Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by the selective and progressive loss of motor neurons. Although many drugs have entered clinical trials, few have shown effectiveness in the treatment of ALS. Other studies have shown that the stimulation of α7 nicotinic acetylcholine receptor (nAChR) can have neuroprotective effects in some models of neurodegenerative disease, as well as prevent glutamate-induced motor neuronal death. However, the effect of α7 nAChR agonists on ALS-associated mutant copper–zinc superoxide dismutase 1 (SOD1) aggregates in motor neurons remains unclear. In the present study, we examined whether α7 nAChR activation had a neuroprotective effect against SOD1G85R-induced toxicity in a cellular model for ALS. We found that α7 nAChR activation by PNU282987, a selective agonist of α7 nAChR, exhibited significant neuroprotective effects against SOD1G85R-induced toxicity via the reduction of intracellular protein aggregates. This reduction also correlated with the activation of autophagy through the AMP-activated protein kinase (AMPK)–mammalian target of rapamycin (mTOR) signaling pathway. Furthermore, the activation of α7 nAChRs was found to increase the biogenesis of lysosomes by inducing translocation of the transcription factor EB (TFEB) into the nucleus. These results support the therapeutic potential of α7 nAChR activation in diseases that are characterized by SOD1G85R aggregates, such as ALS.
Subject terms: Cell death in the nervous system, Molecular neuroscience
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurological disorder that is characterized by muscle weakness and atrophy, paralysis, and eventual death by respiratory failure. Symptoms result from the selective degeneration of upper and lower motor neurons. While 90–95% of ALS cases arise sporadically, 5–10% are familial in nature. In previous studies searching for causal genes associated with familial ALS, copper–zinc superoxide dismutase 1 (SOD1), TAR DNA binding protein of 43 kDa (TDP43), fused in sarcoma (FUS), and optineurin (OPTN) have been identified as playing roles in pathological cascades and phase transitions in sporadic ALS1–7. Among familial ALS patients, mutation in SOD1 is a major autosomal dominant inherited allele associated with disease8.
Several clinical studies suggest a role for glutamate-induced excitotoxicity in familial ALS pathology, although the potential pathogenesis in sporadic ALS is still unclear9–11. Previous experiments have shown that rat spinal cord cultures exposed to long-term low-dose glutamate exhibit selective motor neuronal death12–14. For this reason, riluzole, a glutamate release inhibitor, has been developed as a therapeutic agent for ALS patients, and has been shown to prolong patient life by several months15. Although many drugs have entered into clinical trials, few have demonstrated any effectiveness in the treatment of ALS. Therefore, new prospective drugs are strongly desired in the clinical field.
Nine α (α2–α10) and three β (β2–β4) nicotinic acetylcholine receptor (nAChR) subunits are expressed in the vertebrate brain16. These subunits coassemble to form a family of functionally diverse nAChRs. Among nAChR subunits, the most abundant subtypes in the mammalian nervous system are homomeric α7 nAChRs and heteromeric β2 nAChRs, including α4β2 nAChRs16–19. These receptors are also widely expressed in the dorsal and ventral horns in the spinal cord14,20–22. Loss of nAChRs is associated with a number of disease states, including Alzheimer’s disease, Parkinson’s disease (PD), Lewy body disease, schizophrenia, autism, and attention deficit/hyperactivity disorder (ADHD)23. In addition, accumulating evidence suggests that α7 nAChRs are important targets in the development of therapeutics for PD. We have previously demonstrated that α7 nAChR activation protects against dopaminergic neuronal death in both acute and chronic animal models of PD induced by 6-hydroxydopamine (6-OHDA) and rotenone, respectively24,25.
Nagano et al. have also reported an early decrease in cholinergic input in motor neurons in the spinal cords of patients with ALS26. nAChRs have been implicated in neuroprotection mechanisms against acute cell stress induced by excitotoxicity, namely overactivation of glutamate receptors27. The stimulation of α7 nAChRs by nicotine actually prevented glutamate-induced motor neuronal death14. Therefore, α7 nAChRs represent an important therapeutic target for ALS as well as for PD. Recently, a candidate ALS drug ropinirole (a dopamine D2 and D3 receptor agonist used as an antiparkinsonian drug) has been discovered in drug screenings using ALS patient-derived iPS cells, and is now being evaluated in clinical trials for effectiveness28. Although the mechanistic details are still unknown, there may be commonalities in the pathological mechanisms of PD and ALS. However, the effect of α7 nAChR agonists against mutant SOD1 aggregates in neurons remains unclear. A pathological hallmark of ALS is the presence of cytoplasmic inclusions or protein aggregates in affected motor neurons, suggesting that impairment of protein degradation may play a role in the disease pathology29. SOD1 aggregates are present in both sporadic and familial ALS30,31. In addition, we found that most wild-type SOD1 proteins assume misfolded conformations in cerebrospinal fluid of ALS patients regardless of SOD1 mutation status32. Thus, removal of SOD1 aggregates may be a potential therapeutic approach for ALS treatment. Currently, more than 180 types of SOD1 pathogenic mutations have been identified in ALS patients33. Among those, the pathogenic SOD1G85R mutation has been most frequently studied34–37.
As a strategy to remove mutant SOD1 aggregates, activation of autophagy, a prominent protein degradation pathway, has been reported to be effective in previous studies38. Autophagy is caused by the binding of lysosomes to autophagosomes which are formed by surrounding components to be degraded. Because previous studies have reported that activation of α7 nAChR induces autophagy, we are focusing on autophagy to elucidate the mechanism of the protective effect by activation of α7 nAChR in the present study39,40.
In the present study, we examined whether α7 nAChR activation exhibited neuroprotective effects against SOD1G85R-induced neurotoxicity in a cellular model of ALS.
Results
The α7 nAChR agonist reduces intracellular SOD1G85R aggregates and prevents neurotoxicity of SOD1G85R
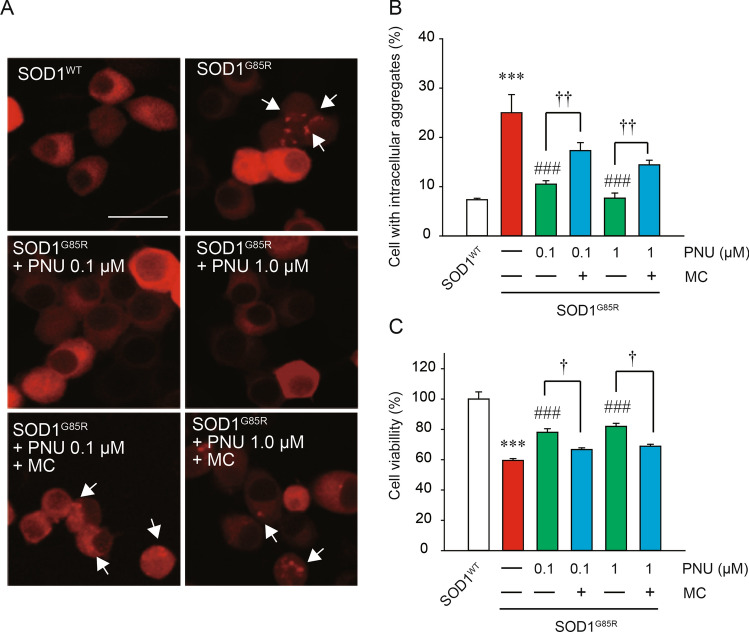
Previously, intracellular aggregate formation was confirmed in mCherry-fused SOD1G85R (hereafter SOD1G85R)-transfected N2a cells, which contributed to cell death via the formation of triton X-100-insoluble aggregates36–38,41. To investigate the effect of α7 nAChR activation on SOD1G85R aggregate formation, we evaluated the number of intracellular aggregates with an Imaging Cytometer after treatment with PNU282987, a selective nAChR agonist39,42 (Fig. 1). SOD1G85R formed intracellular aggregates in approximately 25% of transfected N2a cells. PNU282987 treatment significantly reduced the percentage of cells with intracellular SOD1G85R aggregates (Fig. 1A,B). This reduction in aggregate formation was markedly blocked by methyllycaconitine (MC), an α7 nAChR selective antagonist43,44 (Fig. 1A,B). In order to examine the effect of PNU282987 on SOD1G85R-mediated neurotoxicity, we performed a MTT assay in differentiated N2a cells (Fig. 1C). Although transfection of SOD1WT did not affect cell survival, transfection of SOD1G85R severely induced cell death. PNU282987 prevented SOD1G85R-induced cell death; in addition, MC significantly inhibited the PNU282987-induced neuroprotection (Fig. 1C). Besides, there was no effect of the treatment of MC alone on survival and aggregation (Supplemental Fig. S1). These results suggest that α7 nAChR activation exerts significant neuroprotective effects against SOD1G85R-induced toxicity via the reduction of intracellular protein aggregates. In addition, the expression level of the gene for α7 nAChR was not different in cells transfected with SOD1WT and G85R, which was confirmed by qRT-PCR (Supplemental Fig. S2).
Figure 1.
PNU282987 prevented SOD1G85R-induced neurotoxicity. At 24 h after transfection of each vector into N2a cells, the cells were treated with 0.1 or 1 µM PNU298987 for 24 h in the presence or absence of 20 µM methyllycaconitine (MC) pretreated before 30 min. (A) Representative fluorescent microscopy images. Scale bar: 10 µm. (B) Quantified data of intracellular SOD1 aggregates. (C) The cell viability was measured by MTT assay. Data is expressed as mean ± SEM from three independent experiments. Significance: ***p < 0.001 vs. SOD1WT; ###p < 0.001 vs. SOD1G85R; †p < 0.05, ††p < 0.01 vs SOD1G85R with PNU282987 by One-way ANOVA.
The α7 nAChR agonist exerts neuroprotective effects against SOD1G85R aggregation via activation of autophagy
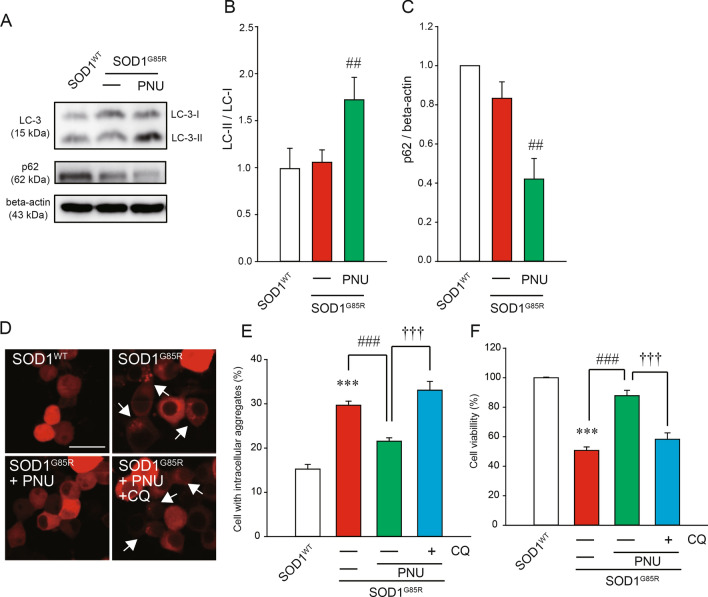
To analyze the mechanism of the reduction of intracellular aggregates upon α7 nAChR activation, we focused on autophagy, a prominent protein degradation pathway. The increase of LC3-II is a marker for the activation of autophagy because LC3-II converted from LC3-I is localized in the membrane of autophagosomes which is essential for autophagy and is proportional to the number of autophagosomes45. In addition, the decrease of p62 is also a marker of autophagy activation because p62 promotes degradation by autophagy and is itself degraded by autophagy46. Western blot analysis showed that PNU282987 treatment increased the formation of LC3-II in N2a cells (Fig. 2A,B). In addition, similar results were confirmed for calculations by taking the ratio of beta-actin (Supplemental Fig. S3). PNU282987 treatment also significantly decreased p62 protein levels (Fig. 2A,C). In addition, chloroquine (CQ), an inhibitor of autophagy, prevented the reduction of cytoplasmic aggregation of SOD1G85R induced by PNU282987 (Fig. 2D,E). To further investigate whether the neuroprotective effects of PNU282987 were associated with autophagy, we performed a MTT assay in the presence of CQ. The protective effect of PNU282987 was significantly inhibited by CQ treatment (Fig. 2F). Besides, there was no effect of the treatment of CQ alone on survival and aggregation (Supplemental Fig. S1). These data suggest that PNU282987 treatment reduces the quantity of subcellular aggregates via the upregulation of autophagy, which prevented SOD1G85R-associated neurotoxicity.
Figure 2.
PNU282987 exerted the activation of autophagy. (A–C) After 24 h of transfection, N2a cells were transfected with SOD1G85R, and then incubated with 1 µM PNU282987. The lysates were analyzed by immunoblotting with anti-LC-3 (A,B) and p62 (A,C) and anti-β-actin antibodies. Levels normalized to the expression of β-actin and quantified based on the band density of SOD1WT. (D–F) N2a cells expressing mCherry-SOD1G85R were treated with 1 µM PNU282987 (PNU) in the presence or absence of 20 nM chloroquine (CQ) pretreated before 30 min. (D) Representative fluorescent microscopy images. Scale bar: 10 µm. (E) Quantified data of intracellular SOD1 aggregates. (F) The cell viability was measured by MTT assay. Data is expressed as mean ± SEM from three independent experiments. Significance: ###p < 0.001 vs. SOD1G85R by One-way ANOVA.
α7 nAChR activation promotes autophagy via Ca2+ influx and the AMP-activated protein kinase (AMPK)-mammalian target of rapamycin (mTOR) signaling pathway
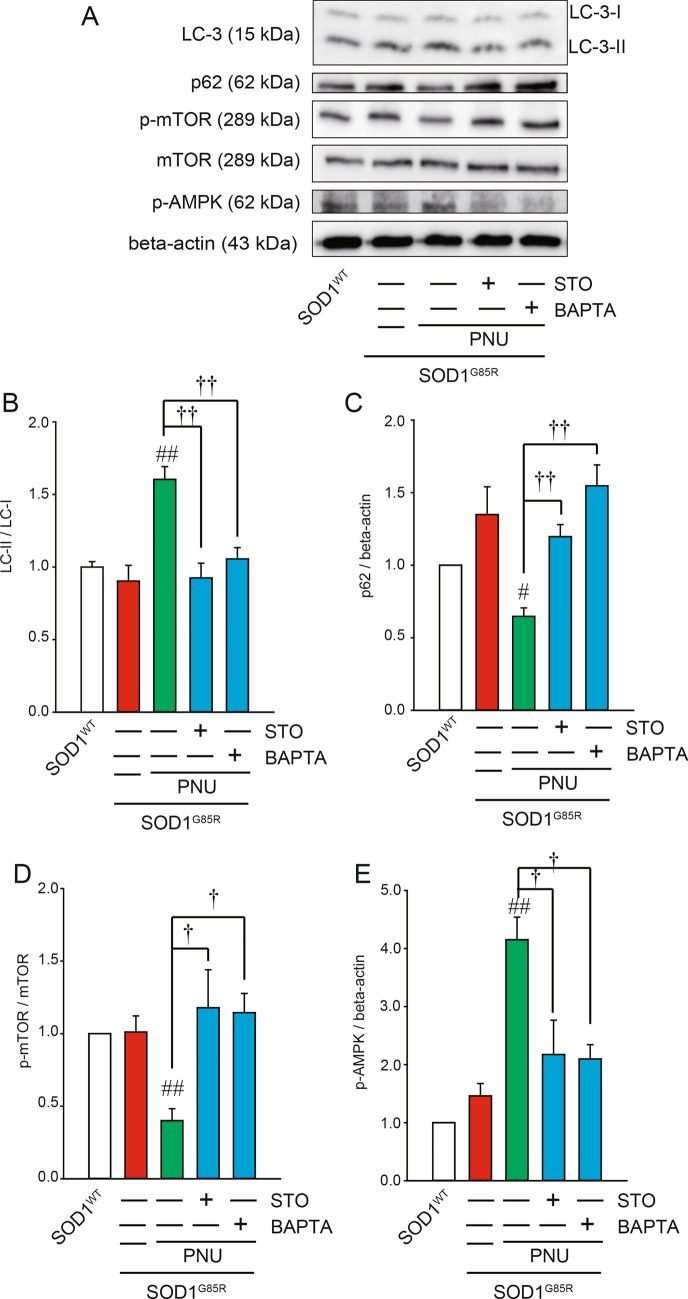
α7 nAChR activation increases intracellular Ca2+ influx through the α7 receptor channel. It is well-established that Ca2+ influx is a regulator of autophagy47. To elucidate the pathways involved in elevated Ca2+ influx after α7 nAChR activation, we used the cell-permeable cytosolic Ca2+ chelator 1,2-bis (2-aminophenoxy) ethane-N,N,N0,N0-tetraacetic acid tetrakis (acetoxymethyl ester) (BAPTA-AM)48,49. Pre-treatment with BAPTA-AM significantly inhibited the formation of LC3-II and the reduction in p62 protein induced by PNU282987 (Fig. 3A–C). CaMKKβ has been proposed as a potential target of cytosolic Ca2+ signaling50, although direct evidence remains elusive. Similar to the effects of BAPTA-AM, the formation of LC3-II and the reduction in p62 induced by PNU282987 were significantly inhibited by STO609, a CaMKKβ-specific inhibitor51,52 (Fig. 3A–C).
Figure 3.
PNU282987-induced Ca2+ influx contributes to autophagy. Immunoblotting analysis of autophagy regulators with a Ca2+ chelator and CaMKK-specific inhibitor. N2a cells expressing SOD1G85R were treated with 1 µM PNU282987 (PNU) in the presence or absence of 1 µM STO609 (STO) or 1 µM BAPTA-AM (BAPTA) pretreated before 30 min. (A) The lysates were analyzed by immunoblotting with antibodies for LC-3, p62, phosphorylated mTOR (p-mTOR), mTOR, phosphorylated AMPK (p-AMPK), β-actin. (B–E) Relative levels normalized by the expression of LC3-I or mTOR or β-actin were quantified, based on the density of SOD1WT. Data is expressed as mean ± SEM from three independent experiments. Significance: #p < 0.05, ##p < 0.01 vs. SOD1G85R; †p < 0.05, ††p < 0.01 vs SOD1G85R with PNU282987 by One-way ANOVA.
The AMP-activated protein kinase (AMPK)–mammalian target of rapamycin (mTOR) signaling pathway is downstream of Ca2+ signaling and plays an important role in the regulation of autophagy in response to different stress conditions53. To further identify the signal transduction pathway mediated by PNU282987, we investigated the AMPK–mTOR pathway. PNU282987 significantly increased AMPK phosphorylation and significantly inhibited mTOR phosphorylation (Fig. 3A,D,E). In addition, BAPTA-AM and STO609 pre-treatment significantly decreased PNU282987-dependent AMPK phosphorylation and restored mTOR phosphorylation (Fig. 3A,D,E), suggesting activation of the AMPK pathway and inhibition of the mTOR pathway occurs via Ca2+ influx following α7 nAChR activation. These results suggest that α7 nAChR activation potentially induces autophagy via Ca2+ influx and signaling through the AMPK and mTOR pathways.
α7 nAChR agonist promotes lysosomal activation via nuclear translocation of transcription factor EB (TFEB)
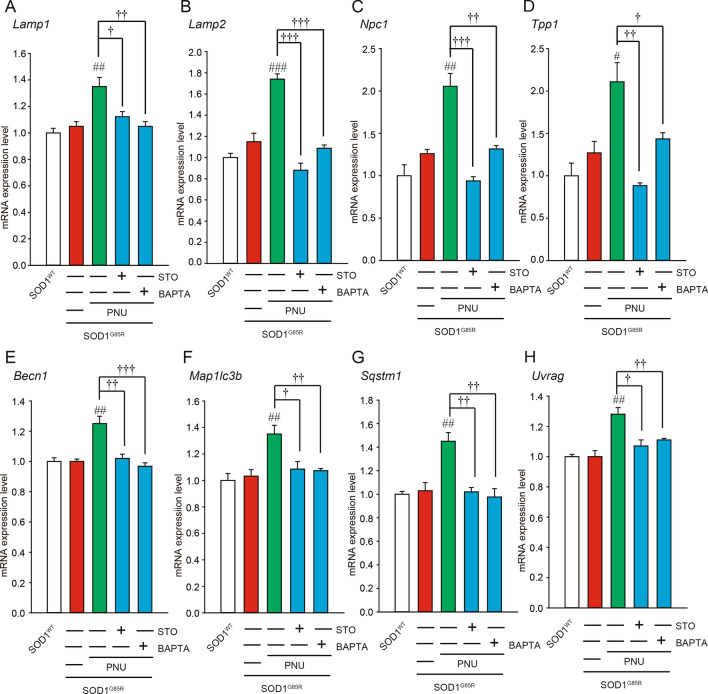
AMPK signaling activates transcription factor EB (TFEB), which is a potential key transcription in the induction of autophagy upon its dephosphorylation and nuclear translocation54. TFEB can enter the nucleus and bind to the E-box of the CLEAR element, which regulates the transcription of genes associated with the biogenesis of lysosomes and autophagosomes55. We first analyzed mRNA levels of lysosomes and autophagy-related genes whose transcription could be regulated by TFEB using qRT-PCR. The mRNA levels of lysosomal-related genes (such as Lamp1, Lamp2, Npc1, and Tpp1) and autophagy-related genes (such as Becn1, Map1lc3b, Sqstm1, and Uvrag) were significantly up-regulated in response to PNU282987 treatment (Fig. 4). Upregulation of these mRNAs was markedly blocked by pre-treatment with BAPTA-AM and STO609 (Fig. 4).
Figure 4.
Effects of PNU282987 on transcription factor EB (TFEB) related mRNA expression. (A–H) N2a cells expressing mCherry-SOD1G85R were treated with 1 µM PNU282987 (PNU) in the presence or absence of 1 µM STO609 (STO) and 1 µM BAPTA-AM (BAPTA) pretreated before 30 min. At 24 h after treatment with PNU, mRNA expressions of Lamp1 (A), Lamp2 (B), Npc1 (C), Tpp1 (D), Becn1 (E), Map1lc3b (F), Sqstm1 (G), Uvrag (H) were analyzed using the SYBR Green-based RT-qPCR assay. The expression levels of mRNA were normalized to the expression levels of β-actin mRNA. Significance: #p < 0.05, ##p < 0.01, ###p < 0.01 vs. SOD1G85R; †p < 0.05, ††p < 0.01, †††p < 0.01 vs SOD1G85R with PNU282987 by One-way ANOVA.
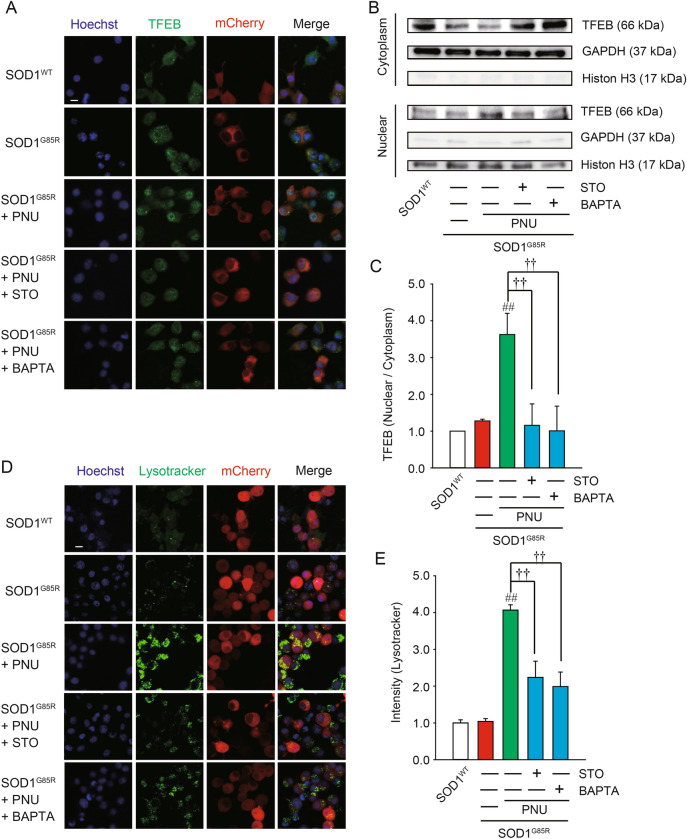
Next, we examined the distribution of TFEB in the SOD1G85R-transfected N2a cells after PNU282987 treatment using immunofluorescence (Fig. 5A). TFEB levels in the nucleus were increased following PNU282987 treatment. Meanwhile, TFEB levels in the cytoplasm were decreased by PNU282987 (Fig. 5A). Western blot also demonstrated that PNU282987 treatment decreased and increased TFEB levels in cytoplasmic and nuclear fractions, respectively (Fig. 5B,C). In addition, nuclear translocation of TFEB induced by PNU282987 was significantly inhibited following pre-treatment with BAPTA-AM and STO609 (Fig. 5B,C). We next determined whether PNU282987 enhances lysosomal activation using immunofluorescence (Fig. 5D,E). PNU282987 promoted lysosomal activation as quantified by fluorescence intensity in LysoTracker-stained cells. The effect of PNU282987 on lysosomal activation was inhibited by pre-treatment with BAPTA-AM and STO609 (Fig. 5D,E). Together, these data suggest that α7 nAChRs activation promotes lysosomal activation via the nuclear transposition of TFEB.
Figure 5.
PNU282987 enhances TFEB activity. N2a cells expressing SOD1G85R were treated with 1 µM PNU282987 (PNU) in the presence or absence of 1 µM STO609 (STO) or 1 µM BAPTA-AM (BAPTA) pretreated before 30 min. (A) Representative fluorescent microscopy images of TFEB. Scale bar: 10 µm. (B) After the fractionation using NE-PER Nuclear and Cytoplasmic Extraction Reagents, the lysates were analyzed by immunoblotting with antibodies for TFEB, GAPDH, and Histone H3. (C) The ratio of nuclear to cytosolic TFEB was calculated and analyzed. (D) Representative fluorescent microscopy images of lysotracker. Scale bar: 10 µm. (E) Quantified data of lysotracker intensity. Significance: ##p < 0.01 vs. SOD1G85R; ††p < 0.01 vs. SOD1G85R with PNU282987 by One-way ANOVA.
Discussion
The goal of the present study was to determine whether activation of α7 nAChR had a neuroprotective effect against SOD1G85R aggregate-induced toxicity in a cellular model of ALS. We demonstrated that PNU282987, an α7 nAChR agonist, induces significant neuroprotective effects via reduction of SOD1G85R intracellular aggregates. This reduction strongly correlated with the activation of autophagy via the AMPK–mTOR signaling pathway. Furthermore, PNU282987 increased lysosomal activation by promoting TFEB translocation into the nucleus. These findings identify α7 nAChR as a novel neuroprotective target against the SOD1G85R aggregate-mediated neurotoxicity. Besides, because of the transient expression of the SOD1 system in this study, there may be a slight change in the gene transfer efficiency. The difference may affect the change in aggregation rate. However, we can always see an increase in aggregation rate in transfected cells with SOD1G85R compared to SOD1WT, therefore, we think that it does not affect the experimental results. In the future, we would like to examine them in a more pathological model using iPS cells rather than this model of transient expression.
Autophagy dysfunction has been implicated in various neurodegenerative diseases, including ALS56,57. The notion that autophagy dysfunction contributes to ALS pathogenesis is strongly supported by the identification of numerous genes associated with familial ALS also involved in regulation of autophagy, including SQSTM158, OPTN4, TBK159, and VCP60. In addition, previous reports also showed that TFEB downregulation induced autophagy defect in spinal motor neurons of patients with SOD1G93A mutations although not confirmed in the mutant SOD1G85A model in this study61. This difference in results may be due to differences in cell type. Therefore, further research is needed on other models. Besides, the presence of intracellular, insoluble inclusions composed of misfolded proteins is a hallmark of ALS pathology29. Therefore, removal of SOD1 aggregates may represent one potential therapeutic approach for ALS treatment.
There are two major pathways for cellular protein degradation: the ubiquitin proteasome system (UPS), and autophagy. Autophagy has been shown to degrade both soluble and aggregated protein substrates that are too large to pass through the pore of the proteasome, such as the toxic SOD1 protein aggregates, while UPS primarily degrades soluble SOD1, suggesting that autophagy regulation is critical for improving ALS pathology57,62.
Treatment of mutant SOD1 transgenic mice with trehalose resulted in increased life span, improved neuronal survival, reduced astrogliosis, and delayed disease onset via activation of autophagy63. Similarly, carbamazepine treatment activated autophagy via the AMPK-ULK1 signaling pathway and promoted the clearance of mutant SOD1 aggregates. Carbamazepine treatment also delayed disease onset and extended life span of SOD1G93A mice64. In our previous studies, autophagy induction has demonstrated beneficial effects in cells harboring pathogenic SOD1 mutations36,37,65. In addition to pharmacological studies, genetic ablation of XBP-1 (X-box-binding protein) in motor neurons of SOD1G85R mice enhanced the clearance of mutant SOD1 aggregates and increased survival via activation of autophagy66. Moreover, bosutinib, which boosts autophagy, can improve the survival of iPS cells-derived motor neurons from patients with familial ALS caused by mutations in SOD167.
Conversely, abnormalities (activations) in autophagy have been observed in numerous neurodegenerative diseases, including ALS68. Pharmacological and genetic modulation of autophagy may result in diverse and even detrimental outcomes to the survival of ALS models; interventions targeting genes including mSOD1, FUS and TDP-4357,62 have shown that it may be necessary to jointly consider the specific effects of each individual mutation, pathology, and possibly other context-dependent influences. These results suggest the need for developing autophagy inducers with higher specificity and lower cytotoxicity based on ALS pathology62. In the present study, PNU282987 exerted neuroprotective effects against SOD1G85R-induced toxicity via autophagy activation. Although it is necessary to explore this finding further with iPS cells and animal models, among candidates of autophagy inducers, α7 nAChR may be a promising candidate.
Our study indicates that PNU282987 decreased mTOR phosphorylation and increased AMPK phosphorylation, and subsequently induced autophagy. The AMPK–mTOR signaling pathway is a downstream target of Ca2+ signaling and plays an important role in the regulation of autophagy in response to different stresses53. In support of this, we showed that AMPK and mTOR phosphorylation were significantly affected by pre-treatment with BAPTA-AM and STO609, indicating activation of the AMPK pathway and inhibition of the mTOR pathway via Ca2+ influx following α7 nAChR activation. In addition, AMPK phosphorylation activates TFEB, which is a potential key transcription for autophagy induction upon its dephosphorylation and nuclear translocation54. It has been reported that under stress conditions or upon loss of function, TDP43 can regulate the nuclear translocation of TFEB in order to promote the transcription of autophagic genes. This indicates that TFEB may play a role in potential strategies for ALS treatment. In the present study, PNU282987 significantly increased the mRNA levels of Lamp1, Lamp2, Npc1, Tpp1, Becn1, Map1lc3b, Sqstm1, and Uvrag the transcription of which could be regulated by TFEB. In addition, PNU282987 significantly increased the TFEB translocation into the nucleus and promoted lysosomal activation. Previously, trehalose, an enhancer of mTOR-independent autophagy, was shown to delay ALS onset and reduce motor neuron loss in SOD1G93A mice69. In contrast, mTOR-dependent activation of autophagy resulted in loss of motor neurons and reduced survival in the same ALS mouse model, which may be due to other physiological functions of mTOR inhibition70. The neuroprotective effects of α7 nAChR activation may be due to pleiotropic effects including other signaling pathways such as FGFR1 and NF-κB, it will be necessary to conduct further studies71–73. In addition, further research is needed on other possibilities for other receptors, such as α4β2 nAChR, the other major nAChR.
ALS is a multifactorial disease encompassing a network of cellular pathways74. Drugs with pleiotropic effects may be practically more effective than drugs with a single effect for patients with ALS. As α7 nAChR activation has various neuroprotective effects including autophagy activation, α7 nAChR activation may possess novel therapeutic potential for ALS. Recently, the usefulness of patient-derived iPS cells as a model for ALS has been demonstrated28,67. Therefore, further research is required at a level closer to the pathological conditions, such as using iPS cells.
Material and methods
Plasmid, cell culture, and transfection
Expression plasmids (pmCherry-N1, Clontech Laboratories Inc., CA, USA) harboring human SOD1 (Wild-type (SOD1WT) or mutant (SOD1G85R)) were prepared as previously reported36,37. Briefly, N2a cells (Mouse Albino neuroblastoma, ECACC, UK) were maintained in Dulbecco’s modified Eagle medium (DMEM, Wako Pure Chemical Industries, Ltd.) containing 10% (v/v) fetal bovine serum (FBS; Thermo Fisher Inc.) under a humidified atmosphere of 5% CO2 at 37 °C. The cells were passaged by trypsinization every 3–4 days. The transient plasmid expression in N2a cells was accomplished with Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Scientific Inc.).
Thiazolyl blue tetrazolium bromide (MTT) assay
N2a cells were seeded at 2.0 × 105 cells/ml in 96-well plates in DMEM containing 10% FBS. Following 24 h of plasmid transfection, the cells were differentiated for 48 h in low glucose (1.0 g/l) DMEM supplemented with 2% FBS and 2 mM N,N-dibutyladenosine 3′,5′-phosphoric acid (dbcAMP; Nacalai Tesque Inc.) with 0.1 or 1 µM PNU282987 (Wako Pure Chemical Industries Ltd.) in the presence or absence of 20 µM MC (Cayman Chemical Ltd.) or 20 nM CQ (Tokyo Chemical Industry Ltd.) pretreated before 30 min. The number of live cells was estimated by Cell Counting Kit-8, following the manufacturer’s instructions (Wako Pure Chemical Industries Ltd.). Briefly, the reagent was added into the wells and the plate was incubated at 37 °C for 4 h. Cell viability was calculated through the detection of the optical density of formazan at 450 nm using GloMax (Promega). A 600 nm wavelength was used as reference.
Aggregation rate analysis
We performed aggregation assay based on previous studies36,37. Briefly, after 24 h of mCherry-fused SOD1WT or SOD1G85R vector transfection, the cells were treated with PNU282987 for 24 h in the presence or absence of 20 µM MC or 20 nM CQ pretreated before 30 min. Subsequently, the cells were washed twice with PBS for 5 min and fixed with 4% paraformaldehyde for 15 min. Fluorescent microscopy images were acquired with a confocal fluorescence microscope (LSM700, Carl Zeiss). For counting the number of aggregates, we used IN Cell Investigator 2200 (GE Healthcare). In each experiment, at least 3000 cells were counted.
Immunoblotting
At 24 h after transfection of each vector into N2a cells, the cells were treated with 1 µM PNU298987 for 24 h in the presence or absence 1 µM STO609 or 1 µM BAPTA-AM pretreated before 30 min. After treatment, the cells were lysed with TNE lysis buffer (50 mM Tris–HCl (pH. 7.4), 150 mM NaCl, 1 mM ethylenediamineteraacetic acid, protease inhibitor cocktail) containing 1% Triton X and then were centrifuged at 15,000×g at 4 °C for 5 min. The supernatant protein sample was collected. Protein concentrations were quantified using a BCA protein assay kit (Thermo Fisher Scientific Inc.) with bovine serum albumin (BSA) as a standard. Lysates were mixed with sample buffer containing 10% 2-mercaptoethanol, and subjected to 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was performed under constant voltage of 200 V at room temperature for 40 min. The separated proteins in polyacrylamide gel were transferred to a PVDF membrane in transfer buffer (0.3% Tris, 1.44% glycine, 20% methanol) under constant voltage of 100 V at 4 °C for 90 min. The membranes were incubated with 5% BSA (Wako) for 60 min, and then with following primary antibodies for overnight: mouse monoclonal antibody against β-actin (1: 2000, Santa Cruz Biotechnology) and rabbit polyclonal antibodies against LC-3 (1:1000, Cell Signaling), p-AMPK (1:1000, Cell Signaling Technology), p-mTOR (1:1000, Cell Signaling Technology), mTOR(1:1000, Cell Signaling Technology), p62 (1:1000, Cell Signaling Technology), TFEB (1:1000, Proteintech). After the primary antibody reaction, the membrane was incubated in the secondary antibody (goat anti-rabbit antibody conjugated with HRP (1:2500, Santa Cruz Biotechnology) or goat anti-mouse HRP antibody conjugated with HRP (1:2500, Santa Cruz Biotechnology)). The membrane was incubated in ECL prime (GE Healthcare, Buckinghamshire, UK) to generate the chemiluminescence from HRP antibodies. The chemiluminescence was detected by Fusion System (Vilber-Lourmat). The band density was measured using ImageJ.
RNA preparation and qRT-PCR
Reverse transcription was performed using the ReverTra Ace qPCR RT Master Mix, in accordance with the manufacturer’s instructions (TOYOBO). qRT-PCR was performed using SYBR Green on a StepOne Real-Time PCR System, in accordance with the manufacturer’s instructions (Life Technologies). The sequences of gene-specific primer sets are shown in Table 1. The expression levels of mRNA were normalized to the expression levels of β-actin mRNA.
Table 1.
Primer pairs used for qRT-PCR.
| Forward | Reverse | |
|---|---|---|
| Lamp1 | 5′tcaaggtggacagtgacaggt3′ | 5′tgactcctcttcctgccaatga3′ |
| Lamp2 | 5′tctccggttaaaggcgcaaag3′ | 5′tcatctcccattctgcataaaggc3′ |
| Npc1 | 5′cgcaatcctgtgtttggtatgg3′ | 5′aagtcatagccgtcctttggg3′ |
| Tpp1 | 5′cccatgttataaggtccccacatcc3′ | 5′ccaagtgcaggctaacagttcc3′ |
| Becn1 | 5′gcggagagattggaccagga3′ | 5′tctccacactcttgagttcgtca3′ |
| Map1lc3b | 5′gtgcctgaccacgtgaacat3′ | 5′tctcactctcgtacacttcgga3′ |
| Sqstm1 | 5′cagatgccagaatcggaaggg3′ | 5′ggactcaatcagccggggat3′ |
| Uvrag | 5′ctgtgtcctgctttgtggtga3′ | 5′tttcattctggttgcgggca3′ |
| Chrna7 | 5′gcccttgatagcacagtacttcg3′ | 5′gatcctggtccacttaggcattt3′ |
| β-actin | 5′cgttgacatccgtaaagacc3′ | 5′gctaggagccagagcagtaa3′ |
Lamp1 lysosome-associated membrane protein 1, Lamp2 lysosome-associated membrane protein 2, Npc1 NPC intracellular cholesterol transporter 1, Tpp1 tripeptidyl peptidase 1, Becn1 Beclin1, Map1lc3b microtubule associated protein 1 light chain 3 beta, Sqstm1 Sequestosome 1, Uvrag UV radiation resistance associated, Chrna7 cholinergic receptor nicotinic alpha 7.
Immunostaining
At 24 h after transfection of each vector into N2a cells, the cells were treated with 1 µM PNU298987 in the presence or absence 1 µM STO609 or 1 µM BAPTA-AM pretreated before 30 min. At 24 h after the treatment, cells were fixed by 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100 diluted in PBS. To block the reaction, the cells were treated with 2% goat serum for 60 min. The cells were incubated with rabbit polyclonal antibody against TFEB (1:200, Proteintech) at 4 °C for overnight. Subsequently, the cells were incubated with secondary antibody (goat anti-rabbit antibody Alexa 488) at room temperature for 1 h. In addition, nuclear staining was performed with Hoechst 33342 (Molecular Probes). Fluorescent microscopy images were acquired with a confocal fluorescence microscope (LSM700, Carl Zeiss).
Lysosomal staining
N2a cells were seeded at 1.0 × 105 cells/ml in CELLview cell culture dishes (Greiner Bio-one). At 24 h after transfection to N2a cells each vector, the cells were treated with 1 µM PNU298987 in the presence or absence 1 µM STO609 or 1 µM BAPTA-AM pretreated before 30 min. At 24 h after the treatment, lysosomes were stained with LysoTracker Green DND-26 (Thermo Fisher Scientific) and nucelei were stained with Hoechst 33342 (Thermo Fisher Scientific), in accordance with the manufacturer’s instructions (Thermo Fisher Scientific). Fluorescent microscopy images were acquired with a confocal fluorescence microscope (LSM700, Carl Zeiss). Image analysis was calculated the fluorescence intensities in each image using ImageJ (thresholding and then brightness measurement) and took the ratio by the number of cells stained with Hoechst.
Cell fractions preparation
At 24 h after transfection of each vector into N2a cells, the cells were treated with 1 µM PNU298987 in the presence or absence 1 µM STO609 or 1 µM BAPTA-AM pretreated before 30 min. At 24 h after the treatment, 1.0 × 106 cells were harvested with trypsin–EDTA and nuclear extraction was performed using NE-PER Nuclear and Cytoplasmic Extraction Reagents, in accordance with the manufacturer’s instructions (Thermo Fisher Scientific). To assess the purity of the fractionation, the cytoplasmic and nuclear fractions were confirmed by immunoblotting using anti-GAPDH (1:1000, MBL Ltd.) as a cytoplasmic marker and anti-Histone H3 (1:1000, Cell Signaling Technology Ltd.) as a nuclear marker.
Statistical analysis
Data are given as the mean ± standard error of the mean (SEM). Significance was determined using the analysis of variance. Further statistical analysis for post hoc comparisons was performed using the Bonferroni/Dunn test (StatView, Abacus). p-values of less than 0.05 were considered to be statistically significant.
Supplementary Information
Acknowledgements
This work was supported by grants from the Takeda Science Foundation, the Japan Society for the Promotion of Science (JSPS) KAKENHI (Grant No. JP17K08311), Grant-in-Aid for Scientific Research on Innovative Areas JSPS KAKENHI (Grant No. JP19H05767A02), Nagai Memorial Research Scholarship from the Pharmaceutical Society of Japan, the API Research Foundation and Smoking Research Foundation.
Author contributions
T.I., M.I. and I.H. designed and organized the project. T.I., T.U., Y.A. and H.K. performed the experiments. T.I., M.I. and I.H. wrote the paper.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Taisei Ito and Masatoshi Inden.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-79189-y.
References
- 1.Rosen DR, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 4.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 5.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr. Opin. Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor JP, Brown RH, Jr, Cleveland DW. Decoding ALS: from genes to mechanism. Nature. 2016;539:197–206. doi: 10.1038/nature20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimori K, et al. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018;24:1579–1589. doi: 10.1038/s41591-018-0140-5. [DOI] [PubMed] [Google Scholar]
- 8.Julien JP. Amyotrophic lateral sclerosis. Unfolding the toxicity of the misfolded. Cell. 2001;104:581–591. doi: 10.1016/S0092-8674(01)00244-6. [DOI] [PubMed] [Google Scholar]
- 9.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole study group. N. Engl. J. Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 10.Couratier P, Hugon J, Sindou P, Vallat JM, Dumas M. Cell culture evidence for neuronal degeneration in amyotrophic lateral sclerosis being linked to glutamate AMPA/kainate receptors. Lancet. 1993;341:265–268. doi: 10.1016/0140-6736(93)92615-Z. [DOI] [PubMed] [Google Scholar]
- 11.Rothstein JD, Van Kammen M, Levey AI, Martin LJ, Kuncl RW. Selective loss of glial glutamate transporter GLT-1 in amyotrophic lateral sclerosis. Ann. Neurol. 1995;38:73–84. doi: 10.1002/ana.410380114. [DOI] [PubMed] [Google Scholar]
- 12.Urushitani M, et al. Mechanism of selective motor neuronal death after exposure of spinal cord to glutamate: involvement of glutamate-induced nitric oxide in motor neuron toxicity and nonmotor neuron protection. Ann. Neurol. 1998;44:796–807. doi: 10.1002/ana.410440514. [DOI] [PubMed] [Google Scholar]
- 13.Kanki R, et al. Effects of mitochondrial dysfunction on glutamate receptor-mediated neurotoxicity in cultured rat spinal motor neurons. Brain Res. 2004;1015:73–81. doi: 10.1016/j.brainres.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 14.Nakamizo T, et al. Stimulation of nicotinic acetylcholine receptors protects motor neurons. Biochem. Biophys. Res. Commun. 2005;330:1285–1289. doi: 10.1016/j.bbrc.2005.03.115. [DOI] [PubMed] [Google Scholar]
- 15.Lucette L, Bensimon G, Leigh PN, Guillet P, Meininger V. Dose-ranging study of riluzole in amyotrophic lateral sclerosis. Amyotrophic lateral sclerosis/riluzole study group II. Lancet. 1996;347:1425–1431. doi: 10.1016/S0140-6736(96)91680-3. [DOI] [PubMed] [Google Scholar]
- 16.Faure P, Tolu S, Valverde S, Naudé J. Role of nicotinic acetylcholine receptors in regulating dopamine neuron activity. Neuroscience. 2014;282C:86–100. doi: 10.1016/j.neuroscience.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 17.Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends Pharmacol. Sci. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Taly A, Corringer PJ, Guedin D, Lestage P, Changeux JP. Nicotinic receptors: allosteric transitions and therapeutic targets in the nervous system. Nat. Rev. Drug Discov. 2009;8:733–750. doi: 10.1038/nrd2927. [DOI] [PubMed] [Google Scholar]
- 19.Rode F, et al. Positive allosteric modulation of α4β2 nAChR agonist induced behaviour. Brain Res. 2012;1458:67–75. doi: 10.1016/j.brainres.2012.03.064. [DOI] [PubMed] [Google Scholar]
- 20.Kiyosawa A, Katsurabayashi S, Akaike N, Pang ZP, Akaike N. Nicotine facilitates glycine release in the rat spinal dorsal horn. J. Physiol. 2001;536:101–110. doi: 10.1111/j.1469-7793.2001.t01-1-00101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth AL, Berg DK. Large clusters of alpha7-containing nicotinic acetylcholine receptors on chick spinal cord neurons. J. Comp. Neurol. 2003;465:195–204. doi: 10.1002/cne.10856. [DOI] [PubMed] [Google Scholar]
- 22.Takeda D, Nakatsuka T, Gu JG, Yoshida M. The activation of nicotinic acetylcholine receptors enhances the inhibitory synaptic transmission in the deep dorsal horn neurons of the adult rat spinal cord. Mol. Pain. 2007;3:26. doi: 10.1186/1744-8069-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimohama S. Nicotinic receptor-mediated neuroprotection in neurodegenerative disease models. Biol. Pharm. Bull. 2009;32:332–336. doi: 10.1248/bpb.32.332. [DOI] [PubMed] [Google Scholar]
- 24.Yanagida T, et al. Synergistic effect of galantamine on nicotine-induced neuroprotection in hemiparkinsonian rat model. Neurosci. Res. 2008;62:254–261. doi: 10.1016/j.neures.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi H, et al. Nicotinic receptor stimulation protects nigral dopaminergic neurons in rotenone-induced Parkinson's disease models. J. Neurosci. Res. 2009;87:576–585. doi: 10.1002/jnr.21869. [DOI] [PubMed] [Google Scholar]
- 26.Nagao M, Misawa H, Kato S, Hirai S. Loss of cholinergic synapses on the spinal motor neurons of amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 1998;57:329–333. doi: 10.1097/00005072-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 27.Kaur J, Rauti R, Nistri A. Nicotine-mediated neuroprotection of rat spinal networks against excitotoxicity. Eur. J. Neurosci. 2018;47:1353–1374. doi: 10.1111/ejn.13950. [DOI] [PubMed] [Google Scholar]
- 28.Okano H, Yasuda D, Fujimori K, Morimoto S, Takahashi S. Ropinirole, a new ALS drug candidate developed using iPSCs. Trends Pharmacol. Sci. 2020;41:99–109. doi: 10.1016/j.tips.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosco DA, et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forsberg K, et al. Novel antibodies reveal inclusions containing non-native SOD1 in sporadic ALS patients. PLoS ONE. 2010;5:e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokuda E, Takei YI, Ohara S, Fujiwara N, Hozumi I, Furukawa Y. Wild-type Cu/Zn-superoxide dismutase is misfolded in cerebrospinal fluid of sporadic amyotrophic lateral sclerosis. Mol. Neurodegener. 2019;14:42. doi: 10.1186/s13024-019-0341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huai J, Zhang Z. Structural properties and interaction partners of familial ALS-associated SOD1 mutants. Front. Neurol. 2019;10:527. doi: 10.3389/fneur.2019.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abel O, Powell JF, Andersen PM, Al-Chalabi A. ALSoD: a user-friendly online bioinformatics tool for amyotrophic lateral sclerosis genetics. Hum. Mutat. 2012;33:1345–1351. doi: 10.1002/humu.22157. [DOI] [PubMed] [Google Scholar]
- 35.Peters OM, Ghasemi M, Jr, Brown RH. Emerging mechanisms of molecular pathology in ALS. J. Clin. Investig. 2015;125:2548. doi: 10.1172/JCI82693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ueda T, et al. The effects of Brazilian green propolis that contains flavonols against mutant copper–zinc superoxide dismutase-mediated toxicity. Sci. Rep. 2017;7:2882. doi: 10.1038/s41598-017-03115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueda T, et al. Effects of gem-dihydroperoxides against mutant copper-zinc superoxide dismutase-mediated neurotoxicity. Mol. Cell Neurosci. 2018;92:177–184. doi: 10.1016/j.mcn.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe S, Hayakawa T, Wakasugi K, Yamanaka K. Cystatin C protects neuronal cells against mutant copper–zinc superoxide dismutase-mediated toxicity. Cell Death Dis. 2014;5:e1497. doi: 10.1038/cddis.2014.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao BZ, et al. Autophagy plays an important role in anti-inflammatory mechanisms stimulated by alpha7 nicotinic acetylcholine receptor. Front. Immunol. 2017;8:553. doi: 10.3389/fimmu.2017.00553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung SY, Huang WP, Liou HC, Fu WM. Autophagy protects neuron from Aβ-induced cytotoxicity. Landes Biosci. 2009;5:502–510. doi: 10.4161/auto.5.4.8096. [DOI] [PubMed] [Google Scholar]
- 41.Oh YK, Shin KS, Yuan J, Kang SJ. Superoxide dismutase 1 mutants related to amyotrophic lateral sclerosis induce endoplasmic stress in neuro2a cells. J. Neurochem. 2008;104:993–1005. doi: 10.1111/j.1471-4159.2007.05053.x. [DOI] [PubMed] [Google Scholar]
- 42.Ke P, Shao BZ, Xu ZO, Chen XW, Wei W, Liu C. Activating α7 nicotinic acetylcholine receptor inhibits NLRP3 inflammasome through regulation of β-arrestin-1. CNS Neurosci. Ther. 2017;23:875–884. doi: 10.1111/cns.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tie K, et al. Histone hypo-acetylation of Sox9 mediates nicotine-induced weak cartilage repair by suppressing BMSC chondrogenic differentiation. Stem Cell Res. Ther. 2018;9:98. doi: 10.1186/s13287-018-0853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Wu M, Lu F, Luo N, He ZP, Yang H. Involvement of α7 nAChR signaling cascade in epigallocatechin gallate suppression of β-amyloid-induced apoptotic cortical neuronal insults. Mol. Neurobiol. 2014;49:66–77. doi: 10.1007/s12035-013-8491-x. [DOI] [PubMed] [Google Scholar]
- 45.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 47.Jing Z, et al. SKF-96365 activates cytoprotective autophagy to delay apoptosis in colorectal cancer cells through inhibition of the calcium/CaMKIIγ/AKT-mediated pathway. Cancer Lett. 2016;372:226–238. doi: 10.1016/j.canlet.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang N, et al. Calcium ion regulation by BAPTA-AM and ruthenium red improved the fertilisation capacity and developmental ability of vitrified bovine oocytes. Sci. Rep. 2017;7:10652. doi: 10.1038/s41598-017-10907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins CS, Mathie A. Effects on K+ currents in rat cerebellar granule neurones of a membrane-permeable analogue of the calcium chelator BAPTA. Br. J. Pharmacol. 1996;118:1772–1778. doi: 10.1111/j.1476-5381.1996.tb15603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scotto Rosato A, et al. TRPML1 links lysosomal calcium to autophagosome biogenesis through the activation of the CaMKKβ/VPS34 pathway. Nat. Commun. 2019;10:5630. doi: 10.1038/s41467-019-13572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu B, et al. Isovitexin-mediated regulation of microglial polarization in lipopolysaccharide-induced neuroinflammation via activation of the CaMKKβ/AMPK-PGC-1α signaling axis. Front. Immunol. 2019;10:2650. doi: 10.3389/fimmu.2019.02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poon AD, et al. Ca2+/calmodulin-dependent protein kinase II and dimethyl sulfoxide affect the sealing frequencies of transected hippocampal neurons. J. Neurosci. Res. 2018;96:1208–1222. doi: 10.1002/jnr.24232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Y, et al. Activation of autophagy through calcium-dependent AMPK/mTOR and PKCθ pathway causes activation of rat hepatic stellate cells under hypoxic stress. FEBS Lett. 2016;590:672–682. doi: 10.1002/1873-3468.12090. [DOI] [PubMed] [Google Scholar]
- 54.Collodet C, et al. AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J. 2019;33:12374–12391. doi: 10.1096/fj.201900841R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu H, et al. Trehalose promotes the survival of random-pattern skin flaps by TFEB mediated autophagy enhancement. Cell Death Dis. 2019;10:483. doi: 10.1038/s41419-019-1704-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beltran S, et al. Network approach identifies Pacer as an autophagy protein involved in ALS pathogenesis. Mol. Neurodegener. 2019;14:14. doi: 10.1186/s13024-019-0313-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Djajadikerta A, Keshri S, Pavel M, Prestil R, Ryan L, Rubinsztein DC. Autophagy induction as a therapeutic strategy for neurodegenerative diseases. J. Mol. Biol. 2020;432:2799–2821. doi: 10.1016/j.jmb.2019.12.035. [DOI] [PubMed] [Google Scholar]
- 58.Fecto F, et al. sSQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 59.Cirulli ET, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson JO, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramesh N, Pandey UB. Autophagy dysregulation in ALS: when protein aggregates get out of hand. Front. Mol. Neurosci. 2017;10:263. doi: 10.3389/fnmol.2017.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, et al. Verapamil ameliorates motor neuron degeneration and improves lifespan in the SOD1G93A mouse model of ALS by enhancing autophagic flux. Aging Dis. 2019;10:1159–1173. doi: 10.14336/AD.2019.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castillo K, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013;9:1308–1320. doi: 10.4161/auto.25188. [DOI] [PubMed] [Google Scholar]
- 64.Zhang JJ, Zhou QM, Chen S, Le WD. Repurposing carbamazepine for the treatment of amyotrophic lateral sclerosis in SOD1-G93A mouse model. CNS Neurosci. Ther. 2018;24:1163–1174. doi: 10.1111/cns.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ueda T, Ito T, Kurita H, Inden M, Hozumi I. p-coumaric acid has protective effects against mutant copper-zinc superoxide dismutase 1 via the activation of autophagy in N2a cells. Int. J. Mol. Sci. 2019;20:2942. doi: 10.3390/ijms20122942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hetz C, et al. XBP-1 deficiency in the nervous system protects against amyotrophic lateral sclerosis by increasing autophagy. Genes Dev. 2009;23:2294–2306. doi: 10.1101/gad.1830709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imamura K, et al. The Src/c-Abl pathway is a potential therapeutic target in amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9:3962. doi: 10.1126/scitranslmed.aaf3962. [DOI] [PubMed] [Google Scholar]
- 68.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy. 2014;10:588–602. doi: 10.4161/auto.27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Switon K, Kotulska K, Janusz-Kaminska A, Zmorzynska J, Jaworski J. Molecular neurobiology of mTOR. Neuroscience. 2017;341:112–153. doi: 10.1016/j.neuroscience.2016.11.017. [DOI] [PubMed] [Google Scholar]
- 71.Morioka N, Hisaoka-Nakashima K, Nakata Y, Akaike A, Shimohama S, Misu Y. Regulation by nicotinic acetylcholine receptors of microglial glutamate transporters: role of microglia in neuroprotection. In: Akaike A, Shimohama S, Misu Y, editors. Nicotinic Acetylcholine Receptor Signaling in Neuroprotection. Berlin: Springer; 2018. pp. 73–88. [PubMed] [Google Scholar]
- 72.Endo F, Yamanaka K. Neuroinflammation in amyotrophic lateral sclerosis. Clin. Neurol. 2014;54:1128–1131. doi: 10.5692/clinicalneurol.54.1128. [DOI] [PubMed] [Google Scholar]
- 73.Patel H, McIntire J, Ryan S, Dunah A, Loring R. Anti-inflammatory effects of astroglial α7 nicotinic acetylcholine receptors are mediated by inhibition of the NF-κB pathway and activation of the Nrf2 pathway. J. Neuroinflamm. 2017;14:192. doi: 10.1186/s12974-017-0967-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dutta K, Patel P, Julien JP. Protective effects of Withania somnifera extract in SOD1G93A mouse model of amyotrophic lateral sclerosis. Exp. Neurol. 2018;309:193–204. doi: 10.1016/j.expneurol.2018.08.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.