Introduction
Malaria led to approximately 228 million cases and 405,000 deaths worldwide in 2018 [1]. Caused by the transmission of Plasmodium parasites by the female anopheles mosquito, malaria poses a risk for almost half of the world’s population, with higher risk of severe disease and mortality in children under the age of five and pregnant women [1]. In addition to the health risk posed by malaria, the disease has a large global economic burden, including the cost of research, distribution of mosquito nets and antimalarials, government spending for in and out patient care, malaria clinics, and lost work due to illness [2–4]. Worldwide efforts to eradicate malaria have led to a decrease in the numbers of mortalities since the early 2000s [5]. However, due to growing resistance to available antimalarials and the lack of an effective, long-lasting vaccine, these numbers have begun to plateau, highlighting the continued need for new, innovative, and affordable methods of treatment [1, 6–9]. Understanding the role of gut microbiota in human health and disease has expanded widely over the last decade, with improved understanding of the ability of gut microbes to alter the immune response to chronic inflammatory and infectious diseases both locally and systemically [10]. Together, the need for novel malaria treatments and the growing knowledge of the role of the gut microbiota in health and disease point to the potential for gut microbiota modulation as a treatment for severe malaria.
Role of gut microbiota in Plasmodium infection
While there is still much to understand before gut microbiota modulation becomes a viable and optimal treatment to prevent severe malaria, recent evidence in both rodent models and human studies have pointed to gut microbiota composition as a factor in disease progression [11, 12]. Villarino et. al. has shown that, when C57BL/6 mice ordered from different vendors are infected with Plasmodium yoelii 17XNL—a nonlethal, rodent-specific strain of Plasmodium—mice exhibited profound differences in infection severity, morbidity, and mortality based on vendor, and that these differences in susceptibility are gut microbiota dependent [13**]. The role of the gut microbiota in Plasmodium severity was further illustrated in research by Morffy Smith et. al. that showed disruption of the gut microbiota in outbred Swiss Webster mice through antibiotic treatment and fecal microbiota transplant resulted in differing susceptibly to and pregnancy outcomes during infection with Plasmodium chabaudi chabaudi—a rodent model for chronic malaria [14].
Additionally, research using mouse models have shown that Plasmodium infections can lead to changes in gut microbiota and gastrointestinal health. Taniguchi et. al. showed that, when C57BL/6 mice were infected with Plasmodium berghei ANKA—a lethal, rodent-specific Plasmodium strain that, in C57BL/6 mice, leads to experimental cerebral malaria (ECM)—changes in intestinal pathology and gut microbiota composition were seen, and these changes correlated with the development of ECM [15]. It has also been shown by Denny et. al. that, in C57BL/6 mice with differing susceptibility to Plasmodium yoelii 17XNL, there is an increase of proinflammatory cells in the lamina propria, prolonged liver damage, and changes in cecal metabolites in mice that exhibit severe malaria upon infection [16]. This report demonstrated shifts in gut bacteria composition in mice regardless of infection severity, yet those changes in gut bacteria composition did not alter susceptibility to future Plasmodium infections. P. yoelii nigeriensis infection also induced changes in gut microbiota, which increased susceptibility to non-typhoid Salmonella infections [17]. Contrary to these rodent models, a paper by Mandal et. al. looking at gut bacteria composition in Kenyan infants found minimal shifts in gut bacteria following a malaria episode and treatment with artemether+lumefantrine [18], illustrating that gut bacterial composition shifts may vary based on host, infection severity, and Plasmodium species.
Several studies have also begun to identify the role of gut microbiota in Plasmodium infection risk and immune response in humans. In a paper by Yooseph et. al., it was shown that there is an association between the composition of stool bacteria composition at the onset of the ensuing malaria transmission season and susceptibility to Plasmodium falciparum infection [19**]. While the gut microbiota was not a good predictor of who would develop a febrile (or symptomatic) infection, it was a good predictor of the likelihood of a P. falciparum infection being established as measured by PCR [19**]. Likewise, Mandal et. al. found variation in the gut microbiota of Kenyan children who had an increased number of malaria episodes rather than a single episode throughout a malaria season [18]. Furthermore, Yilmaz et. al. illustrated that colonization of α-gal producing human gut bacteria E. coli O86:B7 leads to α-gal specific antibodies, and these antibodies are cross-protective against Plasmodium sporozoite infection of hepatocytes in both a mouse model and human populations [20**]. Together, though these human and mouse studies evaluate the role of gut microbiota on the host response to Plasmodium from different viewpoints, they each illustrate the complex relationship between gut microbiota composition and Plasmodium infection.
Immune responses leading to Plasmodium clearance and acquired immunity
Though the immune response necessary to clear Plasmodium and eventually develop acquired immunity is complex due to factors such as large antigenic variation and a complex life cycle [21, 22], it is well established that the production of Plasmodium-specific antibodies are necessary in order to provide at least partial protection from Plasmodium infection [23] (Figure 1). However, protection of Plasmodium-specific antibodies through immune memory only reaches a protective threshold to clinical malaria after years of Plasmodium exposure and can be lost if an individual leaves a malaria endemic region only to return several years later [24, 25]. Studies have shown that it is both a combination of the antibody levels in circulation as well as the number of antigens these antibodies target that can predict protection from clinical malaria [26, 27]. The underlying factors dictating the speed at which an individual gains a protective antibody threshold and develops clinically asymptomatic malaria following Plasmodium infection remain unknown.
Figure 1. Adaptive immune response to Plasmodium.
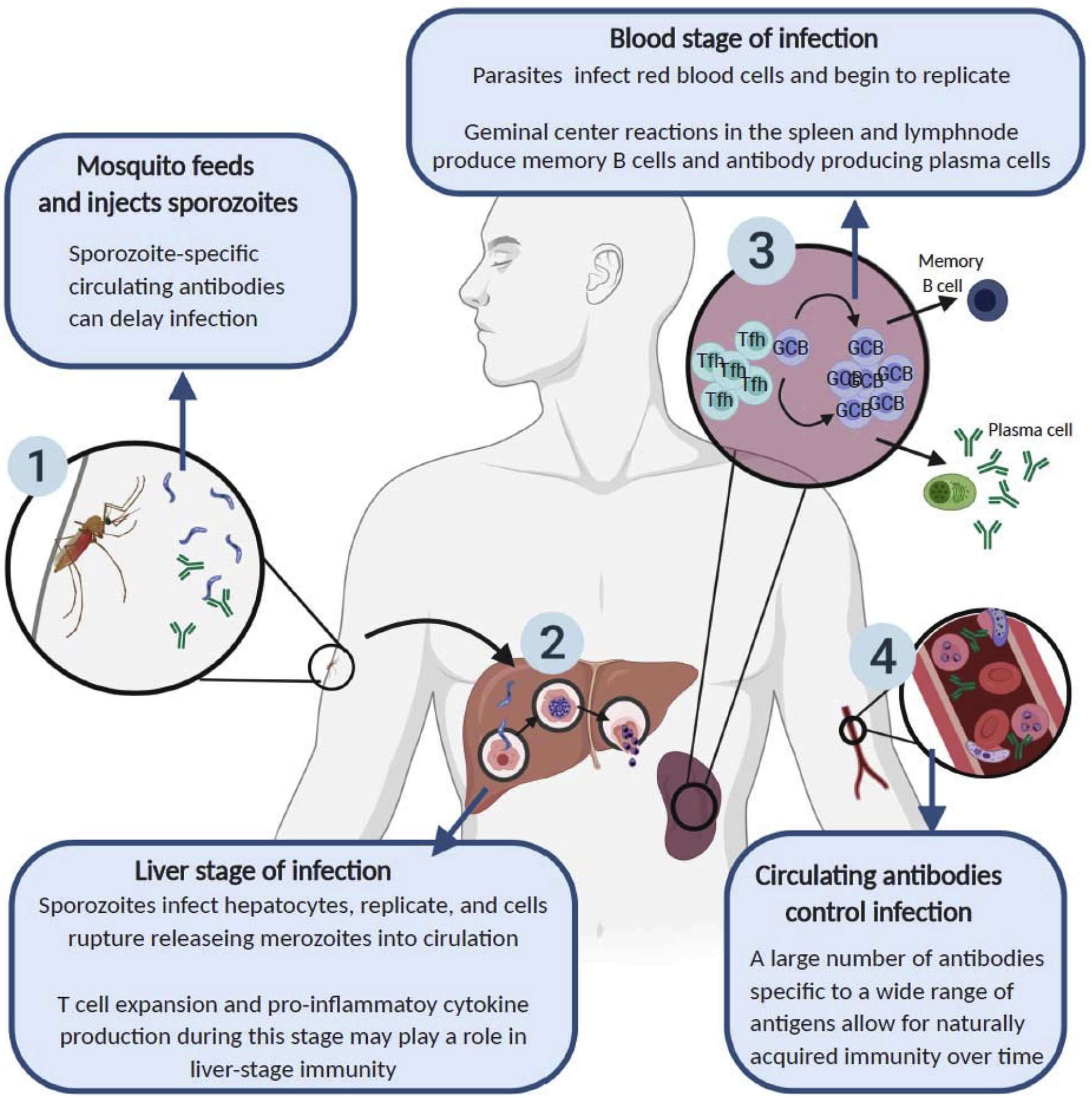
Schematic illustrating the role of the adaptive immune system in controlling a Plasmodium infection from initial parasite inoculation (1) through the liver (2) and control of blood stage of infection (3,4). When a mosquito feeds on a human host, sporozoites are injected into the skin where neutralizing antibodies have been found to immobilize invading sporozoites and delay infection [47, 48]. During the “clinically silent” liver stage of the infection, CD8+ T cells have been observed migrating to the liver to play a role in liver-stage immunity [49], while immune cell expansion and pro-inflammatory immune responses during liver stage play a role in the development of cerebral malaria [50]. Finally, during the blood stage of infection, parasites undergo replication in red blood cells that allows the immune system to recognize Plasmodium antigen and mount an adaptive immune system response, leading to B cell responses that produce high affinity antibodies that enter into circulation, and lead to the elimination of infected red blood cells [51]. Repeated exposures to Plasmodium lead to the development of naturally acquired immunity to clinical malaria over time [25].
In the context of Plasmodium infections, a range of cell types including T follicular helper (Tfh) and germinal center (GC) B cells have been shown to be critical in developing high-affinity, long-lived antibodies in both rodent and human Plasmodium species [28, 29]. It has been shown that disruption of IL-21 signaling produced by Tfh cells leads to sustained high parasitemia in mice infected with P. yoelii and lack of immunity to subsequent infections, and the disruption of Tfh cells in particular leads to an inability of mice to clear the typically self-resolving chronic P. chabaudi infection [29, 30]. Furthermore, impaired Tfh differentiation and thus inefficient GC responses has been shown in severe malaria infection, and these GC responses were restored upon the blockade of inflammatory cytokines [31]. Additional cytokines affecting the humoral immune response to Plasmodium include the presence of IFNγ, which can lead to atypical memory B cells that have reduced effector functions—and B cell intrinsic IL-10—which has been shown to be necessary for GC B cell reaction, efficient antibody responses, and ultimate parasite control and host survival in a murine model [32, 33].
Gut microbiota influence on adaptive immune response
The gut microbiota has a well-established role in the development of a range of immune responses [34]. While a specific mechanism between gut microbiota composition and Plasmodium susceptibility was not identified by Villarino et. al., it was shown that mice resistant to hyperparasitemia following P. yoelii 17XNL infection showed increased numbers of splenic humoral immune cells such as Tfh cells and GC B cells, as well as an increase in Plasmodium-specific antibody titers compared to hyperparasitemia susceptible mice [13**]. Additionally, Yooseph et. al. noted gut microbiota shifts in the age range that children typically develop acquired immunity to Plasmodium [19**], suggesting that age-associated gut microbiota changes may play a previously unidentified role in the ability of an individual to reach a P. falciparium-specific antibody threshold that prevents the development of severe disease. This data leads to the exciting prospect that gut microbiota composition is contributing to the adaptive immune response, thus leading to increased resistance to severe disease and the development of a protective antibody threshold to severe malaria.
Though the specific role of gut microbiota on the systemic immune response to Plasmodium is still being explored, previous data has shown a role of gut microbiota in the development of adaptive immune-mediated protection from extra-GI infections such as influenza and LCMV (Figure 2) [35, 36]. Of note, these studies demonstrated that mice largely devoid of gut microbiota, as a consequence of broad-spectrum antibiotic treatment, have impaired adaptive immune responses. Therefore, it remains less clear how unique compositions of gut microbiota impact adaptive immune responses. To this end, Teng et. al. provided evidence that specific bacteria among the consortium of other gut microbiota can have an effect on the gut-distal immune system [37*]. In the context of an autoimmune arthritis model, they illustrated that gut commensal segmented filamentous bacteria (SFB) led to differentiation of Tfh cells in Peyer’s patches, which then egress and traveled to gut-distal lymphoid tissues where they contributed to GC B cell responses and the production of auto-antibodies (Figure 2) [37*]. In addition to a direct role of gut microbiota on the development of T cells and antibody production, gut microbial products have been shown to have an effect on cytokines such as IL-21 [38, 39], which, as noted above, is essential to the systemic adaptive immune response to Plasmodium.
Figure 2. The known effects of the gut microbiota on the systemic immune system.
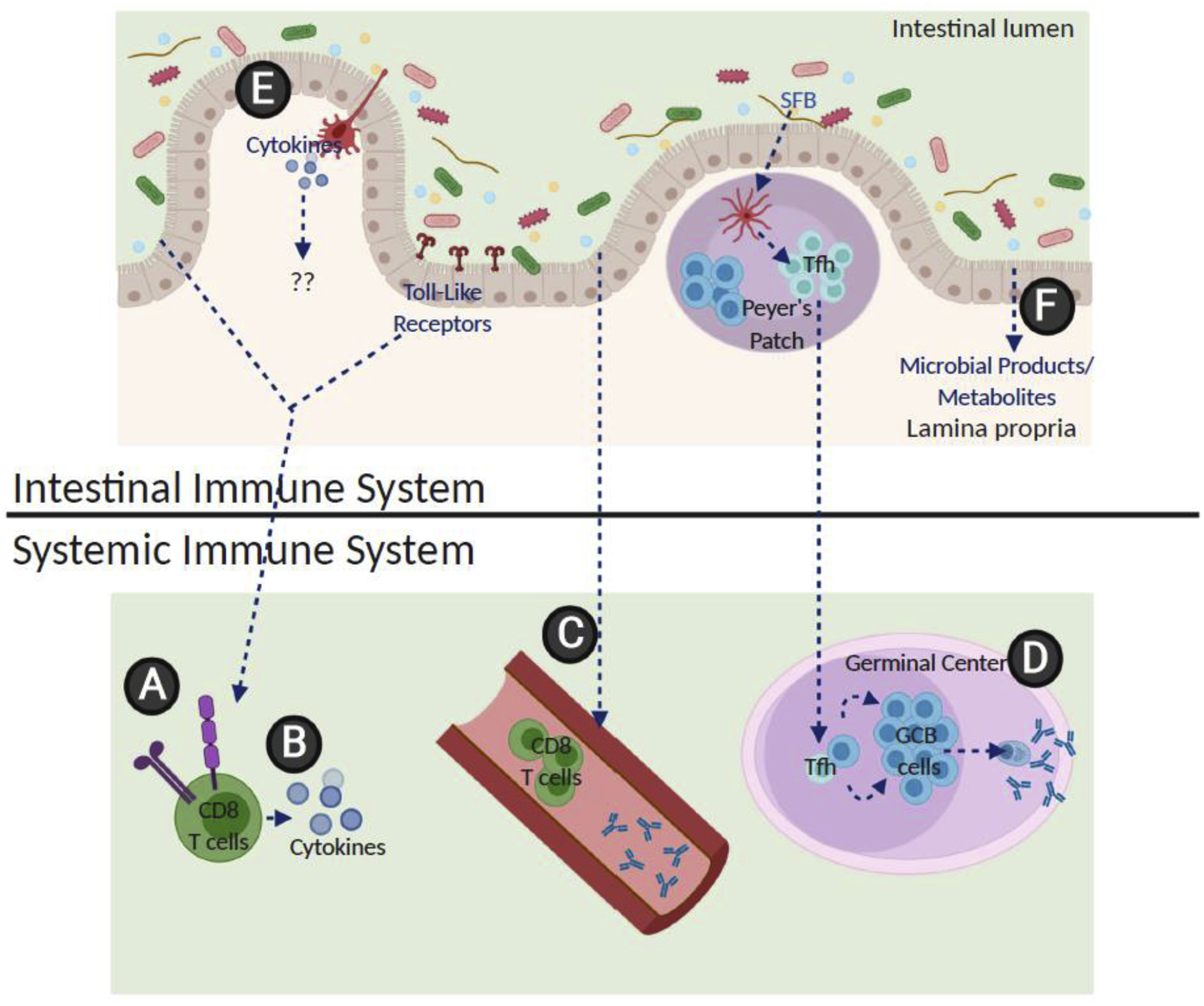
(A) The presence of gut microbes have been found to lead to increased numbers of viral specific CD8+ T cells in the spleen and lungs as well as a decrease in surface inhibitory molecules on these cells during a viral infection [35, 36]. (B) CD8+ T cells also have increased inflammatory cytokine production when the gut microbiota is present, as well as (C) increased numbers of circulating T cells and viral specific antibodies in the blood [35, 36]. (D) Tfh cells primed by SFB in the Peyer’s Patches have been shown to migrate systemically, leading to GC B cell activation and autoantibody production in an autoimmune arthritis model [37*]. (E) While dendritic cells in the lamina propria are known to produce cytokines in response to the gut microbiota and (F) a range of microbial products such as short-chain fatty acids and other metabolites such as PAMPs are known to enter circulation, the exact role these play on the systemic adaptive immune response remain unclear.
Yilmaz et. al. illustrated that bacteria present in the gut microbiota can have an influence on antibody production through cross reactivity, and it is known that a larger breadth of Plasmodium antigens to which an individual produces antibodies leads to fewer cases of clinical malaria [20**, 26, 27]. Together, this data suggests that altering gut microbiota composition could benefit the development of a more efficient adaptive immune response to Plasmodium and lead to increases in Plasmodium antigens that are targeted by the immune response. Though additional research needs to be performed to identify the mechanism by which gut microbiota may be shaping adaptive immunity to Plasmodium, altering gut microbiota composition to modulate adaptive immunity and thus decrease severe or symptomatic malaria remains a potential novel treatment for Plasmodium.
Problems and potential of treating malaria through modulating gut microbiota
Gut microbiota modulation as treatment for diseases, including malaria, has a number of benefits including its low cost. Mathematical modeling used to evaluate the cost-effectiveness of probiotic distribution in sub-Saharan Africa found that probiotic intervention leading to a 2- to 14-fold decrease in incidence of severe malaria would be extremely cost effective for the majority of the region [40]. In addition to being a very low cost therapeutic, probiotic bacteria can be freeze-dried and delivered orally, removing the need for refrigeration or trained medical professionals to provide hands on delivery. Furthermore, gut microbiota supplementation has the potential to prime the host immune system to respond better to Plasmodium rather than targeting the parasite, thus removing the possibility of loss of effectiveness due to Plasmodium resistance. The potential to target gut microbiota as a preventative treatment was illustrated in a study that showed oral symbiotic treatment to target gut microbiota in rural Indian infants provided significant reduction in sepsis-related deaths and unexpectedly a reduction in lower respiratory tract infections [41*].
Though a better understanding of gut microbiota and utilizing the knowledge as a treatment for a range of diseases is an area of research that is important to a variety of fields, there remain important challenges that must be considered as this field moves forward. One of these challenges is identifying the optimal window of treatment for gut microbiota modulation. Villarino et. al. showed that, while there was a significant decrease in malaria susceptibility in mice following Lactobacillus and Bifidobacterium treatment, this only occurred after several weeks of treatment with a broad-spectrum antibiotic cocktail [13**]. Treating otherwise healthy individuals with antibiotics prior to probiotic administration would not only be expensive and encourage the emergence of antibiotic-resistant bacterial strains, but has been shown to negatively affect gut microbiota recovery following probiotic treatment, potentially leading to negative health effects [42]. Still, because gut microbiota of young children—the age most at risk for severe malaria infection and mortality—is very dynamic [43], there likely exists a period where gut microbiota modulation would be most effective.
Finally, even if altering gut microbiota is not sufficient on its own to prevent severe malaria, better understanding how gut microbiota influences the host antibody response toPlasmodium could open the door for gut microbiota modulation in combination with vaccination to improve the efficacy and longevity of a malaria vaccine. Evidence shows that gut microbiota influences antibody responses to a range of vaccinations including the influenza vaccine, meningococcal vaccine, and Infanrix®-hexa combination vaccine [44–46]. Future research targeting gut microbiota modulation in combination with Plasmodium vaccination holds the potential to not only aid in the host immune response to vaccination, but also to expand the breadth of the immune response to vaccinated antigens and provide longer lasting host memory.
Conclusion
Recent research has shown that gut microbiota plays an important role in the severity and outcome of Plasmodium infections in both humans and mice. The specific bacteria driving these differences, the interplay between bacteria and host response, and the mechanism by which severe malaria is being prevented, however, remain to be fully understood. Though more research would need to be done before clinical trials can be pursued, treating young children—the population most at risk of severe malaria and malaria-associate mortality—with probiotics to modulate their microbiome remains an encouraging prospect, and better understanding the interplay between gut microbiota and the host response to Plasmodium will open the door to a range of novel treatment options for malaria.
Highlights.
Gut microbiota is associated with malaria in rodents and humans.
Gut microbiota influences systemic adaptive immune responses.
Probiotic treatment in infants can decrease infection-related mortality.
Acknowledgements
We apologize to the authors who have made important contributions to the subject areas discussed in this article that we were not able to reference owing to space constraints. This work was supported in part by a grant from the National Institute of Allergy and Infectious Disease of the National Institutes of Health (1R01AI123486) to N.W.S. Support provided by the Herman B. Wells Center to N.W.S. was in part from the Riley Children’s Foundation. The project described was supported by the Indiana University Health - Indiana University School of Medicine Strategic Research Initiative. S.D.G.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest:
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.WHO. World Malaria Report 2019.
- 2.Haakenstad A, et al. , Tracking spending on malaria by source in 106 countries, 2000–16: an economic modelling study. Lancet Infect Dis, 2019. 19(7): p. 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallup JL and Sachs JD, The economic burden of malaria. Am J Trop Med Hyg, 2001. 64(1–2 Suppl): p. 85–96. [DOI] [PubMed] [Google Scholar]
- 4.Alonso S, et al. , The economic burden of malaria on households and the health system in a high transmission district of Mozambique. Malar J, 2019. 18(1): p. 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roser M and Ritchie H. Malaria. 2020; Retrieved from: ‘https://ourworldindata.org/malaria’ [Online Resource]].
- 6.Ashley EA, et al. , Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med, 2014. 371(5): p. 411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamilton WL, et al. , Evolution and expansion of multidrug-resistant malaria in southeast Asia: a genomic epidemiology study. Lancet Infect Dis, 2019. 19(9): p. 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menard D and Dondorp A, Antimalarial Drug Resistance: A Threat to Malaria Elimination. Cold Spring Harb Perspect Med, 2017. 7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phyo AP, et al. , Declining Efficacy of Artemisinin Combination Therapy Against P. Falciparum Malaria on the Thai-Myanmar Border (2003–2013): The Role of Parasite Genetic Factors. Clin Infect Dis, 2016. 63(6): p. 784–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durack J and Lynch SV, The gut microbiome: Relationships with disease and opportunities for therapy. J Exp Med, 2019. 216(1): p. 20–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mukherjee D, Chora AF, and Mota MM, Microbiota, a Third Player in the Host-Plasmodium Affair. Trends Parasitol, 2020. 36(1): p. 11–18. [DOI] [PubMed] [Google Scholar]
- 12.Ippolito MM, et al. , Malaria and the Microbiome: A Systematic Review. Clin Infect Dis, 2018. 67(12): p. 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villarino NF, et al. , Composition of the gut microbiota modulates the severity of malaria. Proc Natl Acad Sci U S A, 2016. 113(8): p. 2235–40. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Genetically identical mice ordered from different vendors have profound differences in Plasmodium yoelii infection severity, and that these differences are driven by the gut microbiota.
- 14.Morffy Smith CD, et al. , Composition of the gut microbiota transcends genetic determinants of malaria infection severity and influences pregnancy outcome. EBioMedicine, 2019. 44: p. 639–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi T, et al. , Plasmodium berghei ANKA causes intestinal malaria associated with dysbiosis. Sci Rep, 2015. 5: p. 15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JE, et al. , Differential Sensitivity to Plasmodium yoelii Infection in C57BL/6 Mice Impacts Gut-Liver Axis Homeostasis. Sci Rep, 2019. 9(1): p. 3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooney JP, et al. , Inflammation-associated alterations to the intestinal microbiotareduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci Rep, 2015. 5: p. 14603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal RK, et al. , Longitudinal Analysis of Infant Stool Bacteria Communities Before and After Acute Febrile Malaria and Artemether-Lumefantrine Treatment. J Infect Dis, 2019. 220(4): p. 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yooseph S, et al. , Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics, 2015. 16: p. 631. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Gut microbiota composition prior to the onset of malaria season is a predictor of the risk of a Plasmodium falciparum infection in Malian children.
- 20.Yilmaz B, et al. , Gut microbiota elicits a protective immune response against malaria transmission. Cell, 2014. 159(6): p. 1277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The presence of α-gal antibodies is associated with protection from Plasmodium falciparum in humans, and murine gut colonization with α-gal expressing E. coli O86:B7 results in cross-reactive antibodies that are protective against Plasmodium infection.
- 21.Belachew EB, Immune Response and Evasion Mechanisms of Plasmodium falciparum Parasites. J Immunol Res, 2018. 2018: p. 6529681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long CA and Zavala F, Immune Responses in Malaria. Cold Spring Harb Perspect Med, 2017. 7(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen S, Mc GI, and Carrington S, Gamma-globulin and acquired immunity to human malaria. Nature, 1961. 192: p. 733–7. [DOI] [PubMed] [Google Scholar]
- 24.Crompton PD, et al. , A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci U S A, 2010. 107(15): p. 6958–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss GE, et al. , The Plasmodium falciparum-specific human memory B cell compartment expands gradually with repeated malaria infections. PLoS Pathog, 2010. 6(5): p. e1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osier FH, et al. , Breadth and magnitude of antibody responses to multiple Plasmodium falciparum merozoite antigens are associated with protection from clinical malaria. Infect Immun, 2008. 76(5): p. 2240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogaro SI, et al. , The breadth, but not the magnitude, of circulating memory B cell responses to P. falciparum increases with age/exposure in an area of low transmission. PLoS One, 2011. 6(10): p. e25582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueiredo MM, et al. , T follicular helper cells regulate the activation of B lymphocytes and antibody production during Plasmodium vivax infection. PLoS Pathog, 2017. 13(7): p. e1006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Mazliah D, et al. , Follicular Helper T Cells are Essential for the Elimination of Plasmodium Infection. EBioMedicine, 2017. 24: p. 216–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Mazliah D, et al. , Disruption of IL-21 signaling affects T cell-B cell interactions and abrogates protective humoral immunity to malaria. PLoS Pathog, 2015. 11(3): p. e1004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryg-Cornejo V, et al. , Severe Malaria Infections Impair Germinal Center Responses by Inhibiting T Follicular Helper Cell Differentiation. Cell Rep, 2016. 14(1): p. 68–81. [DOI] [PubMed] [Google Scholar]
- 32.Obeng-Adjei N, et al. , Malaria-induced interferon-gamma drives the expansion of Tbethi atypical memory B cells. PLoS Pathog, 2017. 13(9): p. e1006576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guthmiller JJ, et al. , Cutting Edge: IL-10 Is Essential for the Generation of Germinal Center B Cell Responses and Anti-Plasmodium Humoral Immunity. J Immunol, 2017. 198(2): p. 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamada N and Nunez G, Regulation of the immune system by the resident intestinal bacteria. Gastroenterology, 2014. 146(6): p. 1477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abt MC, et al. , Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity, 2012. 37(1): p. 158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichinohe T, et al. , Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A, 2011. 108(13): p. 5354–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teng F, et al. , Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer’s Patch T Follicular Helper Cells. Immunity, 2016. 44(4): p. 875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]; *The presence of segmented filamentous bacteria in the gut microbiota of mice led to the production of Tfh cell dependent autoantibodies in the systemic lymphoid tissues in an autoimmune arthritis model.
- 38.Cho H, et al. , Microbiota-dependent IL-21 signaling regulates intestinal immune cell homeostasis and immunopathology to infection The Journal of Immunology, 2017. 198(1). [Google Scholar]
- 39.Kim M and Kim CH, Regulation of humoral immunity by gut microbial products. Gut Microbes, 2017. 8(4): p. 392–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greenhalgh S and Chandwani V, Advocating an attack against severe malaria: a cost-effectiveness analysis. BMC Public Health, 2020. 20(1): p. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panigrahi P, et al. , A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature, 2017. 548(7668): p. 407–412. [DOI] [PubMed] [Google Scholar]; *Treatment of Indian infants with Lactobacillus plantarum and fructooligosaccharide (a probiotic and prebiotic respectively) led to a significant decrease in sepsis related death compared to placebo treated infants.
- 42.Suez J, et al. , Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell, 2018. 174(6): p. 1406–1423 e16. [DOI] [PubMed] [Google Scholar]
- 43.Tanaka M and Nakayama J, Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int, 2017. 66(4): p. 515–522. [DOI] [PubMed] [Google Scholar]
- 44.Oh JZ, et al. , TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity, 2014. 41(3): p. 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ciabattini A, et al. , Role of the Microbiota in the Modulation of Vaccine Immune Responses. Front Microbiol, 2019. 10: p. 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynn MA, et al. , Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe, 2018. 23(5): p. 653–660 e5. [DOI] [PubMed] [Google Scholar]
- 47.Barry A, et al. , Functional antibodies against Plasmodium falciparum sporozoites are associated with a longer time to qPCR-detected infection among schoolchildren in Burkina Faso. Wellcome Open Res, 2018. 3: p. 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flores-Garcia Y, et al. , Antibody-Mediated Protection against Plasmodium Sporozoites Begins at the Dermal Inoculation Site. mBio, 2018. 9(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holz LE, Fernandez-Ruiz D, and Heath WR, Protective immunity to liver-stage malaria. Clin Transl Immunology, 2016. 5(10): p. e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato Y, et al. , The Liver-Stage Plasmodium Infection Is a Critical Checkpoint for Development of Experimental Cerebral Malaria. Front Immunol, 2019. 10: p. 2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White NJ, Malaria parasite clearance. Malar J, 2017. 16(1): p. 88. [DOI] [PMC free article] [PubMed] [Google Scholar]


