Abstract
Background:
Transmission dynamics of the infectious disease Corona Virus Disease - 19 (COVID-19) is yet to be understood fully. The study aimed at exploring whether quantitative viral load of COVID-19-infected case indicated by cycle threshold (Ct) value of real-time reverse transcription polymerase chain reaction could predict about transmission pattern in the community.
Materials and Methods:
An observational study was conducted involving 1976 individuals, suspected to be suffering from COVID-19 and contacts, of laboratory confirmed cases from selected districts of Gujarat, India. A total of 138 persons were detected to be positive. Weekly positivity showed an overall increasing trend during the studied weeks. It was observed that only 7% had high, 9% as moderate and rest, 84% had low viral load based on Ct values of real-time RT-PCR.
Results:
Most secondary cases clustered around index cases with high viral load whereas fewer secondary cases clustered around index cases with low viral load. Each index high viral load case transmitted an average of 6.25 secondary cases whereas the same of low viral load transmitted an average of 0.8 case.
Conclusion:
If cases with higher viral load are selectively isolated on detection from the rest of the community along with contact tracing of all individuals, who came in contact with them during the previous 5 days, the quantum of transmission will reduce subsequently. Moreover, health-care workers often get infected while working, probably due to the fact that they often handle cases with higher viral load. The Ct value of all may be provided along with test report to safeguard everybody's health including health-care workers.
Keywords: COVID-19, cycle threshold value, severe acute respiratory syndrome-CoV-2, super-spreader, transmission
INTRODUCTION
India is facing a continuous ascent of the COVID-19 epidemic curve since March 2020 till date. Often there is an argument that India's scenario is much better as the number of new infections is less, the number of deaths also is less compared to that of the western world. There is need to explore the factors underpinning these variations such as universal BCG/OPV vaccination and chloroquine exposure.[1,2,3,4]
COVID-19 is a newly discovered coronavirus which represents the third coronavirus-associated epidemic to emerge from a species leap from wild animals to humans, after severe acute respiratory syndrome (SARS) in 2003, and the Middle East Respiratory Syndrome (MERS) in 2012.[5] Transmission dynamics of COVID-19 are yet to be understood fully. R0 has been defined as the average number of secondary cases of an infectious disease that one case would generate in a completely susceptible population.[6] Estimated basic reproductive number of COVID-19, declared by the WHO (dated January 23, 2020) ranges within 1.4–2.5.[7] A recent review by Liu et al. found the average R0 to be 3.28 and median to be 2.79, which exceed the WHO estimates.[8] It was observed in SARS that a few of the infected people in the community spread most of the infection, whereas most people, although infected, spread it to few people.[9,10,11,12,13,14,15,16]
In addition to R0, scientists use a value called dispersion factor (K), a number indicating the likelihood that a particular disease will spread in clusters.[17] Lower is the K value, more is the transmission that occurs from a small number of people. In 2005, an article in nature estimated that SARS had a K value of 0.16.[17] In a recent publication, K value for COVID-19 was estimated to be as low as 0.1;[18] suggesting that probably about 10% of the cases would spread about 80% of the cases. In view of the above, the current study was conducted to assess the distribution of viral loads among infected cases and whether viral load of COVID-19-infected case indicated by cycle threshold (Ct) value of reverse transcription polymerase chain reaction (RT-PCR) could predict about transmission pattern in the community apart from population mobility and its density.
MATERIALS AND METHODS
Sample collection
The government body had identified certain certified COVID laboratories to diagnose COVID-19 around India. The Indian Council of Medical Research-National Institute of Occupational Health (ICMR-NIOH) was one among the certified centers to execute COVID-19 tests for confirmation and diagnosis. A total of 1976 nasopharyngeal and oropharyngeal swab samples suspected of COVID-19 from Mahisagar and adjacent regions of Gujarat state were provided at ICMR-NIOH by the concerned government health officials during April 2020–May 2020. The samples were collected according to standard procedures and transported using viral transport medium, and accompanied with minimal relevant details such as contact number of the suspected individual, date, and site of sample collection.
Sample analysis
The tests were done by the real-time RT-PCR using standard protocol given by the manufacturer under Principles and Procedure used in laboratory:[19] The real-time fluorescent RT-PCR Kit for Detecting SARS-CoV-2 is a real-time RT-PCR (rRT-PCR) test (BGI Real-Time Fluorescent RT-PCR kit, BGI Biotechnology Co. Ltd, China). The SARS-CoV-2 primer and probe set (s) is designed to detect RNA from the SARS-CoV-2 in the samples. The real-time fluorescent RT-PCR Kit selects a specific target region in the ORF1ab region of SARS-CoV-2 genome. Further, human housekeeping gene β-Actin was worked as the target gene for the internal control. Viral load is considered as an important determinant for transmission and infectiousness of a viral disease from one to other. Here, Ct values were considered as indicator for assessing viral load, which is inversely proportional to the amount of nucleic acid present in a sample.[20,21] Lower Ct values indicate high amounts of targeted nucleic acid, while higher Ct values mean lower (and even too little) amounts of the target nucleic acid. Ct values of all positive samples were noted and categorized as high, medium, and low viral loaded infection as per the NIOH working definition for assessment.
Ct values of the positive samples available from RT-PCR were used for quantifying viral load present in the sample. As an explorative initial approach the Ct values were equally divided into three parts, hence for the purpose of working definition whole range Ct values (within positivity range) were categorized as “high viral load,” “moderate viral load” and “low viral load.” Ct values 17 to <24, 24 to <31, and 31 to 38 were, respectively, categorized as high, moderate, and low viral load.
Secondary case detection
The individuals tested COVID-19 positive, among the received swab samples were contacted via phone. The approximate date of onset of symptom/sample collection (for asymptomatic) and their geographical proximity with the other COVID-19-positive cases were enquired. Secondary cases were identified based on the date of swab collection (among asymptomatic)/development of symptoms (among symptomatic) and the proximity to the nearest COVID-19-positive patient. An individual having tested positive for COVID-19 within the longest incubation period of contacting the COVID-19-positive patient (index case) was regarded as secondary case for the current study. The study was approved both by the Scientific Advisory Committee as well as Institute Ethics Committee prior to initiation of this study.
RESULTS
One thousand nine-hundred and seventy-six naso and oropharyngeal swab samples collected from residents of talukas within and adjacent to Mahisagar district, Gujarat state were processed and of them, 138 samples were detected to be COVID-19 positive with an overall positivity rate of ~7%. Figure 1 shows its week-wise distribution of tested persons along with positive cases. It is evident that there is an increasing trend of COVID-19 infection over the weeks at the studied districts as shown in Figure 2 (dotted line), which is similar to many other places in India.
Figure 1.
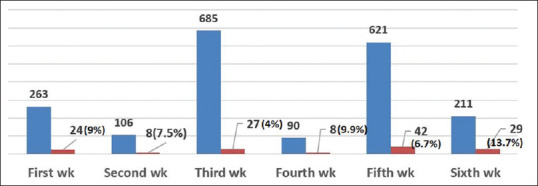
Weekly distribution of COVID-19 samples tested (blue) and positive cases (red)
Figure 2.
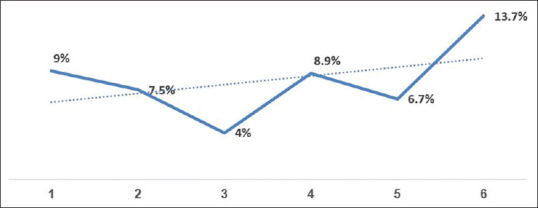
Weekly trend of COVID-19 infection at the selected district
Our study identified 10 (7%) individuals with high viral as per the working definition of this manuscript (Ct values between 17 and <24), while 9% (n = 12) had moderate viral load and remainder 84% (n = 116) had low viral load based on Ct values.
Weekly distribution of infected cases with their viral loads were analyzed, which also showed most infected people were with low viral load and very few were with high and moderate viral loads in the studied weeks. Secondary cases of an index case that develops within the longest incubation period from the occurrence of index case is considered as a valid measure of infectiousness or transmissibility of an infectious disease. It was observed that clustering with several cases occurred around the cases with high viral load in the community whereas very few cases were with that of low viral load. Attempt was made to estimate the secondary infection out of each infected (index) case with high as well as low viral load. We found 8 cases with high viral load who are index cases and there were 50 secondary cases. Similarly, 9 secondary cases were detected out of 11 index cases with low viral load as shown in Table 1. Table 1 shows that each high viral load case transmits 6.25 cases whereas each low load case transmits only 0.8 person. Hence, it may be said that transmission is almost 8 times higher in case of high load case compared to low viral load case.
Table 1.
Secondary cases infected from primary / index case with high viral load
| High viral load |
Low viral load |
|||
|---|---|---|---|---|
| Primary/Index (n=8) | Secondary cases (n=50) | PrimaryIndex (n=11) | Secondary cases (n=9) | |
| Secondary cases per each primary case | 6.25 | 0.8 | ||
DISCUSSION
Analyzing SARS transmission and other infectious disease outbreaks, epidemiologists have found marked heterogeneity in transmission of infectious disease agents where a small fraction of infected patients, “the super spreaders” were observed to contribute to most of the disease transmission events. Rule 20/80, a concept developed through observational and modeling studies have profound implications for infection control which states that 20% of the individuals within any given population are thought to contribute at least 80% of the transmission potential of a pathogen.[22] Predicting and identifying super-spreaders offer significant advantages for infectious disease management and pandemic preparedness plan. Super-spreading events were documented in many infectious diseases such as typhoid, tuberculosis, measles, Ebola hemorrhagic fever, HIV and SARs.[15,23,24,25,26,27,28]
The observation made in this study primarily focuses on what makes an infected case as super spreaders for COVID-19 transmission in the community and a possible marker towards that. Viral load of an infected person at an initial stage (at the time of sample collection) reflects how well a virus is replicating in an infected person which might be an important determining factor for disease transmission in the community apart from population density and its mobility. It has been reported earlier that with reduction in HIV viral load among HIV-infected patients, there are reduced risk of perinatal[29] and sexual transmission of the virus.[30] Another study on healthy volunteers at the National Institutes of Health Clinical Center, USA in 2015 showed that the higher the doses of influenza virus given to healthy volunteers, the worse were their symptoms.[31]
Earlier, researchers from the Méditerranée Infection University Hospital Institute in Marseille, France, also have documented advantages of using Ct values (available from RT-PCR assays) of COVID-19 positive samples in decision-making regarding infectiousness of the cases.[20] Correlation between successful isolation of virus in cell culture and Ct value of quantitative RT-PCR targeting E gene suggested that COVID-19 patients with Ct above 33–34 (using RT-PCR system) are not contagious and thus can be discharged from hospital care or strict confinement for nonhospitalized patients. It was also revealed that patients with Ct values equal or above 34 do not excrete infectious viral particles.
In a hospital-based study with 76 patients of COVID-19 (RT-PCR confirmed), 61% of individuals were classified as mild cases and 39% were classified as severe cases.[32] The mean viral load of severe cases was around 60 times higher than that of mild cases. It was also observed that the Ct values of severe cases remained significantly lower for the first 12 days after onset than those of corresponding mild cases. Mild cases were found to have an early viral clearance, with 90% of these patients repeatedly testing negative on RT-PCR by day 10 post onset. By contrast, all severe cases still tested positive at or beyond day 10 postonset. Overall, the data indicated that patients with severe COVID-19 tend to have a high viral load and a long virus-shedding period. This finding suggests that the viral load of SARS-CoV-2 might be a useful marker for assessing disease severity and prognosis.
In another hospital-based study in China, where the levels of COVID-19 viral load were indicated by the Ct values of RT-PCR assay, it was observed that the Ct values of RT-PCR assays negatively correlated with the probability of progression to severe type of disease in all the patients representing mild-to-moderate severity at admission.[21] It was also observed that the viral load of the sputum specimen in the lower respiratory tract tested at baseline is closely related to the severity of COVID-19. More importantly, patients with a higher baseline viral load were more likely to become severely ill. Findings of these studies support the view of this present study of categorization of viral load using Ct value RT-PCR.
In the light of the current results, a small proportion, i.e., 7% had high viral load, while a larger proportion, i.e., 84% had low viral load. The analysis of secondary cases revealed that virus transmission from the cases with high viral load was almost 8 times higher than the probability of acquiring the infection from cases with low viral load (transmissibility of infection 6.25% vs. 0.8% only). This indicates COVID-19-infected cases with high viral load have the potential of transmitting about 8 times higher number of cases compared to cases with low viral load, which may be viewed as higher or super spreaders in the community. Some other studies also had similar observations (few people spread most as super spreaders and most people spread few) in diseases such as SARS and MERS.[12,16,17] Since transmission of viral infection is related with viral load as observed in other studies,[20,21,29,30,31,33] so the present study could support the view of cases with high viral load are the higher or super-spreader.
Based on the current results, it can be assumed with reasonable certainty that Ct value is a reliable indicator of infectiousness which should be routinely used to assess infection transmitting potential of an individual case and secondly, an infected case with low viral load indicated by high Ct values above 31 can be considered as mildly infectious and further higher Ct value is noninfectious. Similarly, cases with higher viral load indicated by lower Ct values are more infectious. The risk of getting infected with COVID-19 is often higher among health-care workers, probably due to the fact that they spend more time with high viral cases and increase the risk of being exposed to COVID-19-positive cases. If Ct values of the positive patients are routinely communicated to the attending health-care workers, additional care, and attention may be exercised to reduce the transmission from such high viral load individuals and lower the secondary infection among the health-care workers. Future longitudinal multi-centered studies with larger sample size and strict contact tracing of various viral load individuals is required to validate the study observations.
CONCLUSION
Findings of the current study have potentially far reaching implications in strategic decision in the community management of COVID-19 cases and reducing the transmission. This study has shown that 84% of the cases have low viral load and practically will transmit infection to very few of their contacts. If rest 16% could be identified using Ct value and isolate them from the infection control point of view, COVID-19 burden would be potentially reduced subsequently in India. This will reduce concomitant number of hospitalization and deaths, with efficient utilization of hospital services. More clinical, laboratory and mathematical modeling are needed for further understanding towards this.
Financial support and sponsorship
Nil.
Declaration of the Authors
Dr. Kamalesh Sarkar has 32 years' experience and expertise in control of all sorts of locally prevalent communicable and non-communicable diseases. The view and opinions expressed in this article are entirely those of authors.”
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Curtis N, Sparrow A, Ghebreyesus TA, Netea MG. Considering BCG vaccination to reduce the impact of COVID-19. Lancet. 2020;395:1545–6. doi: 10.1016/S0140-6736(20)31025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goswami RP, Mittal DK, Goswami RP. Interaction between malarial transmission and BCG vaccination with COVID-19 incidence in the world map: A changing landscape human immune system? [Last accessed on 2020 Jul 23];Med Rxiv 2020. 2020 doi: https://doi.org/10.1101/2020.04.03.20052563 . [Google Scholar]
- 3.Napoli PE, Nioi M. Global Spread of Coronavirus Disease 2019 and Malaria: An Epidemiological Paradox in the Early Stage of A Pandemic. J Clin Med. 2020;16(9):1138. doi: 10.3390/jcm9041138. doi: 10.3390/jcm9041138. PMID: 32316118; PMCID: PMC7230338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert G, Konstantin C. [Last accessed on 2020 Jul 23];Polio vaccine could beat coronavirus: Dr Robert Gallo, HIV co-discoverer. Financ Express 2020; at shorturl.at/chjlI. [Google Scholar]
- 5.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak-An update on the status. Mil Med Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heesterbeek JA, Dietz K. The concept of Ro in epidemic theory. Stat Neerl. 1996;50:89–110. [Google Scholar]
- 7.Stuart B, Martin C, Supamit C, Vladimir D, Didier H, Youngmee J, et al. Statement on the meeting of the international health regulations (2005) emergency committee regarding the outbreak of novel coronavirus (2019-nCoV) Geneva: World Health Organization (WHO); 2020. [Google Scholar]
- 8.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;13(27):taaa021. doi: 10.1093/jtm/taaa021. doi: 10.1093/jtm/taaa021. PMID: 32052846; PMCID: PMC7074654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauch CT, Lloyd-Smith JO, Coffee MP, Galvani AP. Dynamically modeling SARS and other newly emerging respiratory illnesses: Past, present, and future. Epidemiology. 2005;16:791–801. doi: 10.1097/01.ede.0000181633.80269.4c. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention (CDC). Severe acute respiratory syndrome-Singapore, 2003. MMWR Morb Mortal Wkly Rep. 2003;52:405–11. [PubMed] [Google Scholar]
- 11.Dye C, Gay N. Epidemiology. Modeling the SARS epidemic. Science. 2003;300:1884–5. doi: 10.1126/science.1086925. [DOI] [PubMed] [Google Scholar]
- 12.Kucharski AJ, Althaus CL. The role of superspreading in Middle East respiratory syndrome coronavirus (MERS-CoV) transmission. Euro Surveill. 2015;20:14–8. doi: 10.2807/1560-7917.es2015.20.25.21167. [DOI] [PubMed] [Google Scholar]
- 13.Lipsitch M, Cohen T, Cooper B, Robins JM, Ma S, James L, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300:1966–70. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley S, Fraser C, Donnelly CA, Ghani AC, Abu-Raddad LJ, Hedley AJ, et al. Transmission dynamics of the etiological agent of SARS in Hong Kong: Impact of public health interventions. Science. 2003;300:1961–6. doi: 10.1126/science.1086478. [DOI] [PubMed] [Google Scholar]
- 15.Shen Z, Ning F, Zhou W, He X, Lin C, Chin DP, et al. Superspreading SARS events, Beijing, 2003. Emerg Infect Dis. 2004;10:256–60. doi: 10.3201/eid1002.030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Yu IT, Xu P, Lee JH, Wong TW, Ooi PL, et al. Predicting super spreading events during the 2003 severe acute respiratory syndrome epidemics in Hong Kong and Singapore. Am J Epidemiol. 2004;160:719–28. doi: 10.1093/aje/kwh273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–9. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centre for the Mathematical Modelling of Infectious Diseases COVID-19 Working Group. Endo A, Abbott S, Kucharski AJ, Funk S. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 2020;5:67. doi: 10.12688/wellcomeopenres.15842.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatterjee P, Nagi N, Agarwal A, Das B, Banerjee S, Sarkar S, et al. The 2019 novel coronavirus disease (COVID-19) pandemic: A review of the current evidence. Indian J Med Res. 2020;151:147–59. doi: 10.4103/ijmr.IJMR_519_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–61. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu X, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care. 2020;24:170. doi: 10.1186/s13054-020-02893-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein RA. Super-spreaders in infectious diseases. Int J Infect Dis. 2011;15:e510–3. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks J. The sad and tragic life of Typhoid Mary. CMAJ. 1996;154:915–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen MS, Hoffman IF, Royce RA, Kazembe P, Dyer JR, Daly CC, et al. Reduction of concentration of HIV-1 in semen after treatment of urethritis: Implications for prevention of sexual transmission of HIV-1.AIDSCAP Malawi Research Group. Lancet. 1997;349:1868–73. doi: 10.1016/s0140-6736(97)02190-9. [DOI] [PubMed] [Google Scholar]
- 25.Khan AS, Tshioko FK, Heymann DL, Le Guenno B, Nabeth P, Kerstiëns B, et al. The reemergence of ebola hemorrhagic fever, democratic republic of the congo, 1995.commission de lutte contre les Epidémies à Kikwit. J Infect Dis. 1999;179(Suppl 1):S76–86. doi: 10.1086/514306. [DOI] [PubMed] [Google Scholar]
- 26.Kline SE, Hedemark LL, Davies SF. Outbreak of tuberculosis among regular patrons of a neighborhood bar. N Engl J Med. 1995;333:222–7. doi: 10.1056/NEJM199507273330404. [DOI] [PubMed] [Google Scholar]
- 27.Moss GB, Overbaugh J, Welch M, Reilly M, Bwayo J, Plummer FA, et al. Human immunodeficiency virus DNA in urethral secretions in men: Association with gonococcal urethritis and CD4 cell depletion. J Infect Dis. 1995;172:1469–74. doi: 10.1093/infdis/172.6.1469. [DOI] [PubMed] [Google Scholar]
- 28.Paunio M, Peltola H, Valle M, Davidkin I, Virtanen M, Heinonen OP. Explosive school-based measles outbreak: Intense exposure may have resulted in high risk, even among revaccinees. Am J Epidemiol. 1998;148:1103–10. doi: 10.1093/oxfordjournals.aje.a009588. [DOI] [PubMed] [Google Scholar]
- 29.Ioannidis JP, Abrams EJ, Ammann A, Bulterys M, Goedert JJ, Gray L, et al. Perinatal transmission of human immunodeficiency virus type 1 by pregnant women with RNA virus loads <1000 copies/ml. J Infect Dis. 2001;183:539–45. doi: 10.1086/318530. [DOI] [PubMed] [Google Scholar]
- 30.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Memoli MJ, Czajkowski L, Reed S, Athota R, Bristol T, Proudfoot K, et al. Validation of the wild-type influenza a human challenge model H1N1pdMIST: An a (H1N1) pdm09 dose-finding investigational new drug study. Clin Infect Dis. 2015;60:693–702. doi: 10.1093/cid/ciu924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Yan LM, Wan L, Xiang TX, Le A, Liu JM, et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect Dis. 2020;20:656–7. doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argyropoulos KV, Serrano A, Hu J, Black M, Feng X, Shen G, et al. Association of Initial Viral Load in Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Patients with Outcome and Symptoms. Am J Pathol. 2020:S0002-9440(20)30328-X. doi: 10.1016/j.ajpath.2020.07.001. doi: 10.1016/j.ajpath.2020.07.001. Epub ahead of print. PMID: 32628931; PMCID: PMC7332909. [DOI] [PMC free article] [PubMed] [Google Scholar]


