Abstract
Personalized cancer vaccines are a promising approach for inducing T cell immunity to tumor neoantigens. Using a self-assembling nanoparticle vaccine that links neoantigen peptides to a TLR7/8 agonist (SNP-7/8a), we show how the route and dose alter the magnitude and quality of neoantigen-specific CD8+ T cells. Intravenous vaccination (SNP-IV) induced a higher proportion of TCF1+PD-1+ CD8+ T cells compared to subcutaneous immunization (SNP-SC). Single cell RNA-seq showed that SNP-IV induced stem-like genes (Tcf7, Slamf6, Xcl1) whereas SNP-SC enriched for effector genes (Gzmb, Klrg1, Cx3cr1). Stem-like cells generated by SNP-IV proliferated and differentiated into effector cells upon checkpoint blockade leading to superior antitumor response compared to SNP-SC in a therapeutic model. The duration of antigen presentation by dendritic cells controlled the magnitude and quality of CD8+ T cells. These data demonstrate how to optimize antitumor immunity by modulating vaccine parameters for specific generation of effector or stem-like CD8+ T cells.
INTRODUCTION
The clinical effectiveness of cancer immunotherapy, including checkpoint blockade and adoptive T cell therapy, is based on the magnitude, quality, breadth and ability of T cells to infiltrate tumors1–4. Advances in rapid genomic sequencing has enabled the development of personalized cancer vaccines (PCV) targeting tumor-specific mutations termed neoantigens (neoAg). Early studies in mice5,6 and phase I clinical trials7–10 demonstrate the feasibility of PCV to generate neoAg T cell responses; however, the magnitude of CD8+ T cell responses has been limited to date10.
An important variable in improving CD8+ T cells responses with PCV is the vaccine platform. Accordingly, we developed a self-assembling nanoparticle vaccine platform to standardize the delivery of long peptides (LP)11. At 20–50 nm in size, the nanoparticles allow for efficient drainage via lymphatics and uptake by dendritic cells (DC) that prime CD8+ T cells12. Further, co-delivery of the antigen and TLR7/8 agonist (TLR7/8a) ensures DC maturation via TLR activation in the same cell that has acquired antigen, to achieve optimal presentation to CD8+ T cells. Indeed, the SNP-7/8a vaccine platform generated higher magnitude and breadth of CD8+ T cells with antitumor efficacy when benchmarked against the most commonly used PCV approaches11.
In addition to magnitude, the quality of CD8+ T cell responses can be an important determinant in protection against viral infections and tumors13,14. Studies elucidating the transcriptional and epigenetic heterogeneity of CD8+ T cells has provided important advances in understanding lineage differentiation, proliferative potential and functional capacity. In chronic lymphocytic choriomeningitis virus (LCMV) infection, TCF1 was identified as a critical transcription factor that maintains a precursor population within the exhausted T cell (Tex) pool15–19. Highly expressed in naive CD8+ T cells, TCF1 is downregulated during effector cell differentiation in response to pro-inflammatory cytokines such as IL-12 and type I interferons (IFN-I)20,21. Previously thought to be dysfunctional, a subset of PD-1+ Tex referred to as progenitor exhausted or stem-like cells that express TCF1 remain responsive to checkpoint inhibitors (CPI); they maintain stem cell-like quality by retaining the capacity to proliferate and self-renew22–24. Studies assessing CD8+ tumor infiltrating lymphocytes (TIL) in humans identified gene signatures of stem-like cells including Tcf7 (encoding TCF1), associated with better prognosis and response to checkpoint blockade25–30. Adoptive transfer studies in mice showed that the self-renewing capacity of TIL was regulated by TCF131 whereas cells lacking TCF1 were less effective in controlling tumor growth32. Collectively, these data suggest an important role for TCF1+PD-1+ CD8+ T cells in mediating protection against viral infections and tumors.
Here, we demonstrate how altering the route and dose of vaccination with SNP-7/8a influences the magnitude, transcriptional quality and antitumor capacity of neoAg-specific CD8+ T cells. In addition, we identify the innate mechanisms for how the route of vaccination alters antigen duration and tropism for distinct DC subsets critical for imprinting CD8+ T cell responses.
RESULTS
Route and dose of SNP-7/8a immunization alter the magnitude and quality of neoAg+ CD8+ T cells
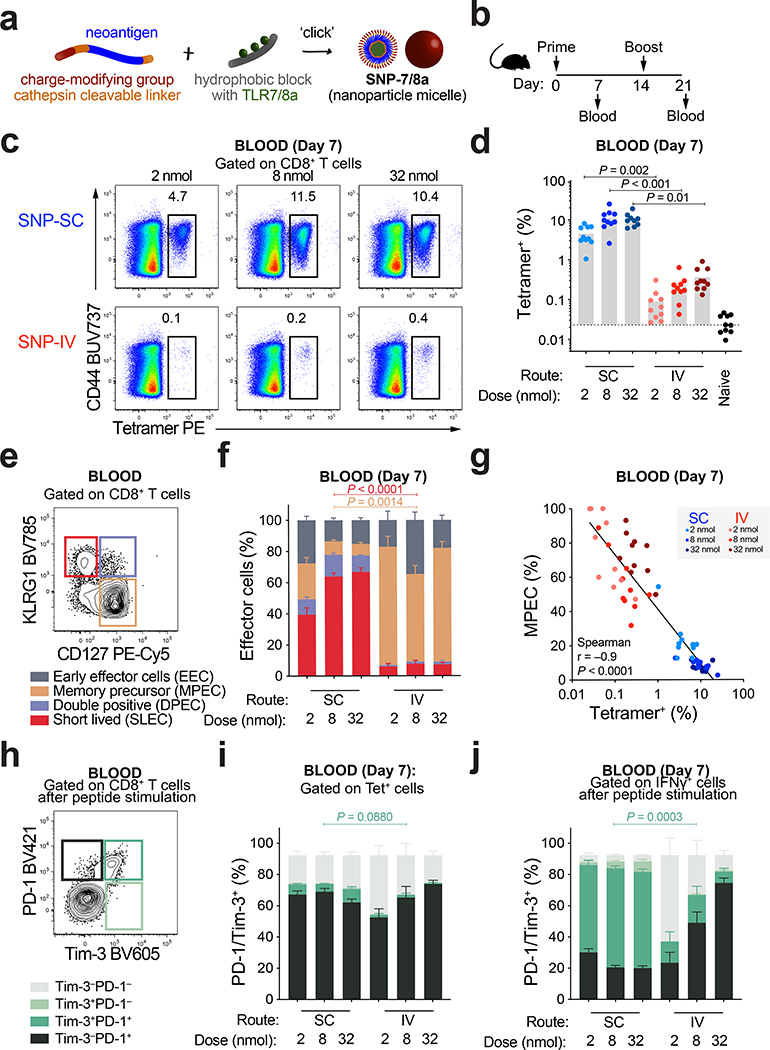
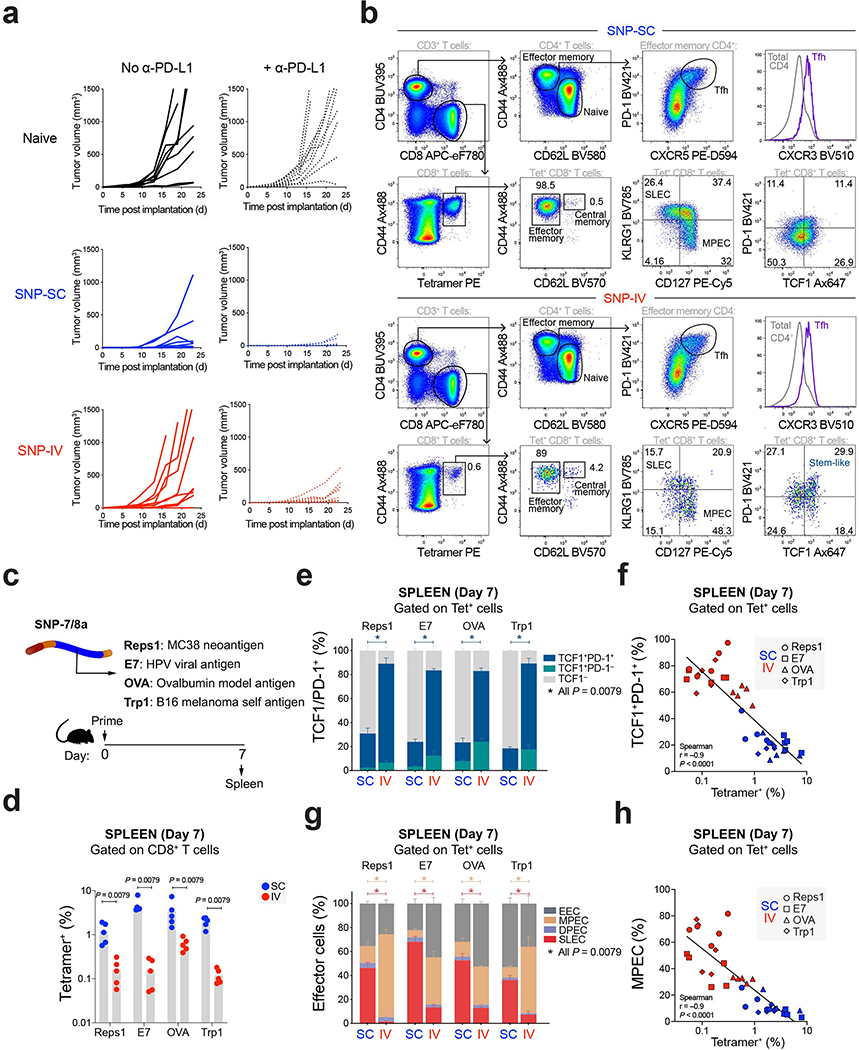
SNP-7/8a is a self-assembling nanoparticle vaccine platform that standardizes the delivery of LP11 (Fig. 1a). Here, we determined how modifying the dose and route of SNP-7/8a immunization influenced CD8+ T cell responses. Mice were vaccinated with SNP-7/8a containing Reps1, an MC38 murine colon carcinoma neoAg (Fig. 1b). Whole blood was collected to measure neoAg-specific CD8+ T cells by tetramer staining (Fig. 1c and Extended Data Fig. 1a) or IFN-γ after re-stimulation with Reps1 peptide (Extended Data Fig. 1b). At a dose of 8 nmol, subcutaneous administration of SNP-7/8a (SNP-SC) generated 20-fold higher neoAg-specific CD8+ T cells compared to IV vaccination (SNP-IV) (Fig. 1d). CD4+ T cells produced IFN-γ at low frequencies as previously described11 (Extended Data Fig. 1c). Together, these data show that the route of vaccination alters the magnitude of the CD8+ T cell response.
Figure 1 |. Route and dose of SNP-7/8a immunization controls the magnitude and phenotype of antigen-specific CD8 T cells.
a, Schematic of peptide–TLR7/8 agonist vaccines that form self-assembling nanoparticles (SNP-7/8a). b, C57BL/6 mice (n = 10) were vaccinated subcutaneously (SC) or intravenously (IV) at 2, 8 or 32 nmol on day 0 and day 14 with SNP-7/8a containing Reps1, an MC38 neoantigen. Whole blood was collected on day 7 and day 21 to measure the frequency of tetramer+ CD8+ T cells. c, Flow cytometric analysis of single cells stained with Reps1 tetramer and CD44 antibody. Numbers indicate percentage of cell population within the gate. d, Bar graphs summarize the frequency of tetramer+ CD8+ T cells from blood (n = 10) on day 7. e, CD8+ T cells were subdivided into memory precursor effector cells (MPEC, tan gate) or short-lived effector cells (SLEC, crimson gate) based on CD127 and KLRG1 expression. f, Bar graphs show proportions of MPEC/SLEC subpopulations in the blood on day 7 (n = 10). g, Frequency of MPEC is negatively correlated to frequency of tetramer+ CD8+ T cells. h, Tetramer+ cells can be subdivided into PD-1+ (black), Tim-3+ (light green) or PD-1+Tim-3+ (dark green) cells. Bar graphs show proportions of PD-1/Tim-3 subpopulations on day 7 (n = 10) of i, tetramer+ cells or j, IFNγ+ cells. Data are representative of two independent experiments. The bars represent the median. Statistics were assessed by Kruskal-Wallis with Dunn’s correction for multiple comparisons (d,f,i,j) and Spearman correlation (g).
Next, we characterized CD8+ memory precursor effector cells (MPEC) or short-lived effector cells (SLEC) based on differential expression of CD127 (IL-7R) and KLRG1 (Fig. 1e). A high proportion of neoAg-specific CD8+ T cells following SNP-SC were SLEC (~60% of tetramer+) whereas SNP-IV cells were primarily MPEC (~60% of tetramer+) (Fig. 1f and Extended Data Fig. 1d). The frequency of MPEC is inversely correlated to the frequency of tetramer+ CD8+ T cells (Fig. 1g and Extended Data Fig. 1e). We then assessed expression of PD-1 and Tim-3, canonical markers of T cell activation, exhaustion or severe dysfunctionality33 (Fig. 1h). Tetramer+ CD8+ T cells from both SNP-SC and SNP-IV mice were PD-1+ but only SC-vaccinated groups expressed low levels of Tim-3 (Fig. 1i and Extended Data Fig. 1f). Peptide re-stimulation markedly increased the expression of PD-1 and Tim-3 following SNP-SC but not SNP-IV (Fig. 1j and Extended Data Fig. 1g). Taken together, the data suggest that the route of vaccination influences the differentiation of neoAg-specific CD8+ T cells.
IV administration of SNP-7/8a generates TCF1+PD-1+ CD8+ T cells with antitumor capacity upon anti-PD-L1 treatment
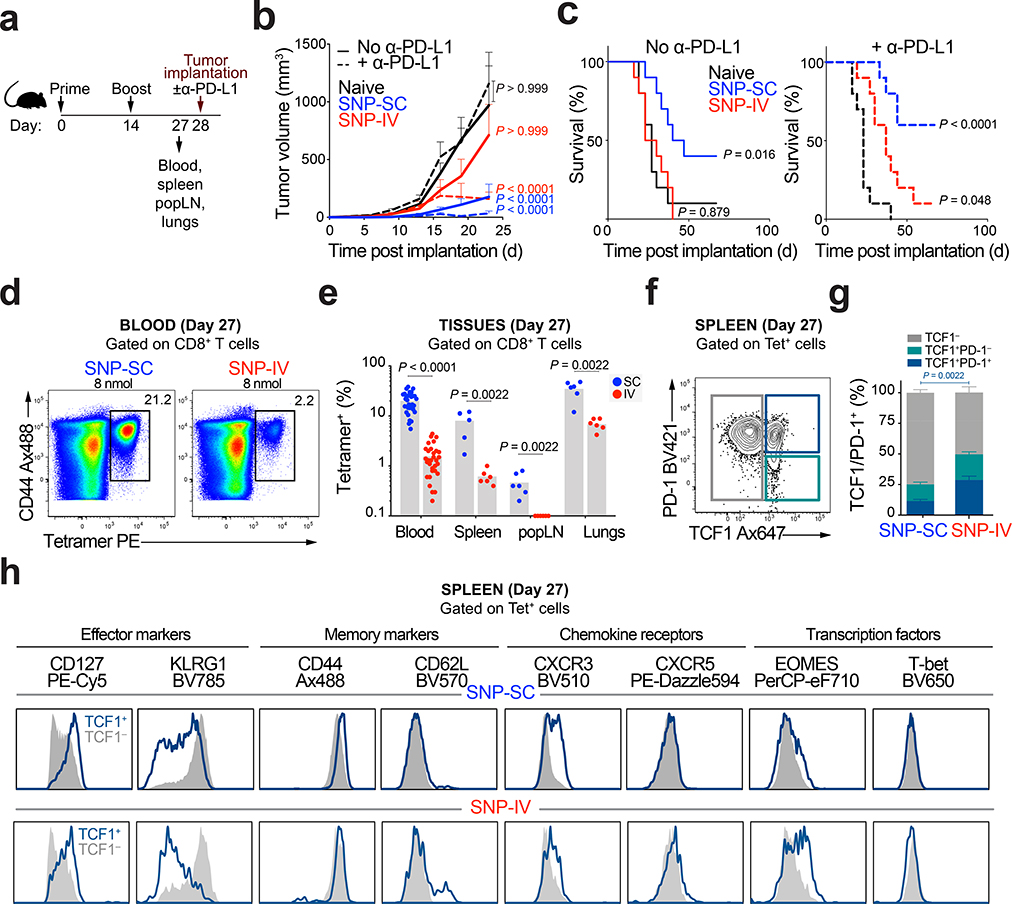
To investigate the antitumor capacity of neoAg-specific CD8+ T cells, mice were challenged with MC38 tumors (Fig. 2a). To evaluate the effect of a checkpoint inhibitor (CPI), mice were also either treated with or without anti-PD-L1. Anti-PD-L1 treatment alone was not sufficient to control tumor growth in unvaccinated mice (Fig. 2b and Extended Data Fig. 2a) or extend survival (Fig. 2c). SNP-SC vaccinated mice significantly controlled tumor growth compared to naive mice (Fig. 2b) independent of CPI treatment. Despite the 10-fold reduction in CD8+ T cell responses (Fig. 2d), SNP-IV vaccinated mice were able to control tumor growth (Fig. 2b) and extend survival compared to naive (Fig. 2c) only when combined with CPI treatment.
Figure 2 |. Intravenous administration of SNP-7/8a generates TCF1+ CD8+ T cells with anti-tumor capacity upon anti-PD-L1 treatment.
a, Mice (n = 16) were vaccinated SC or IV with SNP-7/8a (Reps1). Whole blood, spleens, popliteal lymph nodes (LN) and lungs were collected prior to tumor implantation on day 28. b, Tumor growth curves and c, Kaplan-Meier survival curves of mice unvaccinated (black) or vaccinated with SNP-SC (blue) or SNP-IV (red) with (dotted line) or without α-PD-L1 (solid line) (n = 13). d, Flow cytometric analysis of single cells stained with Reps1 tetramer and CD44 antibody. Numbers indicate percentage of cell population within the gate. e, Frequency of tetramer+ CD8+ T cells from blood (n = 32), spleen, popliteal LN and lungs (n = 6) on day 27 post vaccination. f, Effector CD8+ T cells in the spleen were subdivided into TCF1− (grey), TCF1+PD-1− (teal) or TCF1+PD-1+ (dark blue) populations. g, Bar graphs summarize the frequencies of TCF1 subpopulations in the spleen (n = 6) after SNP-SC or SNP-IV. h, Histograms show differential expression of phenotypic markers expressed by TCF1+ (dark blue) or TCF1− (grey) populations after SNP-SC (top panel) or SNP-IV (bottom panel) as assessed by flow cytometry (concatenated, n = 6). Data are representative of four independent experiments. Mean ± s.e.m. (b,e,g). Statistics were assessed by two-way ANOVA with Bonferroni correction (b), Log-rank test (c) and Mann Whitney test (e,g).
The distribution of tetramer+ CD8+ T cells was assessed at the time of tumor challenge in blood, spleen, popliteal LN (SC vaccine-draining) and the lungs (Fig. 2e). NeoAg-specific CD8+ T cells generated by SNP-SC was significantly higher in all tissues compared to SNP-IV. We detected a population of TCF1+PD-1+ within tetramer+ cells in the spleen (Fig. 2f). Such cells have been described in chronic LCMV15 as a population of exhausted cells that retain proliferative capacity. SNP-IV generated a higher proportion of TCF1+ cells, representing ~50% of tetramer+ CD8+ T cells (Fig. 2g). TCF1+ cells in the SNP-IV group exhibited high CD127 and low KLRG1 (Fig. 2h). TCF1+ CD8+ T cells generated by SNP-IV also expressed proteins upregulated by Tex progenitor cells that retain a stem-like program15,18,27 such as eomesodermin (EOMES) and CXCR3 (Fig. 2h). However, CXCR5 expressed by stem-like cells following chronic LCMV was not detected in TCF1+ CD8+ T cells following SNP-7/8a vaccination. As a staining control, CD4+ T follicular helper (Tfh) cells expressed CXCR5 (Extended Data Fig. 2b). In contrast, TCF1+ CD8+ T cells generated by SNP-SC had high expression of KLRG1 and lower EOMES levels (Fig. 2h). To assess the generalizability of these observations, mice were immunized with SNP-7/8a containing other antigens including E7, OVA and Trp1 (Extended Data Fig. 2c). Similarly, SNP-IV resulted in lower magnitude responses (Extended Data Fig. 2d), but a higher frequency of TCF1+PD-1+ cells (Extended Data Fig. 2e and 2f) and MPEC (Extended Data Fig. 2g and 2h) compared to SNP-SC, regardless of the antigen. These data indicate a phenotypic difference in the neoAg-specific CD8+ T cells generated depending on route of vaccination.
Single cell analysis of neoAg-specific CD8+ T cells identifies stem-like gene signature following SNP-IV
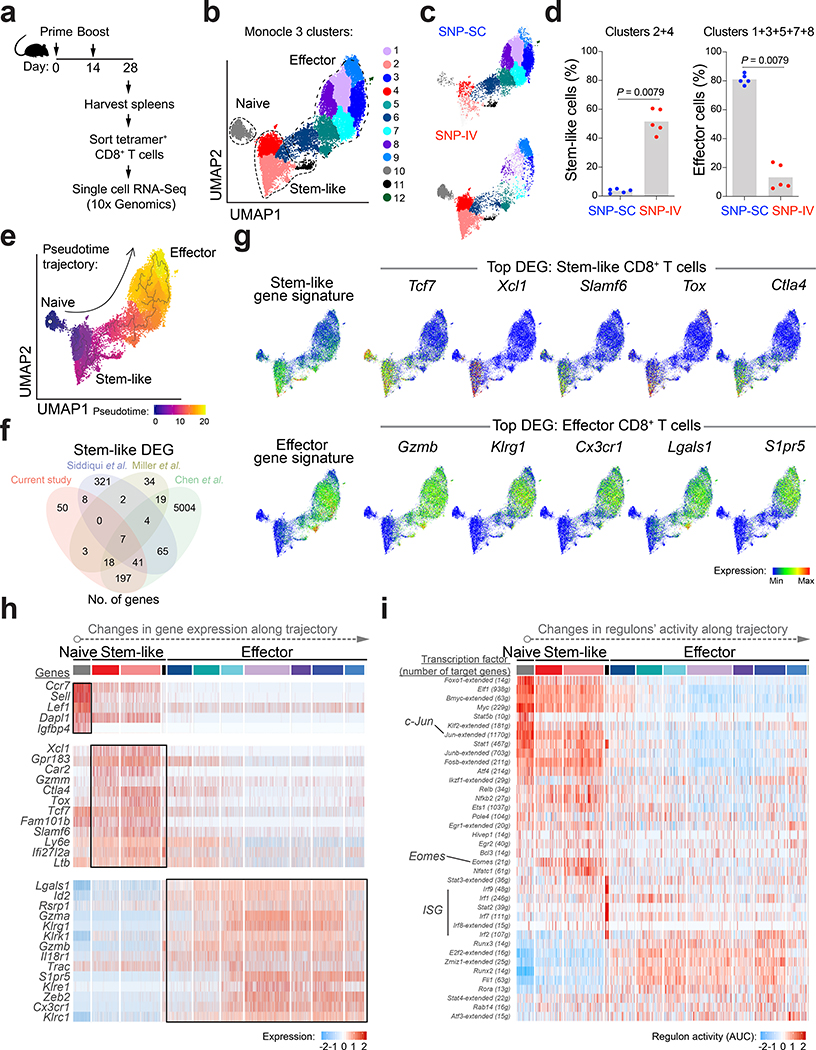
To further characterize the neoAg-specific CD8+ T cells, we sorted tetramer+ cells following SNP-SC and SNP-IV vaccination (Fig. 3a and Extended Data Fig. 3a). Tetramer+ CD8+ T cells from individual animals were barcoded prior to single-cell RNA sequencing (scRNA-seq) using the droplet-based system of 10x Genomics (Extended Data Fig. 3b). Cells were clustered based on gene expression using an unsupervised inference analysis (Monocle 3). The 12 clusters identified were visualized by uniform manifold approximation and projection for dimension reduction (UMAP) algorithm (Fig. 3b). Clustering data showed separation of single cells by route of vaccination (Fig. 3c and Extended Data Fig. 3c). SNP-IV neoAg+ cells were primarily in clusters 2 and 4, which we refer to as “stem-like” cells given that Tcf7 (encoding TCF1) was a significant differentially expressed gene (DEG) in these two clusters (Fig. 3d and Supplementary Table 1). In contrast, SNP-SC neoAg+ cells mainly appeared in the “effector” cluster consisting of clusters 1, 3, 5, 7 and 8 (Fig. 3d). Both stem-like and effector clusters form stable states (high density areas on UMAP)34 (Extended Data Fig. 3d). Cluster 10 represents a small population of naive T cells (Ccr7, Lef1, Sell, Dapl1, Igfbp4) (Extended Data Fig. 3e) that may have contaminated while sorting due to low frequencies with SNP-IV. Excluding naive cells, the T cell receptor (TCR) repertoire is largely diverse following both routes of immunization (Extended Data Fig. 3f). The fraction of stem-like or effector cells within each clone is driven by the route of vaccination, rather than the clonotype identity (Extended Data Fig. 3g).
Figure 3 |. Single-cell analysis of neoAg+ CD8+ T cells by RNA sequencing identifies stem-like gene signature in SNP-IV and effector gene signature in SNP-SC cells.
a, Mice (n = 5) were vaccinated SC or IV with SNP-7/8a (Reps1). Spleens were collected on day 28 and tetramer+ CD8+ T cells were sorted by flow cytometry. Single cell RNA sequencing was performed by 10X Genomics. b, UMAP of sorted neoAg+ CD8+ T cells from spleen. Twelve clusters were generated by Monocle 3 (k = 12) analysis of gene expression. c, Single cells from SNP-IV (Top panel) or SNP-SC (bottom panel) cluster in distinct regions of the UMAP space. d, Bar graphs summarize the frequencies of stem-like cells (clusters 2 and 4, left panel) or effector cells (clusters 1, 3, 5, 7 and 8, right panel) of total tetramer+ CD8+ T cells in the spleen (n = 5). e, Reconstruction of pseudotime trajectory using Monocle 3 algorithm. f, Venn diagram comparing identified differentially expressed genes (DEG) of stem-like cells in this study and three published datasets. g, A list of stem-like or effector gene signatures were overlaid on the UMAP of single cell data. Expression of top DEG of stem-like (top panel) or effector (bottom panel) cells are presented as meaning plots. h, Heatmap of selected DEG expressed in each cluster organized along the pseudotime trajectory. i, Heatmap of significant changes in regulons’ activity as inferred by SCENIC analysis. Statistics were assessed by Mann Whitney test (d).
Next, we reconstructed a developmental trajectory of neoAg-specific CD8+ T cells. The pseudotime analysis showed a trajectory originating from naive T cells, branching into stem-like cells and eventually effector cells (Fig. 3e). We further compared stem-like cells in this study with published datasets from chronic LCMV or tumor models18,22,32, identifying 337 shared genes and 50 unique genes in our dataset (Fig. 3f and Supplementary Table 2). Applying a list of previously described stem-like genes (also referred to as ‘Tex progenitor’) and effector genes (‘Teff’) in the context of LCMV infection18, we confirmed that the stem-like gene signature was highly expressed in clusters 2 and 4, whereas the effector gene signature was more pronounced in clusters 1, 3, 5, 7 and 8 (Fig. 3g).
Consistent with flow cytometry detection of TCF1 in SNP-IV CD8+ T cells (Fig. 2f), the stem-like cluster expressed high levels of Tcf7 (Fig. 3g). These cells also expressed genes associated with T cell exhaustion: Tox, Lag3, and Ctla4, yet upregulated anti-apoptotic genes such as Bcl2 (Supplementary Table 2). As previously observed in stem-like CD8+ T cells from chronic LCMV18,19,35, Slamf6 (encoding Ly108) was highly expressed in the stem-like CD8+ T cells following IV vaccination. Unlike data from LCMV studies, the stem-like CD8+ T cells reported here did not show increased Cxcr5, consistent with the flow cytometric data (Supplementary Table 2 and Extended Data Fig. 3h). The stem-like cluster expressed high levels of Xcl1, which encodes for XCL1 – the ligand for XCR1+ cDC1. The effector cluster expressed genes encoding cytotoxic granules such as Gzma and Gzmb as well as inhibitory receptors such as Klrg1 (Fig. 3h and Supplementary Table 1). A chemokine receptor Cx3cr1 expressed by transitory, effector-like cells in chronic LCMV19,35, is also highly expressed in our effector cluster. Other notable genes that are highly expressed in the effector cluster include Zeb2 (triggers cytotoxic T lymphocytes to adopt terminally differentiated state36), Lgals1 (Galectin-1, a pro-apoptotic molecule37) and S1pr5 (S1PR5, cues exit from lymphoid tissue38) (Fig. 3g and 3h).
To further evaluate cellular states based on expression of transcription factors and target genes, we measured regulons’ activity by performing SCENIC analysis34 (Fig. 3i). Changes in transcriptional network can be detected along the Monocle 3 pseudotime trajectory. Upregulation of Eomes regulon (Eomes and 21 related genes) can be measured in the stem-like cluster, in line with EOMES detected by flow cytometry (Fig. 2h). In contrast, the effector cluster is marked by upregulation of several regulons including Runx3, Runx2, Rora and Fli1 (Fig. 3i). Taken together, at the transcriptional level, the neoAg-specific CD8+ T cells generated by SNP-IV express a stem-like gene signature, whereas SNP-SC cells express an effector gene signature. Further, trajectory inference analysis of scRNA-seq data highlights differences in the developmental states of stem-like and effector CD8+ T cells.
Therapeutic vaccination by SNP-IV controls tumor growth but not SNP-SC
To assess the antitumor capacity of SNP-7/8a in mice with established tumors, MC38 was implanted and vaccination was administered on day 7 (prime) and day 14 (boost) (Fig. 4a). Anti-PD-L1 was given at the time of boost. In this study, the dose of SNP-IV was increased to 32 nmol to generate CD8+ T cells of comparable magnitude to the SNP-SC vaccinated mice (Fig. 4b). Both vaccination routes resulted in ~10% Reps1+ CD8+ T cells (Fig. 4c). Despite similar numbers of neoAg-specific CD8+ T cells, SNP-SC mice could not control tumor growth whereas SNP-IV mice significantly controlled tumor growth compared to naive mice (Fig. 4d). The antitumor efficacy of SNP-IV is antigen-specific and adjuvant-dependent as vaccinating with an irrelevant antigen IV (Fig. 4e) or no adjuvant (SNP with no TLR7/8a) (Extended Data Fig. 4a) was ineffective. The differences in tumor growth is further confirmed in prolonged survival of mice vaccinated with SNP-IV but not SNP-SC (Fig. 4f). Decreasing the SNP-SC dose or reducing to a single vaccination was not effective (Extended Data Fig. 4b). The tumor control (Fig. 4g) and increased survival (Fig. 4h) by SNP-IV were abrogated by depleting CD8+ T cells but not CD4+ T cells or NK cells (Extended Data Fig. 4c and 4d).
Figure 4 |. Therapeutic vaccination with SNP-IV generates neoAg-specific CD8+ T cells with superior anti-tumor capacity.
a, Mice (n = 10) were implanted with MC38, vaccinated with SNP-7/8a (Reps1) and treated with CPI. b,c, Flow analysis of blood stained with tetramer and CD44 antibody (n = 10). d, Tumor growth of SNP-SC or SNP-IV mice (n = 10). e, Average tumor growth and f, survival of Reps1 IV (red), irrelevant IV (maroon), Reps1 SC (blue) and unvaccinated (black) mice. g, Average tumor growth and h, survival of SNP-IV mice treated with isotype (red) or blocking antibodies against CD8+ (black), CD4+ (blue) and NK cells (purple) (n = 10). i, Average tumor growth and j, survival of mice vaccinated with SNP-IV once (blue), vaccinated once and given CPI (green), vaccinated twice (orange), vaccinated twice and given CPI (red) or untreated (black) (n = 10). k, Flow cytometric analysis of spleens stained with tetramer and CD44 antibody. l, Frequency and m, total number of tetramer+ CD8+ T cells on day 21 (n = 3). n, Flow cytometric analysis of spleens stained with TCF1 and Granzyme B. Total number of o, stem-like cells (TCF1+Granzyme B−) and p, effector cells (TCF1− Granzyme B+) on day 21 (n = 3) (unpaired t-test). Data are representative of four independent experiments. The bars represent the median (c) or mean (l,m). Mean ± s.e.m. (e,g,i,o,p). Statistics were assessed by two-way ANOVA (e,g,i), Log-rank test (f,h,j), Mann Whitney test (c) and two-tailed unpaired t-test (l,m,o,p).
To delineate each variable of the SNP-IV treatment regimen, MC38 tumor-bearing mice were vaccinated on day 7 with 32 nmol of SNP containing Reps1 (Fig. 4i). On day 14, groups of mice were either left untreated (“prime”), given CPI (“prime and CPI”), vaccinated again (“prime and boost”), or vaccinated again together with CPI (“prime, boost and CPI”). Indeed, the combination of both vaccinations and CPI was most effective at controlling tumor growth (Fig. 4i) and extending survival (Fig. 4j). Spleens were collected from mice on day 14 (post prime) and day 21 post tumor implantation (Fig. 4k). Without the boost or CPI treatment, the frequency of neoAg-specific CD8+ T cells in the spleen was significantly lower (6-fold) than mice that received a boost and CPI (Fig. 4l). A clear expansion of neoAg-specific CD8+ T cells was observed when enumerating tetramer+ CD8+ T cells in the spleen on day 21 (Fig. 4m). On day 14, approximately 80% of CD8+ T cells were stem-like (TCF1+Granzyme B−) after SNP-IV, compared to 35% following SNP-SC (Fig. 4n). Co-staining with PD-1 confirmed that these cells were high in PD-1 (Extended Data Fig. 4e). By day 21 post SNP-IV, neoAg+ CD8+ T cells from spleens of tumor bearing mice have mostly differentiated to effector cells (TCF1−Granzyme B+) (Fig. 4n). Mice receiving both immunizations and CPI maintained high numbers of stem-like cells in the spleen (Fig. 4o), unlike the other groups that did not receive a boost or CPI. This coincided with a significant 10-fold increase in neoAg+ effector cells expressing Granzyme B in mice that received boost and CPI, compared to mice that received boost with no CPI (Fig. 4p). In contrast, a modest 1.5-fold expansion of effector cells in the spleen was observed following SNP-SC (Extended Data Fig. 4f). Importantly, neoAg-specific CD8+ T cells generated by SNP-SC co-expressed PD-1 and Tim-3 by day 21 (~35%) while SNP-IV cells did not express Tim-3 in the spleen (Extended Data Fig. 4g). On day 21, SNP-IV cells had increased Ki-67 expression in blood and spleen (Extended Data Fig. 4h), indicating an expansion of circulating effector cells. Taken together, these highlight the capacity of stem-like neoAg+ CD8+ T cells generated by SNP-IV to proliferate and replenish effector cells for antitumor responses upon CPI treatment.
Prolonged antigen persistence, DC uptake and activation following SNP-SC but not SNP-IV
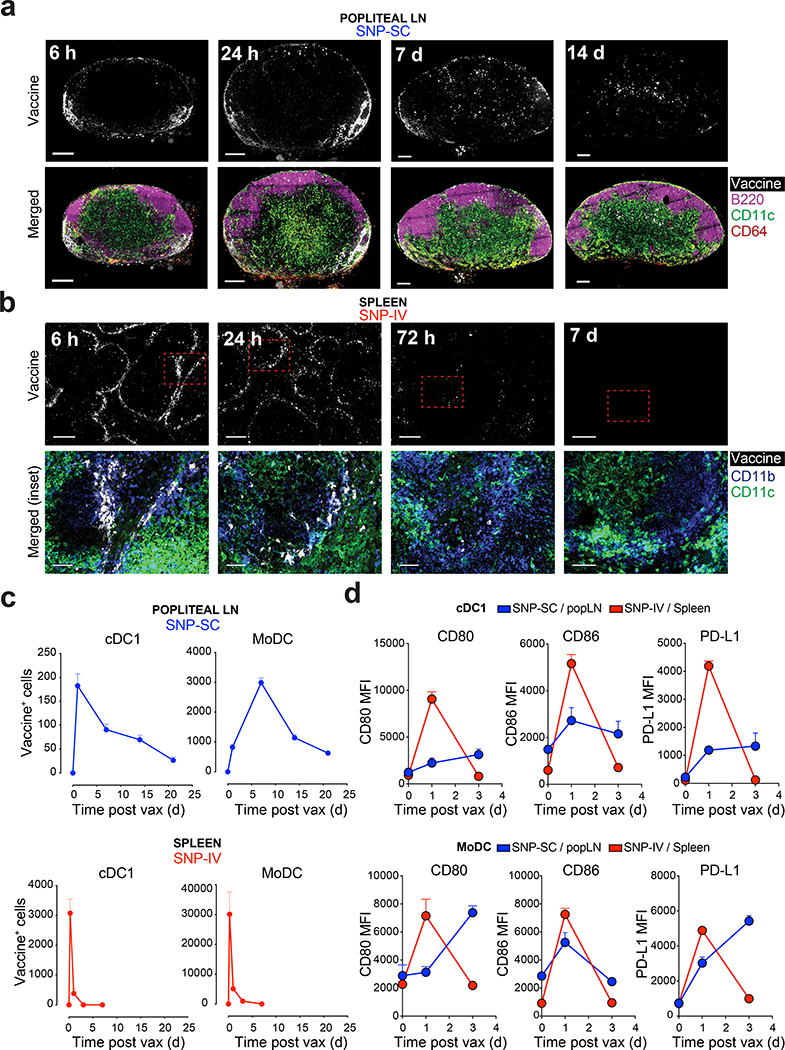
To investigate differences in the mechanism of CD8+ T cell priming, fluorescently labeled SNP-7/8a was used to visualize the distribution and uptake by DC. In vivo imaging of mice revealed that SNP-SC remain localized to the site of vaccination two weeks post vaccination, highlighting a depot effect (Extended Data Fig. 5a). In contrast, SNP-IV resulted in systemic distribution of the vaccine detectable up to 24 hours by in vivo imaging (Extended Data Fig. 5a). LN sections analyzed by confocal microscopy confirmed that vaccine can be detected up to two weeks after SNP-SC, primarily in the T cell zones. (Fig. 5a and Extended Data Fig. 5b). Co-staining with monocyte, macrophage and DC markers revealed co-localization of vaccine with cells expressing CD11b and CD11c (Extended Data Fig. 5c). Following SNP-IV, confocal images of spleen sections show vaccine uptake by CD11b+CD11c+ cells situated in the marginal zones primarily after 6 and 24 hours with minimal vaccine detection after 3 and 7 days (Fig. 5b). Popliteal LN and spleens were harvested for flow cytometric analysis (Extended Data Fig. 5d). Labeled vaccine could be detected in both cDC1 (Lin− MHCII+CD11c+XCR1+) and moDC (Lin−CD64+F4/80+MHCII+CD11c+) in the popliteal LN after SNP-SC as late as 3 weeks after vaccination (Fig. 5c and Extended Data Fig. 5e). Similarly, SNP-IV resulted in uptake by cDC1 and moDC in the spleen. However, this peaked at 6 hours post vaccination (Fig. 5c and Extended Data Fig. 5f). Co-stimulatory molecules CD80 and CD86 as well as the regulatory marker PD-L1 were upregulated in cDC1 and moDC after vaccination by both routes (Fig. 5d). SNP-IV resulted in transient DC activation that peaked at day 1, whereas SNP-SC resulted in prolonged activation of DCs over three days (Fig. 5d). Expressions of CD80, CD86 and the migratory marker CCR7 on moDC were detectable 3 weeks later suggesting continued infiltration of APC into the draining LN (Extended Data Fig. 5g). The frequency of cDC1 in the spleen after SNP-IV peaked at 6 hours and diminished to below baseline levels at 24 hours as assessed by flow cytometry (Extended Data Fig. 5h and 5i). In contrast, the numbers of cDC1 remain high three weeks after SNP-SC in the popliteal LN (Extended Data Fig. 5h and 5i). The differences in presence and activation of cDC1 at the site of priming highlight the potential importance of DC in modulating CD8+ T cell quality after SNP vaccination.
Figure 5 |. Transient vaccine distribution to spleen and activation of migratory cDC1 and moDC in after SNP-IV.
a, Mice (n = 3) were vaccinated with SNP-7/8a labeled with Alexa Fluor 647. Popliteal LNs and spleens were harvested from mice at specified time points. Confocal images of sections from popliteal LNs collected 0 h, 6 h, 24 h, 7 d and 14 d after SC injection with labeled SNP-7/8a. Left panel, vaccine; Right panel, merged. White, vaccine; magenta, B220 (B cells); green, CD11c (moDC or cDC); red, CD64 (monocytes or macrophages). Scale bars, 200 μm. b, Confocal images of sections from spleens collected 0 h, 6 h, 24 h, 3 d and 7 d after IV injection with labeled SNP-7/8a. Left panel, vaccine; Right panel, merged of inset. White, vaccine; blue, CD11b (monocytes, macrophages or cDC2), CD11c (moDC or cDC). Scale bars, 200 μm or 50 μm (inset). c, Popliteal LNs and spleens were harvested, and single cell suspensions were assessed by flow cytometry for detailed identification of DC subsets. Kinetics of vaccine+ cDC1 (XCR1+MHCII+CD11c+; top panels) and moDC (F4/80+CD64+MHCII+CD11c+; bottom panels) in popliteal LN (left panels) or spleen (right panels) after injection with labeled vaccines via SC or IV respectively (n = 3). d, Graphs summarize the mean fluorescence intensity (MFI) of CD80 (left panel), CD86 (middle panel) and PD-L1 (right panel) expressed on cDC1 and moDC in popliteal LN after SNP-SC (blue lines) or in spleen after SNP-IV (red lines) (n = 3). Mean ± s.e.m. (c,d). Data are representative of two independent experiments.
cDC1 and moDC are required for CD8+ T cell priming
To directly address the role of specific APCs in priming CD8+ T cells, wild type (WT), Batf3−/− and Ccr2−/− mice lacking cross-presenting cDC1 and circulating monocytes respectively were immunized with SNP-7/8a (Extended Data Fig. 6a and 6b). The frequency of neoAg-specific CD8+ T cells was significantly decreased following SNP-SC in Batf3−/− (4-fold lower) and Ccr2−/− (20-fold lower) mice (Fig. 6a). As monocytes remain in the spleens of Ccr2−/− mice, we generated bone marrow chimeras of Ccr2DTR mice to conditionally deplete monocytes in peripheral tissues and lymphoid organs (Extended Data Fig. 6c and 6d). Following SNP-IV, the frequency of neoAg-specific CD8+ T cells significantly decreased by diphtheria toxin (DT) treatment of CCR2DTR BM chimera mice (5-fold lower) and Batf3−/− mice (15-fold lower) suggesting dependency on monocytes and cDC1 (Fig. 6b). Mice receiving WT BM with or without DT treatment and mice receiving Ccr2DTR BM without DT treatment generated similar frequencies of neoAg-specific CD8+ T cells after SNP-IV (Extended Data Fig. 6e). Collectively, these data highlight a critical role for not only Batf3-dependent cDC1 but also moDC in priming CD8+ T cells with the SNP-7/8a vaccine.
Figure 6 |. Prolonged antigen presentation by DC drives CD8+ T cell responses after SNP-SC.
Kinetics of neoAg-specific CD8+ T cells following a, SNP-SC or b, SNP-IV in Batf3−/− (triangle), Ccr2−/− (square), Ccr2DTR treated with DT (half square) compared to WT mice (circle) (n = 10). c, IL-12 (left panel) and IFN-α (right panel) were measured by ELISA in supernatants of cultured spleens or popliteal LN following SNP-IV or SNP-SC respectively (n = 3). d, Bar graphs summarize neoAg-specific CD8+ T cells following SNP-SC in WT mice (circle) (n = 8), IL12b−/− (open triangle) (n = 4), Ifnar−/− (open square) (n = 5), Tlr7−/− (open circle) (n = 5). e,f, Flow analysis of splenocytes stained with tetramer and CD44 antibody following SNP-SC in WT (n = 8) or Il12b−/− (n = 4) mice. g,h, CD8+ T cells in the spleen after SNP-SC were subdivided into TCF1− (grey), TCF1+PD-1− (teal) or TCF1+PD-1+ (dark blue). Bar graphs summarize the frequencies of TCF1 subpopulations (n = 4). i, CD45.1 mice (n = 3) were vaccinated with SNP-7/8a (SIINFEKL). 1-, 3- or 7-days post vaccination, naive CD8+ T cells from CD45.2 OT-I mice were labeled and transferred into CD45.1 mice. Four days post transfer, spleens were assessed for cell proliferation. j, Graphs summarize the number of CPDlo cells after SNP-SC (left panel) or SNP-IV (right panel) (n = 3). Data are representative of two independent experiments. The bars represent the median (d) or mean (h). Mean ± s.e.m. (a,b,c,j). Statistics were assessed by Mann Whitney test (a,b,j) or Kruskal-Wallis test with Dunn’s correction for multiple comparisons (d).
As type I IFN (promoting cross-presentation39) and IL-12 (promoting Th1 skewing40) can have critical roles in controlling CD8+ T cell priming, these cytokines were measured. SNP-IV induced transient production of IL-12 and IFN-α which peaked at 6 hours post vaccination in blood and spleen (Fig. 6c and Extended Data Fig. 6f). In contrast, IL-12 was detected up to two weeks after SNP-SC in the popliteal LN (Fig. 6c) consistent with the duration of vaccine detection (Fig. 5a).
To investigate the importance of IL-12 and IFN-α, Il12b−/− and Ifnar−/− mice were vaccinated (Fig. 6d), with Tlr7−/− mice used as a negative control. Both priming and boosting of neoAg-specific CD8+ T cells with SNP-SC or SNP-IV were IFN-α- and TLR7-dependent but IL-12-independent (Fig. 6d). In mice deficient for IFN-α receptor and TLR7, fewer monocytes and cDC1 were in the popliteal LN after SNP-SC than in WT mice (Extended Data Fig. 6g). To assess whether IL-12 affected the quality of neoAg+ CD8+ T cells21, we collected spleens after SNP-SC boost. Similar to blood responses, Il12b−/− mice had comparable tetramer+ CD8+ T cell responses in the spleen as WT mice after SNP-SC (Fig. 6e and Fig. 6f). Further, similar frequencies of TCF1 in both Il12b −/− and WT neoAg+ CD8+ T cells suggest that IL-12 signaling was not mediating TCF1 downregulation (Fig. 6g and 6h). EOMES expression was higher in Il12b −/− compared to WT mice after SNP-SC (Extended Data Fig. 6h).
SNP-SC leads to prolonged antigen retention and presentation in vivo
Based on the data showing prolonged retention of vaccine and innate activation following SC immunization (Fig. 5c and 5d), we hypothesized that antigen persistence mediated the differences in CD8+ cell quality between SC and IV vaccination. Naive CD45.2 OT-I CD8+ T cells were labeled with a fluorescent dye and transferred to CD45.1 congenic mice 1, 3 or 7 days after vaccination with SNP-7/8a delivering the OT-I peptide (SIINFEKL) (Fig. 6i). Spleens were collected 4 days after cell transfer to assess the extent of CD8+ T cell proliferation as an indicator of antigen presence. At all three time points, cell proliferation could be detected, with the largest numbers of OT-I measured being 7 days after SNP-SC vaccination (Fig. 6j). In contrast, SNP-IV resulted in a burst of OT-I expansion when transferred 1 day after vaccination followed by significantly lower cell numbers 3 and 7 days after vaccination (Fig. 6j). Overall, the prolonged persistence of antigen in the popliteal LNs drove T cell differentiation, thus explaining the high magnitude but lower proliferative potential of neoAg-specific CD8+ T cells generated by SNP-SC.
DISCUSSION
In this study, we report how SNP-7/8a dose and route control the magnitude and transcriptional quality of neoAg-specific CD8+ T cells. In the context of cancer immunotherapy, persistence and functional capability are important factors given the chronic nature of cancer. The most striking finding was the demonstration that IV vaccination led to a major difference in the phenotypic and transcriptional quality of the CD8+ T cell response compared to SC vaccination. SNP-7/8a vaccination enables specific generation of stem-like or effector cells by modifying the route of vaccination. We demonstrate how this can be achieved in vivo using CD8+ T cells primed from a polyclonal naive repertoire, without the caveats of TCR-transgenic systems using model antigens. We would note that IV administration of RNA-based PCV has been shown to be more immunogenic than direct LN injection; however the transcriptional quality of CD8+ T cells by this route has not been assessed41,42. Here we clearly demonstrate that neoAg-specific CD8+ T cells generated by SNP-IV have a high proportion of TCF1+PD-1+ stem-like cells and mediate antitumor effects dependent on CPI.
The scRNA-seq data of neoAg+ CD8+ T cells revealed distinct transcriptional landscapes following SNP-SC or SNP-IV, providing insight into the dynamics of CD8+ T cell activation and differentiation following vaccination: naive cells give rise to stem-like cells that can be further differentiated into effector cells. Comparison of clusters revealed several novel genes highly expressed by stem-like CD8+ T cells. Xcl1 expression by stem-like CD8+ T cells suggests a role for XCR1+ cDC143 in licensing their activity in the spleen. In stem-like cells, the expression of Tox, recently described as a crucial regulator of T cell exhaustion44,45, may be beneficial in preventing overstimulation of CD8+ T cells leading to cell death especially in the context of cancer where there is antigen persistence. Interestingly, neoAg-specific CD8+ T cells generated by SNP-IV resemble the stem-like or progenitor exhausted cells described in chronic LCMV infection, whereas cells generated by SNP-SC share genes upregulated in acute LCMV infection15,18,19. Epigenetic analysis, specifically around the Tox locus, may reveal whether vaccine-induced CD8+ T cells resemble a distinct developmental program or share specific features of exhausted cells identified in LCMV46,47. Unlike chronic LCMV where there is persistent antigenic stimulation, we observe stem-like CD8+ T cells following IV vaccination of a peptide-based vaccine where the duration of antigenic delivery peaked at six hours. These data suggest distinct mechanisms of CD8+ T cell priming in vaccination versus natural infection and demonstrate the rapidity by which such cells can be induced by the SNP-7/8a vaccine which will be critical for therapeutic tumor vaccination.
The covalent linking of antigen and adjuvant into a nanoparticle was designed to synchronously deliver the innate stimulation with the antigen for efficient cross-presentation. Several important factors may contribute to the modulation of CD8+ T cell quality upon IV vaccination: location of CD8+ T cell priming, actual dose of vaccine delivered to splenic DC and degree of inflammation. The duration of antigen persistence of up to 3 weeks in the draining LN by SNP-SC contrasted with a short burst of antigen in the spleen by SNP-IV. Despite the brief duration, the spleen remains a large reservoir for DC as noted by the ten-fold increase in absolute numbers of cDC1 or moDC acquiring vaccine at the peak of SNP-IV delivery, compared to numbers of vaccine+ DC in the popliteal LN at the peak of SNP-SC delivery. This is likely the key factor in regulating the magnitude and quality of responses respectively. In a different cancer vaccine model using peptide and incomplete Freund’s adjuvant, antigen persistence also led to dysfunctional CD8+ T cells48. However, it is notable in the prophylactic tumor model presented here, the CD8+ T cells generated by SC vaccination were still highly functional. Confocal images of LN sections showed that, while antigen was retained for 3 weeks in the LN after SC vaccination, the localization of antigen changed: in the first 24 hours, antigen was mostly in the subcapsular sinus, but over the next two weeks, antigen concentrated into the T cell zones. This slow-release effect has been the basis of other rational vaccine design, regulating the kinetics of antigen exposure49. Thus, SNP-SC was efficient at generating effector CD8+ T cells at high numbers. This approach may be valuable for prophylactic cancer vaccines targeting known shared tumor antigens.
Importantly, co-delivery of antigen and adjuvant permits systemic IV administration by limiting the tolerogenic effects of having free peptide taken up by APC in the absence of a danger signal. The short-lived pharmacokinetics of SNP-IV in the spleen likely accounts for the lower magnitude when given at the same dose as SNP-SC. The reduction in numbers of splenic cDC1 between 24 hours and 3 days after IV vaccination may serve as a negative immunoregulatory mechanism. Further, the dependency on moDC for CD8+ T cell responses suggests that these cells either contribute indirectly by providing the required cytokines for T cell activation, by transferring antigen to other DC, or directly by presenting antigens to CD8+ T cells. Following SNP-IV, the frequency of cDC1, the canonical subset for efficient cross-priming, were dramatically reduced. We hypothesize that moDC, generally considered less efficient APC compared to cDC150, may be responsible for providing less-differentiated, more stem-like CD8+ T cells. Future studies will focus on how these distinct DC subsets can modify both the magnitude and quality of CD8+ T cells in the context of vaccination.
In conclusion, this study shows that the route of administration of SNP-7/8a vaccine substantially affected the magnitude and transcriptional quality of neoAg specific CD8+ T cell responses, which had a corresponding impact on functionality and therapeutic outcomes. These findings have implications in the clinical development of therapeutic vaccines for cancer patients.
METHODS
Mice
Female B6 mice (C57Bl/6J), Ccr2−/−, Batf3−/−, Ifnar−/−, Tlr7−/−, Il12b−/− were purchased from Jackson laboratories and maintained in pathogen-free conditions. OT-I Rag2 transgenic mice were purchased from Taconic. CCR2DTR mice51 were bred inhouse. Mice were used in studies when 6–8 weeks old. All animal experiments were performed at the Vaccine Research Center (VRC) at the National Institutes of Health (NIH, USA) with the approval of the Institutional Animal Care and Use Committee (ACUC) at the NIH. Experiments complied with the ethical guidelines set by ACUC and animals were humanely euthanized at defined endpoints.
Generation of bone marrow chimeras
8-week-old recipient mice received 13 Gy of γ-irradiation (2 doses of 6.5 Gy each) prior to IV reconstitution with bone marrow from CCR2DTR mice. Four to eight weeks after reconstitution, successful chimerism was assessed by flow cytometry. Mice were used in studies eight weeks after reconstitution.
In vivo depletions
For cell depletion experiments using neutralizing antibodies, mice were injected with 200 μg/mouse of either anti-CD8β (Clone 53–5.8, BioXcell), anti-CD4 (Clone GK1.5, BioXcell), anti-NK1.1 (Clone PK136, BioXcell) and the respective isotype controls (BioXcell). For cell depletion experiments in conditional knockout mice (Ccr2DTR), 10 ng/g body weight of diphteria toxin (DT) were injected intraperitoneally on day −1, day 1 and day 3 relative to time of vaccination.
Vaccines
SNP conjugate vaccines were produced as previously described11. Briefly, peptide antigens (Genscript, USA) were linked to hydrophobic blocks containing Imidazoquinoline-based TLR7/8a (Avidea Technologies, USA) using a click chemistry reaction. For pharmacokinetics studies, SNP conjugate vaccines were produced by linking Alexa Fluor 647 to hydrophobic blocks.
Immunizations and treatments
Vaccines were prepared in sterile PBS (Gibco) and administered subcutaneously to each footpad (50 μL per site) or intravenously via tail vein injection (200 μL). Animals were treated with 200 μg per mouse of anti-PD-L1 (10F.9G2, BioXcell) in 100 μL of PBS via intraperitoneal injection.
Cells
MC38 cell line was a kind gift from Lélia Delamarre (Genentech, USA). Working cell banks were generated immediately upon receipt and used for tumor experiments. Cells were tested regularly for mycoplasma contamination, and none tested positive throughout the studies.
Tumor implantation
MC38 tumor cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% heat-inactivated fetal calf serum (Atlanta Biologics), 100 U/mL penicillin, 100 μg/mL streptomycin (Gibco), 1x non-essential amino acids (GE Life Sciences) and 1 mM sodium pyruvate (GE Life Sciences). For each tumor implantation, a frozen cell aliquot was thawed and cultured in media at 37°C and 5% CO2, passaged once, and harvested using trypsin EDTA (Gibco). 105 cells in sterile PBS per mouse were implanted subcutaneously on the right flank. Tumors were measured twice a week using digital calipers. Tumor volume was estimated using the formula: [tumor volume = short × short × long / 2]. Animals were euthanized when tumors reached size criteria of 1000 mm3.
Blood and tissue processing
Heparin-treated blood was collected and lysed with ACK lysis buffer (Quality Biologicals). Lungs, liver, kidneys and tumors were harvested in digestion media containing RPMI-1640, 10% fetal calf serum, 50 U/mL DNAse I (Sigma) and 0.2 mg/mL collagenase D (Sigma). Tissues were mechanically disrupted using the respective programs on the gentleMACS dissociator (Miltenyi Biotech, Germany) and incubated at 37°C for 30–45 min in a shaking incubator. Spleens were mechanically disrupted and lysed with ACK lysis buffer. Lymph nodes were mechanically disrupted in BioMasher tubes (Nippi Inc, Japan). All single cell suspensions were filtered through a 40 μm nylon mesh filter plate (EMD Millipore) or 70 μm cell strainer and resuspended in PBS for flow cytometry staining. For ex vivo cultures, cells were resuspended in RPMI-1640 media (GE Life Sceiences) supplemented with 10% heat-inactivated fetal calf serum (Atlanta Biologics), L-Glutamine-Penicillin-Streptomycin (Sigma Aldrich), non-essential amino acids (GE Life Sciences), sodium pyruvate (Hyclone), HEPES buffer (Hyclone) and beta-mercaptoethanol (Sigma).
Peptide restimulation
Cells were cultured in vitro with 2 μg/mL Reps1 peptide antigen (Genscript) and 2μg/mL anti-CD28 (37.51, BD) for 6 h. 10 μg/mL of brefeldin A (BD) was added in the last 4 h.
Flow cytometry
For T cell tetramer analysis, cells were assessed for viability with LIVE/DEAD Fixable Blue (Invitrogen) in PBS containing 50 nM dasatinib (Stemcell Technologies) for 30 min at room temperature, washed and blocked with anti-CD16/CD32 (BD). Cells were then stained with fluorescently conjugated Reps1 (H-2Db AQLANDVVL) tetramer (John Finnigan, Mt Sinai USA) in cell staining buffer (PBS and 2% fetal calf serum) containing dasatinib to enhance staining. Cells were simultaneously stained with the following surface antibodies to: CD8 (53–6.7), PD-1 (29F.A12), CXCR3 (173), CD62L (Mel-14), Tim-3 (RMT3–23), CD44 (IM7), CD39 (Duha59), CD127 (A7R34), NKG2A (16A11), purchased from Biolegend and CD4 (RM4–4), KLRG1 (2F1) and CD103 (M290) purchased from BD. After an hour incubation at 4°C, cells were washed twice in cell staining buffer, fixed and permeabilized using the transcription factor staining buffer set (eBioscience). Cells were stained overnight at 4°C with the following intracellular antibodies to: CD3 (17A2) and Ki-67 (Ki-67) from Biolegend, TCF1 (C63D9) from Cell Signaling, T-bet (O4–46) and Granzyme B (GB11) from BD and Eomes (Dan11mag) from Invitrogen.
For intracellular cytokine analysis after peptide restimulation, cells were assessed for viability with LIVE/DEAD Fixable Blue (Invitrogen) for 10 min at room temperature. Similar to the antibodies used in the tetramer analysis, cells were blocked with anti-FcR antibodies and stained with cell surface antibodies for 20 min at room temperature in cell staining buffer. Cells were fixed and permeabilized using Fix/Perm solution (BD), and subsequently stained with intracellular antibodies to: CD3, IFN-γ (XMG1.2), IL-2 (JES6–5H4) and TNF (MP6-XT22) purchased from BD.
For mononuclear phagocyte uptake analysis, cells were assessed for viability with LIVE/DEAD Fixable Blue (Invitrogen) for 10 min at room temperature. Following FcR blocking, cells were stained with the following surface antibodies to: NK1.1 (PK136), CD19 (1D3), CD3 (145–2C11), Ly6G (1A8), CD45 (30-F11), SiglecH (440c), CD86 (GL1), CD11c (HL3), CD80 (16–10A1), B220 (RA3–6B2), CD64 (X54–5/7.1), CD11b (MI/70) and Ly6C (AL-21) purchased from BD, CCR7 (4B12), MHCII (I-A/I-E, M5/114.15.2), CD169 (3D6.112), XCR1 (ZET) purchased from Biolegend, and CD172a (P84) from Life Technologies.
Cells were acquired on an LSRFortessa X50 (BD) using FACSDiva software (BD) and analyzed on FlowJo software (BD).
OT-I in vivo proliferation assay
Spleens and lymph nodes were harvested from CD45.2 OT-I mice and labeled with Tag-it Violet Proliferation and Cell Tracking Dye (Biolegend) as per manufacturer’s protocol. Labeled cells (2×106) were transferred via retro-orbital intravenous injection to each congenic CD45.1 mouse at different time points after vaccination. Four days after transfer, spleens were harvested from recipient mice for flow cytometric analysis.
In vivo imaging
Whole body imaging of mice following immunization with Alexa Fluor 647-labeled vaccines was performed using IVIS Spectrum (Perkin Elmer).
Cell sorting for scRNA-seq
Spleens from mice vaccinated with SNP via SC or IV injections were collected 2 weeks post boost and processed into a single cell suspension by mechanical dissociation. Splenocytes were stained with Reps1 tetramer and hashtag antibodies (Biolegend Total-Seq-C antibodies 1–5)52. CD8+ T were isolated by fluorescence activated cell sorting (FACS) into 1.5 mL Eppendorf tubes containing staining buffer (2% FBS/PBS). Up to 4×104 cells were sorted per mouse. Cells from mice in both treatment groups with distinct hashtags were mixed to form two pools with an aim for equivalent numbers of cells from each mouse. Each pool of cells was loaded in duplicate in a Chromium single cell sorting system (10X Genomics). Expression and hashtag library construction was performed following Chromium Single Cell VDJ Library protocol with a loading target of 1 ×104 cells per lane. The resulting 4 libraries were pooled prior to sequencing on a Novaseq 6000 S1 chip.
Data processing for scRNA-seq
Raw sequencing files were aligned to the mouse mm10 genome using Cell Ranger software (10X Genomics). Hashtag data was added to each expression library using Seurat V3.1 and demultiplexed using the HTODemux function. After demultiplexing, singlet cells identified by hashtag from each of the libraries were pooled into a single Seurat object. Cells were further filtered by excluding those with greater than 5% mitochondrial genes in their library and fewer than 1000 genes18. 19,368 cells remained after filtering and were used for downstream analysis.
Downstream analysis of scRNA-seq data
Principal component analysis (PCA), uniform manifold approximation and projection (UMAP) for dimension reduction53, clustering the cells (by Leiden method), constructing trajectory and pseudotime analyses54 were performed on UMI counts using Monocle 3 (R package, version 0.2.1)55. Analysis of differentially expressed genes (DEG) and heatmaps were performed on the Log normalized data with Seurat (R package, version 3.1.4)56. The differential expression among the clusters was tested by the non-parametric Wilcoxon rank sum test, followed by Bonferroni correction method (using all features) to calculate adjusted P value. Top DEG were visualized using SeqGeq software (BD). SCENIC pipeline (R package, version 1.1.2.2) was used to construct and score gene regulatory network (regulons) as described previously.34 Each regulon is composed of a transcription factor and its putative target genes. The output of SCENIC is a matrix of regulons’ activity, in which rows correspond to regulons and columns correspond to cells.
TCR analysis of scRNA-seq data
T cell receptor (TCR) libraries were generated independently for all four 10X lanes according to the Chromium Single Cell VDJ library protocol. Raw sequencing files were aligned to the mouse mm10 genome and annotated with their TCR genes by using the cellranger vdj function from the Cell Ranger software suite (10X Genomics). We excluded cells that did not have full length or productive TCR alpha (TRA) or beta (TRB) gene pairs and those with multiple gene pairs (duplicates). Filtered TRA/TRB gene pairs were matched to the cells used for gene expression analysis by 10X barcode and any TRA/TRB pair that was not present in our gene expression data set was removed from further analysis. Clonotype was assigned based on the complementarity-determining region 3 (CDR3) amino acid sequence of the TRA/TRB gene pair. Only clonotypes expressed by more than 100 cells were visualized in the heat map.
Enzyme-linked immunosorbent assay
Serum from whole blood, spleens and popliteal lymph nodes were collected at specified time points after vaccination. Supernatants were collected from single cell suspensions of spleens and LNs that were cultured in vitro for 12 hours in complete RPMI at 37°C. Commercially available ELISA kits were used to measure IL-12p40 (Peprotech) and all subtypes of IFN-α (PBL Assay Science) as per manufacturer’s protocols.
Confocal microscopy
Spleens and popliteal LNs were harvested at indicated times and prepared as previously described57. Briefly, tissues were fixed in periodate-lysine-paraformaldehyde buffer and placed in 30% sucrose in PBS. Tissues were then embedded in OCT medium (Electron Microscopy Sciences), frozen in dry-ice cooled isopentane and sections were cut on a cryostat (Leica Microsystems). Sections were blocked in 5% sera and stained with the following antibodies to: CD11b (M1/70, eBioscience), CD11c (N418, eBioscience), B220 (RA3–6B2, eBioscience), CD64 (polyclonal, R&D Systems), MHC II (I-A/I-E, M5/114.15.2, eBioscience), followed by the relevant secondary antibodies conjugated to fluorophores. Images were acquired using Leica SP8 microscope and analyzed using Imaris software (Bitplane).
Statistical analysis
All results are presented as median with standard deviation. Statistics were assessed by Kruskal-Wallis with Dunn’s correction for multiple comparisons, two-way ANOVA with Bonferroni correction, Log-rank test and Mann Whitney test for immunogenicity (Graphpad Prism).
Data availability
The data that support the findings of this study are available from the corresponding author upon request. The scRNA-seq data have been uploaded to Gene Expression Omnibus (accession number: GSE158240).
Extended Data
Extended Data Fig. 1 |. Route and dose of SNP-7/8a immunization controls the magnitude and phenotype of antigen-specific CD8 T cells.
a, Whole blood was collected on day 21 to measure the frequency of tetramer+ CD8 T cells post boost. Bar graphs summarize the frequency of tetramer+ CD8 T cells from blood (n = 10). b, Bar graphs summarize the frequency of IFNγ+ CD8 T cells from blood (n = 10) on day 21. c, Bar graphs summarize the frequency of IFNγ+ CD4 T cells from blood (n = 10). d, Bar graphs show proportions of MPEC/SLEC subpopulations in the blood (n = 10). e, Frequency of MPECs is negatively correlated to frequency of tetramer+ CD8 T cells. f,g, Bar graphs show proportions of PD-1/Tim-3 subpopulations in the blood (n = 10) of tetramer+ cells (f) or IFNγ+ cells (g). Data are representative of two independent experiments. The bars represent the median. Statistics were assessed by Kruskal-Wallis with Dunn’s correction for multiple comparisons (a,b,d,f,g) and Spearman correlation (e).
Extended Data Fig. 2 |. Intravenous administration of SNP-7/8a generates TCF1+ CD8 T cells with anti-tumor capacity upon anti-PD-L1 treatment.
a, Tumor growth curves of mice unvaccinated (black) or vaccinated with SNP-SC (blue) or SNP-IV (red) with (dotted line) or without α-PD-L1 (solid line) (n = 10). b, Flow cytometric analysis of single cells from spleen (concatenated, n = 6) after SNP-SC (top panel) or SNP-IV (bottom panel). Cells were stained with Reps1 tetramer and other antibodies. Numbers indicate percentage of cell population within the gate. c, Mice were vaccinated with SNP-7/8a containing Reps1, E7, OVA or Trp1 antigens (n = 5). Spleens were collected 7 days post prime. d, Splenocytes were stained with tetramers specific for the respective antigens. Bar graph summarizes the frequencies of antigen-specific CD8 T cells following SNP-SC (blue) or SNP-IV (red). e, Bar graph summarizes the frequencies of TCF1 subpopulations in the spleen (n = 5) after SNP-SC or SNP-IV. f, Frequency of TCF1+PD-1+ cells is negatively correlated to frequency of tetramer+ CD8 T cells. g, Bar graph summarizes the frequencies of early effector cells (EEC, gray), memory precursor effector cells (MPEC, tan), double positive effector cells (DPEC, lilac) and short lived effector cells (SLEC, crimson) in the spleen (n = 5) after SNP-SC or SNP-IV. h, Frequency of MPEC is negatively correlated to frequency of tetramer+ CD8 T cells. Statistics were assessed by Mann Whitney test (d,e,g,) and Spearman correlation (f,h).
Extended Data Fig. 3 |. Single-cell analysis of neoAg+ CD8 T cells by RNA sequencing identifies stem-like gene signature in SNP-IV and effector gene signature in SNP-SC cells.
a, C57BL/6 mice (n = 5) were vaccinated subcutaneously or intravenously at 8 nmol on day 0 and day 14 with SNP-7/8a containing Reps1. Spleens were collected on day 28 and tetramer+ CD8 T cells were sorted by flow cytometry. Flow plots show gating strategy for cell sorting. b, Mice were individually labeled with distinct hashtag oligo-tagged antibodies and pooled for 10x and RNA sequencing. Individual UMAPs show gene expression of each mouse vaccinated SC (left panel) or IV (right panel). c, Bar graph summarizes the frequency of the twelve Monocle 3 clusters that are represented by each vaccination route. d, Density plots to identify stability states corresponding to higher density areas on UMAP, based on 2D kernel density estimation. e, Expression of top differentially expressed genes (DEG) of naïve cells are presented in meaning plots. f, Heatmap summarizes the number of cells that share a clonotype based on paired alpha and beta complementarity-determining region 3 (CDR3) sequences in each individual animal. g, Bar graph shows numbers of stem-like cells (clusters 2 and 4) and effector cells (clusters 1, 3, 5, 7 and 8) in each clonotype from SC or IV vaccinated mice. Only clonotypes expressed by more than 100 cells are represented in the graphs. h, Heatmap of DEG expressed in each cluster organized along the pseudotime trajectory.
Extended Data Fig. 4 |. Therapeutic vaccination with SNP-IV generates neoAg-specific CD8 T cells with superior anti-tumor capacity.
a, Tumor growth of mice treated with SNP-7/8a with (red) or without agonist (gray) (n = 10). b, Average tumor growth of SNP-IV (red), SNP-SC given twice (blue), once on day 7 (dotted blue) or twice at a lower dose (light blue) (n = 10). c, Total numbers of CD8 T cells, CD4 T cells and NK cells and d, frequency of tetramer+ CD8 T cells from blood in mice treated with isotype control antibody (red) or blocking antibodies against CD8β (black), CD4 (blue) or NK1.1 (purple) as assessed by flow cytometry (n = 10). Spleens and tumors were harvested on day 14 (n = 10) and day 21 (n = 3–5). e, Stem-like cells (TCF1+PD-1+; dark blue), f, effector cells (Granzyme B+TCF1−; orange) or g, exhausted cells (PD-1+Tim-3+) of tetramer+ cells were identified by flow cytometry. Bar graphs summarize the frequency of cells in the spleen on day 14 (filled bar) or day 21 (checked bar) (n = 3–10). h, Bar graphs summarize the frequency of Ki-67+ cells in different tissues on day 14 (red bar) or day 21 (checked bar) post SNP-IV (n = 3–10). Data are representative of four independent experiments. The bars represent the median. Statistics were assessed by two-way ANOVA with Bonferroni correction (a,b) and Mann Whitney test (e,f,g,h).
Extended Data Fig. 5 |. Transient vaccine distribution to spleen and activation of migratory cDC1 and moDC in after SNP-IV.
a, Whole body images of mice following SNP-SC or SNP-IV with labeled vaccines. b, Confocal images of LN or spleen sections of an unvaccinated mouse. c, Confocal image of popliteal LN section post SNP-SC. Detailed overlay of additional markers. White, vaccine; red, ERTR7 (stroma); orange, CD11b (monocytes, macrophages or cDC2); CD11c (moDC or cDC). Scale bar, 200 μm or 50 μm (inset). Arrows show co-localization of vaccine and CD11b+CD11c+ cells. d, Gating strategy to identify various populations from popliteal LN and spleen after SNP-SC and SNP-IV: MoDC (red), monocytes (pink), subcapsular sinus macrophages, SCS (gray), red pulp macrophages, RPM (dark gray), cDC1 (maroon), cDC2 (coral). Kinetics of MNPs that are vaccine+ in e, popliteal LNs or f, spleens after SNP-SC or SNP-IV respectively (n = 3). g, Histograms show MFI of CD80, CD86, CCR7 and labeled vaccine in migratory or resident cDC1 or moDC in popliteal LN of naïve (gray) or SNP-SC mice after vaccination (concatenated, n = 3). h,i, Flow cytometric analysis of single cells stained with XCR1 and CD86 after gating on cDC1s in spleens or popliteal LNs of mice post SNP-IV or SNP-SC respectively (n = 3). Data are representative of two independent experiments.
Extended Data Fig. 6 |. Prolonged antigen presentation by DC drives CD8 T cell responses after SNP-SC.
a, WT, Batf3−/− or CCR2−/− mice (n = 10) were vaccinated SC or IV at 8 nmol on day 0 and day 14 with SNP-7/8a (Reps1) (n = 3). b, Total number of cDC1s in spleen and popliteal LN, or monocytes in popliteal LN (right panel) of WT, Batf3−/− or CCR2−/− were measured. c, BM chimeras were performed by irradiating WT CD45.1 mice and transferring BM from CCR2DTR or WT CD45.2 mice. After 8 weeks of proper reconstitution, mice were treated with DT (n = 3). d, Total number of monocytes, cDC1 and cDC2 in spleen of CCR2DTR mice 24 h after DT treatment was measured. e, Kinetics of neoAg-specific CD8 T cell responses in blood of CCR2DTR vaccinated IV without DT treatment, or WT CD45.2 BM chimera vaccinated IV with or without prior DT treatment showed similar responses (n = 8–10). f, Sera were collected after SNP-SC (blue) or SNP-IV (red). IL-12 (left panel) or IFN-α (right panel) were measured by ELISA (n = 3). g, Total number of monocytes and cDC1 in popliteal LN of WT, IFNAR−/− or TLR−/− were measured by flow cytometry. h, Histograms of EOMES gated on tetramer+ cells post SNP-SC in WT or IL-12−/− mice (n = 4). Data are representative of two independent experiments. Statistics were assessed by Mann Whitney test (d) or Kruskal-Wallis with Dunn’s correction for multiple comparisons (g).
Supplementary Material
Acknowledgements
We thank members of the Seder lab for scientific discussions; S. Darko, A. Ransier and D. Douek for scRNA-seq advice; M. Dillon, G. Salbador, S. Rush, L. Gilliam, O. Hernandez, C. Chiedi and D. Scorpio of Translational Research Program (VRC) for their valuable support with animal studies. This work was supported by the Intramural Research Program of the US National Institutes of Health (R.A.S., H.D.H. and J.S.T.), EMBO YIP and Singapore Immunology Network (SIgN) core funding (F.G.).
Footnotes
Competing financial interests
G.M.L., V.L.C., A.S.I. and R.A.S. are listed as inventors on patents describing polymer-based vaccines. G.M.L., V.L.C. and A.S.I. are employees of Avidea Technologies, Inc., which is commercializing polymer-based drug delivery technologies for immunotherapeutic applications.
REFERENCES
- 1.Rosenberg SA & Restifo NP Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schumacher TN & Schreiber RD Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Versluis JM, Long GV & Blank CU Learning from clinical trials of neoadjuvant checkpoint blockade. Nat Med 26, 475–484 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 520, 692–696 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yadav M, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515, 572–576 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Hilf N, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Melief CJM Cancer: Precision T-cell therapy targets tumours. Nature 547, 165–167 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Lynn GM, et al. Peptide-TLR-7/8a conjugate vaccines chemically programmed for nanoparticle self-assembly enhance CD8 T-cell immunity to tumor antigens. Nat Biotechnol 38, 320–332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbarth SC Dendritic cell subsets in T cell programming: location dictates function. Nat Rev Immunol 19, 89–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLane LM, Abdel-Hakeem MS & Wherry EJ CD8 T Cell Exhaustion During Chronic Viral Infection and Cancer. Annu Rev Immunol 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He R, et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature 537, 412–428 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Snell LM, et al. CD8(+) T Cell Priming in Established Chronic Viral Infection Preferentially Directs Differentiation of Memory-like Cells for Sustained Immunity. Immunity 49, 678–694 e675 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z, et al. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity 51, 840–855 e845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hudson WH, et al. Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1(+) Stem-like CD8(+) T Cells during Chronic Infection. Immunity 51, 1043–1058 e1044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu T, et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danilo M, Chennupati V, Silva JG, Siegert S & Held W Suppression of Tcf1 by Inflammatory Cytokines Facilitates Effector CD8 T Cell Differentiation. Cell Rep 22, 2107–2117 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Miller BC, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamphorst AO, et al. Rescue of exhausted CD8 T cells by PD-1-targeted therapies is CD28-dependent. Science 355, 1423–1427 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vodnala SK, et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 176, 775–789 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J Exp Med 215, 2520–2535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sade-Feldman M, et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 175, 998–1013 e1020 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thommen DS, et al. A transcriptionally and functionally distinct PD-1(+) CD8(+) T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat Med 24, 994–1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen CS, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtulus S, et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity 50, 181–194 e186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211 e110 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Wherry EJ T cell exhaustion. Nat Immunol 12, 492–499 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zander R, et al. CD4(+) T Cell Help Is Required for the Formation of a Cytolytic CD8(+) T Cell Subset that Protects against Chronic Infection and Cancer. Immunity 51, 1028–1042 e1024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dominguez CX, et al. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J Exp Med 212, 2041–2056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cedeno-Laurent F & Dimitroff CJ Galectin-1 research in T cell immunity: past, present and future. Clin Immunol 142, 107–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baeyens A, Fang V, Chen C & Schwab SR Exit Strategies: S1P Signaling and T Cell Migration. Trends Immunol 36, 778–787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schiavoni G, Mattei F & Gabriele L Type I Interferons as Stimulators of DC-Mediated Cross-Priming: Impact on Anti-Tumor Response. Front Immunol 4, 483 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trinchieri G Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol 3, 133–146 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Kranz LM, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534, 396–401 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Lewis SM, Williams A & Eisenbarth SC Structure and function of the immune system in the spleen. Sci Immunol 4(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorner BG, et al. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31, 823–833 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alfei F, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Khan O, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beltra JC, et al. Developmental Relationships of Four Exhausted CD8(+) T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity 52, 825–841 e828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hailemichael Y, et al. Persistent antigen at vaccination sites induces tumor-specific CD8(+) T cell sequestration, dysfunction and deletion. Nat Med 19, 465–472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irvine DJ, Swartz MA & Szeto GL Engineering synthetic vaccines using cues from natural immunity. Nat Mater 12, 978–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merad M, Sathe P, Helft J, Miller J & Mortha A The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol 31, 563–604 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hohl TM, et al. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe 6, 470–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoeckius M, et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol 19, 224 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol (2018). [DOI] [PubMed] [Google Scholar]
- 54.Saelens W, Cannoodt R, Todorov H & Saeys Y A comparison of single-cell trajectory inference methods. Nat Biotechnol 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynoso GV, et al. Lymph node conduits transport virions for rapid T cell activation. Nat Immunol 20, 602–612 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request. The scRNA-seq data have been uploaded to Gene Expression Omnibus (accession number: GSE158240).