Abstract
Metabolomics has great potential to process accessible biofluids through high-throughput and quantitative analysis for radiation biomarker screening. This study focused on the potential radiation responsive metabolites in rat plasma and the dose-response relationships. In the discovery stage, 20 male Sprague–Dawley rats were exposed to 0, 1, 3 and 5 Gy of cobalt-60 gamma rays at a dose rate of 1 Gy/min. Plasma samples were collected at 72 h after exposure and analyzed using liquid chromatography mass spectrometry based on non-targeted metabolomics. In the verification stage, 50 additional rats were exposed to 0, 1, 2, 3, 5 and 8 Gy of gamma rays. The concentrations of candidate metabolites were then analyzed using targeted metabolomics methods. Fifteen candidate radiation responsive metabolites were identified as potential radiation metabolite biomarkers. Metabolic pathways, such as linoleic acid metabolism and glycerophospholipid metabolism pathways, were changed after irradiation. Six radiation responsive metabolites, including LysoPC(20:2), LysoPC(20:3), PC(18:0/22:5), L-palmitoylcarnitine, N-acetylornithine and butyrylcarnitine, had good dose-response relationships (R 2 > 0.80). The area under the curve of the panel of the 6 radiation responsive metabolites was 0.923. The radiation exposure metabolomics biomarkers and dose-response curves may have potential for rapid dose assessment and triage in nuclear and radiation accidents.
Keywords: ionizing radiation, metabolomics, biomarker, dose-response, LC-MS
Introduction
Humans can be exposed to ionizing radiation (IR) from several sources, including diagnostic radiography and nuclear medicine, natural background radiation, and nuclear or radiation accidents. The radiological accidents at Fukushima and Chernobyl have reaffirmed the need for a robust method to quickly triage and estimate the radiation biological doses of large numbers of potentially exposed people and help guide appropriate clinical medical response.1,2
Traditional radiobiology dose estimation methods focus on the assessment of DNA damage and repair, chromosomal rearrangement, and mutagenesis.3,4 Chromosomal aberration assays are considered the gold standard for assessing radiation exposure and damage because of the accuracy of biological dose estimation. However, chromosomal assays are labor intensive, time consuming, require well-trained cytogeneticists to score and analyze samples, which may not always be suitable for the large-scale nuclear and radiation emergencies. In addition, cytogenetics methods are not always sensitive enough to detect subtle molecular fluctuations induced by IR prior to cellular and organ damage are expressed or be able to detect alterations in cellular signal pathways.5 Thus, the development of rapid and high-throughput biological method is still an unmet need and crucial for the dose estimation of irradiated populations.
Metabolomics focus on metabolites with molecular weight below 1500 Da in biofluid or tissue samples that can be used to characterize and quantify the concentrations of large numbers of small metabolites simultaneously, thereby achieving a global signature of the changes in metabolite products that can identify physiological conditions even before symptoms appear.6 The non-targeted metabolomics can be utilized to scan the metabolome and reveal patterns that can help identify potential candidate alterations that can be used in a targeted screening panel. The targeted metabolomics focus on specific metabolites or metabolic pathways and could be quantitative analysis. Metabolomics has great potential to contribute to biomarkers screening by being able to identify the molecular targets and related signal pathways induced by radiation, thereby providing more information on the alterations in the cellular physiology.7 Metabolomics methods have been used in many fields, including clinical diagnosis, pharmacokinetic and animal sciences.8-11 As a potential method of radiation biodosimetry, radiation metabolomics can measure the biological response to radiation dose in the past few years.12
IR exposure can potentially alter the complicated networks of cellular and molecular responses that affect metabolic processes and metabolite levels. These metabolites have the potential to become radiation damage biomarkers.13,14 Metabolomics based on mass spectrometry (MS) platforms has shown great potential to explore the biomarkers of radiation damage due to high-throughput and minimally invasive sample collection methods.15,16 Over the last decade, metabolomics studies have been conducted using rodent,17,18 non-human primates,19,20 and humans21,22 to explore IR-induced potential biomarkers. These studies have confirmed a large number of metabolites are altered after exposure to IR. For example, citrulline has been identified in several studies as a potential biomarker of radiation-induced gastrointestinal injury, and decreased levels of plasma citrulline have been reported in irradiated animal models.23-25 Some metabolites may have the potential to be radiation sensitivity biomarkers, but large discrepancies in the reproducibility and the consistency between studies have hindered these efforts. In addition, the dose-response relationships of IR-induced metabolites have not been systematically explored.
In the present study, non-targeted and targeted metabolomics based on liquid chromatography mass spectrometry (LC-MS) were carried out to screen and identify differentially altered metabolites in plasma of rat at 72 h after exposure to cobalt-60 gamma rays. Candidate radiation responsive metabolites that exhibited drastic alterations in rat plasma exposed to IR in the discovery stage were identified. And the concentrations of these metabolites were carefully quantified in a validation phase. Importantly, good dose-response curves of the radiation responsive metabolites were established, which may have significant contribution to develop a high-throughput biodosimetry method for mass population screening.
Materials and Methods
Chemicals
PC(12:0/13:0), PE(12:0/13:0), SM(d18:1/12:0) and Cer(d18:1/12:0) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Stable isotope labeled references were purchased from Cambridge Isotope Laboratories (Andover, MA, USA), including: Glycine (2-13C, 15N), L-Alanine (2,3,3,3-D4), L-Valine (D8), L-Leucine (5,5,5-D3), L-Methionine (methyl-D3), L-Phenylalanine (ring-13C6), L-Tyrosine (ring-13C6), L-Aspartic acid (2,3,3-D3), DL-Glutamic acid (2,4,4-D3), L-Ornithine: HCl (5,5-D2), L-Citrulline (5,5-D2) and L-Arginine (5-13C, 4,4,5,5-D4). Deuterium-labeled carnitine and acylcarnitines (NSK-B set) were purchased from Cambridge Isotope Laboratories (Tewksbury, MA, USA), including 2H3-Acetylcarnitine (C2), 2H3-Propionylcarnitine (C3), 2H3-Butyrylcarnitine (C4), 2H9-Isovalerylcarnitine (C5), 2H3-Octanoylcarnitine (C8), 2H9-Myristoylcarnitine (C14), and 2H3-Palmitoylcarnitine (C16). HPLC-grade ammonium acetate was purchased from Sigma-Aldrich (St. Louis, MO, USA), and HPLC-grade formic acid and acetonitrile were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All solutions were prepared by LC-MS ultra-pure water.
Animals
This study was conducted at the National Institute for Radiological Protection (NIRP), Chinese Center for Disease Control and Prevention. The Animal Care Committee of NIRP approved all animal experiments in this study. A total of 70 male Sprague Dawley (SD) rats (6-8 weeks, 200-250 g, Department of Laboratory Animal Science, Peking University School of Medicine) were used. Twenty rats were randomized into 4 groups in the discovery stage based on non-targeted metabolomics and 50 rats were randomized into 6 groups in the verification stage based on targeted metabolomics.
Irradiation
Rats were exposed by a cobalt-60 gamma ray source at Beijing Radiation Centre, Beijing Normal University. The source radioactivity was 130 TBq, source-sample distance of 83.5 cm, and the homogeneous irradiation field was 50 cm × 50 cm at a dose rate of 1.0 Gy/min.
In the discovery stage, rats were exposed to total body irradiation with 0 (sham irradiation), 1, 3 and 5 Gy for screening the candidate radiation responsive metabolites. In the verification stage, the radiation doses were 0 (sham irradiation), 1, 2, 3, 5 and 8 Gy to establish metabolites dose-response curves. Rats were put back in cages and given food and water after irradiation.
Plasma Sample Separation
Each blood sample (1-2 ml) was collected from the intraorbital canthus venous plexus in ethylenediaminetetra acetic acid (EDTA) vacutainer tubes (Bacton Dickson, USA) at 72 h after irradiation. Blood samples were centrifuged at 3 000 rpm and 4°C for 10 min, and the separated plasma was stored at −80°C until analysis.
Sample Pre-Treatment
Two pre-treatment methods were used to prepare for the chromatographic separation in the following steps. For the reversed phase (RP) mode, plasma samples were thawed on ice at 4°C, and 100 µl plasma was split into a 1.5 ml microcentrifuge tube And 300 µl methanol and 1 ml methyl tert-butyl ether were added and mixed for 15 s, then centrifuged at 12 000 rpm and 4°C for 10 min for protein deposition. An aliquot (400 µl) was dried, then added with 100 µl methanol, and dissolved in ultrasound. For the hydrophilic interaction liquid chromatography (HILIC) mode, 100 µl plasma was split into a 1.5 ml microcentrifuge tube and added with 300 µl acetonitrile, and mixed for 15 s, and centrifuged at 12 000 rpm and 4°C for 10 min for protein deposition. And 100 µl supernatant was transferred into a 200 µl vial insert for analysis.
In the meantime, 10 µl plasma was collected from each sample and pooled as quality control (QC) sample to ensure the data quality. The procedure of the QC sample preparation for LC-MS was carried out in accordance with the above steps.
Non-Targeted Metabolomics Analysis
The chromatographic separation was carried out on the Dionex UltiMate 3000 LC system (Thermo Fisher Scientific, USA) equipped with the RP (HSS T3 column, Waters Corporation, USA) and HILIC (BEH Amide column, Waters Corporation, USA) columns. For the RP separation, mobile phase A consisted of acetonitrile/water (60/40), and mobile phase B consisted of isopropanol/acetonitrile (90/10). Mobile phases A and B were 0.1% formic acid and 10 mmol/l ammonium acetate. The column temperature was 45°C, flow rate was 300 μl/min and injection volume was 1 μl. The RP separation conditions for lipids are shown in Supplemental Table 1. For the HILIC separation, mobile phase A was 0.1% formic acid and 10 mmol/l ammonium acetate in acetonitrile, whereas mobile phase B was 0.1% formic acid and 10 mmol/l ammonium acetate in water. The column temperature was 40°C, flow rate was 300 μl/min and injection volume was 1 μl. The HILIC separation conditions for small polar molecules are shown in Supplemental Table 2.
The Q exactive mass spectrometer (Thermo Fisher Scientific, USA) was applied. The HESI−II spray voltages of positive and negative modes were 3.7 and 3.5 kV, respectively. The temperatures of heated capillary and vaporizer were 320°C and 300°C, respectively. The sheath gas, auxiliary gas and collision gas were nitrogen at the pressure of 30, 10 and 0.2 Pa, respectively. Full mass scan were taken with 70000 resolution, 50 ms maximum isolation time and 50–1500 m/z. The LC-MS platform was equipped with Xcalibur 2.2 SP1.48 software to collect and process the data.
Targeted Metabolomics Analysis
The candidate metabolites were selected based on non-targeted metabolomics analysis in the discovery stage. In the verification stage, the candidate metabolite concentrations were determined using the RP and HILIC columns under ESI+ mode based targeted metabolomics. The RP and HILIC separation conditions are shown in Supplemental Tables 3-4.
In addition, except the candidate metabolites, plasma citrulline concentrations were also determined.
Data Processing and Statistical Analysis
In the discovery stage, the LC-MS data was conducted using Progenesis QI software (Nonlinear Dynamics, Newcastle, UK). Peak alignment and picking were used QC samples as a reference. Normalization was processed by normalizing to all compounds for potential retention time drift and m/z data pairs. Automatic peak picking for the RP and HILIC modes were 0.7-19 and 0.7-13 min respectively. Each feature of the adduct ions (retention time and m/z) were deconvoluted, and the fragmentation patterns were identified in the self-building database containing more than 600 metabolite standards or in the human metabolome database (HMDB, https://hmdb.ca/) and Lipidmaps (https://www.lipidmaps.org/) with an error of less than 10 ppm.
Multivariate statistics, including principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA), were conducted with Simca 14.1 software (Chicago, IL). The parameters R 2 X, R 2 Y and Q 2 were used to evaluate the model quality as well as to prevent over-fitting. Two-tailed Student’s t-test between 5 Gy and 0 Gy group (P < 0.05) and variable importance in projection (VIP) (>1) were used to identify candidate radiation responsive metabolites in this work. The MetaboAnalyst 4.0 and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.genome.jp/kegg/) databases were applied for pathway enrichment analysis to explore the biological function.
In the verification stage, data was processed using Skyline software. In accordance with the exact molecular weight and retention time of the candidate metabolites, the retention time of each metabolite was determined. The metabolites concentration was calculated according to concentration and peak intensities of the known internal standard. The dose-response curves were obtained using SPSS 26.0 software and R 2 was used to evaluate the goodness of fit of the model, which was assessed by Kruskal–Wallis ANOVA test. The receiver operating characteristic (ROC) curve was applied to estimate the classification capacity of metabolites between the exposed and control samples. The level of statistical significance was P < 0.05.
Results
Non-Targeted Metabolomics of Plasma Samples
In the discovery stage, lipids and small polar molecules in plasma samples were separated based on non-targeted metabolomics method. The LC-MS total ion chromatograms of plasma samples on RP (ESI+ and ESI−) and HILIC (ESI+) separation modes for all groups were taken. A total of 13320 ion peaks were identified, including 5204, 4301 and 3815 features in RP (ESI+), RP (ESI−) and HILIC (ESI+) mode, respectively.
QC samples were inserted in plasma samples during the analytical step to maintain the stability of LC-MS platform. The PCA was performed on QC and all samples, and the QC plots were clustered together, which confirmed the stability of the whole process (Supplemental Figure 1).
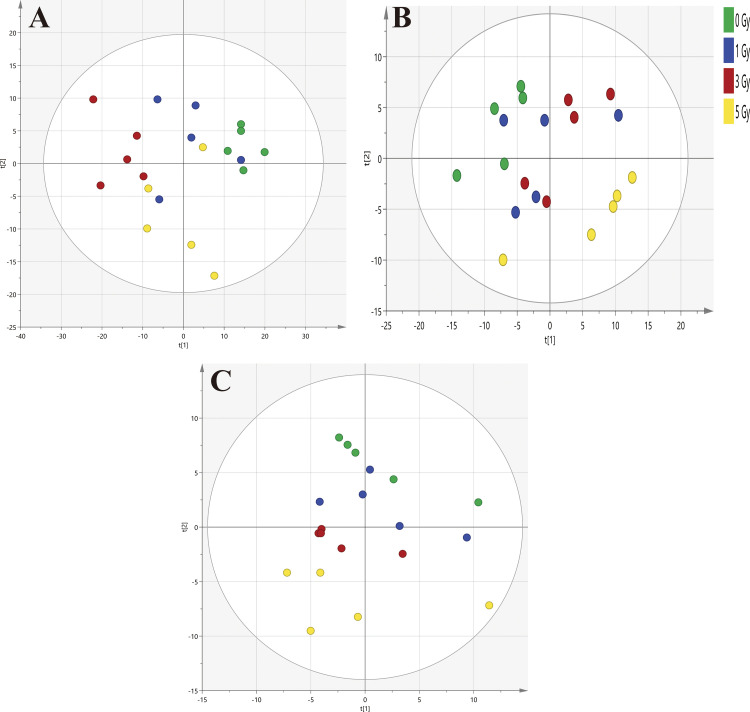
PCA has shown the differences among all groups (Figure 1). PCA score plot displayed the clusters of plasma samples among 1 Gy, 3 Gy, 5 Gy and 0 Gy groups in RP (ESI+) (R 2 X = 0.794, Q 2 = 0.643), RP (ESI−) (R 2 X = 0.702, Q 2 = 0.491) and HILIC (ESI+) (R 2 X = 0.639, Q 2 = 0.363) modes. R2X indicated the describe ability of the variation in x, and Q2 indicated the predictive power of the model. While the cluster region overlaps appeared between the irradiated and 0 Gy groups, separation between 5 Gy and 0 Gy group as well as between 3 Gy and 0 Gy group could be observed, but 1 Gy group could not be clearly distinguish from the other groups.
Figure 1.
Principal component analysis (PCA) score plots. PCA plots demonstrate the clusters of rat plasma samples after 0, 1, 3 and 5 Gy irradiation. Increased separation is evident among the irradiated groups with increased doses and clear separation is evident between 5 Gy and 0 Gy group. A, Reversed phase (ESI+) separation (R 2 X = 0.794, Q 2 = 0.643). B, Reversed phase (ESI−) separation (R 2 X = 0.702, Q 2 = 0.491). C, HILIC (ESI+) separation (R 2 X = 0.639, Q 2 = 0.363).
The OPLS-DA was used for pattern recognition to further explore the differences between 5 Gy and 0 Gy groups. As shown in Supplemental Figure 2(A, B, C), plasma samples between 5 Gy and 0 Gy group were clearly separated in the RP (ESI+) (R 2 Y = 0.973, Q 2 = 0.845), RP (ESI−) (R 2 Y = 0.979, Q 2 = 0.871) and the HILIC (ESI+) (R 2 Y = 0.951, Q 2 = 0.855) modes. The permutation test with 200 iterations was applied to validate the OPLS-DA models with compared between the goodness of fit of the original model and random models. The validation plots indicated that the permuted R 2 and Q 2 points to the left were lower than the original points to the right, and Q 2 regression line was a negative intercept so that the original models were reasonable (Supplemental Figure 2D, E, F).
Candidate Radiation Responsive Metabolites Identification
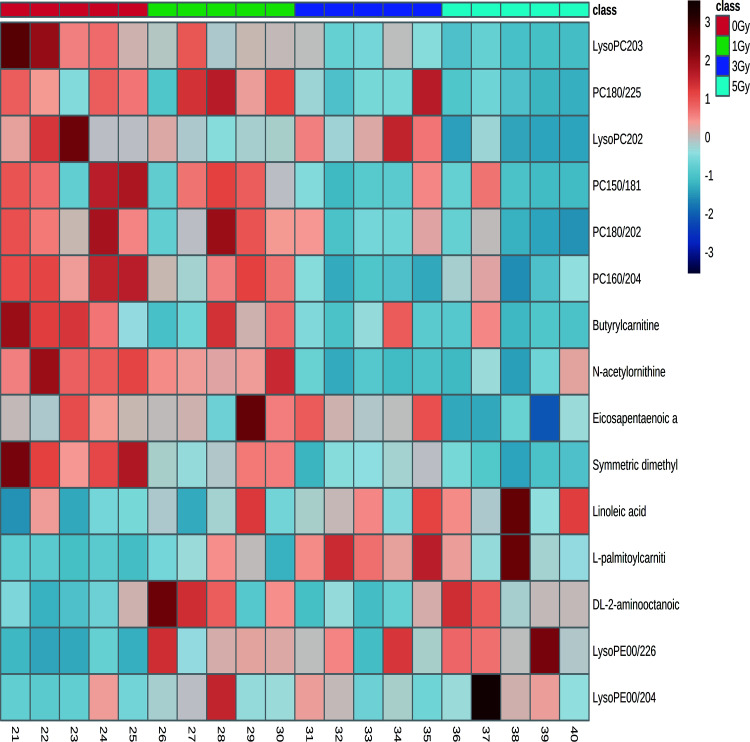
The levels of metabolites were altered significantly after exposure to radiation in rat plasma. Fifteen metabolites were considered as candidate radiation responsive metabolites, which were significant changed between 0 Gy and 5 Gy group (P < 0.05) and VIP value >1, including phosphatidylcholine (PC), lysophosphatidylcholine (LysoPC), lysophosphatidylethanolamine (LysoPE), free fatty acid, amino acid and carnitine. In addition, 8, 2 and 5 candidate metabolites were identified in RP (ESI+), RP (ESI−) and HILIC (ESI+) modes, respectively. Our database and online databases were used to identify these metabolites. For instance, in RP (ESI+) mode, the ion with m/z of 836.6008 and retention time of 14.69 was speculated as C48H86NO8P through the elemental composition and isotopic abundance analyses. Then the fragments of PC(18:0/22:5) (283 and 329) in the HMDB and Lipidmaps database were compared in RP (ESI−) mode to determine that the metabolite was PC(18:0/22:5).
The 15 candidate metabolites, including 5 up-regulated metabolites and 10 down-regulated metabolites, are presented in Table 1, and heatmap of the candidate metabolites is shown in Figure 2. The top 3 up-regulated metabolites based on fold change were LysoPE(0:0/20:4), LysoPE(0:0/22:6) and DL-2-aminooctanoic acid. The top 3 down-regulated metabolites were LysoPC(20:3), PC(18:0/22:5) and LysoPC(20:2).
Table 1.
Candidate Radiation Responsive Metabolites in Rat Plasma After Exposed to 5 Gy Cobalt-60 Gamma Rays.
| Retention time (min) | m/z | Mode | Metabolites | Formula | VIP valuea | p valueb | FDRc | Trend | Fold change |
|---|---|---|---|---|---|---|---|---|---|
| 3.44 | 502.2925 | C18(ESI+) | LysoPE(0:0/20:4) | C25H44NO7P | 1.38 | 0.038 | 0.162 | ↑ | 2.64 |
| 3.24 | 526.2924 | C18(ESI+) | LysoPE(0:0/22:6) | C27H44NO7P | 1.23 | <0.001 | 0.013 | ↑ | 3.41 |
| 4.42 | 546.3549 | C18(ESI+) | LysoPC(20:3) | C28H52NO7P | 1.72 | <0.001 | 0.011 | ↓ | −5.88 |
| 5.37 | 548.3714 | C18(ESI+) | LysoPC(20:2) | C28H54NO7P | 1.20 | 0.001 | 0.023 | ↓ | −2.56 |
| 11.55 | 782.5678 | C18(ESI+) | PC(16:0/20:4) | C44H80NO8P | 1.02 | 0.008 | 0.011 | ↓ | −1.88 |
| 15.30 | 814.6309 | C18(ESI+) | PC(18:0/20:2) | C46H88NO8P | 1.07 | 0.002 | 0.011 | ↓ | −2.00 |
| 14.69 | 836.6143 | C18(ESI+) | PC(18:0/22:5) | C48H86NO8P | 1.38 | <0.001 | 0.011 | ↓ | −3.23 |
| 13.04 | 746.5690 | C18(ESI+) | PC(15:0/18:1) | C41H80NO8P | 1.07 | 0.025 | 0.038 | ↓ | −2.17 |
| 14.93 | 279.2326 | C18(ESI-) | Linoleic acid | C18H32O2 | 1.10 | 0.037 | 0.056 | ↑ | 1.48 |
| 14.16 | 301.2171 | C18(ESI-) | Eicosapentaenoic acid | C20H30O2 | 1.20 | 0.007 | 0.019 | ↓ | −1.42 |
| 1.38 | 400.3414 | HILIC(ESI+) | L-palmitoylcarnitine | C23H45NO4 | 1.25 | 0.013 | 0.056 | ↑ | 1.67 |
| 2.71 | 160.1329 | HILIC(ESI+) | DL-2-aminooctanoic acid | C8H17NO2 | 1.44 | 0.016 | 0.033 | ↑ | 1.74 |
| 6.71 | 203.1500 | HILIC(ESI+) | Symmetric dimethylarginine | C8H18N4O2 | 1.12 | <0.001 | 0.005 | ↓ | −1.25 |
| 6.07 | 175.1075 | HILIC(ESI+) | N-acetylornithine | C7H14N2O3 | 1.26 | 0.002 | 0.011 | ↓ | −1.63 |
| 2.17 | 232.1539 | HILIC(ESI+) | Butyrylcarnitine | C11H21NO4 | 1.42 | 0.008 | 0.025 | ↓ | −1.45 |
Notes. The precise parameters of candidate metabolites based on non-targeted metabolomics.
a VIP value = Variable importance in the projection value.
b T-test, compared with 0 Gy.
c FDR = False discovery rate.
Figure 2.
Heatmap of the candidate radiation responsive metabolites in rat plasma after 0, 1, 3 and 5 Gy irradiation based on non-targeted metabolomics in the discovery stage. The heatmap shows the changes in the abundance of candidate metabolites after irradiation. Metabolites with P < 0.05 and VIP > 1 were selected, and different colors represented the relative concentration (red and blue colors represented high and low concentration, respectively). The conditions of heatmap were as follows: distance measure: pearson; clustering algorithm: ward; color contrast: default; data source: original data; standardization: autoscale features.
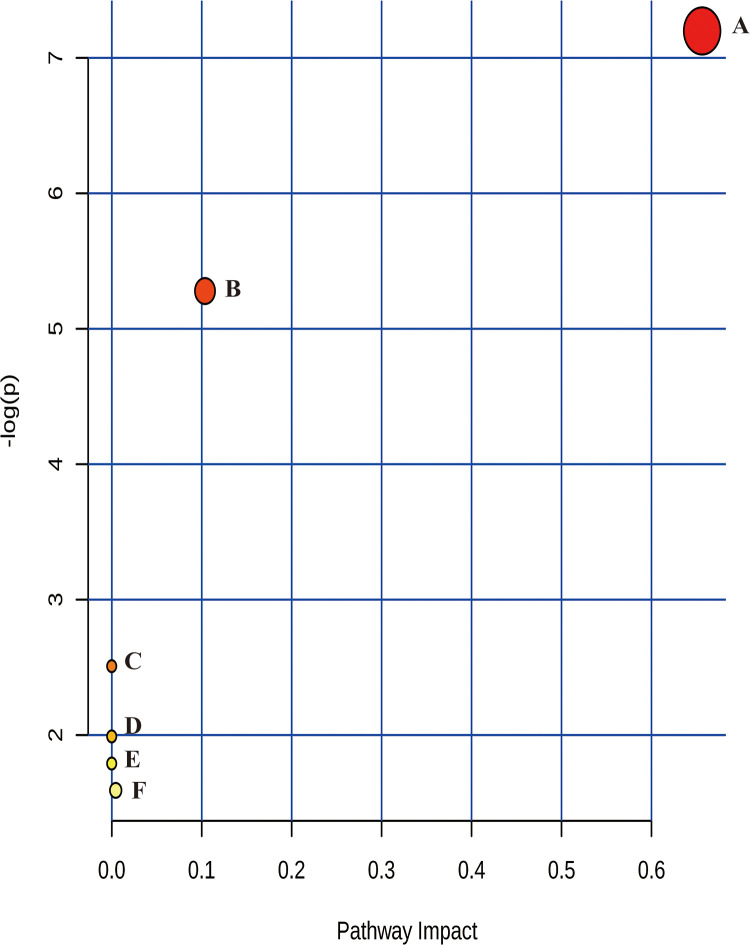
The MetaboAnalyst 4.0 was performed to identify the pathways of the 15 candidate radiation responsive metabolites. As shown in Figure 3 and Supplemental Table 5, 6 metabolic pathways, such as linoleic acid metabolism and glycerophospholipid metabolism, were changed after irradiation.
Figure 3.
Metabolic pathways analysis in rat plasma after exposure to cobalt-60 gamma rays. Significant metabolic pathways related to (A) Linoleic acid metabolism, (B) Glycerophospholipid metabolism, (C) Alpha-Linolenic acid metabolism, (D) Arginine biosynthesis, (E) Arachidonic acid metabolism, (F) Fatty acid degradation. The algorithms were used in MetaboAnalyst were as follow: Over representation analysis: hypergeometric test; pathway topology analysis: relative-betweenness centrality; library: Rattus norvegicus (rat, KEGG).
Concentrations of Candidate Radiation Responsive Metabolites
In the verification stage, the targeted metabolomics method was performed to analysis the concentrations of candidate radiation responsive metabolites selected from discovery phase. Thirteen candidate radiation responsive metabolites (except linoleic acid and eicosapentaenoic acid) had been identified in all groups with RP (ESI+) and HILIC (ESI+) modes. The candidate metabolite concentrations after exposure to 0-8 Gy cobalt-60 gamma rays are shown in Table 2. Compared with the relationship between 5 Gy and 0 Gy group in the discovery stage, 9 of 13 metabolites had the same tendency, including 2 increased metabolites and 7 decreased metabolites. Compared with 0 Gy group, 7 metabolites concentration in 1-8 Gy groups were all increased or decreased. Especially, LysoPC(20:3) and PC(18:0/20:2) concentrations in all irradiated groups were significantly lower than 0 Gy group.
Table 2.
Concentration of Candidate Radiation Responsive Metabolites in Rat Plasma After 0-8 Gy Cobalt-60 Gamma Rays Irradiation Based on Targeted Metabolomics ( ± s ).
| Metabolites | Metabolites concentration (μmol/l) | |||||
|---|---|---|---|---|---|---|
| 0 Gy | 1 Gy | 2 Gy | 3 Gy | 5 Gy | 8 Gy | |
| LysoPE(0:0/22:6) | 0.97 ± 0.27 | 0.78 ± 0.30 | 0.80 ± 0.36 | 0.60 ± 0.20a | 0.78 ± 0.32 | 1.28 ± 0.50 |
| LysoPE(0:0/20:4) | 0.80 ± 0.20 | 0.69 ± 0.14 | 0.64 ± 0.21 | 0.67 ± 0.14a | 0.48 ± 0.14a | 0.64 ± 0.10 |
| LysoPC(20:3) | 3.03 ± 0.59 | 2.49 ± 0.55a | 2.52 ± 0.45a | 2.23 ± 0.26a | 1.60 ± 0.40a | 0.99 ± 0.30a |
| LysoPC(20:2) | 3.08 ± 1.03 | 2.65 ± 0.74 | 2.55 ± 0.74 | 2.98 ± 0.80 | 1.70 ± 0.52a | 1.23 ± 0.43a |
| PC(16:0/20:4) | 44.54 ± 5.45 | 41.12 ± 7.37 | 39.01 ± 7.06 | 41.65 ± 10.25 | 38.10 ± 7.42 | 64.56 ± 8.25a |
| PC(18:0/20:2) | 2.99 ± 0.58 | 2.42 ± 0.65a | 2.22 ± 0.55a | 1.97 ± 0.32a | 1.89 ± 0.41a | 2.26 ± 0.45a |
| PC(18:0/22:5) | 2.99 ± 0.75 | 2.81 ± 0.52 | 3.27 ± 0.90 | 3.23 ± 0.81 | 3.86 ± 0.80a | 7.73 ± 1.27a |
| PC(15:0/18:1) | 3.30 ± 0.10 | 3.17 ± 0.87 | 3.25 ± 0.78 | 3.78 ± 1.24 | 2.90 ± 0.77 | 3.64 ± 0.70 |
| L-palmitoylcarnitine | 0.028 ± 0.006 | 0.026 ± 0.006 | 0.025 ± 0.004 | 0.039 ± 0.014 | 0.038 ± 0.014a | 0.070 ± 0.021a |
| DL-2-aminooctanoic acid | 1.46 ± 0.79 | 1.86 ± 1.58 | 1.70 ± 0.98 | 1.85 ± 1.19 | 1.79 ± 0.98 | 1.59 ± 0.53 |
| Symmetric dimethylarginine | 8.78 ± 0.43 | 7.28 ± 0.89a | 6.66 ± 0.37a | 7.08 ± 1.65 | 8.92 ± 1.93 | 7.16 ± 0.58a |
| N-acetylornithine | 9.34 ± 1.43 | 8.76 ± 2.46 | 8.83 ± 1.12 | 7.40 ± 1.63 | 7.57 ± 1.52a | 6.23 ± 1.58a |
| Butyrylcarnitine | 0.60 ± 0.15 | 0.50 ± 0.13 | 0.49 ± 0.20 | 0.34 ± 0.07a | 0.42 ± 0.15a | 0.24 ± 0.07a |
Notes. Candidate metabolites concentrations in rat plasma at each dose were analyzed in validation phase.
a T-test, compared with 0 Gy, P < 0.05.
Dose-Response Curves of the Radiation Responsive Metabolites
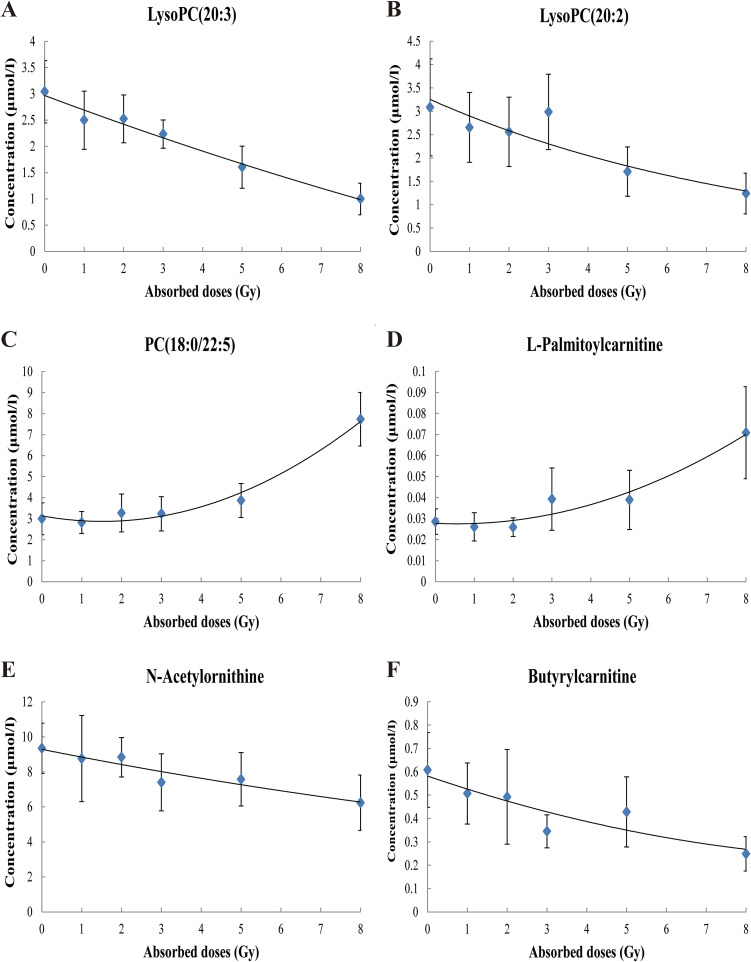
The relationships between concentrations of single metabolite at each dose and absorbed dose were analyzed to establish the dose-response curves. With the absorbed dose of 0-8 Gy Cobalt-60 gamma rays, 9 metabolites concentrations [LysoPE(0:0/20:4), LysoPC(20:3), LysoPC(20:2), PC(18:0/20:2), PC(18:0/22:5), L-palmitoylcarnitine, DL-2-aminooctanoic acid, N-acetylornithine and butyrylcarnitine] had increasing or decreasing trend, and 4 metabolites concentrations [LysoPE(0:0/22:6), PC(16:0/20:4), PC(15:0/18:1) and symmetric dimethylarginine] changed randomly. The concentration of PC(18:0/22:5) and L-palmitoylcarnitine were significantly increased with the increased dose, and the concentration of LysoPC(20:2), LysoPC(20:3), N-acetylornithine and butyrylcarnitine significantly decreased. The dose–response relationships between the metabolite concentration and absorbed doses followed quadratic or exponential models, and the R 2 values were above 0.80 and in the PC(18:0/22:5) model R 2 was 0.981. The regression equations and dose–response curves are shown in Table 3 and Figure 4.
Table 3.
Metabolites Dose-Response Curves of Rat Plasma Exposed to 0-8 Gy Cobalt-60 Gamma Rays.
| Metabolites | Formula a | R2 b | P value c |
|---|---|---|---|
| LysoPC(20:3) | y = 0.0042x2-0.2815x+2.9683 | 0.977 | <0.001 |
| LysoPC(20:2) | y = 3.2537e-0.115x | 0.870 | <0.001 |
| PC(18:0/22:5) | y = 0.1134x2-0.3458x+3.1352 | 0.981 | 0.001 |
| L-palmitoylcarnitine | y = 0.0008x2-0.0009x+0.0278 | 0.945 | <0.001 |
| N-acetylornithine | y = 9.2890e-0.049x | 0.907 | 0.004 |
| Butyrylcarnitine | y = 0.0024x2-0.0582x+0.5819 | 0.817 | <0.001 |
Notes. The dose–response relationship between metabolite concentration and absorbed doses followed quadratic or exponential models.
a y is the metabolite concentration (μmol/l), x is the absorbed dose (Gy).
b R2 value was used to evaluate the goodness of fit of the models.
c Based on Kruskal–Wallis ANOVA test.
Figure 4.
Dose-response curves of the responsive metabolite concentration and the absorbed dose (0-8Gy) of cobalt-60 gamma rays. With the increased dose of 0-8 Gy cobalt-60 gamma rays, the concentrations of PC(18:0/22:5) and L-palmitoylcarnitine significantly increased, and the concentrations of LysoPC(20:2), LysoPC(20:3), N-acetylornithine and butyrylcarnitine significantly decreased. The dose–response relationship between the metabolite concentration and absorbed doses followed quadratic or exponential models. Panels A–F: Dose–response curves of LysoPC(20:3), LysoPC(20:2), PC(18:0/22:5), L-palmitoylcarnitine, N-acetylornithine and butyrylcarnitine.
The coefficients of variation (CVs) of the 6 radiation responsive metabolites at each dose are shown in Supplemental Table 6. The CVs of each metabolite between 0 Gy and irradiated groups no specific increased or decreased trend. CVs ranged from 12.1% to 41.1%, and the majority of them were lower than 35.0%. Typically, LysoPC(20:3) and N-acetylornithine had lower CVs (< 30.0%).
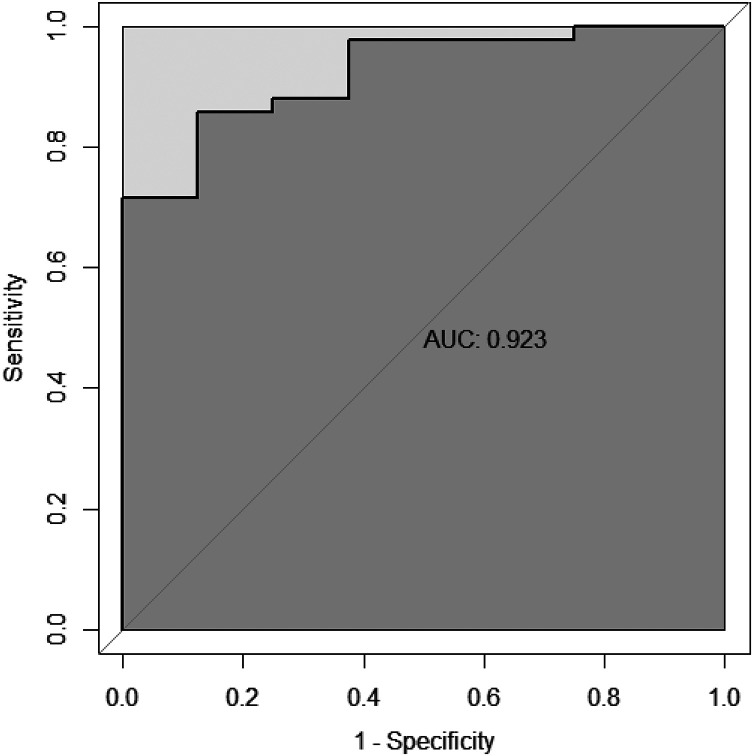
In order to assess the discriminatory capacity of the 6 radiation responsive metabolites aforementioned between irradiated and 0 Gy groups, ROC curve was performed and area under the curve (AUC) was calculated. As shown in Figure 5, AUC of the panel of the 6 radiation responsive metabolites was 0.923, which was higher than those of single metabolites and could evidently distinguish the exposed samples from controls. AUC of the single metabolites were between 0.661 and 0.872 (Supplemental Figure 3).
Figure 5.
ROC curve of the metabolite panel. The discriminatory ability between the exposed samples and controls were analyzed using the ROC curves in the verification stage. The metabolite panel included LysoPC(20:3), LysoPC(20:2), PC(18:0/22:5), L-palmitoylcarnitine, N-acetylornithine and butyrylcarnitine.
Plasma Citrulline
The concentrations were also analyzed based on targeted method. Citrulline concentrations in all irradiated groups decreased, and significantly decreased were found after 3, 5 and 8 Gy irradiation. The dose–response relationship between citrulline concentration and doses followed quadratic models: y = −0.0155x2 −11.337x +111.52, where y is citrulline concentration (μmol/l) and x is dose (Gy). The R 2 value was 0.985 (Supplemental Table 7).
Discussion
In the present study, the non-targeted metabolomics method through LC–MS was performed in rat plasma after cobalt-60 gamma rays total-body irradiation and 15 metabolites were identified as candidate radiation responsive metabolites. The concentration of each candidate metabolites was analyzed by targeted metabolomics methods, and the 0-8 Gy dose–response curves of 6 radiation responsive metabolites were established.
Fifteen metabolites were considered as candidate metabolites in rat plasma after exposure to 5 Gy gamma rays based on non-targeted metabolomics in the discovery stage. In the RP mode, 10 lipids were significant altered, including 4 PCs, 2 LysoPCs, 2 LysoPEs and 2 free fatty acids. Lipids are involved in many cellular structures and perform a variety of functions, which are initially thought to be the formation of cell membranes and storage of energy, and recent studies focus on the roles of lipids in cellular signaling and the inflammatory response.26 Previous studies have reported that PCs, LysoPCs and LysoPEs in serum of rodent and cancer patients changed significantly after exposure to IR and involved in the regulation of crucial signal pathways.27-29 Linoleic acid is reported to be significantly induced by cesium-137 internal exposure in mice serum30 and lung cancer patients’ serum after radiotherapy.31 Eicosapentaenoic acid is not reported to be influenced by IR. In the HILIC model, 3 amino acids and 2 acylcarnitines were identified post-irradiation. Pannkuk has shown that butyrylcarnitine is altered in the nonhuman primate serum 7 days after exposure to gamma rays.32 Symmetric dimethylarginine has changed in rats with cardiovascular disorders after gamma rays irradiation.33 L-palmitoylcarnitine is not found to have a direct relationship with IR, but as a type of fatty acylation, palmitoylation plays an essential role in many biological processes, such as apoptosis and proliferation.34 N-acetylornithine is identified as minor components of deproteinized plasma of humans, and 2-aminooctanoic acid is a fatty acid that modifies a lactoferricin B peptide to improve antimicrobial activity.35 DL-2-aminooctanoic acid and N-acetylornithine were not influenced by IR and should be paid more attention. The effect of IR on apoptosis has been widely reported. In this study, some of candidate metabolites could be attributed to apoptosis, such as PCs, LysoPCs and LysoPEs, which have been proved by previous studies.35 The influence of multiple freeze-thaw cycles on metabolite levels should be considered. Thus, the plasma samples in the present study only underwent two or less freeze thaw cycles. Although the above 15 metabolites in rat plasma changed significantly 72 h after irradiation, further studies should be focused on the multiple time points and other species to verify the applicability of these metabolites.
The above 15 metabolites were used for pathway analysis and 6 metabolic pathways were changed after exposure to cobalt-60 gamma rays, especially in linoleic acid metabolism and glycerophospholipid metabolism. Linoleic acid regulates the cholesterol synthesis, thereby affecting the lipid metabolism. Abnormal linoleic acid metabolism can lead to cell apoptosis and inflammation. Glycerophospholipid, the most abundant phospholipid in living organisms, is an important component of biofilms and bile, and plays an essential role in protein recognition and signal transduction. Glycerophospholipid metabolism disorder may relate to metabolic disease.
The dose-response relationship of radiation responsive metabolites should be carefully assessed to properly evaluate radiation dose for biodosimetry. In the present study, non-targeted and targeted methods were performed to identify potential radiation responsive metabolites to avoid the false positive biomarkers. Good dose-response relationships of 6 radiation responsive metabolites were found, and the corresponding dose-response curves were established. The highest R 2 value was found in PC(18:0/22:5) model, which decreased with the increased doses. Except those of the above 6 metabolites, PC(18:0/20:2) concentrations decreased significantly in all irradiated groups, which may indicate the influence of IR. In the follow-up study, the blind dose test will be performed to validate the applicability and accuracy of dose estimation based on these metabolite dose-response curves.
The variability is another crucial factor to conclude the quality of dose–response curves and evaluate the accuracy of dose estimation based on the metabolite method. In this study, CVs of the 6 radiation responsive metabolites that established the dose-response curves at each dose were analyzed. The changes in the CVs of each metabolite were different. The CVs of LysoPC(20:3) and N-acetylornithine were lower than 30.0% and may be considered as good predictors of radiation biodosimetry. However, the CVs of butyrylcarnitine in 2 Gy group and L-palmitoylcarnitine in 3 Gy group were higher than 35.0%, which may impede the accuracy of dose estimation. These variations may indicate different sensitivity to radiation damage response that may be a limitation of the use of metabolites in triage and dose estimation.
The ROC curves of single metabolites clearly showed high discrimination between the exposed samples and controls. The panel of the 6 radiation responsive metabolites may be potential biomarkers for the rapid classification of radiation injury.
Plasma citrulline may be a potential biomarker of radiation related gastrointestinal tissue damage and epithelial cell loss. Citrulline concentrations are specific to the small intestinal epithelial tissue, which is inversely related to the gastrointestinal injury. The decreased intestinal absorptive function after radiation exposure may result from the loss of intestinal epithelial cells, which form the surface of the absorptive mucosa. The relationship between radiation dose and citrulline concentration has been reported in various animal models.36,37 In the present study, citrulline was not identified as a candidate metabolite through the P and VIP values after exposure to 5 Gy gamma rays based on non-targeted metabolomics. However, the concentrations of citrulline were also analyzed based on the targeted method. Citrulline concentrations decreased with increased doses. Several reports also indicate the decreased levels of citrulline after irradiation.32,38 Actually, knowing the accurate biological dose after radiation or nuclear accidents as soon as possible is good for the irradiated individuals. In this study, we focused on the dose-response relationship of the metabolites 72 h after irradiation. Based on these results, multiple time points will be chosen to explore the time-dependent relationship of radiation responsive metabolites. In addition, the mechanisms of IR-induced metabolic alteration have not been identified yet, but future studies may provide insights into the molecular mechanisms of our model. Although our selected metabolites biomarkers have dose-response relationships, whether these metabolites will have similar trends in humans after irradiation remains unclear. Human radiation metabolomics studies are difficult due to the influence of confounding factors, such as disease, age, gender and diet.39
Conclusions
In summary, the LC-MS platform was used to analyze the effect of IR on metabolites in rat plasma 72 h after cobalt-60 gamma ray exposure. Fifteen metabolites were selected as potential radiation responsive biomarkers. The dose-response curves of 6 metabolites were determined, which may have the potential use in dose estimation or rapid triage. Although these metabolites need further investigation, this study provides additional evidence of the potential of metabolite biomarkers in the determination of radiation exposure in irradiated populations.
Supplemental Material
Supplemental Material, sj-doc-1-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-1-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-2-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-3-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Acknowledgments
The authors thank every researcher in this study.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (81573081).
ORCID iD: Hua Zhao  https://orcid.org/0000-0002-1039-9073
https://orcid.org/0000-0002-1039-9073
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Sproull M, Camphausen K. State-of-the-art advances in radiation biodosimetry for mass casualty events involving radiation exposure. Radiat Res. 2016;186(5):423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Swartz HM, Williams BB, Flood AB. Overview of the principles and practice of biodosimetry. Radiat Environ Biophys. 2014;53(2):221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. IAEA. Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies. Vienna, Austria: IAEA; 2011. [Google Scholar]
- 4. Oya N, Shibamoto Y, Shibata T, Sasai K, Sugiyama T, Abe M. Combined cytokinesis-block micronucleus and chromosomal aberration assay for the evaluation of radiosensitizers at low radiation doses. Int J Radiat Oncol Biol Phys. 1994;30(5):1153–1159. [DOI] [PubMed] [Google Scholar]
- 5. Rothkamm K, Beinke C, Romm H, et al. Comparison of established and emerging biodosimetry assays. Radiat Res. 2013;180(2):111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitsui T, Kira S, Ihara T, et al. Metabolomics approach to male lower urinary tract symptoms: identification of possible biomarkers and potential targets for new treatments. J Urol. 2018;199(5):1312–1318. [DOI] [PubMed] [Google Scholar]
- 7. Menon SS, Uppal M, Randhawa S, et al. Radiation metabolomics: current status and future directions. Front Oncol. 2016;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nalbantoglu S, Abu-Asab M, Suy S, Collins S, Amri H. Metabolomics-based biosignatures of prostate cancer in patients following radiotherapy. OMICS. 2019;23(4):214–223. [DOI] [PubMed] [Google Scholar]
- 9. Murithi JM, Owen ES, Istvan ES, et al. Combining stage specificity and metabolomic profiling to advance antimalarial drug discovery. Cell Chem Biol. 2019;27(2):158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matich EK, Chavez SN, Aga DS, Atilla-Gokcumen GE. Applications of metabolomics in assessing ecological effects of emerging contaminants and pollutants on plants. J Hazard Mater. 2019;373:527–535. [DOI] [PubMed] [Google Scholar]
- 11. Audano M, Maldini M, De Fabiani E, Mitro N, Caruso D. Gender-related metabolomics and lipidomics: from experimental animal models to clinical evidence. J Proteomics. 2018;178:82–91. [DOI] [PubMed] [Google Scholar]
- 12. Coy SL, Cheema AK, Tyburski JB, et al. Radiation metabolomics and its potential in biodosimetry. Int J Radiat Biol. 2011;87(8):802–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Saitoh W, Takada S, Hirao J, et al. Plasma citrulline is a sensitive safety biomarker for small intestinal injury in rats. Toxicol Lett. 2018;295:416–423. [DOI] [PubMed] [Google Scholar]
- 14. Liu C, Gu C, Huang W, et al. Targeted UPLC-MS/MS high-throughput metabolomics approach to assess the purine and pyrimidine metabolism. J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1113:98–106. [DOI] [PubMed] [Google Scholar]
- 15. Haritwal T, Maan K, Rana P, et al. Trichostatin A, an epigenetic modifier, mitigates radiation-induced androphysiological anomalies and metabolite changes in mice as evident from NMR-based metabolomics. Int J Radiat Biol. 2019;95(4):443–451. [DOI] [PubMed] [Google Scholar]
- 16. Laiakis EC, Pannkuk EL, Diaz-Rubio ME, et al. Implications of genotypic differences in the generation of a urinary metabolomics radiation signature. Mutat Res. 2016;788:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang C, Yang J, Nie J. Plasma phospholipid metabolic profiling and biomarkers of rats following radiation exposure based on liquid chromatography-mass spectrometry technique. Biomed Chromatogr. 2009;23(10):1079–1085. [DOI] [PubMed] [Google Scholar]
- 18. Ossetrova NI, Ney PH, Condliffe DP, Krasnopolsky K, Hieber KP. Acute radiation syndrome severity score system in mouse total-body irradiation model. Health Phys. 2016;111(2):134–144. [DOI] [PubMed] [Google Scholar]
- 19. Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJJ. Gas chromatography/mass spectrometry metabolomics of urine and serum from nonhuman primates exposed to ionizing radiation: impacts on the tricarboxylic acid cycle and protein metabolism. J Proteome Res. 2017;16(5):2091–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pannkuk EL, Laiakis EC, Garcia M, Fornace AJJ, Singh VK. Nonhuman primates with acute radiation syndrome: results from a global serum metabolomics study after 7.2 Gy total-body irradiation. Radiat Res. 2018;190(6):576–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang FQ, Li J, Jiang L, et al. Serum-plasma matched metabolomics for comprehensive characterization of benign thyroid nodule and papillary thyroid carcinoma. Int J Cancer. 2019;144(4):868–876. [DOI] [PubMed] [Google Scholar]
- 22. Zhang T, Xu J, Liu Y, Liu J. Metabolomic profiling for identification of potential biomarkers in patients with dermatomyositis. Metabolomics. 2019;15(5):77. [DOI] [PubMed] [Google Scholar]
- 23. Broin PO, Vaitheesvaran B, Saha S, et al. Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury. Int J Radiat Oncol Biol Phys. 2015;91(2):360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones JW, Tudor G, Li F, et al. Citrulline as a biomarker in the murine total-body irradiation model: correlation of circulating and tissue citrulline to small intestine epithelial histopathology. Health Phys. 2015;109(5):452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bujold K, Hauer-Jensen M, Donini O, et al. Citrulline as a biomarker for gastrointestinal-acute radiation syndrome: species differences and experimental condition effects. Radiat Res. 2016;186(1):71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian Y, Xia Z, Li M, et al. The relationship between microwave radiation injury and abnormal lipid metabolism. Chem Phys Lipids. 2019;225:104802. [DOI] [PubMed] [Google Scholar]
- 27. Laiakis EC, Canadell MP, Grilj V, et al. Serum lipidomic analysis from mixed neutron/X-ray radiation fields reveals a hyperlipidemic and pro-inflammatory phenotype. Sci Rep. 2019;9(1):4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park HM, Shin JH, Kim JK, et al. MS-based metabolite profiling reveals time-dependent skin biomarkers in UVB-irradiated mice. Metabolomics. 2014;10(4):663–676. [Google Scholar]
- 29. Jelonek K, Pietrowska M, Ros M, et al. Radiation-induced changes in serum lipidome of head and neck cancer patients. Int J Mol Sci. 2014;15(4):6609–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goudarzi M, Weber WM, Mak TD, et al. Metabolomic and lipidomic analysis of serum from mice exposed to an internal emitter, cesium-137, using a shotgun LC–MSE approach. J Proteome Res. 2014;14(1):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Y, Song X, Zhao X, Zhao X, Xu G. Serum metabolic profiling study of lung cancer using ultra high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2014;966:147–153. [DOI] [PubMed] [Google Scholar]
- 32. Pannkuk EL, Laiakis EC, Authier S, Wong K, Fornace AJJ. Targeted metabolomics of nonhuman primate serum after exposure to ionizing radiation: potential tools for high-throughput biodosimetry. Rsc Adv. 2016;6(56):51192–51202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdel-Magied N, Shedid SM. Impact of zinc oxide nanoparticles on thioredoxin-interacting protein and asymmetric dimethylarginine as biochemical indicators of cardiovascular disorders in gamma-irradiated rats. Environ Toxicol. 2020;35(4):430–442. [DOI] [PubMed] [Google Scholar]
- 34. Chen S, Zhu B, Yin C, et al. Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature. 2017;549(7672):399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Almahboub SA, Narancic T, Devocelle M, et al. Biosynthesis of 2-aminooctanoic acid and its use to terminally modify a lactoferricin B peptide derivative for improved antimicrobial activity. Appl Microbiol Biotechnol. 2018;102(2):789–799. [DOI] [PubMed] [Google Scholar]
- 36. Jones JW, Tudor G, Bennett A, et al. Development and validation of a LC-MS/MS assay for quantitation of plasma citrulline for application to animal models of the acute radiation syndrome across multiple species. Anal Bioanal Chem. 2014;406(19):4663–4675. [DOI] [PubMed] [Google Scholar]
- 37. Goudarzi M, Chauthe S, Strawn SJ, et al. Quantitative metabolomic analysis of urinary citrulline and calcitroic acid in mice after exposure to various types of ionizing radiation. Int J Mol Sci. 2016;17(5):782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones JW, Scott AJ, Tudor G, et al. Identification and quantitation of biomarkers for radiation-induced injury via mass spectrometry. Health Phys. 2014;106(1):106–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawton KA, Berger A, Mitchell M, et al. Analysis of the adult human plasma metabolome. Pharmacogenomics. 2008;9(4):383–397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-doc-1-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-1-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-2-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response
Supplemental Material, sj-tif-3-dos-10.1177_1559325820979570 for Identification of Potential Radiation Responsive Metabolic Biomarkers in Plasma of Rats Exposed to Different Doses of Cobalt-60 Gamma Rays by Hua Zhao, Cong Xi, Mei Tian, Xue Lu, Tian-Jing Cai, Shuang Li, Xue-Lei Tian, Ling Gao, Hai-Xiang Liu, Ke-Hui Liu and Qing-Jie Liu in Dose-Response