Supplemental Digital Content is available in the text.
Keywords: coronavirus disease 2019, hospitalization, intensive care, outcome prediction, pandemic, severe acute respiratory syndrome coronavirus 2
Abstract
Objectives:
To develop an algorithm that predicts an individualized risk of severe coronavirus disease 2019 illness (i.e., ICU admission or death) upon testing positive for coronavirus disease 2019.
Design:
A retrospective cohort study.
Setting:
Cleveland Clinic Health System.
Patients:
Those hospitalized with coronavirus disease 2019 between March 8, 2020, and July 13, 2020.
Interventions:
A temporal coronavirus disease 2019 test positive cut point of June 1 was used to separate the development from validation cohorts. Fine and Gray competing risk regression modeling was performed.
Measurements and Main Results:
The development set contained 4,520 patients who tested positive for coronavirus disease 2019 between March 8, 2020, and May 31, 2020. The validation set contained 3,150 patients who tested positive between June 1 and July 13. Approximately 9% of patients were admitted to the ICU or died of coronavirus disease 2019 within 2 weeks of testing positive. A prediction cut point of 15% was proposed. Those who exceed the cutoff have a 21% chance of future severe coronavirus disease 2019, whereas those who do not have a 96% chance of avoiding the severe coronavirus disease 2019. In addition, application of this decision rule identifies 89% of the population at the very low risk of severe coronavirus disease 2019 (< 4%).
Conclusions:
We have developed and internally validated an algorithm to assess whether someone is at high risk of admission to the ICU or dying from coronavirus disease 2019, should he or she test positive for coronavirus disease 2019. This risk should be a factor in determining resource allocation, protection from less safe working conditions, and prioritization for vaccination.
Most patients diagnosed with coronavirus disease 2019 (COVID-19) do not progress to severe illness, such as the need for ICU admission or death from COVID-19. Proper future resource allocation is essential to preventing undue strain on a healthcare system. Vaccines for COVID-19 are coming, but even when approved, they will not be in sufficient supply to vaccinate everyone on day 1. When deciding whom to vaccinate first, it makes sense to consider, in part, how dangerous COVID-19 would be for the individual should he or she become infected. Those at highest risk for future ICU admission or death might naturally be prioritized after proper consideration of the risk of infection and vaccine efficacy.
Older age (1, 2), smoking (3), diabetes, hypertension, cardiovascular disease, chronic kidney disease, chronic lung disease (4), and cancer (4, 5) have been associated with disease worsening in patients hospitalized with COVID-19. The problem is that these “risk factors” are not very specific, occur in various combinations, and have not been combined into a score applicable to the patient who recently tests positive for COVID-19. Such a scoring system should be optimized for predictive accuracy and then made user-friendly in a way that preserves the accuracy. For example, continuous variables should be left continuous and allowed to have nonlinear effects (6).
We present here a statistical model that can assist with individualized prediction of future ICU admission or death from COVID-19 (i.e., severe COVID-19 illness) for a patient immediately diagnosed with COVID-19. The model was developed and internally validated using data collected from the Cleveland Clinic COVID-19 Registry, which was prospectively created to include all patients being tested for COVID-19 in our integrated healthcare system. The coefficients of the prediction model were expressed as points that can be summed to determine whether someone is at high risk for severe illness (7).
MATERIALS AND METHODS
Patient Selection
We included all patients with COVID-19, regardless of age, who tested positive in the Cleveland Clinic Health System (CCHS) within the United States between March 8, 2020, and July 13, 2020. The CCHS includes greater than 220 outpatient locations and 18 hospitals in Ohio and Florida.
Cleveland Clinic COVID-19 Registry
A Cleveland Clinic enterprisewide research COVID-19 registry was started with initiation of testing capabilities in our organization. Cleveland Clinic Institutional Review Board approval for this study was obtained (#20-283), and a requirement for written Informed Consent was waived. Demographics, comorbidities, travel and COVID-19 exposure history, medications, presenting symptoms, socioeconomic measures, treatment, disease progression, and outcomes were collected. Registry variables were chosen to reflect available literature on COVID-19 disease characterization, progression, and proposed treatments, including medications thought to have benefits through drug-repurposing studies (8). Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was confirmed by laboratory testing using the Centers for Disease Control and Prevention reverse transcription polymerase chain reaction SARS-CoV-2 assay on nasopharyngeal and oropharyngeal swab specimens. Data were extracted via previously validated automated feeds (9) from our electronic health record (Epic, Epic Systems Corporation, Madison, WI) and manually by a study team trained on uniform sources for the study variables. Study data were collected and managed using Research Electronic Data Capture (REDCap) electronic data capture tools hosted at Cleveland Clinic (10, 11). REDCap is a secure, web-based software platform designed to support data capture for research studies, providing: 1) an intuitive interface for validated data capture, 2) audit trails for tracking data manipulation and export procedures, 3) automated export procedures for seamless data downloads to common statistical packages, and 4) procedures for data integration and interoperability with external sources.
Statistical Analysis
The baseline was considered the day the patient was confirmed to be infected with COVID-19. All predictor variable values were recorded as of the baseline day. Baseline data are presented as median (interquartile range [IQR] and number [%]). Continuous variables were compared using the Mann-Whitney U test, and categorical variables were compared using the Chi-square test.
The primary study end point was admission to the ICU or death from COVID-19, whichever occurred first, whereas other-cause death without admission was considered a competing risk (since ICU admission or death from COVID-19 cannot occur afterward). A full multivariable Fine and Gray competing risks regression model was initially constructed based on demographic variables, comorbidities, immunization history, and prescription medications at the time of testing positive. Missing values were imputed using two approaches: 1) using the medians of variables and 2) using multiple imputation by chained equations. The performances of models using each imputation approach were compared. Restricted cubic splines with three knots were applied to continuous variables to relax the linearity assumption. A least absolute shrinkage and selection operator (LASSO) was performed to retain the most predictive features. A 10-fold cross validation method was applied to find the regularization parameter lambda that gave the minimum mean cross-validated concordance index. Predictors with nonzero coefficients following the LASSO procedure were chosen for calculating predicted risk. The final model was internally validated by assessing the discrimination and calibration with 1,000 bootstrap resamples. Discrimination was measured with the time-dependent area under the receiver operating characteristic (ROC) curve (12). Calibration was assessed visually by plotting the statistical model predicted probabilities against the observed event proportions over a series of equally spaced values within the range of the predicted probabilities. The closer the calibration curve lies along the 45° line, the better the calibration. The statistical prediction model was expressed as a nomogram, which allowed points for each variable to be determined from rescaled model coefficients. The performance of risk cutoffs was measured by sensitivity, specificity, negative predictive value, and positive predictive value. Following the internal validation step, the validation dataset was used. We adhered to the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis checklist for reporting the prediction model (13).
RESULTS
Between March 8, 2020, and July 13, 2020, COVID-19 was diagnosed in 7,670 patients. Within about 2 weeks of diagnosis, 619 developed severe COVID-19 (Fig. 1). The long-term cumulative incidence of severe COVID-19 for all who tested positive was about 9%. The risk of severe COVID-19 from 2 weeks after diagnosis until the end of our follow-up increased from only 7.9% to 8.7%, suggesting 2 weeks to be a good end point for the prediction model.
Figure 1.
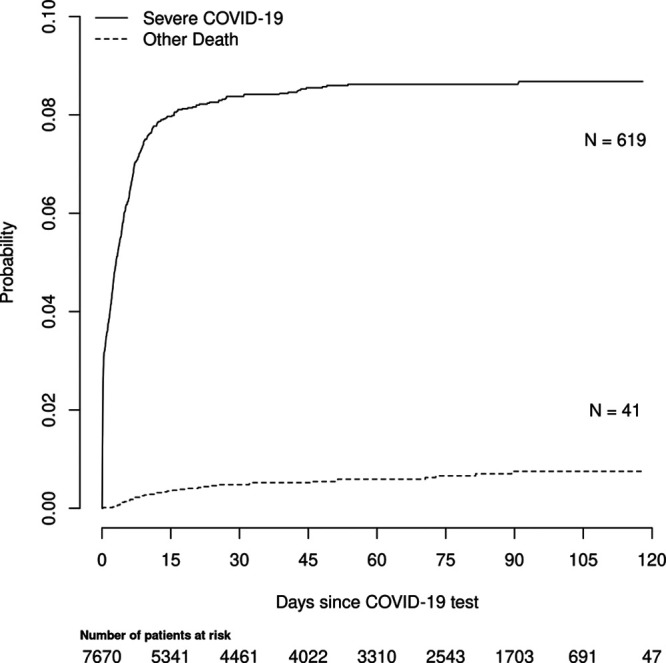
Cumulative incidence of severe coronavirus disease 2019 (COVID-19) for development and test cohorts combined. x-axis is the days from testing positive for COVID-19. Numbers below x-axis legend indicate the number of patients at risk for severe COVID-19 over time. Of the 619 patients who experienced severe illness, 588 were admitted to the ICU, whereas 31 died of COVID-19 without a prior ICU admission.
The demographics of the development and test datasets are presented in Table 1. The statistical prediction model might not be applicable in settings that are grossly different from what is presented in this table. It is noteworthy that several distributions shifted between the time periods, probably reflecting both a change in awareness of the illness and increased capacity for testing.
Not all the variables in the registry were considered for the statistical prediction model. We omitted predictors that could be manipulated, for example, over the counter supplementation and self-reported symptoms. This left the candidate predictors consisting of age, race, body mass index (BMI), household income, gender, documented medical history (e.g., diabetes), immunization history, and prescription medications. The intent was to restrict to those variables that could be verified and not potentially used to “game the system” for access to a vaccine or other resources. We first compared competing risk regression models using the two forms of imputation (medians vs chained equations) for the variables with missing values (Table 1). We found that the bootstrap-corrected, time-dependent areas under the ROC curve were the same (0.781), so we chose the model using medians for practical considerations. A competing risk regression model was constructed, and LASSO was used to simplify the model. This prediction model appears graphically in Figure 2. The following variables were candidate predictors but removed during the LASSO step due to lack of improvement in the performance of the model: history of asthma, cancer, connective tissue disease, inflammatory bowel disease, angiotensin receptor blocker prescription, population per square mile, and population per housing unit. During internal validation, the model performed well at predicting 14-day risk of severe COVID-19 illness, with a time-dependent area under the ROC curve of 0.781 (95% CI, 0.749–0.813). However, the calibration at higher predicted risk levels (above 20%) departs from ideal (see Supplemental Fig. 1, http://links.lww.com/CCX/A454). This suggests that the model may be more useful for discriminating those who will and will not develop severe COVID-19 illness than for providing an absolute level of risk. Therefore, we chose to dichotomize the predicted risk levels of 20% and lower evaluate the prediction model when used as a binary decision rule (i.e., above and below 10%, 15%, and 20% risks). Sensitivity, specificity, positive predictive value, and negative predictive value were calculated, using the test set, at each cutoff. The performance values appear in Table 2. Finally, the points from the nomogram were extracted and summarized in Table 3 so that the model can be easily applied to individual patients. The points for each variable are identified and summed, and then, the final sum is compared with the cutoffs in Table 2 to determine high versus low risk. For example, when using the proposed cutoff of 15%, those who meet the cutoff have a 21% chance of severe illness, while those that do not meet the cutoff have a 96% chance of avoiding severe illness. In addition, application of this decision rule quickly identifies 89% of the population at the very low risk of severe illness (< 4%). Depending on resource availability, a lower risk cutoff could be chosen to define a group with an even lower severe illness. Our tool is freely available online at https://riskcalc.org/SevereCOVID19.
TABLE 1.
Descriptive Statistics for the Development and Test Cohorts
| Variable | Development, March 8 to May 31 | Test, June 1 to July 13 | p |
|---|---|---|---|
| N | 4,520 | 3,150 | |
| Discharged in recent 14 d (%) | 37 (0.8) | 4 (0.6) | 0.001 |
| Demographics | |||
| Race (%) | < 0.001 | ||
| Asian | 47 (1.0) | 28 (0.9) | |
| Black | 1,295 (28.7) | 1,173 (37.2) | |
| Other | 656 (14.5) | 551 (17.5) | |
| White | 2,522 (55.8) | 1,398 (44.4) | |
| Male (%) | 2,082 (46.1) | 1,478 (46.9) | 0.472 |
| Ethnicity (%) | 0.014 | ||
| Hispanic | 470 (10.4) | 384 (12.2) | |
| Non-Hispanic | 3,484 (77.1) | 2,341 (74.3) | |
| Unknown | 566 (12.5) | 425 (13.5) | |
| Smoking (%) | < 0.001 | ||
| Current Smoker | 339 (7.5) | 219 (7.0) | |
| Former Smoker | 1,305 (28.9) | 585 (18.6) | |
| No | 2,273 (50.3) | 1,656 (52.6) | |
| Unknown | 603 (13.3) | 690 (21.9) | |
| Age (median [IQR]) | |||
| Missing: 0.3% | 53.81 (36.22–68.49) | 46.43 (29.66–58.67) | < 0.001 |
| Exposure history | |||
| Exposed to COVID-19? Yes (%) | 3,161 (69.9) | 369 (11.7) | < 0.001 |
| Family member with COVID-19? Yes (%) | 2,781 (61.5) | 328 (10.4) | < 0.001 |
| Presenting symptoms | |||
| Cough? Yes (%) | 3,329 (73.7) | 354 (11.2) | < 0.001 |
| Fever? Yes (%) | 2,673 (59.1) | 258 (8.2) | < 0.001 |
| Fatigue? Yes (%) | 2,723 (60.2) | 278 (8.8) | < 0.001 |
| Sputum production? Yes (%) | 2,006 (44.4) | 219 (7.0) | < 0.001 |
| Flu-like symptoms? Yes (%) | 3,068 (67.9) | 348 (11.0) | < 0.001 |
| Shortness of breath? Yes (%) | 2,149 (47.5) | 230 (7.3) | < 0.001 |
| Diarrhea? Yes (%) | 1,815 (40.2) | 172 (5.5) | < 0.001 |
| Loss of appetite? Yes (%) | 2,289 (50.6) | 223 (7.1) | < 0.001 |
| Vomiting? Yes (%) | 1,289 (28.5) | 151 (4.8) | < 0.001 |
| Comorbidities | |||
| BMI (median [IQR]) | |||
| Missing: 14.5% | 28.69 (28.69–29.17) | 28.69 (28.69–29.99) | 0.576 |
| Chronic obstructive pulmonary disease/emphysema? Yes (%) | 295 (6.5) | 142 (4.5) | < 0.001 |
| Asthma? Yes (%) | 654 (14.5) | 474 (15.0) | 0.502 |
| Diabetes? Yes (%) | 937 (20.7) | 435 (13.8) | < 0.001 |
| Hypertension? Yes (%) | 1,953 (43.2) | 1,008 (32.0) | < 0.001 |
| Coronary artery disease? Yes (%) | 505 (11.2) | 209 (6.6) | < 0.001 |
| Heart failure? Yes (%) | 398 (8.8) | 177 (5.6) | < 0.001 |
| Cancer? Yes (%) | 517 (11.4) | 236 (7.5) | < 0.001 |
| Transplant history? Yes (%) | 29 (0.6) | 19 (0.6) | 0.95 |
| Multiple sclerosis? Yes (%) | 45 (1.0) | 14 (0.4) | 0.01 |
| Connective tissue disease? Yes (%) | 304 (6.7) | 81 (2.6) | < 0.001 |
| Inflammatory bowel disease? Yes (%) | 152 (3.4) | 37 (1.2) | < 0.001 |
| Immunosuppressive disease? Yes (%) | 498 (11.0) | 262 (8.3) | < 0.001 |
| Vaccination history | |||
| Flu shot? Yes (%) | 1,761 (39.0) | 1,237 (39.3) | 0.803 |
| Pneumovax shot? Yes (%) | 764 (16.9) | 473 (15.0) | 0.029 |
| Laboratory findings upon presentation | |||
| Pretesting platelets (median [IQR]) | |||
| Missing: 68.7% | 230.00 (230.00–230.00) | 230.00 (230.00–230.00) | 0.674 |
| Pretesting aspartate amino transferase (median [IQR]) | |||
| Missing: 73.3% | 25.00 (25.00–25.00) | 25.00 (25.00–25.00) | 0.056 |
| Pretesting blood urea nitrogen (median [IQR]) | |||
| Missing: =69.0% | 15.00 (15.00–15.00) | 15.00 (15.00–15.00) | < 0.001 |
| Pretesting chloride (median [IQR]) | |||
| Missing: 69.0% | 100.00 (100.00–100.00) | 100.00 (100.00–100.00) | 0.467 |
| Pretesting creatinine (median [IQR]) | |||
| Missing: 69.0% | 0.93 (0.93–0.93) | 0.93 (0.93–0.93) | 0.653 |
| Pretesting alanine amino transferasea (median [IQR]) | |||
| Missing: 72.9% | 1.32 (1.32–1.32) | 1.32 (1.32–1.32) | 0.02 |
| Pretesting C-reactive protein (median [IQR]) | |||
| Missing: 83.5% | 4.50 (4.50–4.50) | 4.50 (4.50–4.50) | < 0.001 |
| Pretesting hematocrit (median [IQR]) | |||
| Missing: 68.7% | 39.40 (39.40–39.40) | 39.40 (39.40–39.40) | 0.084 |
| Pretesting potassium (median [IQR]) | |||
| Missing: 69.1% | 4.00 (4.00–4.00) | 4.00 (4.00–4.00) | 0.413 |
| Home medications | |||
| Immunosuppressive treatment? Yes (%) | 348 (7.7) | 39 (1.2) | < 0.001 |
| Nonsteroidal anti-inflammatory drugs? Yes (%) | 911 (20.2) | 571 (18.1) | 0.029 |
| Steroids? Yes (%) | 415 (9.2) | 216 (6.9) | < 0.001 |
| Carvedilol? Yes (%) | 115 (2.5) | 52 (1.7) | 0.011 |
| Angiotensin-converting enzyme inhibitor? Yes (%) | 406 (9.0) | 151 (4.8) | < 0.001 |
| Angiotensin receptor blocker? Yes (%) | 279 (6.2) | 131 (4.2) | < 0.001 |
| Melatonin? Yes (%) | 134 (3.0) | 41 (1.3) | < 0.001 |
| Social influencers of health | |||
| Population per km2a (median [IQR]) | |||
| Missing: 0.2% | 3.10 (2.72–3.34) | 3.13 (2.72–3.38) | 0.06 |
| Median income ($1,000, median [IQR]) | |||
| Missing: 0.2% | 54.91 (37.82–73.65) | 48.92 (36.80–71.80) | < 0.001 |
| Population per housing unit (median [IQR]) | |||
| Missing: 0.2% | 2.22 (1.94–2.50) | 2.22 (1.89–2.61) | 0.064 |
BMI = body mass index, COVID-19 = coronavirus disease 2019, IQR = interquartile range.
Figure 2.
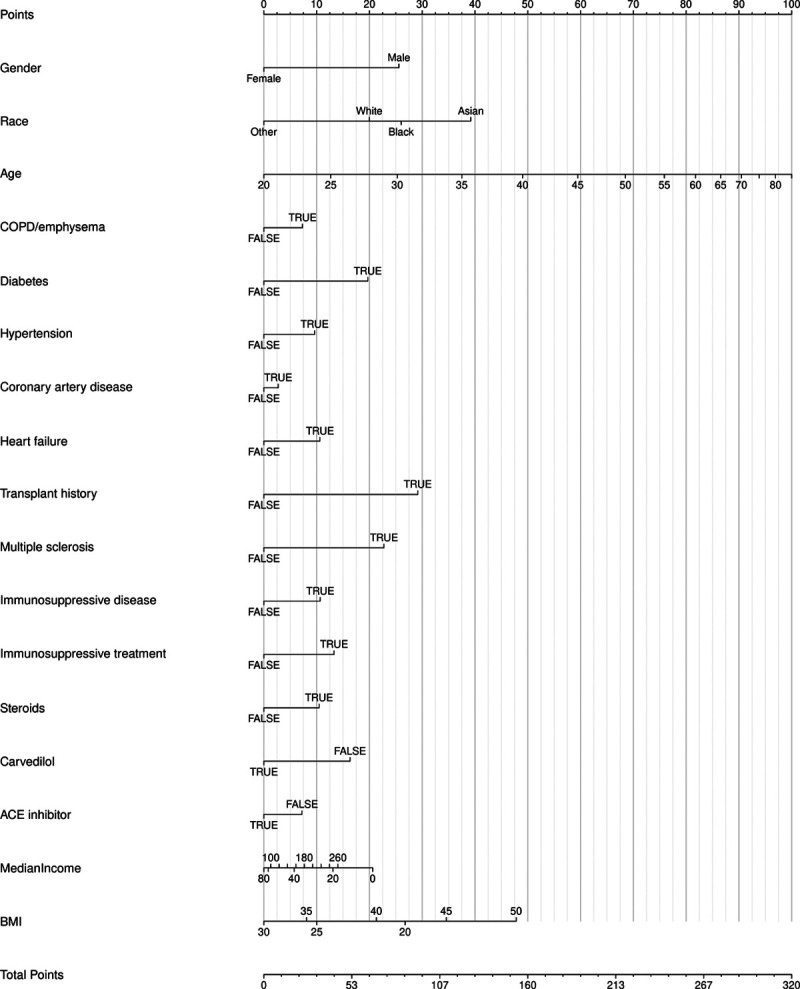
Nomogram for the model predicting severe coronavirus disease 2019. ACE = angiotensin-converting enzyme, BMI = body mass index, COPD = chronic obstructive pulmonary disease.
TABLE 2.
Performance Characteristics of Various Cutoff When Applied to the Test Dataset
| Prediction Model Cutoff Probability (Points) | ||||||
|---|---|---|---|---|---|---|
| Characteristic | 0.05 (140 Points) | 0.1 (169 Points) | 0.15 (186 Points) | 0.2 (199 Points) | Age > 65 | Diabetes |
| Sensitivity | 0.85 | 0.59 | 0.39 | 0.25 | 0.51 | 0.42 |
| Specificity | 0.64 | 0.84 | 0.91 | 0.95 | 0.77 | 0.84 |
| Positive predictive value | 0.13 | 0.18 | 0.21 | 0.23 | 0.16 | 0.18 |
| NPV | 0.99 | 0.97 | 0.96 | 0.95 | 0.95 | 0.95 |
| Proportion declared negative | 0.61 | 0.81 | 0.89 | 0.94 | 0.83 | 0.86 |
| Proportion declared positive | 0.39 | 0.19 | 0.11 | 0.06 | 0.17 | 0.14 |
| Proportion expected for ICU admission | 0.006 | 0.024 | 0.036 | 0.047 | 0.042 | 0.043 |
NPV = negative predictive value.
The choice of cutoff partly depends on the resources available (i.e., what proportion needs to be declared negative). The proportion expected to experience severe illness is estimated as the proportion declared negative (e.g., not vaccinated) times the proportion expected to experience severe illness given declared negative (1-NPV). Making that choice would then dictate the points cutoff to be used for deciding whether a patient meets the cutoff. After the cutoff is chosen, Table 3 may be used for each patient.
TABLE 3.
Points for Each Variable in the Statistical Prediction Model
| Variable | Points |
|---|---|
| Gender = male | 26 |
| Race | |
| Asian | 39 |
| White | 20 |
| Black | 26 |
| Other | 0 |
| Age (yr) | |
| 20 | 0 |
| 30 | 25 |
| 40 | 49 |
| 50 | 68 |
| 60 | 82 |
| 70 | 90 |
| 80 | 97 |
| Chronic obstructive pulmonary disease/emphysema | 7 |
| Diabetes | 20 |
| Hypertension | 10 |
| Coronary artery disease | 3 |
| Heart failure | 11 |
| Transplant history | 29 |
| Multiple sclerosis | 23 |
| Immunosuppressive disease | 11 |
| Immunosuppressive treatment | 13 |
| Steroids | 10 |
| Carvedilol = FALSE | 16 |
| Angiotensin-converting enzyme inhibitor = FALSE | 7 |
| Household income (thousands of dollars) | |
| 0 | 21 |
| 20 | 13 |
| 40 | 6 |
| 60 | 1 |
| 80 | 0 |
| 100 | 1 |
| 120 | 3 |
| 140 | 4 |
| 160 | 6 |
| 180 | 8 |
| 200 | 9 |
| 220 | 11 |
| 240 | 12 |
| 260 | 14 |
| Body mass index | |
| 20 | 27 |
| 25 | 10 |
| 30 | 0 |
| 35 | 8 |
| 40 | 21 |
| 45 | 35 |
| 50 | 48 |
DISCUSSION
Our study provides a simplified method to determine objectively if someone is at high risk for severe COVID-19 illness within 2 weeks of testing positive for COVID-19. The selection of the 2-week prediction horizon follows from Figure 1. It would appear an earlier horizon would not be very definitive, as the risk of severe illness does not clearly taper until approximately day 14. A later time horizon did not seem necessary, since the risk appears to plateau around day 14, and a longer horizon would have fewer patients in the risk set. With the goal to prevent severe COVID-19 related illness requiring ICU admission or death, this tool may be useful for helping to determine who to manage aggressively or vaccinate first and who to protect from settings where social distancing is more challenging (e.g., classrooms). With that use case in mind, we chose not to make the prediction later in the time course (e.g., calculate the prediction at time of hospitalization) when severity of illness predictors might be available (e.g., Acute Physiology and Chronic Health Evaluation scores). One could also argue for an event more severe than ICU admission to be predicted (e.g., mechanical ventilation), but fewer of those events would be available for modeling, and ICU admission regardless of mechanical ventilation support is a serious event.
Perusal of the point allocation from the tool yields some insight. Predictions from the tool are driven heavily by age, race, BMI, and gender, followed by history of diabetes, transplant, and immunosuppressive treatment. Most of the predictors have intuitive effects. Older age (1, 2), underlying diabetes, and chronic obstructive pulmonary disease/emphysema (4) have been previously identified in the literature as predictors of clinical worsening. Much of the previous information about COVID-19 in transplant patients is limited to case reports and anecdotal evidence. There have been documented cases of COVID-19 in patients with a history of kidney (14, 15), liver (16), heart (17), and bone-marrow transplant (18), but with varying outcomes. The clinical course of COVID-19 in transplant patients should be further investigated as the sample size increases. Patients who took carvedilol had a decreased risk of severe disease progression. There have been no clinical studies evaluating the effect of carvedilol in COVID-19 patients; however, based on basic science data and artificial intelligence predictions, beta-adrenergic blockers may be a potential treatment target (19). Since angiotensin-converting enzyme 2 (ACE2) was first identified as the SARS-CoV-2 receptor, speculations abound as to whether the use of ACE inhibitors in patients with COVID-19 would be protective or harmful (20). Thus far, human studies have not shown an increase of ACE2 expression in patients taking ACE inhibitors (21). We recently found no correlation between the use of ACE inhibitors and testing positive for COVID-19 (22), supporting professional societies advocating for their ongoing use in patients already taking them given insufficient evidence to favor discontinuation. More research is needed to explore the potential protective effects of these medications.
Although viewing the nomogram is very useful for the interpretation of how a regression model works, it is important to keep in mind an important assumption that is being made. When examining the effect of a predictor (such as the length of the nomogram axis or the number of points assigned to different values), an assumption is made that all the other predictors are being held constant. This is a hypothetical and artificial situation, of course, since other predictors are likely to be correlated to some degree. For example, moving a medication (such as carvedilol) from off to on likely changes the age of this patient, and perhaps comorbidities. Thus, the addition or subtraction of points from one nomogram axis move may be more than offset in the other direction from other variables that move. This likely explains some counterintuitive findings that result from a regression model.
Our tool appears to be the first such tool for this purpose. It performs well but is not perfect. As with any statistical prediction model, perfect predictive accuracy is not possible. Our cutoff clearly identifies those at high versus low risk for severe illness, but high risk does not connote certain ICU admission or death, and low risk does not prevent ICU admission or death. However, use of this risk tool, as a part of the process, should be better than something very simplistic, such as vaccinate “all senior citizens” or “all those with diabetes.” To illustrate this point, we added the implications of those rules to Table 2. It is easy to see that both rules are dominated in an ROC curve sense by the risk cutoff of 0.1. In addition, those simple rules are dominated by several of the risk cutoffs with respect to positive predictive value (PPV) and negative predictive value (NPV). This analysis shows that risk stems from a constellation of factors, identified as risk factors in other studies, which each need to be considered simultaneously for proper risk determination. For example, using the age 65 cutoff rule would lead to vaccinating 17% of the population, whereas the 15% risk cutoff rule would lead to vaccinating only 11% of the population. In addition, the 15% cutoff rule would reduce the proportion of the population experiencing severe illness, from 4.2% to 3.6%. Granted, our calculations make strong assumptions about the population relative to our COVID-19 testing sample, but the concept is reasonable. In short, fewer resources (e.g., vaccines) are used more efficiently to prevent more ICU admissions or COVID-19 deaths. It should be noted that, although PPV and NPV are pertinent to decision-making policy, they are affected by prevalence, so adjustment is necessary for a setting with different prevalences than ours.
Our study needs to be put in the broader context. Our focus was on the end point of ICU admission or COVID-19 death, and future research should look more broadly. For example, as evidence for chronic complications following COVID-19 infection continues to emerge, analysis of quality-adjusted life years may become warranted. As a limited first step for the issue of vaccination, we assume that the probability that someone should be vaccinated would equal the probability he or she has not already had COVID-19 times the probability he or she will get it in the future times the probability of an ICU or COVID-19 death outcome if infected times the probability that the vaccine will be effective in that individual. Our study helps determine the severe COVID-19 probability and, as such, has numerous limitations. First, it is unclear whether the vaccine will be equally effective in all individuals. For example, it may be that the immune-suppressed or elderly might not benefit as much from a vaccine. However, use of our tool alone, for vaccine eligibility determination, would be assuming that it is equally effective in all people. Once personalized vaccine efficacy becomes available, recommendation tailoring would be feasible. Second, our tool predicts ICU admission or death for the person who now has tested positive. In that sense, it does not consider the potential that some people are more likely to get COVID-19 than others. The risk of getting COVID-19 in the first place should also potentially be considered. It is conceivable that certain subsets of essential workers, or those that live in dense settings (e.g., nursing homes), are more likely to get COVID-19. One way to factor in this risk would be to use a recent published COVID-19 risk calculator (23). At any rate, further refinement regarding the risk of infection would help tailor the allocation of resources. Third, this is not a multicenter study, but it includes all hospitals and outpatient facilities of the CCHS within the United States (> 220 outpatient locations and 18 hospitals in Ohio and Florida) and thus creates robust sampling of the COVID-19 population. It is unknown whether risk of severe COVID-19 illness, after adjustment for the factors in our prediction model, varies based on location. Furthermore, regarding location, while ICU bed availability in our data was not an issue, our findings may not extrapolate well to settings where ICU bed availability is constrained. Fourth, as mentioned above, our tool is not perfectly accurate. However, it provides a much better and far more equitable alternative than assuming everyone is at equal risk of ICU admission or death when compared with a simpler rule, such as, “those older than 65 are at increased risk of admission.”
CONCLUSIONS
We have developed and internally validated an algorithm to assess whether someone is at high risk of severe COVID-19 illness should he or she test positive for COVID-19. This risk should be a factor in determining prioritization for resources and protection from less safe working conditions. Further validation in an external setting is necessary.
Supplementary Material
Footnotes
Dr. Kattan contributed to the conception and design of the work; interpretation of data for the work; and revising the work critically for important intellectual content. Ms. Ji substantially contributed to the design of the work; data analysis; and revising it critically for important intellectual content. Mr. Milinovich, Dr. Adegboye, and Dr. Duggal substantially contributed to the design of the work; data acquisition; and revising it critically for important intellectual content. Drs. Dweik, Khouli, Gordon, and Young substantially contributed to the design of the work; data interpretation; and revising it critically for important intellectual content. Dr. Jehi substantially contributed to the conception and design of the work; interpretation of data for the work; drafting the work; and revising it critically for important intellectual content. All authors approved the final version.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccxjournal).
Supported, in part, by the Cleveland Clinic Lerner Research Institute and National Institutes of Health/National Center for Advancing Translational Sciences UL1TR002548.
The authors have disclosed that they do not have any potential conflicts of interest.
REFERENCES
- 1.Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China. J Infect. 2020; 80:e1–e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020; 395:1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu W, Tao ZW, Lei W, et al. Analysis of factors associated with disease outcomes in hospitalized patients with 2019 novel coronavirus disease. Chin Med J (Engl). 2020; 133:1032–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020; 323:1239–1242 [DOI] [PubMed] [Google Scholar]
- 5.Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: A nationwide analysis in China. Lancet Oncol. 2020; 21:335–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: Issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996; 15:361–387 [DOI] [PubMed] [Google Scholar]
- 7.Kattan MW. Nomograms. Introduction. Semin Urol Oncol. 2002; 20:79–81 [PubMed] [Google Scholar]
- 8.Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020; 6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milinovich A, Kattan MW. Extracting and utilizing electronic health data from epic for research. Ann Transl Med. 2018; 6:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009; 42:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung H, Chiang CT. Estimation methods for time-dependent AUC models with survival data. Canadian J Statistics. 2010; 38:8–26 [Google Scholar]
- 13.Collins GS, Reitsma JB, Altman DG, et al. ; TRIPOD Group. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD statement. The TRIPOD group. Circulation. 2015; 131:211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu L, Xu X, Ma K, et al. “Successful recovery of COVID-19 pneumonia in a renal transplant recipient with long-term immunosuppression.” Am J Transplant. 2020; 20:1859–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee D, Popoola J, Shah S, et al. “COVID-19 infection in kidney transplant recipients.” Kidney Int. 2020; 97:1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhoori S, Rossi RE, Citterio D, et al. “COVID-19 in long-term liver transplant patients: Preliminary experience from an Italian transplant centre in Lombardy.” Lancet Gastroenterol Hepatol. 2020; 5:532–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F, Jie C, Nianguo D. “First cases of COVID-19 in heart transplantation from China.” J Heart Lung Transplant. 2020; 39:496–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Lin H, Wu Y, et al. “COVID-19 in post-transplantation patients-report of two cases.” Am J Transplant. 2020; 20:1879–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heiser K, et al. “Identification of potential treatments for COVID-19 through artificial intelligence-enabled phenomic analysis of human cells infected with SARS-CoV-2.” bioRxiv. 2020 [Google Scholar]
- 20.Hoffmann M, Kleine-Weber H, Schroeder S, et al. “SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor.” Cell. 2020; 181:271–280.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sriram K, Insel PA. “Risks of ACE inhibitor and ARB usage in COVID-19: Evaluating the evidence.” Clinl Pharmacol Ther. 2020; 108:236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta N, Kalra A, Nowacki AS, et al. Association of use of angiotensin-converting enzyme inhibitors and angiotensin ii receptor blockers with testing positive for coronavirus disease 2019 (COVID-19). JAMA Cardiol. 2020; 5:1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jehi L, Ji X, Milinovich A, et al. Individualizing risk prediction for positive COVID-19 testing: Results from 11,672 patients. Chest. 2020; 158:1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


