Abstract
Background
Bone homeostasis is a tightly orchestrated process maintained by osteoblasts and osteoclasts, and a disruption of their steady-state equilibrium can lead to the occurrence of osteoporosis (OP).
Methods
We investigated the differential expression of micro (mi)RNAs in the bone tissues of a postmenopausal osteoporosis rat model induced by ovariectomy (OVX). Real-time PCR was used to verify the differentially expressed miRNAs in bone samples from OP patients and controls. The specific targets of two differentially expressed miRNAs in osteogenic or osteoclast differentiation were determined by bioinformatic prediction, and mRNA and protein detection.
Results
miR-708-5p and miR-708-3p were highly expressed in the bone tissue of OVX rats and OP patients. miR-708-5p negatively regulated osteoblast differentiation in bone marrow mesenchymal stem cells by targeting SMAD specific E3 ubiquitin protein ligase 2, while miR-708-3p positively regulated osteoclast differentiation in bone marrow monocytes by targeting cerebellar degeneration associated protein 1 antisense RNA. miR-708-5p and miR-708-3p were shown to originate from the same precursor miRNA and to have a synergistic effect on the development of osteoporosis with different temporal and spatial patterns.
Conclusion
Our findings provide a referential theoretical basis and targets for the prevention and treatment of osteoporosis.
Keywords: Osteoporosis, miR-708-5p, miR-708-3p, osteoblasts, osteoclasts, differentiation
Introduction
With the advancing mean age of the global population, osteoporosis has become the fourth most prevalent chronic disease worldwide. As a systemic osteopathy, it manifests as a decrease in bone mass, degeneration of the bone microstructure, a decrease in bone strength, and an increase in brittleness, resulting in bones that fracture easily.1 Therefore, the mechanism underlying the development of osteoporosis has become an important health issue requiring exploration. The occurrence of osteoporosis is closely related to osteoblast and osteoclast pathology. Osteoblasts, which are derived from bone marrow mesenchymal stem cells (BMSCs), are responsible for bone formation.2 Conversely, multinucleated osteoclasts, which derive from precursors of macrophages/monocytes, are responsible for bone resorption.3,4 Bone homeostasis depends on the balance between bone resorption, mediated by osteoclasts, and bone formation, mediated by osteoblasts.5 Therefore, it is necessary to elucidate the molecular mechanism of osteoblast and osteoclast differentiation to understand osteoporosis.
The differentiation of BMSCs into osteoblasts is regulated by many signaling pathways, including Wnt, bone morphogenetic protein (BMP)/SMADs, Hedgehog, and Notch signaling pathways. Runt-related transcription factor (Runx)2 is a key regulatory factor associated with osteoblast differentiation and an essential transcription factor in the process of normal bone development and osteogenesis.6 Therefore, it is often used as a marker of osteoblast differentiation. An important signaling pathway involved in osteoclast differentiation is the osteoprotegerin/receptor activator of nuclear factor-κB (RANK)/RANK ligand (RANKL) pathway. The binding of RANKL to RANK activates signal transmission,7 leading to the expression of osteoclast-specific genes such as tartrate-resistant acid phosphatase (TRAP), cathepsin K (CTSK), and matrix metallopeptidase 9 (MMP9).8 The related factors that affect the induced differentiation of osteoblasts and osteoclasts are a potential therapeutic intervention target for osteoporosis prevention.
Osteoblasts and osteoclasts are closely regulated by many factors, such as micro (mi)RNAs.9 Some miRNAs can inhibit osteoblast differentiation by targeting important osteogenic factors. For example, miR-153 reduces osteoblast differentiation by targeting bone morphogenetic protein receptor type II in BMSCs;10 miR-488 negatively regulates psoralen-induced osteoblast differentiation of BMSCs by targeting Runx2;11 miR-340 inhibits osteoclast differentiation by inhibiting the expression of microphthalmia-associated transcription factor (MITF);12 miR-133a participates in the regulation of postmenopausal osteoporosis by promoting osteoclast differentiation;13 and miR-539 promotes osteoblast proliferation, differentiation, and osteoclast apoptosis through the axon-dependent Wnt signaling pathway in a rat model of osteoporosis.14 Therefore, the study of miRNAs is helpful to further understand the pathological mechanism of osteoporosis.
In this study, we sequenced miRNAs expressed in tibial tissue isolated from an ovariectomized (OVX)-induced osteoporosis rat model. We then selected the differentially expressed miRNAs miR-708-5p and miR-708-3p for follow-up analysis to investigate how they regulate the differentiation of osteoblasts and osteoclasts and to provide a new theoretical basis for the intervention and treatment of osteoporosis.
Materials and methods
Ethics statement
The animal care and use protocols were designed in accordance with national and international laws for laboratory animal welfare and experimentation. The protocol was elaborated according to Standard Protocol Items: Recommendations for Animal Research guidelines drawn from the Equator Network (http://www.equator-network.org), and was approved by the Animal Care and Use Committee of Guang'anmen Hospital of China Academy of Traditional Chinese Medicine (Approval No: 2018-087-SQ).
Animals and OVX-induced osteoporosis model
Ten 10-week-old healthy specific pathogen-free female Sprague-Dawley rats were purchased from Viewsolid Biotech (Beijing, China). The rat osteoporosis model was established by ovariectomy (OVX; estrogen withdrawal), as previously described.15,16 Briefly, all rats were kept under standard housing laboratory conditions for 1 week, then randomly divided into two groups: the ovariectomized group (OVX, n = 5) and the sham-operated group (sham, n = 5). At 12 weeks of age, the OVX group underwent OVX (removal of bilateral ovaries) after being anesthetized via intraperitoneal injection of pentobarbital sodium in phosphate-buffered saline (PBS) at 30 mg/kg. For the sham operation group, the same volume of the fat pad was taken out instead of the bilateral ovaries. All animal procedures followed the guidelines of the Chinese Council for Animal Care.
miRNA extraction and sequencing
Only the femoral heads of femur bones were used for miRNA extraction. After 10 weeks of modeling, femur bones were harvested from all rats. Any attached tissue was quickly removed and samples were immediately snap-frozen in liquid nitrogen and stored at −80°C. Total miRNAs were extracted from frozen tissue using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The miRNA concentration, purity, and integrity number were measured using the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) (Appendix 1). T4 RNA ligase was used to add 3′ and 5′ small RNA sequencing ends to total RNA. The product was then amplified by real-time (RT)-PCR (see below) and purified by polyacrylamide gel electrophoresis to produce a small RNA library. The 2000 Bioanalyzer (Agilent Technologies) was used to conduct quantitative and qualitative analysis of the library. The peak fragment length was defined as 130 to 155 nucleotides and the quantitative value was ≥1 fmol as the library was in good condition. Small RNA library samples were sent to Genex Health Co., Ltd. (Beijing, China) for high-throughput deep sequencing analysis.
miRNA prediction and screening
We predicted target genes of candidate miRNAs using miRWalk (http://mirwalk.umm.uni-heidelberg.de/), miRanda (http://www.microrna.org/microrna/home.do), and TargetScan (http://www.targetscan.org/vert_72/). To minimize the probability of introducing false positives and/or negatives, we selected potential targets that were identified by at least two databases.
Bone samples
Ten postmenopausal women undergoing knee replacement because of osteoporotic fracture were enrolled as the experimental group. Another 10 postmenopausal women diagnosed with osteoarthritis in the absence of osteoporosis, according to bone mineral density (BMD) and T-score measurements, were included as the non-osteoporosis group (Appendix 2). None of the participants had a history of other disease including metabolic or endocrine disease, chronic renal failure, chronic liver disease, malignancies, Paget’s disease of bone, or malabsorption syndrome, or had received hormone replacement therapy, anti-resorptive or anabolic agents, oral corticosteroids, or anti-epileptic drugs, or been treated with lithium, heparin, or warfarin. This study was approved by Guang’anmen Hospital. Written informed consent was obtained from all participants.
Osteogenesis induction
Human BMSCs were purchased from Fusheng Industrial Co., Ltd. (Shanghai, China). To induce osteogenic differentiation of BMSCs, cells were cultured in 6-well or 24-well plates under standard culture conditions of 37°C and 5% CO2. They were maintained in osteogenic induction medium (Cyagen, Santa Clara, CA, USA) until they reached 80% confluence, then were differentiated into osteoblasts using osteogenic differentiation-inducing medium (Cyagen) which was composed of 175 mL culture medium, 10% fetal bovine serum, 1% glutamine, 1% penicillin/streptomycin, 0.2% ascorbic acid, 1% β-glycerophosphate, and 0.01% dexamethasone. The cells were induced into osteoblasts for 14 days and the medium was changed every 3 days.
Alizarin red S staining
BMSCs were seeded at a density of 1 × 104 cells/cm2. The culture medium was removed from cells, then they were washed twice with PBS, and fixed with 4% paraformaldehyde for 10 minutes. Then the fixative solution was discarded from samples, they were washed three times with PBS, Alizarin red S (ARS) staining solution (Solarbio, Beijing, China) was added for 30 minutes, and they were washed a further five times with PBS. The extent of osteogenic differentiation was observed under an optical microscope (IX71; Olympus, Tokyo, Japan).
Alkaline phosphatase (ALP) activity assay
BMSCs and induced osteoblasts were fixed with 3.7% formaldehyde and incubated in a staining solution of 0.25% naphthol AS-BI phosphate and 0.75% Fast Blue BB dissolved in 0.1 M Tris buffer (pH 9.3) for 30 minutes and analyzed by absorbance at 405 nm. ALP activity was quantified using a commercial kit according to the manufacturer’s protocol (Cell Biolab, San Diego, CA, USA).
Osteoclast differentiation
Human bone marrow-derived macrophages (BMMs) were purchased from Procell Life Science & Technology (Wuhan, China). They were seeded in 6-well plates (at 5 × 105 cells/well) or 24-well plates (at 3 × 104 cells/well) and cultured in complete medium with 30 ng/ml macrophage colony-stimulating factor (M-CSF) and 50 ng/ml RANKL (R&D Systems, Minneapolis, MN, USA) for 5 days.
TRAP staining
Mature osteoclasts were visualized using TRAP staining, which is a biomarker of osteoclast differentiation. Briefly, the multinucleated osteoclasts were fixed with 3.7% formalin for 10 minutes, permeabilized with 0.1% Triton X-100 for 10 minutes, and then stained with TRAP solution (Sigma-Aldrich, Saint Louis, MO, USA). TRAP+ multinucleated cells containing three or more nuclei were categorized as osteoclasts.
Cell transfection
miR-708-5p, miR-708-5p, and NC mimics were synthesized by GenePharma (Shanghai, China). CDR1a small interfering (si)RNAs were purchased from RiboBio (Ribobio Co., Guangzhou, China). The plasmid pCMV-SMURF2 (RG203417) was obtained from OriGene (OriGene Technologies, Rockville, MD, USA). The pCD5-ciR-CDR1as plasmid was constructed by GENESEED (Geneseed Biotech, Guangzhou, China). BMSCs or BMMs were seeded into six-well plates at 2 × 104 cells/cm2, then grown to about 70% confluence over 24 to 48 hours in complete culture medium. They were transfected with 50 nM miRNA mimics, miRNA inhibitors, or 4 µg plasmids using Lipofectamine2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. At the indicated time after transfection, the cells were lysed for the extraction of RNA and protein.
Luciferase reporter assay
Genomic DNA was extracted from rat BMSCs using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The SMAD specific E3 ubiquitin protein ligase 2 (SMURF2) 3′ untranslated region (UTR; GenBank accession NM_022739.4) containing predicted miRNA binding sites was PCR-amplified from genomic DNA using specific primers. SMURF2 wild-type (wt) primers were: Fw:5′-CCGCTCGAGGAATGCTGACCCCTGCATCT-3′ (bold and italics represent the Xho I site) and Rev: 5′-ATTTGCGGCCGC AAGCGACTCGGCATTTGGTA-3′ (bold and italics represent the Not I site). SMURF2 3′-UTR mutation primers were: Fw: 5′-GTTCAAAACCACGTGTTCAATCCAACT-3′ and Rev: 5′-ACGTGGTTTTGAACTCGAGGAGTCC-3′ (bold and italics represent the mutation sequence). PCR products were cloned into the psiCHECK2 plasmid (Promega) and all constructs were confirmed by sequencing.
293T cells (Cell Resource Center, Peking Union Medical College, Beijing, China) were transferred to 24-well plates at 70% confluence 24 hours before transfection. All transfections were conducted using Lipofectamine2000 (Invitrogen). Firefly luciferase reporter plasmids containing wt or mutant 3′ UTRs (100 ng), the control renilla luciferase plasmid (pRL-CMV, 5 ng) (Promega), and miRNA mimics (20 nM) were co-transfected into cells using Lipofectamine2000 according to the manufacturer’s protocol. After 48 hours, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen). mRNA was reverse-transcribed using the PrimeScript™ RT reagent Kit (TAKARA Bio, Shiga, Japan) according to the manufacturer’s insturctions. miRNAs were reverse-transcribed using specific Bulge-Loop™ miR qRT-PCR primers (Ribobio Co., Ltd., Guangzhou, China). The reverse transcription products were quantified by RT-PCR using SYBR® Premix Ex Taq™ (TAKARA) and the ABI 7500 Real-Time PCR System (Applied Biosystems, Warrington, UK). The thermo-cycling program was set at 95°C for 10 minutes to denature DNA templates, followed by 40 cycles at 95°C for 15 s, 60°C for 30 s, and 72°C for 30 s.
Respective glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and U6 small nuclear RNA genes were used as endogenous normalization controls for mRNAs and miRNAs. Primers used for amplification are listed in Appendix 3.
Western blot analysis
Cells were washed twice with cold PBS and then lysed in lysis buffer (50 mM Tris, pH 7.5, 250 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 2 mM dithiothreitol, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) on ice for 15 minutes. Cell extracts were collected by centrifugation at 15,000 × g at 4°C for 30 minutes, run on 10% SDS-polyacrylamide gel electrophoresis gels, and transferred to polyvinylidene difluoride membranes by electroblotting. The membranes were blocked for 1 hour in a blocking buffer containing 5% powdered milk in Tris-buffered saline and Tween 20. They were then incubated overnight with a primary antibody at 4°C, followed by incubation with a secondary antibody horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (Zhongshan Biotechnology Co., Beijing, China) for 30 minutes at room temperature, and visualized using a chemiluminescence kit (Thermo Scientific, Waltham, MA, USA). Specific antibodies against BMP2 (Abcam, Cambridge, UK, #ab14933), BMP4 (Abcam, #ab39973), RUNX2 (Abcam, #ab76956), SMURF2 (Abcam, #ab53316), TRAP (Abcam, #ab191406), MMP9 (Abcam, #ab38898), cathepsin K (Abcam, #ab187647), and GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA, sc-25778) were used.
Statistical analysis
All data were processed using SPSS version 19.0 software (IBM, Armonk, NY, USA), expressed as means ± SD, and used in the Shapiro–Wilk (W) test to detect normality. For non-normally distributed data, the nonparametric Wilcoxon signed-rank test was used to evaluate statistical differences between groups. When the data were approximately normally distributed, the comparison of multiple group data was performed using one-way analysis of variance, and multiple comparisons of means were analyzed by the least significant difference method.
Results
Analysis of differentially expressed miRNAs
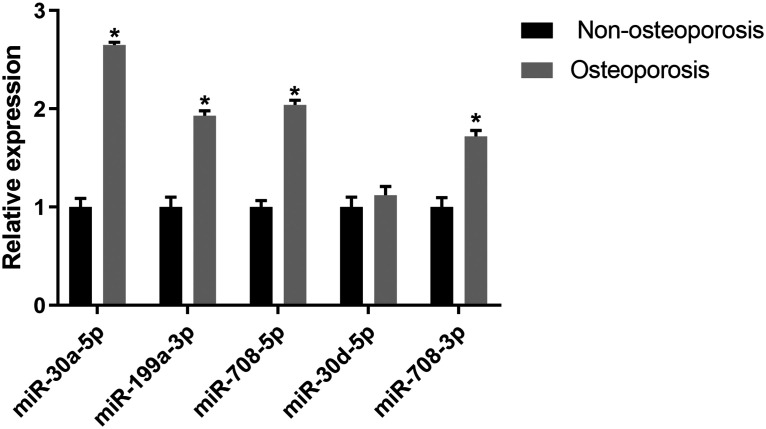
Following miRNA sequencing analysis, we found that 33 miRNAs in the rat model group of osteoporosis were up-regulated compared with the control group (Appendix 4). Table 1 shows the top 10 differentially expressed miRNAs. To verify whether these miRNAs had the same differential expression in osteoporosis patients, we investigated the expression of the top five differentially expressed miRNAs in human bone tissue by qRT-PCR (Figure 1). The expression of miR-30a-5p, miR-199a-3p, miR-708-5p, and miR-708-3p in osteoporosis patients was significantly higher than in the non-osteoporosis group (P < 0.05), which was consistent with findings in the rat model.
Table 1.
miRNAs differentially up-regulated in the rat osteoporosis model.
| miRNA | Fold change | q value |
|---|---|---|
| rno-miR-30a-5p | 12.6518 | 1.03E-21 |
| rno-miR-199a-3p | 8.1760 | 8.13E-15 |
| rno-miR-708-5p | 8.0162 | 5.27E-29 |
| rno-miR-30d-5p | 7.5293 | 1.26E-11 |
| rno-miR-708-3p | 7.6527 | 0.0006281 |
| rno-miR-3588 | 5.7263 | 1.38E-17 |
| rno-miR-26a-5p | 4.2713 | 2.83E-15 |
| rno-miR-21-5p | 4.1733 | 2.58E-07 |
| rno-miR-101b-3p | 3.9173 | 0.00721 |
| rno-miR-664-3p | 3.6283 | 1.82E-17 |
miRNA, microRNA.
Figure 1.
Differences in the expression of corresponding miRNAs in the bone tissue of osteoporosis patients and individuals without osteoporosis by quantitative PCR detection. *P < 0.05 versus non-osteoporosis group.
miRNA, microRNA.
Differential expression patterns of miR-708-5p and miR-708-3p in osteoblasts and osteoclasts
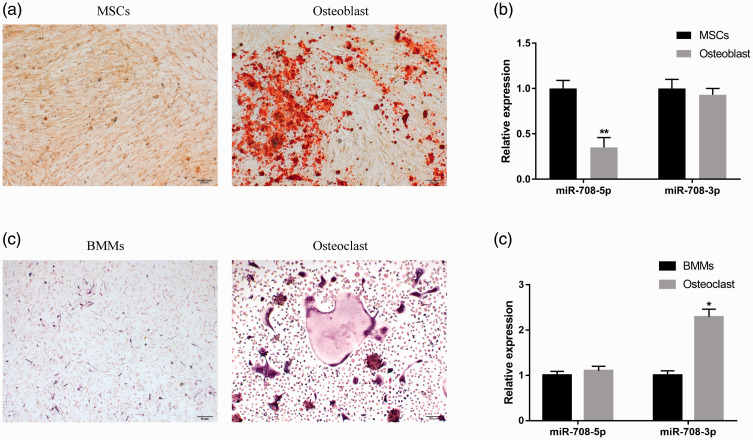
miR-708-5p and miR-708-3p are two mature miRNAs derived from the same precursor miR-708 (pre-miR-708).17 We therefore sought to determine whether differences in homotropism between the two miRNAs had a synergistic or antagonistic effect in osteoporosis. We detected the relative expression of miR-708-5p and miR-708-3p in osteoblast and osteoclast differentiation separately, and found no significant difference in the relative content of miR-708-3p after the osteoblast induction of BMSCs (Figure 2a, b) but a significant down-regulation of miR-708-5p (** P < 0.01, Figure 2b). After the osteoclast differentiation of BMMs (Figure 2c), the expression of miR-708-3p was significantly up-regulated (P < 0.05) but there was no significant difference in miR-708-5p expression (Figure 2d). These results suggest that the functions of miR-708-5p and miR-708-3p in osteoporosis are reflected in the different processes of osteogenesis and osteoclast differentiation.
Figure 2.
The difference in miR-708-5p and miR-708-3p content during osteoblast and osteoclast differentiation. (a) Representative images of Alizarin Red S (ARS) staining of BMSCs after 14 days of osteoblast differentiation. (b) The relative contents of miR-708-5p and 3p before and after BMSC-induced osteogenic differentiation by quantitative PCR. ** P < 0.01 versus BMSCs. (c) Representative TRAP staining of BMMs after 5 days of osteoclast differentiation. BMMs were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) for 5 days. (d) Quantitative PCR was used to detect the difference in miR-708-5p and 3p content before and after BMM-induced osteoclast differentiation. *P < 0.05 versus BMMs.
BMSCs, bone marrow mesenchymal stem cells; BMMs, bone marrow-derived macrophages; TRAP, tartrate-resistant acid phosphatase; M-CSF, macrophage colony-stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand.
miR-708-5p inhibits osteoblast differentiation
To better understand the role of miR-708-5p in the osteoblast differentiation of BMSCs, we overexpressed miR-708-5p mimics by transfecting them into BMSCs during the induction of osteoblast differentiation. Compared with the miR-NC group, osteoblast differentiation of the miR-708-5p mimics group was inhibited (Figure 3a) and ALP activity was significantly decreased (P < 0.05, Figure 3b). mRNA and protein expression levels of osteogenic markers such as BMP2, BMP4, and Runx2 were significantly decreased (P < 0.01, Figure 3c, d), suggesting that miR-708-5p inhibits osteoblast differentiation.
Figure 3.
miR-708-5p negatively regulates the osteoblast differentiation of BMSCs. (a) Alizarin Red S staining (100× magnification) showing that mir-708-5p inhibited the deposition of osteogenic calcium 14 days after BMSC osteoblast induction. (b) Quantitative analysis of ALP activity in BMSCs. mir-708-5p inhibited the ALP activity of osteoblasts. (c, d) Quantitative PCR and western blot analyses showing the effect of miR-708-5p on the mRNA and protein expression of BMP2, BMP4, and Runx2 in osteoblasts. Data are the means ± SD of three independent experiments. *P < 0.05 and ** P < 0.01 versus control.
OSI, osteogenic induction group; CON, non-osteogenic induction group; BMSCs, bone marrow mesenchymal stem cells; ALP, alkaline phosphatase; BMP, bone morphogenetic protein; Runx2, Runt-related transcription factor 2.
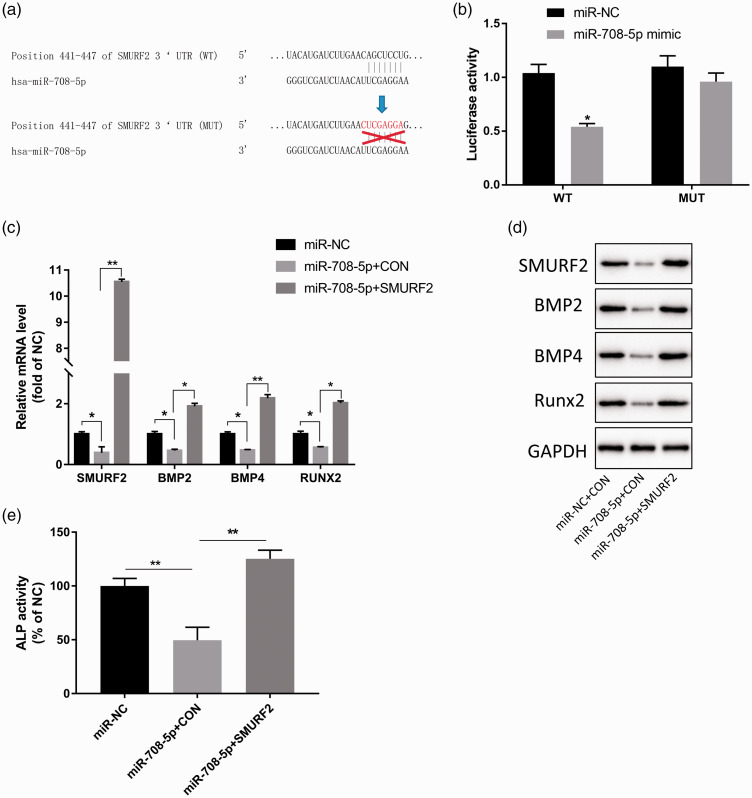
miR-708-5p directly targets SMURF2
To further investigate the molecular mechanism by which miR-708-5p regulates the osteoblast differentiation of BMSCs, we identified its potential target molecules. Multiple target prediction tools, such as miRWalk (http://mirwalk.umm.uni-heidelberg.de/), miRanda (http://www.microrna.org/microrna/home.do), and TargetScan (http://www.targetscan.org/vert_72/), showed that there was a potential binding site for miR-708-5p in the 3′ UTR of SMURF2 (Figure 4a). Additional analysis showed that miR-708-5p mimics had significant inhibitory effects on the luciferase activity of the wt SMURF2 3′ UTR (P < 0.05) but no obvious effects on the luciferase activity of the SMURF2 3′ UTR with a mutated miR-708-5p binding site (Figure 4b). To further explore the mechanism of miR-708-5p/SMURF2, we overexpressed miR-708-5p or SMURF2 by transfecting them during the osteoblast differentiation of BMSCs. miR-708-5p overexpression significantly down-regulated the mRNA and protein expression of osteogenic markers such as BMP2, BMP4, and Runx2 (P < 0.01, Figure 4c, d) and significantly decreased ALP activity (P < 0.01, Figure 4e), while SMURF2 overexpression partially attenuated the down-regulation of miR-708-5p on BMP2, BMP4, and Runx2 expression, as well as the inhibition of ALP activity (P < 0.01, Figure 4c–e). These results suggest that miR-708-5p inhibits osteoblast differentiation by targeting SMURF2.
Figure 4.
miR-708-5p influences the differentiation of osteoblasts by targeting SMURF2. (a) miR-708-5p was predicted to target SMURF2 via TargetScan. (b) miR-708-5p targeting of SMURF2 was verified by dual-luciferase reporter gene assay. (c, d) Quantitative PCR and western blot analyses showing the effect of miR-708-5p and SMURF2 overexpression on the mRNA and protein expression of osteoblast-related markers BMP2, BMP4, and Runx2. (e) Quantitative analysis of the effect of miR-708-5p, miR-708-5p, and SMURF2 overexpression on ALP activity. Data are the means ± SD of three independent experiments. *P < 0.05 and ** P < 0.01 versus control.
SMURF2, SMAD specific E3 ubiquitin protein ligase 2; BMP, bone morphogenetic protein; Runx2, Runt-related transcription factor 2.
The effect of miR-708-3p on osteoclast differentiation
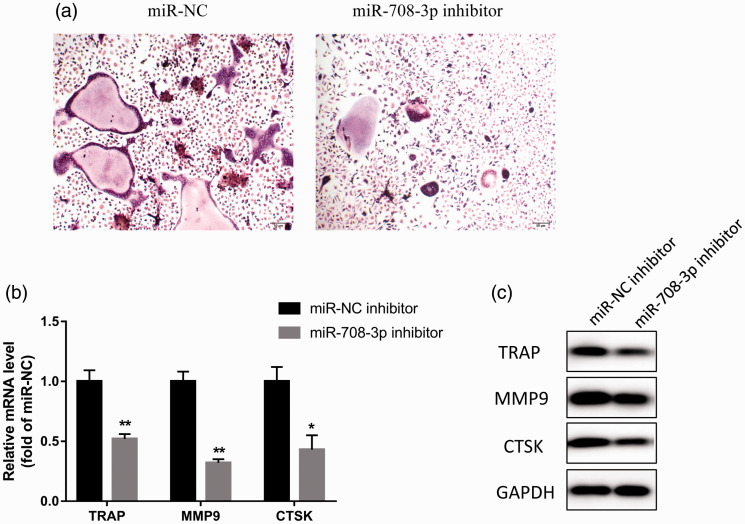
Next, we explored the role of miR-708-3p in the osteoclast differentiation of BMMs. Because miR-708-3p is up-regulated after the osteoclast induction of BMMs (Figure 2d), we transfected miR-708-3p inhibitor into BMMs to investigate its effect on BMM osteoclast differentiation. TRAP staining showed that miR-708-3p inhibitor inhibited osteoclast differentiation compared with the miR-NC inhibitor group (Figure 5a). Furthermore, miR-708-3p inhibitor also significantly inhibited the mRNA and protein expression of osteoclast differentiation markers TRAP, MMP9, and CTSK (P < 0.01, Figure 5b, c). These results show that miR-708-3p inhibitor inhibits osteoclast differentiation, and by extrapolation suggest that miR-708-3p promotes osteoclast differentiation.
Figure 5.
miR-708-3p inhibitor inhibits osteoclast-induced differentiation of BMMs. (a) BMMs were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml) for 5 days. TRAP-stained cells showing the fusion of BMMs on day 5. (b, c) Detection of the effects of mir-708-3p inhibitor on the mRNA and protein expression of TRAP, MMP9, and CTSK during osteoclast differentiation by quantitative PCR and western blot analyses. Data are the means ± SD of three independent experiments. *P < 0.05 and ** P < 0.01 versus control.
BMMs, bone marrow-derived macrophages; M-CSF, macrophage colony-stimulating factor; RANKL, receptor activator of nuclear factor-κB ligand; TRAP, tartrate-resistant acid phosphatase; MMP, matrix metalloproteinase; CTSK, cathepsin K.
Cerebellar degeneration associated protein 1 antisense RNA (CDR1as) is the target gene of miR-708-3p
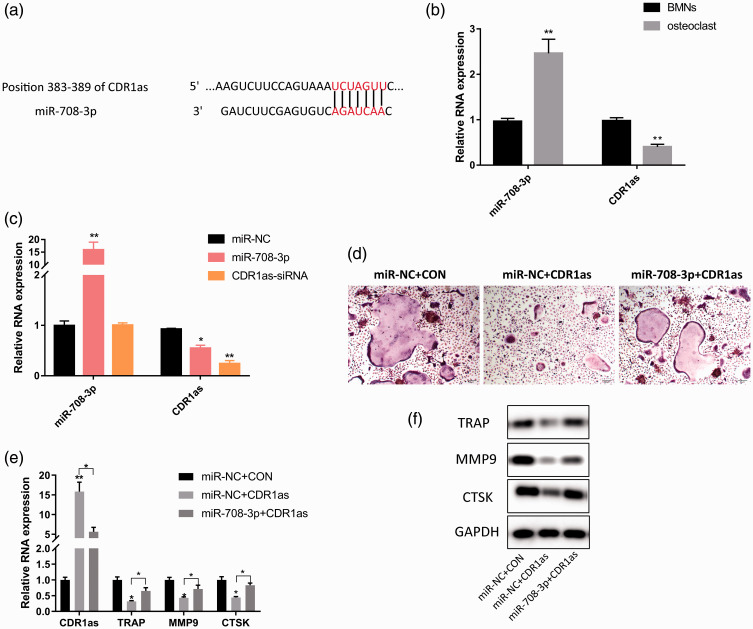
TargetScan showed that CDR1as ranked first as a potential target of miR-708-3p and that its sequence contained a binding site of miR-708-3p (Figure 6a). We next measured the relative content of miR-708-3p and found that it was significantly higher after BMM differentiation into osteoclasts than before, while the relative content of CDR1as was the opposite (P < 0.01, Figure 6b). Transfection of miR-708-3p mimics into BMMs significantly down-regulated the relative expression level of CDR1as (P < 0.01, Figure 6c), which was similar to the findings of the CDR1as siRNA group, revealing the negative regulatory effect of miR-708-3p on CDR1as. To further explore the role of miR-708-3p/CDR1as in osteoclast differentiation, we overexpressed pCD5-ciR-CDR1as plasmids and mir-708-3p mimics in BMMs during osteoclast differentiation. CDR1as overexpression significantly inhibited osteoclast differentiation and down-regulated the mRNA and protein expression of osteoclast markers such as TRAP, MMP9, and CTSK (P < 0.05, Figure 6d, e). However, miR-708-3p overexpression only partially weakened the inhibitory effect of CDR1as on osteoclast differentiation (Figure 6d, e). These results suggest that miR-708-3p affects the differentiation of osteoclasts by the targeted regulation of CDR1as.
Figure 6.
miR-708-3p affects osteoclast differentiation by the targeted regulation of CDR1as. (a) CDR1as is a potential target of miR-708-3p, and its sequence contains miR-708-3p binding sites. (b) Quantitative PCR detection of the relative content of miR-708-3p and CDR1a in BMMs before and after osteoblast induction. (c) Quantitative PCR detection of the inhibitory effect of miR-708-3p mimics and CDR1a siRNA on CDR1as expression. (d) Representative TRAP staining of BMMs after 5 days of osteoclast differentiation. (e) Quantitative PCR analysis of the effects of CDR1as on the relative expression levels of osteoclast-related markers TRAP, MMP9, and CTSK mRNA during osteoclast differentiation. (f) Western blotting analysis of the effect of CDR1as on the relative protein expression levels of TRAP, MMP9, and CTSK during osteoclast differentiation. Data are the means ± SD of three independent experiments. *P < 0.05 and ** P < 0.01 versus control.
CDR1as, cerebellar degeneration associated protein 1 antisense RNA; BMMs, bone marrow-derived macrophages; TRAP, tartrate-resistant acid phosphatase; MMP, matrix metalloproteinase; CTSK, cathepsin K.
Discussion
The growth, development, and metabolism of bone involve two types of cells: osteoblasts, which are responsible for forming new bone tissue, and osteoclasts, which are responsible for absorbing old bone tissue. Through complex interactions between osteoblasts and osteoclasts, the body forms new bone tissue and absorbs old bone tissue in a dynamic balance state to realize the stable maintenance of the skeletal system.18,19 The breakdown of this homeostasis leads to diseases of the skeletal system, such as osteoporosis. The risk of fracture is greatly increased in patients with osteoporosis, and one-quarter of patients with hip fractures were shown to die within 1 year of the fracture.20 Although pyrophosphate and other drugs have been widely used in clinics, they only specifically inhibit bone resorption mediated by osteoclasts, cannot improve the bone-formation ability, and lead to poor bone reconstruction.21 However, there is a lack of more effective treatment options for osteoporosis, so basic research into orthopedics and studying the steady-state balance of the skeletal system will be beneficial.
miRNAs have been found to regulate gene expression in physiological and pathophysiological processes and to be involved in osteoblast and osteoclast differentiation by targeting the expression of key regulatory factors and receptors. In this study, we used the OVX osteoporosis model in rats to show the significant up-regulation of several miRNAs. Dozens of miRNAs are considered to play a role in osteoporosis,22 but the regulatory role of miR-708-5p and miR-708-3p in osteoporosis has rarely been reported. De-La-Cruz-Montoya et al23 showed for the first time that miR-708-5p was up-regulated in peripheral blood mononuclear cells of Mexican women with osteoporosis, and could be used as a marker for osteoporosis. Of note, miR-708-5p and miR-708-3p are processed from the same pre-miRNA, while miR-708-3p usually exists as a passenger strand (miRNA*) and more studies focus on mir-708-5p as a guide-strand. Earlier work suggested that the conservation of passenger-strand miRNAs was lower than that of guide-strand miRNAs, and that their RNA-induced silencing complex was more difficult to form and more readily degraded.24 However, more recent studies indicate that passenger-strand miRNAs play an important role in the regulation of gene expression.24–28 Therefore, whether miR-708-3p has a biological function in osteoblast or osteoclast differentiation is also our focus.
Through in vitro experiments, we found that miR-708-5p was down-regulated in osteoblast differentiation. miR-708-5p overexpression reduced the formation of mineralized tuberculosis (Figure 3a), as well as the expression of BMP2, BMP4, and Runx2 (Figure 3c, d), and ALP activity (Figure 3b). These results clearly indicate that miR-708-5p inhibits the differentiation of osteoblasts.
SMURF2 belongs to the NEDD4 ubiquitin ligase enzyme protein family. All members of this family have a similar molecular structure with a C-terminal HECT domain, an N-terminal C2 domain, and 2 to 4 central WW domains.29 Xu et al.30 showed that SMURF2 is a key factor in osteoblasts that affects homeostasis of the skeletal system. By regulating the activity of SMURF2 in osteoblasts and the interaction between SMAD3 and the vitamin D receptor, it was possible to control bone remodeling and bone density. They also found that conditional knockout of Smurf2 increased RANKL expression, enhanced osteoclast differentiation and bone resorption, and decreased bone mineral density in mice, suggesting that osteoblast and osteoclast differentiation are closely related. In this study, we found that the expression of SMURF2 was targeted by miR-708-5p and that SMURF2 overexpression partially alleviated the inhibitory effect of miR-708-5p on osteoblast differentiation. Therefore, miR-708-5p can inhibit osteoblast differentiation by targeting SMURF2. We also showed that the overexpression of miR-708-3p enhanced osteoclast-specific marker genes such as TRAP, MMP9, and CTSK, and promoted the differentiation of osteoclasts. Through further target gene prediction and experimental verification, we identified CDR1as as the target regulatory gene of miR-708-3p.
CDR1as, a natural antisense transcript of CDR1, was first identified by Hansen in 2011 as a circular RNA (circRNA).31 CircRNAs compete with endogenous RNA to regulate gene transcription, interact with proteins, and encode proteins. CDR1as is one of the most widely studied circRNAs, and has been shown to bind and regulate miRNAs including miR-7,32,33 miR-671,34 miR-1299,35 miR-876-5p,36 and miR-135a.37 Hansen et al.38 reported that human CDR1as contains 74 miRNA-7 binding targets, of which more than 60 are conserved. Therefore, it functions as an miRNA-7 sponge to regulate activity of the miRNA-7 target gene. Yu et al.39 found that CDR1as down-regulation and miRNA-7 up-regulation reduced the proliferation and invasiveness of hepatoma cells. Although most research on CDR1as has focused on tumors,40,41 metabolism,42 and the nervous system,43,44 Li et al.45 reported CDR1as to be up-regulated during osteoblast differentiation. They also found that CDR1as knockout and miR-7 overexpression inhibited ALP activity, ARS staining, and osteogenic gene expression, while in vivo micro computed tomography and histological analysis showed that CDR1as gene knockout reduced bone formation compared with the control group. However, Chen et al.46 observed the up-regulation of CDR1as in BMSCs of patients with steroid-induced osteonecrosis of the femoral head. Their work revealed that CDR1as gene knockout increased osteoblast differentiation and decreased adipogenic differentiation of BMSCs, while CDR1as overexpression decreased osteoblast differentiation and increased adipogenic differentiation of BMSCs. Therefore, current findings on the function of CDR1as in the osteogenic differentiation process are inconsistent and more in-depth research is needed. Our results show that CDR1as is significantly reduced in osteoclast differentiation, and that its overexpression reduces TRAP, MMP9, and CTSK expression and inhibits osteoclast differentiation (Figure 6d, f). These findings are the first, to our knowledge, to show that the miR-708-3p/CDR1as pathway affects the differentiation of osteoclasts.
Our study was limited in that only bone marrow-derived macrophages were used as progenitors for osteoclast formation in vitro. Both peripheral blood monocytes (PBMCs) and bone-derived macrophages are induced into osteoclast-like cells. Rafael et al.23 previously detected the up-regulation of miR-708-5p in peripheral blood mononuclear cells of Mexican osteoporotic women, but we did not observe this in the osteoclast induction of BMMs. Therefore, future work should use PBMCs as the precursor to induce osteoclasts to detect changes in miR-708-5p expression because miRNAs in precursor cells from different sources may have different trends. Another interesting avenue of exploration would be to investigate whether PBMCs from osteoporosis patients have high expression of miR-708-5p, suggesting an inhibitory effect of osteoclast progenitors through miR-708-5p on osteogenesis.
Conclusions
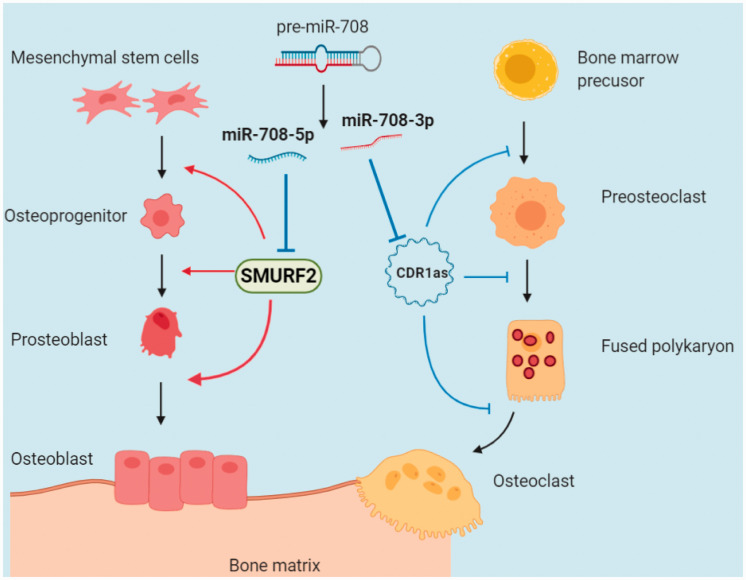
Osteoporosis is the result of an imbalance between osteoblast and osteoclast differentiation. Our results show that miR-708-5p inhibits osteoclast differentiation by targeting SMURF2, while miR-708-3p promotes osteoclast differentiation by regulating the expression of CDR1as. This reveals that miR-708-5p and miR-708-3p affect osteoblast and osteoclast differentiation through separate mechanisms and play a cooperative role with different spatial and temporal patterns in the development of osteoporosis (Figure 7). These findings provide a target and a theoretical basis for the clinical treatment of osteoporosis.
Figure 7.
miR-708-5p and miR-708-3p from the same microRNA precursor accelerate the progression of osteoporosis with different temporal and spatial patterns through their regulatory effects on osteoblast and osteoclast differentiation.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Appendices
Appendix 1. RNA integrity values.
| Sample | ID | RNA (ng/µL) | A260/280 | A260/230 | RIN |
|---|---|---|---|---|---|
| OVX | R201927006 | 162.5 | 1.97 | 1.01 | 5.2 |
| OVX | R201927007 | 290.51 | 1.97 | 1.91 | 6.9 |
| OVX | R201927008 | 209.62 | 2.01 | 1.52 | 8.2 |
| Non-OVX | R201927009 | 280.32 | 1.94 | 1.69 | 6.1 |
| Non-OVX | R201927010 | 184.17 | 1.99 | 1.64 | 7.5 |
| Non-OVX | R201927011 | 220.69 | 1.93 | 1.29 | 6.2 |
| OP | H1 | 174.54 | 1.64 | 0.53 | 8.2 |
| OP | H2 | 84.17 | 1.65 | 0.77 | 5.5 |
| OP | H3 | 270.3 | 1.81 | 1.4 | 6.5 |
| OP | H4 | 130.83 | 1.92 | 1.26 | 6.2 |
| OP | H5 | 159.88 | 1.84 | 0.98 | 6 |
| OP | H6 | 128.42 | 1.78 | 1.41 | 6.7 |
| OP | H7 | 80.1 | 2.09 | 0.82 | 6.6 |
| OP | H8 | 274.5 | 1.9 | 1.52 | 4.8 |
| OP | H9 | 102.66 | 2.06 | 1 | 6.9 |
| OP | H10 | 90.09 | 2.05 | 1.05 | 5.9 |
| Non-OP | H11 | 253.69 | 1.94 | 0.93 | 5.4 |
| Non-OP | H12 | 147.95 | 1.99 | 1.5 | 5.1 |
| Non-OP | H13 | 191.88 | 1.87 | 0.97 | 7 |
| Non-OP | H14 | 178.5 | 2.05 | 1.39 | 5.6 |
| Non-OP | H15 | 204.38 | 1.6 | 1.35 | 6.5 |
| Non-OP | H16 | 141.99 | 1.92 | 0.72 | 6.6 |
| Non-OP | H17 | 111.14 | 2.02 | 1.16 | 5.6 |
| Non-OP | H18 | 274.02 | 1.78 | 0.76 | 6.3 |
| Non-OP | H19 | 196.73 | 2.05 | 0.66 | 6.9 |
| Non-OP | H20 | 268.55 | 2.01 | 0.95 | 7 |
OVX, ovariectomized; OP, osteoporosis; RIN, RNA integrity value.
Appendix 2. Patient characteristics.
| n | Age (years) | T-score | BMD (g/cm2) | |
|---|---|---|---|---|
| Osteoporotic | 10 | 63.16 ± 3.14 | –3.33 ± 0.35 | 0.47 ± 0.07 |
| Non-osteoporotic | 10 | 62.53 ± 7.32 | 0.52 ± 0.47 | 0.84 ± 0.04 |
Data are expressed as the mean ± SD.
BMD, bone mineral density.
Appendix 3. Primer sequences used in qRT-PCR.
| Gene name | Primer sequence (5′–3′) | Reference sequence |
|---|---|---|
| SMURF2 | F: AGAGCTTGGTCCATTGCCTC R: CTGAACCAGGTCTCGCTTGT | NM_022739.4 |
| BMP2 | F: ACTCGAAATTCCCCGTGACCR: CCACTTCCACCACGAATCCA | NM_001200.4 |
| BMP4 | F: TGGGATTCCCGTCCAAGCTATR: AAACGACCATCAGCATTCGG | NM_001202.6 |
| Runx2 | F: CGCCTCACAAACAACCACAGR: TCACTGTGCTGAAGAGGCTG | NM_001024630.4 |
| TRAP | F: CGACGGCAGGGACTGAAGR: AGTCACCCACGGCTACAAAG | NM_001111035.3 |
| MMP9 | F: TCTATGGTCCTCGCCCTGAAR: CATCGTCCACCGGACTCAAA | NM_004994.3 |
| CTSK | F: GACACCCACTGGGAGCTATGR: AACAGGAACCACACTGACCC | NM_000396.4 |
| CDR1as | F: AGGTTTTCTGGTGTCTGCCGR: TGGAAGACGCAGGCTTTTCT | AK094540.1 |
| GAPDH | F: GGTGGTCTCCTCTGACTTCAACAR: GTGGTCGTTGAGGGCAATG | NM_002046 |
| U6 | F: CTCGCTTCGGCAGCACAR: GCGAGCACAGAATTAATACGAC | NG_034215.1 |
Appendix 4. Up-regulated miRNAs in the rat osteoporosis model compared with the sham-operated group (fold-change ≥1).
| miRNA | Fold change | q value |
|---|---|---|
| rno-miR-30a-5p | 12.6518 | 1.03E-21 |
| rno-miR-199a-3p | 8.1760 | 8.13E-15 |
| rno-miR-708-5p | 8.0162 | 5.27E-29 |
| rno-miR-30d-5p | 7.5293 | 1.26E-11 |
| rno-miR-708-3p | 7.6527 | 0.0006281 |
| rno-miR-3588 | 5.7263 | 1.38E-17 |
| rno-miR-26a-5p | 4.2713 | 2.83E-15 |
| rno-miR-21-5p | 4.1733 | 2.58E-07 |
| rno-miR-101b-3p | 3.9173 | 0.00721 |
| rno-miR-664-3p | 3.6283 | 1.82E-17 |
| rno-miR-23b-3p | 3.5165 | 1.16E-11 |
| rno-miR-497-5p | 3.4831 | 1.15E-10 |
| rno-miR-19b-3p | 3.4592 | 7.42E-11 |
| rno-miR-181a-5p | 3.4021 | 8.16E-05 |
| rno-miR-25-3p | 3.3217 | 1.75E-27 |
| rno-miR-674-3p | 3.2876 | 0.00063 |
| rno-miR-93-5p | 3.1834 | 1.42E-18 |
| rno-miR-434-5p | 3.1293 | 9.62E-16 |
| rno-miR-322-5p | 3.6373 | 6.26E-12 |
| rno-miR-101a-3p | 3.0721 | 1.26E-11 |
| rno-miR-24-3p | 3.0268 | 0.000717 |
| rno-miR-10a-3p | 2.8444 | 0.000661 |
| rno-miR-140-5p | 2.6785 | 0.000535 |
| rno-miR-30e-5p | 2.497 | 0.000518 |
| rno-miR-9a-5p | 2.1498 | 0.000415 |
| rno-miR-125a-5p | 1.9628 | 0.000391 |
| rno-miR-194-5p | 1.698 | 0.000315 |
| rno-miR-191a-5p | 1.6571 | 0.000276 |
| rno-miR-103-3p | 1.5955 | 0.000171 |
| rno-miR-598-3p | 1.5924 | 0.000151 |
| rno-miR-17-5p | 1.5838 | 0.000133 |
| rno-miR-598-3p | 1.4839 | 7.17E-05 |
| rno-miR-17-5p | 1.3101 | 4.71E-05 |
miRNA, microRNA.
Footnotes
Authors’ contributions: CRW and SH conceived and designed the present study. RW, JC, HX, HH, SZ, YW, and YF conducted the experiments and collected the data. RW, GX, and SH analyzed and interpreted the data. RW and SH drafted the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interest: The authors declare that there is no conflict of interest.
Funding: This work was supported by the Beijing Traditional Chinese Medicine Technology Development Fund Project [No. JJ2018-59] and the Construction Project of Famous Doctor Studio in Beijing Daxing District [No. DXMY-02-WRR].
ORCID iD: Shengnan Huang https://orcid.org/0000-0003-1099-120X
References
- 1.Maeda SS, Lazaretti-Castro M. An overview on the treatment of postmenopausal osteoporosis. Arq Bras Endocrinol Metabol 2014; 58: 162–171. [DOI] [PubMed] [Google Scholar]
- 2.Niedermair T, Schirner S, Seebröker R, et al. Substance P modulates bone remodeling properties of murine osteoblasts and osteoclasts. Sci Rep 2018; 8: 9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikuta J, Ishii M. Osteoclast migration, differentiation and function: novel therapeutic targets for rheumatic diseases. Rheumatology (Oxford) 2013; 52: 226–234. [DOI] [PubMed] [Google Scholar]
- 4.Teitelbaum SL. Bone resorption by osteoclasts. Science 2000; 289: 1504–1508. [DOI] [PubMed] [Google Scholar]
- 5.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol 2007; 7: 292–304. [DOI] [PubMed] [Google Scholar]
- 6.Vimalraj S, Arumugam B, Miranda PJ, et al. Runx2: Structure, function, and phosphorylation in osteoblast differentiation. Int J Biol Macromol 2015; 78: 202–208. [DOI] [PubMed] [Google Scholar]
- 7.Allison H, McNamara LM. Inhibition of osteoclastogenesis by mechanically stimulated osteoblasts is attenuated during estrogen deficiency. Am J Physiol Cell Physiol 2019; 317: C969–C982. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Kim K, Kim I, et al. Endoplasmic reticulum-bound transcription factor CREBH stimulates RANKL-induced osteoclastogenesis. J Immunol 2018; 200: 1661–1670. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Qiu M, Dou C, et al. MicroRNAs in bone balance and osteoporosis. Drug Dev Res 2015; 76: 235–245. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, LV Q, LV C. MicroRNA-153 suppresses the osteogenic differentiation of human mesenchymal stem cells by targeting bone morphogenetic protein receptor type II. Int J Mol Med 2015; 36: 760–766. [DOI] [PubMed] [Google Scholar]
- 11.Huang Y, Hou Q, Su H, et al. miR‑488 negatively regulates osteogenic differentiation of bone marrow mesenchymal stem cells induced by psoralen by targeting Runx2. Mol Med Rep 2019; 20: 3746–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Zhang J, Shao H, et al. miRNA-340 inhibits osteoclast differentiation via repression of MITF. Biosci Rep 2017; 37: BSR20170302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Zhang W, Huang Y. MiRNA-133a is involved in the regulation of postmenopausal osteoporosis through promoting osteoclast differentiation. Acta Biochim Biophys Sin (Shanghai) 2018; 50: 273–280. [DOI] [PubMed] [Google Scholar]
- 14.Zhu XB, Lin WJ, Lv C, et al. MicroRNA-539 promotes osteoblast proliferation and differentiation and osteoclast apoptosis through the AXNA-dependent Wnt signaling pathway in osteoporotic rats. J Cell Biochem 2018; 119: 8346–8358. [DOI] [PubMed] [Google Scholar]
- 15.Lelovas PP, Xanthos TT, Thoma SE, et al. The laboratory rat as an animal model for osteoporosis research. Comp Med 2008; 58: 424–430. [PMC free article] [PubMed] [Google Scholar]
- 16.Egermann M, Goldhahn J, Schneider E. Animal models for fracture treatment in osteoporosis. Osteoporos Int 2005; 16: S129–S138. [DOI] [PubMed] [Google Scholar]
- 17.Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007; 129: 1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature 2003; 423: 349–355. [DOI] [PubMed] [Google Scholar]
- 19.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature 2003; 423: 337–342. [DOI] [PubMed] [Google Scholar]
- 20.Haleen S, Lutchman L, Mayahi R, et al. Mortality following hip fracture: trends and geographical variations over the last 40 years. Injury 2008; 39: 1157–1163. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqui JA, Partridge NC. Physiological bone remodeling: systemic regulation and growth factor involvement. Physiology (Bethesda) 2016; 31: 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lian JB, Stein GS, Van Wijnen AJ, et al. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 2012; 8: 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De-La-Cruz-Montoya AH, Ramírez-Salazar EG, Martínez-Aguilar MM, et al. Identification of miR-708-5p in peripheral blood monocytes: Potential marker for postmenopausal osteoporosis in Mexican-Mestizo population. Exp Biol Med (Maywood) 2018; 243: 1027–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meijer HA, Smith EM, Bushell M. Regulation of miRNA strand selection: follow the leader. Biochem Soc Trans 2014; 42: 1135–1140. [DOI] [PubMed] [Google Scholar]
- 25.Mah SM, Buske C, Humphries RK, et al. miRNA*: a passenger stranded in RNA-induced silencing complex. Crit Rev Eukaryot Gene Expr 2010; 20: 141–148. [DOI] [PubMed] [Google Scholar]
- 26.Eichner LJ, Perry MC, Dufour CR, et al. miR-378(∗) mediates metabolic shift in breast cancer cells via the PGC-1β/ERRγ transcriptional pathway. Cell Metab 2010; 12: 352–361. [DOI] [PubMed] [Google Scholar]
- 27.Jiang L, Lin C, Song L, et al. MicroRNA-30e* promotes human glioma cell invasiveness in an orthotopic xenotransplantation model by disrupting the NF-κB/IκBα negative feedback loop. J Clin Invest 2012; 122: 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Lee UJ, Kim MN, et al. MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J Biol Chem 2008; 283: 18158–18166. [DOI] [PubMed] [Google Scholar]
- 29.Fu L, Cui CP, Zhang X, et al. The functions and regulation of Smurfs in cancers. Semin Cancer Biol 2019: S1044-579X(19)30423-7. [DOI] [PubMed] [Google Scholar]
- 30.Xu Z, Greenblatt MB, Yan G, et al. SMURF2 regulates bone homeostasis by disrupting SMAD3 interaction with vitamin D receptor in osteoblasts. Nat Commun 2017; 8: 14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495: 384–388. [DOI] [PubMed] [Google Scholar]
- 33.Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17: 205–211. [DOI] [PubMed] [Google Scholar]
- 34.Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol Cell 2017; 66: 22–37.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sang M, Meng L, Liu S, et al. Circular RNA ciRS-7 Maintains metastatic phenotypes as a ceRNA of miR-1299 to target MMPs. Mol Cancer Res 2018; 16: 1665–1675. [DOI] [PubMed] [Google Scholar]
- 36.Sang M, Meng L, Sang Y, et al. Circular RNA ciRS-7 accelerates ESCC progression through acting as a miR-876-5p sponge to enhance MAGE-A family expression. Cancer Lett 2018; 426: 37–46. [DOI] [PubMed] [Google Scholar]
- 37.Li P, Yang X, Yuan W, et al. CircRNA-Cdr1as exerts anti-oncogenic functions in bladder cancer by sponging microRNA-135a. Cell Physiol Biochem 2018; 46: 1606–1616. [DOI] [PubMed] [Google Scholar]
- 38.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013; 73: 5609–5612. [DOI] [PubMed] [Google Scholar]
- 39.Yu L, Gong X, Sun L, et al. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One 2016; 11: e0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Yang W, Gu J, Wang X, et al. Inhibition of circular RNA CDR1as increases chemosensitivity of 5-FU-resistant BC cells through up-regulating miR-7. J Cell Mol Med 2019; 23: 3166–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanniford D, Ulloa-Morales A, Karz A, et al. Epigenetic silencing of CDR1as drives IGF2BP3-mediated melanoma invasion and metastasis. Cancer Cell 2020; 37: 55–70.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stoll L, Sobel J, Rodriguez-Trejo A, et al. Circular RNAs as novel regulators of β-cell functions in normal and disease conditions. Mol Metab 2018; 9: 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495: 333–338. [DOI] [PubMed] [Google Scholar]
- 44.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 2013; 4: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther 2018; 9: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen G, Wang Q, Li Z, et al. Circular RNA CDR1as promotes adipogenic and suppresses osteogenic differentiation of BMSCs in steroid-induced osteonecrosis of the femoral head. Bone 2020; 133: 115258. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.