Abstract
For large-scale vision tasks in biomedical images, the labeled data is often limited to train effective deep models. Active learning is a common solution, where a query suggestion method selects representative unlabeled samples for annotation, and the new labels are used to improve the base model. However, most query suggestion models optimize their learnable parameters only on the limited labeled data and consequently become less effective for the more challenging unlabeled data. To tackle this, we propose a two-stream active query suggestion approach. In addition to the supervised feature extractor, we introduce an unsupervised one optimized on all raw images to capture diverse image features, which can later be improved by fine-tuning on new labels. As a use case, we build an end-to-end active learning framework with our query suggestion method for 3D synapse detection and mitochondria segmentation in connectomics. With the framework, we curate, to our best knowledge, the largest connectomics dataset with dense synapses and mitochondria annotation. On this new dataset, our method outperforms previous state-of-the-art methods by 3.1% for synapse and 3.8% for mitochondria in terms of region-of-interest proposal accuracy. We also apply our method to image classification, where it outperforms previous approaches on CIFAR-10 under the same limited annotation budget. The project page is https://zudi-lin.github.io/projects/#two_stream_active.
Keywords: Active Learning, Connectomics, Object Detection, Semantic Segmentation, Image Classification
1. Introduction
Deep convolutional neural networks (CNNs) have advanced many areas in computer vision. Despite their success, CNNs need a large amount of labeled data to learn their parameters. However, for detection and segmentation tasks, dense annotations can be costly. Further, in the biomedical image domain, annotations need to be conducted by domain experts after years of training. Thus, under the limited annotation budget, it is critical to effectively select a subset of unlabeled data for annotation to train deep learning models.
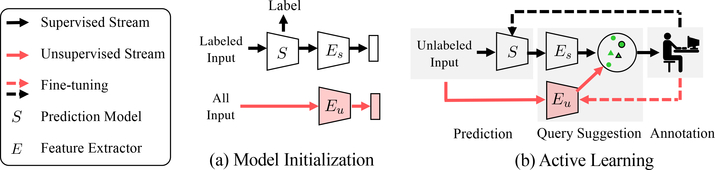
Active learning is a common solution that iteratively improves the prediction model by suggesting informative queries for human annotation to increase labels. There are three main categories of query suggestion approaches that have been explored for CNNs: uncertainty-based [44,42,52], expected model change-based [53], and clustering-based methods [43]. However, all these methods use features extracted from CNNs that are trained on the labeled set (Fig. 1a, →). For example, core-set [45] uses the last feature space before the classification layer to find representative queries, and learning-loss [53] takes multiple features maps to estimate the loss of the model prediction. Therefore, these methods can be biased towards the feature distribution of the small labeled set. Notably, in many biomedical image applications, the labeled dataset is far from representative of the whole dataset due to its vast quantity and great diversity.
Fig. 1.
Two-stream active query suggestion. Active learning methods transform unlabeled data into a feature space to suggest informative queries and improve the base model S. Previous methods optimize their feature extractor (Es) only on the labeled data. We propose a second one (Eu) trained unsupervisedly on all data to capture diverse image features, which can later be updated by fine-tuning with new annotations.
To address this challenge, we propose a two-stream active clustering method to improve query suggestion by introducing an additional unsupervised feature extractor to capture the image statistics of the whole dataset (Fig. 1a,→). During active learning, we combine features extracted by both the supervised and unsupervised streams from the unlabeled data (Fig. 1b). The unsupervised stream can better select representative samples based on image features even when the supervised model makes wrong predictions. Given new annotations, we can further finetune the unsupervised feature extractor to make the embedding space more discriminative. For the clustering module, we show that combining the features from both streams in a hierarchical manner achieves significantly better query suggestion performance than directly concatenating the feature vectors.
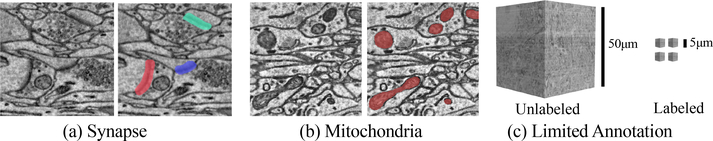
We test our method in the field of connectomics, where the goal is to reconstruct the wiring diagram of neurons to enable new insights into the workings of the brain [26,31]. Recent advances in electron microscopy (EM) allow researchers to collect brain images at nanometer resolution and petabyte scale [21,56]. One crucial task is to detect and segment biological structures like synapses and mitochondria for a deeper understanding of neural anatomy and activation patterns [2] (Fig. 2a–b). However, most labeled connectomics datasets [11,30] are only a few gigavoxels in size, hundreds of times smaller than the unlabeled volume needed for down-stream biological analysis (Fig. 2c).
Fig. 2.
Two essential vision tasks in connectomics: (a) object detection of synapses to quantify the neuronal connectivity strength and (b) semantic segmentation of mitochondria to estimate the neuronal activity level. (c) However, the terabyte-level test data can be 100× larger than the training data, making active learning necessary.
With our two-stream active clustering as the key component, we build an end-to-end framework with a base model and an annotation workflow. Before active learning, our base model achieves state-of-the-art results on public benchmarks. Besides, our annotation workflow reduces interactive error correction time by 26%, as shown by a controlled user study. With this framework, we finished the dense annotation of synapse objects and mitochondria semantic mask for a (50 μm)3 EM image volume (300 gigavoxels) in the rat visual cortex, called EM-R50, which is over 100× larger than existing datasets. For the evaluation of active learning approaches on this connectomics dataset, our method improves the performance of previous state-of-the-art methods by 3.1% for synapses and 3.8% for mitochondria, respectively, in terms of the accuracy of the region-of-interest (ROI) proposals. We further perform ablation studies to examine the importance of different framework components and hyper-parameters. To demonstrate its broader impact, we also benchmark our method on natural image classification (CIFAR-10), which outperforms previous state-of-the-art methods by over 2% under a limited annotation budget ≈ 5% of the total training images.
Contributions.
First, we introduce a novel active learning method that incorporates information from an unsupervised model to improve the effectiveness of query suggestions. Second, our method achieves state-of-the-art results for detection and segmentation tasks on connectomics datasets and image classification on CIFAR-10. Third, we release the code and a densely annotated connectomics dataset (100× bigger than current datasets) to facilitate future researches.
2. Related work
Synapse Detection and Mitochondria Segmentation.
Synapse detection and mitochondria segmentation are two popular tasks in connectomics. Due to the complex shapes, bounding box-based detection [38] and segmentation [12] methods can have poor performance. Thus, most previous works for biomedical vision directly predict the semantic segmentation of the object and generate bounding-box proposals for the detection task via post-processing.
For synapse detection, previous approaches focus on segmenting the synaptic cleft region using hand-crafted image features [25,2,17,19,24,37] or learned features [39]. To further predict synapse polarity, the direction of information transmission among neurons, recent works apply random forest classifiers [23,49], neural networks [18,5,14,34], and combinations [7]. For mitochondria segmentation, earlier works leverage various image processing techniques and manually-designed image features [32,50,30,27,46,36]. Recent methods employ 2D or 3D fully convolutional network architectures [33,6] to regress the semantic mask.
In this paper, we adopt the 3D U-Net [40] model for both synapse detection and mitochondria semantic segmentation. Incorporating recent deep learning techniques including residual blocks [13] and squeeze-and-excitation blocks [16], our model achieves top performance on public connectomics benchmarks.
Active Learning.
Active learning methods iteratively query human annotators to obtain new informative samples to label and then improve the base model. Transductive active learning [54] aims to improve the later step by training the base model on the additional unlabeled data with pseudo labels. The similarity graph among samples [3,35,57] is often used to generate pseudo labels from manual annotations. We focus on the former step to suggest better queries [45], where traditional methods use uncertainty-based sampling [42,52], and diversity-based optimization [9,10]. Tailored for neural networks, recent works explore ideas of maximizing feature coverage [43], margin-based sampling [55], expected error-based selection [53] and adversarial learning [8].
Besides the image classification task, active learning has been applied to object detection for different image domains [4,48,1]. Roy et al. [41] formulates the detection task as a structured prediction with novel margin sampling techniques and Vijayanarasimhan et al. [51] scales up the labeling process with crowdsourcing. Kao et al. [20] proposes location-aware measures for query suggestion. Instead of solely using the feature extractor optimized on the labeled set, our key insights are to improve query suggestions with unsupervised image information and fine-tune the learned feature extractor to distinguish ambiguous samples.
3. Active Learning Framework Overview
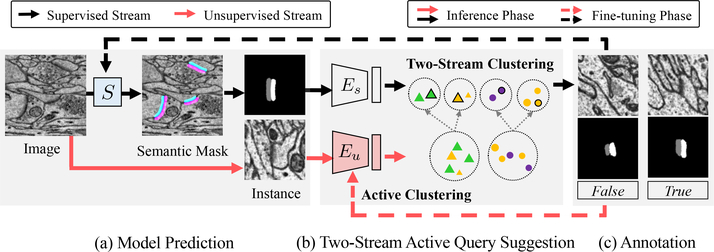
Our active learning framework for large-scale vision tasks in connectomics has three components: base model, query suggestion, and annotation (Fig. 3). We here describe our base model and annotation workflow, leaving the query suggestion method for Sec. 4. Further details are in the supplementary document.
Fig. 3.
Overview of our active learning framework. (a) The base model S predicts semantic masks, which are post-processed to generate ROIs. We align them to the same orientation for better clustering. (b) Our method adds an additional stream of unsupervised feature extracted by Eu. We apply hierarchical clustering to partition the unlabeled data and suggest cluster centers as queries for annotation. (c) Annotators provide True or False annotations for query samples that are used to fine-tune both the based mode S (black dashed line) and the proposed Eu (red dashed line).
Overview.
During active learning on unlabeled images, the base model first predicts dense probability map and generates regions of interest (ROIs). Then the proposed query suggestion method extracts features for all ROIs and suggests queries through the two-stream clustering for annotation. With the new annotation, in addition to fine-tuning the base model, we further fine-tune the proposed query suggestion model to make it more discriminative in query suggestion.
Model Prediction.
The base model handles two tasks: synapse detection and mitochondria segmentation. The irregular shapes make it hard to directly predict 3D bounding boxes for synapses, while the vast volume quantity makes it infeasible to conduct pixel-wise annotation for mitochondria. Therefore, following common practice for biomedical images, we first predict a dense semantic probability map and apply connected component labeling with post-processing to generate ROI proposals. We thus unify two different tasks as judging the correctness of ROIs in active learning. Since finding false positives from proposals is more efficient than locating false negatives in the vast volume, we re-balance the weights between foreground and background pixels to ensure a high recall.
In Fig. 3a, we show an example for synapse detection. Each synapse instance has a pre-synaptic (purple) and a post-synaptic (cyan) segment, and we predict a three-channel probability map representing pre- and post-synaptic regions and their union. We align extracted ROIs to a reference orientation to normalize its rotation variation. To this end, we select the 2D slice with the biggest area from the 3D instance, apply the principal component analysis (PCA) of the mask, and rotate the instance to align its first principal component to the vertical direction. For synapse, we further make sure the pre-synaptic segment (gray) is on the left.
Annotation.
We focus on the correctness of ROIs instead of the pixel-level correctness of the mask. During annotation, an annotator judges the ROI to be correct if the mask within covers more than half of the ground truth mask in this ROI. In practice, thanks to the performance of the base model, annotators find most predicted instances are unambiguously right or wrong. We built a browser-based labeling interface, where annotators can click on each suggested query to change its label, e.g., from True to False (Fig. 3c). For better judgment, we display both image patches and predicted instance masks to annotators. For annotation efficiency, we display the selected query samples in a grouped manner using our clustering results instead of a random order (Sec. 6.3).
4. Two-stream Active Query Suggestion
Compared to previous methods, our two-stream active query suggestion introduces an additional unsupervised feature extractor that is trained on all images to capture dataset statistics. We then cluster unlabeled data with the two-steam features and use cluster centers as query samples for annotation (Sec. 4.1). With new annotations, we further fine-tune the image feature extractor to adjust the feature space for better query suggestions in the next round (Sec. 4.2).
4.1. Two-Stream Clustering
For the task of deciding the correctness of ROI proposals, we use the predicted mask from the base model for the supervised stream and its corresponding raw image for the unsupervised stream. We first apply the feature extractor to reduce the feature dimension for each stream. Then, we fuse the two-stream clustering results to partition the unlabeled data into smaller subsets where the samples share similar features to make the cluster centers more representative.
Feature Extraction Network.
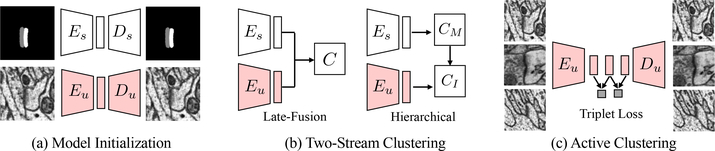
We train the feature extraction model through self-supervision. Specifically, we train a variational auto-encoder (VAE) [22] to regress the input through a bottleneck network structure (Fig. 4a) and use the embedded features as the low-dimensional representations. The VAE model consists of an encoder network E and a decoder network D. We use several convolutional layers for the encoder, followed by a fully connected layer to predict both the mean and standard deviation of the low-dimensional embedding vector. For the decoder, we use deconvolution (or transposed convolution) layers to learn to reconstruct the input image from the embedding vector.
Fig. 4.
Architectures for the two-stream active query suggestion model. (a) For model initialization, we train the supervised (Es) and unsupervised (Eu) feature extractors using VAEs. (b) For two-stream clustering, we compare two design choices to combine Eu and Es features in an either parallel (late-fusion) or hierarchical manner. The block Ci denotes the clustering algorithm. (c) For active clustering, we fine-tune Eu with triplet loss to encourage the learning of discriminative features.
The loss function for the VAE is the sum of the ℓ1 reconstruction loss and the KL-divergence between the distribution of the predicted embedding feature and the standard normal distribution. In practice, we use samples with a fixed patch size where the mask or image is rotated and aligned when training the VAE. We then only use the VAE mean vector as the extracted feature.
Feature Fusion.
Given the extracted image and mask features, we propose two designs of clustering architectures to fuse such two-stream information: late-fusion clustering and hierarchical clustering (Fig. 4b). Inspired by the two-stream architecture designs for video action recognition [47], we design a late-fusion strategy to directly concatenate image and mask features and feed into a clustering module C. We expect that with the combined features from two streams, the clustering method can better distinguish ambiguous samples.
Another strategy is the hierarchical clustering that clusters the mask features and the image features sequentially. The intuition behind the design is that since the embedding spaces for both extractors can be very different (e.g., dimension, and distance scale), the hierarchical approach can alleviate the needs for rebalancing. In the hierarchical clustering, the members of each of the N mask clusters separated in the first round are further divided into M sub-clusters by applying the k-means algorithm on the unsupervised image VAE embedding space, which yields MN clusters in total. We show in Sec. 6.2 that conditioning the image clustering on the mask features can prevent the image features, which are of high dimension than mask features, from dominating the results. Therefore hierarchical clustering can better suggest queries compare to late-fusion.
Query Suggestion.
Given the clustering result from either late-fusion or hierarchical clustering, we run an additional round of clustering (e.g., k-means) with Q clusters and use the samples with minimum distances to each cluster center as queries presented to the annotators. Thus the annotator needs to annotate in total MNQ samples. In the ablation study (Sec. 6.2), we will examine the query suggestion effectiveness and efficiency with different hyper-parameter choices.
4.2. Active Clustering
Since the encoders are learned in an unsupervised manner, we expect that with new annotations, we can improve the encoder to encourage the learning of more discriminative features. Therefore we adaptively adjust the embedding space of the encoder with new labels to make the clustering module active.
Triplet Loss.
We employ the triplet loss [15] to incorporate new label information into the encoder. Suppose that we have a set of labeled positive and negative instances. After randomly select one positive sample xP and one negative sample xN as anchors, we hope that the third sample x becomes close to xP and distant from xN if it is positive, and vice versa. This can encourage the encoders to learn more discriminative features and facilitate query suggestions since samples share the same label are closer while different classes are projected further apart. Following Hoffer et al. [15], we calculate the distances as
| (1) |
where ϕ(x) indicates features extracted from the encoder. We then define the loss function for adjusting the feature extractor as
| (2) |
to minimize dP and maximize dN. Incorporating the triplet loss enables the active adjusting of the feature space to be more discriminative, which can further improve the effectiveness of query suggestion.
4.3. Learning Strategy
Inference Phase.
(Fig. 3, solid line) For both synapses and mitochondria, the base model was initially trained on a small manually labeled volume of size (5μm)3, comparable to public benchmark datasets. We conduct sliding-window prediction for the large test volume of size (50μm)3 and use the connected component algorithm to generate ROI candidates for active learning. The VAE of the mask encoder Es model is trained on the aligned patches with predicted object masks, while the image encoder Eu is trained with image patches uniformly sampled from the whole volume to capture diverse texture information.
Fine-tuning Phase.
(Fig. 3, dashed line) For active clustering, the Eu is initialized by fine-tuning it with labeled patches from the small labeled volume. Then the queries are generated with two-stream hierarchical clustering by successively using the latent spaces of both Es and Eu. After query annotation, we fine-tune the image encoder with new ground truth labels and apply it for future iterations of query suggestion. For non-active clustering, we conduct the same hierarchical clustering but use the original Eu trained under a totally unsupervised setting. In both cases, the new query samples are used to fine-tune and improve the base model as a standard active learning practice.
5. EM-R50 Connectomics Dataset
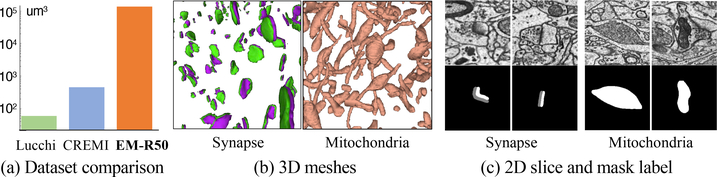
With our two-stream active query suggestion method, we annotated, to the best of our knowledge, the largest 3D EM connectomics image volume datasets with dense synapses object and mitochondria mask annotation. Specifically, we imaged a tissue block from Layer II/III in the primary visual cortex of an adult rat at a resolution of 8×8×30nm3 using a multi-beam scanning electron microscope. After stitching and aligning the images on multi-CPU clusters, we obtained a final volume of 50 μm cube. We also apply deflickering, frame interpolation, and image de-striping techniques to improve image quality.
Annotation Quantity.
All ROIs are annotated by three neuroscience experts, and we take the majority decision mapped back to the volume as the final label. In total, we obtain around 104K synapses, and mitochondria mask that occupies around 7.6% of the voxels. Compared to benchmarks like CREMI [11] and Lucchi [28] in connectomics, EM-R50 dataset is over 150× larger in image volume size, and over 100× larger in terms of the number of synapse instance.
Instance Diversity.
To exhibit instance diversity, we show 3D meshes of all synapses and mitochondria within a subvolume (Fig. 5b). We use representative 2D slices for each 3D ROIs during annotation, and we show the variation of instance shape and orientation (Fig. 5c).
Fig. 5.
EM-R50 connectomics dataset with dense synapse and mitochondria annotation. (a) We compare the size of the densely annotated image volume with other connectomics datasets (log-scale). To visualize the diversity of instance shape and orientation, we show (b) 3D meshes of synapses and mitochondria within a sub-volume, and (c) sample 2D image patches with corresponding mask annotations.
6. Experiments on Connectomics Datasets
We first benchmark our query suggestion method against others on the EM-R50 dataset for the ROI-level accuracy for synapse and mitochondria. Then we examine the design choices of the proposed method and the whole active learning pipeline through ablation studies, public benchmarks, and user studies.
6.1. Comparing with State-of-the-art Methods
Dataset and Metric.
We randomly sample a subset of ROIs for synapses and mitochondria from EM-R50 for the benchmark experiments. The number of samples in the training-test split is 28.7K-10K and 20K-5K for synapse and mitochondria, respectively. We use the ROI proposal accuracy of the base model after fine-tuning as the evaluation metric for active learning methods.
Methods in Comparison.
We compare our method with random uniform sampling, core-set [43], and learning-loss [53] approaches under a two-iteration scenario. For all the methods, after generating queries with a fixed annotation budget (1,280 instances, ≈ 5% of the training set), we use new labels to fine-tune the base network and evaluate the updated network prediction.
Results on Synapse.
The initial accuracy of the network on the active learning test split is 0.811. We fine-tune the network using the instances suggested by different methods from the training set, where it has an initial accuracy of 0.762. To prevent overfitting, we construct mini-batches from the initial labeled volume with a ratio of 0.25. After first-round fine-tuning, the accuracy of the synapse proposals is increased to 0.892 for the test set, which outperforms random uniform sampling, core-set, and learning-loss (Table 1, Round 1). For our method, the new annotations are used to fine-tune the image encoder Eu. After another round of active clustering and fine-tuning the base model, the test accuracy is increased to 0.926, which shows that our method outperforms previous approaches significantly on synapse detection (Table 1, Round 2).
Table 1.
Active learning performance comparison on our EM-R50 connectomics benchmark. Our two-stream query suggestion approach significantly out-perform previous methods in terms of the ROI proposal accuracy (higher is better).
Results on Mitochondria.
Since other structures like lysosome and artifacts caused by tissue staining and imaging look similar to mitochondria, the initial accuracy on the unlabeled ROIs is only 0.57. After applying our approach, the accuracy is improved to 0.809 on the test set, which outperforms core-set [43] method by 4.2% and the learning-loss [53] by 3.8% (Table 1). We observe that the accuracy improvement is relatively small for mitochondria at the second active learning round. This may result from the intrinsic ambiguity of the mitochondria feature, where even human experts are not confident about the label.
Discussion.
Despite the state-of-the-art performance of core-set [43] and learning-loss [53] on natural image benchmarks, those methods are less effective in handling the connectomics tasks due to two reasons. First, both methods use features from the supervised model, which can hardly capture the images features in the large unlabeled volume. Second, for the learning-loss approach, estimating the prediction loss with the global-average-pooling (GAP) module can ignore the useful structure information of objects. Nevertheless, we also compare the CIFAR-10 image classification benchmark (Sec. 7), where the methods are optimally tuned by the authors, for an even fairer comparison.
6.2. Ablation Analysis of Two-Stream Active Query Suggestion
In this part, we validate our design choices of the proposed two-stream active clustering module through ablation studies. Since the goal of query suggestion in active learning is to find the most “representative” samples for annotation, we perform the experiments to evaluate how different hyper-parameter and design choices influence the accuracy of annotation under a limited label budget.
Dataset and Metric.
We use the synapse benchmark dataset above to perform the ablation study. Suppose that after sliding window inference of the detection model, we have N proposed instances with an accuracy of p. Here p is the number of correct predictions over the total number of ROIs. By fixing the annotation budget s, the baseline accuracy is defined by the expectation of the accuracy that can be achieved by random one-by-one annotation, which is . For example, with an initial accuracy of 0.7, randomly annotating 10% of the instances can improve the overall accuracy by 3%, since 70% of the queries are positive, and no errors can be corrected by annotating them. Then for evaluating the proposed methods, after annotating the cluster representatives in the clustering module, we assign the major representative labels to all samples in that cluster and calculate the label accuracy of the ROIs.
Effect of Two-Stream Clustering.
We examine the active learning method accuracy with respect to the number of clusters and clustering architectures. Note that we fix the number of representatives Q = 5. Initially, for the instance proposals generated by the detection model, we assign ‘correct’ labels to all instances, and the accuracy is 0.762. As shown in Table 2, both with manual annotation of 4.46% of the data, random annotation can increase the accuracy by 4.46% × (1 − 0.762) ≈ 0.01, while our clustering module can increase the label accuracy by around 0.08 in absolute value. Besides, combining two-stream information with late fusion performs worse than the ‘mask only’ design. This is because the dimension of image embedding space is 1,000 to achieve reasonable reconstruction performance, which is much larger than the mask embedding space (20). Image embedding tends to dominate the result with direct concatenation and clustering using the same distance metric.
Table 2.
Comparison of design choices for two-stream clustering. We compute the object detection accuracy by assigning the labels of the cluster centers to other cluster members. The number of candidates per cluster, Q, is fixed to 5.
| Description | Random | One-Stream |
Two-Stream |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mask-Only | Image-Only | Late-Fusion | Hierarchical | ||||||||
| Es clusters (N) | - | - | 128 | 256 | 1 | 1 | - | 64 | 128 | 64 | 32 |
| Eu clusters (M) | - | - | 1 | 1 | 128 | 256 | - | 2 | 2 | 4 | 8 |
| Total num. (MN) | - | - | 128 | 256 | 128 | 256 | 256 | 128 | 256 | 256 | 256 |
| Annotation ratio (%) | 2.23 | 4.46 | 2.23 | 4.46 | 2.23 | 4.46 | 4.46 | 2.23 | 4.46 | 4.46 | 4.46 |
| Accuracy | 0.767 | 0.772 | 0.805 | 0.819 | 0.420 | 0.578 | 0.738 | 0.821 | 0.826 | 0.846 | 0.814 |
Effect of Active Clustering.
We examine the effect of active clustering for the feature extractors Eu and Es. There are three choices of the architectures, fine-tuning Es only, fine-tuning Eu only, as well as fine-tuning both Es and Eu. As indicated in Table 3, fine-tuning only Es decreases the accuracy, because add supervision can distort the shape priors learned by the mask VAE; fine-tuning only Eu have a significant improvement over the static hierarchical baseline; fine-tuning both Es and Eu decreases the Eu only performance, which further indicate that the shape information learned in Eu by self-supervision already contains distinguishable information that can be extracted from object masks. Therefore, we only fine-tuning Eu.
Table 3.
Comparison of design choices for active clustering. We show the accuracy w/ or w/o fine-tuning feature extractors. Fine-tuning only Eu shows the best performance while fine-tuning Es can confuse the encoder, which leads to worse performance.
| Active Encoder | None | Es | Eu | Eu and Es |
|---|---|---|---|---|
| Accuracy | 0.846 | 0.830 | 0.880 | 0.871 |
6.3. Ablation Analysis of Active Learning Pipeline
Besides the evaluation of the proposed query suggestion method above, we examine the performance of the other two modules in the whole pipeline.
Model Prediction: Pixel-Level Evaluation.
We provide pixel-level evaluations of the base model2 to show its effectiveness on small benchmark datasets and indicate the necessity of active learning on large datasets. For synaptic cleft, we evaluate on the CREMI Challenge dataset [11], which contains 3 training and 3 test volumes of the size 1250×1250×125 voxels. The results are evaluated by two scores: the average distance of any predicted cleft voxel to its closest ground-truth cleft voxel (ADGT) for penalizing false positives and the average distance of any ground-truth cleft voxel to its closest predicted cleft voxel (ADF) for penalizing false negatives. The final ranking criterion (CREMI score) is the mean of ADGT and ADF over the three test volumes. For mitochondria, we evaluate the model on the Lucchi dataset [30], which contains 1 training and 1 test volumes of size 1024×768×165 voxels. We use the standard VOC score, which is the average of the Jaccard index of the foreground and background pixels.
For synaptic cleft, our proposed model outperforms previous leading methods by 5% and ranks 1st among published results on the public leaderboard (Table 4, left). For mitochondria, our model achieves comparable results to the previous state-of-the-art methods [6,29], with ∼1% difference (Table 4, right). The results suggest that our base model is strong enough to enable a fair comparison of the following active learning methods on the large-scale benchmark dataset.
Table 4.
Pixel-level evaluation on public connectomics datasets. For synapse, ours ranks 1st among results in publications on the CREMI dataset (left). For mitochondria, ours is on-par with state-of-the-art methods on the Lucchi dataset (right).
Model Prediction: Recall.
As objects are sparse in the images, correcting false positive is much easier than finding false negatives. Thus we rebalance the loss and reject batches without foreground pixels with a 95% probability to heavily penalize false negatives as in Heinrich et al. [14]. In return, the instance-level recall for synapses on a fully labeled validation volume is 0.94 (IoU threshold is 0.5), which is adequate for the ROI-based active learning experiments.
Annotation: Query Display Order.
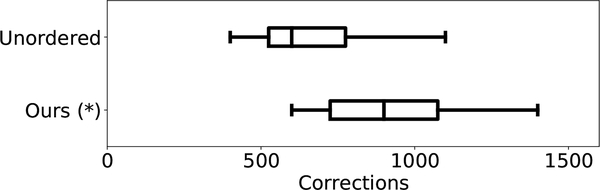
To speed up the annotation, we sort the suggested query samples by their cluster indices and distance from their cluster centers. Such cluster-based query display order potentially allows participants to scan and identify false predictions faster, as patches with similar features are grouped closer than those in a random ordering. For evaluation, we performed a user study with novices as a single factor between-subjects experiment.
From the EM-R50 dataset, we randomly select 2.1K synapses, with 211 are false predictions. We recruited 20 novice participants and asked them to annotate as many synapses as possible within the 30-minute time frame after a 10-min proper instruction on the task. Each participant was randomly assigned to either our clustering method or random ordering of the synapses.
Our clustering method allows study participants to annotate synapse with higher throughput, 930±237 synapses, compared to the random order, 670±224 (Fig. 6). Besides the efficiency improvement, the cluster-based query display order leads to a slight average accuracy improvement: for users with clustering 0.728 ± 0.087 compared to the random ordering with 0.713 ± 0.114.
Fig. 6.
User study on annotation through-put. The box plots show the median and interquartile range of the number of annotated instances in a fixed time frame of 30 minutes.
7. Application to Natural Image Classification
The proposed two-stream active query suggestion can be applied to image classification in the active learning setting. Instead of the predicted mask encoded by a VAE model, we use the class label prediction as the supervised stream feature.
Dataset and Metric.
CIFAR-10 has 60K images of size 32×32 pixels, with 50K for training and 10K for testing. Each image has a label from one of the ten classes. For evaluation, we use the top-1 classification accuracy on the test split.
Methods in Comparison.
We use the same training protocol as Yoo et al. [53] for a fair comparison. For query suggestion methods, we compare with random uniform sampling, core-set [43], and learning-loss [53] approaches. For the active learning pipeline, we run a five-round comparison. We first uniformly sample 1K samples from the training set as the initial pool. After training the classification model, we apply different query suggestion approaches and label additional 1K samples from the unlabeled pool. Then we train the model from scratch again and conduct another round of query suggestion and labeling. We iterate the process until the total number of labeled samples reaches 5K.
Implementation Details.
For classification, we use the same ResNet-18 model as Yoo et al. [53]. During training, we apply data augmentation, including random crop and horizontal flip, and image normalization. During active learning, the number of training epochs is 200, and the mini-batch size is 64. The learning rate of the SGD optimizer is initially 0.1 and decreased to 0.01 after 160 epochs. For indicating the effectiveness of the unsupervised stream, we only use the two-stream clustering module of our query suggestion method. We pre-train the unsupervised stream feature with a VAE on all the training images with a latent dimension of 32. At the clustering phase, we fix the number of clusters at the output space and VAE latent space to be 50 and 20, respectively.
Results.
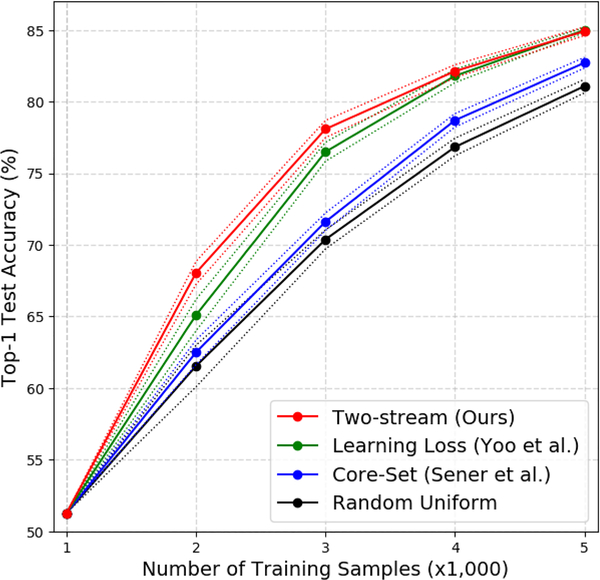
Our proposed method outperforms the random uniform sampling and core-set methods, and is higher or comparable to the recent learning-loss approach (Fig. 7). Empirically, when the number of training samples is around 5% of the whole dataset (i.e., 2K, and 3K out of 50K training images), our method achieves 2–3% improvement upon the learning-loss approach.
Fig. 7.
Active learning results on the CIFAR-10 dataset. The accuracy improvement of our approach over previous state-of-the-art methods is most significant when training with a limited number of samples (2k and 3k out of total 50k images), similar to the annotation budget for EM-R50 (≈ 5%). Mean and standard deviation are estimated from 5 runs. We also show that the accuracy saturates after ten iterations of query suggestion (Fig. S-4 in the supplementary material).
8. Conclusion
In this paper, we demonstrate the effectiveness of our proposed two-stream active query suggestion method for large-scale vision tasks in connectomics under the active learning setting. Besides the state-of-the-art results on the connectomics data, we show its applicability to a natural image classification benchmark. We evaluate each module of our active learning pipeline through public benchmarks, ablation studies, and user studies. As a use case, we build a connectomics dataset from a (50 μm)3 cubic tissue with dense annotation of synapse and mitochondria.
Supplementary Material
Acknowledgments.
This work has been partially supported by NSF award IIS-1835231 and NIH award 5U54CA225088-03.
Footnotes
Please check Sec. S-1 in the supplementary document for model details.
Architecture details are shown in Fig. S-1 in the supplementary document.
References
- 1.Abramson Y, Freund Y: Active learning for visual object detection (2006) 4
- 2.Becker C, Ali K, Knott G, Fua P: Learning context cues for synapse segmentation. IEEE TMI (2013) 3, 4. [DOI] [PubMed] [Google Scholar]
- 3.Belkin M, Niyogi P: Using manifold stucture for partially labeled classification. In: NIPS; (2003) 4 [Google Scholar]
- 4.Bietti A: Active learning for object detection on satellite images. Tech. rep, Technical report, Caltech; (2012) 4 [Google Scholar]
- 5.Buhmann J, Krause R, Lentini RC, Eckstein N, Cook M, Turaga S, Funke J: Synaptic partner prediction from point annotations in insect brains. arXiv preprint arXiv:1806.08205 (2018) 4 [Google Scholar]
- 6.Cheng HC, Varshney A: Volume segmentation using convolutional neural networks with limited training data. In: ICIP (2017) 4, 12 [Google Scholar]
- 7.Dorkenwald S, Schubert PJ, Killinger MF, Urban G, Mikula S, Svara F, Kornfeld J: Automated synaptic connectivity inference for volume electron microscopy. Nature methods 14(4), 435 (2017) 4. [DOI] [PubMed] [Google Scholar]
- 8.Ducoffe M, Precioso F: Adversarial active learning for deep networks: a margin based approach. ICML; (2018) 4 [Google Scholar]
- 9.Dutt Jain S, Grauman K: Active image segmentation propagation. In: CVPR; (2016) 4 [Google Scholar]
- 10.Freytag A, Rodner E, Denzler J: Selecting influential examples: Active learning with expected model output changes. In: ECCV; (2014) 4 [Google Scholar]
- 11.Funke J, Saalfeld S, Bock D, Turaga S, Perlman E: Circuit reconstruction from electron microscopy images. https://cremi.org (2016) 3, 9, 12 [Google Scholar]
- 12.He K, Gkioxari G, Dollár P, Girshick R: Mask r-cnn. In: Proceedings of the IEEE international conference on computer vision pp. 2961–2969 (2017) 4 [Google Scholar]
- 13.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: CVPR; (2016) 4 [Google Scholar]
- 14.Heinrich L, Funke J, Pape C, Nunez-Iglesias J, Saalfeld S: Synaptic cleft segmentation in non-isotropic volume electron microscopy of the complete drosophila brain. MICCAI (2018) 4, 12, 13 [Google Scholar]
- 15.Hoffer E, Ailon N: Deep metric learning using triplet network. In: International Workshop on Similarity-Based Pattern Recognition (2015) 7, 8 [Google Scholar]
- 16.Hu J, Shen L, Sun G: Squeeze-and-excitation networks. In: CVPR; (2018) 4. [DOI] [PubMed] [Google Scholar]
- 17.Huang GB, Plaza S: Identifying synapses using deep and wide multiscale recursive networks. arXiv preprint arXiv:1409.1789 (2014) 4 [Google Scholar]
- 18.Huang GB, Scheffer LK, Plaza SM: Fully-automatic synapse prediction and validation on a large data set. arXiv preprint arXiv:1604.03075 (2016) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagadeesh V, Anderson J, Jones B, Marc R, Fisher S, Manjunath B: Synapse classification and localization in electron micrographs. Pattern Recognition Letters 43, 17–24 (2014) 4 [Google Scholar]
- 20.Kao CC, Lee TY, Sen P, Liu MY: Localization-aware active learning for object detection. ACCV; (2018) 4 [Google Scholar]
- 21.Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, Lee D, Vázquez-Reina A, Kaynig V, Jones TR, et al. :Saturated reconstruction of a volume of neocortex. Cell 162(3), 648–661 (2015) 2 [DOI] [PubMed] [Google Scholar]
- 22.Kingma DP, Welling M: Auto-encoding variational bayes. ICLR; (2013) 6 [Google Scholar]
- 23.Kreshuk A, Funke J, Cardona A, Hamprecht FA: Who is talking to whom: synaptic partner detection in anisotropic volumes of insect brain. In: MICCAI; (2015) 4 [Google Scholar]
- 24.Kreshuk A, Koethe U, Pax E, Bock DD, Hamprecht FA: Automated detection of synapses in serial section transmission electron microscopy image stacks. PloS one 9(2), e87351 (2014) 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreshuk A, Straehle CN, Sommer C, Koethe U, Cantoni M, Knott G, Hamprecht FA: Automated detection and segmentation of synaptic contacts in nearly isotropic serial electron microscopy images. PloS one 6(10), e24899 (2011) 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichtman JW, Sanes JR: Ome sweet ome: what can the genome tell us about the connectome? Current opinion in neurobiology 18(3), 346–353 (2008) 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lucchi A, Li Y, Fua P: Learning for structured prediction using approximate subgradient descent with working sets. In: CVPR; (2013) 4 [Google Scholar]
- 28.Lucchi A, Li Y, Smith K, Fua P: Structured image segmentation using kernelized features. In: ECCV; (2012) 9 [Google Scholar]
- 29.Lucchi A, Márquez-Neila P, Becker C, Li Y, Smith K, Knott G, Fua P: Learning structured models for segmentation of 2d and 3d imagery. IEEE TMI; (2015) 12. [DOI] [PubMed] [Google Scholar]
- 30.Lucchi A, Smith K, Achanta R, Knott G, Fua P: Supervoxel-based segmentation of mitochondria in em image stacks with learned shape features. IEEE TMI (2012) 3, 4, 12 [DOI] [PubMed] [Google Scholar]
- 31.Morgan JL, Lichtman JW: Why not connectomics? Nature methods 10(6), 494 (2013) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narasimha R, Ouyang H, Gray A, McLaughlin SW, Subramaniam S: Automatic joint classification and segmentation of whole cell 3d images. Pattern Recognition (2009) 4 [Google Scholar]
- 33.Oztel I, Yolcu G, Ersoy I, White T, Bunyak F: Mitochondria segmentation in electron microscopy volumes using deep convolutional neural network. In: Bioinformatics and Biomedicine (2017) 4 [Google Scholar]
- 34.Parag T, Berger D, Kamentsky L, Staffler B, Wei D, Helmstaedter M, Lichtman JW, Pfister H: Detecting synapse location and connectivity by signed proximity estimation and pruning with deep nets. arXiv preprint arXiv:1807.02739 (2018) 4 [Google Scholar]
- 35.Parag T, Ciresan DC, Giusti A: Efficient classifier training to minimize false merges in electron microscopy segmentation. In: ICCV; (2015) 4 [Google Scholar]
- 36.Perez AJ, Seyedhosseini M, Deerinck TJ, Bushong EA, Panda S, Tasdizen T, Ellisman MH: A workflow for the automatic segmentation of organelles in electron microscopy image stacks. Frontiers in neuroanatomy 8, 126 (2014) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plaza SM, Parag T, Huang GB, Olbris DJ, Saunders MA, Rivlin PK: Annotating synapses in large EM datasets. arXiv preprint arXiv:1409.1801 (2014) 4 [Google Scholar]
- 38.Ren S, He K, Girshick R, Sun J: Faster r-cnn: Towards real-time object detection with region proposal networks. In: Advances in neural information processing systems. pp. 91–99 (2015) 4 [Google Scholar]
- 39.Roncal WG, Pekala M, Kaynig-Fittkau V, Kleissas DM, Vogelstein JT, Pfister H, Burns R, Vogelstein RJ, Chevillet MA, Hager GD: Vesicle: volumetric evaluation of synaptic interfaces using computer vision at large scale. arXiv preprint arXiv:1403.3724 (2014) 4 [Google Scholar]
- 40.Ronneberger O, Fischer P, Brox T: U-net: Convolutional networks for biomedical image segmentation. In: MICCAI; (2015) 4 [Google Scholar]
- 41.Roy S, Namboodiri VP, Biswas A: Active learning with version spaces for object detection. arXiv preprint arXiv:1611.07285 (2016) 4 [Google Scholar]
- 42.Scheffer T, Decomain C, Wrobel S: Active hidden markov models for information extraction. In: International Symposium on Intelligent Data Analysis (2001) 2, 4 [Google Scholar]
- 43.Sener O, Savarese S: Active learning for convolutional neural networks: A core-set approach. ICLR; (2018) 2, 4, 9, 10, 13 [Google Scholar]
- 44.Settles B: Active learning literature survey. Tech. rep., University of WisconsinMadison Department of Computer Sciences; (2009) 2 [Google Scholar]
- 45.Settles B: Active learning literature survey. 2010. Computer Sciences Technical Report (2014) 2, 4 [Google Scholar]
- 46.Seyedhosseini M, Ellisman MH, Tasdizen T: Segmentation of mitochondria in electron microscopy images using algebraic curves. In: ISBI; pp. 860–863. IEEE (2013) 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simonyan K, Zisserman A: Two-stream convolutional networks for action recognition in videos. In: NIPS; (2014) 6 [Google Scholar]
- 48.Sivaraman S, Trivedi MM: Active learning for on-road vehicle detection: A comparative study. Machine vision and applications (2014) 4 [Google Scholar]
- 49.Staffler B, Berning M, Boergens KM, Gour A, van der Smagt P, Helmstaedter M: Synem, automated synapse detection for connectomics. Elife (2017) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vazquez-Reina A, Gelbart M, Huang D, Lichtman J, Miller E, Pfister H: Segmentation fusion for connectomics. In: ICCV; (2011) 4 [Google Scholar]
- 51.Vijayanarasimhan S, Grauman K: Large-scale live active learning: Training object detectors with crawled data and crowds. IJCV; (2014) 4 [Google Scholar]
- 52.Wang K, Zhang D, Li Y, Zhang R, Lin L: Cost-effective active learning for deep image classification. IEEE TCSVT (2017) 2, 4 [Google Scholar]
- 53.Yoo D, Kweon IS: Learning loss for active learning. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition pp. 93–102 (2019) 2, 4, 9, 10, 13, 14 [Google Scholar]
- 54.Yu K, Bi J, Tresp V: Active learning via transductive experimental design. In: Proceedings of the 23rd international conference on Machine learning pp. 1081–1088 (2006) 4 [Google Scholar]
- 55.Zhang Y, Lease M, Wallace BC: Active discriminative text representation learning. In: AAAI; (2017) 4 [Google Scholar]
- 56.Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, et al. : A complete electron microscopy volume of the brain of adult drosophila melanogaster. Cell (2018) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X, Ghahramani Z, Lafferty JD: Semi-supervised learning using gaussian fields and harmonic functions. In: ICML; (2003) 4 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.