Abstract
Coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is emerging as a global pandemic outbreak. To date, approximately one million deaths and over 32 million cases have been reported. This ongoing pandemic urgently requires an accurate testing device that can be used in the field in a fast manner. Serological assays to detect antibodies have been proven to be a great complement to the standard method of reverse transcription-polymerase chain reaction (RT-PCR), particularly after the second week of infection. We have developed a specific and sensitive immunosensor for immunoglobulin detection produced against SARS-CoV-2. Unlike other lateral flow-based assays (LFAs) involving the utilization of multiple antibodies, we have reported a label-free paper-based electrochemical platform targeting SARS-CoV-2 antibodies without the specific requirement of an antibody. The presence of SARS-CoV-2 antibodies will interrupt the redox conversion of the redox indicator, resulting in a decreased current response. This electrochemical sensor was proven effective in real clinical sera from patients with satisfactory results. In addition, the proposed format was also extended to antigen detection (the spike protein of SARS-CoV-2), which presents new possibilities for diagnosing COVID-19.
Keywords: Paper-based sensors, Electrochemical detection, COVID-19, SARS-CoV-2
1. Introduction
In late 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first discovered and quickly transmitted via humans to humans (World Health Organization 2019). Subsequently, the World Health Organization (World Health Organization 2020) classified this coronavirus disease 2019 (COVID-19) outbreak as a global pandemic. As of now, over 32 million infected cases with approximately 1 million deaths worldwide have been reported (The New York Times 2020). Unfortunately, there is no specific vaccine or drug yet available for this ongoing pandemic; therefore, massive diagnostic devices or biosensors are needed among the international community to reduce the number of undetected cases (Dincer et al., 2019; Peteu 2010; Xu 2016; Xu et al., 2019). Therefore, the early and prompt diagnosis can play a crucial role in making a proper decision for the isolation of infected patients, thus slowing the spread of this infectious disease (Morales-Narváez and Dincer 2020).
Conventionally, the reverse transcription-polymerase chain reaction (RT-PCR)-based test for viral RNA detection is considered the gold-standard method for diagnosing COVID-19. However, the RT-PCR method might not be promptly available or affordable in many countries, as it is generally tested in a centralized laboratory/hospital by skilled personnel. In Thailand, the RT-PCR test was selectively offered to suspected individuals with strict criteria during the initial phase of the pandemic (Nopsopon et al., 2020) due to a paucity of testing supplies (for example, extraction kits and reagents). In addition, limiting factors from RT-PCR (time, cost, sampling errors, specialized handling and transportation, false-negative results in patients with unapparent clinical symptoms, and false-positive results in recovered patients) have also been raised in a recent study (Morales-Narváez and Dincer 2020).
To complement the detection of viral RNA, a few reports have demonstrated lateral flow immunoassay (LFA) platforms with optical detection (colorimetry/fluorescence) for targeting immunoglobulins (IgG, IgM, or IgA) that are produced in response to SARS-CoV-2 during the onset of disease (day four and beyond for IgM and day seven and beyond for IgG (Liu et al., 2020; Chen et al., 2020; Lin et al., 2020). Serological assays to detect these antibodies have proven to be a great complement to RT-PCR, particularly in or after the second week of infection. In addition, serological assays will help assess the immune response to natural infection as well as to vaccination in a population. Although these devices offer a user-friendly and rapid platform, they often exhibit limited sensitivity and require at least two antibodies for detection. To overcome these issues, an electrochemical approach has become an especially well-suited technique to detect SARS-CoV-2 antibodies. This technique offers an excellent capability to discriminate small changes from the recognition event on the electrode surface, thus enabling label-free detection with no need for a single antibody. Technically, such binding events between biomolecules affect the ability of the redox indicator (usually [Fe(CN)6]3-/4-) to reach the electrode surface and consequently its redox conversion (Mayorga-Martinez et al., 2015). Given these properties, this technique can significantly reduce the cost (from multiple antibody requirements) and time (from labeling procedures). Nevertheless, an electrochemical sensor for SARS-CoV-2 antibody detection has rarely been mentioned or explored thus far. Accordingly, we sought to design and fabricate a biosensor for diagnosing COVID-19 based on an electrochemical approach.
The present study demonstrates a paper-based electrochemical platform as a screening tool to detect SARS-CoV-2 immunoglobulins (represented by IgG and IgM). Paper, as a substrate material, was primarily used in this work as it has many advantages (such as low cost, natural abundance, and portability) (Yakoh et al., 2019). Furthermore, the paper itself can be safely disposed of by incineration after use, making it more suitable for infectious disease testing. Herein, the electrochemical paper-based analytical device for diagnosing COVID-19 (COVID-19 ePAD) comprises three parts (working ePAD, counter ePAD, and closing ePAD), which have different functions. In particular, in the test zone of the working ePAD, the SARS-CoV-2 spike protein containing receptor-binding domain (SP RBD) is immobilized to capture incoming SARS-CoV-2 antibodies. For the diagnostic step, the electrochemical response is monitored using the square-wave voltammetry (SWV) technique. In this regard, the SWV response is decreased upon immunocomplex formation. Notably, a series of assays was also performed in real patient sera (both SARS-CoV-2-infected and -uninfected patients), and the results were compared with a commercial standard ELISA method to demonstrate the potential effectiveness of the approach in real-world sample testing. Finally, the proposed platform was additionally extended to the detection of antigen (SARS-CoV-2), exhibiting excellent sensing adaptability for COVID-19 point-of-care (PoC) testing.
2. Materials and methods
2.1. Chemicals and apparatus
Details of the chemical and apparatus are presented in the supporting information.
2.2. Fabrication of the COVID-19 ePAD
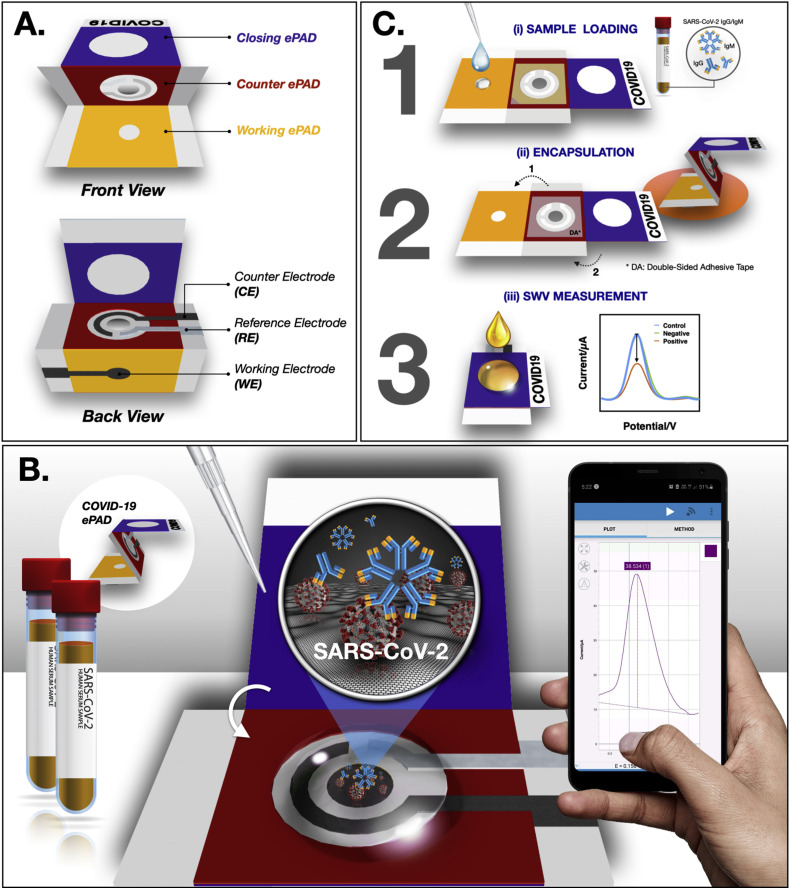
The device was designed using Adobe Illustrator CC (Adobe Systems, USA) and printed using a wax printer (Carrilho et al., 2009; Yakoh et al., 2020). The printed pattern was heated in an oven at 150 °C for 2 min to create a three-dimensional wax barrier. In this work, a single sheet of the COVID-19 ePAD consists of 3 folding layers: a working ePAD, a counter ePAD and a closing ePAD (see Fig. 1 A). The hydrophilic center of each zone was limited by a wax barrier, where the solution could flow through to the test zone at the bottom. Three electrodes were then screen printed at the back of the device and dried in an oven at 55 °C for 30 min. In particular, the working electrode (WE) was printed on the working ePAD, covering the test zone, while the counter electrode (CE) and the reference electrode (RE) were printed on the counter ePAD following the configuration shown in Fig. 1A (back view). The circle center of the counter ePAD was punctured as a hole to allow direct penetration of a solution to the test zone (once the redox solution was dropped). Double-sided adhesive (DA) tape (rectangular-shaped with a circular hole) was attached on both sides of the counter ePAD (front and back) to tighten the stacked layers without requiring an additional PDMS lid. For closing ePAD, it was designed to encapsulate the biohazard sample within the device and prevent it from being exposed to the environment after use.
Fig. 1.
Schematic illustration of the (A) device components, (B) detection principle and (C) detection procedure of the COVID-19 ePAD.
To prepare the COVID-19 ePAD for SARS-CoV-2 antibody detection, the spike protein receptor-binding domain (SP RBD) of SARS-CoV-2 was immobilized on the test zone of the working ePAD (front side) (see Fig. 1B) following the literature (Yakoh et al., 2019). Briefly, the graphene oxide (GO) solution was embedded in a porous structure of a test zone of the working ePAD and allowed to dry at RT. After washing with Milli-Q water, a carboxylic group (–COOH) of GO was activated with 20 mM EDC/40 mM NHS for 1 h and coupled with the SARS-CoV-2 SP RBD for 1 h. After a washing step with 0.01 M PBS (pH 7.4), the test zone was blocked using skim milk for 30 min at RT. The device was stored in the fridge at 4 °C until use. At this point, the device was ready to use.
To prepare a COVID-19 ePAD for SARS-CoV-2 spike protein detection, a similar procedure, as indicated in a previous section, was utilized, with a change from SARS-CoV-2 (SP RBD) immobilization to SARS-CoV-2 IgM immobilization.
2.3. Detection procedure of the COVID-19 ePAD
For the diagnostic step (Figure 1C), 10 μL of a human serum sample containing targeted antibodies is applied to the test zone of the working ePAD and incubated at RT (i). After reaction completed, the test zone is gently washed with 0.01 M PBS (pH 7.4) to remove unbound antibodies. Then, the plastic cover of double-sided adhesive tape attached on both sides of the counter ePAD was detached to enable paper folding. The counter ePAD was manually folded to the working ePAD and stacked with the closing ePAD for further analysis (ii). Notably, this configuration can minimize direct contact with biohazardous fluid and prevent exposure to the environment. For electrochemical detection (iii), a solution of the redox indicator ([Fe(CN)6]3-/4-) will be applied to the closing ePAD. Subsequently, the electrochemical response will be monitored using the square-wave voltammetry (SWV) technique.
Notably, a similar procedure was applied to the detection of the SARS-CoV-2 spike protein (unless stated otherwise).
2.4. Electrochemical measurements
Details of the electrochemical measurement are presented in the supporting information.
2.5. SARS-CoV-2 antibody detection in real human sera
Seventeen clinical sera (7 positive samples and 10 negative samples) were obtained from the Faculty of Medicine, Chulalongkorn University, Thailand. All serum samples were clinically confirmed with a commercial ELISA test kit to assess the IgA and IgG antibodies. The optical density (OD) ratio cutoff for a positive result was 1.1 for IgA and 0.8 for IgG (Kowitdamrong et al., 2020). Four of the negative control samples tested were reactive with anti-hepatitis B surface antigen (anti-HBsAg), anti-hepatitis C virus (anti-HCV), anti-Epstein Barr virus (anti-EBV), anti-Rubella, and anti-cytomegalovirus (anti-CMV), which will be further used for specificity assessment in this study. The study was approved by the Institutional Review Board of the Faculty of Medicine (IRB number 242/63) and the National Blood Center, Thai Red Cross Society (COA No. NBC 5/2020). Experiments were performed in the Chemical and Biological Sections in the Qualified Diagnostic Development Center, Chulalongkorn University by well-trained biosafety practice researchers.
3. Results and discussion
3.1. Design and characterization of the COVID-19 ePAD platform
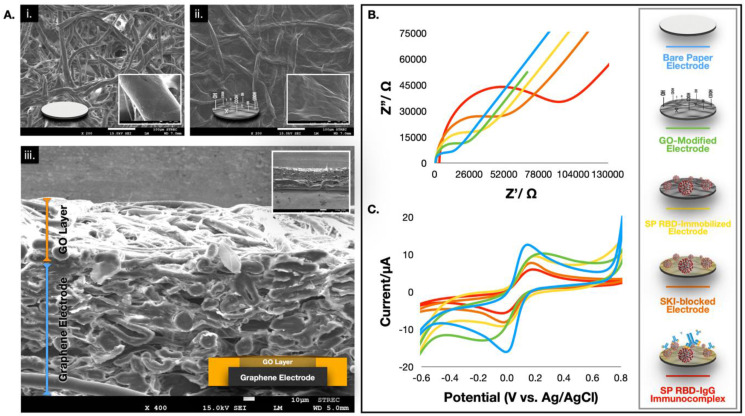
In this section, the conceptual design and characterization of the electrochemical detection of SARS-CoV-2 antibodies (represented by IgG and IgM) are described. The folding paper device comprises three main parts: a working ePAD, a counter ePAD, and a closing ePAD. SP RBD, which binds selectively to SARS-CoV-2 antibodies (both IgG and IgM), was immobilized on the hydrophilic paper zone of the working ePAD through embedded graphene oxide (GO)-EDC/NHS chemistry. Unlike other electrochemical immunosensors, where GO is directly drop-casted on the surface of the working electrode (WE) (Boonkaew et al., 2020; Jampasa et al., 2019; Özcan and Sezgintürk 2016), a layer of film peeling off is usually observed once the surface is rehydrated (by buffers in the system). This observation could be due to the strongly oxygenated and hydrophilic nature of the GO film. Thus, its stability and reproducibility were significantly reduced. Here, we reported a reversed electrode architecture (Fig. 1A) where the GO film was tightly embedded in a front-porous network of the working zone of cellulose paper (note that the WE was printed at the back of the device). This observation was ascribed to the presence of hydroxyl groups exhibited by both paper and GO, which permitted the water-based GO to integrate into the paper network via hydrogen bonding (plus physisorption) and form a stable structure (Cheeveewattanagul et al., 2017). At this stage, the carboxylic (−COOH) termination of GO is ready to activate (via EDC/NHS) and serve as an active site for SP RBD immobilization. We further characterized the morphologies of the proposed structure using SEM, as shown in Fig. 2 A. An unmodified paper device exhibited a porous network of cellulose fibers (panel i), whereas the GO nanosheet was distinctively overspread within the porous structure of the GO-embedded device (panel ii). Additionally, the cross-sectional SEM image (panel iii) clearly exhibited the reversed configuration, where the screen-printed graphene electrode was found at the bottom part of the paper while GO was impregnated over the underlying layer of the graphene working electrode. Additional characterization results from SEM, TEM and laser scanning confocal microscopy techniques are presented in Fig. S1-S3.
Fig. 2.
(A) SEM images of the bare paper (i), the GO modified paper (ii), and its corresponding cross-sectional image (iii). Characterization results of each fabrication step using the EIS (B) and CV (C) techniques. Note that all EIS Nyquist plots were fitted with the Randles equivalent circuit.
Next, the electrochemical behavior of each fabrication step was studied by complementary electroanalytical techniques (electrochemical impedance spectroscopy, EIS and cyclic voltammetry, CV). For EIS data interpretation, real and imaginary impedance components are plotted against one another in the Nyquist plot. It should be noted that each point on the Nyquist plot is the impedance at one frequency, where low-frequency data are on the right side of the plot, and higher frequencies are on the left. The whole of the Nyquist plot contains two parts: the straight diagonal line at lower frequencies indicates a typical diffusion-controlled redox behavior at the planar electrode; the semicircle part at higher frequencies indicates the efficiency of electron transfer from the electrode surface to the redox couple solution (or charge transfer resistance, Rct). For the EIS Nyquist plot shown Fig. 2A, it was observed that the bare paper electrode (blue line) possessed a small charge transfer resistance (small semicircle), which indicates a small obstruction toward redox conversion. When the WE was embedded with GO (green line), the Rct increased. This observation suggests that GO behaves as an insulator, which impedes electron transfer at the interface due to its disrupted sp2 bonding network (Hu et al., 2011). After the SP RBD was immobilized on the activated GO-embedded paper device (yellow line), continued enlargement of the Rct was observed, implying accomplished immobilization of the spike protein, which acts as an obstacle at the interface. Similar electrochemical behavior was also observed when the electrode was blocked with skim milk (SKI) to prevent nonspecific adsorption (orange line). Last, once SARS-CoV-2 IgG was captured with the immobilized SP RBD (red line), the rigidified structure of the antigen-antibody complex substantially forbids the charge transfer of the redox probe, resulting in the largest semicircle. In addition, these results are consistent with the experimental CV results, as indicated in Fig. 2C, whereby a gradual decrease in the peak heights (both oxidation and reduction directions) in each step was observed, suggesting a continued decline in charge transfer kinetics.
3.2. Assay optimization
In the present work, several parameters impacting the performance of COVID-19 ePAD (including the GO concentration, SP RBD concentration, SKI concentration and incubation time) were studied to obtain the maximum sensing efficiency. An optimum concentration for GO, SP RBD, and SKI was 0.4 mg/mL, 10 μg/mL, and 5% (w/v), respectively, with an incubation period of 30 min. Details of the assay optimization are presented in Fig. S4.
3.3. SARS-CoV-2 antibodies detection using the COVID-19 ePAD
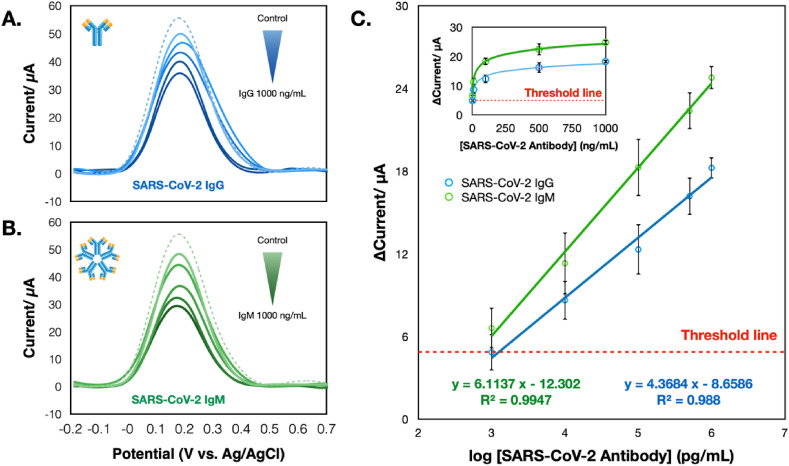
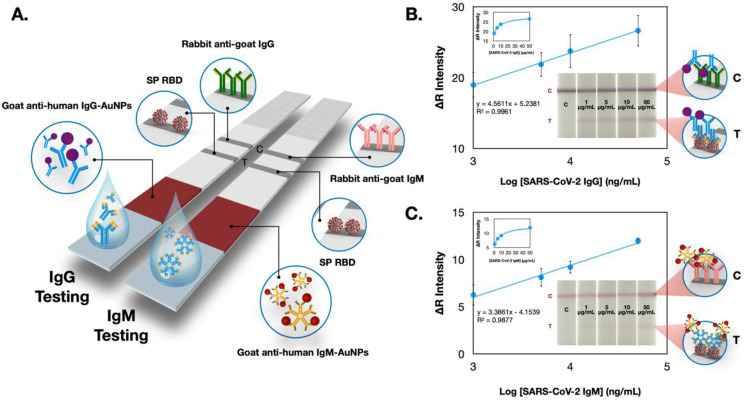
In this regard, the COVID-19 ePAD is intended for the qualitative screening of total SARS-CoV-2 antibodies against the viral protein. Infection with SARS-CoV-2 will initiate an immune response producing circulating immunoglobulin antibodies (both IgG and IgM). Hence, both IgG and IgM were examined individually in this study. From Fig. 3 A and B, both antibodies can be captured by the SP RBD and demonstrate similar performance. Nevertheless, IgM testing revealed a higher sensitivity than IgG testing (Fig. 3C), which could be ascribed to a larger IgM antibody size. Typically, IgM is the largest immunoglobulin, with pentamer units and ten antigen-binding sites (~900 kDa), whereas IgG is a monomer unit with two antigen-binding sites (~150 kDa), which is lighter and smaller (Burrell et al., 2017; Casali 1998; Nezlin 1998). Thus, the more considerable hindrance from IgM (compared with IgG) toward the redox mediator could undoubtedly be observed, as illustrated in the SWV voltammograms. This electrochemical immunosensor exhibited a sensitive response to the presence of SARS-CoV-2 antibodies, where the current proportionally increased with logarithmic concentrations of SARS-CoV-2 IgG and IgM in the range from 1 to 1000 ng/mL (R2 > 0.99). From this logarithmic response, the limit of detection (LOD) values of SARS-CoV-2 IgG and IgM were (3SDblank/slope) 0.96 and 0.14 ng/mL, respectively. Therefore, we estimated the current response based on the LOD and starting point of the calibration plot (~1 ng/mL) as the cutoff threshold for further experiments. Reproducibility evaluated in terms of the % relative standard deviation (RSD) from different ePADs (n = 3) was 4.2% for IgG (1000 ng/mL) and 3.3% for IgM (1000 ng/mL). In addition, we further compared the performance of this COVID-19 ePAD with an in-house colorimetric LFA device prepared using the same batch of immobilized SP RBD (Fig. 4 ). Details of the colorimetric LFA fabrication and detection procedures are presented in the supporting information. Image processing (ImageJ software) was utilized to monitor the color change from the gold nanoparticles (AuNPs). In this regard, the colorimetric LFA platform exhibited the lowest detection limit that could be distinguished by the naked eye at 1 μg/mL for both antibodies (Fig. 4B and C) and was inferior to that of the COVID-19 ePAD. Clearly, a sensitivity enhancement of the COVID-19 ePAD over the traditional LFA was achieved due to the sensitive electrochemical technique and the graphene-based materials utilized in this work. It should be noted that AuNPs 20 nm in size were used for the goat anti-human IgM conjugation, while AuNPs 40 nm in size were used for the goat anti-human IgG conjugation, thereby giving different colors on the T and C lines. Notably, the rigid requirement of multiple antibodies in this colorimetric LFA device significantly amplifies the considerable advantage of the label-free COVID-19 ePAD, in which no antibodies are required for the electrochemical system, thus reducing the overall number of steps (labeling and fabrication steps) and cost of the device. Additionally, the comparison of the analytical performance between this ePAD and other sensors for serological detection of SARS-CoV-2 antibodies published thus far is summarized in Table S1.
Fig. 3.
SWV responses of the COVID-19 ePAD tested with different concentrations of SARS-CoV-2 IgG (A) and SARS-CoV-2 IgM (B) in the presence of 5 mM [Fe(CN)6]3-/4-. (C) A linear relationship between current vs logarithmic concentration of SARS-CoV-2 IgG and IgM and its corresponding relationship between current and concentration of SARS-CoV-2 IgG and IgM. The threshold line was estimated based on an LOD (3SDblank/slope) and the starting point of the calibration plots.
Fig. 4.
(A) Schematic illustration of the in-house LFA colorimetric test strips for detecting SARS-CoV-2 IgG and IgM. (B) and (C) represent the calibration plots of color intensity in the red channel as a function of the logarithmic concentration of SARS-CoV-2 IgG and IgM (n = 3). Illustrated in the insets of each calibration plot are the photographic images of the LFA device after loading different concentrations of IgG and IgM. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The aspect of ePAD stability, which is defined as the retention within 95–105% of the initial response, was also evaluated. By keeping the paper device in the refrigerator at 4 °C before use, the results demonstrated that the COVID-19 ePAD could retain its performance for up to 14 days, indicating satisfactory stability (Fig. S5). All things considered, then it could denote the potential functionality of the proposed platform for the rapid screening of SARS-CoV-2 antibodies.
3.4. Cross-reactivity
Cross-reactivity was initially carried out to investigate any cross-reaction of other antibodies against viral antigens. Four negative controls, which are reactive for anti-HBsAg, anti-HCV, anti-EBV, anti-Rubella, and anti-CMV, were tested with the proposed COVID-19 ePAD, and the results were also compared to those from a commercial ELISA test kit. The cutoff value of the current was approximately set from the LOD and starting point of the working range (~1 ng/mL). This means that the negative control, which had a current difference below the threshold of the LOD, will be inferred as having “no cross-reactivity”. From the experiment, it was found that the presence of nontargeted antibodies does not affect the response of the sensor (Table S2). Therefore, it can be concluded that no cross-reactivity from nontargeted antibodies was observed, indicating the excellent specificity of the developed sensor.
3.5. Clinical samples testing
Finally, to demonstrate the practicality of this COVID-19 ePAD, real clinical serum samples were tested with the prepared ePAD. In this study, 17 serum samples were tested with the ePAD system, in which 7 of these sera were confirmed to be infected with SARS-CoV-2 by a commercial ELISA test kit (the gold standard method for protein detection). The cutoff value was defined by the same threshold mentioned above. A sample that had a relative current difference above the LOD threshold was identified as a positive sample. As shown in Table 1 , 7 human sera from 7 ELISA-confirmed samples were positive. Additionally, 9 serum samples tested to be negative by the ELISA test kit correspondingly tested to be negative by the COVID-19 ePAD, whereas one tested negative control sample was positive by our proposed sensor. This positive result is likely due to the possible SARS-CoV-2 IgM present in the sample, which cannot be detected by ELISA (ELISA responds to only IgA and IgG). Nevertheless, the COVID-19 ePAD can capture all of the immunoglobulins present in the sample (IgG, IgM, and IgA); thus, it may result in a more sensitive response. The statistical Kappa test was also performed to analyze the interrater reliability of qualitative measurements. The analysis data showed a coefficient of 0.881, implying an almost perfect agreement ( > 0.81). In addition, the sensitivity and specificity of the developed COVID-19 ePAD were calculated to be 100% and 90%, respectively (Carvajal and Rowe 2010). Considering all the results described above, it certainly supports the conclusion that the developed COVID-19 ePAD for the serological detection of SARS-CoV-2 antibodies demonstrates potential applicability for qualitative screening of antibodies against SARS-CoV-2. Several advantages, including (i) a small sample volume (10 μL), (ii) no requirements of an antibody, which substantially reduced the overall number of steps and cost, (iii) no tedious labeling procedures (e.g., gold-conjugated antibody), due to the label-free detection, (iv) a compact device that enables it to be remotely used outside the realm of a laboratory, and (v) an encapsulated paper configuration that allows it to be safely disposed and prevents exposure to the environment.
Table 1.
Statistical comparisons between the COVID-19 ePAD and commercial ELISA techniques.
| Commercial ELISA |
||||
|---|---|---|---|---|
| + | – | Total | ||
| COVID-19 ePAD | + | 7 | 1 | 8 |
| – | 0 | 9 | 9 | |
| Total | 7 | 10 | 17 | |
ELISA: enzyme-linked immunosorbent assay, +: positive result, -: negative result.
3.6. SARS-CoV-2 spike protein detection and assay optimization
From the advanced capability of the proposed COVID-19 ePAD system, the scope of the device can be further expanded for direct viral antigen detection. It has been reported in the previous literature that SARS-CoV-2 encodes four proteins, including the spike, envelope, matrix and nucleocapsid proteins. Among these structural proteins, the spike protein is an abundant transmembrane protein of the virus and exhibits high immunogenicity (Seo et al., 2020). Furthermore, the amino acid sequence of this protein possesses diversity from other coronaviruses (Lu et al., 2020), making it a suitable antigen for the selective identification of SARS-CoV-2. Hence, we selected the spike protein as a model antigen for further testing with the COVID-19 ePAD.
Initially, the effect of the concentration of immobilized antibody (SARS-CoV-2 IgM) and incubation time were studied to maximize the electrochemical response (see Fig. S6). Within the studied range of antibody concentrations (0.1, 1, 10, 20, and 50 μg/mL), the highest current was obtained when the IgM concentration of 20 μg/mL was immobilized (Fig. S6A). Likewise, the influence of incubation time for the SARS-CoV-2 spike protein was also investigated from 10 to 60 min. The result showed that the incubation period of 45 min offered the best sensitivity and was then chosen for further experiments (Fig. S6B).
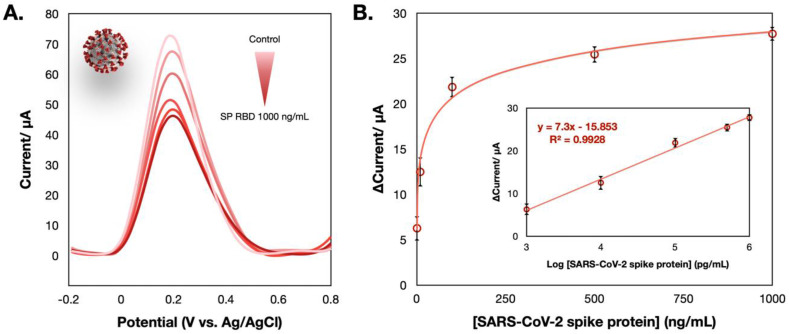
Next, the performance of the electrochemical sensor for detecting the SARS-CoV-2 spike protein was evaluated by the SWV technique. According to the SWV voltammogram presented in Fig. 5 A, the sensor also exhibited a sensitive response to the presence of the SARS-CoV-2 spike protein, where the current response proportionally decreased to the concentration of the SARS-CoV-2 SP RBD. A linear dynamic response toward spike protein sensing was constructed in the range of 1–1000 ng/mL with an LOD of 0.11 ng/mL (3SDblank/slope). Considering this LOD, it is not sufficient to detect the antigen in human nasopharyngeal swab specimens (down to pg/mL). However, the progressive development of SARS-CoV-2 biosensor is achieved among limited sensors that are currently available.
Fig. 5.
(A) SWV responses of the COVID-19 ePAD tested with different concentrations of the SARS-CoV-2 spike protein in the presence of 5 mM [Fe(CN)6]3-/4-. (B) The relationship between current and concentration of SARS-CoV-2 spike protein; the inset shows its corresponding calibration plotted between the current and logarithmic concentration of the SARS-CoV-2 spike protein.
In summary, it can be concluded that the developed electrochemical sensor shows excellent promise for use as a supplementary detection indicator for suspected cases. In addition, using this ePAD format can be extended to a broad application range, allowing for the future development of a point-of-care (POC) diagnostic device for different analytes of interest.
4. Conclusions
Conclusively, we have demonstrated an advantageous paper-based electrochemical platform for diagnosing COVID-19. The sensing scheme relies on the disruption of the redox conversion ([Fe (CN)6]3-/4-) triggered by immunocomplex formation between the captured immunoglobulins produced in response to SARS-CoV-2 in humans with the immobilized spike protein of SARS-CoV-2. Unlike other lateral flow immunoassay (LFA) platforms where multiple antibodies are exploited in charge of the reporter, no antibodies are required in this work as a consequence of the label-free electrochemical system. The fast (30 min) and sensitive detection of SARS-CoV-2 antibodies was recorded with a detection limit of 1 ng/mL, which is more sensitive (3 orders of magnitude) than the colorimetric LFA. In addition, this paper-based device is capable of detecting targeted antibodies in clinical sera from patients and has an acceptable sensitivity and specificity (100% and 90%, respectively). We further expanded the functionality of the paper-based electrochemical platform for the direct detection of the spike protein antigen of SARS-CoV-2. Although the detection limit of this electrochemical sensor for SARS-CoV-2 determination has not yet achieved the detection level in real nasal swab specimens at the present stage of this study, the progressive development of the point-of-care sensor for detecting SARS-CoV-2 is accomplished among limited sensors that are currently available. Furthermore, we believe that the developed rapid serology testing coupled with specific immunoglobulin determination would provide more informative results, thus allowing for better management and treatment in an infected population.
CRediT authorship contribution statement
Abdulhadee Yakoh: Conceptualization, Methodology, Investigation, Writing - original draft, Visualization. Umaporn Pimpitak: Investigation. Sirirat Rengpipat: Resources, Writing - review & editing, Supervision. Nattiya Hirankarn: Validation, Formal analysis, Resources, Writing - review & editing. Orawon Chailapakul: Resources, Writing - review & editing. Sudkate Chaiyo: Conceptualization, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research project is supported by the Second Century Fund (C2F), Chulalongkorn University and partially supported from Innovation Hub of Ageing Society: Diagnostic Platform 2017. Also, the authors would like to thank Chulalongkorn University Technology Center (UTC), and National Research Council of Thailand (NRCT).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112912.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Boonkaew S., Teengam P., Jampasa S., Rengpipat S., Siangproh W., Chailapakul O. Analyst. 2020;145(14):5019–5026. doi: 10.1039/d0an00564a. [DOI] [PubMed] [Google Scholar]
- Burrell C.J., Howard C.R., Murphy F.A. In: Fenner and White's Medical Virology. fifth ed. Burrell C.J., Howard C.R., Murphy F.A., editors. Academic Press; London: 2017. Chapter 6 - adaptive immune responses to infection; pp. 65–76. [Google Scholar]
- Carrilho E., Martinez A.W., Whitesides G.M. Anal. Chem. 2009;81(16):7091–7095. doi: 10.1021/ac901071p. [DOI] [PubMed] [Google Scholar]
- Carvajal D.N., Rowe P.C. Research and statistics. Pediatr. Rev. 2010;31(12):511. doi: 10.1542/pir.31-12-511. [DOI] [PubMed] [Google Scholar]
- Casali P. In: Encyclopedia of Immunology. second ed. Delves P.J., editor. Elsevier; Oxford: 1998. IgM; pp. 1212–1217. [Google Scholar]
- Cheeveewattanagul N., Morales-Narváez E., Hassan A.-R.H.A., Bergua J.F., Surareungchai W., Somasundrum M., Merkoçi A. Adv. Funct. Mater. 2017;27(38):1702741. [Google Scholar]
- Chen Z., Zhang Z., Zhai X., Li Y., Lin L., Zhao H., Bian L., Li P., Yu L., Wu Y., Lin G. Anal. Chem. 2020;92(10):7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- Dincer C., Bruch R., Costa-Rama E., Fernández-Abedul M.T., Merkoçi A., Manz A., Urban G.A., Güder F. Adv. Mater. 2019;31(30):1806739. doi: 10.1002/adma.201806739. [DOI] [PubMed] [Google Scholar]
- Hu Y., Li F., Bai X., Li D., Hua S., Wang K., Niu L. Chem. Commun. 2011;47(6):1743–1745. doi: 10.1039/c0cc04514d. [DOI] [PubMed] [Google Scholar]
- Jampasa S., Lae-ngee P., Patarakul K., Ngamrojanavanich N., Chailapakul O., Rodthongkum N. Biosens. Bioelectron. 2019;142:111539. doi: 10.1016/j.bios.2019.111539. [DOI] [PubMed] [Google Scholar]
- Kowitdamrong E., Puthanakit T., Jantarabenjakul W., Prompetchara E., Suchartlikitwong P., Putcharoen O., Hirankarn N. PloS One. 2020;15(10) doi: 10.1371/journal.pone.0240502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wen D., Wu J., Liu L., Wu W., Fang X., Kong J. Anal. Chem. 2020;92(14):9454–9458. doi: 10.1021/acs.analchem.0c01635. [DOI] [PubMed] [Google Scholar]
- Liu X., Wang J., Xu X., Liao G., Chen Y., Hu C.-H. Emerg. Microb. Infect. 2020;9(1):1269–1274. doi: 10.1080/22221751.2020.1773324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayorga-Martinez C.C., Chamorro-García A., Serrano L., Rivas L., Quesada-Gonzalez D., Altet L., Francino O., Sánchez A., Merkoçi A. J. Mater. Chem. B. 2015;3(26):5166–5171. doi: 10.1039/c5tb00545k. [DOI] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezlin R. In: The Immunoglobulins. Nezlin R., editor. Academic Press; New York: 1998. Chapter 1 - general characteristics of immunoglobulin molecules; pp. 3–73. [Google Scholar]
- Nopsopon T., Pongpirul K., Chotirosniramit K., Hiransuthikul N. medRxiv. 2020 doi: 10.1101/2020.06.24.20139188. 2020.06.24.20139188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan B., Sezgintürk M.K. Talanta. 2016;160:367–374. doi: 10.1016/j.talanta.2016.07.039. [DOI] [PubMed] [Google Scholar]
- Peteu S.F. 2010. CAS 2010 Proceedings (International Semiconductor Conference) pp. 179–190. [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- The New York Times . The New York Times, nytimes.com; 2020. Covid-19 Live Updates: World Nears Grim Milestone of a Million Deaths. [Google Scholar]
- World Health Organization . 2019. Coronavirus Disease (COVID-19) Weekly Epidemiological Update and Weekly Operational Update. [Google Scholar]
- World Health Organization . World Health Organization; 2020. Naming the Coronavirus Disease (COVID-19) and the Virus that Causes it. [Google Scholar]
- Xu K. IEEE Sensor. J. 2016;16(16):6184–6191. [Google Scholar]
- Xu K., Chen Y., Okhai T.A., Snyman L.W. Opt. Mater. Express. 2019;9(10):3985–3997. [Google Scholar]
- Yakoh A., Chaiyo S., Siangproh W., Chailapakul O. ACS Sens. 2019;4(5):1211–1221. doi: 10.1021/acssensors.8b01574. [DOI] [PubMed] [Google Scholar]
- Yakoh A., Siangproh W., Chailapakul O., Ngamrojanavanich N. ACS Appl. Mater. Interfaces. 2020;12(20):22543–22551. doi: 10.1021/acsami.0c03812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.