Abstract
Patient-derived xenografts are high fidelity in vivo tumor models that accurately reflect many key aspects of human cancer. In contrast to either cancer cell lines or genetically engineered mouse models, the utility of PDXs has been limited by the inability to perform targeted genome editing of these tumors. To address this limitation, we have developed methods for CRISPR-Cas9 editing of PDXs using a tightly regulated, inducible Cas9 vector that does not require in vitro culture for selection of transduced cells. We demonstrate the utility of this platform in PDXs (1) to analyze genetic dependencies by targeted gene disruption and (2) to analyze mechanisms of acquired drug resistance by site-specific gene editing using templated homology-directed repair. This flexible system has broad application to other explant models and substantially augments the utility of PDXs as genetically programmable models of human cancer.
INTRODUCTION
Patient-derived xenografts (PDXs) constitute a powerful set of preclinical models for in vivo cancer research, reflecting the spectrum of genomic alterations and therapeutic liabilities of human cancers1-4. These models recapitulate the complex genotypes and intratumoral heterogeneity of their tumors of origin and are not subject to the selective pressure imposed by in vitro cell culture since they are maintained exclusively in vivo5-7. In addition, PDXs have proven to be valuable models of tumor types or genetic alterations for which in vitro models are not readily available8,9. These features have driven the rapid adoption and widespread use of PDXs in preclinical and co-clinical drug development, evaluation of biomarkers and imaging agents, and mechanistic investigation of acquired treatment resistance10-12.
The ability to genetically manipulate cancer models has played an essential role in defining the functional contributions of individual genes and variants to cancer biology and CRISPR-Cas9 has greatly expanded our ability to rapidly perform these studies13,14. CRISPR-Cas9 can be used to disrupt genes through the introduction of frameshift insertions and deletions (indels) by non-homologous end joining (NHEJ) or to precisely alter genomic sequences through homology-directed repair (HDR)15. Combining this technology with in vivo cancer models provides a platform on which to study carcinogenesis and tumor maintenance in a complex environment resembling that of human tumors14.
A diverse array of CRISPR-Cas9 systems have been developed in recent years to perform genome editing of cancer models16. Despite the proven utility of PDXs, application of these systems to in vivo cancer models has been restricted to xenografts of established human and mouse cell lines cultured extensively in vitro or genetically engineered mouse models (GEMMs)13,14. The continuous in vivo passaging of PDXs prevents the use of antibiotic selection methods extensively employed by current CRISPR-Cas9 systems17. While CRISPR-Cas9 vectors with alternative selection methods have been developed18-20, they all lack the complete set of features requisite for use in PDXs, namely 1) a cell surface selection marker, 2) a lentiviral vector with optimized titer, and 3) temporal control of Cas9 expression.
Tight temporal control of Cas9 activity is especially critical for in vivo tumor studies to validate genes required for tumor maintenance and to credential suppressor mutations that may play a role in acquired drug resistance21,22. Several inducible systems have been developed to regulate Cas9 activity at the post-translational level, yet these systems invariably suffer from aberrant or reduced Cas9 activity [reviewed by Gangopadhyay et al.23]. Doxycycline (dox)-inducible expression of Cas9 provides a combination of maximum cutting efficiency in the “on” state while minimizing Cas9 activity in the “off” state through tight transcriptional regulation. However, many current systems are reported to lack complete transcriptional control by dox and are not amenable to use in PDXs because they either rely on inefficient knock-in approaches22,24,25 or employ vectors that exceed the lentiviral packaging limit and consequently result in low viral titers and predictably poor transduction efficiency26,27. These limitations have precluded the application of existing genome editing systems to PDXs.
Given the increasingly central role of PDXs in cancer research, a technological advance to enable inducible CRISPR-Cas9 genome editing of these models would have broad utility to further our understanding of cancer biology and facilitate the development of new therapeutic strategies. We developed pSpCTRE, an all-in-one dox-inducible Cas9 lentiviral vector that encodes CD4T, a compact cell surface selection marker that enables rapid antibody-based selection of transduced PDX cells without intervening in vitro culture. Using this system, we generated a library of SpCTRE-PDXs representing multiple lung cancer subtypes with tightly regulated Cas9 expression. These models can be used to identify in vivo genetic dependencies or, when combined with a DNA repair template, to introduce desired mutations of interest via homology-directed repair (HDR). The methods developed herein significantly enhance the utility of PDXs as genetically programmable models of human cancer.
RESULTS
Development of pSpCTRE
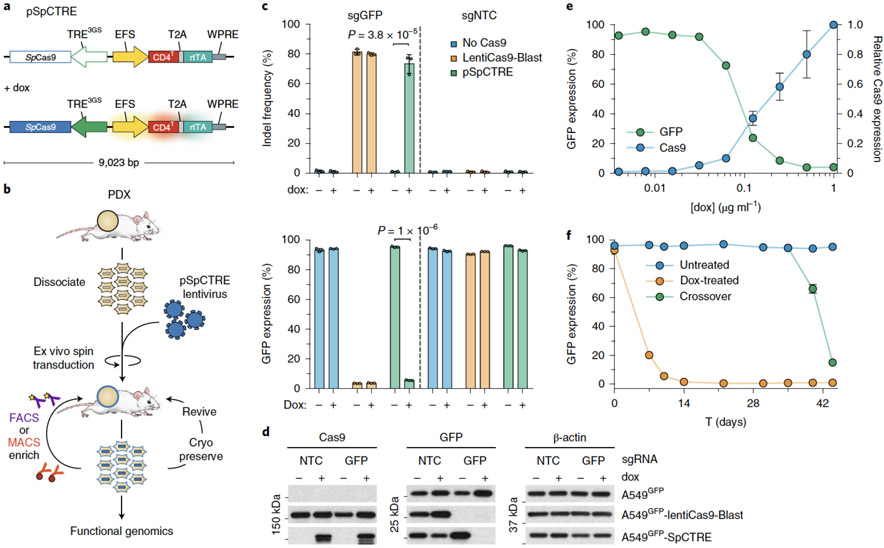
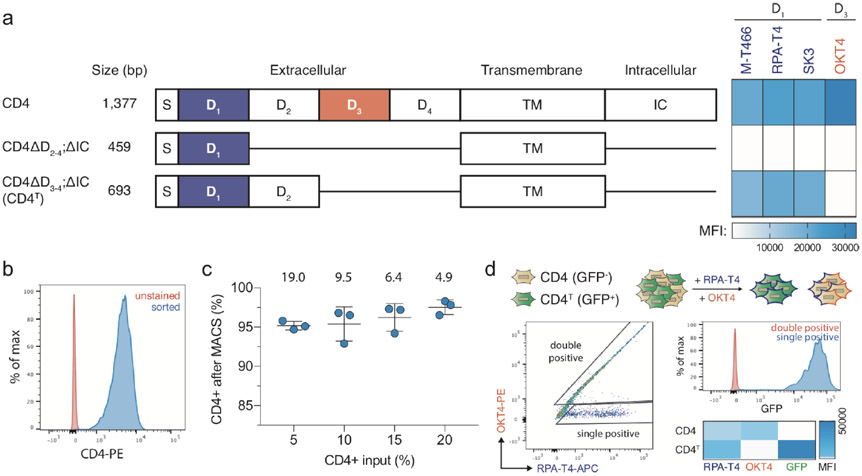
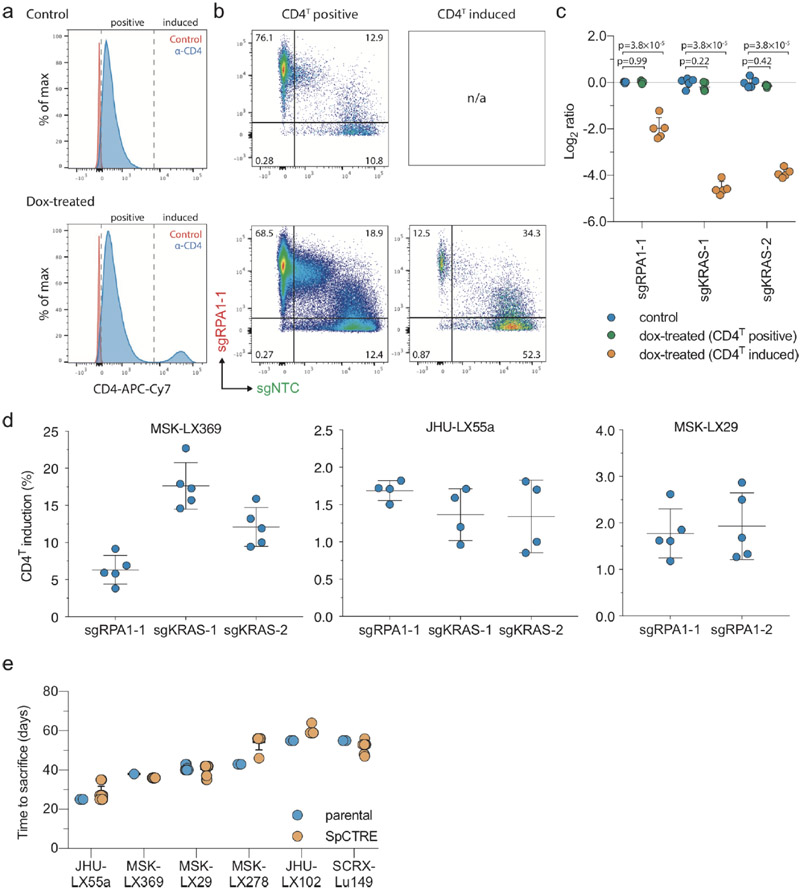
pSpCTRE (S.pyogenes Cas9 – Tet Response Element) is an all-in-one dox-inducible Cas9 lentiviral vector tailored for use in PDXs (Fig. 1a). A constitutively expressed, truncated CD4 (hereafter CD4T) selectable marker enables rapid positive selection of pSpCTRE-transduced cells without prolonged in vitro culture, allowing the system to be employed with PDXs (Fig. 1b). Selectable cell surface markers are particularly useful as they can be enriched through relatively mild methods such as fluorescence activated cell sorting (FACS) or immunomagnetic selection28-30. The diverse range of commercial α-CD4 reagents ensures the availability of bright fluorophores that are spectrally compatible with fluorescent protein markers used in downstream applications. CD4T is a compatible marker for PDXs as immunocompromised mice lack endogenous lymphocytes31 and CD4T lacks the cytoplasmic domain of full length CD4 required for outside-in signal transduction32 (Extended Data Fig. 1a).
Figure 1.
Development and validation of pSpCTRE, a lentiviral Cas9 vector for doxycycline (dox)-inducible genome editing in PDXs. (a) Vector map. Promoter activity and gene expression in the absence (top) or presence (bottom) of dox is depicted as none (outline), active (solid), and induced (glow). CD4T is a minimal, truncated CD4 selectable marker linked to rtTA-V10 by a T2A ribosome skipping sequence. (b) Dissociated tumors are transduced ex vivo with pSpCTRE lentivirus and immediately engrafted in mice without intervening in vitro culture. Resultant SpCTRE PDXs (blue outline) are then subjected to CD4 enrichment, cryopreservation, or functional studies. (c) In vitro genome editing of A549GFP cells lacking Cas9 (blue), with constitutive Cas9 expression from lentiCas9-Blast (orange), or with inducible Cas9 expression from SpCTRE (green) and the indicated sgRNAs. Mean indel frequency (top) and mean GFP expression (bottom) are displayed for n=3 independent cell culture replicates. Data is representative of two independent experiments with similar results. Error bars are SD, with SD < plotting character not drawn. p-values determined between groups by two-way ANOVA. NTC, Nontargeting control (d) Representative western blot from panel c. (e) Dox dose response of A549GFP-SpCTRE cells with sgGFP. Mean Cas9 expression (blue) and mean GFP expression (green) are displayed for n=3 independent cell culture replicates from a single experiment. Error bars are SD, with SD < plotting character not drawn. (f) Long-term culture of A549GFP-SpCTRE cells with sgGFP untreated (blue) or dox-treated (orange). At day 35, previously untreated cells were crossed over to dox media (green). Mean GFP expression is displayed for n=3 independent cell culture replicates from a single experiment. Error bars are SD, with SD < plotting character not drawn. Data for experiments in panels c, e, f, and unmodified gel images for panel d are available as source data (SourceData_Fig1 and Unmodified_Gels_Fig1, respectively).
We designed pSpCTRE to achieve tight temporal control of Cas9 activity and sensitive induction through extensive optimization of its substituent components. The TRE3GS promoter is engineered to eliminate endogenous transcription factor binding sites, thereby minimizing leaky Cas9 expression in the absence of dox33, while the rtTA-V10 variant used displays increased sensitivity to low dox concentrations and may improve dox response in vivo34. Additionally, several studies have shown that relative genome editing efficiency is strongly correlated with Cas9 protein expression, where poorly expressed constructs demonstrate inefficient editing35,36. Therefore, it is imperative to employ an inducible system that can drive sufficient expression of Cas9. To this end, the reverse promoter orientation used in pSpCTRE has been shown to produce significantly higher transgene expression than a similar construct with a forward promoter orientation37.
An added challenge in the design of pSpCTRE is creating a system with all of the necessary components to inducibly express Cas9 yet adequate lentiviral titer to successfully transduce PDXs. Lentiviral titer decreases as a function of proviral genome size38 and the relatively large size of Cas9 has made it challenging to develop high titer CRISPR-Cas9 systems in the past24,39. For this reason, several approaches were taken to reduce the size of pSpCTRE. The optimal composition of CD4T was empirically determined through a deletion series of the extracellular Ig-like domains required for binding of commercial α-CD4 antibodies (Extended Data Fig. 1a). While several α-CD4 antibody epitopes reside within domain D140, we found that both domains D1 and D2 were required for binding of these antibodies. Based on this finding, CD4T is composed of the CD4 signal peptide and extracellular domains D1 and D2 fused to the transmembrane domain (TM) via a flexible linker, with the intracellular portion of the protein omitted entirely. The final coding sequence is approximately half the size of full-length CD4 and can be used with commercial flow cytometry or immunomagnetic sorting reagents (Extended Data Fig. 1b, c). To further minimize vector size, we use the minimal EFS promoter to drive constitutive expression of CD4T and rtTA-V10, linked by a T2A self-cleaving peptide. These optimizations collectively result in a final SpCTRE size of 9,023 bp between the LTRs.
In addition to reducing vector size, the truncated CD4T allows pSpCTRE to be used in the context of CD4-expressing cells, as the former lacks extracellular domains D3 and D4. Therefore, CD4T can be distinguished from full-length CD4 by staining with both domain D1 and D3 specific antibodies (Extended Data Fig. 1d). Cells with full length CD4 will stain with both antibodies and can be easily identified as a double positive population, whereas the cells with CD4T remain single positive.
pSpCTRE Cas9 activity is tightly regulated and comparable to constitutive expression in vitro
We initially examined pSpCTRE editing efficiency through in vitro disruption of GFP in A549 cells expressing GFP (A549GFP). In the absence of dox, A549GFP-SpCTRE cells transduced with sgGFP maintained GFP expression and had no detectable Cas9 protein by western blot (Fig. 1c, d). Conversely, after exposure to dox, A549GFP-SpCTRE cells efficiently edited GFP, similar to A549GFP cells constitutively expressing Cas9 from the lentiCas9-Blast vector (Fig. 1c, d). We observed a dose-dependent increase in Cas9 expression and CD4T induction with concomitant loss of GFP expression with as little as 0.0625 μg/mL dox and a maximum loss of GFP expression at 0.25 μg/mL dox, defining an upper and lower bound for dox response (Fig. 1e).
To more sensitively screen for leaky expression of Cas9 and undesired genome editing, we cultured A549GFP-SpCTRE cells transduced with sgGFP in the absence of dox and observed no decrease in GFP-positive cells over >10 passages. However, these cells retained the capacity to efficiently disrupt GFP upon dox induction after 35 days of continuous culture, conditions that mimic dox-induction in established tumors (Fig. 1f). These data indicate that pSpCTRE is a tightly regulated dox-inducible Cas9 vector that meets our requirements for in vivo use in PDX tumors.
Induced surface expression of CD4T is a reporter for Cas9 expression and genome editing
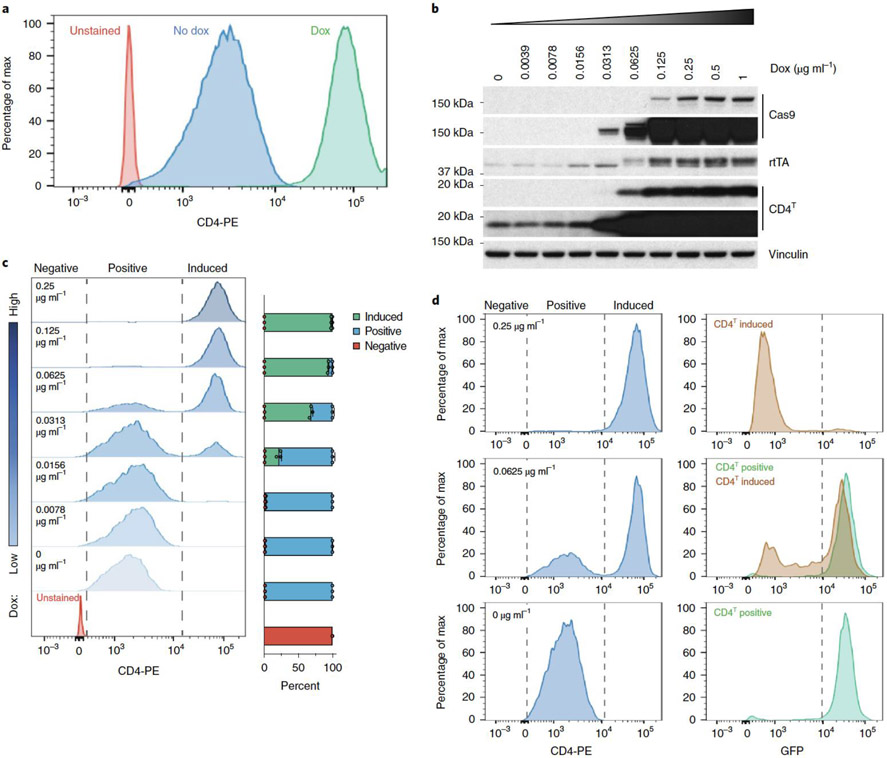
Dox-inducible lentiviral systems are influenced by local regulatory elements of the genomic locus where they randomly integrate, which can lead to heterogeneous and often poor transgene induction33. This problem is accentuated in vivo by lower tumor dox exposure, relative to in vitro culture41. pSpCTRE overcomes this limitation by providing an engineered cell surface reporter of dox-induced transgene expression. This is accomplished by the reverse orientation of the TRE3GS promoter that places it proximal to the constitutive EFS promoter that drives CD4T. This promoter topology is unidirectional (low EFS activity only) in the absence of dox and is strongly bidirectional (both EFS and TRE3GS strongly induced) in the presence of dox, thereby increasing CD4T expression concomitant with Cas9 induction.
This property of pSpCTRE was discovered when we observed an approximately 100-fold increase in CD4T fluorescent signal intensity from dox treated cells (Fig. 2a). We validated by western blot that the change in signal intensity was a result of increased CD4T protein expression and correlated with dox doses that induce Cas9 expression (Fig. 2b). Importantly, we observed that CD4T induced cells are a distinct population that can be easily differentiated from uninduced CD4T positive cells in a mixed population. Rather than a progressive increase in CD4T surface expression at increasing dox concentrations, we see a discrete population of CD4T induced cells emerge (Fig. 2c). We speculate that this stark shift in CD4T surface expression may be caused by a feed forward loop created by a concurrent increase in rtTA-V10 expression to potently drive bidirectional promoter induction. Therefore, there are three distinct populations of cells to note when using pSpCTRE (Fig. 2c): 1) CD4T negative (pSpCTRE negative), 2) CD4T positive (pSpCTRE positive, TRE3GS uninduced), and 3) CD4T induced (pSpCTRE positive, TRE3GS induced).
Figure 2.
Dox induction of EFS promoter activity and CD4T expression above basal levels is a marker for Cas9 expression and genome editing. (a) Flow cytometry analysis of CD4T expression in A549-SpCTRE cells treated with (green) or without (blue) dox. Data is representative of two independent experiments with similar results. (b) Western blot analysis of A549-SpCTRE cells induced with a range of dox concentrations. Arrow indicates specific rtTA band. Data is representative of two independent experiments with similar results. (c) Representative flow cytometry analysis of CD4T expression with the indicated dox concentrations in A549-SpCTRE cells. Mean percentage of cells in the negative, positive, and induced gates is displayed in the bar graph (right) for n=3 independent cell culture replicates from a single experiment. Error bars are SD ,with SD < plotting character not drawn. (d) GFP editing of A549GFP-SpCTRE cells with sgGFP based on gating of CD4T positive or induced populations at the indicated dox doses. Data is representative of n=3 independent cell culture replicates from a single experiment. Data for experiments in panels a, c, d, and unmodified gel images for panel b are available as source data (SourceData_Fig2 and Unmodified_Gels_Fig2, respectively).
Importantly, CD4T induced expression is a robust marker of cells that express Cas9 and are competent to undergo Cas9-mediated genome editing. A549GFP-SpCTRE cells expressing sgGFP efficiently deplete GFP when treated with 0.25 μg/mL dox and all cells within this population induce CD4T (Fig. 2d). However, at a dox concentration of 0.0625 μg/mL, only ~60% of cells induce CD4T. When these cells are gated based on CD4T surface expression, GFP is specifically depleted in the CD4T induced population but not in cells that do not induce CD4T above baseline. Therefore, we can use CD4T induction as a marker to enrich for cells that express Cas9 and exclude cells from analysis that do not undergo genome editing. This feature of pSpCTRE allows for identification of cells with efficient induction of Cas9, enabling the study of heterogeneous tumor cell populations and eliminating the need for selection of single cell clones.
Generating a library of SpCTRE PDXs
Cas9-expressing SpCTRE PDXs can be generated in as few as two in vivo passages. In the first passage, dissociated tumors are subjected to an ex vivo spin transduction and immediately re-engrafted for expansion in vivo. Successfully transduced cells are enriched from the ensuing tumor with α-CD4 positive selection and either cryopreserved or passaged for subsequent use in functional genomic studies.
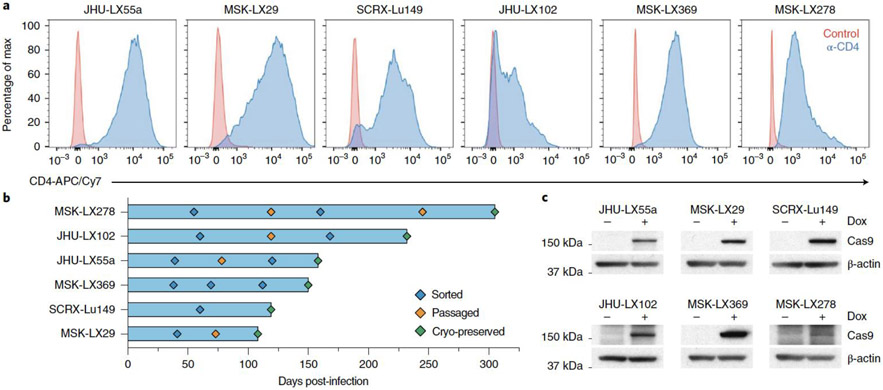
We sought to generate a library of SpCTRE PDXs and transduced 20 PDXs representing multiple lung cancer subtypes with pSpCTRE lentivirus. We were able to detect and enrich CD4T positive cells in six of these models (Fig. 3a). In the 14 models remaining models, no CD4T positive cells were detected, suggesting a lack of pSpCTRE transduction. The median time to establish SpCTRE PDXs following transduction was 154 days (with a range of 108 to 305 days) and one to three in vivo passages were required after transduction (Fig. 3b). Importantly, these SpCTRE PDX tumors express Cas9 exclusively when the mice are administered dox (Fig. 3c).
Figure 3.
Generation of Cas9-expressing PDXs using pSpCTRE. (a) CD4T staining of SpCTRE PDXs successfully transduced with pSpCTRE lentivirus and enriched for CD4T positive cells using FACS. (b) Timeline for SpCTRE PDX model generation. PDXs were 1) sorted to enrich for CD4T positive cells if CD4T positive percentage was below 50% (blue), 2) passaged to expand for additional sorting or cryopreservation (orange), or 3) cryopreserved (green). (c) Western blot analysis from a single experimental replicate of Cas9 expression in the indicated pSpCTRE PDXs from control or dox-treated mice. Data for experiments in panels a-b and unmodified gel images for panel c are available as source data (SourceData_Fig3 and Unmodified_Gels_Fig3, respectively).
Competition assay to identify PDX genetic dependencies
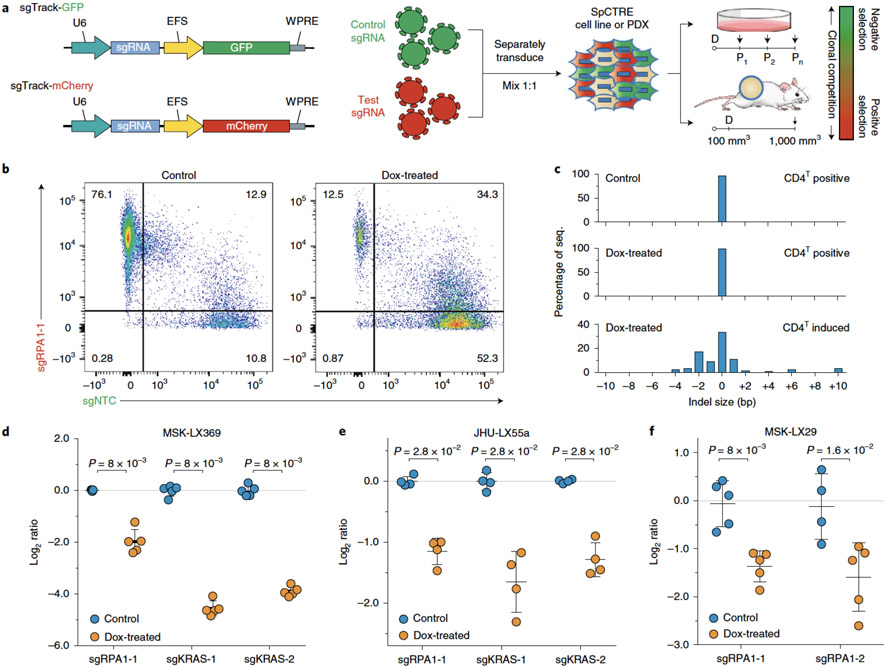
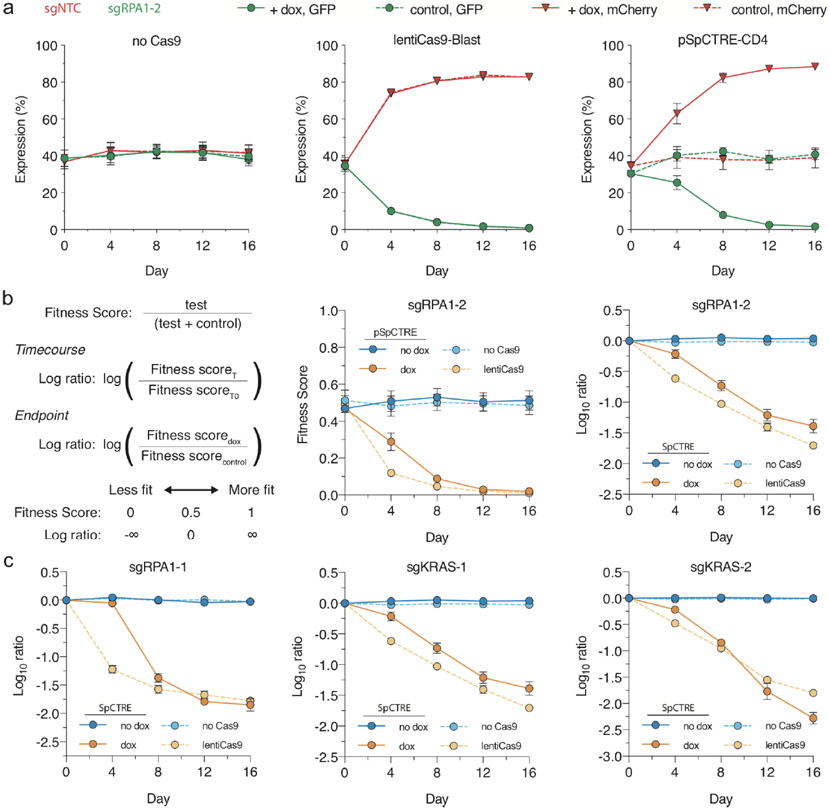
We interrogated genetic dependencies of SpCTRE PDXs using a competition assay, which allows for measurement of gene disruption effects without prior selection for cells transduced with sgRNAs (Fig. 4a). To deliver each sgRNA, we generated a fluorescent reporter lentiviral vector (sgTrack) that constitutively expresses either mCherry or GFP in addition to an sgRNA of interest. We independently transduced SpCTRE PDXs ex vivo with sgTrack vectors encoding a non-targeting control sgRNA or an sgRNA targeting a gene of interest and admixed both populations before engrafting in mice. The fluorescent reporters associated with each sgRNA allowed us to measure the relative abundance of sgRNAs in a population as a proxy for the relative fitness of cells engineered to disrupt a gene of interest. This approach robustly identified differential fitness of cells in vitro, reflected in the reduction of cells carrying an sgRNA targeting the essential gene RPA1 or the KRAS proto-oncogene in a KRASG12S-mutant lung adenocarcinoma cell line (Extended Data Fig. 2a-c).
Figure 4.
Interrogation of genetic dependencies in SpCTRE PDXs using a competition assay. (a) Control or test sgRNAs in sgTrack fluorescent reporter lentiviral vectors are transduced independently into SpCTRE PDXs or cell lines and then admixed and either maintained in culture (if cell lines) or engrafted subcutaneously (if PDXs). Dox addition (D) and flow cytometry analysis (arrows) occurs at the indicated time points and the relative abundance of GFP and mCherry single positive cells is compared between control and dox-treated samples. (b) Flow cytometry analysis of representative tumors from a competition assay with sgRPA1-1 (mCherry) and sgNTC (GFP) in the MSK-LX369 lung adenocarcinoma PDX. Data is representative of n=5 mice (c) RPA1 genome editing of representative tumors from panel b. Analysis was restricted to sgRPA1-1 containing cells from the indicated treatment groups and CD4T gates. (d-f) Competition assays with the indicated sgRNAs in the SpCTRE PDXs (d) MSK-LX369, (e) JHU-LX55a, and (f) MSK-LX29. The log ratio compares the fitness score of dox-treated mice to the fitness score of control mice. Mean log ratio is displayed for n=4 or n=5 mice as indicated. Error bars are SD, with SD < plotting character not drawn. A two-sided Wilcoxon rank sum test was used to determine statistical significance. Data for experiments in panels b-f are available as source data (SourceData_Fig4).
We next performed similar competition assays in vivo to study the effects of single gene disruption on the fitness of three SpCTRE PDXs. Targeting RPA1 in MSK-LX369, JHU-LX55a, and MSK-LX29, we consistently observed a depletion of cells carrying the RPA1 sgRNA and an enrichment of cells carrying a non-targeting control sgRNA in CD4T induced cells from dox-treated mice (Fig. 4b). We verified targeted editing of the RPA1 locus in isolated tumor cells and confirmed the presence of indels exclusively in sgRPA-transduced cells with CD4T induced expression (Fig. 4c). Additionally, we investigated the fitness effects of KRAS gene disruption in two KRAS-mutant lung adenocarcinoma SpCTRE PDXs, MSK-LX369 and JHU-LX55a. We observed a significant depletion of cells harboring KRAS sgRNAs exclusively in CD4T induced cells from dox-treated mice, indicating that these PDXs exhibit reduced fitness in the context of KRAS disruption (Fig. 4d-f and Extended Data Fig. 3a-c) while overall tumor growth rates as measured by time to sacrifice were not radically altered by lentiviral infection, despite robust induction of CD4T (Extended Data Fig. 3d,e). These experiments verify that this system is suited to functionally interrogate PDX gene essentiality in vivo.
Introduction of drug resistance mutations using homology-directed repair
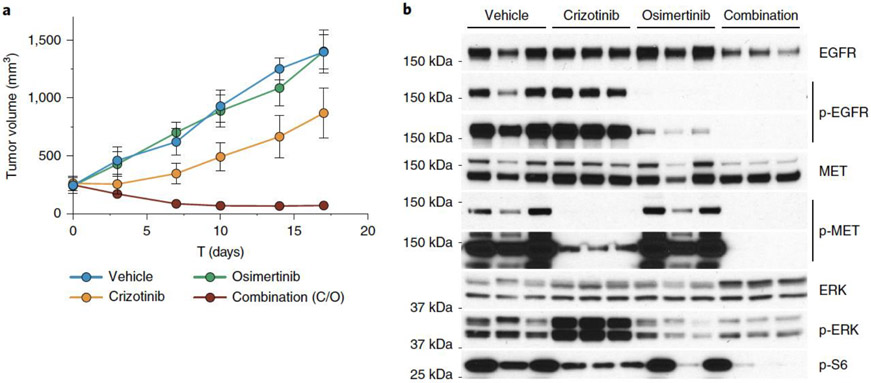
We chose to use SpCTRE PDXs and CRISPR-Cas9 mediated homology-directed repair to explore mechanisms of acquired EGFR inhibitor resistance in lung adenocarcinomas. MSK-LX29 was derived from a patient whose tumor carried a homozygous mutation encoding the EGFRL858R variant and who developed resistance to the 1st generation EGFR inhibitor erlotinib through acquisition of a focal MET amplification. This PDX is resistant to single agent treatment with the 3rd generation EGFR inhibitor osimertinib as well as the MET inhibitor crizotinib but is profoundly and durably sensitive to a crizotinib-osimertinib (C/O) combination therapy (Fig. 5a). Western blot analysis of treated tumors revealed an increase in ERK phosphorylation and incomplete inhibition of S6 phosphorylation in crizotinib-treated tumors, suggesting that MET and EGFR signaling could be buffering in this model (Fig. 5b). We reasoned that restoring EGFR activity by introducing the osimertinib resistance mutation C797S via homology-directed repair would be sufficient to induce resistance of MSK-LX29 to C/O combination therapy.
Figure 5.
Evaluation of EGFR inhibitor combination therapy in a MET amplified PDX (a) MSK-LX29 mean tumor volume for the indicated treatment arms for n=3 mice. Error bars are SD, with SD < plotting character not drawn. C/O, crizotinib/osimertinib combination. (b) Western blot analysis from a single experimental replicate of the tumors treated in panel a. Data for experiments in panel a and unmodified gel images for panel b are available as source data (SourceData_Fig5 and Unmodified_Gels_Fig5, respectively).
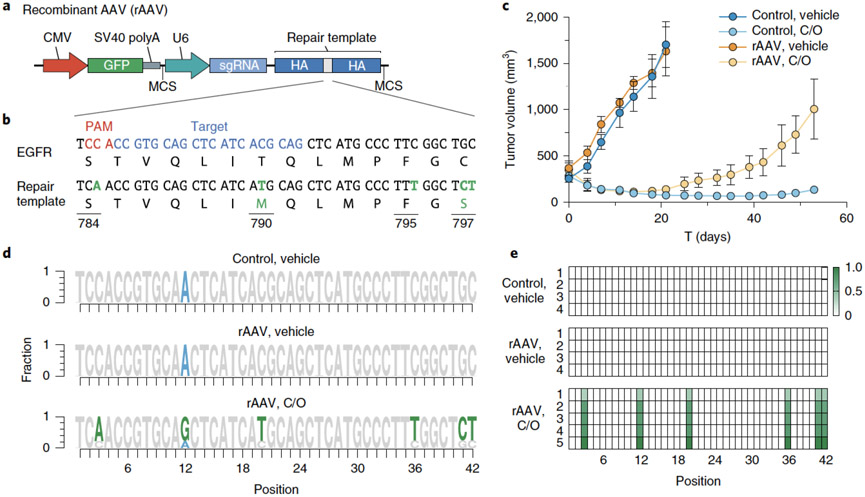
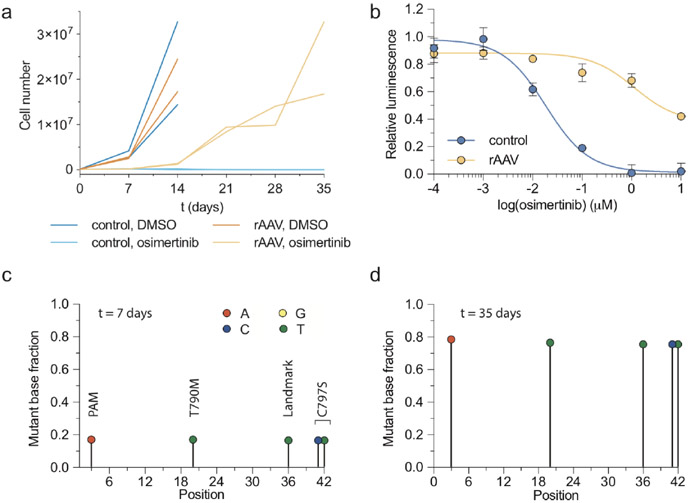
We generated a recombinant adeno-associated virus (rAAV) vector that delivers an sgRNA and a HDR template encoding desired genetic alterations (Fig. 6a). To edit EGFR, this rAAV vector encodes both an sgRNA targeting a sequence proximal to that which encodes the hydrophobic binding pocket of the EGFR kinase domain and a 1.5-kb repair template centered on this region. The repair template encodes nucleotide changes that silently destroy the sgEGFR PAM sequence to prevent Cas9 re-cutting after repair and introduces in cis 1) a T790M 1st generation EGFR inhibitor gatekeeper mutation, 2) a C797S mutation that abolishes activity of 3rd generation covalent EGFR inhibitors and 3) a silent landmark mutation to mark desired nucleotide changes that originate from the repair template (Fig. 6b). Efficient generation of osimertinib resistance in EGFR-mutant PC9 cells using this template was confirmed in vitro (Extended Data Fig. 4).
Figure 6.
Introduction of complex drug resistance mutations in SpCTRE PDXs using recombinant AAV (rAAV). (a) Schematic of rAAV vector that delivers a GFP marker, an sgRNA expressed from a U6 promoter, and a homology-directed repair template that encodes mutations of interest surrounded by homology arms (HA). The U6-sgRNA and repair template are flanked by multiple cloning sites (MCS). (b) sgRNA target site and EGFR T790M/C797S repair template to introduce the indicated nucleotide changes (green). (c) Mean tumor volumes of MSK-LX29-SpCTRE with or without rAAV and treated with a crizotinib/osimertinib (C/O) combination or vehicle for n=5 mice. Error bars are SD, with SD < plotting character not drawn. (d) Sequencing analysis of representative tumors from the indicated treatment groups from panel c. Mutations introduced by rAAV are highlighted (green). MSK-LX29 contains a homozygous SNP (blue) that is removed by the rAAV. (e) Heatmap depicting frequency of rAAV introduced mutations for tumors from the indicated treatment groups from panel c. Data for experiments in panels b-e are available as source data (SourceData_Fig6).
MSK-LX29-SpCTRE cells were infected ex vivo with rAAV and subsequently engrafted into dox-treated mice to induce Cas9 expression and initiate HDR. These tumors were sensitive to C/O combination therapy, which may reflect the latency of Cas9 expression in response to dox treatment or the preference for the NHEJ repair pathway in mammalian cells42,43. However, 2 weeks after the start of treatment MSK-LX29-SpCTRE developed resistance in all five tumors in the rAAV plus C/O treatment group, but in none of the C/O treated tumors not infected with rAAV (Fig. 6c). This observation is consistent with the clonal outgrowth of a subpopulation of cells that successfully introduced the C797S mutation via HDR. Sequencing of all five resistant tumors revealed the presence of the templated C797S mutation (Fig. 6d, e). We conclude that an acquired resistance mutation in EGFR alone is sufficient to restore resistance to crizotinib-osimertinib combination therapy in EGFR mutant lung adenocarcinomas that acquire bypass resistance through MET amplification. More generally, these data confirm that pSpCTRE, in combination with rAAV, can be used to introduce complex template-directed site-specific mutations into PDXs in vivo.
DISCUSSION
The high failure rate for developing new cancer therapeutics can be attributed in part to the extensive use of preclinical models that do not accurately recapitulate many aspects of human tumors. Patient-derived xenografts are high fidelity cancer models that maintain several important features of the tumors from which they were derived, including mutational, gene expression, and epigenetic profiles 6,7,44, and therefore are valuable tools for elucidating novel aspects of tumor biology. However, the use of PDXs in cancer research is limited to a narrow scope of applications that exclude most types of mechanistic experiments due to the technological barriers imposed by continuous in vivo heterotransplantation17. We have developed herein methods for CRISPR-Cas9 genome editing of PDXs while maintaining growth exclusively in vivo.
Although these methods were developed with primary xenografts in mind, this system is readily applicable to other model systems. Unlike mouse organoid cultures, which can be derived from mice engineered with conditional Cas9 expression, human primary organoid explants are subject to many of the same experimental challenges as primary tumor xenografts, suggesting that pSpCTRE constructs would prove particularly useful in this setting. The pSpCTRE cassette may also be useful as the basis for improved general-purpose doxycycline inducible constructs, such as in the context of Col1A1 targeting vectors, which have shown variable degrees of doxycycline control across different adult mouse tissues45,46.
Several aspects of these methods could be improved to enable more complex experimental designs. The current iteration of the clonal competition assay is limited to endpoint analysis of a single genetic perturbation for each in vivo experiment. Future work could address these limitations as needed with new constructs encoding diverse reporters. Secreted luciferases have been used as a surrogate for population size in cell line xenografts and would be equally applicable in this setting47,48. Multiplexing of tens to hundreds of sgRNAs could be made possible by coupling each genetic perturbation to the expression of a unique cell surface protein epitope, significantly increasing the throughput of the system49,50. Similar focused screens of targeted pools of sgRNA may be possible using standard sequencing approaches to quantify changes in sgRNA abundance under different experimental conditions51. Exploration of highly diversified sequence space of a single genomic locus, as previously demonstrated with rAAV in genetically engineered mouse models, would be equally applicable to SpCTRE-PDX52. This initial pSpCTRE construct encodes SpCas9; however, future iterations could employ Cas9 variants with increased fidelity, expanded PAM specificity, or altered enzymatic activity to enable a range of other genetic manipulations in PDXs53-56.
We were able to generate 6 SpCTRE PDXs, however we were unsuccessful in generating SpCTRE PDXs for the remaining 14 PDXs attempted. In each case, the failure was due to a lack of lentiviral transduction as no CD4T positive cells could be detected. Transduction efficiency varies greatly between different cell types57,58 and it is possible that further increasing the titer of pSpCTRE could improve the success rate of transducing PDXs. Additionally, non-viral delivery methods could be explored, such as knock-in to the AAVS1 safe harbor locus, to achieve stable integration of the pSpCTRE cassette.
CD4T was chosen as the cell surface selection marker because PDXs are primarily grown in immunocompromised mice that lack lymphocytes. However, recent methods have been developed to establish PDXs in mice with humanized immune systems for cancer immunotherapy studies59. While CD4T lacks the cytoplasmic domain required for intracellular signaling, we cannot rule out the possibility that it may confound the results of such studies. Accordingly, pSpCTRE could be adapted with an alternative cell surface selectable marker, such as ΔLNGFR29 or huEGFRt30, for these purposes.
Together, these methods constitute a core enabling technology for in vivo functional genomics in PDXs, allowing interrogation of gene essentiality, candidate drug targets, mechanisms of acquired resistance, tumor suppressor function, chemical:genetic interactions, and variants of unknown significance in this tumor model.
METHODS
Patient-derived xenografts (PDXs)
All animal experiments were approved by the Memorial Sloan Kettering Cancer Center (MSKCC) Animal Care and Use Committee. Primary tumors and whole blood samples collected for generation of PDX models were obtained with informed consent from patients under protocols approved by the MSKCC and Johns Hopkins institutional review boards. Subcutaneous flank tumors were generated as described previously11.
Cloning and plasmids
The plasmids generated in this study are available on Addgene as indicated. A list of all primers used in this study is available in Supplementary Table S1.
pSpCTRE (Addgene plasmid # 114010): EFS promoter expresses CD4T-2A-rtTA-V10; TRE-3GS promoter controls tetracycline-inducible expression of S.pyogenes Cas9 as an all-in-one Tet-On system. This vector was derived from the pLVX-TetOne-Puro vector (Clontech), which was digested with XhoI and KpnI to remove the existing rtTA and Puro selection cassettes. The EFS-CD4T-2A-rtTA-WPRE cassette was generated by gene synthesis (IDT) and inserted into the digested vector by Gibson assembly (NEB). Cas9 was PCR amplified and inserted into the multiple cloning site by digestion with EcoRI and BamHI followed by overnight ligation with T4 ligase (NEB).
sgTrack-Gateway (Addgene plasmid # 114011), sgTrack-GFP (Addgene plasmid # 114012) and sgTrack-mCherry (Addgene plasmid # 114013): U6 promoter expresses a single sgRNA; EFS promoter upstream of the Gateway cassette expresses either TurboGFP or mCherry in the respective reporter vectors. These vectors are derived from lentiCRISPRv239, wherein Cas9 was replaced by a Gateway cassette in-frame with the existing 2A-Puro by PCR amplification of the Gateway cassette and Gibson assembly into an XbaI and BamHI digested backbone. Gateway donor vectors with closed TurboGFP and mCherry cDNAs were used to generate the respective reporter vectors using LR clonase (Invitrogen) according to the manufacturer’s protocol. sgRNAs were cloned into these vectors as previously described39.
pAAV-GFP-sgHDR: pAAV-GFP was a gift from John T Gray (Addgene plasmid # 32395)60. A synthetic DNA fragment comprising an SV40 polyA terminator, partial multiple cloning site (BglII, HindIII), the human U6 promoter, EGFR sgRNA, a 1.5 kb EGFR homology directed repair template containing the T790M mutation and destroying the sgEGFR PAM sequence, and a second partial multiple cloning site (ClaI, XhoI, XbaI) was cloned into the NotI and XbaI restriction enzyme sites of pAAV-GFP, replacing the original beta-globin polyA fragment. EGFR C797S coding and silent repair landmark mutations were inserted via site directed mutagenesis using primer pair P1 to generate the rAAV vector.
lentiCas9-Blast (Addgene plasmid # 52962) and lentiGuide-Puro (Addgene plasmid # 52963) were gifts from Feng Zhang. pMD2.G (Addgene plasmid # 12259) and psPAX2 (Addgene plasmid # 12260) were gifts from Didier Trono. pLenti CMV Neo DEST (705-1) (Addgene plasmid # 17392) and pLenti CMV Puro (w118-1) (Addgene plasmid # 17452) were gifts from Eric Campeau and Paul Kaufman61. All Gateway recombination reactions were performed with BP clonase or LR clonase used according to the manufacturer’s protocol (Invitrogen). TurboGFP cDNA was cloned into pLenti CMV Neo DEST (705-1). pDONR221/CD4 was purchased from DNASU (HsCD00413471). CD4ΔD2-4;ΔIC linked to rtTA-V10 by a T2A sequence was generated by gene synthesis (IDT), Gateway adapted by PCR using primer pair P2, and cloned into pDONR221. CD4 domain D2 was then added to this construct to generate CD4ΔD3-4;ΔIC (CD4T) by performing an outward PCR with primer pair P3, extracting domain D2 from pDONR221/CD4 using primer pair P4 and performing a Gibson assembly (NEB) with the PCR products. All CD4 constructs were then cloned into pLenti CMV Puro (w118-1). sgRNAs were cloned into lentiGuide-Puro as previously described39. All transformations were performed in One Shot Stbl3 Chemically Competent cells (Invitrogen). Plasmids were purified using QIAquick Spin Miniprep or Plasmid Plus Midi kits (Qiagen) and digest verified prior to use. All PCRs were performed with Phusion High-Fidelity PCR Master Mix with HF Buffer (NEB).
sgRNA sequences
Target sequences for sgRNAs used in this study are available in Supplementary Table S2. 62-64
Cell culture and lentivirus production
A549, PC9, and HEK293T cells were purchased from ATCC. A549 and PC9 cells were maintained in RPMI-1640 media supplemented with 10% Tet-Free FBS (Gemini) and 1x penicillin/streptomycin (pen/strep, Gibco) and HEK293T cells were maintained in DMEM media supplemented with 10% FBS (Gemini) and 1x pen/strep. All cell lines were verified negative for mycoplasma within 6 months of use. Lentivirus was produced by transfecting HEK293T cells with a 3:2:1 ratio of lentiviral plasmid:psPAX2:pMD2.G with JetPrime transfection reagent (Polyplus) at a 2:1 JetPrime:DNA ratio. Media was changed 24 h after transfection and viral supernatants were collected 72 h after transfection. Viral supernatants were syringe filtered with a 0.45 uM PVDF filter (Millipore) and concentrated approximately 20 fold with Lenti-X Concentrator (Clontech) according to the manufacturer’s protocol. All lentivirus was titrated in A549 cells to control for batch-to-batch variability and to normalize titers between different lentiviral backbones. In vitro lentiviral transductions were performed with 8 μg/mL hexadimethrine bromide (polybrene, Sigma) and at a multiplicity of infection (MOI) of approximately 1, unless otherwise stated.
In vitro validation of pSpCTRE
An A549GFP cell line containing a stably integrated, single-copy TurboGFP gene was generated by transducing A549 with pLenti CMV Neo/TurboGFP lentivirus (MOI 0.3) and single cell FACS sorting using a FACSAria (BD Biosciences). A549GFP cells were subsequently transduced with lentiCas9-Blast or pSpCTRE lentivirus (MOI 0.3) and selected with blasticidin or CD4T single cell FACS sorting, respectively, to generate stable A549GFP Cas9 cell lines. A549GFP Cas9 cells were transduced with lentiGuide-Puro/sgGFP or sgNTC lentivirus and selected with puromycin for 3 days. For analysis of GFP editing efficiency and dox dose response of pSpCTRE, 1 × 105 cells were plated in a 10 cm plate and treated with 0.5 μg/mL doxycycline (dox, Sigma) or a dox dose range, respectively, for 10 days. Cell pellets were collected to perform western blot, Tracking of Indels by Decomposition (TIDE), or flow cytometry analysis as described below. To screen for Cas9 editing in the absence of dox, A549GFP-SpCTRE cells with lentiGuide-Puro/sgGFP were maintained in culture for 6 weeks in RPMI with tet-free FBS. Cells were split when they reached approximately 70% confluence and flow cytometry analysis was performed every 7 days as described below. At day 35, cells not previously treated with dox were split and 0.5 μg/mL dox was added to half of the cells (crossover). In vitro competition assays were performed in A549 cells transduced with lentiCas9-Blast or pSpCTRE lentivirus (MOI 0.3) and selected with blasticidin or CD4T single cell FACS sorting, respectively. A549 Cas9 cell lines were independently transduced with sgTrack lentivirus and, after 3 days, cells containing control and test sgRNAs were mixed with an equal ratio of GFP and mCherry positive cells. Cells were split and flow analysis was performed as described below every 4 days. Results for all experiments represent three independent biological replicates.
Antibodies
A list of antibodies used in this study is available in Supplementary Table S3.
Protein extraction, western blotting and LiCor protein quantification
Whole cell lysates were prepared from frozen cell pellets or flash frozen tumor samples using RIPA lysis buffer with 1x HALT protease inhibitor cocktail (Thermo). Cell pellets were resuspended in 5 volumes of cold lysis buffer and incubated on ice for 10 minutes, followed by sonication for 10 seconds with a 200V microtip sonicator set to 40% amplitude (QSonica, CL 18). Lysates were clarified by centrifugation at 20,000 x g for 10 minutes at 4C. Protein extraction from flash frozen tumor samples was performed as previously described11. Protein was quantified using a BCA protein assay kit (Pierce) and samples were denatured at 70C for 10 minutes in NuPAGE LDS sample buffer with NuPAGE sample reducing agent and then resolved on a 4-12% Bis-Tris gradient gel (Invitrogen). For chemiluminescent detection, gels were wet-transferred to 0.45 μm Immobilon-P PVDF membrane (Millipore) and incubated overnight at 4C with primary antibody diluted in TBS (Fisher) supplemented with 0.1% Tween20 (Fisher) and either 5% BSA (Cell Signaling) or 5% non-fat dry milk (Oxoid). Blots were then incubated at room temperature for 1 hour with the relevant secondary antibody diluted in TBS supplemented with 0.1% Tween20 and 5% non-fat dry milk and then detected using ECL western Blotting Substrate (Pierce). Protein transfer, detection, and quantification using LiCor was performed as previously described11.
Tracing of Indels by Decomposition (TIDE) analysis
Genomic DNA was extracted from cell pellets using the DNeasy Blood & Tissue kit (Qiagen). An approximately 800-bp region centered on the sgRNA cut site was PCR amplified from 50 ng of genomic DNA using primer pair P5 (sgGFP) or P6 (sgRPA1-1) (Supplementary Table S1). Completed PCR reactions were treated with exonuclease I (NEB) according to the manufacturer’s protocol and then purified with the QIAquick PCR purification kit (Qiagen). 20 ng of purified PCR product was Sanger sequenced using an M13-forward primer and chromatograms were analyzed as previously described65.
Flow cytometry analysis for in vitro experiments
A549-SpCTRE or A549GFP-SpCTRE cells for flow cytometry analysis were collected using TrypLE Express (Thermo) according to the manufacturer’s protocol to preserve CD4T cell surface expression. Approximately 1 million cells were resuspended in PBS containing human TruStain FcX (Biolegend) and incubated at 4C for 10 minutes. Cells were stained with the α-CD4 antibody for 30 minutes at 4C, then washed twice with PBS and resuspended in PBS containing 1 μg/mL DAPI. All α-CD4 staining was performed with PE anti-human CD4 antibody clone RPA-T4 (Biolegend), unless otherwise stated. For analysis of GFP expression in CD4 negative cells (i.e. A549GFP-lentiCas9-Blast), approximately 1 million cells were washed twice with PBS and resuspended in PBS containing 1 μg/mL DAPI. All flow analysis was performed on an LSR Fortessa or LSR II (BD Biosciences).
CD4T extracellular domain analysis
A549 cells were co-transfected with a 50:50 mix of each CD4 domain variant and pLenti Neo CMV/TurboGFP using JetPrime transfection reagent (Polyplus) at a 2:1 JetPrime:DNA ratio. Media was changed 24 h after transfection and cells were selected with puromycin for 3 days. Flow cytometry analysis was performed as described above with PE anti-human CD4 antibody clones M-T466 (Miltenyi), RPA-T4, SK3, and OKT4 (Biolegend). Mean fluorescence intensity (MFI) of GFP positive cells was calculated using FlowJo. To compare antibody binding between CD4T and full-length CD4, A549 was transduced with pLenti CMV Puro/CD4 lentivirus and selected with puromycin for 3 days. This cell lines was mixed with A549GFP-SpCTRE and flow cytometry analysis was performed as described above using anti-human CD4 antibodies RPA-T4 (APC) and OKT4 (PE).
Magnetic cell separation (MACS)
A549-SpCTRE cells were spiked into an A549GFP cell suspension at an abundance of 5-20%. This mixture was incubated with CD4 microbeads (Miltenyi) and subjected to immunomagnetic separation with LS columns (Miltenyi) according to the manufacturer’s protocol. Eluted cells were cultured for 3 days to allow for dissociation of magnetic beads and then collected for flow cytometry analysis of CD4T purity, as described above. Results represent three independent biological replicates.
Transduction and enrichment of SpCTRE PDXs
Established PDX tumors were resected and dissociated to a single cell suspension using a gentleMACS tissue dissociator with a human tumor dissociation kit (Miltenyi). Red blood cells were lysed with ACK lysing buffer (Lonza) and mouse stroma cells were subsequently removed by negative immunomagnetic selection using a mouse cell depletion kit (Miltenyi). Cells were then transduced ex vivo with pSpCTRE lentivirus at a relative MOI of 1-6 (based on a functional titer performed in A549) in the presence of 48 μg/mL polybrene in a swinging bucket rotor for 30 min at 800 x g. After transduction, cells were washed twice with PBS to remove the lentivirus and polybrene and engrafted in a 50% Matrigel (BD) mixture into a single flank of 6-8 week old female NSG (Jax) or Nude (Envigo) mice. Resulting tumors were dissociated to a single cell suspension, red blood cells were lysed, and mouse stroma was removed as described above. Cells were prepared for flow cytometry sorting or analysis by resuspending 5-10 million cells (sorting) or 1 million cells (analysis) in FACS buffer (PBS with 2% FBS, 1X pen/strep and 1 mM EDTA) containing human TruStain FcX (Biolegend) and mouse TruStain FcX (anti-mouse CD16/32, Biolegend) and incubating at 4C for 10 minutes. Cells were stained with APC/Cy7 anti-human CD4 (clone RPA-T4, Biolegend) and AlexaFluor647 anti-mouse H-2Kd (clone SF1-1.1, Biolegend) for 30 minutes at 4C. Cells were washed twice with MACS buffer (PBS with 0.5% BSA) and then resuspended in sorting buffer (PBS with 0.5% BSA, 2.5 mM MgCl2, 0.5 mM CaCl2, 1 μg/mL DAPI and 100 U/mL DNaseI, NEB) and incubated for 30 minutes at room temperature before sorting. CD4T positive, H-2Kd negative, DAPI negative cells were collected using a FACSAria (BD Biosciences) and engrafted in a 50% Matrigel mixture into a single flank of a 6-8 week old female NSG or Nude mouse. Fluorescence minus one (FMO) controls were used to identify CD4T positive cells for all experiments66. Resulting tumors were collected and analyzed for CD4T expression as described above. If the tumor was less than 50% CD4T positive, it was sorted again as described and if the tumor was >=50% CD4T positive it was propagated or cryo preserved in RPMI supplemented with 10% FBS, 1x pen/strep, and 10% DMSO. SNP profiles of SpCTRE PDXs were compared to pre-sorted samples using qPCR genotyping of 8 informative SNPs to confirm identity.
In vivo competition assays in SpCTRE PDXs
Established SpCTRE PDX tumors were dissociated to a single cell suspension, red blood cells were lysed, and mouse stroma was removed as described above. Cells were then independently transduced ex vivo with sgTrack lentivirus at a relative MOI of 1 as described above. Each sgTrack transduced SpCTRE PDX was engrafted in a 50% Matrigel mixture into a single flank of one 6-8 week old female NSG or Nude mouse. Resulting tumors were dissociated to a single cell suspension, red blood cells were lysed, and mouse stroma was removed as described above. Cells containing control and test sgRNAs were mixed with an equal ratio of GFP and mCherry positive cells and engrafted in a 50% Matrigel mixture into a single flank of ten 6-8 week old female NSG or Nude mice. Once tumors reached ~100 mm3, mice were randomized to control or dox-treated groups, with dox-treated mice receiving 625 mg/kg doxycycline chow (Envigo). Tumors were collected once they reached 1000 mm3, dissociated to a single cell suspension, and prepared for flow cytometry analysis or sorting as described above. Indel analysis of sorted cells was performed as described above and fitness scores and log ratios were calculated as described in Extended Data Fig. 2b. The Wilcoxon rank sum statistic was used to test if the fitness scores in the dox-treated group were smaller than the fitness scores in the control group.
Recombinant AAV production and validation
Recombinant AAV2/6 pseudotyped virus was produced by the Boston Children’s Hospital Viral Core. PC9 cells transduced with lentiCas9-Blast and selected with blasticidin for 7 days were subsequently transduced with rAAV at an MOI of 2.3. Control and rAAV transduced PC9-lentiCas9-Blast cells were treated with 1 μM osimertinib (Selleck Chemicals) or DMSO (Corning) until they reached confluency, at which time cell pellets were collected for genomic analysis, described below. Osimertinib resistance was confirmed in cells previously transduced with rAAV and selected with osimertinib by seeding 3000 viable cells per well in 100 μL/well media containing a dilution series of osimertinib. Viability was assayed 72 h after plating using CellTiter-Glo (Promega).
In vivo homology-directed repair in MSK-LX29
Crizotinib (LC Laboratories), osimertinib (LC Laboratories), or the combination were formulated in 0.5% hydroxypropyl methylcellulose (HPMC) and given by oral gavage at 25 mg/kg daily for 5 days per week. An MSK-LX29-SpCTRE tumor from a mouse fed with dox chow for 4 weeks prior to dissociation was incubated with the rAAV at an MOI of 1.6 million genomic copies/cell for 1 hour at 37C. Cells were washed twice with PBS and then engrafted in a 50% Matrigel mixture into a single flank of 20 6-8 week old female NSG mice. All mice were administered dox chow at the time of engraftment and treatment groups were randomized to 4 (vehicle groups) or 5 (C/O groups) mice once tumors reached 100 mm3. Tumors were collected once they reached 1000-1500 mm3 and genomic analysis was performed as described below.
Genomic analysis of EGFR homology-directed repair
Genomic DNA was purified from dissociated tumors or cell pellets using the Gentra Puregene Cell kit (Qiagen). A 1,122 bp amplicon, which spans outside the rAAV homology arms to ensure amplification occurs from genomic DNA, was PCR amplified from 200 μg genomic DNA using primer pair P7. The PCR product was then digested with exonuclease I to remove excess primers, column purified using the QIAquick PCR purification kit (Qiagen), and 100 μg was used as the template for a second PCR with primer pair P8 to amplify a 273 bp region centered on the repair site. The second round PCR product was column purified and paired-end Next-Gen sequencing (NGS) was performed by the CCIB DNA Core Facility at Massachusetts General Hospital (Cambridge, MA). NGS sequencing was analyzed using the R statistical computing environment to determine the proportion of reads that underwent rAAV-mediated homology-directed repair.
Statistics & Reproducibility
No statistical method was used to determine sample sizes due to lack of precise information on variability and effect size for each experiment. No data points were excluded from the analyses. Randomization of animals for drug treatment experiments, dosing, and tumor measurement was performed by a technician who was blinded to the experimental hypothesis.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Unmodified gel images for Figures 1, 2, 3, 5, have been provided as Source Data files (Unmodified_Gels_Fig1 - Unmodified_Gels_Fig4). Numerical source data for Main figures 1-6 and Extended Data Figures 1-4 have been provided as Source Data files (SourceData_Fig1 - SourceData_Fig6 and SourceData_ExtendedData_Fig1 - SourceData_ExtendedData_Fig4). All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code Availability
The computer code that supports the findings of this study is available from the corresponding author upon request.
Extended Data
Extended Data Fig. 1 ∣. Truncated CD4T is a size-efficient selectable marker for flow cytometry and immunomagnetic selection.
a, Domain structure of wildtype human CD4 and truncated constructs CD4ΔD2-4;ΔIC and CD4ΔD3-4;ΔIC (hereafter CD4T), where the indicated deletions are replaced by flexible linkers. Heatmap depicts flow cytometry staining intensity of commercially available α-CD4 antibodies, which target the indicated extracellular domains of CD4, to the indicated CD4 constructs. S, signal peptide; TM, transmembrane domain; IC, intracellular domain; MFI, mean fluorescence intensity b, Flow cytometry analysis of A549 cells expressing CD4T and enriched by fluorescence activated cell sorting (FACS). Data is representative of three independent experiments with similar results, c, Immunomagnetic enrichment of A549 cells expressing CD4T, with the indicated enrichment factors (above). Mean percent CD4 positive is displayed for n = 3 independent cell culture replicates. Data is representative of two independent experiments with similar results. Error bars are SD. d, α-CD4 staining strategy with domain D1 and D3 targeting antibodies to differentiate CD4T and full-length CD4 using flow cytometry. A mixture of GFP-negative cells expressing full length CD4 and GFP-positive cells expressing CD4T were stained with the indicated α-CD4 antibody clones. Double positive cells were exclusively GFP-negative (expressing full length CD4) and cells single positive for the domain D1 targeting α-CD4 antibody were exclusively GFP-positive (expressing CD4T). Data is representative of n = 3 independent cell culture replicates from a single experiment. Data for experiments in panels b-d are available as source data (SourceData_ExtendedData_Fig1).
Extended Data Fig. 2 ∣. Competition assay utilizing sgTrack vectors effectively determines fitness effects of gene disruption.
a, Competition assay of sgRPA1-2 in A549 with no Cas9, constitutive Cas9 expression from lentiCas9-Blast, or dox-inducible Cas9 expression from pSpCTRE (left, middle and right panels, respectively). Competition assays were performed with (solid) or without (dashed) dox. Mean fluorescence expression is displayed for n = 3 independent cell culture replicates from a single experiment. Error bars are SD, with SD < plotting character not drawn, b, Formulas for fitness score and log ratio (left). Comparison of fitness score and log ratio analysis methods (right). Mean fitness score or mean log ratio are displayed for n=3 independent cell culture replicates from a single experiment. Error bars are SD, with SD < plotting character not drawn, c, In vitro competition assays in A549 with sgRPA1-1, sgKRAS-1, and sgKRAS-2 (left, middle and right panels, respectively). Log ratio calculations and line assignments are as described in panel b. Mean log ratio is displayed for n = 3 independent cell culture replicates from a single experiment. Error bars are SD, with SD< plotting character not drawn. Data for experiments in panels a-c are available as source data (SourceData_ExtendedData_Fig2).
Extended Data Fig. 3 ∣. CD4T induction in SpCTRE PDXs and gating strategy for in vivo competition assay analysis.
a, Representative flow cytometry analysis of CD4T induction for the competition assay with sgRPA1-1 in MSK-LX369. Data is representative of n=5 mice, b, Flow cytometry analysis of sgRPA1-1 (mCherry) and sgNTC (GFP) in tumors from panel a. Cells are first gated on CD4T positive or CD4T induced populations as shown in panel a. Data is representative of n = 5 mice, c, Log ratio of MSK-LX369 competition assays with the indicated sgRNAs for control or dox-treated mice gated on either CD4T positive or CD4T induced populations. Mean log ratio is displayed for n=5 mice. Error bars are SD, with SD< plotting character not drawn. A two-sided Wilcoxon rank sum test was used to determine statistical significance, d, Percent of CD4T induced cells from dox-treated mice for competition assays with the indicated sgRNAs in the SpCTRE PDXs MSK-LX369, JHU-LX55a, and MSK-LX29 (left, middle, and right panels, respectively). Mean percent CD4 positive cells is displayed for n = 4 or n=5 mice as indicated. Error bars are SD. e, Mean time to sacrifice in days for parental and SpCTRE PDXs for n = 2 to n = 46 mice. Error bars are SD, with SD < plotting character not drawn. Data for experiments in panels a-e are available as source data (SourceData_ExtendedData_Fig3).
Extended Data Fig. 4 ∣. Evaluation of acquired osimertinib resistance driven by rAAV-mediated homology-directed repair in PC9 cells in vitro.
a, Number of PC9 cells under the indicated conditions for n = 2 independent cell culture replicates from a single experiment. Error bars are SD, with SD< plotting character not drawn, b, Osimertinib dose response curves of PC9 cells from the rAAV, osimertinib or control, DMSO treatment arms from panel a. Mean relative luminescence is displayed for n = 3 independent cell culture replicates from a single experiment. Error bars are SD, with SD< plotting character not drawn, c, d, Sequencing analysis of PC9 cells from the rAAV, osimertinib group at (c)t = 7 and (d) t = 35 days for n = 2 biologically independent replicates from panel a. Data for experiments in panels a-d are available as source data (SourceData_ExtendedData_Fig4).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the MSKCC Flow Cytometry core for their technical assistance, members of the Antitumor Assessment Core Facility for assistance with in vivo experiments, and all members of the Rudin lab for critical comments. We thank the Center for Computational and Integrative Biology (CCIB) at Massachusetts General Hospital for the use of the CCIB DNA Core Facility (Cambridge, MA) and the Boston Children’s Hospital Viral Core (Core grant 5P30EY012196, NEI). Supported by NIH U01 CA199215, U24 CA213274, P01 CA129243, and P30 CA008748 (CMR and JTP).
Footnotes
COMPETING INTERESTS
C.M.R. has consulted regarding oncology drug development with AbbVie, Amgen, Ascentage, AstraZeneca, BMS, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Loxo, and PharmaMar, and is on the scientific advisory boards of Elucida, Bridge, and Harpoon. All other authors have no competing interests.
REFERENCES
- 1.Townsend EC et al. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell 29, 574–586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krepler C et al. A Comprehensive Patient-Derived Xenograft Collection Representing the Heterogeneity of Melanoma. Cell Rep 21, 1953–1967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruna A et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell (2016). doi: 10.1016/j.cell.2016.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drapkin BJ et al. Genomic and Functional Fidelity of Small Cell Lung Cancer Patient-Derived Xenografts. Cancer Discovery 8, 600–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel VC et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 69, 3364–3373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirier JT et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene (2015). doi: 10.1038/onc.2015.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo S et al. Molecular Pathology of Patient Tumors, Patient-Derived Xenografts, and Cancer Cell Lines. Cancer Res. 76, 4619–4626 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Puca L et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat Commun 9, 2404 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beshiri ML et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res 24, 4332–4345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao H et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med (2015). doi: 10.1038/nm.3954 [DOI] [PubMed] [Google Scholar]
- 11.Gardner EE et al. Chemosensitive Relapse in Small Cell Lung Cancer Proceeds through an EZH2-SLFN11 Axis. Cancer Cell 31, 286–299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lallo A, Schenk MW, Frese KK, Blackhall F & Dive C Circulating tumor cells and CDX models as a tool for preclinical drug development. Transl Lung Cancer Res 6, 397–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sánchez-Rivera FJ & Jacks T Applications of the CRISPR–Cas9 system in cancer biology. Nat. Rev. Cancer 15, 387–395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura A & Dow LE Modeling Cancer in the CRISPR Era. Annu. Rev. Cancer Biol 2, 111–131 (2018). [Google Scholar]
- 15.Komor AC, Badran AH & Liu DR CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 168, 20–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tschaharganeh DF, Lowe SW, Garippa RJ & Livshits G Using CRISPR/Cas to study gene function and model disease in vivo. FEBS J. 283, 3194–3203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siolas D & Hannon GJ Patient-Derived Tumor Xenografts: Transforming Clinical Samples into Mouse Models. Cancer Res. 73, 5315–5319 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meca-Cortés O et al. CRISPR/Cas9-Mediated Knockin Application in Cell Therapy: A Non-viral Procedure for Bystander Treatment of Glioma in Mice. Mol Ther Nucleic Acids 8, 395–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ablain J, Durand EM, Yang S, Zhou Y & Zon LI A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev. Cell 32, 756–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabadi AM, Ousterout DG, Hilton IB & Gersbach CA Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Research 42, e147–e147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dow LE et al. Conditional Reverse Tet-Transactivator Mouse Strains for the Efficient Induction of TRE-Regulated Transgenes in Mice. PLoS ONE 9, e95236–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dow LE et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 33, 390–394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangopadhyay SA et al. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 58, 234–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González F et al. An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Stem Cell 15, 215–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu M et al. Conditional gene knockout and reconstitution in human iPSCs with an inducible Cas9 system. Stem Cell Research 29, 6–14 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Cao J et al. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Research 60, gkw660–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verma N et al. TET proteins safeguard bivalent promoters from de novo methylation in human embryonic stem cells. Nat. Genet 50, 83–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaines P & Wojchowski DM pIRES-CD4t, a dicistronic expression vector for MACS- or FACS-based selection of transfected cells. Biotech. 26, 683–688 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Bonini C et al. HSV-TK gene transfer into donor lymphocytes for control of allogeneic graft-versus-leukemia. Science 276, 1719–1724 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Wang X et al. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 118, 1255–1263 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirenallur-Shanthappa DK, Ramírez JA & Iritani BM Chapter 5 - Immunodeficient Mice: The Backbone of Patient-Derived Tumor Xenograft Models. Patient Derived Tumor Xenograft Models 57–73 (Elsevier Inc., 2016). doi: 10.1016/B978-0-12-804010-2.00005-9 [DOI] [Google Scholar]
- 32.Koretzky GA Multiple roles of CD4 and CD8 in T cell activation. J. Immunol 185, 2643–2644 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Loew R, Heinz N, Hampf M, Bujard H & Gossen M Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol 10, 81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou X, Vink M, Klaver B, Berkhout B & Das AT Optimization of the Tet-On system for regulated gene expression through viral evolution. Gene Ther 13, 1382–1390 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Kim S, Bae T, Hwang J & Kim J-S Rescue of high-specificity Cas9 variants using sgRNAs with matched 5’ nucleotides. Genome Biol. 18, 218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zafra MP et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat Biotechnol 36, 888–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinz N et al. Retroviral and Transposon-Based Tet-Regulated All-In-One Vectors with Reduced Background Expression and Improved Dynamic Range. Human Gene Therapy 22, 166–176 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Kumar M, Keller B, Makalou N & Sutton RE Systematic Determination of the Packaging Limit of Lentiviral Vectors. Human Gene Therapy 1893–1905 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helling B et al. A specific CD4 epitope bound by tregalizumab mediates activation of regulatory T cells by a unique signaling pathway. Immunol. Cell Biol 93, 396–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cawthorne C, Swindell R, Stratford IJ, Dive C & Welman A Comparison of doxycycline delivery methods for Tet-inducible gene expression in a subcutaneous xenograft model. J Biomol Tech 18, 120–123 (2007). [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Z, Bozzella M, Seluanov A & Gorbunova V Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 7, 1765–1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieber MR The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem 79, 181–211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daniel VC et al. A Primary Xenograft Model of Small-Cell Lung Cancer Reveals Irreversible Changes in Gene Expression Imposed by Culture In vitro. Cancer Res. 69, 3364–3373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dow LE et al. A pipeline for the generation of shRNA transgenic mice. Nat Protoc 7, 374–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beard C, Hochedlinger K, Plath K, Wutz A & Jaenisch R Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Charles JP et al. Monitoring the dynamics of clonal tumour evolution in vivo using secreted luciferases. Nat Commun 5, 3981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rijn S et al. Functional multiplex reporter assay using tagged Gaussia luciferase. Sci Rep 3, 1046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wroblewska A et al. Protein Barcodes Enable High-Dimensional Single-Cell CRISPR Screens. Cell 175, 1141–1155.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poirier JT CRISPR Libraries and Screening. Prog Mol Biol Transl Sci 152, 69–82 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Winters IP et al. Multiplexed in vivo homology-directed repair and tumor barcoding enables parallel quantification of Kras variant oncogenicity. Nat Commun 8, 2053 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kleinstiver BP et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casini A et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nature Publishing Group 36, 265–271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu JH et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sastry L, Johnson T, Hobson MJ, Smucker B & Cornetta K Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Ther 9, 1155–1162 (2002). [DOI] [PubMed] [Google Scholar]
- 58.Zhang B et al. The significance of controlled conditions in lentiviral vector titration and in the use of multiplicity of infection (MOI) for predicting gene transfer events. Genet Vaccines Ther 2, 6 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang M et al. Humanized mice in studying efficacy and mechanisms of PD-1-targeted cancer immunotherapy. FASEB J. 32, 1537–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 60.Gray JT & Zolotukhin S Design and construction of functional AAV vectors. Methods Mol. Biol 807, 25–46 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Campeau E et al. A Versatile Viral System for Expression and Depletion of Proteins in Mammalian Cells. PLoS ONE 4, e6529–18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang T et al. Identification and characterization of essential genes in the human genome. Science 350, 1096–1101 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol (2016). doi: 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brinkman EK, Chen T, Amendola M & van Steensel B Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research 42, e168–e168 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cossarizza A et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol 47, 1584–1797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unmodified gel images for Figures 1, 2, 3, 5, have been provided as Source Data files (Unmodified_Gels_Fig1 - Unmodified_Gels_Fig4). Numerical source data for Main figures 1-6 and Extended Data Figures 1-4 have been provided as Source Data files (SourceData_Fig1 - SourceData_Fig6 and SourceData_ExtendedData_Fig1 - SourceData_ExtendedData_Fig4). All other data supporting the findings of this study are available from the corresponding authors on reasonable request.