Abstract
Background
Glioblastoma stem cells (GSCs) are a subpopulation of glioblastoma (GBM) cells that are critical for tumor invasion and treatment resistance. However, little is known about the function and mechanism of tripartite motif-containing 24 (TRIM24) in GSCs.
Methods
Immunofluorescence, flow cytometry, and western blot analyses were used to evaluate TRIM24 and cluster of differentiation (CD)133 expression profiles in GBM surgical specimens and GSC tumorspheres. Different TRIM24 expression levels in patients’ tumors, as measured by both immunohistochemistry and western blot, were related to their corresponding MRI data. Wound healing, Matrigel invasion, and xenograft immunohistochemistry were conducted to determine GBM cell invasion.
Results
We identified that TRIM24 was coexpressed with CD133 and Nestin in GBM tissues and tumorsphere cells. Limiting dilution assays and xenotransplantation experiments illustrated that knockdown of TRIM24 expression reduced GSC self-renewal capacity and invasive growth. TRIM24 expression levels were positively associated with the volumes of peritumoral T2 weighted image abnormality. Rescue experiments indicated TRIM24 participation in GBM infiltrative dissemination. Chromatin immunoprecipitation, reporter gene assay, PCR, western blot, and immunohistochemistry demonstrated that TRIM24 activated the expression of the pluripotency transcription factor sex determining region Y–box 2 (Sox2) to regulate GBM stemness and invasion in vitro and in vivo. Finally, the close relationship between TRIM24 and Sox2 was validated by testing samples enrolled in our study and exploring external databases.
Conclusions
Our findings uncover essential roles of the TRIM24–Sox2 axis in GBM stemness and invasiveness, suggesting TRIM24 as a potential target for effective GBM management.
Keywords: glioblastoma, invasion, stemness, Sox2, TRIM24
Key Points.
1. TRIM24 regulates GBM stemness via driving Sox2 expression.
2. TRIM24 promotes GBM invasiveness through Sox2.
Importance of the Study.
In the present work, we reveal that TRIM24 is especially enriched in GSCs and regulates GSC properties, including GBM propagation and invasion. Moreover, the clinical and experimental data establish a novel functional mechanism of TRIM24 in cancer stem cells involving activation of Sox2 expression. These findings suggest TRIM24 as an attractive therapeutic target for GBM management.
In the central nervous system, glioblastoma (GBM) is the most common malignant primary tumor, with approximately 6.8% five-year survival rate.1 The dismal prognosis of GBM patients is mainly due to cellular fractions capable of initiating tumor recurrence after multimodal therapies, including surgery, radiation, and chemotherapy.2 These tumor-initiating cells are referred to as GBM stem cells (GSCs), which preferentially reside in a perivascular microenvironment and play pivotal roles in the characteristic invasive growth of GBMs.3 GSCs exhibit features of neural stem cells (NSCs) but display enhanced competency to generate tumors compared with the bulk population within the GBM context.4 The cell fate directions were dictated by transcription factors, chromatin modulators, and associated molecular interactions.5 Induction of GSC differentiation might be an attractive therapeutic approach.
Tripartite motif-containing 24 (TRIM24; also known as transcription intermediary factor [TIF] 1 alpha) is the founding member of the TIF family, which shares a conserved structure with N-terminal tripartite motif (namely a RING [Really Interesting New Gene] domain, 2 B-box zinc fingers, and an associated coiled-coil region) and a C-terminal chromatin-binding unit consisting of tandem plant homeodomain (PHD) finger and bromodomain.6 This unique architecture renders TRIM24 close correlation with several cancers, where TRIM24 may switch its roles between activator and suppressor according to different tumor contexts.7 TRIM24 functions as a liver-specific tumor suppressor in mice,8 whereas it augments androgen receptor signaling in prostate cancer.9 By means of tandem PHD finger and bromodomain, TRIM24 binds specific sites of chromatin to promote breast cancer development.6 However, the functions of TRIM24 in tumors are largely uncovered, especially its molecular mechanisms in cancer stem cells. Interestingly, TRIM24 is readily detected in mouse fetal brains, and its overall levels in the brain reduce markedly after birth, but with exceptionally high expression persisting in the dentate gyrus,10 where NSCs usually reside.4 Here, we show that TRIM24 is especially overexpressed in GSCs and binds chromatin with its bromodomain to activate Sox2 expression, thereby promoting the stemness and invasiveness of GBMs.
Materials and Methods
Tumor Samples
Surgical samples were obtained from patients undergoing resection for newly diagnosed GBMs at the Department of Neurosurgery, Chinese PLA General Hospital. Specimens were classified as GBM isocitrate dehydrogenase (IDH) wildtype (n = 43) and GBM IDH-mutant (n = 8) by 2 pathologists according to World Health Organization (WHO) criteria.11 Written informed consent was obtained from each patient. This study was approved by the institutional review board of Chinese PLA General Hospital and performed in accordance with the Declaration of Helsinki.
Immunofluorescence
Surgical patient GBM sections and cultured cells were blocked with normal goat serum and permeabilized with 0.1% Triton X-100. Primary antibodies used were anti-TRIM24 (Bethyl, A300-815A), anti-TRIM24 (Santa Cruz, sc-271266X), anti-CD31, anti-CD133, anti-Nestin (Proteintech), anti–glial fibrillary acidic protein (GFAP), anti-Sox2 (Cell Signaling), anti–βⅢ tubulin, and anti-CNPase (Abcam). Secondary antibodies used were goat anti-rabbit Alexa Fluor 488 pre-absorbed immunoglobulin (Ig)G (Abcam) and goat anti-mouse Alexa Fluor 594 cross-absorbed IgG (Thermo Fisher). Nuclei were counterstained with Hoechst 33342 (Sigma).
Cell Culture
Fresh tumor samples were dissociated into single cells using StemPro Accutase (Gibco), filtered and resuspended in serum-free medium (DMEM/F12, 1x B-27, 20 ng/mL epidermal growth factor (EGF), 20 ng/mL basic fibroblast growth factor, and 1x GlutaMax) or serum-containing medium (DMEM, 10% fetal bovine serum [FBS]) as described.12 Briefly, GSC subpopulations were isolated by fluorescence-activated cell sorting (FACS) after 6–18 hours recovery in serum-free medium. Fragment crystallizable receptor blocking reagent, cluster of differentiation (CD)133/2 (293C3)–VioBright fluorescein isothiocyanate (FITC), and isotype control IgG2b-VioBright FITC antibodies (Miltenyi) were used according to the instructions. FACS was conducted with a FACSAria flow cytometer (BD Biosciences), and analysis was performed using FlowJo software (Tree Star). The GSC characteristics were validated by serial tumorsphere formation assay, FBS-induced differentiation assay, and in vivo limiting dilution assay. Analysis of mRNA profiles by RNA sequencing and reverse transcription quantitative PCR (RT-qPCR) analyses clustered mesenchymal GSCs (GBM-01, GBM-04, GBM-12, and GBM-37) and proneural GSCs (GBM-19).13 For TRIM24 staining, permeabilized cells were serially incubated with rabbit anti-TRIM24 (1:100) (Bethyl) and phycoerythrin-conjugated goat anti-rabbit IgG (Proteintech), and washed thoroughly between incubation steps. The human GBM cell lines U251 and U87MG and the lentivirus packing cell line 293T were purchased from China Infrastructure of Cell Line Resource and were maintained in DMEM supplemented with 10% FBS. All cell lines were regularly checked for being free of mycoplasma contamination by PCR and culture. The identities of U251, U87MG, and 293T cells were authenticated with short tandem repeat profiling, most recently by Genetic Testing Biotechnology Corporation (Suzhou, China). Only <10 passage cell lines were used in this study.
Immunohistochemistry
Tissue sections were incubated with anti-TRIM24 (Bethyl), anti-human vimentin (Abcam), anti–epidermal growth factor receptor (EGFR), and anti–EGFR variant (v)III (Cell Signaling). Nuclei were counterstained with hematoxylin. The intensity and percentage of positive cells were evaluated. TRIM24 levels were stratified as low expression (no staining or weak staining) and high expression (strong staining) by 2 investigators blinded to clinical data as described.14 All the corresponding fresh samples were also evaluated by western blot and RT-qPCR assays, and the grouping information of TRIM24 levels was consistent.
Lentivirus Infection
The lentiviral vector psi-LVRH1P (Genecopoeia, CSHCTR001-1-LVRH1P) was used to express short hairpin (sh)RNAs. Also purchased from Genecopoeia was the lentiviral vector containing Sox2 open reading frame (ORF) (EX-T2547-Lv105), TRIM24 ORF (EX-T7669-Lv105), EGFRvIII ORF (EX-A8348-Lv105), and empty control (EX-NEG-Lv105). The plasmids of shRNA-resistant wildtype TRIM24 (TRIM24-WT), TRIM24 deleted of the PHD-bromo domain (TRIM24-PHD-BromoΔ), and TRIM24 mutated in bromodomain binding sites (TRIM24-F979A,N980A) were generated by the Phusion site-directed mutagenesis kit (Thermo Fisher). These lentiviral expressing vectors were co-transfected with packing plasmids psPAX2 and pMD2.G (Addgene) (4:3:1) into 293T cells to produce lentivirus. Tumor cells were infected in the presence of 8 µg/mL polybrene (Sigma), and 2 µg/mL puromycin (Sigma) selection was applied.
Tumorsphere Formation Assay
To assess tumorsphere formation capability, cells were seeded at a density of 1 cell per 4 µL serum-free medium in 24-well or 96-well plates. After 10–28 days of culture, tumorspheres consisting of more than 20 cells were scored. For the limiting dilution assay, differently infected cells were seeded in 96-well plates at increasing numbers per well. After 10 days, the percentage of wells not containing tumorspheres was calculated and plotted against plating cell numbers.
Xenotransplantation
Five-week-old female Bagg albino/c nude mice were employed for xenotransplantation experiments and all procedures were conducted in accordance with the protocols approved by the Institutional Animal Care and Use Committee of the Academy of Military Medical Sciences. Tumorsphere cells in 5 µL phosphate buffered saline were stereotactically implanted into the right frontal cortex. Injection coordinates were 1 mm rostral to bregma, 2 mm lateral, and 2.5 mm deep. All transplanted mice were monitored for 90 days or until development of neurologic signs.
Western Blot
Primary antibodies used were anti-TRIM24 (Bethyl), anti-CD133, anti-Nestin, anti-actin (Proteintech), anti-GFAP, anti-Sox2, anti-Notch1, anti–phosphorylated (p)Akt(Ser473), anti-Akt, anti-EGFR, anti–phosphorylated signal transducer and activator of transcription 3 (pSTAT3)(Tyr705), anti-STAT3 (Cell Signaling), anti-pEGFR(Y1173), and inhibitor of DNA binding 1 protein (ID1) (Abcam). After incubation with horseradish peroxidase–labeled secondary antibodies, chemiluminescence and X-ray films were used to develop blots. Each immunoblot was done at least thrice and the signals were quantified with ImageJ software (NIH).
Real-Time PCR Analyses
Total RNA was extracted using RNAiso Plus and reverse transcribed into cDNA by the PrimeScript RT reagent kit (Takara). PCR reactions were performed using TB Green Premix Ex TaqⅡ (Tli RNaseH Plus) (Takara) on an ABI 7500 system. The threshold cycle values for each gene were normalized to the values of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). For validation, the expression values of actin beta (ACTB) were also employed in a similar set of experiments. Primer sequences are listed in Supplementary Table 1.
Published mRNA Expression Datasets
Analyzed as described12 were GBM-TCGA-Firehose Legacy, CGGA-mRNAseq_325 (http://www.cgga.org.cn/), GSE67089,15 and GSE20736.16
Reporter Gene Assay
A lentiviral vector carrying Sox2 promoter luciferase (Genecopoeia, HPRM15202-LvPG04), which also contained the template of secreted alkaline phosphatase for normalization, was co-transfected with packing plasmids into 293T cells to produce lentivirus. Lentivirus supernatants were collected and added to tumor cells, concomitantly with other lentivirus infection if indicated. Three days after infection, a dual luminescence assay kit (Genecopoeia) was used to measure chemiluminescence on a Glomax 20/20 Luminometer (Promega). Each assay was done in triplicate and all the experiments were repeated 3 times.
Chromatin Immunoprecipitation
The precleared chromatin was incubated overnight with 4 µg anti-TRIM24 (Bethyl) or normal rabbit IgG (Millipore). Chromatin immunoprecipitated (ChIPed) DNA was amplified for 30 PCR cycles using specific primers and analyzed by agarose gel electrophoresis. ChIPed DNA was also quantified by real-time PCR analyses. Primer sequences are listed in Supplementary Table 1.
MR Image Analyses
Peritumoral fluid attenuated inversion recovery/T2 weighted image (FLAIR/T2WI) hyperintensity (defined as edema/invasion) and post-contrast T1WI data (enhancement segmentation defined as tumor; region within the tumor without enhancement defined as necrosis) were analyzed by employing 3D Slicer software 4.10.2 (www.slicer.org) as described.17
Wound Healing and Invasion Assays
For wound healing assay, confluent cells were scratched by a sterile 200 µL tip to make a homogeneous wound. Cultures were washed and medium was replaced with serum-free DMEM. Wound borders were captured just after scratches and 24 hours later. Coverage areas were quantified using ImageJ software. For transwell invasion assay, 2.5 × 104 serum-starved cells were seeded into the upper compartment of a Matrigel-coated 8-um pore Boyden chamber and the lower compartment was filled with DMEM containing 10% FBS. After 16–24 hours incubation, the percentage of invaded cells was calculated. For tumorsphere invasion assay, GSC spheroids were embedded into Matrigel matrix (Corning) supplemented with 40% culture medium and 10% FBS, and plated in 96-well plates precoated with Matrigel. Sprouting of tumorspheres was monitored daily and the spread areas were assessed using ImageJ software.
Statistical Analysis
Values in the bar diagrams are shown as means ± SE or ± SD, representing data obtained from at least 3 independent experiments. If normal distribution and homogeneity of variances were both achieved, comparisons among several groups or between 2 groups were performed with a 2-tailed ANOVA test or Student’s t-test. Otherwise, the Mann–Whitney test was employed to assess the differences. The Kaplan–Meier method was employed to draw the survival curves, and the log-rank test was used for significance determination. The correlation between 2 factors was assessed by Pearson correlation analysis if normal distribution of data was fulfilled in both of them. Otherwise, the correlation was presented by the Spearman coefficient value. All statistics were analyzed using SPSS 17.0 software, and P < 0.05 was considered significant.
Results
TRIM24 Is Especially Enriched in GSCs
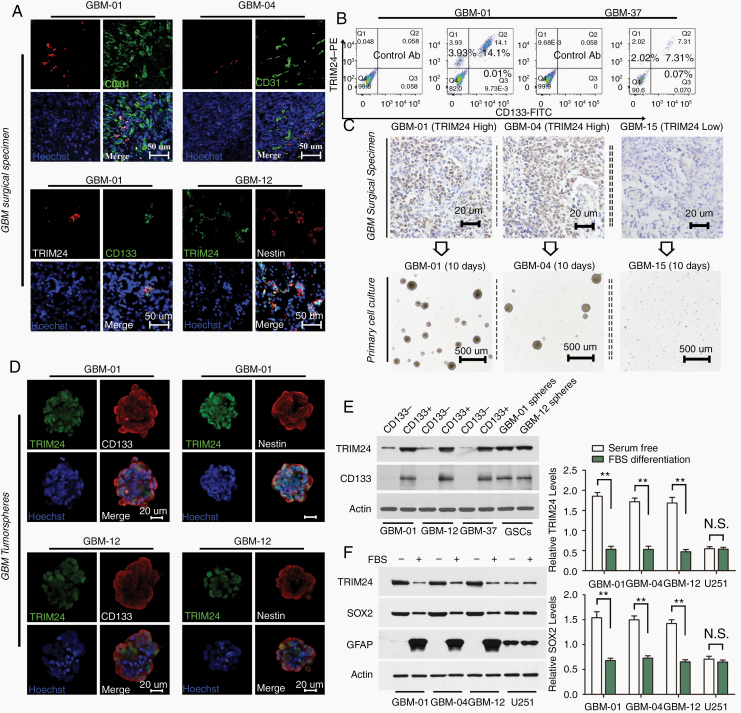
Since GSCs preferentially reside in perivascular niches, to evaluate a possible relationship between TRIM24 and GSCs we labeled GBM surgical specimens with antibodies against TRIM24 and endothelial marker CD31. Immunofluorescence analyses showed strong TRIM24 staining in a fraction of GBM cells in perivascular regions. Besides, TRIM24 was generally coexpressed with stem cell markers, including CD133 and Nestin (Fig. 1A). Flow cytometry analyses confirmed that TRIM24 and CD133 expression overlapped in a fraction of GBM cell population (Fig. 1B).
Fig. 1.
TRIM24 is especially enriched in GSCs. (A) Immunofluorescence analyses were employed to determine TRIM24 expression in the perivascular compartment (blood vessels double stained with CD31) and TRIM24 coexpression with stem cell markers (CD133 and Nestin) in GBM surgical sections. Representative images derived from 15 GBM samples (5 cases per double staining experiment). Scale bars represent 50 µm. (B) Flow cytometry analyses were used to determine the overlap of TRIM24 and CD133 staining in primary isolated GBM cells. Representative results derived from 7 cases. (C) The expression levels and subcellular localization of TRIM24 were evaluated in patient GBM specimens by immunohistochemistry (n = 51). Their corresponding isolated cells were plated in stem cell permissive medium. Scale bars represent 20 µm in the upper panels and 500 µm in the lower panels. (D) Immunofluorescence analyses of TRIM24, CD133, and Nestin staining in tumorsphere cells. Scale bars represent 20 µm. (E) Western blot analyses of FAC-sorted primary cells and GSC tumorspheres. (F) Western blot analyses of GSCs and GBM cell lines cultured in different media for 4 days. **P < 0.01; N.S., not significant.
To assess TRIM24 expression with regard to tumorsphere formation ability, an indicator of self-renewal, we cultured total surgical GBM cells in stem cell permissive medium. Tumor cells of TRIM24 low expressing cases (n = 14) could not be propagated, whereas tumor cells obtained from TRIM24 highly expressed cases (n = 37) were able to propagate at least 5 passages (Fig. 1C). Similar to GBM surgical specimen sections, TRIM24, CD133, and Nestin were coexpressed in tumorsphere cells (Fig. 1D; Supplementary Figure 1). Moreover, tumorsphere cells possessed multilineage differentiation and tumor formation capacity (Supplementary Table 2; Supplementary Figure 2), thus fulfilling the functional definition of GSCs,18 which are frequently identified in malignant gliomas (Supplementary Figure 3).
Western blot analyses demonstrated that FAC-sorted CD133+ cells possessed high TRIM24 expression, compared with CD133 cells (Figure 1E; Supplementary Figure 4B). GSCs growing under differentiation conditions showed remarkable reduction of TRIM24 expression, whereas no significant change in TRIM24 expression levels was observed in GBM cell lines U251 and U87MG, which were continuously cultured in the presence of FBS. Additionally, GSCs and NSCs showed much higher levels of endogenous TRIM24 compared with GBM cell lines and normal astrocytes (Figure 1F; Supplementary Figure 4A and C).
TRIM24 Is Critical for the Stemness of Glioblastoma Cells
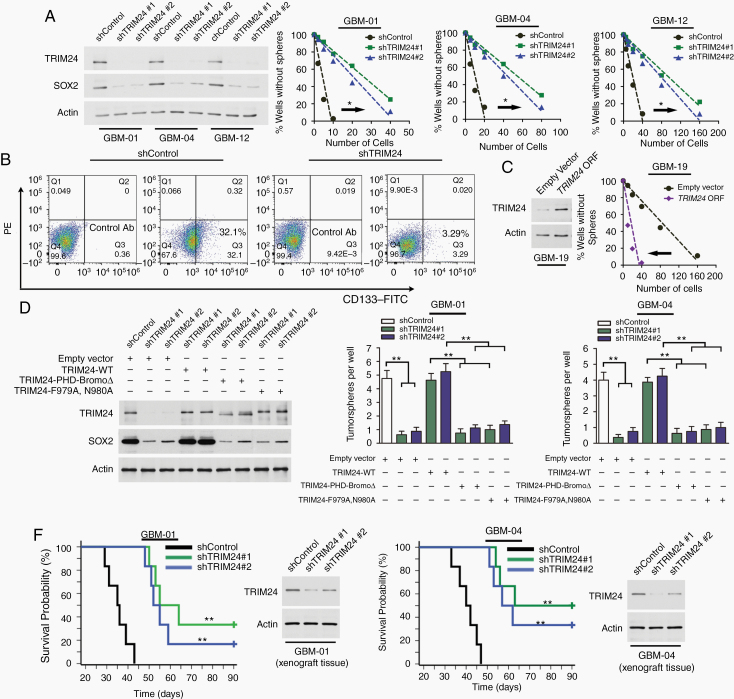
Given that TRIM24 was enriched in GBM cells endowed with GSC properties, we hypothesized that TRIM24 might promote GSC maintenance. Limiting dilution assays showed that knockdown of TRIM24 expression dramatically abrogated tumorsphere formation capability of GSCs (Fig. 2A). TRIM24 knockdown in primary isolated GBM cells from fresh surgical specimens reduced the CD133+ subpopulation (Fig. 2B). On the other hand, overexpression of TRIM24 increased sphere-forming capability of GBM-19 tumorsphere cells and the CD133+ subpopulation of primary isolated GBM-19 cells compared with the control groups (Figure 2C; Supplementary Figure 5). In addition, the crippled tumorsphere formation ability caused by TRIM24 knockdown could be rescued by reintroducing shRNA-resistant TRIM24-WT, but not restored by reexpression of deletion mutant TRIM24-PHD-BromoΔ or disrupted bromodomain mutant TRIM24-F979A,N980A (Fig. 2D). To further interrogate the function of TRIM24 in vivo, we transplanted differently treated GSCs into the brains of nude mice. Compared with nontargeting control GSCs, TRIM24 knockdown impeded tumor formation and greatly prolonged animal survival time (Figure 2E; Supplementary Figure 6).
Fig. 2.
TRIM24 is critical for the stemness of GBM cells. (A) Linear regression analysis was used to evaluate the limiting dilution assays of tumorsphere cells infected with different shRNA expressing lentivirus. *P < 0.05. (B) Flow cytometry analysis of CD133 expression in viable primary isolated GBM-01 cells. Shown are representative results of 3 independent experiments. (C) Limiting dilution analysis of GBM-19 tumorsphere cells infected with TRIM24 ORF containing lentivirus or empty control lentivirus. *P < 0.05. (D) After TRIM24 knockdown and rescue by reexpression of shRNA-resistant wildtype TRIM24 or mutants if indicated, GSCs were seeded and tumorspheres were scored. Representative immunoblots of GSCs derived from GBM-01 cells are shown and similar blots exist in GBM-04 study. Columns, mean; bars, SE; **P < 0.01. (E) Kaplan–Meier method was employed to demonstrate the survival curves of xenograft transplanted nude mice (1000 GSCs per animal, n = 6 per group). The log-rank test was used to determine statistical significance. **P < 0.01 between shControl and shTRIM24 groups.
TRIM24 Impacts GBM Stemness via Driving Sox2 Expression
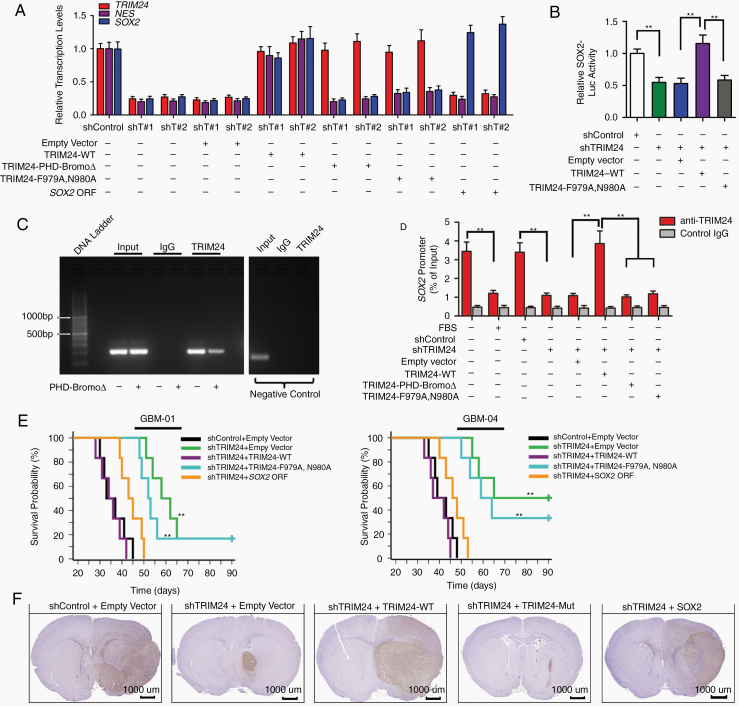
Considering that the bromodomain of TRIM24 has been linked to chromatin binding and gene expression activation,7 we performed RNA sequencing and real-time reverse transcription (RT) PCR analyses, and focused on possible regulators essential for GBM propagation.5 We found that transcriptional levels of Sox2 were significantly downregulated upon TRIM24 knockdown (Figure 3A; Supplementary Figure 7A). Dual luminescence assay demonstrated that TRIM24 knockdown significantly reduced the relative luciferase activity of Sox2 promoter reporter, which was incorporated into the host genome by employing lentiviral infection, and the reduction could be restored by reexpression of TRIM24-WT but not by TRIM24-F979A,N980A (Fig. 3B). On the other hand, TRIM24 overexpression increased the relative luciferase activity of Sox2 promoter reporter using lentiviral infection, whereas no significant change was observed by employing plasmid transfection, further indicating the participation of chromatin histone proteins (Supplementary Figure 7B, C). Moreover, ChIP assays revealed TRIM24-WT binding to Sox2 promoter while TRIM24 mutants did not (Fig. 3C, D). Besides, the binding was downregulated by TRIM24 depletion or FBS-induced differentiation (Figure 3D; Supplementary Figure 7D). Functional analyses demonstrated that the compromised capabilities of in vitro tumorsphere formation and in vivo tumor formation upon TRIM24 depletion could be rescued by reintroduction of Sox2 (Figure 3E; Supplementary Figure 8). These results suggest that TRIM24 activates Sox2 expression to regulate the self-renewal and tumor formation of GSCs. Immunohistochemistry assay using an antibody against human vimentin demonstrated that control GSCs infiltrate both into mouse brain parenchyma and along white matter tracts. Interestingly, TRIM24-downregulated tumor cells formed oval-shaped grafts with well-demarcated edges (Figure 3F; Supplementary Figure 6A, B; Supplementary Figure 9). Immunofluorescence analyses illustrated that TRIM24(+)Nestin(+) GSCs mainly located at the boundary area between the xenograft and normal mouse brain (Supplementary Figure 6C).
Fig. 3.
TRIM24 impacts GBM stemness via driving Sox2 expression. (A) Real-time reverse transcription (RT) PCR analyses of gene expression profiles in GSCs. Shown are relative levels of TRIM24, Nestin (encoded by NES), and Sox2 in GSCs derived from GBM-01 cells. (B) Dual luminescence assay was employed to measure the relative activity of Sox2 promoter after lentiviral infection. Shown are the data of GSCs derived from GBM-01 cells. **P < 0.01. (C) ChIP analysis of the Sox2 promoter using standard PCR in GSCs derived from GBM-01 cells. Shown are gel electrophoresis images using primers corresponding to the site that presents the most binding affinity (-544 to -362 fragment in the Sox2 promoter). The results of negative control KIAA0066 are also presented. (D) Real-time PCR analyses of the ChIPed DNA using primers to amplify the fragment encompassing the -544 to -362 site of Sox2 promoter in GSCs derived from GBM-01 cells. **P < 0.01. (E) Kaplan–Meier method was employed to demonstrate the survival curves of orthotopic transplanted nude mice (1000 GSCs per animal, n = 6 per group). The log-rank test was used to determine statistical significance. P < 0.01 between shTRIM24 + empty vector and shTRIM24 + TRIM24-WT group; P < 0.01 between shTRIM24 + TRIM24-WT and shTRIM24 + TRIM24-F979A,N980A group; P < 0.01 between shTRIM24 + empty vector and shTRIM24 + Sox2 ORF group. **P < 0.01. (F) Differently treated GSCs derived from GBM-01 cells were transplanted into nude mice (1000 cells per animal, n = 5 per group) and all the mice were sacrificed when development of neurologic signs was observed in any group. Representative immunohistochemical images of orthotopic xenografts are shown. Scale bars represent 1 mm. Expanded view of images is presented in Supplementary Figure 9.
TRIM24 Expression Levels Correlate with T2-weighted MRI Abnormality
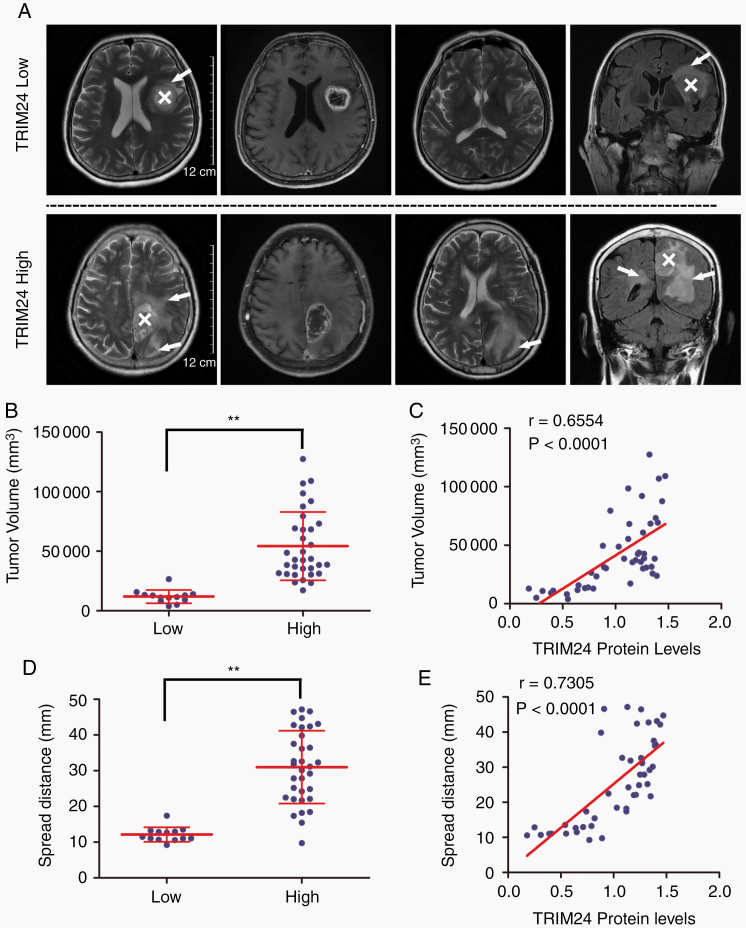
Given that extensive invasive growth is an important factor for poor prognosis of GBM cases and TRIM24 impacts histologic features of xenotransplantation, we attempted to place TRIM24 expression levels in the clinical context. Two independent investigators, who were blinded to the above illustrated in vitro and in vivo experiments, evaluated MRI images of GBM cases enrolled in the study. Regions with peritumoral high T2-weighted signals correspond to edema, likely due to cytokines secreted by invasive tumor cells or leakage from tumor-induced abnormal neovasculature.19 Five cases were excluded from this analysis due to lack of MRI data. GBM cases with high TRIM24 expression (n = 33, all IDH-wildtype cases) presented larger volumes of peritumoral T2WI abnormality compared with low expression cases (n = 13, comprising 6 IDH-wildtype cases and 7 IDH-mutant cases) (Fig. 4A, B). A significant correlation between TRIM24 expression and tumor volume was also identified (Fig. 4C). Besides, similar results were observed by measuring the maximal spread distance in these cases (Fig. 4D, E).
Fig. 4.
TRIM24 expression levels correlate with T2-weighted MRI abnormality. (A) Representative MRI scans of GBM cases with low and high TRIM24 expression levels. White “X” marks the main tumor mass and white arrows indicate invasive regions. Scale bars represent 12 cm. (B) Distribution of peritumoral T2WI abnormality volumes grouped by low (n = 13) and high (n = 33) TRIM24 expression levels. Data are presented as means ± SD. **P < 0.01. (C) Correlation of TRIM24 expression levels (determined by western blot assays) with peritumoral T2WI abnormality volumes. The Pearson coefficient value is presented. (D) Distribution of maximal spread distances grouped by TRIM24 levels. Data are presented as means ± SD. **P < 0.01. (E) Correlation of TRIM24 expression levels (determined by western blot assays) with maximal spread distances. The Pearson coefficient value is presented.
TRIM24 Promotes GBM Invasiveness In Vitro and In Vivo Through Sox2
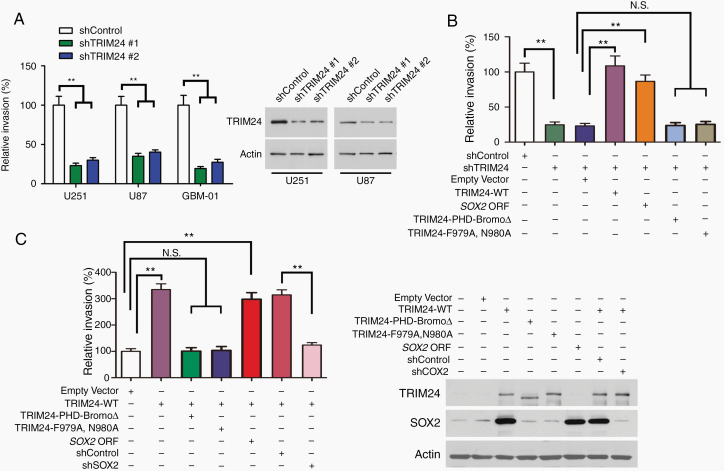
The overexpression of TRIM24 in highly invasive GBMs prompted us to deeply investigate the effects of TRIM24 on cell migration and invasion. Wound healing assays revealed that the migration of bulk GBM cells was suppressed by TRIM24 knockdown or enhanced by TRIM24 upregulation (Supplementary Figure 10). Transwell assays demonstrated that knockdown of TRIM24 expression caused a reduction in the invasion of primary isolated GBM cultures and well-established GBM cell lines (Fig. 5A, B). On the other hand, upregulation of TRIM24 expression increased the invasive ability of GBM cells (Fig. 5C). Then we embedded differently treated tumorspheres into serum-containing Matrigel matrix. The area covered by spread cells was reduced upon TRIM24 knockdown while increased upon TRIM24 overexpression (Supplementary Figure 11). Next, we orthotopically implanted differently infected cells into nude mice. The latency until neurologic symptom onset was drastically shortened in mice bearing TRIM24 upregulated cells (Fig. 6A). Immunohistochemistry analyses of xenograft sections showed that TRIM24 overexpression increased the infiltrative dissemination of GBM cells, illustrated by fingerlike protrusions into brain parenchyma and malignant scatters along white matter tracts (Figure 6B; Supplementary Figure 12). Considering that Sox2 had been reported to enhance the invasion of GSCs and GBM cell lines,12,20 we conducted a series of rescue experiments. Transwell assays showed that the decreased invasion of GBM cells upon TRIM24 knockdown was rescued by reintroducing Sox2 or TRIM24-WT (Fig. 5B). Similarly, the decreased invasion of GSC tumorsphere cells upon TRIM24 knockdown could be rescued by reintroducing Sox2 or TRIM24-WT, but not by TRIM24 mutants (Supplementary Figure 11A). On the other hand, the invasive phenotype of TRIM24 upregulated cells was inhibited by Sox2 silencing in vitro and in vivo (Figure 5C, Figure 6B, Supplementary Figure 11B, Supplementary Figure 12).
Fig. 5.
TRIM24 promotes GBM invasion in vitro through Sox2. (A) Transwell Matrigel invasion assays of primary isolated GBM-01 cells and established GBM cell lines. Values are normalized to shControl group for each cell line. **P < 0.01. (B) Transwell Matrigel invasion assays of primary isolated GBM-01 cells infected with different sets of lentiviruses. **P < 0.01; N.S., not significant. (C) Transwell Matrigel invasion assays of primary isolated GBM-19 cells. **P < 0.01; N.S., not significant.
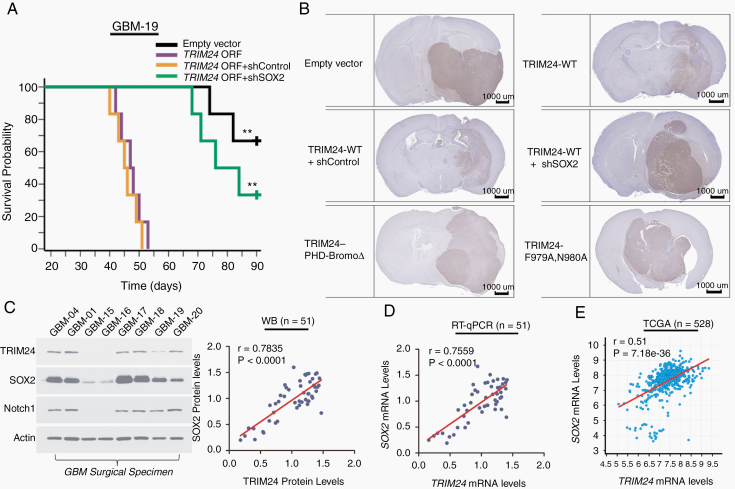
Fig. 6.
TRIM24 promotes GBM invasion in vivo through Sox2 and TRIM24–Sox2 correlation exists in GBM samples. (A) Differently treated GSCs derived from GBM-19 cells were transplanted into nude mice (5000 cells per animal, n = 6 per group). The log-rank test was used to determine differences between Kaplan–Meier survival curves. P < 0.01 between empty vector and TRIM24 ORF group; P < 0.01 between TRIM24 ORF + shControl and TRIM24 ORF + shSox2 group. **P < 0.01. (B) Representative immunohistochemical images of GBM-19 xenografts are shown. Scale bars represent 1 mm. Expanded view of images is presented in Supplementary Figure 12. (C) Western blot analyses of 51 GBM subjects enrolled in the study. The association between TRIM24 and Sox2 protein levels was evaluated by Pearson correlation analyses. Shown are representative blots of 8 subjects. (D) RT-qPCR analyses of TRIM24 and Sox2 mRNA levels in GBM subjects of the study. Their association was evaluated by Pearson correlation analyses. (E) Correlation between TRIM24 and Sox2 mRNA levels in GBM-TCGA-Firehose Legacy using U133 microarray data. The Pearson coefficient value is presented.
TRIM24–Sox2 Correlation Exists in Clinical Context
To further explore the relationship between TRIM24 and Sox2 in the clinical context, we evaluated their expression levels in surgical specimens. Western blot analyses showed a significant correlation between TRIM24 and Sox2 protein levels (Fig. 6C). Their close association was also evident in transcriptional levels by employing RT-qPCR analyses and exploring external databases (Figure 6D, E; Supplementary Figure 13A). Moreover, immunofluorescence analyses demonstrated that TRIM24 and Sox2 staining strongly overlapped in tumor sections, further establishing their close correlation in GBMs (Supplementary Figure 13B).
Discussion
Here, we reveal a novel signaling pathway whereby TRIM24 activates Sox2 expression in GSCs, thus promoting stemness and invasiveness of GBM cells. In embryonic stem cells, TRIM24 acts as a ubiquitin-protein ligase for p53 degradation to antagonize differentiation or shift p53 conformation to avoid cellular transformation.21,22 Moreover, TRIM24 converges with pluripotency transcription factors on multiple enhancers in mouse embryonic stem cells, although the majority of TRIM24 targeted genes are reported to be unlikely dependent on p53.23 In our study, TRIM24 also seems to have p53-independent effects on GSCs, because postsurgical immunohistochemistry indicates that GBM-01 cells express mutant p53, and the bromodomain (not the RING domain) is essential for intact function of TRIM24 as demonstrated by rescue experiments. In chronic myeloid leukemia, TRIM24 is found to be overexpressed in the CD34-positive compartment, which putatively encompasses the leukemia stem cell population.24 Considering the biological similarity between normal stem cells and cancer stem cells,4 even if not mentioning the putative origin of the latter and cellular hierarchies superimposed upon underlying genetic aberrations,25 the roles of TRIM24 in various stem cells might be further explored in the not-so-distant future.
The functional mechanisms of TRIM24 regulation in GSCs can be mediated by Sox2, which is not only a stem cell marker but also a master member in the core set of neurodevelopment transcription factors that are crucial for GSC propagation.5 In the field of cancers, since the weight of a specific signaling or certain pathway differs considerably among diverse tumors, TRIM24 may switch its roles between oncogene and tumor suppressor, thus ultimately dictating the direction in which the balance will be tipped. For example, TRIM24 suppresses development of hepatocellular carcinoma in mice by repressing VL30 retrotransposons.26 On the other hand, TRIM24 promotes progression of breast cancer6 and prostate cancer.9 Nevertheless, TRIM24–Sox2 regulation is unlikely to be dependent on phosphatidylinositol-3 kinase/Akt signaling in GSCs derived from GBM cells (Supplementary Figure 14A).14,27 Besides, TRIM24 participation in EGFR/STAT3/ID1 signaling28 seems largely dependent on continuously activated EGFR status (relative high EGFR expression in GBM-01, GBM-04, and GBM-12, while low EGFR expression in GBM-19) (Supplementary Figure 14B, C). Moreover, we did not observe a significant correlation between TRIM24 and ID1 mRNA levels in external databases (Supplementary Figure 14D and F). In addition, ID1 expression levels increase upon FBS-induced differentiation in GSCs (Supplementary Figure 14E), whereas TRIM24 and Sox2 levels reduce upon differentiation (Figure 1F; Supplementary Figure 4C). Therefore, TRIM24–Sox2 regulation seems unlikely to be dependent on EGFR/STAT3/ID1 signaling in GSCs derived from GBM cells, but they could work cooperatively in GBM cells with highly activated EGFR (especially with EGFRvIII). Interestingly, our findings indicate that GSCs are endowed with escalated levels of endogenous TRIM24, compared with nonstem GBM cells or the bulk population (Figure 1E, F; Supplementary Figure 4). GBMs commonly display remarkable inter- and intratumoral heterogeneity. One possible scenario might be that TRIM24 highly enriched GSCs, acting as pioneers, invade into brain parenchyma to seek out blood vessels or opt for white matter tracts, where Notch1 ligands reside (Fig. 6C).12 Subsequently, the escalated TRIM24 expression tempers to some extent, although it remains higher than normal brain glia, and the residual TRIM24 continues to fuel in situ growth of the bulk population of GBM cells.14 However, high-accuracy in vivo monitoring of exquisitely labeled GSCs would directly illustrate this interesting dynamic evolution. Since Sox2 functions in stem cell maintenance,29 TRIM24–Sox2 regulation might be an important signaling in cancer stem cells. However, experiments using stem cells derived from other body systems are warranted to validate this hypothesis.
Recently, TRIM24 targeting modalities have been explored. Functional TRIM24 degrader has been constructed via conjugation of ineffectual bromodomain and VHL E3 ubiquitin ligases, and has shown therapeutic effect on leukemia.30 Besides, benzimidazolone bromodomain inhibitors show suppressive effect on HeLa cells and myeloma cells.7 These treatment approaches, especially small-molecule inhibitors, might be employed for the management of GBMs, which usually present characteristic infiltrative dissemination and evade traditional therapies due to the blood–brain barrier. In summary, our study establishes the functions of TRIM24 in GBM propagation and invasion through a novel mechanism involving activation of Sox2 expression, suggesting TRIM24 as a potential target for GBM treatment.
Supplementary Material
Acknowledgments
We would like to thank Zhao-Tao Wang from the Second Affiliated Hospital of Guangzhou Medical University for helpful suggestions on cell culture experiments. We also want to thank Yao Chen, Gui-Ling Cai, Qing Liu, and Ai-Xue Huang from the Academy of Military Medical Sciences for helpful assistance in molecular experiments.
Funding
This work was supported by the National Natural Science Foundation of China (81502186 to L.Z., 81571350 to X.Y., 81770857 to D.X.) and the China Postdoctoral Science Foundation (2018M633730 to L.Z.).
Conflict of interest statement. The authors declare no conflict of interest.
Authorship statement. LZ, XY, and DX designed this work; LZ, YY, HC, SF, JL, LC, WF, and GS helped or performed experiments and analyses; YY, HC, SF, JL, LC, WF, GS, and DX helped in preparation of the manuscript; LZ and XY wrote the manuscript.
References
- 1. Ostrom QT, Cioffi G, Gittleman H, et al. . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol. 2019;21(Supplement_5):v1–v100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldape K, Brindle KM, Chesler L, et al. . Challenges to curing primary brain tumours. Nat Rev Clin Oncol. 2019;16(8):509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cuddapah VA, Robel S, Watkins S, Sontheimer H. A neurocentric perspective on glioma invasion. Nat Rev Neurosci. 2014;15(7):455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jung E, Alfonso J, Osswald M, Monyer H, Wick W, Winkler F. Emerging intersections between neuroscience and glioma biology. Nat Neurosci. 2019;22(12):1951–1960. [DOI] [PubMed] [Google Scholar]
- 5. Suvà ML, Rheinbay E, Gillespie SM, et al. . Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157(3):580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai WW, Wang Z, Yiu TT, et al. . TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468(7326):927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Appikonda S, Thakkar KN, Barton MC. Regulation of gene expression in human cancers by TRIM24. Drug Discov Today Technol. 2016;19:57–63. [DOI] [PubMed] [Google Scholar]
- 8. Herquel B, Ouararhni K, Khetchoumian K, et al. . Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2011;108(20):8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Groner AC, Cato L, de Tribolet-Hardy J, et al. . TRIM24 Is an oncogenic transcriptional activator in prostate cancer. Cancer Cell. 2016;29(6):846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Niederreither K, Remboutsika E, Gansmuller A, Losson R, Dollé P. Expression of the transcriptional intermediary factor TIF1alpha during mouse development and in the reproductive organs. Mech Dev. 1999;88(1):111–117. [DOI] [PubMed] [Google Scholar]
- 11. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Xu SL, Duan JJ, et al. . Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1-SOX2 positive-feedback loop. Nat Neurosci. 2019;22(1):91–105. [DOI] [PubMed] [Google Scholar]
- 13. Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin. 2010;60(3): 166–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang LH, Yin AA, Cheng JX, et al. . TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene. 2015;34(5):600–610. [DOI] [PubMed] [Google Scholar]
- 15. Chandran UR, Luthra S, Santana-Santos L, et al. . Gene expression profiling distinguishes proneural glioma stem cells from mesenchymal glioma stem cells. Genom Data. 2015;5:333–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nogueira L, Ruiz-Ontañon P, Vazquez-Barquero A, et al. . Blockade of the NFκB pathway drives differentiating glioblastoma-initiating cells into senescence both in vitro and in vivo. Oncogene. 2011;30(32): 3537–3548. [DOI] [PubMed] [Google Scholar]
- 17. Wangaryattawanich P, Hatami M, Wang J, et al. . Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression-free survival. Neuro Oncol. 2015;17(11):1525–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laug D, Glasgow SM, Deneen B. A glial blueprint for gliomagenesis. Nat Rev Neurosci. 2018;19(7):393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weller M, van den Bent M, Tonn JC, et al. ; European Association for Neuro-Oncology (EANO) Task Force on Gliomas European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017;18(6):e315–e329. [DOI] [PubMed] [Google Scholar]
- 20. Alonso MM, Diez-Valle R, Manterola L, et al. . Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS One. 2011;6(11):e26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain AK, Allton K, Iacovino M, et al. . p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012;10(2):e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rivlin N, Katz S, Doody M, et al. . Rescue of embryonic stem cells from cellular transformation by proteomic stabilization of mutant p53 and conversion into WT conformation. Proc Natl Acad Sci U S A. 2014;111(19):7006–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rafiee MR, Girardot C, Sigismondo G, Krijgsveld J. Expanding the circuitry of pluripotency by selective isolation of chromatin-associated proteins. Mol Cell. 2016;64(3):624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Quintás-Cardama A, Qiu YH, Post SM, et al. . Reverse phase protein array profiling reveals distinct proteomic signatures associated with chronic myeloid leukemia progression and with chronic phase in the CD34-positive compartment. Cancer. 2012;118(21):5283–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herquel B, Ouararhni K, Martianov I, et al. . Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat Struct Mol Biol. 2013;20(3):339–346. [DOI] [PubMed] [Google Scholar]
- 27. Lv D, Jia F, Hou Y, et al. . Histone acetyltransferase KAT6A upregulates PI3K/AKT signaling through TRIM24 binding. Cancer Res. 2017;77(22):6190–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lv D, Li Y, Zhang W, et al. . TRIM24 is an oncogenic transcriptional co-activator of STAT3 in glioblastoma. Nat Commun. 2017;8(1):1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bertolini JA, Favaro R, Zhu Y, et al. . Mapping the global chromatin connectivity network for Sox2 function in neural stem cell maintenance. Cell Stem Cell. 2019;24(3):462–476 e466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gechijian LN, Buckley DL, Lawlor MA, et al. . Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat Chem Biol. 2018;14(4):405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.