Abstract
Nitrogen mustards are among the first modern anticancer chemotherapeutics that are still widely used as non-specific anticancer alkylating agents. While the mechanism of action of mustard drugs involves the generation of DNA interstrand cross-links, the predominant lesions produced by these drugs are nitrogen half-mustard-N7-dG (NHMG) adducts. The bulky major-groove lesion NHMG, if left unrepaired, can be bypassed by translesion synthesis DNA polymerases. However, studies of the translesion synthesis past NHMG have not been reported so far. Here we present the first synthesis of an oligonucleotide containing a site-specific NHMG. We also report kinetic and structural characterization of human DNA polymerase η (polη) bypassing NHMG. The templating NHMG slows dCTP incorporation ~130-fold, while it increases the misincorporation frequency ~10–30-fold, highlighting the promutagenic nature of NHMG. A crystal structure of polη incorporating dCTP opposite NHMG shows a Watson-Crick NHMG:dCTP base pair with a large propeller twist angle. The nitrogen half-mustard moiety fits snugly into an open cleft created by the Arg61-Trp64 loop of polη, suggesting a role of the Arg61-Trp64 loop in accommodating bulky major groove adducts during lesion bypass. Overall, our results presented here provide first insights into translesion synthesis of the major DNA adduct formed by nitrogen mustard drugs.
INTRODUCTION
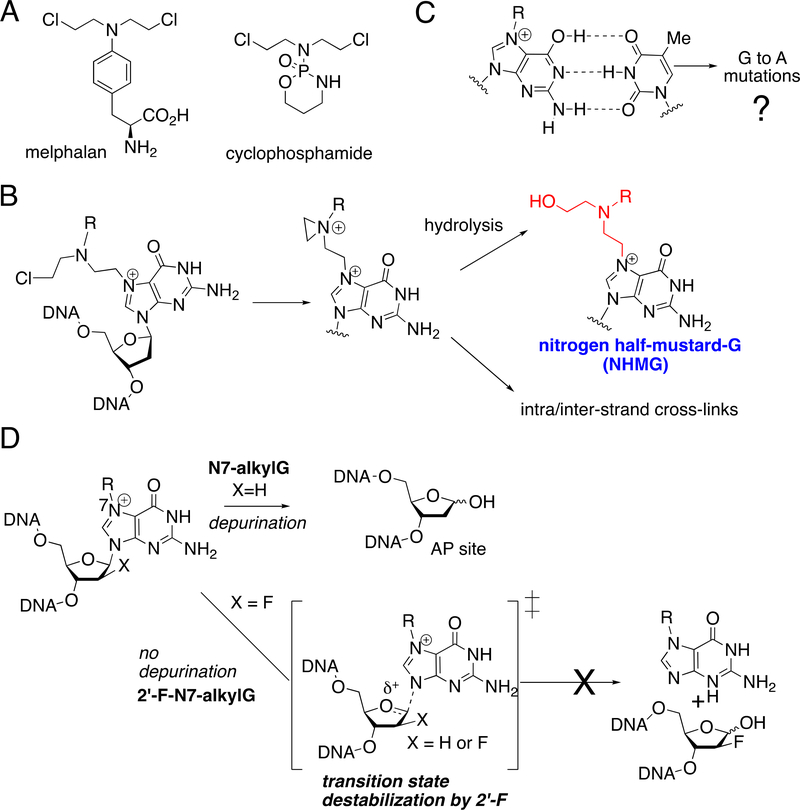
Nitrogen mustards (NM) were among the first anticancer chemotherapeutics developed and have been widely used in the treatment of various blood cancers and solid tumors [1, 2]. Nitrogen mustards such as cyclophosphamide and melphalan (Figure 1A) are used in combination therapies for the treatment of non-Hodgkin’s lymphoma and chronic lymphocytic leukemia. NM are latent electrophiles that generate an aziridinium ion, which can react with a wide variety of nucleophiles including DNA, proteins, and water. The cytotoxicity of NM is believed to be primarily caused by NM-induced DNA interstrand cross-links (ICLs) [3, 4], which can impede DNA strand separation [5]. However, the NM-mediated ICLs represent only a small fraction of DNA lesions formed by the mustard drugs. The major lesions produced by NM including mechlorethamine (>90%), chlorambucil (~100%), and melphalan (~60%) [6, 7] are nitrogen half-mustards-DNA monoadducts, which can be generated by hydrolysis of the initial NM-DNA monoadducts (Figure 1B). The majority of NM-mediated DNA damage occurs at the N7 of guanine to produce nitrogen half-mustard-N7-dG (NHMG) adduct [6], while the N3 of adenine servers as a significant site of DNA alkylation for some NM [8]. The goal of the present work is to delineate the mutagenic potential of the major NM-induced DNA lesion NHMG.
Figure 1. Nitrogen mustards-induced DNA lesions and base pairing properties of N7-alkylG.
(A) Representative nitrogen mustard drugs. (B) Formation of nitrogen half-mustard-dG lesion via hydrolysis of nitrogen mustard-dG monoadduct. (C) Potential promutagenic base pairing of N7-alkylG and thymine. A Watson-Crick-like N7-methyl-G:T base pair has been observed in duplex DNA. (D) Prevention of spontaneous depurination of N7-alkylG by 2′-fluorination.
A prominent drawback of NM-based chemotherapy is the frequent development of secondary cancers in acute myeloid leukemia [6, 9] and the mutagenicity of mustard drugs is related to their ability to form ICLs as well as mono-alkylation adducts [10]. DNA interstrand cross-links generated by bifunctional alkylating agents and platinum-based drugs can be repaired by replication-coupled pathways as well as replication-independent pathways [5, 11–15]. While small N7-alkylG mono-adducts (e.g., N7-methyl-G) are repaired by base excision repair system [16], bulky N7-alkylG mono-adducts (e.g., aflatoxin B1-N7-dG) can be repaired by nucleotide excision repair pathways [17]. If not removed, bulky N7-alkylG mono-adducts can be bypassed by TLS polymerases [17, 18].
Even after several decades of development of the first NM-based alkylating drugs, kinetic and structural studies of DNA polymerase bypassing NHMG have not been reported, thereby significantly limiting our understanding of the miscoding potential of NM-induced DNA damage. Systematic studies of NM-induced DNA lesions have been difficult due to the technical difficulty of preparing the oligonucleotide containing a site-specific incorporated N7-alkylG, which instead rapidly undergoes spontaneous depurination to generate abasic sites. We previously used a 2′-fluorine-mediated transition state destabilization method to incorporate N7-alkylG such as N7MeG and N7-benzyl-G in a site-specific manner [19–22]. Using the 2′-fluorination strategy, we discovered N7-methyl-G (N7MeG) can form Watson-Crick-like base pairing with thymine in the absence of protein contact (Figure 1C and 1D) [20]. Guengerich and colleagues reported N7MeG promotes mutagenic replication by translesion synthesis (TLS) DNA polymerases [23].
Here we describe the first synthesis of a site-specific NHMG-containing oligonucleotide via the 2′-fluorination strategy. In addition, we present kinetic and structural assessment of human DNA polymerase η (polη) bypassing NHMG generated by an N-phenyl nitrogen mustard. Polη was chosen for our studies as it belongs to the Y-family DNA polymerases, which have more spacious active sites than replicative DNA polymerases and thus can accommodate a wide variety of DNA lesions [24, 25]. Polη, which is encoded by the POLH gene in humans, plays a key role in accurate bypass of the UV-induced cyclobutane pyrimidine dimers in vivo [26–29]. Mutations in POLH gene cause the development of human syndrome xeroderma pigmentosum variant (XPV) [30]. Unlike Y-family polymerase κ that is generally blocked by major groove DNA lesions [31], polη is observed to bypass several types of major groove DNA lesions including O6-methylguanine [32], cisplatin-GpG intrastrand cross-links [33, 34], and “unhooked” nitrogen mustards-GG interstrand cross-links [35, 36]. Therefore, our kinetic and structural characterization of polη-mediated bypass of NHMG provides new insights into the promutagenic nature and translesion synthesis of NM-induced DNA lesions
MATERIALS AND METHODS
Preparation of oligonucleotide containing a site specific-incorporated NHMG.
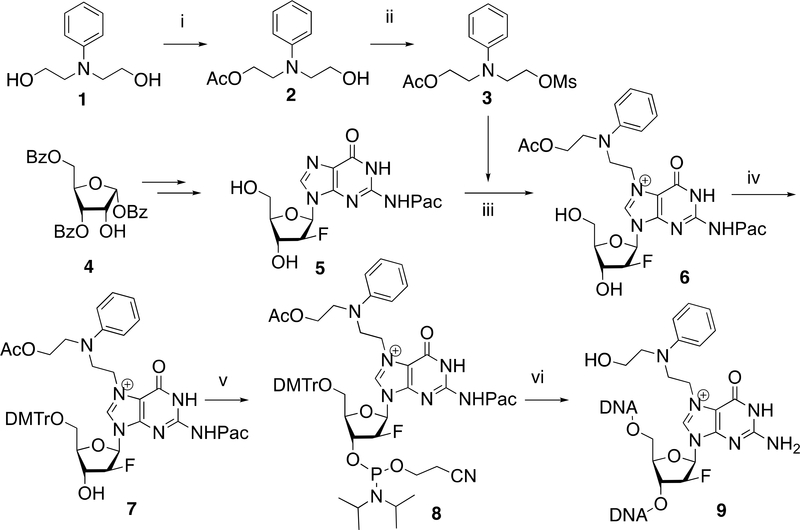
Monoacetylation of commercially available N-phenyldiethanol amine 1 followed by treatment with methanesulfonyl chloride gave mesylated nitrogen half-mustard 3 (Scheme 1). The 2′-fluoro-2′-deoxyguanosine 5 was prepared from a ribose derivative 4 by following the published synthetic procedures [21]. The coupling of the 2′-fluorinated dG 5 with mesylated nitrogen half-mustard 3 took place smoothly in DMF at an ambient temperature to give N7-NHMG adduct 6 in 81% yield. The N7-NHMG 6 was subjected to sequential dimethoxytritylation and phosphatidylation to provide NHMG phosphoramidite 8, which was then incorporated into DNA via the solid phase DNA synthesis and the ultra-mild deprotection to furnish NHMG-modified oligonucleotide 9.
Scheme 1. Preparation of site-specific NHMG-containing oligonucleotide.
Reagents and conditions: i) acetic anhydride, pyridine, 0°C to 25°C, 3 h, 45%; ii) methanesulfonyl chloride, Et3N, CH2Cl2, 25°C, 1 h, 0°C, 79%; iii) DMF, 25°C, 48 h, 81%; iv) DMTrCl, pyridine, 2h, 85%; v) 4,5-dicyanoimidazole, (i-Pr2N)2P(OC2H4CN), CH2Cl2, 25°C, 1 h, 89%; vi) solid phase DNA synthesis and ultra-mild deprotection conditions (K2CO3, MeOH, 25°C).
Synthesis of mono-acetylated N-phenyldiethanolamine (2).
To a solution of N-phenyldiethanolamine 1 (9.1 g, 50 mmol) in pyridine (100 mL) was added acetic anhydride (6 mL, 60 mmol) at 0°C. The resulting mixture was stirred at 25°C for 3 h, quenched with saturated aqueous NaHCO3, and extracted with ethyl acetate (2×200 mL). The resulting organic layer was washed with brine, dried over anhydrous Na2SO4, filtered, concentrated under reduced pressure, and subjected to silica gel column chromatography (hexane–EtOAc, 1:1) to give mono-acetylated N-phenyldiethanolamine 2 (5.1 g, 45%). 1H NMR (400 MHz, CDCl3) δ 7.27 – 7.13 (m, 2H), 6.82 – 6.66 (m, 3H), 4.26 (t, J = 6.0 Hz, 2H), 3.78 (t, J = 5.5 Hz, 2H), 3.62 (t, J = 6.0 Hz, 2H), 3.51 (t, J = 5.5 Hz, 2H), 2.18 (d, J = 8.1 Hz, 1H), 2.01 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 171.05, 147.91, 129.35, 117.46, 113.00, 61.80, 60.12, 54.31, 50.60, 20.85. MS (ESI): m/z = 246.1 [M+Na]
Synthesis of mesylated N-phenyl nitrogen mustard (3).
To a solution of mono-acetylated N-phenyldiethanolamine 2 (4.2 g, 18.83 mmol) and Et3N (8 mL, 56 mmol) in CH2Cl2 (50 mL), was added dropwise methanesulfonyl chloride (3 mL, 37.6 mmol) at 0°C. The resulting mixture was stirred at room temperature for 1 h. The reaction mixture was quenched with saturated aqueous NaHCO3 and extracted with CH2Cl2 (2×150 mL). The organic layer was washed with brine, dried over Na2SO4, filtered, and concentrated under reduced pressure. The resulting residue was purified by column chromatography on silica gel (hexane–EtOAc, 7:3) to give mesylated N-phenyl nitrogen mustard 3 (4.5 g, 79%). 1H NMR (400 MHz, CDCl3) δ 7.27 – 7.15 (m, 2H), 6.79 – 6.64 (m, 3H), 4.33 (t, J = 6.1 Hz, 2H), 4.21 (t, J = 6.1 Hz, 2H), 3.72 (t, J = 6.1 Hz, 2H), 3.62 (t, J = 6.1 Hz, 2H), 2.92 (s, 3H), 2.02 (s, 3H). 13C NMR (100 MHz, CDCl3) δ 170.91, 146.67, 129.60, 117.55, 112.26, 111.94, 66.37, 61.46, 50.20, 50.09, 37.35, 31.54, 20.85. MS (ESI): m/z = 324.1 [M+Na]+. HRMS: calcd. for C13H19NO5S [M+Na]+: 324.0883: found: 324.0876.
Synthesis of 2′-fluoro-2′-deoxy-NHMG nucleoside (6).
To a suspension of N2-phenoxyacetyl (Pac)-2′-fluoro-2′-deoxyguanosine 5 (100 mg, 0.24 mmol) in DMF (5 mL), was added mesylated N-phenyl nitrogen mustard 3 (722 mg, 2.4 mmol). After stirring at room temperature for 48h at 25°C, the reaction mixture was poured into Et2O (100 mL) and the resulting precipitate was collected by filtration. The collected solid residue was purified by silica gel column chromatography with 5–15% MeOH in CH2Cl2 containing 1% Et3N to afford 121 mg (81%) of 2′-fluoro-2′-deoxy NHMG nucleoside 6. 1H NMR (400 MHz, DMSO-d6) δ 9.99 (br, 2H, NHCO), 9.23 (s, 1H, H-8), 7.31 – 7.21 (m, 2H, N-Ph), 7.17 – 7.08 (m, 2H, N-Ph), 6.90 (m, 5H, Pac-Ph), 6.61 (t, J = 7.2 Hz, 1H, N-Ph), 6.36 (dd, J = 15.8, 3.8 Hz, 1H, H-1′), 6.09 – 6.01 (m, 1H, HO-5′), 5.23 (d, J = 15.6 Hz, 1H, H-4′), 5.07 – 4.94 (m, 3H, H-3′ & CH2-Pac), 4.58 (t, J = 7.1 Hz, 2H, N7-CH2-mustard), 4.45 – 4.34 (m, 1H, HO-3′), 4.09 (t, J = 6.2 Hz, 2H, CH2-mustard), 3.93 (q, J = 5.0 Hz, 1H, H-2′), 3.84 (t, J = 7.0 Hz, 2H, CH2-mustard), 3.57 (m, 4H, CH2-mustard & CH2-5′), 1.94 (d, J = 9.8 Hz, 3H, OAc). 13C NMR (100 MHz, DMSO-d6) δ 170.79, 158.46, 148.70, 147.51, 129.97, 129.84, 129.61, 121.77, 121.16, 117.05, 115.00, 114.87, 112.55, 95.88, 93.97, 85.90, 83.79, 83.62, 73.30, 73.07, 68.01, 61.20, 60.96, 50.70, 48.83, 46.21, 21.07, 14.52. MS (ESI): m/z = 625.2 [M]+. HRMS: calcd. for C30H34O8N6F+ [M]+: 625.2417: found: 625.2420.
Synthesis of 5′-O-(4,4′-dimethoxytrityl)-2′-fluoro-2′-deoxy-NHMG (7).
To a pyridine (10 mL) solution of NHMG nucleoside 6 (138 mg, 0.22 mmol), was added 4,4′-dimethoxytrityl chloride (112 mg, 0.33 mmol) and the mixture was stirred at room temperature for 2h. The reaction mixture was diluted with CH2Cl2, washed with saturated aqueous NaHCO3, and brine. The organic layer was dried over anhydrous Na2SO4, filtered, and concentrated. The residue was purified by silica gel chromatography to give 175 mg (86%) of 5′-O-(4,4′-dimethoxytrityl)-2′-fluoro-2′-deoxy-NHMG 7. 1H NMR (400 MHz, CDCl3) δ 9.02 (s, 1H, NHCO), 7.86 (d, J = 2.0 Hz, 1H, H-8), 7.45 – 7.36 (m, 1H), 7.35 – 7.13 (m, 8H), 7.11 – 6.96 (m, 2H), 6.89 – 6.80 (m, 1H), 6.80 – 6.66 (m, 3H), 6.60 (d, J = 8.1 Hz, 1H, H-1′), 5.39 – 5.21 (m, 1H, H-4′), 4.69 (d, J = 16.8 Hz, 1H, 3-H′), 4.64 – 4.45 (m, 2H, N7-CH2-mustard), 4.45–4.23 (m, 3H, CH2-Pac & H-2′), 4.04–3.89 (m, 2H, CH2-mustard), 3.87 (t, J = 5.6 Hz, 2H, CH2-mustard), 3.70 (s, 6H, OCH3-DMTr), 3.44 – 3.24 (m, 4H, CH2-mustard & CH2-5′), 1.89 (s, 2H, CH2-Pac). 13C NMR (100 MHz, CDCl3) δ 170.73, 158.62, 156.80, 149.76, 148.66, 146.04, 144.48, 135.58, 135.53, 130.04, 129.68, 128.16, 127.86, 127.06, 123.69, 122.20, 118.70, 114.70, 113.71, 113.14, 86.71, 86.43, 85.27, 63.50, 61.33, 60.34, 55.16, 50.85, 47.00, 21.00, 20.68, 14.16, 14.07. MS (ESI): m/z = 927.3 [M]+. HRMS: calcd. for C51H52O10N6F+ [M]+: 927.3723: found: 927.3727.
Synthesis of NHMG phosphoramidite (8).
To a solution of 5′-tritylated NHMG nucleoside 7 (93 mg, 0.1 mmol) and dicyanoimidazole (24 mg, 0.2 mmol) in CH2Cl2 (4 mL), was added (2-cyanoethyl-N,N-diisopropylamino)chlorophosphine (42.1 mg, 0.13 mmol) in CH2Cl2. After stirring at room temperature for 1h, the reaction mixture was diluted with CH2Cl2 (50 mL), washed with saturated aqueous NaHCO3, and brine. The organic layer was dried over anhydrous Na2SO4 filtered, and concentrated in vacuo. The residue was purified by silica gel column chromatography with 4–10% MeOH in CH2Cl2 containing 0.5% Et3N to afford 101 mg (90%) of the desired NHMG phosphoramidite 8. 1H NMR (400 MHz, CDCl3) δ 7.95 (d, J = 52.5 Hz, 1H), 7.41 – 7.34 (m, 1H), 7.33 – 7.18 (m, 5H), 7.03 (dt, J = 8.7, 6.3, Hz, 1H), 6.99 – 6.90 (m, 1H), 6.82 – 6.72 (m, 2H), 6.66 (dt, J = 8.3, 4.4 Hz, 1H), 6.62 – 6.46 (m, 2H), 5.43 – 5.18 (m, 1H), 4.79 – 4.63 (m, 1H), 4.52 (s, 1H), 4.23 – 4.13 (m, 1H), 3.97 (dd, J = 7.5, 4.7 Hz, 1H), 3.84 (dt, J = 15.3, 5.4 Hz, 1H), 3.75 (s, 3H), 3.69 – 3.52 (m, 2H), 3.47 – 3.17 (m, 2H), 2.43 (td, J = 6.3, 1.3 Hz, 1H), 1.88 (d, J = 1.4 Hz, 2H), 1.20 – 1.08 (m, 5H). 31P NMR (162 MHz, CDCl3) δ 151.34, 151.2. MS (ESI): m/z = 1127.4 [M]+. HRMS: calcd. for C60H69O11N8F+ [M]+: 1127.4802: found: 1127.4803.
Synthesis of NHMG-containing oligonucleotide (9).
NHMG-containing oligonucleotide was custom-synthesized by Midland Certified Reagent Company Inc. (Midland, TX) via solid phase DNA synthesis using NHMG phosphoramidite 8 and ultramild deprotection conditions (K2CO3 in MeOH, 25°C). The NHMG oligonucleotide was purified by gel filtration, characterized by MALDI-TOF mass spectroscopy, and further purified using denaturing urea-polyacrylamide gel electrophoresis. This NHMG-containing 12-mer (5’-CAT(NHMG)CTCACACT-3’) was used for kinetic and crystallographic studies. 5’-CAT(NHMG)CTCACACT-3’: C125H161O71N42F1P11 Mass calculated [M]: 3745.74; observed [M+1]: 3746.52.
Protein expression and purification.
The catalytic core of human polη (amino acids 1–432) was cloned into pET28a plasmid with NcoI and BamHI restriction enzyme sites. The catalytic domain of polη was used in this study, because its TLS efficiency is comparable to that of the full length polη [29]. E. coli BL21 (DE3) cells transformed with this plasmid were grown at 37°C in LB medium until the OD600 value reached 0.6. Polη expression was induced for eighteen hours at 20°C by adding 0.3 mM isopropyl-β-thiogalactoside. The induced cells were collected by centrifugation at 8,000g for 20 min at 4°C. Proteins were purified by Ni2+-NTA affinity (GE Healthcare), Heparin column and Superdex-75 gel filtration chromatography (GE Healthcare). Purified human polη was concentrated to 15 mg/ml in gel filtration buffer (25 mM Tris, pH 7.5, 300 mM KCl, 10% glycerol and 2 mM dithiothreitol), aliquoted and flash frozen in liquid nitrogen to store at −80°C.
Steady-state kinetics of single nucleotide incorporation opposite a templating NHMG by polη.
Steady-state kinetic parameters for nucleotide insertion opposite NHMG by polη were determined with slight modification of published protocols [37, 38]. The primer for kinetic studies (5´-FAM/AGTGTGAG-3´) was purchased from Integrated DNA Technologies (Coralville, IA). An NHMG-containing template (5´-CAT(NHMG)CTCACACT-3´) was synthesized and purified by Midland Certified Reagent company (Midland, TX). To prepare DNA substrate containing NHMG lesion, the primer and template were annealed in hybridization buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA) at 90°C for 5 min. Polη activities were determined using a reaction mixture containing 40 mM Tris-HCl pH 8.0, 60 mM KCl, 10 mM dithiothreitol, 250 μg/ml bovine serum albumin, 2.5 % glycerol, 5 mM MgCl2, 50–100 nM recessed DNA, and varying concentrations of individual dNTP. To prevent the end-product inhibition and substrate depletion from interfering with accurate velocity measurement, the enzyme concentrations and reaction-time intervals were adjusted for every experiment (less than 20% insertion product formed). The reactions were initiated by the adding the enzyme and stopped with a gel-loading buffer (95% formamide with 20 mM EDTA, 45 mM Tris-borate, 0.1% bromophenol blue, 0.1% xylene cyanol). The quenched reaction samples were separated on 20% denaturing polyacrylamide gels. The gels were analyzed using Typhoon Imager (GE Healthcare) to quantify product formation. The kcat and Km were determined by fitting the reaction rate over dNTP concentrations to the Michaelis-Menten equation. Each experiment was repeated three times to measure the average of the kinetic results. The efficiency of nucleotide insertion was calculated as kcat/Km. The relative frequency of dNTP incorporation opposite NHMG was determined as f = (kcat/Km)[dNTP:NHMG] /(kcat/Km)[dCTP:dG].
Protein crystallization and structure determination.
The ternary polη complex with the templating NHMG was prepared and crystallized with slight modification of published protocols [34, 39]. Purified polη (9 mg/ml) was mixed with a 1.2 molar excess of duplex DNA containing NHMG lesion (primer, 5´-AGTGTGAG-3´ and template, 5´-CAT(NHMG)CTCACACT-3´). Subsequently, a 10-fold molar excess of nonhydrolyzable dCMPNPP (Jena Bioscience) was added to the binary complex of the polη:NHMG duplex DNA. The ternary complex co-crystals with dCMPNPP paired with the templating NHMG were obtained using the hanging drop method in a buffer solution containing 100 mM MES pH 6.0–6.5 and 18% PEG2000 MME. Crystals were harvested and cryoprotected in mother liquor supplemented with 20% glycerol and were flash-frozen in liquid nitrogen. Diffraction data were collected at 100 K at the beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory. The diffraction data were processed using HKL2000 and the scaled set was converted to structure factors by CCP4 [40]. Structures were solved by molecular replacement using polη-dG:dCTP* structure (PDB ID: 4O3N) as a search model. The model was built using COOT [41]and refined using PHENIX [42]. MolProbity in the PHENIX GUI was used to generate Ramachandran plots [43]. All the crystallographic figures were generated using PyMOL.
RESULTS
Single nucleotide insertion opposite templating N7-alkylguanine by human polη.
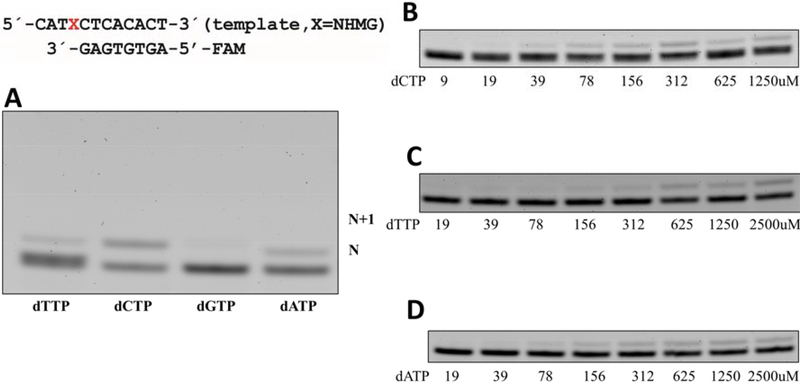
Polη has been shown to efficiently bypass N7-methylG in an error-prone manner [23, 39]. To evaluate the impact of the steric bulkiness of the N7-alkyl group on the mutagenic potential of purine, we performed kinetic studies of polη incorporating nucleotide opposite templating dG, and NHMG. Our steady-state kinetic studies show that the size of the N7-alkyl group significantly influences the catalytic efficiency (kcat/Km) of correct insertion. Published kinetic results show small N7-methyl group moderately (~3-fold; 46 vs. 13.2) decreases the catalytic efficiency for dCTP insertion [39]. Furthermore, NHMG at the templating position decreases the dCTP-insertion efficiency by ~130-fold (kcat/Km: 0.36 vs. 45.6, Table 1 and Figure 2), highlighting the impact of steric bulkiness on the correct insertion opposite N7-alkylG. The Km value of the NHMG:dCTP insertion is ~43-fold greater than that of the dG:dCTP insertion (113.4 vs. 2.7 μM), while its kcat value is 3-fold less than that for the dG:dCTP insertion (40.9×10−3s−1 vs. 120.6×10−3s−1), indicating that the low catalytic efficiency (kcat/Km) for the NHMG:dCTP insertion is mainly driven by increased Km.
Table 1.
Kinetic parameters for nucleotide incorporation opposite N7-alkylG and dG by polη
| template:dNTP | Km (μM) | kcat (10−3s−1) | kcat/Km (10−3s−1μM−1) | f a | replication fidelity |
|---|---|---|---|---|---|
| dG:dCTP | 2.7±0.3 | 120.6±6.1 | 46 | ||
| dG:dTTP | 159.3±2.7 | 74.8±0.9 | 0.47 | 0.01 | 100 |
| dG:dATP | 116.4±8.1 | 35.8±1.0 | 0.31 | 0.007 | 150 |
| MeG:dCTPb | 4.3±0.4 | 56.4±2.7 | 13.2 | ||
| MeG:dTTPb | 52.5±1.7 | 49.3±0.1 | 0.9 | 0.07 | 15 |
| NHMG:dCTP | 113.4±1.4 | 40.9±1.3 | 0.36 | ||
| NHMG:dTTP | 146.3±5.1 | 5.5±0.1 | 0.037 | 0.1 | 10 |
| NHMG:dATP | 156.7±12.5 | 10.4±0.5 | 0.066 | 0.18 | 5 |
Misincorporation frequency: (kcat/Km)[incorrect insertion]/(kcat/Km)[correct insertion].
Data from reference [39]
Replication fidelity: (kcat/Km)[correct insertion]/(kcat/Km)[incorrect insertion]
Figure 2. Incorporation of an individual nucleotide opposite templating NHMG by polη.
(A) A denaturing urea PAGE gel for nucleotide incorporation opposite NHMG by polη. Polη (5 nM), NHMG-containing 12-mer template (50 nM), 8-mer primer (50 nM), and incoming nucleotides (9 to 2,500 μM) were used, and the reaction lasted for 10 min. The formation of an N+1 band was negligible even for dATP, which is a correct insertion for the extension. X denotes NHMG. (B-D) Denaturing gel images for the insertion kinetics of dCTP, dTTP, and dATP.
The introduction of NHMG at the templating position significantly promotes misincorporation by polη. The misincorporation frequency of the dTTP insertion opposite NHMG insertion is 0.1, which is 10-fold greater than that of the dG:dTTP insertion (Table 1). The Km values of the dG:dTTP and NHMG:dTTP insertions are very similar (159 vs. 146 μM), while kcat of the dG:dTTP insertion is ~15-fold greater than that for the NHMG:dTTP insertion (74×10−3s−1 vs. 5×10−3s−1), indicating the error-prone incorporation opposite NHMG is driven by increased kcat.
The catalytic efficiency of dCTP insertion opposite NHMG is only ~10-fold greater than that for dTTP insertion (0.36×10−3s−1μM−1 vs. 0.037×10−3s−1μM−1), while dCTP insertion opposite undamaged dG is ~100-fold more efficient than dTTP insertion opposite dG (46×10−3s−1μM−1 vs. 047×10−3s−1μM−1). This indicates that NHMG decreases the replication fidelity of dTTP insertion ((kcat/Km)[NHMG:dCTP]/ (kcat/Km)[NHMG:dTTP]) by ~10-fold (10 vs. 100, Table 1). In addition, the replication fidelity of dATP insertion opposite NHMG decreases ~30-fold compared to dATP insertion opposite dG (5 vs. 150, Table 1), which demonstrates that the presence of NHMG significantly increases the promutagenicity. Interestingly, dATP insertion opposite NHMG is 2-fold more efficient than dTTP insertion opposite NHMG (0.066 vs. 0.037), whereas the dATP insertion opposite dG is ~1.5-fold less efficient than the dTTP insertion.
Structure of polη incorporating a non-hydrolyzable dCTP analog opposite templating NHMG.
To gain structural insights into polη catalyzing across the major adduct induced by nitrogen mustards, we determined a ternary complex structure of polη incorporating a nonhydrolyzable dCMPNPP (dCTP* hereafter) opposite a templating NHMG in the presence Mn2+ (Figure 3). Extensive attempts for crystallizing the polη-NHMG:dCTP* complex with magnesium ions were not successful. The nonhydrolyzable dCTP* nucleotide was used because it is isosteric to dCTP and its coordination to the active-site metal ions is essentially identical to that of dCTP [34, 44]. The ternary complex was crystallized in the P61 space group with the cell dimension of a = b = 98.9 Å, c = 82.1 Å, α = β = 90.0°, and γ = 120° and one polη was found in the asymmetric unit. The structure, which was refined to 2.69 Å with Rwork = 18.6% and Rfree = 23.8%, displayed the conserved secondary structures and the four characteristic domains (thumb, palm, finger, and little finger) of the Y-family DNA polymerases. The statistics for data collection and the refinement are summarized in Table. 2.
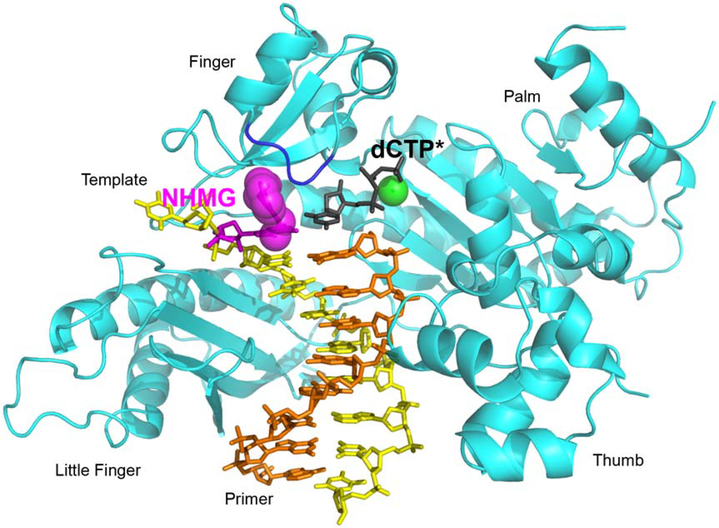
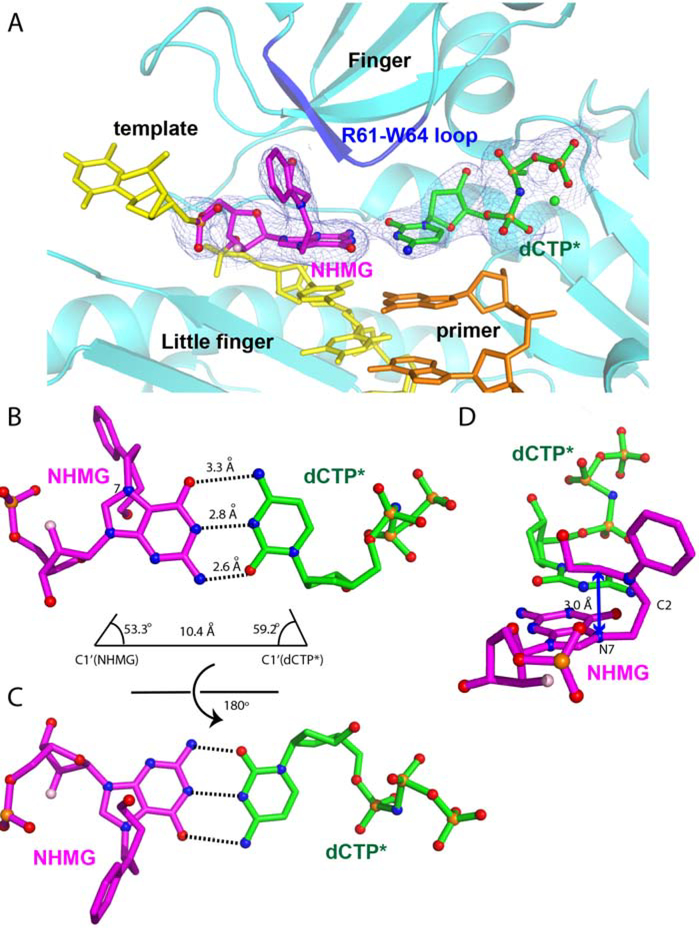
Figure 3. Overall structure of polη incorporating dCTP* opposite a templating NHMG.
NHMG and dCTP* are shown in magenta and black, respectively. The nitrogen half-mustard moiety of the NHMG adduct is shown in magenta spheres. The nucleotide-binding metal ion complexed with dCTP* is shown in a green sphere. The template except for NHMG and the primer are shown in yellow and orange, respectively. The conserved four domains are indicated. The loop containing Arg61, Ser62, Met63, and Trp64 (R61-W64 loop) from the finger domain is colored in blue.
Table 2.
Data Collection and Refinement Statistics.
| PDB CODE | NHMG:C (6V5K) |
|---|---|
| Data Collection | |
| Space group | P61 |
| Cell Constants | |
| a (Å) | 98.911 |
| b | 98.911 |
| c | 82.138 |
| α (°) | 90.00 |
| β | 90.00 |
| γ | 120.00 |
| resolution (Å)a | 50–2.69 (2.73–2.69) |
| Rmerge b (%) | 0.059 (0.569) |
| <I/σ> | 12.4 (1.2) |
| CC1/2 (2.73–2.69) | 0.469 |
| completeness (%) | 99.5 (96.7) |
| redundancy | 10.3 (7.1) |
| Refinement | |
| Rwork c/Rfree d (%) | 18.6/23.8 |
| unique reflections | 12740 |
| Mean B Factor (Å2) | |
| protein | 68.8 |
| ligand | 76.8 |
| Ramachandran Plot | |
| most favored (%) | 94.6 |
| add. allowed (%) | 4.7 |
| RMSD | |
| bond lengths (Å) | 0.010 |
| bond angles (degree) | 1.320 |
Values in parentheses are for the highest resolution shell.
Rmerge = Σ|I-<I>|/ ΣI where I is the integrated intensity of a given reflection.
Rwork = Σ|F(obs)-F(calc)|/ΣF(obs).
Rfree = Σ|F(obs)-F(calc)|/ΣF(obs), calculated using 5% of the data.
The crystal structure of polη in complex with a templating NHMG paired with an incoming dCTP* reveals, for the first time, the base pairing property of a nitrogen mustard-induced DNA lesion in the nascent base pair site of DNA polymerase. A 2Fo-Fc electron density map contoured at 1σ clearly shows the presence of NHMG and dCTP* in the replication site of polη (Figure 4A). NHMG and dCTP* form Watson-Crick base pairing with three hydrogen bonds (Figure 4B and 4C). The distances of interbase hydrogen bonds are 2.6 Å for between N2 of NHMG and O2 of dCTP*, 2.8 Å for between N1 of NHMG and N3 of dCTP*, and 3.3 Å for between O6 of NHMG and N4 of dCTP* (Figure 4B). The distance between the C1´ carbons of NHMG and dCTP* is 10.4 Å and the λ angles are 53.3° (NHMG) and 59.2° (dCTP*), which are similar to the values observed in normal Watson-Crick base pairings [45]. The formation of three hydrogen bonds with Watson-Crick geometry indicates that the keto tautomer of NHMG is involved in the base pairing with dCTP. The propeller twist angle of the purine ring of NHMG and the pyrimidine ring of dCTP* is ~25° (Figure 4D), which is significantly greater than the values observed in normal Watson-Crick base pair. The nitrogen half-mustard moiety bends at the C2 position of NHMG and is positioned on the 5´ side of template strand (Figure 4D). The hydroxyethyl moiety of NHMG is placed above the imidazole ring of NHMG and forms van der Waals interaction with the purine ring of NHMG (Figure 4D). The phenyl ring of the nitrogen half-mustard moiety is almost perpendicular to the purine ring of NHMG. The aromatic moiety of NHMG points toward the Arg61-Trp64 loop in the finger domain.
Figure 4. Close-up view of Polη incorporating dCTP* opposite a templating NHMG.
(A) Base paring of NHMG:dCTP* in the catalytic site of polη. A 2Fo-Fc electron density map contoured at 1σ around the templating NHMG and incoming dCTP* is shown. The Arg61-Trp64 loop is colored in blue. The Mn2+ ion is shown in green sphere. (B) Base pairing properties of NHMG:dCTP*. The Watson-Crick hydrogen bonds and base pairing geometry of NHMG:dCTP* are shown. (C) Base pairing properties of NHMG:dCTP* from another perspective. (D) Conformation of the nitrogen half-mustard moiety of the NMHG adduct. The purine ring of NHMG is distorted about 25° from the plane of the pyrimidine ring of dCTP*.
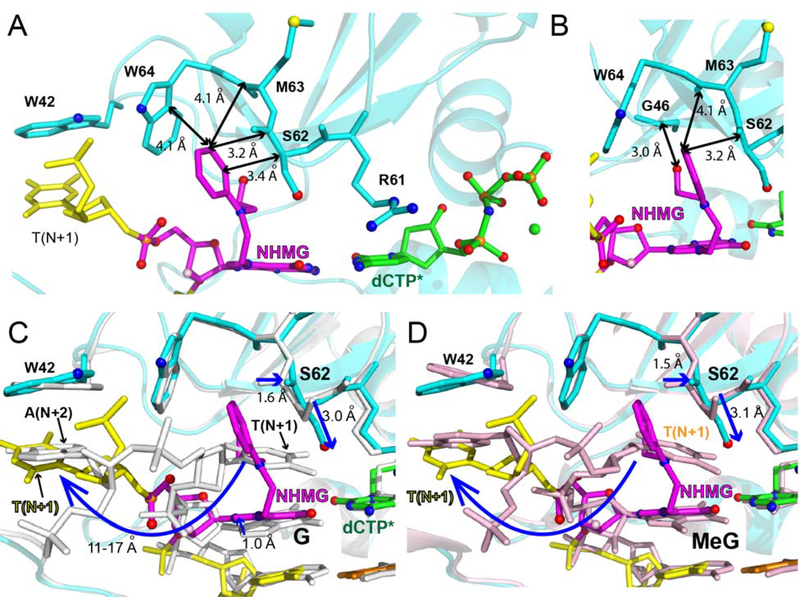
The presence of NHMG at the templating position greatly changes DNA conformation near the lesion, particularly at the binding site of N+1 base of the template strand (Figure 5). The nitrogen half-mustard moiety engages in extensive van der Waals interactions with the Arg61-Trp64 loop of polη (Figure 5), which disrupts the possible stacking interaction between the bases at the N and N+1 positions. The nitrogen half-mustard moiety enters an open cleft generated by the Arg61-Trp64 loop and interacts with Ser62, Met63, and Trp64 (Figure 5A and 5B). In the published polη structure with dG:dCTP (PDB ID:4O3N), the templating dG stacks with an adjacent 5´ base (Figure 5C) [46]. In the NHMG structure, the templating NHMG does not stack with dT on the 5´ side. Instead, the nitrogen half-mustard moiety of NHMG occupies the space where the N+1 template base resides in the dG:dCTP structure, thereby precluding the stacking interaction between the templating base and the 5´ base. The N+1 template dT in the polη-NHMG:dCTP structure swings ~180° from the position in the polη-dG:dCTP structure and points toward the little finger domain of polη, residing where the N+2 template base is positioned in the dG:dCTP structure. This conformational difference suggests that, during the extension across NHMG, the N+1 template dT must undergo a large conformational reorganization to base pair with incoming nucleotide. The bulky NHMG moiety could also interfere the extension step by sterically interacting with the N+1 template dT. The large conformational change of the base 5´ to NHMG may contribute to the lack of extension in polη-mediated bypass of NHMG (Figure 2A). The extension past NHMG could be conducted by other TLS polymerases such as polζ [47], which is specialized to extend distorted base pairs.
Figure 5. Interactions of the templating NHMG with Arg61-Trp64 loop.
(A) Van der Waals interactions of the nitrogen-half mustard moiety with Ser62, Met63, and Trp64. Distances are indicated as double-headed arrows. (B) Van der Waals interactions of the nitrogen-half mustard moiety with Ser62, Met63, and Trp64 from another perspective. (C) Comparison of the conformations of the templating base and the N+1 base in the polη-NHMG:dCTP* structure and the polη-dG:dCTP* structure (PDB code: 4O3N). The conformational shift of the dT at the N+1 position is shown in a curved arrow. The shift of Ser62 is indicated as arrows. (D) Comparison of the conformations of the templating base and the N+1 base in the polη-NHMG:dCTP* structure and the polη-N7MeG:dCTP* structure (PDB code: 6UI2). The conformational shift of the dT at the N+1 base is shown in a curved arrow. The shift of Ser62 is indicated as blue arrows.
The nitrogen half-mustard moiety is oriented toward the 5´ end of the template and almost perpendicular to the purine ring of NHMG. The N-phenyl moiety of NHMG does not engage in stacking interaction with the adjacent bases. The terminal hydroxyl group of the nitrogen mustard monoadduct forms two hydrogen bonds with the backbone carbonyl groups of Gly46 and Ile47 (Figure 5B). To accommodate the arylalkyl moiety of NHMG, the backbone carbonyl group and the sidechain hydroxy group of Ser62 moves by ~1.5 Å and ~3.0 Å, respectively, relative to the positions of Ser62 in the polη-dG:dCTP structure (Figure 5C).
In the catalytic site of the polη-NHMG:dCTP* structure, despite the formation of Watson-Crick NHMG:dCTP base pairing, only the nucleotide-binding metal ion (the B-site metal ion) is observed, which may explain the low catalytic efficiency of the NHMG:dCTP insertion by the enzyme. This is in contrast with published polη-dG:dCTP* [46] and polη-N7MeG:dCTP* [39] structures showing both metal ions in the catalytic site. Superposition of the polη-NHMG:dCTP* structure with the published polη-dG:dCTP* structure reveals that the conformation of the incoming dCTP* is essentially identical (Figure 5C). The conformation of the purine ring of NHMG, however, differs significantly from that of dG, resulting in a greater twist angle value (25°) for NHMG:dCTP* base pair. This canonical but non-optimal base pairing would contribute to the reduced insertion efficiency across NHMG lesion. The favorable interaction of the nitrogen half-mustard moiety with the Arg61-Trp64 loop would lead to a high propeller-twist angle, which could decrease Km for dCTP insertion opposite NHMG and deter binding of metal ion at the metal-A site.
The overlay of the polη-NHMG:dCTP* and published polη-N7MeG:dCTP* (PDB ID: 6UI2) [39] structures reveals that the bulky nitrogen half-mustard group may induce a large conformational change of the templating bases in the N+1 and N+2 positions (Figure 5D). The polη structure with templating N7MeG and incoming dCTP* shows the conformations of DNA and protein are virtually identical to those of the polη-dG:dCTP structure. The N+1 dT engages in π-π stacking with N7MeG, which forms Watson-Crick hydrogen bonds with dCTP with a moderate propeller twist angle (~10°). These indicate the steric bulkiness of the templating N7-alkyl group plays a role in the large conformational reorganization of the template strand.
DISCUSSION
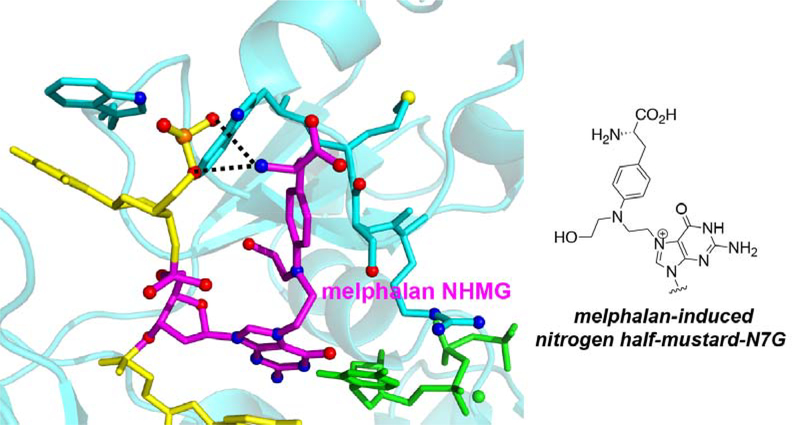
Aromatic nitrogen mustards such as chlorambucil and melphalan are known to produce nitrogen half-mustard-G adducts as major lesions [6, 7]. Our NHMG adduct is structurally very similar to the nitrogen half-mustard-G lesion formed by melphalan. To gain structural insight into how TLS polymerases bypass aromatic nitrogen half-mustard-G adducts, we built a modeled structure where polη incorporates dCTP opposite melphalan-induced NHMG adduct (Figure 6). The modeled structure predicts melphalan-induced NHMG may be accommodated in the replicating base site of polη by generating favorable polar and non-polar interactions. The melphalan moiety would enter the open cleft generated by the Arg61-Trp64 loop and the amino acid moiety of melphalan would make hydrogen bond interactions with the phosphate backbone of the templating DNA and the amino acid residues in Arg61-Trp64 loop. We therefore propose our polη-NHMG:dCTP* structure represents a close approximation of the structure of polη bypassing the aromatic nitrogen mustard-induced NHMG lesions.
Figure 6. Modeled structure of polη bypassing the guanine N7-nitrogen half-mustard adduct formed by melphalan, an aromatic nitrogen mustard.
The melphalan-induced nitrogen half mustard-G lesion is shown in magenta and an incoming dCTP* is shown in green. The amino acid moiety of melphalan-NHMG adduct would form additional hydrogen bonds with the phosphate backbone of template strand and the Arg61-Trp64 loop.
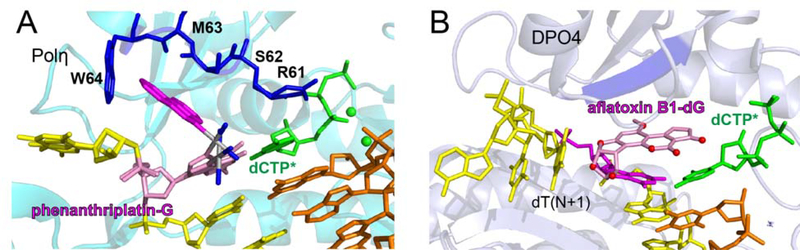
The polη-NHMG:dCTP* complex represents the first crystal structure of DNA polymerase bypassing nitrogen mustard-induced DNA lesions. In contrast, there are published structures of TLS polymerase incorporating nucleotide opposite bulky N7G adducts formed by phenanthriplatin and aflatoxin B1 [48, 49]. Phenanthriplatin is a monofunctional platinum-based anticancer agent with an aromatic phenanthridine moiety. Unlike the NHMG adduct, phenanthriplatin-N7G adduct is efficiently bypassed by polη [48]. Comparison of the polη-NHMG:dCTP* structure with published polη-phenanthriplatin-N7G:dCTP* (PDB ID:4Q8E) structure reveals that the Arg61-Trp64 loop may play a role in accepting bulky major-groove lesions in the enzyme active site (Figure 7A). The overall conformation of the phenanthriplatin-N7G complex is similar to that of the NHMG complex. The phenanthridine moiety of the phenanthriplatin-N7G engages in van der Waals interaction with Ser62 and Trp64, suggesting that the Arg61-Trp64 loop may stabilize bulky N7G lesions at the replicating base pair site. The flexibility of the Arg61-Trp64 loop (Figure 5C) would play an important role in accepting a wide variety of bulky major-groove lesions. In the published polη-phenanthriplatin-N7G:dCTP* structure, unlike in the polη-NHMG:dCTP* structure, both A- and B-site metal ions are present. This is consistent with the efficient incorporation of dCTP opposite the phenanthriplatin-N7G by polη. The crystal structure of Dpo4 incorporating dCTP* opposite aflatoxin B1 (AFB1)-N7G adduct [49] is significantly different from the polη-NHMG:dCTP* structure (Figure 7B). Dpo4 lacks an Arg61-Trp64-like open cleft near the replicating base pair site and has a spacious, solvent accessible active site. The aflatoxin B1 moiety engages in π-π stacking interaction with the modified guanine and incoming dCTP*.
Figure 7. Translesion synthesis DNA polymerase bypassing bulky N7G adducts.
(A) Human polη in complex with templating phenanthriplatin-N7G and incoming dCTP* (PDB code:4Q8E). (B) Solfolobus Solfataricus Dpo4 incorporating dCTP* opposite aflatoxin B1-N7G (PDB code:3PW7).
Our polη-NHMG:dCTP* complex was crystallized in the presence of Mn2+ but not with Mg2+, highlighting the effect of metal cofactor on the crystallization of polymerase-DNA complexes. Mn2+ cofactor has more lenient coordination requirement than Mg2+ and is more tolerant to the changes (e.g., distortion, misalignment, mutation) in the active site of DNA polymerases [50, 51]. For example, in the case of TLS DNA polymerase Dpo4, replacing Mg2+ with Mn2+ dramatically increases the catalytic efficiency of dGTP insertion opposite thymine [52]. In addition, the use of Mn2+ cofactor facilitates the bypass of cyclobutane pyrimidine dimer (CPD) lesion by polη and polι [53]. The fact that polη-NHMG:dCTP* complex was crystallized only with Mn2+ suggests that the presence of templating NHMG causes structural distortion in the active site of polη, which could be tolerated by Mn2+ but not Mg2+.
The bulky nitrogen half-mustard moiety may primarily contribute to the inefficient incorporation of dCTP opposite NHMG by polη. The introduction of the small N7-methylguanine lesion at the templating position only slightly reduces the catalytic efficiency of polη and Solfolobus solfataricus Dpo4 [23], indicating TLS DNA polymerases can readily traverse a small N7-alkylguanine lesion. The presence of NHMG and N7-methylguanine at the templating position increases the frequency of dTTP misincorporation 7 to 10-fold (Table 1), highlighting the promutagenic properties of N7-alkylG lesions. Replication across N7-alkylG by TLS polymerases would, therefore, promote G-to-A mutations, which have been shown to be induced by bulky N7-alkylG lesions in vivo [54]. The high promutagenicity of NHMG lesion suggests the NHMG-induced mutagenesis could be a contributing factor for the development of secondary cancers often found in patients receiving NM-based chemotherapy [6, 9, 10].
N7-alkylG adducts, which exhibit half-lives of several hours to days in duplex DNA [55], can undergo spontaneous depurination to generate abasic sites, which promotes G to T transversions [56]. The primary N7-alkylG lesions with a small alkyl group (e.g. methyl, ethyl) have been considered non-mutagenic [57, 58]. However, recent studies show N7-methylG facilitates dTTP misincorporation opposite the lesion [23, 39]. In addition, the bulky AFB1-N7G adduct has been shown to predominantly induce G to T mutations in primate cells [59], which appears to be caused by intercalation of aflatoxin moiety on the 5´ side of the damaged guanine rather than AFB1-N7G-induced apurinic sites. Indeed, the crystal structure of Dpo4 bound to AFB1-N7G adduct demonstrates the intercalation of the AFB1 moiety above the 5′-face of the modified guanine [49]. Interestingly, nitrogen half-mustard-N7G adducts formed by the acridine half-mustard mutagen ICR-191 strongly induce +1 and +G frame-shift mutations as well as G to A transitions [54, 60], indicating that the N7-alkylG adducts of AFB1 and ICR-191 have different mutagenesis mechanisms. Nevertheless, the observed potent mutagenicity of some bulky N7-alkylG lesions (e.g. ICR-191-N7G) in vivo suggests that the presence of bulky N7-alkylG in DNA may have negative effects on genome integrity. For example, guanine N7-alkylation has been shown to increase the acidity of N1-H of guanine [61, 62], which in turn could destabilize N7-alkylG:C base pairing and promote N7-alkylG:T and N7-alkylG:A mispairs during replication. Bulky N7-alkylG lesions such as AFB1-N7G can be removed by base and/or nucleotide excision DNA repair pathways [17, 63]. If the N7-alkylG lesions are not detected prior to replication, TLS polymerases such as polη could involve in bypassing the lesions. Our studies show the bulky NHMG at the templating position slows catalysis by TLS polymerases, but it increases the misincorporation frequency of dTTP and dATP opposite the lesion, which could contribute to G to A transitions and G to T transversions, respectively.
CONCLUSIONS
The first kinetic and structural studies of NHMG provide insights into how TLS polymerases bypass aromatic NM-induced DNA lesions. We incorporated a 2´-F-mediated transition destabilization strategy to enable a site-specific incorporation of NHMG in DNA for further study. The presence of NHMG at the templating position greatly reduces the dCTP insertion efficiency of polη, suggesting TLS DNA polymerases would slowly catalyze across bulky N7-alkylG mono-adducts formed by aromatic nitrogen mustards (e.g., melphalan, chlorambucil). The templating NHMG increases the misincorporation frequency of dATP ~30-fold, implicating NHMG may promote G to T transversions. A crystal structure of polη-NHMG:dCTP* ternary complex shows a Watson-Crick NHMG:dCTP* base pair with a high degree of propeller twist, which appears to be caused by the insertion of the nitrogen half-mustard moiety of NHMG into an open cleft created by the Arg61-Trp64 loop. This underscores a possible role of the Arg61-Trp64 loop in accommodating bulky major groove N7-alkylG lesions including those produced by aromatic nitrogen mustards. Overall, the results reported here further our understanding of the miscoding potential of the major NM-induced DNA lesions as well as the impact of bulky N7-alkylG on the catalysis by TLS polymerases.
ACKNOWLEDGEMENTS
We are grateful to Dr. Arthur Monzingo for technical assistance. Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at the University of Texas at Austin. Portions of this research were conducted at the Advanced Photon Source with the support of GM/CA. GM/CA@APS has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The Elger 16M detector was funded by an NIH-Office of Research Infrastructure Programs, High-End Instrumentation Grant (1S10OD012289-01A1). The research was supported by the grant from the National Institutes of Health (ES-26676).
Footnotes
Conflict of interest statement. None declared.
ACCESSION NUMBERS
The atomic coordinates of Polη-DNA complexes have been deposited in the Protein Data Bank with the following accession code. PDB ID: 6V5K
REFERENCES
- 1.Singh RK, Kumar S, Prasad DN and Bhardwaj TR (2018) Therapeutic journery of nitrogen mustard as alkylating anticancer agents: Historic to future perspectives. Eur J Med Chem. 151, 401–433 [DOI] [PubMed] [Google Scholar]
- 2.Hurley LH (2002) DNA and its associated processes as targets for cancer therapy. Nat Rev Cancer. 2, 188–200 [DOI] [PubMed] [Google Scholar]
- 3.Hansson J, Lewensohn R, Ringborg U and Nilsson B (1987) Formation and removal of DNA cross-links induced by melphalan and nitrogen mustard in relation to drug-induced cytotoxicity in human melanoma cells. Cancer Res. 47, 2631–2637 [PubMed] [Google Scholar]
- 4.Chen W, Han Y and Peng X (2014) Aromatic nitrogen mustard-based prodrugs: activity, selectivity, and the mechanism of DNA cross-linking. Chemistry. 20, 7410–7418 [DOI] [PubMed] [Google Scholar]
- 5.Deans AJ and West SC (2011) DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Povirk LF and Shuker DE (1994) DNA damage and mutagenesis induced by nitrogen mustards. Mutat Res. 318, 205–226 [DOI] [PubMed] [Google Scholar]
- 7.Mohamed D, Mowaka S, Thomale J and Linscheid MW (2009) Chlorambucil-adducts in DNA analyzed at the oligonucleotide level using HPLC-ESI MS. Chem Res Toxicol. 22, 1435–1446 [DOI] [PubMed] [Google Scholar]
- 8.Bank BB (1992) Studies of chlorambucil-DNA adducts. Biochem Pharmacol. 44, 571–575 [DOI] [PubMed] [Google Scholar]
- 9.Hosing C, Munsell M, Yazji S, Andersson B, Couriel D, de Lima M, Donato M, Gajewski J, Giralt S, Korbling M, Martin T, Ueno NT, Champlin RE and Khouri IF (2002) Risk of therapy-related myelodysplastic syndrome/acute leukemia following high-dose therapy and autologous bone marrow transplantation for non-Hodgkin’s lymphoma. Ann Oncol. 13, 450–459 [DOI] [PubMed] [Google Scholar]
- 10.Sanderson BJ and Shield AJ (1996) Mutagenic damage to mammalian cells by therapeutic alkylating agents. Mutat Res. 355, 41–57 [DOI] [PubMed] [Google Scholar]
- 11.Castano A, Roy U and Scharer OD (2017) Preparation of Stable Nitrogen Mustard DNA Interstrand Cross-Link Analogs for Biochemical and Cell Biological Studies. Methods Enzymol. 591, 415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enoiu M, Jiricny J and Scharer OD (2012) Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 40, 8953–8964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semlow DR, Zhang J, Budzowska M, Drohat AC and Walter JC (2016) Replication-Dependent Unhooking of DNA Interstrand Cross-Links by the NEIL3 Glycosylase. Cell. 167, 498–511 e414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ and Walter JC (2009) The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 326, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knipscheer P, Raschle M, Scharer OD and Walter JC (2012) Replication-coupled DNA interstrand cross-link repair in Xenopus egg extracts. Methods Mol Biol. 920, 221–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo N, Takahashi A, Ono K and Ohnishi T (2010) DNA damage induced by alkylating agents and repair pathways. J Nucleic Acids. 2010, 543531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bedard LL and Massey TE (2006) Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 241, 174–183 [DOI] [PubMed] [Google Scholar]
- 18.Lin YC, Li L, Makarova AV, Burgers PM, Stone MP and Lloyd RS (2014) Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua. J Biol Chem. 289, 18497–18506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koag MC, Kou Y, Ouzon-Shubeita H and Lee S (2014) Transition-state destabilization reveals how human DNA polymerase beta proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 42, 8755–8766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kou Y, Koag MC and Lee S (2015) N7 methylation alters hydrogen-bonding patterns of guanine in duplex DNA. J Am Chem Soc. 137, 14067–14070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, Bowman BR, Ueno Y, Wang S and Verdine GL (2008) Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine. J Am Chem Soc. 130, 11570–11571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kou Y, Koag MC and Lee S (2018) Structural and Kinetic Studies of the Effect of Guanine N7 Alkylation and Metal Cofactors on DNA Replication. Biochemistry. 57, 5105–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Njuma OJ, Su Y and Guengerich FP (2019) The abundant DNA adduct N (7)-methyl deoxyguanosine contributes to miscoding during replication by human DNA polymerase eta. J Biol Chem. 294, 10253–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman MF and Woodgate R (2013) Translesion DNA polymerases. Cold Spring Harb Perspect Biol. 5, a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling H, Boudsocq F, Woodgate R and Yang W (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 107, 91–102 [DOI] [PubMed] [Google Scholar]
- 26.Yoon JH, Prakash L and Prakash S (2009) Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci U S A. 106, 18219–18224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling H, Boudsocq F, Plosky BS, Woodgate R and Yang W (2003) Replication of a cis-syn thymine dimer at atomic resolution. Nature. 424, 1083–1087 [DOI] [PubMed] [Google Scholar]
- 28.Washington MT, Johnson RE, Prakash S and Prakash L (2000) Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase eta. Proc Natl Acad Sci U S A. 97, 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F and Yang W (2010) Structure and mechanism of human DNA polymerase eta. Nature. 465, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K and Hanaoka F (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 31.Stern HR, Sefcikova J, Chaparro VE and Beuning PJ (2019) Mammalian DNA Polymerase Kappa Activity and Specificity. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patra A, Zhang Q, Guengerich FP and Egli M (2016) Mechanisms of Insertion of dCTP and dTTP Opposite the DNA Lesion O6-Methyl-2’-deoxyguanosine by Human DNA Polymerase eta. J Biol Chem. 291, 24304–24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP and Carell T (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science. 318, 967–970 [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Biertumpfel C, Gregory MT, Hua YJ, Hanaoka F and Yang W (2012) Structural basis of human DNA polymerase eta-mediated chemoresistance to cisplatin. Proc Natl Acad Sci U S A. 109, 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho TV and Scharer OD (2010) Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 51, 552–566 [DOI] [PubMed] [Google Scholar]
- 36.Ho TV, Guainazzi A, Derkunt SB, Enoiu M and Scharer OD (2011) Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 39, 7455–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koag MC, Jung H and Lee S (2019) Mutagenic Replication of the Major Oxidative Adenine Lesion 7,8-Dihydro-8-oxoadenine by Human DNA Polymerases. J Am Chem Soc. 141, 4584–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koag MC, Jung H and Lee S (2020) Mutagenesis mechanism of the major oxidative adenine lesion 7,8-dihydro-8-oxoadenine. Nucleic Acids Res. 48, 5119–5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koag MC, Jung H, Kou Y and Lee S (2019) Bypass of the Major Alkylative DNA Lesion by Human DNA Polymerase eta. Molecules 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, N. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 41.Emsley P and Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK and Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 43.Davis IW, Murray LW, Richardson JS and Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC and Wilson SH (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vaisman A, Frank EG, Iwai S, Ohashi E, Ohmori H, Hanaoka F and Woodgate R (2003) Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase iota. DNA Repair (Amst). 2, 991–1006 [DOI] [PubMed] [Google Scholar]
- 46.Patra A, Nagy LD, Zhang Q, Su Y, Muller L, Guengerich FP and Egli M (2014) Kinetics, structure, and mechanism of 8-Oxo-7,8-dihydro-2’-deoxyguanosine bypass by human DNA polymerase eta. J Biol Chem. 289, 16867–16882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma S, Helchowski CM and Canman CE (2013) The roles of DNA polymerase ζ and the Y family DNA polymerases in promoting or preventing genome instability. Mutat Res. 743–744, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregory MT, Park GY, Johnstone TC, Lee YS, Yang W and Lippard SJ (2014) Structural and mechanistic studies of polymerase eta bypass of phenanthriplatin DNA damage. Proc Natl Acad Sci U S A. 111, 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banerjee S, Brown KL, Egli M and Stone MP (2011) Bypass of aflatoxin B1 adducts by the Sulfolobus solfataricus DNA polymerase IV. J Am Chem Soc. 133, 12556–12568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Junop MS and Haniford DB (1996) Multiple roles for divalent metal ions in DNA transposition: distinct stages of Tn10 transposition have different Mg2+ requirements. EMBO J. 15, 2547–2555 [PMC free article] [PubMed] [Google Scholar]
- 51.Villani G, Tanguy Le Gac N, Wasungu L, Burnouf D, Fuchs RP and Boehmer PE (2002) Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucleic Acids Res. 30, 3323–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaisman A, Ling H, Woodgate R and Yang W (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank EG and Woodgate R (2007) Increased catalytic activity and altered fidelity of human DNA polymerase iota in the presence of manganese. J Biol Chem. 282, 24689–24696 [DOI] [PubMed] [Google Scholar]
- 54.Sahasrabudhe SR, Luo X and Humayun MZ (1990) Induction of G.C to A.T transitions by the acridine half-mustard ICR-191 supports a mispairing mechanism for mutagenesis by some bulky mutagens. Biochemistry. 29, 10899–10905 [DOI] [PubMed] [Google Scholar]
- 55.Gates KS, Nooner T and Dutta S (2004) Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 17, 839–856 [DOI] [PubMed] [Google Scholar]
- 56.Boiteux S and Guillet M (2004) Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst). 3, 1–12 [DOI] [PubMed] [Google Scholar]
- 57.Boysen G, Pachkowski BF, Nakamura J and Swenberg JA (2009) The formation and biological significance of N7-guanine adducts. Mutat Res. 678, 76–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Philippin G, Cadet J, Gasparutto D, Mazon G and Fuchs RP (2014) Ethylene oxide and propylene oxide derived N7-alkylguanine adducts are bypassed accurately in vivo. DNA Repair (Amst). 22, 133–136 [DOI] [PubMed] [Google Scholar]
- 59.Bailey EA, Iyer RS, Stone MP, Harris TM and Essigmann JM (1996) Mutational properties of the primary aflatoxin B1-DNA adduct. Proc Natl Acad Sci U S A. 93, 1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sahasrabudhe SR, Luo X and Humayun MZ (1991) Specificity of base substitutions induced by the acridine mutagen ICR-191: mispairing by guanine N7 adducts as a mutagenic mechanism. Genetics. 129, 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lawley PD and Brookes P (1961) Acidic dissociation of 7:9-dialkylguanines and its possible relation to mutagenic properties of alkylating agents. Nature. 192, 1081–1082 [DOI] [PubMed] [Google Scholar]
- 62.Sowers LC, Shaw BR, Veigl ML and Sedwick WD (1987) DNA base modification: ionized base pairs and mutagenesis. Mutat Res. 177, 201–218 [DOI] [PubMed] [Google Scholar]
- 63.De Alencar TA, Leitao AC and Lage C (2005) Nitrogen mustard- and half-mustard-induced damage in Escherichia coli requires different DNA repair pathways. Mutat Res. 582, 105–115 [DOI] [PubMed] [Google Scholar]