Abstract
Cholesterol is a major carbon source for Mycobacterium tuberculosis (Mtb) during infection, and cholesterol utilization plays a significant role in persistence and virulence within host macrophages. Elucidating the mechanism by which cholesterol is degraded may permit identification of new therapeutic targets. Here, we characterized EchA19 (Rv3516), an enoyl-CoA hydratase involved in cholesterol side chain catabolism. Steady-state kinetics assays demonstrated that EchA19 preferentially hydrates cholesterol enoyl-CoA metabolite 3-oxo-chol-4,22-diene-24-oyl-CoA, an intermediate of side chain β-oxidation. In addition, succinyl-CoA, a downstream catabolite of propionyl-CoA that forms during cholesterol degradation, covalently modifies targeted mycobacterial proteins, including EchA19. Inspection of a 1.9 Å resolution X-ray crystallography structure of Mtb EchA19 suggests that succinylation of Lys132 and Lys139 may perturb enzymatic activity by modifying the entrance to the substrate binding site. Treatment of EchA19 with succinyl-CoA revealed that these two residues are hotspots for succinylation. Replacement of these specific lysine residues with negatively-charged glutamate reduced the rate of catalytic hydration of 3-oxo-chol-4,22-diene-24-oyl-CoA by EchA19, as does succinylation of EchA19. Our findings suggest that succinylation is a negative feedback regulator of cholesterol metabolism, thereby adding another layer of complexity to Mtb physiology in the host. These regulatory pathways are potential non-catabolic targets for antimicrobial drugs.
Keywords: Mycobacterium tuberculosis, cholesterol, succinylation, hydratase
Graphical Abstract
Cholesterol is a major carbon source for Mycobacterium tuberculosis (Mtb) during infection, and cholesterol metabolism is linked to energy production, virulence, and pathogenesis.1–2 In addition, transcriptional profiling has demonstrated that cholesterol regulates a subset of the Mtb genome, including genes involved in lipid metabolism.3 Disruption of several of these genes leads to growth and persistence defects in mouse models of infection,3–5 which suggests that cholesterol utilization in Mtb has the potential to be a new therapeutic target.6
In efforts to target the cholesterol catabolic pathway, our laboratory is working on elucidating its biochemical steps, focusing specifically on the degradation of the cholesterol side chain. The side chain is degraded through a series of steps that are analogous to those involved in the β-oxidation of fatty acids and are catalyzed by gene products regulated by the KstR1 repressor.7 Specifically, side chain degradation involves three multistep β-oxidation cycles, each of which produces a steroid intermediate with a truncated side chain and either acetyl-CoA or propionyl-CoA.8 We previously characterized and assigned the functions of several enzymes involved in these cycles.3, 9–14 The first oxidation step in each cycle is catalyzed by one of three KstR1-regulated acyl-CoA dehydrogenases (ACADs):10 ChsE1-ChsE2 (Rv3544c-Rv3543c), ChsE3 (Rv3573c), and ChsE4-ChsE5 (Rv3504-Rv3505). ChsE4-ChsE5 operates in the first cycle, utilizing 3-oxo-cholest-4-en-26-oyl-CoA (3-OCS-CoA); in the second cycle, ChsE3 catalyzes the dehydrogenation of 3-oxo-chol-4-en-24-oyl-CoA (3-OCO-CoA); and in the last cycle, ChsE1-ChsE2 oxidizes 3-oxo-4-pregnene-20-carboxyl-CoA. Both ChsE1-ChsE2 and ChsE4-ChsE5 form unique α2β2 heterotetramers and are thus structurally different from the ACADs produced by humans.11 ChsE3 adopts a similar structure in which the α and β domains are fused into a single polypeptide chain.15 Enoyl-CoA hydratases catalyze the second step in each β-oxidation cycle, that is, hydration of the unsaturated enoyl-CoA product of the ACAD reaction. Although β-oxidation is typically accomplished by members of the crotonase (S)-enoyl-CoA hydratase family (which is structurally distinct from the (R)-enoyl-CoA/MaoC hydratase family), the MaoC-like enoyl-CoA hydratase ChsH1-ChsH2 catalyzes hydration of branched enoyl-CoA steroid intermediate 3-oxo-4,17-pregnadiene-20-carboxyl-CoA (3-OPDC-CoA) in the third β-oxidation cycle in Mtb.9, 16 Work aimed at assignment of the hydratases that function in the first and second β-oxidation cycles is ongoing in our laboratory.
One candidate for the second cycle is the enoyl-CoA hydratase EchA19, which is encoded by the KstR1 regulon and is annotated to have both hydratase and 3-cis- to 2-trans-enoyl-CoA isomerase activity.17 Protein sequence alignments suggest that EchA19 possesses the canonical crotonase protein fold (which is used by enoyl-CoA hydratases involved in mitochondrial and peroxisomal β-oxidation)18–20 and that EchA19 has the same structure as the Mycobacterium marinum homolog (PDB:4F47).
Post-translational covalent modification of amino acid side chains is known to influence an array of protein functions, including protein–protein interactions, enzymatic activity, and protein localization and stability.21–24 Recently reported evidence suggests that the enzymes that regulate cholesterol side chain degradation undergo post-translational modifications. For example, the thiolase responsible for generating acetyl- and propionyl-CoA, FadA5, is regulated by reversible disulfide bond between two pairs of cysteines in the active site.14 Specifically, we have shown that the catalytic activity of FadA5 depends on the mycothiol redox potential in Mtb, which varies in response to oxidative stress encountered in host macrophages. In addition, other investigators have used whole-cell proteomic analyses to show that the Mtb proteome is highly succinylated, with protein targets having been found in various metabolic processes, including central carbon metabolism, lipid metabolism, and cholesterol metabolism.25–26 Nineteen KstR1-regulated enzymes, including EchA19, were identified in the succinylome analysis. The high level of succinylation on KstR1 enzymes suggests that steroid catabolism may be regulated by this post-translational modification, although its precise role in cholesterol metabolism has not been elucidated. Because complete degradation of the cholesterol side chain yields androstenedione, acetyl-CoA, and propionyl-CoA and because succinyl-CoA forms from propionyl-CoA via methylmalonyl-CoA, we hypothesized that succinylation of KstR1-regulon enzymes might regulate the generation of propionyl-CoA, a toxic metabolite, from cholesterol.
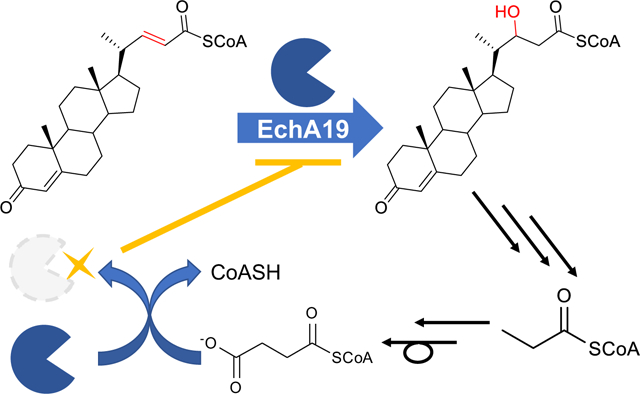
In the current study, we sought to establish whether EchA19 functions in steroid side chain catabolism and to identify its preferred steroid substrate. In addition, we investigated whether succinylation of the Lys132 and Lys139 residues of EchA19 alters the enzyme’s activity and whether the NAD+-dependent sirtuin Rv1151c catalyzes the desuccinylation of succinylated EchA19. Herein, we demonstrate that EchA19 specifically catalyzes the hydration of 3-oxo-chol-4,22-diene-24-oyl-CoA (3-OCDO-CoA) to form 3-oxo-chol-4-ene-22-hydroxy-24-oyl-CoA (22-HOCO-CoA) and that the catalytic activity of EchA19 is depressed by succinylation of both Lys132 and Lys139.
Results and Discussion
The cholesterol side chain catabolic pathway comprises three β-oxidation cycles involving KstR1-regulated enzymes and produces both methyl-branched (in cycles one and three) and straight-chain (in cycle two) steroid intermediates. We previously determined the substrate specificities of the three KstR1 ACADs (Scheme 1).10 Additionally, we demonstrated that the enoyl-CoA hydratase ChsH1-ChsH2 hydrates methyl-branched steroid 3-OPDC-CoA and thereby functions in the second step of the third cycle.9 There are two additional enoyl-CoA hydratases in the KstR1 regulon: EchA19 and Rv3538. Rv3538 is predicted to be a MaoC-like enoyl-CoA hydratase, like ChsH1-ChsH2. EchA19, in contrast, belongs to the crotonase superfamily of hydratases and possesses a different structural architecture. The structural differences among the steroid intermediates and among the potential enzymes that degrade them, along with the activity of ChsH1-ChsH2, led us to hypothesize that the EchA19 utilizes straight-chain steroid intermediate 3-OCDO-CoA and functions in the second β-oxidation cycle.
Scheme 1.
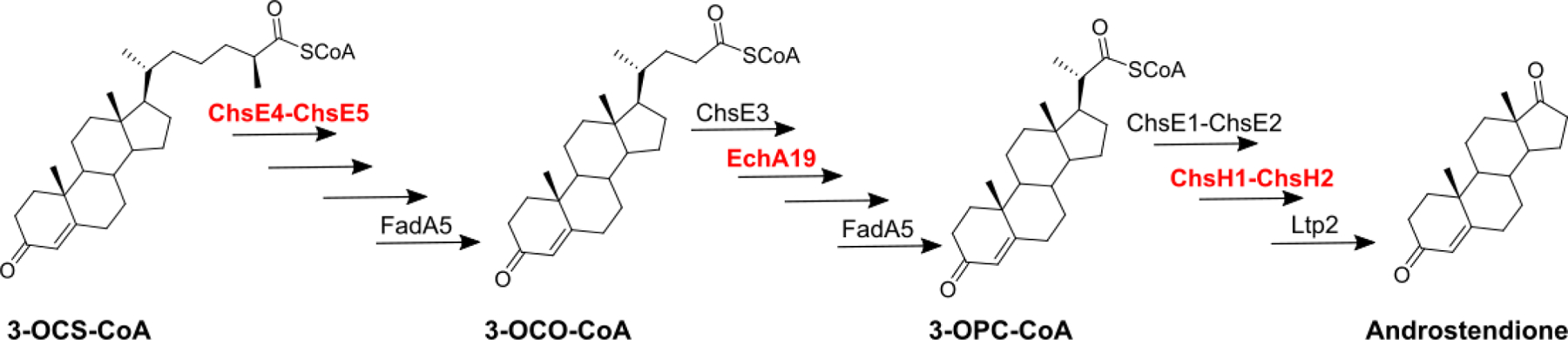
Cholesterol side chain catabolism involves three β-oxidation cycles.
Moreover, the potential additional layer of regulatory complexity that post-translational modifications introduce into the cholesterol catabolic pathway25, 27 prompted us to investigate how such modifications might affect EchA19 activity. Specifically, although various studies have demonstrated the effects of acetylation on metabolism in Mtb,28–32 the biochemical consequences of succinylation have yet to be fully investigated. Because succinyl-CoA is a downstream intermediate of cholesterol catabolism, we reasoned that succinylation could function as a negative feedback regulator. Therefore, we sought to investigate the activity of EchA19, assign its role in the cholesterol catabolic pathway in Mtb and to elucidate the effects of succinylation on EchA19 activity toward its preferred steroid substrate.
Assignment of EchA19 as an enoyl-CoA hydratase that preferentially hydrates cholesterol enoyl-CoA.
KstR1-regulated enzymes are annotated as catalyzing β-oxidation of fatty acids but demonstrate a preference for cholesterol-derived substrates over straight-chain fatty acids.9–10 To determine whether EchA19 conformed to this paradigm, we assayed its catalytic activity with four different substrates: straight-chain fatty acid octenoyl-CoA and three cholesterol catabolites: (E)-3-OCDS-CoA, 3-OCDO-CoA, and 3-OPDC-CoA. The hydration reaction was monitored spectrophotometrically.
(E) −3-OCDS-CoA was prepared enzymatically from a synthetically produced mixture of (R/S)-3-OCS-CoA. Because ChsE4-ChsE5 oxidizes only (S)-3-OCS-CoA, MCR was included in the reaction mixture to epimerize the 3-OCS-CoA.13 3-OCDO-CoA and 3-OPDC-CoA were prepared synthetically as previously described.9–10
First, by demonstrating turnover of octenoyl-CoA, we established that EchA19 is an enoyl-CoA hydratase (Table 1). Then we determined its substrate specificity by testing it with the three cholesterol catabolites. EchA19 exhibited strict specificity for the unbranched side-chain catabolite, 3-OCDO-CoA (Table 1 and Scheme 2). Comparison of the rates of EchA19-catalyzed hydration of octenoyl-CoA and 3-OCDO-CoA revealed that the kcat/Km for 3-OCDO-CoA was 89 times higher than for octenoyl-CoA, clearly establishing EchA19 as a hydratase that functions in the second β-oxidation cycle (Scheme 1). The steady-state activity profile is clearly hyperbolic (Figure S1). The 3-OCDO-CoA reaction was analyzed at equilibrium by LC-UV-MS and the 3-OCDO-CoA:22-HOCO-CoA substrate: hydrated product ratio was 30:70. On the basis of structural and sequence alignments, Srivastava et al. predicted that Mtb EchA19 possesses 3-cis to 2-trans enoyl-CoA isomerase activity; however, we did not investigate whether EchA19 catalyzes isomerization, because the cholesterol side chain catabolic pathway in Mtb is not predicted to produce unconjugated enoyl-CoA metabolites.
Table 1.
Catalytic rate constants for wild type and mutant EchA19.a
| Enzyme | substrate | Km (μM) | kcat (s−1) | kcat/Km (M−1 s−1) | Specific Activity (μmol min−1 mg−1)b |
|---|---|---|---|---|---|
| EchA19wt | octenoyl-CoA | 3.4 ± 1.8 | 1.2 ± 0.1 | (3.5 ± 0.1) × 105 | ND |
| EchA19wt | 3-OCDO-CoA | 5.8 ± 1.9 | 181.2 ± 18.7 | (31.2 ± 7.6) × 106 | 18.2 ± 5.0 |
| EchA19K132E | 3-OCDO-CoA | ND | ND | ND | 2.2 ± 0.7 |
| EchA19K139E | 3-OCDO-CoA | ND | ND | ND | 2.4 ± 0.7 |
| EchA19K132E&K139E | 3-OCDO-CoA | ND | ND | ND | 1.2 ± 0.5 |
| Succinylated EchA19wt | 3-OCDO-CoA | 8.0 ± 2.7 | 45.7 ± 5.6 | (5.7 ± 1.4) × 106 | 8.6 ± 0.8 |
| EchA19wt | 3-OCDS-CoA | NA | NA | NA | NA |
| EchA19wt | 3-OPDC-CoA | NA | NA | NA | NA |
Rate constants are averages of three technical replicates. Errors are standard deviations of nonlinear fits to the Michaelis–Menten equation. NA, no activity; no product could be detected with 40 μM substrate and 300 pM enzyme. ND, not determined. Where Km and kcat were not determined, values could not be calculated, because at concentrations required to measure saturated activity, the substrates formed aggregates that prevented accurate monitoring of enzymatic activity.
Specific activity (μmol min−1 mg−1) was determined by calculating the rate of product formation when 1 nM of enzyme was added to 10 μM substrate (3-OCDO-CoA). The error bars are the standard deviations of the calculated rates for three or four independent experiments.
Scheme 2.
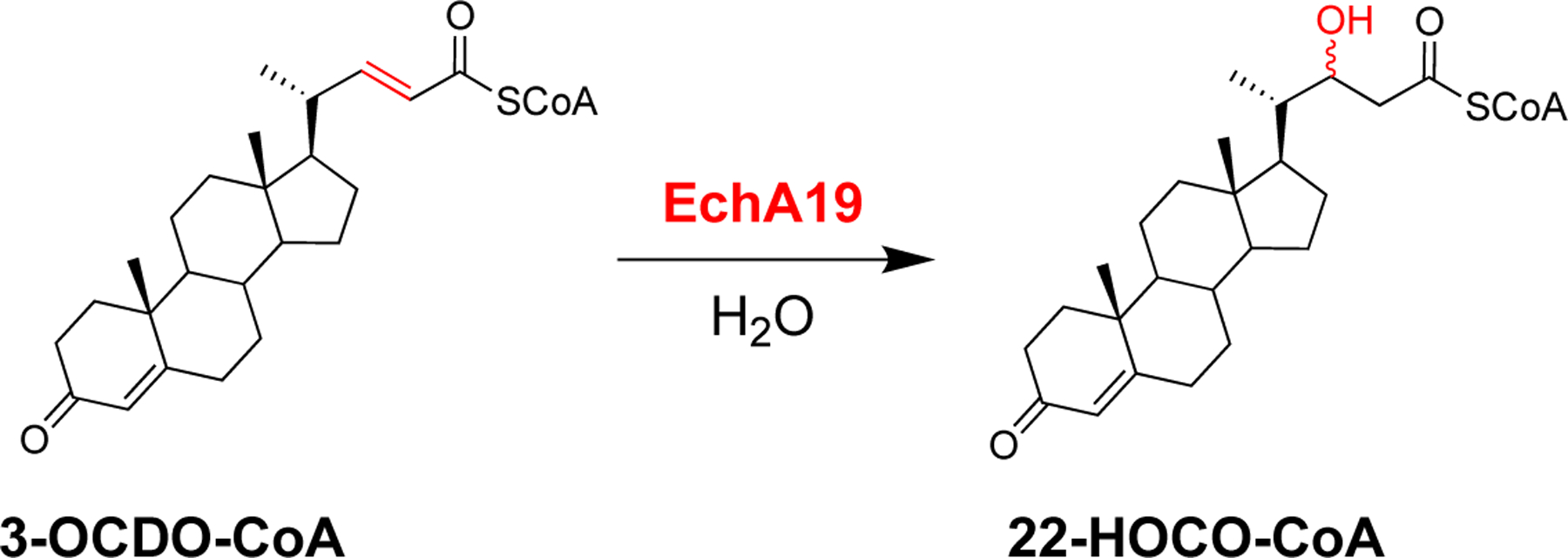
EchA19 catalyzes hydration of 3-OCDO-CoA to form 22-HOCO-CoA.
Homotrimer formation by Mtb EchA19.
We obtained an X-ray crystal structure of EchA19 at a resolution of 1.9 Å (Table 2), which revealed the structure to be nearly identical to that of the M. marinum EchA19 orthologue previously deposited in the Protein Data Bank (PDB ID 4F47). EchA19 forms a homotrimeric complex with three complete active sites (Figure 1A), and each monomer possesses the canonical ββα protein fold of the crotonase superfamily.18 To investigate the interactions needed for substrate binding and catalysis, we modeled the substrate analog, 3-OCDO-CoA into the active site of Mtb EchA19 (Figure 1B,C). Structural superposition of EchA19 with the rat mitochondrial bifunctional enoyl-CoA hydratase (PDB ID 2DUB) revealed that the catalytic glutamate residues and the substrate stabilization residues are conserved.19–20 The monomer–monomer interfaces of EchA19 form elongated tunnels that accommodate the aliphatic side chain of 3-OCO-CoA, and the phosphates of the CoA moiety face the solvent. At the end of each tunnel is a cleft that houses the cyclic steroid backbone of 3-OCO-CoA. 3-OCO-CoA binding is stabilized by hydrogen-bonding interactions between the Gly114, Ala64, Leu68, Ala25, Ala28, and Lys69 residues of monomer B (cyan) and the carbonyl oxygen at the 24-position of 3-OCO-CoA and the adenine ring of CoA (Figure 1C). Phe252′ of monomer C (purple) stabilizes the adenine ring of the adenosine through pi-stacking interactions. Lys69 and Lys255′ of monomer C form salt bridges with the 3′-phosphates of CoA (Figure 1C). This predicted binding conformation suggests that the aliphatic side chain of 3-OCO-CoA is located such that active site Glu117 and Glu137 residues are properly positioned to coordinate a water molecule that assists in the hydration of the α,β-alkene of the preferred enoyl-CoA substrate, 3-OCDO-CoA (Figure 1D).
Table 2.
Crystallography data collection and refinement statistics for Mtb EchA19
| Parameter | Valuea |
|---|---|
| Wavelength (Å) | 0.97934 |
| Resolution range (Å) | 37.71–1.915 (1.983–1.915) |
| Space group | P 3 2 1 |
| Unit cell | a = b = 75.413 Å, c = 68.89 Å, α = β = 90°, γ = 120° |
| Total reflections | 344811 (12826) |
| Unique reflections | 17862 (1755) |
| Multiplicity | 19.3 (14.8) |
| Completeness (%) | 99.88 (100.00) |
| Mean (I) (sigma) | 19.4 (2.4) |
| Wilson B-factor (Å2) | 35.94 |
| Rmerge | 0.078 (0.867) |
| Rmeas | 0.080 (0.898) |
| Rpim | 0.018 (0.231) |
| CC1/2 | 0.999 (0.931) |
| Reflections used in refinement | 17845 (1755) |
| Reflections used for Rfree | 861 (95) |
| Rwork | 0.1810 (0.2788) |
| Rfree | 0.2342 (0.3856) |
| Number of nonhydrogen atoms | 1825 |
| Macromolecules | 1748 |
| Ligands | 1 |
| Solvent | 76 |
| Protein residues | 237 |
| RMS (bonds) (Å) | 0.011 |
| RMS (angles) (deg) | 1.06 |
| Ramachandran favored (%) | 96.57 |
| Ramachandran allowed (%) | 2.58 |
| Ramachandran outliers (%) | 0.86 |
| Rotamer outliers (%) | 1.16 |
| Clashscore | 2.01 |
| Average B-factor Å2 | 56.23 |
| Macromolecules | 56.44 |
| Ligands | 40.65 |
| Solvent | 51.65 |
| Number of TLS groups | 4 |
| PDB ID | 6WYI |
Statistics for the highest-resolution shell are shown in parentheses.
Figure 1. Mtb enoyl-CoA hydratase EchA19 is a homotrimer with conserved active site residues and CoA binding residues and large binding pockets to accommodate cholesterol metabolite substrates.
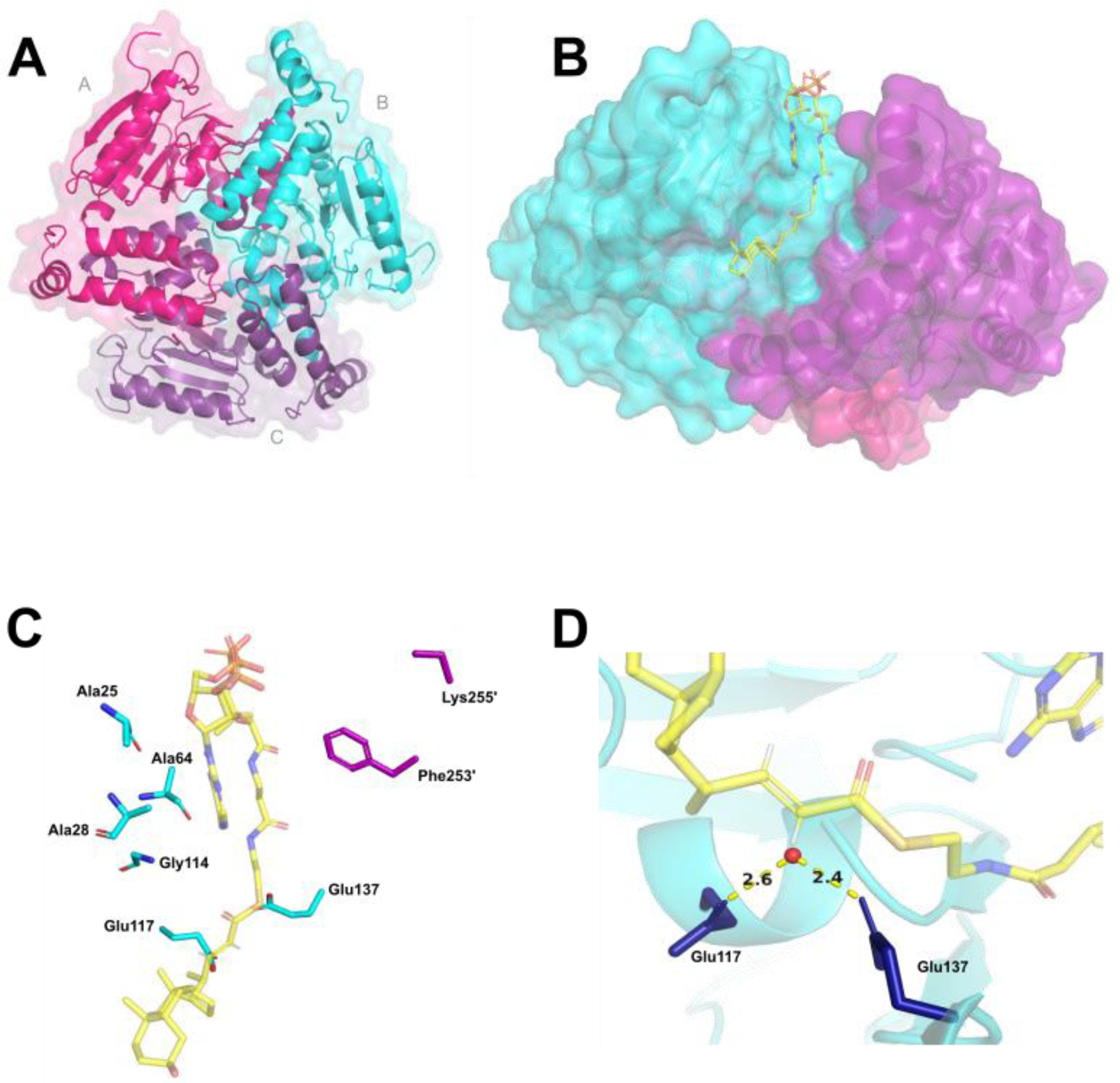
A) KstR1-regulated EchA19 is a homotrimer (monomer A, magenta; monomer B, cyan; monomer C, purple) with three substrate binding pockets formed by the monomer–monomer interfaces. Each monomer possesses the canonical αββ protein fold of the crotonase-like family. B) 3-OCDO-CoA is modeled into an active site of Mtb EchA19. 3-OCDO-CoA adopts an elongated conformation within the active site (monomers B and C are shown). C) Conserved active site residues (Glu117 and Glu137) and substrate interacting residues in the substrate binding pocket are Ala25, Ala28, Ala64, Leu68, Lys69, and Gly114 (which belong to monomer B) and Lys255’ and Phe253’ (which belong to monomer C). The Leu68 and Lys69 residues are located in a disordered region of the Mtb EchA19 crystal structure (Gly65–Arg89). The shortened side chain of Lys255’ is the result of incomplete electron density for this residue in the protein structure. D) Catalytic residues Glu117 and Glu137 coordinate a water molecule (depicted as a red circle), positioning it for addition to the alkene of 3-OCDO-CoA.
Members of the crotonase superfamily of enoyl-CoA hydratases are generally used by bacteria and eukaryotes for the hydration of fatty acid enoyl-CoA metabolites.33–34 Interestingly, EchA19 utilizes straight-chain steroid enoyl-CoA metabolite 3-OCDO-CoA, whose aliphatic side chain is structurally like those of fatty acid enoyl-CoA metabolites. Conversely, the other enoyl-CoA hydratases in the cholesterol side chain catabolic pathway (ChsH1-ChsH2 and Rv3538) possess a unique MaoC-like protein fold. ChsH1-ChsH2 hydrates α-methyl-branched steroid enoyl-CoA 3-OPDC-CoA. The α-methyl branch structure of the 3-OCDS-CoA substrate and the predicted MaoC-like motif of Rv3538 suggest that Rv3538 catalyzes hydration of 3-OCDS-CoA in the first β-oxidation cycle of the pathway. These assignments suggest that structural differences determine substrate specificity.
Susceptibility of Mtb EchA19 Lys132 and Lys139 to succinylation.
Whole-cell proteomic analysis has shown that succinylation of the proteome occurs extensively in Mtb25 and that several cholesterol catabolic enzymes, including EchA19, are succinylated. EchA19 is succinylated at Lys132 and Lys139. Although the cellular concentration of succinyl-CoA will depend on the cell’s metabolic state, the succinyl-CoA concentration in the mitochondrial matrix is estimated to be 0.1–0.6 mM, which is sufficient to non-enzymatically succinylate protein lysines.35–36 Additionally, non-enzymatic succinylation can occur at pH 4–8, a range that overlaps with the intracellular pH (7.2) of Mtb in host macrophages.37–38 Non-enzymatic succinylation is attributed to succinyl-CoA’s higher reactive acylation potential relative to those of other transferred groups, including acetyl-CoA.37, 39–42 The reactivity of succinyl-CoA is postulated to result from its terminal carboxylate and its propensity to self-hydrolyze into free CoASH to form highly reactive succinic anhydride at physiological pH.37 Thus, succinylation of Mtb EchA19 may occur non-enzymatically in the cell.
We utilized protein mutagenesis and immunoblot analysis to investigate the extent to which Lys132 and Lys139 are required for the overall succinylation state of Mtb EchA19. To block succinylation at these two residues, we generated single lysine-to-glutamate mutants EchA19K132E and EchA19K132E and the double mutant EchA19K132E&K139E. These mutants also serve to model the negative charge introduced upon succinylation, although the side chains are at a shorter distance from the polypeptide backbone than those in the succinylated wild-type protein. Recombinantly expressed and purified EchA19wt and the lysine mutants were non-enzymatically succinylated with 50 μM succinyl-CoA at pH 7.2 and pH 8.1, and then the extent of succinylation was monitored with a polyclonal pan anti-succinyllysine antibody. After 60 minutes, succinylation of EchA19wt was only detected at pH 8.1. The single Lys132 and Lys139 mutants were succinylated to a lesser extent than EchA19wt. However, mutation of both Lys132 and Lys139 resulted in nearly complete abolishment of Mtb EchA19 succinylation (Figure 2A). An experiment with EchA19wt that was expressed in E. coli but not exposed to succinyl-CoA revealed that the in vivo concentration of succinyl-CoA in E. coli during recombinant expression was insufficient to succinylate EchA19 to detectable levels.
Figure 2. EchA19 enzymes are succinylated at sites near the substrate binding pockets.
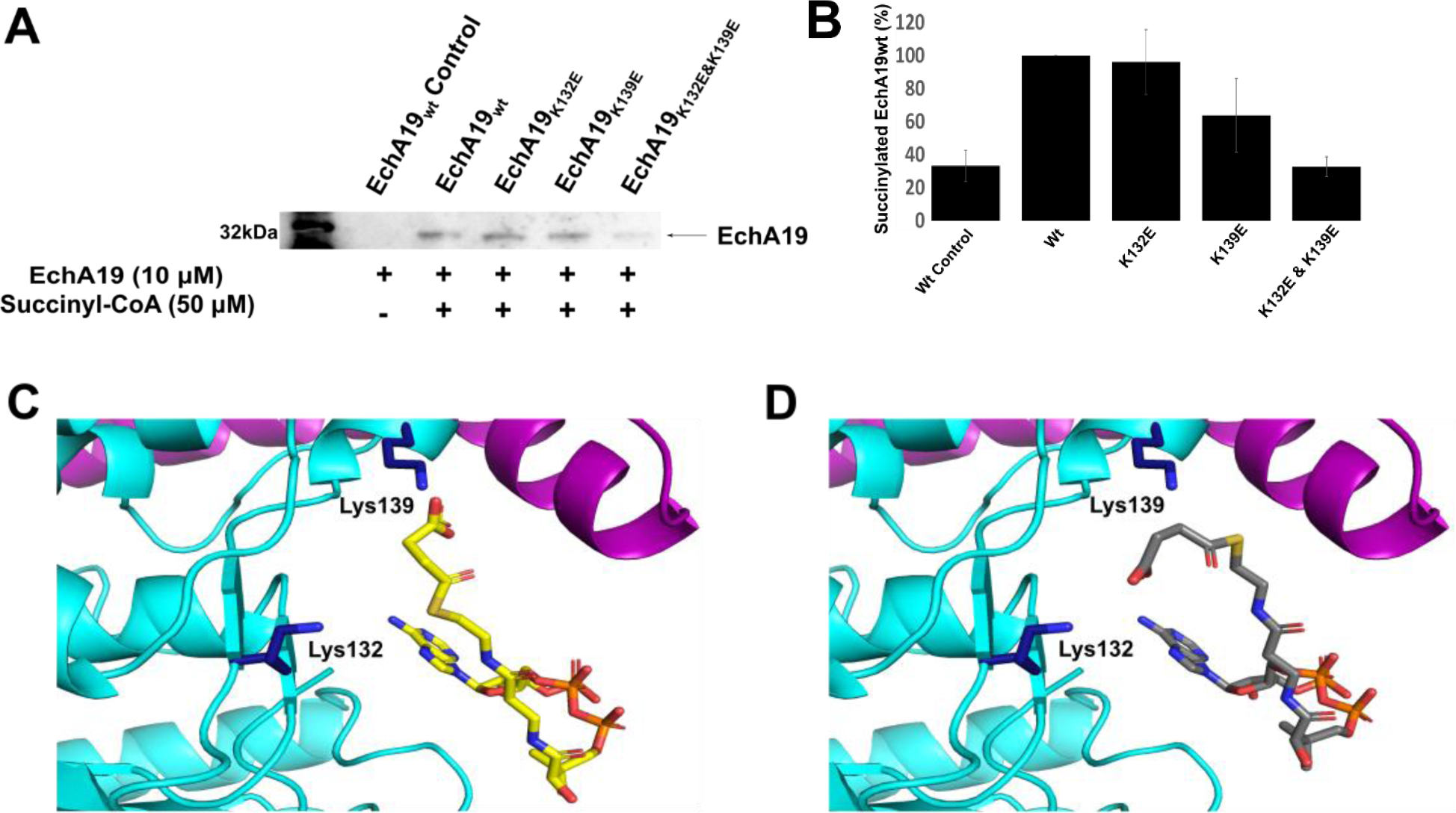
A) Recombinant EchA19wt and three lysine mutants (EchA19K132E, EchA19K139E, and EchA19K132E&K139E) were incubated with 50 μM succinyl-CoA at pH 8.1 for 60 min, and the extent of succinylation was monitored with a pan anti-succinyllysine antibody. B) Densitometry analysis of blots from three replicates of experiment shown in (A). Intensity was scaled to 100% for succinylated EchA19wt. Errors shown are the standard error of measurement. C) and D) The active site of EchA19 with succinyl-CoA modeled in the CoA pocket identified in the 3-OCDO-CoA-bound model (Figure 1). Lys132 and Lys139 (dark blue) are located near the CoA binding pocket of EchA19.
There are 13 lysine residues in each Mtb EchA19 monomer, and consistent with prior proteomic analysis, we demonstrated that Lys132 and Lys139 were most reactive. Their hyper-reactivity is consistent with their location at the entrance to the substrate binding site (Figure 2C, 2D). We hypothesize that the CoA moiety of succinyl-CoA may bind in the EchA19 CoA pocket, thereby increasing the effective concentration of succinyl-CoA and enhancing the rate of its reaction with the proximal ε-amino moieties of Lys132 and Lys139.40 The coenzyme A moiety of succinyl-CoA was modelled into the coenzyme A binding site. There are 2 possible conformations (yellow: conformation 1; gray: conformation 2, Figure 2C, 2D). The amino moieties of Lys132 and Lys139 are located near the thioester of the succinyl-CoA. In conformation 1, Lys139 can form a salt bridge with the terminal carbonyl and Lys132 can be succinylated by the second carbonyl. In conformation 2, Lys 132 can form a salt bridge and Lys 139 is succinylated. Therefore, non-enzymatic succinylation of EchA19 at Lys132 and Lys139 may be due to self-succinylation.
In addition to non-enzymatic or self-succinylation, there may be as-yet-unidentified succinyl transferases present in Mtb. The Mtb genome encodes 30 CoA transferases, including the well-characterized MtPat (Rv0998). Although these enzymes are annotated as acetyl-CoA transferases, some of them may catalyze the transfer of other acyl groups. Rv2170 transfers succinyl-, propionyl-, and acetyl-CoA to isocitrate dehydrogenase lysines.43 Additionally, Rv0802c contains a GCN5-related N-acetyltransferase motif and co-crystallizes with succinyl-CoA.44
Effect of succinylation on Mtb EchA19 activity.
In the Mtb EchA19 structure, Lys132 and Lys139 are located at the entrance to the substrate binding site in a gate-like conformation, and their side chains extend into the solvent channel that leads to the active site (Figure 2C and 2D). Modeling suggests that succinylation at these specific sites disrupts substrate binding and subsequent catalysis (Figure 2C and 2D).
The lysine-to-glutamate EchA19 mutants mimicked succinylation, owing to the inversion of charge. Therefore, we investigated the effects of these introduced negative charges on the activity of EchA19 toward its preferred substrate, 3-OCDO-CoA (Table 1). We observed that the specific activity values for both EchA19K132E and EchA19K139E were lower than the value for EchA19wt, but reduction in activity was greatest for the double mutant (EchA19K132E&K139E); the rate for this mutant was nearly 1/16 that of the Wt). Thus, we inferred that succinylation of Mtb EchA19 has important consequences for enzymatic activity and suppresses cholesterol catabolism. As a further test, we measured the catalytic activity of EchA19wt(succ) and found that the kcat/Km was reduced 5.5-fold compared to EchA19wt, primarily a consequence of a reduced kcat. The specific activity of EchA19wt(succ) was half that of EchA19wt. Comparison of these rates suggests that EchA19wt succinylation is approximately 50% complete under the conditions utilized. Small-angle X-ray scattering analysis did not reveal any structural differences between EchA19wt and EchA19K139E (Table S3 and Figure S2)
Evaluation of succinylated EchA19wt as a substrate for Rv1151c.
The Mtb genome encodes one known deacylase, the NAD+-dependent sirtuin Rv1151c. Like other members of the sirtuin superfamily, which demonstrate activity with acylated proteins,42, 45–46 Rv1151c catalyzes deacetylation, depropionylation, or desuccinylation of various proteins, including those involved in central carbon metabolism and cholesterol C and D ring catabolism.25, 28–29, 47 Therefore, we investigated whether succinylated EchA19wt was a substrate for Rv1151c. Succinylated EchA19wt was incubated with Rv1151c, and desuccinylation was monitored over 2 h. Succinylation is decreased 85% after 1 hour compared to the control and no further decrease was observed after an additional hour (Figure 3). Thus, succinylated EchA19wt is a substrate for Rv1151c. Because Rv1151c activity depends on the presence of NAD+ as a second substrate and because the NADH/NAD+ ratio increases as a consequence of cholesterol β-oxidation, the NADH/NAD+ potential may provide yet another post-translational regulatory mechanism for cholesterol catabolism in Mtb.
Figure 3. Succinylated Mtb EchA19 is a substrate for the sirtuin Rv1151c.
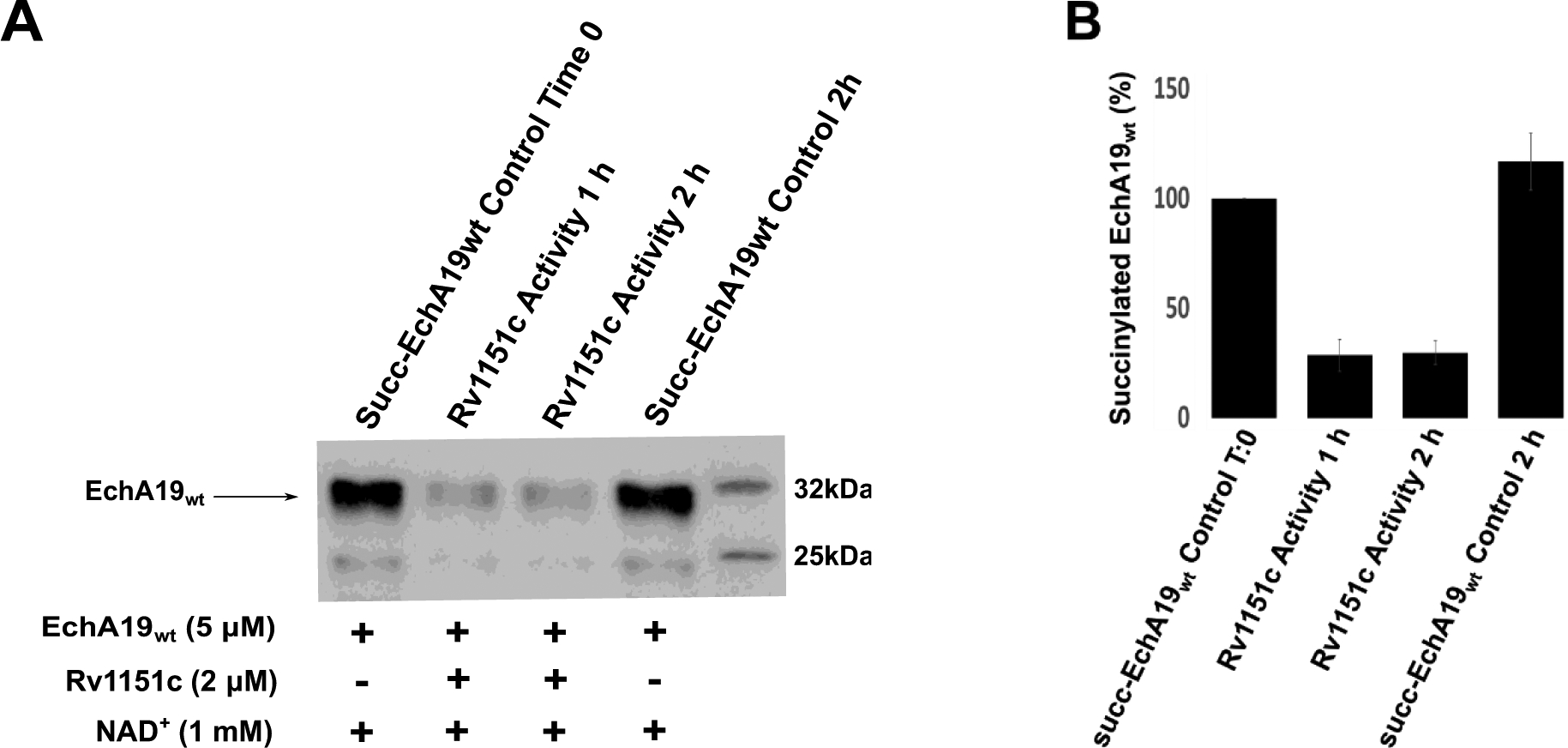
A) Succinylated EchA19wt (5 μM) was incubated with 2 μM Rv1151c in the presence of 1 mM NAD+ in a solution containing 25 mM Tris-HCl (pH 7.5), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, and 0.1% BSA. Desuccinylase activity catalyzed by Rv1151c was monitored as a function of time over 2 h, and remaining succinylated EchA19 was detected with a pan anti-succinyllysine antibody. B) Densitometry analysis of blots from three replicates of experiment shown in (A). Intensity was scaled to 100% for succinylated EchA19wt. Intensity was scaled to 100% for succinylated EchA19wt. Errors shown are the standard error of measurement.
Conclusion
The ability of Mtb to survive and persist in the harsh host environment is a tribute to its ability to adapt to changing environmental conditions. In this study, we identified EchA19 as the enoyl-CoA hydratase that catalyzes the hydration of 3-OCDO-CoA, and we found that the activity of this enzyme is negatively regulated by reversible succinylation. Two other enzymes involved in cholesterol side chain catabolism—ChsE4-ChsE5 (the ACAD in the first β-oxidation cycle) and ChsH1-ChsH2 (an enoyl-CoA hydratase in the third β-oxidation cycle)—are also succinylated,25–26 suggesting that all three β-oxidation cycles are post-translationally regulated (Figure 4). Furthermore, enzymes involved in the degradation of the steroid rings carry the succinylation mark (FadA6 [Rv3556c], FadD3 [Rv3561], FadE32 [Rv3563], HsaC [Rv3568c], HsaD [Rv3569c], and Rv3570c).25, 48 Employment of a downstream byproduct (succinyl-CoA) as a post-translational modifier to reduce catalytic activity suggests that succinylation operates as a negative feedback mechanism. The multivariate molecular cross-talk modalities in the cholesterol catabolic pathway highlight a sophisticated metabolic network with multiple layers of regulation, all of which must be considered in attempts to develop new antimycobacterial therapeutics.48
Figure 4. Succinylation provides an additional layer of metabolic regulation for cholesterol utilization in Mtb.
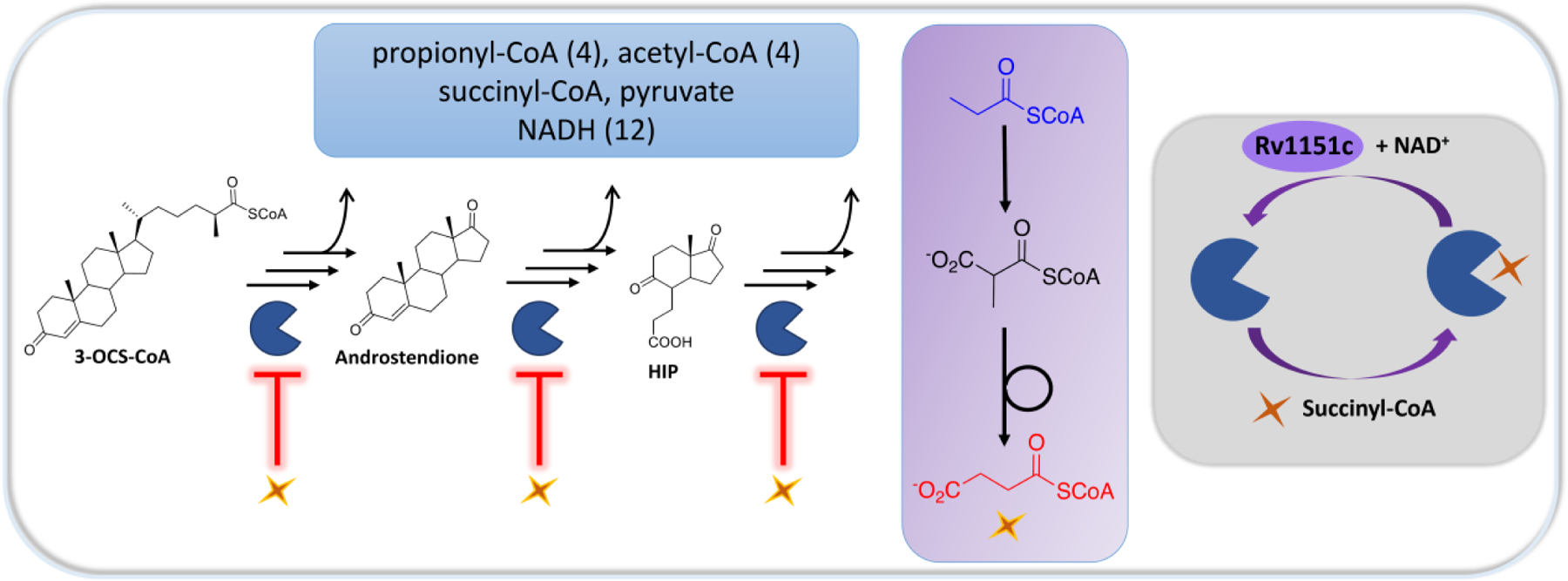
During cholesterol catabolism in Mtb, the cellular NADH pool increases, and toxic propionyl-CoA is produced (blue box). Succinyl-CoA is produced by C and D ring metabolism and by carboxylation of propionyl-CoA to form methylmalonyl-CoA followed by isomerization (purple box). The cholesterol catabolic enzyme EchA19 is post-translationally regulated by succinylation, which reduces the enzyme’s activity, suggesting that a negative feedback mechanism regulates cholesterol catabolism on the basis of the levels of propionyl-CoA and succinyl-CoA produced (gray box). Succinylation of EchA19 is reversed by Rv1151c. Several other enzymes at each stage of cholesterol catabolism have known sites of succinylation,25 and the catalytic activities of these enzymes are postulated to be similarly regulated by this modification. HIP: 3aα‐H‐4α(3’‐propanoate)‐7aβ‐methylhexahydro‐1,5‐indanedione.
Methods
Materials, Bacterial Strains, Media, and General Methods.
The pET28B expression vector and ligation-independent cloning T4 polymerase were purchased from Novagen (Madison, WI). Polyclonal rabbit pan anti-succinyllysine primary antibody was purchased from PTM Biolabs (Chicago, IL). HindIII and NdeI restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA). XL1 Blue and BL21 (DE3) Escherichia coli cells, iProof high-fidelity DNA polymerase, and horseradish peroxidase–conjugated goat anti-rabbit secondary antibody were purchased from BioRad (Hercules, CA). A QuikChange site-directed mutagenesis kit and DpnI was purchased from Agilent (Santa Clara, CA). Kanamycin, ampicillin, and isopropyl β-d-1-thiogalactopyranoside were purchased from GoldBio (St. Louis, MO). His NTA resin was purchased from Qiagen (Germantown, MD). A HiLoad 16/60 Superdex 200 prep-grade GE column was purchased from GE Healthcare Biosciences Corp. (Piscataway, NJ). MgCl2, ATP, free CoA, LB medium, HEPES, TAPS, and Tris-HCl were purchased from Fisher Scientific (Pittsburgh, PA). Ferrocenium hexaflurophosphate, bovine serum albumin (BSA) and 2,3-octenoic acid were purchased from Sigma Aldrich (St. Louis, MO). Tryptone and yeast extract were purchased from Becton Dickinson (Franklin Lakes, NJ). 4,22-stigmastadiene-3-one was purchased from MP Biochemicals (Solon, OH). Primary crystallization screening kits were purchased from Hampton Research (Aliso Viejo, CA). Mtb genomic DNA was extracted from wild-type (WT) Mtb H37Rv bacteria culture. Traditional PCR and site-direct mutagenesis oligonucleotides were synthesized by IDT Inc. (Coralville, IA). Plasmid DNA sequencing was conducted by Genewiz (South Plainfield, NJ). LC-MS analysis was conducted on an Agilent UPLC/MS instrument with diode array and single quadrupole detector. Proteins were purified on a Knauer Azura fast protein LC system. Matrix-assisted laser-desorption time-of-flight (MALDI-TOF) MS was conducted on a Bruker Autoflex II TOF/TOF for MALDI-TOF. Bacteria cells were lysed with a Constant Systems TS series benchtop cell disruptor (Kennesaw, GA). Fatty acid and cholesterol substrates were purified using a Shimadzu (Somerset, NJ) HPLC system. Steady-state kinetics assays were conducted on a Shimadzu UV2550 UV–vis spectrophotometer. 2xYT medium is composed of 16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl. Recombinantly expressed proteins were purified in the buffers described below.
α-Methyl-CoA racemase (MCR) buffers.
MCR Buffer A (lysis/washing): 20 mM Tris-HCl (pH 8), 300 mM NaCl, 10 mM imidazole. MCR Buffer B (elution): 20 mM Tris-HCl (pH 8), 300 mM NaCl, 500 mM imidazole. MCR Buffer C (dialysis/storage): 20 mM Tris-HCl (pH 8), 300 mM NaCl, supplemented with 5% glycerol.
Buffers for EchA19wt and succinylation mutants.
EchA19 Buffer A (lysis/washing): 20 mM Tris-HCl (pH 8), 300 mM NaCl. EchA19 Buffer B (elution): 20 mM Tris-HCl (pH 9), 300 mM NaCl (pH 9), 500 mM imidazole. EchA19 Buffer C (storage): 20 mM Tris-HCl (pH 8), 300 mM NaCl, supplemented with 5% glycerol.
Rv1151c buffers.
Rv1151c Buffer A (lysis/washing): 50 mM NaH2PO4 (pH 8, 300 mM NaCl, 20 mM imidazole. Rv1151c Buffer B (elution): 50 mM NaH2PO4 (pH 8), 300 mM NaCl, 500 mM imidazole. Rv1151c Buffer C (dialysis/storage): 50 mM Tris-HCl (pH 7.5), 150 mM NaCl.
Expression Plasmid Construction.
Mtb genomic DNA was used as a template for PCR amplification of ChsE4-ChsE5 (Rv3504-Rv3505), MCR (Rv1143), EchA19 (Rv3516), FadD17 (Rv3506), and Rv1151c (Rv1151c). The constructs used in this work are listed in Table S1 with the primers listed in Table S2 for either traditional or ligation-independent cloning, as described below.
Traditional cloning.
The pET28 vector was used to generate N-terminal His-tagged constructs for ChsE4-ChsE5, EchA19 (wild type and mutants), FadD17, and Rv1151c. Gene inserts were PCR-amplified by means of the iProof high-fidelity DNA polymerase protocol. Gene inserts and the pET28B vector were digested with NdeI and HindIII restriction enzymes for 6 h at 37 °C, and the enzymes were subsequently heat-inactivated for 20 min at 80 °C. The vector was also treated with calf alkaline phosphatase to prevent self-ligation. The digested gene inserts and vector were gel-purified and then ligated with T4 ligase at 37 °C for 14 h. Ligation reactions were transformed into XL1 Blue E. coli cells and selected on kanamycin (50 μg/mL) LB medium. Plasmids were amplified in and purified from XL1 Blue E. coli cells, and gene insertion was confirmed by sequencing of the full gene across insertion sites.
Ligation-independent cloning.
The ligation-independent cloning vector 2BT was used to generate N-terminal His-tagged constructs for MCR expression. Ligation reactions were transformed into XL1 Blue E. coli cells and selected on ampicillin (100 μg/mL) LB medium. Plasmids were amplified in and purified from XL1 Blue E. coli cells, and gene insertion was confirmed by sequencing the full gene across insertion sites.
Site-directed mutagenesis.
EchA19 mutants (K132E, K139E, K132&K139) were generated using the Agilent site-directed mutagenesis protocol. The mutagenic primer sequences are shown in Table S2. The pET28B (EchA19wt) expression vector was used as a template for generation of the pET28B (EchA19K132E) and pET28B (EchA19K139E) plasmids. The pET28B(EchA19K139E) expression vector was used as the template for pET28B (EchA19K132E&K139E) plasmid generation. After PCR amplification, the parental DNA was digested with 1 μL of DpnI (10 U/μL). The DpnI-treated DNA (2 μL) was transformed into XL1 Blue E. coli cells for plasmid amplification and purification. Mutagenesis was confirmed by DNA sequencing of the coding sequence of the expression plasmids.
Protein Expression and Purification.
ChsE4-ChsE5 and FadD17 were expressed and purified as previously described.10, 12 All other proteins were expressed and purified by means of the following general procedure, unless indicated otherwise. A 7 mL culture (supplemented with 50 μg/mL kanamycin or 100 μg/mL ampicillin [final concentrations]) was used to inoculate 1 L of 2xYT medium, and the 1 L culture was incubated at 37 °C until it reached an OD600 of 0.8 (about 3 h). The incubation temperature of the 1 L culture was lowered to 25 °C, and the culture was allowed to equilibrate for 20 min. Isopropyl β-d-1-thiogalactopyranoside (final concentration, 400 μM) was added, and the cell pellet was harvested after 20h.
The bacterial cultures were pelleted at 4870 g for 20 min at 4 °C. Buffer A (40–100 mL) was added to the cell pellet, and the cells were lysed with a cell disruptor. Homogenates were subjected to ultracentrifugation at 18600 xg for 1 h at 4 °C. The protein-containing supernatant was loaded onto an IMAC His Bind·Resin (5 mL), which was washed with Buffer A and eluted with Buffer B. Fractions were collected and dialyzed overnight against Buffer C with 10 kDa molecular weight cutoff MWCO dialysis tubing.
After dialysis, the protein solution was concentrated by ultrafiltration (MWCO 10 kDa) to less than 2 mL, and the protein was purified by size-exclusion chromatography on a Superdex 200 column pre-equilibrated with Buffer C. Protein-containing fractions were analyzed with 12% SDS PAGE gel. The fractions were then concentrated, and protein concentrations were determined by means of UV spectroscopy at 280 nm using the following extinction coefficients: ε280nm (ChsE4) = 68,995 M−1 cm−1, ε280nm(ChsE5) = 36,565 M−1 cm−1, ε280nm(EchA19) = 17,210 M−1 cm−1, ε280nm(MCR) = 57,410 M−1cm−1, ε280nm(FadD17) = 56,840 M−1 cm−1, and ε280nm(Rv1151c) = 51,700 M−1 cm−1. Protein purities were determined by SDS-PAGE (Figure S3) and identities were confirmed by means of tryptic digestion and MALDI-TOF MS.
Synthesis of Cholesterol-CoA Substrates.
The protocols for synthesis of the saturated cholesterol substrates 3-OCO-CoA and 3-OCS-CoA and unsaturated cholesterol substrate 3-OPDC-CoA have been described previously.9–10 Octenoyl-CoA was synthesized from octenoic acid by means of the mixed anhydride method, as previously described.9
3-OCDS-CoA.
3-OCDS-CoA was synthesized enzymatically with the acyl-CoA dehydrogenase ChsE4-ChsE5 and MCR.10, 13 Specifically, an isomeric mixture of (R) and (S) 3-OCS-CoA (1 mM) was incubated with ChsE4-ChsE5 (10 μM) and MCR (10 nM) with the electron acceptor ferrocenium hexafluorophosphate (250 μM) in 100 mM TAPS buffer (pH 8.5) for 5 h at 25 °C. The progress of the reaction was monitored by MALDI-TOF MS. The product was purified by HPLC on a semiprep C18 column (Phenomenex: Torrance, CA) with 10 mM ammonium acetate containing acetonitrile (linear gradient from 5% to 100% over 50 min).
3-OCDO-CoA.
20-Formyl-pregn-4-en-3-one was obtained by ozonolysis of 4,22-stigmastadiene-3-one with reductive workup.49 To a cooled (0 °C on ice) solution of NaH (30 mg, 1.25 mmol) in 5 mL of THF, tert-butyl diethylphosphonoacetate (315 mg, 1.25 mmol) was added dropwise. After the reaction was stirred for 5 min, a solution of 20-formyl-pregn-4-en-3-one (275 mg, 0.8 mmol) in 5 mL of THF was added, and the mixture was allowed to warm to 25 °C and then stirred for an additional 12 h. The THF was removed by rotary evaporation, and the resulting solid residue was dissolved in 5 mL of TFA and 5 mL of CH2Cl2 and stirred at 25 °C for 3 h. The TFA and CH2Cl2 were evaporated, and the residue was purified by flash chromatography with 1:4 EtOAc/hexane as the eluent to yield 3-OCDO-acid (180 mg, 61%). 3-OCDO-CoA was then synthesized enzymatically from the 3-OCDO-acid using the steroid-CoA ligase FadD17. Specifically, 3-OCDO-acid (0.66 mM) was incubated with FadD17 (0.5 μM) for 1 h at 30 °C in a reaction buffer containing 1.1 mM free CoA, 2.5 mM ATP, 10 mM MgCl2, and 100 mM HEPES (pH 8.0). The product was purified on an HPLC system equipped with a semiprep C18 column (Phenomenex) with 10 mM ammonium acetate containing acetonitrile (linear gradient from 5% to 100% over 50 min) as the eluent.
Non-enzymatic Succinylation of Proteins and Immunoblot Analysis.
WT and mutant EchA19 (10 μM) were incubated with 50 μM succinyl-CoA in 100 mM Tris-HCl (pH 8.1) at 25 °C. At the indicated reaction time, an aliquot of the reaction mixture was added to SDS loading dye (final concentration, 1× SDS 6% β-mercaptoethanol). The reaction/SDS aliquots were boiled for 10 min, allowed to cool, and then loaded onto 12% SDS PAGE gel, with a total of 200 ng of protein added to each well. The gels were run at 70 V for 30 min and then at 135 V for 1.5 h. The gels were then transferred to pre-activated (with methanol) polyvinylidene difluoride membrane at a constant voltage of 30V for 16h at 4 °C; the transfer buffer contained 25 mM Tris (pH 8.3), 190 mM glycine, and 20% methanol. After transfer, the membrane was blocked with TBST buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.5 mM KCl, 0.1% Tween 20) supplemented with 5% (w/v) non-fat milk for 1 h at rt with rocking. The membrane was then incubated with TBST buffer supplemented with 5% non-fat milk and pan anti-succinyllysine antibody (1:1000 dilution) for 16 h at 4 °C with rocking. The membrane was washed in TSBT for 10 min with rocking (3 times) and then incubated with horseradish peroxidase–conjugated goat anti-rabbit antibody (1:3000 dilution) for 1 h at rt with rocking. The membrane was subsequently washed with TBST buffer for 10 min (3 times). Blots were developed with chemiluminescence enhancer solution using a ChemiDoc instrument (BioRad: Hercules, CA).
Hydratase Assay.
The activities of WT and mutant EchA19 were assayed with octenoyl-CoA, 3-OPDC-CoA, 3-OCDO-CoA, and 3-OCDS-CoA as substrates in 100 mM HEPES (pH 7.4) and 3 mM ethylenediaminetetraacetic acid (EDTA). Inclusion of EDTA was essential to eliminate a contaminating phosphatase activity that could not be removed through repeated chromatography. Enzymatic activity was monitored at substrate concentrations ranging from 2 to 100 μM (cholesterol substrates) or 2 to 300 μM (octenoyl-CoA). Reactions were monitored at 263 nm, and initial velocities were determined upon addition of one of the following enzymes: EchA19wt (89 nM with octenoyl-CoA or 300 pM with 3-OCDS-CoA), EchA19K132E (500 pM), EchA19K139E (1 nM), EchA19K132E&K139E (3 nM), or succinylated EchA19wt (1 nM). A change in extinction coefficient (ε263nm) value of 6700 M−1 cm−1, which corresponds to α,β-unsaturation of the enoyl-CoA substrates, was used to quantify product formation.
LC-MS Analysis.
Cholesterol substrates and products were detected with a Kinetex EVO C18 column (2.6 μm, 100 Å, 100 × 2.1 mm; Phenomenex) with Solvent A1 (H2O containing 10 mM ammonium acetate, pH 7) and Solvent B2 (95:5 CH3CN/H2O containing 10 mM ammonium acetate, pH 7) using the following separation parameters: t = 0–3 min, B2 = 5–30%; t = 3–28 min, B2 = 30–55%; t = 28–33 min, B2 = 55–95%; t = 33–34 min, B2 = 95–99%; the flow rate was 0.40 mL/min, and the temperature was 35 °C. The MS method was as follows: —instrument state, TOF 2 GHz ExtDyn 3200; calibration with a small-molecule reference mix (3 reference masses); acquisition, ESI m/z = 100–3200, 1 Hz, centroid mode; Vcap, 4000 V; desolvation gas flow rate, 11 L/min; desolvation gas temperature, 300 °C; nebulizer pressure, 45 psig; fragmentor voltage, 175 V; detector voltage, 780 V. The diode-array detector (DAD) method parameters were as follows: 205–500 nm, 2 nm slit, 2 nm step, 1 mAU threshold, DAD1-A 215 ± 5 nm; DAD1-B 260 ± 5 nm; reference 500 ± 25 nm, acquisition, 5 Hz. The injection volume was 10.0 μL (T = 5 °C). The identity of the 3-OCDO-CoA hydration product at reaction equilibrium was confirmed by high resolution LC-MS analysis. 22-HOCO-CoA ([MH+]/z, UV260 30%): 1137.3660 (calc) 1137.3567 (meas); 3-OCDO-CoA ([MH+]/z, UV260 70%): 1119.3554 (calc), 1119.3454 (meas).
Rv1151c Desuccinylase Activity.
EchA19wt (20 μM) was incubated with 100 μM succinyl-CoA for 60 min, and then the reaction mixture was buffer exchanged against EchA19 storage buffer 20 mM Tris-HCl (pH 8), 300 mM NaCl with a spin centricon (MWCO 10 kDa) to remove excess succinyl-CoA; with a final succinyl-CoA concentration of 1 pM. The resulting protein (5 μM) was incubated for 2 h with Rv1151c (2 μM) in a solution containing 25 mM Tris-HCl (pH 7.5), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.1% BSA, and 1 mM NAD+. An aliquot of the reaction mixture was added to SDS loading dye (final concentration, 1× SDS) containing 6% β-mercaptoethanol. Reactions were monitored by means of immunoblot analysis, as described above. Two hundred nanograms of protein was loaded in each well.
X-Ray Crystallization, Data Collection and Structure Determination.
Potential conditions for crystallization of EchA19wt (13 mg mL−1) were screened with the following 96-well crystallization screens: NeXtal (NeXtal Biotechnologies), Crystal Screen, and Index (Hampton Research). Hit conditions were optimized by means of the hanging drop method in 24-well trays. Crystals were grown in a solution of 0.2 M MgCl2 and 0.1 M Tris (pH 8.5) supplemented with 25% PEG3350 and cryocooled in a cryoprotecting solution containing 5% glycerol. Data were collected at the FMX beamline at NSLS-II (Brookhaven National Laboratory, Upton, NY) and processed using the autoPROC software package.50 The structure was solved through molecular replacement using Phaser,51 with 4F47 as the search model. Refinement was carried out with the PHENIX suite52 with several cycles of manual model building using Coot.53 Data collection and refinement statistics are summarized in Table 2. The electron density of Mtb EchA19Wt was of sufficient quality to build most protein residues (Glu2-Ala64, Ile90-Arg263). Mtb EchA19 crystallized in the P231 space group, with one monomer of the protein in the asymmetric unit. The biological assembly is (believed to be) a trimer, and thus includes two symmetry-related copies of the molecule.
Supplementary Material
Acknowledgments
The X-ray and small-angle X-ray scattering experiments were conducted using resources of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under contract no. DE-SC0012704. Beamline LiX is supported primarily by the National Institutes of Health (NIH); by the National Institute of General Medical Sciences (NIGMS), through a Biomedical Technology Research Resource P41 grant (P41GM111244); and by the DOE Office of Biological and Environmental Research (KP1605010). This research is funded by an NIH grant (RO1AI134054) to N.S.S. A.C.B. acknowledges support from the Turner Fellow Program (Stony Brook University), Molecular Pharmacology Training Program (NIGMS T32GM007518), and the Chemical Biology Training Program (NIGMS T32GM092714). Small-angle X-ray scattering experiments were conducted by Xiaoxi Yu of the Department of Chemistry, Stony Brook University. We thank all members of the Sampson and Garcia-Diaz laboratories for insightful discussion and support.
Footnotes
Supporting Information Additional information on expression constructs, small-angle X-ray scattering, and immunoblots is available in the Supporting Information section. This information is available free of charge on the ACS Publications website.
Accession codes
UniProt accession IDs for proteins used in this work: EchA19, O53561; MCR, O06543; Rv1151c, P9WGG3; ChsE4-ChsE5, I6YCA3-I6Y3Q0; FadD17, O53551. The structure factors and atomic coordinates for Mtb EchA19 have been deposited into the Protein Data Bank with accession code 6WYI. Authors will release the atomic coordinates and experimental data upon article publication.
Conflicts of interest: The authors declare they have no conflicts of interest with the contents of this article.
References
- 1.Lee W; VanderVen BC; Fahey RJ; Russell DG, Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 2013, 288, 6788–6800 DOI 10.1074/jbc.M112.445056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Carvalho LP; Fischer SM; Marrero J; Nathan C; Ehrt S; Rhee KY, Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol 2010, 17, 1122–1131 DOI 10.1016/j.chembiol.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 3.Nesbitt NM; Yang X; Fontan P; Kolesnikova I; Smith I; Sampson NS; Dubnau E, A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect Immun 2010, 78, 275–282 DOI 10.1128/IAI.00893-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey AK; Sassetti CM, Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 2008, 105, 4376–4380 DOI 10.1073/pnas.0711159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang JC; Harik NS; Liao RP; Sherman DR, Identification of mycobacterial genes that alter growth and pathology in macrophages and in mice. J Infect Dis 2007, 196, 788–795 DOI 10.1086/520089 [DOI] [PubMed] [Google Scholar]
- 6.Yuan T; Sampson NS, Hit generation in TB drug discovery: from genome to granuloma. Chem Rev 2018, 118, 1887–1916 DOI 10.1021/acs.chemrev.7b00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kendall SL; Withers M; Soffair CN; Moreland NJ; Gurcha S; Sidders B; Frita R; Ten Bokum A; Besra GS; Lott JS; Stoker NG, A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol Microbiol 2007, 65, 684–699 DOI 10.1111/j.1365-2958.2007.05827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Geize R; Yam K; Heuser T; Wilbrink MH; Hara H; Anderton MC; Sim E; Dijkhuizen L; Davies JE; Mohn WW; Eltis LD, A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc Natl Acad Sci U S A 2007, 104, 1947–1952 DOI 10.1073/pnas.0605728104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang M; Guja KE; Thomas ST; Garcia-Diaz M; Sampson NS, A distinct MaoC-like enoyl-CoA hydratase architecture mediates cholesterol catabolism in Mycobacterium tuberculosis. ACS Chem Biol 2014, 9, 2632–2645 DOI 10.1021/cb500232h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang M; Lu R; Guja KE; Wipperman MF; St Clair JR; Bonds AC; Garcia-Diaz M; Sampson NS, Unraveling cholesterol catabolism in Mycobacterium tuberculosis: ChsE4-ChsE5 alpha2beta2 acyl-CoA dehydrogenase intiates beta-oxidation of 3-oxo-cholest-4-en-26-oyl CoA. ACS Infect Dis 2015, 1, 110–125 DOI 10.1021/id500033m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wipperman MF; Yang M; Thomas ST; Sampson NS, Shrinking the FadE proteome of Mycobacterium tuberculosis: insights into cholesterol metabolism through identification of an alpha2beta2 heterotetrameric acyl coenzyme A dehydrogenase family. J Bacteriol 2013, 195, 4331–4341 DOI 10.1128/JB.00502-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaefer CM; Lu R; Nesbitt NM; Schiebel J; Sampson NS; Kisker C, FadA5 a thiolase from Mycobacterium tuberculosis: a steroid-binding pocket reveals the potential for drug development against tuberculosis. Structure 2015, 23, 21–33 DOI 10.1016/j.str.2014.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu R; Schmitz W; Sampson NS, alpha-Methyl acyl CoA racemase provides Mycobacterium tuberculosis catabolic access to cholesterol ester. Biochemistry 2015, 54, 5669–5672 DOI 10.1021/acs.biochem.5b00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R; Schaefer CM; Nesbitt NM; Kuper J; Kisker C; Sampson NS, Catabolism of the cholesterol side chain in Mycobacterium tuberculosis Is controlled by a redox-sensitive thiol switch. ACS Infect Dis 2017, 3, 666–675 DOI 10.1021/acsinfecdis.7b00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruprecht A; Maddox J; Stirling AJ; Visaggio N; Seah SY, Characterization of novel acyl coenzyme A dehydrogenases involved in bacterial steroid degradation. J Bacteriol 2015, 197, 1360–1367 DOI 10.1128/JB.02420-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan T; Yang M; Gehring K; Sampson NS, Mycobacterium tuberculosis exploits a heterohexameric enoyl-CoA hydratase retro-aldolase complex for cholesterol catabolism. Biochemistry 2019, 58, 4224–4235 DOI 10.1021/acs.biochem.9b00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava S; Chaudhary S; Thukral L; Shi C; Gupta RD; Gupta R; Priyadarshan K; Vats A; Haque AS; Sankaranarayanan R; Natarajan VT; Sharma R; Aldrich CC; Gokhale RS, Unsaturated lipid assimilation by mycobacteria requires auxiliary cis-trans enoyl CoA isomerase. Chem Biol 2015, 22, 1577–1587 DOI 10.1016/j.chembiol.2015.10.009 [DOI] [PubMed] [Google Scholar]
- 18.Hamed RB; Batchelar ET; Clifton IJ; Schofield CJ, Mechanisms and structures of crotonase superfamily enzymes--how nature controls enolate and oxyanion reactivity. Cell Mol Life Sci 2008, 65, 2507–2527 DOI 10.1007/s00018-008-8082-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel CK; Mathieu M; Zeelen JP; Hiltunen JK; Wierenga RK, Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket. Embo j 1996, 15, 5135–5145 [PMC free article] [PubMed] [Google Scholar]
- 20.Engel CK; Kiema TR; Hiltunen JK; Wierenga RK, The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain fatty acid-CoA molecule. J Mol Biol 1998, 275, 847–859 DOI 10.1006/jmbi.1997.1491 [DOI] [PubMed] [Google Scholar]
- 21.Kochanowski K; Sauer U; Noor E, Posttranslational regulation of microbial metabolism. Curr Opin Microbiol 2015, 27, 10–17 DOI 10.1016/j.mib.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 22.Pisithkul T; Patel NM; Amador-Noguez D, Post-translational modifications as key regulators of bacterial metabolic fluxes. Curr Opin Microbiol 2015, 24, 29–37 DOI 10.1016/j.mib.2014.12.006 [DOI] [PubMed] [Google Scholar]
- 23.Xie Z; Dai J; Dai L; Tan M; Cheng Z; Wu Y; Boeke JD; Zhao Y, Lysine succinylation and lysine malonylation in histones. Mol Cell Proteomics 2012, 11, 100–107 DOI 10.1074/mcp.M111.015875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hentchel KL; Escalante-Semerena JC, Acylation of biomolecules in prokaryotes: a widespread strategy for the control of biological function and metabolic stress. Microbiol Mol Biol Rev 2015, 79, 321–346 DOI 10.1128/MMBR.00020-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang M; Wang Y; Chen Y; Cheng Z; Gu J; Deng J; Bi L; Chen C; Mo R; Wang X; Ge F, Succinylome analysis reveals the involvement of lysine succinylation in metabolism in pathogenic Mycobacterium tuberculosis. Mol Cell Proteomics 2015, 14, 796–811 DOI 10.1074/mcp.M114.045922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie L; Liu W; Li Q; Chen S; Xu M; Huang Q; Zeng J; Zhou M; Xie J, First succinyl-proteome profiling of extensively drug-resistant Mycobacterium tuberculosis revealed involvement of succinylation in cellular physiology. J Proteome Res 2015, 14, 107–119 DOI 10.1021/pr500859a [DOI] [PubMed] [Google Scholar]
- 27.Liu F; Yang M; Wang X; Yang S; Gu J; Zhou J; Zhang XE; Deng J; Ge F, Acetylome analysis reveals diverse functions of lysine acetylation in Mycobacterium tuberculosis. Mol Cell Proteomics 2014, 13, 3352–3366 DOI 10.1074/mcp.M114.041962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nambi S; Gupta K; Bhattacharyya M; Ramakrishnan P; Ravikumar V; Siddiqui N; Thomas AT; Visweswariah SS, Cyclic AMP-dependent protein lysine acylation in mycobacteria regulates fatty acid and propionate metabolism. J Biol Chem 2013, 288, 14114–14124 DOI 10.1074/jbc.M113.463992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H; Hegde SS; Blanchard JS, Reversible acetylation and inactivation of Mycobacterium tuberculosis acetyl-CoA synthetase is dependent on cAMP. Biochemistry 2011, 50, 5883–5892 DOI 10.1021/bi200156t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayden JD; Brown LR; Gunawardena HP; Perkowski EF; Chen X; Braunstein M, Reversible acetylation regulates acetate and propionate metabolism in Mycobacterium smegmatis. Microbiology 2013, 159, 1986–1999 DOI 10.1099/mic.0.068585-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nambi S; Basu N; Visweswariah SS, cAMP-regulated protein lysine acetylases in mycobacteria. J Biol Chem 2010, 285, 24313–24323 DOI 10.1074/jbc.M110.118398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nambi S; Badireddy S; Visweswariah SS; Anand GS, Cyclic AMP-induced conformational changes in mycobacterial protein acetyltransferases. J Biol Chem 2012, 287, 18115–18129 DOI 10.1074/jbc.M111.328112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterson RM; Hill RL, Enoyl coenzyme A hydratase (crotonase). Catalytic properties of crotonase and its possible regulatory role in fatty acid oxidation. J Biol Chem 1972, 247, 5258–5265 [PubMed] [Google Scholar]
- 34.Willadsen P; Eggerer H, Substrate stereochemistry of the enoyl-CoA hydratase reaction. Eur J Biochem 1975, 54, 247–252 DOI 10.1111/j.1432-1033.1975.tb04134.x [DOI] [PubMed] [Google Scholar]
- 35.Hansford RG; Johnson RN, The steady state concentrations of coenzyme A-SH and coenzyme A thioester, citrate, and isocitrate during tricarboxylate cycle oxidations in rabbit heart mitochondria. J Biol Chem 1975, 250, 8361–8375 [PubMed] [Google Scholar]
- 36.Wagner GR; Payne RM, Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem 2013, 288, 29036–29045 DOI 10.1074/jbc.M113.486753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner GR; Bhatt DP; O’Connell TM; Thompson JW; Dubois LG; Backos DS; Yang H; Mitchell GA; Ilkayeva OR; Stevens RD; Grimsrud PA; Hirschey MD, A class of reactive acyl-CoA species reveals the non-enzymatic origins of protein acylation. Cell Metab 2017, 25, 823–837 e8 DOI 10.1016/j.cmet.2017.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vandal OH; Pierini LM; Schnappinger D; Nathan CF; Ehrt S, A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat Med 2008, 14, 849–854 DOI 10.1038/nm.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagner GR; Hirschey MD, Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol Cell 2014, 54, 5–16 DOI 10.1016/j.molcel.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harmel R; Fiedler D, Features and regulation of non-enzymatic post-translational modifications. Nat Chem Biol 2018, 14, 244–252 DOI 10.1038/nchembio.2575 [DOI] [PubMed] [Google Scholar]
- 41.Kulkarni RA; Worth AJ; Zengeya TT; Shrimp JH; Garlick JM; Roberts AM; Montgomery DC; Sourbier C; Gibbs BK; Mesaros C; Tsai YC; Das S; Chan KC; Zhou M; Andresson T; Weissman AM; Linehan WM; Blair IA; Snyder NW; Meier JL, Discovering targets of non-enzymatic acylation by thioester reactivity profiling. Cell Chem Biol 2017, 24, 231–242 DOI 10.1016/j.chembiol.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simic Z; Weiwad M; Schierhorn A; Steegborn C; Schutkowski M, The varepsilon-amino group of protein lysine residues Is highly susceptible to nonenzymatic acylation by several physiological acyl-CoA thioesters. ChemBioChem 2015, 16, 2337–2347 DOI 10.1002/cbic.201500364 [DOI] [PubMed] [Google Scholar]
- 43.Lee W; VanderVen BC; Walker S; Russell DG, Novel protein acetyltransferase, Rv2170, modulates carbon and energy metabolism in Mycobacterium tuberculosis. Sci Rep 2017, 7, 72 DOI 10.1038/s41598-017-00067-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vetting MW; Errey JC; Blanchard JS, Rv0802c from Mycobacterium tuberculosis: the first structure of a succinyltransferase with the GNAT fold. Acta Crystallogr Sect F Struct Biol Cryst Commun 2008, 64, 978–985 DOI 10.1107/S1744309108031679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du J; Zhou Y; Su X; Yu JJ; Khan S; Jiang H; Kim J; Woo J; Kim JH; Choi BH; He B; Chen W; Zhang S; Cerione RA; Auwerx J; Hao Q; Lin H, Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 2011, 334, 806–809 DOI 10.1126/science.1207861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J; Chen Y; Tishkoff DX; Peng C; Tan M; Dai L; Xie Z; Zhang Y; Zwaans BM; Skinner ME; Lombard DB; Zhao Y, SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 2013, 50, 919–930 DOI 10.1016/j.molcel.2013.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gu J; Deng JY; Li R; Wei H; Zhang Z; Zhou Y; Zhang Y; Zhang XE, Cloning and characterization of NAD-dependent protein deacetylase (Rv1151c) from Mycobacterium tuberculosis. Biochemistry (Mosc) 2009, 74, 743–748 [DOI] [PubMed] [Google Scholar]
- 48.Bonds AC; Sampson NS, More than cholesterol catabolism: regulatory vulnerabilities in Mycobacterium tuberculosis. Curr Opin Chem Biol 2018, 44, 39–46 DOI 10.1016/j.cbpa.2018.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas ST; VanderVen BC; Sherman DR; Russell DG; Sampson NS, Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J Biol Chem 2011, 286, 43668–43678 DOI 10.1074/jbc.M111.313643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vonrhein C; Flensburg C; Keller P; Sharff A; Smart O; Paciorek W; Womack T; Bricogne G, Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr 2011, 67, 293–302 DOI 10.1107/s0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ; Grosse-Kunstleve RW; Adams PD; Winn MD; Storoni LC; Read RJ, Phaser crystallographic software. J Appl Crystallogr 2007, 40, 658–674 DOI 10.1107/s0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zwart PH; Afonine PV; Grosse-Kunstleve RW; Hung LW; Ioerger TR; McCoy AJ; McKee E; Moriarty NW; Read RJ; Sacchettini JC; Sauter NK; Storoni LC; Terwilliger TC; Adams PD, Automated structure solution with the PHENIX suite. Methods Mol Biol 2008, 426, 419–435 DOI 10.1007/978-1-60327-058-8_28 [DOI] [PubMed] [Google Scholar]
- 53.Emsley P; Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004, 60, 2126–2132 DOI 10.1107/s0907444904019158 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


