Abstract
Electrochemical biosensors combine the selectivity of electrochemical signal transducers with the specificity of biomolecular recognition strategies. Although they have been broadly studied in different areas of diagnostics, they are not yet fully commercialized. During the COVID-19 pandemic, electrochemical platforms have shown the potential to address significant limitations of conventional diagnostic platforms, including accuracy, affordability, and portability. The advantages of electrochemical platforms make them a strong candidate for rapid point-of-care detection of SARS-CoV-2 infection by targeting not only viral RNA but antigens and antibodies. Herein, we reviewed advancements in electrochemical biosensing platforms towards the detection of SARS-CoV-2 through studying similar viruses.
Keywords: Pathogen diagnostics, Coronaviruses, Nucleic acid–based detection, Serological testing, Recognition element, Electrochemical signal
1. Introduction
This review perspective discusses the most recent advancements in electrochemical diagnostic methods to combat the current viral pandemic. The massive number of publications on COVID-19 diagnostics motivated this review intending to encourage the development of effective point-of-care approaches based on electrochemical biosensing. We dedicate our work to the healthcare providers and the front-line workers whose roles can be assisted through more efficient methods for point-of-care diagnostics during the pandemic.
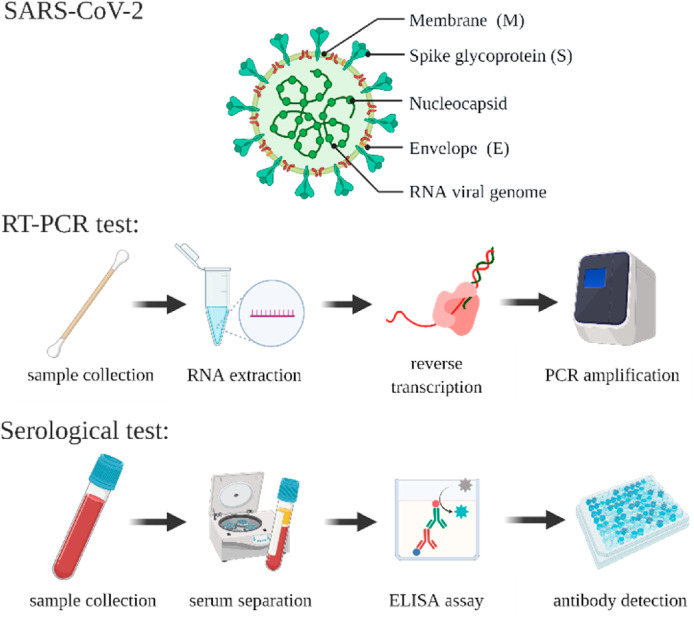
One of the main challenges during the COVID-19 pandemic is an urgent need for improved pathogen diagnostic techniques (Cesewski and Johnson 2020; Uhteg et al. 2020). Accurate and widespread testing is essential for the containment of SARS-CoV-2, facilitating efficient contact tracing and necessary treatment (Qin et al. 2020; Shen et al. 2020). However, restricted by supply-chain shortages and limited accredited laboratories, the implementation of adequate testing regimes has been substandard in various countries (Germany 2020; Moatti 2020). Conventional detection platforms such as polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) create and perpetuate these issues as these laboratory-based techniques often require trained personnel to perform multiple time-consuming steps, using large volumes of expensive reagents (Scheme 1 ). The complicated nature of these tests makes them unsuitable for rapid large-scale diagnostics, restricting the availability and distribution of COVID-19 tests (Feng et al. 2020). Table 1 summarizes the advantages and limitations of existing diagnostic methods.
Scheme 1.
(top) The Novel Coronavirus SARS-CoV-2 illustrated with its components, including the surface proteins and viral RNA. Illustration of various steps to perform (middle) RT-PCR, and (bottom) ELISA-serological tests.
Table 1.
Comparison of electrochemical and conventional pathogen detection platforms.
Improved assays have achieved results in less than 1 h.
In particular, reverse-transcriptase polymerase chain reaction (RT-PCR) is the gold standard for diagnosis of COVID-19, as recommended by the World Health Organization (WHO) and the American Center for Disease Control (CDC) (Control and Prevention 2020; Organization 2020). The testing workflow consists of the reverse transcription of viral RNA to complementary DNA strands, followed by PCR-based amplification of specific regions on the complementary DNA (Scheme 1, middle). In the case of SARS-related viral genomes, there are three main conserved target sequences: (1) RNA-dependent RNA polymerase gene (RdRp gene), (2) envelope protein gene (E gene), and (3) nucleocapsid protein gene (N gene) (Corman et al. 2020a; Udugama et al. 2020). A recommended RT-PCR protocol initially developed by Corman et al. (2020b) was a combination of an E gene assay and a SARS-CoV-2-selective RdRp gene assay with detection limits of 3.9 and 3.6 copies per reaction, respectively. Despite the high sensitivity of this method, reports studying the assessment of COVID-19 diagnostic methods found that these tests can be susceptible to false negative rates due to insufficient viral genetic material in the sample as well as laboratory and transportation error (Huang et al. 2020; Jung et al. 2016; Kubina and Dziedzic 2020; Li et al. 2020a; Santiago 2020; Velay et al. 2020; Xie et al. 2020).
The superior sensitivity and limit of detection (LOD) of a diagnostic platform depend on the infectious dose and the minimum viral load of the SARS-CoV-2 virus. The infectious dose of a virus is the number of virus particles that are enough to infect 50% of a given population (Schröder 2020), and the viral load is the number of virus particles in an infected individual (Walsh et al. 2020). For SARS-CoV-2, the infectious dose is unknown, and the viral load is highly variable (Schröder 2020; Walsh et al. 2020). As a result, defining the minimum detection limit for the diagnostic platform is challenging. Until the virological levels are concretely known, comparative studies are an effective means of determining the required detection limits for SARS-CoV-2 diagnostic tests. Current ‘best-in-class’ diagnostic tests have detection limits around 100 copies/mL (Arnaout et al. 2020).
In general, the aforementioned limitations establish an immediate need for more accurate, affordable, and portable tests that are easy-to-use at large scales. This review explores electrochemical diagnostic tests as an alternative to conventional pathogen detection methods by studying recent advancements in the field of biosensing.
2. Electrochemical biosensors for pathogen diagnostics
Electrochemical sensors, analogous to glucose meters (Scheme 2 ), attempt to address the need for rapid, sensitive, low-cost, and easy-to-operate detection strategies (Huy et al. 2011; Layqah and Eissa 2019). By employing a signal transducer, based on the direct quantifiable electronic response of an electrochemical reaction on the electrode, these sensors can provide signal selectivity in complex media such as whole blood, unlike optical- or mass-based detection methods (Thévenot et al. 2001). The signal is defined as an increase (signal-on) or a decrease (signal-off) in electrochemical response of the electrode by the target concentration elevation, which is measured either through the electron transfer (CA, CV, SWV) or the electron transfer resistance (EIS) (Xiao et al. 2005). Furthermore, the immobilization of biomolecular recognition elements on the electrode’s surface adds specificity to the target, required in complex media. In this regard, electrochemical (bio)sensors have become an ideal tool for the direct detection of biomarkers related to, for example, pathogens in human specimens (Cesewski and Johnson 2020).
Scheme 2.
Electrochemical biosensors can potentially assess various human specimens including finger-prick blood, nasopharyngeal, and saliva samples, for target analytes. The platforms possess (1) a readout device (2) a biosensing chip with sample delivery system and electronic leads to transfer the response of the electrodes to the readout device, (3) electrodes with the corresponding recognition elements of complementary oligonucleotides, antibodies, antigens to detect the target oligonucleotides, antigens, and antibodies, respectively, with a quantifiable current response proportional to the analyte concentration.
In addition to sensitivity and specificity, widespread sensing issues, such as high costs, large reagent volumes, and lengthy analysis times, can also be addressed with the electrochemical platforms. These favourable characteristics can be optimized through a multitude of strategies, including modification of the electrode materials, recognition probes, immobilization methods, and redox labels. For example, using the redox label ferrocene leads to high target affinity, whereas using methylene blue enhances the long-term stability (Kang et al. 2009). Additionally, nanotechnology has had a significant impact on all electrochemical biosensors. Specifically, nanoparticles can be used as electrochemical labels, or to increase the electron-transfer efficiency or the solution-interface surface area (Jianrong et al. 2004). For example, Khater et al., Lee et al., and Steinmetz et al. used gold nanoparticles to improve the response of their sensors when detecting viral pathogens (Khater et al. 2019; Lee et al. 2019; Steinmetz et al. 2019). The use of nanotechnology is in accordance with the recent trend of miniaturization and enhancing portability in biosensors (da Silva et al. 2017; Khater et al. 2017). Similarly, advancements in isothermal amplification methods have led to the development of ultrasensitive electrochemical sensors to detect various pathogens (de la Escosura-Muñiz et al. 2016; Yu et al. 2015). Improving parameters, such as portability while maximizing sensitivity, is vital during the COVID-19 pandemic and can be achieved using these novel approaches (Cesewski and Johnson 2020). A multitude of novel testing platforms is reviewed here and summarized in Table 2 .
Table 2.
Classification of electrochemical biosensors studied in this review: SWV: square wave voltammetry; CV: cyclic voltammetry; EIS: electrical impedance spectroscopy; CA: chronoamperometry; IV: Influenza virus.
| Authors | Target | Signal Output | Recognition Element | Limit of Detection (LOD) | Linear Range | |
|---|---|---|---|---|---|---|
| Nucleic Acid Detection | Abad-Valle et al. (2005) | SARS-CoV (30-bp of genome) | SWV signal-on | Thiol-modified target complementary DNA probe | 6 pM | 0.102–5.10 nM |
| (Martínez-Paredes et al. 2009) | SARS-CoV (30-bp of genome) | CV signal-on | Alkaline phosphatase labelled target complementary DNA probe | 2.5 pM (2.5 pmol/L) | 2.5–50 pmol/L | |
| Díaz-González et al. (2008) | SARS-CoV (30-bp of genome) | CA signal-on | complementary DNA probe (electrostatic immobilization) | 0.5 nM | 20–200 pM | |
| Malecka et al. (2016) | Avian IV H5N1 | SWV signal-off | Thiolated ssDNA probe | 1 pM | Not Reported | |
| Chung et al. (2011) | Type A IV | CV signal-off | Biotinylated Complementary DNA probe | (85.1 fM) 8.51 × 10-14 M | 1 × 10-13 - 1 × 10-10 M |
|
| Ju et al. (2003) | HBV | CV signal-on | HBV complementary ss-DNA fragments | 10-21 mole of the original fragment | Not Reported | |
| Jampasa et al. (2014) | HPV | SWV signal-off | Anthraquinone-labelled pyrrolidinyl peptide nucleic acid probe | 4 nM | 0.02–12 μM | |
| Viral Antigen Detection | Hassen et al. (2011) | Type A IV | EIS signal-on | Polyclonal antibodies | 8 ng/mL | 0–64 ng/mL |
| Han et al. (2016) | H1N1, H5N1, H7N9 IV | CA signal-on | Target specific capture antibodies | 1 pg/mL | 1–10 ng/mL | |
| Singh et al. (2017) | H1N1 IV | CA signal-on | Target specific capture antibodies | 0.5 PFU/mL | 1 - 1 × 104 PFU/mL | |
| Layqah and Eissa (2019) | MERS-CoV and H–CoV | SWV signal-on | MERS-CoV specific and human coronavirus antibodies | 0.4 pg/mL and 1.0 pg/mL | 10-3 – 102 ng/mL and 10-2 – 104 ng/mL | |
| Seo et al. (2020) | SARS-CoV-2 | FET | SARS-CoV-2 spike antibody | 1 fg/mL and 1.6 × 101 PFU/mL | 1.6 × 101 -1.6 × 104 PFU/mL | |
| Antibody Detection | Jarocka et al. (2014) | Anti-hemagglutinin antibodies | EIS signal-on | His-tagged hemagglutinin and anti-his antibodies | 2.1 pg/mL | 4–20 pg/mL |
| Mikuła et al. (2018) | Anti-hemagglutinin antibodies | SWV signal-off | Recombinant his-tagged hemagglutinin | Not Reported | Not Reported |
SARS-CoV-2 can be detected in multiple ways: the genetic material of the virus, viral antigens, or serological antibody testing (Huang et al. 2009; Huy et al. 2011; Morales-Narváez and Dincer 2020). A significant contributor to the severity of the pandemic was the lack of knowledge of SARS-CoV-2 and the progression of the infection. Throughout the pandemic, scientific understanding of the virus has grown exponentially, particularly by drawing similarities between the novel SARS-CoV-2 and physically and genetically similar viruses, such as SARS-CoV, MERS-CoV, or influenza. For example, one of SARS-CoV-2’s entry mechanisms was determined from the existing research on SARS-CoV’s (Rabaan et al. 2020). This practice can be further employed for the mechanisms of detection. By studying established platforms capable of detecting well-studied viruses, more efficient methods for detecting SARS-CoV-2 can be designed (Al Johani and Hajeer 2016; Andersen et al. 2020; Dudas et al. 2018; Wu et al. 2020).
3. RNA/DNA detection for coronaviruses
Nucleic acid-based electrochemical sensors (genosensors) use synthetic oligonucleotide constructs as recognition elements, with specific sequences complementary to the target DNA or RNA (Abad-Valle et al. 2005; González-López and Abedul 2020). If present, the target analyte will hybridize to the recognition element immobilized on the electrode’s surface. This hybridization induces a quantifiable change proportional to the analyte concentration and is detected using electrochemical signal transducers (Liu et al. 2009; Yu et al. 2017; Zhou et al. 2014). To act as a reasonable alternative to RT-PCR-based assays, electrochemical genosensors must have a comparable sensitivity, which could be tunable through the various optimization strategies discussed above.
Electrochemical approaches, for the detection of SARS-CoV nucleic acids, constituted of probe strands that are complementary to a portion of the target genome, which are immobilized on the surface of an electrode. Some earlier works such as Abad-Valle et al. (2005) and Martínez-Paredes et al. (2009) developed similar electrochemical genosensors targeting the 30-base-pair sequences of the genome of SARS-associated coronavirus on the surface of 100-nm gold-sputtered films (Abad-Valle et al. 2005) and gold nanostructured screen-printed carbon electrodes (Martínez-Paredes et al. 2009). The targets were further conjugated to alkaline phosphatase-labelled streptavidin (AP-SA) through biotinylation, following hybridization with the complementary probes on the electrode’s surface. The biotinylated targets acted as a substrate for the enzymatic reaction with AP-SA (Scheme 3 -A). The reactions generate an electrochemical signal (Table 2, Scheme 3-A’ and 3-A’’), which passes through the probe to the electrode, ensuring selectivity against mismatches (Scheme 3-A’’’). Both platforms exhibited low picomolar detection limits of 6 pM and 2.5 pM, respectively (Abad-Valle et al. 2005; Martínez-Paredes et al. 2009). Despite requiring an additional modification step for the target, by employing the unconventional gold electrodes smaller sample volumes could be used. These platforms evidently address a key concern of traditional nucleic-acid detection assays while reducing the overall cost and achieving low detection limits comparable to PCR-based tests. Díaz-González et al. also suggested the use of an Au(I) complex, sodium aurothiomalate, to label the same 30-base-pair target. The labelled target would then hybridize with probes electrostatically immobilized on the polylysine-modified electrode surface (Scheme 3-B). Through this platform, hydrogen evolution, catalyzed by the gold label, causes a current proportional to probe hybridization (a signal-on assay, Scheme 3-B’), with selectivity against a three-base mismatch. This sensor was compared to a similar one with an alkaline phosphatase-label and displayed a higher limit of detection, 0.5 nM to the 8 pM of the alternative label. Despite its higher detection limit, the sodium aurothiomalate-labelled biosensor boasted a significant reduction in the analysis time, approximately half of the time required by the alkaline phosphate labelled sensor (Díaz-González et al. 2008). This study raises the question of property priority; is it permissible, in certain circumstances, to have reduced sensitivity in favour of low analysis time?
Scheme 3.
Brief illustration of the electrochemical biosensing platforms through the assay format and electrochemical responses for the detection of RNA/DNA. Panels A to F represent the assay formats for various platforms discussed in this study; A’ to F’ and A’’ show the type of electrochemical responses and how it varies with target concentration ((a) and (b) represents either with target (+target) or without target (–target)); A‴, C’’, D’’ and F’’ are shown in their original scale representing the details on calibration curves and selectivity responses.
In another attempt, Malecka et al. (2016) and Chung et al. (2011) developed label-free genosensor platforms to detect different strains of the Influenza virus by directly targeting the nucleic acid sequences of the virus without any modifications to the target. Malecka et al. (2016) designed and compared two platforms, one on gold disc electrodes and another on gold screen-printed electrodes; both platforms were immobilized with the same manner using a thiol-gold binding approach (Scheme 3-C). Chung et al. (2011) prepared an avidin-modified glassy carbon electrode functionalized with biotinylated DNA probes using an avidin-biotin immobilization approach (Scheme 3-D). For all of these platforms, the electrochemical signal was generated by the redox-active marker [Fe(CN)6]3-/4-, which was added to the testing solution to distinguish the lower current signal with the target DNA or RNA hybridization from the higher current signal without (Table 2, Scheme 3-C’, 3-D’ and 3-D’’). In the case of gold-based electrodes designed by Malecka et al., the RNA transcripts could be specifically detectable at the minimum concentration of 1 pM without the need for further amplification (Scheme 3-C’’). However, the refolding of the RNA transcripts could negatively impact the accuracy of the sensor (Malecka et al. 2016). Using avidin/biotin-modified glassy carbon electrode, a lower limit of detection of 85.1 fM (8.51 × 10-14 M) was achieved by Chung et al. (Scheme 3-D’’) (Chung et al. 2011). Conclusively, these platforms offer more straightforward approaches compared to the previous ones discussed due to the absence of a target modification step and the label-free nature of their signaling mechanism (i.e. using redox-active marker [Fe(CN)6]3-/4- instead).
Ju et al. (2003) also proposed a label-free biosensor with the hybridization approach for the detection of hepatitis B virus (HBV) DNA as the product of PCR. They covalently immobilized the single-stranded HBV-DNA fragments on the surface of a gold electrode modified with a thioglycolic acid monolayer (Pividori et al. 2000). The detection was performed through hybridization of the target DNA to the complementary sequence, where di(2,2′-bipyridine)osmium (III) ([Os(bpy)2Cl2]+) acted as the electroactive marker, similar to the [Fe(CN)6]3-/4- marker (Table 2, Scheme 3-E and 3-E’). The resultant sensor demonstrated a higher signal in the presence of the hybridization process (Scheme 3-E’). In this case, a sensitivity of 5 × 103 HBV copies, equivalent to 8.3 × 10-21 moles of original genomic fragments, was achieved. Jampasa et al. (2014) developed another type of label-free genosensor capable of detecting the human papillomavirus (HPV) using the redox label anthraquinone (AQ) attached to the free end of the probes immobilized to the surface. The probes were made of 14-mer pyrrolidinyl PNA (peptide nucleic acid) constructs (Püschl et al. 2000). Through the cross-linking of amino groups, these constructs were covalently immobilized on the screen-printed carbon electrodes modified with chitosan (CHT). Once hybridized to the complementary 14-nucleotide targeted region of the HPV specific gene, the electrochemical signal of AQ decreased as the result of the increased rigidity of the duplexes on the surface compared to single-strand probes, which limits the electron transfer between the redox moiety and electrode surface (Table 2, Scheme 3-F, 3-F’ and 3-F’’). The resultant genosensor achieved a linear range of 0.02 to 12.0 μM and a limit of detection of 4 nM (Scheme 3-F’’). The main advantage of Jampasa et al.‘s method is the use of pyrrolidinyl PNA probes, which possess the pseudo-peptide backbone and boast an improved binding affinity to DNA and RNA in comparison to DNA or PNA (Nielsen et al. 1994) probes, ensuring the elevated sensitivity of the platform.
Commercially available electrochemical genosensors are primarily a combination of PCR with microfluidic systems, such as the ePlex platform by GenMark Diagnostics. ePlex is capable of detecting a variety of respiratory pathogens, including human coronavirus, in a multiplexing fashion in one single-test. This platform features sample dispensing, nucleic acid extraction, amplification, and detection steps with an automated fluidic transport system. During the PCR assay, target single-stranded amplicons are generated. These amplicons are partially hybridized to ferrocene-conjugated signal probes and partially to their complementary capture probes, previously immobilized to the gold electrode surface. As a result of hybridization, the ferrocene redox moiety generates a signal when subjected to a potential and in contact with the electrode surface. The platform possesses multiple gold electrodes immobilized with various capture probes, each complementary to the amplicon of a specific pathogen, allowing a single sample to be tested for multiple pathogens in parallel (Schmitz and Tang 2018).
Most recently, the electrochemical detection of SARS-CoV-2 genetic materials using smartphone as a portable signal transducer has been reported.
4. Antibody and antigen detection for coronaviruses
In addition to the genetic material, viral proteins and antibodies can serve as disease-specific biomarkers. The recognition strategies for the detection of these molecules are far more diverse than for the DNA/RNA molecules. They include not only nucleic acid-based recognition probes, such as aptamer-binding proteins, but also antibodies, proteins, and their peptide-derivatives. In this case, the target protein binding affinity and the specificity of the reaction are the main focuses when dealing with target detection in complex media (Chambers et al. 2008; Lin et al. 2016; Mahshid et al. 2019). Similar to genosensors, these sensors use an electrical signal transducer to quantify a concentration-proportional change induced by a chemical reaction, specifically an immunochemical reaction (Cristea et al. 2015).
4.1. Virus detection using antibodies
The direct detection of pathogens through electrochemical biosensing using viral antibodies as the recognition element is a potential candidate to address the challenges of current methods. As with nucleic-acid based sensors, such platforms must achieve sensitivities comparable to RT-PCR assays. Indeed, the detection platforms for viruses such as influenza A virus (IAV), as a well-studied pathogen with established sensing techniques (Hassen et al. 2011; Tepeli and Ülkü 2018), can serve as practical models for the detection of SARS-CoV-2.
Hassen et al. (2011) have developed an electrochemical impedance spectroscopy (EIS)-based sensor, which consists of gold electrodes immobilized with an IAV antibody-neutravidin-thiol complex for label-free detection of IAV (H3N2). The attachment of the target virus to the surface-immobilized antibodies results in a quantifiable increase in the electrical impedance measured on the electrode surface (Table 2 and Scheme 4 -A). The sensor was capable of detecting IAV in PBS (phosphate-buffered saline) at low concentrations, down to 8 ng/mL, despite the presence of interfering species (Scheme 4-A’ and 4-A’’). However, as the detection method relies on small changes in the electrical resistivity of the surface monolayer, the ability to sensitively detect analytes in undiluted media becomes challenging for this technique.
Scheme 4.
Brief illustration of the electrochemical biosensing platforms the assay format and electrochemical responses for the detection antibody and antigen. Panels A to E represent the assay formats for various platforms discussed in this study; A’ to E’ show the type of electrochemical responses and how it varies with target concentration ((a) and (b) represents either with target (+target) or without target (–target)); A’’, B’’, C’’, C‴, D’’, D‴, and E’’ are shown in their original scale representing the details on calibration curves, multiplexing, and selectivity responses.
Three subtypes of IAV, H1N1, H5N1, and H7N9, were chosen as the target for a multi-virus microfluidic-based electrochemical immunosensor, developed by Han et al. (2016). The device was designed with one inlet and three outlets, each outlet channel containing a sensor region with different virus-specific capture-antibodies. ZnO nanorods were grown in the sensor regions of the PDMS-microfluidic device hanging over the electrodes and were electrostatically immobilized with capture antibodies. To electrochemically detect the three virus subtypes, they used a modified sandwich ELISA assay with the detection antibodies conjugated to horseradish peroxidase (HRP). When the detection antibodies bind to target viruses on the sensor regions, the applied potential causes HRP to generate electrons through the oxidation of 3,3’,5,5’-tetramethylbenzidine in solution (Scheme 4-B). The resultant current was recorded by the amperometric measurements with a limit of detection of 1 pg/mL for all analytes (Table 2, Scheme 4-B’and 4-B’’).
Singh et al. (2017) also established an immunosensor combining electrochemical and microfluidic techniques and by taking advantage of the improved conductivity and favourable structure of a reduced graphene (rGO) coated electrode. H1N1 specific monoclonal antibodies were immobilized on the rGO surface through the direct interaction of antibody amino groups and carboxyl groups of the graphene. The response of virus detection was recorded through amperometric measurements using the [Fe(CN)6]3-/4- solution (Table 2, Scheme 4-C and 4-C’). The resultant device demonstrated highly specific and selective detection of influenza virus H1N1, with a limit of detection of 0.5 PFU/mL (Scheme 4-C’’ and 4-C’’’).
Layqah and Eissa (2019) developed an electrochemical competition-based immunosensor for MERS-CoV on a gold nanoparticle-modified electrode with recombinant spike protein S1 immobilized on the surface. The prepared electrodes were incubated in a solution of human coronavirus antibodies and different dilutions of free MERS-CoV. The target virus detection is based on the competition for available antibodies between the free MERS-CoV and the immobilized S1 protein, in which the signal is measured using the [Fe(CN)6]3-/4- redox-active marker in the testing solution (when surface is covered with the antibody the access of redox marker is reduced resulting in decrease in the current) (Table 2, Scheme 4-D and 4-D’). A similar platform was also developed for targeting the human coronavirus, HCoV (Scheme 4-D’’ and 4-D’’’). This platform, together with the two above electrochemical immunosensors, confer the specificities required to operate in undiluted samples. The high specificities are due to their antibody-recognition strategies, as well as the choice of signal transduction mechanisms, such as CA, CV, and SWV.
To detect the SARS-CoV-2 virus using antibodies, Seo et al. (2020) developed an immunosensor with a field-effect transistor (FET) as the signal transducer using graphene sheets immobilized with antibodies against spike proteins. The immobilization was done by coupling the antibodies with 1-pyrenebutyric acid N-hydroxysuccinimide on the graphene sheet. The combination of the selectivity of antibodies and the ultrasensitive FET-based detection platform yielded a simple and highly responsive SARS-CoV-2 biosensor. The sensor achieved a limit of detection of 1 fg/mL and 100 fg/ml against SARS-CoV-2 spike proteins in PBS and clinical transport media, respectively. The response of such a device was also recorded in culture medium with a detection limit of 16 PFU/mL, as well as in clinical samples with a minimum of 242 copies/mL without any preprocessing or preparation.
4.2. Antibody detection using antigens/proteins
Serological testing works through the detection of antibodies in bodily fluids, namely blood, and through a larger diagnostic window can be used to detect past and present infections, even in asymptomatic individuals (Meyer et al. 2014). The ability to detect antibodies against viruses in asymptomatic and recovered individuals is crucial for accurate epidemiology, understanding herd immunity, and determining transmission trends and prevalence (Hoffman et al. 2020). Serological testing was used during the original SARS outbreak, the MERS epidemic, and is currently being studied for the SARS-CoV-2 pandemic. The two globulin proteins involved in the immune response to these viruses are the IgM and IgG antibodies. Following SARS infection, the IgM antibody can be detected within 3-6 days, and the IgG antibody can be detected in patients’ blood after 8 days (Li et al. 2020b). Earlier studies on electrochemical detection of similar viruses demonstrated the use of viral proteins, including the spike surface glycoprotein (S) and nucleocapsid protein (N), as recognition elements to the viral antibodies.
Jarocka et al. (2014) developed an EIS-based platform to optimize the sensitivity of existing sensors for the detection of anti-hemagglutinin antibodies, a frequently targeted antibody for influenza viruses. A glassy carbon electrode was modified with carboxylic groups and covalently immobilized with protein A. Subsequently, by interacting with the bonded protein A, anti-His IgG monoclonal antibodies were immobilized on the electrode and free to bind recombinant His-tagged hemagglutinin, as the recognition complex (Scheme 4-E and 4-E’). In the presence of the redox marker, [Fe(CN)6]3-/4-, the recognition complex combined with the target anti-hemagglutinin antibodies produces an increase in the electron transfer resistance that is not affected by the interference of the control antibody, resulting in a limit of detection of 2.1 pg/mL (Table 2, and Scheme 4-E’’) The applicability of this biosensor was verified using dilutions of vaccinated hen sera in the concentration range of 7 × 103 to 7 × 107, which showed almost 104 times more sensitivity compared to ELISA. The same research group later developed a similar platform for the detection of anti-hemagglutinin antibodies against swine originated H1N1. They used the same His-tagged hemagglutinin (His6-H5 HA) as the recognition element, which was covalently attached to a 4-mercaptobutanol-modified gold electrode. The recognition element was immobilized in an oriented fashion using dipyrromethene (DPM)-Cu (II) complex as a histidine tag surface-receptor. The DPM-Cu (II) complex also served as the electroactive marker, relying on the Cu(II)/Cu(I) redox current to generate a response measured through square wave voltammetry (Table 2). This sensor was assessed using vaccinated mice sera samples at various dilutions. Antibodies were detected at sera dilutions from 1 × 108 to 1 × 109 fold, which is approximately 107 times more sensitive than standard ELISA tests (Mikuła et al. 2018). Despite the high sensitivities, the success of these tests is based on the immune response to the virus, not the virus itself. As a result, these tests would largely serve a supplemental role in diagnostics (Kasetsirikul et al. 2020).
The current platforms for serological testing of SARS-CoV-2 have illustrated the use of conventional ELISA (Amanat et al. 2020; Freeman et al. 2020; Yan et al. 2020; Zhang et al. 2020), lateral flow immunoassay (LFA) (Cassaniti et al. 2020; Hoffman et al. 2020), or (electro)chemiluminescent immunoassay techniques (Hörber et al. 2020). These platforms use either the S- or N- proteins as recognition elements of viral antibodies. For instance, Li et al. (2020b) presented an LFA platform with gold nanoparticles and recombinant S-proteins to detect both anti-SARS-CoV-2-IgM and anti-SARS-CoV-2-IgG with a sensitivity of 88.66% and a specificity of 90.63%.
As with nucleic acid detection (ePlex platform by GenMark Diagnostics), an electrochemiluminescence-based sensor was developed and commercialized by Roche diagnostics to detect SARS-CoV-2 antibodies (Hörber et al. 2020). The Roche diagnostic platform involves streptavidin-biotin complexes with ruthenium electrochemiluminescence, where biotinylated antibodies, ruthenium labelled antibodies, and streptavidin particles combine with the sample to form a complex (Gassner and Jung 2014). When subjected to an electrical potential, this complex becomes excited and emits light (Sano and Tatsumi 1996). Compared with ELISA and chemiluminescence assays, the Roche platform shows similar and promising specificity, albeit with a slightly lower sensitivity (Hörber et al. 2020).
However, currently, none of these platforms have employed the use of electrochemical biosensors with electrochemical readout strategies for the detection of SARS-CoV-2 antibodies.
5. Closing remarks
Throughout the COVID-19 pandemic, the role of testing and the degree of implementation has been widely debated. Early and widespread testing for SARS-CoV-2 has proven to reduce mortality rates and improve contact tracing. However, the value of testing is directly linked to the availability and accuracy of diagnostic tests. As concerns grow regarding the application of RT-PCR and ELISA-based testing, particularly the complexity of the assays during inventory shortages, the development of easy-to-use alternative platforms is encouraged with specific attention paid to sensitivity and simplicity.
Electrochemical-based sensors remediate the majority of the abovementioned concerns while maintaining the required sensitivity for diagnostic tests. Primarily defined by the signal transduction methods, a diverse range of analytes can be detected with high effectiveness, depending on the associated biorecognition element. The broad variability of electrochemical-based devices is useful in pathogen diagnostics, as different analytes appear at different times throughout the progress of an infection. Advances in electrochemical detection of viral nucleic acids have demonstrated high sensitivities and low detection limits for various viruses. Exploring different electrode materials and redox markers allows for improved sensor properties, often reaching and surpassing the effectiveness of RT-PCR-based assays while simultaneously rectifying broader issues, such as long analysis time and high costs. The detection of SARS-CoV-2 antigens over nucleic acids has the advantages of an earlier detection window and more straightforward sample preparation, often at the expense of sensitivity. Using electrochemical concepts and microfluidic devices combines the portability with sensitivity and selectivity of the detection of pathogenic antigens at the laboratory-levels in real-life samples. Serological testing is vital for a complete understanding of the epidemiology of a pandemic and the response of the immune system to a pathogen. Electrochemical sensors used for the detection of antibodies have demonstrated trail-blazing sensitivities, outmatching ELISA-based assays dramatically. In addition to detecting pathogens in real-life samples, immunosensors were shown to be capable of distinguishing coronaviruses similar to SARS-CoV-2, proposing an apparent alternative to traditional ELISA assays.
Despite the promising results, the majority of studies explored in this perspective were focused on the detection of viruses analogous to SARS-CoV-2. As a result, further work must be done to determine the efficiency of electrochemical platforms for the detection of SARS-CoV-2, in particular. Additionally, the diversity of analytes, pathogens, and detection mechanisms studied makes it challenging to compare the results within the field of electrochemical sensing directly and between novel and traditional diagnostic methods. The continual efflux of studies and an ever-growing understanding of the pandemic must be monitored and applied to existing electrochemical concepts to develop and commercialize alternative electrochemical-based SARS-CoV-2 diagnostic tests.
CRediT authorship contribution statement
Sahar Sadat Mahshid: Conceptualization, Investigation, Visualization, Resources, Writing - original draft, Writing - review & editing, Supervision. Sarah Elizabeth Flynn: Investigation, Visualization, Writing - original draft. Sara Mahshid: Conceptualization, Resources, Writing - review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We dedicate our work to the front-line workers and healthcare providers whose roles can be assisted through more efficient methods for point-of-care diagnostics during the pandemic. The authors thank the Natural Science and Engineering Research Council of Canada (NSERC, G247765), and MI4 Seed Fund Grant (250611) for financial support.
References
- Abad-Valle P., Fernández-Abedul M.T., Costa-García A. Genosensor on gold films with enzymatic electrochemical detection of a SARS virus sequence. Biosens. Bioelectron. 2005;20(11):2251–2260. doi: 10.1016/j.bios.2004.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abduljalil J.M. New microbes and new infections; 2020. Laboratory Diagnosis of SARS-CoV-2: Available Approaches and Limitations; p. 100713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afsahi S., Lerner M.B., Goldstein J.M., Lee J., Tang X., Bagarozzi D.A., Jr., Pan D., Locascio L., Walker A., Barron F. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018;100:85–88. doi: 10.1016/j.bios.2017.08.051. [DOI] [PubMed] [Google Scholar]
- Al Johani S., Hajeer A.H. MERS-CoV diagnosis: an update. J. Infect.Public Health. 2016;9(3):216–219. doi: 10.1016/j.jiph.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020:1–4. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout R., Lee R.A., Lee G.R., Callahan C., Yen C.F., Smith K.P., Arora R., Kirby J.E. SARS-CoV2 testing: the limit of detection matters. bioRxiv : the preprint server for biology. 2020;2002:131144. 2020.2006. [Google Scholar]
- Bhalla N., Pan Y., Yang Z., Payam A.F. Opportunities and challenges for biosensors and nanoscale Analytical tools for pandemics: COVID-19. ACS Nano. 2020;14(7):7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter L.J., Garner L.V., Smoot J.W., Li Y., Zhou Q., Saveson C.J., Sasso J.M., Gregg A.C., Soares D.J., Beskid T.R. ACS Publications; 2020. Assay Techniques and Test Development for COVID-19 Diagnosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassaniti I., Novazzi F., Giardina F., Salinaro F., Sachs M., Perlini S., Bruno R., Mojoli F., Baldanti F. Performance of VivaDiag COVID‐19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID‐19 in acute patients referring to emergency room department. J. Med. Virol. 2020;92(10):1724–1727. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesewski E., Johnson B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020:112214. doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers J.P., Arulanandam B.P., Matta L.L., Weis A., Valdes J.J. Texas Univ at San Antonio Dept of Biology; 2008. Biosensor Recognition Elements. [PubMed] [Google Scholar]
- Chung D.-J., Kim K.-C., Choi S.-H. Electrochemical DNA biosensor based on avidin–biotin conjugation for influenza virus (type A) detection. Appl. Surf. Sci. 2011;257(22):9390–9396. [Google Scholar]
- Control C.f.D., Prevention CDC 2019-novel coronavirus (2019-nCoV) real-time RT-PCR diagnostic panel. ReVision. 2020;3:30. [Google Scholar]
- Corman V., Bleicker T., Brünink S., Drosten C., Zambon M., Organization W.H. World Health Organization; Geneva: 2020. Diagnostic Detection of Wuhan Coronavirus 2019 by Real-Time RT-PCR. January 13. [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea C., Florea A., Tertiș M., Săndulescu R. IntechOpen; 2015. Immunosensors. Biosensors-Micro and Nanoscale Applications. [Google Scholar]
- D'Cruz R.J., Currier A.W., Sampson V.B. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Front. Cell. Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva E.T.S.G., Souto D.E.P., Barragan J.T.C., Giarola J.de F., de Moraes A.C.M., Kubota L.T. Electrochemical biosensors in point-of-care devices: recent advances and future trends. ChemElectroChem. 2017;4(4):778–794. [Google Scholar]
- de la Escosura-Muñiz A., Baptista-Pires L., Serrano L., Altet L., Francino O., Sánchez A., Merkoçi A. Magnetic bead/gold nanoparticle double-labeled primers for electrochemical detection of isothermal amplified leishmania DNA. Small. 2016;12(2):205–213. doi: 10.1002/smll.201502350. [DOI] [PubMed] [Google Scholar]
- Díaz-González M., de la Escosura-Muñiz A., González-García M.B., Costa-García A. DNA hybridization biosensors using polylysine modified SPCEs. Biosens. Bioelectron. 2008;23(9):1340–1346. doi: 10.1016/j.bios.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G., Carvalho L.M., Rambaut A., Bedford T. MERS-CoV spillover at the camel-human interface. Elife. 2018;7 doi: 10.7554/eLife.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., Newbigging A.M., Le C., Pang B., Peng H., Cao Y., Wu J., Abbas G., Song J., Wang D.-B., Cui M., Tao J., Tyrrell D.L., Zhang X.-E., Zhang H., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- Freeman B., Lester S., Mills L., Rasheed M.A.U., Moye S., Abiona O., Hutchinson G.B., Morales-Betoulle M., Krapinunaya I., Gibbons A. bioRxiv; 2020. Validation of a SARS-CoV-2 Spike Protein ELISA for Use in Contact Investigations and Serosurveillance. [Google Scholar]
- Gassner D., Jung R. First fully automated immunoassay for anti-Müllerian hormone. Clin. Chem. Lab. Med. 2014;52(8):1143–1152. doi: 10.1515/cclm-2014-0022. [DOI] [PubMed] [Google Scholar]
- Germany L. The COVID-19 testing debacle. Nat. Biotechnol. 2020;38(6) doi: 10.1038/s41587-020-0575-3. 653-653. [DOI] [PubMed] [Google Scholar]
- González-López A., Abedul M.T.F. Elsevier; 2020. Genosensor on Gold Films with Enzymatic Electrochemical Detection of a SARS Virus Sequence. Laboratory Methods in Dynamic Electroanalysis; pp. 213–220. [Google Scholar]
- Han J.-H., Lee D., Chew C.H.C., Kim T., Pak J.J. A multi-virus detectable microfluidic electrochemical immunosensor for simultaneous detection of H1N1, H5N1, and H7N9 virus using ZnO nanorods for sensitivity enhancement. Sensor. Actuator. B Chem. 2016;228:36–42. [Google Scholar]
- Hassen W.M., Duplan V., Frost E., Dubowski J.J. Quantitation of influenza A virus in the presence of extraneous protein using electrochemical impedance spectroscopy. Electrochim. Acta. 2011;56(24):8325–8328. [Google Scholar]
- Hoffman T., Nissen K., Krambrich J., Rönnberg B., Akaberi D., Esmaeilzadeh M., Salaneck E., Lindahl J., Lundkvist Å. Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2. Infect. Ecol. Epidemiol. 2020;10(1):1754538. doi: 10.1080/20008686.2020.1754538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörber S., Soldo J., Relker L., Jürgens S., Guther J., Peter S., Lehmann R., Peter A. Evaluation of three fully-automated SARS-CoV-2 antibody assays. Clin. Chem. Lab. Med. 2020;58(12):2113–2120. doi: 10.1515/cclm-2020-0975. [DOI] [PubMed] [Google Scholar]
- Huang J.C., Chang Y.-F., Chen K.-H., Su L.-C., Lee C.-W., Chen C.-C., Chen Y.-M.A., Chou C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009;25(2):320–325. doi: 10.1016/j.bios.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., Hu X., Chen J., Liu B. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295(1):22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huy T.Q., Hanh N.T.H., Thuy N.T., Van Chung P., Nga P.T., Tuan M.A. A novel biosensor based on serum antibody immobilization for rapid detection of viral antigens. Talanta. 2011;86:271–277. doi: 10.1016/j.talanta.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jampasa S., Wonsawat W., Rodthongkum N., Siangproh W., Yanatatsaneejit P., Vilaivan T., Chailapakul O. Electrochemical detection of human papillomavirus DNA type 16 using a pyrrolidinyl peptide nucleic acid probe immobilized on screen-printed carbon electrodes. Biosens. Bioelectron. 2014;54:428–434. doi: 10.1016/j.bios.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Jarocka U., Sawicka R., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecki J., Radecka H. Electrochemical immunosensor for detection of antibodies against influenza A virus H5N1 in hen serum. Biosens. Bioelectron. 2014;55:301–306. doi: 10.1016/j.bios.2013.12.030. [DOI] [PubMed] [Google Scholar]
- Jianrong C., Yuqing M., Nongyue H., Xiaohua W., Sijiao L. Nanotechnology and biosensors. Biotechnol. Adv. 2004;22(7):505–518. doi: 10.1016/j.biotechadv.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Ju H.-X., Ye Y.-K., Zhao J.-H., Zhu Y.-L. Hybridization biosensor using di(2,2′-bipyridine)osmium (III) as electrochemical indicator for detection of polymerase chain reaction product of hepatitis B virus DNA. Anal. Biochem. 2003;313(2):255–261. doi: 10.1016/s0003-2697(02)00625-5. [DOI] [PubMed] [Google Scholar]
- Jung I.Y., You J.B., Choi B.R., Kim J.S., Lee H.K., Jang B., Jeong H.S., Lee K., Im S.G., Lee H. A highly sensitive molecular detection platform for robust and facile diagnosis of Middle East respiratory syndrome (MERS) corona virus. Advanced healthcare materials. 2016;5(17):2168–2173. doi: 10.1002/adhm.201600334. [DOI] [PubMed] [Google Scholar]
- Kang D., Zuo X., Yang R., Xia F., Plaxco K.W., White R.J. Comparing the properties of electrochemical-based DNA sensors employing different redox tags. Anal. Chem. 2009;81(21):9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasetsirikul S., Umer M., Soda N., Sreejith K.R., Shiddiky M.J.A., Nguyen N.-T. Detection of the SARS-CoV-2 humanized antibody with paper-based ELISA. Analyst. 2020;145(23):7680–7686. doi: 10.1039/d0an01609h. [DOI] [PubMed] [Google Scholar]
- Khater M., de la Escosura-Muñiz A., Merkoçi A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017;93:72–86. doi: 10.1016/j.bios.2016.09.091. [DOI] [PubMed] [Google Scholar]
- Khater M., de la Escosura-Muñiz A., Quesada-González D., Merkoçi A. Electrochemical detection of plant virus using gold nanoparticle-modified electrodes. Anal. Chim. Acta. 2019;1046:123–131. doi: 10.1016/j.aca.2018.09.031. [DOI] [PubMed] [Google Scholar]
- Koczula K.M., Gallotta A. Lateral flow assays. Essays Biochem. 2016;60(1):111–120. doi: 10.1042/EBC20150012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács A., Palásti P., Veréb D., Bozsik B., Palkó A., Kincses Z.T. The sensitivity and specificity of chest CT in the diagnosis of COVID-19. Eur. Radiol. 2020:1–6. doi: 10.1007/s00330-020-07347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubina R., Dziedzic A. Molecular and serological tests for COVID-19 a comparative review of SARS-CoV-2 coronavirus laboratory and point-of-care diagnostics. Diagnostics. 2020;10(6):434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layqah L.A., Eissa S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Microchimica Acta. 2019;186(4):224. doi: 10.1007/s00604-019-3345-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Ahn J.-H., Park S.Y., Kim G.-H., Kim J., Kim T.-H., Nam I., Park C., Lee M.-H. Recent advances in AIV biosensors composed of nanobio hybrid material. Micromachines. 2018;9(12):651. doi: 10.3390/mi9120651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Park S.Y., Jang H., Kim G.-H., Lee Y., Park C., Mohammadniaei M., Lee M.-H., Min J. Fabrication of electrochemical biosensor consisted of multi-functional DNA structure/porous au nanoparticle for avian influenza virus (H5N1) in chicken serum. Mater. Sci. Eng. C. 2019;99:511–519. doi: 10.1016/j.msec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT‐PCR testing of SARS‐CoV‐2 for hospitalized patients clinically diagnosed with COVID‐19. J. Med. Virol. 2020;92:903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yi Y., Luo X., Xiong N., Liu Y., Li S., Sun R., Wang Y., Hu B., Chen W. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J. Med. Virol. 2020;92:1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R.R.X., Bonanni A. The potential of electrochemistry for the detection of coronavirus-induced infections. Trac. Trends Anal. Chem. 2020;133:116081. doi: 10.1016/j.trac.2020.116081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Song P., Zhou G., Zuo X., Aldalbahi A., Lou X., Shi J., Fan C. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 2016;11(7):1244–1263. doi: 10.1038/nprot.2016.071. [DOI] [PubMed] [Google Scholar]
- Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109(5):1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahshid S.S., Dabdoub A. Development of a novel electrochemical immuno-biosensor for circulating biomarkers of the inner ear. Biosens. Bioelectron. 2020:112369. doi: 10.1016/j.bios.2020.112369. [DOI] [PubMed] [Google Scholar]
- Mahshid S.S., Mahshid S., Vallée-Bélisle A., Kelley S.O. Peptide-mediated electrochemical steric hindrance assay for one-step detection of HIV antibodies. Anal. Chem. 2019;91(8):4943–4947. doi: 10.1021/acs.analchem.9b00648. [DOI] [PubMed] [Google Scholar]
- Malecka K., Stachyra A., Góra-Sochacka A., Sirko A., Zagórski-Ostoja W., Radecka H., Radecki J. Electrochemical genosensor based on disc and screen printed gold electrodes for detection of specific DNA and RNA sequences derived from Avian Influenza Virus H5N1. Sensor. Actuator. B Chem. 2016;224:290–297. [Google Scholar]
- Martínez-Paredes G., González‐García M.B., Costa‐García A. Genosensor for SARS virus detection based on gold nanostructured screen‐printed carbon electrodes. Electroanalysis: Int. J.Devoted.Fund.Practical Aspect.Electroanalysis. 2009;21(3‐5):379–385. [Google Scholar]
- Merckx J., Wali R., Schiller I., Caya C., Gore G.C., Chartrand C., Dendukuri N., Papenburg J. Diagnostic accuracy of novel and traditional rapid tests for influenza infection compared with reverse transcriptase polymerase chain reaction: a systematic review and meta-analysis. Ann. Intern. Med. 2017;167(6):394–409. doi: 10.7326/M17-0848. [DOI] [PubMed] [Google Scholar]
- Meyer B., Drosten C., Müller M.A. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–183. doi: 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuła E., Silva C.E., Kopera E., Zdanowski K., Radecki J., Radecka H. Highly sensitive electrochemical biosensor based on redox - active monolayer for detection of anti-hemagglutinin antibodies against swine-origin influenza virus H1N1 in sera of vaccinated mice. BMC Vet. Res. 2018;14(1):328. doi: 10.1186/s12917-018-1668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moatti J.-P. The French response to COVID-19: intrinsic difficulties at the interface of science, public health, and policy. The Lancet Public Health. 2020;5(5):e255. doi: 10.1016/S2468-2667(20)30087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. Biosensors and Bioelectronics; 2020. The impact of biosensing in a pandemic outbreak: COVID-19; p. 112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P.E., Egholm M., Buchardt O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjugate Chem. 1994;5(1):3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- Organization W.H. World Health Organization; 2020. Laboratory Testing for Coronavirus Disease 2019 (COVID-19) in Suspected Human Cases: Interim Guidance, 2 March 2020. [Google Scholar]
- Pallett S.J., Rayment M., Patel A., Fitzgerald-Smith S.A., Denny S.J., Charani E., Mai A.L., Gilmour K.C., Hatcher J., Scott C. Point-of-care serological assays for delayed SARS-CoV-2 case identification among health-care workers in the UK: a prospective multicentre cohort study. The Lancet Respiratory Medicine. 2020;8(9):885–894. doi: 10.1016/S2213-2600(20)30315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pividori M., Merkoci A., Alegret S. Electrochemical genosensor design: immobilisation of oligonucleotides onto transducer surfaces and detection methods. Biosens. Bioelectron. 2000;15(5–6):291–303. doi: 10.1016/s0956-5663(00)00071-3. [DOI] [PubMed] [Google Scholar]
- Püschl A., Tedeschi T., Nielsen P.E. Pyrrolidine PNA: A novel conformationally restricted PNA analogue. Org. Lett. 2000;2(26):4161–4163. doi: 10.1021/ol000300l. [DOI] [PubMed] [Google Scholar]
- Qin Z., Peng R., Baravik I.K., Liu X. Fighting COVID-19: integrated micro- and nanosystems for viral infection diagnostics. Matter. 2020;3(3):628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Inf. Med. 2020;28(2):174–184. [PubMed] [Google Scholar]
- Sano M., Tatsumi N. Electro chemiluminescence immunoassay. Rinsho Byori. 1996;44(11):1076–1079. [PubMed] [Google Scholar]
- Santiago I. Trends and innovations in biosensors for COVID‐19 mass testing. Chembiochem : Eur. J.Chem. Biol. 2020;21:1–11. doi: 10.1002/cbic.202000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J.E., Tang Y.-W. The GenMark ePlex®: another weapon in the syndromic arsenal for infection diagnosis. Future Microbiol. 2018;13(16):1697–1708. doi: 10.2217/fmb-2018-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder I. COVID-19: a risk assessment perspective. J. Chem. Health Saf. 2020;27(3):160–169. doi: 10.1021/acs.chas.0c00035. [DOI] [PubMed] [Google Scholar]
- Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- Shen M., Zhou Y., Ye J., Abdullah Al-maskri A.A., Kang Y., Zeng S., Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. Journal of Pharmaceutical Analysis. 2020;10(2):97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Hong S., Jang J. Label-free detection of influenza viruses using a reduced graphene oxide-based electrochemical immunosensor integrated with a microfluidic platform. Sci. Rep. 2017;7(1):42771. doi: 10.1038/srep42771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Lima D., Viana A.G., Fujiwara S.T., Pessôa C.A., Etto R.M., Wohnrath K. A sensitive label-free impedimetric DNA biosensor based on silsesquioxane-functionalized gold nanoparticles for Zika Virus detection. Biosens. Bioelectron. 2019;141:111351. doi: 10.1016/j.bios.2019.111351. [DOI] [PubMed] [Google Scholar]
- Tepeli Y., Ülkü A. Electrochemical biosensors for influenza virus a detection: the potential of adaptation of these devices to POC systems. Sensor. Actuator. B Chem. 2018;254:377–384. [Google Scholar]
- Thévenot D.R., Toth K., Durst R.A., Wilson G.S. Electrochemical biosensors: recommended definitions and classification. Anal. Lett. 2001;34(5):635–659. doi: 10.1016/s0956-5663(01)00115-4. [DOI] [PubMed] [Google Scholar]
- Turner A.P.F. Biosensors: sense and sensibility. Chem. Soc. Rev. 2013;42(8):3184–3196. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y., Chen H., Mubareka S., Gubbay J.B., Chan W.C. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- Uhteg K., Jarrett J., Richards M., Howard C., Morehead E., Geahr M., Gluck L., Hanlon A., Ellis B., Kaur H. Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays. J. Clin. Virol. 2020:104384. doi: 10.1016/j.jcv.2020.104384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen A., Veen J., Arends H.A., Koets M. In: Handbook of Immunoassay Technologies. Vashist S.K., Luong J.H.T., editors. Academic Press; 2018. Chapter 7 - lateral flow immunoassays; pp. 157–182. [Google Scholar]
- Velay A., Gallais F., Benotmane I., Wendling M.J., Danion F., Collange O., De Sèze J., Schmidt-Mutter C., Schneider F., Bilbault P. Evaluation of the performance of SARS-CoV-2 serological tools and their positioning in COVID-19 diagnostic strategies. Diagn. Microbiol. Infect. Dis. 2020;98(4):115181. doi: 10.1016/j.diagmicrobio.2020.115181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P., Ahern S., Carty P.G., O'Brien K.K., O'Murchu E., O'Neill M., Smith S.M., Ryan M., Harrington P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J. Infect. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Piorek B.D., Plaxco K.W., Heeger A.J. A reagentless signal-on architecture for electronic, aptamer-based sensors via target-induced strand displacement. J. Am. Chem. Soc. 2005;127(51):17990–17991. doi: 10.1021/ja056555h. [DOI] [PubMed] [Google Scholar]
- Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Radiology; 2020. Chest CT for Typical 2019-nCoV Pneumonia: Relationship to Negative RT-PCR Testing; p. 200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Chang L., Wang L. Laboratory testing of SARS‐CoV, MERS‐CoV, and SARS‐CoV‐2 (2019‐nCoV): current status, challenges, and countermeasures. Rev. Med. Virol. 2020;30(3) doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes N., Al-Sadeq D.W., Al-Jighefee H., Younes S., Al-Jamal O., Daas H.I., Yassine H.M., Nasrallah G.K. Challenges in laboratory diagnosis of the novel coronavirus SARS-CoV-2. Viruses. 2020;12(6):582. doi: 10.3390/v12060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.L.L., Maslova A., Hsing I.M. Rational design of electrochemical DNA biosensors for Point‐of‐Care applications. ChemElectroChem. 2017;4(4):795–805. [Google Scholar]
- Yu Y., Chen Z., Jian W., Sun D., Zhang B., Li X., Yao M. Ultrasensitive electrochemical detection of avian influenza A (H7N9) virus DNA based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzyme nanowires. Biosens. Bioelectron. 2015;64:566–571. doi: 10.1016/j.bios.2014.09.080. [DOI] [PubMed] [Google Scholar]
- Zari N., Mohammedi H., Amine A., Ennaji M. DNA hydrolysis and voltammetric determination of guanine and adenine using different electrodes. Anal. Lett. 2007;40(9):1698–1713. [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microb. Infect. 2020;9(1):386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen W., Smith E., Manji R., Schron D., Berry G.J. Clinical evaluation of three sample-to-answer platforms for detection of SARS-CoV-2. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Jimmy Huang P.-J., Ding J., Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139(11):2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]