Abstract
MAPK pathways regulate different responses yet can share common components. Although core regulators of MAPK pathways are well known, new pathway regulators continue to be identified. Overexpression screens can uncover new roles for genes in biological processes and are well suited to identify essential genes that cannot be evaluated by gene deletion analysis. In this study, a genome-wide screen was performed to identify genes that, when overexpressed, induce a reporter (FUS1-HIS3) that responds to ERK-type pathways (Mating and filamentous growth or fMAPK) but not p38-type pathways (HOG) in yeast. Approximately 4500 plasmids overexpressing individual yeast genes were introduced into strains containing the reporter by high-throughput transformation. Candidate genes were identified by measuring growth as a readout of reporter activity. Fourteen genes were identified and validated by re-testing: two were metabolic controls (HIS3, ATR1), five had established roles in regulating ERK-type pathways (STE4, STE7, BMH1, BMH2, MIG2) and seven represent potentially new regulators of MAPK signaling (RRN6, CIN5, MRS6, KAR2, TFA1, RSC3, RGT2). MRS6 encodes a Rab escort protein and effector of the TOR pathway that plays a role in nutrient signaling. MRS6 overexpression stimulated invasive growth and phosphorylation of the ERK-type fMAPK, Kss1. Overexpression of MRS6 reduced the osmotolerance of cells and phosphorylation of the p38/HOG MAPK, Hog1. Mrs6 interacted with the PAK kinase Ste20 and MAPKK Ste7 by two-hybrid analysis. Based on these results, Mrs6 may selectively propagate an ERK-dependent signal. Identifying new regulators of MAPK pathways may provide new insights into signal integration among core cellular processes and the execution of pathway-specific responses.
Subject terms: Functional genomics, Fungal genomics
Introduction
During cell differentiation, cells specialize into specific types by the action of signal transduction pathways. Mitogen-activated protein kinase (MAPK) pathways control numerous responses, including cell differentiation, proliferation, cell migration, and apoptosis1,2. MAPK pathways control diverse responses by regulating the expression of a large number of target genes. There are four types of MAPK pathways: RAF-MEK-ERK1/2, JNK1/2/3, p38α/β/γ/δ, and ERK51,3. Remarkably, these pathways can share common components, which leads to proper cross-talk in normal settings and unregulated cross-talk in the disease state. Mis-regulation of MAPK signaling leads to inappropriate responses, such as cancers and problems with immune system function4–6. Due to the crucial roles of MAPK pathways in regulating fundamental cellular processes, they remain the focus of investigation by many labs and are a focus for therapeutic targeting7–12.
MAPK pathways are evolutionarily conserved signaling modules in eukaryotes, and fundamental insights into MAPK pathway regulation have come from studies in many systems. The budding yeast Saccharomyces cerevisiae is a unicellular organism that has been extensively used as a model for studying signaling pathways7,13–19. Like in other eukaryotes, yeast utilizes ERK-type and p38-type MAPK pathways20,21. One ERK-type pathway mediates the response to nutrient-limiting conditions that permit filamentous (pseudohyphal/invasive) growth, a fungal-type foraging response resulting in the formation of chains of elongated interconnected cells22,23. This pathway functions through a set of kinases that function in a tandem series: p21 activated [PAK] Ste20 (MAPKKKK), Ste11 (MAPKKK), Ste7 (MAPKK), and Kss1 (MAPK)24,25. A second ERK-type pathway in yeast controls the mating of haploid cells through an almost identical set of kinases: Ste20 (PAK), Ste11 (MAPKKK), Ste7 (MAPKK), and Fus3 and Kss1 (MAPK). Two MAPKs, Fus3, and Kss1, function in mating and filamentous growth pathways, respectively. It has been shown that the deletion of KSS1 causes a reduction in agar penetration26, a phenotype called invasive growth that is related to filamentous growth22, while it has little effect on mating efficiency27. In contrast, deletion of FUS3 allows cells to penetrate the agar more vigorously26 while they cause a moderate decrease in mating efficiency27. This and other data support the idea that one MAPK promotes invasive/filamentous growth (Kss1), and while another mainly functions to regulating mating (Fus3). Surprisingly, the elimination of both MAPKs results in more agar penetration, which identified an inhibitory role for the unphosphorylated form of Kss1 regulating filamentous growth26–29.
A p38-type pathway, the high osmolarity glycerol response (HOG) pathway, allows the response to hyperosmotic conditions through Pbs2 (MAPKK) and Hog1 (MAPK)30–32. One branch of this pathway shares components with the mating and fMAPK pathways33. Specifically, Ste20 and Ste11 function to regulate Pbs2 and Hog1. Therefore, MAPK pathways in yeast can share some common components despite the fact that the pathways induce different transcriptional and morphogenetic responses.
In pathogens, the filamentation response is critical for host-cell attachment, invasion into tissues, and virulence34. In S. cerevisiae haploid cells, filamentous growth is triggered by growth in a non-preferred carbon source. The response is regulated by multiple signal transduction pathways35,36, including the RAS-cAMP-PKA pathway23,37–39 and the filamentous growth MAPK pathway (fMAPK)25. These pathways induce target genes that reorganize cell polarity, the cell cycle, and cell adhesion to bring about a new cell type40–43. The signaling mucin Msb2 operates at the head of the fMAPK pathway, and through the adaptor protein, Sho1, regulates MAPK activity by interaction with the Ras-homology (Rho)-type GTPase Cdc42. Sho1 interacts with Msb2 and Ste11 and functions in both the fMAPK and HOG pathways33,44,45. Cdc42 is an essential gene that is required for the maintenance of cell polarity and signaling. Human homolog Cdc42 is 81% identical to the yeast protein46–51. Cdc42 regulates the fMAPK pathway by interacting with Ste2025,26,40.
Several mechanisms that promote insulation have been described. One mechanism involves scaffolds, such as Ste552–54 and Pbs245. Ste5 activates Fus3 by forming a multi-kinase complex that joins the Ste11, Ste7, and Fus3 kinases52,55,56. Pbs2 regulates the HOG pathway by being activated through two different branches, SLN1-SSK1 and Sho145. Another mechanism that is employed to maintain specificity involves cross-pathway inhibition. In this case, a transcription factor for the filamentation pathway, Tec1, is phosphorylated by Fus3, which leads to its turnover by a ubiquitin ligase complex57,58. An intriguing challenge, therefore, is to understand how pathways that share elements establish and maintain their identity59,60.
The core regulators of the fMAPK pathway (MAPKKK- > MAPKK- > MAPK) are well known, and several proteins have been identified that regulate the fMAPK pathway at or above the level of Cdc42. However, some proteins that regulate the fMAPK pathway may remain unidentified. For example, genome-wide screens have recently identified new proteins that regulate the fMAPK pathway61. Loss-of-function studies also have identified a broad set of genes that contribute to filamentous growth. Nevertheless, no single genetic approach can be expected to yield comprehensive results, and in this light, gene overexpression screens have proven to be an effective complement to gene deletion analysis62,63. Analysis of filamentation phenotypes from gene overexpression collections continues to provide a more comprehensive understanding of pseudohyphal growth regulation. We, therefore, performed an overexpression screen to identify new regulators of ERK-type pathways in yeast. Among the genes identified was a new pathway regulator, Mrs6, that when overexpressed stimulates the fMAPK pathway but not the HOG pathway. Since many of the new regulators identified have homologs in other eukaryotes, including humans, investigation of fMAPK pathway regulators provides a foundation for understanding MAPK pathway regulation in general. This may contribute to the development of new therapeutic targets in related species of fungal pathogens and can be linked to other signaling systems in higher organisms, with implications in the understanding and treatment of human disease.
Results
A genome-wide screen in yeast identifies new regulators of ERK-type MAPK pathways
Three MAPK pathways in yeast require a subset of common components, including the Rho-type GTPase Cdc42, PAK Ste20, and MAPKKK Ste11, yet the pathways induce different responses [Fig. 1,64,65]. A genetic screen was performed to identify regulators of ERK-type MAPK pathways in yeast. An ordered collection of overexpression plasmids66 was examined for the induction of a MAPK pathway-dependent growth reporter [Fig. 2A, FUS1-HIS3,67,68]. The reporter provides a readout of two ERK-type MAPK pathways, mating and fMAPK (Fig. 1). However, cells lacking an intact mating pathway were evaluated (ste4Δ), which biases reporter activity towards the fMAPK pathway69.
Figure 1.
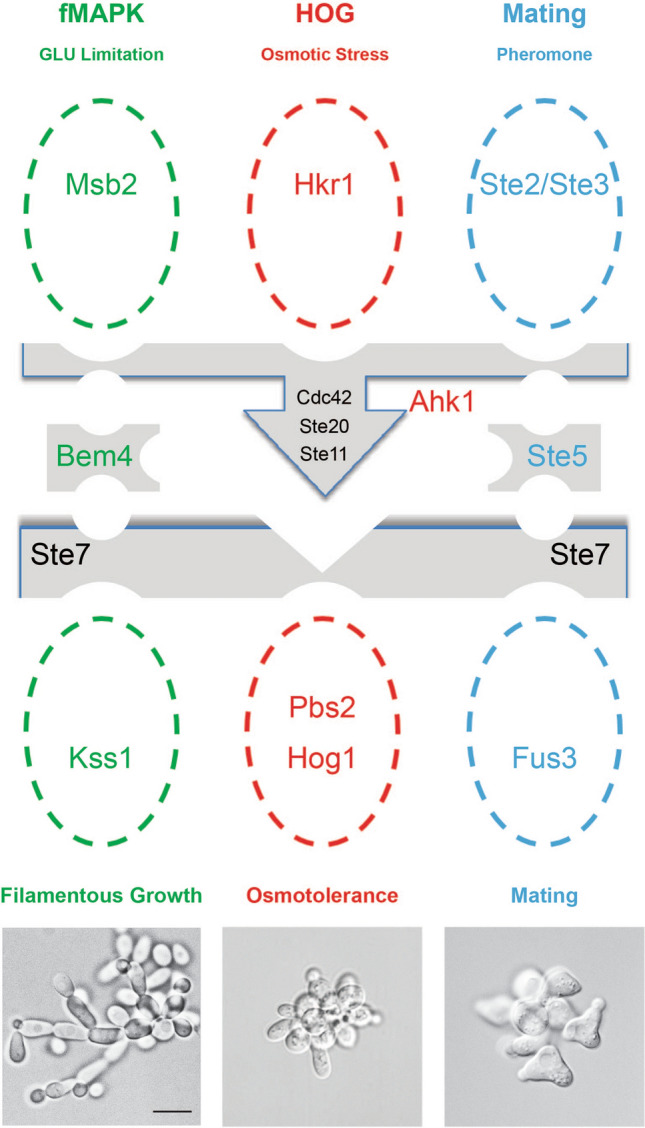
Three MAPK pathways in yeast share a subset of common components. Common components are shown in black, and pathway specific proteins are shown in color for the fMAPK (red), HOG (green), and mating (blue) pathways. Each pathway has a scaffold-type adaptor, Bem470, Pbs245 and Ahk171, and Ste552, and a specific MAP kinase. Cells undergo filamentous growth under nutrient-limiting conditions (left), cells do not change their morphology when exposed to YEP-GAL + 1.0 M KCl salt (middle), and YEP-GAL + 1 mg/ml α-factor stimulates an elongated cell shape or shmoo (right). Scale bar, 10 μm.
Figure 2.
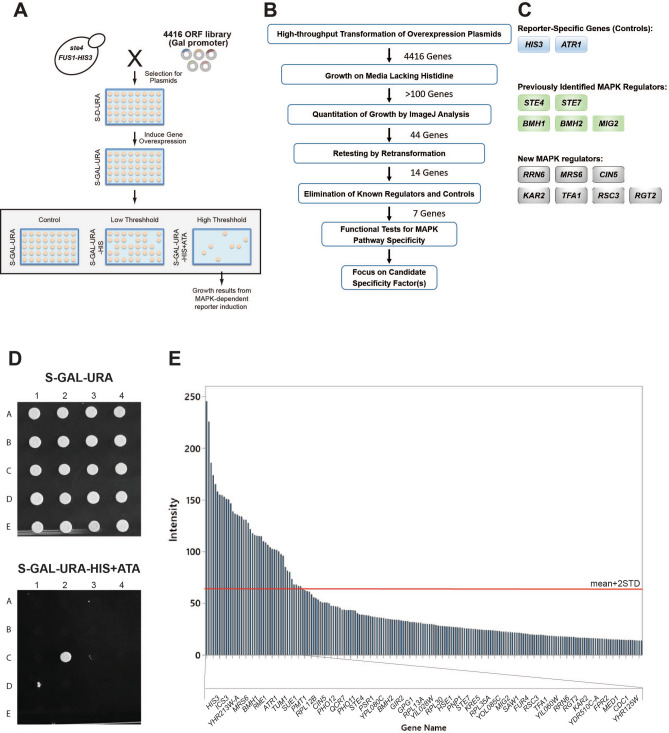
Genome-wide overexpression screen for new MAPK pathway regulatory proteins. (A) Diagram of the overexpression screen. An ordered collection of 4416 ORF overexpression plasmids covering ~ 80% of yeast genome controlled by an inducible (pGAL1) promoter (circles66) was introduced into a ste4 FUS1-HIS3 strain (PC999) by high-throughput transformation. Transformants were generated by a microtiter plate method and pinned onto S-D-URA to select for plasmids. Overexpression of genes was accomplished by pinning colonies from S-D-URA to S-GAL-URA medium to induce overexpression of the genes. On the following day, cells were pinned to low threshold and high threshold (containing ATA, a competitive inhibitor of the His3 enzyme) media to identify genes that induce a MAPK pathway-dependent growth reporter (FUS1-HIS3) on media lacking histidine. The genes, which could overcome ATA, were identified as the candidates that, when overexpressed, can turn the pathway up (colored spots). (B) Pipeline for identifying functionally relevant MAPK pathway regulators. 44 genes were identified and prioritized for further analysis. The validation screen identified 14 genes from the initial screen. (C) The list of 14 genes that induced the MAPK pathway-dependent reporter, FUS1-HIS3, when overexpressed. Genes fell into three categories (see Table 1 for more details). (D) Example of a portion of one plate from the overexpression screen (the full screen is available in Table S4). The colony growing in the lower panel, C2, overexpresses MRS6. (E) The graph shows the results of the top genes identified by overexpression. Colony growth on S-GAL-URA-HIS + ATA resulting from reporter (FUS1-HIS3) expression was measured by ImageJ analysis. Growth based on spot intensity and determined and plotted in the graph. The top 200 genes are shown. Forty-four genes passed a cut-off of mean + 2STD (red bar) and are labelled here.
Specifically, high-throughput transformation was used to introduce 4416 plasmids into a wild-type yeast strain (Fig. 2A)69 containing the FUS1-HIS3 reporter. Gene overexpression was induced by galactose, as the plasmid library is designed to induce gene expression by the strong pGAL1 promoter. Galactose is also a non-preferred carbon source that stimulates filamentous growth. In this way, gene induction occurred under the same conditions that induce filamentous growth. Specifically, colonies were transferred in 96-well format from S-D-URA media to S-GAL-URA media to induce overexpression of the genes. After 24 h, cells were pinned from S-GAL-URA to S-GAL-URA (as a control for growth), S-GAL-URA-HIS, and S-GAL-URA-HIS containing 3-amino-1, 2, 4-triazole (ATA) media (10 µl of 2 M in 25 ml plates). ATA is a competitive inhibitor of the His3 enzyme, and its inclusion allows for selection for high levels of reporter activity 72. Genes that inhibited growth on S-GAL-URA-HIS (Fig. 2A, Low Threshold) may, when overexpressed, dampen reporter activity and will be discussed elsewhere. Genes that induce growth on S-GAL-URA-HIS + ATA media may stimulate MAPK pathway activity due to transcriptional up-regulation of the growth reporter (Fig. 2A). These genes, in principle, have the potential to encode new MAPK pathway regulatory proteins.
A scheme was employed to identify relevant MAPK pathway regulators, represented by a flowchart (Fig. 2B). In the initial screen, > 100 genes were identified that showed some growth on S-GAL-URA-HIS + ATA media (Fig. 2D, Table S4). To quantitatively assess differences in growth, colony size was measured by ImageJ analysis [Table S2, 73]. By applying a rigorous cut-off of the mean + 2SD, 44 genes were identified that showed elevated MAPK reporter activity when overexpressed (Fig. 2B,E). To independently validate genes identified by the screen, plasmids containing candidate genes were re-transformed into wild-type cells and re-tested for reporter activity. Fourteen genes passed this validation step (Fig. 2, B and C; see Table S3 for the raw data).
The genes that passed the above criteria fell into three categories (Fig. 2C, Table 1). The first category was metabolic controls. Two controls were identified, HIS3, which allows growth on media lacking histidine74, and ATR1, which encodes a multidrug efflux pump that confers ATA resistance72. The second category was known regulators of MAPK pathways. These included STE4, which regulates the mating pathway and complemented the signaling defect of the ste4 mutant75; STE7, the MAPKK that regulates the mating and fMAPK pathways76; BMH1 and BMH2, which are members of the 14-3-3 family of proteins and are established regulators of the fMAPK pathway 7, and MIG2 a transcriptional repressor77 that has been implicated in fMAPK pathway regulation78. Not all components of the fMAPK pathway were identified: STE20, STE50, and STE11 were not present in the collection; MSB2 and CDC42 would not be expected to be identified as C-terminal fusions of the proteins, which occur in the library, are not functional in the fMAPK pathway; OPY2 was identified but fell below the threshold for statistical significance, and TEC1 does not induce the growth reporter. In its unphosphorylated form, the MAPK Kss1 would also not be expected to activate the reporter and may not be identified for this reason26,28,29. SHO1, BEM4, and STE12 were present in the collection but did not induce the reporter for reasons that have not been explored. The third category was potentially new MAPK pathway regulators. These included RRN6, MRS6, CIN5, KAR2, TFA1, RSC3, and RGT2 (Fig. 2C, Table 1).
Table 1.
Functional classification of MAPK pathway regulatory genes identified by gene overexpression alongside human homologs.
| Genes | Standard name | Name description | Normalized growth intensity in WT[a] | Normalized growth intensity in msb2Δ[a] | Molecular Function[b] | Biological Process[b,c,d] | Human Homolog[c] |
|---|---|---|---|---|---|---|---|
| YOR202W | HIS3 | HIStidine | 22[e] | 22 | Allows cells to grow on media lacking histidine | Catalyzes the sixth step in histidine biosynthesis | NA |
| YOR212W | STE4 | STErile | 20 | 20 | beta subunit of the first identified heterotrimeric G-protein | pheromone-dependent signal transduction involved in conjugation with cellular fusion, invasive growth in response to glucose limitation, regulation of transposition, RNA-mediated, chemotropism | G protein subunit beta 1(GNB1), GNB2, GNB5, GNB4, GNB3 |
| YDL159W | STE7 | STErile | 18 | 13 | MAP kinase kinase activity | MAPK cascade involved in cell wall organization or biogenesis, signal transduction involved in filamentous growth, invasive growth in response to glucose limitation, protein phosphorylation | mitogen-activated protein kinase kinase 2 (MAP2K2),MAP2K1 |
| YER177W | BMH1 | Brain Modulosignalin Homolog | 18 | 9 | RNA polymerase II activating transcription factor binding, DNA replication origin binding, phosphoserine binding | DNA damage checkpoint, signal transduction involved in filamentous growth, glycogen metabolic process,fungal-type cell wall chitin biosynthetic process, negative regulation of apoptotic process, pseudohyphal growth, negative regulation of transcription from RNA polymerase II promoter, Ras protein signal transduction | YWHAE, YWHAZ, YWHAB, SFN, YWHAG, YWHAH, YWHAQ |
| YDR099W | BMH2 | Brain Modulosignalin Homolog | 16 | 2 | DNA replication origin binding, phosphoserine binding | DNA damage checkpoint, signal transduction involved in filamentous growth, glycogen metabolic process,fungal-type cell wall chitin biosynthetic process, negative regulation of apoptotic process, pseudohyphal growth, negative regulation of apoptotic process | YWHAE, YWHAB, SFN, YWHAG, YWHAH, YWHAQ |
| YGL209W | MIG2 | Multicopy Inhibitor of Galactose gene expression | 16 | 6 | Zinc finger transcriptional repressor | cooperates with Mig1p in glucose-induced gene repression; under low glucose conditions relocalizes to mitochondrion, where it interacts with Ups1p, antagonizes mitochondrial fission factor Dnm1p, indicative of a role in mitochondrial fusion or regulating morphology; regulates filamentous growth in response to glucose depletion; activated in stochastic pulses of nuclear localization in response to low glucose | EGR1 [f] |
| YBL014C | RRN6 | Regulation of RNA polymerase I | 16 | 2 | RNA polymerase | Component of the core factor (CF) rDNA transcription factor complex; CF is required for transcription of 35S rRNA genes by RNA polymerase I and is composed of Rrn6p, Rrn7p, and Rrn11p | NA |
| YOR370C | MRS6 | Mitochondrial RNA Splicing 5 | 15 | 2 | Rab geranylgeranyltransferase activity, Rab GTPase binding | protein targeting to membrane, ER to Golgi vesicle-mediated transport, protein geranylgeranylation, activation of GTPase activity | CHM Rab escort protein (CHM), CHML |
| YOR028C | CIN5 | Chromosome INstability | 14 | 2 | Basic leucine zipper (bZIP) transcription factor of the yAP-1 family | physically interacts with the Tup1-Cyc8 complex and recruits Tup1p to its targets; mediates pleiotropic drug resistance and salt tolerance; nuclearly localized under oxidative stress and sequestered in the cytoplasm by Lot6p under reducing conditions | NA |
| YJL034W | KAR2 | KARyogamy | 14 | 4 | ATPase activity, unfolded protein binding | karyogamy involved in conjugation with cellular fusion, response to unfolded protein, SRP-dependent cotranslational protein targeting to membrane, translocation, fungal-type cell wall beta-glucan biosynthetic process | heat shock protein family A (Hsp70) member 5 (HSPA5) |
| YKL028W | TFA1 | Transcription Factor a, subunit 1 | 13 | < 0.1 | TFIIE large subunit; RNA polymerase II core binding | involved in recruitment of RNA polymerase II to the promoter, activation of TFIIH, and promoter opening | general transcription factor IIE subunit 1 (GTF2E1) |
| YDR303C | RSC3 | Remodel the Structure of Chromatin | 13 | < 0.1 | Component of the RSC chromatin remodeling complex | essential gene required for maintenance of proper ploidy and regulation of ribosomal protein genes and the cell wall/stress response; RSC3 has a paralog, RSC30, that arose from the whole genome duplication | NA |
| YDL138W | RGT2 | Restores Glucose Transport | 12 | < 0.1 | Plasma membrane high glucose sensor that regulates glucose transport | low affinity sesnor that contains 12 predicted transmembrane segments and a long C-terminal tail required for hexose transporter induction; phosphorylation of the tail by Yck1p/Yck2p facilitates binding to the HXT co-repressors, Mth1p and Std1p; RGT2 has a paralog, SNF3, that arose from the whole genome duplication | solute carrier family 2 member 8 (SLC2A8), SLC2A10, SLC2A12 |
| YML116W | ATR1 | AminoTriazole Resistance | 11 | < 0.1 | required for resistance to aminotriazole and 4-nitroquinoline-N-oxide | Multidrug efflux pump of the major facilitator superfamily; ATR1 has a paralog, YMR279C, that arose from the whole genome duplication; protein abundance increases in response to DNA replication stress | NA |
[a] Spot intensity was measured by ImageJ analysis and was normalized to wild-type values (see Table S2 for the raw data).
[b] Data comes from SGD (https://www.yeastgenome.org/).
[c] Data comes from Database Integration Tools (MARRVEL, Gene2Function, monarch INITIATIVE, ALLIANCE of GENOME RESOURCES, NCBI).
[d] Not all biological processes are mentioned.
[e] Growth intensity rates of the canididates validated in wild type is normalized to msb2Δ mutant.
[f] Not all homologs are listed here.
To explore the characteristics of the genes identified by the screen, we used gene ontology (GO) annotations and database integration tools to identify the molecular and biological roles of proteins and determine whether they had mammalian homologs79–84. Many of the identified genes had human homologs with established functions in diverse biological processes (Table 1). These included BMH1, BMH2 7, TFA185, MRS686,87, and KAR288. Moreover, the screen identified several essential genes (MRS6, KAR2, TFA1, and RRN6), and a set of paralogs (BMH1 and BMH2), which might be missed in whole-genome deletion screens.
Many signaling pathways can influence the activity of the fMAPK pathway. One mechanism for this regulatory input comes from the regulation of the expression of the MSB2 gene89. MSB2 encodes the mucin-type glycoprotein that regulates the fMAPK pathway69. To determine whether these genes fall above or below Msb2 in their ability to stimulate MAPK pathway activity, candidates from the screen were examined for overexpression-dependent bypass the signaling defect of the msb2Δ mutant. A subset of the genes tested restored signaling in the msb2Δ mutant (Table 1, see Table S3 for the raw data), which indicates that they function below the level of Msb2 in the MAPK pathway. We were interested in new regulators that, when overexpressed, bypass the signaling defect of the msb2Δ mutant (MRS6, RRN6, and KAR2), because of their potential to modulate MAPK pathway activity directly.
Examining the role of new MAPK pathway regulators in polarity reorganization during filamentous growth
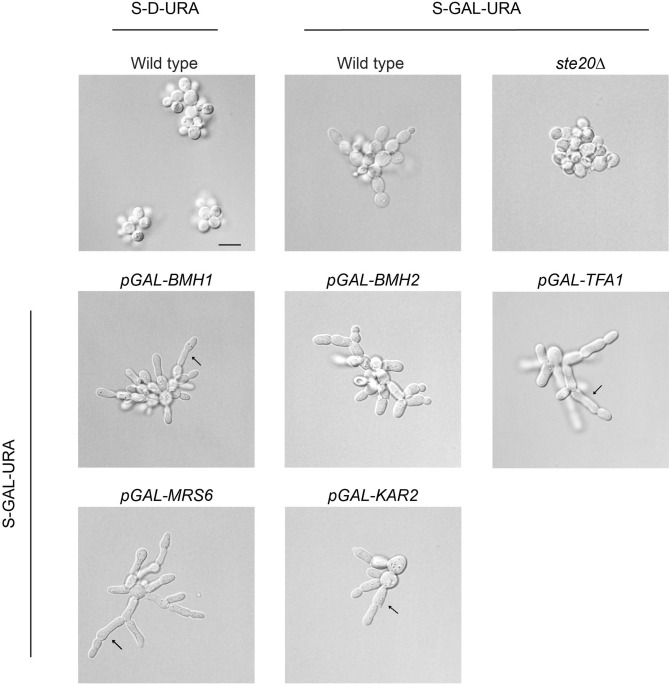
During filamentous growth, yeast cells produce an elongated cell morphology, which results from hyper-polarized growth90. Hyperpolarized growth is caused by the fMAPK pathway22, which induces the expression of genes that cause a delay in the G1 and G2/M phases of the cell cycle91,92. The single-cell invasive growth assay24 was used to examine the polarized growth of a subset of candidate genes identified in the screen. Wild-type cells exist in the yeast form when grown in glucose (Fig. 3, S-D-URA) and undergo filamentous growth when grown in the non-preferred carbon source galactose (Fig. 3, S-GAL-URA). Cells lacking an intact fMAPK pathway are defective for filamentous growth by this assay (Fig. 3, ste20Δ). Overexpression of MRS6, BMH1, BMH2, KAR2, and TFA1 induced hyperpolarized growth. Specifically, the cells were longer and had irregular morphologies (Fig. 3, arrows). This phenotype is distinct from activation of the HOG pathway, which shares components with the fMAPK pathway but does not induce a morphogenetic change when activated21.
Figure 3.
Morphological analysis of cells overexpressing genes that stimulate MAPK pathway signaling. (A) Cell morphology of the indicated strains by the single-cell invasive growth assay by DIC microscopy at 100X magnification. Scale bar, 10 μm. As controls, wild-type cells were grown in glucose (Glu, S-D) and galactose (S-Gal) media, and the ste20Δ mutant was grown in S-Gal media. Overexpression of MRS6, BMH1, BMH2, KAR2, and TFA1 induced hyperpolarized morphologies. Arrows show elongated cells making chains of filaments.
Analysis of fMAPK pathway regulatory proteins by functional tests for MAPK pathways
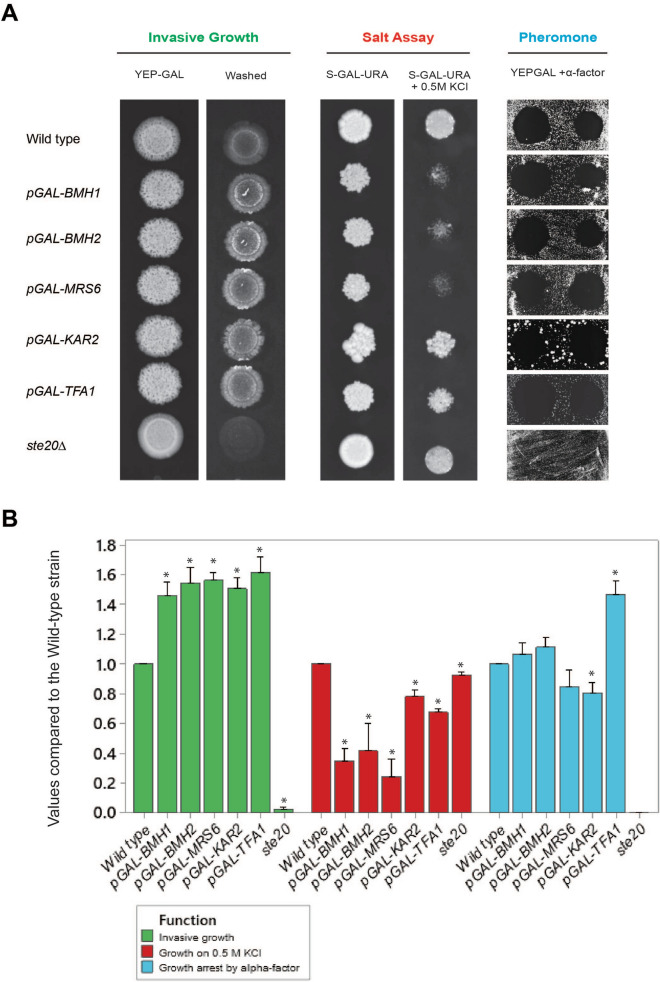
Three MAPK pathways in yeast share components, including the Rho-type GTPase Cdc42, PAK Ste20, and MAPKKK Ste11 (Fig. 1, 64,65). As a complementary approach to assess the role of overexpression of MRS6, BMH1, BMH2, KAR2, and TFA1 on these pathways, functional tests were performed that provide a readout of the three MAPK pathways. The plate-washing assay (PWA) measures the invasion of cells into the agar, which can be revealed by washing plates in a stream of water, and which is dependent on an intact fMAPK pathway25. Salt sensitivity was used to measure the activity of the HOG pathway45,93, and growth arrest by α-factor (halo assay) was used to measure the activity of the mating pathway94. Growth on galactose (YEP-GAL) resulted in hyper-invasive growth for each of the candidate genes tested by the PWA (Fig. 4A, green). Overexpression of BMH1, BMH2, and MRS6 caused a growth defect on media containing salt (Fig. 4A, red). This result was interesting because the fMAPK pathway functions antagonistically with the HOG pathway 95. Thus, it is plausible that elevated activation of the fMAPK pathway by overexpression of these genes might result in a dampened HOG response. Overexpression of these genes did not result in a defect in halo formation (Fig. 4A, blue).
Figure 4.
Phenotypic analysis of the role of overexpression of selected candidates on MAPK pathway activity. (A) Wild-type cells (PC6810) containing the indicated plasmids were grown in S-D-URA for 16 h and spotted onto the indicated media. For the PWA, cells were spotted onto YEP-GAL medium for 96 h. The plate was photographed (YEP-GAL), washed in a stream of water, and photographed again (Washed). To assess salt sensitivity, cells were spotted on S-GAL-URA and S-GAL-URA + 0.5 M KCl media for 72 h at 30 °C. To determine sensitivity to α-factor, cells were spread onto S-GAL-URA plates. 10 μl and 3 μl drops of 1 mg/ml α-factor were applied to the plates followed by incubation for 48 h. (B) Plot shows the quantified data for the invasive growth, salt assay, and pheromone. Values normalized to wild type (WT) values, which were set to a value of 1. Bars represent the average of at least three independent experiments. Error bars represent the standard deviation between trials. Asterisks indicate significant differences compared to the wild-type strain for the same condition (p-value < 0.01 by Student’s t-test).
Quantifying the data also supports the idea that some genes turn fMAPK pathway up, and turn HOG down (Fig. 4B). These results support the idea that MRS6, BMH1, BMH2, KAR2, and TFA1 stimulate the activity of the fMAPK pathway and might potentially play a specific role in that pathway when these genes are overexpressed.
Mrs6 overexpression stimulates the fMAPK pathway and dampens the HOG pathway
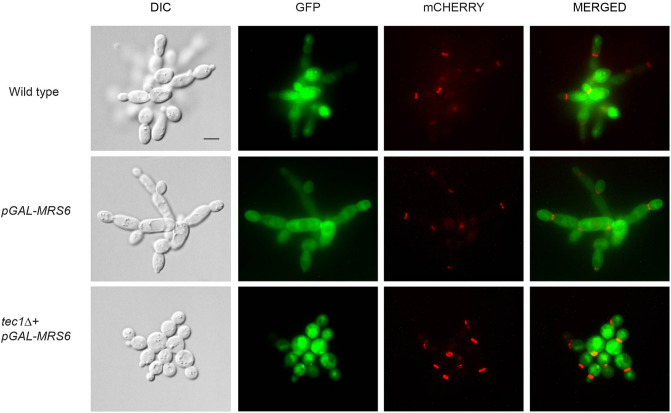
We focused on MRS6 because it was one of the strongest hits from the screen (Fig. 2D, the spot represents MRS6). Overexpression of MRS6 also strongly induced polarized growth (Fig. 3), induced hyper-invasive growth (Fig. 4, green), and dampened the HOG pathway (Fig. 4, red). Mrs6 is also an essential protein and, when overexpressed, bypassed the signaling defect of the msb2 mutant (Table 1). Mrs6 is a Rab escort protein96 and has recently been identified as a modulator of the activity of the TOR pathway97. We confirmed that overexpression of MRS6 induces hyperpolarized growth (Fig. 3). To determine whether this results from problems in cell polarity, we examined cells by fluorescence microscopy for defects in the localization of polarity proteins GFP-Cdc42 and septin Cdc3-mCHERRY 98. The localization of these proteins was normal in cells overexpressing MRS6, which indicates that Mrs6 does not promote cell elongation solely by perturbing proper cell morphogenesis. Interestingly, the elongated cell morphology seen in cells overexpressing MRS6 was dependent on the fMAPK pathway, as it was not seen in the tec1Δ mutant, which lacks a key transcription factor for the pathway40 (Fig. 5). Additional examples of the morphology of these strains can be seen over a time-course experiment (Movies S1–S3). Taken together, these results provide support for a role for Mrs6 in positively regulating the fMAPK pathway.
Figure 5.
The localization of GFP-Cdc42 and the septin (by Cdc3-mCHERRY) in cells overexpressing MRS6 with and without the transcription factor Tec1 were examined by fluorescence microscopy. Wild-type cells, and cells overexpressing MRS6 in wild-type cells and the tec1Δ mutant cells were grown for 16 h in media [0.67% YNB without ammonium sulfate, 0.1% monosodium glutamate (MSG), 2% dextrose, 1 X amino acid stock without uracil, 0.36 mg/ml gent]. Cells were grown to mid-log phase for 6 h and photographed by fluorescence microscopy utilizing the GFP, Rhodamine, and DIC filter sets. Scale bar, 5 microns.
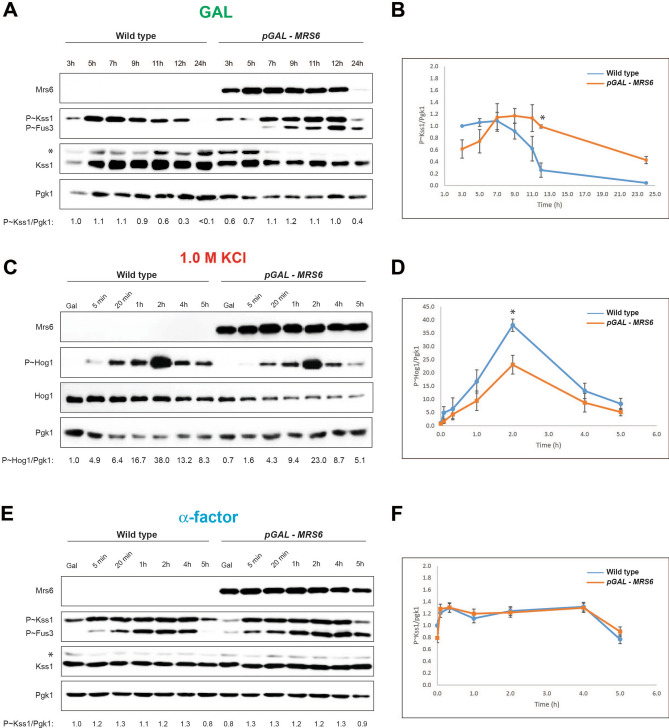
To explore the role of Mrs6 in regulating MAPK pathways, the phosphorylation of MAP kinases was examined, which provides a diagnostic readout of their activities. Based on immunoblot analysis, we typically see a > 100-fold increase in Mrs6 protein levels upon overexpression by the pGAL1 promoter (Fig. 6A). This induction is similar to what has been reported for other proteins driven by that promoter 66,99. Using anti phospho p44-42 antibodies that detect the phosphorylated MAP kinases, Kss1 and Fus3, we found that overexpression of MRS6 induced phosphorylation of Kss1 (Fig. 6A, P ~ Kss1). In comparison to wild-type cells, where the levels of P ~ Kss1 increased after 3 h growth in Gal and decreased after 7 h, overexpression of MRS6 caused a delay in the phosphorylation of Kss1, which was sustained until 12 h and then decreased (Fig. 6B). This result indicates that MRS6 alters the kinetics of the fMAPK pathway in a manner that might be expected to promote cell elongation during filamentous growth.
Figure 6.
Impact of overexpression of MRS6 on the fMAPK, HOG, and mating pathways. Wild-type cells (PC6810) and cells overexpressing MRS6 (PC7447) were examined under conditions that induce MAPK pathway signaling. Cell extracts were evaluated by MAP kinase phosphorylation by immunoblot (IB) analysis. One example is shown for panels A, C, and E (raw data is shown in Fig. S3 A, B, and C, respectively). For panels B, D, and F, the data represent the average of at least three independent experiments. Error bars indicate the standard error of mean between trials (Asterisks, p-values < 0.05 by student’s t-test). (A) Cells were grown in the non-preferred carbon
source galactose (YEP-GAL) for the times indicated. Cell extracts were examined by IB analysis for P ~ Kss1 and P ~ Fus3 by p44/42 antibodies. Mrs6 proteins were detected at ~ 91 kDa. Total Kss1 levels and Pgk1 (loading control, ~ 45 kDa) also were assessed. The ratio of P ~ Kss1 to Pgk1 normalized to wild-type values, which were set to a value of 1. (B) Graph visualizes the ratio of P ~ Kss1 to Pgk1 for wild-type and pGAL-MRS6. (C) Cells were pre-grown in YEP-GAL for 4 h following by growing in YEP-GAL medium containing 1.0 M KCl to examine P ~ Hog1. (D) Graph showing P ~ Hog1 to Pgk1 ratios, normalized to wild-type values, which were set to a value of 1. (E) Phosphorylation of Kss1 and Fus3 in response to pheromone. Cells were grown in YEP-GAL for 4 h, and incubated in YEP-GAL medium containing 1 mg/ml α-factor for the times indicated. Fus3 bands run in the same size as a degradation product of MRS6. (F) Graph showing P ~ Kss1 to Pgk1, normalized to wild-type values, which were set to a value of 1.
By comparison, the terminal MAP kinase in the HOG cascade, Hog1, was under-phosphorylated in response to salt due to MRS6 overexpression (Fig. 6C). Immunoblot data indicated that overexpression of MRS6 caused a modest reduction in HOG pathway activity (Fig. 6D). These results match with the fact that overexpression of MRS6 caused a growth defect on high-osmolarity media (Fig. 4). Given that the fMAPK and HOG pathways can function antagonistically95, our results suggest that MRS6 may be a specific regulator of the fMAPK pathway. Overexpression of MRS6 did not have a dramatic effect on the mating pathway (Fig. 6, E and F). The main MAP kinase for the mating pathway, Fus3, is phosphorylated in response to pheromone. Although Fus3 phosphorylation was similar between wild-type cells and cells overexpressing MRS6, Fus3 migration overlapped with a degradation product of Mrs6 and was not used for quantitation. Therefore, MRS6 overexpression led specifically to phosphorylation (activation) of the MAP kinase Kss1, which is consistent with a specific role for the protein in regulating the fMAPK pathway.
Mrs6 interacts with the protein kinases Ste20 and Ste7
To define how Mrs6 might specifically regulate the fMAPK pathway, genetic suppression analysis was performed. Genetic suppression analysis can allow the ordering of proteins into a pathway using gain- and loss-of-function alleles. pGAL-MRS6 was introduced into mutants that lack fMAPK pathway components. Reporter induction by overexpression of Mrs6 was compared in cells lacking components of the fMAPK pathway (Fig. 1). We looked at many components of fMAPK, including the msb2Δ, sho1Δ, opy2Δ, ste20Δ, bem4Δ, ste50Δ, and ste11Δ mutants. The results showed that Mrs6 overexpression partially bypassed the signaling defect of the sho1Δ mutant but not the ste11Δ mutant (Fig. S1, data shown for sho1Δ and ste11Δ). This experiment indicates that Mrs6 regulates the fMAPK pathway at or above the level of Ste11 in the fMAPK pathway. We also noticed that overexpression of MRS6 induced a growth defect. The growth defect was separate from its induction of the fMAPK pathway, as it was seen in cells lacking fMAPK pathway components (Fig. S2). Interestingly, diploid strains heterozygous for MRS6 also have a growth defect100.
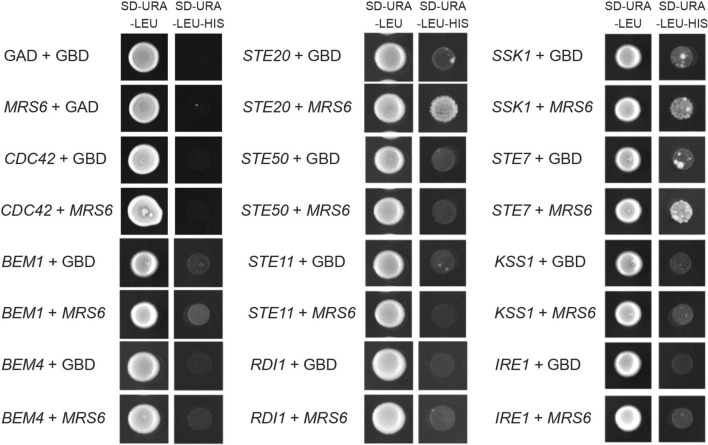
To further define how Mrs6 regulates the fMAPK pathway, we analyzed the ability of Mrs6 to interact with fMAPK components by the two-hybrid system101. Two-hybrid analysis can identify protein interactions in vivo by reconstitution of the binding and activation domains of fusion proteins to the Gal4 transcription factor, evaluated by a growth reporter 101. Two-hybrid analysis has proven to be a useful tool in detecting interactions in many biological systems, including the isolated domains of interacting proteins102,103. The gene encoding Mrs6 was cloned into a two-hybrid vector (bait) and probed for interactions with a panel of proteins that regulate MAP kinase pathways. The analysis identified a robust interaction between Mrs6 and Ste20 (Fig. 7). Two-hybrid analysis also identified an interaction between Mrs6 and Ste7. Also, we saw a very weak positive signal for the Ssk1 protein. Mrs6 did not associate with other components of fMAPK by two-hybrid analysis. Therefore, the two-hybrid analysis may provide an explanation for how MRS6 promotes fMAPK signaling, which includes the kinases Ste20, Ste11, Ste7, and Kss1, but not the HOG pathway, which includes the kinases Ste20, Ste11, Pbs2, and Hog1.
Figure 7.
Two-hybrid analysis between Mrs6 and proteins that regulate fMAPK pathway. In the panels, GAD refers to pGAD-C1, and GBD refers to pGBDU-C1. Cells were grown on S-D-URA-LEU to maintain selection for the bait and prey plasmids. Growth on medium lacking histidine (S-D-URA-LEU-HIS) displayed an interaction of Mrs6 with MAPKKK kinase Ste20, and an interaction between Ste7 and Mrs6. Based on two-hybrid analysis, Mrs6 did not associate with other components of fMAPK.
Discussion
MAPK pathways regulate diverse cellular responses and are controlled by an expanding repertoire of regulatory proteins. In this study, we uncovered new regulators of an ERK-type MAPK pathway in yeast. We screened a S. cerevisiae library of covering 80% of the genome for genes that, when overexpressed, induce a MAPK-dependent growth reporter. Overexpression screens can uncover new roles for genes in biological processes and are well suited to identify roles for essential genes that cannot be evaluated by deletion analysis. In this study, we identified 12 regulatory genes of the MAPK pathway and two metabolic controls. The seven new genes identified in this study as MAPK pathway regulators provide a platform for exploring how a different cellular processes connect to and regulate MAPK pathways. We followed up on one of these candidates and showed by a combination of genetic and biochemical approaches that Mrs6 regulates the MAPK pathway that controls filamentous growth. The phenotypic difference between a wild-type strain and the indicated overexpression plasmids was apparent by reporter activity (Fig. 2), the elongated morphology of cells compared to wild-type cells (Fig. 3) and hyper-invasive growth by the plate-washing assay (Fig. 4A). These outcomes demonstrate that filamentous growth resulting from overexpression of these genes occurs under conditions (nutrient poor, galactose) when the pathway is active. Whether they also induce pathway activity under basal (nutrient-rich) conditions has not been tested. Furthermore, we found out Mrs6 interacts with kinases that regulate that pathway and might play a role in pathway specificity.
MRS6 is an essential gene that may regulate the fMAPK pathway in several ways. One way might be through its role in regulating protein trafficking. Rab-type GTPases regulate protein trafficking104–106. Mrs6 is a Rab escort protein (REP) that makes a complex with the Rab GTPases Ypt1, Sec4, Ypt6, Vps21107–109. Given that Cdc42 is itself a component of the exocyst complex110,111, it is possible that Mrs6 might contribute to the delivery of Cdc42 or other fMAPK pathway components to the plasma membrane. Similarly, Mrs6 may regulate the assembly of the fMAPK signaling complex and/or its function in the secretory pathway. Ypt1 also regulates the UPR by promoting the decay of HAC1 RNA112. Interestingly, Msb2 and the fMAPK pathway are regulated by the UPR113. Perhaps some of the regulators identified in this study connect the MAPK pathway to the UPR pathway. In a related study, we found that BMH1 and BMH2 showed a connection to the UPR but not MRS6 (Jamalzadeh et. al, unpublished data).
Mrs6 functions with members of the Rab family of GTPases, which control vesicle trafficking in the secretory pathway114. In particular, Mrs6 is a Rab-escort protein that promotes lipid modification (prenylation) of the Rab GTPase Ypt1 by the Bet2 and Bet4 geranylgeranyltransferase complex II96. Mrs6 specifically facilitates geranylgeranylation for the prenylation of Ypt1 at the Golgi through the Bet2 and Bet3 enzymes115. Mrs6 might regulate the fMAPK pathway through its role in regulating Rab prenylation. Ypt1 and the Bet proteins are essential proteins that cannot be readily analyzed by deletion analysis. We found that Bet proteins, Bet3, and the Rab GTPase, Ypt1, when overexpressed did not impact fMAPK pathway activity (Table S4, see labeled genes). Mrs6 has also been identified as a modulator of the TOR pathway by interacting with the transcription factor Sfp197,116. Sfp1, when overexpressed, did not impact the activity of the fMAPK pathway (Table S4, see labeled genes). However, interestingly, overexpression of SFP1 stimulates filamentous growth117. Thus, Mrs6 may have a separate function in regulating the fMAPK pathway than its role in Rab or TOR pathway regulation. In the fMAPK pathway, the Rho-type GTPase Cdc42 is modified by lipid geranyl groups (by Cdc43); thus, Mrs6 may impact the lipid modification of Cdc42. However, Mrs6 did not associate with Cdc42 by two-hybrid analysis.
Two-hybrid analysis showed that Mrs6 interacts with Ste20. Ste20 is the PAK kinase that regulates the fMAPK pathway25,118,119. Ste20 is recruited by a complex containing Cdc42 and Cdc24 to the membrane120. Thus, Mrs6 may regulate the fMAPK pathway by promoting the plamsa membrane recruitment or activation of Ste20. Mrs6 also interacts with Ste7 (a MAPKK). Given that MRS6 specifically stimulates the fMAPK pathway, Mrs6 might facilitate interactions among members of the kinase cascade. In support of this possibility, overexpression of MRS6 dampened the activity of the HOG pathway. Alternatively, Mrs6 may interact with Ste20 in one complex and Ste7 in another complex. Future studies will be required to determine how these interactions promote fMAPK pathway induction.
Signal transduction pathways operate in different ways with vastly different kinetics5. The activation kinetics of signaling pathways are crucial to determine the nature of the biological response. The fMAPK pathway operates with slower kinetics compared to the mating and HOG pathways [121, this study]. The kinetics of activation has probably been fine-tuned for the filamentous growth response. Overexpression of MRS6 increases fMAPK pathway activity (see Fig. 6B). By examining the kinetics of the fMAPK pathway, we show that overexpression of MRS6 extends the amount of time the pathway is active. The interactions between Mrs6 with Ste20 and Ste7 might extend pathway activity. Phenotypically, this may augment the MAPK-dependent cell-cycle delays, resulting in hyper-invasive growth, which we also observe upon MRS6 overexpression.
In a separate study, MRS6 was shown to regulate the TORC1 pathway through SFP1 to control ribosome biogenesis97,116. TOR is a master regulatory pathway of cell growth and nutrient sensing122. TOR’s activator, GOLPH3, has been identified recently as an oncogene in many human cancers123. Hence, the identification of Mrs6 as a key regulator of the fMAPK pathway in yeast raises the possibility that REP1/REP2 may link fMAPK signaling to the TOR pathway and to the secretory system in higher organisms.
In mammalian cells, MRS6 homolog encoded by CHM, which is the human Rab escort proteins REP1/CHM or REP2/CHML and share 50% sequence identity with Mrs686. CHM is a disease of the retina, which causes progressive vision loss12. Furthermore, REP1/CHM has been shown to regulate the epidermal growth factor receptor (EGFR) through the transcription factor STAT3. EGRF is also a major regulator of the Grb-SOS-RAS-MEK-ERK pathway, which is commonly misregulated in cancer cells 124. Given that EGFR also signals through RAS-MEK-ERK125,126, our screen may have identified a new and general regulator of ERK-type MAPK pathways.
Materials and methods
Strains and plasmids
Strains used in the study are listed in Table S1. Strains were cultured in yeast extract and peptone (YEP) media (1% yeast extract and 2% bactopeptone) with a source of carbon [2% glucose (D) or 2% galactose (GAL)] for growth in liquid culture or 2% agar for growth in semi-solid agar media. All experiments were carried out at 30 °C unless otherwise specified. Synthetic complete (S) medium was used for maintaining selection for plasmids. Bacterial cultures of Escherichia coli were proliferated in LB + CARB media (carbenicillin) by standard methods 127. The pRS plasmids (pRS315 and pRS316) have been described 128. To construct two-hybrid plasmids, plasmids pGAD-C1 and pGBDU-C1 were used 129.
Analysis of a gene overexpression collection for altered activity of a MAPK pathway-dependent growth reporter
A microtiter-based high throughput transformation method130 was used to introduce a collection of ~ 4500 plasmids, each overexpressing a different yeast gene66 into strain (PC999). Transformants were screened for Msb2-HA secretion as described89 and the activity of the fMAPK pathway in this study. Specifically, transformants were pinned onto S-D-URA to select for plasmids. Colonies that grew onto S-D-URA were then pinned to S-GAL-URA to induce gene overexpression. From S-GAL-URA, cells were pinned onto S-GAL-URA, S-GAL-URA-HIS, and S-GAL-URA-HIS + ATA to identify positive regulators of the fMAPK pathway. Colonies that grew on S-GAL-URA-HIS + ATA media resulted from elevated fMAPK pathway activity due to the up-regulation of the growth reporter (FUS1-HIS3).
Genome-wide screen and data analysis
The growth of 4416 genes was examined from 46 plates (raw data is available in Table S4, see labeled genes for hits). Not all of the genes from the collection were analyzed. This may have resulted because of the failure of some plasmids to be transformed and contamination on several plates. ImageJ analysis (https://imagej.nih.gov/ij/) was used to quantify colony growth. Images of the plates from the screen were converted to 8-bit and inverted. A threshold adjustment was performed, followed by analysis by the DNA microarray plugin to measure spot intensity for each colony (Table S2). Outputs from ImageJ were saved as cvs format for additional analysis.
A MATLAB script was written to identify growth that was statistically significant. A cut-off of mean + 2STD identified the top 3% of genes that, when overexpressed, showed growth that was above background. Validation of candidates was performed by re-transformation of plasmids containing genes, by standard transformations 131, into a wild-type strain (PC6021) and testing for reporter induction by growth on S-GAL-URA-HIS + ATA media (Table S3). The same plasmids were also transformed into the msb2Δ mutant (PC3209) to determine the bypass of that regulator of the pathway (Table S3). Database Integration Tools were used for further describing the identified genes’ characteristics and their orthologs in a concise manner79–84.
Microscopy
Differential interference contrast (DIC) microscopy was performed at 100X using an Axioplan 2 fluorescent microscope (Zeiss) with a Plan-Apochromat 100X/1.4 (oil) objective (N.A. 1.4) (coverslip 0.17) (Zeiss). Digital images were obtained with the Axiocam MRm camera (Zeiss) and Axiovision 4.4 software (Zeiss) was used for image acquisition. Adobe Photoshop was used for brightness and contrast adjustments. Polarized cells were assigned by examining cells over multiple focal planes by DIC.
Localization and fluorescence microscopy
Wild-type cells with integrated Cdc3-mCherry that also contained pGFP-Cdc42 and either pRS316 (PC7589) or pGAL-MRS6 (PC7590), and tec1Δ cells with integrated Cdc3-mCherry and contained pGFP-Cdc42 and pGAL-MRS6 (PC7592) were examined by fluorescence microscopy. Plasmids were selected on S-D-URA + Geneticin (Cat#11811-031) semi-sold agar media [2% agar, 0.67% YNB without ammonium sulfate, 0.1% monosodium glutamate (MSG), 2% dextrose, 1 X amino acid stock without uracil, 0.36 mg/ml Gent]132 at 30◦C. Samples were grown for 16 h in S-D-URA + 0.1% MSG + 0.36 mg/ml Gent. Five hundred microliters of each culture was collected by centrifugation, washed twice in distilled water and transferred to 10 ml of YEP-GAL + MSG + Gent media. Cells were grown for 6 h. The mid-log phase samples were washed twice with water, and cells were examined by fluorescence microscopy at 100X utilizing GFP, Rhodamine, and DIC filter sets using an Axioplan 2 fluorescent microscope (Zeiss). Cells were examined at serial sections on the plane of the Z-axis. Brightness and contrast were adjusted to reduce background using Adobe Photoshop.
Time-lapse fluorescence microscopy was performed as described132. Cells were grown for 16 h in in S-D-URA + 0.1% MSG + 0.36 mg/ml Gent at 30 °C. Approximately 800 µl of YEP-GAL + 1% agarose was placed on 12 mm Nunc glass base dishes (150,680, Thermo Scientific, Waltham, MA). One thousand microliters of cells were washed in water and resuspended in YEP-GAL media. 25 µl of cells were placed underneath of the agarose pad by gently lifting the pad with a scalpel. The plate was incubated for 30 min at 30◦C for stabilizing the cells. Cells were examined by a Zeiss 710 confocal microscope equipped with a Plan-Apochromat 40x/1.4 Oil DIC M27 for 5 h with 10 min intervals. Serial sections were examined in the plane of the Z-axis.
Functional assays for MAPK pathway activity
Cell morphology was assessed by the single-cell invasive growth assay 24. Invasive growth was assessed by the PWA 25. For the PWA, equal concentrations of cells were spotted onto YEP-GAL media. The activity of the HOG pathway was assessed by growth on high-osmolarity media. Equal concentrations of cells were spotted onto S-GAL-URA and S-GAL-URA + 0.5 M KCl media. Halo assays were performed as described 133. Cells were spotted onto YEP-GAL media followed by spotting 3 μl and 10 μl α-factor (1 mg/ml) on the plate. Plates were incubated at 30 °C and photographed at 24 h and 48 h. The single-cell invasive growth assay was performed as described 24.
Phospho-immunoblot analysis
Phosphorylation of different MAP kinases in response to different stimuli was examined as described 134,135. Cells were grown to mid-log phase from a saturated culture in YEP-D or YEP-GAL media for 4 h. Cells were washed and sub-cultured into YEP-GAL, YEP-GAL with 1.0 M KCl, or YEP-GAL with α-factor (1 mg/ml). Cells were collected at various times by centrifugation, washed once, and stored at -80 ˚C. Proteins were extracted by trichloroacetic acid precipitation (TCA) and resuspended in 0.15 ml sample buffer by heating to 90 °C. Protein samples were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide). Proteins were transferred from polyacrylamide to nitrocellulose membranes (AmershamTM ProtranTM Premium 0.45 μm NC, GE Healthcare Life sciences, 10600003) by electrotransfer (Bio-Rad laboratories Inc.). Membranes were blocked with 5% BSA in 1X TBST (10 mM TRIS–HCl pH 8, 150 mM NaCl, 0.05% Tween 20).
Phosphorylation of mating and fMAPK pathways (P ~ Kss1 and P ~ Fus3) was investigated with p44-42 antibody (Cell Signaling Technology, Danvers, MA, 4370) at a dilution of 1:10,000 to detect ERK-type MAP kinases. Phosphorylated Hog1 was detected using a 1:10,000 dilution of α-phospho p38 antibody (Santa Cruz Biotechnology, Santa Cruz CA; #yC-20). Total Kss1 was detected with α-Kss1 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA; #6775) at a 1:5,000 dilution. Total Hog1 was detected with α-Hog1 antibodies at a 1:5,000 dilution and Pgk1 was detected using mouse monoclonal antibodies at a 1:5,000 dilution (Novex, 459250). Membranes were incubated 16 h with primary antibodies in 1X TBST with 5% BSA at 4 °C. Control membranes were incubated 16 h in Pgk1 antibodies in 1X TBST with 5% non-fat dried milk at 4 °C. To detect the primary antibodies, secondary antibodies of goat anti-rabbit IgG-HRP at a 1:10,000 dilution (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, 111–035-144), and Goat α-mouse secondary (Bio-Rad Laboratories, Hercules, CA, 1706516) at a 1:5,000 dilution were used within milk as blocking buffer. The pGAL-MRS6 plasmid encodes a Mrs6-HA-HIS-Protein A fusion protein, which can be detected with the abovementioned antibodies. Blots were visualized by chemiluminescence using a Bio-Rad ChemiDoc XRS + system (Bio-Rad, 1708265). Image Lab Software (Bio-Rad, Inc.) was applied to analyze the band intensity.
Genetic suppression analysis
Control (pRS316) and pGAL-MRS6 plasmids were transformed into wild-type strain (PC538) and MAPK pathway mutants. These included msb2Δ (PC3209), sho1Δ (PC5692), opy2Δ (PC3752), ste20Δ (PC5692), bem4Δ (PC3551), ste50Δ (PC610), and ste11Δ (PC3861) mutants. Cells were grown on S-GAL-URA and S-GAL-URA-HIS to evaluate growth, which we infer to represent bypass of the mutant phenotype.
Cloning the MRS6 gene into two-hybrid plasmids
The MRS6 gene was cloned into the pGAD-C1 and pGBDU-C1 vectors in the following way. The MRS6 gene was amplified by PCR using the forward primer 5′-ATGCATCGATATGTTAAGTCCTGAACGTAGACC-3′ and reverse primer 5′-ATGCGTCGACTCATATCTCCATTTCACCTACAAATTC-3′. The PCR product was purified with QIAquick PCR Purification Kit, Qiagen, CA#28106. The PCR product and pGAD-C1 vector were digested with ClaI (5′-ATCGAT-3′, New England BioLabs Inc., MA, CA#R0197S) and SalI (5′-GTCGAC-3′, New England Biolabs Inc., MA, CA#R3138S) restriction enzymes. Digested insert and vector DNAs were run on a 1% agarose gel containing ethidium bromide. Bands were extracted from the gel using the QIAquick Gel Extraction Kit, Qiagen (CA#28704). A quick Ligation Kit (New England Biolabs Inc., MA, CA#M200l) was used for ligating the insert and vector. The ligation mixture was transformed into E. coli (One-Shot MAX Efficiency DH5α-T1 Competent Cells, ThermoFisher, CA# 12297016), followed by plating on LB + Carb plates. The plates were incubated at 37 °C for 24 h. Transformants were confirmed by digestion with ClaI and SalI. Plasmids were sequenced at the Roswell Park Sequencing facility (Roswell Park Cancer Institute, Buffalo, NY).
Two-hybrid assay
Two-hybrid constructs (pGBDU-C1 bait and pGAD-C1 prey) and empty vectors as controls were introduced into strain PJ694A (PC284)129 using the lithium acetate transformation standard protocols136. Transformants were selected on S-D media lacking uracil (URA) and leucine (LEU) to maintain selection for plasmids. Protein–protein interactions were screened by spotting cells onto S-D-URA-LEU media that was also lacking histidine (HIS) and containing ATA. Growth in this media results from the induction of a two-hybrid transcriptional reporter as the readout of protein–protein interactions.
Supplementary information
Acknowledgments
Thanks to Heather Dionne for performing the screen; to Alan Siegal for his assistance with the time-lapse experiments. Thanks to previous and current members of the Cullen laboratory, especially Dr. Beatriz Gonzalez and Aditi Prabhakar, for technical assistance and insightful discussions throughout the course of this work. This work was supported by a grant from the NIH (GM098629).
Abbreviations
- ATA
3-Amino-1, 2, 4-triazole
- CARB
Carbenicillin
- CHM
Choroideremia
- CHML
Choroideremia-like
- D
Dextrose
- DIC
Differential interference contrast
- E. coli
Escherichia coli
- ERK
Extracellular-signal-regulated kinase
- GAL
Galactose
- GLU
Glucose
- GAD
Gal4 activation domain
- GBD
Gal4 binding domain
- Gent
Geneticin
- GO
Gene ontology
- GTP
Guanine nucleotide triphosphate
- HIS
Histidine
- HOG
High osmolarity glycerol response
- LEU
Leucine
- MAPK
Mitogen-activated protein kinase
- MAPKKK
Mitogen-activated protein kinase kinase kinase
- MEKK
Mitogen-activated protein kinase kinase kinase
- MEK
Mitogen-activated protein kinase kinase
- MSG
Monosodium glutamate
- PAK
P21-activated protein kinase
- PWA
Plate washing assay
- RabGDI
RabGDPdissociation inhibitor
- RabGGTase
Rab geranylgeranyl transferase
- REP
Rab escort protein
- Rho
Ras homology
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- STD
Standard deviation
- S
Synthetic
- TCA
Trichloroacetic acid
- URA
Uracil
- WT
Wild type
- YEP
Yeast extract and peptone
Author contributions
S.J. designed and performed experiments, analyzed the data, and wrote the paper. A.N.P. performed experiments. P.J.C. designed experiments and wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
is available for this paper at 10.1038/s41598-020-78470-4.
References
- 1.Cicenas J, et al. JNK, p38, ERK, and SGK1 Inhibitors in Cancer. Cancers (Basel). 2017 doi: 10.3390/cancers10010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, et al. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduction. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412. [DOI] [PubMed] [Google Scholar]
- 3.Peng Q, et al. Mitogen-activated protein kinase signaling pathway in oral cancer. Oncol. Lett. 2018;15:1379–1388. doi: 10.3892/ol.2017.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papa S, Choy PM, Bubici C. The ERK and JNK pathways in the regulation of metabolic reprogramming. Oncogene. 2019;38:2223–2240. doi: 10.1038/s41388-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raman M, Chen W, Cobb M. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 6.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–8023. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 7.Roberts RL, Mosch HU, Fink GR. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/S0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- 8.de Dios CH, Roman E, Monge RA, Pla J. The role of MAPK signal transduction pathways in the response to oxidative stress in the fungal pathogen Candida albicans: implications in virulence. Curr. Protein Pept. Sci. 2010;11:693–703. doi: 10.2174/138920310794557655. [DOI] [PubMed] [Google Scholar]
- 9.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: an update. Arch. Toxicol. 2015;89:867–882. doi: 10.1007/s00204-015-1472-2. [DOI] [PubMed] [Google Scholar]
- 10.McGivern N, et al. Activation of MAPK signalling results in resistance to saracatinib (AZD0530) in ovarian cancer. Oncotarget. 2018;9:4722–4736. doi: 10.18632/oncotarget.23524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smalley I, Smalley KSM. ERK inhibition: a new front in the War against MAPK pathway-driven cancers? Cancer Discov. 2018;8:140–142. doi: 10.1158/2159-8290.CD-17-1355. [DOI] [PubMed] [Google Scholar]
- 12.Alory C, Balch WE. Organization of the Rab-GDI/CHM superfamily: the functional basis for choroideremia disease. Traffic. 2001;2:532–543. doi: 10.1034/j.1600-0854.2001.20803.x. [DOI] [PubMed] [Google Scholar]
- 13.McCaffrey G, Clay FJ, Kelsay K, Sprague GF., Jr Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:2680–2690. doi: 10.1128/MCB.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brizzio V, et al. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:609–626. doi: 10.1091/mbc.10.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwartz MA, Madhani HD. Principles of map kinase signaling specificity in Saccharomyces cerevisiae. Annu. Rev Genet. 2004;38:725–748. doi: 10.1146/annurev.genet.39.073003.112634. [DOI] [PubMed] [Google Scholar]
- 16.Karunanithi S, et al. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS ONE. 2012;7:e32294. doi: 10.1371/journal.pone.0032294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno T, Masuda Y, Irie K. The Saccharomyces cerevisiae AMPK, Snf1, negatively regulates the Hog1 MAPK pathway in ER stress response. Plos Genet. 2015;11:e1005491. doi: 10.1371/journal.pgen.1005491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasserre JP, et al. Yeast as a system for modeling mitochondrial disease mechanisms and discovering therapies. Dis. Model Mech. 2015;8:509–526. doi: 10.1242/dmm.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.19Dohlman, H. G. & Slessareva, J. E. Pheromone signaling pathways in yeast. Sci. STKE : Signal Transduction Knowl. Environ.2006, cm6 (2006). [DOI] [PubMed]
- 20.Martin SG. Molecular mechanisms of chemotropism and cell fusion in unicellular fungi. J. Cell Sci. 2019 doi: 10.1242/jcs.230706. [DOI] [PubMed] [Google Scholar]
- 21.Saito H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 2010;13:677–683. doi: 10.1016/j.mib.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 23.Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-R. [DOI] [PubMed] [Google Scholar]
- 24.Cullen PJ, Sprague GF., Jr Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Gene Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 26.Cook JG, Bardwell L, Thorner J. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 27.Madhani HD, Styles CA, Fink GR. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/S0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 28.Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc. Natl. Acad. Sci. 1998;95:15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bardwell L, et al. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Gene Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewster JL, de Valoir T, Dwyer ND, Winter E, Gustin MC. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 31.Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 1994;14:4135–4144. doi: 10.1128/MCB.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev.: MMBR. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 34.Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bharucha N, et al. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol. Biol. Cell. 2008;19:2708–2717. doi: 10.1091/mbc.e07-11-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin R, Dobry CJ, McCown PJ, Kumar A. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell. 2008;19:284–296. doi: 10.1091/mbc.e07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harashima T, Heitman J. The Galpha protein Gpa2 controls yeast differentiation by interacting with kelch repeat proteins that mimic Gbeta subunits. Mol. Cell. 2002;10:163–173. doi: 10.1016/S1097-2765(02)00569-5. [DOI] [PubMed] [Google Scholar]
- 38.Pan X, Heitman J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Mol. Cell. Biol. 2002;22:3981–3993. doi: 10.1128/MCB.22.12.3981-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosch HU, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhani HD, Fink GR. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 41.Madhani HD. Interplay of intrinsic and extrinsic signals in yeast differentiation. Proc. Natl. Acad. Sci. 2000;97:13461–13463. doi: 10.1073/pnas.011511198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kron SJ. Filamentous growth in budding yeast. Trends Microbiol. 1997;5:450–454. doi: 10.1016/S0966-842X(97)01131-1. [DOI] [PubMed] [Google Scholar]
- 43.Pan X, Harashima T, Heitman J. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 2000;3:567–572. doi: 10.1016/S1369-5274(00)00142-9. [DOI] [PubMed] [Google Scholar]
- 44.Tatebayashi K, et al. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 2006;25:3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 46.Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000;113 (Pt 3):365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- 48.Kachroo AH, et al. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/MMBR.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziman M, et al. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol. Biol. Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chol K-Y, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 53.Printen JA, Sprague GF. Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marcus S, Polverino A, Barr M, Wigler M. Complexes between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc. Natl. Acad. Sci. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zarrinpar A, Bhattacharyya RP, Nittler MP, Lim WA. Sho1 and Pbs2 act as coscaffolds linking components in the yeast high osmolarity MAP kinase pathway. Mol. Cell. 2004;14:825–832. doi: 10.1016/j.molcel.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharyya RP, et al. The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 57.Bao MZ, Schwartz MA, Cantin GT, Yates JR, III, Madhani HD. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 2004;119:991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 58.Chou S, Huang L, Liu H. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 2004;119:981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 59.Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–686. doi: 10.1126/science.1198701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witzel F, Maddison L, Bluthgen N. How scaffolds shape MAPK signaling: what we know and opportunities for systems approaches. Front. Physiol. 2012;3:475. doi: 10.3389/fphys.2012.00475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chavel CA, Caccamise LM, Li BY, Cullen PJ. Global regulation of a differentiation MAPK pathway in yeast. Genetics. 2014;198:1309–1320. doi: 10.1534/genetics.114.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sopko R, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 63.64Douglas, A. C. et al. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda)2, 1279–1289. http://doi.org/10.1534/g3.112.003400 (2012). [DOI] [PMC free article] [PubMed]
- 64.Roberts CJ, et al. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 2000;287:873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- 65.Maleri S, et al. Persistent activation by constitutive Ste7 promotes Kss1-mediated invasive growth but fails to support Fus3-dependent mating in yeast. Mol. Cell. Biol. 2004;24:9221–9238. doi: 10.1128/MCB.24.20.9221-9238.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gelperin DM, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Gene Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCaffrey G, Clay FJ, Kelsay K, Sprague G. Identification and regulation of a gene required for cell fusion during mating of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2680–2690. doi: 10.1128/MCB.7.8.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horecka J, Sprague GF., Jr Use of imidazoleglycerolphosphate dehydratase (His3) as a biological reporter in yeast. Methods Enzymol. 2000;326:107–119. doi: 10.1016/s0076-6879(00)26049-7. [DOI] [PubMed] [Google Scholar]
- 69.Cullen PJ, et al. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 2004;18:1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pitoniak A, et al. Cdc42p-interacting protein Bem4p regulates the filamentous-growth mitogen-activated protein kinase pathway. Mol. Cell Biol. 2015;35:417–436. doi: 10.1128/Mcb.00850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishimura A, et al. Scaffold protein Ahk1, which associates with Hkr1, Sho1, Ste11, and Pbs2, inhibits cross talk signaling from the Hkr1 osmosensor to the Kss1 mitogen-activated protein kinase. Mol. Cell. Biol. 2016;36:1109–1123. doi: 10.1128/MCB.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanazawa S, Driscoll M, Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol. Cell. Biol. 1988;8:664–673. doi: 10.1128/MCB.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alifano P, et al. Histidine biosynthetic pathway and genes: structure, regulation, and evolution. Microbiol. Rev. 1996;60:44–69. doi: 10.1128/MR.60.1.44-69.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barr MM, Tu H, Van Aelst L, Wigler M. Identification of Ste4 as a potential regulator of Byr2 in the sexual response pathway of Schizosaccharomyces pombe. Mol. Cell Biol. 1996;16:5597–5603. doi: 10.1128/mcb.16.10.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neiman AM, Herskowitz I. Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc. Natl. Acad. Sci. USA. 1994;91:3398–3402. doi: 10.1073/pnas.91.8.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lutfiyya LL, et al. Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics. 1998;150:1377–1391. doi: 10.1093/genetics/150.4.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Karunanithi S, Cullen PJ. The filamentous growth MAPK pathway responds to glucose starvation through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics. 2012;192:869–887. doi: 10.1534/genetics.112.142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J, et al. MARRVEL: integration of human and model organism genetic resources to facilitate functional annotation of the human genome. Am. J. Human Genet. 2017;100:843–853. doi: 10.1016/j.ajhg.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.81Skrzypek, M. S. & Hirschman, J. Using the Saccharomyces Genome Database (SGD) for analysis of genomic information. Current protocols in bioinformatics35, 1.20. 21–21.20. 23 (2011). [DOI] [PMC free article] [PubMed]
- 81.Coordinators NR. Database resources of the national center for biotechnology information. Nucl. Acids Res. 2018;46:D8. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alliance of Genome Resources Portal Unified model organism research platform. Nucl. Acids Res. 2020;48:D650–D658. doi: 10.1093/nar/gkz813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mungall CJ, et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucl. Acids Res. 2017;45:D712–D722. doi: 10.1093/nar/gkw1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.85Hu, Y., Comjean, A., Mohr, S. E., Perrimon, N. & Consortium, F. Gene2Function: an integrated online resource for gene function discovery. G3: Genes, Genomes, Genetics7, 2855–2858 (2017). [DOI] [PMC free article] [PubMed]
- 85.Feaver WJ, et al. Yeast TFIIE. Cloning, expression, and homology to vertebrate proteins. J. Biol. Chem. 1994;269:27549–27553. [PubMed] [Google Scholar]
- 86.Alory C, Balch WE. Molecular evolution of the Rab-escort-protein/guanine-nucleotide-dissociation-inhibitor superfamily. Mol. Biol. Cell. 2003;14:3857–3867. doi: 10.1091/mbc.e03-04-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alory C, Balch WE. Molecular basis for Rab prenylation. J. Cell Biol. 2000;150:89–103. doi: 10.1083/jcb.150.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 89.Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. Multiple signals converge on a differentiation MAPK pathway. Plos Genet. 2010;6:e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Madhani HD, Galitski T, Lander ES, Fink GR. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA. 1999;96:12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loeb JD, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Maeda T, Wurgler-Murphy SM, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 94.Sprague GF, Jr, Blair LC, Thorner J. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu. Rev. Microbiol. 1983;37:623–660. doi: 10.1146/annurev.mi.37.100183.003203. [DOI] [PubMed] [Google Scholar]
- 95.Adhikari H, Cullen PJ. Metabolic respiration induces AMPK- and Ire1p-dependent activation of the p38-Type HOG MAPK pathway. Plos Genet. 2014;10:e1004734. doi: 10.1371/journal.pgen.1004734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benito-Moreno RM, Miaczynska M, Bauer BE, Schweyen RJ, Ragnini A. Mrs6p, the yeast homologue of the mammalian choroideraemia protein: immunological evidence for its function as the Ypt1p Rab escort protein. Curr. Genet. 1994;27:23–25. doi: 10.1007/BF00326574. [DOI] [PubMed] [Google Scholar]
- 97.Singh J, Tyers M. A Rab escort protein integrates the secretion system with TOR signaling and ribosome biogenesis. Gene Dev. 2009;23:1944–1958. doi: 10.1101/gad.1804409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park H-O, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moriya H. Quantitative nature of overexpression experiments. Mol. Biol. Cell. 2015;26:3932–3939. doi: 10.1091/mbc.E15-07-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ryan O, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 101.Fields S, Song O-K. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 102.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J. Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Allen JB, Walberg MW, Edwards MC, Elledge SJ. Finding prospective partners in the library: the two-hybrid system and phage display find a match. Trends Biochem. Sci. 1995;20:511–516. doi: 10.1016/S0968-0004(00)89119-7. [DOI] [PubMed] [Google Scholar]
- 104.Bauer BE, et al. Amino- and carboxy-terminal domains of the yeast Rab escort protein are both required for binding of Ypt small G proteins. Mol. Biol. Cell. 1996;7:1521–1533. doi: 10.1091/mbc.7.10.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miaczynska M, et al. The yeast Rab escort protein binds intracellular membranes in vivo and in vitro. J. Biol. Chem. 1997;272:16972–16977. doi: 10.1074/jbc.272.27.16972. [DOI] [PubMed] [Google Scholar]
- 106.Lamber EP, Siedenburg AC, Barr FA. Rab regulation by GEFs and GAPs during membrane traffic. Curr. Opin. Cell Biol. 2019;59:34–39. doi: 10.1016/j.ceb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 107.Sidorovitch V, Niculae A, Kan N, Ceacareanu AC, Alexandrov K. Expression of mammalian Rab Escort protein-1 and -2 in yeast Saccharomyces cerevisiae. Protein Expr. Purif. 2002;26:50–58. doi: 10.1016/S1046-5928(02)00506-5. [DOI] [PubMed] [Google Scholar]
- 108.Fujimura K, Tanaka K, Nakano A, Toh-e A. The Saccharomyces cerevisiae MSI4 gene encodes the yeast counterpart of component A of Rab geranylgeranyltransferase. J. Biol. Chem. 1994;269:9205–9212. [PubMed] [Google Scholar]
- 109.Bialek-Wyrzykowska U, et al. Low levels of Ypt protein prenylation cause vesicle polarization defects and thermosensitive growth that can be suppressed by genes involved in cell wall maintenance. Mol. Microbiol. 2000;35:1295–1311. doi: 10.1046/j.1365-2958.2000.01782.x. [DOI] [PubMed] [Google Scholar]
- 110.TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. The EMBO journal. 1996;15:6483–6494. doi: 10.1002/j.1460-2075.1996.tb01039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang X, et al. Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem. 2001;276:46745–46750. doi: 10.1074/jbc.M107464200. [DOI] [PubMed] [Google Scholar]
- 112.Tsvetanova NG, Riordan DP, Brown PO. The yeast Rab GTPase Ypt1 modulates unfolded protein response dynamics by regulating the stability of HAC1 RNA. PLoS Genet. 2012;8:e1002862. doi: 10.1371/journal.pgen.1002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adhikari H, et al. Role of the unfolded protein response in regulating the mucin-dependent filamentous-growth mitogen-activated protein kinase pathway. Mol. Cell. Biol. 2015;35:1414–1432. doi: 10.1128/MCB.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas LL, Fromme JC. Extensive GTPase crosstalk regulates Golgi trafficking and maturation. Curr. Opin. Cell Biol. 2020;65:1–7. doi: 10.1016/j.ceb.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Witter DJ, Poulter CD. Yeast geranylgeranyltransferase type-II: steady state kinetic studies of the recombinant enzyme. Biochemistry. 1996;35:10454–10463. doi: 10.1021/bi960500y. [DOI] [PubMed] [Google Scholar]
- 116.Lempiainen H, et al. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell. 2009;33:704–716. doi: 10.1016/j.molcel.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 117.Shively CA, et al. Genetic networks inducing invasive growth in Saccharomyces cerevisiae identified through systematic genome-wide overexpression. Genetics. 2013;193:1297–1310. doi: 10.1534/genetics.112.147876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Peter M, Neiman A, Park H, Van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. doi: 10.1002/j.1460-2075.1996.tb01096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Leberer E, et al. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.123Pesce, G. et al. Cell-to-cell variability in the yeast pheromone response: Cytoplasmic microtubule function stabilizes signal generation and promotes accurate fate choice. bioRxiv, 093195 (2016).
- 121.125Basu, S. et al. Functions for Cdc42p BEM Adaptors in Regulating a Differentiation-Type MAP Kinase Pathway. Mol Biol Cell, mbcE19080441. http://doi.org/10.1091/mbc.E19-08-0441 (2020). [DOI] [PMC free article] [PubMed]
- 122.Kunkel J, Luo X, Capaldi AP. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat. Commun. 2019;10:3558. doi: 10.1038/s41467-019-11540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scott KL, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459:1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yuan ZL, et al. Central role of the threonine residue within the p+1 loop of receptor tyrosine kinase in STAT3 constitutive phosphorylation in metastatic cancer cells. Mol. Cell Biol. 2004;24:9390–9400. doi: 10.1128/MCB.24.21.9390-9400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zebisch A, et al. Signaling through RAS-RAF-MEK-ERK: from basics to bedside. Curr. Med. Chem. 2007;14:601–623. doi: 10.2174/092986707780059670. [DOI] [PubMed] [Google Scholar]
- 126.Li L, et al. The Ras/Raf/MEK/ERK signaling pathway and its role in the occurrence and development of HCC. Oncol. Lett. 2016;12:3045–3050. doi: 10.3892/ol.2016.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.131Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular cloning: a laboratory manual. (Cold spring harbor laboratory press, 1989).
- 128.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gietz RD, Schiestl RH. Microtiter plate transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:5–8. doi: 10.1038/nprot.2007.16. [DOI] [PubMed] [Google Scholar]
- 131.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 132.Prabhakar A, Chow J, Siegel AJ, Cullen PJ. Regulation of intrinsic polarity establishment by a differentiation-type MAPK pathway. J. Cell Sci. 2020 doi: 10.1242/jcs.241513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jenness D, Goldman B, Hartwell L. Saccharomyces cerevisiae mutants unresponsive to alpha-factor pheromone: alpha-factor binding and extragenic suppression. Mol. Cell Biol. 1987;7:1311–1319. doi: 10.1128/MCB.7.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Basu S, et al. Spatial landmarks regulate a Cdc42-dependent MAPK pathway to control differentiation and the response to positional compromise. Proc. Natl. Acad. Sci. USA. 2016;113:E2019–2028. doi: 10.1073/pnas.1522679113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lee MJ, Dohlman HG. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr. Biol. 2008;18:211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.140Gietz, R. D. & Woods, R. A. in Methods in enzymology Vol. 350 87–96 (Elsevier, 2002). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.