Abstract
Homologous recombination (HR) is initiated by double-strand break (DSB) resection, during which DSBs are processed by nucleases to generate 3' single-strand DNA. DSB resection is initiated by CtIP and Mre11 followed by long-range resection by Dna2 and Exo1 in Saccharomyces cerevisiae. To analyze the relative contribution of four nucleases, CtIP, Mre11, Dna2 and Exo1, to DSB resection, we disrupted genes encoding these nucleases in chicken DT40 cells. CtIP and Dna2 are required for DSB resection, whereas Exo1 is dispensable even in the absence of Dna2, which observation agrees with no developmental defect in Exo1-deficient mice. Despite the critical role of Mre11 in DSB resection in S. cerevisiae, loss of Mre11 only modestly impairs DSB resection in DT40 cells. To further test the role of CtIP and Mre11 in other species, we conditionally disrupted CtIP and MRE11 genes in the human TK6 B cell line. As with DT40 cells, CtIP contributes to DSB resection considerably more significantly than Mre11 in TK6 cells. Considering the critical role of Mre11 in HR, this study suggests that Mre11 is involved in a mechanism other than DSB resection. In summary, CtIP and Dna2 are sufficient for DSB resection to ensure efficient DSB repair by HR.
Introduction
DNA double-strand breaks (DSBs) are the most dangerous DNA damage, as a single unrepaired DSB can trigger apoptosis. DSBs are generated during physiological replication and induced by ionizing-radiation. DSBs are repaired by two major DSB-repair pathways, homologous recombination (HR) and non-homologous end-joining (NHEJ). The choice of DSB-repair pathway depends on the cell-cycle phase and the DNA-damaging agent (Symington & Gautier 2011). HR repairs DSBs in the S to G2 phases, whereas NHEJ operates in all the cell phases. HR is more prominent than NHEJ in the repair of DSBs occurring during DNA replication (Hochegger et al. 2006; Qing et al. 2011) and is essential for cellular proliferation. Indeed, loss of critical HR factors, including CtIP, Mre11 and Rad51, causes mortality due to severe genome instability (Yamazoe et al. 2004; Nakamura et al. 2010; Hoa et al. 2015).
HR is carried out in a series of steps, beginning with the 5'-to-3' strand resection of DSBs, which is called DSB resection (reviewed in Stracker & Petrini 2011; Symington & Gautier 2011). The resulting 3'-over-hang is coated with a single-strand DNA binding protein, replication protein A (RPA). RPA is subsequently replaced with polymerized Rad51 recombinase, which polymerization results in the formation of subnuclear Rad51 foci. Polymerized Rad51 performs homology search and strand invasion into intact homologous sequences leading to formation of D-loop and Holliday junction structures. Biochemical and genetic studies have shown that in Saccharomyces cerevisiae (S. cerevisiae), DSB resection is initiated by Mre11 nuclease, which physically associates with Rad50 and Xrs2 (the MRX complex). The MRX complex and Sae2 are the orthologs of human Mre11/ Rad50/Nbs1 (the MRN complex) and CtIP, respectively. Yeast MRX plays an important role in initiating HR by removing up to a few hundred nucleotides from the 5' end. In S. cerevisiae, this short-range resection is sufficient for efficient HR (Mimitou & Symington 2008; Zhu et al. 2008). Sae2/CtIP is required for the initiation of DSB resection by the MRX complex (Sartori et al. 2007; Nicolette et al. 2010). The resulting short 3'-overhangs are further processed by two alternative pathways involving Dna2 and Exo1 nucleases (Gravel et al. 2008; Mimitou & Symington 2008; Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010). This process is called long-range resection and generates more than 10 kb 3'-overhangs (Mimitou & Symington 2008; Zhu et al. 2008). DSB resection by Dna2 requires DNA unwinding by Sgs1, the RecQ DNA helicase in S. cerevisiae (Zhu et al. 2008).
The following data suggest that the contribution of the four nucleases to DSB resection may differ distinctly between S. cerevisiae and vertebrate cells. Nuclease-dead mre11 mutants have a significantly milder phenotype during mitosis than do null-mre11 mutants in S. cerevisiae (Bressan et al. 1998; Moreau et al. 1999; Lewis et al. 2004; Krogh et al. 2005). By sharp contrast, nuclease-deficient MRE11−/H129N mice phenocopied Mre11-null-deficient MRE11−/− mice, including embryonic lethality associated with marked genome instability (Buis et al. 2008). The phenotypic similarity between MRE11−/H129N and MRE11−/− mice has been interpreted as evidence of the critical role played by MRN in DSB resection. Another difference is that although short-range resection by the MRX is sufficient for efficient HR in S. cerevisiae (Mimitou & Symington 2008; Zhu et al. 2008), long-range resection by Dna2 seems to play an essential role in HR in mammalian and chicken cell lines (Peng et al. 2012; Hoa et al. 2015). Exo1 might play a minor role, as evidenced by the fact that Exo1-deficient mice develop normally (Schaetzlein et al. 2013). The relative contributions of vertebrate Mre11, CtIP, Dna2 and Exo1 to DSB resection have not yet been defined in any mammalian cell lines, as previous studies have relied on the incomplete depletion of the relevant proteins by siRNA. Furthermore, the role of Mre11-nuclease activity has not yet been defined in human cells.
We previously reported that depletion of Mre11, Rad50 or Nbs1 does not compromise the formation of Rad51 foci at ionizing-radiation-induced DSB sites in the chicken DT40 B cell line (Yamaguchi-Iwai et al. 1999; Nakahara et al. 2009). Also, we previously showed that CtIP and Dna2 are required for efficient DSB resection, in which CtIP physically associates with Dna2 and recruits it to DSB sites (Hoa et al. 2015). We here quantitated the relative contributions of CtIP, Dna2, Exo1 and Mre11 to DSB resection in DT40 cells. Contrary to the current view (Stracker & Petrini 2011), CtIP plays the dominant role in DSB resection over Mre11. The dominant role of CtIP over Mre11 in the initiation of DSB resection is also seen in the human TK6 B cell line (Honma et al. 2003). Inactivation of the nuclease activity associated with CtIP has no detectable effect on DSB resection in DT40 or TK6 cells as previously reported for mammalian cells (Makharashvili et al. 2014; Wang et al. 2014). Exo1 has a very limited role in DSB resection even in the absence of Dna2 in DT40 cells. In summary, CtIP and Dna2 are sufficient for efficient DSB resection with CtIP being responsible for loading Dna2 onto DSB sites in the chicken DT40 cell line. Considering the critical role of Mre11 in DSB repair by HR, the current data suggest that Mre11 may play an important role in a step other than DSB resection in HR.
Results
Generation of nuclease-deficient DT40 cells
We previously created CtIP-deficient (CtIP−/−/−), DNA2−/− and MRE11−/− DT40 cells (Table 1). To selectively inactivate the nuclease activity of Dna2, we previously inserted the D245A mutation into the endogenous DNA2 gene and generated DNA2−/D245A cells. Note that all the mutants are unable to proliferate and were conditionally generated using transgenes under the control of the tetracycline-repressible promoter. To comprehensively understand the roles of DSB resection nucleases in DT40 cells, we here created the following mutants (Table 1). We selectively inactivated the nuclease activity of Mre11 (Fig. S1 in Supporting Information) and CtIP (Fig. S2 in Supporting Information), selectively inactivated the DNA-helicase activity of Dna2 (K623E) (Masuda-Sasa et al. 2006) (Fig. S3 in Supporting Information), and disrupted the EXO1 gene in wild-type and DNA2−/D245A cells (Fig. S4 in Supporting Information).
Table 1.
List of cell lines used in this study
| Genotype | Name of cell line and species | † | Marker genes | Reference |
|---|---|---|---|---|
| MRE11loxP/loxP | Human TK6 (TSCER2) | 4 | puroR, neoR, hygroR | ‡ |
| MRE11WT/WT | Human TK6 (TSCER2) | hygroR, neoR | ||
| MRE11loxP/WT | Human TK6 (TSCER2) | 4 | puroR, neoR, hygroR | |
| MRE11loxP/H129N | Human TK6 (TSCER2) | 4 | puroR, neoR, hygroR | |
| MRE11−/H129N | Chicken DT40 | – | ||
| EXO1−/− | Chicken DT40 | bsrR, hisR | ||
| DNA2−/D245A/EXO1−/− | Chicken DT40 | 1.5 | puroR, neoR, bsrR, hisR | |
| DNA2−/K623E | Chicken DT40 | 1.5 | puroR, neoR | |
| CtIPWT/WT | Human TK6 (TSCER2) | puroR, hygroR | ||
| CtIPND/ND (ND: N181A/R185A) | Human TK6 (TSCER2) | puroR, neoR | ||
| CtIPmAID/mAID | Human TK6 (TSCER2) | 1/3 | neoR, hisR | |
| RAD54−/− | Human TK6 (TSCER2) | puroR, neoR | Keka et al. (2015) | |
| MRE11−/− | ALTD2 | – | Purchased from ATCC, USA. ALTD2 is transformed with SV40 T antigen (Kobayashi et al. 2010). | |
| NBS1ΔN § | GM07166 | – | Kobayashi et al. (2010) | |
| MRE11−/− reconstituted with a MRE11 transgene | ATLD2-MRE11 | neoR | ||
| NBS1ΔN reconstituted with a NBS1 transgene § | GM07166-NBS1 | hygroR | ||
| MRE11−/− | Chicken DT40 | hisR, bsrR | Yamaguchi-Iwai et al. (1999) | |
| KU70−/−/MRE11−/− | Chicken DT40 | 3 | hisR, bsrR | Yamaguchi-Iwai et al. (1999) |
| KU70−/− | Chicken DT40 | hisR, bsrR | Takata et al. (1998) | |
| RAD50−/− | Chicken DT40 | 3 | puroR, neoR | Nakahara et al. (2009) |
| NBS1−/−/− | Chicken DT40 | 3 | puroR, neoR, hygroR | Nakahara et al. (2009) |
| CtIP−/−/ND (ND: N183A/R187A) | Chicken DT40 | 1 | hisR, bsrR | Hoa et al. (2015) |
| CtIP−/−/− | Chicken DT40 | puroR, bsrR, hisR | Nakamura et al. (2010) | |
| DNA2−/− | Chicken DT40 | 1.5 | puroR, neoR, hisR | Hoa et al. (2015) |
| BRCA2−/− | Chicken DT40 | puroR, neoR | Qing et al. (2011) | |
| RECQL1−/− | Chicken DT40 | hisR, bsrR | Wang et al. (2003) | |
| BLM−/−/WRN−/− | Chicken DT40 | hisR, bsrR, puroR, neoR | Imamura et al. (2002) | |
| RECQL4−/−/− | Chicken DT40 | 1.5 | hisR, bsrR, puroR | Abe et al. (2011) |
| RECQL4−/−/− + GFP-hRECQL4 (1–496) | Chicken DT40 | hisR, bsrR, puroR | Abe et al. (2011) | |
| RECQL5−/− | Chicken DT40 | hisR, bsrR | Wang et al. (2003) |
At the indicated days after the conditional inactivation of relevant genes, ionizing-radiation was carried out to measure Rad51 focus formation.
This study. TSCER2 is a cell line derived from TK6 and carries a substrate DNA for the analysis of HR (Honma et al. 2003).
The genotype is not clear. The cells express an N-terminal truncated Nbs1 protein.
To selectively inactivate the nuclease activity of Mre11, we inserted the H129N mutation into the endogenous MRE11 gene and generated MRE11−/H129N cells (Fig. S1 in Supporting Information). Expression of the mutated MRE11 mRNA was confirmed by reverse-transcription PCR (RT-PCR) (Fig. S1C in Supporting Information). MRE11−/H129N cells were capable of proliferating, with the length of a doubling time increasing from 8 to 21 h (Fig. S1D in Supporting Information and Table 2). To inactivate the endonuclease activity of CtIP, we inserted the N183A/R187A mutations (Wang et al. 2014) into the wild-type allelic gene of CtIP−/−/+ cells (Nakamura et al. 2010) (Fig. S2 in Supporting Information, Hoa et al. 2015). The resulting CtIP−/−/ND cells (ND: nuclease-dead) cells were able to proliferate with kinetics comparable to that of CtIP−/−/+ cells (Table 2). To inactivate the DNA-helicase activity of Dna2, we inserted the K623E mutation into one of the two allelic DNA2 genes and generated DNA2−/K623E cells, which carried a wild-type DNA2 transgene under the control of tetracycline-repressible promoter (the tet-DNA2 transgene) (Fig. S3 in Supporting Information). We checked the expression of the DNA2K623E allelic gene by RT-PCR (Fig. S3B in Supporting Information). Like DNA−/− and DNA2−D245A cells, DNA2−/K623E cells were unable to proliferate and stopped dividing at 2 days after the addition of doxycycline, a tetracyline analogue (Fig. S3C in Supporting Information). After repression of the tet-DNA2 transgene, DNA2−/K623E cells showed a great increase in the number of spontaneous chromosomal breaks very similar to that of DNA2−/− and DNA2−/D245A clones (Fig. S3D in Supporting Information, Table 3). Thus, the helicase and nuclease activities of Dna2 equally contribute to genome maintenance during the cell cycle. To investigate a functional overlap between Dna2 and Exo1, the two nucleases involved in long-range DSB resection in S. cerevisiae (Gravel et al. 2008; Mimitou & Symington 2008; Zhu et al. 2008; Cejka et al. 2010; Niu et al. 2010), we disrupted the EXO1 gene in wild-type and DNA2−/D245A cells (Fig. S4 in Supporting Information). Remarkably, disruption of the EXO1 gene had no detectable impact on cellular proliferation (Fig. S4D in Supporting Information) or spontaneous chromosomal breaks (Fig. S4E in Supporting Information, Table 3) in DNA2−/D245A cells. This observation indicates that Exo1 might not play an important role in DSB resection in DT40 cells.
Table 2.
Summary of the phenotype of DT40 and TK6 mutants analyzed by this study
| Genotype | The name of cell line | The method of conditional inactivation | Cell growth (doubling time) |
|---|---|---|---|
| Wild-type | TK6 | Proliferating (14 h) | |
| MRE11loxP/loxP | TK6 | Tamoxifen | Lethal |
| MRE11WT/WT | TK6 | Proliferating (14 h) | |
| MRE111oxP/WT | TK6 | Tamoxifen | Proliferating (14 h) |
| MRE111oxP/H129N | TK6 | Tamoxifen | Lethal |
| CtIPWT/WT | TK6 | Proliferating (14 h) | |
| CtIPND/ND (ND: N181A/R185A) | TK6 | Proliferating (14 h) | |
| CtIpmAID/mAID | TK6 | Auxin | Lethal |
| RAD54−/− | TK6 | Proliferating (15.5 h) | |
| MRE11−/− | ATLD2 | Proliferating | |
| NBS1ΔN† | GM07166 | Proliferating | |
| MRE11−/− reconstituted with a MRE11 transgene | ATLD2-MRE11 | Proliferating | |
| NBS1ΔN reconstituted with a NBS1 transgene† | NBS1ΔN-NBS1 | Proliferating | |
| Wild-type | DT40 | Proliferating (8 h) | |
| EXO1−/− | DT40 | Proliferating (8 h) | |
| DNA2−/K623E | DT40 | Doxycycline | Lethal |
| DNA2−/D245A/EXO1−/− | DT40 | Doxycycline | Lethal |
| CtIP−/−/− | DT40 | Doxycycline | Lethal |
| CtIP−/−/ND | DT40 | Doxycycline | Proliferating |
| MRE11−/H129N | DT40 | Proliferating (21 h) |
The genotype is not clear. The cells express an N-terminal truncated Nbs1 protein.
Table 3.
Spontaneous chromosomal aberrations per 60 cells
| Cell lines | Days after conditional inactivation | Treatment for conditional inactivation | Chromatid | Isochromatid | Exchanged | Total aberration (±SE)† |
|---|---|---|---|---|---|---|
| DT40 | Wild-type cells (Hoa et al. 2015) | |||||
| No | 1.1 | 0 | 0 | 1.1 ± 1.0 | ||
| MRE11−/H129N cells | ||||||
| No | 2.8 | 1.9 | 0.9 | 5.6 ± 2.3 | ||
| EXO1−/− DT40 cells | ||||||
| No | 1.1 | 0 | 0 | 1.1 ± 1.0 | ||
| DNA2−/− cells (Hoa et al. 2015) | ||||||
| Day 0 | No | 1.2 | 0.0 | 0.0 | 1.2 ± 1.0 | |
| Day 1 | Doxycycline | 2.4 | 1.2 | 0.0 | 3.6 ± 1.9 | |
| Day 2 | 6.0 | 1.2 | 1.2 | 8.4 ± 2.9 | ||
| Day 3 | 19.2 | 18 | 2.4 | 39.6 ± 6.3 | ||
| DNA2−/D245A cells (Hoa et al. 2015) | ||||||
| Day 0 | No | 1.2 | 1.2 | 0.0 | 2.4 ± 1.5 | |
| Day 1 | Doxycycline | 3.8 | 0.9 | 0.0 | 4.8 ± 1.6 | |
| Day 2 | 4.8 | 1.2 | 0.0 | 6.0 ± 2.4 | ||
| Day 3 | 20.4 | 14.4 | 1.2 | 36.0 ± 6.0 | ||
| DNA2−/K623E cells | ||||||
| Day 0 | No | 1.2 | 0.0 | 0.0 | 1.2 ± 1.0 | |
| Day 1 | Doxycycline | 4.2 | 1.5 | 0.3 | 6.0 ± 1.6 | |
| Day 2 | 3.6 | 3.6 | 0.0 | 7.2 ± 2.7 | ||
| Day 3 | 13.2 | 18.0 | 3.6 | 34.8 ± 5.9 | ||
| DNA2−/D245A/EX01−/− cells | ||||||
| Day 0 | No | 1.0 | 1.0 | 0.0 | 2.0 ± 1.4 | |
| Day 1 | Doxycycline | 3.6 | 0.9 | 0.0 | 4.5 ± 1.6 | |
| Day 2 | 4.8 | 2.4 | 1.2 | 8.4 ± 2.6 | ||
| Day 3 | 15.0 | 16.9 | 3.8 | 35.7 ± 5.9 | ||
| TK6 | Wild-type cells | |||||
| No | 1.5 | 0 | 0 | 1.5 ± 1.2 | ||
| MRE11−/− cells | ||||||
| Day 0 | No | 2.1 | 0.0 | 0.0 | 2.1 ± 1.3 | |
| Day 4 | Tamoxifen | 1.8 | 0.0 | 0.0 | 1.8 ± 1.1 | |
| Day 5 | 2.3 | 3.0 | 3.8 | 9.0 ± 2.2 | ||
| Day 6 | 7.6 | 13.1 | 21.8 | 42.5 ± 5.7 | ||
| MRE11−/Hl29N cells | ||||||
| Day 0 | No | 1.5 | 0.0 | 0.6 | 2.1 ± 1.2 | |
| Day 4 | Tamoxifen | 1.7 | 0.0 | 0.3 | 2.0 ± 1.3 | |
| Day 5 | 5.9 | 4.6 | 1.4 | 11.8 ± 3.1 | ||
| Day 6 | 18.3 | 4.7 | 1.8 | 24.8 ± 4.5 | ||
| CtIPmAID/mAID cells | ||||||
| Day 0 | No | 5 | 8 | 0.0 | 13 ± 3.5 | |
| Day 1 | Auxin | 22 | 27 | 2 | 51 ± 5.6 | |
| CtIPWT/WT cells | ||||||
| No | 2.1 | 0.0 | 0.6 | 3.0 ± 1.6 | ||
| CtIPND/ND cells | ||||||
| No | 2.1 | 0.0 | 0.6 | 3.0 ± 1.6 |
Data are presented as the number of aberrations per 60 cells. The actual number of cells analyzed was 60 for each genotype. Defining the number of cells analyzed and total chromosomal aberrations as N and x, respectively, we calculated the number of total aberrations per cell ± SE as , based on the Poisson distribution of spontaneous chromosomal aberrations previously observed (Sonoda et al. 1998). ‘Days’ indicates the duration of treatment with either doxycycline, tamoxifen or auxin.
Proficient ionizing-radiation-induced Rad51 focus formation in DT40 cells deficient in Mre11, Rad50 and Nbs1
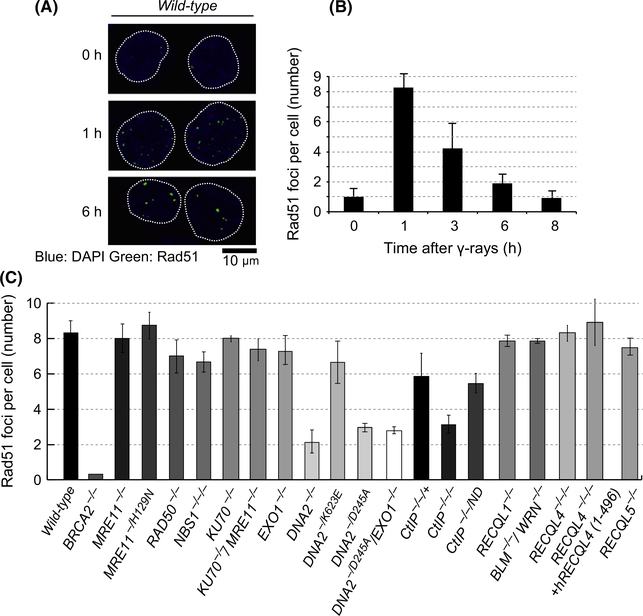
We measured the kinetics of Rad51 focus formation after exposure of DT40 cells to ionizing-radiation. We counted all the Rad51 foci without distinguishing foci having strong fluorescent signals from foci having faint signals. In wild-type cells, the number of Rad51 foci reached a peak 1 h after the radiation and decreased thereafter (Fig. 1A,B), with the intensity of detectable individual foci increasing from 1 to 6 h after ionizing-radiation (Fig. 1A). As a negative control, we used BRCA2−/− cells (Fig. 1C). The depletion of BRCA2 gene greatly diminished Rad51 focus formation (Qing et al. 2011). We evaluated DSB resection mainly by counting Rad51 foci at 1 h after ionizing-radiation. This is because Rad51 foci are very bright and thus clearly detectable even at this very early time point. It should be noted that Rad51 and RPA foci at the later time points after the 1 h might represent D-loop and Holliday junction structures rather than resected DSBs because a decrease in the number of Rad51 foci after the 1 h suggests that HR begins to complete the repair of DSBs at the later time points.
Figure 1.
Loss of CtIP and Dna2, but not loss of Exo1 or Mre11, causes a decrease in Rad51 focus formation in gamma-irradiated DT40 cells. (A) Rad51 focus formation kinetics of wild-type chicken DT40 cells at the indicated time after exposure to 2-Gy ionizing-radiation. (B) Representative images of Rad51 foci in wild-type DT40 cells at the indicated times after gamma-radiation. (C) The average number of Rad51 foci per cell of the indicated genotypes. Cells were analyzed 1 h after the ionizing-radiation. (A and C) The average number of Rad51 foci per cell was calculated from at least 100 cells. Note that we counted all foci including faint ones. Error bars are plotted for standard deviation (Buis et al.) from three independent experiments.
We first analyzed short-range DSB resection involving CtIP and the MRN complex. Earlier, we conditionally generated MRE11−/−, RAD50−/− and NBS1−/−/− DT40 cells (Yamaguchi-Iwai et al. 1999; Nakahara et al. 2009). We measured Rad51 foci 3 days after the addition of either doxycycline (for MRE11−/− and RAD50−/−) or tamoxifen (for NBS1−/−/−), when MRE11−/−, RAD50−/− and NBS1−/−/− DT40 cells are able to proliferate with normal kinetics (Yamaguchi-Iwai et al. 1999; Nakahara et al. 2009). The number of Rad51 foci in the three mutant cells was very similar to that of wild-type cells (Fig. 1C). Nuclease-deficient MRE11−/H129N cells, which are able to proliferate, also showed intact Rad51 focus formation. Thus, the proficient Rad51 focus formation of MRE11−/− cells is not due to residual Mre11 activity in the conditional MRE11−/− cells. These observations stand in marked contrast to the results seen in the CtIP−/−/− DT40 mutant, which showed a very severe defect in ionizing-radiation-induced Rad51 focus formation (Nakamura et al. 2010; Hoa et al. 2015) (Fig. 1C). As indicated by a previous study for mammalian cells (Makharashvili et al. 2014), CtIP does not contribute to DSB resection as a nuclease in DT40 cells, as nuclease-deficient CtIP−/−/ND and CtIP−/−/+ cells showed indistinguishable Rad51 focus formation (Fig. 1C).
Ku70/Ku80, a heterodimer protein essential for initiation of NHEJ, prevents DSB resection by Dna2 and Exo1 in the absence of functional Mre11 in yeast species (Tomita et al. 2003; Mimitou & Symington 2010; Shim et al. 2010). We thus analyzed DSB resection in KU70−/−/MRE11−/− cells. The loss of Ku70 increases the efficiency of HR-mediated DSB repair during DNA replication in DT40 cells (Trujillo & Sung 2001; Hochegger et al. 2006). Nonetheless, KU70−/− and KU70−/−/MRE11−/− cells showed the same Rad51 focus formation as wild-type cells (Fig. 1C), suggesting that neither Ku nor Mre11 affects DSB resection in DT40 cells.
We next analyzed the long-range DSB resection by Dna2 and Exo1. We previously reported that DNA2−/− and DNA2−/D245A cells are severely deficient in ionizing-radiation-induced Rad51 focus formation (Hoa et al. 2015). By contrast, the inactivation of Exo1 has no detectable impact on Rad51 focus formation even in the DNA2−/D245A background (Fig. 1C). In summary, the roles played by CtIP and Dna2 are considerably more important to DSB resection than are the roles of Exo1 and the Mre11/Rad50/Nbs1 complex in DT40 cells.
We finally analyzed the involvement of DNA helicases in DSB resection, as S. cerevisiae Dna2 requires the RecQ-family DNA helicase, Sgs1, for DSB resection both in vivo and in vitro (Zhu et al. 2008; Cejka et al. 2010). DNA2−/K623E cells retain the capability of forming Rad51 foci, which finding agrees with the helicase-dead mutant of S. cerevisiae (Zhu et al. 2008). There are five RecQ ortholog genes in mammalian and chicken cells, including RECQL1, BLM (RECQL2), WRN (RECQL3), RECQL4 and RECQL5 (Seki et al. 2008). We measured ionizing-radiation-induced Rad51 focus formation in RECQL1−/−, BLM−/−/WRN−/−, RECQL4−/−/− and RECQL5−/− DT40 cells (Table 1). Note that RECQL4−/−/− cells are mortal and carry a human RECQL4 transgene under the control of tetracycline-repressible promoter. We also analyzed RECQL4−/−/− + GFP-hRECQL4 (1–496) cells (Abe et al. 2011), where a RECQL4 transgene carrying the N-terminal half (1–496 amino acids) is expressed and rescues the mortality of RECQL4−/−/− cells (Table 1). None of the mutants shows a defect in Rad51 focus formation (Fig. 1C). In summary, the requirement of DNA helicases for DSB resection is different between S. cerevisiae and DT40 cells, as the latter cells do not require the RecQ orthologs despite the dominant role for the Dna2 nuclease in DSB resection.
Generation of Mre11 mutant cells from the human TK6 B cell line
No significant contribution of Mre11 to DSB resection in DT40 cells is very surprising. To investigate the reproducibility of this result in human cells, we conditionally disrupted the MRE11 and CtIP genes in the TK6 cell line. The TK6 B cell line has been widely used by the governments of developed countries to evaluate the genotoxicity of industrial chemical compounds (Zhang et al. 1995; Kirkland et al. 2007). We did the conditional disruption by inserting a full-length MRE11 cDNA flanked by the LoxP signals at both ends into the first exon of the endogenous MRE11 allelic genes in the TK6 cell line and generated MRE11loxP/loxP TK6 cells (Fig. S5A, left, and B in Supporting Information). As a control, we inserted a full-length MRE11 cDNA without having the LoxP signals (MRE11WT) into the first exon of the endogenous MRE11 allelic genes and generated MRE11WT/WT TK6 cells (Fig. S5A, right in Supporting Information). We then stably transfected a DNA construct expressing the chimeric Cre recombinase associated with the estrogen receptor (ER-Cre) (Peng et al. 2012). We added tamoxifen to activate ER-Cre, leading to the generation of MRE11−/− cells from the MRE11loxP/loxP cells. After the addition of tamoxifen, the Mre11 protein decreased in its expression over time and was undetectable on day three (Fig. S5C in Supporting Information).
To selectively inactivate the nuclease activity of the MRE11 gene, we inserted a full-length MRE11 cDNA carrying the H129N mutation into the wild-type (+) MRE11 allelic gene of the MRE11loxP/+ TK6 cells and generated MRE11loxP/H129N cells (Fig. S6A in Supporting Information). We also inserted a full-length wild-type MRE11 cDNA and generated MRE11loxP/WT cells. Note that the cDNAs are expressed by the endogenous MRE11-gene promoter in MRE11loxP/H129N and MRE11loxP/WT cells. We then stably expressed ER-Cre recombinase, and exposed MRE11loxP/H129N and MRE11loxP/WT cells to tamoxifen, leading to the generation of MRE11 −/H129N and MRE11−/WT cells. RT-PCR followed by direct nucleotide sequencing showed that the number of MRE11H129N transcripts was more than 20 times higher than the number of MRE11loxP transcripts 4 days after the addition of tamoxifen to MRE11loxP/ H129N cells (Fig. S6C in Supporting Information). Western blot analysis shows that the amount of Mre11 protein in MRE11loxP/H129N and MRE11loxP/WT cells is comparable to that of endogenous Mre11 protein in wild-type cells (Fig. S6D, left in Supporting Information). Using tamoxifen treatment, the total amount of Mre11 protein in MRE11−/H129N and MRE11−/WT cells is reduced to an approximately half of that in wild-type cells (Fig. S6D, right, and E in Supporting Information).
The proliferation kinetics of MRE11−/− TK6 cells slowed 6 days after the addition of tamoxifen (Fig. S5D in Supporting Information). The number of spontaneously arising chromosomal aberrations increased (Table 3), as seen in DT40 cells deficient in Mre11, Rad50 or Nbs1 (Yamaguchi-Iwai et al. 1999; Nakahara et al. 2009). As with the MRE11−/− TK6 cells, the MRE11−/H129N TK6 cells stopped proliferation 6 days after the addition of tamoxifen (Fig. S6F in Supporting Information), exhibiting spontaneous chromosomal aberrations (Table 3). The spontaneous chromosomal aberrations in MRE11−/H129N cells are distinctly different from those in MRE11−/− cells (Table 3), suggesting that both non-catalytic and catalytic roles of Mre11 may contribute to maintenance of genomic DNA during the cell cycle.
Generation of CtIP mutant cells from the human TK6 B cell line
We conditionally disrupted the CtIP gene in TK6 cells using the auxin-inducible degron (AID) system (Nishimura et al. 2009). We used the AID system, as depletion of CtIP might immediately compromise cellular viability and, thus, synchronous quick depletion of CtIP protein would be required for accurate phenotypic analysis. We inserted the mini-AID (mAID) tag combined with GFP tag and the terminal codon (mAID-GFP tag) into the intronic sequences 5' of the last exon in the two CtIP allelic genes (Fig. S7A in Supporting Information) and confirmed the targeting events by genomic PCR (Fig. S7B in Supporting Information). The resulting CtIPmAID/mAID TK6 cells expressed a 60% lower amount of CtIP carrying the mAID-GFP tag when compared with the amount of endogenous CtIP in wild-type cells (Fig. S7C in Supporting Information). We also stably expressed the SCF F-box protein, OsTIR1, which recognizes the mAID tag (Kubota et al. 2013). We exposed the resulting CtIPmAID/mAID TK6 cells to the auxin hormone and observed an over 90% decrease in the amount of CtIP-mAID chimeric protein at 8 h (Fig. S7C in Supporting Information). CtIPmAID/mAID TK6 cells stopped proliferating at 24 h after the addition of auxin (Fig. S7D in Supporting Information).
To generate endonuclease-deficient CtIP, we inserted either wild-type CtIP cDNA or a mutant cDNA carrying the N181A/R185A mutations (nuclease-dead (ND) mutations)into the first exon of the endogenous CtIP gene (Fig. S8 in Supporting Information). CtIPND transcripts were expressed in both CtIP+/ND and CtIPND/ND cells (Fig. S8C in Supporting Information). The amount of CtIP protein expressed in the resulting CtIPWT/WT and CtIPND/ND cells was only 20% of that expressed in wild-type cells (Fig. S8D in Supporting Information). These cells proliferated with the same kinetics as wild-type cells (Table 2). In summary, both the null mutation and nuclease deficiency of Mre11 cause mortality, whereas only null mutation but not nuclease deficiency of CtIP causes mortality in the TK6 B cell line.
MRE11−/H129N and MRE11−/− TK6 cells retain the capability of resecting DSB ends
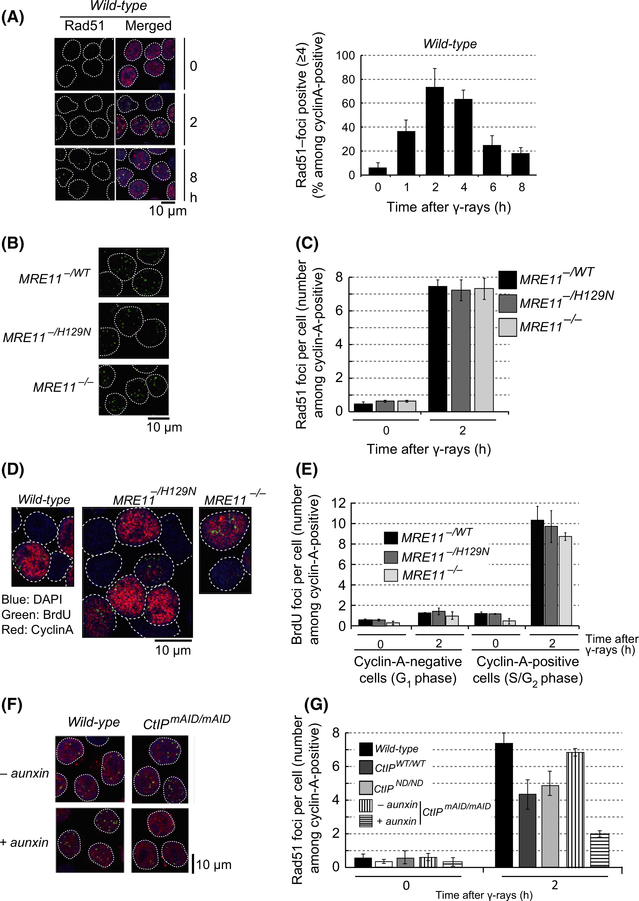
We monitored the kinetics of Rad51 focus formation afterionizing-radiation in MRE11−/−, MRE11−/H129N, CtIPmAID/mAID, CtIPWT/WT and CtIPND/ND TK6 cells. Note that we counted all detectable foci, including faint fluorescent signals (Fig. 2A, left panels). Rad51 focus formation was hardly detectable in nonirradiated cells, and peaked 2 h after ionizing-radiation in wild-type TK6 cells (Fig. 2A, right graph). We measured the number of Rad51 foci at 0 and 2 h after ionizing-radiation, focusing on cells in the S/G2 phase expressing cyclin A (Fig. 2B,C). We took the measurement 4 days after the addition of tamoxifen, when the Mre11 mutant cells were still capable of proliferating with normal kinetics (Figs S5D and S6F in Supporting Information). Like MRE11−/− and MRE11−/H129N DT40 cells, MRE11−/− and MRE11−/H129N TK6 cells were still capable of forming Rad51-foci. To confirm the poor contribution of Mre11-nuclease activity to DSB resection, we assessed single-strand formation at DSB sites by counting BrdU foci. To this end, we labeled the whole genomic DNA with BrdU, exposed the cells to ionizing-radiation, and stained the fixed cells with anti-BrdU antibody, which binds BrdU only on resected single-strand DNA but not on intact duplex DNA. We counted BrdU foci 2 h after ionizing-radiation in cyclin-A-positive (S/G2 phase) cells. The TK6 cells showed BrdU foci only in γ-irradiated S/ G2 cells (Fig. 2D,E). We did not count the foci of the RPA single-strand binding protein due to background signals even in G1 -phase cells (Barton et al. 2014). The number of ionizing-radiation-induced BrdU foci in the MRE11−/H129N and MRE11−/− cells was similar to that of the MRE11−/WT cells, which agrees with the results for Rad51 focus formation (Fig. 2B,C). We therefore conclude that the nuclease activity of Mre11 is largely dispensable for DSB resection in the human and chicken B cell lines.
Figure 2.
MRE11−/H129N and MRE11−/− TK6 cells retain the capability of performing DSB resection. (A) Representative images of Rad51 foci (green) in wild-type TK6 cells (left). Nuclei stained with red fluorescence indicate cyclin-A-positive cells. The histogram shows kinetics of Rad51 focus formation in wild-type TK6 cells at the indicated times after 0.5-Gy gamma-ray exposure (right). Note that we counted all foci including faint ones. Rad51-positive cells are defined as cells showing not less than four Rad51 foci. (B) Representative images of Rad51 foci at 2 h after exposure of the indicated genotypes to 0.5-Gy ionizing-radiation. (C) The average number of Rad51 foci per cyclin-A-positive cell for the indicated genotype at 0 and 2 h after 0.5-Gy ionizing-radiation. (D) Representative images of BrdU foci for the indicated cells. (E) The average number of BrdU foci per cell in cyclin-A-positive or cyclin-A-negative cells at the indicated times after 0.5-Gy ionizing-radiation. (F) Representative images of Rad51 foci in the indicated genotypes. CtIPmAID/mAID-conditional mutant cells were irradiated at 6 h and analyzed at 8 h after the treatment of cells with auxin. (G) The average number of Rad51 foci per cyclin-A-positive cell for indicated CtIP mutants at 0 and 2 h after 0.5-Gy ionizing-radiation. Dotted lines in (A), (B), (D) and (F) indicate the nuclear envelop contours. Error bars in (A), (C), (E) and (G) are plotted for SD from three independent experiments. At least 100 cells were analyzed in each experiment.
We exposed CtIPmAID/mAID cells to ionizing-radiation and counted Rad51 foci at 8 h after the addition of auxin, when the cells are capable of proliferating (Fig. S7D in Supporting Information). CtIP-depleted TK6 cells showed poor induction of Rad51 foci (Figs 2F,G and S7E in Supporting Information), as is also seen with CtIP-deficient DT40 cells (Fig. 1C). Likewise, CtIPWT/WT cells, where expression of CtIP was reduced to 20% of that of wild-type cells (Fig. S8D in Supporting Information), also showed an approximately 30%–40% decrease in the number of ionizing-radiation-induced Rad51 foci (Fig. 2G). We therefore conclude that the contribution of Mre11 to DSB resection is significantly smaller than that of CtIP, which is in marked contrast to the requirement of both Sae2/CtIP and Mre11 for efficient DSB resection in S. cerevisiae (Sartori et al. 2007; Nicolette et al. 2010). The endonuclease activity of CtIP is dispensable for DSB resection as Rad51 focus formation was very similar between CtIPND/ND and CtIPWT/WT cells (Fig. 2G). Thus, noncatalytic role for CtIP plays an important role in DSB resection in both DT40 and TK6 cell lines, as previously indicated (Makharashvili et al. 2014; Hoa et al. 2015).
Efficient Rad51 focus formation in Mre11-depleted U2OS, and Mre11- and Nbs1-deficient human cell lines
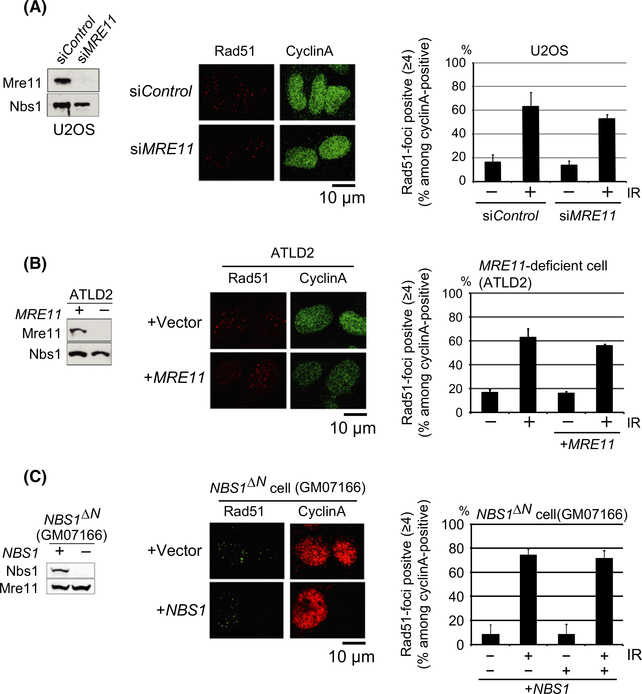
We analyzed ionizing-radiation-induced Rad51 focus formation in other three human cell lines, U2OS and two cell lines established from patients suffered from hereditary diseases, human ataxia-telangiectasia-like disorder (ATLD2) deficient in Mre11 (Stewart et al. 1999) and Nijmegen breakage syndrome (NBS) deficient in Nbs1. Note that ATLD2 and NBS1 cells are transformed with SV40 T antigen (Table 1). We also analyzed isogenic control cells, where expression of the defective proteins was restored by MRE11 and NBS1 transgenes (Ito et al. 1999; Uziel et al. 2003; Kobayashi et al. 2009). We counted all the Rad51 foci in cyclin-A-positive cells, including faint ones. Mre11-depleted U2OS cells showed the same level of Rad51 focus formation as the control U2OS cells (Fig. 3A). As with the MRE11−/− TK6 and NBS1−/− DT40 cells (Figs 1C and 2C), the ATLD2 and NBS1 cells exhibited relatively efficient Rad51 focus formation 4 h after ionizing-radiation (Fig. 3B,C). In summary, the MRN complex does not play a very important role in DSB resection in all human cell lines tested here.
Figure 3.
Mre11- and Nbs1-deficient human cell lines are capable of inducing Rad51 foci. (A, B) Western blotting of Mre11 for whole cell extract prepared from U2OS cells treated with siMRE11 (A, left) and ATLD2 cells (B, left). Representative images of Rad51 foci (red) at 4 h after 6-Gy ionizing-radiation in siMRE11-treated U2OS (A, middle) and Mre11-deficient ATLD2 (B, middle) cells. The histograms show the percentage of cells representing at least four Rad51 foci (Rad51-positive cells) among cyclin-A-positive cells (green) in U2OS cells treated with siMRE11 (A, right) and ATLD2 cells (B, right). ‘+MRE11’ indicates ATLD2 cells reconstituted with a MRE11 transgene. (C) Western blotting of Nbs1 for whole cell extract prepared from Nbs1-deficient (GM07166) cells and the cells reconstituted with a NBS1 transgene (+NBS1) (left). Representative images of Rad51 foci (green) at 4 h after 6-Gy ionizing-radiation in GM07166 cells (middle). The histogram shows the percentage of cells representing at least four Rad51 foci (Rad51-positive cells) among cyclin-A-positive cells (red) in GM07166 cells ( −NBS1) and GM07166 cells reconstituted with wild-type NBS1 transgene (+NBS1) (right). (A–C) Cells were fixed at zero (IR− ) and 4 h (IR+) after 6 Gy of ionizing-radiation (IR) treatment. Error bars are plotted for SD of three independent experiments. At least 100 cells in total were counted for the individual three experiments.
Defective HR-mediated DSB repair in MRE11−/H129N and MRE11−/− cells
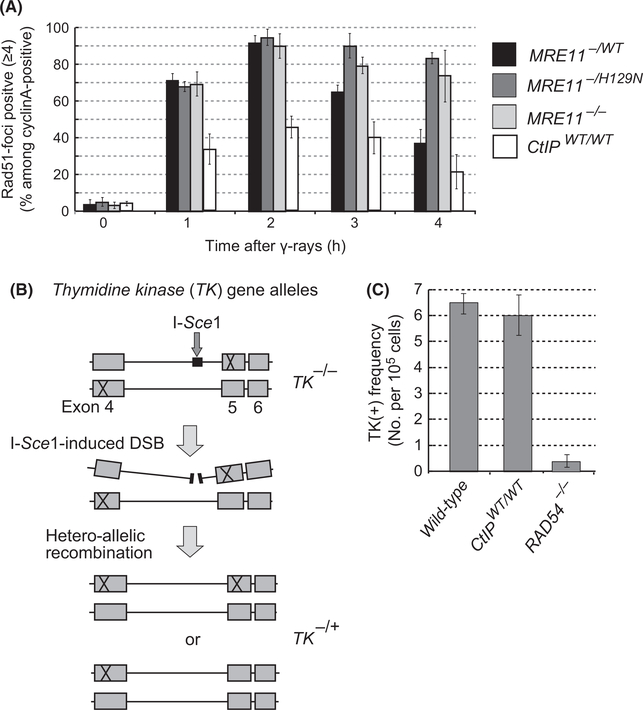
A large number of studies have indicated the critical role of Mre11 in HR (Stracker & Petrini 2011). To assess the capability of the Mre11-deficient TK6 cells to repair ionizing-radiation-induced DSBs, we monitored the kinetics of Rad51 focus formation over time after ionizing-radiation. Although the number of Rad51 foci was very similar among MRE11−/WT, MRE11−/H129N and MRE11−/− TK6 cells at 1 and 2 h after ionizing-radiation, the latter two mutants showed a significant delay in the resolution of Rad51 foci in comparison with MRE11−/WT cells at the later time points (Fig. 4A). This observation is consistent with the data that MRE11−/− DT40 cells are severely defective in HR-dependent repair of ionizing-radiation-induced DSBs despite efficient induction of Rad51 foci by ionizing-radiation (Fig. 1C) (Yamaguchi-Iwai et al. 1999). We conclude that Mre11 nuclease may contribute to HR through a mechanism other than DSB resection. Remarkably, although CtIPWT/WT cells showed an approximately 40% decrease in the Rad51 focus formation (Fig. 2G), they retained the capability of HR to efficiently perform DSB repair, as the number of Rad51 foci decreased from 2 to 4 h after ionizing-radiation in the same manner as MRE11−/WT cells (Fig. 4A). To assess the capability of HR to repair DSBs, we measured heteroallelic recombination frequency between the thymidine kinase (TK) allelic genes carrying mutations at different sites (Honma et al. 2003; Keka et al. 2015) (Fig. 4B). The HR frequency of HR-deficient RAD54−/− cells (Table 1) was 15 times smaller than that of wild-type cells (Fig. 4C). Although the number of ionizing-radiation-induced Rad51 foci in CtIPWT/WT cells was only 60% of that in wild-type cells, the frequency of heteroallelic recombination was very similar between CtIPWT/WT and wild-type cells. One possible explanation of this result is that the half reduced Rad51-foci are sufficient for efficient heteroallelic HR, which is likely to require more intensive homology search when compared to HR between two sister chromatids. In summary, Mre11 significantly contributes to HR-dependent DSB repair presumably through a mechanism other than DSB resection.
Figure 4.
A 40% decrease in DSB resection does not compromise the capability of HR to perform heteroallelic recombination in CtIPWT/WT TK6 cells. (A) Kinetics of Rad51 focus formation after 0.5-Gy ionizing-radiation treatment for the indicated TK6 cell lines. Error bars are plotted for SD from three independent experiments. At least 100 cells were analyzed in each experiment. (B) Schematic diagram showing the repair of an I-SceI-induced double-strand break (DSB) in the endogenous thymidine kinase (TK) locus. Compound heterozygous TK−/− cells carry an I-SceI restriction-enzyme site in intronic sequences of a TK allelic gene. The site of mutations in the TK allelic genes is marked as ‘X’ at exon 4 and exon 5. HR-mediated DSB repair associated with crossover (upper panel of TK−/+) or without crossover (lower panel of TK−/+) would yield TK−/+ clones from TK−/− cells. The number of TK−/+ clones was measured by counting the number of HAT-resistant colonies. (C) Histogram represents the frequency of DSB-repair events (y-axis) in the indicated genotypes (x-axis). Error bars indicate SD of more than three independent experiments.
Discussion
This study is the first comprehensive genetic study to accurately measure the relative contribution of the four nucleases, CtIP, Dna2, Exo1 and Mre11, to DSB resection in a single cell line, DT40 cells. We showed that CtIP and Dna2 are sufficient for DSB resection to ensure efficient HR. CtIP plays the significantly more prominent role in DSB resection than Mre11. Thus, Mre11 plays a critical role in HR through a mechanism other than DSB resection. The nuclease-deficient CtIP is capable of fully performing DSB resection, indicating that the role played by CtIP is noncatalytic. A previous report indicated the noncatalytic role of CtIP in DSB resection (Makharashvili et al. 2014). We previously showed that Dna2 focus formation at induced DSB sites requires CtIP but not its nuclease activity (Hoa et al. 2015). These observations suggest that the noncatalytic role of CtIP is to recruit Dna2 to DSB sites facilitating the long-range resection. Exo1 does not contribute to DSB resection even in the absence of Dna2. The modest contribution of Exo1 to DSB resection agrees with the fact that Exo1-deficient mice develop normally despite the embryonic mortality of various HR-deficient mice (Schaetzlein et al. 2013). In summary, only Dna2 contributes to DSB resection as a nuclease among the four nucleases in DT40 cells.
We found that although the nuclease- and helicase-dead mutants of DNA2 are both lethal, only the nuclease-dead mutant showed a strong decrease in Rad51 focus formation (Fig. 1C). The finding supports the idea that the lethality of the helicase- as well as nuclease-dead mutant might be due to a defect in a mechanism other than HR such as Okazaki fragment maturation during DNA replication. In fact, the suppression of lethality in the S. cerevisiae Dna2 mutant by over-expression of Fen-1, a nuclease involved in the Okazaki fragment maturation (Budd et al. 2011), implies an essential role for Dna2 in DNA replication in DT40 cells. However, it is hard to exclude a defect in HR as a critical cause of the lethality in DNA2−/K623E DT40 cells. This is because a defect in the Okazaki fragment maturation would cause a great increase in the amount of recombinogenic substrates and strongly stimulate HR. As a consequence, the inactivation of HR in a Fen-1 mutant causes a synthetic lethality in both Schizosaccharomyces pombe and S. cerevisiae (Debrauwere et al. 2001; Akamatsu et al. 2003). The roles of the vertebrate Dna2 helicase activity in HR and DNA replication are an important issue for future studies.
The data that Mre11-deficient human and chicken cells still retain the capability of performing DSB resection challenge the current view of Mre11. A large number of studies have shown the important role of Mre11 in DSB resection. Most of the studies seem to have counted the number of only bright Rad51 and RPA foci, and have founded reduced focus formation in Mre11-depleted cells. However, no study has measured the relative contribution of nucleases to DSB resection, as the previous studies set up an arbitrary threshold of brightness when they identified subnuclear foci, and thus, it is impossible to directly compare the number of foci in one study with that in other studies. Another problem is that most studies only partially depleted DSB resection nucleases using siRNA. To overcome these problems, we analyzed gene-disrupted clones and set up a simple criterion for identifying subnuclear foci, counting every focus formation. Moreover, we counted a very early time point, 1 and 2 h after ionizing-radiation, which early-time focus formation may represent resected DNA but not its association with intact homologous sequences to form Holliday junctions. We here examined the effect of Mre11 depletion in the DT40 (Fig. 1), TK6 (Fig. 2) and U2OS (Fig. 3) cell lines and also examined ATLD2 cells (Fig. 3). We found that Mre11 contributes only little to DSB resection in the various cell lines. The data suggest that the absence of Mre11 does not show the detectable defect in DSB resection to ensure efficient HR at least in some cell lines.
The conserved MRX/MRN complex has been believed to be the major regulator of DSB resection in mammalian cells as well as in S. cerevisiae. The importance of the nuclease activity of the mammalian MRN complex is indicated by Buis et al. (Buis et al. 2008), who found that nuclease-deficient MRE11−/H129N mice phenocopied mice with null mutation of Mre11 (MRE11−/− mice). Both mutant mice exhibited a very severe phenotype, including early embryonic lethality and dramatic genomic instability (Buis et al. 2008). Likewise, MRE11−/− and MRE11−/H129N TK6 cells showed a similar phenotype: cellular mortality associated with dramatic genomic instability (Table 3). Because of the critical role played by yeast MRX in DSB resection, the phenotypic similarity between the MRE11−/H129N and MRE11−/− mice has been interpreted as an evidence of a dominant role for MRN in DSB resection in mammalian cells. It should be noted that the MRE11−/H129N and MRE11−/− mice showed only a ∼50% decrease in X-ray-induced Rad51 focus formation, compared with wild-type controls (Buis et al. 2008). Likewise, a recent study directly measuring the length of resected 3' single-strand overhangs also shows only approximately a 50%–70% decrease in DSB resection when Mre11 is depleted by siRNA (Zhou et al. 2014). It is unlikely that such modest decreases in DSB resection significantly decrease HR efficiency, as CtIPWT/WT cells showing an approximately 40% decrease in the Rad51 focus formation fully retain the capability of HR to perform DSB repair by heteroallelic recombination (Fig. 4C). Hence, although Mre11 facilitates DSB resection to some extent, as indicated by a number of studies, its limited contribution to DSB resection may not account for the very severe defect in DSB repair by HR in Mre11-deficient cells (Yamaguchi-Iwai et al. 1999; Shibata et al. 2014). We propose that Mre11 contributes to HR-mediated DSB repair in a mechanism other than DSB resection. Besides DSB resection, there are several possible roles of Mre11 (MRN complex) in DSB repair by HR. First, Rad50, a component of the MRN complex, belongs to the structural maintenance of chromosomes (SMC) family members including cohesin and condensin (Kinoshita et al. 2009). Accordingly, the MRN complex has been proposed to tether two ends of a single DSB site, thereby facilitating DSB repair by HR (de Jager et al. 2001; Nakai et al. 2011). Second, MRN complex facilitates DSB repair probably by eliminating second structures at DSB sites as well as chemical adducts such as the topoisomerase-cleavage complex (Lobachev et al. 2002). This idea is supported by the following findings. First, although S. cerevisiae Mre11 significantly contributes to cellular tolerance of ionizing-radiation, which induces DSBs associated with nonphysiological chemical modification, Mre11 is dispensable for HR-dependent repair of DSBs induced by the HO restriction enzyme (Llorente & Symington 2004; Hartsuiker et al. 2009). Second, Mre11 eliminates topoisomerase adducts covalently bound to DSB ends (Neale et al. 2005; Hartsuiker et al. 2009; Lee et al. 2012; Cannavo & Cejka 2014). In addition to the elimination of chemical adducts from DSB ends, the nuclease activity of Mre11 contributes to genome stability by opening hairpin-capped ends and maintaining the progression of replication forks in S. cerevisiae (Paull & Gellert 1998; Trujillo & Sung 2001; Lobachev et al. 2002). The nuclease activity of Mre11 also contributes to the maintenance of replication fork progression, as the DNA-polymerase-α/δ-inhibitor, aphidicolin, induced higher number of chromosome breaks in mitotic chromosome spreads in MRE11−/H129N and MRE11−/− mice than in the control mice (Buis et al. 2008). The catalytic and noncatalytic activities of Mre11 stabilize DNA replication forks when they are stalled at damaged template strands (Tittel-Elmer et al. 2009; Schlacher et al. 2011). Future studies are required to define the key role played by Mre11 in genome maintenance other than DSB resection.
Experimental procedures
Cell lines and culture conditions
All cell lines used in this study are shown in Table 1. The DT40 cell line, derived from chicken B lymphoma (Buerstedde & Takeda 1991), was cultured as previously described (Sonoda et al. 1998). Cells were incubated at 39.5 °C with 5% CO2 in a RPMI-1640 medium (Nacalai Tesque, Japan) with glutamine (11875; Invitrogen, USA) supplemented with 1% chicken serum (GIBCO-BRL, Grand Island, NY, USA), 10% heat-inactivated fetal bovine serum (FBS) (100–106; Gemini Bio-Products, West Sacramento, CA, USA), 50 μM beta-mercaptoethanol (Nacalai Tesque), 50 U/mL penicillin and 50 μg/ mL streptomycin (Nacalai Tesque). Doxycycline (Nacalai Tesque) was added at a final concentration of 100 ng/mL to inactivate the expression of the tetracycline-repressible promoter.
The human lymphoblastoid cell lines, TSCE5 and TSCER2, derived from the TK6 cell line with an I-SceI inserted into the TK locus (Honma et al. 2003), were cultured as previously described (Honma et al. 2003). Cells were grown in a RPMI-1640 medium (Nacalai Tesque) supplemented with 5% heat-inactivated horse serum (Life Technologies, USA), 200 μg/mL sodium pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin (Nacalai Tesque) and maintained at 37 °C in a 5% CO2 atmosphere with 100% humidity. We added tamoxifen (Sigma, USA), an estrogen antagonist, to the medium at a final concentration of 200 nM to activate the estrogen receptor (ER-Cre) and generated the MRE11−/− gene from MREllloxP/loxP cells and the MRE11−/H129N gene from MRE11loxP/H129N cells.
Human fibroblast U2OS, ATLD2 and GM07166 cells were obtained from the American Type Culture Collection (ATCC). These cell lines were maintained at 37 °C with 5% CO2 in DMEM medium (Nacalai Tesque) supplemented with 10% FBS (Gemini Bio-Products). Note that ATLD2 cell lines are transformed with SV40 DNA virus (Kobayashi et al. 2010).
Cell counting and cell-cycle analysis
Cells were counted and cell-cycle analysis was carried out using LSRFortessa (BD Biosciences, USA). Immediately before being counted, cells were mixed with reference beads (Polyscience Inc., USA) and propidium iodide. For cell-cycle analysis, cells fixed with 70% ethanol were treated with RNase A and then stained with propidium iodide and analyzed using LSRFortessa. FlowJo software was used to process the data.
Chromosome aberration analysis in mitotic chromosome spreads
DT40 and TK6 cells were treated with 0.1 μg/mL colcemid (Invitrogen) for 3 h. Cells were suspended in 75 mM potassium chloride for 15 min, washed with Carnoy’s solution (a 3: 1 mixture of methanol and acetic acid), dropped on slides and stained with a 5% Giemsa solution for 10 min.
Assay of HR-dependent repair of I-SceI-induced DSBs
A TK6-derived cell line (TSCER2) that is compound heterozygous for point mutations in both exon 4 and exon 5 of the thymidine kinase gene (TK−/−) was used to measure the frequency of HR events as described previously (Honma et al. 2003). TK6 cells transfected with 2 μg of I-SceI expression vector using Neon were incubated for 48 h without antibiotics and then seeded on hypoxanthine–aminopterin-thymidine (HAT) medium (Yasui et al. 2014).
siRNA knockdown assay
A cocktail of three siRNA oligonucleotides targeting human MRE11 with two thymidine residues (dTdT) at the 3' end of the sequence was purchased from B-Bridge International Inc. (Sunnyvale, CA). These siRNA oligonucleotides correspond to nucleotides 912–930, 1171–1189 and 1397–1415 of the human MRE11 gene (Accession Number: NM_005590). The sequences are as follows: siMRE11-1 (sense: 5'-GAUGAGAA CUCUUGGUUUATT-3'), siMRE11-2 (sense: 5'-GGAAGA UGAAUAUGCAUAATT-3') and siMRE11-3 (sense: 5'-GG ACUAUAGUGGAGGUUUUTT-3'). Each siRNA oligonucleotide included in the cocktail was separately available and used for preliminary experiments. To verify sequence-specific effectiveness of the MRE11-siRNAs, we also used negative control siRNAs (NC-siRNA; B-Bridge International Inc., USA) that have no significant homology with any known sequence in the human genome. Transfection was carried out using Lipofectamine 2000 (Life Technologies), according to the specified protocol. After transfection, U2OS cells were cultured in fresh media for 48 h before being harvested for Western blotting and immunostaining assays.
Transfection for TK6 cells
Both TALEN- and Cas9-mediated gene knockin was carried out using the Neon system (Invitrogen). The target sequences of TALEN and CRISPR are shown in supplementary information. TK6 cells (4 × 106 cells) suspended in 100 μL of solution R were electroporated with 6 μg of TALEN/pX330-Cas9 and 2 μg of each targeting vector (1350-V, 10-ms, 3-pulse, 100-μL Neon tip). After being incubated for 48 h in RPMI-1640 with 10% horse serum, the transfectants were selected in RPMI-1640 containing antibiotics and 5% horse serum.
Immunostaining and microscopic analysis
Cells were fixed with either 4% paraformaldehyde (Nacalai Tesque) for 10 min at room temperature for chicken DT40 and human TK6 cells or cold methanol on ice for human fibroblast cells. Fixed cells then were permeabilized with either 0.1% Nonidet P-40 (Nacalai Tesque) in PBS for 15 min for the chicken DT40 cells or 0.5% Triton X-100 (Sigma-Aldrich, USA) in PBS for 20 min at room temperature for the human TK6 and on ice for the fibroblasts. Permeabilized cells were incubated with blocking solution 5% BSA in PBS for 1 h at room temperature for human cells and 3% BSA in PBST for 1 at 37 °C for chicken DT40 cells. Fixed cells were treated with the following primary antibodies on slide glass: anti-Rad51, anti-BrdU and anti-cyclin A. Cells were then treated with the following secondary antibodies: Alexa Fluor 488-conjugated anti-mice IgG (1/1000; Molecular Probes) for Rad51 staining (Figs 2A–C,F,G, 3C, 4A and S7E in Supporting Information) and BrdU staining (Fig. 2D,E), or Alexa Flour 594-conjugated anti-mice IgG (1/1000, Molecular Probes) for Rad51 staining (Fig. 3A,B), or Alexa Flour 488-conjugated anti-rabbit IgG (1/1000; Life Technologies) for Rad51 staining (Fig. 1) and cyclin A staining (Fig. 3A,B), or Alexa Flour 594-conjugated anti-rabbit IgG (1/1000; Invitrogen) for cyclin A staining (Figs 2, 3C, 4A and S7E in Supporting Information). For BrdU staining, cells were incubated with BrdU (final concentration 10 μg/mL) for two cell-cycle times before fixation.
To analyze DSB-end resection in S/G2-phase cells, antibody against the protein cyclin A (rabbit polyclonal, 1/100; Santa Cruz Biotechnology, Inc., USA) was used as a biomarker to exclude G1-phase cells from the count. Images were taken with a confocal microscope (TCS SP8; Leica Microsystems Inc., Germany) and analyzed by Leica Application Suite (LAS X) software (Leica Microsystems Inc.) using the same settings for all samples. Z-stack digital images were collected optically at every 0.6 μm depth (z-step size). Optical sections (steps) were projected using the maximum-intensity projection. We also distinguish cells in the G1, S and G2 phases by measuring the intensity of DAPI staining in the nucleus of individual fixed cells.
Antibodies
Antibodies used were as follows: anti-CtIP rabbit polyclonal (1/1000; Bethyl, USA), anti-Mre11 mouse monoclonal (1/1000; GeneTex, USA), anti-NBS1 mouse monoclonal (1/ 1000; GeneTex), anti-actin mouse monoclonal (1/5000; Sigma-Aldrich), anti-Rad51 mouse polyclonal (1/500; Abnova, Taiwan), anti-Rad51 rabbit polyclonal (1/1000; Bio Academia, Japan), anti-BrdU mouse monoclonal (1/100; BD Pharmingen, USA), anti-BrdU rat polyclonal (1/250; Abcam, UK), anti-cyclin A rabbit polyclonal (1/100; Santa Cruz Biotechnology); Alexa Fluor 488-conjugated anti-mice IgG (1/1000; Molecular Probes), Alexa Flour 594-conjugated anti-mice IgG (1/1000; Molecular Probes), Alexa Fluor 594-conjugated anti-rabbit (1/1000; Invitrogen) and Alexa Flour 488-conjugated anti-rabbit IgG (1/1000; Life Technologies).
Supplementary Material
Data S1 Supplementary materials
Figure S1 Generation of the conditional MRE11−/H129N mutant in chicken DT40 cells.
Figure S2 Generation of the CtIP−/−/ND mutant from CtIP−/−/+/tet-CtIP chicken DT40 cells.
Figure S3 Generation of the conditional DNA2−/K623E mutant in chicken DT40 cells.
Figure S4 Generation and phenotypic analysis of the EXO1 mutants in chicken DT40 cells.
Figure S5 Generation of the conditional MRE11−/− mutant and MRE11WT/WT cells from human TK6 cells.
Figure S6 Generation of the conditional MRE11−/H129N mutant in human TK6 cells.
Figure S7 Generation of the conditional CtIP mutant (CtIP-mAID/mAID) in human TK6 cells.
Figure S8 Generation of CtIPWT/WT and CtIPND/ND mutants in human TK6 cells.
Table S1 Primer list used in this study.
Acknowledgements
We are grateful to P. Jeggo, J. Tainer, T. Paull and S. Gasser for his constructive comments on the manuscript. We also thank A. Noguchi and A. Kobayashi for their technical assistance and the laboratory members for their stimulating discussion. FACS analysis and DNA sequencing were carried out at the Medical Research Support Center, Graduate School of Medicine, Kyoto University. This work was supported by a Grant-in-Aid from the Ministry of Education, Science, Sport and Culture to J. K., K. K., T. N., M. K., S. T. and H. S.
Footnotes
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher’s web site:
References
- Abe T, Yoshimura A, Hosono Y, Tada S, Seki M & Enomoto T (2011) The N-terminal region of RECQL4 lacking the helicase domain is both essential and sufficient for the viability of vertebrate cells. Role of the N-terminal region of RECQL4 in cells. Biochim. Biophys. Acta 1813, 473–479. [DOI] [PubMed] [Google Scholar]
- Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H & Iwasaki H (2003) Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl Acad. Sci. USA 100, 15770–15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton O, Naumann SC, Diemer-Biehs R, Kunzel J, Steinlage M, Conrad S, Makharashvili N, Wang J, Feng L, Lopez BS, Paull TT, Chen J, Jeggo PA & Lobrich M (2014) Polo-like kinase 3 regulates CtIP during DNA double-strand break repair in G1. J. Cell Biol 206, 877–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE & Petrini JH (1998) Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics 150, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd ME, Antoshechkin IA, Reis C, Wold BJ & Campbell JL (2011) Inviability of a DNA2 deletion mutant is due to the DNA damage checkpoint. Cell Cycle 10, 1690–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde JM & Takeda S (1991) Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67, 179–188. [DOI] [PubMed] [Google Scholar]
- Buis J, Wu YP, Deng YB, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S & Ferguson DO (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135,85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E & Cejka P (2014) Sae2 promotes dsDNA endonuclease activity within Mre11–Rad50–Xrs2 to resect DNA breaks. Nature 514, 122–125. [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL & Kowalczykowski SC (2010) DNA end resection by Dna2–Sgs1–RPA and its stimulation by Top3–Rmi1 and Mre11–Rad50–Xrs2. Nature 467, 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere H, Loeillet S, Lin W, Lopes J & Nicolas A (2001) Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl Acad. Sci. USA 98, 8263–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C & Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22, 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ & Carr AM (2009) Distinct requirements for the Rad32(Mre11) nuclease and Ctp1 (CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol. Cell 33, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Kobayashi J, Omura M, Hirakawa M, Yang SH, Komatsu K, Paull TT, Takeda S & Sasanuma H (2015) BRCA1 and CtIP are both required to recruit Dna2 at double-strand breaks in homologous recombination. PLoS ONE 10, e0124495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G & Takeda S (2006) Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J 25, 1305–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M, Izumi M, Sakuraba M, Tadokoro S, Sakamoto H, Wang W, Yatagai F & Hayashi M (2003) Deletion, rearrangement, and gene conversion; genetic consequences of chromosomal double-strand breaks in human cells. Environ. Mol. Mutagen 42, 288–298. [DOI] [PubMed] [Google Scholar]
- Imamura O, Fujita K, Itoh C, Takeda S, Furuichi Y & Matsumoto T (2002) Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene 21, 954–963. [DOI] [PubMed] [Google Scholar]
- Ito A, Tauchi H, Kobayashi J, Morishima K, Nakamura A, Hirokawa Y, Matsuura S, Ito K & Komatsu K (1999) Expression of full-length NBS1 protein restores normal radiation responses in cells from Nijmegen breakage syndrome patients. Biochem. Biophys. Res. Commun 265, 716–721. [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R & Wyman C (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 8, 1129–1135. [DOI] [PubMed] [Google Scholar]
- Keka IS, Mohiuddin, Maede Y, Rahman MM, Sakuma T, Honma M, Yamamoto T, Takeda S & Sasanuma H (2015) Smarcal1 promotes double-strand-break repair by nonhomologous end-joining. Nucleic Acids Res 43, 6359–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita E, van der Linden E, Sanchez H & Wyman C (2009) RAD50, an SMC family member with multiple roles in DNA break repair: how does ATP affect function? Chromosome Res 17, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland D, Pfuhler S, Tweats D et al. (2007) How to reduce false positive results when undertaking in vitro genotoxicity testing and thus avoid unnecessary follow-up animal tests: report of an ECVAM Workshop. Mutat. Res 628, 31–55. [DOI] [PubMed] [Google Scholar]
- Kobayashi J, Kato A, Ota Y, Ohba R & Komatsu K (2010) Bisbenzamidine derivative, pentamidine represses DNA damage response through inhibition of histone H2A acetylation. Mol. Cancer 9, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Chen B, Burma S, Tashiro S, Matsuura S, Tanimoto K, Chen DJ & Komatsu K (2009) Histone H2AX participates the DNA damage-induced ATM activation through interaction with NBS1. Biochem. Biophys. Res. Commun 380, 752–757. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A & Symington LS (2005) Mutations in Mre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11–Rad50–Xrs2 complex stability in addition to nuclease activity. Genetics 171, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT & Donaldson AD (2013) The Elg1 replication factor C-like complex functions in PCNA unloading during DNA replication. Mol. Cell 50, 273–280. [DOI] [PubMed] [Google Scholar]
- Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, Morris NJ, Jackson GH, Cockell SJ, Tainer JA & Austin CA (2012) MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biol. Open 1, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P & Resnick MA (2004) Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics 166, 1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B & Symington LS (2004) The Mre11 nuclease is not required for 5' to 3' resection at multiple HO-induced double-strand breaks. Mol. Cell. Biol 24, 9682–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA & Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108, 183–193. [DOI] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, Wu X & Paull TT (2014) Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol. Cell 54, 1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Imamura O & Campbell JL (2006) Biochemical analysis of human Dna2. Nucleic Acids Res 34, 1865–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP & Symington LS (2008) Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP & Symington LS (2010) Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J 29, 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR & Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell. Biol 19, 556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara M, Sonoda E, Nojima K, Sale JE, Takenaka K, Kikuchi K, Taniguchi Y, Nakamura K, Sumitomo Y, Bree RT, Lowndes NF & Takeda S (2009) Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig V in chicken B lymphocytes. PLoS Genet 5, e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W, Westmoreland J, Yeh E, Bloom K & Resnick MA (2011) Chromosome integrity at a double-strand break requires exonuclease 1 and MRX. DNA Repair 10, 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, Ogi T, Takeda S & Taniguchi Y (2010) Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet 6, e1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J & Keeney S (2005) Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE & Paull TT (2010) Mre11–Rad50–Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol 17, 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T & Kanemaki M (2009) An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. [DOI] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, Ira G & Sung P (2010) Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467, 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT & Gellert M (1998) The 3' to 5' exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979. [DOI] [PubMed] [Google Scholar]
- Peng G, Dai H, Zhang W, Hsieh HJ, Pan MR, Park YY, Tsai RY, Bedrosian I, Lee JS, Ira G & Lin SY (2012) Human nuclease/helicase DNA2 alleviates replication stress by promoting DNA end resection. Cancer Res 72, 2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y, Yamazoe M, Hirota K, Dejsuphong D, Sakai W, Yamamoto KN, Bishop DK, Wu XH & Takeda S (2011) The epistatic relationship between BRCA2 and the other RAD51 mediators in homologous recombination. PLoS Genet 7, e1002148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bar-tek J, Baer R, Lukas J & Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450, 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaetzlein S, Chahwan R, Avdievich E, Roa S, Wei K, Eoff RL, Sellers RS, Clark AB, Kunkel TA, Scharff MD & Edelmann W (2013) Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc. Natl Acad. Sci. USA 110, E2470–E2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H & Jasin M (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Otsuki M, Ishii Y, Tada S & Enomoto T (2008) RecQ family helicases in genome stability — lessons from gene disruption studies in DT40. Cell Cycle 7, 2472–2478. [DOI] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS et al. (2014) DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53,7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim EY, Chung WH, Nicolette ML, Zhang Y, Davis M, Zhu Z, Paull TT, Ira G & Lee SE (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J 29, 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y & Takeda S (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NGJ, Raams A, Byrd PJ, Petrini JHJ & Taylor AMR (1999) The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99, 577–587. [DOI] [PubMed] [Google Scholar]
- Stracker TH & Petrini JHJ (2011) The MRE11 complex: starting from the ends. Nat. Rev. Mol. Cell Biol 12, 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS & Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu. Rev. Genet 45, 247–271. [DOI] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Sonoda E, Morrison C, Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A & Takeda S (1998) Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P & Cobb JA (2009) The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J 28, 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, Iwasaki H, Mizuno K, Ohta K, Uritani M, Ushimaru T., Yoshinaga K & Ueno M. (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol. Cell. Biol 23, 5186–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM & Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem 276, 35458–35464. [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L & Shiloh Y (2003) Requirement of the MRN complex for ATM activation by DNA damage. EMBO J 22, 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HL, Li YJ, Truong LN, Shi LDZ, Hwang PYH, He J, Do J, Cho MJ, Li HZ, Negrete A, Shiloach J, Berns MW, Shen BH, Chen LC & Wu XH (2014) CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell 54, 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WS, Seki M, Narita YA, Nakagawa T, Yoshimura A, Otsuki M, Kawabe Y, Tada S, Yagi H, Ishii Y & Enomoto T (2003) Functional relation among RecQ family Helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell. Biol 23, 3527–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Sonoda E, Sasaki MS, Morrison C, Haraguchi T, Hiraoka Y, Yamashita YM, Yagi T, Takata M, Price C, Kakazu N & Takeda S (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazoe M, Sonoda E, Hochegger H & Takeda S (2004) Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair (Amst.) 3, 1175–1185. [DOI] [PubMed] [Google Scholar]
- Yasui M, Kanemaru Y, Kamoshita N, Suzuki T, Arakawa T & Honma M (2014) Tracing the fates of site-specifically introduced DNA adducts in the human genome. DNA Repair 15,11–20. [DOI] [PubMed] [Google Scholar]
- Zhang LS, Honma M, Hayashi M, Suzuki T, Matsuoka A & Sofuni T (1995) A comparative-study of TK6 human lymphoblastoid and L5178y mouse lymphoma cell lines in the in vitro micronucleus test. Mutat. Res 347, 105–115. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G & Paull TT (2014) Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res 42, e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE & Ira G (2008) Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 Supplementary materials
Figure S1 Generation of the conditional MRE11−/H129N mutant in chicken DT40 cells.
Figure S2 Generation of the CtIP−/−/ND mutant from CtIP−/−/+/tet-CtIP chicken DT40 cells.
Figure S3 Generation of the conditional DNA2−/K623E mutant in chicken DT40 cells.
Figure S4 Generation and phenotypic analysis of the EXO1 mutants in chicken DT40 cells.
Figure S5 Generation of the conditional MRE11−/− mutant and MRE11WT/WT cells from human TK6 cells.
Figure S6 Generation of the conditional MRE11−/H129N mutant in human TK6 cells.
Figure S7 Generation of the conditional CtIP mutant (CtIP-mAID/mAID) in human TK6 cells.
Figure S8 Generation of CtIPWT/WT and CtIPND/ND mutants in human TK6 cells.
Table S1 Primer list used in this study.