Abstract
Alphaviruses are transmitted by an arthropod vector to a vertebrate host. The disease pathologies, cellular environments, immune responses, and host factors are very different in these organisms. Yet, the virus is able to infect, replicate, and assemble into new particles in these two animals using one set of genetic instructions. The balance between conserved mechanisms and unique strategies during virus assembly is critical for fitness of the virus. In this review, we discuss new findings in receptor binding, polyprotein topology, nucleocapsid core formation, and particle budding that have emerged in the last five years and share opinions on how these new findings might answer some questions regarding alphavirus structure and assembly.
Introduction
Alphaviruses are enveloped viruses that are transmitted by arthropod vectors to vertebrate hosts [1–3]. Alphaviruses can be divided into two groups based on disease outcome. The arthralgia-causing viruses, including Chikungunya virus (CHIKV) and Ross River virus (RRV), which cause fever, rash, and polyarthritis, are often endemic to certain areas, and can trigger large scale epidemics [4,5]. The second group of alphaviruses is neuroinvasive, includes Venezuelan equine encephalitis virus (VEEV) and Eastern equine encephalitis virus (EEEV), and frequently causes encephalitis and death in a range of mammalian species [6]. Despite having different disease outcomes, the two groups of alphaviruses are very similar in their genome organization, replication, assembly pathway, and virion structure [7••,8,9].
The goal of this review is to highlight recent findings in alphavirus structure and assembly and to discuss some unanswered questions. As we cannot cover every aspect of this topic in this review due to space limitations, we will refer readers to several recent reviews that provide details on subjects that we do not address.
The virion: what is in it and what is needed to make it
The alphavirus genome is a positive-sense, single-stranded RNA that encodes four nonstructural proteins and six structural proteins. The structural proteins are produced from a subgenomic mRNA or second open-reading frame. The proteins produced are capsid (CP), E3, E2, 6K, transframe (TF), and E1, and are involved in assembly of an infectious virion; however, not all of these proteins need to be produced or incorporated into a virion for it to be infectious. The alphavirus virion consists of three concentric layers: the viral genome surrounded by the CP (the nucleocapsid core), a host-derived lipid bilayer, and 80 trimeric glycoprotein spikes embedded in the lipid bilayer (Figure 1a). The spikes are trimers of E2 and E1 heterodimers that bind to the cellular receptor and mediate fusion between the viral and endosomal membranes. Multiple receptors for alphaviruses have been identified [2,3,10].
Figure 1.
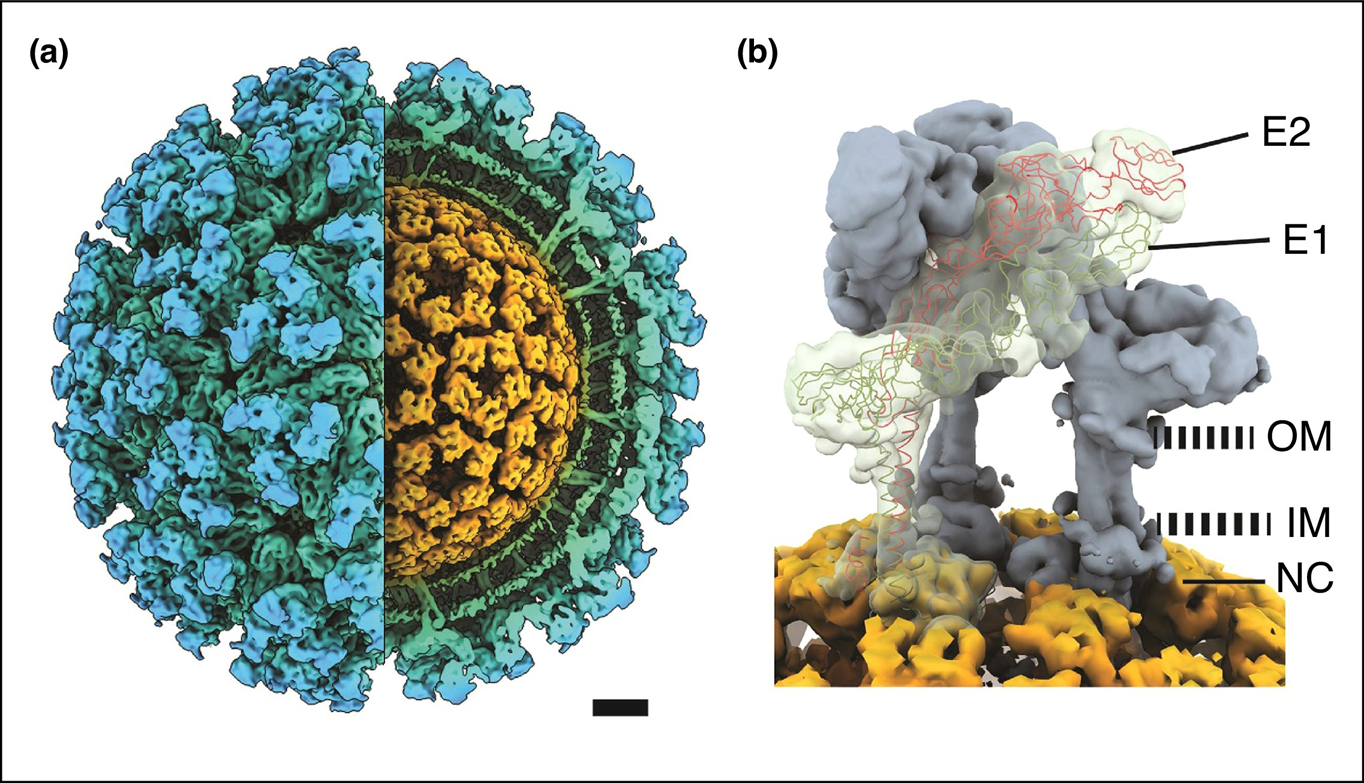
Overview of alphavirus structure.
(a) A cutaway isosurface representation of the alphavirus Sindbis viewed along two-fold axis. Structure determined by cryo-EM and 3D reconstruction. The virion consists of three concentric layers: the glycoprotein spikes (blue) embedded in the lipid bilayer (green), and the nucleocapsid core (orange) which consists of the capsid protein and RNA genome. Both glycoprotein and the core are arranged in a T = 4 surface lattice. (b) Each spike is a trimer of E1 and E2 heterodimers. Shown are two heterodimers in gray and one with the atomic trace. The cytoplasmic domain of E2 interacts with capsid protein. OM, outer membrane. IM, inner membrane. Scale bar, 5 nm.
Alphaviruses are unique enveloped viruses because the nucleocapsid core and glycoprotein spikes are symmetrically aligned in a T = 4 arrangement [7••,8,11]. The E2 protein has a 33-amino acid domain that is embedded in the lipid bilayer and interacts with the hydrophobic pocket of CP (Figure 1b). Initial structures suggested that each of the 240 E2 proteins interact with each of the 240 CPs and that these interactions facilitate the symmetrical matching between the two protein layers [12–16]. One-to-one interactions are energetically favorable, however during virion assembly, the nucleocapsid core consists of hexamers and pentamers and the spikes are trimers. From an assembly perspective, this suggests that the interacting components are not symmetrically matched and that the virus must undergo additional steps to ensure E2 and CP do interact. From a functional perspective, the advantages of a symmetrical match, such as transmission or an internal checkpoint during particle assembly and disassembly, remain unclear.
The structural proteins E3, 6K, and TF have roles in particle assembly, but do not need to be present in a virus particle for it to be infectious. E3 aids in the heterodimerization and transport of the E2–E1 glycoprotein spikes from the endoplasmic reticulum (ER) to the plasma membrane. E3 presumably acts as a clamp that holds the E2–E1 dimer together to prevent premature disassembly as the dimer travels through the host secretory pathway and is exposed to low pH [17–19]. TF is produced from a programmed ribosomal frameshifting event during translation of 6K [20], resulting in a different amino acid sequence after the frameshifting slip site. The N-termini of 6K and TF are identical but the C-termini are unique [20,21]. 6K and TF are dispensable in tissue culture, but are required for robust infection in animals. Studies with Sindbis, CHIKV, RRV, and VEEV [21–25] showed that TF is a virulence factor. Although the mechanistic roles of 6K and TF have not been fully determined, palmitoylation of TF has been shown to be important for virus assembly [21] and TF has been shown to antagonize interferon production during early infection [26]. 6K has been proposed to function as a viroporin [27,28]. A schematic of alphavirus replication and assembly and the cellular locations of the different viral proteins are shown in Figure 2; we refer you to Jose et al. [29•] for more details.
Figure 2.
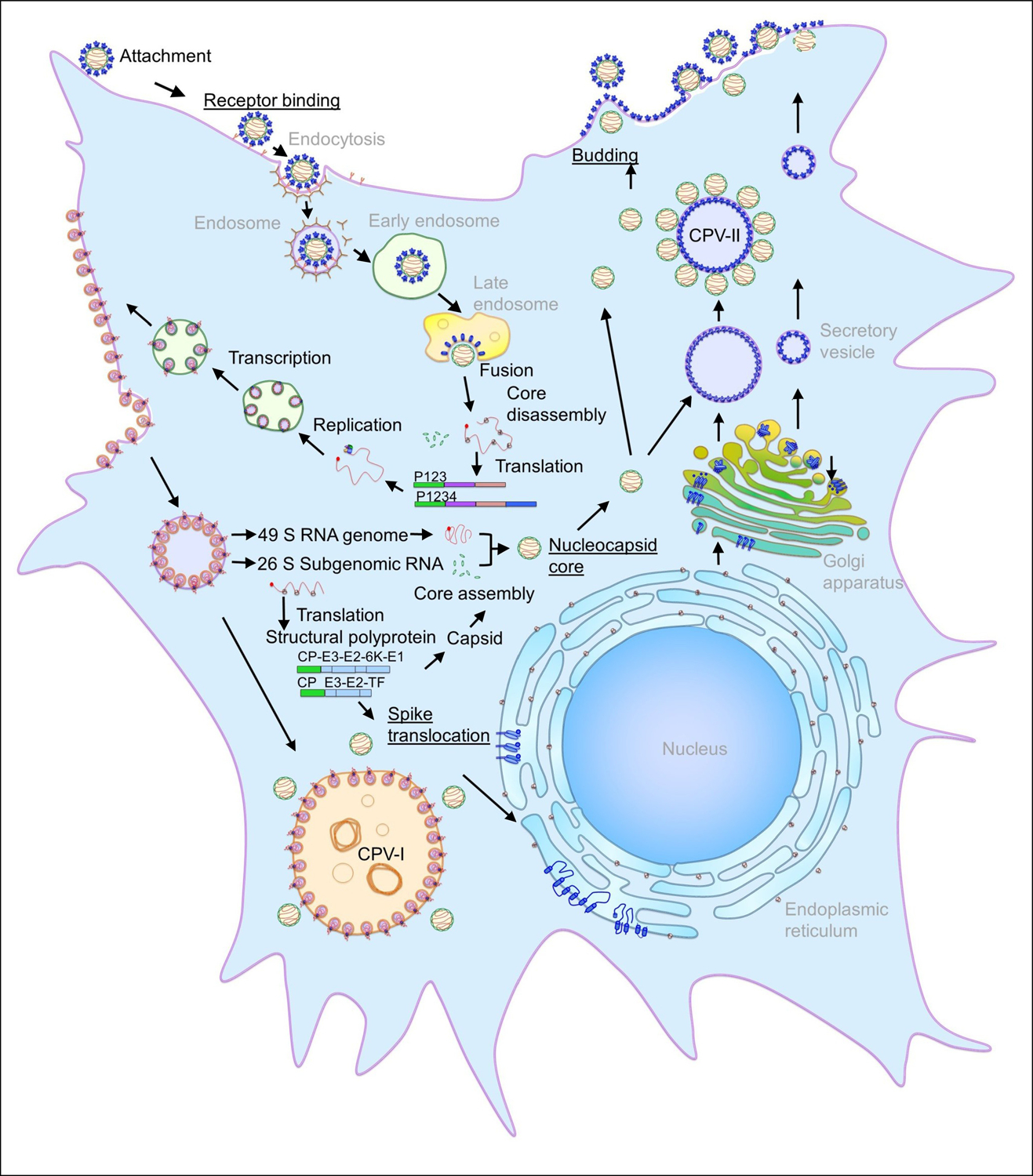
Alphavirus replication and assembly in a vertebrate cell.
Alphavirus particles enter the host cell through receptor-mediated endocytosis and the low pH environment triggers viral and host membrane fusion followed by core disassembly. The viral genome is replicated and subsequently the core and the spikes are assembled in two independent pathways. Particles are released from the cell by budding from the plasma membrane. The steps of the replication and assembly pathway are labeled in black, the steps discussed in this review are underlined, and host processes/components are in gray. Figure adapted from Jose et al. Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. (2017) mBio 8:e02294–16 [29•] and with permission from the authors.
Mxra8: first atomic structure of a receptor-alphavirus particle complex
Many receptors for alphaviruses have been identified in both mammalian and arthropod hosts [2,3,10] and neutralizing antibodies have been mapped to alphavirus particles [7••,8,30], but the atomic level interactions between receptor and particle were unknown. In 2018, Zhang et al. [31] identified the cell adhesion molecule Mxra8 as a receptor specific for the entry of arthritogenic alphaviruses, including CHIKV and RRV; this receptor was not used by the encephalitic viruses. Zhang et al. [32••] showed attenuated infectivity in mice upon infection with a recombinant CHIKV with reduced binding to Mxra8 and enhanced CHIKV infectivity in transgenic flies expressing Mxra8.
The crystal structures of the Mxra8 receptor, which is a novel two-domain immunoglobulin-like receptor, alone and in complex with the virion were solved independently by two groups and provided the first atomic structure of a receptor with an alphavirus particle (Figure 3) [32••,33••]. Although the two groups had similar findings, they used different nomenclatures to describe the Mxra8 domains and the regions on E2–E1 where the receptor bound. In this review, we follow the nomenclature of Basore et al. [32••] because this group published several additional papers using this nomenclature. Mxra8 forms a unique structure than previously described two-domain immunoglobulin-like receptors in that the full domain 1 (D1) is inserted between two fragments of domain 2 (D2) forming two hinge loops between these domains which are held together by an interdomain disulfide bond.
Figure 3.
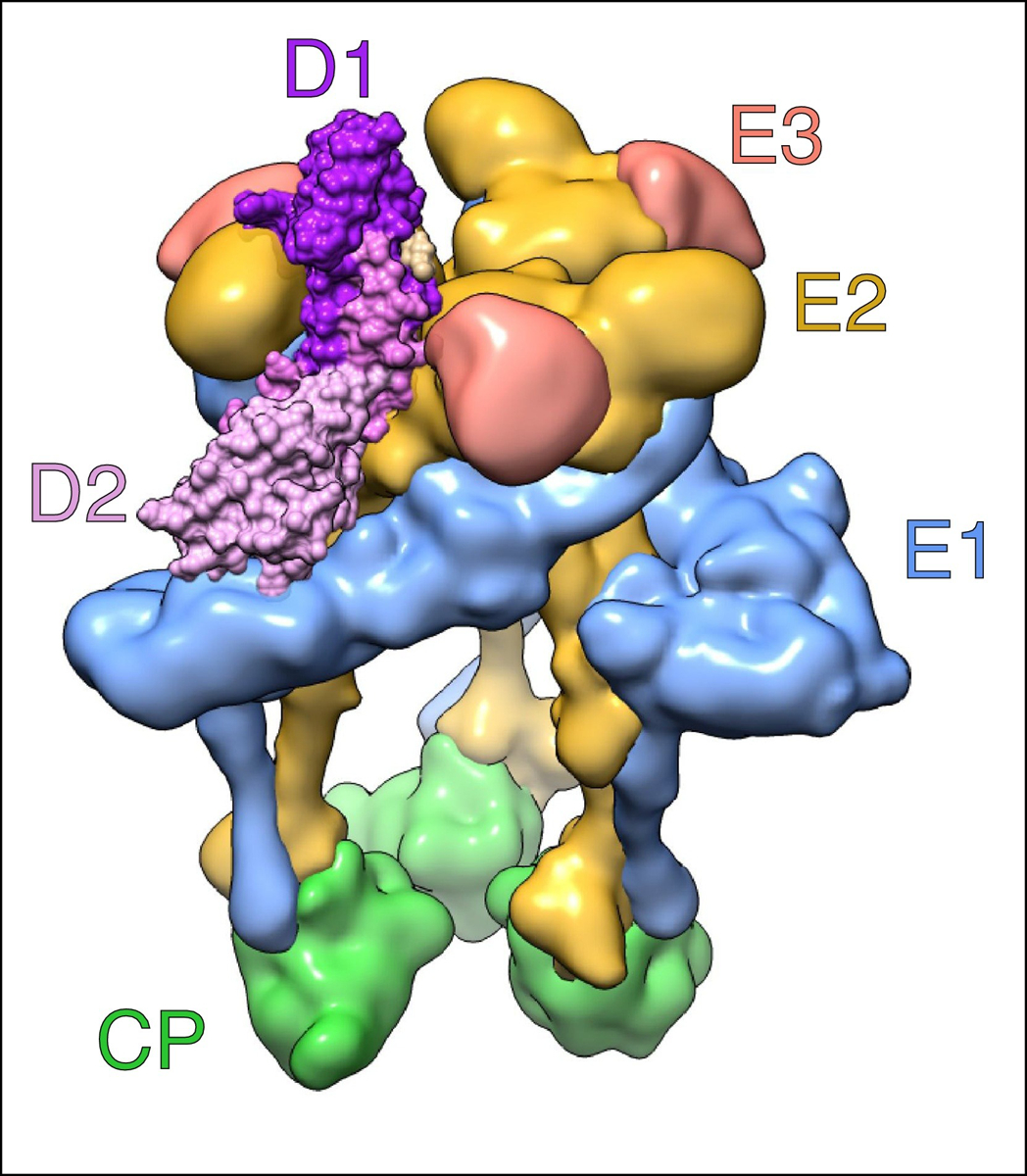
Mxra8 bound to CHIKV.
Side view of a CHIKV spike complexed with its Mxra8 receptor. The viral proteins are CP (green), E1 (light blue), E2 (yellow), and E3 (pink). The two domains of Mxra8, D1 and D2, are shown in dark purple and light purple, respectively. Interestingly, the receptor binds to both E2 and E1 and intercalates itself between adjacent E2–E1 heterodimers in the spike complex. The nomenclature is based on Basore et al. [32••] and is consistent with findings from Song et al. [33••]. EMD-9395 with PDB-6NK6 were used to construct the figure in Chimera [70].
Previously, it was thought that only E2 was involved in receptor binding [34], but both E1 and E2 interact with the two domains of Mxra8. Mxra8 is deeply embedded in a cleft or ‘canyon’ between the two E1–E2 heterodimers of a single CHIKV spike trimer, and Mxra8 makes most of its contacts at the distal end of one E1–E2 heterodimer, which is referred to as ‘wrapped’. Contacts are also made at the adjacent ‘intraspike’ heterodimer on the same spike trimer and an adjacent ‘interspike’ heterodimer of the neighboring spike trimer [32••]. Overall, Mxra8 adopts a 3:3 binding motif with a single spike trimer in CHIKV [33••]. However, depending on the presence or absence of E3 on the trimer, two classes of binding sites have been identified. Mxra8 occupies both high-affinity and low-affinity binding sites on CHIK virus-like particles (VLPs) lacking E3, but occupies only a single high-affinity binding site when E3 is present because retention of E3 blocks the low-affinity binding sites [32••]. In addition to the two domains D1 and D2, Mxra8 has a 48-residue stalk region, a 23-residue transmembrane region, and an 81-residue cytoplasmic tail. Truncations and mutations to the stalk region of Mxra8 diminishes binding; thus, the stalk region of Mxra8 also plays a crucial role in viral binding and entry [33••].
Not all species use Mxra8 for entry. Kim et al. [35] identified a 15-amino acid insertion in the Mxra8 receptor of a Bovinae species that prevents alphaviruses from binding by sterically hindering interaction with E2 residues. Interestingly, a mosquito ortholog of Mxra8 does not exist, indicating that other unidentified host factors must be involved in the binding and entry of alphaviruses in mosquito cells.
This new information on Mxra8 may contribute to the development of novel vaccines, inhibitors that block virus entry, and broadly neutralizing antibodies that target arthritogenic alphaviruses. Nevertheless, questions regarding the mechanism of alphavirus infection post-entry and post-fusion remain. Upon alphavirus binding and entry, alphaviruses are in an early endosome and the low pH mediates viral-host membrane fusion. E1 undergoes a permanent conformational change and forms a fusogenic trimer. It is also unclear how many receptors the virus needs to bind to for entry, how many E1 trimers are necessary for fusion, and what conformational changes E2 undergoes in the endosome before/during fusion. To begin to answer these questions, Cao et al. [36] determined the low resolution structure of the Sindbis virus in a liposome at low pH. With advances in cryo-electron microscopy (cryo-EM), the different stages of fusion and the movement and conformational changes that E2 undergoes during entry may be identified.
Two topologies: connection to frameshifting
The canonical model for the topological arrangement of the alphavirus structural polyprotein, in which E2 and 6K each have two transmembrane domains, is shown as Old Model in top panel (Figure 4). There are, however, inconsistencies between the virus structure and this topology. First, structural studies show that E2 has only one transmembrane domain and a cytoplasmic domain that interacts with CP on the interior side of the lipid bilayer [7••]. Although it has been speculated that a second TM domain within E2 is extruded from the membrane during processing [37], energetic predictions of transmembrane helices suggest that this predicted second transmembrane helix may fail to undergo trans-locon-mediated membrane integration in the first place [38,39•]. Second, the predicted hydrophobic region of 6 K is 35 amino acids in length (based on Sindbis virus numbering and similar to that of other alphaviruses [21]), which is too short to form two complete helices. Third, the palmitoylated residues on TF are present before the programmed ribosomal frameshift site; therefore, they should also be present in 6K. However, these residues are not palmitoylated in 6K, possibly because of self-regulation or because these cysteine residues are not on the cytoplasmic side of the membrane [21].
Figure 4.
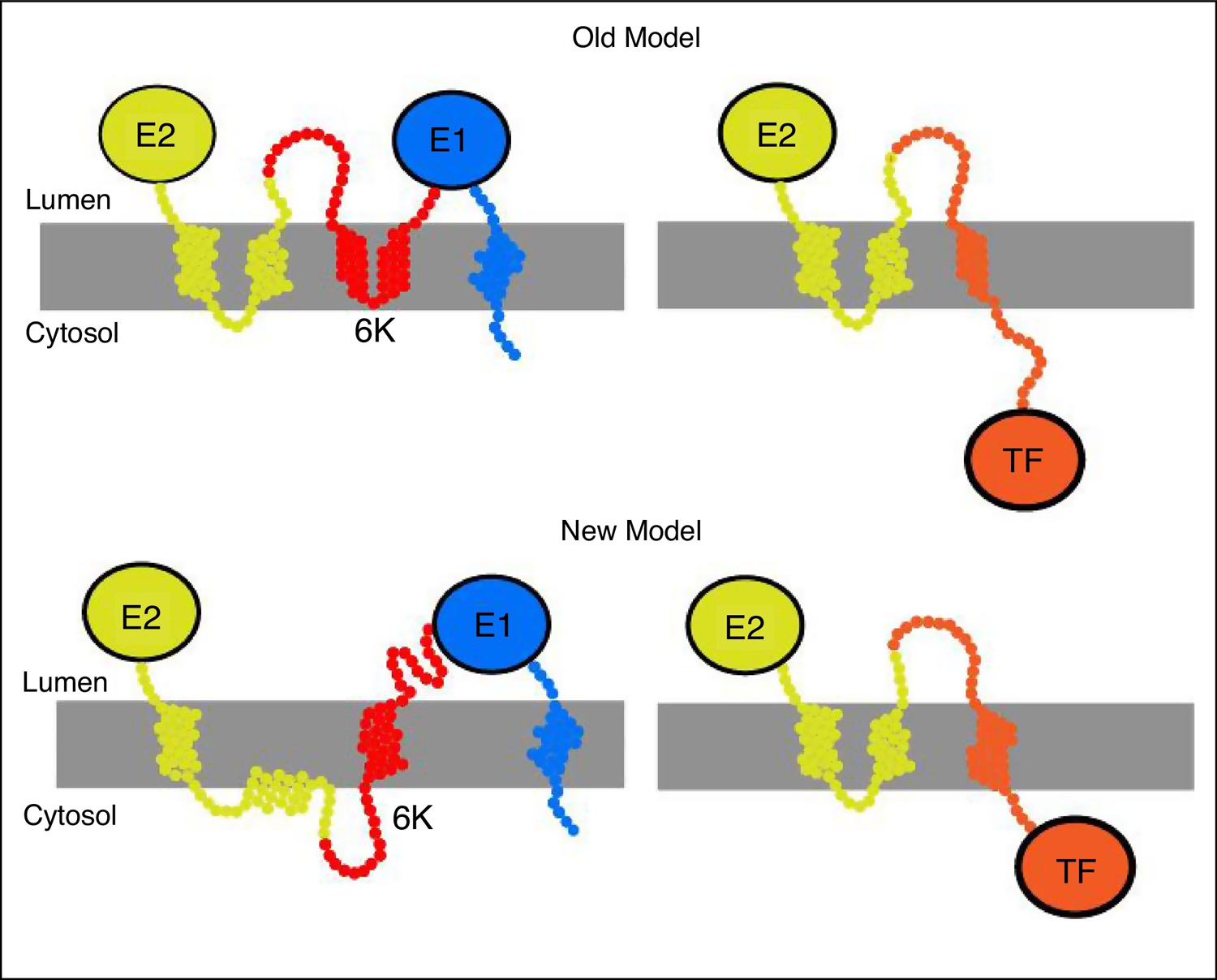
Topological models of the alphavirus structural polyprotein.
The top panel shows the ‘old’ model in which E2 and 6 K each have two transmembrane helices and TF contains a single transmembrane helix. The bottom panel shows the ‘new’ model in which E2 contains a single transmembrane helix, the cytoplasmic domain of E2 is in the cytoplasm, and 6 K contains a single transmembrane helix. When the cytoplasmic domain of E2 enters the membrane, programmed ribosomal frameshifting occurs and TF is produced. This model is based on Figure 3 in Harrington et al. [39•].
Recent work by Harrington et al. [39•] evaluated the topology of the structural polyprotein in relation to TF frameshifting and found that there are two topological conformations of the nascent form of the alphavirus structural polyprotein within the ER. Upon production of 6K, both E2 and 6K have one transmembrane domain each. When the cytoplasmic domain of E2 enters the membrane bilayer, this causes tension on the ribosome, promotes ribosomal frameshifting, and causes TF production. In this situation, the orientation of TF is inverted relative to 6K, and the cysteine residues can be palmitoylated (Figure 4), shown as New Model in the bottom panel. Palmitoylation of TF is important for its incorporation into particles [21] and for its role as a virulence factor [26].
Although TF is produced in low amounts relative to 6K, it is more abundant in released virions [20]. TF is present in lower stoichiometric amounts relative to CP, E2, and E1, but its location in the virion and its oligomeric state is unknown. Although several functions of 6K have been proposed [21,27], its role in virus assembly remains unclear. The function of TF as a virulence factor may depend on its role in structure and assembly and vice versa. It is important to understand the regulation of the proteins that are not incorporated into the virion to identify their roles in infectious particle assembly.
Why are cores not closed spherical capsids?
The ‘perfect’ nature of the alphavirus particle, which is an icosahedral core with spikes aligned together and no external host proteins in the glycoprotein layer, should make alphavirus particles ideal candidates for high resolution structures. However, only a handful of alphavirus structures have resolutions better than 5Å [7••,8,9,40]. Heterogeneity of the glycan structures and conformations, as well as the unknown location of TF could limit the resolution. In addition, the core and its lack of interactions with the E2 cytoplasmic domain could contribute to an imperfect virion with internal heterogeneity.
Traditionally, the nucleocapsid core of alphaviruses has been thought to be composed of a viral genome surrounded by 240 copies of CP arranged in a closed shell with T = 4 icosahedral symmetry [41]. However, both closed complete and open incomplete shells of RRV cores exist [42•,43], and these structural differences have been observed in virus particles purified from both vertebrate and mosquito cells. The incomplete cores were observed when only the cores were used in the reconstruction process, instead of using the glycoprotein layer or the entire particle. The interpretation was that the symmetry of the glycoprotein in the particle dominated the analysis, especially when imposing icosahedral symmetry [42•]. As of now, it is unknown whether closed cores, open cores, or both are biologically active. An incomplete core may be more sensitive to the environment and may assist in the disassembly of the virus upon entry into a new cell [43].
Reconstructions of the flavivirus Kunjin have also shown different results depending on whether the entire particle or only the core region was used for reconstruction and icosahedral averaging was imposed [44]. Therkelsen et al. [44] found that organization of the nucleocapsid core changes during particle maturation. In addition, in both immature and mature virions, the densities of the glycoprotein shell and inner membrane leaflet at the distal pole of the virion were perturbed or missing [44]. The authors proposed that the different interactions in the immature versus mature virions may be important for assembly and budding. It is becoming clear that selecting only the best particles and imposing symmetry to get the best structures results in a significant loss of information.
The highest resolution cryo-EM structures do not provide information regarding CP interactions with viral RNA [7••,8,9,40] or the proteins incorporated into the virions [45,46]. With higher resolution structures, structures of different classes, and structures of assembly intermediates becoming available, we can begin to determine the steps of core and virion assembly and disassembly. The complex interactions that occur between the nucleocapsid core and the cytoplasmic domain of E2 have led to the hypothesis that these interactions cause a conformational change in the core that primes it for disassembly in a newly infected cell [7••,16,47]. This disassembly likely occurs at the threefold axes because there are only weak stabilizing interactions between the subunits. It has also been proposed that the disassembly is mediated by the low pH environment of the endosome during entry because the intrasubunit-stabilizing interactions are electrostatic [7••,15].
Budding: more than E2–CP interactions?
During budding, nucleocapsid cores that form in the cytoplasm interact with the cytoplasmic domain of E2 at the plasma membrane [48]. With the discovery of partially open cores, it remains to be determined how these particles assemble and bud from the membrane. For example, it is not yet known whether these cores form core-E2 interactions or whether their assembly mechanism differs from that of closed core assembly. Although many studies have modeled the interaction of CP with the cytoplasmic domain of E2 [12–16], it is unclear how many of these interactions are necessary for budding to occur [7••,15]. Furthermore, because the most detailed alphavirus structures have been produced by imposing icosahedral symmetry, any differences in the interactions between CP and the cytoplasmic domain of E2 within a particle would not be identified. Because the core is often incomplete, it is unlikely that there would be 240 equivalent E2–CP interactions within a particle. Molecular dynamic simulations have shown that nucleocapsid core-mediated budding results in particles of uniform size and morphology [49•], an advantage of nucleocapsid core mediated budding.
Previous work can now be reconciled with new structural findings. It has been observed that CP interacts with the viral genome in the cytoplasm without forming a complete closed core [50,51]. CP likely interacts with E2 at the plasma membrane and in cytopathic vesicles II (CPV-II), intracellular, virus-induced vesicles (Figure 2) [29•,52]. In multiple Semliki Forest virus mutants, CP can be detected in the virus particle but cannot form nucleocapsid cores in the cytoplasm [50–53]. It has recently been shown in CHIKV, Semliki Forest, and Sindbis viruses that CP is not required for assembly or budding of infectious virus particles [54,55]. When the genomes of these viruses lack the CP sequence, infectious microvesicles (iMVs) containing glycoproteins and genomes are released from the plasma membranes of infected insect and mammalian cells. The iMVs are pleomorphic, have a significantly lower titer than the wild-type virus [54], and are analogous to the subviral particles produced by flaviviruses [56].
Interestingly, multiple studies have shown differences in the assembly and budding of viruses in vertebrate and invertebrate cells; for example, a viral mutant is able to form nucleocapsid cores in mosquito cells, but not in mammalian cells. It has also been demonstrated that internal budding in CPV-II is more common in mosquito cells than in mammalian cells [29•,57]. In addition, it has been shown that the organization of viral proteins and RNA in replication complexes differs between mammalian and mosquito cells and that novel replication complexes form in mosquito cells [29•].
The scission machinery that releases budding particles remains unknown for alphaviruses. Alphaviruses do not use the host endosomal sorting complexes required for transport (ESCRT) system [58], but it is unclear if another host system is used or if the virus encodes for its own viral scission machinery (e.g. the Influenza M2 protein) [59]. Analysis of mutations in 6 K and TF suggest that they could function in viral budding [60–62]. A neutralizing antibody that interferes with CHIK virus budding, but not entry, has recently been identified. This antibody reduces the number of released particles by crosslinking the glycoprotein spikes at the plasma membrane [63•]. Alphaviruses can also spread via cell-to-cell transmission [64,65], and this mechanism is reviewed in Brown et al. [66].
Conclusion
Structure and function go hand in hand. The structure of the released virion is the final metastable form of the particle, and to get to that conformation, many regulated events must occur. Similarly, when a virus enters a cell to initiate an infection, there is a systematic process for disassembly. With the recent advances in high-resolution structural biology, it is a great time to focus on the assembly intermediates that allow a virion to form and the disassembly intermediates that form during infection. New and much improved tools for single-particle structural biology [67,68] and advances in sample preparation and data acquisition for high resolution cryo-tomography [69] make it possible to describe the structure of assembly and disassembly intermediates. Identifying the intermediates and determining their cellular locations will guide our hypotheses for cellular studies. Structural and cellular data will connect the dots between structure and function, which has tremendous implications for vaccine development, drug design, evolutionary biology, and the use of viral particles as biological templates.
Acknowledgements
We thank J. Jose, and R.J. Kuhn for providing Figure 2, H. Harrington for providing Figure 4, and our lab, collaborators, and members of the Indiana University Virology group for valuable discussions. We apologize to the colleagues whose contributions were not discussed or cited due to space constraints.
Funding
This review did not receive any specific grants from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Conflict of interest statement Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Weaver SC, Winegar R, Manger ID, Forrester NL: Alphaviruses: population genetics and determinants of emergence. Antiviral Res 2012, 94:242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn RJ: Chapter 22: togaviradae In Fields’ Virology. Edited by Knipe DM, Howley PM. Lippincott Williams & Wilkins; 2013:629–650. [Google Scholar]
- 3.Griffin DE: Chapter 23: alphaviruses In Fields’ Virology. Edited by Knipe DM, Howley PM. Lippincott Williams & Wilkins; 2013:652–770. [Google Scholar]
- 4.Silva LA, Dermody TS: Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J Clin Invest 2017, 127:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X, Tharmarajah K, Taylor A: Ross river virus disease clinical presentation, pathogenesis and current therapeutic strategies. Microbes Infect 2017, 19:496–504. [DOI] [PubMed] [Google Scholar]
- 6.Yu YA, Bolton M: A case of eastern equine encephalitis. Clin Pediatr (Phila) 2019, 58:245–246. [DOI] [PubMed] [Google Scholar]
- 7.••.Hasan SS, Sun C, Kim AS, Watanabe Y, Chen CL, Klose T, Buda G, Crispin M, Diamond MS, Klimstra WB et al. : Cryo-EM structures of eastern equine encephalitis virus reveal mechanisms of virus disassembly and antibody neutralization. Cell Rep 2018, 25:3136–3147 e3135. [DOI] [PMC free article] [PubMed] [Google Scholar]; This high-resolution cryo-EM structure of EEEV provides the most detailed information to date on the conformation of the cytoplasmic domain of E2 with CP. Modeling of the N-terminal residues of CP into the structure suggests how CP may interact with viral RNA during assembly and with the ribosome during disassembly. Additionally, this structure provides insights into receptor binding and immune evasion by the virus.
- 8.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG: Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization. eLife 2013, 2: e00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, Wang M, Zhu D, Sun Z, Ma J, Wang J, Kong L, Wang S, Liu Z, Wei L et al. : Implication for alphavirus host-cell entry and assembly indicated by a 3.5A resolution cryo-EM structure. Nat Commun 2018, 9:5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnierle BS: Cellular attachment and entry factors for Chikungunya virus. Viruses 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng RH, Kuhn RJ, Olson NH, Rossmann MG, Choi HK, Smith TJ, Baker TS: Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 1995, 80:621–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Lindqvist B, Garoff H, von Bonsdorff CH, Liljestrom P: A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J 1994, 13:4204–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Owen KE, Kuhn RJ: Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 1997, 230:187–196. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Owen KE, Choi HK, Lee H, Lu G, Wengler G, Brown DT, Rossmann MG, Kuhn RJ: Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 1996, 4:531–541. [DOI] [PubMed] [Google Scholar]
- 15.Tang J, Jose J, Chipman P, Zhang W, Kuhn RJ, Baker TS: Molecular links between the E2 envelope glycoprotein and nucleocapsid core in Sindbis virus. J Mol Biol 2011, 414:442–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snyder JE, Berrios CJ, Edwards TJ, Jose J, Perera R, Kuhn RJ: Probing the early temporal and spatial interaction of the Sindbis virus capsid and E2 proteins with reverse genetics. J Virol 2012, 86:12372–12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder AJ, Mukhopadhyay S: The alphavirus E3 glycoprotein functions in a clade-specific manner. J Virol 2012, 86:13609–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchime O, Fields W, Kielian M: The role of E3 in pH protection during alphavirus assembly and exit. J Virol 2013, 87:10255–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fields W, Kielian M: Interactions involved in pH protection of the alphavirus fusion protein. Virology 2015, 486:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth AE, Chung BY, Fleeton MN, Atkins JF: Discovery of frameshifting in alphavirus 6K resolves a 20-year enigma. Virol J 2008, 5:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramsey J, Mukhopadhyay S: Disentangling the frames, the state of research on the alphavirus 6K and TF Proteins. Viruses 2017, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder JE, Kulcsar KA, Schultz KL, Riley CP, Neary JT, Marr S, Jose J, Griffin DE, Kuhn RJ: Functional characterization of the alphavirus TF protein. J Virol 2013, 87:8511–8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kendra JA, de la Fuente C, Brahms A, Woodson C, Bell TM, Chen B, Khan YA, Jacobs JL, Kehn-Hall K, Dinman JD: Ablation of programmed-1 ribosomal frameshifting in Venezuelan equine encephalitis virus results in attenuated neuropathogenicity. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hallengard D, Kakoulidou M, Lulla A, Kummerer BM, Johansson DX, Mutso M, Lulla V, Fazakerley JK, Roques P, Le Grand R et al. : Novel attenuated chikungunya vaccine candidates elicit protective immunity in C57BL/6 mice. J Virol 2014, 88:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor A, Melton JV, Herrero LJ, Thaa B, Karo-Astover L, Gage PW, Nelson MA, Sheng KC, Lidbury BA, Ewart GD et al. : Effects of an in-frame deletion of the 6k gene locus from the genome of ross river virus. J Virol 2016, 90:4150–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers KJ, Jones-Burrage S, Maury W, Mukhopadhyay S: TF protein of Sindbis virus antagonizes host type I interferon responses in a palmitoylation-dependent manner. Virology 2020, 542:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dey D, Siddiqui SI, Mamidi P, Ghosh S, Kumar CS, Chattopadhyay S, Ghosh S, Banerjee M: The effect of amantadine on an ion channel protein from Chikungunya virus. PLoS Negl Trop Dis 2019, 13:e0007548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Xie S, Sun B: Viral proteins function as ion channels. Biochim Biophys Acta 2011, 1808:510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.•.Jose J, Taylor AB, Kuhn RJ: Spatial and temporal analysis of alphavirus replication and assembly in mammalian and mosquito cells. mBio 2017, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This side-by-side study of alphavirus infection in mammalian and insect cells demonstrates the differences in the mechanism and timing of viral replication, which provides insights into the different types of alphavirus infections in different organisms.
- 30.Davis NL, Pence DF, Meyer WJ, Schmaljohn AL, Johnston RE: Alternative forms of a strain-specific neutralizing antigenic site on the Sindbis virus E2 glycoprotein. Virology 1987, 161:101–108. [DOI] [PubMed] [Google Scholar]
- 31.Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, Rimkunas R, Fong RH, Lin H, Poddar S et al. : Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 2018, 557:570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.••.Basore K, Kim AS, Nelson CA, Zhang R, Smith BK, Uranga C, Vang L, Cheng M, Gross ML, Smith J et al. : Cryo-EM structure of Chikungunya virus in complex with the Mxra8 receptor. Cell 2019, 177:1725–1737 e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors solved the X-ray crystal structure of mouse Mxra8 (2.2 Å) alone and the cryo-EM structures of Mxra8 bound to either CHIK VLPs (4.06 Å) or infectious viruses (4.99Å). These structures revealed two binding modes with fully mature VLPs and provided the first atomic details of a receptor–virus interaction. This article complements the Song et al. study.
- 33.••.Song H, Zhao Z, Chai Y, Jin X, Li C, Yuan F, Liu S, Gao Z, Wang H, Song J et al. : Molecular basis of arthritogenic alphavirus receptor MXRA8 binding to Chikungunya virus envelope protein. Cell 2019, 177:1714–1724 e1712. [DOI] [PubMed] [Google Scholar]; The authors report the crystal structures of mouse Mxra8 (2.4 Å) and the human Mxra8 (hMxra8)-CHIKV E protein complex (3.49Å) and cryo-EM structures of hMXRA8 bound to CHIK VLPs (8.9 Å). This article complements the Basore et al. study.
- 34.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA: Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468:709–712. [DOI] [PubMed] [Google Scholar]
- 35.Kim AS, Zimmerman O, Fox JM, Nelson CA, Basore K, Zhang R, Durnell L, Desai C, Bullock C, Deem SL et al. : An evolutionary insertion in the Mxra8 receptor-binding site confers resistance to alphavirus infection and pathogenesis. Cell Host Microbe 2020, 27:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao S, Zhang W: Characterization of an early-stage fusion intermediate of Sindbis virus using cryoelectron microscopy. Proc Natl Acad Sci U S A 2013, 110:13362–13367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liljestrom P, Garoff H: Internally located cleavable signal sequences direct the formation of Semliki forest virus membrane proteins from a polyprotein precursor. J Virol 1991, 65:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G: Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 2007, 450:1026–1030. [DOI] [PubMed] [Google Scholar]
- 39.•.Harrington HR, Zimmer MH, Chamness LM, Nash V, Penn WD, Miller TF 3rd, Mukhopadhyay S, Schlebach JP: Cotranslational folding stimulates programmed ribosomal frameshifting in the alphavirus structural polyprotein. J Biol Chem 2020, 295:6798–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identify two competing topologies for the alphavirus structural polyprotein, which involve frameshifting and production of TF.
- 40.Zhang R, Hryc CF, Cong Y, Liu X, Jakana J, Gorchakov R, Baker ML, Weaver SC, Chiu W: 4.4 A cryo-EM structure of an enveloped alphavirus Venezuelan equine encephalitis virus. EMBO J 2011, 30:3854–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendes A, Kuhn RJ: Alphavirus nucleocapsid packaging and assembly. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.•.Wang JC, Chen C, Rayaprolu V, Mukhopadhyay S, Zlotnick A: Self-assembly of an alphavirus core-like particle is distinguished by strong intersubunit association energy and structural defects. ACS Nano 2015, 9:8898–8906. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show, for the first time, that alphavirus nucleocapsid cores are not closed capsids.
- 43.Wang JC, Mukhopadhyay S, Zlotnick A: Geometric defects and Icosahedral viruses. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Therkelsen MD, Klose T, Vago F, Jiang W, Rossmann MG, Kuhn RJ: Flaviviruses have imperfect icosahedral symmetry. Proc Natl Acad Sci U S A 2018, 115:11608–11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sokoloski KJ, Nease LM, May NA, Gebhart NN, Jones CE, Morrison TE, Hardy RW: Identification of interactions between Sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog 2017, 13:e1006473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokoloski KJ, Snyder AJ, Liu NH, Hayes CA, Mukhopadhyay S, Hardy RW: Encapsidation of host-derived factors correlates with enhanced infectivity of Sindbis virus. J Virol 2013, 87:12216–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paredes A, Alwell-Warda K, Weaver SC, Chiu W, Watowich SJ: Structure of isolated nucleocapsids from Venezuelan equine encephalitis virus and implications for assembly and disassembly of enveloped virus. J Virol 2003, 77:659–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Acheson NH, Tamm I: Replication of Semliki forest virus: an electron microscopic study. Virology 1967, 32:128–143. [DOI] [PubMed] [Google Scholar]
- 49.•.Lazaro GR, Mukhopadhyay S, Hagan MF: Why enveloped viruses need cores-the contribution of a nucleocapsid core to viral budding. Biophys J 2018, 114:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses simulations to show differences in the morphologies and sizes of released particles upon glycoprotein-directed budding versus nucleocapsid-directed budding.
- 50.Forsell K, Griffiths G, Garoff H: Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J 1996, 15:6495–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forsell K, Xing L, Kozlovska T, Cheng RH, Garoff H: Membrane proteins organize a symmetrical virus. EMBO J 2000, 19:5081–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soonsawad P, Xing L, Milla E, Espinoza JM, Kawano M, Marko M, Hsieh C, Furukawa H, Kawasaki M, Weerachatyanukul W et al. : Structural evidence of glycoprotein assembly in cellular membrane compartments prior to alphavirus budding. J Virol 2010, 84:11145–11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skoging U, Vihinen M, Nilsson L, Liljestrom P: Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 1996, 4:519–529. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Guillen M, Gabev E, Quetglas JI, Casales E, Ballesteros-Briones MC, Poutou J, Aranda A, Martisova E, Bezunartea J, Ondiviela M et al. : Capsid-deficient alphaviruses generate propagative infectious microvesicles at the plasma membrane. Cell Mol Life Sci 2016, 73:3897–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang YN, Deng CL, Li JQ, Li N, Zhang QY, Ye HQ, Yuan ZM, Zhang B: Infectious Chikungunya Virus (CHIKV) with a complete capsid deletion: a new approach for a CHIKV vaccine. J Virol 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lindenbach BD, Rice CM: Molecular biology of flaviviruses. Adv Virus Res 2003, 59:23–61. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez R, Lee H, Nelson C, Brown DT: A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J Virol 2000, 74:4220–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taylor GM, Hanson PI, Kielian M: Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J Virol 2007, 81:13631–13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossman JS, Jing X, Leser GP, Lamb RA: Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142:902–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaedigk-Nitschko K, Ding MX, Levy MA, Schlesinger MJ: Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology 1990, 175:282–291. [DOI] [PubMed] [Google Scholar]
- 61.Gaedigk-Nitschko K, Schlesinger MJ: The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 1990, 175:274–281. [DOI] [PubMed] [Google Scholar]
- 62.Lusa S, Garoff H, Liljestrom P: Fate of the 6K membrane protein of Semliki forest virus during virus assembly. Virology 1991, 185:843–846. [DOI] [PubMed] [Google Scholar]
- 63.•.Jin J, Galaz-Montoya JG, Sherman MB, Sun SY, Goldsmith CS, O’Toole ET, Ackerman L, Carlson LA, Weaver SC, Chiu W et al. : Neutralizing antibodies inhibit Chikungunya virus budding at the plasma membrane. Cell Host Microbe 2018, 24:417–428 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors identify a neutralizing antibody that crosslinks spikes at the plasma membrane, and consequently, inhibits budding.
- 64.Martinez MG, Kielian M: Intercellular extensions are induced by the alphavirus structural proteins and mediate virus transmission. PLoS Pathog 2016, 12:e1006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martinez MG, Snapp EL, Perumal GS, Macaluso FP, Kielian M: Imaging the alphavirus exit pathway. J Virol 2014, 88:6922–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown RS, Wan JJ, Kielian M: The alphavirus exit pathway: what we know and what we wish we knew. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bai XC, McMullan G, Scheres SH: How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 2015, 40:49–57. [DOI] [PubMed] [Google Scholar]
- 68.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y: Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 2013, 10:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang P: Advances in cryo-electron tomography and subtomogram averaging and classification. Curr Opin Struct Biol 2019, 58:249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE: UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004, 25:1605–1612. [DOI] [PubMed] [Google Scholar]


