Abstract
Resting state functional connectivity refers to the temporal correlations between spontaneous hemodynamic signals obtained using functional magnetic resonance imaging. This technique has demonstrated that the structure and dynamics of identifiable networks are altered in psychiatric and neurological disease states. Thus, resting state network organizations can be used as a diagnostic, or prognostic recovery indicator. However, much about the physiological basis of this technique is unknown. Thus, providing a translational bridge to an optimal animal model, the macaque, in which invasive circuit manipulations are possible, is of utmost importance. Current approaches to resting state measurements in macaques face unique challenges associated with signal-to-noise, the need for contrast agents limiting translatability, and within-subject designs. These limitations can, in principle, be overcome through ultra-high magnetic fields. However, imaging at magnetic fields above 7T has yet to be adapted for fMRI in macaques. Here, we demonstrate that the combination of high channel count transmitter and receiver arrays, optimized pulse sequences, and careful anesthesia regimens, allows for detailed single-subject resting state analysis at high resolutions using a 10.5 Tesla scanner. In this study, we uncover thirty spatially detailed resting state components that are highly robust across individual macaques and closely resemble the quality and findings of connectomes from large human datasets. This detailed map of the rsfMRI ‘macaque connectome’ will be the basis for future neurobiological circuit manipulation work, providing valuable biological insights into human connectomics.
Keywords: Functional connectivity, Rhesus macaque, Resting-state, Spontaneous activity, Functional MRI (fMRI)
1. Introduction
Resting state functional connectivity refers to the temporal correlations between spontaneous blood oxygenation level dependent (BOLD) signals obtained using functional magnetic resonance imaging (fMRI). These fluctuations were first noted in motor cortex (Biswal et al., 1995), but since then many other large-scale networks of correlated temporal patterns in the resting brain have been identified (Biswal et al., 2010; Heuvel and Pol, 2010; Smith et al., 2013; Wang et al., 2009). It was clear even before the advent of fMRI that functional networks at multiple spatial and temporal scales are embedded in the mammalian brain (Essen and Maunsell, 1983; Felleman and Essen, 1991); many years of fMRI neuroimaging have catalogued the spatial and, to some degree, dynamic organization (Allen et al., 2014; Hindriks et al., 2016; Ju and Bassett, 2020; Hutchison et al., 2013; Hutchison et al., 2013) of these networks. These different networks have distinct temporal properties (~0.01 - ~0.1 Hz fluctuations) and persist through different states such as sleep or anesthesia. Moreover, they are generally consistent across subjects (Smith et al., 2013, 2009) and to some degree generalize across species (Li et al., 2013; Jonckers et al., 2011).
As a method, characterization of resting state networks has proven invaluable for psychiatry (Floris et al., 2018; Konova et al., 2015; Hulvershorn et al., 2013), basic neuroscience (Fox and Raichle, 2007; Glasser et al., 2016; Smith et al., 2013), and psychology (Laird et al., 2011). In particular, it offers the ability to simultaneously characterize networks across the whole brain within a short period of time, and without the need to engage the subject in order to collect the data, as opposed to classical task-based experimental designs. These brain networks (and any individual variations) can then be studied across development, aging or disease and provide a high-throughput approach to both basic and clinical human neuroscience.
For several reasons, the resting state technique is a particularly important one to implement in the macaque monkey model. First, macaques offer a critical intermediary between humans and rodent models and have gross neuroanatomy that is highly homologous to that of humans (Hutchison and Everling, 2012; Mars et al., 2016; Petrides and Pandya, 2002; Watson and Platt, 2012; Heilbronner et al., 2016). Second, macaques are a valuable model for many complex behaviors and cognitive processes, and thus potentially for psychiatric diseases. Third, macaques offer the opportunity to perform types of invasive manipulations, including microstimulation, chemogenetics (Galvan et al., 2019; Nagai et al., 2016; Raper et al., 2019; Upright et al., 2018; Cushnie et al., 2020) and optogenetics (Galvan et al., 2017; Khateeb et al., 2019), which are impossible or of limited applicability in humans. Fourth, additional measurement techniques that are rare or impossible in humans, like single-unit electrophysiological recordings and neuroanatomical tract-tracing using invasive tracers, have allowed for careful validation of non-invasive neuroimaging tools such as fMRI and the BOLD response (Logothetis, 2003; Logothetis et al., 2001) or diffusion-weighted-imaging for tractography (dMRI) (Bakker et al., 2012; Markov et al., 2012). Thus, findings in nonhuman animal models constrain theory and experimentation in human neuroscience. On the other hand, investigations using non-invasive tools in humans can guide us in designing and conducting experiments to elucidate the basic mechanism of those findings using invasive tools in animals.
Macaques, however, do have a critical disadvantage as a model organism for resting state fMRI (rsfMRI). That is, until now, our ability to collect high resolution, high signal-to-noise ratio (SNR) whole brain data in macaques has been extremely difficult. This is a multifaceted problem resulting mainly from the macaques’ small head size (~6 % of the human brain volume), the use of standard field strength magnets, the challenges in doing alert studies (especially at high resolutions) and the unavailability of commercial high channel count surface coils arrays. Moreover, work on macaques is costly and labor intensive. Thus, sample sizes in classical macaque experiments are generally low (usually 2-4 subjects per experiment), which contradicts the notion of generalization and out-of-sample prediction in human studies. That, in turn, means the utility of neuroimaging in macaques would be greatly enhanced if we could develop ways to improve SNR and contrast-to-noise ratio (CNR) dramatically.
One tool to improve SNR is using exogenous blood-pool contrast agents that induce strong susceptibility gradients in their vicinity. Such agents include monocrystalline iron oxide nanoparticles (MION), or similar more modern agents (Ferumoxytol, Senirem, Feraheme) as a contrast agent (Moeller et al., 2009; Tsao et al., 2003; Vanduffel et al., 2001). However, this approach has several disadvantages: (1) the signal then reflects blood volume changes (Leite et al., 2002) rather than the typical blood oxygenation level dependent (BOLD) response, complicating its translatability to typical human studies, (2) it has a slightly slower hemodynamic response (Leite et al., 2002; Pelekanos et al., 2020), and (3) it can accumulate in the brain and causes potentially long-lasting health disadvantages (Li et al., 2012; Mustafa and Mohammed-Rasheed, 2019). The SNR problem can to some degree also be overcome with larger datasets (i.e. many hours of data collection) but this is expensive and impractical and is accompanied by unavoidable risks to the subjects if general anesthetics are utilized.
One way to overcome the sensitivity problem of acquiring high resolution fMRI is to use ultra-high field strengths in combination with high channel count receiver arrays (Lagore et al., 2020; Uğurbil et al., 2019), an approach which has proven extremely productive, to this end, in human studies (Martino et al., 2018, 2011; Ugurbil, 2014; Uğurbil et al., 2019, 2019, 2003; Yacoub et al., 2008, 2001; Zimmermann et al., 2011). Increasing magnetic fields not only provide enhanced SNR but also lead to higher BOLD contrast, especially for the microvascular BOLD contributions that produce mapping signals with higher fidelity to sites of neuronal activity (Uğurbil, 2018; Ugurbil, 2016) . Thus, increasing the magnetic field produces multiplicative gains in BOLD based fMRI studies. Because of these advantages and an increasing number of solutions introduced to tackle challenges confronted in imaging at such magnetic fields, the ultrahigh magnetic field of 7 Tesla has emerged as the preferred platform for non-clinical fMRI in humans (Uğurbil, 2018). Following this trend, recently, efforts have been undertaken to significantly surpass 7 Tesla in magnetic field strength and introduce magnetic fields greater than 10 Tesla.
While some macaque studies focused on resting state functional magnetic resonance imaging (rsfMRI) (Gilbert et al., 2016; Goulas et al., 2017; Hutchison et al., 2011; Hutchison et al., 2013; Schaeffer et al., 2018; Vincent et al., 2009, 2007) to date, to our knowledge, resolutions in macaque rsfMRI continue to be greater than 1 mm3 as well as longer than 1 second.
Here we explore the advantages of 10.5T neuroimaging for rsfMRI in the macaque. combined with a specialized 32 Rx and 8 Tx channel coil (Lagore et al., 2020), implemented on a 10.5T human scanner with a commercial console (Siemens). We demonstrate robust, within subject and group ‘connectome’ style BOLD-based rsfMRI data in 6 lightly anesthetized macaques (for a similar approach at 3T see Autio et al., 2020). Our approach is extremely robust and allows us to repeatedly and reliably measure intrinsic BOLD signals in macaques at high spatial and temporal resolution and with high SNR. Our findings, extending previous approaches, show robust resting state networks similar to human findings that are consistent between macaques and, importantly, identifiable robustly in the individual. Equally important, the components we identify adhere extremely well to anatomical architecture, as well as showing homogeneous sensitivity to cortex and subcortical regions. Our findings demonstrate the general utility that 10.5T imaging has for in vivo intrinsic BOLD investigations in macaques and non human primates in general.
2. Materials and methods
2.1. Animal preparation
We obtained data from 6 adult macaque monkeys (Macaca fascicularis: 4 female, and Macaca mulatta, 1 male and 1 female). Weights ranged from 3.0 kg to 9.0 kg. Experimental procedures were carried out in accordance with University of Minnesota Institutional Animal Care and Use Committee approval and in accord with the National Institute of Health standards for the care and use of non human primates. All subjects were fed ad libitum and pair housed within a light and temperature controlled colony room. Animals were not water restricted. None of the subjects had any prior implant or cranial surgery. Animals were fasted for 14-16 hours prior to imaging.
On scanning days, anesthesia was first induced by intramuscular injection of atropine (0.5 mg/kg) and ketamine hydrochloride (7.5 mg/kg). Subjects were transported to the scanner anteroom and intubated using an endotracheal tube. Initial anesthesia was maintained using 1.5-2% isoflurane mixed with oxygen (1L/m during intubation and 2L/m during scanning to compensate for the 12m length of the tubing used).
Subjects were placed onto a custom-built coil bed with integrated head fixation by placing stereotactic ear bars into the ear canals. The position of the animal corresponds to the sphinx position. For functional imaging, the isoflurane level was lowered to 1%. Experiments were performed with animals freely breathing. Additionally, for Macaca fascicularis subjects only, an initial bolus injection of 1.5ug/kg fentanyl was administered IV followed by a continuous administration of 3ug/kg/hr using a syringe pump. Rectal temperature (~99.6F), respiration (10-15 breaths/min), end-tidal CO2 (25-40), electrocardiogram (70-150 bpm) and SpO2 (>90%) were monitored using an MRI compatible monitor (IRADIMED 3880 MRI Monitor, USA). Temperature was maintained using a circulating water bath as well as chemical heating pads and padding for thermal insulation. Anesthesia was used to eliminate motion effects and physiological stress as well as allow the imaging of surgically naive subjects. Isoflurane is a vasodilator (Farber et al., 1997) and modulates cerebrovascular activity (Vincent et al., 2007). However, previous studies have shown successful synchronous BOLD fluctuations and resting state activity in macaques as well as mice and rats under isoflurane (Hutchison et al., 2011; Hutchison et al., 2013; Okada et al., 2018; Vincent et al., 2009, 2007).
2.2. Data acquisition
All data were acquired on a passively shielded 10.5 Tesla, 88 cm diameter clear bore magnet coupled to Siemens gradients (“SC72” body gradients operating at a slew rate of 200 mT/m/s, and 70 mT/m maximal strength) and electronics (Magnetom 10.5T Plus) (Siemens, Erlangen, Germany). Within the gradient set and the bore-liner, the space available for subject insertion had a 60 cm diameter.
The 10.5T system operates on the E-line (E12U) platform which is directly comparable to clinical platforms (3T Prisma/Skyra, 7T Terra). As such, the user interface and pulse sequences were identical to those running on clinical platforms. A custom in-house built and designed RF coil with an 8-channel transmit/receive end-loaded dipole array of 18 cm length (individually) combined with a close-fitting 16-channel loop receive array head cap, and an 8-channel loop receive array of 50 × 100mm size located under the chin (Lagore et al., 2020). The size of 14 individual receive loops of the head cap was 37 mm with 2 larger ear loops of 80 mm - all receiver loops were arranged in an overlapping configuration for nearest neighbor decoupling. The resulting combined 32 receive channels were used for all experiments and supported 3-fold acceleration in phase encoding direction. The coil holder was designed to be a semi-stereotaxic instrument holding the head of the animal in a centered sphinx position via customized ear bars. The receive elements were modelled to adhere as close to the surface of the animal’s skulls as possible. Transmit phases, for the individual transmit channels were fine-tuned for excitation uniformity for one representative mid-sized animal and the calculated phases were then used for all subsequent acquisitions. Magnetic field homogenization (B0 shimming) was performed using a customized field of view with the Siemens internal 3D mapping routines. Multiple iterations of the shims (using the adjusted FOV shim parameters) were performed and further fine adjustment performed manually on each animal. Third order shim elements were ignored for these procedures.
In all animals a B1 + (transmit B1) fieldmap was acquired using a vendor provided flip angle mapping sequence (example result in Sup. Fig. 9) and then power calibrated for each individual. Following B1 + transmit calibration, 3-5 averages (23 minutes) of a T1 weighted magnetization prepared rapid acquisition gradient echo protocol (3D MP-RAGE) were acquired for anatomical processing (TR = 3300 ms, TE = 3.56 ms, TI = 1140 ms, flip angle = 5°, slices = 256, matrix = 320 × 260, acquisition voxel size = 0.5 × 0.5 × 0.5 mm3). Images were acquired using in plane acceleration GRAPPA = 2. A resolution and FOV matched T2 weighted 3D turbo spin echo sequence (variable flip angle) was run to facilitate B1 inhomogeneity correction.
Before the start of the functional data acquisition, five images were acquired in both phase encoding directions (R/L, L/R or F/H, H/F as appropriate) for offline EPI distortion correction. For four monkeys, six runs of 700 continuous 2D multiband (MB) EPI (Moeller et al., 2010; Setsompop et al., 2012; Uğurbil et al., 2013) functional volumes (TR = 1110 ms; TE = 17.6 ms; flip angle = 60°, slices = 58, matrix = 108 × 154; FOV = 81 × 115.5 mm2; acquisition voxel size = 0.75 × 0.75 × 0.75 mm3) were acquired. Images were acquired with a left-right phase encoding direction using in plane acceleration factor GRAPPA = 3, partial Fourier = 7/8th, and MB or simultaneous multislice factor = 2. For the two other monkeys, five runs of 700 continuous 2D MB EPI functional volumes were acquired (TR = 1200 ms; TE = 17.6 ms; flip angle = 60°, slices = 58, matrix = 106 × 212; FOV = 79.5 × 159 mm2; acquisition voxel size = 0.75 × 0.75 × 0.75 mm3). Images were scanned in foot-head phase encoding direction using in-plane acceleration factor GRAPPA = 3, partial Fourier = 7/8th and MB = 2. The change in sequence parameters was made after upgrading to the 10.5 Plus (E12U) platform which brought with it software changes that made the left right phase encoding more favorable. Since macaques were scanned in sphinx positions, the orientations noted here are what is consistent with a (head first supine) typical human brain study (in terms of gradients) but translate differently to the actual macaque orientation. While left / right orientation is the same, foot-head is actually anterior-posterior in the monkey.
2.3. Image preprocessing
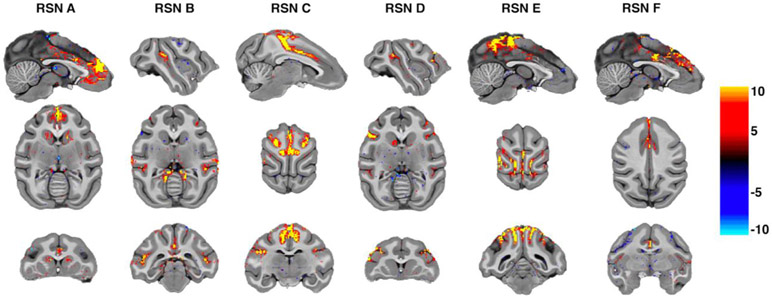
Image processing was performed using a custom pipeline relying on FSL (Jenkinson et al., 2012), ANTs (Avants et al., 2014, 2011), AFNI (Cox, 1996) and a heavily modified CONN (Whitfield-Gabrieli and Nieto-Castanon, 2012) toolbox. Images were first motion corrected using mcflirt (registration to the first image). Motion parameters never exceeded 0.5 mm or 0.4° rotation except in one run of one animal, which was discarded (the animal experienced anesthesia induced emesis during the last run). Images were slice time corrected and EPI distortion corrected using topup. High magnetic fields and large matrix sizes are commonly associated with severe EPI distortions. Supplementary Fig 1 shows an overview of the extent of the distortion as well as the result of the corrected warps. Anatomical images were nonlinearly warped into the NMT (Seidlitz et al., 2018) template using ANTs and 3DQwarp in AFNI. The distortion correction, motion correction and normalization were performed using a single sinc interpolation. Images were spatially smoothed (FWHM = 2 mm), linear detrended, denoised using a linear regression approach including heart rate and respiration, as well as a 5 component nuisance regressor of the masked white matter and cerebrospinal fluid and band-pass filtering (0.008 - 0.09 Hz) (Hallquist et al., 2013). While we chose here to present the data as is typically done in resting state applications, to demonstrate the raw SNR and overall BOLD sensitivity we show an unsmoothed example on the individual macaques brain in Fig. 1. A raw tSNR map of one animal can be found in Sup. Fig. 10.
Fig. 1.
Independent components data in individual subject space. Displayed are 6 RSNs in one unsmoothed macaque dataset processed without spatial smoothing. For this visualization thresholds were set to 4,−4. The results show robust results in the unsmoothed volumes at native resolution demonstrating our high SNR regime. Networks adhere to the cortical ribbon topography without any registration performed.
2.4. Independent component analysis
We used group ICA to uncover ordered reproducible components across subjects. This approach uncovers a set of interpretable group components. Algorithms were used as implemented in the GIFT (Calhoun et al., 2001) software package. Our preprocessing pipeline produced one nuisance corrected concatenated time course of all runs per subject. The temporal dimension of the concatenated time course was then reduced using principal component analysis (PCA). Spatial resting state components were then estimated using the Infomax algorithm approach with the standard GIFT parameters. Time-courses and spatial maps associated with each component were then back projected as implemented in the group-ICA tools in GIFT. Since the process of dimensionality reduction and model selection are somewhat arbitrary (one needs to specify the number of components to estimate), there is no real consensus on how many components to extract. It has been noted that there is no best dimensionality for the underlying neurophysiology of multiple distributed systems (Cole et al., 2010). There are always multiple valid solutions, but we argue that a higher model order (Abou-Elseoud et al., 2010; Kiviniemi et al., 2009; Smith et al., 2009) than previously used (Hutchison et al., 2011; Hutchison et al., 2013; Vincent et al., 2007) is beneficial in macaques. While one can compartmentalize the brain into networks with associations to visual, motor, sensory, and auditory processing, the goal in macaque work is usually shifted towards finer-grain coding to allow for better cross-method comparison. Previous papers have not ignored this but have rather been limited by data quality (e.g., a high dimensional ICA decomposition cannot be obtained from a short rsfMRI run at standard field strength using high resolution). Multiple criteria (Jafri et al., 2008; Li et al., 2007; Zuo et al., 2010) currently exist for optimally selecting the number of independent components for a given dataset. The minimum description length criterion, however, yielded a mean estimation of 703 (SD = 69) independent components for our dataset. To obtain a manageable number of components and to allow for easier comparison with human and previous macaque datasets, 40 components were extracted. To test for the reliability of the decomposition the ICASSO (Li et al., 2007) toolbox was used and ICA iterated 20 times. Lastly, mean group independent components were scaled to empirically derived z-scores. These z-scores are an approximation of the temporal correlation between each voxel and the associated components time-course. A threshold of +/−2 was used as a lower limit of functional connectivity for visualization.
2.5. Identification of resting state networks and visualization
Components resulting from the independent component analysis were manually inspected and labeled according to anatomical and functional locations. Since ICA can be extremely noise sensitive, some extracted components are typically associated with motion, scanner noise artifacts or physiological confounds. In our dataset, 6 out of 40 components were discarded as they showed noisy, nonspecific, low correlation activation patterns or correspondence to large veins. Data was =visualized in volume space (custom code written in c++ and openGL) or projected to the surface reconstruction of the NMT template (Seidlitz et al., 2018) using AFNI (Cox, 1996) and SUMA (Saad et al., 2004; Saad and Reynolds, 2012).
2.6. Single subject ICA
Since studies in macaque usually have low numbers of subjects, and because subtle individual features of resting-state connectivity can be lost in group analysis, we also performed single subject ICA (example of ICA components extracted from data not subjected to spatial smoothing in Fig. 1) Parameters were matched to those of the group ICA.
2.7. Independent component network connectivity
Temporal correlations between components resulting from spatial ICA analysis as performed here can be high (Calhoun et al., 2003), since our approach to ICA maximizes the statistical independence in space while reducing the dimensionality of the temporal domain. To explore the relationship between the independent components from a temporal perspective we performed functional network connectivity analysis (FNC) (Jafri et al., 2008) between our resulting ICs. Our ICs were further clustered using a purely data driven hierarchical clustering procedure (nonspatial complete-linkage clustering). FNC then analyzes the entire set of connections between pairs of networks in terms of the within- and between- network connectivity sets. Resulting F-statistics were used to test statistically significant connections (connection threshold p<0.05 p-uncorrected, cluster threshold p<0.05 p-FDR corrected using MVPA omnibus test).
3. Results
3.1. Quantitative image metrics
Since this study presents results from a novel instrument (10.5T) we report simple image metrics. Supplementary Fig. 10 presents representative raw tSNR metrics from one functional run of one subject. Next, we evaluated the mean fraction of outliers per fMRI volume for all subjects (AFNI 3dToutcount with alpha = 0.001). A mean of 4.4851 × 10−4 with a standard deviation of 9.8672 × 10−4 voxels were selected as outliers. Additionally, we investigated the physiological noise regime of our data by computing functional network connectivity on the unsmoothed data while including 1-5 acquired rsfMRI runs incrementally. Supplementary Fig. 11 demonstrates that z-scored connectivity values increase as a function of runs included in the analysis indicating that our data is not noise dominated by physiological noise.
3.2. Group resting state networks
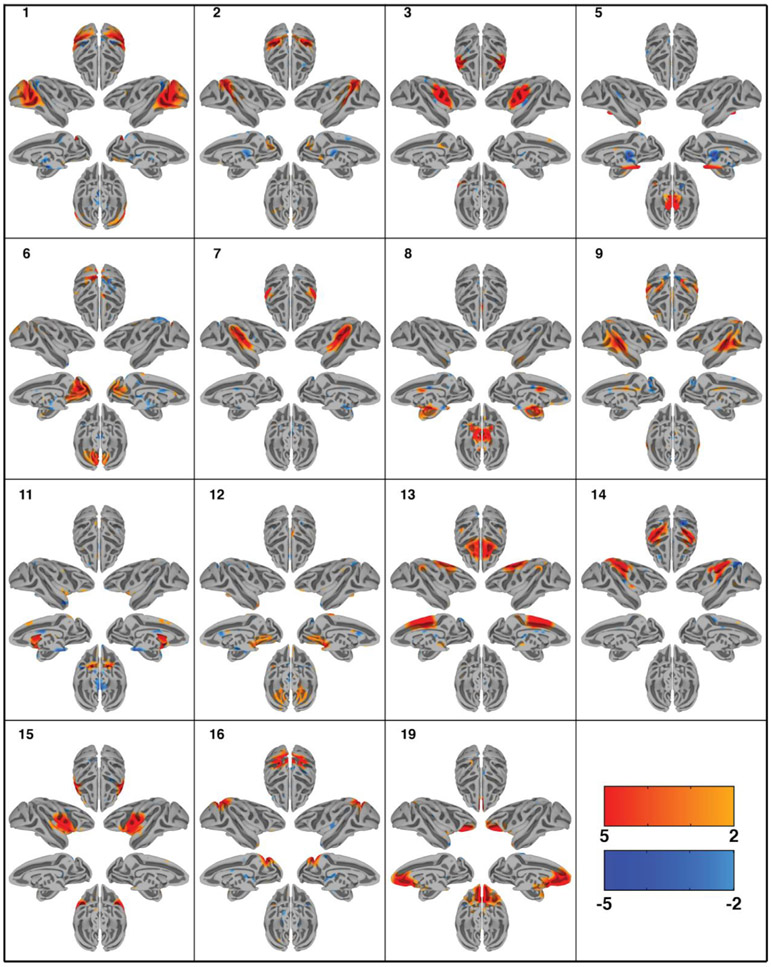
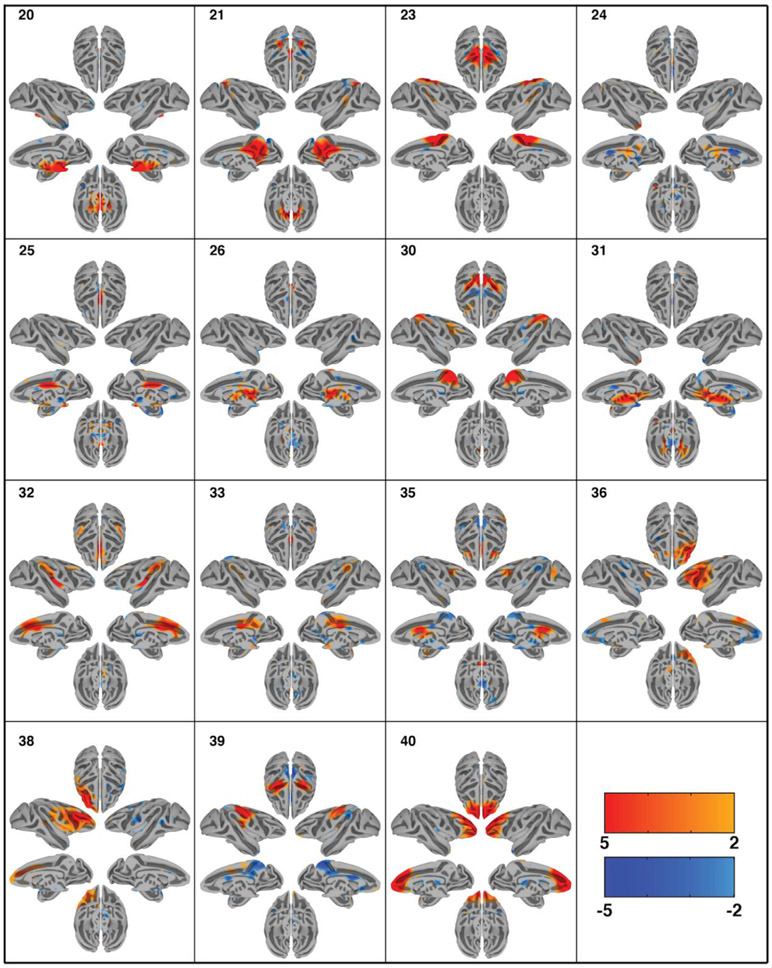
Group-ICA decomposed the dataset of 6 monkeys into 40 independent components as specified. ICASSO returned a stability index of 0.96 (SD = 0.04) demonstrating that the components are close to orthogonal clusters and highly consistent across multiple ICA iterations. Out of the 40 extracted components, 30 were deemed physiologically relevant, containing more than one anatomical area, adhering to the gray matter as well as displaying a reasonable heavy-tailed frequency distribution. The spatial maps of the resting state networks (RSNs) obtained via ICA analysis are shown in Figs. 2 and 3. The order of components is non informative. The computed components account for 56.39% of the data’s variance. An overview of the cortical coverage, as well as the overlap of the components can be found in supplementary Fig. 2. The resulting 30 RSNs are described below following anatomical classification from the Saleem and Logothetis atlas (Saleem and Logothetis, 2012):
Fig. 2.
Cortical surface representation of the first 15 resting state networks (RSNs) identified using group independent component analysis (GICA) in 6 macaque monkeys. Overlayed color maps represent thresholded z-scores. Individual subjects were normalized to the NMT template. Each component shows the medial and lateral view of each hemisphere independently as well as the dorsal and ventral view of the hemispheres combined.
Fig. 3.
Cortical surface representation of the second 15 resting state networks (RSNs) identified using group independent component analysis (GICA) in 6 macaque monkeys. Overlayed color maps represent thresholded z-scores. Individual subjects were normalized to the NMT template. Each component shows the medial and lateral view of each hemisphere independently as well as the dorsal and ventral view of the hemispheres combined.
Network 1 (lateral occipital): This RSN is mainly in the lateral occipital cortex. It covers portions of early visual areas V1, V2, and V4. The network also includes anticorrelations to regions within the intraparietal sulcus.
Network 2 (dorsal superior temporal sulcus): This RSN includes the areas surrounding the dorsal portion of the STS, including areas 7a, FST, MT, and MST.
Network 3 (somatosensory): This RSN is primarily composed of somatosensory cortical regions, extending somewhat into the primary motor cortex as well. Anticorrelations with insula and lateral intraparietal area are observed.
Network 5 (pulvinar-pons-cerebellum): This RSN is primarily subcortical and includes portions of the dorsal thalamus, particularly the medial and lateral pulvinar, much of the pons, and part of the cerebellum.
Network 6 (medial occipital): This RSN covers medial areas of the occipital lobe. It is primarily composed of area V2, but extends into aspects of areas V6 and, more rostrally, 7m.
Network 7 (lateral and cingulate sulci): This RSN follows the lateral sulcus closely, and thus covers primary auditory cortex, some auditory association areas, area 7op, and large portions of the insula. Although poorly visible on the surface, this network has substantial connectivity with cingulate areas 23c and 24c.
Network 8 (ventromedial subcortex): This RSN covers a large swath of the ventromedial subcortical surface of the brain, including amygdala, basal forebrain, and hippocampus. Cortically, correlations in anterior cingulate cortex 24a/b as well as anticorrelations in nearby 23a/b are observed.
Network 9 (middle STS-dlPFC-cingulate): This RSN covers the middle portions (dorsal-ventrally) of the superior temporal sulcus. This covers many areas associated with multisensory processing, including TPO, TAa, TEO, PGa, and some of the insula. The network also includes portions of the anterior and posterior cingulate gyrus, the outer edges of the intraparietal sulcus, and the dorsolateral prefrontal cortex (specifically areas 46 and 8a). Anticorrelations are seen with visual areas V2 and V6.
Network 11 (striatal-medial cortical): This RSN covers the entire striatum, as well as some medial cortical areas, including precuneus, V6, and dorsomedial frontal areas 6M and 8M. Additional connectivity to orbitofrontal cortex (area 13) and anticorrelations with frontal pole and midbrain are present.
Network 12 (cerebellar-frontal pole): This RSN covers the cerebellum and ventromedial frontal pole.
Network 13 (dorsomedial cortex): This RSN covers a large portion of the dorsomedial cortex, including areas of the frontal lobe (medial areas 9, 8, 6, 4, and 24) and parietal lobe (somatosensory cortices, precuneus, and are 23).
Network 14 (intraparietal sulcus): This RSN covers much of the intraparietal sulcus, including LIP, VIP, and MIP. Rostrally, it extends into the insula.
Network 15 (insula): This RSN is mostly situated in the insula and adjacent operculum. Dorsally, it bleeds into the somatosensory and premotor cortices; ventrally and rostrally, it bleeds into the ventrolateral prefrontal cortex.
Network 16 (caudal dorsomedial cortex): This RSN is in the dorsal parietal and occipital lobes. It includes portions of V1, V3, V4, V6, precuneus, Opt, LIP, and PEa.
Network 19 (orbitofrontal cortex): This RSN covers nearly all of the orbitofrontal cortex, spanning from orbitofrontal regions of the frontal pole to ventral insula regions of the orbitofrontal cortex. The network extends somewhat dorsally into the medial prefrontal cortex (mainly area 32).
Network 20 (midbrain): This RSN is centered in the midbrain, but extends into portions of the thalamus, hypothalamus, cerebellum, basal forebrain, and pons.
Network 21 (posteromedial cortex): This RSN covers a large portion of the posteromedial cortex, including posterior cingulate areas 23 and 31, parietal area precuneus, and, caudally, some of V6 and V2. There are smaller lateral regions in this network as well: portions of TPO, 7a, and LIP are present. Finally, much of the intraparietal sulcus shows anticorrelations to this RSN.
Network 23 (sensorimotor): This RSN is situated centrally and dorsomedially, spanning the border between the frontal lobe and parietal lobe. Thus, it includes primary motor cortex, primary somatosensory cortex, supplementary motor area, premotor cortex, anterior and posterior cingulate cortices, and PE.
Network 24 (lower brainstem): This RSN is situated in the pons and medulla.
Network 25 (middle-caudal cingulate): This RSN is mainly situated in the caudal anterior cingulate cortex (area 24’, sometimes called the midcingulate cortex) and posterior cingulate cortex (areas 23 and 31).
Network 26 (dorsal thalamus): This RSN is situated in the dorsal portion of the thalamus.
Network 30 (dorsal parietal cortex): This RSN is located in the dorsal parietal cortex, and covers the intraparietal sulcus, including areas LIPv, LIPd, area 5 and MIP. It extends towards the medial wall, covering areas V6, precuneus, and parts of the posterior cingulate cortex.
Network 31 (ventral thalamus): This RSN is located in the ventral thalamus, with some extension into the midbrain.
Network 32 (anterior cingulate-insula): This RSN covers the entire anterior cingulate cortex (area 24), much of the insula, and a small portion of the lateral parietal cortex (7op and PFG).
Network 33 (posterior cingulate-lateral parietal cortices): This RSN covers the posterior cingulate cortex (areas 23 and 31) as well as lateral parietal areas 7op and Tpt. There is also a small area of correlation in the temporal pole.
Network 35 (dorsolateral-cingulate-thalamus): This RSN includes areas in the dorsolateral prefrontal cortex such as area 8 as well as the anterior cingulate cortex area 24 and some rostral thalamus. Anticorrelations to posterior cingulate cortex and precuneus regions are observed.
Network 36 (lateral prefrontal right): This RSN covers the right lateral prefrontal cortex around the principle and arcuate sulcus. Anticorrelations with the medial prefrontal cortex, such as area 9 and 25, as well as part of the anterior cingulate cortex are observed.
Network 38 (lateral prefrontal left): This RSN covers the lateral left prefrontal cortex around the principle and arcuate sulcus.
Network 39 (sensorimotor 2): This RSN covers both sides of the central sulcus, and thus involves both motor and somatosensory cortices. The network also includes anticorrelations of posterior cingulate cortex and more caudal somatosensory areas.
Network 40 (frontal pole): This RSN is mainly in the frontal pole. Caudally, it extends into the dorsal prefrontal cortex and anterior cingulate cortex.
3.3. Individual resting state networks
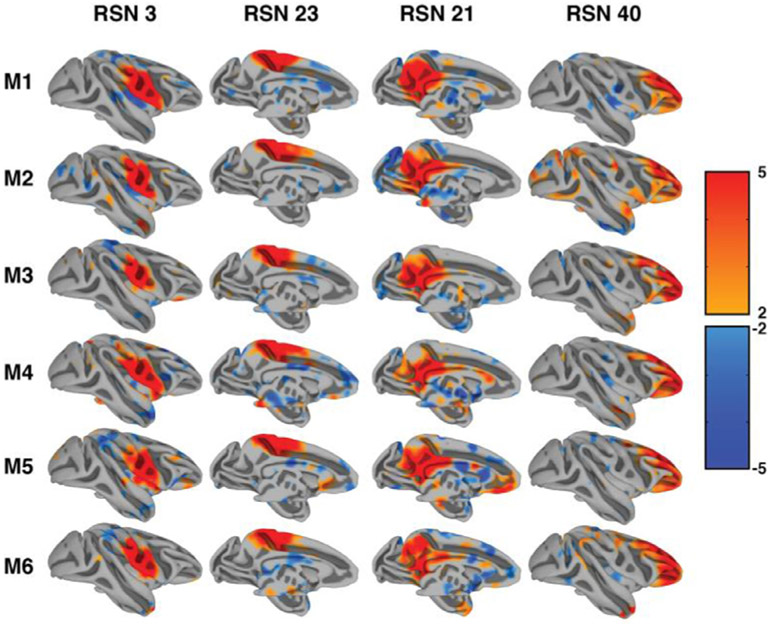
Since group analysis can obscure individual anatomical and functional details, we present all individual ICA results in supplementary Figs. 3-8. Fig. 4 shows the consistency of our single subject independent component results in all subjects for four representative components. Note the consistency in spatial location at the same threshold. This finding demonstrates the consistency and data quality of our imaging approach and shows that our group level findings are highly representative of the individual monkey.
Fig. 4.
Consistency of single subject resting state networks. The figure demonstrates the same four resting state components in all six monkeys. Thresholds were kept consistent with group analysis results.
Projection to surface space can additionally obscure results and is generally ill-suited for use in individual animal physiological use. Fig. 5. shows 7 of the 30 components displayed from one representative single subject volume space projected onto the NMT template. The results demonstrate the close correspondence of the components with anatomical and functional landmarks as well as specificity to both cortical and subcortical regions.
Fig. 5.
Example resting state networks (RSNs) of one macaque following single-subject independent component analysis (ICA). Images are normalized to the NMT space and overlaid onto the NMT-template. Overlaid color maps represent thresholded z-scores. Arbitrary slices were selected to demonstrate the diversity of components from subcortical to cortical components as well as the detailed anatomical correspondence.
3.4. Independent component network connectivity
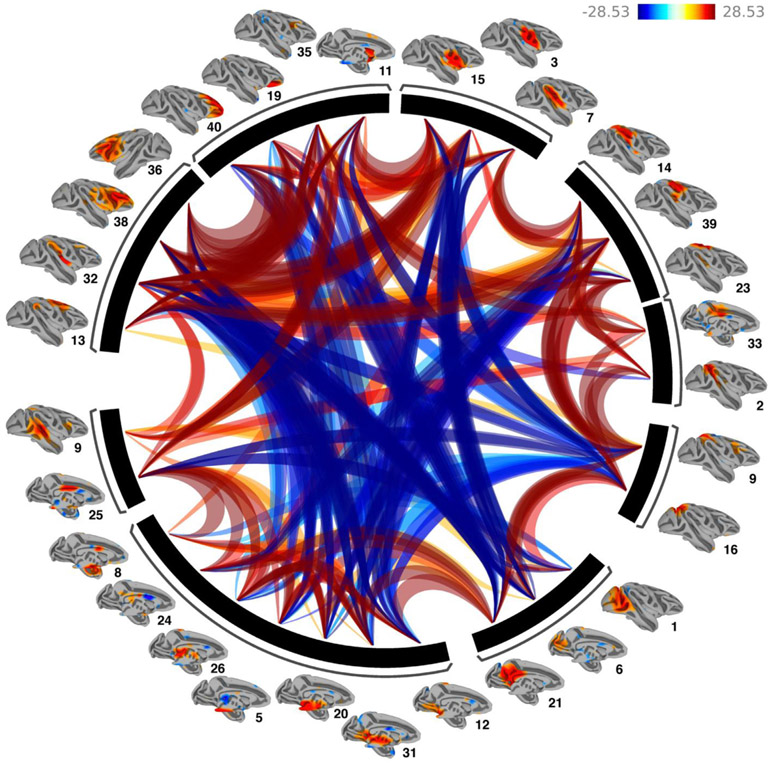
Data driven hierarchical clustering of the independent component time courses revealed 9 clusters of components, with strong positive functional connectivity within cluster and between adjacent clusters, and strong negative functional connectivity between non-adjacent clusters. Even though clustering was not based on spatial information, the resulting adjacencies reveal anatomical neighboring relationships (Fig. 6). Clusters fall into the following categories: 1. Subcortical components, 2. Prefrontal components, 3. Visual components, 4. Parietal components, 5. Superior-temporal components and 6. Motor components.
Fig. 6.
Functional Network Connectivity of the 30 extracted Independent components. Lines portray significant connections between the networks and color indicates positive (red) and negative (blue) functional connectivity. Networks were clustered using hierarchical clustering. Results demonstrate strong positive within cluster network connections and primarily negative between cluster connections.
4. Discussion
Here we present the first demonstration of connectome style quality resting state fMRI data in the macaque at ultra-high magnetic fields significantly beyond 7T. The delineated RSNs closely resemble the quality and findings of connectomes from large human datasets, providing a detailed map of the rsfMRI ‘macaque connectome’ as a basis for future interventional work.
Resting state functional connectivity patterns are an extremely important tool used in current neuroscientific research into the origin of mental disease states (Bai et al., 2011; Lawrie et al., 2002; Lustig et al., 2003; Quigley et al., 2001; Xie et al., 2012; Zilverstand et al., 2018). Yet, much of this technique’s physiological basis is not well understood. Since macaques offer an extremely promising translational model (Heilbronner and Chafee, 2019; Krubitzer and Huffman, 2000; Consortium et al., 2020; Hutchison et al., 2011; Hutchison and Everling, 2012; Milham et al., 2018), a detailed map of the rsfMRI ‘macaque connectome’ is needed to build the basis for future interventional work. While human rsfMRI studies have made significant advances in the ability to acquire highly sensitive and informative images of functional connectivity (Essen et al., 2012; Glasser et al., 2016; Harms et al., 2018), including at relatively high spatial and temporal resolutions (Moeller et al., 2010; Vu et al., 2016), similar non-invasive neuroimaging advances in the macaque have not been as widespread or productive.
Several barriers have previously prevented such translations from being successful. Since invasive work, such as single unit recording, anatomical dissection, or chemo- and optogenetic manipulations in the macaque is usually done on the individual subject level at high spatial and temporal scales, extremely robust within individual rsfMRI maps are needed. In vivo experiments using very high magnetic fields facilitate such high spatial-temporal resolutions and overall sensitivity across the entire brain. Next, high channel count coils with sufficient parallel imaging performance, specifically designed for macaque setups (Autio et al. 2020; Autio et al. this issue), are also essential for achieving the translational sensitivity and spatial-temporal resolutions. The use of parallel imaging is essential for reducing the volume acquisition times and hence increasing the temporal sampling rate of the fMRI time series. However, this results in a spatially non-uniform increase in the noise of the measurement through the so called g-factor noise (Pruessmann et al., 1999). The g-factor noise is mitigated with higher channel counts and independently with increasing magnetic fields (Ohliger et al., 2003; Wiesinger et al., 2006, 2004). Thus, the two approaches employed together, as in this 10.5T study, work synergistically to provide multiplicative gains, enabling high accelerations with suppressed g-factor noise (Uğurbil et al., 2019).
Further, even though human studies are typically completed within a couple of hours and macaque studies have the luxury of extending well beyond this to increase overall sensitivity, extending session duration can still be problematic in terms of physiological (and therefore fMRI) stability. For this reason, shorter scans with higher SNR are still desirable.
Another approach to achieving sensitivity with shorter scan durations is the use of contrast agents, which greatly improves lower field studies. However, the use of contrast agents has significant limitations as well, such as: 1) It does not measure intrinsic BOLD signals (which is what the vast majority of human studies use), 2) studying temporal dynamics or paradigms other than block designs is much more difficult, and 3) there are risks associated with its use and it is hence not used in non-clinical human studies.
While implementation on a human-based scanner is not absolutely necessary, it does provide a convenient avenue by which imaging methods and sequences routinely used in the human can be applied to the macaques (Autio et al. this issue). Finally, while obtaining high resolution functional images of the alert monkey would be more ideal than the low anesthesia strategy we employed here, this is still an enormous technical challenge that we and others are working to overcome. Despite this, our data demonstrate 10.5T coupled with high channel count arrays can achieve submillimeter acquisitions using the advanced accelerated sequences developed for the Human Connectome Project (Uğurbil et al., 2013). Further, the setup described here could be used for other applications, such as task fMRI.
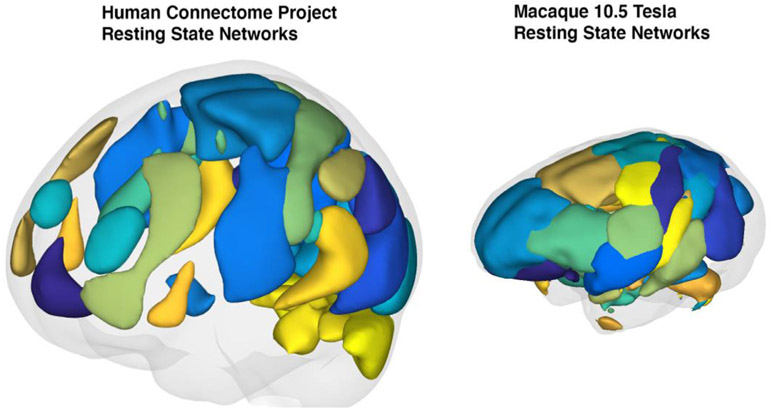
The macaque connectome has been studied previously (Hutchison et al., 2011) using the ultrahigh magnetic field of 7 Tesla with 1.3 × 1.3 × 1.5 mm3 resolution and 3 mm FWHM spatial smoothing, revealing 11 macaque RSNs using group ICA analysis. Here, following this earlier work (Hutchison et al., 2011) we report the first whole brain (0.75 mm3 isotropic at ~1.2s TR, smoothed at 2mm) comprehensive group independent component analysis in macaques at 10.5T. Although our data was of sufficient quality to produce RSNs without spatial smoothing , in this first 10.5T study, we employed spatial smoothing following coming practices employed in the resting state fMRI literature. Our approach identified 30 reproducible RSNs covering the entire macaque cortex as well as subcortical contributions throughout the thalamus, brainstem and cerebellum. The RSNs we were able to identify closely resemble higher model order RSNs identified in large sample sizes of human experiments (Abou-Elseoud et al., 2010; Kiviniemi et al., 2009; Li et al., 2007; Smith et al., 2013, 2012, 2009). In these findings, high model order RSNs are a tradeoff between the number of components extracted and the detectability of unique networks observed. Extremely high model order in ICA produces a larger number of networks but fails to uncover additional brain regions involved in a network. Our networks represent an intermediary between extremely high model orders (Moeller et al., 2009) and more conservative approaches (Hutchison et al., 2011), covering nearly the entire brain (see Fig. 7 and supplementary Fig. 2) while uncovering networks that include long range functional connections. Using this approach, we were able to demonstrate remarkable reproducibility of the RSN decomposition even in the individual animal without the use of contrast agents. This achievement is of utmost importance when trying to understand and model the relationship between real anatomical neural connections only obtainable invasively and functional connectivity based on neuroimaging. Our RSN findings reveal very similar network organization between humans and macaques at the mesoscopic level (see Fig. 7 for a qualitative comparison). We observe RSNs in all of the networks that are commonly identified in human experiments (Beckmann et al., 2005; Jafri et al., 2008; Smith et al., 2013, 2009). For example, we identify multiple networks that resemble the commonly described fronto-temporal, default-mode, fronto-parietal, cerebellar, visual, somato-motor, intraparietal, lateral prefrontal, basal ganglia, insular, and cingular networks (Fig. 7).
Fig. 7.
Qualitative comparison between resting state networks identified from the Human Connectome Project database (supplied by the CONN toolbox and Connectome Workbench) and our six subject macaque group resting state networks. Individual regions of interest were smoothed using a 10 mm spherical kernel interpolated in 3D. Gray shading represents a reconstruction of the estimated pial surface.
Previous work found 20 stimulus evoked components (Moeller et al., 2009) using contrast agents or 11 high quality intrinsic components (Hutchison et al., 2011). Extending this work, we were able to resolve previously not observed (Hutchison et al., 2011; Moeller et al., 2009; Vincent et al., 2009, 2007) lateralized dorsolateral prefrontal components (RSN 36,38) that have readily been described in the human literature and attributed to language, memory and cognitive attentional processes (Beckmann et al., 2005; Jafri et al., 2008; Smith et al., 2009). Our lateralized network does not include the parietal contribution present in the human literature, however. Another set of components that have previously remained elusive (Vincent et al., 2009, 2007) in the macaques are the dorso medial and orbital contributions of the prefrontal cortex. In our findings (RSN 19, 40) these were readily present. It is difficult to assess why these findings have previously not been reported, but SNR limitations as well as susceptibility artefacts close to the orbits and sinuses have likely contributed to the problem. One difference in our findings with the human literature is the absence of a large component covering the primary visual cortex’s caudal extent. Our visual cortex related RSNs (RSN 1,6) cover both medial and lateral portions of V1, V2 but miss the occipital pole. This is likely the result of our coil having the lowest sensitivity in visual cortex due to the macaque’s sphinx position. Another difference in our findings compared to previous macaque rsfMRI studies (Hutchison et al., 2011; Moeller et al., 2009; Vincent et al., 2009, 2007), but in agreement with high model order RSN studies (Abou-Elseoud et al., 2010; Kiviniemi et al., 2009) as well as clustering approaches (Leech et al., 2012, 2011), is the fractioning of RSNs that are associated with the default mode network. In our findings multiple RSNs (21, 26, 30, 31, 33, 39) show at least partial overlap with posteromedial cortex. Another finding that has previously not been reported is the detailed delineation of the cingulate cortex that can be observed in our RSNs. RSNs (5,8,9,11,13,21,24,25,32,33,35,39) all exhibit involvement of the cingulate cortex partitioning the anatomical areas 23a,b,c and 24a,b,c. While some of these networks demonstrate overlap in their cingulate cortex involvement, unique partitioning is directly observable in others (RSNs 5,24,25,26,35). Again, it is unclear why these effects were not previously found but inhomogeneous excitation from local transmitters or low SNR in medial regions further away from the scalp (especially true if headposts are used), and, compared to our study, lower BOLD contrast, could contribute to this omission.
To address one of our motivations, the robust identification of RSNs within the individual subject, single subject ICA using the same model order of 40 was performed and assessed for reproducibility and compared to the group results. Compared to previous studies (Hutchison et al., 2011) we found remarkable reproducibility for most of the 30 resulting RSNs. First, using the same thresholds as in the group-ICA resulted in more widespread diffuse activity, yet the pattern and anatomical correspondence of the main group-ICA findings were largely preserved in the individual. It is difficult to compare our current approach to previous findings since we used diffeomorphic transformations for normalization that likely resulted in better anatomical correspondence and acquired significantly more data in an individual subject.
Taken together, we have achieved a highly detailed, individually robust delineation of RSNs in the macaque that closely resembles the quality and findings of connectomes from large human datasets, further demonstrating the benefits of using extremely-high magnetic fields in vivo.
One countering recent argument and effort (Consortium et al., 2020; Milham et al., 2018) that has seen high translational success has been the cross laboratory pooling of macaque fMRI datasets. We argue that our rationale of advocating high field macaque neuroimaging does not negate or change the need for such databases, but rather aims at providing data and solutions for a different problem. The big data consortium approach allows for the study of general principles derived from rsfMRI data in macaques such as physiological states (Xu et al., 2019) (anesthetized vs. awake) or macroscale area organizations (Xu et al., 2018) within the species. These are incredibly important efforts that can also serve an important translational role. What these approaches cannot provide, and where we believe extremely-high field comes into play, is measuring the effects of within subject manipulation using invasive tools. If we are interested in figuring out how structural connections and their electrical and chemical changes enable functional connectivity in rsfMRI and behavior, disease, or developmental processes, resting state neuroimaging at high magnetic fields coupled with within-subject invasive manipulations will pave the way.
5. Conclusions
To our knowledge, this is the first detailed demonstration of a macaque connectome (or HCP) style resting state dataset, which has been available for human applications for around 10 years. While previous macaque resting state studies have demonstrated typical RSNs, none have achieved whole brain sub-millimeter acquisitions with an approximate 1 sec temporal resolution, let alone doing it using only intrinsic BOLD signals and with a scan duration of around an hour. Data sets such as the current one allow for more straightforward translations of macaque data to the human for the purposes of understanding brain connectivity in individuals and how it is altered in disease.
Supplementary Material
Acknowledgements
We thank Steve Jungst for continuing support with our coils and hardware setup. We thank Hannah Lee, Jen Holmberg, Adriana Cushnie, Tanya Casta, and Megan Monko for support with animal care and data acquisition. We thank Research Animal Resources at UMN, especially Whitney McGee and Anne Merley, for helping us implement new and improved anesthesia protocols.
Funding statement
This work was supported by NIH grants P30 DA048742 (to AZ, SRH, BYH and JZ), RF1 MH116978 (to EY), R01 DA038615 (to BYH), U01 EB025144 (to KU), by a P41 EB027061 (to KU, EY, NH, GA, BYH and JZ), R01 MH118257 (to SRH), an NINDS R01 NS081118 and P50 NS098573 Udall center to NH by an award from MNFutures to BYH, from the Digital Technologies Initiative to JZ, and BYH, from the Templeton Foundation to BYH, a Young Investigator Award from the Brain & Behavior Research Foundation to SRH, a Medical Discovery Team on Addiction Pilot Grant to SRH and BYH, and a UMN AIRP award to JZ, BYH, SRH and AZ.
Footnotes
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2020.117349.
Bibliography
- Abou-Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V, 2010. The effect of model order selection in group PICA. Hum. Brain Mapp 31. doi: 10.1002/hbm.20929, NA-NA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD, 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autio JA, Glasser MF, Ose T, Donahue CJ, Bastiani M, Ohno M, Kawabata Y, Urushibata Y, Murata K, Nishigori K, Yamaguchi M, Hori Y, Yoshida A, Go Y, Coalson TS, Jbabdi S, Sotiropoulos SN, Kennedy H, Smith S, Essen DCV, Hayashi T, 2020. Towards HCP-Style macaque connectomes: 24-Channel 3T multi-array coil, MRI sequences and preprocessing. Neuroimage 215, 116800. doi: 10.1016/j.neuroimage.2020.116800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC, 2014. The Insight ToolKit image registration framework. Front. Neuroinform 8 (44). doi: 10.3389/fninf.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC, 2011. An Open Source Multivariate Framework for n-Tissue Segmentation with Evaluation on Public Data. Neuroinformatics 9, 381–400. doi: 10.1007/s12021-011-9109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Xie C, Watson DR, Shi Y, Yuan Y, Wang Y, Yue C, Teng Y, Wu D, Zhang Z, 2011. Aberrant hippocampal subregion networks associated with the classifications of aMCI subjects: a longitudinal resting-state study. Plos One 6, e29288. doi: 10.1371/journal.pone.0029288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker R, Wachtler T, Diesmann M, 2012. CoCoMac 2.0 and the future of tract-tracing databases. Front. Neuroinform 6 (30). doi: 10.3389/fninf.2012.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM, 2005. Investigations into resting-state connectivity using independent component analysis. Philos. Trans. R. Soc. B Biol. Sci 360, 1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS, 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med 34, 537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski A-M, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kötter R, Li S-J, Lin C-P, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SARB, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng G-J, Veijola J, Villringer A, Walter M, Wang L, Weng X-C, Whitfield-Gabrieli S, Williamson P, Windischberger C, Zang Y-F, Zhang H-Y, Castellanos FX, Milham MP, 2010. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. USA 107, 4734–4739. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ, 2001. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp 14, 140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushnie AK, El-Nahal HG, Bohlen MO, May PJ, Basso MA, Grimaldi P, Wang MZ, Velasco E.M.F., de, Sommer MA, Heilbronner SR, 2020. Using rAAV2-retro in rhesus macaques: Promise and caveats for circuit manipulation. J. Neurosci. Methods 345, 108859. doi: 10.1016/j.jneumeth.2020.108859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DM, Smith SM, Beckmann CF, 2010. Advances and pitfalls in the analysis and interpretation of resting-state FMRI data. Front. Syst. Neurosci 4 (8). doi: 10.3389/fnsys.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, T., Prim DE, PRIME-D GCW, Milham M, Petkov CI, Margulies DS, Schroeder CE, Basso MA, Belin P, Fair DA, Fox A, Kastner S, Mars RB, Messinger A, Poirier C, Vanduffel W, Essen DCV, Alvand A, Becker Y, Hamed SB, Benn A, Bodin C, Boretius S, Cagna B, Coulon O, El-Gohary SH, Evrard H, Forkel SJ, Friedrich P, Froudist-Walsh S, Garza-Villarreal EA, Gao Y, Gozzi A, Grigis A, Hartig R, Hayashi T, Heuer K, Howells H, Ardesch DJ, Jarraya B, Jarrett W, Jedema HP, Kagan I, Kelly C, Kennedy H, Klink PC, Kwok SC, Leech R, Liu X, Madan C, Madushanka W, Majka P, Mallon A-M, Marche K, Meguerditchian A, Menon RS, Merchant H, Mitchell A, Nenning K-H, Nikolaidis A, Ortiz-Rios M, Pagani M, Pareek V, Prescott M, Procyk E, Rajimehr R, Rautu I-S, Raz A, Roe AW, Rossi-Pool R, Roumazeilles L, Sakai T, Sallet J, García-Saldivar P, Sato C, Sawiak S, Schiffer M, Schwiedrzik CM, Seidlitz J, Sein J, Shen Z, Shmuel A, Silva AC, Simone L, Sirmpilatze N, Sliwa J, Smallwood J, Tasserie J, Schotten M.T., de, Toro R, Trapeau R, Uhrig L, Vezoli J, Wang Z, Wells S, Williams B, Xu T, Xu AG, Yacoub E, Zhan M, Ai L, Amiez C, Balezeau F, Baxter MG, Blezer ELA, Brochier T, Chen A, Croxson PL, Damatac CG, Dehaene S, Everling S, Fleysher L, Freiwald W, Griffiths TD, Guedj C, Hadj-Bouziane F, Harel N, Hiba B, Jung B, Koo B, Laland KN, Leopold DA, Lindenfors P, Meunier M, Mok K, Morrison JH, Nacef J, Nagy J, Pinsk M, Reader SM, Roelfsema PR, Rudko DA, Rushworth MFS, Russ BE, Schmid MC, Sullivan EL, Thiele A, Todorov OS, Tsao D, Ungerleider L, Wilson CRE, Ye FQ, Zarco W, Zhou Y, 2020. Accelerating the evolution of nonhuman primate neuroimaging. Neuron 105, 600–603. doi: 10.1016/j.neuron.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res 29, 162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Essen DCV, Maunsell JHR, 1983. Hierarchical organization and functional streams in the visual cortex. Trends Neurosci. 6, 370–375. doi: 10.1016/0166-2236(83)90167-4. [DOI] [Google Scholar]
- Essen DCV, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Penna SD, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E, Consortium, W.-M.H., 2012. The Human Connectome Project: A data acquisition perspective. Neuroimage 62, 2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber NE, Harkin CP, Niedfeldt J, Hudetz AG, Kampine JP, Schmeling WT, 1997. Region-specific and agent-specific dilation of intracerebral microvessels by volatile anesthetics in rat brain slices. Anesthesiology 87, 1191–1198. doi: 10.1097/00000542-199711000-00024. [DOI] [PubMed] [Google Scholar]
- Felleman DJ, Essen DCV, 1991. Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47. doi: 10.1093/cercor/1.1.1. [DOI] [PubMed] [Google Scholar]
- Floris DL, Lai M-C, Nath T, Milham MP, Martino AD, 2018. Network-specific sex differentiation of intrinsic brain function in males with autism. Mol. Autism 9 (17). doi: 10.1186/s13229-018-0192-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME, 2007. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci 8, 700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Galvan A, Raper J, Hu X, Paré J, Bonaventura J, Richie CT, Michaelides M, Mueller SAL, Roseboom PH, Oler JA, Kalin NH, Hall RA, Smith Y, 2019. Ultrastructural localization of DREADDs in monkeys. Eur. J. Neurosci 50, 2801–2813. doi: 10.1111/ejn.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvan A, Stauffer WR, Acker L, El-Shamayleh Y, Inoue K-I, Ohayon S, Schmid MC, 2017. Nonhuman primate optogenetics: recent advances and future directions. J. Neurosci. Official J. Soc. Neurosci 37, 10894–10903. doi: 10.1523/jneurosci.1839-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert KM, Gati JS, Barker K, Everling S, Menon RS, 2016. Optimized parallel transmit and receive radiofrequency coil for ultrahigh-field MRI of monkeys. NeuroImage 125, 153–161. doi: 10.1016/j.neuroimage.2015.10.048. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Smith SM, Marcus DS, Andersson JLR, Auerbach EJ, Behrens TEJ, Coalson TS, Harms MP, Jenkinson M, Moeller S, Robinson EC, Sotiropoulos SN, Xu J, Yacoub E, Ugurbil K, Essen DCV, 2016. The Human Connectome Project’s neuroimaging approach. Nat. Neurosci 19, 1175–1187. doi: 10.1038/nn.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas A, Stiers P, Hutchison RM, Everling S, Petrides M, Margulies DS, 2017. Intrinsic functional architecture of the macaque dorsal and ventral lateral frontal cortex. J. Neurophysiol 117, 1084–1099. doi: 10.1152/jn.00486.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallquist MN, Hwang K, Luna B, 2013. The nuisance of nuisance regression: spectral misspecification in a common approach to resting-state fMRI preprocessing reintroduces noise and obscures functional connectivity. Neuroimage 82, 208–225. doi: 10.1016/j.neuroimage.2013.05.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MP, Somerville LH, Ances BM, Andersson J, Barch DM, Bastiani M, Bookheimer SY, Brown TB, Buckner RL, Burgess GC, Coalson TS, Chappell MA, Dapretto M, Douaud G, Fischl B, Glasser MF, Greve DN, Hodge C, Jamison KW, Jbabdi S, Kandala S, Li X, Mair RW, Mangia S, Marcus D, Mascali D, Moeller S, Nichols TE, Robinson EC, Salat DH, Smith SM, Sotiropoulos SN, Terpstra M, Thomas KM, Tisdall MD, Ugurbil K, van der Kouwe A, Woods RP, Zöllei L, Essen DCV, Yacoub E, 2018. Extending the human connectome project across ages: imaging protocols for the lifespan development and aging projects. Neuroimage 183, 972–984. doi: 10.1016/j.neuroimage.2018.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronner SR, Chafee MV, 2019. Learning how neurons fail inside of networks: nonhuman primates provide critical data for psychiatry. Neuron 102, 21–26. doi: 10.1016/j.neuron.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilbronner SR, Rodriguez-Romaguera J, Quirk GJ, Groenewegen HJ, Haber SN, 2016. Circuit-Based Corticostriatal Homologies Between Rat and Primate. Biol. Psychiat 80, 509–521. doi: 10.1016/j.biopsych.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Pol HEH, 2010. Exploring the brain network: A review on resting-state fMRI functional connectivity. Eur. Neuropsychopharm 20, 519–534. doi: 10.1016/j.euroneuro.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Hindriks R, Adhikari MH, Murayama Y, Ganzetti M, Mantini D, Logothetis NK, Deco G, 2016. Can sliding-window correlations reveal dynamic functional connectivity in resting-state fMRI? Neuroimage 127, 242–256. doi: 10.1016/j.neuroimage.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulvershorn LA, Mennes M, Castellanos FX, Martino AD, Milham MP, Hummer TA, Roy AK, 2013. Abnormal amygdala functional connectivity associated with emotional lability in children with attention-deficit/hyperactivity disorder. J. Am. Acad. Child Psy 53. doi: 10.1016/j.jaac.2013.11.012, 351–61.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Everling S, 2012. Monkey in the middle: why non-human primates are needed to bridge the gap in resting-state investigations. Front. Neuroanat 6 (29). doi: 10.3389/fnana.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Leung LS, Mirsattari SM, Gati JS, Menon RS, Everling S, 2011. Resting-state networks in the macaque at 7T. Neuroimage 56, 1546–1555. doi: 10.1016/j.neuroimage.2011.02.063. [DOI] [PubMed] [Google Scholar]
- Hutchison RM Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Penna SD, Duyn JH, Glover GH, Gonzalez-Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, Pasquale de, F., Sporns O, Walter M, Chang C, 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80, 360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS, 2013. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum. Brain Mapp 34, 2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD, 2008. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage 39, 1666–1681. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM, 2012. FSL. Neuroimage 62, 782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Jonckers E, Audekerke JV, Visscher GD, der Linden AV, Verhoye M, 2011. Functional connectivity fMRI of the rodent brain: comparison of functional connectivity networks in rat and mouse. Plos One 6, e18876. doi: 10.1371/journal.pone.0018876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H, Bassett DS, 2020. Dynamic representations in networked neural systems. Nat. Neurosci 1–10. doi: 10.1038/s41593-020-0653-3. [DOI] [PubMed] [Google Scholar]
- Khateeb K, Griggs DJ, Sabes PN, Yazdan-Shahmorad A, 2019. Convection enhanced delivery of optogenetic adeno-associated viral vector to the cortex of rhesus macaque under guidance of online MRI images. J. Vis. Exp doi: 10.3791/59232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Starck T, Remes J, Long X, Nikkinen J, Haapea M, Veijola J, Moilanen I, Isohanni M, Zang Y-F, Tervonen O, 2009. Functional segmentation of the brain cortex using high model order group PICA. Hum. Brain Mapp 30, 3865–3886. doi: 10.1002/hbm.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konova AB, Moeller SJ, Tomasi D, Goldstein RZ, 2015. Effects of chronic and acute stimulants on brain functional connectivity hubs. Brain Res. 1628, 147–156. doi: 10.1016/j.brainres.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krubitzer L, Huffman KJ, 2000. Arealization of the neocortex in mammals: genetic and epigenetic contributions to the phenotype. Brain Behav. Evol 55, 322–335. doi: 10.1159/000006667. [DOI] [PubMed] [Google Scholar]
- Lagore R, Moeller S, Delabarre L, Grant A, Radder J, Zimmermann J, Ugurbil K, Yacoub E, Harel N, Adriany G, 2020. An 8 channel dipole transceive and 24 loop receive array for non-human primate head imaging at 10.5T. In: Proceedings of the 28th International Social for Magnetic Resonance in Medicine Scientific Meeting ISMRM. [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT, 2011. Behavioral interpretations of intrinsic connectivity networks. J. Cognit. Neurosci 23, 4022–4037. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie SM, Buechel C, Whalley HC, Frith CD, Friston KJ, Johnstone EC, 2002. Reduced frontotemporal functional connectivity in schizophrenia associated with auditory hallucinations. Biol. Psychiat 51, 1008–1011. doi: 10.1016/s0006-3223(02)01316-1. [DOI] [PubMed] [Google Scholar]
- Leech R, Braga R, Sharp DJ, 2012. Echoes of the brain within the posterior cingulate cortex. J. Neurosci 32, 215–222. doi: 10.1523/jneurosci.3689-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leech R, Kamourieh S, Beckmann CF, Sharp DJ, 2011. Fractionating the default mode network: distinct contributions of the ventral and dorsal posterior cingulate cortex to cognitive control. J. Neurosci 31, 3217–3224. doi: 10.1523/jneurosci.5626-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite FP, Tsao D, Vanduffel W, Fize D, Sasaki Y, Wald LL, Dale AM, Kwong KK, Orban GA, Rosen BR, Tootell R, Mandeville JB, 2002. Repeated fMRI using iron oxide contrast agent in awake, behaving macaques at 3 tesla. NeuroImage 16, 283–294. doi: 10.1006/nimg.2002.1110. [DOI] [PubMed] [Google Scholar]
- Li L, Hu X, Preuss TM, Glasser MF, Damen FW, Qiu Y, Rilling J, 2013. Mapping putative hubs in human, chimpanzee and rhesus macaque connectomes via diffusion tractography. Neuroimage 80, 462–474. doi: 10.1016/j.neuroimage.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Kim HS, Tian L, Yu MK, Jon S, Moon WK, 2012. Comparison of two ultrasmall superparamagnetic iron oxides on cytotoxicity and mr imaging of tumors. Theranostics 2, 76–85. doi: 10.7150/thno.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-O, Adalı T, Calhoun VD, 2007. Estimating the number of independent components for functional magnetic resonance imaging data. Hum. Brain Mapp 28, 1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, 2003. The underpinnings of the BOLD functional magnetic resonance imaging signal. J. Neurosci 23, 3963–3971. doi: 10.1523/jneurosci.23-10-03963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A, 2001. Neuro-physiological investigation of the basis of the fMRI signal. Nature 412 (150). doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Lustig C, Snyder AZ, Bhakta M, O’Brien KC, McAvoy M, Raichle ME, Morris JC, Buckner RL, 2003. Functional deactivations: change with age and dementia of the Alzheimer type. Proc. Natl. Acad. Sci 100, 14504–14509. doi: 10.1073/pnas.2235925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov NT, Ercsey-Ravasz MM, Gomes ARR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey-Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, Essen DCV, Kennedy H, 2012. A weighted and directed interareal connectivity matrix for macaque cerebral cortex Cereb. Cortex New York N Y: 1991 24, 17–36. doi: 10.1093/cercor/bhs270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars RB, Verhagen L, Gladwin TE, Neubert F-X, Sallet J, Rushworth MFS, 2016. Comparing brains by matching connectivity profiles. Neurosci. Biobehav. Rev 60, 90–97. doi: 10.1016/j.neubiorev.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino FD, Esposito F, van de Moortele P-F, Harel N, Formisano E, Goebel R, Ugurbil K, Yacoub E, 2011. Whole brain high-resolution functional imaging at ultra high magnetic fields: An application to the analysis of resting state networks. Neuroimage 57, 1031–1044. doi: 10.1016/j.neuroimage.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino FD, Yacoub E, Kemper V, Moerel M, Uludağ K, Weerd PD, Ugurbil K, Goebel R, Formisano E, 2018. The impact of ultra-high field MRI on cognitive and computational neuroimaging. Neuroimage 168, 366–382. doi: 10.1016/j.neuroimage.2017.03.060. [DOI] [PubMed] [Google Scholar]
- Milham MP, Ai L, Koo B, Xu T, Amiez C, Balezeau F, Baxter MG, Blezer ELA, Brochier T, Chen A, Croxson PL, Damatac CG, Dehaene S, Everling S, Fair DA, Fleysher L, Freiwald W, Froudist-Walsh S, Griffiths TD, Guedj C, Hadj-Bouziane F, Hamed SB, Harel N, Hiba B, Jarraya B, Jung B, Kastner S, Klink PC, Kwok SC, Laland KN, Leopold DA, Lindenfors P, Mars RB, Menon RS, Messinger A, Meunier M, Mok K, Morrison JH, Nacef J, Nagy J, Rios MO, Petkov CI, Pinsk M, Poirier C, Procyk E, Rajimehr R, Reader SM, Roelfsema PR, Rudko DA, Rushworth MFS, Russ BE, Sallet J, Schmid MC, Schwiedrzik CM, Seidlitz J, Sein J, Shmuel A, Sullivan EL, Ungerleider L, Thiele A, Todorov OS, Tsao D, Wang Z, Wilson CRE, Yacoub E, Ye FQ, Zarco W, Zhou Y, Margulies DS, Schroeder CE, 2018. An open resource for non-human primate imaging. Neuron 100, 61–74. doi: 10.1016/j.neuron.2018.08.039, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Nallasamy N, Tsao DY, Freiwald WA, 2009. Functional connectivity of the macaque brain across stimulus and arousal states. J. Neurosci. Official J. Soc. Neurosci 29, 5897–5909. doi: 10.1523/jneurosci.0220-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Uğurbil K, 2010. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med 63, 1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa TA, Mohammed-Rasheed MA, 2019. Accumulation and cytotoxicity assessment of TAT-IONPs on cancerous mammalian cells. Anim. Biotechnol 1–6. doi: 10.1080/10495398.2019.1658595. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Kikuchi E, Lerchner W, Inoue K, Ji B, Eldridge MAG, Kaneko H, Kimura Y, Oh-Nishi A, Hori Y, Kato Y, Hirabayashi T, Fujimoto A, Kumata K, Zhang M-R, Aoki I, Suhara T, Higuchi M, Takada M, Richmond BJ, Minamimoto T, 2016. PET imaging-guided chemogenetic silencing reveals a critical role of primate rostromedial caudate in reward evaluation. Nat. Commun 7, 13605. doi: 10.1038/ncomms13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohliger MA, Grant AK, Sodickson DK, 2003. Ultimate intrinsic signal-to-noise ratio for parallel MRI: Electromagnetic field considerations. Magn. Reson. Med 50, 1018–1030. doi: 10.1002/mrm.10597. [DOI] [PubMed] [Google Scholar]
- Okada S, Bartelle BB, Li N, Breton-Provencher V, Lee JJ, Rodriguez E, Melican J, Sur M, Jasanoff A, 2018. Calcium-dependent molecular fMRI using a magnetic nanosensor. Nat. Nanotechnol 13, 473–477. doi: 10.1038/s41565-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelekanos V, Mok RM, Joly O, Ainsworth M, Kyriazis D, Kelly MG, Bell AH, Kriegeskorte N, 2020. Rapid event-related, BOLD fMRI, non-human primates (NHP): choose two out of three. Sci. Rep-uk 10, 7485. doi: 10.1038/s41598-020-64376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrides M, Pandya DN, 2002. Comparative cytoarchitectonic analysis of the human and the macaque ventrolateral prefrontal cortex and corticocortical connection patterns in the monkey: ventrolateral prefrontal cortex in human and monkey. Eur. J. Neurosci 16, 291–310. doi: 10.1046/j.1460-9568.2001.02090.x. [DOI] [PubMed] [Google Scholar]
- Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P, 1999. SENSE: sensitivity encoding for fast MRI. Magn. Reson. Med 42, 952–962. doi: . [DOI] [PubMed] [Google Scholar]
- Quigley M, Cordes D, Wendt G, Turski P, Moritz C, Haughton V, Meyerand ME, 2001. Effect of focal and nonfocal cerebral lesions on functional connectivity studied with MR imaging. Ajnr Am. J. Neuroradiol 22, 294–300. [PMC free article] [PubMed] [Google Scholar]
- Raper J, Murphy L, Richardson R, Romm Z, Kovacs-Balint Z, Payne C, Galvan A, 2019. Chemogenetic inhibition of the amygdala modulates emotional behavior expression in infant rhesus monkeys. Eneuro 6. doi: 10.1523/eneuro.0360-19.2019, ENEURO.0360-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, 2012. SUMA. Neuroimage 62, 768–773. doi: 10.1016/j.neuroimage.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall B, Japee S, Cox RW, 2004. SUMA: An interface for surface-based intra- and inter-subject analysis with AFNI. In: Proceedings of the 2nd IEEE International Symposium Biomed Imaging Nano Macro Ieee Cat 04ex821, pp. 1510–1513. doi: 10.1109/isbi.2004.1398837. [DOI] [Google Scholar]
- Saleem KS, Logothetis NK, 2012. A Combined MRI and Histology Atlas of the Rhesus Monkey Brain in Stereotaxic Coordinates. Academic Press. [Google Scholar]
- Schaeffer DJ, Johnston KD, Gilbert KM, Gati JS, Menon RS, Everling S, 2018. In vivo manganese tract tracing of frontal eye fields in rhesus macaques with ultra-high field MRI: Comparison with DWI tractography. Neuroimage 181, 211–218. doi: 10.1016/j.neuroimage.2018.06.072. [DOI] [PubMed] [Google Scholar]
- Seidlitz J, Sponheim C, Glen D, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, Messinger A, 2018. A population MRI brain template and analysis tools for the macaque. NeuroImage 170. doi: 10.1016/j.neuroimage.2017.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL, 2012. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn. Reson. Med 67. doi: 10.1002/mrm.23097, 1210–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF, 2009. Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. USA 106. doi: 10.1073/pnas.0905267106, 13040–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Essen DCV, Feinberg DA, Yacoub ES, Ugurbil K, 2012. Temporally-independent functional modes of spontaneous brain activity. Proc. Natl. Acad. Sci. USA 109. doi: 10.1073/pnas.1121329109, 3131–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Vidaurre D, Beckmann CF, Glasser MF, Jenkinson M, Miller KL, Nichols TE, Robinson EC, Salimi-Khorshidi G, Woolrich MW, Barch DM, Uğurbil K, Essen DCV, 2013. Functional connectomics from resting-state fMRI. Trends Cogn. Sci 17. doi: 10.1016/j.tics.2013.09.016, 666–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao DY, Freiwald WA, Knutsen TA, Mandeville JB, Tootell RBH, 2003. Faces and objects in macaque cerebral cortex. Nat. Neurosci 6, 989–995. doi: 10.1038/nn1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uğurbil K, 2018. Imaging at ultrahigh magnetic fields: History, challenges, and solutions. Neuroimage 168, 7–32. doi: 10.1016/j.neuroimage.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurbil K, 2016. What is feasible with imaging human brain function and connectivity using functional magnetic resonance imaging. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci 371, 20150361. doi: 10.1098/rstb.2015.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugurbil K, 2014. Magnetic resonance imaging at ultrahigh fields. IEEE Trans. Bio-med. Eng 61, 1364–1379. doi: 10.1109/tbme.2014.2313619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uğurbil K, Adriany G, Andersen P, Chen W, Garwood M, Gruetter R, Henry P-G, Kim S-G, Lieu H, Tkac I, Vaughan T, Moortele P-FVD, Yacoub E, Zhu X-H, 2003. Ultrahigh field magnetic resonance imaging and spectroscopy. Magn. Reson. Imaging 21, 1263–1281. doi: 10.1016/j.mri.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Uğurbil K, Auerbach E, Moeller S, Grant A, Wu X, Moortele P.-F.V.de, Olman C, DelaBarre L, Schillak S, Radder J, Lagore R, Adriany G, 2019. Brain imaging with improved acceleration and SNR at 7 Tesla obtained with 64-channel receive array. Magn. Reson. Med 82, 495–509. doi: 10.1002/mrm.27695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uğurbil K, Xu J, Auerbach EJ, Moeller S, Vu AT, Duarte-Carvajalino JM, Lenglet C, Wu X, Schmitter S, Moortele P.F.V.de, Strupp J, Sapiro G, Martino FD, Wang D, Harel N, Garwood M, Chen L, Feinberg DA, Smith SM, Miller KL, Sotiropoulos SN, Jbabdi S, Andersson JLR, Behrens TEJ, Glasser MF, Essen DCV, Yacoub E, Consortium, W.-M.H., 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. Neuroimage 80, 80–104. doi: 10.1016/j.neuroimage.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upright NA, Brookshire SW, Schnebelen W, Damatac CG, Hof PR, Browning PGF, Croxson PL, Rudebeck PH, Baxter MG, 2018. Behavioral effect of chemogenetic inhibition is directly related to receptor transduction levels in rhesus monkeys. J. Neurosci. Official J. Soc. Neurosci 38, 7969–7975. doi: 10.1523/jneurosci.1422-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanduffel W, Fize D, Mandeville JB, Nelissen K, Hecke PV, Rosen BR, Tootell RBH, Orban GA, 2001. Visual motion processing investigated using contrast agent-enhanced fMRI in awake behaving monkeys. Neuron 32, 565–577. doi: 10.1016/s0896-6273(01)00502-5. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Kahn I, Essen DCV, Buckner RL, 2009. Functional connectivity of the macaque posterior parahippocampal cortex. J. Neurophysiol 103, 793–800. doi: 10.1152/jn.00546.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Essen DCV, Zempel JM, Snyder LH, Corbetta M, Raichle ME, 2007. Intrinsic functional architecture in the anaesthetized monkey brain. Nature 447, 83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Vu AT, Jamison K, Glasser MF, Smith SM, Coalson T, Moeller S, Auerbach EJ, Uğurbil K, Yacoub E, 2016. Tradeoffs in pushing the spatial resolution of fMRI for the 7T human connectome project. Neuroimage 154, 23–32. doi: 10.1016/j.neuroimage.2016.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Zang Y, Yang H, Tang H, Gong Q, Chen Z, Zhu C, He Y, 2009. Parcellation-dependent small-world brain functional networks: a resting-state fMRI study. Hum. Brain Mapp 30. doi: 10.1002/hbm.20623, 1511–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson KK, Platt ML, 2012. Of mice and monkeys: using non-human primate models to bridge mouse- and human-based investigations of autism spectrum disorders. J. Neurodev. Disord 4 (21). doi: 10.1186/1866-1955-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S, Nieto-Castanon A, 2012. Conn : a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2, 125–141. doi: 10.1089/brain.2012.0073. [DOI] [PubMed] [Google Scholar]
- Wiesinger F, Moortele PV, de Adriany G, Zanche ND, Ugurbil K, Pruessmann KP, 2006. Potential and feasibility of parallel MRI at high field. NMR Biomed. 19, 368–378. doi: 10.1002/nbm.1050. [DOI] [PubMed] [Google Scholar]
- Wiesinger F, Moortele PV, de Adriany G, Zanche ND, Ugurbil K, Pruessmann KP, 2004. Parallel imaging performance as a function of field strength—An experimental investigation using electrodynamic scaling. Magn. Reson. Med 52, 953–964. doi: 10.1002/mrm.20281. [DOI] [PubMed] [Google Scholar]
- Xie C, Bai F, Yu H, Shi Y, Yuan Y, Chen Gang, Li W, Chen Guangyu, Zhang Z, Li S-J, 2012. Abnormal insula functional network is associated with episodic memory decline in amnestic mild cognitive impairment. Neuroimage 63. doi: 10.1016/j.neuroimage.2012.06.062, 320–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Falchier A, Sullivan EL, Linn G, Ramirez JSB, Ross D, Feczko E, Opitz A, Bagley J, Sturgeon D, Earl E, Miranda-Domínguez O, Perrone A, Craddock RC, Schroeder CE, Colcombe S, Fair DA, Milham MP, 2018. Delineating the macroscale areal organization of the macaque cortex in vivo. Cell Rep. 23, 429–441. doi: 10.1016/j.celrep.2018.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Sturgeon D, Ramirez JSB, Froudist-Walsh S, Margulies DS, Schroeder CE, Fair DA, Milham MP, 2019. Inter-individual variability of functional connectivity in awake and anesthetized rhesus monkeys. Biol. Psychiatry Cognit. Neurosci. Neuroimaging doi: 10.1016/j.bpsc.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub E, Harel N, Uğurbil K, 2008. High-field fMRI unveils orientation columns in humans. Proc. Natl. Acad. Sci 105, 10607–10612. doi: 10.1073/pnas.0804110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoub E, Shmuel A, Pfeuffer J, Moortele PVD, Adriany G, Andersen P, Vaughan JT, Merkle H, Ugurbil K, Hu X, 2001. Imaging brain function in humans at 7 Tesla. Magn. Reson. Med 45, 588–594. doi: 10.1002/mrm.1080. [DOI] [PubMed] [Google Scholar]
- Zilverstand A, Huang AS, Alia-Klein N, Goldstein RZ, 2018. Neuroimaging impaired response inhibition and salience attribution in human drug addiction: a systematic review. Neuron 98, 886–903. doi: 10.1016/j.neuron.2018.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Goebel R, Martino FD, van de Moortele P-F, Feinberg D, Adriany G, Chaimow D, Shmuel A, Uğurbil K, Yacoub E, 2011. Mapping the organization of axis of motion selective features in human area MT using high-field fMRI. Plos One 6, e28716. doi: 10.1371/journal.pone.0028716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X-N, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP, 2010. Reliable intrinsic connectivity networks: Test–retest evaluation using ICA and dual regression approach. Neuroimage 49, 2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.