Abstract
Scleroproteins are an important category of proteins within the human body that adopt filamentous, elongated conformations in contrast with typical globular proteins. These include keratin, collagen, and elastin, which often serve a common mechanical function in structural support of cells and tissues. Genetic mutations alter these proteins, disrupting their functions and causing diseases. Computational characterization of these mutations has proven to be extremely valuable in identifying the intricate structure-function relationships of scleroproteins from the molecular scale up, especially if combined with multiscale experimental analysis and the synthesis of model proteins to test specific structure-function relationships. In this work, we review numerous critical diseases that are related to keratin, collagen, and elastin, and through several case studies, we propose ways of extensively utilizing multiscale modeling, from atomistic to coarse-grained molecular dynamics simulations, to uncover the molecular origins for some of these diseases and to aid in the development of novel cures and therapies. As case studies, we examine the effects of the genetic disease Epidermolytic Hyperkeratosis (EHK) on the structure and aggregation of keratins 1 and 10; we propose models to understand the diseases of Osteogenesis Imperfecta (OI) and Alport syndrome (AS) that affect the mechanical and aggregation properties of collagen; and we develop atomistic molecular dynamics and elastic network models of elastin to determine the role of mutations in diseases such as Cutis Laxa and Supravalvular Aortic Stenosis on elastin’s structure and molecular conformational motions and implications for assembly.
Keywords: Keratin, Collagen, Elastin, Molecular Dynamics, Human Disease Phenotypes, Mechanics, Mechanobiology, Mutation, Defect, Failure, Elasticity
1. Introduction
Keratin, collagen, and elastin belong to an important category of proteins within the human body, known as fibrous proteins or scleroproteins, which adopt filamentous, elongated conformations in contrast with typical globular proteins [1, 2]. They typically consist of tandemly repeating units of amino acid sequences, such as the heptad repeats in keratins [3], the G-X-Y motifs in collagen [4], and the V-P-G-V-G motifs in elastin [5]. These fibers provide structural support to cells and tissues due to their unique mechanical properties conferred by the repeating motifs and the resultant secondary structures, as well as their tendency to aggregate and form functional filaments [2]. Examples of these filaments include the helical coiled-coil bundles of keratins, the triple-helical fibers of collagen I, and the elastin microfibrils formed through coacervation and crosslinking. Given the remarkable importance of these proteins as structural elements, it is of no surprise that many disease phenotypes arise as a result of mutations in these proteins that lead to misaggregation that diminishes or completely disrupts their ability to form fibrillar structures, thereby dramatically reducing their mechanical functions. Due to the molecular nature of these disruptions, experimental characterization of diseases related to fibrous proteins has been significantly assisted by the proliferation of computational studies that are able to probe the intricate structure-function relationships of these scleroproteins on the molecular scale. Here, we review the molecular origins of numerous critical diseases that are related to keratin, collagen, and elastin, through several case studies. We propose ways of extensively utilizing multiscale modeling, from atomistic to coarse-grained molecular dynamics simulations, to uncover the underling molecular mechanisms for some of these diseases, helping to pave the way towards novel cures and therapies.
1.1. Keratin-related Diseases
The human skin, a complex multilayered structure encompassing almost the entirety of the human body, provides crucial biological functions, such as regulating the egress of body fluids, preventing the entry of foreign substances, and maintaining the body’s temperature [6, 7]. To perform those functions, cells known as keratinocytes undergo cellular differentiation and outward migration, leading to the formation of the final layer of the skin, known as the stratum corneum (SC) [8, 9], which consists of a dead flat layer of corneocytes. In healthy human skin, the primary structural components in the SC corneocytes are keratins [9], which are long strands of proteins with a high molecular weight, accounting for the major portion of the total protein content in the epidermis [8]. Importantly, many inherited skin disorders have been found to explicitly link with mutations in keratin molecular structures [10], and have been listed in the Human Intermediate Filament database [9].
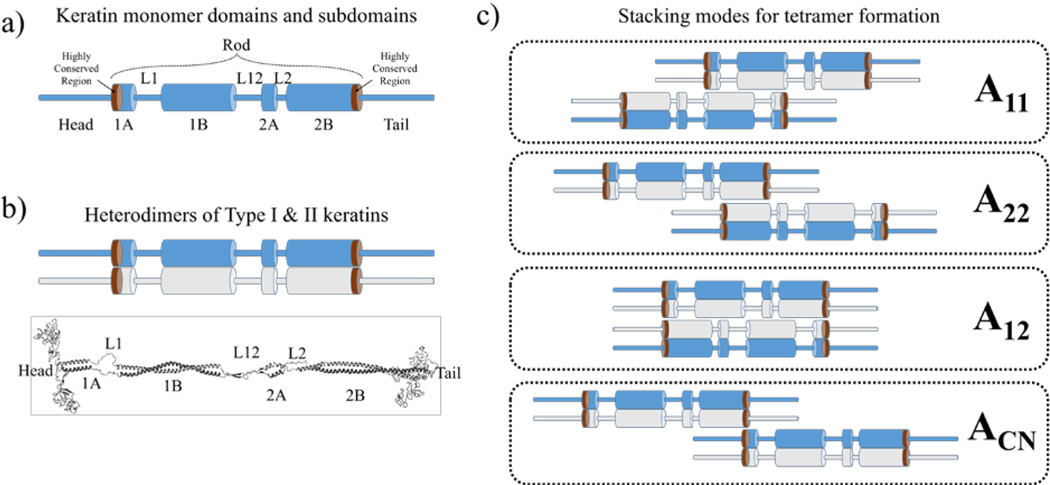
Keratins in the human skin are rich in glutamic acid, serine, leucine, and glycine [9, 11]. The primary structure of keratins consists of chains of these amino acids with high sequence identity although the exact number and sequence of amino acids vary slightly. The amino acid sequences also display a periodicity in the arrangement of the residues, consisting of heptad repeats of (a-b-c-d-e-f-g)n such that residues a and d are typically hydrophobic and e and g are charged [3]. The precise amino acid sequence exerts immense influence on the functions, structures, and properties of keratins [12]. The secondary structure of keratins consists of multiple domains and subdomains, as depicted in Figure 1a. Each keratin monomer consists of head, rod, and tail domains. The head and tail domains are largely non-helical and globular structures with some content of β-turns [13, 14]. The rod domain is further divided into subdomains of right-handed α-helical segments (1A, 1B, 2A, and 2B) connected by strands of non-helical linkers (L1, L12, and L2).
Figure 1.
(a) Schematics of a keratin monomer secondary structure at the molecular scale. (b) Parallel coiled-coil heterodimers of keratin Types I (blue) and II (grey) stylized from the proposed tertiary structure of heterodimeric coiled-coil keratins (bottom). (c) Different stacking modes of keratin tetramers during the formation of keratin filaments. Thin cylinders denote random coils, thick cylinders denote α-helices, and brown regions denote the highly-conserved regions of keratins.
The tertiary structures of keratin filaments consist of Type I and II keratins assembled as left-handed coiled-coil heterodimers [15] as shown in Figure 1b, where the hydrophobic residues a and d from each keratin monomer are wrapped up within the coil, forming an amphipathic structure with hydrophilic residues on the surface. Homodimers of either type are highly unstable, and thus degrade swiftly, while the stability of heterodimers helps maintain a balanced ratio between Types I and II [10]. This is due to Type I keratins being acidic, while Type II keratins are neutral to basic. Finally, unit length filaments (ULFs) are formed through bundling the heterodimers through four different modes of antiparallel stacking [16, 17] as illustrated in Figure 1c. These stacking modes were identified after crosslinks were induced between human K1 and K10 epidermal keratin chains and the linked residues were isolated through trypsin digestion [16]. The precise reason that ULFs have these stacking modes are still unknown but it is speculated that these can lead to spatially efficient packing into a lattice structure [18] through electrostatic interactions between charged amino acids on their surfaces [11].
Epidermolysis Bullosa Simplex (EBS), Epidermolytic Palmoplantar Keratoderma (EPPK), and Epidermolytic Hyperkeratosis (EHK) are examples of inherited skin disorders that are explicitly linked to keratin mutations [15]. EBS and EHK display remarkable similarities in that the most frequent mutations occur in the highly-conserved regions in segments 1A and 2B (also known as mutation ‘hot-spots’) as highlighted in Figure 1a. For EBS, mutations occur in both basal keratin 5 and 14 (K5 and K14), while EHK is a result of mutations in both suprabasal keratin 1 and 10 (K1 and K10), which are the predominant keratins found in human SC [11]. As there are 28 Type I and 26 Type II intermediate filament proteins, nomenclature of these keratins have been standardized according to the three categories of (1) epithelial keratins/genes, (2) hair keratins/genes, and (3) keratin pseudogenes, taking into account historical precedence [9, 19]. Moreover, one of the most frequently observed mutations in EHK is the substitution of the positively-charged arginine residue in position 156 of K10 by six other residues [9]. Three of these are polar residues: cysteine, histidine, and serine; while the other three are hydrophobic residues: proline, leucine, and glycine. For ease of reference, the substitutions are simplified as three-letter protein codes, namely p.ARG156CYS, p.ARG156HIS, p.ARG156SER, p.ARG156PRO, p.ARG156LEU, and p.ARG156GLY respectively. Characteristics of EHK include skin redness coupled with broad thickening of the topmost layer of the skin and severe blistering [17]. These mutations also result in keratins failing to assemble into filament structures, leading to malformed clumps of keratins [12, 16].
The exact molecular mechanisms behind the inability of the keratins to assemble after mutations have not been fully understood. Through experimental observations, Steinart et al. [16] proposed a surface lattice model of K1 and K10 packing and suggested that there likely exists a recurring pattern of ten to eleven residues overlapping between the highly conserved 1A and 2B segments, implying that any mutations in these regions may result in significant interference with the structural integrity of keratin filaments. Similarly, through differential interference contrast microscopy and rheological assays, Ma et al. [20] characterized keratin 14 (K14) mutations involved in EBS and found that mutations significantly decreased the keratins’ ability to bundle and weakened the network’s mechanical properties. Furthermore, structural characteristics of various mutated keratins have been studied by computer simulations [21, 22]. For instance, molecular dynamics (MD) modelling by Smith et al. [22] on twenty-two mutations in segment 1A of keratin 5 (K5) and K14 responsible for EBS showed that structural distortions due to these mutations correlate with clinical severity, and the distortions vary according to different mutations.
1.2. Collagen-related Diseases
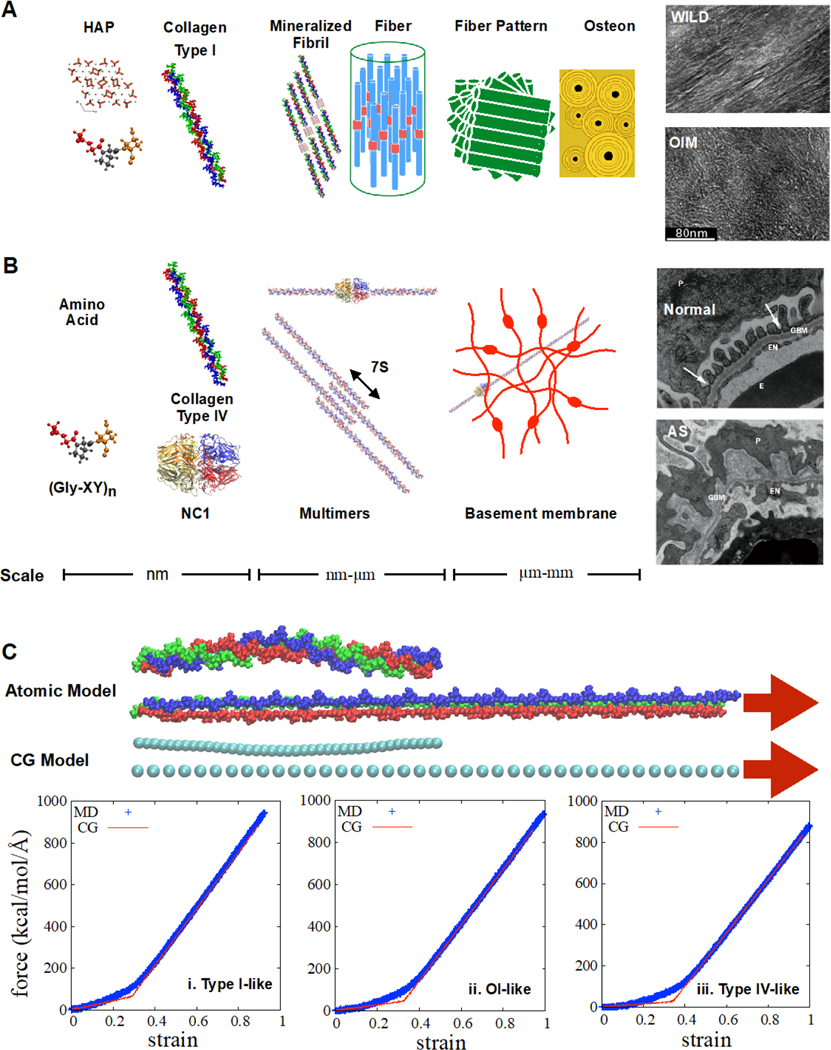
Collagen is one of the most abundant proteins in human body that confers mechanical stability, elasticity, and integrity to bio-organisms in combination with other important materials such as hydroxyapatite (HAP) minerals and elastin [23–25]. The basic building block of collagen molecules is a Gly-X-Y amino acid triplet. Repeated triplets form long chain of polypeptides, and three strands of polypeptides assemble into a triple helical tropocollagen molecule. This tropocollagen molecule can be assembled further into two main types of collagen assemblies as shown in Figure 2a and b [26]: fibrillar (Type I-like, I, II, III, V, and XI) and non-fibrillar networks (Type IV-like). Type I collagen molecules form ordered fibers where the axial length is 67 nm and this is known as the D period [27]. The unit cell consists of five molecular segments with an overlap region (0.46D ~ 31 nm) and a gap region (0.54D ~ 36 nm), forming a right-handed twist with the neighboring fibrils [28]. The collagen I fiber is a main structural component of the muscular-skeletal components such as tendons, ligaments, and bone with the other building blocks of HAP mineral nano-plates (2–5nm thick, 15–55 nm long and 5–25nm wide) [25, 29, 30]. On the other hand, type IV collagen is non-fibrillar and mostly found in the basal lamina, forming a structurally complex membrane basement with laminins which has filtration properties in capillaries and glomeruli [31].
Figure 2.
a) Hierarchical structures of bone and TEM images of mineralized collagens in wild type and OIM mice adapted from [32] licensed under CC BY 3.0 from Elsevier. b) Hierarchical structure of glomeruli in kidney and TEM images of 9-week-old normal glomeruli and 9-week-old Alport syndrome glomeruli, adapted by permission from Macmillan Publishers Ltd: Nature Reviews Nephrology [33], copyright (2013). P: Podocyte, EN: Endothelium GBM: glomerular basement membrane. c) Atomistic model and bilinear spring CG model of (GPO)13 normal (i) and mutated (ii and iii) collagen molecules with strain rates of 10 m/s and time steps of 1 fs. The difference between panels ii and iii is the location of the glycine deletion. The mutation is located at the center of all 3 chains in (ii) and is spatially separated in (iii) to mimic the similar conditions of OI and normal collagen IV. The force-strain behavior is well captured with a bilinear spring model.
A single point mutation of amino acids in these collagens can cause serious genetic diseases. Considering their portion in the total collagen molecules, it is intriguing that such small changes can have significant influence on the function and stability of the organs that contain collagen. Osteogenesis imperfecta (OI), also known as brittle bone syndrome, is a result of mutations in collagen I and it is characterized by significant bone fragility. Due to the sophisticated hierarchical structure of bone, it is still elusive how such a small alteration can significantly affect the entire system. Several abnormalities such as differences in mineralization, collagen crosslinking, fiber diameter, and mineral crystallinity are reported [32, 34, 35]. There is no known complete cure for this disease but some medicines such as bisphosphonate or treatment with vibration can mitigate the symptoms [36, 37].
Missense mutations in collagen I, such as replacing glycine with another amino acid that has a bulkier side chain, can cause serious malfunctions in the mechanical properties of bone due to tissue disorders [38]. In contrast, missing glycines in the Gly-X-Y triplet is considered to be vital for non-fibrillar collagen IV and these are termed natural interruptions [39]. There are more than 20 natural interruptions in the entire collagen IV sequence [40] where six different chains of α1 to α6 have been identified in the collagen IV family. These chains are composed of three main domains: a short 7s domain at the N-terminal, a long collagenous domain, and a non-collagenous (NC1) domain at the C-terminal, as shown in Figure 2b [41]. Alport syndrome (AS) is a genetic disorder associated with mutations in collagen type IV. Most patients with Alport syndrome have X-linked inheritance of mutation in the gene responsible for encoding the collagen IV α5 chain, which results in thinning or detaching of the glomerular basement membrane (GBM), as shown in Figure 2b. The main feature of X-linked AS is the reduction or total loss of α3α4α5 triple helices, while some mutations with less severe AS still allow for the formation of α3α4α5 [42].
Those mutations may result in incorrect structural assembly of the α3α4α5 heterotrimer and cause them to degrade easily, although the detailed mechanisms are still not clear. The natural interruptions perturb the fibrillar packing but allow dense networks to form by reducing the fibril’s bending and tensile stiffness. Glycine missense mutations (i.e. mutations related to AS) near these natural interruptions can reduce the stability of triple helices further or weaken the mechanical properties of collagen IV networks [43]. However, these molecular details remain very elusive in terms of experimental characterization, therefore integrative atomistic and coarse-grained modeling can be performed together with detailed experiments to shed light on the mechanics of the tissues and the detailed molecular mechanisms that are caused by diseases.
1.3. Elastin-related Diseases
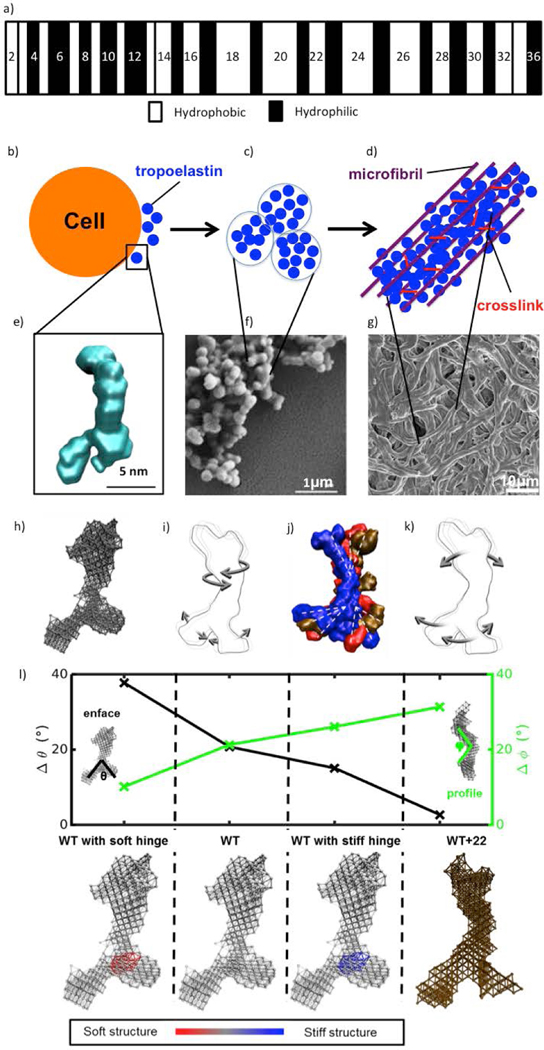
Elastin is an extracellular matrix protein that forms an essential component of elastic fibers which confer reversible deformability, recoil and resilience to connective and vascular tissue, including the skin, arteries, heart and the lungs [44, 45]. Native elastin is a durable constitutive material that is insoluble and extensively cross-linked [44], and thus difficult to isolate. Tropoelastin is the soluble, monomeric building block of the elastin protein. It is encoded by a single gene (ELN) with 34 exons that may generate multiple isoforms depending on alternative splicing patterns [46]. A mature molecular structure has the molecular weight of approximately 60 kDa [46]. Tropoelastin is composed of alternating hydrophilic domains rich in lysine and alanine residues and hydrophobic domains, rich in valine, proline and glycine residues, often occurring in repeats of VPGVG, in single amino-acid code (Figure 3a).
Figure 3.
(a) Structure of human tropoelastin with alternating hydrophobic and hydrophilic domains, adapted from [47], Copyright (2005), with permission from Elsevier. (b) Tropoelastin is secreted from the cell, (c) self-assembles into a coacervate form, whereupon it is (d) crosslinked and deposited onto a microfibrillar scaffold, forming a growing elastic fiber. (e) Tropoelastin structure based on small-angle X-ray scattering, adapted from [48] with permission from the National Academy of Science, USA. (f) Scanning electron micrograph of coacervates of tropoelastin, adapted from [49], Copyright (2011), with permission from Elsevier. (g) Scanning electron micrograph of elastic fiber purified from human skin biopsies, adapted from [50] with permission from John Wiley and Sons. (h) Elastic network model of a SAXS-based structure of tropoelastin. (i) Wild type (WT) tropoelastin global dynamics, depicting the twist of the N-terminal scissors-like flexion of the hinge and foot regions. (j) Representative solution shapes of wild type tropoelastin showing conformations consistent with the range of molecular motions defined by the elastic network model. (k) Domain motion of WT+22 tropoelastin, characterized by the N-terminal bend along the perpendicular axis of the body and the parallel motion between the hinge region and the foot region. (l) Change in representative angles for four models: WT, mutant tropoelastin (WT+22), WT with a stiffened hinge region representative of alpha helix–to–beta sheet transition (WT with stiff hinge), and WT with a softened hinge region as an alternate control (WT with soft hinge). Corresponding model representations are presented below. The color bar indicates relative stiffness in the structure. (h-l) are adapted from [51] licensed under CC BY-NC 4.0 from AAAS.
It is a dynamic, highly disordered molecule, containing a high content of labile β-turns, poly-proline II helix (PPII) and disordered structure, as well as stable α-helices principally in polyalanine cross-linking domains [52, 53]. Tropoelastin interacts with cell-surface receptors, assisting cellular functions, and stimulates cell responses, including chemotaxis, proliferation and cell adhesion [54]. Of particular importance for these functions is domain 36, the final capping domain of the molecule that contains an important cell-interactive RKRK sequence [55, 56].
Elastogenesis is the elastic fiber assembly process by which tropoelastin is deposited onto a microfibrilar scaffold, forming the growing elastic fiber. Microfibrils consist of fibrillin, microfibril associated glycoproteins (MAGPs), fibulins, and EMILIN-1. Microfibrils and their components, which include fibrillin, microfibril associated glycoproteins (MAGPs), fibulins, and EMILIN-1, assist in lysyl oxidase (LOX) cross-linking and alignment of tropoelastin clusters onto the growing fiber [57]. First, tropoelastin is secreted from elastogenic cells (Figure 3b), such as smooth muscle cells and fibroblasts, coalescing on the cell surface, where it begins to be cross-linked by the enzyme lysyl oxidase, a process facilitated by fibulins. In parallel to the cross-linking process, the monomers aggregate together through coacervation (Figure 3c, f). In vitro coacervation has been observed, showing a sequential assembly of growing spherical clusters of tropoelastin [49, 58]. The growing aggregates of tropoelastin are eventually transferred to microfibrils in the extracellular environment, bound to the cell surface through integrins. The elastin aggregates extend on the microfibril scaffold where further cross-linking takes place (Figure 3d, g).
Until recently, the molecular structure of tropoelastin has remained a mystery due to its large size and disordered nature, and insoluble, highly cross-linked state in vivo. After the elucidation of the human tropoelastin-encoding gene, recombinant full-length tropoelastin expression was realized, opening the path to probe the complete molecular structure [48, 59]. Recent small-angle X-ray (SAXS) and neutron scattering (SANS) studies captured the distinguishable global shape of tropoelastin [48] (Figure 3e). It was found that tropoelastin is an asymmetric molecule, with an extended, coil-like, N-terminal region, supported by a hinge that props a bridge linking the N-terminal to the cell-interactive C-terminal contained in the exposed foot region [48].
A number of diseases, including cutis laxa, emphysema, and supravalvular aortic stenosis are associated with mutations within or improper function of elastic fibers [60]. Cutis laxa includes a collection of disorders characterized by loose, wrinkled skin, affecting both the appearance and the protective function of tissues in the face, hands, feet, joints and torso [61, 62]. Autosomal dominant congenital cutis laxa (ADCL), in particular, is associated with mutations in the elastin (ELN) gene. It is genetically heterogeneous and shows clinical variability. In addition to loose, inelastic skin, ADCL may include gastrointestinal diverticula, hernia, and genital prolapsem [62]. Rare manifestations are pulmonary artery stenosis, aortic aneurysm, bronchiectasis, and emphysema [61, 63]. The majority of ADCL mutations are due to frame-shift mutations in the ELN gene at the 3’-end, resulting in an extended missense peptide sequence at the carboxy-terminal end of secreted tropoelastin [64]. Mutant tropoelastin is deficient in fibrillin binding and has enhanced self-association properties [65]. After mutant tropoelastin is secreted it is incorporated into elastin fibers, forming elastic structures with reduced stiffness and increased compliance [66], and yielding observed disease phenotypes. A major challenge for understanding the mechanisms of operation of cutis laxa and other ELN-related diseases is to determine how the known mutations affect the structure of the tropoelastin molecule, thereby influencing exposed binding domains, mechanical properties and assembly.
2. Simulation Methodologies
2.1. Atomistic Modeling
Molecular dynamics (MD) simulations are immensely powerful and useful tools for determining numerical solutions for classical many-body problems that commonly lack analytical solutions. In atomistic MD simulations, atoms are assumed to be point masses that interact with each other according to classical Newtonian equations of motion [67, 68], where Euler equations, Hamilton’s quaternions, and Lagrange’s method of geometric constraints, can be incorporated depending on the complexity of the problem at hand [69]. Classical MD deals with the atoms or molecules based on their interactions where the electronic structures of atoms are not changed significantly, e.g., excluding bond breaking and forming. Based on this classical description, the Hamiltonian, H, is the sum of the potential energy, U, and kinetic energy, K, of a system that has N number of particles. The K and U are usually functions of coordinates qi and momenta pi of each atom i, where the variables in bold refer to vectors. Under given volume and interatomic potential U(qi), the equations of motion are solved by numerical integration such as velocityVerlet or Gear method. Then, the total Hamiltonian, H = K(pi)+U(qi), is naturally conserved with a proper time step during the time integration (NVE ensemble). Since temperature and pressure are common macroscopic variables in experiments, the NVT (canonical) and NPT (isobaric-isothermal) ensembles, where the equations of motion are generally derived from the extended Hamiltonian systems including thermostat and barostat, are frequently adopted in MD simulations.
To describe the properties of a model system correctly, the potential, U, plays a key role in defining their behaviors and therefore, a broad range of MD potentials have been developed to characterize numerous types of interatomic interactions, which are able to describe biological, chemical, mechanical, and physical properties of many materials. Through a comparatively simple mathematical formulation, one of the most fundamental of these potential energy functions is the Lennard-Jones (LJ) potential which could accurately model the properties of several low-density gases, such as argon [70, 71]. Moreover, a more significant usage of the LJ potential is to couple it to other interatomic potentials to implement dispersive interactions. This allows a much broader range of materials to be modeled [72–75] by approximating the interactions between chemically dissimilar atoms with the Lorentz-Berthelot mixing rules [67]. Even though these mathematical formulations have proven to be suitable for a broad range of applications, simulations of biological phenomena typically demand greater complexity, especially when modeling chemical species that have complex bonding structures, such as those in carbon and silicon compounds. Highly intricate potential energy functions have been derived, optimized, and validated against Density Functional Theory (DFT) and experimental results, including the Adaptive Intermolecular Reactive Empirical Bond Order (AIREBO) potential [72], the Tersoff potential [76], and the large family of CHARMM [77] and AMBER [78] potentials to name just a few. Here, for instance, the Amber03 force field [79] was used to parameterize the keratins, as implemented in the GROMACS 4.6.5 software package [80]. This force field has been widely used to simulate peptides, proteins, and helical coiled-coils such as collagen [81–85]. The force field is comprised of five separate energy terms, namely, the bond stretching, angle bending, torsional, van der Waals (vdW), and electrostatic interactions. Explicitly,
where Kr and Kθ are the bond and angle stiffnesses; r and θ the bond length and bond angle; req and θeq the equilibrium bond length and bond angle; Vn the dihedral stiffness; φ the dihedral angle; γ the phase angle; Rij the distance between two non-bonded atoms; Aij and Bij the constants for non-bonded interactions; q the charge of the atoms; ε the dielectric constant.
2.2. Coarse-grained Modeling
Even though highly detailed characteristics of atoms can be determined from MD simulations, significant computational costs have to be incurred for systems that extend beyond millions of atoms because of their intrinsically short time step, typically around 1 or 2 fs. This might be infeasible even with current computational hardware, especially for biological materials, as adequate sampling of configurational space for large systems becomes prohibitive. An alternative to a brute force approach is to employ a simplification of interatomic details through coarse-graining. Coarse-grained (CG) models are efficient and computationally economical techniques for investigating the response, property, and/or behavior of interest in systems with spatiotemporal scales that are not accessible to numerical methods that capture phenomena on a much smaller scale, such as MD simulations. Moreover, hierarchical “handshaking” can be established between the various scales, leading to progressively multiscale transitions to determine each system’s nano-, meso-, and macro-scale structure-property-function relationships. By adopting this “finer-trains-coarser” approach, the ability to characterize and understand biological phenomena from the bottom-up is greatly fortified through the incorporation of a comprehensive theoretical foundation to predict and characterize multiple properties in these biological systems.
One of the more widely used CG models is a physics-based approach where experimental data or quantum-mechanical calculations are used to parameterize CG forcefields. Due to this similarity with parameterizing fully atomistic MD forcefields, physics-based CG forcefields can usually be assimilated into existing MD software conveniently, thus ensuring excellent portability for wider usage. Amongst these numerous CG methods, the MARTINI forcefield [86, 87] is widely used for modeling biological systems and materials. From its initial parameterization to model lipids [88], the MARTINI methodology has been standardized subsequently for the systematic reproduction of the free energies of hydration, vaporization, and partitioning of polar and apolar phases for a wide variety of chemical compounds such as hexadecane, chloroform, ether, and octanol [86]. The MARTINI forcefield clusters atoms into four main types of polar (P), nonpolar (N), apolar (C), and charged (Q) CG beads where four heavy atoms and their corresponding hydrogen atoms are modelled as one CG bead. Even though this is a comparatively basic representation of molecules, this model has been parameterized and validated for a broad range of biomaterials, such as polymers and block co-polymers such as PEG and PEO [89], polysaccharides [90], proteins [91, 92], and DNA [93]. A major downside is that only tertiary structures of proteins, DNA, and RNA can be determined as the secondary structures have to be constrained via an elastic network, therefore a MARTINI model cannot capture conformational changes in the secondary structure.
However, structural details in MARTINI models can be reinforced through the construction of appropriate elastic network models (ENMs) [51]. ENMs are most useful in capturing global domain motions of loops and subdomains, as these phenomena occur at longer time scales and larger length scales inaccessible to MD. Such approaches have been instrumental in identifying protein functions in ion gating and self-assembly (further references can be found in [94]). ENMs simplify a structure by modeling it as a network of masses interacting through a simple harmonic potential, creating a coupled bead-spring system [94]. Nodes correspond to equidistant points on a grid fit to atomistic or small-angle X-ray scattering derived geometries. Molecular normal mode analysis is based on the assumption that a molecule will oscillate about an equilibrium configuration as a result of thermally induced fluctuations at the atomic scale. Mathematically, the molecule is modeled as a system containing N interacting sites with coordinates qi. The assumption in normal mode analysis maintains that the starting structure is an equilibrium configuration, such that are coordinates in the equilibrium configuration. The potential energy near equilibrium is approximated as:
The terms are components of the Hessian matrix. Considering the molecule as a system of classically interacting particles, the equation of motion is:
where M is the mass matrix, q are the coordinate components and H is the Hessian matrix. The equation of motion can be written as:
assuming a solution of the form
This is a generalized eigenvalue problem, where the solution set, uk, with 1 ≤ k ≤ 3N, and squared frequencies, ω2, represent the eigenvectors and eigenvalues, respectively. The eigenvectors of the mass-weighted Hessian matrix form an orthonormal basis set defining the normal modes of the molecule. The energy of a mode is directly proportional to the eigenvalue, or the squared frequency of motion, λk = ω2k. From normal mode analysis, low-frequency modes are most accessible and least energetically expensive.
Dissipative particle dynamics (DPD) is an alternative physics-based method of coarse-graining [95, 96] where a combination of a dissipative force, a random force, and a conservative linear repulsive force are exerted on the CG beads. The corresponding chemical identity of each CG bead is enforced by the conservative force which mimics excluded-volume interactions while the system’s temperature is maintained by a thermostat comprising of the dissipative and random forces. Furthermore, bonds between the CG beads are usually soft Hookean springs or the finitely extensible nonlinear elastic (FENE) model [97]. DPD simulations have been broadly used to model the properties of proteins [98–100], polymers [101, 102], complex fluids [103], and biomembranes [104]. For instance, ultraviolet inks for 3D inkjet printing in nanoscale additive manufacturing were modelled with DPD [102] to demonstrate that sodium dodecyl sulphate is an effective surfactant for reducing the average size of polyethylene glycol and polystyrene agglomerates, thereby enhancing their uniformity. Another study employed DPD to identify experimental parameters that are critical for the processing of bioinspired silk fibres [99]. To design hierarchical materials in a predictive manner, the co-polymer’s ideal lengths were determined and the ratios of the hydrophobic and hydrophilic domains in the co-polymer are optimized. It was verified experimentally that fibers with excellent mechanical properties were formed. This model was extended further to determine how fibrillation was influenced by terminal modification of the peptides [100] and their results demonstrated that decreasing the hydrophobicity in combination with increasing the molecular weight at the terminals augmented the alignment of the peptide chains under shear. Despite a reduction in extensibility and toughness, the mechanical strength and elasticity were enhanced.
3. Computational Characterization of Disease Phenotypes
3.1. Keratin
Fully atomistic molecular dynamics (FAMD) and coarse-grain molecular dynamics (CGMD) simulation methods have successfully used to reveal the dynamics of different types of proteins such as HIV-1 protease at different timescales [105, 106]. These methods can also be applied to determine the molecular mechanisms behind poor assembly of mutated K1 and K10. In this case study, we perform both FAMD and CGMD simulations on the six ARG mutations in segment 1A of K10. Through FAMD simulations, we characterize the evolution and equilibration of the secondary and tertiary structures of these mutated segments. In order to simulate the binding of the overlapping regions of keratin segments, which is on a time-scale beyond the reach of FAMD simulations, cluster analyses [107] are performed on the trajectories of the FAMD simulations and the most probable equilibrated structures are coarse-grained. Subsequently, the binding energies between segments 1A and 2B of each mutation are analyzed by using CGMD. As mentioned in Section 1.1, mutations related to EHK result in poor binding of keratins, leading to the inability to assemble into filament structures. The present case study provides fundamental understanding of the effects of different mutations in highly conserved regions of keratins on the structural characteristics and aggregation dynamics, which may lead to potential therapeutic treatments for EHK as this disorder currently has no known cure beyond amelioration of its symptoms. Details of the FAMD and CGMD models and simulation procedures are provided in Section 1 of the Supporting Information.
3.1.1. Atomistic modeling
Hydrophobic substitutions of the ARG156 residue in segment 1A have marked effects on the structural stability of the entire segment. Table 1 collates the structural data averaged over the last 10 ns of the equilibrations in comparison with the wild-type segments.
Table 1.
RMSD after equilibration and number of α-helical residues averaged over the last 10 ns of the equilibration run together with their corresponding standard deviations. Further comparisons with the wild-type keratin were made by calculating the RMSD after backbone alignments with the mutated keratin structures from cluster analyses of the equilibration trajectories, which is given in the second column. Final two columns are the average total interaction energy and the corresponding percentage decrease after mutations from CGMD simulations. These are also averaged over the last 10ns of the simulations.
| RMSD after Cluster Analysis and Alignment (nm) | Number of α-Helical Residues | Average Total Interaction Energy (kJ/mol) | Percentage Decrease in Total Interaction Energy (%) | |
|---|---|---|---|---|
| Wild-Type | ----- | 61 ± 1 | −5,427 | ----- |
| p.ARG156PRO | 0.36 | 56 ± 3 | −4,873 | 10.2 |
| p.ARG156LEU | 0.23 | 58 ± 2 | −4,874 | 10.2 |
| p.ARG156GLY | 0.16 | 58 ± 2 | −4,227 | 22.1 |
| p.ARG156CYS | 0.08 | 60 ± 2 | −5,397 | 0.6 |
| p.ARG156HIS | 0.07 | 60 ± 2 | −5,086 | 6.3 |
| p.ARG156SER | 0.06 | 61 ± 2 | −4,255 | 21.6 |
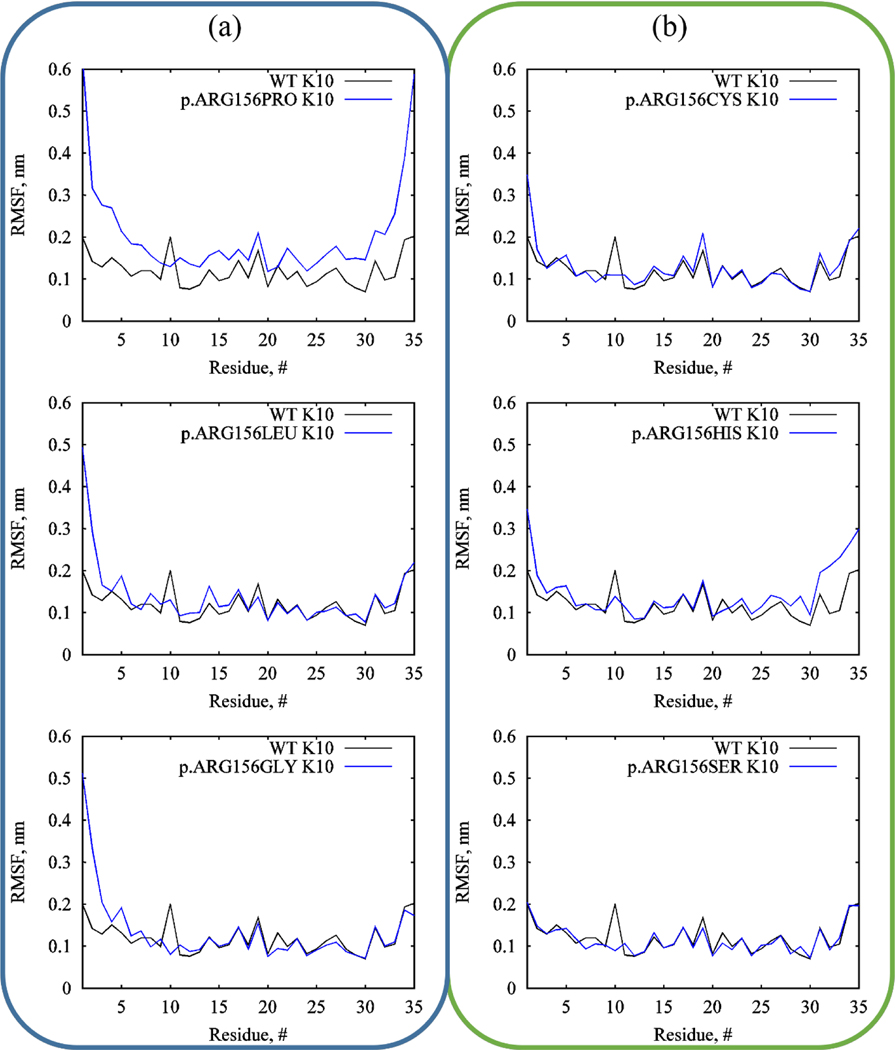
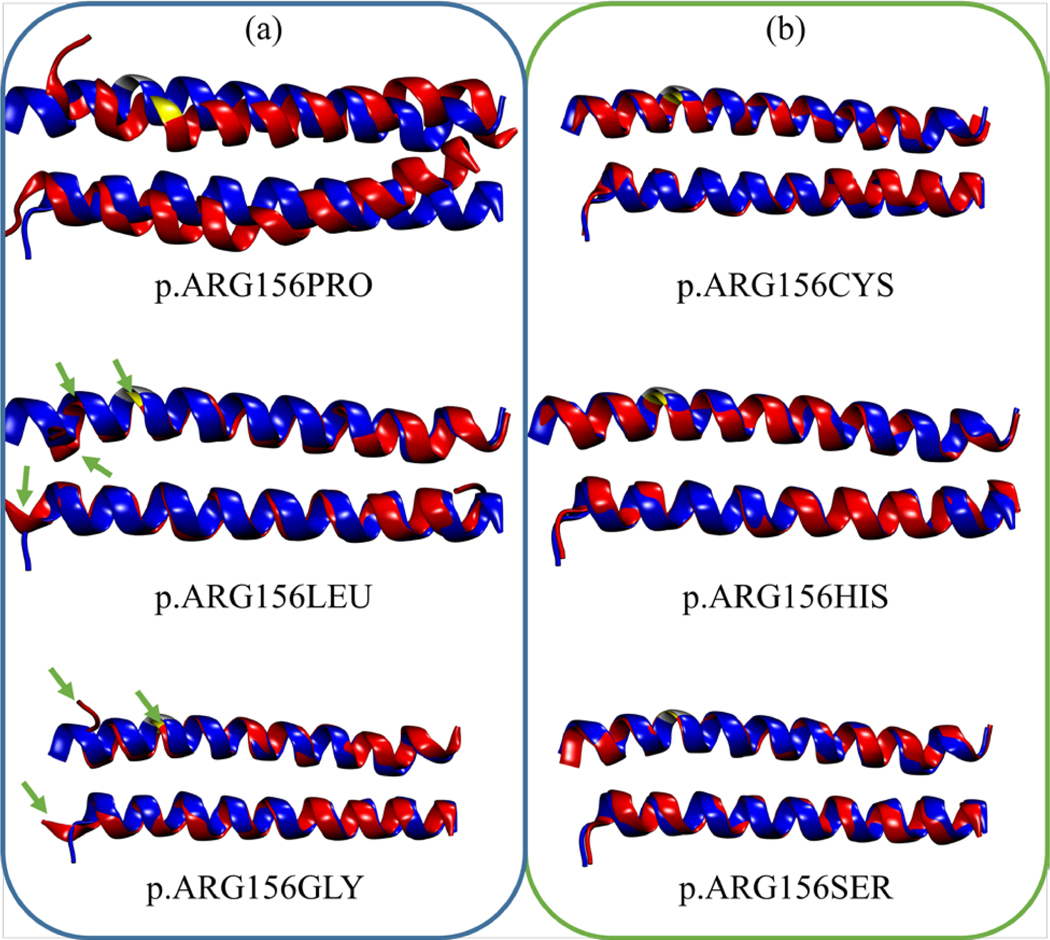
As mentioned in Section 1, protein coiled-coil formation is due to the burying of hydrophobic residues between the coils away from water solvents. For K1/K10 segment 1A, the ARG156 residue directly faces into the water solvent, as illustrated in Figure S1a. In contrast, hydrophobic substitutions immediately result in the destabilization of the coiled-coil structure as the keratins attempt to re-fold their tertiary structure. Evidence for this is provided in Figure 4, where the average root-mean-square fluctuation (RMSF) of the backbone atoms in each residue of K10 is plotted. There are 35 residues in segment 1A of K10, and the substituted ARG residue is the tenth residue in K10. From Figure 4a, it can be observed that the hydrophobic residues (PRO, LEU, GLY) induced significant fluctuations in all residues at the nearest ends (residues 1 to 9), whereas these were largely stable for polar substitutions (CYS, HIS, SER) in Figure 4b. These fluctuations are most apparent for residues 1 to 5, where the RMSF for these residues in hydrophobic mutations can be almost double that for wild-type and hydrophilic mutations. PRO substitutions underwent the most dramatic fluctuations, affecting the entire length of K10. This is due to the fact that PRO residues have low propensity to form helices, as well as steric hindrance of the PRO sidechain with the backbone atoms of the preceding turn within the helix [108, 109]. As shown in Table 1, substitutions with hydrophobic residues also resulted in more decreased α-helical content than polar residues. The average number of α-helical residues in wild-type keratins is 61 ± 1. This number dipped for PRO, LEU, and GLY hydrophobic substitutions, whereas it was relatively stable for polar substitutions of CYS, HIS, and SER.
Figure 4.
Evolution of the protein backbone RMSF of each residue in K10 over 20 ns for (a) hydrophobic and (b) polar mutations in comparison with the wild-type segment. The substituted ARG residue is the tenth residue in K10.
The combined effects of significant increases in the RMSF with the decreases in the α-helical content due to hydrophobic substitutions can be ascertained visually, as illustrated in Figure 5. Cluster analyses [107] were performed on the trajectories of the equilibration run in GROMACS to determine the conformations with the highest number of transitions, i.e. conformations which have been accessed the most number of times throughout the simulation. Thereafter, the backbone atoms in mutated keratins were aligned with the wild-type keratin and their RMSD were determined. On the one hand, it can be observed that hydrophobic residues tend to produce a localized kink in the region of mutation, and this is especially dramatic for PRO substitutions due to the reasons aforementioned. This leads to much larger RMSD values after alignment with the wild-type, as seen in the first column of Table 1. On the other hand, polar substitutions produced negligible kinking, largely retaining similar conformations with the wild-type, resulting in very small RMSD values after alignment. Clearly, local structural distortions could be a significant factor behind the in vitro observation of misaggregation of mutated keratin bundles. To deepen our understanding of the origin and driving force behind this misaggregation, we performed CGMD on much longer time-scales to determine the interaction energies during the aggregation process.
Figure 5.
Alignment of the backbone atoms in mutated K1/K10 segment 1A (red cartoons) after cluster analyses of the equilibration trajectory for (a) hydrophobic and (b) polar mutations with the wild-type segment (blue cartoons). For greater clarity in the structural changes for the p.ARG156LEU and p.ARG156GLY mutations, green arrows denote regions that are highly perturbed after the hydrophobic mutations and equilibration.
3.1.2. Coarse-grained Modeling
The vdW, Coulombic, and total interaction energies between segments 1A and 2B were determined over 200 ns and these data were averaged over four different simulations for each mutation together with the application of the Savitzky-Golay smoothing filter, as shown in Figure S2. There were no significant variations in the short-range vdW interaction energies for both hydrophobic and polar mutations. However, substitutions with hydrophobic residues led to extensive reductions in inter-segment long-range Coulombic interactions, while substitutions with polar residues only resulted in slight reductions in the Coulombic interactions. As shown in Table 1 and Figure S2, for hydrophobic mutations, this resulted in decreases of more than 10% in the average total interaction energy between segments 1A and 2B. The underlying reasons for the differences are that hydrophobic substitutions of the positively-charged arginine residue greatly reduced long-range electrostatic interactions in the highly polar region at the end of segment 2B; while the varying degrees of polarity in cysteine, histidine and serine substitutions ensured that the weakening was lessened to a great extent, though the effects were still apparent, especially for serine mutations due to their relatively small sidechains.
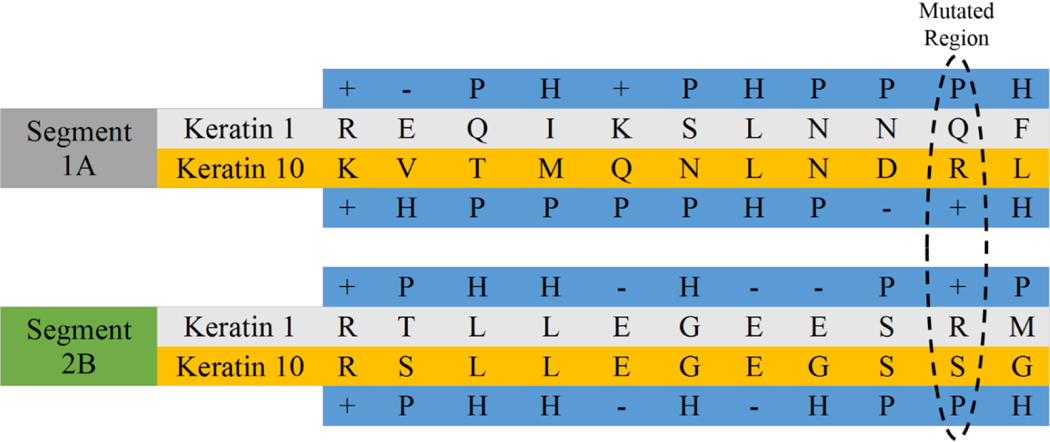
To analyze this issue, the amino acid sequences for segments 1A and 2B are listed in Figure 6. It is shown that hydrophobic mutations of ARG156 strongly perturb the mutated region dominated by the polar interactions, leading to the reduction in the long-range electrostatic interactions. By summing up both of these interactions, we see that the total interaction energies in Figure S2 point to the fact that a single mutation in the ‘hot-spot’ is able to cause significant losses of intersegment cohesiveness along the overlapping regions, thus potentially leading to filament misaggregation observed in experiments [12, 16]. This is especially true for substitutions with hydrophobic residues.
Figure 6.
Standard single-letter amino acid sequences for the first eleven residues of segment 1A and last eleven residues of segment 2B of wild-type K1/K10. Blue highlights describe whether the amino acids are positively-charged (+), negatively-charged (−), polar (P), or hydrophobic (H). Stippled line shows the amino acids overlapping with the mutated ARG156 in K10.
3.1.3. Discussion and Implications
From the Human Intermediate Filament Database [9], the vast majority of mutations in K10 can be found in segment 1A, especially in the ARG156 hotspot. Moreover, the most frequent substitutions are of HIS, followed by CYS, with the other mutations being rare. The origins of these keratin mutations have a genetic basis, where mutations of a single base to different bases in various positions of the KRT10 gene result in different expressions of keratin 10 segment 1A [110]. The remarkably high frequency of HIS and CYS substitutions observed in literature [9] correlates well with our findings that these substitutions result in binding energies which are largely similar to that of wild-type keratins, implying that these substitutions are more energetically favorable and thus increase the relative probability of such mutations. Furthermore, there is a broad spectrum of phenotypic expressions of EHK, where even the same residue substitution can result in a different level of phenotypic severity and different expression [110–113], leading to significant variation and tailoring of therapies and disease management for each individual. This phenotype spectrum also implies that keratins have multiple functions, some of which are not yet well characterized and understood. Hence, deepening the understanding of phenotypic traits and keratin functions could be made possible through untangling the complex interactions involved.
It is known that changes in keratin’s secondary and tertiary structures are strongly related to the side chain properties (size, acidic, basic, neutral) of mutated residues and how they interact with other biomolecules in their close environment, which are the key drivers behind the aggregation and clumping caused by mutations [110]. In the present study, by isolating localized atomic interactions, we identified the molecular traits associated with different residues and conformations. Our results can be useful in identifying different classes of phenotypic expressions observed in experiments, enabling better prediction of disease symptoms, especially since these symptoms vary with individuals and change with age and different therapies. For instance, our models predict that the lower binding affinity due to hydrophobic mutations presents a greater probability of severe disease phenotypes. Otherwise, it is likely that another confounding mechanism dominates, such as the expression of other keratin types like keratin 2 or 9 which may reinforce the functions of defective K1/K10. Further numerical studies may be needed to identify such confounding mechanisms in order to correlate with the phenotypes determined experimentally.
Current treatments of this type of genetic diseases are confined to management through daily skin care, or oral ingestion and skin application of various retinoids. However, there are promising future prospects of more comprehensive treatments through gene regulation by topical delivery of siRNA-based spherical nucleic acid nanoparticles [114]. While this is still under intensive research, the existing symptomatic relief of these diseases could be further strengthened by targeting the specific keratin mutations presented in our study. The most notable success in this form of therapy is the treatment of specific mutations in cystic fibrosis [115, 116]. High-throughput screening of small molecules has successfully identified molecules which can correct or improve the functions of both deletion and point substitution mutations in the cystic fibrosis transmembrane conductance regulator protein even when this protein is misfolded as a result of these mutations. Our model is a crucial step towards identifying similar drugs which can restore the binding affinity of segments 1A and 2B, leading to corrections and/or improvements in the structural and mechanical properties in the keratin filaments.
3.2. Collagen
Fully atomistic and CG MD simulations are utilized to understand the mechanical properties of type I collagen molecule and fibrils. Details of the FAMD and CGMD models and simulation procedures are provided in Section 2 of the Supporting Information. Like other biological materials, the deformation and stress-strain behaviors of collagen molecules are strongly affected by their hydrogen bond networks during the deformation [117]. The stress-strain curve of a single collagen (GPO)13 homotrimer shows hyper-elastic behavior, as shown in Figure 2c. The initial low Young’s modulus is a result of rotation and unwinding of the triple helix, while the subsequent high modulus is due to stiffening as the backbone stretches. The transition from low modulus to high modulus depends on the loading rate and hydration [118, 119]. The Young’s modulus under a strain of 0.1 converged to around 4 GPa when the tensile loading rate is lower than 1 m/s. When the collagen I molecule is dehydrated, it becomes stiffer and less tough due to reduced energy dissipation by the water environment. Atomistic simulations of fibril-level deformation and mechanical properties are prohibitive due to the extremely large system sizes. Therefore, CG models are developed to understand fibril level deformation and mechanics [120–122]. Unlike other CG methods such as the MARTINI forcefield [92], previous CG models for fibrils are developed for mechanical properties without considering thermodynamic properties such as free energy. These CG models capture the nonlinear stress–strain behaviors by adopting bilinear spring constants derived from atomistic MD simulation (examples are shown in Figure 2c). A group of amino acids is represented by a bead and each bead is connected to others with a bilinear spring. The overall behavior of a single molecule is described very well with this simple model. The spring constant includes hydration and loading rate effects from the atomistic MD conditions. One may include the loading rate effects into the CG models by considering the instant relative velocities between the beads. In the CG modeling of collagen assembly, the interchain friction is usually oversimplified because the Lenard-Jones (LJ) type potential is usually adapted to determine the non-bonded interactions and it cannot capture the friction arising from variations in geometries, which might play an important role in toughening and impact tolerance.
Mutations related to the OI disease have been studied with various atomistic MD simulations. Previous studies have discussed the effects of mutation-induced structural changes, including kinks, locally unfolded helix, and disturbed hydrogen bonds at the atomic scale [123–125]. However, the assembly process of nano-fibrils is still elusive, hence a reliable CG forcefield tuned for thermodynamics properties is required to model the self-assembly of collagen. On the other hand, the CG models for mechanical properties of the mutated collagen molecules can be developed with in same manner of bilinear spring models to determine the mechanical behaviors under given geometries. For example, the force-strain behaviors by deleting 6th glycine in the center of collagen molecule of (GPO)13 show reduction in the stiffness, and the bilinear spring model can still capture the overall mechanical behavior, as shown in Figure 2c(ii).
Srinivasan et al. have studied the molecular structures and stress-strain relationship in collagen IV due to mutations related to AS [126], although the selected sequences do not include natural interruptions. Results from peptide models in experiments [127, 128] show that the natural interruptions can decrease the stability of triple helical structure but the changes strongly depend on the specific sequences, indicating that the natural interruptions are not randomly distributed but nature may have designed them through evolution. Also, the mutations of X-linked AS are dispersed in α5 chain without distinct hot spots [40], which is consistent with distributions of natural interruptions, and 75% of Gly missense mutations related AS are located within a distance of 20 residues away from the natural interruptions [43]. Figure 2c(iii) shows the force-strain behavior of (GPO)13 with distributed natural interruptions. The three chains of the collagen molecule have different locations of Gly missing with 9 amino acids in spatial separation, showing softened behavior compared to the other two samples. The bilinear spring CG model also can capture the behaviors properly. Although the detailed geometries of the networks still need to be addressed, CG and atomistic MD can shed light on the effects of AS mutations or natural interruptions in collagen IV on the mechanical properties of the basement membrane.
3.3. Elastin
Mutations within the elastin gene (ELN) have been shown to result in a series of diseases, including cutis laxa, supervalvular aortic stenosis and emphezyma [60]. The phenotypes share a common thread as they ultimately stem from the disruption in the molecular structure and consequent effects on mechanical function and elastic fiber assembly, starting at the single molecule level. Therefore, it is crucial to understand the structure-function relationship within the mutated elastin molecule to identify the mechanisms of these diseases and thereafter to design treatment strategies targeted to the molecular scale. Though traditionally thought to be a fully disordered protein, tropoelastin has a defined shape in solution [51, 129], and conserved sequence elements across mammalian species suggest that specific regions of the molecule are tied to distinct structural functionality [130–132]. Elastic fiber formation begins with tropoelastin association, followed by molecular deposition and cross-linking [133]. Linking molecular scale changes and implications for higher-order assembly may be critical for understanding disease etiology.
To consider single-molecule kinetics and implications for assembly of tropoelastin, elastin network models were developed, yielding descriptions of concerted molecular motions (Figure 3h). These collective motions are intrinsically accessible to the molecule and are governed by the representative native fold [51]. The wild type (WT) elastic network model displays a scissors-like motion in the lower region of the molecule, and a twist in the upper region of the molecule (Figure 3i). A lock-and-key mechanism is identified, suggesting a predisposition to a head-to-tail, chain-like assembly pattern of tropoelastin into higher order structures [51]. The scissor-like motion involves the hinge region, producing a high stress concentration in the region that is effectively strengthened through clamping imposed during cross-linking. The dynamics results provide a good qualitative match to SAXS snapshots (Figure 3j). This data supports and redefines the role played by the C-terminal and the hinge region of tropoelastin in cellular and intra-molecular binding [51, 129]. On the other hand, the mutant WT+22, which introduces a normally spliced out sequence corresponding to exon 22, displays significantly different dynamics from WT (Figure 3k). The hinge and foot regions of the WT+22 molecule move in parallel, while its coil region bends along a perpendicular axis, which may shield intermolecular regions of contact and reduce the likelihood of head-to-tail assembly [51]. The WT+22 mutant also displays an increase in beta sheet secondary structure in the hinge region compared to the WT [51], suggesting changes in local stiffness within the molecular structure. It is found that local stiffness in fact plays an important role in the molecule’s global dynamic behavior (Figure 3l). We propose that similar analysis may be performed on disease-associated tropoelastin isoforms to uncover disease etiology at the molecular scale.
Elastin (ELN) based diseases have not been well-characterized through computational approaches to date. Based on recent progress in computational modeling of tropoelastin, modeling presents a useful avenue to understand the structural underpinnings of elastin (ELN) based diseases. Full-atomistic molecular models of tropoelastin and related mutations will be key for determining molecular mechanisms of these diseases in the future.
4. Conclusions
Through a series of reviews and case studies, we examined the role of numerous critical diseases that are disruptive to the normal functions of scleroproteins, including keratin, collagen, and elastin. We also analyzed the importance of multiscale modeling, from atomistic to coarse-grained molecular dynamics simulations in complementing experimental studies to unearth the molecular effects of genetic diseases. In particular, our case studies focus on Epidermolytic Hyperkeratosis (EHK), and the structure and aggregation of keratins 1 and 10; the diseases of Osteogenesis Imperfecta (OI) and Alport syndrome (AS) that affect the mechanical and aggregation properties of collagen; and the role of mutations on elastin’s structure and molecular conformational motions. These diseases are currently debilitating yet incurable, therefore we are hopeful that integrative computational and experimental approaches may help to develop novel cures and therapies.
Supplementary Material
Acknowledgments
The authors, J. Yeo, G.S. Jung, A. Tarakanova, F.J. Martín-Martínez, Z. Qin, and M.J. Buehler, acknowledge support from the US Department of Defense, Office of Naval Research (N00014-16-1-233) and the National Institutes of Health (U01 EB014976). Part of this work was supported by the A*STAR Computational Resource Centre through the use of its high-performance computing facilities.
References
- [1].Ferraro V, Anton M, Santé-Lhoutellier V. The “sisters” α-helices of collagen, elastin and keratin recovered from animal by-products: Functionality, bioactivity and trends of application. Trends in Food Science & Technology. 2016;51(Supplement C):65–75. [Google Scholar]
- [2].Parry DAD, Squire JM. Fibrous Proteins: New Structural and Functional Aspects Revealed Advances in Protein Chemistry: Academic Press; 2005, p. 1–10. [DOI] [PubMed] [Google Scholar]
- [3].Mason JM, Arndt KM. Coiled Coil Domains: Stability, Specificity, and Biological Implications. ChemBioChem. 2004;5(2):170–6. [DOI] [PubMed] [Google Scholar]
- [4].Khoshnoodi J, Pedchenko V, Hudson B. Mammalian Collagen IV. Microscopy research and technique. 2008;71(5):357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Muiznieks LD, Weiss AS, Keeley FW. Structural disorder and dynamics of elastin. Biochemistry and Cell Biology. 2010;88(2):239–50. [DOI] [PubMed] [Google Scholar]
- [6].Madison KC. Barrier Function of the Skin: “La Raison d’Etre” of the Epidermis. J Investig Dermatol. 2003;121(2):231–41. [DOI] [PubMed] [Google Scholar]
- [7].Proksch E, Brandner JM, Jensen J-M. The skin: an indispensable barrier. Experimental Dermatology. 2008;17(12):1063–72. [DOI] [PubMed] [Google Scholar]
- [8].Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6(4):328–40. [DOI] [PubMed] [Google Scholar]
- [9].Szeverenyi I, Cassidy AJ, Chung CW, Lee BTK, Common JEA, Ogg SC, et al. The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Human Mutation. 2008;29(3):351–60. [DOI] [PubMed] [Google Scholar]
- [10].Lane EB, McLean WHI. Keratins and skin disorders. Journal of Pathology. 2004;204(4):355–66. [DOI] [PubMed] [Google Scholar]
- [11].Bragulla HH, Homberger DG. Structure and functions of keratin proteins in simple, stratified, keratinized and cornified epithelia. Journal of Anatomy. 2009;214(4):516–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fuchs E The cytoskeleton and disease: genetic disorders of intermediate filaments. Annual review of genetics. 1996;30(1):197–231. [DOI] [PubMed] [Google Scholar]
- [13].Rafik ME, Doucet J, Briki F. The intermediate filament architecture as determined by X-ray diffraction modeling of hard α-keratin. Biophys J. 2004;86(6):3893–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Steinert PM, Steven AC, Roop DR. The molecular biology of intermediate filaments. Cell. 1985;42(2):411–9. [DOI] [PubMed] [Google Scholar]
- [15].Crick FHC. Is α-Keratin a Coiled Coil? Nature. 1952;170(4334):882–3. [DOI] [PubMed] [Google Scholar]
- [16].Steinert PM, Marekov LN, Fraser RDB, Parry DAD. Keratin Intermediate Filament Structure: Crosslinking Studies Yield Quantitative Information on Molecular Dimensions and Mechanism of Assembly. J Mol Biol. 1993;230(2):436–52. [DOI] [PubMed] [Google Scholar]
- [17].Chamcheu JC, Siddiqui IA, Syed DN, Adhami VM, Liovic M, Mukhtar H. Keratin gene mutations in disorders of human skin and its appendages. Archives of biochemistry and biophysics. 2011;508(2):123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fraser RDB, Parry DAD. The three-dimensional structure of trichocyte (hard α-) keratin intermediate filaments: Features of the molecular packing deduced from the sites of induced crosslinks. Journal of Structural Biology. 2005;151(2):171–81. [DOI] [PubMed] [Google Scholar]
- [19].Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, et al. New consensus nomenclature for mammalian keratins. The Journal of Cell Biology. 2006;174(2):169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ma L, Yamada S, Wirtz D, Coulombe PA. A ‘hot-spot’ mutation alters the mechanical properties of keratin filament networks. Nat Cell Biol. 2001;3(5):503–6. [DOI] [PubMed] [Google Scholar]
- [21].Danciulescu C, Nick B, Wortmann F-J. Structural Stability of Wild Type and Mutated α-Keratin Fragments: Molecular Dynamics and Free Energy Calculations. Biomacromolecules. 2004;5(6):2165–75. [DOI] [PubMed] [Google Scholar]
- [22].Smith TA, Steinert PM, Parry DAD. Modeling effects of mutations in coiled-coil structures: Case study using epidermolysis bullosa simplex mutations in segment 1a of k5/k14 intermediate filaments. Proteins: Structure, Function, and Bioinformatics. 2004;55(4):1043–52. [DOI] [PubMed] [Google Scholar]
- [23].Kadler KE, Baldock C, Bella J, Boot-Handford RP. Collagens at a glance. J Cell Sci. 2007;120(12):1955. [DOI] [PubMed] [Google Scholar]
- [24].Kielty CM, Sherratt MJ, Shuttleworth CA. Elastic fibres. J Cell Sci. 2002;115(14):2817–28. [DOI] [PubMed] [Google Scholar]
- [25].Olszta MJ, Cheng X, Jee SS, Kumar R, Kim Y-Y, Kaufman MJ, et al. Bone structure and formation: A new perspective. Materials Science and Engineering: R: Reports. 2007;58(3–5):77–116. [Google Scholar]
- [26].Jung GS, Buehler MJ. Multiscale Modeling of Muscular-Skeletal Systems. Annu Rev Biomed Eng. 2017;19(1):435–57. [DOI] [PubMed] [Google Scholar]
- [27].Petruska JA, Hodge AJ. A SUBUNIT MODEL FOR THE TROPOCOLLAGEN MACROMOLECULE. Proc Natl Acad Sci USA. 1964;51(5):871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Orgel JPRO Irving TC, Miller A Wess TJ. Microfibrillar structure of type I collagen in situ. Proceedings of the National Academy of Sciences. 2006;103(24):9001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nudelman F, Pieterse K, George A, Bomans PHH, Friedrich H, Brylka LJ, et al. The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. 2010:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fratzl P, Fratzl-Zelman N, Klaushofer K, Vogl G, Koller K. Nucleation and growth of mineral crystals in bone studied by small-angle X-ray scattering. Calcified Tissue International. 1991;48(6):407–13. [DOI] [PubMed] [Google Scholar]
- [31].Yurchenco PD, Smirnov S, Mathus T. Analysis of basement membrane self-assembly and cellular interactions with native and recombinant glycoproteins Methods Cell Biol: Academic Press; 2002, p. 111–44. [DOI] [PubMed] [Google Scholar]
- [32].Vanleene M, Porter A, Guillot P-V, Boyde A, Oyen M, Shefelbine S. Ultra-structural defects cause low bone matrix stiffness despite high mineralization in osteogenesis imperfecta mice. Bone. 2012;50(6):1317–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kruegel J, Rubel D, Gross O. Alport syndrome[mdash]insights from basic and clinical research. Nat Rev Nephrol. 2013;9(3):170–8. [DOI] [PubMed] [Google Scholar]
- [34].Vetter U, Weis MA, Mörike M, Eanes ED, Eyre DR. Collagen crosslinks and mineral crystallinity in bone of patients with osteogenesis imperfecta. J Bone Miner Res. 1993;8(2):133–7. [DOI] [PubMed] [Google Scholar]
- [35].Cassella JP, Ali SY. Abnormal collagen and mineral formation in osteogenesis imperfecta. Bone Miner. 1992;17(2):123–8. [DOI] [PubMed] [Google Scholar]
- [36].Harrington J, Sochett E, Howard A. Update on the Evaluation and Treatment of Osteogenesis Imperfecta. Pediatr Clin North Am. 2014;61(6):1243–57. [DOI] [PubMed] [Google Scholar]
- [37].Dwan K, Phillipi CA, Steiner RD, Basel D. Bisphosphonate therapy for osteogenesis imperfecta. Cochrane Database of Systematic Reviews. 2016(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Myllyharju J, Kivirikko KI. Collagens, modifying enzymes and their mutations in humans, flies and worms. Trends Genet. 2004;20(1):33–43. [DOI] [PubMed] [Google Scholar]
- [39].Brodsky B, Thiagarajan G, Madhan B, Kar K. Triple-helical peptides: An approach to collagen conformation, stability, and self-association. Biopolymers. 2008;89(5):345–53. [DOI] [PubMed] [Google Scholar]
- [40].Hudson BG, Reeders ST, Tryggvason K. Type IV collagen: structure, gene organization, and role in human diseases. Molecular basis of Goodpasture and Alport syndromes and diffuse leiomyomatosis. J Biol Chem. 1993;268(35):26033–6. [PubMed] [Google Scholar]
- [41].Hudson BG, Tryggvason K, Sundaramoorthy M, Neilson EG. Alport’s Syndrome, Goodpasture’s Syndrome, and Type IV Collagen. New Engl J Med. 2003;348(25):2543–56. [DOI] [PubMed] [Google Scholar]
- [42].Jais JP, Knebelmann B, Giatras I, Marchi MD, Rizzoni G, Renieri A, et al. X-linked Alport Syndrome: Natural History in 195 Families and Genotype- Phenotype Correlations in Males. J Am Soc Nephrol. 2000;11(4):649–57. [DOI] [PubMed] [Google Scholar]
- [43].Hwang ES, Brodsky B. Folding Delay and Structural Perturbations Caused by Type IV Collagen Natural Interruptions and Nearby Gly Missense Mutations. J Biol Chem. 2012;287(6):4368–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Debelle L, Alix AJP. The structures of elastins and their function. Biochimie. 1999;81(10):981–94. [DOI] [PubMed] [Google Scholar]
- [45].Wise SG, Mithieux SM, Weiss AS. Engineered Tropoelastin and Elastin-Based Biomaterials. Adv Protein Chem Str. 2009;78:1–24. [DOI] [PubMed] [Google Scholar]
- [46].Wise SG, Weiss AS. Tropoelastin. International Journal of Biochemistry & Cell Biology. 2009;41(3):494–7. [DOI] [PubMed] [Google Scholar]
- [47].Wise SG, Mithieux SM, Raftery MJ, Weiss AS. Specificity in the coacervation of tropoelastin: solvent exposed lysines. Journal of Structural Biology. 2005;149(3):273–81. [DOI] [PubMed] [Google Scholar]
- [48].Baldock C, Oberhauser AF, Ma L, Lammie D, Siegler V, Mithieux SM, et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. P Natl Acad Sci USA. 2011;108(11):4322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yeo GC, Keeley FW, Weiss AS. Coacervation of tropoelastin. Advances in Colloid and Interface Science. 2011;167:94–103. [DOI] [PubMed] [Google Scholar]
- [50].Schmelzer CEH, Jung MC, Wohlrab J, Neubert RHH, Heinz A. Does human leukocyte elastase degrade intact skin elastin? Febs J. 2012;279(22):4191–200. [DOI] [PubMed] [Google Scholar]
- [51].Yeo GC, Tarakanova A, Baldock C, Wise SG, Buehler MJ, Weiss AS. Subtle balance of tropoelastin molecular shape and flexibility regulates dynamics and hierarchical assembly. Sci Adv. 2016;2(2):e1501145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tamburro AM, Bochicchio B, Pepe A. Dissection of human tropoelastin: exon-by-exon chemical synthesis and related conformational studies. Biochemistry. 2003;42(45):13347–62. [DOI] [PubMed] [Google Scholar]
- [53].Muiznieks LD, Weiss AS. Flexibility in the solution structure of human tropoelastin. Biochemistry. 2007;46(27):8196–205. [DOI] [PubMed] [Google Scholar]
- [54].Rodgers UR, Weiss AS. Cellular interactions with elastin. Pathol Biol. 2005;53(7):390–8. [DOI] [PubMed] [Google Scholar]
- [55].Bax DV, Rodgers UR, Bilek MMM, Weiss AS. Cell Adhesion to Tropoelastin Is Mediated via the C-terminal GRKRK Motif and Integrin alpha(V)beta(3). Journal of Biological Chemistry. 2009;284(42):28616–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Broekelmann TJ, Kozel BA, Ishibashi H, Werneck CC, Keeley FW, Zhang LJ, et al. Tropoelastin interacts with cell-surface glycosaminoglycans via its COOH-terminal domain. Journal of Biological Chemistry. 2005;280(49):40939–47. [DOI] [PubMed] [Google Scholar]
- [57].Wagenseil JE, Mecham RP. New insights into elastic fiber assembly. Birth Defects Res C Embryo Today. 2007;81(4):229–40. [DOI] [PubMed] [Google Scholar]
- [58].Tu YD, Wise SG, Weiss AS. Stages in tropoelastin coalescence during synthetic elastin hydrogel formation. Micron. 2010;41(3):268–72. [DOI] [PubMed] [Google Scholar]
- [59].Martin SL, Vrhovski B, Weiss AS. Total Synthesis and Expression in Escherichia-Coli of a Gene Encoding Human Tropoelastin. Gene. 1995;154(2):159–66. [DOI] [PubMed] [Google Scholar]
- [60].Vrhovski B, Weiss AS. Biochemistry of tropoelastin. Eur J Biochem. 1998;258(1):1–18. [DOI] [PubMed] [Google Scholar]
- [61].Online Mendelian Inheritance in Man. 2016 [cited 2016 September 2016]; Gene description: 130160. ELASTIN; ELN]. Available from: http://omim.org/entry/130160?search=cutis%20laxa&highlight=laxa%20cuti
- [62].Royce PM, Steinmann B. Connective tissue and its heritable disorders molecular, genetic, and medical aspects. 2nd ed. New York; Chichester: Wiley, 2002, p. 992 p. [Google Scholar]
- [63].Graul-Neumann LM, Hausser I, Essayie M, Rauch A, Kraus C. Highly variable cutis laxa resulting from a dominant splicing mutation of the elastin gene. Am J Med Genet A. 2008;146A(8):977–83. [DOI] [PubMed] [Google Scholar]
- [64].Uitto J, Li Q, Urban Z. The complexity of elastic fibre biogenesis in the skin--a perspective to the clinical heterogeneity of cutis laxa. Exp Dermatol. 2013;22(2):88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Callewaert B, Renard M, Hucthagowder V, Albrecht B, Hausser I, Blair E, et al. New Insights into the Pathogenesis of Autosomal-Dominant Cutis Laxa with Report of Five ELN Mutations. Hum Mutat. 2011;32(4):445–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hu Q, Shifren A, Sens C, Choi J, Szabo Z, Starcher BC, et al. Mechanisms of emphysema in autosomal dominant cutis laxa. Matrix Biol. 2010;29(7):621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Allen MP, Tildesley DJ. Computer simulation of liquids. New York: Oxford University Press; 1989. [Google Scholar]
- [68].Rapaport DC. The art of molecular dynamics simulation. 2nd ed. Cambridge: Cambridge University Press; 2004. [Google Scholar]
- [69].Goldstein H, Poole CP, Safko JL. Classical mechanics, 3rd ed. San Francisco: Addison Wesley; 2002. [Google Scholar]
- [70].Verlet L Computer “Experiments” on Classical Fluids. I. Thermodynamical Properties of Lennard-Jones Molecules. Physical Review. 1967;159(1):98–103. [Google Scholar]
- [71].Hansen J-P, Verlet L. Phase Transitions of the Lennard-Jones System. Physical Review. 1969;184(1):151–61. [Google Scholar]
- [72].Stuart SJ, Tutein AB, Harrison JA. A reactive potential for hydrocarbons with intermolecular interactions. J Chem Phys. 2000;112(14):6472–86. [Google Scholar]
- [73].Qian D, Yu M-F, Ruoff RS, Wagner GJ, Liu WK. Mechanics of carbon nanotubes. Applied Mechanics Reviews. 2002;55(6):495–533. [Google Scholar]
- [74].Becker CA, Hoyt JJ, Buta D, Asta M. Crystal-melt interface stresses: Atomistic simulation calculations for a Lennard-Jones binary alloy, Stillinger-Weber Si, and embedded atom method Ni. Physical Review E. 2007;75(6):061610. [DOI] [PubMed] [Google Scholar]
- [75].Jang I, Phillips R, Sinnott SB. Study of C3H5+ ion deposition on polystyrene and polyethylene surfaces using molecular dynamics simulations. J Appl Phys. 2002;92(6):3363–7. [Google Scholar]
- [76].Tersoff J New empirical approach for the structure and energy of covalent systems. Physical Review B: Condensed Matter and Materials Physics. 1988;37(12):6991–7000. [DOI] [PubMed] [Google Scholar]
- [77].Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, Groot BLd, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nature Methods. 2016;14(1):71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, et al. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117(19):5179–97. [Google Scholar]
- [79].Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. [DOI] [PubMed] [Google Scholar]
- [80].Pronk S, Páll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29(7):845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Matthes D, de Groot BL. Secondary Structure Propensities in Peptide Folding Simulations: A Systematic Comparison of Molecular Mechanics Interaction Schemes. Biophys J. 2009;97(2):599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ribeiro AAST, Ortiz V Energy Propagation and Network Energetic Coupling in Proteins. J Phys Chem B. 2015;119(5):1835–46. [DOI] [PubMed] [Google Scholar]
- [83].Suárez E, Díaz N, Suárez D. Entropic Control of the Relative Stability of Triple-helical Collagen Peptide Models. J Phys Chem B. 2008;112(47):15248–55. [DOI] [PubMed] [Google Scholar]
- [84].Cheng Y, Koh L-D, Wang F, Li D, Ji B, Yeo J, et al. Carbon nanoscroll-silk crystallite hybrid structures with controllable hydration and mechanical properties. Nanoscale. 2017;9(26):9181–9. [DOI] [PubMed] [Google Scholar]
- [85].Yeo J, Cheng Y, Han YT, Zhang Y, Guan G, Zhang Y-W. Adsorption and Conformational Evolution of Alpha-Helical BSA Segments on Graphene: A Molecular Dynamics Study. International Journal of Applied Mechanics. 2016;08(02):1650021. [Google Scholar]
- [86].Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J Phys Chem B. 2007;111(27):7812–24. [DOI] [PubMed] [Google Scholar]
- [87].Marrink SJ, Tieleman DP. Perspective on the Martini model. Chem Soc Rev. 2013;42(16):6801–22. [DOI] [PubMed] [Google Scholar]
- [88].Marrink SJ, De Vries AH, Mark AE. Coarse grained model for semiquantitative lipid simulations. The Journal of Physical Chemistry B. 2004;108(2):750–60. [Google Scholar]
- [89].Lee H, de Vries AH, Marrink S-J, Pastor RW. A coarse-grained model for polyethylene oxide and polyethylene glycol: conformation and hydrodynamics. The journal of physical chemistry B. 2009;113(40):13186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].López CA, Rzepiela AJ, De Vries AH, Dijkhuizen L, Hünenberger PH, Marrink SJ. Martini coarse-grained force field: extension to carbohydrates. Journal of Chemical Theory and Computation. 2009;5(12):3195–210. [DOI] [PubMed] [Google Scholar]
- [91].Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink S-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. Journal of Chemical Theory and Computation. 2008;4(5):819–34. [DOI] [PubMed] [Google Scholar]
- [92].Gautieri A, Russo A, Vesentini S, Redaelli A, Buehler MJ. Coarse-Grained Model of Collagen Molecules Using an Extended MARTINI Force Field. Journal of Chemical Theory and Computation. 2010;6(4):1210–8. [Google Scholar]
- [93].Uusitalo JJ, Ingólfsson HI, Akhshi P, Tieleman DP, Marrink SJ. Martini coarse-grained force field: extension to DNA. Journal of chemical theory and computation. 2015;11(8):3932–45. [DOI] [PubMed] [Google Scholar]
- [94].Bahar I, Lezon TR, Bakan A, Shrivastava IH. Normal Mode Analysis of Biomolecular Structures: Functional Mechanisms of Membrane Proteins. Chem Rev. 2010;110(3):1463–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hoogerbrugge P, Koelman J. Simulating microscopic hydrodynamic phenomena with dissipative particle dynamics. EPL (Europhysics Letters). 1992;19(3):155. [Google Scholar]
- [96].Espanol P, Warren P. Statistical mechanics of dissipative particle dynamics. EPL (Europhysics Letters). 1995;30(4):191. [Google Scholar]
- [97].Kremer K, Grest GS. Dynamics of entangled linear polymer melts: A molecular - dynamics simulation. The Journal of Chemical Physics. 1990;92(8):5057–86. [Google Scholar]
- [98].Peter EK, Lykov K, Pivkin IV. A polarizable coarse-grained protein model for dissipative particle dynamics. Physical Chemistry Chemical Physics. 2015;17(37):24452–61. [DOI] [PubMed] [Google Scholar]
- [99].Lin S, Ryu S, Tokareva O, Gronau G, Jacobsen MM, Huang W, et al. Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres. Nat Commun. 2015;6:6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jacobsen MM, Tokareva OS, Ebrahimi D, Huang W, Ling S, Dinjaski N, et al. Effect of Terminal Modification on the Molecular Assembly and Mechanical Properties of Protein-Based Block Copolymers. Macromolecular Bioscience.1700095-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Moeendarbary E, Ng T, Zangeneh M. Dissipative particle dynamics in soft matter and polymeric applications—A review. International Journal of Applied Mechanics. 2010;2(01):161–90. [Google Scholar]
- [102].Aphinyan S, Geethalakshmi K, Yeo J, Shakouri A, Ng TY. Numerical characterization of ultraviolet ink fluid agglomeration and the surfactant effect in nanoinkjet printing. Polymers for Advanced Technologies. 2017. [Google Scholar]
- [103].Moeendarbary E, Ng T, Zangeneh M. Dissipative particle dynamics: introduction, methodology and complex fluid applications—a review. International Journal of Applied Mechanics. 2009;1(04):737–63. [Google Scholar]
- [104].Guigas G, Morozova D, Weiss M. Exploring membrane and protein dynamics with dissipative particle dynamics. Advances in protein chemistry and structural biology. 2011;85:143–82. [DOI] [PubMed] [Google Scholar]
- [105].Li D, Liu MS, Ji B, Hwang K, Huang Y. Coarse-grained molecular dynamics of ligands binding into protein: The case of HIV-1 protease inhibitors. The Journal of Chemical Physics. 2009;130(21):215102. [DOI] [PubMed] [Google Scholar]
- [106].Cheng Y, Li D, Ji B, Shi X, Gao H. Structure-based design of carbon nanotubes as HIV-1 protease inhibitors: Atomistic and coarse-grained simulations. Journal of Molecular Graphics and Modelling. 2010;29(2):171–7. [DOI] [PubMed] [Google Scholar]
- [107].Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Peptide Folding: When Simulation Meets Experiment. Angewandte Chemie International Edition. 1999;38(1–2):236–40. [Google Scholar]
- [108].Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys J. 1998;75(1):422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Richardson JS. The Anatomy and Taxonomy of Protein Structure In: C.B. Anfinsen JTE, Frederic MR, eds. Advances in Protein Chemistry: Academic Press; 1981, p. 167–339. [DOI] [PubMed] [Google Scholar]
- [110].Arin MJ, Oji V, Emmert S, Hausser I, Traupe H, Krieg T, et al. Expanding the keratin mutation database: novel and recurrent mutations and genotype–phenotype correlations in 28 patients with epidermolytic ichthyosis. British Journal of Dermatology. 2011;164(2):442–7. [DOI] [PubMed] [Google Scholar]
- [111].Haruna K, Suga Y, Mizuno Y, Hasegawa T, Kourou K, Matsuba S, et al. R156C mutation of keratin 10 causes mild form of epidermolytic hyperkeratosis. Journal of Dermatology. 2007;34(8):545–8. [DOI] [PubMed] [Google Scholar]
- [112].Sun X, Ma L, Xie Y, Zhu X. Keratin 1 and keratin 10 mutations causing epidermolytic hyperkeratosis in Chinese patients. Journal of Dermatological Science. 2002;29(3):195–200. [DOI] [PubMed] [Google Scholar]
- [113].Smith FD. The Molecular Genetics of Keratin Disorders. Am J Clin Dermatol. 2003;4(5):347–64. [DOI] [PubMed] [Google Scholar]
- [114].Zheng D, Giljohann DA, Chen DL, Massich MD, Wang X-Q, Iordanov H, et al. Topical delivery of siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation. Proceedings of the National Academy of Sciences. 2012;109(30):11975–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kerem E Mutation specific therapy in CF. Paediatric Respiratory Reviews. 2006;7, Supplement 1:S166–S9. [DOI] [PubMed] [Google Scholar]
- [116].Thursfield RM, Davies JC. Genotype-specific small-molecule therapy for cystic fibrosis. Breathe. 2013;9(3):176–86. [Google Scholar]
- [117].Jung GS, Qin Z, Buehler MJ. Mechanical Properties and Failure of Biopolymers: Atomistic Reactions to Macroscale Response Polymer Mechanochemistry. Cham: Springer Berlin Heidelberg Springer International Publishing; 2015, p. 317–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Hierarchical Structure and Nanomechanics of Collagen Microfibrils from the Atomistic Scale Up. Nano Lett 2011, p. 757–66. [DOI] [PubMed] [Google Scholar]
- [119].Gautieri A, Buehler MJ, Redaelli A. Deformation rate controls elasticity and unfolding pathway of single tropocollagen molecules. Journal of the Mechanical Behavior of Biomedical Materials. 2009;2(2):130–7. [DOI] [PubMed] [Google Scholar]
- [120].Buehler MJ. Atomistic and continuum modeling of mechanical properties of collagen: Elasticity, fracture, and self-assembly. J Mater Res. 2011;21(08):1947–61. [Google Scholar]
- [121].Depalle B, Qin Z, Shefelbine SJ, Buehler MJ. Influence of cross-link structure, density and mechanical properties in the mesoscale deformation mechanisms of collagen fibrils. Journal of the Mechanical Behavior of Biomedical Materials. 2015;52:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Depalle B, Qin Z, Shefelbine SJ, Buehler MJ. Large Deformation Mechanisms, Plasticity, and Failure of an Individual Collagen Fibril With Different Mineral Content. J Bone Miner Res. 2016;31(2):380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Osteogenesis imperfecta mutations lead to local tropocollagen unfolding and disruption of H-bond network. RSC Adv. 2012;2:3890–6. [Google Scholar]
- [124].Chang S-W, Shefelbine SJ, Buehler MJ. Structural and Mechanical Differences between Collagen Homo- and Heterotrimers: Relevance for the Molecular Origin of Brittle Bone Disease. Biophys J. 2012;102(3):640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Gautieri A, Uzel S, Vesentini S, Redaelli A, Buehler MJ. Molecular and Mesoscale Mechanisms of Osteogenesis Imperfecta Disease in Collagen Fibrils. Biophys J. 2009;97(3):857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Srinivasan M, Uzel SGM, Gautieri A, Keten S, Buehler MJ. Alport Syndrome mutations in type IV tropocollagen alter molecular structure and nanomechanical properties. J Struct Biol. 2009;168(3):503–10. [DOI] [PubMed] [Google Scholar]
- [127].Mohs A, Popiel M, Li Y, Baum J, Brodsky B. Conformational Features of a Natural Break in the Type IV Collagen Gly-X-Y Repeat. J Biol Chem. 2006;281(25):17197–202. [DOI] [PubMed] [Google Scholar]
- [128].Sun X, Chai Y, Wang Q, Liu H, Wang S, Xiao J. A Natural Interruption Displays Higher Global Stability and Local Conformational Flexibility than a Similar Gly Mutation Sequence in Collagen Mimic Peptides. Biochemistry. 2015;54(39):6106–13. [DOI] [PubMed] [Google Scholar]
- [129].Baldock C, Oberhauser AF, Ma L, Lammie D, Siegler V, Mithieux SM, et al. Shape of tropoelastin, the highly extensible protein that controls human tissue elasticity. Proceedings of the National Academy of Sciences. 2011;108(11):4322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Hirano E, Knutsen RH, Sugitani H, Ciliberto CH, Mecham RP. Functional rescue of elastin insufficiency in mice by the human elastin gene - Implications for mouse models of human disease. Circulation Research. 2007;101(5):523–31. [DOI] [PubMed] [Google Scholar]
- [131].Piontkivska H, Zhang Y, Green E, Elnitski L. Multi-species sequence comparison reveals dynamic evolution of the elastin gene that has involved purifying selection and lineage-specific insertions/deletions. BMC Genomics. 2004;5(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Bashir MM, Indik Z, Yeh HS, Ornstein-Goldstein N, Rosenbloom J, Abrams WR, et al. Characterization of the complete human elastin gene. The Journal of Biological Chemistry. 1989;264(15):8887–91. [PubMed] [Google Scholar]
- [133].Wise SG, Weiss AS. Tropoelastin. International Journal of Biochemistry & Cell Biology. 2009;41:494–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.