Abstract
Purpose
To define genetic variants associated with variable severity of X-linked progressive retinal atrophy 1 (XLPRA1) caused by a five-nucleotide deletion in canine RPGR exon ORF15.
Methods
A genome-wide association study (GWAS) was performed in XLPRA1 phenotype informative pedigree. Whole genome sequencing (WGS) was used for mutational analysis of genes within the candidate genomic region. Retinas of normal and mutant dogs were used for gene expression, gene structure, and RNA duplex analyses.
Results
GWAS followed by haplotype phasing identified an approximately 4.6 Mb candidate genomic interval on CFA31 containing seven protein-coding genes expressed in retina (ROBO1, ROBO2, RBM11, NRIP1, HSPA13, SAMSN1, and USP25). Furthermore, we identified and characterized two novel lncRNAs, ROBO1-AS and ROBO2-AS, that display overlapping gene organization with axon guidance pathway genes ROBO1 and ROBO2, respectively, producing sense-antisense gene pairs. Notably, ROBO1-AS and ROBO2-AS act in cis to form lncRNA/mRNA duplexes with ROBO1 and ROBO2, respectively, suggesting important roles for these lncRNAs in the ROBO regulatory network. A subsequent WGS identified candidate genes within the genomic region on CFA31 that might be implicated in modifying severity of XLPRA1. This approach led to discovery of genetic variants in ROBO1, ROBO1-AS, ROBO2-AS, and USP25 that are strongly associated with the XLPRA1 moderate phenotype.
Conclusions
The study provides new insights into the genetic basis of phenotypic variation in severity of RPGRorf15-associated retinal degeneration. Our findings suggest an important role for ROBO pathways in disease progression further expanding on our previously reported changes of ROBO1 expression in XLPRA1 retinas.
Keywords: X-linked retinal degeneration, genetic variants, genotype-phenotype correlation, animal disease model
Retinitis pigmentosa (RP) is a large heterogeneous grouping of inherited retinal degenerative diseases leading to photoreceptor cell death and blindness.1 As a class, X-linked RP (XLRP) comprise some of the most severe diseases, and accounts for 10% to 20% of all RP cases.2 Although six disease loci have been mapped on the X-chromosome (RP2, RP3, RP6, RP23, RP24, and RP34; https://sph.uth.edu/retnet/disease.htm) approximately 75% of XLRP cases map to the RP3 locus3,4 that encodes the disease causative gene retinitis pigmentosa GTPase regulator (RPGR).5,6 RPGR-XLRP demonstrates considerable allelic heterogeneity, and more than 350 different RPGR sequence variants have been identified to date in patients with XLRP (http://rpgr.hgu.mrc.ac.uk).
The RPGR gene is subject to alternative splicing and two major isoforms are expressed in the retina.5,7,8 The constitutive RPGR isoform is encoded by exons 1 through 19, whereas the RPGRorf15 variant is encoded by exons 1 through 14 and terminates with a large alternative purine-rich ORF15 exon derived from the extension of exon 15 into intron 15. The RPGRorf15 plays a critical role in retinal function and viability, as multiple disease-causing mutations in the ORF15 exon have been identified in humans, dogs, and mice.6,9,10 Furthermore, gene augmentation therapy with RPGRorf15 preserves photoreceptors function and prevents their degeneration in animal disease models.11,12
Extensive phenotypic diversity is observed between patients with different RPGR mutations, and even between patients in families with the same mutation.13,14 The molecular basis of such clinical variability in XLRP is poorly understood. It might be partially due to RPGR allelic heterogeneity13 but it can also reflect environmental or genetic background influences. Genetic variants contributing to background effects are termed “modifiers,” and these may alter the penetrance or expressivity of a pathogenic variant.13,15–18 It has been previously shown that the presence of common variant in RPGR-interacting protein NPHP5 (I393N) is associated with more severe disease in patients with XLRP with mutations in RPGR exons 1 to 14,13 suggesting that variation in NPHP5 may serve as modifier for the disease. However, to date, there were no genetic variant(s) reported to be associated with severity of retinal degeneration caused by mutations in the RPGR exon ORF15.
Clinically relevant naturally occurring canine models of X-linked progressive retinal atrophy (XLPRA) have proven useful in exploring molecular mechanisms of XLRP and testing therapeutic strategies.12,19–22 Using the XLPRA1 model of RPGRorf15-XLRP,9 the present study seeks to provide new insights into the genetic basis of phenotypic variation in disease severity. The disease is caused by a microdeletion in RGPR exon ORF15 (del1028–1032) that produces a premature stop resulting in a C-terminal truncation of 230 residues. The mutant protein has a shortened Glu-Gly-rich acidic domain and lacks the basic ORF15 C-terminal domain known to interact with nucleophosmin23 and tubulin tyrosine ligase like-5.24 Affected dogs have normal photoreceptor morphogenesis, after which there is gradual but progressive degeneration and eventual vision loss.25 The mutant retinas display mislocalized opsin, upregulation of pro-inflammatory genes expression early in disease, and remodeling defects in the outer retina, correlated with depletion of axon guidance receptor ROBO1 in rod synaptic terminals.19–22
Similar to human patients with RPGR mutations, XLPRA1-affected dogs display broad phenotypic variability in disease severity and progression.25 The original research colony developed to map and characterize the disease was established by outcrossing a single XLPRA1 affected male to unrelated normal females free of known inherited retinal degenerations,26 and subsequently expanded by intercrosses/backcrosses to dogs of known genotypes at the XLPRA1 locus. Because of distinct phenotypic differences in disease severity25 we hypothesized that the severity of the photoreceptor degeneration in affected dogs can be influenced by the action of modifier genes. However, analysis of 12 gene-candidates encoding RPGR interacting proteins as well as proteins essential for ciliary trafficking (RPGRIP1, RPGRIP1L, RANBP2, NPM1, PDE6D, NPHP5, ABCA4, DFNB31, RAB8A, RAB11B, CEP290, and CC2D2A) resulted in their exclusion as potential genetic disease modifiers.20,27
To identify genetic variants associated with disease severity, in this study, we conducted a genome-wide association study (GWAS) in the XLPRA1 phenotype informative pedigree and identified a candidate genomic interval on CFA31 containing potential genetic modifiers. As a majority of genes within the genomic region on CFA31 had not yet been annotated in the dog genome, we carried out a detailed characterization of these genes. A subsequent whole genome sequencing and genotype-phenotype association study demonstrated strong association between genetic variants in four genes (roundabout guidance receptor 1 [ROBO1], ROBO1 antisense RNA [ROBO1-AS], ROBO2 antisense RNA [ROBO2-AS], and ubiquitin specific peptidase 25 [USP25]) within the candidate genomic interval on CFA31 and disease severity.
Materials and Methods
Ethics Statement
The research was conducted in full compliance and strict accordance with the Association for Research in Vision and Ophthalmology (ARVO) Resolution on the Use of Animals in Ophthalmic and Vision Research. All the studies have been approved by the University of Pennsylvania Institutional Animal Care and Use Committee (IACUC).
Pedigree Resources
Details of the origin and composition of the XLPRA1 colony have been previously published.20,27 Briefly, the colony was established by outcrossing a single XLPRA1-affected male Siberian husky to unrelated healthy female beagles shown to be free from inherited retinal degeneration based on test breeding to known homozygous affected dogs with other autosomal recessive diseases.26 The carrier progeny were subsequently mated with mixed breed or other purebred dogs of varied genetic background to produce informative hemizygous or homozygous affected males or females, respectively, and heterozygous females. All dogs were maintained under specific and standard conditions where all animals have the same exposure to cyclic light (12 hours: 12 hours light-dark cycle), receive the same diet, and have the same medical procedures and vaccinations. A subset of the colony, consisting of 45 dogs (affected for RPGR mutation [N = 29] and normal [N = 16]), was selected for the studies (Supplementary Fig. S1). Twenty-four of the total 29 XLPRA1 affected dogs were the same used in prior analyses of potential candidate disease modifier genes,20,27 and were included based on the results of serial clinical assessment of retinal disease status using indirect ophthalmoscopy, electroretinography (ERG), and high resolution optical microscopy of plastic embedded retinal tissues. The remaining five affected dogs were added to the study based on the clinical assessment of retinal disease status and ERG. All colony dogs were maintained under identical conditions, including diet, exposure to cyclic light, and medications/vaccination, at the Retinal Disease Studies (RDS) facility, University of Pennsylvania. Morphologic criteria were used to establish grades of disease severity taking into account the animal's age, degree, and extent of disease.25 Three disease phenotypes were defined: (1) mild = degeneration present only in periphery after 1.5 years of age or later, (2) moderate = peripheral retinal degeneration develops between 11 and 15 months of age, and (3) severe = photoreceptor degeneration stage 2 or more advanced, present both centrally and peripherally before 11 months of age).25
Study Samples
The study used archival DNA samples from normal and XLPRA1 dogs as well as retinal samples mostly remaining from the previously published studies.20,25,27,28 Sections of retinal tissues embedded in an epoxy resin were prepared in our previous study.25 Canine brain tissue was collected and stored at −80°C. The total RNA from mouse retina (1 month old, C57BL/6 strain) was a kind gift from Dr. Helen Léger (University of Pennsylvania).
RNA Extraction and cDNA Synthesis
Total RNA was isolated from canine tissues using a modified TRIzol and single chloroform extraction protocol as previously described.21 First strand cDNA for real-time PCR was synthesized using the High Capacity RNA-to-cDNA kit (Applied BioSystems, Foster City, CA, USA) following the manufacturer's recommendations. First strand cDNA for full length transcripts analysis was gained using the ThermoScript RT-PCR System (Life Technologies, Carlsbad, CA, USA).
Genome-wide Association Mapping and Haplotype Analysis
Genomic DNA from 16 control and 29 XLPRA1 affected cases (15 dogs with severe, 12 dogs with moderate, and 2 dogs with mild disease phenotype) was genotyped on the Illumina CanineHD BeadChip (Illumina, San Diego, CA, USA) that contains 173,662 single nucleotide variants (SNVs). The data set was filtered for genotyping rates (< 95%), low minor allele frequencies (< 0.05), and deviation from Hardy Weinberg equilibrium (P < 10−6). The GenABEL package was used to carry out the GWAS,29 applying a mixed model approach. The files for the haplotype phasing program BEAGLE was prepared with PLINK version 1.9.30 Haplotypes within the highest associated locus were reconstructed using BEAGLE version 3.0,31 phasing the whole chromosome using the default options and 1000 iterations. The same procedure was used in the second iteration, with the additional SNVs added using a text editor.
RNA-Seq Analysis
To have a visual reference of retina-expressed genes, we used the RNA-seq data generated by our group as a part of the previous study.32 Briefly, the reads were mapped to the CanFam3.1 canine reference genome assembly using STAR version 2.7.33 The SAM file obtained was then converted to BAM and the reads were sorted using Samtools.34 The mapped reads were visualized with Integrated Genome Viewer (https://software.broadinstitute.org/software/igv/).
Whole Genome Sequencing and Variants Comparison
Four Illumina TruSeq PCR-free DNA libraries were prepared, with an insert size of 350 base pairs (bps). The sequenced dogs selected were two severe cases (H2 and H38), and two moderate cases (H29 and H59; Supplementary Fig. S1). After collecting HiSeq2500 paired-end reads (2 × 150 bp), the fastq files were created using Casava version 1.8. A total of 854,110,100 150 bp paired-end reads were collected for the 4 dogs (489,897,687 and 364,212,413 for the moderate and severe cases, respectively). The reads were mapped to the CanFam3.1 using Burrows-Wheeler Aligner version 0.5.9-r1635 using the default settings. The SAM file generated was converted to BAM, and the reads were sorted using Samtools.34 The average coverage was 15.12 for the moderate cases and 11.33 for the severe cases.
The GATK version 2.4.9 program36 was used for variant calling within the candidate interval, using the “HaplotypeCaller” module. The variant data for each sample were obtained in variant call format (vcf, version 4.0). Variant filtration followed the best practice documentation of GATK version 4. SnpEff software37 and the CanFam3.1 assembly were used to predict the functional effects of the variants. Then, each group of dogs (moderate and severe) was filtered against the other in order to detect variants exclusive for both moderate cases. Each variant was carefully checked against the annotation.
Rapid Amplification of cDNA Ends
The 5′- and 3′-Rapid Amplification of cDNA Ends (RACE) were performed with the RNA-ligase–mediated RACE (RLM-RACE) system (Thermo Fisher Scientific, Rockford, IL, USA), according to the manufacturer's recommendations and using gene-specific primers listed in Supplementary Table S1.
Amplification of Long PCR Fragments
Long-range PCR was performed using GoTaq Long PCR Master Mix (Promega, Madison, WI, USA). Corresponding primers sequences are listed in Supplementary Table S1. All novel transcript data generated in this study have been deposited to the National Center for Biotechnology Information (NCBI) GenBank database and Accession Numbers are listed in Supplementary Tables S1 and S2.
Genotyping
Candidate genetic variants were genotyped in XLPRA1 pedigree using direct sequencing of PCR products. Fisher's exact test (two-sided) was used for statistical analysis (P ≤ 0.05). For in silico genotyping, we searched through the .vcf file containing the SNV called from the whole genome sequencing (WGS) data of 800 dogs of various breeds part of the studies carried out by the Dog Biomedical Variant Database Consortium.38 The SNVs were extracted with a simple bash script and formatted with a text editor.
Ribonuclease Protection Assay
Total RNA (approximately 5–6 µg) from 16 week old canine retina was orderly digested with TURBO DNAase (Thermo Fisher Scientific) for 30 minutes at 16°C and RNase A/T1 Mix (Thermo Fisher Scientific) for 1 hour and 30 minutes at 16°C to remove all the genomic DNA contamination and single-strand RNAs. RNA was purified after each step with the RNA Clean & Concentrator kit. The cDNA from endogenous double-strand RNAs (dsRNA) was produced using the ThermoScript RT-PCR System (Life Technologies) and the mixture of three gene-specific primers. The reaction conditions involved two cycles of heating at 82°C for 1 minute, 65°C for 1 minute, 60°C for 30 seconds, and 57°C for 30 minutes. The double-stranded cDNA was amplified in 25 ul PCR reaction system. After 35-cycle amplification, the products were checked by electrophoresis on 5% polyacrylamide gel (PAAG) with ethidium bromide staining. Primer sequences are listed in Supplementary Table S1. Notes: Total RNA used for these experiments, was isolated from canine retina under mild/nondenaturing conditions to preserve prospective natural RNA duplexes.
Relative Quantification (ddCt) Assay
Gene expression was determined by quantitative real-time PCR (qRT-PCR) in 16 week old normal (N = 3) and XLPRA1-affected retinas (N = 3–6). Each group analyzed included at least three animals. Real-time PCR was performed in a total volume of 25 µL in 96-well microwell plates on the Applied Biosystems 7500 Real-Time PCR System. All PCRs were performed in triplicate using cDNA generated from 20 ng DNAase-treated RNA. The SYBR green platform was used for gene expression analysis using a primer concentration of 0.2 µM. The TATA-box binding protein (TBP) gene expression level was used to normalize the cDNA templates.20 Primers used are listed in Supplementary Table S1. Amplification data were analyzed with the 7500 Software version 2.0.1 (Applied Biosystems). Unpaired t-test was used for statistical analysis. Genes with P < 0.05 and fold changes (FCs) > +/− 2 were considered differentially expressed.
Sequence Analysis Tools
Possible impact of an amino acid substitution on the structure and function of proteins was evaluated with the PolyPhen-2 online software (http://genetics.bwh.harvard.edu/pph2/).
The IntaRNA program39 was used for the prediction of RNA-RNA interactions (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp).
Results
GWAS Detects a Candidate Genomic Interval on CFA31
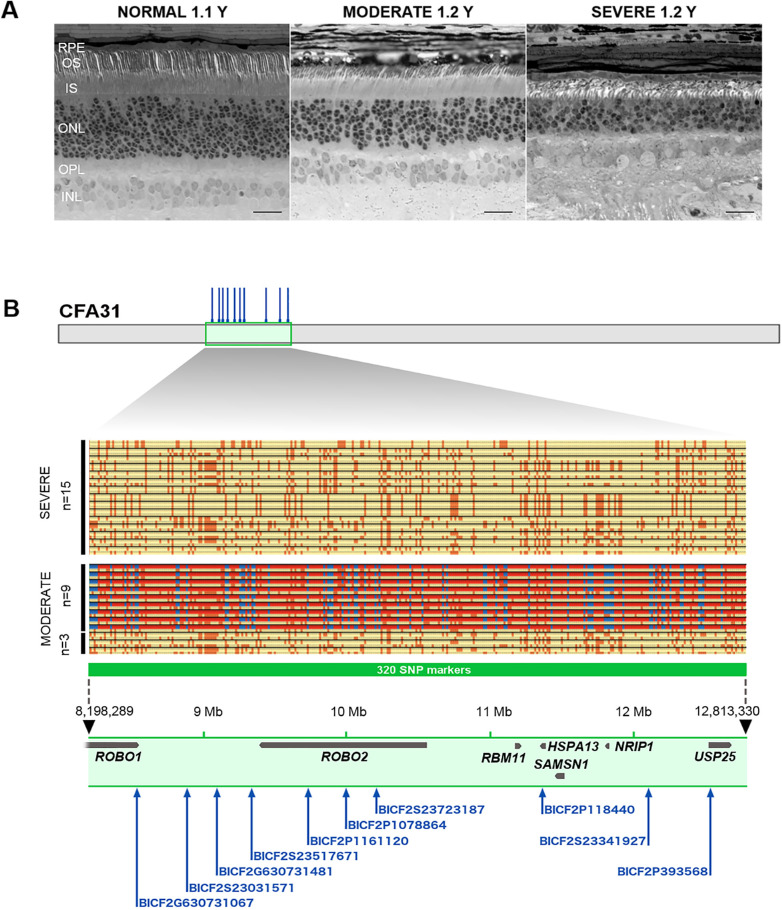
To map genomic loci containing candidate modifiers that affect distinct XLPRA1 phenotypes (Fig. 1A and Supplementary Fig. S1), we performed GWAS analysis in the disease phenotype informative pedigree (Supplementary Fig. S1). A cohort of 16 controls and 29 XLPRA1 cases was genotyped on Illumina CanineHD BeadChips. After quality control, 132,218 SNVs remained for association analysis in GenABEL. The calculated genomic inflation factor (lambda) of 1.38 indicated a degree of stratification, therefore a mixed model approach was used for the association analysis. As an additional control, an association study was performed to replicate the mapping of the XLPRA1 disease causative locus on canine chromosome X.9 The 10 most highly associated SNVs reached a P value of 6.69 × 10−8 and were located on CFAX between 31,161,359 (BICF2P479827) and 34,529,510 (BICF2P1287979) bp (Supplementary Fig. S2A, S2B). Considering that RPGR gene is positioned on CFAX between 33,056,371 and 33,105,037 bp, the XLPRA1 causative locus was reliably remapped.
Figure 1.
Variability in XLPRA1 disease progression rate and fine-mapping of the critical region on CFA31. (A) Representative images of normal and XLPRA1 retinas with moderate and severe phenotype. Examination of 1 µm sections from archival tissues embedded in epoxy resin shows differences in severity of photoreceptor degeneration in mutant retinas from littermates H81 (moderate) and H78 (severe). When compared to normal, disease samples display reduction of ONL thickness of approximately 25% and 50% in H81 (moderate) and H78 (severe), respectively. IS and OS of remaining photoreceptors are shortened and disorganized as disease progresses. RPE, retinal pigment epithelium; OS, outer segment; IS, inner segment; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer. Scale bar is 20 µm. (B) Results of the phasing of CFA31. A 320 SNV markers haplotype h1, spanning a genomic region of approximately 4.6 Mb (highlighted in green), has been identified in 9 of the 12 cases with a “moderate” phenotype, and in none of the 15 dogs with a “severe” phenotype. In all nine dogs, the haplotype is heterozygous (marked in red and blue). The critical interval on CFA31 between 8,198,289 and 12,813,330 bp is marked by black arrow heads. Seven retina-expressed genes (ROBO1, ROBO2, RBM11, NRIP1, HSPA13, SAMSN1, and USP25), along with their positions within the interval are indicated. The blue upward arrows represent the 10 best trait-associated SNVs in GWAS, where BICF2G630731481 and BICF2P393568 are the markers with the highest association (P values of 7.16e-5). Note: positions are given according CanFam3.1 dog genome assembly.
Next, we carried out the association analysis, using 15 dogs with severe (group 1, “control”) and 12 dogs with moderate (group 2, “cases”) disease phenotypes. Although the remaining dogs were not a part of either group, they were left in the dataset for calculation of minor allele frequencies and Hardy-Weinberg equilibrium. As the group sizes were small and stratified, the GWAS analysis did not reach significance after Bonferroni correction. Nonetheless, we identified a single locus on CFA31 with notable trend toward association with the XLPRA1 phenotype (Supplementary Fig. S2C, S2D). Notably, the peak associated with the region showed a log (P value) just below the very strict Bonferroni correction threshold. To define the precise boundaries of this critical interval, SNV data from CFA31 were phased. The haplotype h1, composed of 320 SNVs linked to this locus, is positioned on CFA31 between 8,198,289 and 12,813,330 bp (see Fig. 1B and Supplementary Dataset S1). This haplotype was found in XLPRA1 pedigree in a subset of affected dogs as well as in five clinically normal breeding dogs that passed it on to their offspring (see Supplementary Dataset S1). Thereafter, two XLPRA1 phenotypes (severe and moderate, see Fig. 1A and Supplementary Fig. S1) were tested for their haplotype association. The haplotype h1 was present in heterozygous state in 9 of 12 affected dogs with moderate disease phenotype (P < 0.02, Fisher's exact test) but absent in any of the 15 dogs with severe disease phenotype (see Supplementary Dataset S1). After this last step, we considered the CFA31 4.6 Mb interval CFA31:31,161,359 to 34,529,510 as a suitable candidate region for the moderate phenotype.
To examine this genomic interval for retinal genes expression, we used RNA-seq data from adult normal dog retinas (GSE9763832). RNA-seq data were visually inspected using the Integrative Genomics Viewer (IGV) software. Seven protein-coding genes (roundabout guidance receptor 1 and 2 [ROBO1 and ROBO2, respectively], RNA binding motif protein 11 [RBM11], nuclear receptor interacting protein 1 [NRIP1], heat shock protein family A member 13 [HSPA13], SAM domain SH3 domain and nuclear localization signals 1 [SAMSN1], and ubiquitin specific peptidase 25 [USP25]) were found to be expressed in this approximately 4.6 Mb interval, with ROBO1 and ROBO2 genes occupying approximately half of the locus (see Fig. 1B).
Characterization of the Genes Within the Genomic Interval on CFA31
The accurate exon-intron structure of the seven genes within the CFA31 region is required for the downstream mutational analysis. Of the mapped seven genes within the genomic region, only two, ROBO1 and ROBO2, were previously characterized in the normal dog retina.22 Therefore, we first examined the exon-intron structure of the remaining five transcripts (RBM11, NRIP1, HSPA13, SAMSN1, and USP25) using IGV software. The structure of the four transcripts (RBM11, HSPA13, SAMSN1, and USP25) was further confirmed by sequencing of corresponding PCR products (Supplementary Fig. S2E). As the sequencing of NRIP1 PCR products presented a major challenge due to high guanine and cytosine (GC) content, we chose to use its Ensembl transcript as a reference sequence for downstream mutational analysis. Brief characterization of the transcripts within the genomic interval is given in Supplementary Tables S2 and S3. Predicted structure for sufficiently long open reading frames (ORFs) was based on the similarity to the known human and mouse orthologous sequences.
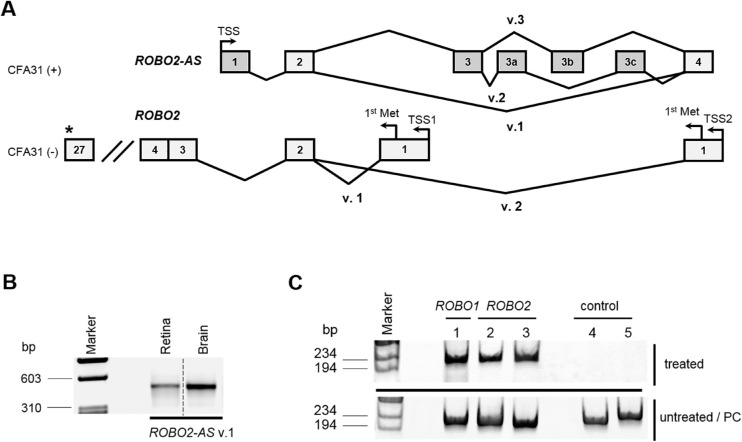
By comprehensively exploring the gene expression landscape in the CFA31 region we also identified two novel lncRNAs, ROBO1-AS and ROBO2-AS, that overlapped with canine ROBO1 and ROBO2, respectively, but were transcribed in the antisense direction. We first discovered ROBO2-AS in the course of experimental verification of the expression of testis-derived EST CX986571 (CFA31: 9,715,744 to 10,176,010) in normal adult dog retina. Of four CX986571 exons, three were found to be expressed in the retina. RACE analysis to identify 5' and 3' ends of the transcript and computational evaluation of its coding potential revealed that it is actually a noncoding natural antisense transcript. Overall, three alternatively spliced transcriptional variants of ROBO2-AS (v.1 [composed of 3 exons], v.2 [6 exons], and v.3 [5 exons], schematically shown on Fig. 2A) were found in the retina. These variants have a common start in the second intron of ROBO2 as well as common second and last exons with 100% antisense homology to the ROBO2 v.2 second exon and the first exon, respectively. Exonic organization of the ROBO2-AS v.1 was validated by RT-PCR and expression of this transcript was found in both, the retina and brain (Fig. 2B). ROBO2-AS v.2 and v.3 have predicted structure supported by two PCR products with overlap in the common exon 3. To confirm on a preliminary basis that sense-antisense organization of ROBO2/ROBO2-AS genes is conserved between species, we examined the presence of orthologous Robo2-AS in mouse retina. The identified transcript (Supplementary Fig. S2F) displayed structural and sequencing homology to canine ROBO2-AS v.1 and formed a sense-antisense overlap with mouse Robo2.
Figure 2.
Structural organization of lncRNA ROBO2-AS and its functional analysis in the retina. (A) Exon structure and alternative transcripts of the sense-antisense ROBO2 / ROBO2-AS gene pair. ROBO2-AS produces three transcripts through alternative splicing (v.1 [exons 1, 2, and 4], v.2 [exons 1 to 3, 3a, 3c, and 4], and v.3 [exons 1 to 3, 3b, and 4]) (top). Two transcript variants of ROBO2 (27 coding exons each) have common 26 exons but differ in their first coding exon (bottom). (B) Expression of lncRNA ROBO2-AS v.1 in normal canine retina (24 weeks) and brain (1.1 years). Note: In order to present the data in a streamlined way the irrelevant lanes were spliced out from the 12-wells agarose gel (indicated by dashed lines). (C) Results of the RPA. Upper panel: Presence of lncRNA/mRNA duplex between the ROBO1-AS/ROBO1 (lane 1 [exons 10 to 11 of ROBO1] and ROBO2-AS/ROBO2 [lanes 2 and 3: exon 2 and exons 1 and 2 of ROBO2 v.2, respectively) were validated by RT-PCR in cDNA produced from nuclease treated RNA, lanes 4 and 5 show negative controls (genomic and ssRNA, respectively). Lower panel: Positive control (PC) experiment where the same analysis was repeated in cDNA produced from untreated RNA.
Canine lncRNA ROBO1-AS was identified subsequently by screening the ROBO1 exons in antisense direction using 5′RACE. Three alternative 3′-ends of ROBO1-AS were identified by 3′RACE. The assembly of ROBO1-AS transcript variants v.1, v.2, and v.3 was done through overlap of 5′RACE and 3′RACE products. Briefly, all three variants are predicted to have 100% antisense homology to part of the ROBO1 mRNA where ROBO1-AS v.1, v.2, and v.3 overlap ROBO1 exons 8 to 13, 5 to 13, and 4 to 13, respectively. Brief characterization and accession numbers of ROBO1-AS and ROBO2-AS transcripts are given in Supplementary Tables S2 and S3.
The overlapping organization of ROBO genes to produce sense-antisense gene pairs, ROBO1/ROBO1-AS and ROBO2/ROBO2-AS suggests that cis-antisense RNAs ROBO1-AS and ROBO2-AS may act as key regulators of ROBO1 and ROBO2 transcriptional or translational output. To examine the interaction between ROBO1-AS and ROBO1 mRNA, as well as the interaction between ROBO2-AS and ROBO2 mRNA, we performed a ribonuclease protection assay (RPA) on total RNA extracted from 16 weeks old normal canine retina. RPA showed that sense/antisense RNA duplex formation occurred between the two strands, resulting in protection (see Fig. 2C).
Last, we examined whether the expression profile of the genes within the CFA31 region is impaired in predisease XLPRA1 retinas. To this end, we evaluated expression of ROBO1, ROBO2, ROBO2-AS, RBM11, NRIP1, HSPA13, SAMSN1, and USP25 in 16 weeks old mutant retinas using real-time PCR. At this time of age, when the retina is structurally normal,25 there were no differences in expression of these eight genes in disease compared to normal control (data not shown). Because of the lack of unique exons and exon-exon junctions distinguishing ROBO1-AS from ROBO1 we could not accurately determine expression levels of ROBO1-AS.
Mutational Analysis of the Genes in the Genomic Interval on CFA31
To further assess if sequence variants of ROBO1, ROBO1-AS, ROBO2, ROBO2-AS, RBM11, NRIP1, HSPA13, SAMSN1, and USP25 contribute to variation in XLPRA1 phenotype, we carried out mutational analysis of these genes in XLPRA1 affected dogs. To this end, DNAs from two affected dogs with a severe phenotype (H2 and H38 <negative for h1>) and two affected dogs with a moderate phenotype (H29 and H59 <carriers for h1>) were used for the initial analysis (see Supplementary Fig. S1). Exons of ROBO1/ROBO1-AS, ROBO2/ROBO2-AS, RBM11, NRIP1, HSPA13, SAMSN1, and USP25 genes were analyzed by WGS (see Materials and Methods for details). Given the large number of sequence variants identified using WGS, we applied high-stringency filters for narrowing down the search. Specifically, in search for genetic determinants that have same distribution as the haplotype h1, variants in each group of dogs (moderate [H29 and H59] and severe [H2 and H38]) were filtered against the other in order to detect variants exclusive for both moderate cases. Next, we compared and filtered SNVs as well as small indels called in both groups. The total of 8515 SNVs and small indels were found in the interval. These variants were then filtered according to two criteria: (1) filtering out the variants present in the severe cases, and (2) filtering out the variant not present in both moderate cases. Of 1752 filtered variants (listed in Supplementary Dataset S2) exclusive for both moderate cases, 4 SNVs (shown in Table 1) were found in exons of ROBO1 (rs8523 and rs8509), ROBO1-AS (rs8523), ROBO2-AS (ss1350), and USP25 (rs8524) and were further used in association studies. None of the filtered variants were found within predicted ORF of the ROBO2, RBM11, NRIP1, HSPA13, and SAMSN1 genes.
Table 1.
Characterization of Allelic Variants in ROBO1/ROBO1-AS, ROBO2-AS, and USP25
| Transcript | SNV Position | Allele 1 | Allele 2 | AA Change | Accession Numbera | Study ID |
|---|---|---|---|---|---|---|
| ROBO1 | CFA31: 8515350 | C | T | A434A | rs852373101 | rs8523b |
| CFA31: 8556222 | A | T | P1110P | rs850923081 | rs8509 | |
| ROBO1-AS | CFA31: 8515350 | G | A | rs852373101 | rs8523b | |
| ROBO2-AS | CFA31:10176026 | C | G | ss1350332223581 | ss1350 | |
| USP25 | CFA31: 12651217 | G | A | R615H | rs852430218 | rs8524 |
Note: SNVs positions are given according CanFam3.1 dog reference genome assembly.
Variants accession numbers were obtained from Ensembl genome database [www.ensembl.org/] and European Variation Archive [https://www.ebi.ac.uk/eva/] (<rs> and <ss>, respectively).
Prevalence of rs8523, rs8509, ss1350, and rs8524 in Dogs With Moderate Disease Phenotype
Genotype frequencies of the ROBO1 (rs8523 and rs8509), ROBO1-AS (rs8523), ROBO2-AS (ss1350), and USP25 (rs8524) sequence variants were analyzed in the XLPRA1-affected dogs and evaluated for association with moderate and severe phenotypes (Table 2). All four examined variants showed strong association with moderate XLPRA1 phenotype (P < 0.02). We found that rs8509, ss1350, and rs8524 variants fully cosegregate with the haplotype h1 from the GWAS-identified modifier locus. The rs8523 variant was present in all 9 rs8509/ss1350/rs8524 positive dogs, but also found in an additional 3 dogs (1 with moderate and 2 with severe XLPRA1 phenotype) that did not harbor the rs8509, ss1350, or rs8524 variants.
Table 2.
Distribution of ROBO1/ROBO1-AS (rs8523), ROBO1 (rs8509), ROBO2-AS (ss1350), and USP25 (rs8524) SNVs in XLPRA1 Pedigree
| Sample ID | rs8523 | rs8509 | ss1350 | rs8524 |
|---|---|---|---|---|
| Severe | ||||
| H2 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H64 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H104 | 1, 2 | 1, 1 | 1, 1 | 1, 1 |
| H105 | 1, 2 | 1, 1 | 1, 1 | 1, 1 |
| H78 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H79 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H82 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H143 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H35 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H38 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H71 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H72 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H73 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H118 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H497 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| Moderate | ||||
| H29 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H31 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H130 | 1, 2 | 1, 1 | 1, 1 | 1, 1 |
| H131 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H81 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H208 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H59 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H201 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H202 | 1, 1 | 1, 1 | 1, 1 | 1, 1 |
| H498 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H499 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
| H500 | 1, 2 | 1, 2 | 1, 2 | 1, 2 |
Note: 1 = Allele 1 (wild type); 2 = Allele 2 (sequence variation).
We further examined whether rs8523, rs8509, ss1350, and rs8524 are a part of the haplotype h1. To this end, these four variants were merged with a subset of the SNV dataset used for GWAS and first round of phasing (CFA31 8,106,389 to 12,877,782). This second dataset was again phased using the BEAGLE software. Two XLPRA1 phenotypes, severe and moderate, were tested once more for association with each haplotype identified. For a second time, we found significant association (P < 0.02) of the major haplotype with a moderate XLPRA1 disease phenotype (shown in Supplementary Dataset S3). The haplotype identified in nine dogs with moderate disease phenotype matched to the haplotype h1 described above but modified by rs8523, rs8509, ss1350, and rs8524 variants, hereafter referred to as h1R. Notably, the h1R haplotype included ROBO1/ROBO1-AS rs8523, ROBO1 rs8509, ROBO2-AS ss1350, and USP25 rs8524, supporting a potential role for these variants in the genetic architecture of the disease.
Distribution of rs8523, rs8509, ss1350, and rs8524 Sequencing Variants
We next determined if the ROBO1/ROBO1-AS, ROBO2-AS, and USP25 sequence variants were subject to positive or negative selection in dog populations. For this, we queried the Dog Biomedical Variant Database Consortium, which has WGS data from domesticated dogs worldwide.38 Using in silico genotyping, we determined that allelic frequency of rs8523, rs8509, ss1350, and rs8524 in this heterogeneous dog population (N = 800, 126 breeds) was 16.2%, 16.8%, 5.3%, and 5.4%, respectively, as shown in Table 3. No significant differences were found between observed and expected genotype frequencies, excluding the possibility that rs8523, rs8509, ss1350, and rs8524 are deleterious alleles.
Table 3.
Allele and Genotype Frequencies of ROBO1/ROBO1-AS (rs8523), ROBO1 (rs8509), ROBO2-AS (ss1350), and USP25 (rs8524) Variants in a Heterogenic Dog Population
| Genotype, N (Genotype Frequency O/E) | |||||||
|---|---|---|---|---|---|---|---|
| SNV | Dogs, N | 1,1 | 1,2 | 2,2 | P Value* | Allele 1 Frequency, % | Allele 2 Frequency, % |
| rs8523 | 799 | 587 (0.73/0.70) | 165 (0.21/0.27) | 47 (0.06/0.03) | 0.66 | 83.8 | 16.2 |
| rs8509 | 796 | 581 (0.73/0.70) | 163 (0.21/0.27) | 52 (0.06/0.03) | 0.66 | 83.2 | 16.8 |
| ss1350 | 798 | 725 (0.91/0.89) | 61 (0.08/0.10) | 12 (0.01/0.01) | 0.88 | 94.7 | 5.3 |
| rs8524 | 800 | 724 (0.91/0.89) | 66 (0.08/0.10) | 10 (0.01/0.01) | 0.49 | 94.6 | 5.4 |
Note: *The χ2 test was used to compare observed (O) and expected (E) genotype frequencies.
Both rs8523 and rs8509 are synonymous substitution in ROBO1 (p. A434A and p. P1110P, respectively) and whereas rs8524 is the missense change in USP25 (p. R615H) it is predicted to be benign by the PolyPhen2 software. The functional significance of rs8523 and ss1350 requires further clarification as these sequence variants may have an impact on ROBO1-AS and ROBO2-AS interaction with the cis-target transcripts (ROBO1 and ROBO2, respectively) as well as with their potential trans-targets that are yet to be identified.
Association of ROBO1-AS rs8523 and ROBO2-AS ss1350 With Reduced Immune Response
The intensity of the immune response has been shown to contribute to neurodegeneration in retinal pathologies.40–42 As previous studies demonstrated the ability of some lncRNAs to modulate the host immune response,43–45 we examined whether ROBO1-AS rs8523 and ROBO2-AS ss1350 may be associated with variation in immune response in affected retinas. To this end, we performed a gene expression experiment in pre-disease 16 week old mutant retinas that differ by rs8523/ss1350 genotype (rs8523−/−/ss1350−/− [16w-group 1] and rs8523−/+/ss1350−/+ [16w-group 2]), using a subset of the immune response genes previously shown to be upregulated very early in XLPRA1 disease.20,21 Although at 16 weeks, XLPRA1 retinas are still morphologically normal and the disease severity phenotype is yet unknown, one can expect that adaptive changes in the immune response in this early time point may ameliorate further disease progression.
We found that although the entire subset of immune response genes studied (TLR4, P2RX7, IL1B, GFAP, IL4, FGF, and LIF) was significantly upregulated in both 16w-groups compared to normal control (P < 0.05), but the increase of gene expression in the 16w-group 2 (rs8523/ss1350−/+) was of considerably lower magnitude (Table 4). These results point to a potential value of rs8523 and ss1350 as predictive markers of modulated immune reaction in the retina, although the precise mechanism underlying such modulation remains to be defined.
Table 4.
Comparative Analysis of Gene Expression in Two Study Groups
| Relative Expression vs | ||
|---|---|---|
| Normal Control, Fold Changes | ||
| 16w-Group 1 | 16w-Group 2 | |
| Gene | rs8523−/−/ss1350−/− | rs8523−/+/ss1350−/+ |
| TLR4 | 13.1 | 7.6 |
| P2RX7 | 4.1 | 2.2 |
| IL1B | 29.2 | 14.8 |
| GFAP | 9.9 | 5.3 |
| IL4 | 14.2 | 8.8 |
| FGF | 5.3 | 3.4 |
| LIF | 4.5 | 3.4 |
Discussion
Identifying the genetic factors that contribute to interindividual differences in disease is critical for understanding genotype-phenotype correlations and for developing appropriate targeted therapies. In this study, we have examined genotype–phenotype correlations in a naturally occurring large animal model of RPGR-XLRP using a GWAS approach and targeted genetic testing.
The XLPRA1 study pedigree is a small and inbred affected population. Although these limitations affected the P value of the signal and the pattern of the QQ-plot (Supplementary Fig. S2D), a combined approach of GWAS and phasing allowed us to pinpoint an approximately 4.6 Mb candidate genomic interval on CFA31 that contained candidate modifier genes of XLPRA1 and to identify a haplotype segregating in a statistically significant manner between severe and moderate cases. We found that the genomic region on CFA31 contains seven protein-coding genes (ROBO1, ROBO2, RBM11, NRIP1, HSPA13, SAMSN1, and USP25) and most notably, two novel lncRNAs ROBO1-AS and ROBO2-AS that display overlapping gene organization with axon guidance receptor genes ROBO1 and ROBO2, respectively.
Next, we evaluated sequence variations in the genes within the CFA31 region and their putative association with disease phenotype. We found strong association between the genetic variants in ROBO1/ROBO1-AS (rs8523), ROBO1 (rs8509), ROBO2-AS (ss1350), and USP25 (rs8524) and moderate disease phenotype both individually and as an intrinsic part of the reconstructed haplotype h1R. Genotype distribution of rs8523, rs8509, ss1350, and rs8524 in dog population further confirmed our initial hypothesis that these variants are not rare mutations accumulated in XLPRA1 pedigree due to the breeding process but are relatively common DNA polymorphisms. However, this fact cannot disqualify either of these variants to be a potential modifier as genetic polymorphisms may exert an impact on disease phenotype in the presence of disease causal mutation.15
Having identified a list of four genetic variants, we speculate that not each of these sequence variants might impact the expressivity of disease phenotypes. ROBO1 synonymous substitutions (rs8509 and rs8523) may not have an effect on ROBO1 protein functions. On the other hand, ss1350 and rs8523 variants are more complicated to interpret as ss1350 is a part of ROBO2-AS and rs8523 is a part of both ROBO1 and ROBO1-AS transcripts. Both ROBO1-AS and ROBO2-AS are newly described lncRNAs and their exact role in retinal function is not established yet.
In this study, we demonstrated ROBO1-AS and ROBO2-AS act in cis to form lncRNA/mRNA duplexes with ROBO1 and ROBO2, suggesting a contribution of these natural antisense transcripts to regulatory complexity of axon guidance ROBO pathway. Still, the underlying basis for such interactions remains to be further characterized as duplex RNA between sense and antisense transcripts may bring about a variety of outcomes, including changes in mRNA stability and translation, modulation of mRNA nuclear transport, splicing, and editing.46,47 Taking into consideration the important role of ROBO signaling for neuronal cell functions, it is not surprising that the activity of ROBO receptors is highly controlled at all possible levels, including gene expression, splicing, mRNA stability, translation, post-translational modification, and cell-type specific cleavage.48–50 Whereas the ROBO pathway is best known for mediating axon repulsion in the developing nervous system,48,51 results of our recent study highlight the role of ROBO signaling in adult mammalian retina.22 We showed that in fully developed normal canine retinas, ROBO1 protein is predominantly expressed in photoreceptors and depletion of ROBO1 levels in rod synaptic terminals strongly correlates with the remodeling of axonal and dendritic processes in the outer retina of XLPRA1 dogs. We have also found prominent expression of ROBO2 in the subset of amacrine and ganglion cells, suggesting a range of functions for this receptor in the retinal neurons.22 One cannot exclude the possibility that changes in regulation of ROBO levels by natural antisense transcripts may play a role in retinal remodeling associated with XLPRA1 progression. This assumption will be addressed in future studies.
Recent evidence increasingly implicates lncRNAs in the critical regulation of multiple biological processes through epigenetic regulation, chromatin remodeling, effects on gene and protein expression, cellular transport, cell differentiation, organ or tissue development, metabolic processes, and chromosome dynamics.52–54 Therefore, one might surmise that genetic variants or dysregulation of lncRNAs can exert broad effects on health and/or disease. For instance, our data point to the potential regulatory effect of ROBO1-AS and/or ROBO2-AS on variation in immune response in mutant retinas, either directly or indirectly.
Specifically, we identified substantial differences in expression levels of seven immune response genes in pre-disease mutant retinas (with unknown yet disease severity phenotype) that harbor the ROBO1-AS rs8523 and ROBO2-AS ss1350 variant. It has been shown that some lncRNAs can modulate immune response through direct regulation of immune cell differentiation and function.43–45 On the other hand, lncRNAs are known to participate in almost every aspect of visual maintenance and impairment.54 In retinal disorders, neuroinflammation is an adaptive response to tissue stress, regulated by retina-resident nonimmune and immune cells, which are in constant communication with retinal neurons to sense the release of damage-associated signals. In pathological conditions, this tight communication between cells mediate the magnitude of inflammatory responses in the retina.
How lncRNAs ROBO1-AS and ROBO2-AS are being involved into regulation of physiological and pathological responses of retinal cells can be critical for understanding the pathogenesis of RPGR-associated retinal degeneration. Thus, whereas the relationship between ROBO1-AS rs8523 and ROBO2-AS ss1350 and the resultant XLPRA1 phenotype is clearly provocative, further functional studies will be required to mechanistically determine how these variants may modulate expressivity of XLPRA1.
Regarding the USP25 missense variant (rs8524) predicted to be benign, we suggest that it may not lead to a substantial effect on USP25 protein functions. Although USP25 was shown to be involved in regulation of the immune pathways in non-retinal cells,55,56 the role of this protein in the retina remains poorly understood57 and would require further examination.
Overall, our results demonstrate a genetic association of ROBO1/ROBO1-AS rs8523, ROBO1 rs8509, ROBO2-AS ss1350, and USP25 rs8524 variants with milder disease phenotype in XLPRA1 dogs. These findings open up new opportunities to explore whether genetic variants in corresponding human orthologous genes have a prognostic value in individuals with RPGR-XLRP with a range of disease severity. In addition, our data suggest a complex connection between ROBO pathways and the disease progression and provide a basis for future mechanistic studies that will inform the molecular events responsible for complex regulation of ROBO1-AS and ROBO2-AS signaling in the retina, and provide new insights into potential therapeutic strategies for retinal degeneration.
Supplementary Material
Acknowledgments
The authors thank Leslie King and Jacob G. Appelbaum for helpful discussions and comments, Joseph Krupiak and Jacqueline Wivel for assistance with archival tissues database, and Helen Leger for mouse RNA sample. We acknowledge collaborators of the Dog Biomedical Variant Database Consortium for sharing dog genome sequence data.
Supported by grants EY-06855, EY-17549, the Foundation Fighting Blindness, the Van Sloun Fund for Canine Genetic Research, Hope for Vision, and is partially supported by the Vision Research Center (P30-EY001583).
Disclosure: T. Appelbaum, None; L. Murgiano, None; D. Becker, None; E. Santana, None; G.D. Aguirre, None
References
- 1. Sahel J, Bonnel S, Mrejen S, Paques M. Retinitis pigmentosa and other dystrophies. Dev Ophthalmol. 2010; 47: 160–167. [DOI] [PubMed] [Google Scholar]
- 2. Breuer DK, Yashar BM, Filippova E, et al.. A comprehensive mutation analysis of RP2 and RPGR in a North American cohort of families with X-linked retinitis pigmentosa. Am J Hum Genet. 2002; 70: 1545–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ott J, Bhattacharya S, Chen JD, et al.. Localizing multiple X chromosome-linked retinitis pigmentosa loci using multilocus homogeneity tests. Proc Natl Acad Sci USA. 1990; 87: 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teague PW, Aldred MA, Jay M, et al.. Heterogeneity analysis in 40 X-linked retinitis pigmentosa families. Am J Hum Genet. 1994; 55: 105–111. [PMC free article] [PubMed] [Google Scholar]
- 5. Meindl A, Dry K, Herrmann K, et al.. A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat Genet. 1996; 13: 35–42. [DOI] [PubMed] [Google Scholar]
- 6. Vervoort R, Lennon A, Bird AC, et al.. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat Genet. 2000; 25: 462–466. [DOI] [PubMed] [Google Scholar]
- 7. Ferreira PA. Insights into X-linked retinitis pigmentosa type 3, allied diseases and underlying pathomechanisms. Hum Mol Genet. 2005; 14(Spec No. 2): R259–R267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wright AF, Shu X.. Focus on molecules: RPGR. Exp Eye Res. 2007; 85: 1–2. [DOI] [PubMed] [Google Scholar]
- 9. Zhang Q, Acland GM, Wu WX, et al.. Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum Mol Genet. 2002; 11: 993–1003. [DOI] [PubMed] [Google Scholar]
- 10. Thompson DA, Khan NW, Othman MI, et al.. Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One. 2012; 7: e35865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Z, Hiriyanna S, Qian H, et al.. A long-term efficacy study of gene replacement therapy for RPGR-associated retinal degeneration. Hum Mol Genet. 2015; 24: 3956–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beltran WA, Cideciyan AV, Lewin AS, et al.. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci USA. 2012; 109: 2132–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fahim AT, Bowne SJ, Sullivan LS, et al.. Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One. 2011; 6: e23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Charng J, Cideciyan AV, Jacobson SG, et al.. Variegated yet non-random rod and cone photoreceptor disease patterns in RPGR-ORF15-associated retinal degeneration. Hum Mol Genet. 2016; 25: 5444–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013; 132: 1077–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danciger M, Ogando D, Yang H, et al.. Genetic modifiers of retinal degeneration in the rd3 mouse. Invest Ophthalmol Vis Sci. 2008; 49: 2863–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Khanna H, Davis EE, Murga-Zamalloa CA, et al.. A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat Genet. 2009; 41: 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riordan JD, Nadeau JH. From peas to disease: modifier genes, network resilience, and the genetics of health. Am J Hum Genet. 2017; 101: 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beltran WA, Acland GM, Aguirre GD. Age-dependent disease expression determines remodeling of the retinal mosaic in carriers of RPGR exon ORF15 mutations. Invest Ophthalmol Vis Sci. 2009; 50: 3985–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Appelbaum T, Becker D, Santana E, Aguirre GD. Molecular studies of phenotype variation in canine RPGR-XLPRA1. Mol Vis. 2016; 22: 319–331. [PMC free article] [PubMed] [Google Scholar]
- 21. Appelbaum T, Santana E, Aguirre GD. Strong upregulation of inflammatory genes accompanies photoreceptor demise in canine models of retinal degeneration. PLoS One. 2017; 12: e0177224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Appelbaum T, Santana E, Aguirre GD. Critical decrease in the level of axon guidance receptor ROBO1 in rod synaptic terminals is followed by axon retraction. Invest Ophthalmol Vis Sci. 2020; 61: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shu X, Fry AM, Tulloch B, et al.. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet. 2005; 14: 1183–1197. [DOI] [PubMed] [Google Scholar]
- 24. Sun X, Park JH, Gumerson J, et al.. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc Natl Acad Sci USA. 2016; 113: E2925–E2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zeiss CJ, Acland GM, Aguirre GD. Retinal pathology of canine X-linked progressive retinal atrophy, the locus homologue of RP3. Invest Ophthalmol Vis Sci. 1999; 40: 3292–3304. [PubMed] [Google Scholar]
- 26. Acland GM, Blanton SH, Hershfield B, Aguirre GD. XLPRA: a canine retinal degeneration inherited as an X-linked trait. Am J Med Genet. 1994; 52: 27–33. [DOI] [PubMed] [Google Scholar]
- 27. Guyon R, Pearce-Kelling SE, Zeiss CJ, Acland GM, Aguirre GD. Analysis of six candidate genes as potential modifiers of disease expression in canine XLPRA1, a model for human X-linked retinitis pigmentosa 3. Mol Vis. 2007; 13: 1094–1105. [PMC free article] [PubMed] [Google Scholar]
- 28. Genini S, Beltran WA, Aguirre GD. Up-regulation of tumor necrosis factor superfamily genes in early phases of photoreceptor degeneration. PLoS One. 2013; 8: e85408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007; 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- 30. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Browning SR, Browning BL.. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007; 81: 1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sudharsan R, Beiting DP, Aguirre GD, Beltran WA. Involvement of innate immune system in late stages of inherited photoreceptor degeneration. Sci Rep. 2017; 7: 17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dobin A, Davis CA, Schlesinger F, et al.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li H, Handsaker B, Wysoker A, et al.. The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McKenna A, Hanna M, Banks E, et al.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cingolani P, Platts A, Wangle L, et al.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jagannathan V, Drogemuller C, Leeb T, Dog Biomedical Variant Database Consortium (DBVDC). A comprehensive biomedical variant catalogue based on whole genome sequences of 582 dogs and eight wolves. Anim Genet. 2019; 50: 695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017; 45: W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rashid K, Akhtar-Schaefer I, Langmann T. Microglia in retinal degeneration. Front Immunol. 2019; 10: 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whitcup SM, Nussenblatt RB, Lightman SL, Hollander DA. Inflammation in retinal disease. Int J Inflam. 2013; 2013: 724648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao L, Zabel MK, Wang X, et al.. Microglial phagocytosis of living photoreceptors contributes to inherited retinal degeneration. EMBO Mol Med. 2015; 7: 1179–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y, Cao X.. Long noncoding RNAs in innate immunity. Cell Mol Immunol. 2016; 13: 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017; 18: 962–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heward JA, Lindsay MA.. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014; 35: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saberi F, Kamali M, Najafi A, Yazdanparast A, Moghaddam MM. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett. 2016; 21: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Faghihi MA, Wahlestedt C.. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009; 10: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016; 143: 3037–3044. [DOI] [PubMed] [Google Scholar]
- 49. Coleman HA, Labrador JP, Chance RK, Bashaw GJ. The Adam family metalloprotease Kuzbanian regulates the cleavage of the roundabout receptor to control axon repulsion at the midline. Development. 2010; 137: 2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang T, Huang H, Shao Q, Yee S, Majumder T, Liu G. miR-92 suppresses Robo1 translation to modulate slit sensitivity in commissural axon guidance. Cell Rep. 2018; 24: 2694–2708.e6. [DOI] [PubMed] [Google Scholar]
- 51. Bashaw GJ, Klein R.. Signaling from axon guidance receptors. Cold Spring Harb Perspect Biol. 2010; 2: a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018; 46: 2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen X, Yan CC, Zhang X, You ZH. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2017; 18: 558–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wan P, Su W, Zhuo Y. Precise long non-coding RNA modulation in visual maintenance and impairment. J Med Genet. 2017; 54: 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhong B, Liu X, Wang X, et al.. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal. 2013; 6: ra35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lin D, Zhang M, Zhang MX, et al.. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc Natl Acad Sci USA. 2015; 112: 11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Esquerdo M, Grau-Bové X, Garanto A, et al.. Expression atlas of the deubiquitinating enzymes in the adult mouse retina, their evolutionary diversification and phenotypic roles. PLoS One. 2016; 11: e0150364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.