Abstract
BACKGROUND.
Polygenic risk scores comprising established susceptibility variants have shown to be informative classifiers for several complex diseases including prostate cancer. For prostate cancer it is unknown if inclusion of genetic markers that have so far not been associated with prostate cancer risk at a genome-wide significant level will improve disease prediction.
METHODS.
We built polygenic risk scores in a large training set comprising over 25,000 individuals. Initially 65 established prostate cancer susceptibility variants were selected. After LD pruning additional variants were prioritized based on their association with prostate cancer. Six-fold cross validation was performed to assess genetic risk scores and optimize the number of additional variants to be included. The final model was evaluated in an independent study population including 1,370 cases and 1,239 controls.
RESULTS.
The polygenic risk score with 65 established susceptibility variants provided an area under the curve (AUC) of 0.67. Adding an additional 68 novel variants significantly increased the AUC to 0.68 (P = 0.0012) and the net reclassification index with 0.21 (P = 8.5 E-08). All novel variants were located in genomic regions established as associated with prostate cancer risk.
CONCLUSIONS.
Inclusion of additional genetic variants from established prostate cancer susceptibility regions improves disease prediction.
Keywords: prostate cancer, polygenic risk score, risk prediction
INTRODUCTION
After extensive genotyping efforts in several large international consortia a total of 100 prostate cancer susceptibility loci have been identified to date [1]. Overall, these established single nucleotide polymorphisms (SNPs) have been estimated to explain 33% of the familial risk of prostate cancer. Individually, these SNPs accounted for a modest part of disease risk and their ability to individually discriminate between prostate cancer cases and unaffected controls was limited. However, by combining established risk SNPs in polygenic risk scores the predictive performance of prostate cancer risk has proven to be substantial [2–6]. Eeles and coworkers showed, by using a polygenic risk score consisting of 68 established prostate cancer risk SNPs, that men in the top 1% of the risk distribution have more than four-fold increased risk for prostate cancer compared with those with average risk [7]. This effect size is similar to that conferred by deleterious BRCA1 [8,9] and BRCA2 [10–12] mutation carriers, undergoing targeted screening in clinical trials [13]. Moreover, a polygenic risk score including 35 established prostate cancer risk SNPs has recently shown to decrease the number of biopsies by 23% at a cost of 3% fewer cases detected in a Swedish cohort of men that underwent biopsy of the prostate [14].
Several studies of complex diseases [15] have shown that by including SNPs that do not individually achieve genome-wide significant levels, an increasing proportion of heritability can be explained. Regarding prostate cancer, two studies that applied this approach have provided inconsistent results; in one study a modest increase in predictive performance was observed by relaxing inclusion thresholds of available SNPs [16], while the other study reported no significant improvement in prediction capacity [17]. These observations are in line with results from a recent methodological study concluding that power (i.e., sample size) is of importance in order to improve the predictive capacity of polygenic risk scores [18].
The aim of this study was to explore the possibility to improve predictive performance of a polygenic risk score, comprised only of established prostate cancer risk SNPs, by including additional genetic markers that have so far not been associated with prostate cancer risk at a genome-wide significant level. Utilizing over 13,000 cases and 14,000 controls from the PRACTICAL consortium with genotypes available from a collection of approximately 83,000 SNPs distributed across the genome, we developed a cross-validated polygenic score and assessed its predictive performance in an independent population.
MATERIAL AND METHODS
Study Subjects - PRACTICAL
The international prostate cancer genetics consortium (PRACTICAL) is a part of the Collaborative Oncological Gene-environment Study (COGS [19]) with three other cancer genetics consortia (breast, ovarian, and BRCA1/2 mutation carriers). The initial aim of the COGS project was to design an Illumina Custom Infimum array (iCOGS), where each consortium nominated SNPs in three categories: markers indicated by genome-wide association studies (GWAS), fine-mapping of already established susceptibility regions, and candidate genes. PRACTICAL’s part of COGS has been described in detail previously [7]. In this study we included all 85,278 SNPs (out of totally 211,155 on the iCOGS chip) that were nominated by PRACTICAL, specifically chosen to be relevant for prostate cancer. The main component of these markers (74,001 SNPs) was selected based on a meta-analysis of four prostate cancer GWAS. An additional 13,739 SNPs were selected to fine-map already established prostate cancer susceptibility regions and 1,398 SNPs were selected to explore candidate genes in key biological pathways (including hormone metabolism, HOX class of genes, the cell cycle and DNA repair). After quality control assessment 82,895 SNPs remained for analysis in the present study.
The iCOGS chip was genotyped in approximately 50,000 subjects in 32 sub-populations of PRACTICAL. Most of these studies represent either population-based cases-control studies or nested case-control studies. To avoid overfitting, we excluded 5,293 subjects that were previously used in GWAS analysis for nomination of SNPs to the iCOGS chip. Furthermore, we restricted this study to only include men of European ancestry below 70 years-old at time of study enrollment. The age limit was selected to create a study population that could potentially benefit from prostate cancer screening. Finally, each included sub-study that did not contribute both prostate cancer cases and controls was excluded, resulting in a final study sample comprising 13,532 cases and 14,242 controls from 21 different studies (Table I).
TABLE I.
Study Populations From PRACTICAL Included in Risk Prediction Analysis
| Study | Country | No. cases | No. controls |
|---|---|---|---|
| Training data - Internal validation samples | |||
| CPCS1 | Denmark | 454 | 2122 |
| CPCS2 | Denmark | 212 | 621 |
| FHCRC | USA | 696 | 662 |
| MAYO | USA | 584 | 348 |
| MCCS | Australia | 1525 | 1179 |
| QLD | Australia | 156 | 60 |
| ProtecT | UK | 1509 | 1473 |
| STHM1 | Sweden | 1413 | 1522 |
| UKGPCS | UK | 2351 | 2184 |
| Training data - Other samples | |||
| CAPS | Sweden | 299 | 147 |
| EPIC | EU | 586 | 1001 |
| EPIC-Norfolk | UK | 177 | 361 |
| ESTHER | Germany | 255 | 265 |
| MEC | USA | 308 | 298 |
| MOFFITT | USA | 297 | 81 |
| PCMUS | Bulgaria | 84 | 92 |
| Poland | Poland | 283 | 259 |
| ProMPT | UK | 109 | 2 |
| ULM | Germany | 495 | 176 |
| UTAH | USA | 360 | 150 |
| Total (training data) | 12153 | 13003 | |
| External test sample | |||
| SEARCH | UK | 1370 | 1239 |
| Total | 13532 | 14242 | |
Prediction Model
The prediction model that we aimed to optimize for discrimination between prostate cancer cases and controls was a logistic regression with two polygenic risk scores (sum of risk alleles, weighted by the log OR of each SNP), one containing 65 established risk SNPs and the second including novel markers. The development of this model was exclusively performed in a training set and thereafter evaluated in an independent test set. The UK Study of Epidemiology and Risk factors in Cancer Heredity (SEARCH) with 1,370 cases and 1,239 controls was chosen as the external test set, since it is a population-based study and has a similar structure as our target population, i.e. men aged <70 years with European ancestry. The rest of the data (i.e., excluding SEARCH) with 12,153 cases and 13,003 controls were used to train the model.
The primary goals with the training set were to determine the optimal number of novel SNPs that should be added to the model, aside from already known risk markers, in order to optimize the predictive capacity, and to determine weights (log odds ratios) for SNPs included in the two polygenic risk scores. To avoid over fitting when the optimal genetic profile for risk prediction was developed we used six large sub-populations in the training data for internal validation: Australia (MCCS + QLD), Denmark (CPCS1 + 2), PROTECT, UKGPCS, STHM1, and USA (FHCRC + MAYO). The model development procedure was implemented as follows:
Individual associations of each SNP with prostate cancer were assessed in logistic regression, assuming a log-additive genetic model and adjusting for population stratification (six principal components), setting one internal validation population aside.
Linkage Disequilibrium (LD) - based pruning was performed, i.e., one SNP with lowest P-value (in step1) was selected from each LD-block (r2> = 0.2).
A genetic risk score was created with 65 established risk SNPs (54 directly genotyped and 11 surrogates with r2 > 0.8). Log odds ratios obtained in step 1 were used as weights.
From the LD-pruned list, novel SNPs were added one by one, ordered by P-value (starting with lowest), into a second risk score. The two risk scores were fitted as covariates in a logistic regression model.
Every time a SNP was added to the second risk score in step 4, the prediction performance of the model was assessed in the internal validation population that was set aside, by the area under the receiver operator characteristic (ROC) curve (AUC).
We iterated between steps 1–5 six times, each time setting aside different validation data.
Among the six models (one per excluded internal validation set) in the training data with the same number of added SNPs to the polygenic risk score, the mean value of the predictions was calculated. The maximum of this average AUC was used to determine the optimal number of top ranked SNPs to include in the final prediction model. In order to construct the final risk score for this model, weights were obtained and SNPs were ranked according to P-values, using results from logistic regressions (as before, adjusting for six principal components on an LD-pruned set of SNPs) on the complete training set.
In the external test data (SEARCH), the association between prostate cancer and the two risk scores (both treated as linear predictors) was evaluated using a logistic regression model. Both improvement in AUC (DeLong nonparametric test [20]) and the continuous version of Net Reclassification Index (NRI) [21,22] were used to evaluate the final prediction model in the test data, when the novel SNP risk score was added to the established risk score. Finally, the association between prostate cancer and ordered categories of the final polygenic risk score (containing both established and novel SNPs together) was assessed with logistic regression.
RESULTS
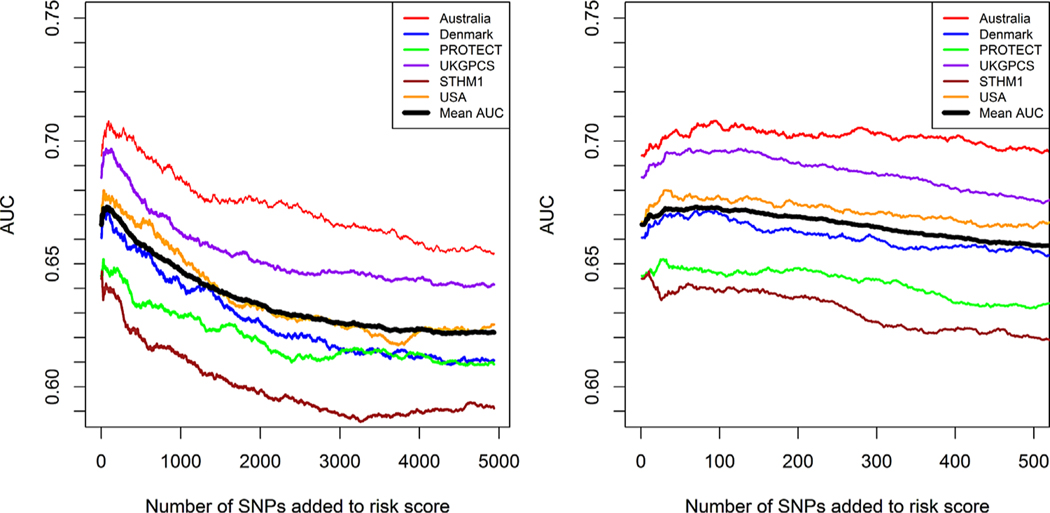
In the initial training step, performing cross-validated SNP selection, substantial variation in the predictive performance of the polygenic scores was observed between the different internal validation study samples (Fig. 1). In the model with the risk score containing only the 65 established risk SNPs, AUC values ranged between 0.64 and 0.69 for the different populations. In general we observed improved predictive performance, with a maximum increase in AUC of approximately 0.01, when additional SNPs were added to the model. Using the mean AUC of the six internal validation samples, the optimal predictive capacity in the training data was obtained when 68 SNPs were added to the model (Fig. 1, Supplementary Table). Of note, each of the additional 68 selected SNPs was located in a genomic region already established as associated with prostate cancer risk.
Fig. 1.
Prediction performance in different study populations included in the training set. The left plot shows prediction performance when up to 5,000 novel SNPs are added to the prediction model. The right plot is zoomed in on the part where the predictions increase. The black line corresponds to the mean AUC.
In the external independent test sample (SEARCH) strong statistical association was observed both between the genetic score composed of the 65 established SNPs (P = 5.0E-16) and the score composed of the additional 68 SNPs (P = 2.5E-10). The AUC increased significantly (P = 0.0012) from 0.67, using only established risk SNPs, to 0.68 when the optimal genetic risk profile derived from the training set was added (Table II). Furthermore, the optimal model including the 68 additional SNPs showed a significantly improved NRI, both in cases and controls separately and overall, compared with the model with only established risk SNPs (Table II). Stratifying individuals by their genetic profile resulted in a linear trend of increasing ORs (Table III). Compared with the reference category (40–60%), individuals with lowest risk scores (lowest 5% percentile) had 84% decreased risk of prostate cancer, while individuals with highest risk scores (highest 5% percentile) had a four-fold increased relative risk of prostate cancer.
TABLE II.
Contrasting an Established Risk Score with an Addition of 68 Prostate Cancer Associated Genetic Variants
| Cases | Controls | Established | Established + top68 | P | |
|---|---|---|---|---|---|
| AUC | 1370 | 1239 | 0.67(0.65–0.69) | 0.68(0.66–0.7) | 0.0012 |
| NRI | Reclassified upwards | Reclassified downwards | NRI (95% CI) | P | |
| Cases | 0.55 | 0.45 | 0.09(0.04–0.15) | 0.00051 | |
| Controls | 0.44 | 0.56 | 0.12(0.06–0.17) | 4.32E-05 | |
| All | - | - | 0.21(0.13–0.29) | 8.46E-08 |
TABLE III.
Risk Distribution in Different Percentiles of a Genetic Risk Score, Containing Both 65 Established and 68 Novel SNPs, Evaluated in an External Test Sample
| Percentiles (%) | OR (95% CI) | P value |
|---|---|---|
| 0–5 | 0.16 (0.1,0.27) | 4.43e-12 |
| 5–10 | 0.52 (0.35,0.77) | 0.0012 |
| 10–20 | 0.41 (0.3,0.56) | 2.85e-08 |
| 20–30 | 0.82 (0.61,1.1) | 0.18 |
| 30–40 | 0.92 (0.69,1.24) | 0.60 |
| 40–60 | 1.00(ref) | – |
| 60–70 | 1.36 (1.01,1.84) | 0.046 |
| 70–80 | 1.6 (1.18,2.16) | 0.0026 |
| 80–90 | 2.58 (1.86,3.56) | 9.66e-09 |
| 90–95 | 2.37 (1.56,3.6) | 5.07e-05 |
| 95–100 | 4.00 (2.51,6.39) | 6.5e-09 |
DISCUSSION
Individual prostate cancer risk profiling, using polygenic risk scores, has the potential to facilitate risk stratification in targeted screening and prevention programs. In this study we optimized a polygenic risk score starting with a set of established prostate cancer susceptibility variants and then included additional variants until maximum discrimination was achieved. We observed substantial variations in predictive capacity between our training populations with AUC values ranging between 0.65 and 0.71. This may suggest that the utility of the derived risk score varies between different populations. However, the diverse ascertainment schemes applied in the included studies most likely explain the observed difference in discrimination between the study populations. Final assessment of the derived polygenic risk model was performed in the SEARCH population. SEARCH is a population-based case-control study ascertained in UK utilizing regional cancer- and population registers. In this population the optimal model gave an AUC of 0.68 and we argue that this is a representative assessment that can be generalized to other European populations.
Interestingly, a similar trend in AUC values could be observed when novel SNPs were added to the prediction models when different validation samples (except for STHM1) in the training data were used. An initial increase in the AUC, peaking at around 68 added markers, followed by a decline when more markers were added. Furthermore, the 68 top associated novel SNPs in the training sample significantly improved predictions in the independent SEARCH study. Thus, the iCOGS chip is enriched with additional genetic variants that improve prostate cancer risk prediction in addition to already established susceptibility variants. Of note, all 68 non-established variants included in the optimal prediction model were located in already established prostate cancer susceptibility regions. This suggests that fine-mapping of established regions may reveal additional genetic variants that are independently associated with prostate cancer risk.
It has been argued that a prediction model should at minimum have an AUC of at least 0.75 in order to have any benefits in a screening context [23]. The rather modest improvement (0.01 increase in AUC) in predictions that we demonstrated by adding 68 novel SNPs, suggests that there is noise (false positives) in our data that needs to be removed in order to improve the predictive capacity. In addition, it is most likely that the iCOGS chip contains genetic markers further down in the P-value ranked list that are truly associated with prostate cancer risk that could further improve the prediction capacity if included in the polygenic score. Therefore, future efforts assessing even larger study populations with genome-wide data hold promise to further improve polygenic risk prediction of prostate cancer. However, the risk stratification achieved in our independent test sample (SEARCH) already shows that we can identify sub-groups in the tails of the risk score distributions with extremely high and low risk of prostate cancer, which may be useful in a prostate cancer screening setting. Furthermore, by combining a genetic risk score with established biomarkers, such as PSA level and family history of disease, would probably improve predictions substantially [5,24,25]. Prostate cancer intervention studies addressing this possibility are warranted.
A strength with our study is the large sample size of the training data set, which is known to be an important factor regarding the accuracy of genetic scores for individual prediction [24]. We found that the risk score containing 68 novel SNPs was significantly associated with prostate cancer in our external test sample (SEARCH), adjusting for already known markers. In a similar study [17] that used GWAS data from a much smaller study population no improvement in risk prediction was achieved by adding additional variants to established markers, probably due to limited statistical power [18]. Another advantage with our study was the enrichment of prostate cancer risk variants on the iCOGS chip. Most variants on the chip were selected based on previous prostate cancer GWAS studies, therefore we can expect a larger proportion of truly associated variants on the iCOGS chip compared to a general genome-wide array.
CONCLUSION
In summary, we have derived a polygenic prediction model that improves discrimination between prostate cancer cases and controls. Further studies exploring the utility of polygenic risk stratification in screening and prevention programs are warranted.
Supplementary Material
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
Full list of participants of The PRACTICAL Consortium is provided in the Supplementary material.
REFERENCES
- 1.Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z, Saunders E, Leongamornlert D, Lindstrom S, Jugurnauth-Little S, Dadaev T, Tymrakiewicz M, Stram DO, Rand K, Wan P, Stram A, Sheng X, Pooler LC, Park K, Xia L, Tyrer J, Kolonel LN, Le Marchand L, Hoover RN, Machiela MJ, Yeager M, Burdette L, Chung CC, Hutchinson A, Yu K, Goh C, Ahmed M, Govindasami K, Guy M, Tammela TL, Auvinen A, Wahlfors T, Schleutker J, Visakorpi T, Leinonen KA, Xu J, Aly M, Donovan J, Travis RC, Key TJ, Siddiq A, Canzian F, Khaw KT, Takahashi A, Kubo M, Pharoah P, Pashayan N, Weischer M, Nordestgaard BG, Nielsen SF, Klarskov P, Roder MA, Iversen P, Thibodeau SN, McDonnell SK, Schaid DJ, Stanford JL, Kolb S, Holt S, Knudsen B, Coll AH, Gapstur SM, Diver WR, Stevens VL, Maier C, Luedeke M, Herkommer K, Rinckleb AE, Strom SS, Pettaway C, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, Tay E, Truelove A, Niwa S, Chokkalingam AP, Cannon-Albright L, Cybulski C, Wokolorczyk D, Kluzniak W, Park J, Sellers T, Lin HY, Isaacs WB, Partin AW, Brenner H, Dieffenbach AK, Stegmaier C, Chen C, Giovannucci EL, Ma J, Stampfer M, Penny KL, Mucci L, John EM, Ingles SA, Kittles RA, Murphy AB, Pandha H, Michael A, Kierzek AM, Blot W, Signorello LB, Zheng W, Albanes D, Virtamo J, Weinstein S. Nemesure B, Carpten J, Leske C, Wu SY, Hennis A, Kibel AS, Rybicki BA, Neslund-Dudas C, Hsing AW, Chu L, Goodman PJ, Klein EA, Zheng SL, Batra J, Clements J, Spurdle A, Teixeira MR, Paulo P, Maia S, Slavov C, Kaneva R, Mitev V, Witte JS, Casey G, Gillanders EM, Seminara D, Riboli E, Hamdy FC, Coetzee GA, Li Q, Freedman ML, Hunter DJ, Muir K, Gronberg H, Neal DE, Southey M, Giles GG, Severi G, Breast, Prostate Cancer Cohort C, Consortium P, Consortium C, Consortium G-OE, Cook MB, Nakagawa H, Wiklund F, Kraft P, Chanock SJ, Henderson BE, Easton DF, Eeles RA, Haiman CA . A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet 2014;46(10):1103–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Balter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Gronberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med 2008;358-(9):910–919. [DOI] [PubMed] [Google Scholar]
- 3.Johansson M, Holmstrom B, Hinchliffe SR, Bergh A, Stenman UH, Hallmans G, Wiklund F, Stattin P. Combining 33 genetic variants with prostate-specific antigen for prediction of prostate cancer: Longitudinal study. Int J Cancer 2012;130(1):129–137. [DOI] [PubMed] [Google Scholar]
- 4.Sun J, Kader AK, Hsu FC, Kim ST, Zhu Y, Turner AR, Jin T, Zhang Z, Adolfsson J, Wiklund F, Zheng SL, Isaacs WB, Gronberg H, Xu J. Inherited genetic markers discovered to date are able to identify a significant number of men at considerably elevated risk for prostate cancer. Prostate 2011;71(4)):421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindstrom S, Schumacher FR, Cox D, Travis RC, Albanes D, Allen NE, Andriole G, Berndt SI, Boeing H, Bueno-de-Mesquita HB, Crawford ED, Diver WR, Gaziano JM, Giles GG, Giovannucci E, Gonzalez CA, Henderson B, Hunter DJ, Johansson M, Kolonel LN, Ma J, Le Marchand L, Pala V, Stampfer M, Stram DO, Thun MJ, Tjonneland A, Trichopoulos D, Virtamo J, Weinstein SJ, Willett WC, Yeager M, Hayes RB, Severi G, Haiman CA, Chanock SJ, Kraft P. Common genetic variants in prostate cancer risk prediction-results from the NCI Breast and Prostate Cancer Cohort Consortium (BPC3). Cancer Epidemiol Biomarkers Prev 2012;21(3):437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amin Al Olama A, Benlloch S, Antoniou AC, Giles GG, Severi G, Neal D, Hamdy FC, Donovan JL, Muir K, Schleutker J, Henderson BE, Haiman C, Schumacher FR, Pashayan N, Pharoah PD, Ostrander EA, Stanford JL, Batra J, Clements JA, Chambers SK, Weischer M, Nordestgaard BG, Ingles SA, Sorensen KD, Orntoft TF, Park JY, Cybulski C, Maier C, Doerk T, Dickinson JL, Cannon-Albright L, Brenner H, Rebbeck TR, Zeigler-Johnson C, Habuchi T, Thibodeau SN, Cooney K, Chappuis PO, Hutter P, Kaneva RP, Foulkes WD, Zeegers MP, Lu YJ, Zhang HW, Stephenson R, Cox A, Southey MC, Spurdle AB, FitzGerald L, Leongamornlert D, Saunders E, Tymrakiewicz M, Guy M, Dadaev T, Little SJ, Govindasami K, Sawyer E, Wilkinson R, Herkommer K, Hopper JL, Lophatonanon A, Rinckleb AE, Kote-Jarai Z, Eeles RA, Easton DF. Risk analysis of prostate cancer in PRACTICAL, a multinational consortium, using 25 known prostate cancer susceptibility loci. Cancer Epidemiol Biomarkers Prev 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, Initiative CO-CRUG-E, Australian Prostate Cancer B, Oncology UKGPCSCBAoUSSo, Collaborators UKPS, Consortium P, Kote-Jarai Z, Easton DF . Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 2013;45(4): 385–391, 391 e381–e382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, Goh C, Govindasami K, Guy M, O’Brien L, Sawyer E, Hall A, Wilkinson R, Easton D, Collaborators U, Goldgar D, Eeles R, Kote-Jarai Z. Germline BRCA1 mutations increase prostate cancer risk. Br J Cancer 2012;106(10):1697–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson D, Easton DF, Breast Cancer Linkage C. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst 2002;94(18):1358–1365. [DOI] [PubMed] [Google Scholar]
- 10.Breast Cancer Linkage C Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 1999;91(15):1310–1316. [DOI] [PubMed] [Google Scholar]
- 11.Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, Guy M, Edwards S, O’Brien L, Sawyer E, Hall A, Wilkinson R, Dadaev T, Goh C, Easton D, Collaborators U, Goldgar D, Eeles R. BRCA2 is a moderate penetrance gene contributing to young-onset prostate cancer: Implications for genetic testing in prostate cancer patients. Br J Cancer 2011;105-(8):1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Asperen CJ, Brohet RM, Meijers-Heijboer EJ, Hoogerbrugge N, Verhoef S, Vasen HF, Ausems MG, Menko FH, Gomez Garcia EB, Klijn JG, Hogervorst FB, van Houwelingen JC, van’t Veer LJ, Rookus MA, van Leeuwen FE, Netherlands Collaborative Group on Hereditary Breast C. Cancer risks in BRCA2 families: Estimates for sites other than breast and ovary. J Medical Genet 2005;42(9):711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bancroft EK, Page EC, Castro E, Lilja H, Vickers A, Sjoberg D, Assel M, Foster CS, Mitchell G, Drew K, Maehle L, Axcrona K, Evans DG, Bulman B, Eccles D, McBride D, van Asperen C, Vasen H, Kiemeney LA, Ringelberg J, Cybulski C, Wokolorczyk D, Selkirk C, Hulick PJ, Bojesen A, Skytte AB, Lam J, Taylor L, Oldenburg R, Cremers R, Verhaegh G, van Zelst-Stams WA, Oosterwijk JC, Blanco I, Salinas M, Cook J, Rosario DJ, Buys S, Conner T, Ausems MG, Ong KR, Hoffman J, Domchek S, Powers J, Teixeira MR, Maia S, Foulkes WD, Taherian N, Ruijs M, Helderman-van den Enden AT, Izatt L, Davidson R, Adank MA, Walker L, Schmutzler R, Tucker K, Kirk J, Hodgson S, Harris M, Douglas F, Lindeman GJ, Zgajnar J, Tischkowitz M, Clowes VE, Susman R, Ramon y Cajal T, Patcher N, Gadea N, Spigelman A, van Os T, Liljegren A, Side L, Brewer C, Brady AF, Donaldson A, Stefansdottir V, Friedman E, Chen-Shtoyerman R, Amor DJ, Copakova L, Barwell J, Giri VN, Murthy V, Nicolai N, Teo SH, Greenhalgh L, Strom S, Henderson A, McGrath J, Gallagher D, Aaronson N, Ardern-Jones A, Bangma C, Dearnaley D, Costello P, Eyfjord J, Rothwell J, Falconer A, Gronberg H, Hamdy FC, Johannsson O, Khoo V, Kote-Jarai Z, Lubinski J, Axcrona U, Melia J, McKinley J, Mitra AV, Moynihan C, Rennert G, Suri M, Wilson P, Killick E, Collaborators I, Moss S, Eeles RA. Targeted prostate cancer screening in BRCA1 and BRCA2 mutation carriers: Results from the initial screening round of the IMPACT study. Eur Urol 2014;66(3):489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D’Amato M, Adolfsson J, Gronberg H. Polygenic risk score improves prostate cancer risk prediction: Results from the Stockholm-1 cohort study. Eur Urol 2011;60(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460(7256):748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witte JS, Hoffmann TJ. Polygenic modeling of genome-wide association studies: An application to prostate and breast cancer. Omics : J Integrat Biol 2011;15(6):393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machiela MJ, Chen CY, Chen C, Chanock SJ, Hunter DJ, Kraft P. Evaluation of polygenic risk scores for predicting breast and prostate cancer risk. Genet Epidemiol 2011;35(6):506–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013;9(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. http://ec.europa.eu/research/health/medical-research/cancer/fp7-projects/cogs_en.html.
- 20.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988;44-(3):837–845. [PubMed] [Google Scholar]
- 21.Pencina MJ, D’Agostino RB Sr., D’Agostino RB Jr., Vasan RS. Evaluating the added predictive ability of a new marker: From area under the ROC curve to reclassification and beyond. Stat Med 2008;27(2):157–172. discussion 207–112. [DOI] [PubMed] [Google Scholar]
- 22.Pencina MJ, D’Agostino RB Sr., Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 2011;30(1):11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssens AC, Moonesinghe R, Yang Q, Steyerberg EW, van Duijn CM, Khoury MJ. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med 2007;9(8):528–535. [DOI] [PubMed] [Google Scholar]
- 24.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet 2013;45(4):400–405 405e401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.So HC, Kwan JS, Cherny SS, Sham PC. Risk prediction of complex diseases from family history and known susceptibility loci, with applications for cancer screening. Am J Hum Genet 2011;88(5):548–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.