Abstract
Synthetic biology strives to reliably control cellular behavior, typically in the form of user-designed interactions of biological components to produce a predetermined output. Engineered circuit components are frequently derived from natural sources and are therefore often hampered by inadvertent interactions with host machinery, most notably within the host central dogma. Reliable and predictable gene circuits require the targeted reduction or elimination of these undesirable interactions to mitigate negative consequences on host fitness and develop context-independent bioactivities. Here, we review recent advances in biological orthogonalization, namely the insulation of researcher-dictated bioactivities from host processes, with a focus on systematic developments that may culminate in the creation of an orthogonal central dogma and novel cellular functions.
Keywords: Bioengineering, directed evolution, biological orthogonalization, genetic circuit design, synthetic biology
Engineering gene circuits
Natural gene circuits rely on regulatory factors to govern cellular activities, leveraging elements of both circuit insulation and interaction to afford the necessary response(s) [1]. These natural gene circuits are maintained through discrete functional modules, which allow for both robustness and plasticity in adapting to changing environmental conditions [2]. Over the past two decades, components of natural gene circuits have been isolated and repurposed to develop increasingly complex engineered gene circuits [3], wherein multiple user-defined inputs may act in concert to produce the desired outputs [4]. Increasing circuit complexity has been driven by the incremental realization of synthetic biology goals: controlled gene expression [5], genetic code expansion [6] and development of synthetic cells [7]. In turn, these and other accomplishments have catalysed significant research into more complex genetic circuitry that continue to co-opt host cell machinery and resources for researcher-dictated bioactivities.
Historically, engineered gene circuits have been implemented at the transcriptional and post-transcriptional levels [4,5,8–11], integrating RNAs [9–11], proteins [5,8], or combinations thereof [12]. Engineered circuits, therefore, rely heavily on elements within the central dogma of the host to generate the necessary biomolecules, which can inadvertently reduce host fitness by depleting essential resources or by enforcing non-native functions onto pre-existing machinery [12–19]. Efforts to improve engineered circuit performance and overcome pleiotropic consequences on the host have focused on limiting component overproduction by affecting transcription or translation rates [12]. While this is an important starting point for circuit optimization, these ad hoc approaches may similarly disrupt the balance of native cellular components [14]. Furthermore, empirical approaches for improving synthetic circuits and their components become particularly challenging when circuits grow in complexity, as the combinations of possible modifications grows exponentially [13].
To date, bottlenecks in engineering more complex gene circuits have stemmed from two broad factors: (i) engineered circuits typically repurpose components that are optimized to function within their native contexts, and which are not immediately suited for user-defined objectives, such as high levels of basal expression in an off-state [20], and (ii) their performance is often limited by incompatibility of components, such as cross-talk or toxicity in the chosen host [21]. Insulation of these components from host processes (Biological Orthogonalization; Box 1) has been described as early as the 1960s [22] (Figure 1) and has significantly improved the predictability of engineered circuits by limiting known instances of cellular cross-talk [5]. An orthogonal and user-controlled paralogue [23] of the central dogma may therefore continue to improve the reliability of engineered circuits, and enable increasingly numerous elements to co-exist alongside host components without adverse cellular effects. In this review, we discuss prior studies aimed at segregating synthetic and host elements, and take inspiration from native biological systems to selectively partition, compartmentalize, or orthogonalize native and/or engineered components to enable specific goals. As modern engineered gene circuits continue to rely heavily on the host central dogma, we discuss these developments with a focus on their integration to furnish an orthogonal central dogma, wherein elements of information maintenance, transfer, and translation have been modified and/or insulated from host interactions.
Box 1. Biological Orthogonalization.
The term “orthogonal” or “orthogonality” in synthetic biology describes the inability of two or more biomolecules, similar in composition and/or function, to interact with one another or affect their respective substrates. For example, two proteases may be mutually orthogonal if they are unable to cleave one another’s respective substrates, or two aminoacyl-tRNA synthetases that do not cross aminoacylate their non-cognate tRNA. By necessity, all biomolecules within a cell should not interact directly with one another. This allows key processes such as those within the central dogma to perform discrete functions (replication, transcription and translation) while remaining intrinsically linked as each function is dependent on the prior stages. As it relates to the development of engineered gene circuits, the necessary degree of orthogonality depends on the user-defined objectives. For example, it may be necessary to orthogonally control the transcription of multiple genes using independent promoters in an engineered gene circuit. Here, transcription factors (TFs) with known mutual orthogonality in their respective operator sequences may be sufficient. Conversely, more complex goals such as the large-scale implementation of expanded genetic codes will undoubtedly require a larger repertoire of orthogonal elements.
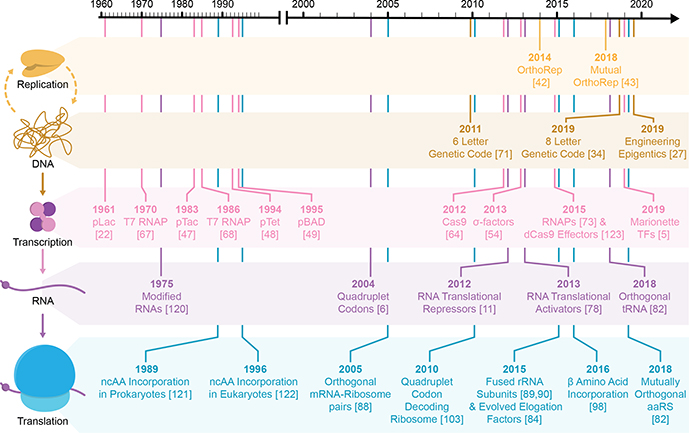
Fig. 1 |. Key Figure| Timeline of Developing Orthogonal Elements.
Schematic representation of key orthogonal elements discovered through investigation of natural biological systems, biomolecular engineering, and/or directed evolution. Elements are segregated by colour according to their order within the central dogma: genetic information storage and replication (yellow/orange), transcription (pink/purple) and translation (blue) [121–123]. In each case, the year of their first literature report is provided.
Orthogonal genetic information-storage and replication
Insulation via non-canonical nucleobases:
Engineered gene circuits can be introduced into a host chassis as standalone episomes [24] or integrated into the host genome [25]. As inadvertent recognition of engineered sequences by host components may affect circuit performance in unpredictable ways, mechanisms that insulate exogenous genetic information from host machinery would limit reliance on the host for replication or gene expression [26]. Epigenetic modification of nucleic acids is among the most common mechanisms exploited by natural biological systems to insulate specific cellular processes. Taking inspiration from these natural systems, Khalil and colleagues recently established an orthogonal methodology to control gene transcription in eukaryotic cells through epigenetic signalling [27]. This system uses a modified nucleobase commonly found in prokaryotes but absent in eukaryotic genomes, N6-methyldeoxyadenosine (m6dA), by porting the requisite methyltransferase and transcription factors, thereby enabling efficient and orthogonal information storage and propagation (Figure 2A).
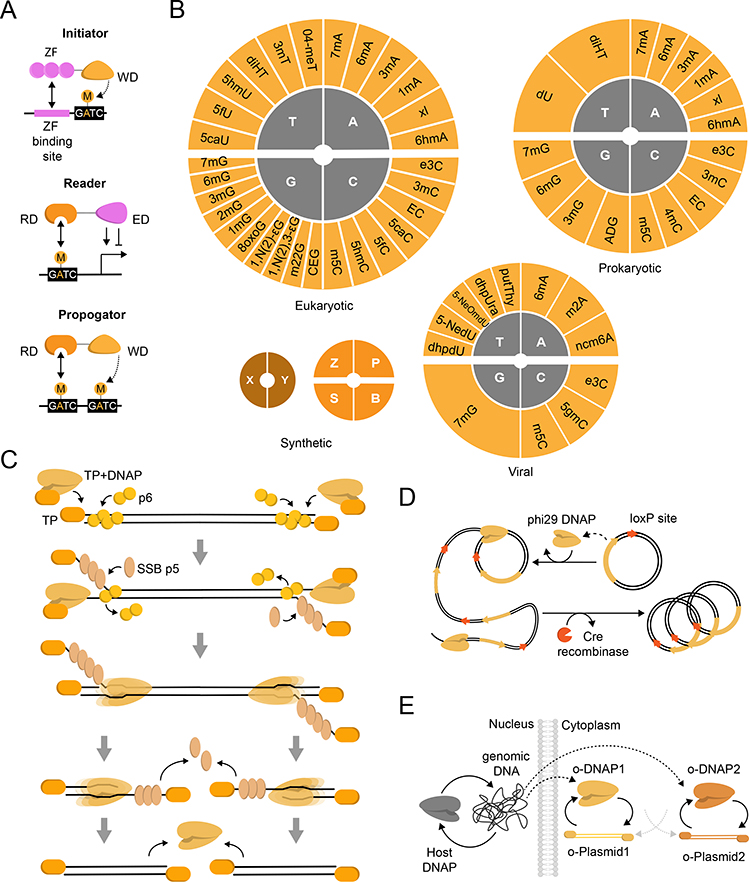
Fig. 2 |. Orthogonal Genetic Information.
Schematic representation of components fulfilling aspects of orthogonal genetic information maintenance and propagation. Grayscale elements represent canonical or host-associated components. A) A system for programable DNA modification composed of two main elements: a writer domain (WD) and reader domain (RD). These can be used to propagate DNA modification and affect gene expression by incorporating an effector domain (ED). B) Naturally-occurring nucleic acid modifications [28], which have been expanded with synthetic nucleobases: X–Y rely on hydrophobic interactions, whereas Z–P and S–B use hydrogen bonding reminiscent of natural Watson-Crick base pairing. C) An overview of linear φ29 bacteriophage genome replication [39]. D) An orthogonal transcription and translation-coupled DNA replication system (TTcDR) consisting of a circular DNA containing the φ29-DNAP gene and a loxP site. φ29-DNAP initiates rolling-circle replication on circular DNA, yielding two complementary ssDNA of tandem repeating sequence. Cre recombinase acts on loxP sites to produce circular products [40]. E) The yeast-derived orthogonal replication system OrthoRep consists of mutually orthogonal cytosolic DNAP-plasmid pairs. Separation by the nuclear envelope prevents crosstalk with the host genome or DNAP(s) [44].
A great diversity of DNA modifications exist in natural genomes [28] (Figure 2B), wherein the abundance of non-canonical nucleobases may make their genetic information innately orthogonal to the host machinery. For example, the genomes of a eukaryotic multicellular organism may be identical in sequence but produce distinct gene expression patterns through alternative epigenetic profiles [27]. Such modifications may also provide protective functions to cellular circuits. Bacteriophage genomes can have partial or complete substitution of nucleobases by modified variants [26], which are thought to resist endonuclease-mediated digestion upon infection. However, incorporation of non-canonical nucleobases may require the action of dedicated polymerases to facilitate replication and propagation [29]. Modified nucleotides in bacteriophage are generated primarily through enzymatic modification of cellular precursors [26], however some bacteriophage components can introduce secondary nucleotide modifications following replication [26,30]. It may therefore be possible to exploit these modifications to program additional layers of orthogonal information maintenance in non-natural hosts.
The observation of non-canonical nucleobases in natural systems has motivated significant interest in incorporating synthetic nucleotide pairs into cellular DNA, where such efforts could enable genetic code expansion through increased sequence diversity. Recent work to expand the genetic code has recently resulted in accretion from the canonical four to six [31–33] or eight [34] synthetic nucleobase codes, thereby increasing the possible information density while reducing the potential for nucleobase-host component interactions (Figure 2B). While these systems have expanded the repertoire of viable codons in vivo for non-canonical amino acid (ncAA) incorporation [33], they require in vitro synthesis of (deoxy)nucleoside triphosphates for cellular uptake to maintain noncanonical information transfer [35]. Similarly, non-canonical sugars in nucleic acids have served as a basis for artificial information maintenance and transfer using evolved polymerases [36]. Finally, nucleic acid phosphate backbones are known to be extensively modified to phosphorothioate across diverse bacterial genera [37], an interesting example of an artificial innovation that was later discovered to occur naturally in biological systems. Recent work by Holliger and colleagues extended this observation by creating alkyl phosphonate nucleic acids, showcasing how such modifications can give rise to novel biomolecular interactions in evolved aptamers [38].
Orthogonal DNA replication
Whereas non-canonical nucleobases may limit interactions with host machinery, a complementary mechanism leverages orthogonal replication of genetic information. Among well-studied systems, orthogonal replication of the lytic Bacillus φ29 bacteriophage genome is perhaps the most well understood, and used as a model for protein-primed replication [39]. φ29 DNA replication is initiated asynchronously at both ends of the linear genome, wherein a terminal protein (TP)-DNA polymerase (DNAP) heterodimer recognizes the protein-capped replication origins (Figure 2C). The DNAP then dissociates from the TP and continues processive elongation coupled to strand displacement. Continuous elongation by two DNAPs gives rise to the complete duplication of the parental strands [39]. Taking inspiration from natural viral replication systems, φ29 bacteriophage components have enabled minimally self-replicating DNA systems in vitro (Figure 2D). Ichihashi and colleagues developed a minimal transcription- and translation-coupled DNA replication (TTcDR) system wherein a plasmid encoding its own DNAP replicates by rolling circle amplification [40]. Following amplification, Cre recombinase loxP sites facilitate re-circularization of the nascent DNA, with rounds of directed evolution employed to improve circularization efficiency by 60-fold. Danelon and colleagues [7] recently reported the self-replication of a linear DNA flanked by φ29 bacteriophage replication origins in liposome-based synthetic cells. While both systems demonstrate the capacity of φ29 bacteriophage components to support an orthogonal DNA replication modality in vitro, their extension to cellular systems has not yet been demonstrated, suggesting the potential toxicity of φ29 protein production in vivo.
Natural auxiliary genetic information has been noted beyond bacteriophage systems, including cytosolic plasmids, mitochondrial, and chloroplast genomes. The latter two genomes exist through symbiotic relationships to their host and have evolved to have share components of their replication machinery. A minimal mitochondrial DNA replication system has been recapitulated in vitro, consisting of only of two protein factors: DNA polymerase POL𝛾 and TWINKLE helicase [41]. As above, this system has not yet been extended to a heterologous host, likely a reflection of slow polymerase elongation rate (280 bp/min) and a requirement of two asymmetric origins of replication.
Recently, an orthogonal DNA replication (OrthoRep) system has been established in yeast to realize cellular mutation rates beyond the error catastrophe threshold while mitigating negative consequences on host fitness (Figure 2E) [42–44]. OrthoRep builds upon the native cytoplasmic plasmids of Kluveromyces lactis to enable orthogonal DNA replication, wherein an orthogonal DNAP is generated from the host genome using native machinery and exclusively replicates the cognate cytoplasmic plasmid. Critically, OrthoRep operates exclusively in the cytoplasm whereas host genome replication occurs in the nucleus, and plasmid propagation relies on protein-primed replication. This system has been elaborated to generate two mutually orthogonal and host-independent DNAPs [43], has strain-independent DNA replication activity [45] and has been used to evolve robust drug resistance and improved enzymatic capabilities [46]. Together, this system highlights the utility of leveraging both non-canonical initiation and compartmentalization to facilitate orthogonal DNA replication.
Orthogonal transcriptional regulation
Small molecule inducible transcription factors
Beyond the development and maintenance of orthogonal DNA, mechanisms for insulating transcriptional and regulatory elements have historically served as the foundation for synthetically controlling gene expression, building upon pioneering studies of inducible systems [47–49]. Rules governing model inducer-responsive transcriptional units have been established and natural regulatory components have been exploited through engineering and directed evolution to yield synthetic paralogues more suited to their new contexts [5]. Inducible systems classically consist of three components: a protein transcription factor (TF), a DNA motif specifically bound by the TF, and a non-toxic, cell-permeable small molecule that promotes or inhibits the binding of a TF to its cognate DNA motif (Figure 3A). Though not typically identified as orthogonal systems during their development, the success of small molecule-inducible systems has derived from significant efforts to insulate their functionality from host machinery and to limit crosstalk with pre-existing components [5]. This includes the development of non-metabolizable (e.g. Isopropyl β-D-1-thiogalactopyranoside; IPTG) or non-toxic (e.g. anhydrotetracycline; aTc) inducers, elimination of native pathways for inducer detection or catabolism (e.g. lactose and arabinose catabolism), or modulation of TFs to mitigate pleiotropic effects (e.g. IPTG and arabinose crosstalk) [50]. Additionally, to overcome undesirable characteristics that had been previously identified in prokaryotic inducible systems (i.e., noise, low dynamic range and poor sensitivity), Voigt and colleagues developed a panel of twelve mutually orthogonal TFs using an in vitro compartmentalized self-replication (CSR) directed evolution strategy, providing a robust set of orthogonal transcriptional regulators [5].
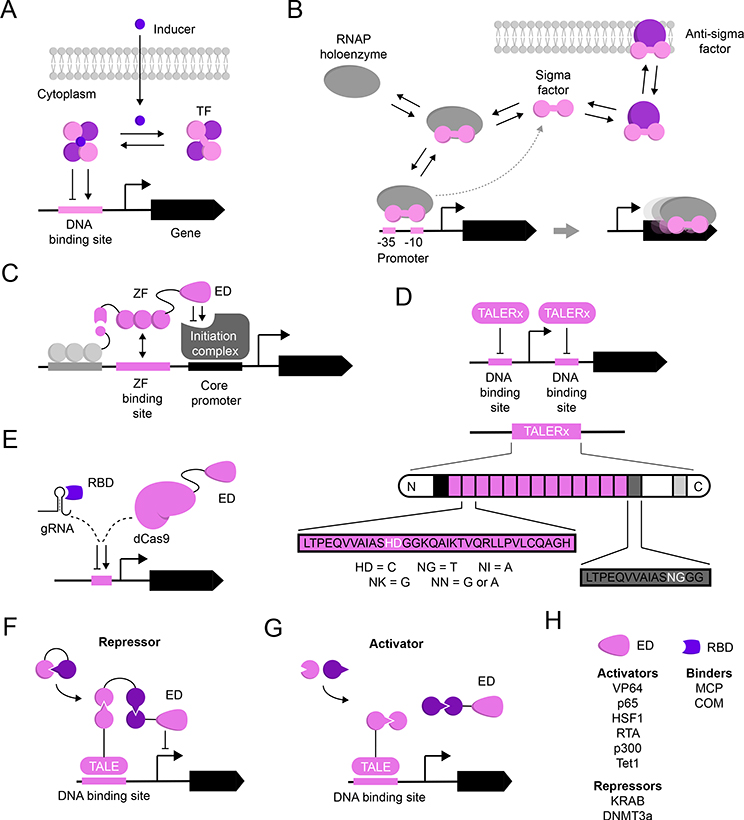
Fig. 3 |. Orthogonal Transcription Components.
Schematic representation of components fulfilling aspects of orthogonal transcription. Grayscale elements represent canonical or host-associated components. A) Small molecule-inducible transcription factors (TF): transcriptional units consist of a TF which binds to its cognate promoter in a small molecule-dependant manner [5]. B) Extracytoplasmic sigma (σ-factors): membrane-anchored anti-σ-factors bind their cognate σ-factor to inhibit association with the host RNAP holoenzyme. Following an external stimulus, the σ-factor dissociates from its anti-σ-factor, thereby allowing for σ-RNAP interaction and initiating transcription from its cognate promoter [54]. C) Zinc fingers (ZFs) are modular domains found in many eukaryotic TFs that make sequence-specific contacts with cognate DNA binding sites. Synthetic ZFs have been employed as programable TFs to define transcriptional states by fusing it with an effector domain (ED) [58]. D) TALERx is a programable TF platform based on transcription activator-like effectors (TALEs). Recognition is imparted by two residues in a repeating peptide sequences (pink rectangles with black outline) and correspond to specific nucleotide sequence in a designed DNA sequence [59]. E) Engineered CRISPR/dCas9-based transcriptional regulators: an engineered guide RNA (gRNA) enables dCas9 binding to the DNA sequence of interest. Either the protein (dCas9) or the gRNA can be modified to include additional functionality by recruiting trans-activating proteins [124]. F) De novo designed logic gates based on heterodimeric protein-protein interaction pairs [66]. These modular heterodimers can be used to generate transcriptional repressors (F) or activators (G). H) Effector domains and RNA-binding domains used to control gene expression [124].
Sigma factors
Natural and engineered TFs often regulate transcriptional outcomes via occlusion of effectors [51]. However, some natural effectors can orthogonally regulate transcriptional output using more direct modalities, and have been employed to control gene expression in vivo. Bacterial sigma factors (σ-factors) and anti-σ-factors are essential components of RNA polymerases (RNAPs), which are employed to differentially affect genomic promoters under variable environmental conditions [52,53]. Natively, the membrane-anchored anti-σ-factors bind their cognate σ-factor to inhibit association with the host RNAP holoenzyme. Following an external stimulus, the anti-σ-factor dissociates from its cognate σ-factor, thereby allowing for σ-RNAP interaction and initiating transcription from its cognate promoter (Figure 3B) [54]. While many stimuli remain unknown, numerous σ-factors show mutual and host orthogonality in Escherichia coli [54,55]. Of these, extracytoplasmic function (ECF) σ-factors are the smallest and most diverse in terms of sequence [56,57]. Rhodius and colleagues investigated the mutual orthogonality of 86 ECF σ-factors from phylogenetically diverse organisms in E. coli [54]. Following significant promoter optimization of components derived from GC rich genomes, 20 σ-factor-promoter pairs were found to be mutually orthogonal. Notably, ECF σ-factor and promoter chimeras expanded this panel to 24 mutually orthogonal pairs. Finally, anti-σ-factors can be used as threshold-gates to minimize leaky expression from orthogonal σ-factor-dependent promoters [54].
Programable transcription factors
Whereas steric occlusion of transcriptional machinery is common regulatory modality in prokaryotic systems, many natural and engineered eukaryotic systems will often recruit specific factors to defined DNA motifs to affect transcriptional regulation [51]. Programable TFs have historically required the expression of orthogonal DNA-binding protein modules to regulate transcriptional circuits, incorporating zinc fingers (ZFs) (Figure 3C) [58] or transcription activator-like effectors (TALEs) (Figure 3D) [59] fused to relevant activating [60–63] or repressive [64] effector domains (EDs). In the last decade, the nuclease-null CRISPR-Cas (dCas) proteins have seen tremendous success for increasingly orthogonal regulation of gene expression, and benefit from utilization of a single protein component alongside engineered guide RNAs (gRNA) (Figure 3E). Programmable dCas-based TFs have served as the basis for multi-input transcriptional enhancers and repressors in mammalian cells [65], owing to the relative ease in programmability as compared to ZFs and TALEs.
Mirroring the utility of orthogonal dCas/sgRNA homologs in eukaryotic gene regulation, Baker and colleagues developed a new approach for orthogonal regulation of gene expression in vivo through de novo protein-protein interaction logic gates [66] (Figure 3F-G). Importantly, these logic gates implement 2-component modules that are mutually orthogonal heterodimeric pairs, have tuneable binding affinities, and associate cooperatively to limit sensitivity to stoichiometric input changes. As these heterodimeric pairs interact selectively, this method will likely see facile integration into existing technologies for regulation by small molecules, sigma-factors, CRISPRs, TALEs, ZFs and/or other effector domains (Figure 3H).
Orthogonal RNAPs and non-canonical nucleobases:
Naturally-derived and engineered prokaryotic and eukaryotic TFs rely on the host transcriptional machinery to regulate gene expression. Compared to these multicomponent systems, bacteriophage-derived RNAPs provide singular polypeptide components that are naturally orthogonal and do not require supplementary cellular factors to affect transcription. T7 RNAP has proven to be particularly versatile due to its specific recognition of a short 17-bp promoter [67,68], with extensive prokaryotic, eukaryotic, and in vitro applications. While additional mutually orthogonal bacteriophage-derived RNAPs are known [69], Ellington and colleagues bolstered this set using compartmentalized partnered replication (CPR) to afford a panel of mutually orthogonal T7 RNAP variants [70]. Additionally, the versatility of bacteriophage RNAPs has been extended to the polymerization of orthogonal nucleobases in live cells [71,72]. Wildtype T7 RNAP has been used to transcribe mRNAs and tRNAs containing the unnatural nucleotides dNaM (X) and dTPT3 (Y) for an expanded six letter genetic alphabet in vivo [71]. The engineered T7-FAL (Y639F, H784A, P266L) variant [73] can support transcription of the eight letter “hachimoji” code in vitro [34], whereas wildtype T7 RNAP can support the polymerization of all but one nucleotide, failing to incorporate riboS opposite deoxyriboB [34]. The extensibility of orthogonal RNAPs to accommodate orthogonal synthetic DNA marks a significant milestone in multi-layered orthogonality, and future efforts will likely continue to engineer and/or evolve RNAPs to improve their transcription capabilities for non-canonical nucleobases.
Orthogonal translation
Post-transcriptional regulation
Following transcription, mRNAs can be differentially affected via higher order regulatory elements in natural biological systems [74–76], which may influence the functionality of engineered gene circuits. A recent surge in the detection and functional assessment of RNA modifications has revealed roles in dynamic regulation of mRNAs (Figure 4A) [77], suggesting that future engineering efforts may co-opt these modifications to further insulate or affect synthetic circuits. Additionally, regulatory small RNAs (sRNAs) can have profound impacts on mRNA translation (Figure 4B, C). As sRNAs operate via canonical Watson-Crick base-pairing, they offer an attractive engineering modality to orthogonally control protein abundance. Indeed, this principle has been recently abstracted to generate synthetic orthogonal RNA regulators of translation in an effort to improve control over protein expression [10,11,78,79]. Recently, Green, Yin and colleagues developed 15 mutually orthogonal sRNA–mRNA pairs with a dynamic range of up to 300–fold [10]. These devices enable rapid, multi-input Boolean logic gates in E. coli that do not require translation of the actuator to function, as is the case in protein-based cellular logic. Similar mechanisms have previously been demonstrated [11,78,79], all of which selectively alter the mRNA 5’ UTR secondary and/or tertiary structure to modulate RBS or start codon availability for translational initiation.
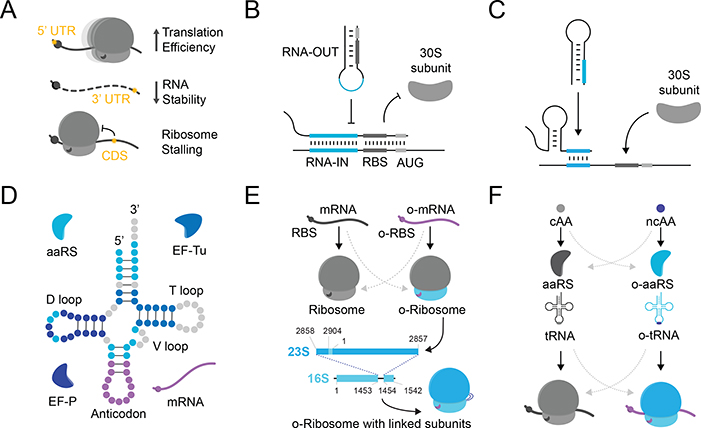
Fig. 4 |. Orthogonal Translation Components.
Schematic representation of components fulfilling aspects of orthogonal translation and regulation. Grayscale elements represent canonical or host-associated components. A) Modified nucleobases found in mRNA can influence translation efficiency and/or mRNA stability [75]. B) RNA-based translational repressors can be programmed to restrict ribosome initiation in vivo [11]. C) RNA-based translational activators can be engineered to liberate ribosome translation initiation sites in vivo [78]. D) Efforts to engineer tRNAs have affected interactions with various protein-translation factors, further orthogonalizing these biomolecules from host machinery [125]. E) Orthogonal ribosomes, rely on an o-RBS/o-antiRBS interaction, can be used to selectively direct ribosome initiation to an orthogonal transcript. Covalent linkage of circularly-permutated rRNAs further improves cellular orthogonality [88,89]. F) Genetic code expansion via ncAAs: to incorporate ncAAs into proteins, the host is supplemented with an orthogonal aminoacyl-tRNA synthetase (o-aaRS) that selectively charges a cognate orthogonal (o-tRNA) with the ncAA.
Orthogonal tRNA engineering
The extensive bioactivities required for tRNA maturation have made their effectors targets for engineering and directed evolution (Figure 4D), and have been aimed largely at improving ncAA incorporation in vivo [80]. Broad genetic code expansion efforts have seen the development of orthogonal aminoacyl-tRNA synthetase (aaRS)–tRNA pairs to incorporate ncAAs across diverse prokaryotic and eukaryotic cellular systems [81]. Orthogonal aaRS enzymes are well suited to discriminate against native cellular components as each native aaRS-tRNA pair is mutually orthogonal, permitting their directed evolution to catalyse ncAA incorporation. Recently, directed evolution of tRNAs was shown to improve mutual orthogonality for aaRSs in E. coli through manipulation of the variable loop region, a known determinant of aaRS–tRNA interactions [82]. Rational engineering of the elongation factor Tu (EF-Tu)–tRNA interface can similarly improve ncAA incorporation in vivo by improving interactions necessary to shuttle ncAA-charged tRNAs to the ribosome [83]. The elongation factor P (EF-P) has been suggested as a candidate for further improvement [84]. These extensive developments have been collated to significantly expand the amino acid diversity in vivo by incorporating up to three unique ncAAs using multiple approaches for codon reassignment [85]. Future efforts, which will likely integrate recently demonstrated modes for codon expansion using synthetic bases [86] and mutually orthogonal aaRS-tRNA pairs [87], are expected to enable a larger repertoire of ncAAs to be incorporated into a contiguous polypeptide chain.
Dedicated, mRNA-specific orthogonal ribosomes
The ribosome translates the complete cellular protein repertoire and serves as a signalling nexus to affect cell fate decisions and growth dynamics. Accordingly, inadvertent effects on translation from engineered genetic circuits can have devastating consequences on cellular fitness [14]. To mitigate these effects, a specific and orthogonal ribosome (o-ribosome) population can be generated to exclusively translate researcher-defined orthogonal mRNAs (o-mRNAs) (Figure 4E). Orthogonal translation has been developed in prokaryotes and integrates an altered, orthogonal anti-ribosome binding site (o-antiRBS) in the 16S rRNA to selectively initiate protein translation using a corresponding o-RBS [88]. However, both the orthogonal and native 16S rRNAs can freely associate with wild-type 23S rRNAs, leading to the production of heterogenous ribosome populations in vivo. Pioneering work has addressed this limitation by covalently linking circularly-permutated rRNAs to yield a contiguous, orthogonal ribosome (Figure 4E) [89,90]. More recently, the association of covalently-linked ribosomal subunits with their native counterparts has been minimized by optimizing the inter-subunit linker, facilitating the directed evolution of enzymatic capabilities that are inaccessible to otherwise native o-ribosomes [91,92].
The development of increasingly orthogonalized ribosomes in vivo may enable expanded functions or alternative mechanisms of translation. In E. coli, only three codons are natively used to initiate protein: AUG (83%), GUG (14%) and UUG (3%) [93–95], yet 47 of the 64 native codons may also support translation initiation [96]. Ribosomes engineered or evolved to leverage the remaining codons for translational initiation may therefore impart further degrees of orthogonality in vivo. Furthermore, improved modes of orthogonal translation catalyze the incorporation of diverse, non-canonical amino acids in cellular proteins. There has been significant progress towards this goal over nearly 30 years, with the development of orthogonal aaRSs (o-aaRS) that selectively charge cognate tRNAs with >150 unnatural amino acids (Figure 4F) [97,98]. Historically, these methods co-opt poorly used stop codons (e.g. amber/ochre stop), but have suffered from low efficiencies due to competition with native termination factors [99,100]. To fulfil the potential of non-canonical amino acid incorporation, complete genome modification [101], de novo synthesis of recoded genomes [102], expanded four-base (quadruplet) codons [6,103–105] and synthetic nucleobases [86] have been successfully used to reassign codons in vivo.
Retroactivity and resource allocation
Limiting the impact of engineered circuits on the expression and function of endogenous genes or proteins has been a major area of research. Specifically, minimizing the burden of engineered circuits on both the host cell and on one another can have profound impacts on their functionality. Discrete modules, typically characterized in isolation, will often fail to function in concert in novel user-defined contexts. This failure is often attributed to: 1) off-target effects between circuit components and/or with host components [106,107], 2) unbalanced flux through a signalling cascade, wherein intermediate products are overproduced above the necessary concentrations [108–111], and/or 3) unbalanced host resource allocation to generate the requisite biomolecules, whose production can be coupled due to resource competition (TFs, RNAPs, Ribosomes, metabolites) [13]. Collectively, these factors often manifest as a reduction in host growth rate [19], which can exacerbate engineered circuit function by changing protein dilution [112] or translation rates [113]. Strategies to mitigate these outcomes have incorporated buffering loops that function at more rapid kinetics than the engineered circuits [108], including sRNA-based [12] and phosphorylation-based circuit capacitors [114]. The continued implementation of these or related strategies in higher order engineered circuits will undoubtedly be necessary to support their increasingly complex designs.
Conclusions and future perspective
While significant effort has been devoted to the creation of orthogonal components that alleviate interactions with the host, these innovations have not yet coalesced into a contiguous and orthogonal central dogma (see Outstanding Questions). A major contributing factor has been the development and characterization of orthogonal parts required to achieve a desired outcome, exclusively insulating the necessary elements. For example, the selective control of gene transcription using orthogonal TFs has become routine and typically requires a small number of components [5], whereas the incorporation of numerous ncAAs in vivo remains a major challenge due to crosstalk with host machinery requiring host engineering [115,116]. Nonetheless, combining orthogonal modalities at multiple nodes of the central dogma has greatly improved circuit performance for otherwise intractable bioactivities. Numerous strategies have improved ncAA incorporation efficiency with orthogonal aaRS–tRNA pairs while limiting host fitness consequences [117]: controlling o-ribosomes with inducible TFs [89,103], developing complete transcription–translation networks that integrate T7 RNAP, o-mRNA and o-ribosome [118], or even decoding cellular codons bearing synthetic nucleobases [72].
Outstanding Questions Box.
Three orthogonal pairs of nucleobases can be replicated and transcribed in vitro, with one pair functioning in vivo. Combining all three artificial base pairs would exceed the diversity of possible codon combinations in extant biological nucleic acids. Accordingly, how close are we to genes that are exclusively encoded by orthogonal synthetic nucleotides? Is additional biosynthetic machinery needed to realize this goal?
Given that entire organisms (e.g. E. coli) can function with a few hundred transcription factors, how many independent transcriptional regulators are needed to build elaborate and complex synthetic behavior that can mirror natural processes?
Will future synthetic genetic circuit development move towards highly programable components (e.g. CRISPR-based systems), harness the potential orthogonality of heterologous components, rely on de novo modular elements, or a combination thereof?
Can polypeptide translation truly be made orthogonal from the host cell given the number of biomolecules involved? Would true orthogonalization require physical separation (e.g. designer membranes, compartmentalization) or can it be achieved more directly?
Orthogonal aminoacyl-tRNA synthetase(aaRS)/tRNA pairs have enabled non-canonical amino acid (ncAAs) incorporation in living cells, often using recoded organisms that eliminate specific codon-amino acid assignments. How many novel or recoded codons can be used simultaneously in a living cell and what factors limit this number? What enabling technologies are needed to expand the genetic code beyond three ncAAs in a contiguous polypeptide in living cells?
Current synthetic biology efforts still rely heavily on host replication and translation components, yet significant effort has been devoted to limit this dependence. What prompts the first study to bring all levels of orthogonality together within the central dogma?
Furthermore, host dependence has restricted the utility of many orthogonal components to a small subset of host organisms. Whereas some standalone orthogonal elements (e.g. T7 RNAP) do not rely on the host machinery and have seen utility in model organisms spanning the tree of life [118–120], most components have been developed to take advantage of host-specific phenomena to direct increasingly orthogonal bioactivities (e.g. RBS/antiRBS complementarity to afford o-ribosomes) [88]. To date, bacterial systems enable the greatest repertoire of orthogonal elements spanning both transcription and translation, while orthogonal DNA replication mechanisms have comparatively lagged. Conversely, robust methods for orthogonal DNA replication and transcription have been described in eukaryotic organisms, whereas orthogonal translation has not seen nearly the same degree of utility as in prokaryotes. It should be noted, however, that these seminal contributions have relied on biological insight or researcher-guided evolution to afford the requisite bioactivities. In the future, it may be possible to develop orthogonal components de novo, bolstering the available repertoire of parts and providing access to otherwise intractable cellular functions.
The orthogonalization of engineered gene circuits has reflected a necessity in achieving key bioengineering goals. A fully orthogonal central dogma, therefore, may be required to fulfil increasingly complex user-defined behaviour not found in nature. Indeed, the design of advanced cellular circuits to sense, process and actuate in response to a diverse set of input signals will rely not only robust and predictable module behaviour, but also the development of guidelines to connect discrete modules for higher order information maintenance and transfer. Progress towards this goal has relied on the accretion of elements and insulation strategies over many decades and from various fields, often limited in their attribution of orthogonal behaviour by field-specific jargon or engineering goals. We believe that viewing these components through the lens of orthogonality, whether natural or engineered, most notably as it pertains to insulation at all stages of the central dogma, will catalyse significant progress towards attaining a fully orthogonal, cellular central dogma and enable truly modular and predictable engineered biological systems.
Highlights.
Development of fully synthetic nucleobase pairs that faithfully interact in living cells, and their applications in creating semisynthetic organisms with expanded and orthogonal information-carrying capacity.
Harnessing naturally occurring and mutually orthogonal DNA replication systems to enable replication of target genes. Highly error-prone variations on these systems enable robust directed evolution of biomolecules.
Engineering and directed evolution of mutually orthogonal transcription factors that operate with high dynamic range, low background, and respond to a wide repertoire of stimuli in vivo.
Recent developments in in vivo orthogonal protein translation including: orthogonal RBS–orthogonal antiRBS pairs, covalently linked rRNA subunits to discover novel enzymatic capabilities, improved incorporation of non-canonical amino acids and decoding quadruplet codons.
Acknowledgements
This work was supported by the National Institutes of Health Director’s Early Independence Award (DP5-OD-024590), the NASA Exobiology Program (NNH17ZDA001NEXO), and the Broad Institute of MIT and Harvard.
Glossary
- aaRS
An aminoacyl-tRNA synthase, also known as a tRNA-ligase, is an enzyme that covalently attaches an amino acid to a tRNA
- Biological orthogonalization
The purposeful insulation of biomolecules from host components, aimed at limiting undesirable cross-talk with cellular biomolecules or environmental stimuli.
- Codon
A sequence of three nucleotides in a DNA or RNA molecule that forms a translatable unit within the genetic code
- DNAP
An enzyme that polymerizes deoxyribonucleotides to afford DNA
- EF-P
Elongation factor P is a prokaryotic factor that improves the efficiency of initial peptide bond formation during the transition from initiation to elongation
- EF-Tu
Elongation factor Tu is a prokaryotic factor that regulates aminoacyl-tRNA selection en route to the ribosome
- Engineered gene circuit
A synthetic collective of interactions using naturally-derived, engineered and/or evolved biomolecules (DNA, RNA, proteins, and/or small molecules) that cumulatively affect a user-defined process or outcome
- Genetic code expansion
The addition of synthetic nucleic acids or ncAAs to living biological systems to supplement their information-carrying capacities, often via an increased number of unique and functional codons
- Host
As it pertains to synthetic biology, a host is the cell in which exogenous genetic information has been introduced
- mRNA
A messenger RNA contains an open reading frame encoding a protein
- Natural gene circuit
The natural collective of interactions using cellular factors (DNA, RNA, proteins, and/or small molecules) that exist within natural biological systems and are integrated to affect a specific process or outcome
- ncAA
An unnatural amino acid not found in the 20 proteogenic amino acids commonly found in biological systems
- Orthogonal translation
The partitioning of ribosomal pools within a cell, wherein a discrete population of the ribosomes exclusively initiate the translation of researcher-defined orthogonal mRNAs
- Quadruplet codon
A sequence of four nucleotides in a DNA or RNA molecule that forms a translatable unit within an expanded genetic code
- RNAP
An enzyme that polymerizes ribonucleotides to afford RNA during transcription, often uses a DNA sequence as the template
- Synthetic cell
A broad term that includes an artificial/engineered particle that imitates one or many functions of a natural biological cell, typically defined as biologically-active material enclosed in a membrane
- Synthetic nucleobases
Nucleobases synthesized in vitro and not found in natural biological systems
- T7
Refers to T7 bacteriophage, a lytic virus that infects bacteria
- tRNA
A transfer RNA is a non-coding 70–90 nucleotide RNA molecule that decodes codons in mRNAs and is used by the ribosome to translate mRNA sequences to the corresponding proteins
- UTR
The untranslated region of an mRNA are sequences flanking the open reading frame defining the protein sequence
Footnotes
Conflicts of Interest
AC and AHB declare no conflicts of Interest.
References
- 1.Müller IE et al. (2019) Gene networks that compensate for crosstalk with crosstalk. Nature Communications 10, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hartwell LH et al. (1999) From molecular to modular cell biology. Nature 402, C47–C52 [DOI] [PubMed] [Google Scholar]
- 3.Cameron DE et al. (2014) A brief history of synthetic biology. Nat Rev Microbiol 12, 381–390 [DOI] [PubMed] [Google Scholar]
- 4.Brophy JAN and Voigt CA (2014) Principles of genetic circuit design. Nature Methods 11, 508–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer AJ et al. (2019) Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology 15, 196–204 [DOI] [PubMed] [Google Scholar]
- 6.Anderson JC et al. (2004) An expanded genetic code with a functional quadruplet codon. PNAS 101, 7566–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Nies P et al. (2018) Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nature Communications 9, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nielsen AAK et al. (2016) Genetic circuit design automation. Science 352, [DOI] [PubMed] [Google Scholar]
- 9.Carlson PD et al. (2018) De novo Design of Translational RNA Repressors. bioRxiv DOI: 10.1101/501767 [DOI] [Google Scholar]
- 10.Kim J et al. (2019) De novo-designed translation-repressing riboregulators for multi-input cellular logic. Nature Chemical Biology 15, 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutalik VK et al. (2012) Rationally designed families of orthogonal RNA regulators of translation. Nature Chemical Biology 8, 447–454 [DOI] [PubMed] [Google Scholar]
- 12.Ceroni F et al. (2018) Burden-driven feedback control of gene expression. Nature methods 15, 387. [DOI] [PubMed] [Google Scholar]
- 13.Gyorgy A et al. (2015) Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J. 109, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Y et al. (2017) Resource Competition Shapes the Response of Genetic Circuits. ACS Synth. Biol. 6, 1263–1272 [DOI] [PubMed] [Google Scholar]
- 15.Kurland CG and Dong H (1996) Bacterial growth inhibition by overproduction of protein. Mol. Microbiol. 21, 1–4 [DOI] [PubMed] [Google Scholar]
- 16.Sleight SC and Sauro HM (2013) Visualization of evolutionary stability dynamics and competitive fitness of Escherichia coli engineered with randomized multigene circuits. ACS Synth Biol 2, 519–528 [DOI] [PubMed] [Google Scholar]
- 17.Cardinale S and Arkin AP (2012) Contextualizing context for synthetic biology – identifying causes of failure of synthetic biological systems. Biotechnology Journal 7, 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moser F et al. (2012) Genetic circuit performance under conditions relevant for industrial bioreactors. ACS Synth Biol 1, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borkowski O et al. (2016) Overloaded and stressed: whole-cell considerations for bacterial synthetic biology. Current Opinion in Microbiology 33, 123–130 [DOI] [PubMed] [Google Scholar]
- 20.Haseltine EL and Arnold FH (2007) Synthetic Gene Circuits: Design with Directed Evolution. Annual Review of Biophysics and Biomolecular Structure 36, 1–19 [DOI] [PubMed] [Google Scholar]
- 21.Ellefson JW et al. (2014) Directed evolution of genetic parts and circuits by compartmentalized partnered replication. Nature Biotechnology 32, 97–101 [DOI] [PubMed] [Google Scholar]
- 22.Jacob F and Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. Journal of Molecular Biology 3, 318–356 [DOI] [PubMed] [Google Scholar]
- 23.Liu CC et al. (2018) Toward an Orthogonal Central Dogma. Nat Chem Biol 14, 103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kittleson JT et al. (2011) Rapid optimization of gene dosage in E. coli using DIAL strains. J Biol Eng 5, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaidukov L et al. (2018) A multi-landing pad DNA integration platform for mammalian cell engineering. Nucleic Acids Res. 46, 4072–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weigele P and Raleigh EA (2016) Biosynthesis and Function of Modified Bases in Bacteria and Their Viruses. Chem. Rev. 116, 12655–12687 [DOI] [PubMed] [Google Scholar]
- 27.Park M et al. (2019) Engineering Epigenetic Regulation Using Synthetic Read-Write Modules. Cell 176, 227–238.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sood AJ et al. (2019) DNAmod: the DNA modification database. Journal of Cheminformatics 11, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren RAJ (1980) Modified Bases in Bacteriophage DNAs. Annual Review of Microbiology 34, 137–158 [DOI] [PubMed] [Google Scholar]
- 30.Gommers-Ampt JH and Borst P (1995) Hypermodified bases in DNA. FASEB J. 9, 1034–1042 [DOI] [PubMed] [Google Scholar]
- 31.Yang Z et al. (2011) Amplification, Mutation, and Sequencing of a Six-Letter Synthetic Genetic System. J. Am. Chem. Soc. 133, 15105–15112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhami K et al. (2014) Systematic exploration of a class of hydrophobic unnatural base pairs yields multiple new candidates for the expansion of the genetic alphabet. Nucleic Acids Res 42, 10235–10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirao I and Kimoto M (2012) Unnatural base pair systems toward the expansion of the genetic alphabet in the central dogma. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci 88, 345–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshika S et al. (2019) Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 363, 884–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feldman AW et al. (2018) A Tool for the Import of Natural and Unnatural Nucleoside Triphosphates into Bacteria. J. Am. Chem. Soc. 140, 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro VB et al. (2012) Synthetic genetic polymers capable of heredity and evolution. Science 336, 341–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L et al. (2011) DNA phosphorothioation is widespread and quantized in bacterial genomes. PNAS 108, 2963–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arangundy-Franklin S et al. (2019) A synthetic genetic polymer with an uncharged backbone chemistry based on alkyl phosphonate nucleic acids. Nature Chemistry 11, 533–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salas M et al. (2016) DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication. Front Mol Biosci 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakatani Y et al. (2018) Self-replication of circular DNA by a self-encoded DNA polymerase through rolling-circle replication and recombination. Scientific Reports 8, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korhonen JA et al. (2004) Reconstitution of a minimal mtDNA replisome in vitro. The EMBO Journal 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ravikumar A et al. (2014) An orthogonal DNA replication system in yeast. Nat. Chem. Biol. 10, 175–177 [DOI] [PubMed] [Google Scholar]
- 43.Arzumanyan GA et al. (2018) Mutually Orthogonal DNA Replication Systems In Vivo. ACS Synth. Biol. 7, 1722–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ravikumar A et al. (2018) Scalable, Continuous Evolution of Genes at Mutation Rates above Genomic Error Thresholds. Cell 175, 1946–1957.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Javanpour AA and Liu CC (2019) Genetic Compatibility and Extensibility of Orthogonal Replication. ACS Synth Biol 8, 1249–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong Z et al. (2020) Automated continuous evolution of proteins in vivo, Synthetic Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Boer HA et al. (1983) The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U.S.A. 80, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skerra A (1994) Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene 151, 131–135 [DOI] [PubMed] [Google Scholar]
- 49.Guzman LM et al. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SK et al. (2007) Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl. Environ. Microbiol. 73, 5711–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hook-Barnard IG and Hinton DM (2007) Transcription Initiation by Mix and Match Elements: Flexibility for Polymerase Binding to Bacterial Promoters. Gene Regul Syst Bio 1, 275–293 [PMC free article] [PubMed] [Google Scholar]
- 52.Helmann JD (1999) Anti-sigma factors. Curr. Opin. Microbiol. 2, 135–141 [DOI] [PubMed] [Google Scholar]
- 53.Campbell EA et al. (2008) Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol. 11, 121–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhodius VA et al. (2013) Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol Syst Biol 9, 702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bervoets I et al. (2018) A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res. 46, 2133–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Helmann JD (2002) The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46, 47–110 [DOI] [PubMed] [Google Scholar]
- 57.Staroń A et al. (2009) The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74, 557–581 [DOI] [PubMed] [Google Scholar]
- 58.Khalil AS et al. (2012) A Synthetic Biology Framework for Programming Eukaryotic Transcription Functions. Cell 150, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Y et al. (2015) Modular construction of mammalian gene circuits using TALE transcriptional repressors. Nat Chem Biol 11, 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mali P et al. (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavez A et al. (2015) Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hilton IB et al. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu X et al. (2016) A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2, 16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilbert LA et al. (2013) CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y et al. (2016) Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nature Methods 13, 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z et al. (2020) De novo design of protein logic gates. Science 368, 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chamberlin M et al. (1970) New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature 228, 227–231 [DOI] [PubMed] [Google Scholar]
- 68.Studier FW and Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 69.Temme K et al. (2012) Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Research 40, 8773–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer AJ et al. (2015) Directed Evolution of a Panel of Orthogonal T7 RNA Polymerase Variants for in Vivo or in Vitro Synthetic Circuitry. ACS Synth. Biol. 4, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y et al. (2017) A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feldman AW et al. (2019) Optimization of Replication, Transcription, and Translation in a Semi-Synthetic Organism. J. Am. Chem. Soc. 141, 10644–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyer AJ et al. (2015) Transcription yield of fully 2’-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res. 43, 7480–7488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tejada-Arranz A et al. (2020) Bacterial RNA Degradosomes: Molecular Machines under Tight Control. Trends in Biochemical Sciences 45, 42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao BS et al. (2017) Post-transcriptional gene regulation by mRNA modifications. Nature Reviews Molecular Cell Biology 18, 31–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filipowicz W et al. (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nature reviews.Genetics 9, 102. [DOI] [PubMed] [Google Scholar]
- 77.Zaccara S et al. (2019) Reading, writing and erasing mRNA methylation. Nature Reviews Molecular Cell Biology 20, 608–624 [DOI] [PubMed] [Google Scholar]
- 78.Isaacs FJ et al. (2004) Engineered riboregulators enable post-transcriptional control of gene expression. Nature Biotechnology 22, 841–847 [DOI] [PubMed] [Google Scholar]
- 79.Callura JM et al. (2012) Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. U.S.A. 109, 5850–5855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoffman KS et al. (2018) Versatility of Synthetic tRNAs in Genetic Code Expansion. Genes (Basel) 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melnikov S et al. (2012) One core, two shells: bacterial and eukaryotic ribosomes. Nature Structural & Molecular Biology 19, 560–567 [DOI] [PubMed] [Google Scholar]
- 82.Willis JCW and Chin JW (2018) Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nature Chem 10, 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fan C et al. (2015) Rationally evolving tRNAPyl for efficient incorporation of noncanonical amino acids. Nucleic Acids Res. 43, e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hammerling MJ et al. (2020) Strategies for in vitro engineering of the translation machinery. Nucleic Acids Res 48, 1068–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Italia JS et al. (2019) Mutually Orthogonal Nonsense-Suppression Systems and Conjugation Chemistries for Precise Protein Labeling at up to Three Distinct Sites. J. Am. Chem. Soc. 141, 6204–6212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fischer EC et al. (2020) New codons for efficient production of unnatural proteins in a semisynthetic organism. Nat Chem Biol DOI: 10.1038/s41589-020-0507-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cervettini D et al. (2020) Rapid discovery and evolution of orthogonal aminoacyl-tRNA synthetase–tRNA pairs. Nat Biotechnol DOI: 10.1038/s41587-020-0479-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rackham O and Chin JW (2005) A network of orthogonal ribosome·mRNA pairs. Nature Chemical Biology 1, 159–166 [DOI] [PubMed] [Google Scholar]
- 89.Orelle C et al. (2015) Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 [DOI] [PubMed] [Google Scholar]
- 90.Fried SD et al. (2015) Ribosome Subunit Stapling for Orthogonal Translation in E. coli. Angew Chem Int Ed Engl 54, 12791–12794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmied WH et al. (2018) Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature 564, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Carlson ED et al. (2019) Engineered ribosomes with tethered subunits for expanding biological function. Nature Communications 10, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Adams JM and Capecchi MR (1966) N-formylmethionyl-sRNA as the initiator of protein synthesis. Proc Natl Acad Sci U S A 55, 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nirenberg M and Leder P (1964) RNA CODEWORDS AND PROTEIN SYNTHESIS. THE EFFECT OF TRINUCLEOTIDES UPON THE BINDING OF SRNA TO RIBOSOMES. Science 145, 1399–1407 [DOI] [PubMed] [Google Scholar]
- 95.Blattner FR et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277, 1453–1462 [DOI] [PubMed] [Google Scholar]
- 96.Hecht A et al. (2017) Measurements of translation initiation from all 64 codons in E. coli. Nucleic Acids Research 45, 3615–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dumas A et al. (2015) Designing logical codon reassignment – Expanding the chemistry in biology. Chem. Sci. 6, 50–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Melo Czekster C et al. (2016) In Vivo Biosynthesis of a β-Amino Acid-Containing Protein. J. Am. Chem. Soc. 138, 5194–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu CC and Schultz PG (2010) Adding New Chemistries to the Genetic Code. Annual Review of Biochemistry 79, 413–444 [DOI] [PubMed] [Google Scholar]
- 100.Hoesl MG and Budisa N (2012) Recent advances in genetic code engineering in Escherichia coli. Curr. Opin. Biotechnol. 23, 751–757 [DOI] [PubMed] [Google Scholar]
- 101.Lajoie MJ et al. (2013) Genomically recoded organisms expand biological functions. Science 342, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Fredens J et al. (2019) Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neumann H et al. (2010) Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 [DOI] [PubMed] [Google Scholar]
- 104.Niu W et al. (2013) An Expanded Genetic Code in Mammalian Cells with a Functional Quadruplet Codon. ACS Chem. Biol. 8, 1640–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang K et al. (2014) Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nature Chemistry 6, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Halleran AD and Murray RM (2018) Cell-Free and In Vivo Characterization of Lux, Las, and Rpa Quorum Activation Systems in E. coli. ACS Synth. Biol 7, 752–755 [DOI] [PubMed] [Google Scholar]
- 107.Meyer AJ et al. (2019) Escherichia coli “Marionette” strains with 12 highly optimized small-molecule sensors. Nature Chemical Biology 15, 196–204 [DOI] [PubMed] [Google Scholar]
- 108.Grunberg TW and Del Vecchio D (2020) Modular Analysis and Design of Biological Circuits. Current Opinion in Biotechnology 63, 41–47 [DOI] [PubMed] [Google Scholar]
- 109.Jayanthi S et al. (2013) Retroactivity Controls the Temporal Dynamics of Gene Transcription. ACS Synth. Biol. 2, 431–441 [DOI] [PubMed] [Google Scholar]
- 110.Lee T-H and Maheshri N (2012) A regulatory role for repeated decoy transcription factor binding sites in target gene expression. Molecular Systems Biology 8, 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Del Vecchio D et al. (2008) Modular cell biology: retroactivity and insulation. Molecular Systems Biology 4, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tan C et al. (2009) Emergent bistability by a growth-modulating positive feedback circuit. Nature Chemical Biology 5, 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klumpp S and Hwa T (2014) Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Current Opinion in Biotechnology 28, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mishra D et al. (2014) A load driver device for engineering modularity in biological networks. Nature Biotechnology 32, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson DBF et al. (2011) RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 7, 779–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hong SH et al. (2014) Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth Biol 3, 398–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bryson DI et al. (2017) Continuous directed evolution of aminoacyl-tRNA synthetases. Nature Chemical Biology 13, 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.An W and Chin JW (2009) Synthesis of orthogonal transcription-translation networks. Proceedings of the National Academy of Sciences 106, 8477–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dower K and Rosbash M (2002) T7 RNA polymerase-directed transcripts are processed in yeast and link 3’ end formation to mRNA nuclear export. RNA 8, 686–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lieber A et al. (1989) High level gene expression in mammalian cells by a nuclear T7-phase RNA polymerase. Nucleic Acids Res 17, 8485–8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wei C-M et al. (1975) Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 4, 379–386 [DOI] [PubMed] [Google Scholar]
- 122.Forchhammer K et al. (1989) Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342, 453–456 [DOI] [PubMed] [Google Scholar]
- 123.Low SC and Berry MJ (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 21, 203–208 [PubMed] [Google Scholar]
- 124.Komor AC et al. (2017) CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mukai T et al. (2017) Rewriting the Genetic Code. Annual Review of Microbiology 71, 557–577 [DOI] [PMC free article] [PubMed] [Google Scholar]