SUMMARY
Ribosome movement is not always smooth and is rather often impeded. For ribosome pauses, fundamental issues remain to be addressed, including where ribosomes pause on mRNAs, what kind of RNA/amino acid sequence causes this pause, and the physiological significance of this attenuation of protein synthesis. Here, we survey the positions of ribosome collisions caused by ribosome pauses in humans and zebrafish using modified ribosome profiling. Collided ribosomes, i.e., disomes, emerge at various sites: Pro-Pro/Gly/Asp motifs; Arg-X-Lys motifs; stop codons; and 3’ untranslated regions. The electrostatic interaction between the charged nascent chain and the ribosome exit tunnel determines the eIF5A-mediated disome rescue at the Pro-Pro sites. In particular, XBP1u, a precursor of endoplasmic reticulum (ER)-stress-responsive transcription factor, shows striking queues of collided ribosomes and thus acts as a degradation substrate by ribosome-associated quality control. Our results provide insight into the causes and consequences of ribosome pause by dissecting collided ribosomes.
In Brief
Han et al. survey ribosome collision sites across the transcriptome by sequencing disome footprints in humans and zebrafish and find that inefficient contexts for elongation, termination, and recycling of ribosomes contributes to the collision of ribosomes. They also uncover XBP1u as an endogenous target of ribosome-associated quality control.
Graphical Abstract
INTRODUCTION
Translation, which serves as the interface between nucleic acids and amino acids in the central dogma of life, is regulated at multiple steps. Among these steps, translation elongation, a process in which ribosomes travel along mRNAs to synthesize peptides by decoding codons, is interrupted by a wide range of factors: downstream secondary RNA structures (Dana and Tuller, 2012; Charneski and Hurst, 2013; Pop et al., 2014; Ibrahim et al., 2018; Sharma et al., 2019); amino acid/tRNA availability (Dana and Tuller, 2012; Hussmann et al., 2015; Weinberg et al., 2016; Zhang et al., 2017; Dao Duc and Song, 2018; Diament et al., 2018; Mohammad et al., 2019; Sharma et al., 2019); and the presence of nascent peptide chains within the ribosome exit tunnel (Lu and Deutsch, 2008; Brandman et al., 2012; Dana and Tuller, 2012; Charneski and Hurst, 2013; Weinberg et al., 2016; Dao Duc and Song, 2018; Nakatogawa and Ito, 2002; Yanagitani et al., 2011). The movement of ribosomes across the transcriptome can be directly assessed by ribosome profiling, in which ribosome-protected mRNA fragments are generated by RNase treatment and assessed by deep sequencing (Ingolia et al., 2009; Brar and Weissman, 2015). Indeed, this approach has revealed ribosome pauses under a wide range of conditions (Buskirk and Green, 2017).
Ribosome pause has long been speculated to cause ribosome queueing. Although ribosome profiling has revealed ribosome queueing under extreme conditions (e.g., upon perturbation of the translation machinery and depletion of charged tRNAs; Lareau et al., 2014; Young et al., 2015; Woolstenhulme et al., 2015; Schuller et al., 2017; Darnell et al., 2018; Mohammad et al., 2019; Wu et al., 2019a), ribosome queueing has not been observed in naive cells (Ingolia et al., 2011). Whether the lack of stacked ribosomes under native conditions originates from the technical limitations of regular ribosome profiling or the possibility that too few ribosomes are loaded onto an mRNA to form a ribosome queue is unknown.
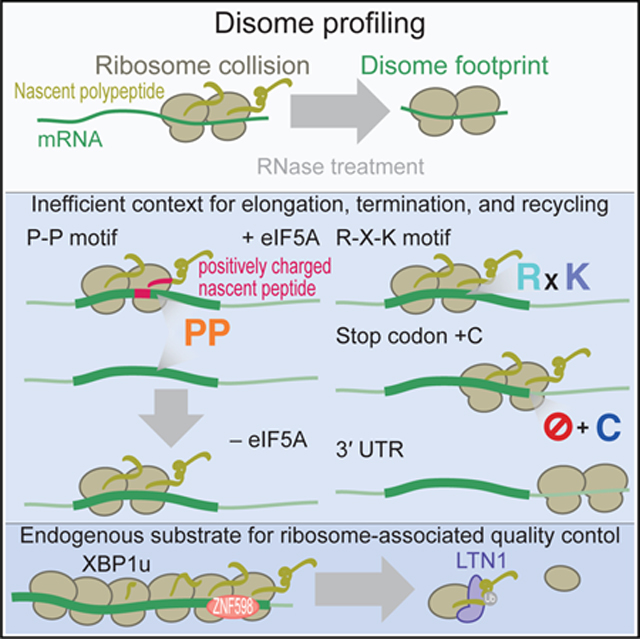
Cells should avoid ribosome traffic jams because the arrested ribosomes could be the source of truncated and deleterious proteins. Indeed, ribosome arrest undergoes surveillance in cells by ribosome-associated quality control (RQC) for nascent polypeptide degradation. In this system, the collided ribosome (also called the di-ribosome or disome) has been proposed as a characteristic sensor for arrested translation (Simms et al., 2017; Juszkiewicz et al., 2018; Ikeuchi et al., 2019). The disome interface recruits the E3 ligase ZNF598 (a yeast Hel2 homolog) for ubiquitination of the ribosomal proteins eS10/uS10/eS3 (Sundaramoorthy et al., 2017; Garzia et al., 2017; Juszkiewicz and Hegde, 2017) in mammals and uS10/eS3 in yeast (Matsuo et al., 2017). This ubiquitination triggers remodeling of the arrested ribosomes and splits them into two subunits via RQC trigger (Rqt) 2, Rqt3, and Rqt4 (Slh1, Cue3, and yKR023W, respectively, in yeast; Matsuo et al., 2017). On the split 60S subunit, the nascent chain is polyubiquitinated by the E3 ligase LTN1 for proteasomal degradation (Bengtson and Joazeiro, 2010; Brandman et al., 2012; Defenouillère et al., 2013; Shao et al., 2013). The addition of a C-terminal alanine-threonine (CAT) tail to the nascent chain (CATylation) further facilitates protein degradation (Shen et al., 2015; Osuna et al., 2017; Kostova et al., 2017; Sitron and Brandman, 2019). Although the pivotal role of RQC has been illustrated by neurodegeneration in mice in the absence of LTN1 (Chu et al., 2009), the endogenous targets of these systems remain largely unknown.
In this study, we probed ribosome collisions by the genome-wide survey of disomes in humans and zebrafish with modified ribosome profiling. Unlike the results of disome profiling conducted in yeast (Guydosh and Green, 2014; Diament et al., 2018), the clear accumulation of disome footprints at discrete codons showed that unique amino acid motifs and the nonoptimal context of stop codons are contributing factors for ribosome pauses. Furthermore, long ribosome queues (~5 ribosomes) were observed in vertebrates under normal conditions. Moreover, disome profiling suggests that post-termination ribosomes along the 3’ untranslated region (UTR) could obstruct ribosomes that follow during the elongation process. We found that severe ribosome traffic jams on X-box binding protein 1 (XBP1), a stress response transcription factor (Hetz and Papa, 2018), induce the degradation of undesirable proteins by RQC in the absence of stress. Our approach provides a versatile framework for the investigation of ribosome traffic jams in cells.
RESULTS
Disome Footprints Define Ribosome Pause Sites
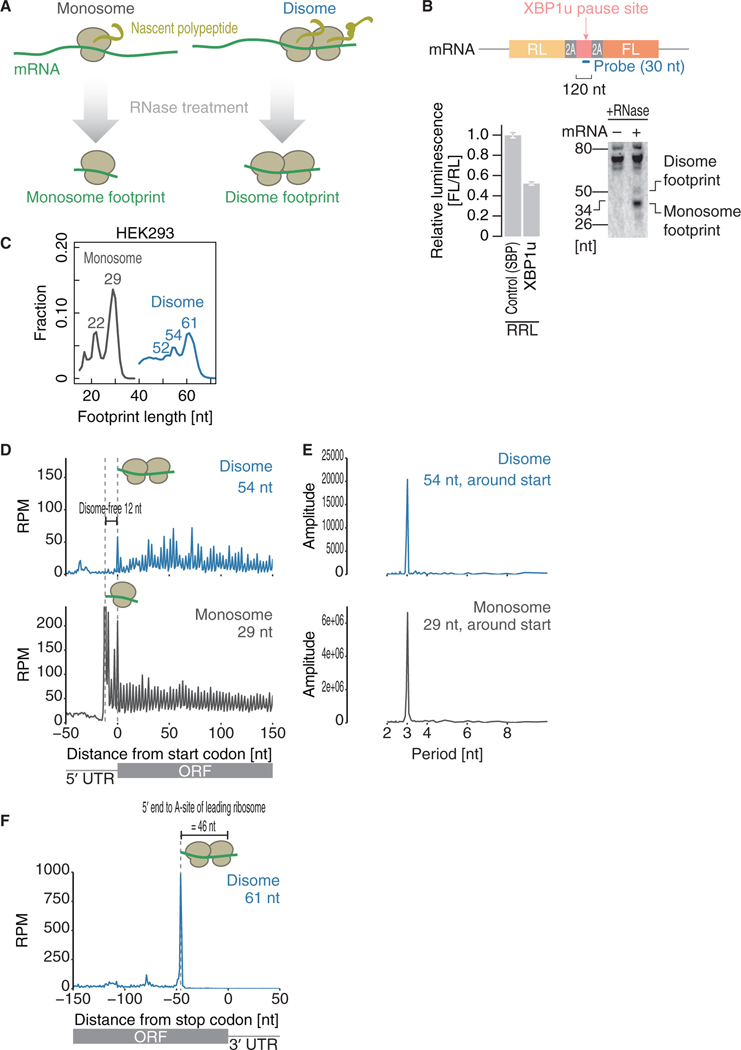
To determine the positions of ribosome traffic jams on mRNAs in humans, we modified the ribosome profiling technique. In typical ribosome profiling (hereafter termed “monosome” profiling), 17- to 34-nt-long RNA fragments generated by nuclease digestion are isolated (Figure 1A). In the modified method (or “disome” profiling), we focused on collided ribosomes that can be represented by two ribosomes as a unit. As tightly packed ribosome pairs were reported to cover 40- to 65-nt mRNA fragments (Wolin and Walter, 1988; Guydosh and Green, 2014; Subramaniam et al., 2014; Diament et al., 2018; Juszkiewicz et al., 2018; Ikeuchi et al., 2019), the lengths of disome fragments were expected to be in the same range.
Figure 1. Disome Footprints Reflect the Collision of Ribosomes.
(A) Schematic representation of disome profiling.
(B) Northern blot of footprints generated by stalled ribosomes on the XBP1u pause site in rabbit reticulocyte lysate (RRL) (bottom right). Schematic representations of a reporter and designed probe are shown (top). As a control, the streptavidin-binding peptide (SBP)-tag was replaced with the XBP1u pause site. The XBP1u pause site and control SBP-tag were sandwiched by self-cleavage 2A sequences. Thus, the readout from upstream Renilla luciferase (RL) and downstream firefly luciferase (FL) could directly reflect the net translation and avoid cotranslational protein degradation, if any, in the cell-free translation system. Translation attenuation by the XBP1u pause site verified the ribosome stalling (bottom left). Data represent the mean and SD (n = 3).
(C) Distribution of the lengths of fragments generated by monosome profiling (regular ribosome profiling, gray line) and disome profiling (blue line) in HEK293 cells.
(D and F) Metagene analysis showing the 5’ ends of fragments of the indicated read lengths around start codons (D) and stop codons (F). The position of the first nucleotide in the start or stop codon is set as 0.
(E) Discrete Fourier transform of reads showing their periodicity around start codons.
RPM, reads per million mapped reads.
See also Figures S1 and S2.
This hypothesis was validated by northern blotting of ribosome-protected RNA fragments generated by stalled ribosomes. An mRNA reporter containing the XBP1u pause site (Yanagitani et al., 2009, 2011; Kanda et al., 2016; Shanmuganathan et al., 2019; Figure 1B, top; also see below and legend for details of reporter design) was subjected to in vitro translation with rabbit reticulocyte lysate to induce ribosome collision, which was verified by the attenuation of protein synthesis (Figure 1B, bottom left). We designed a northern blot probe to hybridize with the ribosome footprints originating from the pause site and indeed observed an RNA fragment with a length of ~60 nt, almost twice the length of the ~30-nt monosome footprint (Figure 1B, bottom right). Therefore, to monitor the human disome in vivo, we generated deep-sequencing libraries from 50- to 80-nt mRNA fragments generated by the RNase digestion of mRNA-ribosome complexes from HEK293 cells (Figure S1A). As a reference, monosome profiling libraries were also prepared from the same samples.
In our pilot experiments, a few specific sites derived from 28S rRNA fragments overpopulated the deep-sequencing libraries (Figures S1B and S1C, top). These contaminants are typically generated during RNase digestion, which partially cleaves rRNAs within ribosomes (Ingolia et al., 2012; Miettinen and Björklund, 2015; McGlincy and Ingolia, 2017; Gerashchenko and Gladyshev, 2017), and escape the rRNA depletion step with hybridizing oligonucleotides. To maximize the disome footprint sequencing space in the experiments, we optimized the protocol by introducing Cas9 treatment to degrade specific sequences in the libraries (i.e., depletion of abundant sequences by hybridization or DASH; Gu et al., 2016). Cas9 programmed with designed guide RNAs (gRNAs) (Figure S1D) was used to remove the contaminating 28S rRNA fragments from the libraries (Figure S1C, bottom), enabling a prominent increase in the number of reads usable for downstream analysis (Figure S1E). Similar improvement following Cas9 treatment was also observed in monosome profiling (Figure S1F).
Disome footprints showed the hallmarks of ribosomes in the elongation process. The populations of mRNA fragments protected by disomes were found to be 52, 54, and 61 nt in length (Figure 1C, blue line), as expected if two consecutive monosome footprints were joined from tail to head. Similar to observations from monosome profiling, discrete Fourier transform showed a 3-nt periodicity in the disome footprints across the coding sequences (CDSs) (Figures 1D and 1E), which reflects the codon-wise movement of ribosomes.
Unlike the general monosome footprints (Figure 1D, bottom), the disome footprints showed no clear accumulation of the first 12 nt surrounding the start codons by metagene analysis (Figure 1D, top). As discussed in an earlier study (Guydosh and Green, 2014), the scanning ribosome (or initiating 80S ribosome) at the start codon may require this additional region of mRNA; thus, the formation of these fragments would be blocked until the downstream ribosome proceeds forward (Figure S1G). Indeed, the footprints of scanning pre-initiation complexes contain longer fragments than those of the elongating ribosomes (Archer et al., 2016). Similar disome-free regions were obtained in previous studies in yeast, although the disome-free area in yeast was longer (~25 nt; Guydosh and Green, 2014). This inconsistency in the length of the disome-free area may be due to the structural differences in ribosomes between mammals and yeast (Anger et al., 2013).
In contrast to the 54-nt disome fragments, which were distributed across the CDSs (Figure 1D), the longer 61-nt disome fragments were predominantly found around stop codons (Figure 1F; note the difference in the vertical axis scale compared with that in Figure 1D, top), with the 5’ end of the fragments located 46 nt upstream of stop codons. These observations represent a scenario in which a leading ribosome pauses at the stop codon and the trailing ribosome collides into it. Thus, the distance from the 5’ ends to stop codons (46 nt) corresponds to the A-site position in the leading ribosome in the disome and matches the size of one ribosome (~30 nt) plus the distance from the A-sites to the 5’ ends (15 nt) observed in monosome profiling. Similar A-site distances were observed in the 52- and 54-nt footprints (Figure S2A). This distance is valuable for addressing the A-site positions of the leading ribosomes at the sub-codon resolution in disome profiling.
Because the two populations of monosome footprints (with peaks at 22 and 29 nt; Figure 1C, gray line) were reported to reflect A-site tRNA accommodation (Wu et al., 2019a), we reasoned that a similar explanation applies to the differential disome footprints. Cycloheximide (CHX), which is used to trap disomes, blocks ribosome translocation, allows A-sites to be occupied by tRNAs (Budkevich et al., 2011; Garreau de Loubresse et al., 2014), and then results in 29-nt footprints. On the other hand, the peptidyl transferase inhibitor anisomycin (ANS) (Garreau de Loubresse et al., 2014) induces the drop-off of A-site tRNAs from ribosomes and eventually leads to the trimming of 7 nt from 3’ ends by RNase (thus resulting in 22-nt footprints; Lareau et al., 2014; Wu et al., 2019a). Similar to the reported drug sensitivity in monosome footprints, disome profiling with ANS treatment induced greater accumulation of short 52- and 54-nt footprints than CHX treatment (Figure S2B). Moreover, consistent with the variable 3’ ends of monosome footprints between 22 and 29 nt long, the major differences in the long and short disome footprints were at the 3’ ends (Figures 1F, S2A, and S2C). Thus, the most straightforward interpretation of these results is that the presence of A-site tRNA in the leading ribosome of disomes extended the footprints, as reported for monosome footprints (Wu et al., 2019a).
The disome occupancy at the codons on which the A-sites of the leading ribosome lay was calculated to evaluate the pause site. This measurement approach was robust because of its high reproducibility regardless of the drug used to trap the disome on mRNA (Figure S2D). Taken together, these data suggest disome profiling as a platform to survey the landscape of collided ribosomes.
Precise Annotation of Pause Sites in Reported Translational Attenuator Sequences
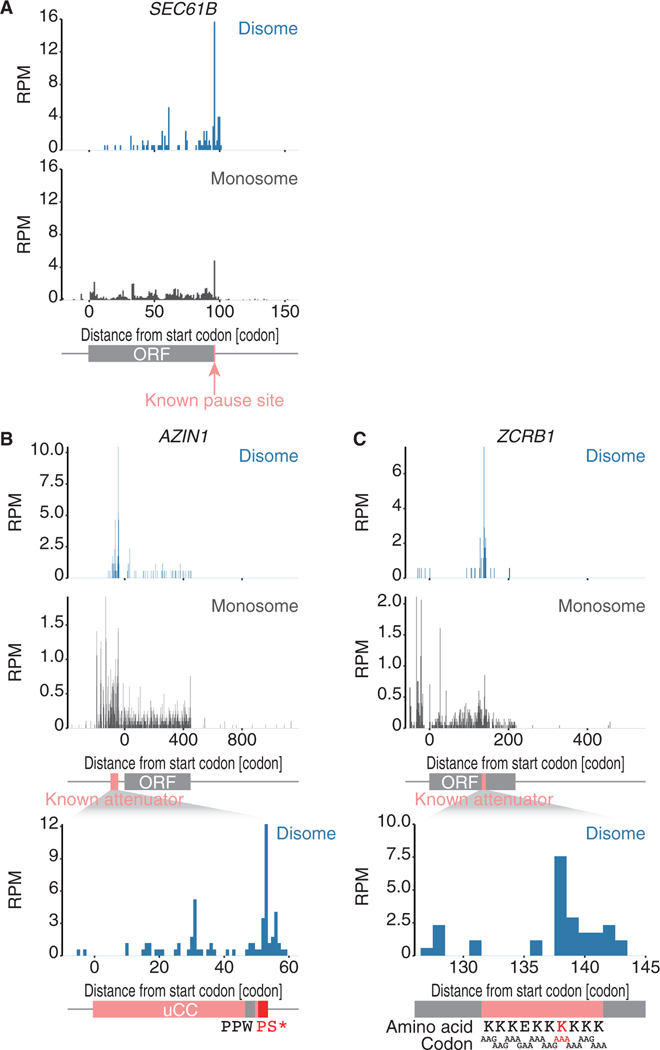
Disome profiling enabled us to revisit the limited number of translational attenuator sites annotated by earlier studies at a sub-codon resolution. SEC61B, a tail-anchored (TA) protein, is one example; its protein synthesis is paused before completion to provide enough time for the recruitment of the membrane insertion machinery (Mariappan et al., 2010). Disome profiling revealed the ribosome collision at its stop codon (Figure 2A), the same site previously reported (Mariappan et al., 2010; Ingolia et al., 2011).
Figure 2. Precise Annotation of Ribosome Pause Sites in Reported Translation Attenuators.
(A–C) The read distribution along indicated mRNAs is depicted, showing the A-site positions in a monosome (bottom) and in the leading ribosome of a disome (top). The known pause site in SEC61B (Mariappan et al., 2010; A) and the known attenuator sequences in AZIN1 (Ivanov et al., 2018; B) and ZCRB1 (Arthur et al., 2015; C) are highlighted in pink.
The second example is the upstream conserved coding region (uCC) (Ivanov et al., 2008, 2018) located in the 5’ UTR of the mRNA of antienzyme inhibitor 1 (AZIN1), a regulator of cellular polyamine synthesis. Paused ribosomes during uCC translation have been proposed to be a roadblock to inhibit the translation from the main downstream open reading frames (ORFs) and to increase the duration time of the scanning ribosome to allow the recognition of near-cognate start sites in the uCC (Ivanov et al., 2018). The uCC in AZIN1 exhibited a marked pile-up of disome footprints (Figure 2B). Notably, although an earlier study reported that the Pro-Pro-Trp motif and Pro-Ser-stop motif are potential pause sites in the uCC (Ivanov et al., 2018), ribosome collision was found only at the PS-stop motif (Figure 2B, bottom).
The third example of ribosome collision focuses on the ZCRB1 and MTDH mRNAs (Arthur et al., 2015). These transcripts have attenuator motifs attributed to adenosine-rich sequences to pause ribosomes; however, no clear evidence of ribosome pause has been provided (Arthur et al., 2015). Our disome profiling showed an apparent ribosome collision in the middle of the motif (Figures 2C and S3A). Even with the homopolymeric sequences in the motif, ribosome collision did not occur randomly but rather at specific codon positions.
Disome Pause Sites Are Widespread across the Transcriptome
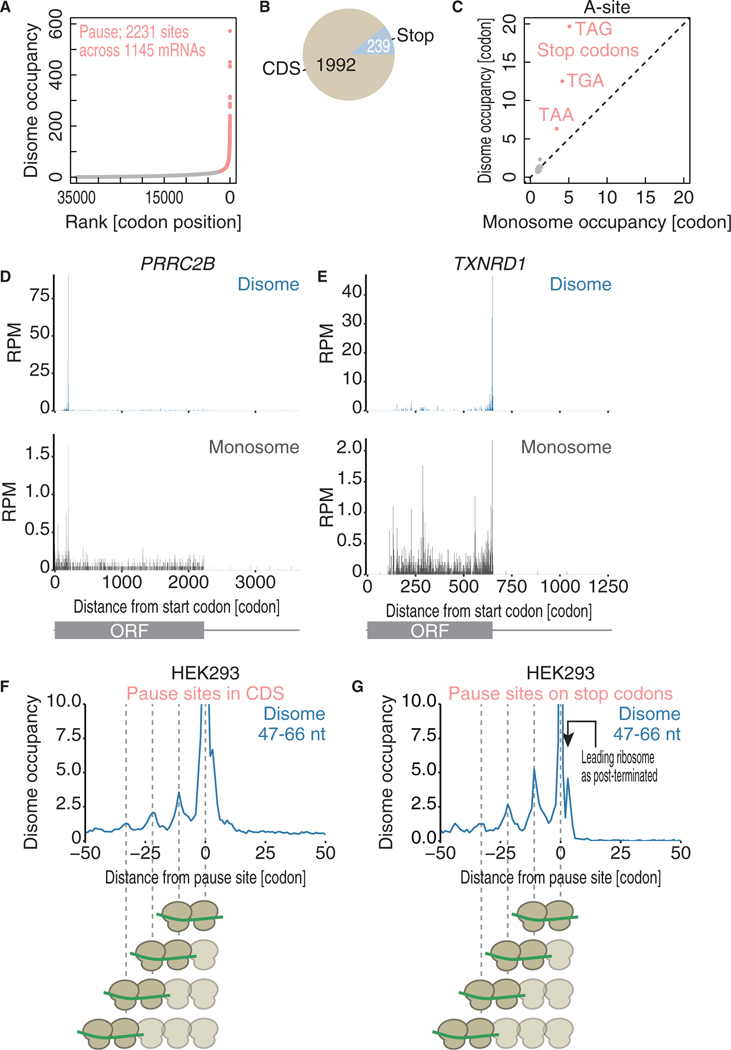
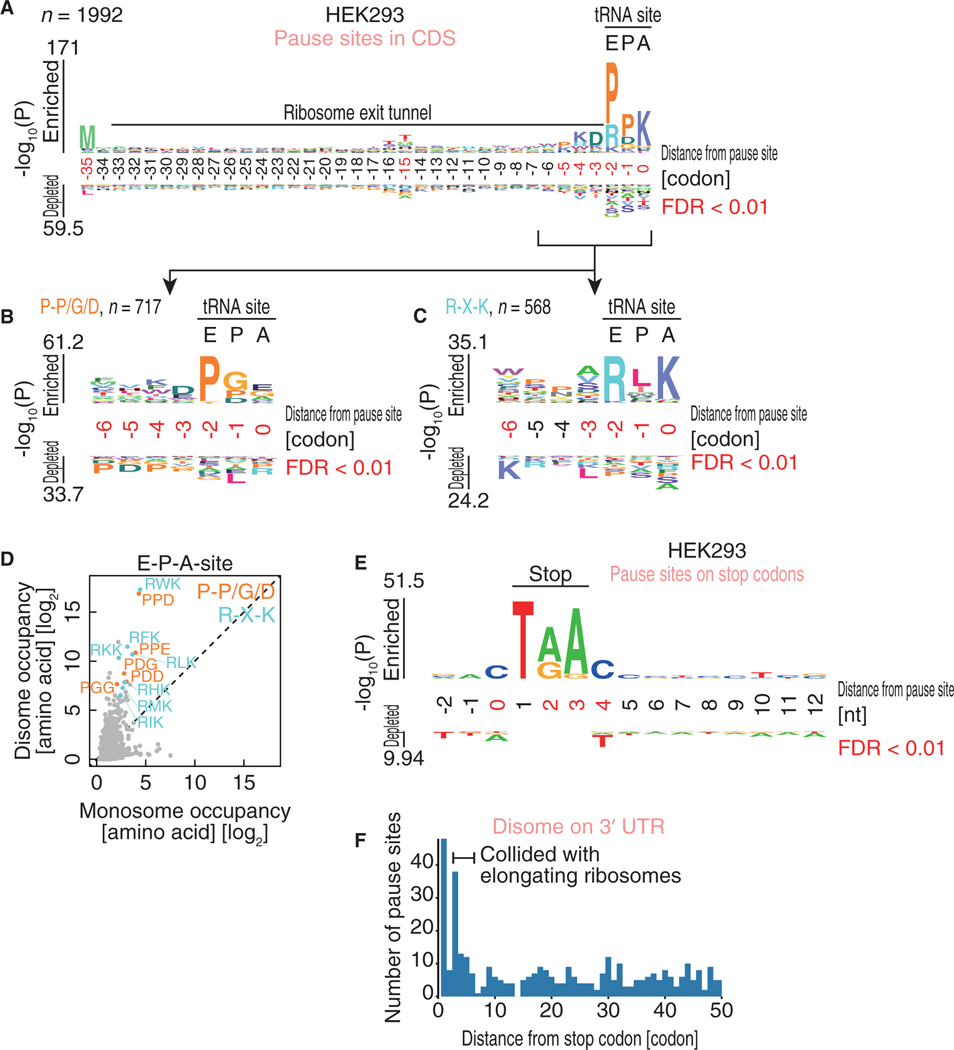
The precise assignments within the known translation attenuators identified by disome profiling led us to conduct an unbiased transcriptome-wide analysis of pronounced pause sites with collided ribosomes (see the STAR Methods section for details). Here, we defined more than two thousand ribosome collision sites (2,231 disome pause sites in 1,145 genes; Figure 3A; Table S1). Given the number of genes (10,315) surveyed in the analysis, we estimated that ~11% of genes have at least one ribosome collision site. The ribosome collision sites could be mechanistically sub-grouped into CDS and stop codon sites, reflecting the regulation of elongation and termination, respectively. Given the number of codons in mRNAs, stop codons were significantly enriched in ribosome collision sites (Figure 3B; p < 2.2 × 10−16; hypergeometric test). The overall disome codon occupancy along tRNA binding sites also showed that ribosomes decoding stop codons at the A-site are prevalent roadblocks for trailing ribosomes (Figure 3C).
Figure 3. Ribosome Collision Sites Are Found in a Wide Range of mRNAs.
(A) Calculated disome occupancies across codons. The collision sites, which are defined as codons with disome occupancies larger than the mean + SD, arehighlighted in pink.
(B) Pie chart indicating the positions of disome pause sites in mRNAs.
(C) Monosome and disome codon occupancies on A-site codons.
(D and E) The read distributions along the PRRC2B (D) and TXNRD1 (E) mRNAs are depicted, showing the A-site position in the monosome (bottom) and in the leading ribosome of the disome (top).
(F and G) Metagene analysis of disome reads around collision sites in the CDS (F) and stop codons (G). The A-site position in the leading ribosome is shown.
The distinct ribosome collision sites were exemplified in the representative mRNAs PRRC2B and TXNRD1 (Figures 3D and 3E); strikingly, disome profiling enabled the enrichment of the paused ribosome fraction and depletion of the fraction under typical elongation/termination conditions, which dominates monosome profiling. Conversely, regular monosome profiling may miss a fraction of stalled ribosomes, as collided ribosomes are relatively resistant to RNase and are not digested into monosomes as previously predicted (Diament et al., 2018).
Next, we investigated the functional implication of the genes associated with ribosome collisions and observed that genes involved in a subset of terms (helicase activity, microtubule cytoskeleton organization and biogenesis, lipid transport, etc.) are more likely to have disomes. In contrast, translation of ribosomal genes proceeded smoothly without ribosome traffic jams (Figure S3B).
Disome Pause Sites Have Long Ribosome Queues
Although disome footprints were generated from two consecutive ribosomes, disome profiling revealed an even longer queue of protein synthesis machinery. Strikingly, the relative disome occupancies around the disome pause sites in the CDS and at stop codons showed three upstream peaks every 11 codons (33 nt), which was consistent with the size of a single ribosome (Figures 3F and 3G). Such a long queue of ribosomes was observed for the DDX39B (Figure S3C) and XBP1u mRNAs (Figure 7A). These ribosome queues around disome pause sites were also observed by monosome profiling (Figures S3D and S3E), although they were less clear than those in the disome profiles. Ribosome queueing upstream of stop codons was also supported by unbiased metagene analysis around stop codons (Figure S2A, top), in which a discrete Fourier transform showed a periodicity of 30 nt–the size of a ribosome (Figure S2E). These observations strongly support a model in which the long pause of a ribosome at a specific site causes a long queue of stalled ribosomes even under normal conditions.
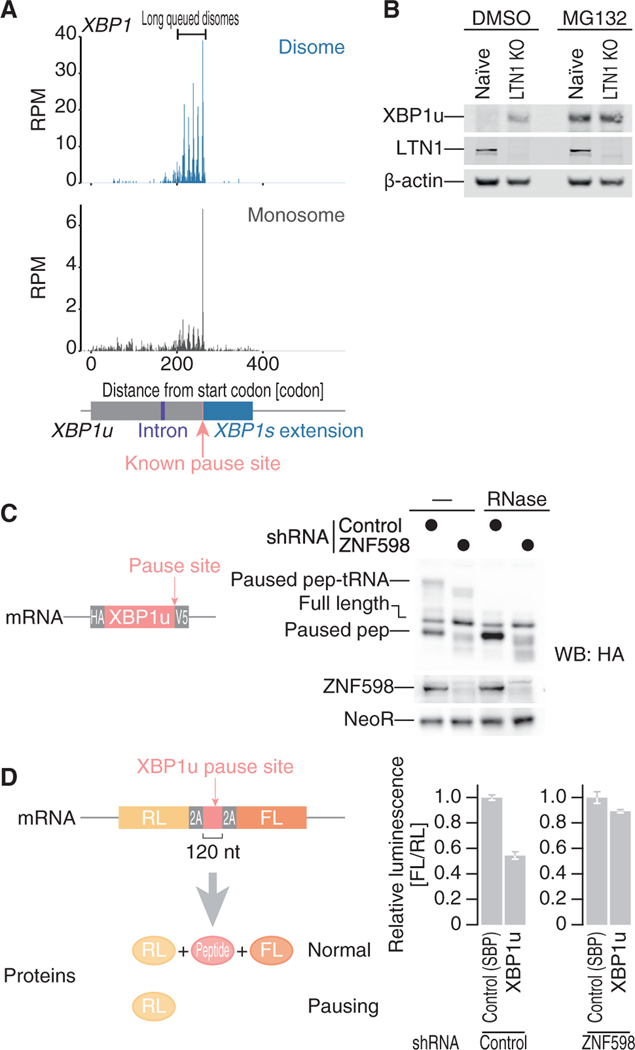
Figure 7. Collided Ribosomes on XBP1u Are Rescued by the RQC Pathway.
(A) The read distributions along human XBP1u mRNA are depicted, showing the A-site positions in the monosome (bottom) and in the leading ribosome of the disome (top). The known ribosome pause site (Yanagitani et al., 2011) is highlighted in pink.
(B) Western blot for the indicated proteins in naive or LTN1 knockout HEK293 cells.
(C) Western blot for proteins expressed from a reporter plasmid (left) in ZNF598 knockdown and control cells. Proteins with or without RNase treatment were separated by neutral PAGE to detect the peptidyl-tRNAs.
(D) Luciferase assay with the reporter with an XBP1u pause site sandwiched between self-cleavage 2A tags (left; same as Figure 1B) in ZNF598 knockdown and control cells. Data represent the mean and SD (n = 3).
See also Figure S7.
Motifs Associated with Ribosome Collisions during Elongation
To reveal the determinants of ribosome collision, we investigated whether the general rates of initiation and elongation could define the ribosome collision frequency. In theory, the high rate of translation initiation–the general limiting step in the overall translation process (Morisaki et al., 2016; Wu et al., 2016; Yan et al., 2016; Wang et al., 2016)–increases the number of ribosomes on mRNAs and thus may lead to ribosome collision. However, ribosome collision was not correlated with translation efficiency (Figure S4A), which was calculated by the over-/underrepresentation of the number of monosome footprints over RNA sequencing (RNA-seq) reads and is generally considered to be the translation initiation rate (Weinberg et al., 2016).
Generally, the rate of ribosome movement along mRNAs has been thought to be shaped by tRNA availability (Dana and Tuller, 2012; Hussmann et al., 2015; Weinberg et al., 2016; Zhang et al., 2017; Dao Duc and Song, 2018; Diament et al., 2018; Mohammad et al., 2019; Sharma et al., 2019). Thus, the cognate tRNA abundance as measured by demethylase (DM)-tRNA-seq (Zheng et al., 2015) was compared to disome occupancy on each codon. However, we did not observe a correlation (Figure S4B). In addition to the tRNA supply, the codon usage frequency–the demand for tRNA–also showed a poor correlation with the disome occupancy (Figure S4C). Thus, more specific contexts, which consume even more time than overall translation initiation and elongation, may determine ribosome collisions in cells.
Indeed, the motifs around ribosome collision sites potentially explain the origins of ribosome traffic jams. Ribosome collision sites found in the CDS showed a strong Pro-Pro-Lys tendency at the E-P-A-site of the leading ribosome (Figure 4A). To separate the combinatorial effects of the multiple motifs, we further clustered the ribosome collision sites and found that the motifs clearly fell into two amino acid sequences: Pro-Pro/Gly/Asp at E-P-sites (Figure 4B) and Arg-X-Lys at E-P-A-sites (Figure 4C). The former motif at E-P-sites has a unique conformation of the nascent chain and is incompatible with the peptidyl transferase center (PTC) (Doerfel et al., 2015; Melnikov et al., 2016a) and thus may cause ribosome pause. In contrast, the latter motif was not well characterized in earlier studies. Importantly, again, ribosome collision on those motifs was underestimated by monosome profiling; disome occupancy on the tri-peptide motif at E-P-A-sites strikingly highlighted the pause on the identified motifs (Figure 4D).
Figure 4. The Motifs Associated with Ribosome Collision.
(A–C) Amino acid sequences enriched in ribosome collision sites found in the CDS (A). The motifs are further sub-clustered (B and C). The codon position relative to the A-site in the leading ribosome is shown.
(D) Monosome and disome occupancies on tri-peptides at E-P-A-sites. Amino acids corresponding to Pro-Pro/Gly/Asp and Arg-X-Lys are highlighted in orange and light blue, respectively.
(E) Nucleic acid enrichment around ribosome collision sites on stop codons. Nucleic acids from the stop codon (where 1 indicates the first nucleotide in the stop codon) are depicted.
(F) Histogram of the number of disome pause sites found in the 3’ UTR along with the distance from the stop codon.
The logos in (A)–(C) and (E) were drawn with kpLogo (Wu and Bartel, 2017). The positions with significantly enriched sequences are highlighted in red.
See also Figure S4.
In contrast to the clear motif found in the PTC, the amino acids held within the ribosome exit tunnel did not show any apparent trends (Figure 4A). Although electrostatic interaction of the negatively charged ribosome exit tunnel with the stretch of positive charges in the nascent chain has been reported to modulate elongation (Lu and Deutsch, 2008; Brandman et al., 2012; Dana and Tuller, 2012; Charneski and Hurst, 2013; Weinberg et al., 2016; Dao Duc and Song, 2018), enrichment of a positive net charge was not observed in nascent chain segments of ribosome collision sites (Figure S4D). Another characteristic of the nascent chain that may influence the elongation rate is its hydrophobicity (Dao Duc and Song, 2018); however, this property did not explain ribosome collision (Figure S4E).
In addition to amino acids, RNA with a strong secondary structure has been recognized as another roadblock for elongating ribosomes (Dana and Tuller, 2012; Charneski and Hurst, 2013; Pop et al., 2014; Ibrahim et al., 2018; Sharma et al., 2019). However, no RNA structure probed by dimethyl sulfide (DMS)-seq (Rouskin et al., 2014) was found around ribosome collision sites (Figure S4F). Thus, although these characteristics of amino acids and nucleic acids may fine-tune ribosome movement, they may not generate a long enough duration for the leading ribosome to be caught by the trailing ribosomes in HEK293 cells.
Motifs Associated with Ribosome Collisions during Termination
Unlike PTC motifs found in the CDS, these regions in stop codons with ribosome collisions showed no clear enrichment (Figure S4G). Instead, we observed a trend in nucleotides tilted toward a C after the stop codon (+4 position; Figure 4E). This motif could be attributed to the mechanism of stop codon recognition by eRF1; the +4 purine nucleotide adopts a preferential conformation induced by eRF1 as it interacts with base G626 of 18S rRNA (Brown et al., 2015) and is thus frequently found downstream of ORFs in eukaryotes (Brown et al., 1990). Thus, inefficient translation termination with a C pyrimidine at the +4 site (McCaughan et al., 1995) would cause the collision of upstream translating ribosomes with paused ribosomes at the stop codon that had not swiftly dissociated.
Disomes with Post-termination Ribosomes
During our analysis of disome accumulation around stop codons, we found a population of disomes spanning the CDS and 3’ UTR (Figures 3G and S2A). Because the A-sites of leading ribosomes lay outside of the CDS, we argued that those disomes represent post-termination ribosomes. Indeed, the disome footprints originating from the 3’ UTR did not show 3-nt periodicity, ruling out the possibility that disomes in the 3’ UTR are undergoing active elongation (Figure S4H). The shorter disome and monosome footprints from the 3’ UTR (Figure S4I) also supported the hypothesis that the leading ribosomes do not possess tRNA in the decoding center. Here, we expanded our survey of disome pause sites along the 3’ UTR and found the enrichment of disome footprint fragments 3–5 codons (9–15 nt) downstream of stop codons (Figure 4F). Given the accumulation of post-termination ribosomes at stop codons and along the 3’ UTR found in regular monosome profiling (Figure S4I, bottom; Guydosh and Green, 2014; Young et al., 2015; Mills et al., 2016; Guydosh and Green, 2017; Sudmant et al., 2018), our data suggested that ribosomes that fail to dissociate after termination and leak into the 3’ UTR could be roadblocks for upcoming ribosomes.
Conservation of Ribosome Collision Sites in Vertebrates
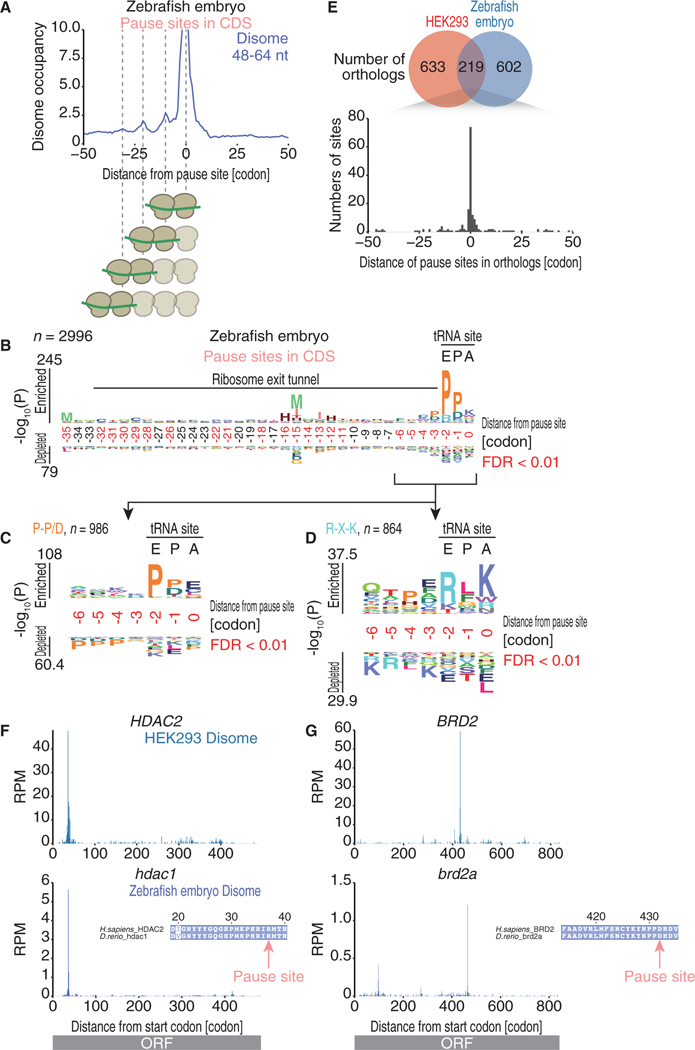
To understand the basis of ribosome collisions across vertebrates, we conducted disome profiling in zebrafish embryos. Identical to the observations made in human cells, zebrafish disome footprint fragments were twice the length of monosome footprint fragments and exhibited a 3-nt periodicity (Figures S5A–S5C), the first 12 nt of the CDS were disome free (Figure S5B), and a 44-nt stretch to the A-site of the leading ribosome was determined by reads upstream of stop codons (Figure S5D). The zebrafish disomes also showed ribosome pauses at the known pause site in Sec61b (Figure S5E). Furthermore, we surveyed de novo pause sites and found them located across CDS and on stop codons in a distribution similar to that in HEK293 cells (Figures S5F and S5G).
Similar to the results of human disome profiling (Figure 3F), long queues of ribosomes formed upstream of disome pause sites in the zebrafish embryo disome profile (Figure 5A). The identical amino acids enriched at the E-P-A-site of the disome pause sites suggest a common basis for elongation regulation in vertebrates (Figures 5B–5D). We observed a fraction of the disome pause sites found in the orthologous mRNAs of the two species (Figure 5E, top). The shared orthologs exhibited ribosome collisions at the exact same sites (Figure 5E, bottom), as exemplified by HDAC2/hdac1 and BRD2/brd2a (Figures 5F and 5G). The conservation of ribosome collision between these two species suggests a commonality in the mechanistic basis of ribosome pause among vertebrates.
Figure 5. The Conservation of Ribosome Collisions between Humans and Zebrafish.
(A) Metagene analysis of zebrafish disome reads around ribosome collision sites in the CDS.
(B–D) Same logo as that in Figures 4A–4C showing zebrafish ribosome collision sites in the CDS.
(E) Venn diagram showing the overlap of ribosome collisions identified in humans and zebrafish orthologs (top). Distribution of the distance between the collision sites of different orthologs is shown (bottom).
(F and G) Disome footprint distributions in human and zebrafish orthologs (HDAC2/hdac1, F; BRD2/brd2a, G) are depicted, showing the A-site position in the leading ribosome of the disome.
The Positively Charged Nascent Chain Determines the Susceptibility of eIF5A-Mediated Ribosome Rescue at the Pro-Pro Sites
The Pro-Pro motifs found in our disome profiling (Figures 4B and 5C) are widely recognized as a characteristic sequence on which eIF5A facilitates the elongation (Gutierrez et al., 2013; Melnikov et al., 2016b; Schmidt et al., 2016; Schuller et al., 2017; Pelechano and Alepuz, 2017; Manjunath et al., 2019). The high level of eIF5A mRNA translation in HEK293 cells suggested that eIF5A was not the limiting factor causing the buildup of disomes on the Pro-Pro motifs (Figure S6A). Thus, we reasoned that ribosome collisions on the identified proline stretches may form independently of eIF5A.
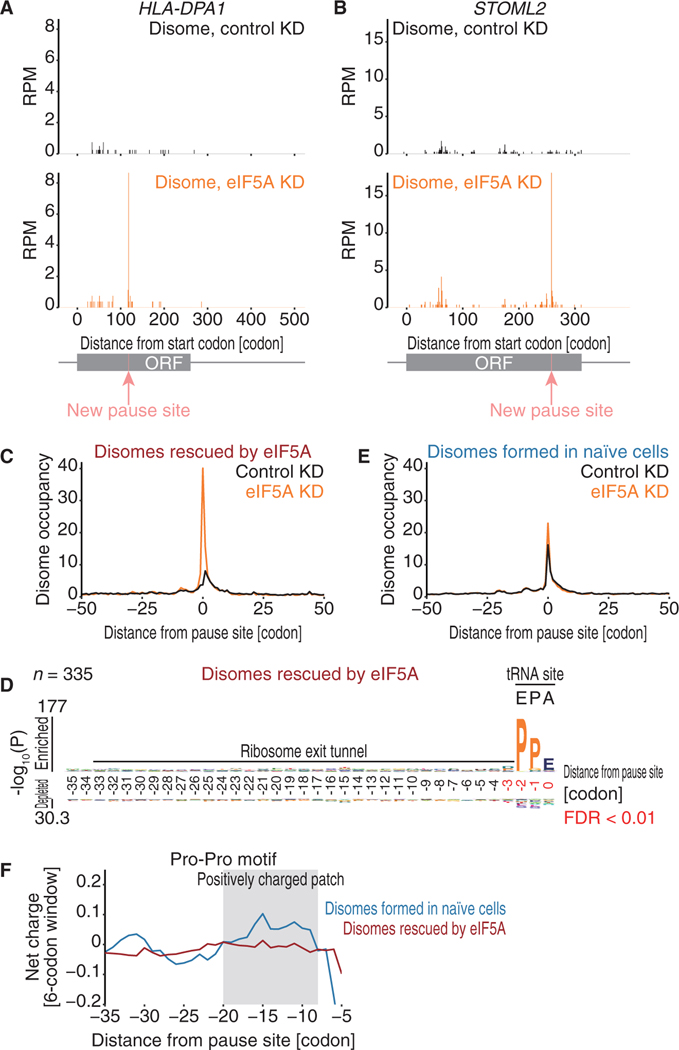
To understand the impact of eIF5A on the ribosome collision landscape, we performed disome profiling with eIF5A knockdown (Figure S6B). Upon eIF5A depletion, we clearly observed the emergence of novel disomes not observed in control cells (Figures 6A–6C; Table S3), indicating that eIF5A otherwise prevents these ribosome collisions. The motif analysis revealed Pro-Pro motifs at the E-P-site, consistent with the earlier reports (Figure 6D; Gutierrez et al., 2013; Melnikov et al., 2016b; Schmidt et al., 2016; Schuller et al., 2017; Pelechano and Alepuz, 2017; Manjunath et al., 2019). In contrast, the ribosome collision sites that formed even in the presence of eIF5A (pause sites used in Figures 3 and 4) were less susceptible to eIF5A depletion (Figure 6E).
Figure 6. Disomes on Pro-Pro Motifs Bearing Positively Charged Nascent Chains Escape from Rescue by eIF5A.
(A and B) The read distributions along human HLADPA1 (A) and STOML2 (B) mRNA from control (top) and eIF5A knockdown (bottom) samples are depicted, showing the A-site positions in the leading ribosome of the disome. The newly identified ribosome pause sites are highlighted in pink.
(C and E) Metagene analysis of disome reads around collision sites observed upon eIF5A knockdown (C) and found in naive cells (pause sites used in Figures 3 and 4; E).
(D) Amino acid sequences enriched in ribosome collision sites observed upon eIF5A knockdown.
(F) Net charge in the nascent chain around ribosome collision sites observed upon eIF5A knockdown and found in naive cells (pause sites used in Figures 3 and 4).
Because essentially the same motifs at the E-P-site were associated with ribosome collisions irrespective of eIF5A presence, we hypothesized that an additional factor outside the PTC determines the impact of eIF5A on ribosome procession. To ensure comparable analysis, here, we restricted the analysis to disomes on the Pro-Pro motif at the E-P-site. We found that the disomes formed in the presence of eIF5A (or found in naive cells) tend to have positively charged nascent peptides within the ribosome exit tunnel (Figure 6F). On the other hand, eIF5A-rescued disomes contained neutral nascent peptides in charge. Thus, although the peptidyl transfer of unfavorably orientated Pro-Pro in the PTC could be facilitated by eIF5A, the electrostatic interaction between the ribosome exit tunnel and the nascent chain conferred resistance to the effect of eIF5A. We note that the impact of positively charged nascent peptides was masked in the meta-analysis shown in Figure S4D, because multiple contexts were averaged. Our disome profiling revealed that the combination of the two factors (ProPro motif and positively charged nascent peptide) provides a stronger barrier to ribosome movement.
XBP1u Protein Was Degraded by RQC
We further traced the fates of the disomes that escape from the aid of eIF5A and constitutively form in cells. We searched for mRNAs that contain significant ribosome queues upstream of disome pause sites. XBP1u emerged as the mRNA with the most prominent ribosome queueing features (Figure 7A). We observed five sharp disome peaks (representing 6 ribosomes queued in a row) at 11-codon intervals, and the highest peak in the disome footprint occurred at the reported ribosome pause site (Yanagitani et al., 2011; Figure 7A), at which the PTC of the ribosome is distorted by the unique conformation of the nascent chain within the ribosome exit tunnel (Shanmuganathan et al., 2019).
Because ribosome collisions drive RQC (Simms et al., 2017; Juszkiewicz et al., 2018; Ikeuchi et al., 2019), the long queue of ribosome on the XBP1u mRNA led us to investigate whether the XBP1u protein is subject to RQC. For this purpose, we knocked out LTN1 (Figure S6C), an E3 ligase that triggers nascent chain ubiquitination and subsequent proteasomal degradation (Inada, 2017). Because the XBP1u protein is rapidly degraded through the ubiquitin-proteasome pathway (Lee et al., 2003; Yoshida et al., 2006), no protein accumulation was observed (Figure 7B). In contrast, LTN1 depletion (or treatment with the proteasome inhibitor MG132) rescued the protein from degradation (Figure 7B).
The RQC pathway is initiated by the recruitment of the E3 ligase ZNF598 (yeast Hel2homolog) to collided ribosomes (Simms et al., 2017; Juszkiewicz et al., 2018; Ikeuchi et al., 2019), which is then followed by the dissociation of the stalled ribosome, leaving paused peptidyl-tRNA in the 60S subunit. When ZNF598 does not recognize the collided ribosomes, the paused ribosomes read through the pause site and translate the downstream regions (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017; Matsuo et al., 2017; Juszkiewicz et al., 2018). We showed that the XBP1u pause site phenocopied the reported fate of the RQC target. ZNF598 knockdown alleviated the accumulation of paused peptidyl-tRNA, observed as the slowly migrating bands that disappeared with RNase treatment, and produced more full-length proteins than observed in control cells (Figure 7C).
To quantitatively monitor readthrough of the pause site, we constructed a reporter containing in-frame N-terminal Renilla and C-terminal firefly luciferase ORFs and the XBP1u pause sequence inserted in the middle (Figure 7D, left). Viral 2A sequences, which induce self-cleavage of peptide bonds during translation (de Felipe et al., 2006; Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017), flanking the XBP1u pause site liberated the individual luciferase proteins. This mRNA reporter ensures the readout from net translation and does not reflect cotranslational protein degradation by RQC, which is restricted only in the middle peptide flanked by the 2A sequence in this construct (Juszkiewicz and Hegde, 2017; Sundaramoorthy et al., 2017). Indeed, readthrough of the pause site in this reporter was restored in ZNF598 knockdown cells (Figure 7D, right). Considering the data together, we concluded that ribosomes queued at the XBP1u pause site are subject to RQC.
DISCUSSION
XBP1u translation is essential to tether the mRNA-ribosome-nascent chain complex to the endoplasmic reticulum (ER) membrane until the unfolded protein response (UPR) is induced (Yanagitani et al., 2009, 2011; Kanda et al., 2016; Shanmuganathan et al., 2019), which leads to unconventional splicing of XBP1u mRNA anchored on the ER (Yoshida et al., 2001; Calfon et al., 2002) and a frameshift to produce the XBP1s protein, a mature stress-responsive transcription factor. However, the protein product of the unspliced form of XBP1 is undesirable under normal conditions (Yoshida et al., 2006). Thus, cells harness the RQC system to degrade the unnecessary precursor form of the transcription factor in the absence of stress. Notably, the intron of XBP1u is not covered by stacked ribosomes (Figure 7A), thus allowing its splicing upon ER stress without steric hindrance by ribosomes.
Great progress in understanding the molecular mechanism of RQC and harnessing the artificial reporter system with programmed pause sites has been made in recent years (Dimitrova et al., 2009; Brandman et al., 2012; Simms et al., 2017; Matsuo et al., 2017; Sundaramoorthy et al., 2017; Garzia et al., 2017; Juszkiewicz and Hegde, 2017; Kostova et al., 2017; Juszkiewicz et al., 2018; Kuroha et al., 2018; Verma et al., 2018; Winz et al., 2019; Ikeuchi et al., 2019). In contrast, the endogenous substrate of the RQC system is unknown. Here, by identifying long ribosome queueing via disome profiling, we found the XBP1u pause site as an endogenous substrate of RQC. Given the recently reported examples of mitochondria targeting proteins exhibiting translation termination failure (Wu et al., 2019b) and misfolded proteins during membrane insertion (Lakshminarayan et al., 2020), the list of endogenous RQC targets will expand. Notably, given the link between RQC and the stresses in mitochondria (Wu et al., 2019b) and ER (Higgins et al., 2015) to RQC, cellular stress appears to be a key driver for RQC. In contrast, our findings of XBP1u in the regular, nonstressed growth condition provided a complementary and unique example of RQC targets.
Ribosome stacking is not rigid but is rather flexible, probably due to diverse cellular factors, such as the kinetics of translation initiation, elongation, termination, and recycling, which consequently determine the number of ribosomes in a queue. Indeed, in zebrafish embryos, we observed the buildup of disomes at the xbp1u pause site (Figure S7A) and the inhibition of downstream translation by the pause site sequence on reporter mRNAs (Figures S7B–S7D). However, the queueing of ribosomes upstream of the xbp1u pause site was not obvious (Figure S7A). Therefore, we would not be surprised if the number of queued ribosomes differs across cell types and species.
The sorting of ribosomes for RQC and ribosome queueing should be related but distinct issues. Whereas we found that the long queue on XBP1u is rescued by RQC, ribosome collisions alone were not enough to predict the RQC substrate; readthrough of ribosome collision sites in ZNF598 knockdown cells was not observed in monosome profiling (Figure S6D), and endogenous MTDH, which showed a remarkable pileup of disomes (Figure S3A), was not rescued by LTN1 knockout (Figure S6E). Although the disome may provide a platform for ZNF598 recruitment to drive RQC, additional determinants might enhance the reaction efficiency in human cells.
In addition to its role in quality control, ribosome pauses are suggested to facilitate cotranslational nascent protein folding (Gloge et al., 2014; Wilson et al., 2016; Collart and Weiss, 2020). As cotranslational chaperone recruitment to domains during synthesis was recently reported to be coupled with pause (Stein et al., 2019), we observed that a fraction of ribosome collision sites resided within domains (Figures S6F and S6G). In yeast, the association of the ribosome collision with the ends of domains (Figure S6H) correlated with the recruitment of TRiC chaperone complex to the nascent chain, which is facilitated by a reduced elongation rate (Stein et al., 2019). The programmed ribosome pause at the site where domain synthesis is largely completed may be a strategy for protein folding in humans as well.
Although the speed of codon decoding by tRNA is widely expected to be a rate-limiting step of elongation, our disome profiling suggested that the difficulty of the peptidyl transfer could define the rate of ribosome movement, as illustrated by the clear amino acid motifs at the PTC (e.g., Pro-Pro/Gly/Asp). The Arg-X-Lys motifs found in both humans and zebrafish were coincident with the amino acid context specifically stalled by macrolide antibiotics, such as erythromycin, telithromycin, and azithromycin, in bacteria (Sothiselvam et al., 2014; Davis et al., 2014). A subsequent study reported that this context per se was an inefficient motif for peptide bond formation, even without the antibiotics (Sothiselvam et al., 2016). Disome profiling will expand our knowledge regarding the in vivo kinetics of ribosome movement and the fates of mRNAs and proteins defined by cotranslational events.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Shintaro Iwasaki (shintaro.iwasaki@riken.jp).
Material Availability
The materials generated in this study will be distributed upon request. There are restrictions to the availability due to Material Transfer Agreement (MTA).
Data and Code Availability
The accession numbers for monosome profiling and disome profiling of HEK293 cells and zebrafish reported in this paper are GEO: GSE145723 and GEO: GSE133392. All custom scripts of R used for data analysis are available upon request. Original images used for the figures have been deposited to Mendeley Data: https://doi.org/10.17632/9y9bpn4w87.1.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HEK293 cells were cultured in DMEM + GlutaMAX-I (Thermo Fisher Scientific) with 10% FBS at 5% CO2 at 37°C.
LTN1 knockout cells were generated using CRISPR-Cas9 approaches (Ran et al., 2013). The guide sequence used to target the LTN1 locus was 5’-ATTCCACCACAACCTAACCATGG-3’ (Figure S6C) and was cloned into the BbsI restriction sites in the pSpCas9(BB)-2A-Puro (PX459) plasmid. HEK293T cells were transfected with the plasmid and selected with 10 μg/ml puromycin. Single-cell clones were then isolated by a limiting dilution approach and screened for LTN1 expression by western blotting.
ZNF598 knockdown cells were generated using shRNA-expressing lentivirus (Matsuo et al., 2017). The shRNA against ZNF598 (designed to express the 5’-GCCAGTTGCCGTCGTCGTTAAT-3’ gRNA (Matsuo et al., 2017)) was cloned into the HpaI/XhoI restriction sites in the pll3.7 plasmid with substitution of its GFP gene with a puromycin resistance gene. HEK293T cells infected with shZNF598-expressing lentivirus were selected with 10 μg/ml puromycin.
The sex of HEK293 cells is female.
Zebrafish
Zebrafish AB strain was raised and maintained at 28.5°C under standard laboratory conditions according to Animal Experiment Protocol (2018–46) at Kyoto Sangyo University. Fertilized eggs were obtained by natural bleeding of the AB strain.
METHOD DETAILS
Northern blotting
Reporter mRNAs (see below for details) were prepared as previously described (Iwasaki et al., 2016, 2019) and translated in a rabbit reticulocyte lysate system (Promega) according to the manufacturer’s instructions. Luminescence was detected with a dual-luciferase reporter assay system (Promega) and GloMax (Promega).
The reaction was diluted in 300 μl of lysis buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, 1 mM dithiothreitol (DTT), 100 μg/ml CHX, and 1% Triton X-100] and treated with 20 units of micrococcal nuclease (TaKaRa) at 25 °C for 45 min. Nuclease digestion was stopped by the addition of 20 μl of 0.1 M ethylene glycol tetraacetic acid (EGTA). Ribosome recovery and RNA extraction were performed as previously described (McGlincy and Ingolia, 2017). RNA containing ribosome footprints was separated by polyacrylamide gel electrophoresis (PAGE) in 1 × TBE buffer (89 mM Tris, 89 mM boric acid, and 2 mM ethylenediaminetetraacetic acid), and transferred to a Biodyne B pre-cut modified nylon membrane (Thermo Fisher Scientific) using 1 × TBE buffer. RNA was immobilized by ultraviolet cross-linking.
To prepare the IRDye800-conjugated probe, the complementary sequence surrounding the pause site of XBP1u (5’-GGUCAA AACGAAUGAGUUCAUUAAUGGCUU-3’) was transcribed using a T7-Scribe Standard RNA IVT Kit (CELLSCRIPT) with 1.25 mM 5-azido-C3-UTP (Jena Bioscience). The azido-containing transcript was labeled with IRDye 800CW DBCO (LI-COR) and purified by PAGE. Hybridization was performed using PerfectHyb Plus Hybridization Buffer (Sigma-Aldrich) at 42°C overnight. Signals were detected in an Odyssey CLx Infrared Imaging System (LI-COR).
siRNA knockdown
For knockdown, HEK293 cells were transfected with siRNAs targeting eIF5A1 (Dharmacon On-Target plus, L-015739–00-0005) or control siRNAs (Dharmacon, D-001810–01-05) with Lipofectamine RNAiMAX (Thermo Fisher Scientific) for 72 h before cell lysis.
Monosome and disome profiling
Library preparations
HEK293 cell disome and monosome profiling libraries were prepared as described previously (McGlincy and Ingolia, 2017). CHX (Sigma-Aldrich) and ANS (Wako Chemicals) (both 100 μg/ml) were included in the lysis buffer. Protected RNA fragments ranging from 17–34 nt and 50–80 nt were gel-excised for monosome and disome profiling, respectively.
Fifty to sixty zebrafish embryos were snap frozen at the sphere stage in liquid nitrogen and lysed with lysis buffer (McGlincy and Ingolia, 2017). These embryos were injected with mRNA libraries of GFP reporters containing various codon-tag sequences. Detailed information about the library and related results will be described elsewhere (Y.M. and S.I., unpublished data). To isolate ribosomes, an S-400 HR gel filtration spin column (GE Healthcare) was used instead of sucrose cushion ultracentrifugation. RNA fragments ranging from 26–34 nt and 50–80 nt were selected for monosome and disome profiling, respectively.
PCR-amplified DNA libraries were further treated with gRNA-programmed Cas9 protein. To prepare gRNAs by in vitro transcription, template DNAs were amplified by PCR (PrimeSTAR Max, TaKaRa) with the 4 overlapping DNA oligos (three scaffolding oligos and one target-specific oligo) listed below:
scaffold oligo DNAs:
gRNA scaffold 1 (T7 promoter): 5’-TGACTAATACGACTCACTATAGG-3’,
gRNA scaffold 2: 5’-AAAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTAAACTTGCTATGC TGTTTCCAGC-3’, and
gRNA scaffold 3: 5’-AAAAAAAGCACCGACTCGGTGC-3’;
target-specific oligo DNAs:
Disome g1: 5’-TAATACGACTCACTATAGGCCGGTCGCGGCGCACCGCCGGTTTAAGAGCTATGCTGGAAACAGCATAGCAAG TTTAAATAAGG-3’,
Disome g2: 5’-TAATACGACTCACTATAGGTCGCCGAATCCCGGGGCCGAGTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTT TAAATAAGG-3’,
Disome g3: 5’-TAATACGACTCACTATAGGAGGCCTCTCCAGTCCGCCGAGTTTAAGAGCTATGCTGGAAACAGCATAGCAAGTT TAAATAAGG-3’,
Monosome g1: 5’- TGACTAATACGACTCACTATAGGCGGAGGATTCAACCCGGCGGGTTTAAGAGCTATGCTGGAAACAGCA TAGCAAGTTTAAATAAGG-3’,
Monosome g2: 5’-TGACTAATACGACTCACTATAGGACGCCGGCGCGCCCCCGCGGTTTAAGAGCTATGCTGGAAACAGCATA GCAAGTTTAAATAAGG-3’,
Monosome g3: 5’- TGACTAATACGACTCACTATAGGGCCGGGCCACCCCTCCCAGTTTAAGAGCTATGCTGGAAACAGCATAG CAAGTTTAAATAAGG-3’,
Monosome g4: 5’- TGACTAATACGACTCACTATAGGCCCGGGCGGGTCGCGCCGTCGTTTAAGAGCTATGCTGGAAACAGCAT AGCAAGTTTAAATAAGG-3’,
Monosome g5: 5’-TGACTAATACGACTCACTATAGGCCGGCCGAGGTGGGATCCCGGTTTAAGAGCTATGCTGGAAACAGCATA GCAAGTTTAAATAAGG-3’,
Monosome g6: 5’-TGACTAATACGACTCACTATAGGACGGGCCGGTGGTGCGCCCTGTTTAAGAGCTATGCTGGAAACAGCATA GCAAGTTTAAATAAGG-3’,
Monosome g7: 5’-TGACTAATACGACTCACTATAGGCGCTTCTGGCGCCAAGCGCCGTTTAAGAGCTATGCTGGAAACAGCATA GCAAGTTTAAATAAGG-3’,
Monosome g8: 5’-TGACTAATACGACTCACTATAGGTTGGTGACTCTAGATAACCTGTTTAAGAGCTATGCTGGAAACAGCATAG CAAGTTTAAATAAGG-3’,
Monosome g9: 5’-TGACTAATACGACTCACTATAGGCGCTCAGACAGGCGTAGCCCGTTTAAGAGCTATGCTGGAAACAGCAT AGCAAGTTTAAATAAGG-3’, and
Monosome g10: 5’-TAATACGACTCACTATAGGGCCCAAGTCCTTCTGATCGGTTTAAGAGCTATGCTGGAAACAGCATAGCAA GTTTAAATAAGG-3’.
Purified DNAs were used for in vitro transcription with a T7-Scribe Standard RNA IVT Kit (CELLSCRIPT). To generate the disome profiling library, an RNA-protein complex was formed with 1 μM Cas9 protein (New England Biolabs) and 1 μM gRNA pool (3 gRNAs in total) in 4 × Cas9 Reaction Buffer (New England Biolabs) at 25°C for 10 min in a 5 μl reaction volume. The reaction mixture was directly added to 15 μl of DNA library containing 0.17 pmol DNA (thus diluting the Cas9 Reaction Buffer to 1 ×) and incubated at 37°C for 30 min. To generate the monosome profiling library, 2 μM Cas9 protein and 2 μM gRNA pool (10 gRNAs in total) were preincubated and used to generate 0.28 pmol of DNA library. To quench the reaction, gRNAs were degraded by the addition of 0.5 μl of RNase I (10 U/μl, Epicenter) and incubation at 37°C for 10 min, and then Cas9 proteins were digested with 0.5 μl of proteinase K (~20 mg/ml, Roche) at 37°C for 10 min. DNA was further gel-excised and sequenced with the HiSeq4000 platform (Illumina).
Data analysis
Data were processed as previously described (Ingolia et al., 2012; Iwasaki et al., 2016). Disome and monosome occupancies were calculated as the ratio of reads at given codons to the average reads per codon on the transcript. The disome pause sites were defined as codons with disome occupancies larger than the mean + SD. Gene Ontology analysis was performed with iPAGE (Goodarzi et al., 2009). The highest disome occupancy was used as a representative value for the analysis. CHX-treated libraries were used unless otherwise indicated. The eIF5A knockdown-specific disome pause sites were defined as codons with disome occupancies larger than the mean + SD in eIF5A knockdown sample and with read counts fewer than three in the control knockdown.
For calculation of the translation efficiencies, the read number obtained by ribosome profiling within each CDS were normalized by the read number obtained by RNA-seq (GSE126298) (Hia et al., 2019) using the DESeq package (Anders and Huber, 2010). Reads corresponding to the first and last five codons of each CDS were omitted.
The tRNA abundance was estimated by DM-tRNA-seq of samples from HEK293T cells (GSE66550) (Zheng et al., 2015). Reads mapped to each tRNA (GtRNAdb) (Chan and Lowe, 2016) were counted. To estimated cognate tRNA abundances, wobble parameters (Lim and Curran, 2001) were used for individual codons as previously reported (Weinberg et al., 2016). Codon usage was followed as previously published (Puigbò, 2008)
The motifs associated with ribosome collision sites were illustrated with kpLogo (http://kplogo.wi.mit.edu) (Wu and Bartel, 2017). The ribosome collision sites were further clustered by GibbsCluster (http://www.cbs.dtu.dk/services/GibbsCluster-2.0/) (Andreatta et al., 2017) and were then visualized with kpLogo.
The net charge (Lehninger pKa scale) and hydrophobicity (Kyte-Doolittle scale) of the nascent chain within a 6 amino acid window was calculated with the Peptides package in R. The annotated domains of peptides were obtained from the UniProt database.
The in vivo secondary structure was estimated with the published DMS-seq data from K562 cells (GSE45803) (Rouskin et al., 2014). For calculation of the DMS-seq value, the raw read counts of each nucleotide were normalized to the most reactive base within the indicated window encompassing a pause site, and the normalized counts of the in vivo sample were then compared to those of the denatured sample. We excluded transcripts with an average of 15 or fewer reads over the A or C position in the given window.
The definition of orthologs from Homologene (NCBI) was used. Human codon positions corresponding to zebrafish disome pause sites were found by pairwise alignment, and the distance to the disome pause site in HEK293 cells was calculated.
All custom scripts used in this study are available upon request.
DNA constructs
psiCHECK2-2A-3xFLAG-XBP1u_pause-2A and 2A-3xFLAG-SBP-2A
DNA fragments with the following sequence were inserted between the Renilla luciferase (hRluc) and firefly luciferase (luc+) ORFs in the psiCHECK2-EIF2S3 5’ UTR (Iwasaki et al., 2016) to generate an in-frame ORF containing both luciferase ORFs:
2A-3xFLAG-XBP1u_pause-2A: 5’-GGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACC CTGGACCTGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGCCTGGAGGAGCTCCCAGAGGTCTACCCAGAAGGACCCAGTTCCTTACCAGCCTCCCTTTCTCTGTCAGTGGGGACGTCATCAGCCAAGCTGGAAGCCATTAATGAACTCATTCGTTTTGACCGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAG GAGAACCCTGGACCT-3’ and
2A-3xFLAG-SBP-2A: 5’-GGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAACCCTGGACCTGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGACGAGAAAACCACCGGCTGGCGGGGAGGCCACGTGGTGGAAGGGCTGGCAGGCGAGCTGGAACAGCTGCGGGCCAGACTGGAACACCACCCCCAGGGCCAGAGAGAGCCTAGCGGCGGAGGAGGAAGCGGAGCTACTAACTTCAGCCTGCTGAAGCAGGCTGGAGACGTGGAGGAGAA CCCTGGACCT-3’.
PCR products generated from the plasmids and the primers (5’-TGACTAATACGACTCACTATAGG-3’ and 5’-TGTATCTTATC ATGTCTGCTCGAAG-3’) were used as templates for in vitro transcription.
pcDNA3.1(+)-HA-XBP1u-V5
DNA fragments with following sequence were inserted to pcDNA3.1(+) at the NheI-ApaI restriction sites:
5’-gccgccaccATGGGATACCCATACGACGTCCCAGACTACGCGAAGCTTGGTACCGGATCCGAATTTgtggtggtggcagccgcgccgaacccggccgacgggacccctaaagttctgcttctgtcggggcagcccgcctccgccgccggagccccggccggccaggccctgccgctcatggtgccagcccagagaggggccagcccggaggcagcgagcggggggctgccccaggcgcgcaagcgacagcgcctcacgcacctgagccccgaggagaaggcgctgaggaggaaactgaaaaacagagtagcagctcagactgccagagatcgaaagaaggctcgaatgagtgagctggaacagcaagtggtagatttagaagaagagaaccaaaaacttttgctagaaaatcagcttttacgagagaaaactcatggccttgtagttgagaaccaggagttaagacagcgcttggggatggatgccctggttgctgaagaggaggcggaagccaaggggaatgaagtgaggccagtggccgggtctgctgagtccgcagcactcagactacgtgcacctctgcagcaggtgcaggcccagttgtcacccctccagaacatctccccatggattctggcggtattgactcttcagattcagagtctgatatcctgttgggcattctggacaacttggacccagtcatgttcttcaaatgcccttccccagagcctgccagcctggaggagctcccagaggtctacccagaaggacccagttccttaccagcctccctttctctgtcagtggggacgtcatcagccaagctggaagccattaatgaacGAATTCGATATCACCGGTCTCGAGTCTAGAGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGTAA-3’,
where the lowercase italic letters indicate the XBP1u CDS, the uppercase letters indicate the HA/V5 tag sequence, and the bold letters indicate the multicloning site sequence.
pCS2+sfGFP-zXbp1opt-suv39h1a 3’ UTR
ADNA fragment encoding the sfGFPOR Flacking as top codon and containing EcoRI-XhoIsite satits3’ end was synthesized by the Thermo Fisher Gene Art Strings service and inserted into the NcoI-XhoI sites in pCS2+EGFP-suv39h1a (Mishima and Tomari, 2016) to replace the EGFP ORF (pCS2+sfGFP-suv39h1a). DNA oligos corresponding to the zebrafish xbp1 C-terminal pause sequence or its mutant, as shown below, were annealed and inserted into the XhoI-XbaI sites in pCS2+sfGFP-suv39h1a. The pause sequence was codon-optimized to the zebrafish genome to minimize the effect of synonymous codon choice on mRNA stability (Mishima and Tomari, 2016).
y750zxbppauseOptRIXIwtFW: 5’-aattcCAGGAGAGCAAGTACCTGCCTCCGCACCTGCAGCTGTGGGGCCCGCACCAGCTGAG CTGGAAGCCCCTGATGAACTGAc-3’,
y750zxbppauseOptRIXIwtRV: 5’-tcgagTCAGTTCATCAGGGGCTTCCAGCTCAGCTGGTGCGGGCCCCACAGCTGCAGGTGCGGAGGCAGGTACTTGCTCTCCTGg-3’,
y751zxbppauseOptRIXImutFW: 5’-aattcCAGGAGAGCAAGTACCTGCCTCCGCACgcaCAGCTGTGGGGCCCGCACCAGCTGAG CgcaAAGCCCCTGATGAACTGAc-’ and
y751zxbppauseOptRIXImutRV: 5’-tcgagTCAGTTCATCAGGGGCTTtgcGCTCAGCTGGTGCGGGCCCCACAGCTGtgcGTGCGG AGGCAGGTACTTGCTCTCCTGg-3’
Reporter assay in HEK293 cells
Reporter mRNAs were prepared and transfected as previously described (Iwasaki et al., 2016, 2019). Luminescence was detected with a dual-luciferase reporter assay system (Promega) and GloMax (Promega).
Neutral PAGE
For neutral PAGE, cells were lysed with passive lysis buffer (Promega) and centrifuged at 13,500 × g for 1 min. Supernatants were collected, and equal amounts of total protein were used as protein samples. For RNase(+) samples, RNase A (QIAGEN) was added at a final concentration of 0.05 μg/ml and incubated on ice for 20 min; for RNase(−) samples, Milli-Q water was added instead. After incubation, NuPAGE sample buffer [200 mM Tris-HCl (pH 6.8), 8% w/v SDS, 40% glycerol, 0.04% BPB, and 100 mM DTT] was added and heated at 65°C for 10 min. Proteins were separated by 15% PAGE under neutral pH conditions (pH 6.8) for 4 h with a 50 mA constant current in MES-SDS buffer (1 M MES, 1 M Tris base, 69.3 mM SDS, and 20.5 mM EDTA) and were transferred to a PVDF membrane (Millipore, IPVH00010).
Western blot analysis
To inhibit proteasome, cells were treated with 0.5 μM MG132 (Wako Chemicals) dissolved in DMSO for 6 h, before lysis.
Anti-XBP1 [Cell Signaling Technology (CST), 12782], anti-LTN1 (Abcam, ab104375), anti-β-actin [Medical & Biological Laboratories (MBL), M177–3], anti-HA-Peroxidase (Roche, 12013819001), anti-ZNF598 (Novus Biologicals, NBP1–84658), anti-neomycin phosphotransferase II (Millipore, 06–747), anti-GFP (MBL, 598), anti-MTDH (CST, 14065), anti-eIF5A (BD Biosciences, 611976), and anti-α-tubulin (Sigma-Aldrich, T6074) primary antibodies were used for western blotting. To generate the western blot shown in Figures 7B and S6E, IRDye680- or IRDye800CW-conjugated secondary antibodies (LI-COR, 925–68070/71 and 926–32210/11, respectively) were used to detect proteins, and images were acquired in an Odyssey CLx Infrared Imaging System (LI-COR).
To generate the western blot shown in Figures S6B and 7C, HRP-linked anti-rabbit IgG antibodies (GE Healthcare, NA934) were used for detection and chemiluminescence images were acquired with a LAS 4000 mini (GE Healthcare). To detect zebrafish embryo proteins (Figure S7C) by western blotting, HRP-conjugated anti-rabbit IgG (MBL, 458) and HRP-conjugated anti-mouse IgG (MBL, 330) antibodies were used. Signals were detected with Lumina Forte (Merck Millipore) and an Amersham Imager (GE Healthcare).
Zebrafish microinjection
Capped and polyadenylated sfGFP-Xbp1 mRNAs were synthesized in vitro as described previously (Mishima and Tomari, 2016) and injected into 1-cell stage embryos at a concentration of 50 ng/μl.
QUANTIFICATION AND STATISTICAL ANALYSIS
For Figures 1B and 7D, data represent the mean and SD (n = 3). In Figures 4A–4C, 4E, 5B–5D, 6D, and S4G, false discovery rate (FDR) was calculated with kpLogo pakage (http://kplogo.wi.mit.edu) (Wu and Bartel, 2017). Other statistical analyses were performed on R. In Figures S2D and S4A–S4C, r and ρ stand for Pearson’s and Spearman’s correlations, respectively. Stop codon enrichments in ribosome collision sites (Figure 3B) was calculated by hypergeometric test. In Figure S6D, the significance of the difference of the two distributions was calculated with Wilcoxon’s test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-XBP1 antibody | Cell Signaling Technology | Cat# 12782; RRID: AB_2687943 |
| Rabbit polyclonal anti-LTN1 antibody | Abcam | Cat# ab104375; RRID: AB_10711159 |
| Mouse monoclonal anti-β-actin antibody | Medical & Biological Laboratories | Cat# M177-3; RRID: AB_10697039 |
| Rat monoclonal anti-HA conjugated with Peroxidase | Roche | Cat# 12013819001; RRID: AB_390917 |
| Rabbit polyclonal anti-ZNF598 antibody | Novus Biologicals | Cat# NBP1-84658; RRID: AB_11039996 |
| Rabbit polyclonal anti-neomycin phosphotransferase II antibody | Millipore | Cat# 06-747; RRID: AB_310234 |
| Rabbit polyclonal anti-GFP antibody | Medical & Biological Laboratories | Cat# 598; RRID: AB_591816 |
| Rabbit monoclonal anti-MTDH antibody | Cell Signaling Technology | Cat# 14065; RRID: AB_2798381 |
| Mouse monoclonal anti-eIF5A antibody | BD Biosciences | Cat# 611976; RRID: AB_399397 |
| Mouse monoclonal anti-α-tubulin antibody | Sigma-Aldrich | Cat# T6074; RRID: AB_477582 |
| Goat polyclonal anti-mouse IgG conjugated with IRDye680 | LI-COR | Cat# 925-68070; RRID: AB_2651128 |
| Goat polyclonal anti-rabbit IgG conjugated with IRDye680 | LI-COR | Cat# 925-68071; RRID: AB_2721181 |
| Goat polyclonal anti-mouse IgG conjugated with IRDye800CW |
LI-COR | Cat# 926-32210; RRID: AB_621842 |
| Goat polyclonal anti-rabbit IgG conjugated with IRDye800CW |
LI-COR | Cat# 926-32211; RRID: AB_621843 |
| Donkey polyclonal anti-rabbit IgG antibody conjugated with HRP | GE Healthcare | Cat# NA934; RRID: AB_772206 |
| Goat polyclonal anti-rabbit IgG antibody conjugated with HRP | Medical & Biological Laboratories | Cat# 458; RRID: AB_2827722 |
| Goat polyclonal anti-mouse IgG antibody conjugated with HRP | Medical & Biological Laboratories | Cat# 330; RRID: AB_2650507 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MG132 | Wako Chemicals | Cat# 135-18453 |
| 5-azido-C3-UTP | Jena Bioscience | Cat# NU-157S |
| IRDye 800CW DBCO | LI-COR | Cat# 929-50000 |
| CHX | Sigma-Aldrich | Cat# C4859-1ML |
| ANS | Wako Chemicals | Cat# 017-16861 |
| Cas9 protein and Cas9 Reaction Buffer | New England Biolabs | Cat# M0386T |
| RNase I | Epicenter | Cat# N6901K |
| Proteinase K | Roche | Cat# 3115887001 |
| RNase A | QIAGEN | Cat# 19101 |
| Micrococcal Nuclease | TaKaRa | Cat# 2910A |
| PrimeSTAR Max | TaKaRa | Cat# R045 |
| Critical Commercial Assays | ||
| Rabbit reticulocyte lysate system, nuclease treated | Promega | Cat# L4960 |
| Dual-luciferase reporter assay system | Promega | Cat# E1910 |
| Biodyne B pre-cut modified nylon membrane | Thermo Fisher Scientific | Cat# 77016 |
| T7-Scribe Standard RNA IVT Kit | CELLSCRIPT | Cat# C-AS3107 |
| illustra MicroSpin S-400 HR Column | GE Healthcare | Cat# 27-5140-01 |
| Passive lysis buffer | Promega | Cat# E194A |
| PVDF membrane | Millipore | Cat# IPVH00010 |
| PerfectHyb Plus Hybridization Buffer | Sigma-Aldrich | Cat# H7033-50ML |
| Deposited Data | ||
| DM-tRNA-seq | Zheng et al., 2015 | GEO: GSE66550 |
| Monosome and disome profiling of HEK293 cells, raw and processed data | This study | GEO: GSE145723 |
| Monosome and disome profiling of zebrafish embryos, raw and processed data | This study | GEO: GSE133392 |
| Monosome profiling and RNA-Seq, raw and processed data | Hia et al., 2019 | GEO: GSE126298 |
| DMS-seq data from K562 cells | Rouskin et al., 2014 | GEO: GSE45803 |
| Original images used for the figures | This study | Mendeley Data: https://doi.org/10.17632/9y9bpn4w87.1 |
| Experimental Models: Cell Lines | ||
| HEK293 | American Type Culture Collection (ATCC) | N/A |
| HEK293T | American Type Culture Collection (ATCC) | N/A |
| HEK293T, control shRNA expressed | Matsuo et al., 2017 | N/A |
| HEK293T, ZNF598 shRNA expressed | Matsuo et al., 2017 | N/A |
| HEK293T, LTN1 knocked out | This study | N/A |
| Experimental Models: Organisms/Strains | ||
| Zebrafish AB strain | Zebrafish International Resource Center (ZRC) | ZDB-GENO-960809-7 |
| Oligonucleotides | ||
| ON-TARGETplus Human EIF5A siRNA | Dharmacon | Cat# L-015739-00-0005 |
| ON-TARGETplus Non-targeting Control siRNAs | Dharmacon | Cat# D-001810-01-05 |
| Primers for gRNA preparation | This study | See STAR Methods |
| Recombinant DNA | ||
| psiCHECK2-2A-3xFLAG-XBP1u_pause-2A and 2A-3xFLAG-SBP-2A | This study | N/A |
| pcDNA3.1(+)-HA-XBP1u-V5 | This study | N/A |
| pCS2+sfGFP-zXbp1opt-suv39h1a 3’ UTR | This study | N/A |
| pSpCas9(BB)-2A-Puro (PX459) | Ran et al., 2013 | Addgene, Cat# 48139 |
| Software and Algorithms | ||
| iPAGE | Goodarzi et al., 2009 | https://tavazoielab.c2b2.columbia.edu/iPAGE/ |
| DESeq | Anders and Huber, 2010 | https://bioconductor.org/packages/release/bioc/html/DESeq.html |
| kpLogo | Wu and Bartel, 2017 | http://kplogo.wi.mit.edu |
| GibbsCluster | Andreatta et al., 2017 | http://www.cbs.dtu.dk/services/GibbsCluster-2.0/ |
Highlights.
Disome profiling reveals widespread ribosome collisions in vertebrates
Ribosomes are in queues at Pro-Pro/Gly/Asp, Arg-X-Lys, stop codons, and 3’ UTRs
The positively charged nascent chain weakens the eIF5A-mediated rescue of disomes
The stalled disomes on XBP1u mRNA are an endogenous substrate of RQC
ACKNOWLEDGMENTS
We thank all the members of the Iwasaki laboratory for constructive discussion, technical help, and critical reading of the manuscript. S.I. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas “nascent chain biology” (JP17H05679) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT); a Grant-in-Aid for Young Scientists (A) (JP17H04998) from the Japan Society for the Promotion of Science (JSPS); the Pioneering Projects (“Cellular Evolution”) and the Aging Project from RIKEN; and the Takeda Science Foundation. M.Y. was supported by a Grant-in-Aid for Scientific Research (S) (JP19H05640) from JSPS and a Grant-in-Aid for Scientific Research on Innovative Areas (JP18H05503) from MEXT. Y.M. was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (“nascent chain biology”; JP17H05662) from MEXT and a Grantin-Aid for Scientific Research (B) (JP18H02370) from JSPS. T.I. was supported by a Grant-in-Aid for Scientific Research (A) (JP18H03977) from JSPS and by the Japan Agency for Medical Research and Development (AMED) (JP19gm1110010). DNA libraries were sequenced by the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by an NIH S10 OD018174 Instrumentation Grant. Computations were supported by Manabu Ishii, Itoshi Nikaido, and the Bioinformatics Analysis Environment Service on the RIKEN Cloud at RIKEN ACCC. Y.S. was a JSPS Research Fellow (PD) (JP19J00920).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107610.
REFERENCES
- Anders S, and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta M, Alvarez B, and Nielsen M. (2017). GibbsCluster: unsupervised clustering and alignment of peptide sequences. Nucleic Acids Res. 45 (W1), W458–W463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, and Beckmann R. (2013). Structures of the human and Drosophila 80S ribosome. Nature 497, 80–85. [DOI] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, and Preiss T. (2016). Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574. [DOI] [PubMed] [Google Scholar]
- Arthur L, Pavlovic-Djuranovic S, Smith-Koutmou K, Green R, Szczesny P, and Djuranovic S. (2015). Translational control by lysine-encoding A-rich sequences. Sci. Adv. 1, e1500154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtson MH, and Joazeiro CA (2010). Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, et al. (2012). A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, and Weissman JS (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 16, 651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CM, Stockwell PA, Trotman CN, and Tate WP (1990). Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 18, 6339–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Shao S, Murray J, Hegde RS, and Ramakrishnan V. (2015). Structural basis for stop codon recognition in eukaryotes. Nature 524, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, and Spahn CM (2011). Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol. Cell 44, 214–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk AR, and Green R. (2017). Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, and Ron D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Chan PP, and Lowe TM (2016). GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 44 (D1), D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, and Hurst LD (2013). Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol. 11, e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Hong NA, Masuda CA, Jenkins BV, Nelms KA, Goodnow CC, Glynne RJ, Wu H, Masliah E, Joazeiro CA, and Kay SA (2009). A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl. Acad. Sci. USA 106, 2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, and Weiss B. (2020). Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res. 48, 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, and Tuller T. (2012). Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput. Biol. 8, e1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao Duc K, and Song YS (2018). The impact of ribosomal interference, codon usage, and exit tunnel interactions on translation elongation rate variation. PLoS Genet. 14, e1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell AM, Subramaniam AR, and O’Shea EK (2018). Translational control through differential ribosome pausing during amino acid limitation in mammalian cells. Mol. Cell 71, 229–243.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Gohara DW, and Yap MN (2014). Sequence selectivity of macrolide-induced translational attenuation. Proc. Natl. Acad. Sci. USA 111, 15379–15384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P, Luke GA, Hughes LE, Gani D, Halpin C, and Ryan MD (2006). E unum pluribus: multiple proteins from a self-processing polyprotein. Trends Biotechnol. 24, 68–75. [DOI] [PubMed] [Google Scholar]
- Defenouillère Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, and Fromont-Racine M. (2013). Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl. Acad. Sci. USA 110, 5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diament A, Feldman A, Schochet E, Kupiec M, Arava Y, and Tuller T. (2018). The extent of ribosome queuing in budding yeast. PLoS Comput. Biol. 14, e1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova LN, Kuroha K, Tatematsu T, and Inada T. (2009). Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kubyshkin V, Starosta AL, Wilson DN, Budisa N, and Rodnina MV (2015). Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J. Am. Chem. Soc. 137, 12997–13006. [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, and Yusupov M. (2014). Structural basis for the inhibition of the eukaryotic ribosome. Nature 513, 517–522. [DOI] [PubMed] [Google Scholar]
- Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T, and Sonenberg N. (2017). The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 8, 16056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, and Gladyshev VN (2017). Ribonuclease selection for ribosome profiling. Nucleic Acids Res. 45, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, and Bukau B. (2014). Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol. 24, 24–33. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, and Tavazoie S. (2009). Revealing global regulatory perturbations across human cancers. Mol. Cell 36, 900–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Crawford ED, O’Donovan BD, Wilson MR, Chow ED, Retallack H, and DeRisi JL (2016). Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 17, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, and Dever TE (2013). eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, and Green R. (2014). Dom34 rescues ribosomes in 3’ untranslated regions. Cell 156, 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh NR, and Green R. (2017). Translation of poly(A) tails leads to precise mRNA cleavage. RNA 23, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, and Papa FR (2018). The unfolded protein response and cell fate control. Mol. Cell 69, 169–181. [DOI] [PubMed] [Google Scholar]
- Hia F, Yang SF, Shichino Y, Yoshinaga M, Murakawa Y, Vandenbon A, Fukao A, Fujiwara T, Landthaler M, Natsume T, et al. (2019). Codon bias confers stability to human mRNAs. EMBO Rep. 20, e48220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, et al. (2015). The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell 59, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, and Press WH (2015). Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 11, e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim F, Maragkakis M, Alexiou P, and Mourelatos Z. (2018). Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 25, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K, Tesina P, Matsuo Y, Sugiyama T, Cheng J, Saeki Y, Tanaka K, Becker T, Beckmann R, and Inada T. (2019). Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, e100276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada T. (2017). The ribosome as a platform for mRNA and nascent polypeptide quality control. Trends Biochem. Sci. 42, 5–15. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, and Weissman JS (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, and Weissman JS (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, and Atkins JF (2008). uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc. Natl. Acad. Sci. USA 105, 10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Shin BS, Loughran G, Tzani I, Young-Baird SK, Cao C, Atkins JF, and Dever TE (2018). Polyamine control of translation elongation regulates start site selection on antizyme inhibitor mRNA via ribosome queuing. Mol. Cell 70, 254–264.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Floor SN, and Ingolia NT (2016). Rocaglates convert DEADbox protein eIF4A into a sequence-selective translational repressor. Nature 534, 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Iwasaki W, Takahashi M, Sakamoto A, Watanabe C, Shichino Y, Floor SN, Fujiwara K, Mito M, Dodo K, et al. (2019). The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol. Cell 73, 738–748.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, and Hegde RS (2017). Initiation of quality control during poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell 65, 743–750.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juszkiewicz S, Chandrasekaran V, Lin Z, Kraatz S, Ramakrishnan V, and Hegde RS (2018). ZNF598 is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda S, Yanagitani K, Yokota Y, Esaki Y, and Kohno K. (2016). Autonomous translational pausing is required for XBP1u mRNA recruitment to the ER via the SRP pathway. Proc. Natl. Acad. Sci. USA 113, E5886–E5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova KK, Hickey KL, Osuna BA, Hussmann JA, Frost A, Weinberg DE, and Weissman JS (2017). CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha K, Zinoviev A, Hellen CUT, and Pestova TV (2018). Release of ubiquitinated and non-ubiquitinated nascent chains from stalled mammalian ribosomal complexes by ANKZF1 and Ptrh1. Mol. Cell 72, 286–302.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshminarayan R, Phillips BP, Binnian IL, Gomez-Navarro N, Escudero-Urquijo N, Warren AJ, and Miller EA (2020). Pre-emptive quality control of a misfolded membrane protein by ribosome-driven effects. Curr. Biol. 30, 854–864.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, and Brown PO (2014). Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife 3, e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Iwakoshi NN, Anderson KC, and Glimcher LH (2003). Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA 100, 9946–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim VI, and Curran JF (2001). Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA 7, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, and Deutsch C. (2008). Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath H, Zhang H, Rehfeld F, Han J, Chang TC, and Mendell JT (2019). Suppression of ribosomal pausing by eIF5A is necessary to maintain the fidelity of start codon selection. Cell Rep. 29, 3134–3146.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, and Hegde RS (2010). A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature 466, 1120–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K, et al. (2017). Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 8, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughan KK, Brown CM, Dalphin ME, Berry MJ, and Tate WP (1995). Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl. Acad. Sci. USA 92, 5431–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy NJ, and Ingolia NT (2017). Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Rigger L, Neuner S, Shin BS, Yusupova G, Dever TE, Micura R, and Yusupov M. (2016a). Molecular insights into protein synthesis with proline residues. EMBO Rep. 17, 1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov S, Mailliot J, Shin BS, Rigger L, Yusupova G, Micura R, Dever TE, and Yusupov M. (2016b). Crystal structure of hypusine-containing translation factor eIF5A bound to a rotated eukaryotic ribosome. J. Mol. Biol. 428, 3570–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen TP, and Björklund M. (2015). Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3’ untranslated regions. Nucleic Acids Res. 43, 1019–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EW, Wangen J, Green R, and Ingolia NT (2016). Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, and Tomari Y. (2016). Codon usage and 3’ UTR length determine maternal mRNA stability in zebrafish. Mol. Cell 61, 874–885. [DOI] [PubMed] [Google Scholar]
- Mohammad F, Green R, and Buskirk AR (2019). A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife 8, e42591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Lyon K, DeLuca KF, DeLuca JG, English BP, Zhang Z, Lavis LD, Grimm JB, Viswanathan S, Looger LL, et al. (2016). Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429. [DOI] [PubMed] [Google Scholar]
- Nakatogawa H, and Ito K. (2002). The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636. [DOI] [PubMed] [Google Scholar]
- Osuna BA, Howard CJ, Kc S, Frost A, and Weinberg DE (2017). In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing. eLife 6, e27949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, and Alepuz P. (2017). eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 45, 7326–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, and Koller D. (2014). Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 10, 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P; Bravo IG, Garcia-Vallvé S. (2008). E-CAI: a novel server to estimate an expected value of Codon Adaptation Index (eCAI). BMC Bioinformatics 9, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, and Weissman JS (2014). Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C, Becker T, Heuer A, Braunger K, Shanmuganathan V, Pech M, Berninghausen O, Wilson DN, and Beckmann R. (2016). Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 44, 1944–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, and Green R. (2017). eIF5A functions globally in translation elongation and termination. Mol. Cell 66, 194–205.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmuganathan V, Schiller N, Magoulopoulou A, Cheng J, Braunger K, Cymer F, Berninghausen O, Beatrix B, Kohno K, von Heijne G, and Beckmann R. (2019). Structural and mutational analysis of the ribosomearresting human XBP1u. eLife 8, e46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, von der Malsburg K, and Hegde RS (2013). Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell 50, 637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AK, Sormanni P, Ahmed N, Ciryam P, Friedrich UA, Kramer G, and O’Brien EP (2019). A chemical kinetic basis for measuring translation initiation and elongation rates from ribosome profiling data. PLoS Comput. Biol. 15, e1007070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. (2015). Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, and Zaher HS (2017). Ribosome collision is critical for quality control during No-go decay. Mol. Cell 68, 361–373.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, and Brandman O. (2019). CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat. Struct. Mol. Biol. 26, 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, Atkinson GC, Brauer A, Remm M, Tenson T, Schulten K, et al. (2014). Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc. Natl. Acad. Sci. USA 111, 9804–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sothiselvam S, Neuner S, Rigger L, Klepacki D, Micura R, Vázquez-Laslop N, and Mankin AS (2016). Binding of macrolide antibiotics leads to ribosomal selection against specific substrates based on their charge and size. Cell Rep. 16, 1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein KC, Kriel A, and Frydman J. (2019). Nascent polypeptide domain topology and elongation rate direct the cotranslational hierarchy of Hsp70 and TRiC/CCT. Mol. Cell 75, 1117–1130.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, and O’Shea EK (2014). An integrated approach reveals regulatory controls on bacterial translation elongation. Cell 159, 1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Lee H, Dominguez D, Heiman M, and Burge CB (2018). Widespread accumulation of ribosome-associated isolated 3’ UTRs in neuronal cell populations of the aging brain. Cell Rep. 25, 2447–2456.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, and Bennett EJ (2017). ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell 65, 751–760.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Reichermeier KM, Burroughs AM, Oania RS, Reitsma JM, Aravind L, and Deshaies RJ (2018). Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Han B, Zhou R, and Zhuang X. (2016). Real-time imaging of translation on single mRNA transcripts in live cells. Cell 165, 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg DE, Shah P, Eichhorn SW, Hussmann JA, Plotkin JB, and Bartel DP (2016). Improved ribosome-footprint and mRNA measurements provide insights into dynamics and regulation of yeast translation. Cell Rep. 14, 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Arenz S, and Beckmann R. (2016). Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr. Opin. Struct. Biol. 37, 123–133. [DOI] [PubMed] [Google Scholar]
- Winz ML, Peil L, Turowski TW, Rappsilber J, and Tollervey D. (2019). Molecular interactions between Hel2 and RNA supporting ribosome-associated quality control. Nat. Commun. 10, 563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, and Walter P. (1988). Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7, 3559–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Guydosh NR, Green R, and Buskirk AR (2015). High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, and Bartel DP (2017). kpLogo: positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 45 (W1), W534–W538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Eliscovich C, Yoon YJ, and Singer RH (2016). Translation dynamics of single mRNAs in live cells and neurons. Science 352, 1430–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Zinshteyn B, Wehner KA, and Green R. (2019a). High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol. Cell 73, 959–970.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Tantray I, Lim J, Chen S, Li Y, Davis Z, Sitron C, Dong J, Gispert S, Auburger G, et al. (2019b). MISTERMINATE mechanistically links mitochondrial dysfunction with proteostasis failure. Mol. Cell 75, 835–848.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Hoek TA, Vale RD, and Tanenbaum ME (2016). Dynamics of translation of single mRNA molecules in vivo. Cell 165, 976–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, and Kohno K. (2009). Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol. Cell 34, 191–200. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, and Kohno K. (2011). Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science 331, 586–589. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, and Mori K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Oku M, Suzuki M, and Mori K. (2006). pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J. Cell Biol. 172, 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, and Green R. (2015). Rli1/ABCE1 recycles terminating ribosomes and controls translation reinitiation in 3’UTRs in vivo. Cell 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Hu H, Zhou J, He X, Jiang T, and Zeng J. (2017). Analysis of ribosome stalling and translation elongation dynamics by deep learning. Cell Syst. 5, 212–220.e6. [DOI] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, and Pan T. (2015). Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for monosome profiling and disome profiling of HEK293 cells and zebrafish reported in this paper are GEO: GSE145723 and GEO: GSE133392. All custom scripts of R used for data analysis are available upon request. Original images used for the figures have been deposited to Mendeley Data: https://doi.org/10.17632/9y9bpn4w87.1.