Abstract
Mammalian polychlorinated biphenyl (PCB) metabolism has not been systematically explored with nontarget high-resolution mass spectrometry (Nt-HRMS). Here we investigated the importance of the gut microbiome in PCB biotransformation by Nt-HRMS analysis of feces from conventional (CV) and germ-free (GF) adult female mice exposed to a single oral dose of an environmental PCB mixture (6 mg/kg or 30 mg/kg in corn oil). Feces were collected for 24 h after PCB administration, PCB metabolites were extracted from pooled samples, and the extracts were analyzed by Nt-HRMS. Twelve classes of PCB metabolites were detected in the feces from CV mice, including PCB sulfates (PCB sulfates), hydroxylated PCB sulfates (OH-PCB sulfates), PCB sulfonates (PCB sulfonates), and hydroxylated methyl sulfone PCBs (OH-MeSO2-PCBs) reported previously. We also observed eight additional PCB metabolite classes that were tentatively identified as hydroxylated PCBs (OH-PCBs), dihydroxylated PCBs (diOH-PCBs), monomethoxylated dihydroxylated PCBs (MeO-OH-PCBs), methoxylated PCB sulfates (MeO-PCB sulfates), mono- to tetra-hydroxylated PCB quinones ((OH)x-Q, x=1-4), and hydroxylated polychlorinated benzofurans (OH-PCDF). Most metabolite classes were also detected in the feces from GF mice, except for MeO-OH-PCBs, OH-MeSO2-PCBs, and OH-PCDFs. Semi-quantitative analyses demonstrate that relative PCB metabolite levels increased with increasing dose and were higher in CV than GF mice, except for PCB sulfates and MeO-PCB sulfates, which were higher in GF mice. These findings demonstrate that the gut microbiome plays a direct or indirect role in the absorption, distribution, metabolism, or excretion of PCB metabolites, which in turn may affect toxic outcomes following PCB exposure.
Keywords: enterotype, feces, PCB metabolites, Mus musculus, nontarget high-resolution mass spectrometry
Capsule:
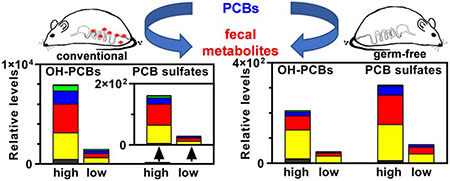
Female conventional and germ-free mice eliminate up to twelve classes of PCB metabolites with the feces following oral exposure to an environmental PCB mixture in a dose- and enterotype-dependent manner.
Graphical Abstract
1. Introduction
Polychlorinated biphenyls (PCBs) are persistent organic pollutants (ATSDR, 2000). Their production was banned worldwide because of environmental and human health concerns. Despite their persistence in the environment, PCB congeners undergo biotransformation in mammals, resulting in the formation of potentially toxic metabolites (Dhakal et al., 2018; Grimm et al., 2015b; Kania-Korwel and Lehmler, 2016). Although there are species differences in the metabolism of PCBs, the overall metabolic pathways show similarities between humans and other species, including mice (Kania-Korwel and Lehmler, 2016; Lehmler et al., 2010). PCBs are initially oxidized by mammalian cytochrome P450 enzymes, either by direct insertion of an oxygen atom into an aromatic C-H bond or via arene oxide intermediates that rearrange to hydroxylated PCBs (OH-PCBs). Subsequently, OH-PCBs are conjugated to sulfate or glucuronide metabolites. There are a total of 837 possible congeners for each of these metabolite classes (Dhakal et al., 2018). Alternatively, PCB arene oxides are conjugated with glutathione. The resulting glutathione conjugates are further metabolized to methyl sulfone PCBs (MeSO2-PCBs). These PCB metabolite classes can be further metabolized, for example, to hydroxylated sulfate or glucuronide conjugates, methylated conjugates of dihydroxylated metabolites, hydroxylated MeSO2-PCBs, or PCB quinone metabolites. PCB metabolites of lower chlorinated PCBs appear to be readily excreted with the urine, whereas the metabolites of higher chlorinated are typically excreted with the feces (Birnbaum, 1985).
Mammals are exposed environmentally to PCB mixtures containing over 100 individual congeners, which undergo congener specific metabolism to diverse PCB metabolites (Dhakal et al., 2018; Grimm et al., 2015b; Kania-Korwel and Lehmler, 2016). The resulting PCB metabolite mixtures can be highly complex and, in humans, are typically studied using serum (Bergman et al., 1994; Grimm et al., 2017; Koh et al., 2016) or, more recently, in urine (Haga et al., 2018; Quinete et al., 2016). Characterizing the complex PCB metabolite mixtures in laboratory animals and humans is important, considering the well-documented toxicities of PCB metabolites (Grimm et al., 2015b; Liu et al., 2020). Unfortunately, only a limited number of OH-PCBs and a few other PCB metabolites (e.g., PCB sulfates) are typically quantified in animal and epidemiological studies because only a small number of analytical standards are available. Nontarget high-resolution mass spectrometry (Nt-HRMS) approaches are a powerful alternative to study the complex PCB metabolite profiles present in laboratory animals, wildlife, and humans. For example, we recently detected hundreds of PCB metabolites in the serum of polar bears and discovered previously unidentified PCB metabolite classes using Nt-HRMS (Liu et al., 2018). In the same study, we confirmed that conventional (CV) mice excrete many of the same PCB metabolites in feces following exposure to the Fox River Mixture, an environmental PCB mixture associate with neurotoxic outcomes in rodent models (Dreiem et al., 2010; Kostyniak et al., 2005; Powers et al., 2006; Sable et al., 2009; Sable et al., 2006).
Exposure to PCBs, such as the Fox River Mixture, can alter the composition and function of the intestinal microbiome (Cheng et al., 2018; Choi et al., 2013; Lim et al., 2020; Petriello et al., 2018). The microbiome also contributes to the reductive metabolism and deconjugation of xenobiotics, including environmental contaminants (Claus et al., 2016; Klaassen and Cui, 2015). Similar to other environmental pollutants (Li et al., 2017), it is likely that the microbiome plays a yet unexplored role in the disposition of PCB and their metabolites in rodent models and humans. Early studies demonstrate that the intestinal microbiome plays a role in the metabolism of PCB glutathione adducts to methyl sulfone PCBs (Bakke et al., 1982; Brandt et al., 1982; Gustafsson et al., 1981). Moreover, sulfate and glutathione conjugates of OH-PCBs are excreted into the intestinal content, where they may be subject to deconjugation reactions. The resulting OH-PCBs may undergo enterohepatic circulation, thus increasing the half-life of these potentially toxic metabolites (Roberts et al., 2002). It is unknown how the intestinal microbiome alters the PCB metabolite profile in feces or affects toxicity by potentially altering PCB toxicokinetics.
Here, we report a semi-quantitative analysis of the PCB metabolite profiles in feces from CV and germ-free (GF) mice, sampled during a previously described toxicity study (Cheng et al., 2018; Liu et al., 2018). The goals were to use an Nt-HRMS approach to explore dose- and enterotype-dependent differences in the relative levels of PCB metabolite classes in feces and to examine the presence of new PCB metabolites. These fundamental studies lay the groundwork for future research investigating the role of dysbiosis of the microbiome in the disposition and toxicity of PCBs.
2. Materials and Methods
2.1. Animal maintenance and exposure to PCBs
The Fox River Mixture approximates human PCB exposures from the consumption of fish from the PCB contaminated Fox River in Wisconsin, USA (Kostyniak et al., 2005). This environmental PCB mixture was prepared by mixing Aroclor 1242, Aroclor 1248, Aroclor 1254, and Aroclor 1260 at a weight ratio of 35:35:15:15 and authenticated as reported previously (Cheng et al., 2018; Liu et al., 2018). Female GF and CV mice on the C57BL/6 background were housed according to guidelines from the Association for Assessment and Accreditation of Laboratory Animal Care International in the animal facility at the University of Washington. The study was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Washington.
As described previously, the initial breeding colony of GF mice on the C57BL/6 background was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill, NC) (Cheng et al., 2018; Liu et al., 2018). All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines, and the animal studies were approved by the Institutional Animal Care and Use Committee at the University of Washington (Seattle, WA, USA). The GF mice were fed a laboratory autoclaved rodent diet (LabDiet #5010); LabDiet, St. Louis, MO, USA) and nonacidified autoclaved water, and maintained on autoclaved Enrich-N’Pure bedding (Andersons, Maumee, OH, USA). Germ-free maintenance colonies were housed in germ-free isolators, and the germ-free status was routinely monitored through fecal pellet culture and 16S rDNA qPCR of universal bacteria. Chemical treatment was performed after germ-free mice were transferred in sterile conditions to the Techniplast caging system. The germ-free status of the GF mice was further confirmed through 16S rDNA qPCR analysis at the end of the experiment. In addition, specific pathogen-free female C57BL/6J mice (CV mice) were purchased from the Jackson Laboratory (Bar Harbor, Maine) and acclimated to the animal facility at the University of Washington for one week before beginning PCB exposure. The CV mice were maintained analogous to the GF mice in the sterile Techniplast caging system on autoclaved Enrich-N’Pure bedding with autoclaved rodent diet and nonacidified water.
At 90 days of age, female GF mice (mice without a microbiome) and CV mice (mice with a microbiome) were exposed by oral gavage to corn oil (vehicle control), low-dose Fox River Mixture (LD, 6 mg/kg), or high-dose Fox River Mixture (HD, 30 mg/kg) in corn oil (Cheng et al., 2018; Liu et al., 2018) with each group containing 6-8 individual mice. Fecal samples were collected 24 h after PCB administration and stored at −80°C in glass vials. The average amount of feces samples from each exposure groups were gut microbiome dependent, i.e., the GF mice produced more feces than CV mice in the same experimental period (CVcontrol: 0.62 g/mouse, n = 8; CVLD: 0.63 g/mouse, n = 8; CVHD: 0.56 g/mouse, n = 8; GFcontrol: 0.88 g/mouse, n = 6; GFLD: 0.93 g/mouse, n = 6; GFHD: 0.91 g/mouse, n = 7).
2.2. Sample extraction and instrument analysis
Feces samples were pooled across animals from the same exposure group (i.e., conventional mice exposed to the high dose [CVHD], low dose [CVLD], or vehicle [CVcontrol], and germ-free mice exposed to the high dose [GFHD], low dose [GFLD], or vehicle [GFcontrol)]) to generate one large pooled sample by grinding in a mortar and, thus, to minimize the intra- and inter-individual variability of the feces samples analyzed. Two hundred milligrams of the pooled feces samples were homogenized in 2 mL of water with TissueRuptor (Qiagen, Hilden, Germany) for 30 seconds (Liu et al., 2018). Two solvent blanks without feces (i.e., method blanks) were extracted in parallel to control for any background contamination. 3-F,4′PCB 3 sulfate (sulfuric acid mono-(4′-chloro-3′-fluoro-biphenyl-4-yl ammonium salt; 5 ng in acetonitrile) was spiked to each homogenate as internal standard (Dhakal et al., 2012), 6 mL of acetonitrile was added, and the samples were mixed and centrifuged at 1,800 g for 5 min. Supernatants were transferred to new glass vials. The pellets were re-extracted with 4 mL of fresh acetonitrile/water mixture (3:1, v/v), and, after inverting and centrifugation, the supernatant was combined with the above extract. The combined supernatants (~12 mL) plus a 2 mL acetonitrile rinse solution were loaded onto a methanol-preconditioned HybridSPE cartridge (500 mg/6mL, Sigma-Aldrich, St. Louis, MO, USA) for phospholipid and protein removal before being subjected to a stir bar-sorptive extraction (SBSE) process as described in the following. Briefly, HybridSPE extracts were concentrated with high purity nitrogen gas at 35°C to ~ 1.75 mL in a 15 mL glass tube. The sample was then transferred to a 20 mL glass vial, and 200 μL of ACN:H2O = 1:1 (v/v) and 1 mL of H2O were used successively to rinse the sample tube. Sodium carbonate (0.05 M) and water were used to make up the total volume to 10 mL (pH 10.5). After adding 0.09 g of tetrabutylammonium bromide (TBABr), a magnetic stir bar, and ten 10 mm-long PES capillary segments (UltraPES0.7, 0.7 mm outer diameter, 150 μm wall thickness; 3M Deutschland GmbH, Wuppertal, Germany), the samples were sealed and placed on a magnetic stirrer (700 rpm) for 36 h at room temperature. All PES segments were then removed, rinsed with water, and immersed in 1 mL methanol for 30 min for analyte desorption. After removing the PES capillaries, the extracts were dried with high purity nitrogen gas, reconstituted in 100 μL of water:methanol = 1:1 (v/v) and filtered.
An Orbitrap Elite™ hybrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) coupled with an Accela HPLC was used for the analyses of the extracts, as described (Liu et al., 2018). An Xselect CSH C18 XP column (130 Å pore size, 2.5 μm particle size, 3 mm inner diameter, 150 mm length; Waters, Milford, MA) was employed with a mobile flow rate and pressure of 0.5 mL/min and 400-900 bar, respectively. Water and methanol were used as mobile phase A and B, respectively. Both mobile phases contained 2 mM ammonium acetate and 2 mM 1-methyl piperidine (pH of mobile phase A = 9.8). The HPLC gradient program was as follows: starting at 38% B, increased linearly to 50% B between 0.17 and 2.67 min, then to 99% B at 41 min, held for 6 min, and returning to 38% B with a hold for 6 min before the next injection. The sample injection volume was 20 μL of the final extract, with instrumental blanks (i.e., 50% H2O/50% MeOH) used to monitor for carryover.
The Orbitrap was operated in negative electrospray ionization mode. The source parameters, including capillary voltage, capillary temperature, vaporizer temperature, sheath gas, auxiliary gas, and sweep gas, were 5 kV, 275 °C, 325 °C, 40, 5, and 2 (arbitrary units), respectively. Analyses were performed in the full scan mode (m/z=150-1500, Orbitrap resolution 120,000). These analyses were performed in parallel with our extensive characterization of PCB metabolites in the polar bear serum (Liu et al., 2018), where an additional structural characterization analysis (i.e., MS/MS) was performed following full-scan. Before performing any analysis, the Orbitrap was calibrated to ensure the high resolution and high mass accuracy, and serial OH-PCBs and PCB sulfate standard solutions were injected to evaluate instrumental and method sensitivity, as detailed in our previous polar bear study (Liu et al., 2018).
2.3. Identification of PCB metabolites
A suspect screening list with the molecular ion formulas and m/z values of potential mono- to deca-chlorinated PCB metabolites was developed based on the known composition of the Fox River Mixture (Cheng et al., 2018; Kostyniak et al., 2005; Liu et al., 2018) and published studies to identify PCB metabolites in the feces of PCB exposed mice (Dhakal et al., 2012; Grimm et al., 2015b; Liu et al., 2018). Chromatographic peak extraction against m/z values of each potential PCB metabolites was performed manually with the CVHD and GFHD sample using Xcalibur software (version 1.0.51.0, Thermo Scientific), and important parameters in the extraction process included mass tolerance (±5 ppm), mass precision decimals (5), and corrected isotopic patterns (e.g., the spectral peak height follows M:(M+2):(M+4)=100:97:31 for trichlorinated OH-PCBs). Whenever a potential PCB metabolite was discovered, control mice sample and method blanks were checked for possible false positive and/or background contamination. This approach allowed us to assign a confidence level of 4 (Schymanski et al., 2014) to all metabolites within a metabolite class (Scheme 1 and Figure 1). In our parallel study (Liu et al., 2018), some of the metabolites were further identified with a confidence level of 3 based on the MS/MS analysis of their molecular ions (i.e., the molecular formula was certain, but the position of chlorine and functional group substitutions could not be confirmed).
Scheme 1.
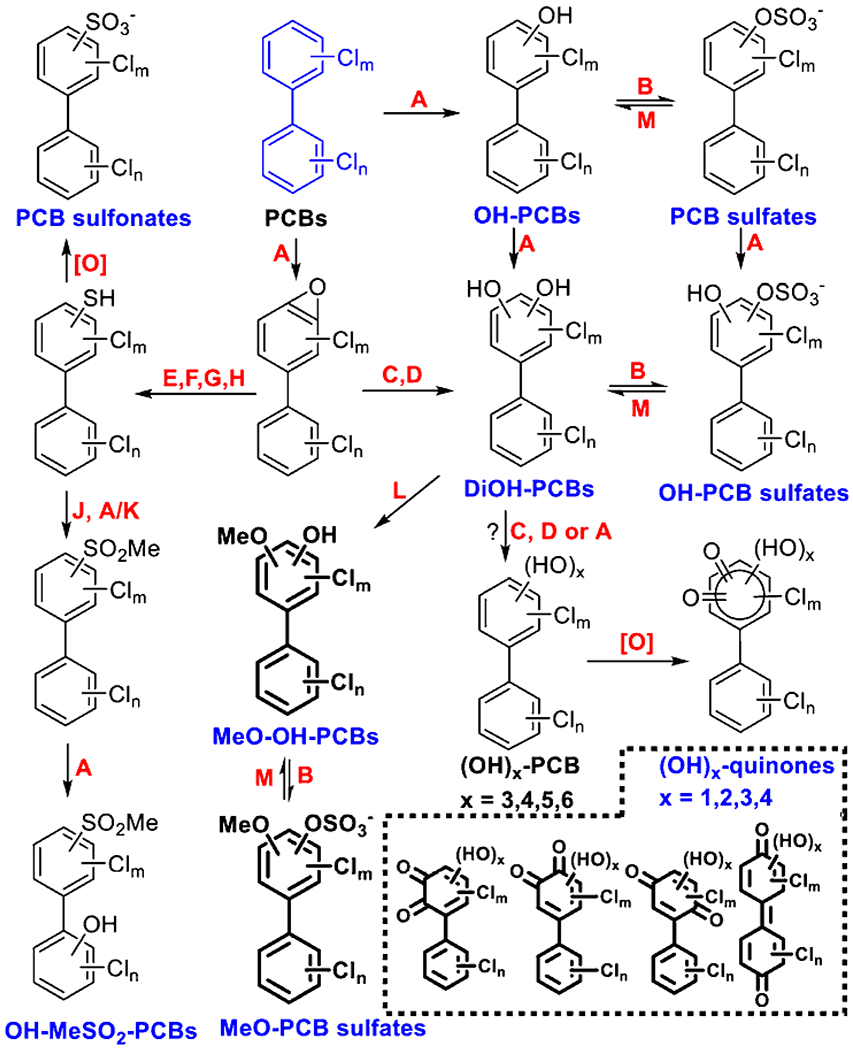
Mice metabolize and excrete at least twelve distinct classes of PCB me-tabolites with the feces, based on the semi-quantitative characterization of the fecal PCB metabolite profiles in CV and GF mice (Dhakal et al., 2012; Grimm et al., 2015b; Kania-Korwel and Lehmler, 2016; Liu et al., 2018). Only representative structures are shown; the position of the substituents is unknown. Metabolites that were detected in our study are indicated with a blue label. Newly detected PCB metabolites are shown as bold structures. A: Cytochrome P450; B: Sulfotransferase; C: Epoxide hydrolase; D: Dihydrodiol dehydrogenase; E: Glutathione-S-transferase; F: γ-glutamyl transpeptidase; G: Cysteinylglycinase; H: Cysteine S-conjugate β-lyase; I: Cysteine S-conjugate N-acetyltransferase; J: Thiol-S-methyltransferase; K: Flavin containing monooxygenases; L: Catechol O-methyltransferase; M: Sulfatase.
Figure 1.
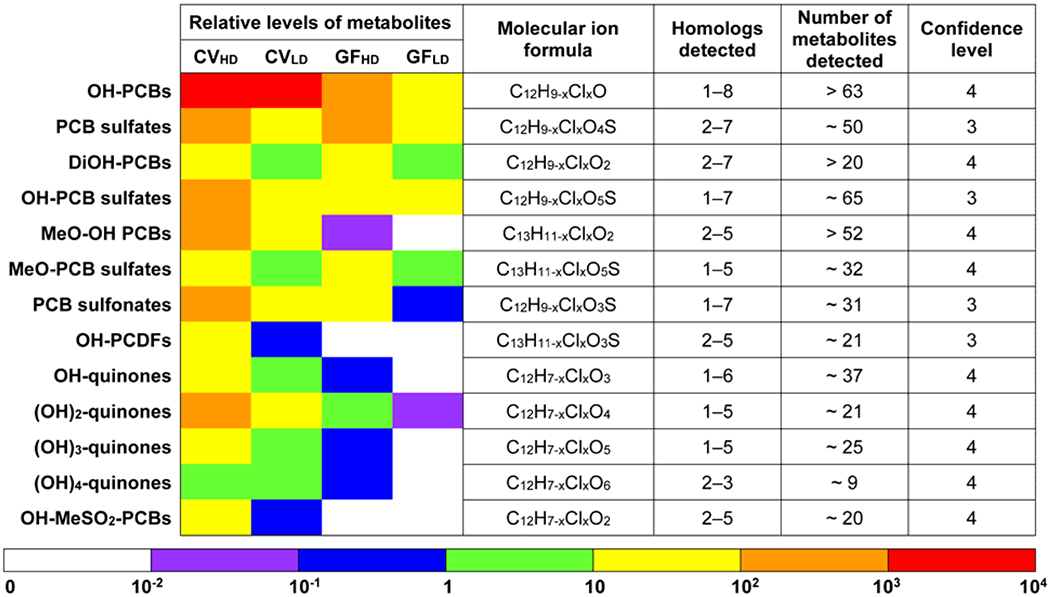
A heatmap-style comparison of the relative levels of the sum of the PCB metabolites (Pmetabolites) and a summary of the PCB metabolites detected in feces from CV and GF mice exposed orally to the Fox River Mixture. Confidence levels were assigned as described (Schymanski et al., 2014). Briefly, a confidence level of 3 represents tentative PCB metabolites with possible structures but an unconfirmed position of substituents. A confidence level of 4 describes PCB metabolites identified based on the molecular formula and the isotope pattern.
2.4. Semi-quantitative analysis of PCB metabolites
The Nt-HRMS chromatograms of suspect PCB metabolites were extracted with Xcalibur software (version 1.0.51.0, Thermo Scientific), and only peaks with matching isotope patterns and ring double bond (RDB) equivalents were integrated (details see Table S1). For each PCB metabolite class (i.e., PCB metabolites with the same functional groups) or each metabolite homolog (i.e., PCB metabolite isomers with the same chlorine numbers), peak areas of all detected metabolites were summed up and normalized to the peak area of the internal standard, 3-F,4′PCB 3 sulfate, and the resulting relative metabolite levels are reported as Σmetabolites. These Σmetabolite values obtained from the extract of each pooled feces sample were then adjusted with the average amount of feces that was produced by one mouse in each exposure group in the 24 h exposure period, and the resulting data were used for group comparison.
Chromatograms showed that some hydroxylated PCB metabolites (i.e., OH-PCBs, dihydroxylated PCBs (DiOH-PCBs), and monomethoxylated DiOH-PCBs (MeO-OH-PCBs)) co-eluted with a sulfate metabolite (i.e., PCB sulfate, hydroxylated PCB sulfates (OH-PCB sulfate), and methoxylated PCB sulfates (MeO-PCB sulfates)). Thus, the hydroxylated fragment ions from the corresponding sulfate metabolite would have contributed to the peak area of these hydroxylated metabolites. We, therefore, used two approaches to correct for the contribution of these fragment ions to the peak area of each affected hydroxylated PCB metabolites: (1) subtracting the estimated hydroxylated metabolite peak area resulting from the fragmentation of the co-eluting sulfate metabolites. The estimated peak area was obtained by multiplying the sum of the peak area of the co-eluting PCB sulfate metabolites by an adjustment factor derived from the internal standard signals in the same sample, [3-F,4′PCB 3-O]−/[3-F,4′PCB 3 sulfate]− (see Table S1 and S2). (2) Alternatively, we made the correction by excluding hydroxylated PCB metabolite peaks that co-eluted with a sulfate metabolite (see data in parentheses in Table S1). The raw data, including the raw instrument data, for individual PCB metabolites and total peak areas for all metabolite classes, are available free of charge through Iowa Research Online (Li et al., 2020).
3. Results and Discussion
3.1. Identification of PCB metabolites eliminated with the feces
As shown in Scheme 1 and Figure 1, we tentatively identified and proposed 12 classes of PCB metabolites in mice feces, including the four PCB metabolite classes that were identified (with a confidence level of 3) and reported in our study comparing the fecal PCB metabolites of CV mice to PCB metabolites present in polar bear serum (Liu et al., 2018), including PCB sulfates, hydroxylated PCB sulfates (OH-PCB sulfates), PCB sulfonates, and hydroxylated methyl sulfone PCBs (OH-MeSO2-PCBs) (Scheme 1). Here, we identify eight additional PCB metabolite classes in the feces from mice exposed to the Fox River Mixture, with a confidence level of 3 or 4, including OH-PCBs, DiOH-PCBs, MeO-OH-PCBs, MeO-PCB sulfates, and mono- to tetrahydroxylated PCB quinones ((OH)n-Q, n=1-4) (Scheme 1). None of the 12 classes were detected in any control mice groups or method blanks.
All 12 PCB metabolite classes were detected in feces from CV mice, and detailed information are as follows: PCB sulfates (C12H9-nClnSO4−, n=2-7; ~50 total analytes), OH-PCB sulfates (C12H9-nClnSO5−, n=1-7; ~65 total analytes), PCB sulfonates (C12H9-nClnSO3−, n=1-7; ~31 total analytes), OH-MeSO2-PCBs (C13H11-nClnSO3, n=2-5; ~21 total analytes), OH-PCBs (C12H9-nClnO, n=1-8; > 63 total analytes), DiOH-PCBs (C12H9-nClnO2, n=2-7; > 20 total analytes), MeO-OH-PCBs (C13H11-nClnO2, n=2-5; > 52 total analytes), MeO-PCB sulfates (C13H11-nClnSO5−, n=1-5; ~32 total analytes), OH-Q (C12H7-nClnO3, n=1-6, ~37 analytes), (OH)2-Q (C12H7-nClnO4, n=1-5, ~21 analytes), (OH)3-Q (C12H7-nClnO5, n=1-5, ~25 analytes), and (OH)4-Q (C12H7-nClnO6, n=2-3, ~9 analytes). Most, but not all PCB metabolite classes present in feces from CV mice were also observed in GF mice (Table S1).
The formation of mono-hydroxylated metabolites and the corresponding sulfate and glucuronide conjugates is well documented across species, including humans (Grimm et al., 2017; Lucier et al., 1978; Wu et al., 2020); however, the excretion of these metabolites with the feces has received limited attention. The formation of DiOH-PCBs and their conjugates has also been observed in diverse animal models (Dhakal et al., 2012; Grimm et al., 2015a; Haraguchi et al., 2004). It is unknown if these metabolites are formed in vivo by oxidation of mono-hydroxylated PCBs, as indicated by in vitro metabolism studies (Lu et al., 2013; McLean et al., 1996), or by oxidation of the mono-sulfated or mono-glucuronidated metabolite, as suggested by an in vivo disposition study (Grimm et al., 2015a). There is also evidence that hydroxylated-methoxylated PCB metabolites are formed in mammals (Ariyoshi et al., 1997; Goto et al., 1974; Hutzinger et al., 1974). Although this has not been shown experimentally, indirect evidence indicates that these metabolites are formed by methylation of a PCB catechol metabolite by catechol-O-methyltransferase (COMT) (Sadeghi-Aliabadi et al., 2007).
Chromatograms and mass spectra showing, for example, the presence of trichlorinated mono- and dihydroxylated quinones in feces from CV mice exposure to 30 mg/kg of the Fox River Mixture are shown in Figure 2. The formation of reactive and toxic PCB quinone metabolites from DiOH-PCBs has been demonstrated in vitro (Amaro et al., 1996). There is also evidence that PCB quinones are formed in rats exposed to PCB 52 (2,2′,5,5′-tetrachlorobiphenyl) (Lin et al., 2000), a major congener present in the Fox River Mixture (Kostyniak et al., 2005). Thus, it is not unexpected that hydroxylated PCB quinones are formed in vivo, for example, via the oxidation of polyhydroxylated PCB metabolites; however, additional experiments are needed to confirm the structure of this new class of PCB metabolites.
Figure 2.
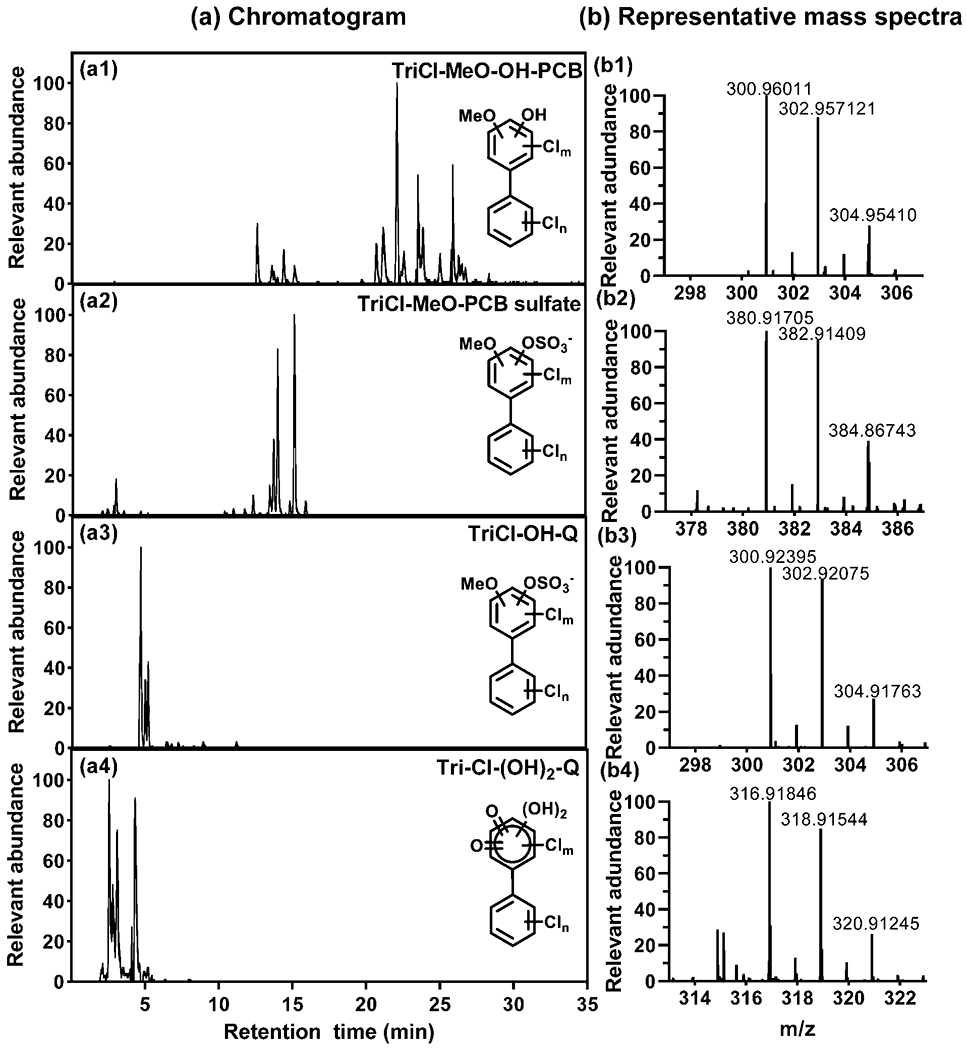
Representative chromatograms showing the presence of several trichlorinated (a1) MeO-OH-PCB, (a2) MeO-PCB sulfate, (a3) OH-quinones, and (a4) (OH)2-quinones in feces from female CV mice exposed to the high dose (30 mg/kg) of the Fox River mixture. Representative mass spectra with the molecular ions are shown for each metabolite class (panels b1-b4). Only representative structures are shown; the position of the substituents is unknown.
Traces of metabolites with the general formula C12H7-nClnO2 (n=2-5, ~21 analytes) were detected in feces from the CV but not the GF mice. These metabolites were tentatively identified as hydroxylated polychlorinated benzofurans (OH-PCDFs, Figure S2). OH-PCDFs can be formed from 2-chloro-2′-hydroxy PCBs during gas chromatographic-mass spectrometry (GC-MS) analysis on electron impact (EI) mode (Li et al., 2009). However, this possible formation pathway is unlikely in vivo because the oxidation of PCBs in the ortho position is, at best, a minor metabolism pathway in mammals (McLean et al., 1996; Wu et al., 2013). Besides, the collision energy of the Nt-HRMS analyses is much lower compared to gas chromatographic analyses, thus making the formation of breakdown products during the analysis unlikely. It is more probable that OH-PCDF metabolites are formed from polychlorinated benzofurans (PCDF) impurities that are present in Aroclor 1254 (Johnson et al., 2008; Morita et al., 1978).
PCB glucuronides were not detected in any feces samples. In contrast, PCB glucuronides have been observed in rat and human urine samples (Dhakal et al., 2012; Grimm et al., 2017). A PCB 11 glucuronide was detected in the bile of rats exposed intravenously to a PCB 11 sulfate (Grimm et al., 2015a). Moreover, rat UDP-glucuronosyltransferases (UGTs) are also known to metabolize OH-PCBs to PCB glucuronides in vitro (Daidoji et al., 2005; Tampal et al., 2002) and modulate the OH-PCB and DiOH-PCB profiles in vivo (Haraguchi et al., 2004). We detected a hydroxylated PCB glucuronide in urine from mice exposed orally to a PCB 91 (2,2’,3,4’,6-pentachlorobiphenyl) (Wu et al., 2020). It is possible that PCB glucuronides were not eliminated with the feces of CV mice because they are rapidly deconjugated to OH-PCBs by microbial glucuronidases in the gastrointestinal tract. Alternatively, these metabolites were not extracted onto the polyethersulfone stir bars because of their high polarity.
We also did not observe glutathione adducts or metabolites from the mercapturic acid pathway, despite that some PCB mercapturic acids can be eliminated with rat feces (Dhakal et al., 2012). Although we detected OH-MeSO2-PCBs, the presumed intermediate metabolites were not detected in our analysis. It is not surprising that we did not observe OH-MeSO2-PCBs in GF mice because the gut microbiome plays a role in the formation of MeSO2-PCBs, putative precursors on OH-MeSO2-PCBs, in rodents (Brandt et al., 1982; Gustafsson et al., 1981).
3.2. Relative levels of PCB metabolites in the feces
GF mice have an altered hepatic expression of xenobiotic processing genes, including genes involved in the metabolism of PCBs (Selwyn et al., 2015). Besides, GF mice cannot conjugate host metabolites, such as sulfated and glucuronidated PCB metabolites, in the lumen of the gastrointestinal tract due to the absence of bacterial deconjugating enzymes. We semi-quantitatively determined the levels of the different metabolite classes, as the sum of the relative peak areas (Σmetabolites) adjusted for feces weight, to explore how differences in host and microbial metabolism affect the excretion of PCB metabolites.
Overall, the approach used to calculate the Σmetabolites for hydroxylated PCB metabolites (i.e., OH-PCBs, DiOH-PCBs, and MeO-OH-PCBs) did not change the trends observed and discussed in the manuscript. Using either of the two peak area correction methods, OH-PCBs were the major metabolites in CV mice compared to other metabolite classes, irrespective of dose and enterotype (Figure 3). The Σmetabolites for different metabolite classes were typically 4- to 19-fold higher in the high vs. low PCB exposure group of CV mice (Table S2), which is consistent with the 5-fold difference in the PCB dose. Other major metabolite classes were PCB sulfates, DiOH-PCBs, and OH-PCB sulfates. Besides, PCB sulfonates, metabolites that we reported for the first time in the serum from polar bears (Liu et al., 2018), were also major PCB metabolites in mice. MeO-OH-PCBs, MeO-PCB sulfates, (OH)n-quinones (n=1-4), and OH-PCDFs were relatively minor metabolites. Interestingly, compared to other class of PCB metabolites, the homolog profiles of the novel (OH)n-quinone metabolites showed a shift to lower chlorinated metabolites. Mono to trichlorinated (OH)n-quinones accounted for > 72% of the relative levels of these four metabolite classes. In contrast, OH-PCB metabolites with ≤ three chlorine substituents accounted for approximately 40% of the ΣOH-PCBs in CV mice. For comparison, the Fox River Mixture contains 18.5 % of mono- to trichlorinated PCBs. The detection of mostly lower chlorinated (OH)n-quinone metabolites is consistent with the faster metabolism of lower than higher chlorinated PCBs (Lucier et al., 1978; Matthews and Anderson, 1975).
Figure 3.
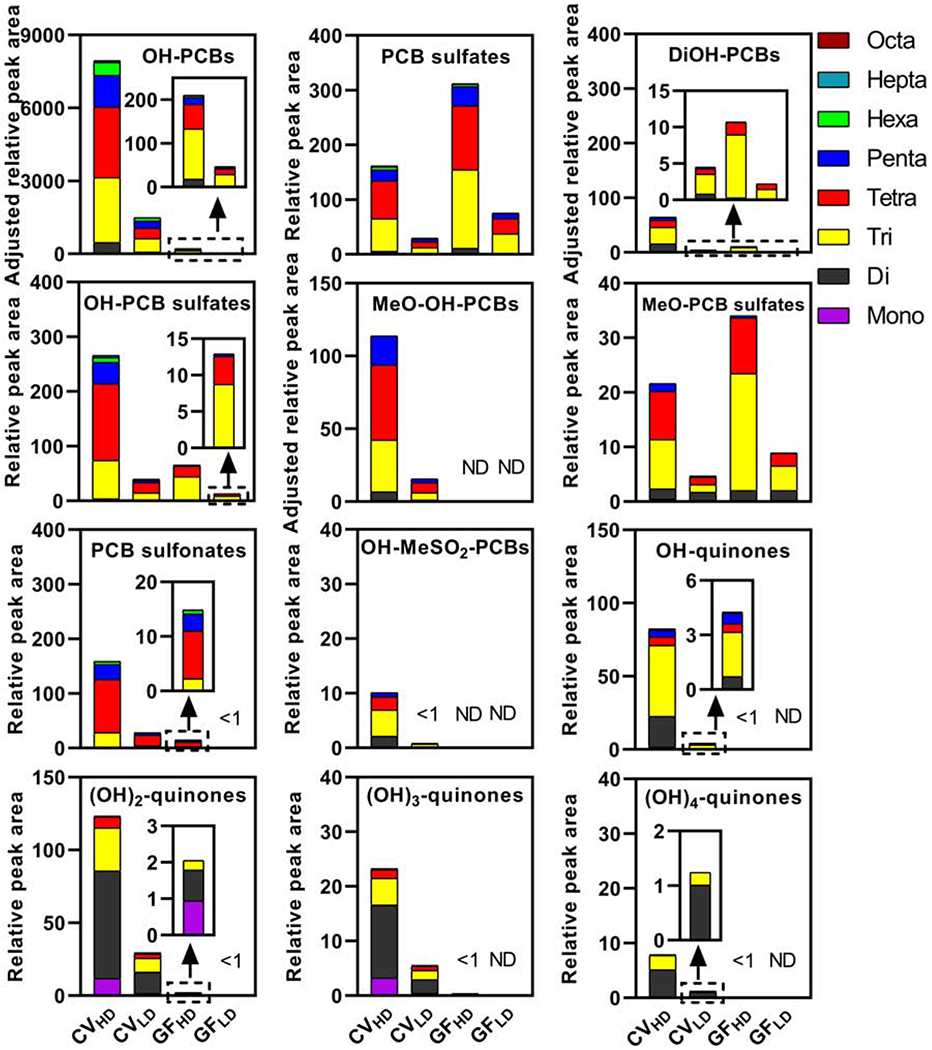
The sum of the relative peak area of selected major PCB metabolites in the 24 h exposure period shows dose- and gut microbiome-dependent differences in feces samples from CV and GF mice. Data are the sum of the peak areas of PCB metabolites adjusted by the internal standard and the feces weight to account for the different amount of feces excreted over 24 h by CV vs. GF mice. For the relative peak areas of metabolites by class and homolog group, see Table S1–2.
In GF mice, PCB sulfates, OH-PCB sulfates, and MeO-PCB sulfates were the major metabolite detected in feces (Figure 3). The total levels of the metabolite classes were typically higher in the high vs. low exposure group of GF mice, consistent with the difference in the PCB dose administered to the GF mice. Interestingly, the ΣPCB sulfates and ΣMeO-PCB sulfates were 2- and 1.7-fold higher in GF compared to CV mice in the high and low PCB exposure groups, respectively. The levels of ΣOH-PCB, ΣDiOH-PCB, ΣMeO-OH-PCB, and, to a lesser extent, ΣOH-PCB sulfate metabolites were more elevated in CV compared to GF mice. The putative quinone and OH-PCDF metabolites were typically not detected in feces from GF mice.
PCB sulfonate levels showed dose- and gut microbiome-dependent differences (Table S2). The ΣPCB sulfonates differed by 5.7- and 16-fold between the high and low exposure groups in CV and GF mice, respectively. Levels of these metabolites were 11-times higher in CV than GF mice in the high exposure group, whereas this difference was 29-fold in the low exposure group. The differences in the ΣPCB sulfonate levels by enterotype suggest that gut microbial metabolism plays a role in forming this novel class of PCB metabolites. At the same time, the dose-dependent differences in both CV and GF mice may be the result of differences in host metabolism. This interpretation of our findings is consistent with the observation that PCBs induce the hepatic expression of cytochrome P450 enzymes in a dose-dependent manner in mice (Lim et al., 2020; Robertson et al., 1984). However, it remains unclear how the sulfonate metabolites are formed in mice.
The differences in the relative levels of the Σmetabolites (Figure 3), ultimately, translate into different metabolite profiles when comparing PCB metabolite classes (Figure 1, Table S3) and homologs of a specific metabolite class between CV and GF mice (Figure S1). In contrast, PCB metabolite profiles are very similar when comparing the low to the high PCB exposure groups for each enterotype (Table S3). The pronounced differences between CV and GF mice may be due to physiological differences between GF and CV mice, including differences in host and microbial PCB metabolism. We demonstrated that exposure to the Fox River Mixture differentially affects the hepatic expression of xenobiotic processing genes (Lim et al., 2020). For example, CV showed a more pronounced induction of cytochrome P450 enzymes involved in the metabolism of PCBs in the liver than GF mice. This observation is consistent with the typically lower levels of PCB metabolites in GF compared to CV mice. Our previous study of the liver transcriptome also revealed a PCB-mediated upregulation of sulfotransferases in GF mice (Lim et al., 2020), which may explain the higher levels of different sulfate metabolites in GF compared to CV mice. Moreover, microbial sulfatase activity likely contributes to the deconjugation of some PCB sulfate classes to the corresponding (Di)OH-PCBs (Cheng et al., 2018), thus resulting in higher levels of hydroxylated metabolites in the CV than the GF mice. Besides, other physiological differences, such as the larger cecum and the higher intestinal motility of GF than CV mice, as well as differences in the composition of the gastrointestinal content (Nicklas et al., 2015), may explain the different profiles and relative levels of PCB metabolites in the fecal pellet from GF and CV mice. Indeed, GF mice produced a large amount of feces than CV mice in the 24 h study duration.
4. Conclusions
Our results demonstrate differences in the fecal excretion of PCB metabolites that depend on the presence or absence of the gut microbiome. Because PCBs alter the composition and function of the microbiome, it is likely that dysbiosis of the microbiome alters the excretion of PCB metabolites and, consequently, may affect the disposition and, ultimately, toxicity of PCBs in rodent and humans. Although some structures of new metabolites require further confirmation, and we only performed a semi-quantitative analysis, OH-PCBs appear to be major metabolites in the feces of CV but not GF mice. Since the Σmetabolites for most metabolite classes are lower in GF than CV mice and the hepatic activity of many drug-metabolizing enzymes is lower in GF than CV mice, we hypothesize that the host metabolism of PCBs may be less extensive in GF than CV mice. Alternatively, the absorption, distribution, or excretion of PCBs and their metabolites may fundamentally differ between GF and CV mice due to physiological differences, a possibility that warrants further attention. Our results also reveal that some sulfated metabolites are more readily excreted with the feces in GF mice, possibly due to the absence of gut microbial metabolism. In contrast, the sulfated PCB metabolites are deconjugated in the gastrointestinal tract of CV mice, resulting in OH-PCB metabolites that undergo enterohepatic circulation. As a result, OH-PCBs are expected to have a longer half-life in the CV than GF mice. These findings demonstrate for the first time that the gut microbiome plays a role in the distribution, metabolism, and excretion of hydroxylated PCB metabolites and their conjugates. Further studies are, therefore, needed to assess how the microbiome affects the toxicokinetics and toxicodynamics of PCBs in mice and humans.
Supplementary Material
Highlights.
PCB metabolites were identified with non-target high-resolution mass spectrometry
Feces from mice exposed to a PCB mixture contained twelve PCB metabolite classes
PCB metabolite levels were typically higher in conventional than germ-free mice
Levels of some sulfated metabolites were higher in germ-free than conventional mice
Some PCB metabolite classes were not detected in conventional mice
Acknowledgments
The authors would like to thank Dr. Lihua Cheng (University of Washington) for help with the animal study.
Funding sources
The research reported in this publication was supported by the National Institute Of Environmental Health Sciences and the National Institute of General Medical Sciences of the National Institutes of Health under Award Numbers ES005605 [HJL], ES0007033 [JYC], ES013661 [HJL], ES019487 [JYC], ES025708 [JYC], ES031098 [HJL, JYC], and GM111381 [JYC]. Additional support was provided by the University of Washington Sheldon Murphy Endowment [JYC]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interests: The authors declare no competing financial interests.
Appendix A.: Supplementary data
Supplementary data to this article can be found openly available in Iowa Research Online at https://doi.org/10.25820/data.006119 (Li et al., 2020).
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Amaro AR, Oakley GG, Bauer U, Spielmann HP, Robertson LW, 1996. Metabolic activation of PCBs to quinones: Reactivity toward nitrogen and sulfUr nucleophiles and influence of superoxide dismutase. Chem. Res. Toxicol 9, 623–629. [DOI] [PubMed] [Google Scholar]
- Ariyoshi N, Koga N, Yoshimura H, Oguri K, 1997. Metabolism of 2,4,5,2′,4′,5′-hexachlorobiphenyl (PCB153) in guinea pig. Xenobiotica 27, 973–983. [DOI] [PubMed] [Google Scholar]
- ATSDR, 2000. Toxicological Profile for Polychlorinated Biphenyls (PCBs). https://www.atsdr.cdc.gov/toxprofiles/tp.asp?id=142&tid=26. [PubMed]
- Bakke JE, Bergman AL, Larsen GL, 1982. Metabolism of 2,4′,5-trichlorobiphenyl by the mercapturic acid pathway. Science 217, 645–647. [DOI] [PubMed] [Google Scholar]
- Bergman Å, Klasson-Wehler E, Kuroki H, 1994. Selective retention of hydroxylated PCB metabolites in blood. Environ. Health Perspect 102, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum LS, 1985. The role of structure in the disposition of halogenated aromatic xenobiotics. Environ. Health Perspect 61, 11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt I, Klasson-Wehler E, Rafter J, Bergman Å, 1982. Metabolism of 2,4′,5-trichlorobiphenyl: tissue concentrations of methylsulphonyl-2,4′,5-trichlorobiphenyl in germ-free and conventional mice. Toxicology Letters 12, 273–280. [DOI] [PubMed] [Google Scholar]
- Cheng SL, Li X, Lehmler H-J, Phillips B, Shen D, Cui JY, 2018. Gut microbiota modulates interactions between polychlorinated biphenyls and bile acid homeostasis. Toxicol. Sci 166, 269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M, 2013. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ. Health Perspect 121, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus SP, Guillou H, Ellero-Simatos S, 2016. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2, 16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daidoji T, Gozu K, Iwano H, Inoue H, Yokota H, 2005. UDP-glucuronosyltransferase isoforms catalyzing glucuronidation of hydroxy-polychlorinated biphenyls in rat. Drug Metab. Dispos 33, 1466–1476. [DOI] [PubMed] [Google Scholar]
- Dhakal K, Gadupudi GS, Lehmler HJ, Ludewig G, Duffel MW, Robertson LW, 2018. Sources and toxicities of phenolic polychlorinated biphenyls (OH-PCBs). Environ. Sci. Pollut. Res. Int 25, 16277–16290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal K, He X, Lehmler H-J, Teesch LM, Duffel MW, Robertson LW, 2012. Identification of sulfated metabolites of 4-chlorobiphenyl (PCB3) in the serum and urine of male rats. Chem. Res. Toxicol 25, 2796–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreiem A, Okoniewski RJ, Brosch KO, Miller VM, Seegal RF, 2010. Polychlorinated biphenyls and polybrominated diphenyl ethers alter striatal dopamine neurochemistry in synaptosomes from developing rats in an additive manner. Toxicol. Sci 118, 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Sugiura K, Hattori M, Miyagawa T, Okamura M, 1974. Metabolism of 2,3,5,6-tetrachlorobiphenyl-14C and 2,3,4,5,6-pentachlorobiphenyl-14C in the rat. Chemosphere 5, 233–238. [Google Scholar]
- Grimm FA, He X, Teesch LM, Lehmler HJ, Robertson LW, Duffel MW, 2015a. Tissue distribution, metabolism, and excretion of 3,3′-dichloro-4′-sulfooxy-biphenyl in the rat. Environ. Sci. Technol 49, 8087–8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Hu D, Kania-Korwel I, Lehmler HJ, Ludewig G, Hornbuckle KC, Duffel MW, Bergman A, Robertson LW, 2015b. Metabolism and metabolites of polychlorinated biphenyls. Crit. Rev. Toxicol 45, 245–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm FA, Lehmler HJ, Koh WX, DeWall J, Teesch LM, Hornbuckle KC, Thorne PS, Robertson LW, Duffel MW, 2017. Identification of a sulfate metabolite of PCB 11 in human serum. Environ. Int 98, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson JA, Rafter JJ, Bakke JE, Gustafsson BE, 1981. The effect of intestinal microflora on the enterohepatic circulation of mercapturic acid pathway metabolites. Nutr Cancer 2, 224–231. [DOI] [PubMed] [Google Scholar]
- Haga Y, Suzuki M, Matsumura C, Okuno T, Tsurukawa M, Fujimori K, Kannan N, Weber R, Nakano T, 2018. Monitoring OH-PCBs in PCB transport worker’s urine as a non-invasive exposure assessment tool. Environ. Sci. Poll. Res 25, 16446–16454. [DOI] [PubMed] [Google Scholar]
- Haraguchi K, Kato Y, Koga N, Degawa M, 2004. Metabolism of polychlorinated biphenyls by Gunn rats: Identification and serum retention of catechol metabolites. Chem. Res. Toxicol 17, 1684–1691. [DOI] [PubMed] [Google Scholar]
- Hutzinger O, Jamieson WD, Safe S, Paulmann L, Ammon R, 1974. Identification of metabolic dechlorination of highly chlorinated biphenyl in rabbit. Nature 252, 698–699. [DOI] [PubMed] [Google Scholar]
- Johnson GW, Hansen LG, Hamilton MC, Fowler B, Hermanson MH, 2008. PCB, PCDD and PCDF congener profiles in two types of Aroclor 1254. Environ. Toxicol. Pharmacol 25, 156–163. [DOI] [PubMed] [Google Scholar]
- Kania-Korwel I, Lehmler HJ, 2016. Chiral polychlorinated biphenyls: absorption, metabolism and excretion-a review. Environ. Sci. Pollut. Res. Int 23, 2042–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY, 2015. Review: Mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab. Dispos 43, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh WX, Hornbuckle KC, Marek RF, Wang K, Thorne PS, 2016. Hydroxylated polychlorinated biphenyls in human sera from adolescents and their mothers living in two U.S. Midwestern communities. Chemosphere 147, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyniak PJ, Hansen LG, Widholm JJ, Fitzpatrick RD, Olson JR, Helferich JL, Kim KH, Sable HJK, Seegal RF, Pessah IN, Schantz SL, 2005. Formulation and characterization of an experimental PCB mixture designed to mimic human exposure from contaminated fish. Toxicol. Sci 88, 400–411. [DOI] [PubMed] [Google Scholar]
- Lehmler H-J, Harrad SJ, Huhnerfuss H, Kania-Korwel I, Lee CM, Lu Z, Wong CS, 2010. Chiral polychlorinated biphenyl transport, metabolism, and distribution: A review. Environ. Sci. Technol 44, 2757–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Lee S, Cade S, Kuo LJ, Schultz IR, Bhatt DK, Prasad B, Bammler TK, Cui JY, 2017. Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals PBDEs. Drug Metab. Dispos 45, 1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Robertson LW, Lehmler H-J, 2009. Electron ionization mass spectral fragmentation study of sulfation derivatives of polychlorinated biphenyls. Chem. Cent. J 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JJ, Li X, Lehmler H-J, Wang D, Gu H, Cui JY, 2020. Gut microbiome critically impacts PCB-induced changes in metabolic fingerprints and the hepatic transcriptome in mice. Toxicol. Sci 177, 168–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PH, Sangaiah R, Ranasinghe A, Upton PB, La DK, Gold A, Swenberg JA, 2000. Formation of quinonoid-derived protein adducts in the liver and brain of Sprague-Dawley rats treated with 2,2′,5, 5′-tetrachlorobiphenyl. Chem. Res. Toxicol 13, 710–718. [DOI] [PubMed] [Google Scholar]
- Liu J, Tan Y, Song E, Song Y, 2020. A Critical Review of Polychlorinated Biphenyls Metabolism, Metabolites, and Their Correlation with Oxidative Stress. Chemical Research in Toxicology 33, 2022–2042. [DOI] [PubMed] [Google Scholar]
- Liu Y, Richardson ES, Derocher AE, Lunn NJ, Lehmler HJ, Li X, Zhang Y, Cui JY, Cheng L, Martin JW, 2018. Hundreds of unrecognized halogenated contaminants discovered in polar bear serum. Angew. Chem. Int. Ed. Engl 57, 16401–16406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Kania-Korwel I, Lehmler HJ, Wong CS, 2013. Stereoselective formation of mono- and dihydroxylated polychlorinated biphenyls by rat cytochrome P450 2B1. Environ. Sci. Technol 47, 12184–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucier GW, McDaniel OS, Schiller CM, Matthews HB, 1978. Structural requirements for the accumulation of chlorinated biphenyl metabolites in the fetal rat intestine. Drug Metab. Dispos 6, 584–590. [PubMed] [Google Scholar]
- Matthews HB, Anderson MW, 1975. Effect of chlorination on the distribution and excretion of polychlorinated biphenyls. Drug Metab. Dispos 3, 371–380. [PubMed] [Google Scholar]
- McLean MR, Bauer U, Amaro AR, Robertson LW, 1996. Identification of catechol and hydroquinone metabolites of 4-monochlorobiphenyl. Chem. Res. Toxicol 9, 158–164. [DOI] [PubMed] [Google Scholar]
- Morita M, Nakagawa J, Rappe C, 1978. Polychlorinated dibenzofuran (PCDF) formation from PCB mixture by heat and oxygen. Bull Environ Contam Toxicol 19, 665–670. [DOI] [PubMed] [Google Scholar]
- Nicklas W, Keubler L, Bleich A, 2015. Maintaining and monitoring the defined microbiota status of gnotobiotic rodents. ILAR J 56, 241–249. [DOI] [PubMed] [Google Scholar]
- Petriello MC, Hoffman JB, Vsevolozhskaya O, Morris AJ, Hennig B, 2018. Dioxin-like PCB 126 increases intestinal inflammation and disrupts gut microbiota and metabolic homeostasis. Environ. Pollut 242, 1022–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers BE, Widholm JJ, Lasky RE, Schantz SL, 2006. Auditory deficits in rats exposed to an environmental PCB mixture during development. Toxicol. Sci 89, 415–422. [DOI] [PubMed] [Google Scholar]
- Quinete N, Esser A, Kraus T, Schettgen T, 2016. Determination of hydroxylated polychlorinated biphenyls (OH-PCBs) in human urine in a highly occupationally exposed German cohort: New prospects for urinary biomarkers of PCB exposure. Environ Int 97, 171–179. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Magnusson BM, Burczynski FJ, Weiss M, 2002. Enterohepatic Circulation. Clinical Pharmacokinetics 41, 751–790. [DOI] [PubMed] [Google Scholar]
- Robertson LW, Parkinson A, Bandiera S, Lambert I, Merrill J, Safe SH, 1984. PCBs and PBBs: Biologic and toxic effects on C57BL/6J and DBA/2J inbred mice. Toxicology 31, 191–206. [DOI] [PubMed] [Google Scholar]
- Sable HJK, Eubig PA, Powers BE, Wang VC, Schantz SL, 2009. Developmental exposure to PCBs and/or MeHg: Effects on a differential reinforcement of low rates (DRL) operant task before and after amphetamine drug challenge. Neurotoxicol. Teratol 31, 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sable HJK, Powers BE, Wang VC, Widholm JJ, Schantz SL, 2006. Alterations in DRH and DRL performance in rats developmentally exposed to an environmental PCB mixture. Neurotoxicol. Teratol 28, 548–556. [DOI] [PubMed] [Google Scholar]
- Sadeghi-Aliabadi H, Chan K, Lehmler H-J, Robertson LW, O’Brien PJ, 2007. Molecular cytotoxic mechanisms of catecholic polychlorinated biphenyl metabolites in isolated rat hepatocytes. Chem.-Biol. Interact 167, 184–192. [DOI] [PubMed] [Google Scholar]
- Schymanski EL, Jeon J, Gulde R, Fenner K, Ruff M, Singer HP, Hollender J, 2014. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol 48, 2097–2098. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY, 2015. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci 147, 84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampal N, Lehmler H-J, Espandiari P, Malmberg T, Robertson LW, 2002. Glucuronidation of hydroxylated polychlorinated biphenyls (PCBs). Chem. Res. Toxicol 15, 1259–1266. [DOI] [PubMed] [Google Scholar]
- Wu X, Duffel M, Lehmler H-J, 2013. Oxidation of polychlorinated biphenyls by liver tissue slices from phenobarbital-pretreated mice is congener-specific and atropselective. Chem. Res. Toxicol 26, 1642–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhai G, Schnoor JL, Lehmler H-J, 2020. Atropselective disposition of 2,2′,3,4′,6-pentachlorobiphenyl (PCB 91) and identification of its metabolites in mice with liver-specific deletion of cytochrome P450 reductase. Chem. Res. Toxicol 33, 1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


