Abstract
Background
Glioblastoma (GBM) stemlike cells (GSCs) are thought to be responsible for the maintenance and aggressiveness of GBM, the most common primary brain tumor in adults. This study aims at elucidating the involvement of deregulations within the imprinted delta-like homolog 1 gene‒type III iodothyronine deiodinase gene (DLK-DIO3) region on chromosome 14q32 in GBM pathogenesis.
Methods
Real-time PCR analyses were performed on GSCs and GBM tissues. Methylation analyses, gene expression, and reverse-phase protein array profiles were used to investigate the tumor suppressor function of the maternally expressed 3 gene (MEG3).
Results
Loss of expression of genes and noncoding RNAs within the DLK1-DIO3 region was observed in GSCs and GBM tissues compared with normal brain. This downregulation is mainly mediated by epigenetic silencing. Kaplan–Meier analysis indicated that low expression of MEG3 and MEG8 long noncoding (lnc)RNAs significantly correlated with short survival in GBM patients. MEG3 restoration impairs tumorigenic abilities of GSCs in vitro by inhibiting cell growth, migration, and colony formation and decreases in vivo tumor growth, reducing infiltrative growth. These effects were associated with modulation of genes involved in cell adhesion and epithelial-to-mesenchymal transition (EMT).
Conclusion
In GBM, MEG3 acts as a tumor suppressor mainly regulating cell adhesion, EMT, and cell proliferation, thus providing a potential candidate for novel GBM therapies.
Keywords: cancer stem cells, chromosome 14q32, glioblastoma, MEG3 lncRNA
Key Points.
1. The expression of imprinted DLK-DIO3 region at chromosome 14q32 is downregulated in GSCs and GBM tissues compared with normal brain, mainly by epigenetic silencing.
2. Downregulation of MEG3 and MEG8 lncRNAs significantly correlates with a poor clinical outcome.
3. MEG3 functions as a tumor suppressor by regulating genes involved in cell adhesion and EMT.
Importance of the Study.
Despite current multimodal therapies, treatment of GBM poses a significant clinical challenge, and the life expectancy of GBM patients remains dismal. LncRNAs have emerged as important players in cancer. The present study provides evidence of the role of lncRNAs in GBM pathobiology. Our data show that MEG3 is involved in a complex network that regulates cell adhesion, DNA damage, cell proliferation, and stemness by targeting a multitude of genes. The analysis of biological processes affected by MEG3, coupled with functional in vitro and in vivo assays, identifies known as well as unexplored pathways that can be targeted by innovative therapies.
Glioblastoma (GBM) is the most frequent and aggressive primary adult brain tumor, with a 5.1% survival rate at 5 years.1 Even with the current multimodal therapy, the mean survival of GBM patients is still approximately 15 months. Because of its extremely unfavorable prognosis, it is imperative to develop more effective therapeutic strategies for this cancer.
The identification of GBM initiating stemlike cells (GSCs) has introduced a new paradigm in therapy, since these cells are likely to comprehend a population uniquely able to support tumor growth and should represent a primary therapeutic target.2 Analyzing a collection of patient-derived GSC lines by complementary molecular approaches, we previously identified 2 GSC clusters: one characterized by a proneural-like phenotype (GSf-like) and the other showing a mesenchymal-like phenotype (GSr-like).3 Furthermore, by investigating the GSC micro (mi)RNA profiles, we identified a set of 3 miRNAs able to discriminate GSf- and GSr-like GSC phenotypes as well as mesenchymal and proneural GBM patient subtypes with different clinical outcomes.4 In the same study we also noticed that, independently of GSC phenotypes, many of the miRNAs mapping to region q32.2 of chromosome 14 were downregulated compared with normal neural stem cells of both adult and fetal origin. This region, spanning from delta-like homolog 1 (DLK1) to type III iodothyronine deiodinase (DIO3), is subject to imprinting and plays an important role in development. Patients with imprinting defects in the DLK1-DIO3 region suffer from a range of abnormalities, including growth delay, skeletal malformations, developmental delay/intellectual disability, and even perinatal death.5,6
The 14q32 imprinted region is regulated by 2 differentially methylated regions (DMRs), the paternally methylated imprinting control region, called the InterGenic (IG)-DMR, and the somatic DMR within the maternally expressed 3 (MEG3) promoter.7
This region contains the paternally expressed imprinted genes DLK1, DIO3, and retrotransposon-like 1 (RTL1), the maternally expressed imprinted long noncoding (lnc)RNAs (MEG3, MEG8, MEG9 and LINC00524, and anti-sense RTL1), a large miRNA cluster (53 miRNAs), and 2 families of small nucleolar RNAs (SNORDs; SNORD113 and SNORD114 coding for 9 and 31 SNORDs, respectively).
Correlation between methylation patterns in the MEG3-DMR promoter and expression of noncoding (nc)RNAs and several miRNAs in the 14q32 cluster has been suggested.5 Deregulated expression of the ncRNAs from this region has been implicated in several human malignancies, including gliomas.8 Particularly, either the loss or the downregulation of MEG3 lncRNA is reported in an expanding list of human tumors of nervous origin, including neuroblastomas,9 meningiomas,10 and gliomas.8
While several studies recently explored the mode of action of MEG3 and its association with transcription factors and chromatin, we focused our attention on the cellular processes and pathways regulated by MEG3 lncRNA in the context of GBM pathophysiology.
Materials and Methods
Cell Cultures
GSCs were isolated from surgical samples of adult patients who underwent craniotomy at the Institute of Neurosurgery, Catholic University of Rome, upon patient informed consent and approval by the local ethical committee.11 Information of human adult neural stem cell (NSC) lines and human neural progenitor cell (HNPC) lines are described in the Supplementary Methods.
Real-Time PCR
Total RNAs were extracted from cells using TRIzol reagent (Life Technologies) and from microdissected paraffin embedded GBM sections using the miRNeasy FFPE kit (Qiagen) and reverse transcribed by Moloney murine leukemia virus reverse transcriptase (Life Technologies). Real-time (RT) PCR was performed with SYBR Green Master Mix in the StepOnePlus Real-Time PCR System (Applied Biosystems). Primers are listed in Supplementary Table 1.
Methylation Analysis
Methylation-specific multiple ligation probe amplification (MS-MLPA) analysis was performed by using the ME032 UPD7-UPD14 A1 Kit (MRC-Holland) according to manufacturer’s instructions. The assay we used does not evaluate the methylation status of DLK1 or RTL1.
Methylation profile (Infinium Methylation 450K, Illumina) was performed by Genomix4life (Laboratory of Molecular Medicine and Genomics, University of Salerno). The detailed methods are described in the Supplementary Methods.
Array-Based Comparative Genomic Hybridization
Array comparative genomic hybridization (array-CGH) was performed on GBM tissues using SurePrint G3 Human CGH Microarrays, 8x60K (Agilent) according to manufacturer’s instructions and scanned on a DNA Microarray Scanner (Agilent). Feature extracted data were then analyzed using CytoGenomics Software (Agilent).
Plasmid Constructs and Lentivirus Infection
The MEG3 variant 1 (NR_002766), from HNPCs was cloned in a GFP lentiviral vector. Primers used were: forward 5′-agcccctagcgcagacggcgga-3′ and reverse 5′-ttgttaagacaggaaacacatttattgagagcac-3′. Short hairpin (sh)MEG3 lentiviral green fluorescent protein (GFP) vectors (TL320132) were from OriGene Technologies. Lentiviral particles were produced in 293T packaging cell line and infection performed as previously described.12 After infection, GFP fluorescence was evaluated by FACSCanto (BD Biosciences) and GFP-positive cells were flow sorted by FACS ARIA (BD Biosciences).
Cell Growth, Migration, and Colony Formation Assays
Detailed information on viability assay, cell proliferation, motility, and colony formation ability are described in the Supplementary Methods.
Intracranial and Subcutaneous Implantation of Glioma Stemlike Cells in Immunocompromised Mice and Analysis of Brain Xenografts
Experiments involving animals were approved by the Ethical Committee of the Istituto Superiore di Sanità, Rome, Italy. For intracranial implantation, 4- to 6-week-old nonobese diabetic severe combined immunodeficient mice (Charles River) were implanted intracranially with 2 × 105 GFP-MEG3 or empty vector transduced GSC#1 cells (Supplementary Methods).13 For subcutaneous implantation, 4- to 6-week-old Hsd:athymic nude mice (Charles River) were implanted subcutaneously with 5 × 105 GSC#61 cells either overexpressing MEG3 or carrying the empty vector (see Supplementary Methods).
Gene Array
Total RNA, extracted from MEG3 and empty vector transduced cells, was labeled and hybridized to the Agilent-019118 array for miRNAs (Agilent) and Affymetrix GeneChip1.0ST array (Affymetrix) following the manufacturer’s instructions. Hybridization values were normalized by the RMA (robust multi-array average) method.
Reverse-Phase Protein Arrays
Cell lysates for reverse-phase protein lysate microarray (RPPA) analysis were performed as previously described.14 The list of antibodies selected for RPPA analysis is available in Supplementary Table 2. See the Supplementary Methods for details.
Statistical Methods
Detailed information of all the statistical analyses performed is described in the Supplementary Methods.
Results
Dysregulation of DLK1-DIO3 Transcripts in GBM and GSCs: Expression of MEG3 and MEG8 lncRNAs Is Associated to GBM Patient Overall Survival
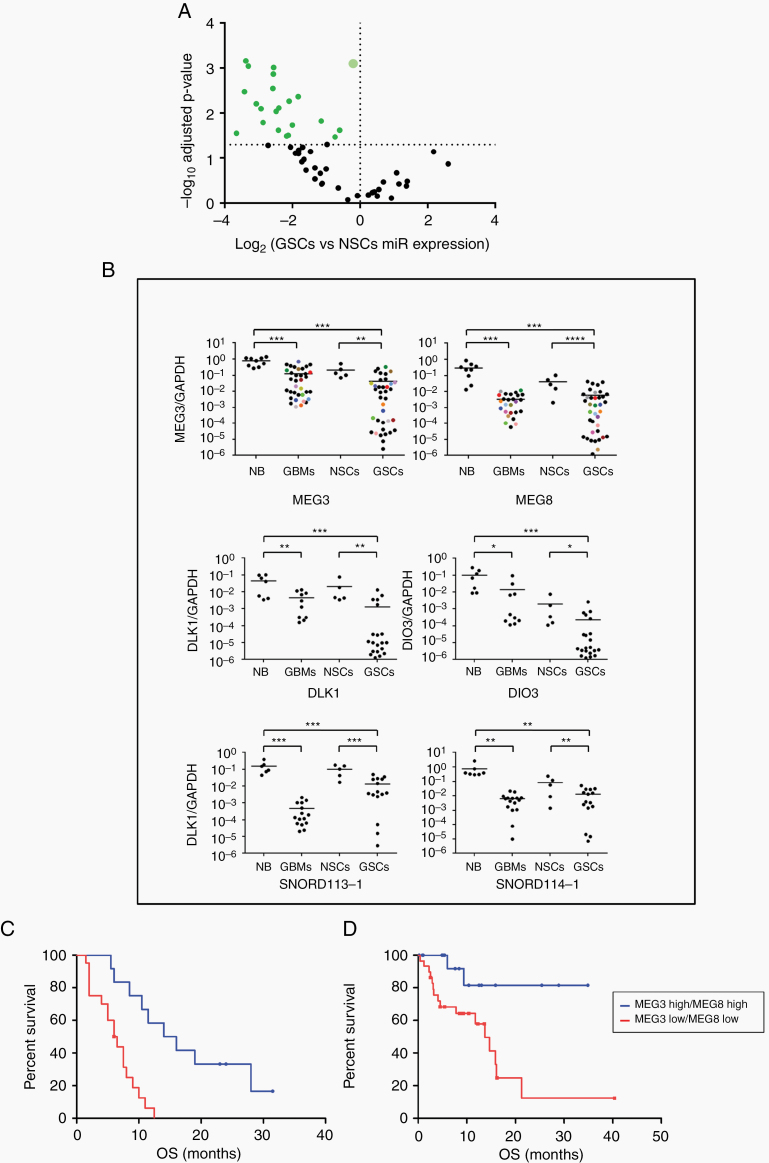
By investigating the miRNA profiles by gene expression microarrays, we found that miRNAs from a large cluster on chromosome 14q32 were significantly downregulated in GSCs compared with normal NSCs (P < 0.05, Student’s t-test) (Supplementary Table 3, Supplementary Figure 1A and Figure 1A). MiRNA microarray results were validated by RT-quantitative (q)PCR on an independent set of GSC lines (n = 10) (Supplementary Figure 1B). Analyzing the expression of genes and ncRNAs located on the 14q32 imprinted region by RT-qPCR, we found that the DLK1 and DIO3 genes, SNORD113-1, SNORD114-1, MEG3, and MEG8 were significantly downregulated in GSCs and tumor tissue derived from GBM patients compared with normal brain and with NSCs (Figure 1B). Noteworthy, MEG3 and MEG8 expression levels are directly correlated in both GSCs and GBM (Spearman correlation coefficients 0.81 and 0.75, respectively; P < 0.0001) (Supplementary Figure 2). Moreover, MEG3 expression, although significantly lower than in normal brains, appears to be heterogeneous among GSCs (coefficient of variation [CV] 0.53 in normal brain and 1.86 in GSCs, CV ratio 3.51) (Figure 1B). These results suggested that deregulated expression of the transcripts contained in the DLK1-DIO3 region could be involved in GBM pathogenesis.
Fig. 1.
Expression of gene transcripts from DLK1-DIO3 region: MEG3 and MEG8 downregulation correlates with patient overall survival. (A) Volcano-plot representing the expression level of miRNAs on chromosome 14q32 from GSCs (n = 9) compared with normal neural stem cells (NSCs) from both adult (olfactory bulb) and fetal origin (n = 3). The bigger plot represents the 8 miRNA samples with the same value (miRs -433, -432*, -380*, 323–5p, -300, -453, -496, -412). P-values are based on Student’s t-test and adjusted by Bonferroni correction. (B) Real time RT-PCR analysis performed on normal brain samples (NB), GBM tissues (GBM), NSCs, and GSCs. The colored points in MEG3 and MEG8 panels represent GBM tissue and GSCs derived from the same patient. Samples were run in duplicate and normalized with glyceraldehyde 3-phosphate dehydrogenase. No statistically significant differences (P = 0.30) were observed among paired samples. (C-D) Kaplan–Meier survival curves: MEG3 and MEG8 downregulation significantly correlates with a poor clinical outcome in our cohort of patients (C; P = 0.0002) and in patients from the glioblastoma database of TCGA (D; P = 0.0066).
Since lncRNAs have been implicated in glioma progression, we analyzed more in detail the heterogeneity of MEG3 and MEG8 expression in GSCs.
Thirty-five GSCs derived from GBM surgical specimens were assessed for the expression of MEG3 and MEG8 by RT-PCR. Clinical and pathological features are summarized in Supplementary Table 4. Survival analysis adjusted for relevant predictors as well as multivariate analysis for survival are shown in Supplementary Figure 3. Based on the expression value of MEG3 (median = 0.044) and MEG8 (median = 0.0063), GSCs were classified into 4 subgroups (ie, MEG3 high and MEG8 high; MEG3 high and MEG8 low; MEG3 low and MEG8 high; MEG3 low and MEG8 low). Kaplan–Meier analysis showed that patients whose tumors expressed low levels of both MEG3 and MEG8 had significantly shorter overall survival than patients with high expression of both MEG3 and MEG8 (P = 0.0002; hazard ratio [HR], 0.1818; 95% CI: 0.07422–0.4452; Figure 1C). Kaplan–Meier survival curves of MEG3 and MEG8 individually evaluated as prognostic factors did not significantly correlate with clinical outcome (Supplementary Figure 4). Our results were confirmed by an analysis performed on the database of The Cancer Genome Atlas (TCGA), using the University of California Santa Cruz Xena software (https://xena.ucsc.edu/). After downloading raw data and grouping the GBM into low and high expression groups, we found that patients with high expression of both MEG3 and MEG8 had a better prognosis compared with those showing low expression of both lncRNAs (n = 51; P = 0.0066; HR, 0.2700; 95% CI: 0.1049–0.6945; Figure 1D). Further, in silico analysis of the other ncRNAs within the DLK1-DIO3 region was not performed due to the limited availability of data on these transcripts in the database of TCGA.
Epigenetic Modifications Are the Main Regulator of Chromosome 14q32 Gene Expression in GSCs
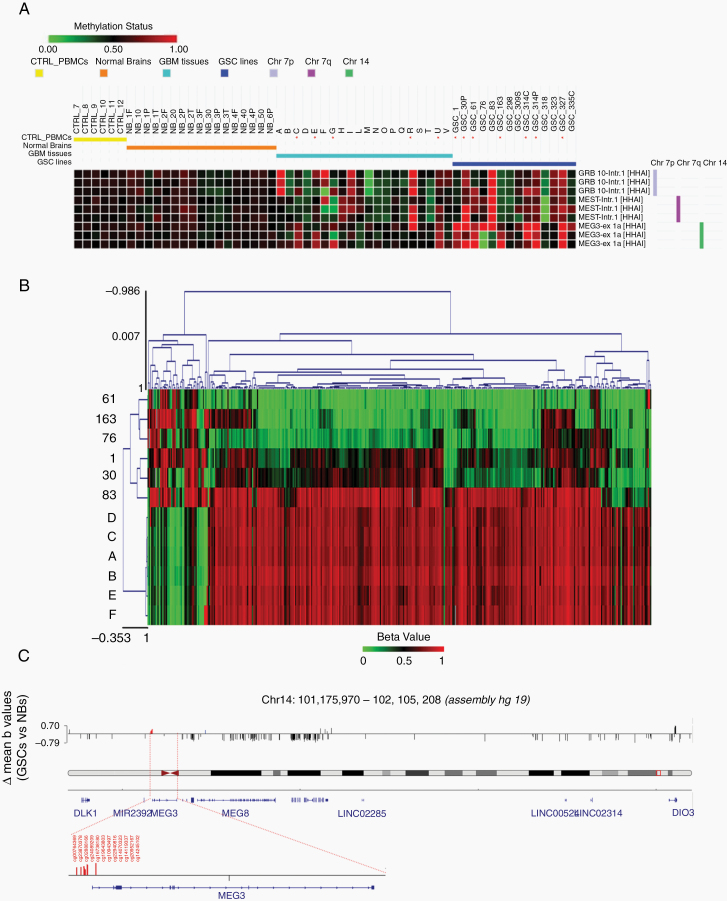
Multiple mechanisms may contribute to nullizygosity of the DLK1-DIO3 region in tumors, including genomic deletions, epigenetic modifications, or a combination of the two. To determine the type of derangement of this region in our cohort, we performed MS-MLPA analysis of 14 GSC lines, 20 GBM tissue specimens, and 6 normal brain tissues. Samples from normal brain and peripheral blood displayed the same monoallelic methylation pattern at the MEG3 locus (Figure 2A).
Fig. 2.
Analysis of methylation pattern of DLK1-DIO3 region. (A) Annotated heatmap of chromosome 7 and 14 probes informative for methylation status. MS-MLPA analysis was performed on HhaI digested DNA from 20 GBM tissue specimens, 14 GSC, 6 normal brains (NB) and 6 peripheral blood mononuclear cell (PBMC) samples from normal controls (CTRL). For NBs, the region of origin of the sample is shown: F = frontal, O = occipital, P = parietal, T = temporal; for some samples, multiple regions were examined. Probes are named by gene and position (Intr = intron, ex = exon). For further information about probe genomic positions, please refer to MRC-Holland Salsa MS-MLPA probemix ME032-A1 UPD7/UPD14—description version 05;10–12–2014. Hypermethylated samples are marked with an asterisk (*). (B) Heatmap generated starting from the beta values of the CpGs associated to the DLK1-DIO3 region, considering Illumina array 450k methylation data. The samples (A to F: Normal Brains; #1, #30p, #61, #76, #83 and #163: GSC lines) have been clustered using the Euclidean distance metric. (C) Integrative Genomics Viewer (IGV) screenshot of all significantly differentially methylated Infinium probes (P < 0.01) in the DLK1-DIO3 genomic region. In red are highlighted the 12 CpG probes within MEG3-DMR, while in blue is highlighted one CpG probe (cg16126137) within the putative MEG8-DMR.
Five out of the 20 GBMs (25%) displayed hypermethylation of MEG3. Hypermethylation was detected for at least 1 of the 3 methylation-sensitive MEG3 probes included in the MLPA kit. In 3 of these (C, E, and U), one MEG3 allele was deleted, as demonstrated by MS-MLPA analysis and confirmed by array-CGH (Supplementary Figure 5A, B). The deletion likely involved the maternal allele, since the allele retained in these samples was hypermethylated. The other 2 samples with MEG3 hypermethylation (G and R) did not show MEG3 copy number changes.
Of the 14 GSCs, 7 (50%) were hypermethylated at the 3 MEG3 cytosine-phosphate-guanine (CpG) islands analyzed. The discrepancy between tumors and cell lines could be due to the presence of a mixed cell population in tumors or to a consequence of prolonged in vitro culture.
However, the methylation pattern was similar between matched pairs of GBM samples and cell lines (Supplementary Figure 5C) and was consistent in most of the GSCs analyzed at early and late passages (ie, up to 10 and more than 20 passages, respectively) except for GSC#76, which displays differences limited to a single MEG3 probe. Overall, our data indicate that maintenance of GSCs in vitro did not affect the methylation status of MEG3 (Supplementary Figure 5D).
Since the data obtained with MLPA were limited to a restricted number of probes, we analyzed the methylation profile in 6 GSC lines (#61, #30p, #83, #163, #1, and #76) established from primary GBM specimens as well as in 6 human normal brain tissue samples (Infinium Methylation 450K, Illumina). The 6 GSCs were selected based on their MEG3 expression levels, GSCs #30p and #163 were chosen among the lowest expressing cells, GSCs #1 and #76 among the medium, and GSCs #61 and #83 among the highest (Supplementary Table 5).
Methylation profiles clearly separated GSCs and normal brain samples into discrete groups (Figure 2B and Supplementary Figure 6). Particularly, the analysis revealed a significant hypermethylation (P < 0.01) of 12 Infinium probes within the MEG3-DMR and of 1 probe (cg16126137) within the putative MEG8-DMR15 in all GSC samples analyzed compared with normal brain (Figure 2C and Supplementary Table 6). Nine of the 12 probes were present in the database of TCGA and covaried (first principal component explaining 73.8% of cumulative variance), implying a combined regulation of these 12 CpG methylation sites (Supplementary Table 7).
Starting from the methylation profiling obtained with Illumina array 450K, we considered the probes annotated on the genes of interest and extracted the associated beta values. These data were used to create the heatmaps, and Euclidean Distance Metric was applied to cluster the samples (Supplementary Figure 7). The obtained heatmaps confirmed a clear distribution of the samples between the 2 groups, and showed a higher methylation level for probes annotated on the MEG3 gene of the GSC samples compared with normal brain.
Interestingly, unlike the other GSC lines analyzed, GSC #76 showed poor level of MEG3 expression and low level of methylation (Supplementary Figure 8A). To assess whether other epigenetic modifications, such as histone acetylation, take part in MEG3 silencing, we treated the 6 profiled GSC lines with valproic acid (VPA), an inhibitor of histone deacetylases (classes I and IIa). VPA treatment resulted in a 1.3- to 2-fold increase of MEG3 levels in GSCs #1, #30p, #163, #61, and #83 while, in GSC #76 lacking constitutive MEG3 hypermethylation, the expression of MEG3 was 7-fold increased following VPA treatment (Supplementary Figure 8B), suggesting that diverse epigenetic modifications may contribute to MEG3 silencing.
An inverse correlation between MEG3 expression and methylation level (r = −0.68; P < 0.01) was observed indeed only in GSCs #1, #30p, #163, #61, and #83 (Supplementary Figure 8C).
Restoration of MEG3 Impairs Tumorigenic Properties of GSCs In Vitro and In Vivo
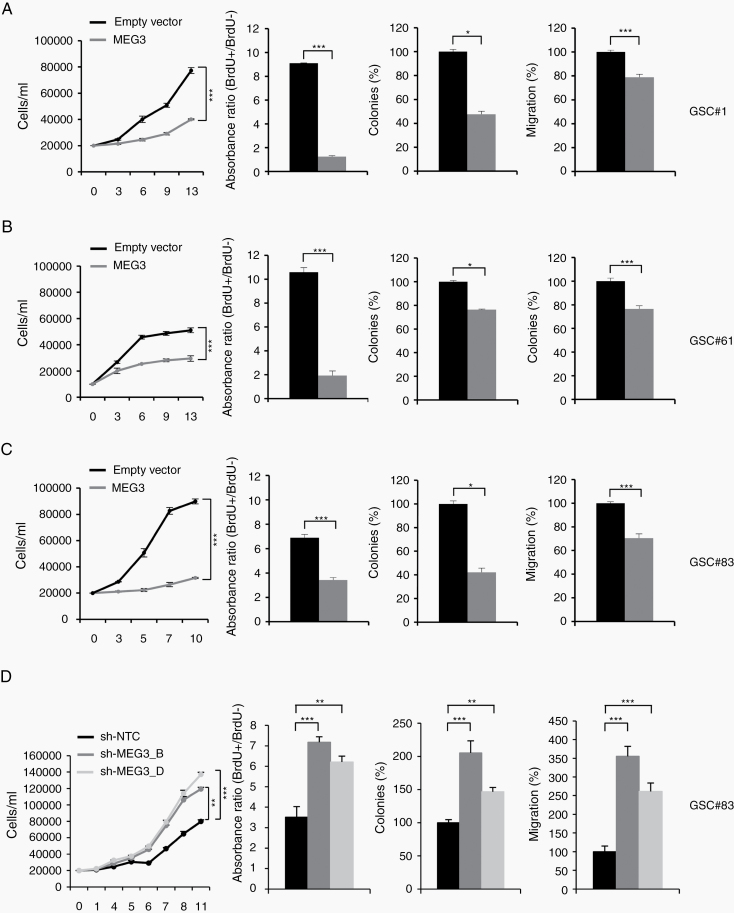
Since restoration of the whole DLK1-DIO3 genomic region was not feasible and accumulating evidence shows that MEG3 deregulation might be involved in gliomagenesis, we decided to focus our attention on MEG3 and performed enforced reexpression of MEG3 in GSCs. To this end we overexpressed MEG3 (NR_002766) in 3 GSC lines (#1, #61, #83), chosen for different MEG3 expression levels, by transducing a lentiviral vector carrying MEG3 and GFP as reporter genes used to select cells by flow cytometry sorting. LncRNA restoration was confirmed by RT-PCR (Supplementary Figure 9A).
Ectopic expression of MEG3 induced a significant, stable decrease in the growth rate of all GSC lines tested (Figure 3). In detail, MEG3 significantly reduced bromodeoxyuridine (BrdU) incorporation showing a decreased progression in the cell cycle through the S phase. Moreover, MEG3-GSCs formed significantly fewer colonies compared with empty vector transduced cells (Figure 3). The motility of MEG3-transduced GSCs was dramatically reduced (Figure 3). Thus, MEG3 restoration resulted in a considerable inhibition of proliferation, migration, and colony formation of all the GSCs tested.
Fig. 3.
MEG3 tuning modulates cell growth, migration and clonogenic abilities of GSCs. Functional in vitro assays on (A) GSC#1 (B) GSC#61, and (C) GSC#83 cell lines transduced with either empty or MEG3 vectors. Growth curves of GSCs (left panels). Points and range lines at each day represent mean and SD of at least 2 independent experiments in triplicate. Two-way analysis of variance for repeated measures was performed on the whole set of data. BrdU incorporation after 72 h pulse in empty vector and MEG3 transduced GSC lines (center-left panels). Absorbance (450–550 nm) has been shown as ratio between BrdU + and BrdU− wells. Values are reported as mean ± SD from 2 independent experiments in triplicate. Analysis of efficiency in colony formation of GSCs after transduction with MEG3 (center-right panels). Percent colony number values from 2 independent experiments in duplicate were calculated over the correspondent empty vector and are shown as mean ± SD for each GSC line. Analysis of variance demonstrated a significant effect of the restoration of MEG3 on the colony-forming ability of GSCs. Analysis of migration efficiency in GSCs transduced with MEG3 48 h after induction (right panels). Values are reported as percent relative to control vector and shown as mean ± SD from 2 independent experiments in duplicate. (D) Functional in vitro assays on GSC#83 transduced with sh-NTC, sh-MEG3_B or sh-MEG3_D vectors.
These results were implemented by MEG3 silencing. To this end, 2 shMEG3 lentiviral GFP constructs were transduced in the MEG3 highly expressing line GSC#83. MEG3 silencing, verified by RT-PCR (Supplementary Figure 9B), significantly increased proliferation, migration, and colony formation of GSC#83 (Figure 3D).
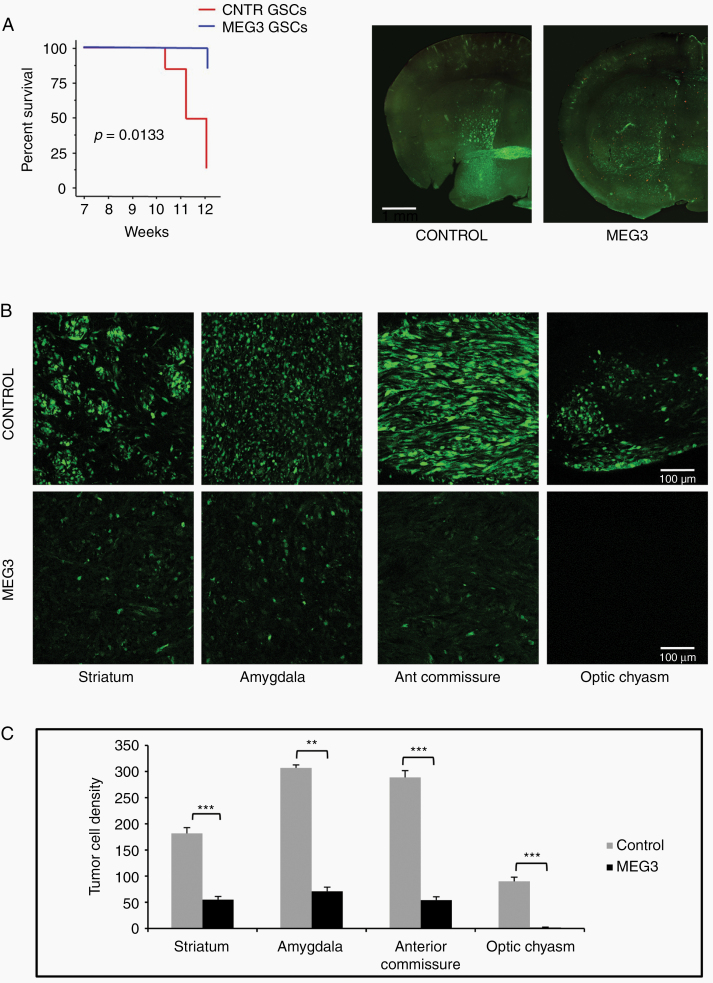
Following orthotopic injection into immunocompromised mice, patient-derived GSCs generate highly infiltrative tumors that closely reproduce the parent neoplasm.14 Therefore, we utilized this model to test in vivo the effects of MEG3 overexpression on the growth of brain tumors. Kaplan–Meier analysis of mice outcome showed that mice grafted with MEG3-GSCs did significantly better than those grafted with control GSCs (n = 12, P = 0.0133 log-rank test) (Figure 4A). At 12 weeks after grafting, mice grafted with control GSCs (n = 6) harbored tumors that invaded extensively the striatum, piriform cortex, and amygdaloid area and spread through the corpus callosum, anterior commissure, optic chiasm, septal nuclei, and fimbria hippocampus. In mice grafted with MEG3-GSCs (n = 6), the degree of brain invasion was highly reduced, as demonstrated by the significant reduction of tumor cell density in the striatum, amygdaloid area, anterior commissure, and optic chiasm (Figure 4B, C).
Fig. 4.
Effect of MEG3 over-expression on the growth of brain xenografts of GFP expressing GSC #1. (A) Kaplan–Meier analysis of mice with brain grafts of GSCs (left). Mice grafted with MEG3 GSCs showed weight loss (>20% of initial weight) or neurological signs later than those grafted with control GSCs (n = 12; P = 0.0133, log rank test). Coronal sections of brain across the grafting site in a control and MEG3 mouse (right). (B) The density of tumor cells spreading in the striatum, amygdala, anterior commissure, and optic chiasm is highly reduced in xenografts of MEG3 overexpressing cells. (C) Graphs showing results of tumor cell counts in the brain regions analyzed; **P < 0.01; ***P < 0.001.
As a second in vivo model, we used subcutaneous grafts of GSC#61 cells either overexpressing MEG3 or carrying an empty vector (CNTR).
We found that 8 weeks after inoculation, mice injected with MEG3 GSC#61 cells developed significantly smaller tumors (Supplementary Figure 10A, B). Tumor proliferation, as assessed by Ki67 staining, which ranged between 76% and 85% in CNTR GSC#61 xenografts, was dramatically lower in MEG3 GSC#61 xenografts (range 14–35%). MEG3 GSC#61 tumors also showed expression of glial fibrillary acidic protein in 16–26% of cells, which was completely absent in CNTR GSC#61 subcutaneous xenografts, indicating an astrocyte differentiation (Supplementary Figure 10C).
To confirm that MEG3 reactivation may inhibit tumor growth, in vivo severe combined immunodeficient mice (n = 12) were grafted onto the right striatum either with untreated GFP + GSC#1 cells (n = 8) or with GFP + GSC#1 cells pre-treated in vitro with VPA (4 mM; n = 4), as epigenetic drug that has been administered for the prevention or treatment of seizure disorder in GBM patients.16 One week after surgery, the mice were either treated with saline i.p. (0.2 mL, b.i.d. for 5 d/wk over 3 wk) (n = 4) or treated with VPA i.p. (n = 8; 200 mg /kg in 0.2 mL, b.i.d. for 5 d/wk over 3 wk). Beginning and duration of i.p. treatment were chosen to maximize VPA passage across the damaged blood–brain barrier in the grafted brain area.17 Mice were sacrificed at 8 weeks after grafting.
Results are shown in Supplementary Figure 11. In mice treated with VPA i.p., the brain xenografts were significantly smaller than in saline-treated controls (P = 0.017; unpaired Student’s t-test). In vitro exposure of GFP + GSC#1 to VPA further inhibited tumor growth, whereby mice grafted with VPA pretreated GFP + GSC#1 cells that received additional VPA i.p. after surgery developed significantly smaller tumor than mice treated with VPA i.p. only (P = 0.003; unpaired Student’s t-test) (Supplementary Figure 11A, B). Immunostaining with Ki67 showed that cell proliferation was 24.4 ± 2.1% (mean ± SEM), 13.1 ± 1.4%, and 7.9 ± 0.9% in saline-treated controls, VPA i.p. treated mice, and VPA i.p. treated mice grafted with VPA pretreated GPF GSC#1 cells, respectively. Thus, treatment with VPA i.p. reduced significantly tumor cell proliferation compared with saline (P < 0.001; unpaired Student’s t-test), and pretreating the tumor cells with VPA further inhibited cell proliferation (P < 0.01; unpaired Student’s t-test) compared with VPA i.p. only (Supplementary Figure 11C). This experiment suggests that epigenetic silencing of MEG3 is important for activation of the glioma malignancy, and those epigenetic targeting drugs such as VPA can inhibit tumor growth.
Tumor-Suppressor Function of MEG3 Involved Cell Adhesion Signaling Pathways
To further investigate the molecular mechanism underlying the tumor-suppressor function of MEG3, we analyzed gene expression profiles of GSCs transduced with MEG3 or with an empty vector. In detail, we selected GSC lines #1 and #61, characterized by a “pro-neural-” and “mesenchymal”-like signature,3 respectively, as prototype GSCs with opposing basal expression of MEG3.
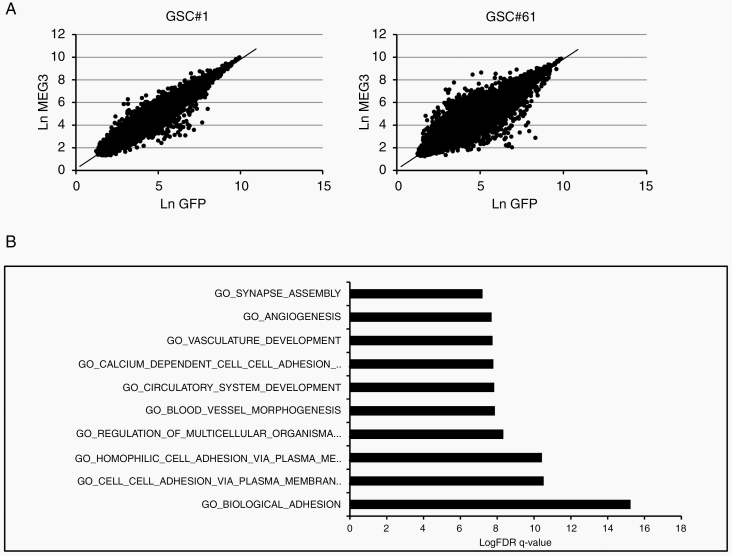
In order to identify the most deviating genes from what was expected by the shared tissue attractor, we computed for both cell lines the linear regression equation, linking the logarithm expression level in control GSCs with the expression levels of MEG3-GSC transduced cells. Both cell lines gave rise to robust and statistically significant models allowing us to predict the “expected” expression value for each gene in MEG3-transduced GSC based on its corresponding levels in control cells. The 2 linear equations were: MEG3 (expected) = 0.097 + 0.974*GFP (GSC line #61), and MEG3 (expected) = 0.137 + 0.968*GFP (GSC line #1). The 2 regression models gave rise to Pearson correlation coefficients equal to 0.98 and 0.87 for line #1 and line #61, respectively (Figure 5A).
Fig. 5.
Effect of MEG3 overexpression on gene expression and signaling pathways. (A) Linear regression equation between the log expression level in control GSCs (ln-GFP) with MEG3-GSC (ln-MEG3) cells evaluated for GSC #1 (left) and for GSC #61 (right) lines. (B) Pathway enrichment analysis examined by GSEA of highly modulated genes in both GSC#1 and #61 overexpressing MEG3.
The genes most deviating from the regression lines are those more affected by MEG3 reexpression: we opted for a threshold equal to 2 standard deviations of the residuals (difference of observed and expected expression values) in order to select potentially interesting genes. We selected only those genes that significantly deviate from their expected value in both lines.
The genes selected were then analyzed for pathway enrichment analysis by gene set enrichment analysis (GSEA) online tool; http://www.broadinstitute.org/gsea/index.jsp), setting Gene Ontology biological process terms and Kyoto Encyclopedia of Genes and Genomes pathways. The most significantly enriched pathways were related mainly to cell adhesion (Figure 5B and Supplementary Table 8).
On the other hand, previous studies showed that MEG3 regulates EMT, cell cycle, DNA damage, and Wnt pathways in several human malignancies.18,19 Since our data showed that enforced MEG3 reexpression inhibited cell growth, migration, and colony-forming ability of GSCs, we sought to investigate the effects of MEG3 reintegration on GSC signaling. We selected a set of endpoints related to these pathways (Supplementary Table 2) and measured their expression by RPPA in control and MEG3-transduced GSCs.
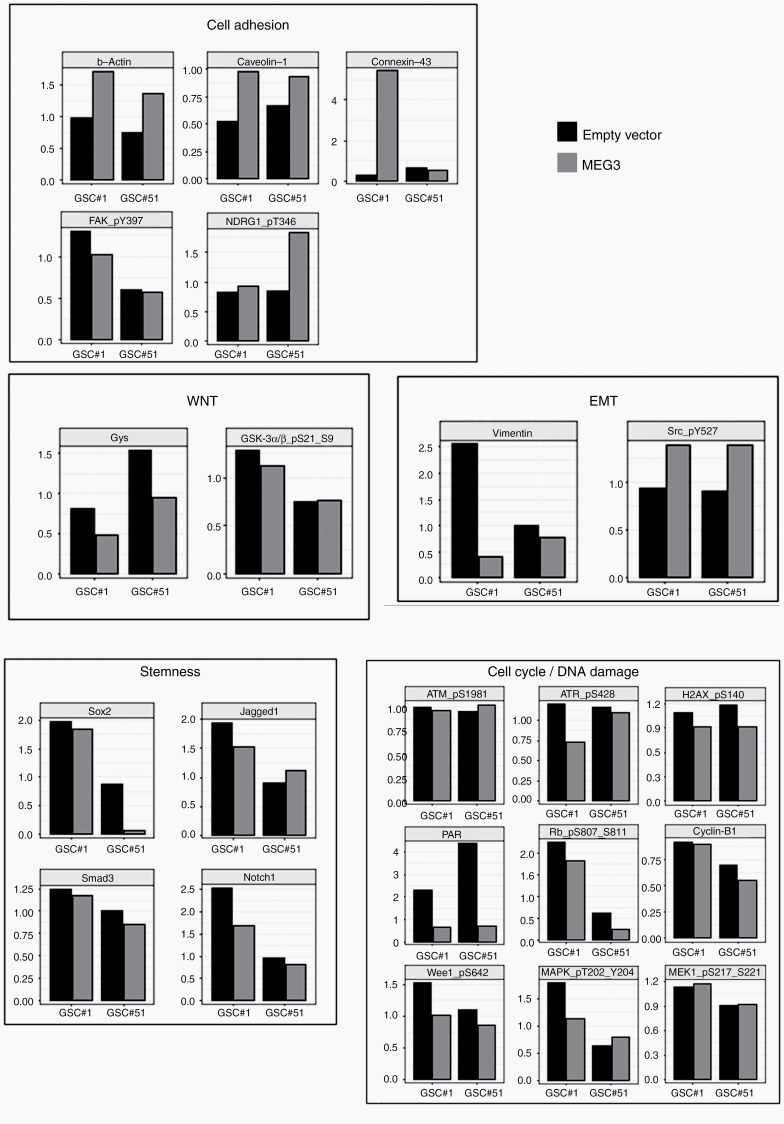
In line with a reinstatement of the epithelial program, we found that MEG3 induced a marked downmodulation of vimentin along with an increased β-actin content and inhibitory phosphorylation of Src (Src_pY527). Furthermore, MEG3 restoration caused a reduction of total and active focal adhesion kinase (FAK_pY397) as well as an increase in caveolin-120 and connexin-4321 levels together with activated N-myc downstream regulated gene 1 (NDRG1_pT346), the latter previously shown to suppress metastasis and cell migration in a colorectal cancer model.22 Intriguingly, RPPA levels of total glycogen synthase were decreased by MEG3, pointing to an active inhibition by GSK3, which ultimately targets and activates NDRG1. We did not detect significant changes in the levels of key players of the Wnt pathway.
Along similar lines, reexpression of MEG3 by GSCs was associated with a decrease of DNA damage-related components and targets such as total (but not phosphorylated) ataxia telangiectasia mutated kinase, activated ataxia telangiectasia and Rad3-related protein (ATR_pS428), poly(ADP-ribose) polymerase, and H2AX-gamma_pS140. Despite a reduction in total and inactive Wee1 (Wee1_pS642) following MEG3 overexpression, we found that readouts of cell proliferation such as cyclin-B1, MEK1_pS217_S221, MAPK_pT202_Y204, and Rb_pS807_S811 were not affected by MEG3. Therefore, the downregulation of DNA damage markers may correlate with the absence of an active proliferative state of GSCs after transduction with MEG3. Finally, we found that MEG3 reexpression might contribute to the reduction of the levels of some endpoints involved in stemness, such as sex determining region Y–box 2 (Sox2), Notch, and transforming growth factor beta (TGF-β) family members (Figure 6 and Supplementary Figure 12).
Fig. 6.
Targeted pathway analysis on GSCs. Normalized, relative protein expression levels as measured by RPPA analysis on control and MEG3-overexpressing GSC#1 and GSC#61. Barplots of selected, functional protein endpoints are grouped by pathway, as reported in Supplementary Table 2.
Discussion
Loss of expression of loci contained in the DLK1-DIO3 region on chromosome 14q32 is a frequent event in several types of cancers7 and is mainly mediated by epigenetic silencing.5 Here we report that deregulated expression of genes and several noncoding RNAs within this region is a common event in tumor samples and GSCs derived from GBM patients compared with normal brain.
Importantly, the overall survival of GBM patients with low expression of both MEG3 and MEG8 was significantly lower than that of patients with high expression of both lncRNAs. These data were confirmed on a cohort of GBM patients available from the database of TCGA.
Our analysis showed that downregulation of genes within the DLK1-DIO3 region in GBM is mainly mediated by epigenetic alterations and, particularly, by hypermethylation within the differentially methylated region MEG3-DMR as described in the other tumors.23
Since MEG3 dysregulation has been implicated in gliomagenesis,24 we decided to study the functional impact of MEG3 restoration on GSCs. Interestingly, we found that MEG3 inhibited cell growth, migration, and colony-forming ability of GSCs in vitro and significantly decreased the growth of GSC-derived tumors in vivo, as demonstrated by the significant reduction of cell density in several brain regions that are usually invaded by GSCs.
A number of studies have shown lncRNAs as crucial components of complex gene regulatory networks by regulating gene expression at the transcriptional, posttranscriptional, and epigenetic levels. Some of them can serve as scaffolds to regulate protein-protein interaction, decoys binding microRNAs, or guides to recruit epigenetic regulators on chromatin.
Previous studies in colon and brain cancer first demonstrated a function of MEG3 as a tumor suppressor through activation of p53, leading to increased p53 protein levels and stimulation of p53-dependent transcription.8,25
Recently, MEG3 has been shown to interact with components of the polycomb repressive complex 2 (PRC2), an epigenetic regulator involved in transcriptional repression. Acting in tandem with PRC2 components, MEG3 negatively regulates the activity of a set of genes of the TGF-β pathways, and suppresses c-Met gene transcription.26–28
Besides regulating 2 of the most potent EMT inducers by transcriptional repression, MEG3 inhibits EMT by acting as competing endogenous RNA (ceRNA).
The cross talk of ceRNAs with other RNA species (eg, miRNA sharing) suggests that ceRNAs may regulate diverse biological processes. Therefore, disruption of axes involving ceRNAs and miRNAs could represent a critical step in cancer pathogenesis.29
MEG3 inhibits cell proliferation, migration, and invasion and induces apoptosis by acting as ceRNA of miR-19a30 in glioma cells and of miR-499-5p in melanoma,31 by regulating E-cadherin and forkhead box O1 expression through competitive binding to miR-9 in esophageal cancer,32 and by sponging miR-421, upregulating E-cadherin and downregulating zinc finger E-box-binding homeobox 1 (ZEB1), ZEB2, and vimentin in breast cancer.33
A further functional role of MEG3 has been revealed from studies on liver functions and vascular endothelium whereby MEG3 acts as an RNA scaffold.34,35
Altogether, data available in the literature underline the molecular complexity of MEG3 interactome, pointing to multiple modes of action in several and diverse biological processes, in a cell-specific manner.
GSEA revealed cell adhesion and EMT as principal biological processes affected by MEG3 ectopic expression in GSCs. MEG3 restoration predominantly increased the expression of protocadherin (PCDH)-beta genes. PCDHs are a group of cell-cell adhesion molecules, belonging to the cadherin family, predominantly expressed in the nervous system, and having a major role in regulating neuronal function.36,37
Recently, PCDHs have been reported to be broadly involved in tumor suppression,38–42 and epigenetic dysregulation of several PCDHs has been observed in a variety of tumors.43,44
Besides PCDHs, MEG3 restoration modulated the expression of matrix gamma-linolenic acid protein,45,46 periostin,47,48 and versican,49 which play a major role in glioma invasion, angiogenesis, and migration and significantly contribute to malignant progression.
The complexity of MEG3 activity is further highlighted by the diversity of RPPA endpoint activated. In line with the MEG3-dependent reinstatement of an epithelial program, we found that MEG3 induced a marked downmodulation of vimentin in GSCs along with an increased β-actin content coupled to elevated inhibitory Src phosphorylation. Furthermore, MEG3 restoration caused (i) a reduction of total and active FAK, (ii) an increase in connexin-4321 and caveolin-1, the latter being a negative regulator of GBM growth,20 and (iii) upmodulation of activated NDRG1, a metastasis suppressor.22
Notably, enforced expression of MEG3 in GSCs caused a decrease of DNA damage-related players, which correlates with the absence of readouts of cell proliferation by RPPA analysis. Finally, we found that MEG3 reexpression reduced the levels of some proteins involved in stemness (ie, Sox2 and Notch and TGF-β family members).
It should be emphasized that although the regulation by MEG3 of the endpoints analyzed is heterogeneous in different cell lines, in both cell lines tested MEG3 functions as a tumor suppressor regulating the same pathways. The inconsistencies of the individual analytes indicates that a similar effect can be produced in each GSC line through different effectors belonging to the same pathway, depending on the specific complex network of interactions of the cell line. This hypothesis is likely for a tumor like GBM, which is highly heterogeneous at the inter- and intratumor level.
Overall, our data confirm and expand on those present in the literature and show that lncRNA MEG3 is involved in a complex network regulating diverse signaling pathways. Nonetheless, the molecular mechanisms governing the tumor suppressor activity of MEG3 in GBM remain elusive.
We hypothesize that other members of the DLK1-DIO3 region may participate in the regulation of processes involving MEG3 and that aberrant expression of this cluster may trigger cell growth and proliferation while modulating cell adhesion, thus promoting EMT and ultimately contributing to GBM pathogenesis.
Supplementary Material
Funding
This work was supported by the Italian Ministry of Health (RF-2016–02361089 to L.R.V.) and the Italian Association for Cancer Research (IG 2019 Id.23154 to R.P.).
Conflict of interest statement. None disclosed.
Authorship statement. G.M. and L.R.V. (conceived and designed the study); M.Bu. and G.C. (GSC cultures and in vitro assays); A.G. and V.L. (molecular data analysis); M.M. (clinical data analysis); QG.D’A. and R.P. (brain xenografts and data analysis, provided surgical specimens); S.G. (confocal microscopy); G.G. (analysis of Methylation profile data); A.N., S.S., and M.G. (MLPA experiments and data analysis); Giu.M. (array CGH experiments); M.S. (proteomic experiments); technical support: R.I. and A.B.; data interpretations and manuscript writing: G.M., L.R.V., A.G., M.S., M.Bi., and R.P.; figure assembly: L.R.V., V.L., M.S., and M.M.; financial support: L.R.V. and M.Bi. All the authors revised the manuscript.
References
- 1. Ostrom QT, Gittleman H, Xu J, et al. . CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016;18(suppl_5):v1–v75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eramo A, Ricci-Vitiani L, Zeuner A, et al. . Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13(7):1238–1241. [DOI] [PubMed] [Google Scholar]
- 3. Marziali G, Signore M, Buccarelli M, et al. . Metabolic/proteomic signature defines two glioblastoma subtypes with different clinical outcome. Sci Rep. 2016;6:21557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marziali G, Buccarelli M, Giuliani A, et al. . A three-microRNA signature identifies two subtypes of glioblastoma patients with different clinical outcomes. Mol Oncol. 2017;11(9):1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kagami M, O’Sullivan MJ, Green AJ, et al. . The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 2010;6(6):e1000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kagami M, Sekita Y, Nishimura G, et al. . Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet. 2008;40(2):237–242. [DOI] [PubMed] [Google Scholar]
- 7. Benetatos L, Vartholomatos G, Hatzimichael E. DLK1-DIO3 imprinted cluster in induced pluripotency: landscape in the mist. Cell Mol Life Sci. 2014;71(22):4421–4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113(6):1868–1874. [DOI] [PubMed] [Google Scholar]
- 9. Astuti D, Latif F, Wagner K, et al. . Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br J Cancer. 2005;92(8):1574–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X, Gejman R, Mahta A, et al. . Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res. 2010;70(6):2350–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pallini R, Ricci-Vitiani L, Banna GL, et al. . Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14(24):8205–8212. [DOI] [PubMed] [Google Scholar]
- 12. Ricci-Vitiani L, Pedini F, Mollinari C, et al. . Absence of caspase 8 and high expression of PED protect primitive neural cells from cell death. J Exp Med. 2004;200(10):1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ricci-Vitiani L, Pallini R, Biffoni M, et al. . Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–828. [DOI] [PubMed] [Google Scholar]
- 14. Signore M, Pelacchi F, di Martino S, et al. . Combined PDK1 and CHK1 inhibition is required to kill glioblastoma stem-like cells in vitro and in vivo. Cell Death Dis. 2014;5:e1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Court F, Tayama C, Romanelli V, et al. . Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24(4):554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tsai HC, Wei KC, Tsai CN, et al. . Effect of valproic acid on the outcome of glioblastoma multiforme. Br J Neurosurg. 2012;26(3):347–354. [DOI] [PubMed] [Google Scholar]
- 17. Haas TL, Sciuto MR, Brunetto L, et al. . Integrin alpha7 is a functional marker and potential therapeutic target in glioblastoma. Cell Stem Cell. 2017;21(1):35–50 e39. [DOI] [PubMed] [Google Scholar]
- 18. He Y, Luo Y, Liang B, Ye L, Lu G, He W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget. 2017;8(42):73282–73295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an oncogenic long non-coding RNA in different cancers. Pathol Oncol Res. 2019;25(3):859–874. [DOI] [PubMed] [Google Scholar]
- 20. Quann K, Gonzales DM, Mercier I, et al. . Caveolin-1 is a negative regulator of tumor growth in glioblastoma and modulates chemosensitivity to temozolomide. Cell Cycle. 2013;12(10):1510–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tittarelli A, Guerrero I, Tempio F, et al. . Overexpression of connexin 43 reduces melanoma proliferative and metastatic capacity. Br J Cancer. 2016;115(9):e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mi L, Zhu F, Yang X, et al. . The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene. 2017;36(30):4323–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48(3):R45–R53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kiang KM, Zhang XQ, Leung GK. Long non-coding RNAs: the key players in glioma pathogenesis. Cancers (Basel). 2015;7(3):1406–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Benetatos L, Vartholomatos G, Hatzimichael E. MEG3 imprinted gene contribution in tumorigenesis. Int J Cancer. 2011;129(4):773–779. [DOI] [PubMed] [Google Scholar]
- 26. Mondal T, Subhash S, Vaid R, et al. . MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Commun. 2015;6:7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaneko S, Bonasio R, Saldaña-Meyer R, et al. . Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53(2):290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iyer S, Modali SD, Agarwal SK. Long noncoding RNA MEG3 is an epigenetic determinant of oncogenic signaling in functional pancreatic neuroendocrine tumor cells. Mol Cell Biol. 2017;37(22):e00278–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qin N, Tong GF, Sun LW, Xu XL. Long Noncoding RNA MEG3 suppresses glioma cell proliferation, migration, and invasion by acting as a competing endogenous RNA of miR-19a. Oncol Res. 2017;25(9):1471–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Long J, Pi X. lncRNA-MEG3 suppresses the proliferation and invasion of melanoma by regulating CYLD expression mediated by sponging miR-499-5p. Biomed Res Int. 2018;2018:2086564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dong Z, Zhang A, Liu S, et al. . Aberrant methylation-mediated silencing of lncRNA MEG3 Functions as a ceRNA in esophageal cancer. Mol Cancer Res. 2017;15(7):800–810. [DOI] [PubMed] [Google Scholar]
- 33. Zhang W, Shi S, Jiang J, Li X, Lu H, Ren F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed Pharmacother. 2017;91:312–319. [DOI] [PubMed] [Google Scholar]
- 34. Shihabudeen Haider Ali MS, Cheng X, Moran M, et al. . LncRNA Meg3 protects endothelial function by regulating the DNA damage response. Nucleic Acids Res. 2019;47(3):1505–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y, Wu J, Liangpunsakul S, Wang L. Long non-coding RNA in liver metabolism and disease: current status. Liver Res. 2017;1(3):163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol Rev. 2012;92(2):597–634. [DOI] [PubMed] [Google Scholar]
- 37. Redies C, Hertel N, Hübner CA. Cadherins and neuropsychiatric disorders. Brain Res. 2012;1470:130–144. [DOI] [PubMed] [Google Scholar]
- 38. Yu JS, Koujak S, Nagase S, et al. . PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene. 2008;27(34):4657–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhong X, Zhu Y, Mao J, Zhang J, Zheng S. Frequent epigenetic silencing of PCDH10 by methylation in human colorectal cancer. J Cancer Res Clin Oncol. 2013;139(3):485–490. [DOI] [PubMed] [Google Scholar]
- 40. Hu X, Sui X, Li L, et al. . Protocadherin 17 acts as a tumour suppressor inducing tumour cell apoptosis and autophagy, and is frequently methylated in gastric and colorectal cancers. J Pathol. 2013;229(1): 62–73. [DOI] [PubMed] [Google Scholar]
- 41. Imoto I, Izumi H, Yokoi S, et al. . Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66(9):4617–4626. [DOI] [PubMed] [Google Scholar]
- 42. Zhu P, Lv J, Yang Z, et al. . Protocadherin 9 inhibits epithelial-mesenchymal transition and cell migration through activating GSK-3β in hepatocellular carcinoma. Biochem Biophys Res Commun. 2014;452(3):567–574. [DOI] [PubMed] [Google Scholar]
- 43. Dallosso AR, Hancock AL, Szemes M, et al. . Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009;5(11):e1000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Asada K, Abe M, Ushijima T. Clinical application of the CpG island methylator phenotype to prognostic diagnosis in neuroblastomas. J Hum Genet. 2013;58(7):428–433. [DOI] [PubMed] [Google Scholar]
- 45. Martini M, Cenci T, D’Alessandris GQ, et al. . Epigenetic silencing of Id4 identifies a glioblastoma subgroup with a better prognosis as a consequence of an inhibition of angiogenesis. Cancer. 2013;119(5):1004–1012. [DOI] [PubMed] [Google Scholar]
- 46. Mertsch S, Schurgers LJ, Weber K, Paulus W, Senner V. Matrix gla protein (MGP): an overexpressed and migration-promoting mesenchymal component in glioblastoma. BMC Cancer. 2009;9:302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Butcher JT, Norris RA, Hoffman S, Mjaatvedt CH, Markwald RR. Periostin promotes atrioventricular mesenchyme matrix invasion and remodeling mediated by integrin signaling through Rho/PI 3-kinase. Dev Biol. 2007;302(1):256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park SY, Piao Y, Jeong KJ, Dong J, de Groot JF. Periostin (POSTN) regulates tumor resistance to antiangiogenic therapy in glioma models. Mol Cancer Ther. 2016;15(9):2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Touab M, Villena J, Barranco C, Arumí-Uría M, Bassols A. Versican is differentially expressed in human melanoma and may play a role in tumor development. Am J Pathol. 2002;160(2):549–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.