Abstract
Myosin II is the motor protein responsible for contractility in muscle and nonmuscle cells1. The molecule has two identical heads attached to an elongated tail, and can exist in two conformations: 10S and 6S, named for their sedimentation coefficients2,3. The 6S conformation has an extended tail and assembles into polymeric filaments, which pull on actin filaments to generate force and motion. In 10S myosin, the tail is folded into three segments and the heads bend back and interact with each other and the tail3-7, creating a compact conformation, in which ATPase activity, actin activation and filament assembly are all highly inhibited7,8. This switched-off structure appears to function as a key energy-conserving storage molecule in muscle and nonmuscle cells9-12, which can be activated to form functional filaments as needed13 – but the mechanism of its inhibition is not understood. Here we have solved the structure of smooth muscle 10S myosin to a global resolution of 4.3 Å by cryo-EM, revealing near-atomic level details of its structure for the first time. The reconstruction provides a new understanding of the head and tail regions of the molecule and of the key intramolecular contacts that cause inhibition. Our results suggest an atomic model for the off-state of myosin II, for its activation and unfolding by phosphorylation, and for understanding the clustering of disease-causing mutations near sites of intramolecular interaction.
Myosin II consists of pairs of heavy chains (HCs), essential light chains (ELCs), and regulatory light chains (RLCs) that combine to form the two-headed molecule and α-helical, coiled-coil tail (Fig. 1a). Each head comprises a motor domain (MD) and regulatory domain (RD), containing one RLC and one ELC. In the 10S structure, inhibition occurs through: (i) interaction of the actin-binding region of one head (the blocked head, BH) with the ATP-binding region of the other (free head, FH)6,14, forming an “interacting-heads motif” (IHM); and (ii) head interactions with the three segments of the tail (seg1, seg2 and seg3) and of the tail with itself4,7. A similar IHM structure occurs in thick filaments, contributing to the relaxed state of striated muscle7,15, but intramolecular interactions with segs 2 and 3 are absent as the molecules are extended. The 10S structure is in equilibrium with thick filaments in smooth and nonmuscle cells, regulated by phosphorylation of its RLCs. Phosphorylation promotes unfolding to the extended (6S) structure5,10,16, which forms thick filaments that interact with actin to produce contractility and regulate actin dynamics. Filaments depolymerize to the 10S form when the RLCs are dephosphorylated13. Mutations in the heads and tail impair function and cause muscle and other diseases17. The 10S conformation has been conserved throughout animal evolution18, indicating its fundamental importance to cell function. Previous studies of 10S myosin have been limited to 20 Å resolution, leaving many unknowns concerning its structure and function4,6,7,13. Our cryo-EM reconstruction provides novel insights, in near-atomic detail, into the structure of the 10S molecule, the molecular basis of inhibition and activation, and the mechanism of disease.
Fig. 1. 3D reconstruction of 10S myosin.
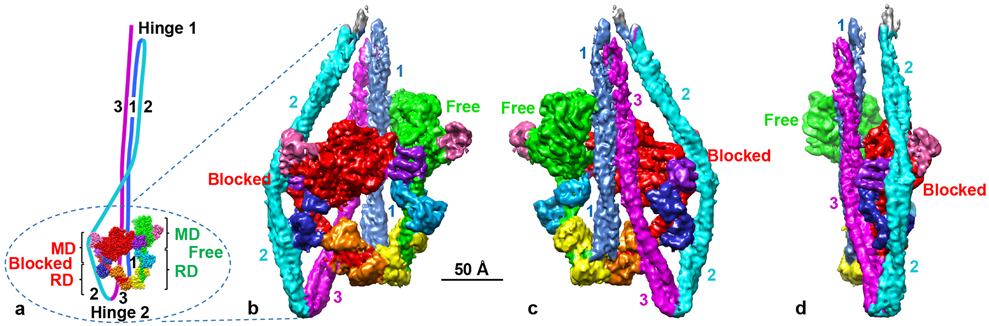
a. Overview of 10S molecule, showing two heads and folded tail. Heads comprise N-terminal halves of HCs (red, green), ELCs (blue, cyan), and RLCs (orange, yellow). C-terminal halves of HCs combine to form the coiled-coil tail, folded at two hinges, creating tail segments 1, 2 and 3. Heads interact to form interacting-heads motif, with “blocked” and “free” heads. Ellipse shows region of molecule in reconstruction; upper portion is excluded due to flexibility. MD, RD: motor and regulatory domains. SH3 and converter domains of MDs are pink and purple respectively. b, c, d. Front, back and side views of reconstruction, using same color scheme (map, based on 168,613 particles, shown at contour level 0.0125—see Methods). See Supplementary Video 1.
Cryo-EM structure of 10S myosin II
Class averages of cryo-imaged molecules showed multiple views of the 10S conformation, with evidence of secondary structure in the heads and clear density for all three tail segments (Extended Data Fig. 1; Methods). The refined reconstruction (EMD-22145; resolution range ~4-9 Å) confirmed this appearance, revealing secondary structure in the MDs (including side-chain detail), the light chains, and the individual α-helices of the tail (Figs. 1b-d, Extended Data Figs. 1b, 2; Extended Data Table 1). These features, observed here in intact myosin II, have been seen previously only in X-ray structures of its separate components. The two heads interact with each other through their motor domains. Seg1 of the tail (subfragment 2, or S21) exits the heads at the junction of the two RDs, crosses the BH, reverses direction at hinge 1 (not seen in the map due to flexibility), where it becomes seg2. Seg2 passes around the edge of the BH and reverses direction at hinge 2, becoming seg3, which crosses the BH parallel to, but resolved from, seg1 (Fig. 1).
We interpreted the structure by rigidly fitting the motor and regulatory domains of a two-headed myosin fragment (PDB 1i8414) independently into the EM map, and then refining the fit (Fig. 2a; Methods). We fitted and refined the tail in a similar way, using the α-carbon backbone of the subfragment-2 coiled-coil (PDB 2FXM)19. The refined model (PDB 6XE9; Figs. 2a, b, Extended Data Figs. 2c-g) revealed novel detail of the heads, the head-tail junction, the tail, and the intramolecular interfaces that clamp the molecule in the off-state.
Fig. 2. Fitting of atomic model to map (a) and locations of mutations (b, c).
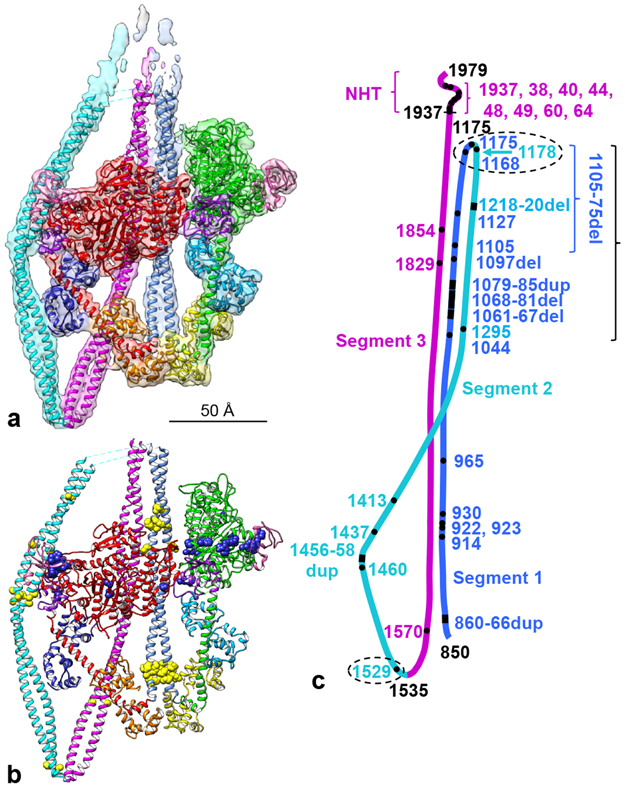
a. Cryo-EM map from Fig. 1 (contour level 0.0125) fitted with refined atomic model (PDB 6XE9; Supplementary Video 2). b. Refined model (Supplementary Video 3), showing locations of disease-causing mutations in smooth and nonmuscle myosin II (Extended Data Table 2) plotted onto the atomic model (MD mutations in blue, tail mutations in yellow). c. Locations of all tail mutations (heads omitted for clarity). Ellipses, mutations that could affect folding at hinges; long bracket, clustering of mutations in the three tail segments presumed to interact with each other in the 10S structure; these may affect stability of the folded molecule. Numbers are amino acid positions: black, ends of the coiled-coil (850-1937), the two hinges (1175, 1535), and the C-terminus (1979); amino acids ~1938-1979 form a non-helical tailpiece (NHT); colored numbers are positions of point, duplication or deletion mutations. See also Extended Data Fig. 9 and Supplementary Video 4.
Structure of the heads.
The reconstruction and the fitted model show that the two MDs are essentially identical in structure (Extended Data Fig. 3a). Similar location and structure of the converter domains suggests that the MDs are in the same pre-powerstroke, ADP.Pi nucleotide state20. In contrast, the RDs make quite different angles with their MDs, due to different flexing in the pliant region of the head (near L790; Extended Data Fig. 3b,c14). Similar differential flexing of the BH and FH occurs in the thick filament IHM21, suggesting that it is a fundamental feature of the motif. It enables the heads, interacting asymmetrically at their MD interface, to come together at their C-termini and attach to S2 without requiring significant unwinding of its coiled coil (Extended Data Fig. 3d). Our structure does not support a recent low-resolution thick filament model in which BH flexing is much smaller than we find22. We also observe another point of flexibility enabling the heads to attach to S2 without requiring its uncoiling: a straightening of the FH RD between the ELC and RLC, which brings the C-termini of the heads ~ 7 Å closer together (Extended Data Fig. 3e,f; cf.23). In each RD, the density for the α-helical backbone, to which the ELCs and RLCs bind, was continuous, with little evidence for melting in the BH suggested from flexible fitting to a previous 20 Å resolution filament structure21,24.
Structure of the head-tail junction.
Flexing about the head-tail junction is essential for myosin function, but the structure of this region is unknown, as this flexibility has made it impossible to study by X-ray crystallography. Our map clearly shows the junction, which starts with an abrupt bend (“hook”) at the C-terminal end of the RD HC23,25. The hook leads to invariant proline P849 and the start of the coiled-coil tail (Extended Data Fig. 4). The map densities suggest that both hooks are α-helical, with no major melting suggested from low resolution filament models21,24. The hook angles are similar in both heads (~90°; Extended Data Figs. 3e, 4), and show no evidence for differences proposed as a source of compliance facilitating connection to the tail in the IHM23,25. As discussed, this compliance apparently occurs at the pliant point and between the light chains in the RD. The α-helices of seg1 lead directly from the hooks, with possible confined melting around P849 (~K848-Q852; Extended Data Fig. 4). We see no evidence for dissociation of the first eight heptads of each S2 HC into individual α-helices, nor their binding to the surface of the heads26. Instead, the helices apparently associate with each other from the start of S2 (~V853), though with a slightly longer helical pitch and less order in the first few residues (Extended Data Fig. 5b; next section). This may also contribute to relief of stress in the IHM (see above), and could be important in generating the off-state27. Lower stability of the coiled-coil at the start of S2 is suggested by less ordering in X-ray crystal structures19,28 and in our density map (Extended Data Fig. 2).
Structure and path of the tail.
The organization of the tail in 10S myosin has not previously been observed at high resolution4,7,14. Our reconstruction clearly resolves the two α-helical densities, which twist around each other in a left-handed supercoil29 (Figs. 2, Extended Data Figs. 2 and 5). The pitch of the coiled-coil varies substantially along its length (Extended Data Fig. 5), as also observed in thick filaments30. While some portions have fairly constant crossover distances (~ 70 Å), the start of seg3 shows that the two helices run roughly parallel for ~ 100 Å before normal coiling resumes (Extended Data Fig. 5a-c). This disruption to the canonical structure may relate to skip residue Q1592, located in this section of seg3, or to head-head interaction27. Similar disruption occurs in regions containing skip residues in isolated tail fragments31 and thick filaments30. The tail shows a sharp bend at hinge 2 (~E15354; Extended Data Fig. 5d), which is not explained by associated skip residues or predicted coiled-coil weakening4,32. The density map suggests almost continuous coiling of the α-helices about each other at the hinge, with local melting at the immediate site of bending. Seg2 appears to be relatively straight, except for a gentle bend at the BH MD (residues ~1462-1472; Extended Data Fig. 5b), which may be facilitated by predicted coiled-coil weakening in this region32.
The locations of segments 1 and 3 on the BH were a surprise. The IHM in thick filaments15 shows that S2 bends after leaving the heads15,19,21, then crosses the BH ‘mesa’ 33 along a path of positive charge over the center of the head, thought to interact with negative charge on S219. This charge complementarity21,34 is thought to be important in the off-state of thick filaments15,33. However, the path of seg1 in our map is unambiguously different (Extended Data Fig. 6). After leaving the heads, it follows a straight course, crossing over the edge of the BH, to the right of seg1 in the filament IHM (as viewed in Extended Data Fig. 6a, c). In fact, it is seg3 in the 10S molecule that coincides with seg1 of the filament map. There are thus two variants of the IHM, with different S2 lateral positions on the BH: one for thick filaments and the other for 10S molecules. S2 in heavy meromyosin (HMM), lacking segments 2 and 3, has the same position as that in 10S myosin4. Interactions with other myosin molecules in thick filaments may contribute to S2’s different position and bent conformation in the polymer compared with the monomer.
Interactions producing the off-state
Our map shows striking new detail of multiple intramolecular contacts, which we suggest clamp 10S myosin in the inhibited state (Fig. 3, Supplementary Table 1). These occur in the same regions suggested by our previous negative stain reconstruction7, and we use the interaction nomenclature defined in that work. We analyzed these putative interactions using UCSF Chimera (Methods). Because of the limited resolution of the reconstruction, we consider them potential rather than definitive, at least in the lower resolution regions.
Fig 3. Intramolecular interactions in the 10S atomic model.
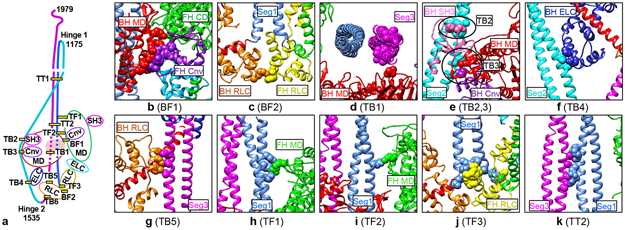
a. Cartoon of 10S structure showing locations of interactions (nomenclature from 7; B=BH, F=FH, T=tail). Interaction TB6 is discussed in Extended Data Fig. 8. b-k. Detail of interactions (see Supplementary Table 1). See a and Fig. 2a, b for color scheme. All views from front of molecule, except d (from bottom) and g (from side); parts of molecule removed for clarity. Cnv=converter, CD=catalytic domain. See Supplementary Video 5.
Head-head interactions.
The density map shows clear contact between the BH and FH MDs, involving the actin-binding surface of the BH and the catalytic and converter domains of the FH (Figs. 1, 2). Specifically, loop I365-N381 and helix T382-L390 of the BH lie near helix E727-Y734 and loop E735-D748 of the FH converter (interaction BF1, Fig. 3b, Supplementary Table 1). BH helix V395-L403 lies over helix F727-Y734 of the FH converter and helix I153-D167 of the FH catalytic domain. These contacts broadly support previous work at 20 Å resolution14,21, but precisely locate the potential interacting residues. However, we do not observe any contact between the BH MD and the FH ELC, thought previously to form a significant interface of the IHM14 (Fig. 3b, Supplementary Table 1). These BH-FH interactions likely contribute to inhibition by hindering motions of the FH converter required for ATPase activity and by blocking BH binding to actin6.
Blocked-head–tail interactions.
The BH appears to be locked down by three interactions with the tail not previously seen at high resolution6,7. 1. As segment 2 travels from the top of the molecule, it passes through a groove on the edge of the BH, making contact at ~L1431-D1436 with helix K72-D74 of the SH3 domain and at ~Q1445-L1452 with ~R718 and L766 in/near the converter (TB2, TB3, Fig. 3e). Strikingly, in this location seg2 physically blocks movement of the BH converter required for phosphate release (Extended Data Fig. 7a-d), directly explaining the inhibition of ATP turnover by the BH through trapping of ATP hydrolysis products in the active site7,8. 2. The tail (~L1494-L1498) next contacts helix D (E67-A77) of the ELC N-lobe (interaction TB4), potentially stabilizing the 10S conformation (Fig. 3f). 3. After a hairpin bend at hinge 2, seg3 (~A1577-R1584) contacts helix E of the BH RLC (E99-A107) (TB5, Fig. 3g) and comes close to RD α-helix Q817-V824. It then crosses the BH MD, parallel to and contacting seg1 (~L1628-E1647 in seg3 with R910-M925 in seg1; TT2, Fig. 3k). Strikingly, seg1 and seg3 both appear to “hover” above the surface of the BH (interaction TB1, Fig. 3d), contacting it only between ~L1604-E1612 of seg3 and loop L450-F460 of the BH MD. It has previously been assumed that there are multiple interactions of segments 1 and 3 with the surface of the BH. With the ~ 8 Å gap between these tail segments and the BH, contacts are apparently few. Weak electrostatic interaction (5-10 Å; 35) might occur over these longer ranges, consistent with weak, salt-sensitive binding of S2 to myosin heads in solution33.
RLC-RLC interaction.
The RLCs approach within ~4-4.5 Å of each other at the base of their N-terminal lobes, where helix A and the A-B linker in the BH and helix D and the A-B linker in the FH come together (BF2, Fig. 3c), strengthening previous suggestions of RLC-RLC interaction14,21,36, thought to be important in regulating smooth and skeletal muscle activity37.
Free-head–tail interaction.
The FH contacts the tail at three sites. The CM loop (T404-K420) and loop 2 (K626-T658), in the actin-binding interface, form contacts TF1 and TF2 with seg1 (M925-A941; Fig. 3h-i), as previously proposed for thick filaments19,21, sterically inhibiting FH-actin binding (Extended Data Fig. 7h-k). And helix A in the N-lobe of the FH RLC fills the gap between the two α-helices at the origin of seg1 (L850-Q856), with multiple potential interactions with the tail (TF3, Fig. 3j), likely impacting regulatory movements of this light chain.
Mechanism of inhibition and activation.
We conclude that these multiple contacts together pin the myosin II molecule in its 10S conformation. Both heads are in the pre-powerstroke, ADP.Pi state, with hydrolysis products trapped by inhibition of their converter movements, conserving ATP. HMM, lacking segments 2 and 3, is ten times more active than 10S myosin8, supporting the inhibitory role played by these segments7. Actin-interaction loops are blocked in both heads through head-head (BH) and head-tail (FH) interactions, while unfolding of the tail is inhibited due to its multiple intramolecular interactions. The result is complete inhibition of the molecule in the 10S, dephosphorylated state. The 10S structure is activated by phosphorylation of S19 on the RLCs, leading to breaking of the inhibitory interactions. A possible mechanism for activation based on the atomic model, including visualization for the first time of the N-terminal 24 residues of the BH RLC, is presented in Extended Data Fig. 8.
The 10S structure and myosin mutations
A number of human diseases are connected to mutations in smooth and nonmuscle myosin II HCs, including breast and prostate cancer, blood diseases and smooth muscle dysfunction17,38. These myosins, which both form the 10S structure5,39, play essential roles in smooth and nonmuscle cellular functions17. Could alteration of the 10S conformation contribute to these diseases? We aligned the smooth and nonmuscle sequences (Supplementary Table 2), then mapped the mutations onto the smooth muscle HC (Extended Data Table 2) and the 10S atomic model (Fig. 2b). We considered the potential effects of mutations based on their proximity to (potential impact on interaction with) other regions of the molecule in the folded structure.
Most FH mutations have no relation to the 10S conformation, being distant from any intramolecular contacts (Figs. 2b, Extended Data Fig. 9a). These might impact head function directly, independently of 10S folding. Three exceptions are R253, R507 and R731 (smooth muscle numbering), in the interface with the BH (Extended Data Table 2, Extended Data Fig. 9a), which might impact head-head interaction; a BH mutation in the interface (K386) may have a similar effect. The situation is different for the BH, which has multiple mutation sites close to seg2. One group stands out, clustered where seg2 wraps around the BH, especially residues in and near the SH3 and converter domains, some coming within 10 Å of seg2, and some < 4 Å (Figs. 2b, c, Extended Data Fig. 9b). While some may directly affect ATPase activity, their proximity to seg2 suggests they could alter BH-seg2 interaction and thus stability of the 10S structure. Mutations in seg2 also occur in this region, and could similarly affect this contact. If seg2/converter interaction inhibits BH ATPase activity (see earlier), mutations in seg2 might disrupt this inhibition.
There are also multiple mutations that might affect tail-tail interaction. These include mutations in seg1 that coincide with interaction site TT2, potentially impacting interaction with seg3 (Figs. 3a, k, Extended Data Fig. 9d), and a cluster in the upper part of the tail (not included in the reconstruction), where the tail segments appear tightly apposed4 (Fig. 2c). These mutations, occurring in all three segments, may impact the stability of the 10S structure. Mutations near the two hinges could affect folding, and those in the non-helical tailpiece, near the assembly competence domain40, could impact filament formation after activation. Mutation of R1570 in seg3, part of the proposed binding site for the BH PD (Extended Data Fig. 8b, c, i), could impact folding or phosphorylation (Extended Data Fig. 9c).
This structure-mutation analysis parallels that on the IHM of cardiac thick filaments in relation to hypertrophic cardiomyopathy, where disease correlated with mutations in the head-head interface, which were proposed to destabilize head-head interaction33,41. Our analysis suggests that intramolecular interfaces may be important for smooth and nonmuscle myosin II diseases, in this case mostly related to interactions involving the tail.
Conclusion
Our cryo-EM reconstruction provides the first near-atomic-level insights into the structure of intact myosin II in the 10S conformation and the physical basis of its inhibition, correcting and integrating previous structural and solution studies. Our model for the off-state, and for its activation and unfolding, provides a framework for further testing functional mechanisms and for understanding how mutations may cause disease, with implications for drug design.
Methods
Protein preparation:
Smooth muscle myosin II was purified in the dephosphorylated state42 from adult turkey (Meleagris gallopavo) gizzard, obtained from a local turkey farm. 10 μl aliquots (30 μM,15 mg/ml) were flash-frozen in liquid nitrogen and stored at −80°C. Smooth muscle myosin has similar structural and functional properties to nonmuscle myosin II, both forming essentially identical 10S structures5,39. Conclusions from our reconstruction are therefore also relevant to nonmuscle myosin II.
Specimen optimization:
Negative staining was used to check specimen purity, quality and grid loading. A thawed aliquot of myosin II was diluted to 10 nM in 0.15 M NaAc, 1 mM EGTA, 2.5 mM MgCl2, 0.5 mM ATP, 10 mM MOPS, pH 7.5, generating the 10S structure7. The 10S conformation is formed by weak interactions that are easily disrupted by EM preparative conditions18,43. Molecules were therefore crosslinked in solution at room temperature for 1 min with 0.1% glutaraldehyde followed by quenching with 100 mM Tris pH 8.0, immediately before grid preparation7,18,39. This stabilizes the 10S conformation, without substantially altering the structure of the molecules7,39,43,44. 5 μL of the diluted and crosslinked sample were applied to a carbon-coated EM grid and negatively stained with 1% (w/v) uranyl acetate7,45. Grids, pretreated with UV light to optimize stain spreading39,46, were imaged on an FEI Tecnai Spirit transmission electron microscope at 120 kV with a 2K x 2K CCD camera.
Cryo-EM:
A thawed aliquot of smooth muscle myosin was freshly diluted to 0.2 μM and crosslinked for 1 min as described above. A 3 μL droplet was applied to a freshly glow-discharged ultrathin 0.2 nm carbon film on a lacey carbon 300 mesh copper support grid (Ted Pella). Glow discharge was carried out for 60 s at 25 mA on a PELCO easiGlow (Ted Pella). The grid was blotted for 6 s at pressure setting 5 then vitrified in liquid ethane cooled by liquid nitrogen, using a Vitrobot Mark IV (FEI/ThermoFisher) operated at 10°C and 95% relative humidity.
Data acquisition:
Grids were screened on a Talos Arctica (ThermoFisher) cryo-EM at 200 kV. Optimal regions of selected grids were then imaged (SerialEM 3.8.047) on a Titan Krios transmission electron microscope (Thermo Fisher) at 300 kV, using a K3 direct electron detector (Gatan) in counting mode and a Gatan GIF Quantum energy filter with a slit width of 20 eV. 10,950 movies were collected at a nominal magnification of 105,000 X in nanoprobe EFTEM super-resolution mode yielding a physical pixel size of 0.83 Å at the specimen level. Each movie contained 27 frames over 1.6 s exposure time, with a dose rate of 18.45 e-/pix/s and total accumulated dose of 43.3 e-/Å2. The nominal defocus range was set from −1.2 to −3.5 μm.
Data Processing:
Each frame in a movie was aligned to correct beam-induced motion using MotionCor248 with 5 × 5 patches and a B-factor of 150, and then summed with a binned pixel size of 0.83 Å/pixel. The defocus of each image was determined using CTFFIND449. The subsequent image processing was carried out using RELION 3.050. 10,604 images were selected for further image processing after removing images with defocus larger than 3.5 μm. 10S myosin molecules consist of a folded tail region, which is flexible, and relatively rigid heads4,7. Only the structure of the head portion was investigated in this study and windowed out in the center of a particle box7. 3,218 particles were manually picked and subjected to 2D classification. Good class averages were selected as templates for automated particle picking from the images. 1,765,220 particles were picked and extracted at 3.32 Å/pixel with a box size of 120 × 120 pixels. To clean up the data set, multiple rounds of 2D classification were performed to remove particles in thick ice, contaminated ice or dirt, resulting in 260,360 particles in 43 class averages showing secondary structural features. A 3D reconstruction of 10S myosin from a previous negative stain study7 was low-pass filtered to 50 Å and used as the starting model to perform unsupervised 3D classification. Six classes of 3D reconstruction were generated with particle numbers/resolutions of, respectively: class 1, 43,641/12.8 Å; class 2, 68,336/9.9 Å; class 3, 17,484/15.9 Å; class 4, 30,621/14.2 Å; class 5, 50,151/11.7 Å; class 6, 50,126/11.4 Å. The reconstructions of the 6 classes showed that the portions of tail segments 1, 2 and 3 included in the reconstruction were flexible compared with the heads. Class 2, 5 and 6 exhibited similar structural features and resolution, and therefore were combined for 3D refinement. A soft mask (5-pixel extension, 6-pixel soft cosine edge), enclosing the entire 10S structure, was created for 3D refinement and post-processing. The combined data set, containing 168,613 particles, was re-extracted and re-centered using a box size of 300 × 300 pixels and pixel size of 1.328 Å, and then subjected to 3D refinement, CTF refinement and polishing. The resolution of the refined 3D reconstruction was estimated to be 4.3 Å based on the gold-standard Fourier shell correlation (FSC) 0.143 criterion (Extended Data Fig. 1b, Extended Data Table 1) using the above soft mask. To visualize the map, the modulation transfer function (MTF) of the detector and a B-factor were implemented to the map after refinement. An estimated local resolution map, calculated using ResMap51, showed highest resolution in the motor domains of the two heads, with lower resolution in the regulatory domains and tail segments, consistent with the results of 3D classification.
Atomic fitting:
A refined atomic model of the interacting heads motif14, based on chicken gizzard smooth muscle myosin (PDB: 1I846), was docked into the EM density map by rigid body fitting using UCSF Chimera 1.1452. The two motor domains, the two regulatory domain α-helices, and the four light chains were each docked individually. To fit the tail regions of the reconstruction, human β-myosin subfragment 2 (PDB: 2FXM19) was used to create a homology model for the turkey gizzard smooth muscle myosin amino acid sequence (residues 853-954) using the SWISS-MODEL server. This model was docked into segment 1 by rigid body fitting using UCSF Chimera. There is currently no atomic model for segments 2 or 3. These two segments were modelled using the α-carbon backbone (poly-A model) of segment 1 (i.e. after removing side chains). The residues were then numbered by assigning E1535 to hinge 24. This assignment is based on measurements made from negative stain 2D class averages and is therefore subject to uncertainty, which could be 2-3 amino acids (3-4.5 Å) either side of 15354. The resulting model (consisting of the two heavy chains and four light chains) was subjected to multiple cycles of real-space refinement using Phenix 1.17.153 followed by manual modification of the model in Coot 0.8.9.2 EL54. The map was sharpened using the auto-sharpen map tool in Phenix. The resulting map was used to improve the fitting of the model, as some of the bulky side chains became resolved. A real space refinement was done with the improved model and the resulting PDB was deposited.
Figures were made using UCSF Chimera52 (contour levels shown in figure legends) and PyMOL (www.pymol.org).
Extended Data
Extended Data Fig. 1. Imaging and processing of 10S myosin II.
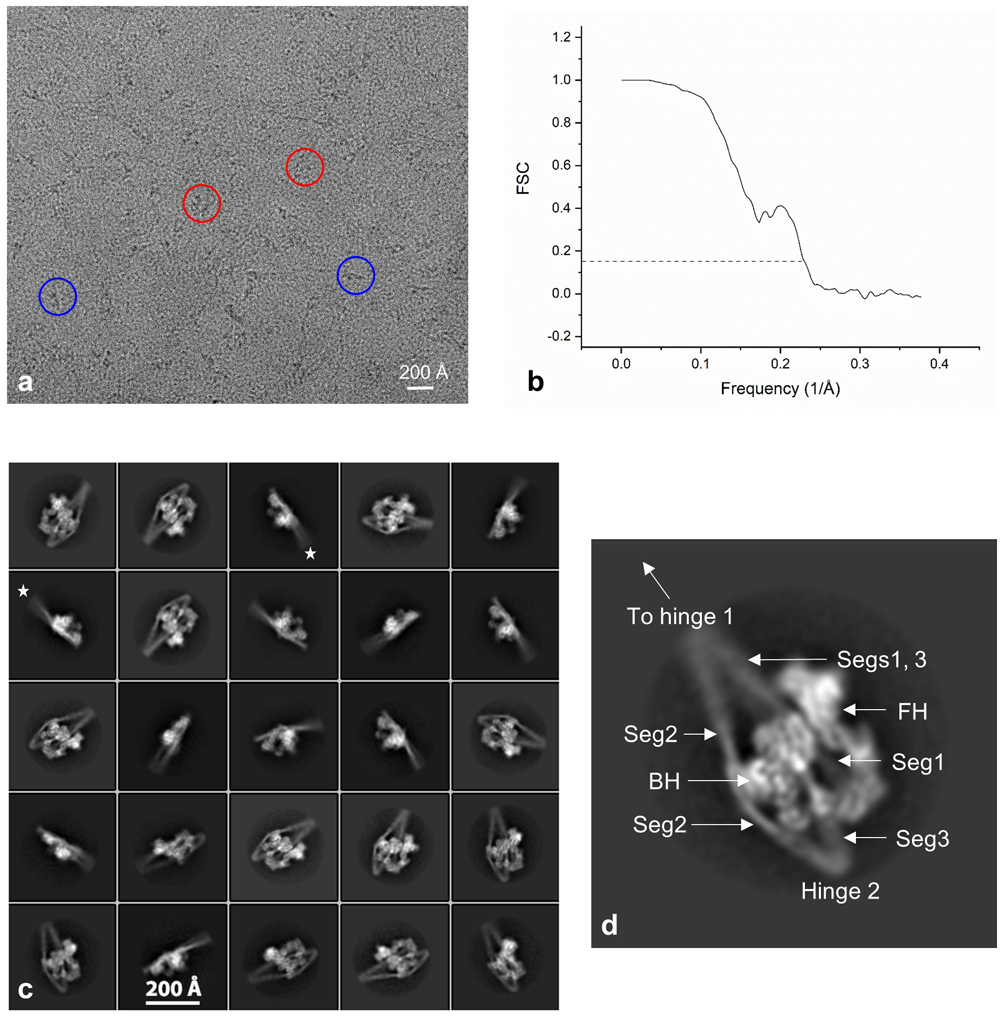
a. Raw cryo-EM image. Circles indicate individual molecules; red, face view; blue, edge view. This is one of 10,950 micrographs recorded at 300 kV on the Titan Krios, and is representative of those showing the particles most clearly. A preliminary set of 400 micrographs from a different set of grids was first recorded on a Talos Arctica at 200 kV, producing a similar, initial reconstruction, at 9 Å resolution. b. Gold-standard Fourier shell correlation (FSC) curve using half maps; global resolution estimate is 4.3 Å by the FSC 0.143 criterion. c. Typical 2D class averages of 10S myosin (25 of a total of 43 good class averages, representing a total of 260,360 particles). Edge views show poor definition of the longer end of the tail (stars), corresponding to mobility in the upper part of the reconstruction (Extended Data Fig. 2). d. Enlarged class average showing main features in reconstruction. BH/FH = blocked/free head; Seg = segment number.
Extended Data Fig. 2. Resolution and atomic fitting of 10S structure.
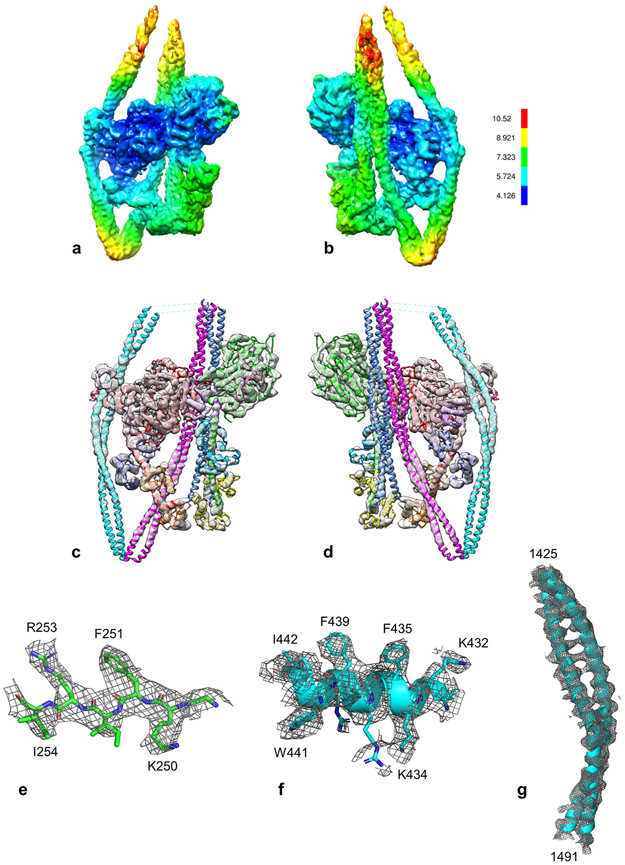
a, b. Front and rear views of density map (contour level 0.0125), showing estimated local resolution according to RESMAP51. Resolution of the heads is highest in the MDs (especially the BH), and lower in the RDs (especially the FH), corresponding to local regions of varied mobility. Resolution in the tail is best where it is stabilized by contacts with other domains (seg2 with the BH MD and ELC), especially in the specific α-helix making the contact. The tail regions at the top are noisy and of low density (Extended Data Fig. 1c). c, d. Docking of the refined model to the map shown at high contour cutoff (0.025; cf. Fig. 2a), revealing clear secondary structure and quality of fit (front, rear views respectively). e-g. Fitting to show map quality. e. β-strand 249-254 of the BH MD, showing side-chain density. f. α-helix 431-443 in the BH MD, showing side-chain density g. Coiled-coil in seg2 contacting the BH MD (1425-1491), showing 5.4 Å α-helical pitch (cf. d).
Extended Data Fig. 3. Comparison of free and blocked heads.
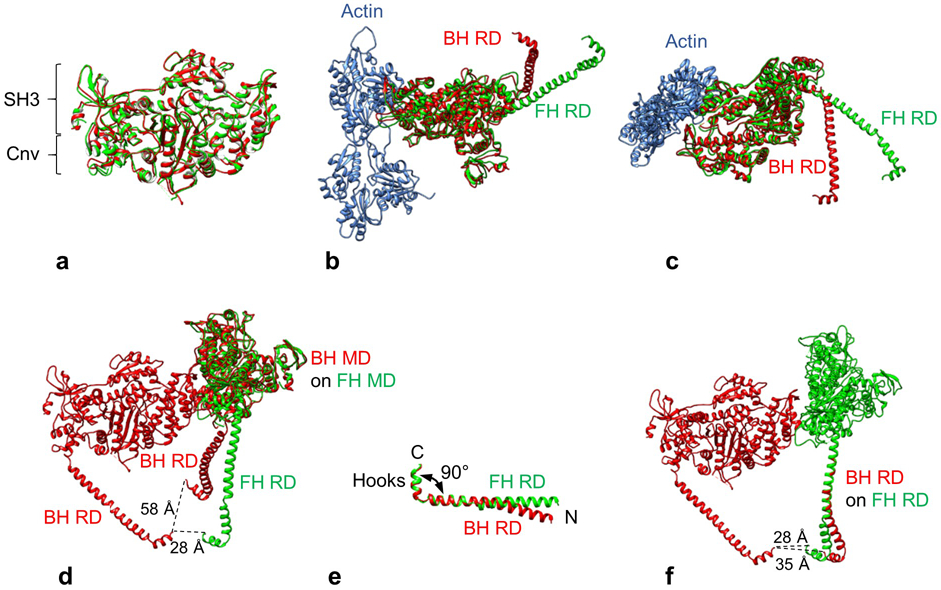
a. Superposition of BH (red) and FH MDs (green) using Matchmaker in Chimera. There is an almost perfect match, including the converter domains (Cnv), which show no more than ~5 Å movement, suggesting that the two heads are in the same biochemical state. Orientation of heads is that seen in front view of BH. b, c. Alignment of MDs of BH and FH, oriented as attached to actin in rigor state. There is a large difference in angle between the BH and FH RDs with respect to their MDs in both longitudinal (b) and azimuthal planes (c) (defined with respect to plane of filament sliding)14. BH and FH were aligned by superposing their MDs on the MD (not shown) of mammalian actomyosin in the rigor state (PDB 5H53). RLCs and ELCs have been removed for clarity. d. Face-view of IHM shows how C-termini of the two heads (P849), at bottom of RDs, come within ~28 Å of each other where they meet S2 (not shown). This proximity depends on the differential flexing of the RDs with respect to the MDs in the two heads. If the FH had the BH RD angle, the C-termini in the IHM would be too far apart (~ 58 Å) to join to the 20 Å-diameter S2 without its substantial uncoiling. The angle of the FH RD is the major structural difference that brings the C-termini of the two heads close enough together to make their simultaneous attachment to S2 possible. Comparison of RD angles was made by superposing the BH and FH MDs. e. Comparison of isolated regulatory domain structures in BH and FH (LCs omitted for clarity) after aligning residues K823-P849 (the left half of the molecule). The C-terminal hooks make similar ~90° angles with the RD helix. There is a small difference in angle between the FH and BH helices in the N-terminal half of the RD HC. f. Comparison of regulatory domain structures within IHM. The BH RD was superimposed on the FH RD in the N-terminal half. The straighter course of the FH RD brings the FH and BH C-termini that attach to S2 closer together by ~7 Å, facilitating attachment to S2 without any substantial unwinding of the coiled coil. This flexibility in the RD, bringing the FH C-terminus closer to the BH, thus aids in formation of the IHM, along with the different angles of the RDs with respect to the MDs seen in b-d.
Extended Data Fig. 4. Structure of head-tail junction.
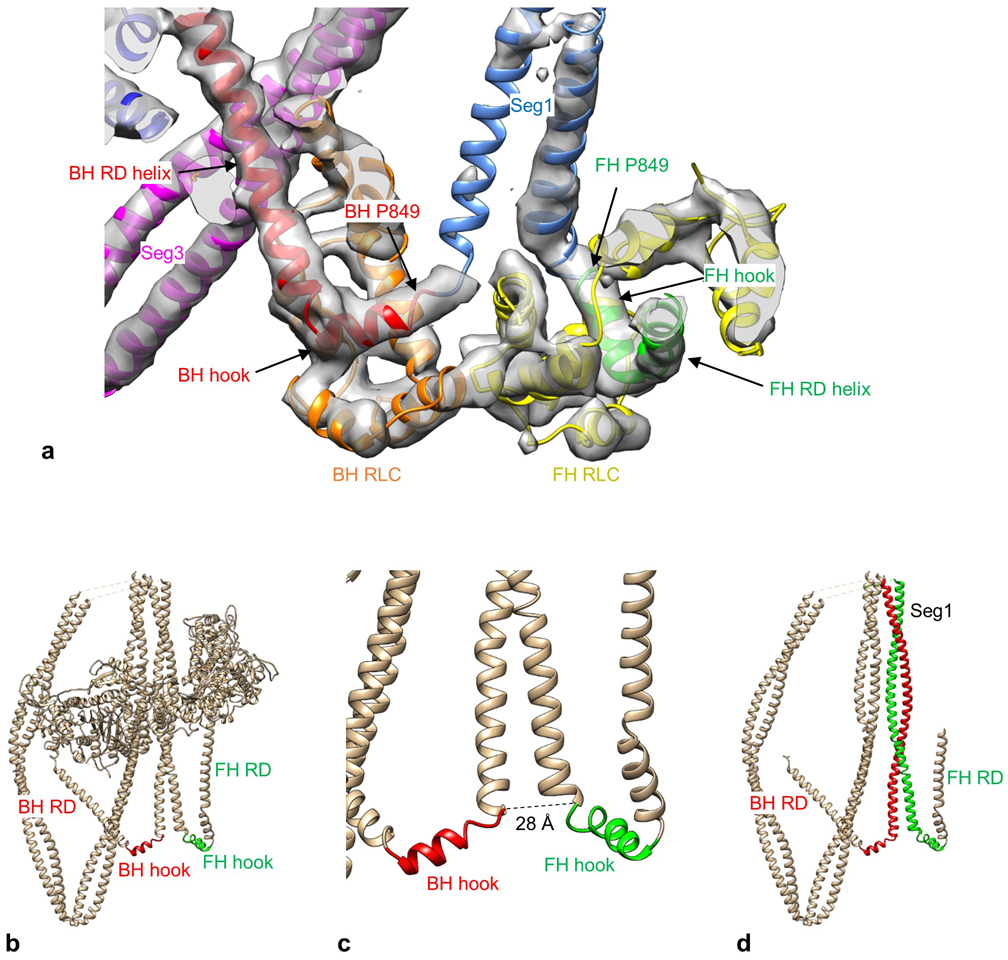
a. Atomic model fitted into the electron density map (contour level 0.021) in the region of the head-tail junction. The map shows tubes of density for the two hooks, which form 90° bends with the α-helical HC of the RDs. Such tubular density is characteristic of α-helices in other parts of the structure. P849 is the invariant proline that marks the junction between each head and the tail. View is from front. b. IHM front-view showing hooks at C-terminal end of each RD. ELCs and RLCs removed for clarity. c. Enlargement of hook region showing ~28 Å distance between invariant prolines at the C-terminus of each head HC. d. IHM face-view showing α-helical backbone of BH and FH RDs, hooks, and coiled-coil tail regions. Seg1 α-helices (red, green) continue from BH and FH hooks.
Extended Data Fig. 5. Variations in the coiled-coil of 10S myosin.
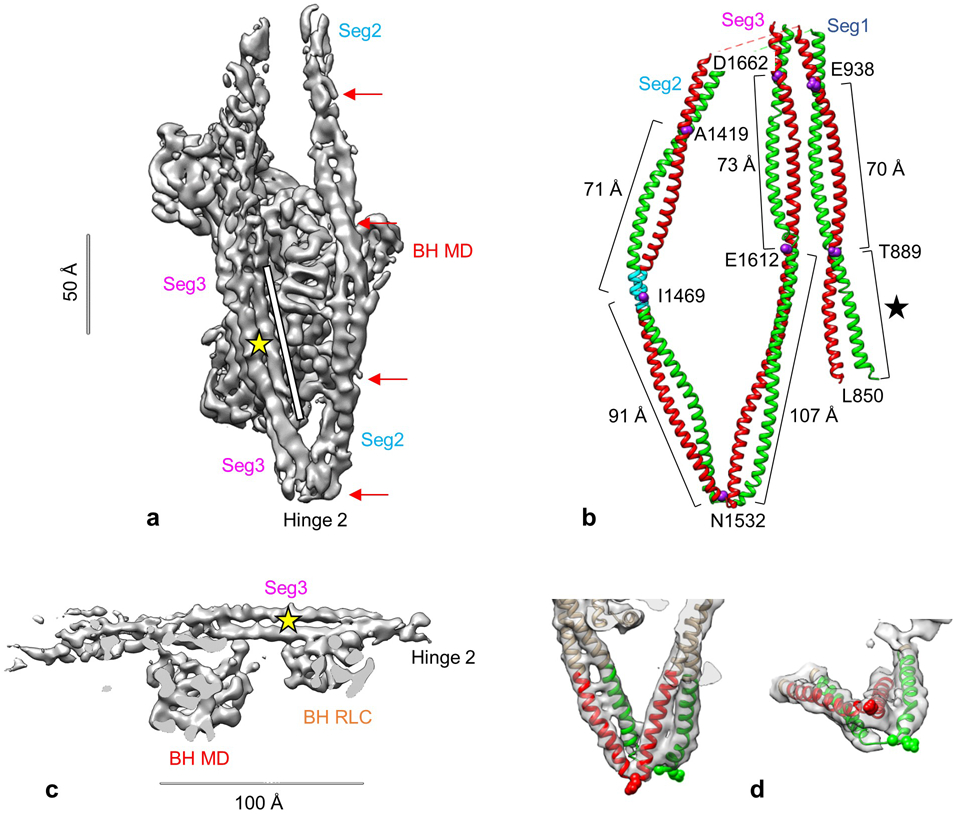
a. Oblique view of back of reconstruction. Red arrows indicate relatively regular crossovers of coiled-coil (~60-75 Å apart) as segment 2 wraps around the BH MD, and a long, parallel (untwisted) stretch in segment 3 after leaving hinge 2 (~100 Å; white bar). b. Atomic model of tail showing approximate distance between crossovers in coiled coil. Purple residues (numbered) are estimated to be at crossovers as seen in this face view, with distances between them shown. E1535-E1612 is an almost straight, non-coiled region of the tail, especially the green chain. The two heads would connect to the bottom of seg1 at L850. L850-T889 represents the first ~ 5 heptads of S2 (black star), which appear to associate with each other from the start, though with a slightly longer helical pitch in the first few residues. The two α-helices are shown in different colors for clarity. Their specific connection to the BH or FH is unknown (apart from seg1) due to lack of continuity of density in the top half of the reconstruction. Blue region in seg2 is M1462-K1472 (see text). c. Side-view of reconstruction showing the long stretch of untwisted coiled-coil in segment 3, running over the BH. Yellow star here and in a shows position of skip residue 1592. d. Hinge 2, in face- and end-views, showing continuity of coiling of the two α-helices about each other through this sharp bend, with local melting of α-helices likely at the bend. Glu1535 (spheres) is thought to mark the hinge point, although uncertainty in measurement of negative stain images means that the hinge could occur 2-3 amino acids either side of 15354. For this reason, numbering of amino acids in segs 2 and 3 in the atomic model is uncertain to the same degree. Density maps shown at contour level of 0.016.
Extended Data Fig. 6. Comparison of segment 1 position in filament and 10S molecule.
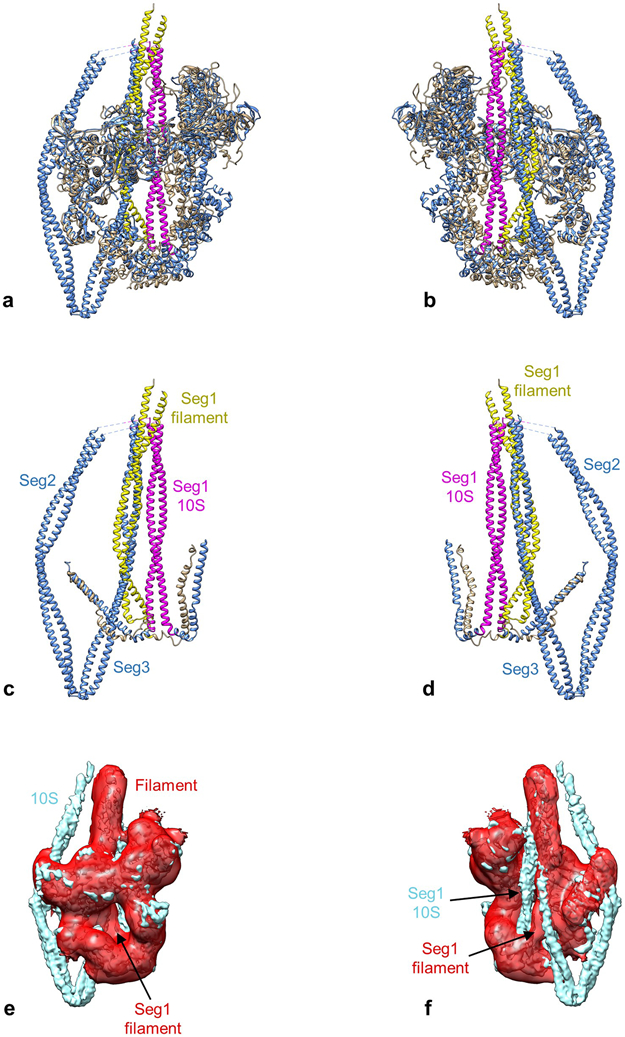
The different position of seg1 in the IHM of the tarantula filament and the 10S molecule is illustrated by comparing the PDBs of the best fits (tarantula filament, PDB 3JBH; 10S molecule, PDB 6XE9 [this work]). a, b. Front and rear views of IHM in which the head regions (filament, tan; 10S, blue) have been superposed (using Matchmaker in Chimera). c, d. Same views as for a, b, but with heads removed for clarity. Seg1 is yellow for the filament (bent conformation) and pink for the molecule (straight). The two segments run in different positions, ~ 20 Å apart, center to center. Strikingly, the position occupied by seg1 in the filament is taken by seg3 in the 10S molecule (yellow/blue overlap). e, f. The different seg1 positions are also clear when the filament (red) and 10S (blue) maps are compared. Front and rear views confirm that seg1’s in the two reconstructions are laterally displaced from each other. Similar results are obtained for two independent filament maps: EMD-1950 (shown)36 and EMD-651255.
Extended Data Fig. 7. Mechanisms of converter domain and actin-binding inhibition in BH and FH.
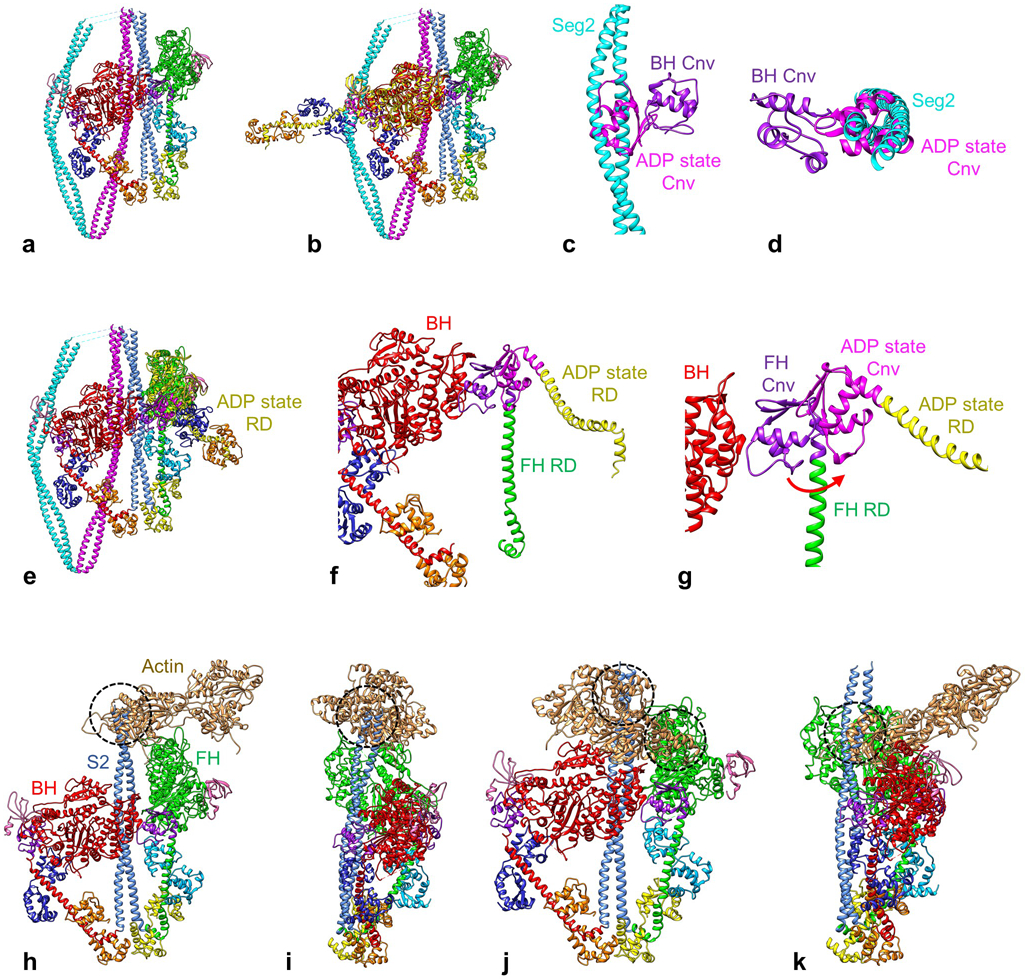
a-g. Converter domain inhibition. a. Atomic model of 10S structure. Both heads are in the ADP.Pi state (see text). b. 10S model, with a myosin head in ADP state (PDB 3I5F, yellow HC) superimposed on the BH by matching motor domains. The RD in this nucleotide state is straight and the Cnv (pink) is in a very different location from the BH Cnv (purple), clearly clashing with seg2. c, d. Detail of this clash in front and end views, with other parts of molecule removed. This comparison shows that for the BH to lose its Pi (going to the ADP state), its Cnv will clash with seg2. We conclude that seg2 acts as a physical barrier, preventing this transition and inhibiting BH ATP turnover. e. 10S model, with ADP-state head (PDB 3I5F) superposed on the FH MD. f, g. Detail of e, with LCs removed for clarity. The ADP-state RD and its Cnv (pink) are to the right (red arrow) of the ADP.Pi-state FH. This comparison suggests that for the FH to lose its Pi, the interaction of its Cnv with the BH MD must be broken. We propose that this is inhibited in the 10S structure by the strength of this interaction (BF), reinforced by FH interactions with the tail (TF1, TF2 and TF3). Together these interactions would prevent the ADP.Pi→ADP transition and inhibit FH ATP turnover. h-k. Actin-binding inhibition. The BH and FH were attached to actin by superposing their MDs on the MD (not shown) of mammalian actomyosin in the rigor state (PDB 5H53), as described in Extended Data Fig. 3. Segments 2 and 3 were removed for clarity. The modeling shows that attachment via both BH and FH is inhibited due to major steric clashes of other parts of the 10S structure with actin. h. Inhibition of binding via the FH. Front view of IHM shows that S2 clashes (dashed circle) with actin (2 monomers shown). i. Rotated 90° around vertical axis with respect to h. j. Inhibition of binding via the BH. Front view of IHM shows that S2 and the FH both clash with actin (dashed circles). k. Rotated 90° around vertical axis with respect to j.
Extended Data Fig. 8. Proposed mechanism of 10S myosin inhibition and activation, based on the atomic model and MD simulations.
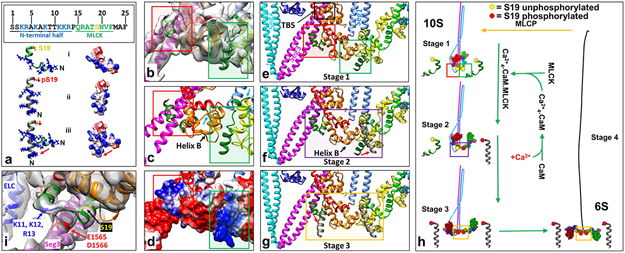
Smooth and nonmuscle myosin IIs are activated by phosphorylation of their RLCs on S19, leading to breaking of the 10S intramolecular interactions, unfolding to the extended structure, and assembly into functional filaments. Our atomic model indicates a possible mechanism.
Our previous work suggested that the single interaction most critical to the folded conformation is that occurring between seg3 and the BH RD7, and we noted how proximity of BH RLC S19 might regulate this interaction. Our atomic model suggests that seg3 in fact contacts the BH RLC at two sites. One is the C-lobe (TB5, Fig. 3a, g, and e above). The other involves the 24-residue N-terminal extension of the RLC, the phosphorylation domain (PD56), containing S19 (interaction TB6, Fig 3a). The PD is not observed in structures of the myosin head, but has been modeled by molecular dynamics simulations (a(i), dephosphorylated PD, ribbon and surface charge depictions; red negative, blue positive; upper box, PD sequence, MLCK binding site green; S19, yellow; N-terminal half positively charged)56. Our EM map reveals significant density (b, red rectangle), extending from F25, that fits this PD (b shows best fit of model from a(i) to BH PD density) and lies over seg3, below TB5 (red rectangles in b, showing fitting; c, model based on fit; e, zoomed-out model). In the atomic model (c), interaction occurs between positively charged residues of the PD N-terminal half and a negatively charged patch (~1560-1572) in seg37,43 (d, red rectangle; surface charge depiction; red, negatively charged; blue, positively charged; see interaction TB6 in Supplementary Table 1), which could strengthen TB5 (e). There is also significant density for a portion of the FH PD (b, green rectangle), which fits residues 20-24, while the positively charged N-terminal half (a) fits weak density near to negatively charged residues of BH RLC helix B (b-e, green rectangles). This would strengthen interaction BF2 between the RLCs. These interactions involving the RLCs, especially the BH PD with seg3, appear to be the key features creating the off-state, supported by the other interactions already described.
The structural basis of unfolding upon S19 phosphorylation remains unknown due to the absence of the PD in previous structures. The apparent PD densities we observe suggest the following model (h). Phosphorylation appears to occur first on the FH, then the BH7. EPR and molecular dynamics simulations suggest that phosphorylation causes straightening and stiffening of the PD56 (a: i. dephosphorylated, ii. phosphorylated, iii. transition, dephosphorylated → phosphorylated). When the unphosphorylated PDs (compact in our map; e, h, stage 1 in the activation sequence) are replaced by the phosphorylated (straightened) conformations (grey helices in f, g, using PD structures from a(ii)), the FH PD interaction with BH RLC helix B is removed (due to straightening and to the reduction in positive charge), which could weaken the RLC-RLC and thus head-head interaction (f, FH RLC phosphorylated, purple rectangle)36, releasing the FH, while retaining the folded tail structure (h, stage 2). When the BH is also phosphorylated, straightening/stiffening of its PD, and reduction in its positive charge, breaks its interaction with seg3 (g, red arrow, yellow rectangle; h, stage 3). With weakening of these interactions, seg2 could dissociate from the BH MD and ELC, leading to complete unfolding to the 6S structure (h, stage 4). In support of this proposal, replacement of charged amino acids near S19 in the RLC PD showed that unfolding upon phosphorylation may be due to net charge reduction of the PDs57. This physical model suggests that the two PDs with their phosphorylation sites, and the associated regions of seg3, represent a localized structural confluence in which the key events of activation and deactivation take place (the “phosphorylation zone”, e-g).
We tested the PD structure suggested by the MD simulations (in the case of the BH) by examining the sharpened map in this region and manually creating a model with the PD sequence to best fit the map using Coot (panel (i) above; viewing angle changed slightly from b to best show density and model features). The density clearly suggests a short helix followed by a loop and a second helix, with density present for the entire length of the PD. This is the first time that the PD has been directly visualized, as it is disordered in isolated myosin heads. We suggest that it is the binding of the PD to seg3 (occurring only in the 10S structure) that makes this visualization possible. The atomic model based on this fitting broadly supports the bent, helix-loop-helix conformation suggested by the MD simulations of the unphosphorylated PD (a(i)). The model (panel i) suggests that basic residues K11, K12, and R13, close to acidic residues D1565 and E1566 in seg3, electrostatically hold seg3 in the folded conformation—the most crucial interaction of the 10S structure—and in close proximity to the regulatory S19. MD simulations suggest that phosphorylation creates a salt bridge between phosphorylated S19 and R16, which causes the PD loop to become α-helical, straightening and stiffening the PD as a whole56. As discussed above, we propose that it is this straightening, and the reduction in positive charge of the PD, that cause the dissociation of seg3 from the PD, leading to unfolding and activation of the 10S structure as a whole (a, g, h). From the model it is not clear whether the BH RLC would be fully available for binding by MLCK in the 10S structure. Importantly, even if sufficiently exposed, the interaction of K11-R13 with seg3 could slow binding by MLCK, as these residues are also involved in MLCK substrate recognition58. If such hindrance occurs, this would be consistent with the proposal7 that BH phosphorylation, occurring after FH phosphorylation (h), is the final, required step for activation and unfolding.
Extended Data Fig. 9. Locations of disease-causing mutations in 10S structure.
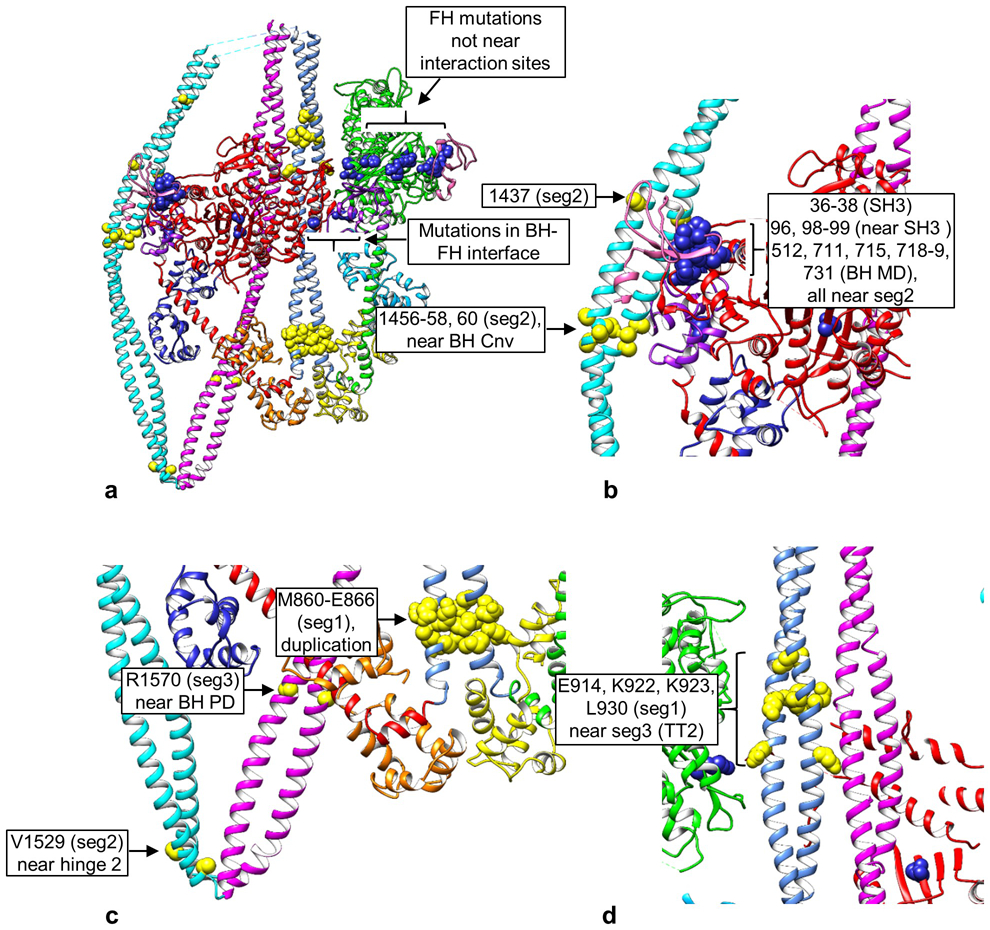
a. Overview of 10S molecule showing distribution of mutations, yellow in tail, blue in heads. b-d. Enlargements of mutation regions. b. Proximity of BH converter (Cnv, purple) and SH3 domain (pink) to seg2 mutations, and of BH SH3 and MD mutations to seg2. c. Mutations in segs 1, 2, and 3. V1529 is near hinge 2 and could impact hinge function. R1570 is part of the proposed interaction region of the BH RLC PD on seg3, and could impact RLC function. M860-E866 is a duplication, which could translate seg1, impacting its interactions downstream. d. Mutations in seg1 coincident with TT2 interaction. See Fig. 2b, Extended Data Table 2.
Extended Data Table 1.
Cryo-EM data collection, refinement and validation statistics.
| 10S smooth muscle myosin (EMDB-22145) (PDB 6XE9) |
|
|---|---|
| Data collection and processing | |
| Magnification | 105,000x |
| Voltage (kV) | 300 |
| Electron exposure (e–/Å2) | 43 |
| Defocus range (μm) | −1.2 to −3.5 |
| Pixel size (Å) | 0.415 |
| Symmetry imposed | C1 |
| Initial particle images (no.) | 1,765,220 |
| Final particle images (no.) | 260,360 |
| Map resolution (Å) | 4.3 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 4.1 – 9.5 |
| Refinement | |
| Initial model used (PDB code) | 1i84, 2FXM |
| Model resolution (Å) | 4.4 |
| FSC threshold | 0.143 |
| Model resolution range (Å) | 4.0-4.6 |
| Map sharpening B factor (Å2) | 40 |
| Model composition | |
| Non-hydrogen atoms | 22,081 |
| Protein residues | 2,949 |
| Ligands | 0 |
| B factors (Å2) | |
| Protein | 257.18 |
| Ligand | -- |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.03 |
| Bond angles (°) | 0.488 |
| Validation | |
| MolProbity score | 2.19 |
| Clashscore | 20.98 |
| Poor rotamers (%) | 0.77 |
| Ramachandran plot | |
| Favored (%) | 94.60 |
| Allowed (%) | 5.40 |
| Disallowed (%) | 0.00 |
Extended Data Table 2. Locations of disease-causing mutations in smooth and nonmuscle myosin II.
The table lists mutations in nonmuscle myosin IIA, IIB, IIC and smooth muscle myosin that have been associated with disease, together with regions of the molecule in close proximity to the mutations. The table and Figs. 2b, c demonstrate that many of the mutations occur close to regions we have identified as sites of intramolecular interaction in the 10S molecule. This suggests that mutation could alter the structure or stability of the folded conformation (see text).
| Mutated residue* | Gene/myosin type† | MYH11 equivalent‡ | Location§ | Potential interaction partner(s)‖ | Ref. |
|---|---|---|---|---|---|
| W33, V34, P35 | MYH9/NMII-A | W36, V37, P38 | SH3 | BH-Seg2/FH-none | 17 |
| N93, A95, S96 | MYH9/NMII-A | N96, A98, S99 | MD, near SH3 | BH-Seg2/FH-none | 17 |
| N97 | MYH10/NMII-B | N96 | MD, near SH3 | BH-Seg2/FH-none | 59 |
| S120 | MYH14/NMII-C | S99 | MD, near SH3 | BH-Seg2/FH-none | 17 |
| S237 (DAN) | MYH11/SMM | S245 | Switch 1 | BH-none/FH-none | 38 |
| R247 (MUS) | MYH11/SMM | R253 | Near switch 1 | BH-Seg3/FH-BH CM loop | 60 |
| K373 | MYH9/NMII-A | K386 | MD | BH-FH/FH-none | 17 |
| G376 | MYH14/NMII-C | G363 | MD | BH-none/FH-none | 17 |
| R501 | MYH11/SMM | R507 | Switch 2 | BH-none/FH-BH C-loop | 61 |
| W512 (DAN) | MYH11/SMM | W512 | Relay loop | BH-Seg2/FH-none | 62 |
| R702, R705, Q706 | MYH9/NMII-A | R715, R718, Q719 | Near SH1 helix | BH-Seg2/FH-none | 17 |
| R709 | MYH10/NMII-B | R715 | Near SH1 helix | BH-Seg2/FH-none | 59 |
| R718 | MYH9/NMII-A | R731 | Converter | BH-none/FH-BH C-loop | 17 |
| L722 | MYH14/NMII-C | L711 | Near SH1 helix | BH-Seg2/FH-none | 59 |
| R726 | MYH14/NMII-C | R715 | Near SH1 helix | BH-Seg2/FH-none | 17 |
| M847-E853dup | MYH9/NMII-A | M860-E866dup | Seg1 | Seg1-none | 17 |
| E908 | MYH10/NMII-B | E914 | Seg1, ring 1¶ | Seg1-Seg3/Seg1-FH | 17 |
| K910 | MYH9/NMII-A | K923 | Seg1, near ring 1,2¶ | Seg1-Seg3/Seg1 none | 17 |
| R933 | MYH14/NMII-C | K922 | Seg1, near ring 1,2¶ | Seg1-Seg3/Seg1-FH | 17 |
| V941 | MYH14/NMII-C | L930 | Seg1, near ring 2¶ | Seg1-Seg3/Seg1-FH | 63 |
| L976 | MYH14/NMII-C | L965 | Seg1 | Seg1-Seg3 | 17 |
| K1044 | MYH11/SMM | K1044 | Seg1, upper# | Seg1-Segs2,3** | 61 |
| K1048-E1054del | MYH9/NMII-A | K1061-E1067del | Seg1, upper# | Seg1-Segs2,3** | 17 |
| G1055-Q1068del | MYH9/NMII-A | G1068-Q1081del | Seg1, upper# | Seg1-Segs2,3** | 17 |
| E1066-A1072del/dup | MYH9/NMII-A | E1079-A1085dup | Seg1, upper# | Seg1-Segs2,3** | 17 |
| E1084del | MYH9/NMII-A | E1097del | Seg1, upper# | Seg1-Segs2,3** | 17 |
| V1092-R1162del | MYH9/NMII-A | L1105-R1175del | Seg1, upper# | Seg1-Segs2,3** | 17 |
| S1114 | MYH9/NMII-A | S1127¶ | Seg1, upper# | Seg1-Segs2,3** | 17 |
| T1155, R1162, R1165 | MYH9/NMII-A | T1168, R1175, R1178 | Hinge 1# | Segs1,2-Seg3** | 17 |
| T1162 | MYH10/NMII-B | T1168 | Hinge 1# | Seg1-Segs2,3** | 17 |
| L1205-Q1207del | MYH9/NMII-A | L1218-Q1220del | Seg2, upper# | Seg2-Segs1,3** | 17 |
| L1287 (DAN) | MYH11/SMM | V1295 | Seg2, upper# | Seg2-Segs1,3** | 38 |
| R1400 | MYH9/NMII-A | Q1413 | Seg2, lower | None | 17 |
| D1424 | MYH9/NMII-A | D1437 | Seg2, lower | Seg2-BH SH3 | 17 |
| Q1443-K1445dup | MYH9/NMII-A | Q1456-K1458dup | Seg2, lower | Seg2-BH converter | 17 |
| D1447 | MYH9/NMII-A | D1460 | Seg2, lower | Seg2-BH converter | 17 |
| V1516 | MYH9/NMII-A | V1529 | Hinge 2 | None | 17 |
| R1557 | MYH9/NMII-A | R1570 | Seg3, near BH | Seg3-BH RLC PD | 17 |
| 11816 | MYH9/NMII-A | 11829 | Seg3, upper# | Seg3-Segs1,2** | 17 |
| E1841 | MYH9/NMII-A | D1854 | Seg3, upper# | Seg3-Segs1,2** | 17 |
| G1924, D1925, P1927, | MYH9/NMII-A | G1937, N1938, P1940, | Seg3, non-helical tailpiece# | None | 17 |
| V1930, R1933, M1934, | MYH9/NMII-A | A1944, R1948, M1949, | Seg3, non-helical tailpiece# | None | 17 |
| D1941, E1945 | MYH9/NMII-A | D1960, E1964 | Seg3, non-helical tailpiece# | None | 17 |
Location of mutation in nonmuscle or smooth muscle myosin II sequence. DAN, zebrafish; MUS, mouse; all others, human.
Myosin heavy chain gene/myosin type (nonmuscle myosin: NMII-A, B, C; smooth muscle myosin: SMM).
Equivalent residue number in chicken smooth muscle myosin (MYH11) after sequence alignment (Supplementary Table 2).
Location is the approximate domain location (for heads) or position in tail. Mutations can occur in FH and BH and in both chains of the tail.
The first symbol shows which head (BH, FH) or segment (1, 2, 3) contains the mutation; the second symbol, after the hyphen, shows the nearby domains/potential interaction partners. After the “/”, the same notation applies to the other head or segment. Because the two heads have different environments, some mutations might affect BH but not FH interactions and vice versa. These potential interactions are mapped onto the 10S structure in Fig. 2b, c and Extended Data Fig. 9. BH, FH and segments are color-coded to correspond to Fig. 2b, c. Our criteria for “nearby domains/potential interaction partners” are distances of 3.5 to ~ 10 Å. ~1/3 of these are in the 3.5-5 Å range; mutation sites > 5 Å from a partner (not close enough for actual contact) would still impact local conformation and possibly alter nearby interactions.
Rings 1 and 2 refer to negatively charged regions (905-916 and 932-946, respectively) on segment 119.
Although the upper regions of segments 1, 2 and 3 are not in the reconstruction, they lie close together in negative stain images4, and appear likely to interact with each other.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health grants AR072036, AR067279, and HL139883 (RC), and HL075030, HL111696 and HL142853 (MI), and a University of Texas STARs PLUS Award (MI). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Drs. Karthikeyan Subramanian, Christna Ouch, William Royer, Chen Xu, Christl Gaubitz, and Nicholas Stone for advice and discussion on fitting and refinement, Kang Kang Song and Chen Xu for cryo-EM imaging, Lorenzo Alamo and Antonio Pinto for advice on homology modeling, and Michel Espinoza-Fonseca for providing the atomic models of the RLC NTEs. Cryo-EM imaging was carried out in the Massachusetts Facility for High-Resolution Electron Cryo-Microscopy at the University of Massachusetts Medical School. The Titan Krios was purchased with a grant from the Massachusetts Life Sciences Center capital fund.
Footnotes
Competing interests. The authors declare no competing interests.
Supplementary Information is available for this paper (two tables, five videos).
Reprints and permissions information is available at www.nature.com/reprints.
Data availability. Structural data that support the findings of this study on the structure of 10S myosin II have been deposited in the Worldwide Protein Data Bank (wwPDB) under accession codes EMD-22145 (the EM density map) and PDB 6XE9 (the atomic model). PDB data used to build the initial model were PDB 1i84 and PDB 2FXM.
References
- 1.Geeves MA & Holmes KC Structural mechanism of muscle contraction. Annu. Rev. Biochem 68, 687–728 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Suzuki H, Kamata T, Onishi H & Watanabe S Adenosine triphosphate-induced reversible change in the conformation of chicken gizzard myosin and heavy meromyosin. J Biochem 91, 1699–1705 (1982). [DOI] [PubMed] [Google Scholar]
- 3.Trybus KM, Huiatt TW & Lowey S A bent monomeric conformation of myosin from smooth muscle. Proc Natl Acad Sci U S A 79, 6151–6155 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess SA et al. Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state. J. Mol. Biol 372, 1165–1178 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Craig R, Smith R & Kendrick-Jones J Light-chain phosphorylation controls the conformation of vertebrate non-muscle and smooth muscle myosin molecules. Nature 302, 436–439 (1983). [DOI] [PubMed] [Google Scholar]
- 6.Wendt T, Taylor D, Trybus KM & Taylor K Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. U. S. A 98, 4361–4366 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S et al. The central role of the tail in switching off 10S myosin II activity. J Gen Physiol 151, 1081–1093, doi: 10.1085/jgp.201912431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross RA, Jackson AP, Citi S, Kendrick-Jones J & Bagshaw CR Active site trapping of nucleotide by smooth and non-muscle myosins. J. Mol. Biol 203, 173–181 (1988). [DOI] [PubMed] [Google Scholar]
- 9.Kiboku T et al. Nonmuscle myosin II folds into a 10S form via two portions of tail for dynamic subcellular localization. Genes Cells 18, 90–109, doi: 10.1111/gtc.12021 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Liu X et al. Effect of ATP and regulatory light-chain phosphorylation on the polymerization of mammalian nonmuscle myosin II. Proc Natl Acad Sci U S A 114, E6516–E6525, doi: 10.1073/pnas.1702375114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breckenridge MT, Dulyaninova NG & Egelhoff TT Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell 20, 338–347, doi: 10.1091/mbc.E08-04-0372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Milton DL et al. Direct evidence for functional smooth muscle myosin II in the 10S self-inhibited monomeric conformation in airway smooth muscle cells. Proc Natl Acad Sci U S A 108, 1421–1426, doi: 10.1073/pnas.1011784108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross RA What is 10S myosin for? J. Muscle Res. Cell Motil 9, 108–110 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Wendt T, Taylor D & Taylor K Refined model of the 10S conformation of smooth muscle myosin by cryo-electron microscopy 3D image reconstruction. J. Mol. Biol 329, 963–972 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Woodhead JL et al. Atomic model of a myosin filament in the relaxed state. Nature 436, 1195–1199 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Trybus KM & Lowey S Conformational states of smooth muscle myosin. Effects of light chain phosphorylation and ionic strength. J. Biol. Chem 259, 8564–8571 (1984). [PubMed] [Google Scholar]
- 17.Ma X & Adelstein RS The role of vertebrate nonmuscle Myosin II in development and human disease. Bioarchitecture 4, 88–102, doi: 10.4161/bioa.29766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KH et al. Interacting-heads motif has been conserved as a mechanism of myosin II inhibition since before the origin of animals. Proc Natl Acad Sci U S A 115, E1991–E2000, doi: 10.1073/pnas.1715247115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blankenfeldt W, Thoma NH, Wray JS, Gautel M & Schlichting I Crystal structures of human cardiac beta-myosin II S2-Delta provide insight into the functional role of the S2 subfragment. Proc. Natl. Acad. Sci. U. S. A 103, 17713–17717 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez R, Freyzon Y, Trybus KM & Cohen C Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre- power stroke state. Cell 94, 559–571 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Alamo L et al. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J Mol Biol 428, 1142–1164, doi: 10.1016/j.jmb.2016.01.027 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robert-Paganin J, Auguin D & Houdusse A Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat Commun 9, 4019, doi: 10.1038/s41467-018-06191-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pylypenko O & Houdusse AM Essential “ankle” in the myosin lever arm. Proc Natl Acad Sci U S A 108, 5–6, doi: 10.1073/pnas.1017676108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson RL et al. Deciphering the super relaxed state of human beta-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc Natl Acad Sci U S A 115, E8143–E8152, doi: 10.1073/pnas.1809540115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JH et al. Visualizing key hinges and a potential major source of compliance in the lever arm of myosin. Proc Natl Acad Sci U S A 108, 114–119, doi: 10.1073/pnas.1016288107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rimm DL, Sinard JH & Pollard TD Location of the head-tail junction of myosin. J Cell Biol 108, 1783–1789 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tama F, Feig M, Liu J, Brooks CL III & Taylor KA The requirement for mechanical coupling between head and S2 domains in smooth muscle myosin ATPase regulation and its implications for dimeric motor function. J. Mol. Biol 345, 837–854 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Brown JH et al. An unstable head-rod junction may promote folding into the compact off-state conformation of regulated myosins. J. Mol. Biol 375, 1434–1443, doi:S0022–2836(07)01570–7 [pii]; 10.1016/j.jmb.2007.11.071 [doi] (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolfson DN The design of coiled-coil structures and assemblies. Adv. Protein Chem 70, 79–112 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Hu Z, Taylor DW, Reedy MK, Edwards RJ & Taylor KA Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 A resolution. Sci Adv 2, e1600058, doi: 10.1126/sciadv.1600058 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor KC et al. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc Natl Acad Sci U S A 112, E3806–3815, doi: 10.1073/pnas.1505813112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olney JJ, Sellers JR & Cremo CR Structure and function of the 10 S conformation of smooth muscle myosin. J. Biol. Chem 271, 20375–20384 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Nag S et al. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat Struct Mol Biol 24, 525–533, doi: 10.1038/nsmb.3408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodhead JL & Craig R The mesa trail and the interacting heads motif of myosin II. Arch Biochem Biophys 680, 108228, doi: 10.1016/j.abb.2019.108228 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou HX & Pang X Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Chem Rev 118, 1691–1741, doi: 10.1021/acs.chemrev.7b00305 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brito R et al. A molecular model of phosphorylation-based activation and potentiation of tarantula muscle thick filaments. J. Mol. Biol 414, 44–61, doi:S0022–2836(11)01023–0 [pii]; 10.1016/j.jmb.2011.09.017 [doi] (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li XD, Saito J, Ikebe R, Mabuchi K & Ikebe M The interaction between the regulatory light chain domains on two heads is critical for regulation of smooth muscle myosin. Biochemistry 39, 2254–2260 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Abrams J et al. Graded effects of unregulated smooth muscle myosin on intestinal architecture, intestinal motility and vascular function in zebrafish. Dis Model Mech 9, 529–540, doi: 10.1242/dmm.023309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung HS, Komatsu S, Ikebe M & Craig R Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell 19, 3234–3242, doi: 10.1091/mbc.E08-02-0206 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dulyaninova NG & Bresnick AR The heavy chain has its day: regulation of myosin-II assembly. Bioarchitecture 3, 77–85, doi:26133 [pii] (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alamo L et al. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. Elife 6, doi: 10.7554/eLife.24634 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods and Extended Data references
- 42.Ikebe M & Hartshorne DJ Effects of Ca2+ on the conformation and enzymatic activity of smooth muscle myosin. J. Biol. Chem 260, 13146–13153 (1985). [PubMed] [Google Scholar]
- 43.Jung HS et al. Role of the tail in the regulated state of myosin 2. J Mol Biol 408, 863–878, doi: 10.1016/j.jmb.2011.03.019 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urnavicius L et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446, doi: 10.1126/science.aaa4080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takizawa Y et al. While the revolution will not be crystallized, biochemistry reigns supreme. Protein Sci 26, 69–81, doi: 10.1002/pro.3054 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burgess SA, Walker ML, Thirumurugan K, Trinick J & Knight PJ Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol 147, 247–258 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Mastronarde DN Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152, 36–51, doi: 10.1016/j.jsb.2005.07.007 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65, doi: 10.1038/nmeth.2727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544, doi: 10.1107/S2059798318006551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang S, Woodhead JL, Zhao FQ, Sulbaran G & Craig R An approach to improve the resolution of helical filaments with a large axial rise and flexible subunits. J Struct Biol 193, 45–54, doi: 10.1016/j.jsb.2015.11.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espinoza-Fonseca LM, Kast D & Thomas DD Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J 93, 2083–2090, doi: 10.1529/biophysj.106.095802 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sweeney HL, Yang Z, Zhi G, Stull JT & Trybus KM Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc. Natl. Acad. Sci. U. S. A 91, 1490–1494 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gallagher PJ, Herring BP & Stull JT Myosin light chain kinases. J. Muscle Res. Cell Motil 18, 1–16 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Kim KY, Kovacs M, Kawamoto S, Sellers JR & Adelstein RS Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J Biol Chem 280, 22769–22775, doi: 10.1074/jbc.M503488200 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Huang J et al. Genetic approaches to identify pathological limitations in aortic smooth muscle contraction. PLoS One 13, e0193769, doi: 10.1371/journal.pone.0193769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alhopuro P et al. Unregulated smooth-muscle myosin in human intestinal neoplasia. Proc Natl Acad Sci U S A 105, 5513–5518, doi: 10.1073/pnas.0801213105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wallace KN et al. Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev Cell 8, 717–726, doi: 10.1016/j.devcel.2005.02.015 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Almutawa W et al. The R941L mutation in MYH14 disrupts mitochondrial fission and associates with peripheral neuropathy. EBioMedicine 45, 379–392, doi: 10.1016/j.ebiom.2019.06.018 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


