Key Points
Question
Can an end-to-end deep learning network model be used to identify patients with stage IV epidermal growth factor receptor (EGFR) variant–positive non–small cell lung cancer who will not benefit from EGFR–tyrosine kinase inhibitor (TKI) therapy?
Findings
In this diagnostic/prognostic study of 342 patients receiving EGFR-TKI therapy, a bidirectional generative adversarial network model demonstrated a 36% reduction in the progression-free survival of patients at high risk for rapid progression but no significant difference in the progression-free survival between these patients and those receiving first-line chemotherapy. The proposed deep learning semantic signature eliminated all manual interventions required while using previous radiomics methods and had a better prognostic performance.
Meaning
An end-to-end clinically applicable approach is promising for quantitatively identifying the benefit of EGFR-TKI therapy.
Abstract
Importance
An end-to-end efficacy evaluation approach for identifying progression risk after epidermal growth factor receptor (EGFR)–tyrosine kinase inhibitor (TKI) therapy in patients with stage IV EGFR variant–positive non–small cell lung cancer (NSCLC) is lacking.
Objective
To propose a clinically applicable large-scale bidirectional generative adversarial network for predicting the efficacy of EGFR-TKI therapy in patients with NSCLC.
Design, Setting, and Participants
This diagnostic/prognostic study enrolled 465 patients from January 1, 2010, to August 1, 2017, with follow-up from February 1, 2010, to June 1, 2020. A deep learning (DL) semantic signature to predict progression-free survival (PFS) was constructed in the training cohort, validated in 2 external validation and 2 control cohorts, and compared with the radiomics signature.
Exposures
An end-to-end bidirectional generative adversarial network framework was designed to predict the progression risk in patients with NSCLC.
Main Outcomes and Measures
The primary end point was PFS, considering the time from the initiation of therapy to the date of recurrence, confirmed disease progression, or death.
Results
A total of 342 patients with stage IV EGFR variant–positive NSCLC receiving EGFR-TKI therapy met the inclusion criteria. Of these, 145 patients from 2 of the hospitals (n = 117 and 28) formed a training cohort (mean [SD] age, 61 [11] years; 87 [60.0%] female), and the patients from 2 other hospitals comprised 2 external validation cohorts (validation cohort 1: n = 101; mean [SD] age, 57 [12] years; 60 [59.4%] female; and validation cohort 2: n = 96, mean [SD] age, 58 [9] years; 55 [57.3%] female). Fifty-six patients with advanced-stage EGFR variant–positive NSCLC (mean [SD] age, 52 [11] years; 26 [46.4%] female) and 67 patients with advanced-stage EGFR wild-type NSCLC (mean [SD] age, 54 [10] years; 10 [15.0%] female) who received first-line chemotherapy were included. A total of 90 (26%) receiving EGFR-TKI therapy with a high risk of rapid disease progression were identified (median [range] PFS, 7.3 [1.4-32.0] months in the training cohort, 5.0 [0.6-34.6] months in validation cohort 1, and 6.4 [1.8-20.1] months, in validation cohort 2) using the DL semantic signature.The PFS decreased by 36% (hazard ratio, 2.13; 95% CI, 1.30-3.49; P < .001) compared with that in other patients (median [range] PFS, 11.5 [1.5-64.2] months in the training cohort, 10.9 [1.1-50.5] in validation cohort 1, and 8.9 [0.8-40.6] months in validation cohort 2. No significant differences were observed when comparing the PFS of high-risk patients receiving EGFR-TKI therapy with the chemotherapy cohorts (median PFS, 6.9 vs 4.4 months; P = .08). In terms of predicting the tumor progression risk after EGFR-TKI therapy, clinical decisions based on the DL semantic signature led to better survival outcomes than those based on radiomics signature across all risk probabilities by the decision curve analysis.
Conclusions and Relevance
This diagnostic/prognostic study provides a clinically applicable approach for identifying patients with stage IV EGFR variant–positive NSCLC who are not likely to benefit from EGFR-TKI therapy. The end-to-end DL-derived semantic features eliminated all manual interventions required while using previous radiomics methods and have a better prognostic performance.
This diagnostic/prognostic study proposes a clinically applicable large-scale bidirectional generative adversarial network for predicting the efficacy of epidermal growth factor receptor–tyrosine kinase inhibitor therapy in patients with non–small cell lung cancer.
Introduction
Epidermal growth factor receptor (EGFR)–tyrosine kinase inhibitor (TKI) therapy plays an important role in treating clinical stage IV EGFR (OMIM 131550) variant–positive non–small cell lung cancer (NSCLC).1,2 Previous studies3,4 have found that EGFR-TKIs, such as erlotinib, gefitinib, and icotinib, promote longer progression-free survival (PFS) in patients with EGFR variant–positive NSCLC than conventional chemotherapy. On the basis of these results, the current clinical guidelines recommend EGFR-TKI therapy in patients with stage IV EGFR variant–positive NSCLC.5
Despite potential benefits, studies6,7 have reported that 30% of patients with stage IV EGFR variant–positive NSCLC nevertheless developed rapid tumor progression after EGFR-TKI therapy, and TKIs were deemed ineffective or less effective in these patients.4,8 Currently, there are no known biomarkers predictive of which patients might not benefit from TKIs and are thus at risk for rapid disease progression. Next-generation TKIs (osimertinib) or intercalated therapy may be considered in patients with EGFR T790M variants or those more likely to develop tumor progression.9,10,11 Recent clinical studies12,13,14 have suggested that the efficacy of EGFR-TKIs should be evaluated before clinical decision-making and alternative treatments potentially prioritized in patients at high risk for rapid tumor progression.
However, in current clinical practice, no clear strategies exist for assessing future risk status after EGFR-TKI therapy in patients with stage IV EGFR variant–positive NSCLC. The primary method remains surveillance using chest computed tomography (CT) in search of macroscopic changes in disease. Recent studies15,16 have suggested potential utility of CT-based radiomics-derived predictors of prognosis in patients with stage IV EGFR variant–positive NSCLC. However, clinical translation of radiomics can be limiting because of multiple steps, such as manual segmentation of tumor boundaries, feature extraction, feature selection, and model development procedures.17 Bias might arise from interobserver variations in determining tumor boundaries and different methods used to decode radiomic features. Furthermore, layer-by-layer region of interest labeling is labor-intensive; thus, it is difficult to meet the clinical real-time requirements. A clinically applicable, end-to-end approach to more precisely stratify patients with stage IV EGFR variant–positive NSCLC who are likely responders and nonresponders to EGFR-TKI therapy could help to individualize care for this disease.
Recent developments in deep learning (DL) provide the potential to mitigate such clinical challenges.18,19 A generative adversarial network (GAN) has been reported to be useful in simulating real images and thereby bolstering learning image characteristics.20 Recent modifications to self-supervised representation learning with a GAN, the so-called large-scale bidirectional generative adversarial network (BigBiGAN), have resulted in state-of-the-art performances in generating high-quality, mimic images and automatically recognizing high-dimensional semantic features.21 Studies22,23 have found that semantic features extracted by the BigBiGAN framework can boost model performance in detecting coronavirus disease 2019 pneumonia and brain abnormalities.
We propose an end-to-end BigBiGAN-based DL approach to predicting the efficacy of EGFR-TKI therapy in patients presenting with stage IV EGFR variant–positive NSCLC without requiring manual tumor volume delineation or feature engineering procedures of traditional radiomics. We hypothesized that a DL-based representation framework could identify significant CT-based image features associated with disease progression in patients with stage IV EGFR variant–positive NSCLC and thereby contribute to identifying patient cohorts more likely to benefit from EGFR-TKI therapy.
Methods
Study Design
The flowchart of this study is shown in Figure 1. First, 120-dimensional DL semantic features were extracted by the BigBiGAN framework to identify the CT features in individual patients with stage IV EGFR variant–positive NSCLC. Second, a semantic signature was proposed using the least absolute shrinkage and selection operator (LASSO) Cox proportional hazards regression method to distinguish the risk of progression after EGFR-TKI therapy in patients in the training cohort, which was subsequently validated in multiple external validation cohorts. Third, 2 control cohorts of patients with advanced-stage (stage III-IV) EGFR variant–positive and EGFR wild-type NSCLC who received first-line chemotherapy were respectively enrolled to compare survival benefits. This diagnostic/prognostic study was approved by the institutional review boards of the participating institutions, which waived the need for informed consent. All data were deidentified. This study followed the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) reporting guidelines.24
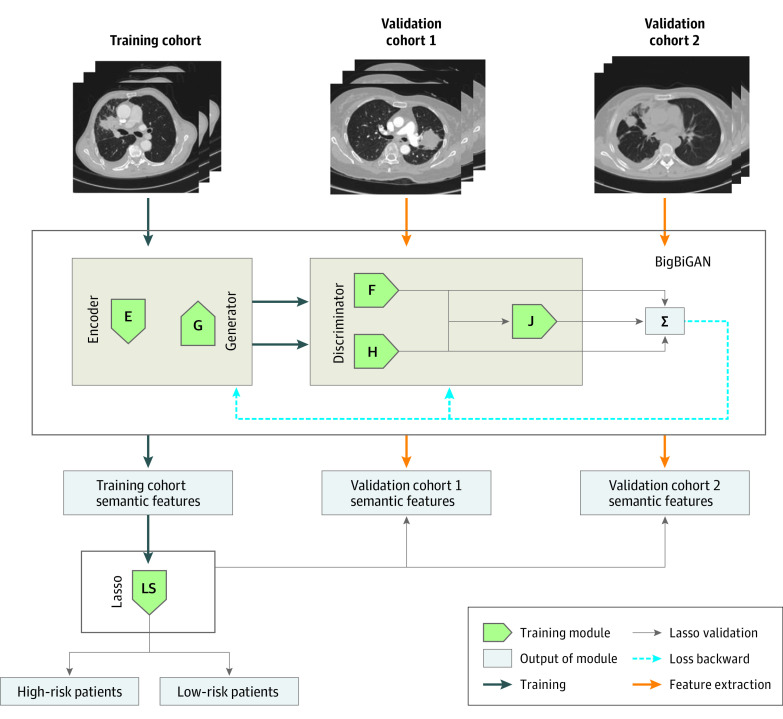
Figure 1. Flowchart of the Bidirectional Generative Adversarial Network (BigBiGAN) Training, Deep Learning Image Semantic Feature Extraction, and Construction and Validation of the Semantic Signature.
The training cohort was input into the encoder (E) module of the BigBiGAN for semantic feature extraction. The generator (G) module was used to mimic the original image from a random vector. Then the data pair of the original and mimic image was input into the F module of the discriminator, and the data pair of semantic features and random vector was input into the H module for loss calculation. When the loss of the model (J) was minimum, the semantic features of the training cohort were extracted for least absolute shrinkage and selection operator (LASSO) signature construction, and the signature was then tested on the 2 external validation cohorts. Σ indicates the sum of the loss.
Patients
From January 1, 2010, to August 1, 2017, we retrospectively enrolled patients with clinical stage IV EGFR variant–positive NSCLC treated with EGFR-TKI therapy and patients with advanced-stage NSCLC treated with first-line chemotherapy (without EGFR-TKIs) from 6 hospitals. For the EGFR-TKI therapy cohorts, the inclusion criteria were as follows. Experienced practitioners (N.N.N., M.Z., J.S., N.W., W.L., and Z.L.) collected CT scans and clinical data from each participating hospital. Clinical data elements included sex, age, smoking status, performance status score, histopathologic subtype (adenocarcinoma, squamous cell carcinoma, or others), EGFR variant subtype, and the administered therapeutic regimen. Patients diagnosed with clinical stage IV NSCLC according to the American Joint Committee on Cancer guidelines were older than 20 years, had a confirmed EGFR variant (EGFR19 exon deletion, exon 21 L858R substitution, or other EGFR variant subtype), and had received EGFR-TKI therapy according to current clinical guidelines. Patients with a history of systemic anticancer therapy for advanced disease or resection were excluded. Patients obtained contrast enhanced and/or noncontrast CT before EGFR-TKI administration. Detailed CT protocols from each included hospital are listed in eAppendix 1 in the Supplement. To reduce the impact of the variation in images from different sources of CT scanners, all input images were standardized (eAppendix 1 in the Supplement).
In addition, 2 control cohorts of patients treated with first-line chemotherapy (non–EGFR-TKI) were included according to the following criteria: patients had been diagnosed with clinical advanced-stage NSCLC, had the above-mentioned complete demographic characteristics and histopathologic subtype, and had an EGFR variant test result (EGFR variant–positive or EGFR wild-type NSCLC). All clinical treatments in this study were conducted on a voluntary basis.
The patients receiving EGFR-TKI therapy at 2 of the hospitals were used as the training cohort, and patients from 2 other hospitals served as the 2 external validation cohorts. The control cohorts of patients with advanced-stage EGFR variant–positive and EGFR wild-type NSCLC treated with first-line chemotherapy served to compare survival benefits.
The Response Evaluation Criteria in Solid Tumors 1.1 standard was used to determine tumor response on CT examinations. The primary end point in this study was PFS (time from the initiation of therapy to the date of recurrence); confirmed disease progression; or death. The occurrence of relapse or progression was defined as the development of a new lesion or progression of the primary lesion. Patients were censored at the date of death from other causes or at the last visit (for those who were living without documented disease progression).
DL Semantic Features
The patient’s original whole CT images were used as the input for the BigBiGAN framework. As shown in Figure 1, the CT images were first input into the encoder module to learn the semantic characteristics of the images, and then, the generator module generated the mimic images. In this study, we used only CT slices that contained 1 or more lung tumors to reduce unnecessary learning. Second, the image data pair (real and generated images) and semantic feature data pair (learned from the real image and random) were input into the discriminator to calculate the loss of each module. By continuous self-supervised training of the BigBiGAN, 120-dimensional semantic features representative of the specific image characteristics were extracted from each CT image. The model was trained on the training cohort until the loss was minimum and kept stable. Then we used the model with the loss at the minimum state to test the 2 external validation cohorts. To ensure reproducibility, all the source codes are open access (eAppendix 1 in the Supplement) and implemented in the free Google Colab (Google Research).
DL Semantic Signature
On the basis of the 120-dimensional semantic feature vector of each image obtained by the BigBiGAN model, for all CT slices from the same patient, we sequentially integrated the semantic feature vectors into a matrix. Finally, we calculated the mean value of each feature on all CT layers per patient. The averaged 120-dimensional semantic features of each patient in the training cohort were input into LASSO Cox proportional hazards regression to construct a semantic signature.
On the basis of the semantic signature score of each patient by LASSO, X-tile25 was used to obtain the optimal cutoff threshold to stratify the patients in the training cohort into the high-progression-risk group after EGFR-TKI therapy and the low-progression-risk group. Kaplan-Meier curves were plotted to present differences in PFS between different risk groups, and the log-rank test was used to evaluate differences between groups. Subsequently, the signature and the cutoff threshold were validated in the 2 external validation EGFR-TKI cohorts. Time-dependent receiver operating characteristic (ROC) curve analysis was used to evaluate the prognostic accuracy of the semantic signature in the 3 cohorts by the cutoffs of 10 and 12 months.
To further validate the semantic signature proposed in this study, we compared the radiomics prognostic signature for the efficacy evaluation of EGFR-TKIs. On the basis of radiomic features provided by the previous radiomics study,15 we compared the performance of the semantic signature proposed in this study with the radiomics signature in fitting the patients’ actual survival benefit. A more detailed introduction of the radiomics signature15 is presented in eAppendix 1 in the Supplement. The decision curve analysis and clinical impact curve analysis were used to evaluate the performances of the 2 signatures in actual clinical application.
Statistical Analysis
Statistical analysis was performed using R software, version 3.2.3 (R Foundation for Statistical Computing). Independent, unpaired, 2-tailed t tests were used to compare the demographics of different groups for continuous variables. The Fisher exact test was used to identify associations between categorical variables. The PFS predicted using the semantic signature in the different progression risk groups was compared using hazard ratios (HRs). Time-dependent ROC curve analysis was performed using the package survivalROC. The decision curve analysis and clinical impact curve analysis results were plotted by dca.R and rmda, respectively. P < .05 (2-tailed) was considered statistically significant.
Results
Patients
A total of 342 patients with stage IV EGFR variant–positive NSCLC receiving EGFR-TKI therapy met the inclusion criteria. Of these, 145 patients from 2 of the hospitals (n = 117 and 28) formed a training cohort (mean [SD] age, 61 [11] years; 87 [60.0%] female), and the patients from 2 other hospitals comprised 2 external validation cohorts (validation cohort 1: n = 101; mean [SD] age, 57 [12] years; 60 [59.4%] female; and validation cohort 2: n = 96, mean [SD] age, 58 [9] years; 55 [57.3%] female). Fifty-six patients with advanced-stage EGFR variant–positive NSCLC (mean [SD] age, 52 [11] years; 26 [46.4%] female) and 67 patients with advanced-stage EGFR wild-type NSCLC (mean [SD] age, 54 [10] years; 10 [15.0%] female) who received first-line chemotherapy were included. The demographics and clinicopathologic data of the enrolled patients are presented in the Table. Detailed treatment scheme is presented in eAppendix 1 in the Supplement.
Table. Demographic and Histopathologic Characteristics of Study Patientsa .
| Characteristic | EGFR-TKI therapy cohorts | Chemotherapy cohorts | |||
|---|---|---|---|---|---|
| Training cohort (n = 145) | Validation cohort 1 (n = 101) | Validation cohort 2 (n = 96) | EGFR variant positive (n = 56) | EGFR wild type (n = 67) | |
| No. of CT sections | 3481 | 1855 | 868 | NA | NA |
| Sex | |||||
| Male | 58 (40.0) | 41 (40.6) | 41 (42.7) | 30 (53.6) | 57 (85.1) |
| Female | 87 (60.0) | 60 (59.4) | 55 (57.3) | 26 (46.4) | 10 (14.9) |
| Age, y | |||||
| ≤65 | 84 (57.9) | 66 (65.3) | 66 (68.8) | 43 (76.8) | 60 (89.6) |
| >65 | 61 (42.1) | 35 (34.7) | 30 (31.2) | 13 (23.2) | 7 (10.4) |
| PS score | |||||
| ≥2 | 100 (69.0) | 68 (46.9) | 62 (64.6) | 43 (76.8) | 39 (58.2) |
| <2 | 45 (31.0) | 33 (53.1) | 34 (35.4) | 13 (23.2) | 28 (41.8) |
| PFS, median (SD), mo | 10.1 (15.0) | 9.2 (9.0) | 8.2 (7.9) | 4.5 (4.7) | 3.6 (3.6) |
| EGFR variant | |||||
| EGFR 19Del | 72 (49.7) | 60 (59.4) | 41 (42.7) | 21 (37.5) | NA |
| EGFR 21L858R | 59 (40.7) | 35 (34.6) | 48 (50.0) | 30 (53.6) | NA |
| Other | 14 (9.6) | 6 (6.0) | 7 (7.3) | 5 (8.9) | NA |
| Tobacco use | |||||
| Smoker | 60 (41.3) | 21 (20.8) | 17 (17.7) | 14 (25.0) | 55 (82.1) |
| Nonsmoker | 85 (58.7) | 80 (79.2) | 79 (82.3) | 42 (75.0) | 12 (17.9) |
| Histopathology | |||||
| Adenocarcinoma | 135 (93.1) | 99 (98.0) | 92 (95.8) | 31 (55.4) | 12 (17.9) |
| SCC | 9 (6.2) | 1 (1.0) | 3 (3.1) | 25 (44.6) | 55 (82.1) |
| Other | 1 (0.7) | 1 (1.0) | 1 (1.1) | 0 | 0 |
Abbreviations: EGFR, epidermal growth factor receptor; CT, computed tomography; NA, not applicable; PFS, progression-free survival; PS, performance status; SCC, squamous cell carcinoma; TKI, tyrosine kinase inhibitor.
Data are presented as number (percentage) of patients unless otherwise indicated.
The median (range) follow-up periods were 15.9 (1.7-68.0) months in the training cohort, 13.5 (1.5-57.0) months in external validation cohort 1, and 11.8 (2.0-55.0) months in external validation cohort 2, and 313 of 342 patients (91.5%) in the EGFR-TKI cohorts experienced tumor progression (29 patients, censored data). No significant difference in PFS among the 3 cohorts (median [range] PFS, 9.9 [1.4-64.2] months in the training cohort, 9.2 [0.6-50.5] months in validation cohort 1, and 8.2 [0.8-40.6] months in validation cohort 2; Kruskal-Wallis H test, P = .20) was found. Furthermore, no significant differences in patient demographics among all 3 cohorts presented in the Table were found. Two patients receiving chemotherapy were censored during the follow-up; in 1 case, treatment was discontinued because of serious adverse reactions. The median (range) PFS were 3.6 (1.0-16.3) in months for EGFR wild-type NSCLC and 4.5 (0.9-25.9) months in EGFR variant-positive NSCLC.
DL Semantic Features and Signature
A total of 6204 CT sections that contained lung tumor(s) from the 342 patients in the EGFR-TKI therapy cohorts were included. In the last epoch of the training of the BigBiGAN framework, we extracted the semantic features when the network loss was minimum.
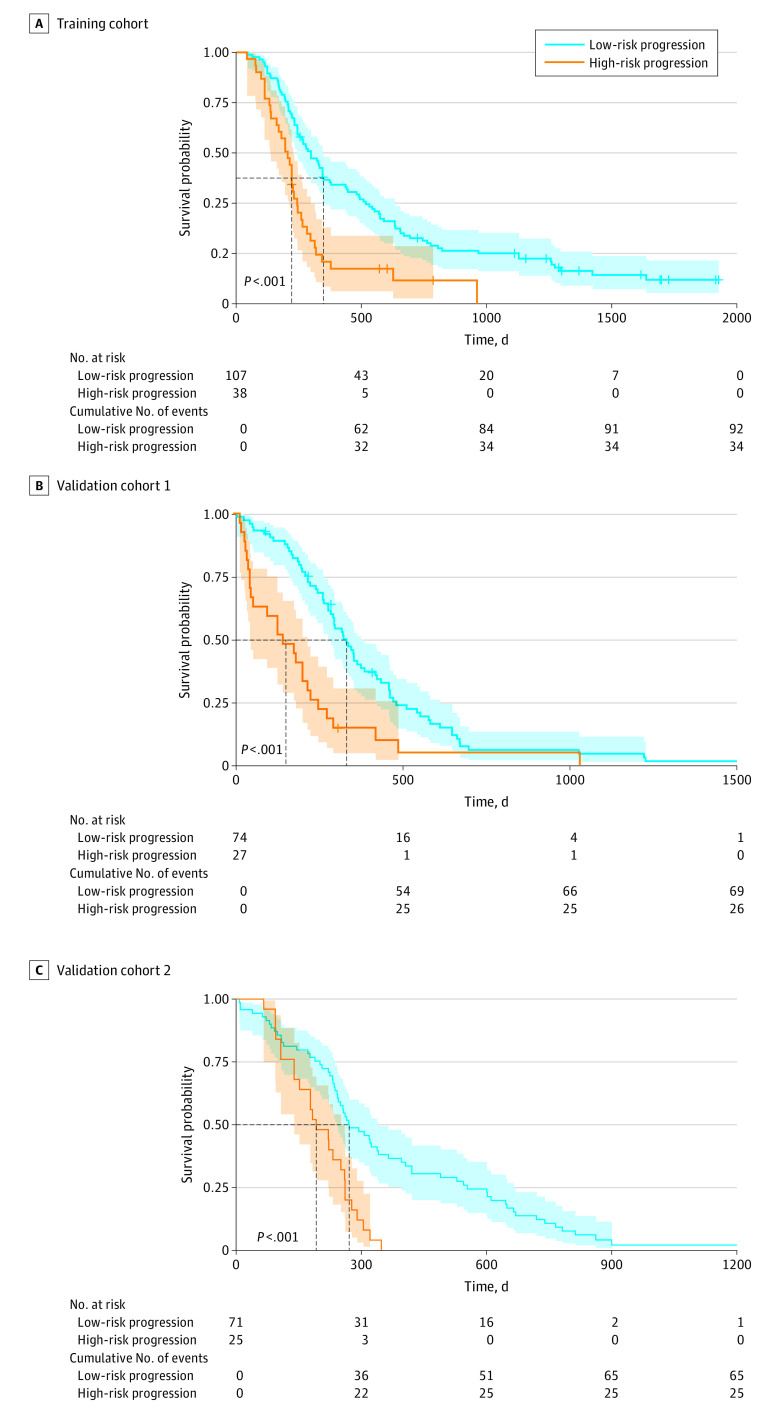
The 145 × 120–dimensional feature matrix obtained from the training cohort was input into the LASSO Cox proportional hazards regression model to construct the semantic prognostic signature. The coefficients of the 18 significant semantic features calculated by LASSO Cox proportional hazards regression are listed in eAppendix 2 in the Supplement. On the basis of the semantic signature score calculated for the patients in the training cohort, a cutoff threshold of −0.67 was obtained by X-tile to stratify the patients into low-progression-risk (107 patients; median [range] PFS, 11.5 [1.5-64.2] months) and high-progression-risk (38 patients; median [range] PFS, 7.3 [1.4-32.0] months) groups. A significant difference in PFS was found between the 2 groups (HR, 2.13; 95% CI, 1.30-3.49; P < .001). The Kaplan-Meier curves are presented in Figure 2A.
Figure 2. Kaplan-Meier Curves of Patients in the 3 Epidermal Growth Factor Receptor–Tyrosine Kinase Inhibitor Therapy Cohorts Stratified as High Risk and Low Risk of Rapid Progression Using the Deep Learning Semantic Signature Proposed in this Study.
Shaded areas indicate 95% CIs.
When applying the signature to the external validation cohorts, the median (range) PFS was 10.9 (1.1-50.5) months for the low-risk group (74 patients) and 5.0 (0.6-34.6) months for the high-risk group (27 patients) in validation cohort 1. Significant differences in PFS were found between the 2 risk groups (HR, 2.35; 95% 15 CI, 1.31–4.21; P < .001). Regarding validation cohort 2, a significant PFS difference was found between the low-risk (71 patients; median [range], 8.9 [0.8-40.6] months) and high-risk (25 patients; median [range], 6.4 [1.8-20.1] months) groups (HR, 2.32; 95% CI, 1.28–4.20; P < .001). The Kaplan-Meier curves are presented in Figure 2B and C.
Time-dependent ROC analysis indicated that the area under the curves of the survival benefits predicted by the semantic signature in the training cohort were 0.78 (95% CI, 0.75-0.80) under the 12-month cutoff and 0.75 (95% CI, 0.71-0.79) under the 10-month cutoff. The prediction area under the curves in external validation cohort 1 was 0.73 (95% CI, 0.70-0.77) under the 12-month cutoff and 0.72 (95% CI, 0.68-0.76) under the 10-month cutoff. In external validation cohort 2, the prediction area under the curves was 0.75 (95% CI, 0.72-0.80) under the 12-month cutoff and 0.70 (95% CI, 0.66-0.74) under the 10-month cutoff (eFigure 1 in the Supplement).
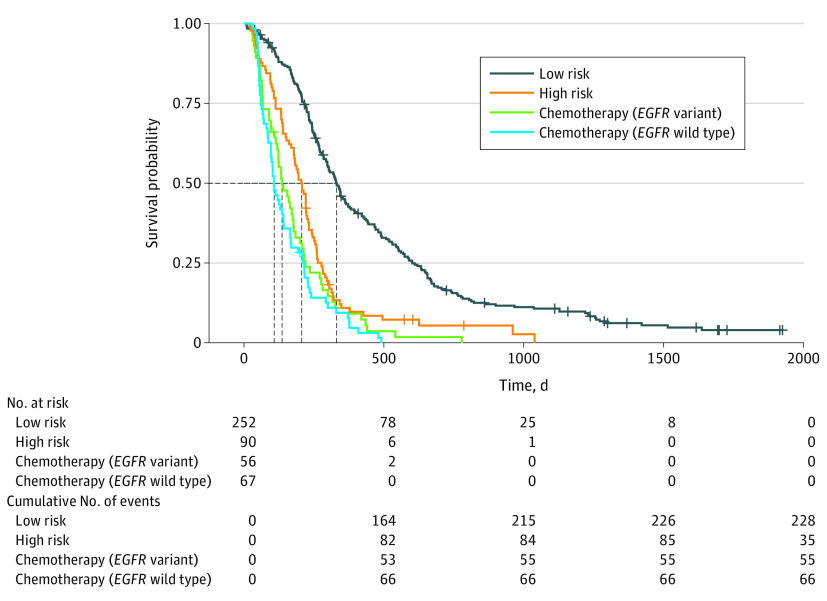
The results indicated that the median (range) PFS was 10.8 (0.8-64.2) months for the low-progression-risk group, 6.9 (0.6-34.6) months for the high-progression-risk group, 4.5 (0.9-25.9) months for the EGFR variant–positive chemotherapy group, and 3.6 (1.0-16.3) months for the EGFR wild-type chemotherapy group. No significant differences in PFS were found between the high-progression-risk patients in the EGFR-TKI cohort and the patients with EGFR variant–positive or EGFR wild-type NSCLC receiving chemotherapy (median PFS, 6.9 vs 4.4 months; P = .08). However, the survival benefit was significantly better in the low-progression-risk patients in the EGFR-TKI cohort than in the other 3 groups (median PFS, 10.8 vs 4.4 months; P < .001) (Figure 3). A more detailed comparison of these 4 groups is presented in eAppendix 3 in the Supplement.
Figure 3. Kaplan-Meier Curves of Patients With Low Progression Risk and High Progression Risk Who Received Epidermal Growth Factor Receptor–Tyrosine Kinase Inhibitor and Patients With EGFR Variant–Positive and EGFR Wild-Type Non–Small Cell Lung Cancer Who Received First-Line Chemotherapy.
The dotted lines represent the median time of progression-free survival of patients in each cohort.
Comparison With Radiomics Signatures
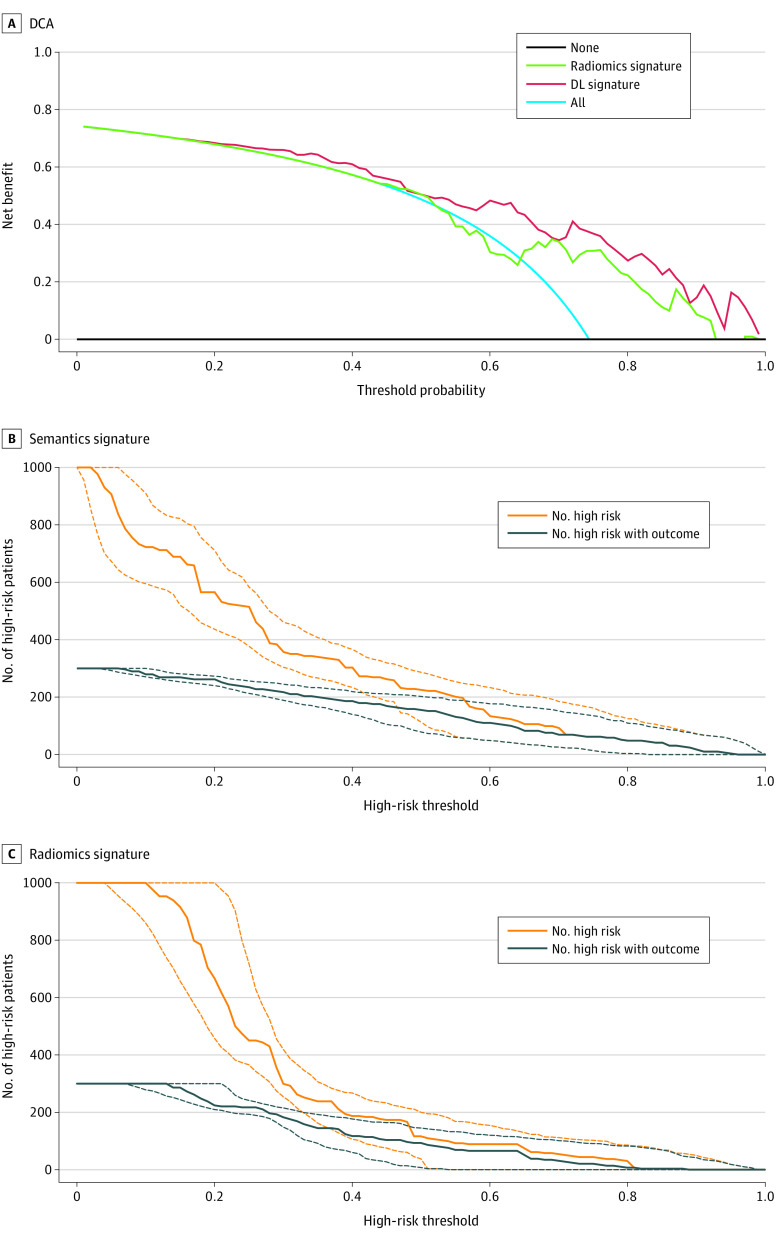
The decision curve analysis indicated that when the semantic signature is used to predict the risk of progression after EGFR-TKI therapy, patients could obtain better clinical benefits across all risk probabilities (Figure 4A). Furthermore, the comparison of clinical impact curves between the 2 signatures indicated that the predictions by the semantic signature of which patients were at high risk for progression were consistent with who actually experienced disease progression when the risk threshold was 0.7; this value was 0.8 for the radiomics signature (Figure 4B and C).
Figure 4. Decision Curve Analysis (DCA) and Clinical Impact Curve Analysis (CIA).
The orange line in the CIA plots indicates the number of patients predicted by deep learning (DL) to be at high risk for disease progression, and the blue line indicates the actual number at high risk for progression. The prediction result of the DL signature is consistent with that of patients who actually progressed when the high-risk probability was higher than 0.7.
Discussion
This diagnostic/prognostic study examined an end-to-end, CT-based DL approach to predict efficacy of EGFR-TKI therapy in patients with stage IV EGFR variant–positive NSCLC based on disease course. The study used a state-of-the-art representation learning framework to build a DL semantic prognosis signature from pretherapy CT images of patients with NSCLC. The proposed semantic signature suggested that 30% of patients were less likely to benefit from EGFR-TKI therapy. Patients with CT-based semantic features that were considered low risk for progression (74% of all patients) had a median (range) PFS of 327 (0.8-64.2) days compared with those with high-risk sematic features, who demonstrated a median (range) PFS of 208 (0.6-34.6) days. In addition, no significant difference in survival was found between patients with high-risk semantic features and who received first-line chemotherapy (without EGFR-TKI therapy: median PFS, 6.9 vs 4.4 months; P = .08).
Compared with radiomics, the approach used in this study not only avoids human intervention procedures requiring manual region of interest segmentation and predesigning heterogeneous features, but it also appears to be associated with better survival prognostic performance. Finally, the open-access source code provided in this study enables interested readers to reproduce the results of this study.
In a recent study8 on EGFR-TKI therapy, approximately 30% of patients failed to benefit from TKI drugs, with earlier disease progression in these patients. On the basis of DL semantic signature, patients predicted to have high risk of rapid progression accounted for 26% of all patients and a PFS that was similar to patients with advanced-stage NSCLC who received conventional chemotherapy. Furthermore, the clinical impact curve analysis curves indicated that when using the DL semantic signature to identify patients at high risk of progression, the predicted result was consistent with the patient’s actual tumor progression when the probability was higher than 0.7. This result had good agreement with the report of previous clinical studies,8,15 in which 30% of patients had rapid progression after EGFR-TKI therapy. These findings suggest potential use of a DL-based framework for more precisely stratifying patients with NSCLC who are more likely to benefit from EGFR-TKI therapy.
There have been many challenges to developing clinically applicable tools to evaluate the efficacy of EGFR-TKI therapy.26 Although potential image-based methods have been reported,15,16,26,27 important hurdles that relate to clinical operability and real-time requirements in clinics exist. Manual delineation of tumor boundary can create bias and can be labor-intensive, especially with multiple lung lesions.28 In this study, a DL-based approach was used to identify significant semantic features without requiring tumor margin delineation. Thus, unlike the more labor-intensive radiomics-based image predictors, an end-to-end DL approach to generate CT-based biomarkers to stratify patients who are more likely to benefit from EGFR-TKI therapy can facilitate more rapid clinical translation. Direct comparisons between the image features extracted by DL networks and those extracted by radiomics are currently lacking. These results also suggest that DL-based semantic features achieve better performance than radiomics in predicting the efficacy of EGFR-TKI therapy.
Chemotherapy remains the mainstay therapy for advanced-stage EGFR wild-type NSCLC. In this study, one potential reason for worse PFS in the EGFR wild-type group may be the rate of squamous cell carcinoma pathology, which was 82% in this group. Prior studies29,30 have suggested that the efficacy of chemotherapy in patients with lung adenocarcinoma is superior to that in patients with other pathologic subtypes. However, studies31,32 have not found this to be the case for EGFR-TKI therapy, prompting a clinical need to better stratify patients with EGFR variant–positive NSCLC for EGFR-TKI therapy efficacy.
Limitations
This study has limitations. The efficacy of osimertinib was much better than that of previous drugs, such as erlotinib, gefitinib, and icotinib, in this study. More patients receiving osimertinib should be included in future studies to clarify the risk of progression according to different TKI drugs. In addition, this study used CT images that only included lung tumors to train the model and thus reduce unnecessary calculations; therefore, screening of CT sections is needed. Also, there is currently no specific definition of the DL semantic features, and we will further explore the image encoding process corresponding to the DL semantic features to explain the specific image characteristics reflected by these features.
Conclusions
We describe an end-to-end CT-based DL approach to stratify patients who are more likely to benefit from EGFR-TKI therapy and contribute to precision in therapeutics in NSCLC. Unlike the more labor-intensive radiomics-based image predictors, an end-to-end DL approach to generate CT-based biomarkers can help toward more rapid clinical translation.
eAppendix 1. Open Access Source Code, CT Protocols of the Included Hospitals, Treatment Details and Follow-up, and the Radiomics Signature We Used for Comparison
eAppendix 2. Patient Enrollment and Construction and Performance of the Signature
eAppendix 3. Comparison of EGFR-TKI and Chemotherapy
References
- 1.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31(8):1070-1080. doi: 10.1200/JCO.2012.43.3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly K, Altorki NK, Eberhardt WE, et al. Adjuvant erlotinib versus placebo in patients with stage IB-IIIA non-small-cell lung cancer (RADIANT): a randomized, double-blind, phase III trial. J Clin Oncol. 2015;33(34):4007-4014. doi: 10.1200/JCO.2015.61.8918 [DOI] [PubMed] [Google Scholar]
- 3.Gao G, Ren S, Li A, et al. Epidermal growth factor receptor-tyrosine kinase inhibitor therapy is effective as first-line treatment of advanced non-small-cell lung cancer with mutated EGFR: a meta-analysis from six phase III randomized controlled trials. Int J Cancer. 2012;131(5):E822-E829. doi: 10.1002/ijc.27396 [DOI] [PubMed] [Google Scholar]
- 4.Soria JC, Wu YL, Nakagawa K, et al. Gefitinib plus chemotherapy versus placebo plus chemotherapy in EGFR-mutation-positive non-small-cell lung cancer after progression on first-line gefitinib (IMPRESS): a phase 3 randomised trial. Lancet Oncol. 2015;16(8):990-998. doi: 10.1016/S1470-2045(15)00121-7 [DOI] [PubMed] [Google Scholar]
- 5.Ettinger DS, Aisner DL, Wood DE, et al. NCCN guidelines insights: non-small cell lung cancer, Version 5.2018. J Natl Compr Canc Netw. 2018;16(7):807-821. doi: 10.6004/jnccn.2018.0062 [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi T, Ando M, Asami K, et al. Randomized phase III trial of erlotinib versus docetaxel as second- or third-line therapy in patients with advanced non-small-cell lung cancer: Docetaxel and Erlotinib Lung Cancer Trial (DELTA). J Clin Oncol. 2014;32(18):1902-1908. doi: 10.1200/JCO.2013.52.4694 [DOI] [PubMed] [Google Scholar]
- 7.Spigel DR, Edelman MJ, O’Byrne K, et al. Results from the phase III randomized trial of onartuzumab plus erlotinib versus erlotinib in previously treated stage IIIB or IV non-small-cell lung cancer: METLung. J Clin Oncol. 2017;35(4):412-420. doi: 10.1200/JCO.2016.69.2160 [DOI] [PubMed] [Google Scholar]
- 8.Barnet MB, O’Toole S, Horvath LG, et al. EGFR-co-mutated advanced NSCLC and response to egfr tyrosine kinase inhibitors. J Thorac Oncol. 2017;12(3):585-590. doi: 10.1016/j.jtho.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 9.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28(4):784-790. doi: 10.1093/annonc/mdx017 [DOI] [PubMed] [Google Scholar]
- 10.Wu YL, Lee JS, Thongprasert S, et al. Intercalated combination of chemotherapy and erlotinib for patients with advanced stage non-small-cell lung cancer (FASTACT-2): a randomised, double-blind trial. Lancet Oncol. 2013;14(8):777-786. doi: 10.1016/S1470-2045(13)70254-7 [DOI] [PubMed] [Google Scholar]
- 11.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16(9):e447-e459. doi: 10.1016/S1470-2045(15)00246-6 [DOI] [PubMed] [Google Scholar]
- 12.Crystal AS, Shaw AT, Sequist LV, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346(6216):1480-1486. doi: 10.1126/science.1254721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soucheray M, Capelletti M, Pulido I, et al. Intratumoral heterogeneity in EGFR-Mutant NSCLC results in divergent resistance mechanisms in response to EGFR tyrosine kinase inhibition. Cancer Res. 2015;75(20):4372-4383. doi: 10.1158/0008-5472.CAN-15-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou C, Yao LD. Strategies to improve outcomes of patients with EGRF-mutant non-small cell lung cancer: review of the literature. J Thorac Oncol. 2016;11(2):174-186. doi: 10.1016/j.jtho.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 15.Song J, Shi J, Dong D, et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res. 2018;24(15):3583-3592. doi: 10.1158/1078-0432.CCR-17-2507 [DOI] [PubMed] [Google Scholar]
- 16.Li H, Zhang R, Wang S, et al. CT-based radiomic signature as a prognostic factor in stage IV ALK-positive non-small-cell lung cancer treated with TKI crizotinib: a proof-of-concept study. Front Oncol. 2020;10:57. doi: 10.3389/fonc.2020.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150-R166. doi: 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559-1567. doi: 10.1038/s41591-018-0177-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao W, Yang J, Ni B, et al. Toward automatic prediction of EGFR mutation status in pulmonary adenocarcinoma with 3D deep learning. Cancer Med. 2019;8(7):3532-3543. doi: 10.1002/cam4.2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal. 2019;58:101552. doi: 10.1016/j.media.2019.101552 [DOI] [PubMed] [Google Scholar]
- 21.Donahue J, Simonyan K Large scale adversarial representation learning. Paper presented at: Advances in Neural Information Processing Systems; December 8, 2019; Vancouver, Canada. [Google Scholar]
- 22.Mozafari M, Reddy L, VanRullen R Reconstructing natural scenes from fMRI patterns using BigBiGAN. Cornell University Library. May 17, 2020. Accessed November 4, 2020. https://arxiv.org/abs/2001.11761
- 23.Song J, Wang H, Liu Y, et al. End-to-end automatic differentiation of the coronavirus disease 2019 (COVID-19) from viral pneumonia based on chest CT. Eur J Nucl Med Mol Imaging. 2020;47(11):2516-2524. doi: 10.1007/s00259-020-04929-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015;350:g7594. doi: 10.1136/bmj.g7594 [DOI] [PubMed] [Google Scholar]
- 25.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252-7259. doi: 10.1158/1078-0432.CCR-04-0713 [DOI] [PubMed] [Google Scholar]
- 26.O’Connor JP, Jackson A, Asselin MC, Buckley DL, Parker GJ, Jayson GC. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 2008;9(8):766-776. doi: 10.1016/S1470-2045(08)70196-7 [DOI] [PubMed] [Google Scholar]
- 27.Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749-762. doi: 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 28.Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46(13):2638-2655. doi: 10.1007/s00259-019-04391-8 [DOI] [PubMed] [Google Scholar]
- 29.Dahlberg SE, Sandler AB, Brahmer JR, Schiller JH, Johnson DH. Clinical course of advanced non-small-cell lung cancer patients experiencing hypertension during treatment with bevacizumab in combination with carboplatin and paclitaxel on ECOG 4599. J Clin Oncol. 2010;28(6):949-954. doi: 10.1200/JCO.2009.25.4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scagliotti GV, Parikh P, von Pawel J, et al. Phase III study comparing cisplatin plus gemcitabine with cisplatin plus pemetrexed in chemotherapy-naive patients with advanced-stage non-small-cell lung cancer. J Clin Oncol. 2008;26(21):3543-3551. doi: 10.1200/JCO.2007.15.0375 [DOI] [PubMed] [Google Scholar]
- 31.Kawaguchi T, Ando M, Asami K, et al. Randomized phase III trial of erlotinib versus docetaxel as second-or third-line therapy in patients with advanced non–small-cell lung cancer: Docetaxel and Erlotinib Lung Cancer Trial (DELTA). J Clin Oncol. 2014;32(18):1902-1908. doi: 10.1200/JCO.2013.52.4694 [DOI] [PubMed] [Google Scholar]
- 32.Chiu CH, Chou TY, Chiang CL, Tsai CM. Should EGFR mutations be tested in advanced lung squamous cell carcinomas to guide frontline treatment? Cancer Chemother Pharmacol. 2014;74(4):661-665. doi: 10.1007/s00280-014-2536-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix 1. Open Access Source Code, CT Protocols of the Included Hospitals, Treatment Details and Follow-up, and the Radiomics Signature We Used for Comparison
eAppendix 2. Patient Enrollment and Construction and Performance of the Signature
eAppendix 3. Comparison of EGFR-TKI and Chemotherapy