Abstract
Background:
Several available data suggest the association between specific molecular subtypes and BRCA1/2 mutational status. Previous investigations showed the association between BRCA1/2 pathogenic variants (PVs) in specific genomic regions and phenotypic variations of cancer relative risk, while the role of PV type and location in determining the breast cancer (BC) phenotypic features remains still unclear. The aim of this research was to describe the germline BRCA1/2 PVs in triple-negative breast cancer (TNBC) versus luminal-like BC and their potential leverage on BC phenotype.
Patients & methods:
We retrospectively collected and analyzed all clinical information of 531 patients with BC genetically tested for germline BRCA1/2 PVs by Next-Generation Sequencing analysis at University Hospital Policlinico “P. Giaccone” of Palermo (Sicily) from January 2016 to February 2020.
Results:
Our results corroborate the evidence that BRCA1-related tumors often have a profile which resembles the TNBC subtype, whereas BRCA2-associated tumors have a profile that resembles luminal-like BC, especially the Luminal B subtype. Interestingly, our findings suggest that the PVs identified in TNBC were not largely overlapping with those in luminal-like tumors. Differences in the frequency of two PVs potentially associated with different molecular tumor subtypes were observed. BRCA1-633delC was detected with relatively higher prevalence in patients with TNBC, whereas BRCA2-1466delT was found mainly in Luminal B tumors, but in no TNBC patient.
Conclusion:
Future studies examining the type and location of BRCA1/2 PVs within different molecular subtypes are required to verify our hypothesis and could provide an interesting insight into the complex topic of genotype–phenotype correlations. Additionally, a more in-depth understanding of the potential correlations between BRCA PVs and clinical and phenotypic features of hereditary BC syndrome patients could be the key to develop better strategies of prevention and surveillance in BRCA-positive carriers without disease.
Keywords: BRCA1, BRCA2, breast cancer, genetic testing, germline pathogenic variants, luminal-like breast cancer, triple-negative breast cancer
Introduction
The critical role of BRCA1 and BRCA2 genes in breast cancer (BC) and ovarian cancer (OC) was discovered for the first time in 1994 and 1995.1,2 Since then, the characteristics of BRCA1/2-related BCs have been well investigated. The recent diffusion of high-throughput next-generation sequencing technologies has provided a deep insight into the genetics and molecular biology of these tumors,3 shedding light on their biological heterogeneity. Over the years, the eligibility criteria for BRCA1/2 genetic testing have been expanded and updated.4,5 BRCA1 and 2, in addition to being considered indicators for cancer risk assessment, have become also biomarkers of predictive utility. Several other genes involved in the homologous recombination (HR) pathway have been evaluated,6–10 and the knowledge on BRCA-mediated tumor phenotypes continues to evolve. In this context, marked by great progress and change in the field of genetic testing, the study and characterization of pathogenic variants (PVs) in BRCA1/2 genes are becoming increasingly important for BC and OC screening and prevention.
Germline PVs detected in BRCA1/2 genes are the primary causes of inherited breast tumors and confer an approximately 70% of lifetime risk in mutation carriers.11,12 PVs are detected in 5–10% of all women with BC, and the prevalence increases to 10–20% in triple-negative breast cancer (TNBC).13,14 TNBC is a particularly proliferative and aggressive subtype because the absence of actionable targets results in poor prognosis and unfavorable outcomes for the patients.15,16
Several available data suggest the association between specific molecular subtypes and BRCA1/2 mutational status. BRCA1/2 PV carriers have been shown to harbor TNBC more frequently, and this association is strongest of all in BRCA1-related BC.17,18
Studies about the molecular and clinicopathological characteristics of patients with BRCA1- versus BRCA2-related BC showed significant and distinct features. Approximately 70% of BCs developing in BRCA1 PV carriers are TNBCs, which tend to have higher histological grade than BRCA2 PV carriers, and are most frequently invasive ductal carcinomas. Conversely, approximately 75% of BCs in BRCA2 PV carriers are estrogen-receptor (ER)-positive, more often Luminal B. ER-positive BCs which develop in BRCA PV carriers are lobular or ductal type, and have higher histologic grade than sporadic ER-positive BCs.19,20
These differences highlight the heterogeneity in BC biology and molecular phenotype among tumors related to different germline BRCA1/2 PVs. Although the molecular and clinicopathological characteristics of BRCA1/2-related BCs have been well investigated, the available studies on the genotype–phenotype correlations are predominantly based on the involvement of the BRCA1 or BRCA2 gene. To date, the role of PV type and its location in determining the BC phenotypic features still remains unclear. Previous investigations showed the association between BRCA1/2 PVs in specific genomic regions and phenotypic variations of cancer relative risks.21 Further and deeper information on BRCA PVs could help us better understand the impact of genotype on breast tumor phenotype. Finally, some studies revealed the existence of a close correlation between specific BRCA1/2 PVs and variations of BC and OC relative risks, by identifying specific putative breast cancer cluster regions (BCCRs) and ovarian cancer cluster regions (OCCRs) located on the coding DNA sequences of BRCA1 and BRCA2 genes.21,22
Based on a Breast Cancer BRCA System database retrospectively collected at University Hospital Policlinico “P. Giaccone” of Palermo, the aim of this research was to describe the type and gene location of germline BRCA1/2 PVs in TNBC versus other BC molecular subtypes (Luminal A, Luminal B and HER2-enriched) and their potential leverage on BC phenotype.
Patients and methods
Study population
We carried out a retrospective cohort study at the “Sicilian Regional Center for the Prevention, Diagnosis and Treatment of Rare and Heredo-Familial Tumors” of the Section of Medical Oncology of University Hospital Policlinico “P. Giaccone” of Palermo. We have collected and analyzed all information regarding all women diagnosed with primary BC who underwent to germline BRCA1/2 genetic testing from January 2016 to February 2020.
Genetic counseling was performed by a multidisciplinary team consisting mainly of an oncologist, a geneticist and a psychologist. Information about personal and familial history of cancer, family geographical origin, age of cancer diagnosis, histological tumor subtype, molecular phenotype and disease stages (I–IV), was anonymously recorded for all patients who previously provided a written informed consent. The study (G-Land 2017, approval number: 01-03-2017) was approved by the ethical committee (Comitato Etico Palermo 1) of the University-affiliated hospital A.O.U.P. “P. Giaccone” of Palermo. The ER, progesterone-receptor (PgR), HER2-receptor status (HER2), Ki67 status, and histological grade (Grades I, II, and III) of the primary tumors were reported in local pathology testing of diagnostic core biopsies or tumor resections for clinical use. Based on histological grade and biomarker expression, invasive tumors were categorized as Luminal A-like (LA = ER/PR+ and HER2–, histological grade 1 or 2), Luminal B-like (LB = ER/PR+ and HER2+, or ER/PR+, HER2–, and grade 3), HER2 enriched (E) (ER/PR– and HER2+) or triple negative (TN = ER–, PR– and HER2–).23
The patients were selected for mutational screening based on probability rate of carrying PV assessed by the BRCAPRO genetic risk prediction model24 and according to the previously established criteria by the Italian Association of Medical Oncology (AIOM) (https://www.aiom.it/linee-guida-aiom-neoplasie-della-mammella-2019/). These criteria are based on personal and family history and age of cancer onset, in order to identify individuals at high risk of harboring a PV in the Hereditary Breast and Ovarian Cancer (HBOC) predisposition genes. The AIOM every year updates its guidelines for identifying the individuals who should receive BRCA genetic testing, and in 2016 AIOM included in the population to be genetically tested woman with TNBC <60 years, regardless of family history.
The following criteria were used to select patients to be genetically tested for germline BRCA1/2 PVs: (i) Personal history of (a) male with BC; (b) women with BC and OC; (c) woman with BC <36 years; (d) woman with TNBC <60 years; (e) woman with bilateral BC <50 years; (ii) Personal history of BC <50 years and at least one first-degree relative with: (a) BC <50 years; (b) non-mucinous and non-borderline OC at any age; (c) bilateral BC; (d) male BC; (iii) Personal history of BC >50 years and family history of BC or OC in two or more relatives who have a first-degree relationship with each other (including one who has a first-degree relationship with her); (iv) Family history of known pathogenic variant in a predisposing gene (https://www.aiom.it/linee-guida-aiom-neoplasie-della-mammella-2019/).
The BRCA test result was considered informative when a pathogenic or likely pathogenic variant was identified in an individual. Conversely, BRCA test result was considered not informative when no pathogenic or likely pathogenic variant was identified but its presence could not be excluded, or a variant of uncertain significance (VUS) to which it was not possible to attribute a risk value was detected.
Patients harboring a germline PV in BRCA1/2 genes were directed to enhanced screening programs and/or risk-reducing surgical strategies by a professional with expertise in cancer genetics. Targeted BRCA1/2 testing was proposed and extended to the first-degree family members of BRCA-variant patients, after providing informed consent.
Sample collection and next-generation sequencing analysis for BRCA1/2 genes
Peripheral blood samples were collected from BC patients. Genomic DNA was isolated from the peripheral blood using the DNeasy® Blood Kit (QIAGEN, Hilden, Germany), quantified by Qubit®3.0 fluorometer (Thermofisher Scientific, Waltham, MA, USA) and its quality was assessed by using 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). We used 4 ng of DNA to prepare the barcoded library using BRCA Screen kit (4bases SA) that has allowed us to investigate all the exons of BRCA1 (NM_007300.3) and BRCA2 (NM_000059.3) genes. The kit consists of three multiplex PCR primer pools. We used 20 ng of DNA per primer pool for multiplex PCR amplification, followed by ligation of a barcode and purification with Agentcourt AMPureXP reagent (Beckman Coulter, Beverly, MA, USA). The quantity and the quality of prepared libraries were evaluated using Qubit®3.0 fluorometer (Thermofisher Scientific, Waltham, MA, USA) and Agilent 2100 Bioanalyzer on-chip electrophoresis (Agilent Technologies, Santa Clara, CA), respectively, as previously described.25 Subsequently, libraries were mixed in an equimolar ratio and emulsion PCR was performed with the Ion OneTouch OT2 System (Thermofisher Scientific, Waltham, MA, USA) using Ion 520 & Ion 530 Kit-OT2 (Thermofisher Scientific, Waltham, MA, USA). Finally, sequencing was performed with Ion 520 Chip (Thermofisher Scientific, Waltham, MA, USA) using Ion Torrent S5 (Thermofisher Scientific, Waltham, MA, USA) instrument. The sequencing data were analyzed with Amplicon Suite (SmartSeq s.r.l., Novara, Italy) and Ion Reporter Software v.5.12 (Thermofisher Scientific, Waltham, MA, USA).
Sanger sequencing
Pathogenic variants of BRCA1/2 genes were confirmed by Sanger sequencing using a BigDye Therminator 3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) and read through the 3130×l Genetic Analyzer (Applied Biosystems, Foster City, CA, USA), according to the manufacturers’ protocols.
CNV analysis by multiplex ligation-dependent probe amplification analysis
The presence of large genomic rearrangements (LGRs) was additionally tested by Multiplex ligation-dependent probe amplification (MLPA), using the SALSA MLPA probemix P002-C2 for BRCA1 gene and SALSA MLPA Probemix P090 for BRCA2 gene according to the manufacturer’s instructions (MRC–Holland, Amsterdam, the Netherlands). Probe amplification products were analyzed by capillary electrophoresis using ABI 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). Results were analyzed by GeneMapper™ Software Version 3.5 (Applied Biosystems, Foster City, CA, USA) to determine peak heights and areas, and fragment sizes in base pairs (bp), as described previously.26,27 Positive results were confirmed with an additional analysis using the same kit on a second blood sample.
Genetic variant classification
The BRCA genetic variants were screened based on the classification criteria developed by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) consortium (https://enigmaconsortium.org/) and according to IARC recommendations,28 using a system of division into five classes: benign (class I), likely benign (class II), VUS (class III), likely pathogenic (class IV), and pathogenic (class V). Several databases, such as ClinVar, BRCA Exchange, LOVD, were used for the search and classification of BRCA variants. The positions of the variants on BRCA1 and BRCA2 genes were obtained and graphically represented using the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics.29,30
The BRCA PVs identified in BRCA-positive carriers were named according to the systematic nomenclature of The Breast Cancer Information Core (BIC) database (http://research.nhgri.nih.gov/bic/),31 and to the recommendations for the description of sequence variants established by the Human Genome Variation Society (HGVS). HGVS nomenclature was authorized by the HGVS, Human Variome Project, and the Human Genome Organization.32
Statistical analysis
Clinicopathological variables and prevalence of BRCA1/2 PVs were evaluated for each subgroup of patients. The comparison between subgroups was made with Fisher’s Exact test. p-values <0.05 were considered significant.
Statistical analyses were conducted using IBM SPSS Statistics for Windows Version 23.0 (IBM Corporation, Armonk, NY, USA).
Results
Distribution of molecular subtypes
Between 1 January 2016 and 28 February 2020, 531 patients with BC who met eligibility criteria for BRCA1/2 gene testing were included in a retrospective analysis. Mutational screening was offered at the “Regional Center for the prevention, diagnosis and treatment of rare and heredo-familial tumors of adults” of the Section of Medical Oncology of the University Hospital Policlinico “P. Giaccone” of Palermo, according to national guidelines (see section Patients and Methods). All women were tested for BRCA1 and BRCA2 germline PVs, after appropriate genetic counseling.
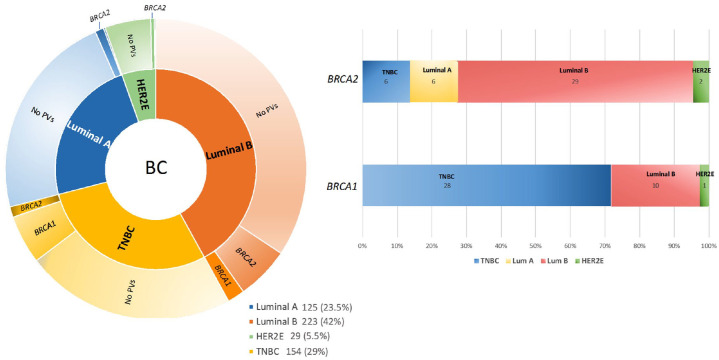
The distribution of BC molecular subtypes involved 125 (23.5%) Luminal A (LA), 223 (42%) Luminal B (LB), 29 (5.5%) HER2-enriched (HER2E), and 154 (29%) TNBC.
Breast cancer patients with BRCA1/2 pathogenic variants
In total, 83 out of 531 BC patients (15.6%) resulted positive for BRCA1/2 PVs; 39 (47%) were BRCA1-positive, 43 (51.8%) were BRCA2-positive, and one patient (1.2%) showed double heterozygosity for BRCA1 and BRCA2 PVs. Among BRCA1-positive patients, 28 (71.9%) had a TNBC, 10 (25.6%) a LB tumor, one (2.5%) HER2E and none LA. Among BRCA2-positive tumors, 29 (67.5%) were LB, six (13.9%) TNBC, six (13.9%) LA and two (4.7%) HER2E (Figure 1).
Figure 1.
Number of breast cancer patients genetically tested for BRCA1/2 PVs (Jan 2016–Feb 2020).
(a) Distribution of molecular subtypes in the study population; (b) Prevalence of molecular subtypes in BRCA1- and BRCA2-positive breast cancer patients. The numbers reported inside each box indicate the patient number, excluding the LB breast cancer patient showing double heterozygosity for BRCA1 and BRCA2 PVs.
Tumors from patients with BRCA1 PVs were predominantly TNBCs (p = 0.0001) and tumors with BRCA2 PVs were mainly LB/HER2-negative (p = 0.0014) (Table 1).
Table 1.
BRCA1/2 PV detection rate in Luminal A, Luminal B, HER2-enriched and TNBC patients.
| Total | BRCA1 | BRCA2 | DH BRCA1-BRCA2 PVs | Absence of PVs | p-value * | |
|---|---|---|---|---|---|---|
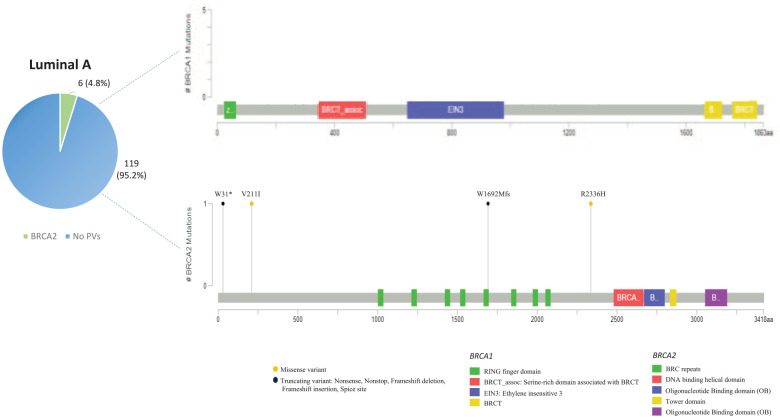
| Luminal A | 125 | 0 (0%) | 6 (4.8%) | 0 (0%) | 119 (95.2%) | p = 0.213 |
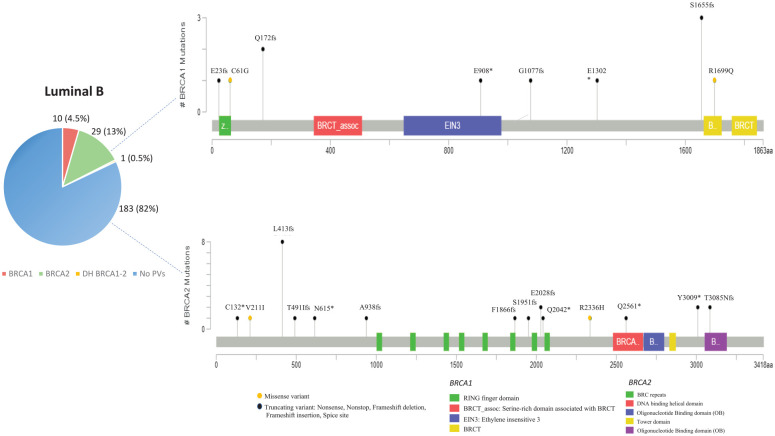
| Luminal B | 223 | 10 (4.5%) | 29 (13%) | 1 (0.5%) | 183 (82%) | p = 0.0014 |
| HER2E | 29 | 1 (3.4%) | 2 (6.8%) | 0 (0%) | 26 (89.8%) | p = 1.00 |
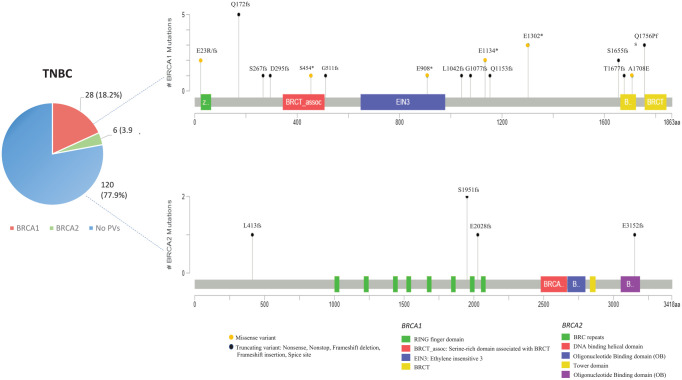
| TNBC | 154 | 28 (18.2%) | 6 (3.9%) | 0 (0%) | 120 (77.9%) | p = 0.0001 |
Comparison BRCA1 PV versus BRCA2 PV versus BRCA1/2 w.t.
DH, Double Heterozygosity; HER2E, Her2-enriched; TNBC, Triple Negative Breast Cancer.
Type and gene location of pathogenic variants of triple-negative versus luminal-like breast cancers
Our study was also aimed to evaluate the typology and gene location of germline BRCA1 and BRCA2 PVs in triple-negative versus luminal-like BCs, in order to investigate potential associations between specific PVs and tumor phenotype.
Based on the classification criteria developed by the ENIGMA consortium (https://enigmaconsortium.org/) and according to the IARC recommendations,28 mutational analysis revealed that 45 PVs were present in a total of 83 patients with BRCA1/2-related BC. Mutational screening showed that 23 (51.1%) out of 45 observed PVs were detected in TNBCs, 18 of which were in BRCA1 gene and five in BRCA2 gene (Table 2), whereas 33 PVs were found in luminal-like BCs, eight of which in BRCA1 and 25 in BRCA2, and, finally, three were observed in HER2E BCs, one of which in BRCA1 and two in BRCA2 (Table 3). Fourteen of 45 total PVs are reported two folds in Tables 2 and 3, beacause they were observed in different molecular subtypes of BC.
Table 2.
BRCA1/2 pathogenic variants in TNBCs.
| TNBC | ||||||
|---|---|---|---|---|---|---|
| Gene | Type of PV | HGVS Nomenclature | BIC Nomenclature | Protein change | No. families | No. PV carriers (patients and family members) |
| BRCA1 | Deletion | c.514del | 633delC | p.Gln172fs | 5 (14.5%) | 15 |
| BRCA1 | SNV | c.3904G>T | 4023G>T | p.Glu1302Ter | 3 (9.1%) | 10 |
| BRCA1 | Duplication | c.5266dupC | 5382insC | p.Gln1756Profs | 3 (9.1%) | 5 |
| BRCA1 | Deletion | c.4964_4982del | 5083del19 | p.Ser1655fs | 2 (6%) | 4 |
| BRCA1 | SNV | c.3400G>T | 3519G>T | p.Glu1134Ter | 2 (6%) | 2 |
| BRCA1 | Deletion | c.798_799del | 916delTT | p.Ser267fs | 1 (2.9%) | 9 |
| BRCA1 | Deletion | c.1360_1361del | 1479delAG | p.Glu453_Ser454insTer | 1 (2.9%) | 7 |
| BRCA1 | Deletion | c.3228_3229del | 3347delAG | p.Gly1077fs | 1 (2.9%) | 6 |
| BRCA1 | Deletion | c.1531del | / | / | 1 (2.9%) | 6 |
| BRCA1 | Deletion | c.5030_5033del | 5147del4 | p.Thr1677fs | 1 (2.9%) | 4 |
| BRCA1 | Duplication | c.66dupA | 185insA | p.Glu23Argfs | 1 (2.9%) | 3 |
| BRCA1 | SNV | c.5123C>A | 5242C>A | p.Ala1708Glu | 1 (2.9%) | 1 |
| BRCA1 | Deletion | c.3266del | 3385delT | p.Leu1089fs | 1 (2.9%) | 1 |
| BRCA1 | Deletion | c.3599_3600del | 3718delAG | p.Gln1200Argfs | 1 (2.9%) | 1 |
| BRCA1 | Deletion | c.882del | 1001delA | p.Asp295fs | 1 (2.9%) | 1 |
| BRCA1 | SNV | c.2722G>T | 2841G>T | p.Glu908Ter | 1 (2.9%) | 1 |
| BRCA1 | Deletion | c.66_67del | 185_186delAG | p.Glu23fs | 1 (2.9%) | 1 |
| BRCA1 | LGR | c.-232_4675del | / | / | 1 (2.9%) | 1 |
| BRCA2 | Deletion | c.5851_5854del | 6076del4 | p.Ser1951fs | 2 (6%) | 4 |
| BRCA2 | SNV | c.8954-15T>G | / | / | 1 (2.9%) | 4 |
| BRCA2 | Deletion | c.1238del | 1466delT | p.Leu413fs | 1 (2.9%) | 1 |
| BRCA2 | Deletion | c.9455_9456del | 9683delAG | p.Glu3152fs | 1 (2.9%) | 1 |
| BRCA2 | Deletion | c.6082_6086del | 6310del5 | p.Glu2028fs | 1 (2.9%) | 1 |
Abbreviations: LGR, large genomic rearrangement; PV, pathogenic variant; SNV, single nucleotide variant.
Table 3.
BRCA1/2 pathogenic variants in luminal-like and HER2E patients.
| Luminal B | ||||||
|---|---|---|---|---|---|---|
| Gene | Type of PV | HGVS Nomenclature | BIC Nomenclature | Protein change | No. families | No. PV carriers (patients and family members) |
| BRCA1 | Deletion | c.4964_4982del | 5083del19 | p.Ser1655fs | 3 (7.5%) | 7 |
| BRCA1 | Deletion | c.514del | 633delC | p.Gln172fs | 2 (5%) | 2 |
| BRCA1 | SNV | c.2722G>T | 2841G>T | p.Glu908Ter | 1 (2.4%) | 2 |
| BRCA1 | SNV | c.5096G>A | 5215G>A | p.Arg1699Gln | 1 (2.4%) | 1 |
| BRCA1 | Deletion | c.3228_3229del | 3347delAG | p.Gly1077fs | 1 (2.4%) | 1 |
| BRCA1 | Deletion | c.66_67del | 185_186delAG | p.Glu23fs | 1 (2.4%) | 1 |
| BRCA1 | SNV | c.3904G>T | 4023G>T | p.Glu1302Ter | 1 (2.4%) | 1 |
| BRCA2 | Deletion | c.1238del | 1466delT | p.Leu413fs | 8 (19.5%) | 15 |
| BRCA2 | Deletion | c.9026_9030del | 9254del5 | p.Tyr3009fs | 2 (5%) | 4 |
| BRCA2 | Deletion | c.6082_6086del | 6310del5 | p.Glu2028fs | 2 (5%) | 4 |
| BRCA2 | SNV | c.476-2A>G | IVS5-2A>G | / | 2 (5%) | 4 |
| BRCA2 | Duplication | c.9253dup | 9481insA | p.Thr3085Asnfs | 2 (5%) | 2 |
| BRCA2 | SNV | c.631G>A | 859G>A | p.Val211Ile | 1 (2.4%) | 3 |
| BRCA2 | Deletion | c.5851_5854del | 6076del4 | p.Ser1951fs | 1 (2.4%) | 3 |
| BRCA2 | SNV | c.8754+4A>G | IVS21+4A>G | / | 1 (2.4%) | 3 |
| BRCA2 | SNV | c.8632+2T>C | / | / | 1 (2.4%) | 2 |
| BRCA2 | SNV | c.6124C>T | 6352C>T | p.Gln2042Ter | 1 (2.4%) | 2 |
| BRCA2 | SNV | c.7681C>T | 7909C>T | p.Gln2561Ter | 1 (2.4%) | 2 |
| BRCA2 | Deletion | c.2808_2811del | 3036del4 | p.Ala938Profs | 1 (2.4%) | 2 |
| BRCA2 | Duplication | c.1842dup | 2070insT | p.Asn615Terfs | 1 (2.4%) | 1 |
| BRCA2 | SNV | c.7007G>A | 7235G>A | p.Arg2336His | 1 (2.4%) | 1 |
| BRCA2 | Deletion | c.1472del | 1700delC | p.Thr491Ilefs18 | 1 (2.4%) | 1 |
| BRCA2 | SNV | c.396T>A | 624T>A | p.Cys132Ter | 1 (2.4%) | 1 |
| BRCA2 | Deletion | c.5595_5596del | 5823delAT | p.Phe1866fs | 1 (2.4%) | 1 |
| BRCA2 | SNV | c.8487+1G>A | IVS19+1G>A | / | 1 (2.4%) | 1 |
| BRCA1/BRCA2 | SNV | c.181T>G* | 300T>G* | p.Cys61Gly | 1 (2.4%) | 4 |
| SNV | c.8331+2T>C* | IVS18+2T>C* | / | |||
| Luminal A | ||||||
| Gene | Type of PV | HGVS Nomenclature | BIC Nomenclature | Protein change | No. families | No. PV carriers (patients and family members) |
| BRCA2 | SNV | c.631G>A | 859G>A | p.Val211Ile | 1 (16.67%) | 3 |
| BRCA2 | SNV | c.8487+1G>A | IVS19+1G>A | / | 1 (16.67%) | 2 |
| BRCA2 | SNV | c.93G>A | 321G>A | p.Trp31Ter | 1 (16.67%) | 1 |
| BRCA2 | SNV | c.7007G>A | 7235G>A | p.Arg2336His | 1 (16.67%) | 1 |
| BRCA2 | Duplication | c.5073dup | 5301insA | p.Trp1692Metfs | 1 (16.67%) | 1 |
| BRCA2 | SNV | c.8754+4A>G | IVS21+4A>G | / | 1 (16.67%) | 1 |
| HER2E | ||||||
| Gene | Type of PV | HGVS Nomenclature | BIC Nomenclature | Protein change | No. families | No. PV carriers (patients and family members) |
| BRCA1 | Duplication | c.5266dupC | 5382insC | p.Gln1756Profs | 1 (33.3%) | 2 |
| BRCA2 | Deletion | c.5073del | 5301delA | p.Lys1691fs | 1 (33.3%) | 1 |
| BRCA2 | Deletion | c.7679-7680del | 7907delTT | p.Phe2560fs | 1 (33.3%) | 1 |
Abbreviations: PV, Pathogenic Variant; SNV, Single Nucleotide Variant.
These PVs are present in one proband showing double heterozygosity for BRCA1 and BRCA2 PVs.
The most frequent PV identified in most of the analyzed BRCA1-positive TNBCs is named BRCA1-633delC (HGVS nomenclature: c.514del; p.Gln172fs) and involves the deletion of one nucleotide containing a cytosine (C) in BRCA1 exon 8, which causes a frameshift, resulting in the substitution of the amino acid glutamine with asparagine at codon 172, creation of a premature stop codon at position 62 of the new reading frame, and formation of a truncated or absent BRCA1 protein.33,34 This alteration was detected in five Sicilian families, involving a total of 15 PV carriers (five probands and 10 family members). The second most recurrent BRCA1 PV associated with TNBCs and found in three families for a total of 10 PV carriers (three probands and seven family members) is named BRCA1-4023G>T (HGVS nomenclature: c.3904G>T; p.Glu1302Ter). This variant involves the substitution of one nucleotide containing a guanine (G) with another containing a thymine (T) in the BRCA1 exon 11, causing the change of glutamic acid with a premature stop codon at codon 1302, resulting in the formation of a truncated BRCA1 protein.35 In general, most PVs detected in the BRCA1 gene of TNBC patients showed a low prevalence. Also, the few identified BRCA2 PVs were detected with a lower frequency in TNBC patients. Therefore, no association between specific BRCA1/2 PVs and TNBC in Sicilian study population was observed (Figure 2).
Figure 2.
Lollipop plots showing the distribution and frequency of BRCA1 and BRCA2 PVs identified in TNBC patients. The plots were obtained by the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics (GenBank Reference BRCA1: NM_007294 and GenBank Reference BRCA2: NM_000059). The Intronic Variant Sequences (IVS) are not shown in the lollipop plots. The lollipop height indicates the frequency of BRCA1/2 PVs in different molecular subgroups of our study cohort.
Concerning the distribution of BRCA1/2 PVs in patients with luminal-like BC, the most frequent PV, named BRCA2-1466delT (HGVS nomenclature: c.1238del; p.Leu413fs), was observed in the BRCA2 gene of LB molecular subtypes and detected in a total of 15 PV carriers (eight probands and seven family members). This variant involves the deletion of one nucleotide containing a thymine (T) in the BRCA2 exon 10, which causes a frameshift resulting in the change of a Leucine with a Histidine at codon 413, formation of a premature termination codon and loss of the normal protein function.36
Interestingly, the most common Sicilian founder variant named BRCA1-5083del19 (HGVS nomenclature: c.4964_4982del; p.Ser1655fs)27 showed a low prevalence both in TNBCs (6%) and luminal-like BCs (7.5% in LB tumors), as this PV was detected only in two and three families, respectively.
In general, most of the PVs found in patients with luminal-like BC are localized on BRCA2 gene. No BRCA1 PV was observed in patients harboring LA molecular subtypes. In the same way as TNBC patients, most PVs detected in BRCA1 and BRCA2 genes of LA and LB molecular subtypes showed a low prevalence in the Sicilian population, suggesting the absence of a significant association between specific BRCA1/2 PVs and luminal-like tumors (Figures 3 and 4).
Figure 3.
Lollipop plots showing the distribution and frequency of BRCA1 and BRCA2 PVs identified in luminal B breast cancer patients. The plots were obtained by the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics (GenBank Reference BRCA1: NM_007294 and GenBank Reference BRCA2: NM_000059). The Intronic Variant Sequences (IVS) are not shown in the lollipop plots. The lollipop height indicates the frequency of BRCA1/2 PVs in different molecular subgroups of our study cohort.
Figure 4.
Lollipop plots showing the distribution and frequency of BRCA1 and BRCA2 PVs identified in luminal A breast cancer patients. The plots were obtained by the informatic tool Mutation Mapper-cBioPortal for Cancer Genomics (GenBank Reference BRCA1: NM_007294 and GenBank Reference BRCA2: NM_000059). The Intronic Variant Sequences (IVS) are not shown in the lollipop plots. The lollipop height indicates the frequency of BRCA1/2 PVs in different molecular subgroups of our study cohort.
As regards the gene location of BRCA1 and BRCA2 PVs detected in TNBC patients of the Sicilian study cohort, most PVs have been shown to be localized inside three hypothetical cluster regions present in the BRCA1 protein structure which include the RING domain at the N-terminus, region encoded by exon 11, and BRCT domain near the C-terminus (Supplemental material Figure 1). Ten (55.5%) out of 18 BRCA1 PVs were detected in exon 11 of TNBC patients (nucleotides: 916-4023; codons: 267-1302), whereas four were in the sequence corresponding to the BRCT repeats (nucleotides: 5083-5382; codons: 1655-1756) and only two in the RING domain (nucleotide: 185; codon: 23). More than half (11) of BRCA1 PVs were frameshift mutations, whereas three were nonsense and three missense. Only a LGR involving a deletion (c.-232_4675del) ranging from exon 1 to exon 15 of BRCA1 gene was detected in one TNBC patient. Almost all BRCA2 PVs (four) observed in TNBCs were frameshift mutations distributed along the entire gene sequence (Supplemental material Figure 1).
Although BRCA1 PVs are poorly represented in patients with luminal-like BC, these few variants are equally distributed into three putative cluster regions containing the RING domain, region encoded by exon 11, and BRCT domain, as already observed in TNBC patients. Conversely, most PVs observed in patients with luminal-like BC were mainly localized inside three other putative cluster regions present in the BRCA2 protein structure, which include the PALB2 binding site at the N-terminus, BRC repeats (located within the exon 11), and DNA binding helical domain near the C-terminus (Supplemental material Figure 2). Six (24%) out of 25 BRCA2 PVs were detected in exon 11 (mainly within the BRC repeats) of patients with luminal-like BC (nucleotides: 3036-6352; codons: 938-2042), whereas three were in exon 10 (nucleotides: 1466-2070; codons: 413-615) and three in the DNA binding helical domain (nucleotides: 7909-9481; codons: 2561-3085). More than a third (nine) of BRCA2 PVs were frameshift mutations, seven were intronic variants (IVS), whereas five were nonsense and four missense. Half of the BRCA1 PVs (four) observed in LB cancer patients were frameshift mutations distributed along the entire gene sequence (Supplemental material Figure 2).
Association between BRCA1/2 pathogenic variants and clinical variables
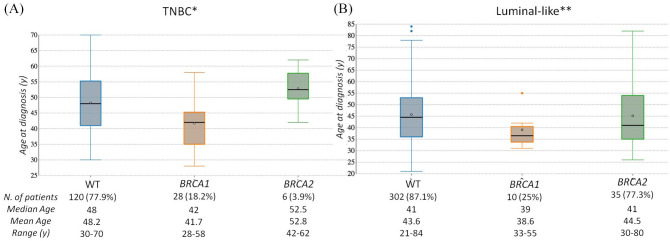
In the TNBC subgroup, the vast majority (82.4%) of BRCA-carriers were premenopausal at BC diagnosis (before the age of 50 years), with mean age of 43.7 years (median: 43). A statistically significant difference in mean age between BRCA1 and BRCA2 PV carriers is evident. The mean age at BC diagnosis of BRCA1 PV carriers was 41.7 years (median: 42; range: 28–58 years), 52.8 years (median 52.5; range: 42–62 years) for BRCA2 PV carriers, and 48.2 years (median 48; range: 30–70 years) for subjects with no BRCA1/2 PV. On average, patients with TNBC and BRCA1 PVs developed BC 6.45 years earlier than non-carrier individuals (p < 0.001), and 11.1 years earlier than BRCA2 PV carriers (p < 0.001) (Figure 5a). Prevalence of PVs was 12/34 (35.3%) in the age group ⩽40 years, 16/34 (47.1%) in age range of 41–50 years, 5/34 (14.7%) in 51–60 years, and 1/34 (2.9%) in subjects with age >60 years (Table 4).
Figure 5.
Boxplots showing difference in age at diagnosis among women without BRCA1/2 PVs versus women with BRCA1 or BRCA2 PV. (a) TNBC subgroup; (b) Luminal-like BC subgroup.
*WT versus BRCA1 p < 0.001; WT versus BRCA2 p = 0.26; BRCA1 versus BRCA2 p < 0.001; **WT versus BRCA1 p = 0.053; WT versus BRCA2 p = 0.94; BRCA1 versus BRCA2 p = 0.78.
Table 4.
Baseline characteristics and clinicopathological information of Triple-Negative and Luminal-like BC patients.
| TNBC* | Luminal-like** | *p-value | **p-value | |||
|---|---|---|---|---|---|---|
| WT | BRCA1/2 | WT | BRCA1/2 | |||
| Number of patients (502) | 120 (77.9%) | 34 (22.1%) | 302 (86.8%) | 46 (13.2%) | - | - |
| Age at diagnosis (y): | ||||||
| Median | 48 | 43 | 41 | 40 | 0.013 | 0.308 |
| Mean | 48.25 | 43.7 | 43.6 | 42.8 | ||
| Range | 30–70 | 28–62 | 21–84 | 30–80 | ||
| Age groups (y) | ||||||
| ⩽ 40 | 30 (25%) | 12 (35.3%) | 140 (46.3%) | 27 (58%) | 0.135 | 0.580 |
| 41–50 | 43 (35.8%) | 16 (47.1%) | 86 (28.4%) | 9 (20%) | ||
| 51–60 | 36 (30%) | 5 (14.7%) | 49 (16.2%) | 6 (13%) | ||
| >60 | 11 (9.2%) | 1 (2.9%) | 27 (9.1%) | 4 (9%) | ||
| Histological Subtype | ||||||
| Ductal | 119 (99.2%) | 33 (97%) | 237 (78.5%) | 36 (78.2%) | 0.337 | 0.700 |
| Lobular | 0 (%) | 0 (%) | 32 (10.6%) | 7 (15.2%) | ||
| Others | 1 (0.8%) | 1 (3%) | 30 (9.9%) | 3 (6.6%) | ||
| unknown | \ | \ | 3 (1%) | \ | ||
| ER (%) | ||||||
| ⩽20 | 14 (4.6%) | 8 (17.4%) | - | 0.001 | ||
| >20 | 267 (88.5%) | 34 (73.9%) | ||||
| unknown | \ | \ | 21 (6.9%) | 4 (8.7%) | ||
| PR (%) | ||||||
| ⩽20 | 60 (19.9%) | 19 (41.3%) | - | 0.007 | ||
| >20 | 211 (69.9%) | 23 (50%) | ||||
| unknown | \ | \ | 31 (10.2%) | 4 (8.7%) | ||
| HER2 (%) | ||||||
| pos | \ | \ | 63 (20.9%) | 4 (8.7%) | - | 0.048 |
| neg | 220 (72.8%) | 42 (91.3%) | ||||
| unknown | 19 (6.3%) | / | ||||
| Ki-67 (%) | ||||||
| <20 | 10 (8.3%) | 2 (5.9%) | 105 (34.8%) | 6 (13%) | 0.854 | <0.001 |
| 20–50 | 34 (28.4%) | 9 (26.5%) | 120 (39.7%) | 21 (45.7%) | ||
| >50 | 76 (63.3%) | 23 (67.6) | 39 (12.9%) | 15 (32.6%) | ||
| unknown | \ | \ | 38 (12.6%) | 4 (8.7%) | ||
| Histological grade | ||||||
| G1 | 4 (3.3) | 1 (2.9%) | 43 (14.3%) | 1 (2.2%) | 0.882 | <0.001 |
| G2 | 18 (15%) | 4 (11.8%) | 141 (46.7%) | 14 (30.4%) | ||
| G3 | 98 (81.7%) | 29 (85.3%) | 84 (27.8%) | 26 (56.5%) | ||
| unknown | \ | \ | 34 (11.2%) | 5 (10.9%) | ||
| Tumor size (T) | ||||||
| T1 | 74 (61.7%) | 18 (53%) | 142 (47.1%) | 19 (41.3%) | 0.802 | 0.920 |
| T2 | 34 (28.3%) | 12 (35.3%) | 77 (25.6%) | 10 (21.7%) | ||
| T3 | 10 (8.3%) | 3 (8.8%) | 5 (1.7%) | / | ||
| T4 | 2 (1.7%) | 1 (2.9%) | 4 (1.4%) | 1 (2.2%) | ||
| unknown | \ | \ | 73 (24.2%) | 16 (34.8%) | ||
| Axillary nodal involvement (N) | ||||||
| N0 | 88 (73.3%) | 13 (38.2%) | 126 (41.7%) | 16 (34.8%) | 0.002 | 0.016 |
| N1 | 22 (18.3%) | 14 (41.3%) | 66 (21.9%) | 10 (21.7%) | ||
| N2 | 8 (6.7%) | 6 (17.6%) | 13 (4.3%) | 2 (4.4%) | ||
| N3 | 2 (1.7%) | 1 (2.9%) | 4 (1.3%) | 4 (8.7%) | ||
| unknown | \ | \ | 93 (30.8%) | 14 (30.4%) | ||
| Bilateral | ||||||
| Yes | 12 (10%) | 5 (14.7%) | 66 (21.8%) | 13 (28.2%) | 0.439 | 0.425 |
| No | 108 (90%) | 29 (85.3%) | 236 (78.2%) | 33 (71.8%) | ||
| Median age at diagnosis (y) | ||||||
| Primary tumor | 48 | 40 | 48 | 41 | 0.033 | 0.0474 |
| Secondary tumor | 56 | 50 | 53 | 52 | ||
| Time between 1st and 2nd tumors (y) | ||||||
| Median | 6.5 | 10 | 3 | 4 | 0.389 | 0.465 |
Comparison TNBC WT versus BRCA1/2; **Comparison luminal-like WT versus BRCA1/2.
In the luminal-like subgroup, the mean age at BC diagnosis for BRCA-positive carriers was 43.75 years (median 40), 39.1 years (median: 36.5; range: 31–55 years) for BRCA1 PV carriers, and 45.1 years (median: 41; range: 26–82 years) for BRCA2 PV carriers. Individuals with no BRCA1/2 PV showed mean age at BC diagnosis of 45.7 years, with a wider range (21–84 years; median: 44.5). On average, patients with luminal-like BRCA1-PVs developed BC 6.6 years earlier than non-carrier individuals (p = 0.0538), and 6 years earlier than BRCA2 PV carriers (p = 0.78) (Figure 5b).
Prevalence of PVs was 27/46 (58%) in the age group ⩽40 years, 9/46 (20%) in age range of 41–50 years, 6/46 (13%) in 51–60 years, and 4/46 (9%) in subjects with age >60 years (Table 4).
Significant clinicopathological differences between BRCA PV carriers and non-carrier BC patients were observed (Table 4). In the luminal-like subgroup, BRCA-positive BCs were more likely associated to lower ER (p = 0.001) and PR expression (p = 0.007), and were more frequently HER2-negative (p = 0.048). BRCA PV carriers had a high proliferation rate (Ki-67%; p = <0.001) and higher histological grade (Grade III versus I/II) than non-carriers (p = <0.001). In either subgroup, TNBCs and luminal-like tumors, patients with BRCA PV more likely had an axillary nodal involvement (p = 0.002 and p = 0.016, respectively), while no significant differences were observed in tumor size (T) (p = 0.802 and p = 0.920, respectively).
All TNBC and most luminal-like BC patients showed ductal histotype, without statistically significant differences between BRCA PV carriers and non-carriers (p = 0.337 and p = 0.7, respectively).
Contralateral breast tumors occurred in 96 (19.1%) patients: five (14.7%) out of 34 BRCA1/2 PV carriers with TNBC, 13 (28.2%) out of 46 BRCA1/2 PV carriers with luminal-like BC, and 78 (18.4%) out of 422 BRCA-negative patients, including 12 (10%) out of 120 TNBCs and 66 (21.8%) out of 302 luminal-like tumors. Controlateral tumors in BRCA1/2-positive TNBC patients were diagnosed at a younger age (50 years) than non-carriers (56 years) (p = 0.033). In patients with luminal-like BC, the difference between median age of BRCA-carriers and non-carriers was lower (52 versus 53 years).
Median time to contralateral BC was 10 years in BRCA-positive TNBC patients and 6.5 years in BRCA-negative TNBC patients (p = 0.389). In the luminal-like subgroup, the median time between the first and second tumor was shorter both in BRCA-positive (4 years) and BRCA-negative patients (3 years) (p = 0.465). Overall, the median time of onset of bilateral tumors was lower in luminal-like than TNBC patients. Clinicopathological characteristics of Triple-Negative and Luminal-like BC patients are presented in Table 4.
Discussion
Breast cancer is a heterogeneous disease at the genetic, histological, molecular and clinical level, showing a wide variability in the prognosis, treatment and patient outcomes. This heterogeneous nature is highlighted from recent advances in genetic and genomic fields. In recent years, an increasing amount of new information on germline PVs in cancer susceptibility genes has been collected,37 determining a substantial increase in the request and indication of genetic testing for cancer risk assessment27 and requiring physicians to integrate this information into strategies of prevention, surveillance and treatment decision making.38
Germline BRCA1/2 PVs confer high risk of developing BC, increased more than fourfold compared with the general population. Multiple germline or somatic mutations in other genes involved in homologous recombination deficiency (HRD),6 such as PALB2, CHEK2, ATM, RAD51, ATR, CHK1 and WEE1, have been recently observed, but at relatively low frequencies, occurring in 4–6% of BC patients, and with lower lifetime risk than BRCA1/2 PVs.39–41 However, the evidence regarding other BC susceptibility genes is still limited and additional studies are needed to better define their role. Therefore, despite variations in the prevalence among different ethnic groups and geographical zones,42–44 inherited PVs in BRCA1/2 genes are confirmed as the most frequent in BC. Integrating all genetic knowledge into surveillance and prevention strategies and patient care is the crucial aim of physicians.
The germline PVs in BRCA1/2 genes are associated with an increased risk of developing cancer for each molecular subtype of BC, defined by estrogen, progesterone and HER2 receptor status. However, it was demonstrated that BRCA1-related tumors have often a profile which resembles the TNBC subtype, whereas BRCA2-associated tumors have a profile that resembles luminal B or, less frequently, luminal A tumor subtypes.45 These differences point to a heterogeneity in BC biology and molecular phenotype among tumors related to different germline BRCA1 and BRCA2 PVs.19
Previous research has indicated that structural and functional changes of mutated proteins caused by different BRCA1 PVs are not identical and can lead to various tumor phenotypes.22 However, several studies were mainly focused on the impact of PVs located in different exons of the BRCA1 or BRCA2 genes and on phenotypic variations of cancer relative risks.46,47 Rebbeck et al.21 investigated whether the type and location of BRCA1/2 PVs were associated with the variation in BC and OC risk, showing that patients carrying BRCA1 PVs within exon 11 had different disease phenotypes than patients carrying BRCA1 PVs in other gene loci. Similarly, different PVs in specific genomic regions were associated with variability in BC and OC risk.21,47–49 Also murine models of different mutations in BRCA1/2 suggested that a genotype–phenotype correlation exist.49,50 However, how molecular phenotypes differ by type, function and location of BRCA1/2 PVs has not been fully investigated.
In this study, we screened 531 patients with BC for germline PVs in BRCA1/2 genes according to national guidelines. We detected 45 BRCA1/2 PVs in 83 BC patients. TNBC has been shown to be the molecular subgroup where BRCA1/2 PVs are found more frequently (22.1%), followed by Luminal B tumors (18%), whereas BRCA1/2 alterations are less frequent in HER2E (10.2%) and Luminal A (4.8%) BC patients. We confirmed a significant association between TNBC and BRCA1 PVs and between Luminal B tumors and BRCA2 PVs.
Confirming the previous findings, in our patient cohort, breast tumors with BRCA PVs occur in younger women. BRCA1 PV carriers, in addition to a higher predisposition toward the onset of TNBC, developed BC earlier than BRCA2 PV carriers and non-carrier individuals, and the difference in age at diagnosis between BRCA1 and BRCA2 PV carriers is greater in TNBC patients.
The findings in this study indicate that tumors present in BRCA1/2 PV carriers were differentially associated with several prognostic factors compared with non-carriers. The tumors in BRCA1/2-positive patients showed a higher proportion of Ki67-positive cells, a higher histological grade and an axillary nodal involvement. In the luminal-like subgroup, BRCA-positive BCs were more likely associated with low ER, PR and HER2 expression. Furthermore, differences were observed between PV carriers and non-carriers in the presence of bilateral tumors. In BRCA-positive patients, controlateral BC was more common and had a lower median time to second tumor development in luminal-like BC compared with TNBC and BRCA-negative patients.
Concerning the variant type, although no significant association between specific BRCA1/2 PVs and TNBC or luminal-like tumors in the study cohort was observed, a phenotypic variation of BC in patients with different BRCA1/2 PV type seems to be detectable. Most of the TNBC-associated BRCA1 PVs and luminal-like BC-associated BRCA2 PVs were frameshift mutations for both molecular subtypes. However, a significant percentage of pathogenic IVS was detected in BRCA2 gene of luminal-like BC patients. Interestingly, differences in the frequency of two PVs potentially associated with TNBC and luminal-like tumors, respectively, were observed. BRCA1-633delC was detected with higher prevalence in TNBC patients (five families, including 15 PV carriers) and only in two families with LB tumors, whereas BRCA2-1466delT was found in eight families (including 15 PV carriers) with LB tumors, but in no TNBC patient.
The BRCA1-633delC emerged as a PV type related to TNBC diagnosed at younger age and featuring poor prognostic factors, such as high proliferation rate and nuclear grade.
The BRCA2-1466delT was more likely associated to HER2-negative BC with higher ER expression (range 70–95%), in patients who carried a high proportion of bilateral breast tumors.51 In addition, as BRCA1-633delC has been infrequently observed in other Italian regions or in the world, this PV could be further investigated for a possible founder effect specific for the Sicilian population.27
Understanding the mutational background underlying the phenotype of each tumor may have not only prognostic, but also preventive and therapeutic implications. The four surrogate intrinsic subtypes are the most important criteria for clinical decisions and imply distinct treatment approaches. Systemic therapies are routinely selected through a few well-established biomarkers of response, including tumor ER and PgR expression, and amplification or overexpression of tumor HER2. Although advances in molecular profiling and genetic expression studies have identified different subtypes of TNBC, making it increasingly heterogeneous, primary TNBC continues to be typically treated as a single disease, due to the absence of specific drivers.52
In TNBC, chemotherapy is the standard treatment option and typically involves the use of anthracycline and taxane. For BRCA-associated TNBC, the platinum-based agents and PARP inhibitors, such as olaparib and talazoparib, showed a particular efficacy.53
We hypothesized that also among BRCA-related TNBCs there is a marked genetic heterogeneity, which could define the phenotype of BCs associated with mutations. This could affect the natural history of these tumors and make them potential candidates for different treatment options.
In our cohort, the patients with BRCA1-633delC were found to be less chemosensitive than those harboring BRCA1-4023G>T and BRCA1-5382insC, who showed, instead, a more chemosensitive and prolonged survival benefit. Our observations should be interpreted cautiously due to the limited number of patients in each subgroup. Nevertheless, our investigation suggests that chemosensitivity in TNBC patients may widely vary in the same molecular phenotype of the tumor. Conversely, no correlation between different responses to treatment and different mutation type/localization was observed in patients with luminal-like tumor.
BRCA testing, that was previously used solely to predict the risk of future cancers and drive surgical treatments, could acquire, in the future, an additional significance for treatment response and resistance.
Referring to the work by Rebbeck and colleagues,21 we tried to identify the regions for which the variant site could define a possible genotype/phenotypic effect related to TNBC risk or luminal-like tumors in the Sicilian population. In particular, we have observed that most of the BRCA1 PVs (55.5%) and BRCA2 PVs (24%) detected in TNBC and luminal-like patients, respectively, were located within exon 11, which represents the majority of the coding sequence of both genes and is generally considered a “coldspot” for missense PVs.54 In addition to the region encoded by exon 11, other two BRCA1 protein regions, RING domain at the N-terminus and BRCT domain, seem to be involved, to a lesser extent, in the TNBC risk, confirming the crucial role in tumor suppression played by these structural components.54,55 Our data did not allow us to define new regions other than those already known in the literature, such as the OCCRs or BCCRs. The heterogeneous distribution of PVs and their low prevalence in TNBC patients could reflect the genetic heterogeneity of the Sicilian population, probably determined by the colonization of this island of Mediterranean Sea by many and different peoples throughout history.
Although this study adds significant and useful information to the current knowledge in the field, it does however, show some potential limitations. Our study is a retrospective analysis of BC patients who were referred to the genetic counseling service for testing of the BRCA1 and BRCA2 genes. Thus, the BRCA non-carrier control group may not be a fair representation of sporadic cancers. In addition, our results need to be confirmed by future studies, which prospectively test for BRCA1/2 mutations in BC patients, in order to minimize the possibility of selection bias.
In conclusion, our results corroborate the evidence that BRCA1-related tumors have often a profile which resembles the TNBC subtype, whereas BRCA2-associated tumors have a profile that resembles luminal-like BCs, especially the luminal B tumor subtypes.
Previous studies showed the association between type and location of BRCA1/2 PVs and phenotypic variations of cancer relative risks. The findings from this study suggest that, although no clear association between specific BRCA1/2 PVs and TNBC or luminal-like tumors was observed, the pathogenic variants identified in TNBC were not largely overlapping with those detected in luminal-like tumors. Future studies examining the type and location of BRCA1/2 PVs within the molecular subtype are required to verify this hypothesis, and could offer an interesting insight into the complex topic of genotype–phenotype correlations.
Supplemental Material
Supplemental material, sj-pdf-1-tam-10.1177_1758835920975326 for BRCA1/2 pathogenic variants in triple-negative versus luminal-like breast cancers: genotype–phenotype correlation in a cohort of 531 patients by Lorena Incorvaia, Daniele Fanale, Marco Bono, Valentina Calò, Alessia Fiorino, Chiara Brando, Lidia Rita Corsini, Sofia Cutaia, Daniela Cancelliere, Alessia Pivetti, Clarissa Filorizzo, Maria La Mantia, Nadia Barraco, Stefania Cusenza, Giuseppe Badalamenti, Antonio Russo and Viviana Bazan in Therapeutic Advances in Medical Oncology
Acknowledgments
All authors thank Dr. Chiara Drago for the language English revision.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author contributions: Conceptualization, L.I., D.F., A.R. and V.B.; genetic counselling D.F., V.C.; sample collection and gene testing, M.B., D.C., A.F., N.B., and A.P.; data curation and analysis, L.I., D.F., C.B., S.C., G.B., L.R.C, M.L., and S.C; writing L.I. and D.F.; supervision, A.R., G.B. and V.B. All authors have read and agreed to the published version of the manuscript.
Supplemental material: Supplemental material for this article is available online.
ORCID iDs: Clarissa Filorizzo  https://orcid.org/0000-0002-0164-6992
https://orcid.org/0000-0002-0164-6992
Antonio Russo  https://orcid.org/0000-0002-4370-2008
https://orcid.org/0000-0002-4370-2008
Contributor Information
Lorena Incorvaia, Section of Medical Oncology, Department of Biomedicine, Neuroscience and Advanced Diagnostics (Bi.N.D.), University of Palermo, Palermo, Italy.
Daniele Fanale, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Marco Bono, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Valentina Calò, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Alessia Fiorino, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Chiara Brando, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Lidia Rita Corsini, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Sofia Cutaia, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Daniela Cancelliere, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Alessia Pivetti, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Clarissa Filorizzo, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Maria La Mantia, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Nadia Barraco, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Stefania Cusenza, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Giuseppe Badalamenti, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Palermo, Italy.
Antonio Russo, Section of Medical Oncology, Department of Surgical, Oncological and Oral Sciences, University of Palermo, Via del Vespro 129, Palermo, 90127, Italy.
Viviana Bazan, Section of Medical Oncology, Department of Biomedicine, Neuroscience and Advanced Diagnostics (Bi.N.D.), University of Palermo, Palermo, Italy.
References
- 1. Miki Y, Swensen J, Shattuck-Eidens D, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994; 266: 66–71. [DOI] [PubMed] [Google Scholar]
- 2. Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature 1995; 378: 789–792. [DOI] [PubMed] [Google Scholar]
- 3. Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016; 17: 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tung NM, Boughey JC, Pierce LJ, et al. Management of hereditary breast cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline. J Clin Oncol 2020; JCO.20.00299. [DOI] [PubMed] [Google Scholar]
- 5. Yadav S, Couch FJ. Germline genetic testing for breast cancer risk: the past, present, and future. Am Soc Clin Oncol Educ Book 2019; 39: 61–74. [DOI] [PubMed] [Google Scholar]
- 6. Easton DF, Pharoah PDP, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med 2015; 372: 2243–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371: 1650–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002; 31: 55–59. [DOI] [PubMed] [Google Scholar]
- 9. Weischer M, Bojesen SE, Ellervik C, et al. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: meta-analyses of 26,000 patient cases and 27,000 controls. J Clin Oncol 2008; 26: 542–548. [DOI] [PubMed] [Google Scholar]
- 10. Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 2006; 38: 873–875. [DOI] [PubMed] [Google Scholar]
- 11. Kuchenbaecker KB, Hopper JL, Barnes DR, et al. Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017; 317: 2402. [DOI] [PubMed] [Google Scholar]
- 12. Hartmann LC, Longo DL, Lindor NM. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N Engl J Med 2016; 374: 454–468. [DOI] [PubMed] [Google Scholar]
- 13. Arun BK, Lu KH, Litton JK, et al. Prospective evaluation of universal BRCA testing for women with triple-negative breast cancer. JNCI Cancer Spectrum 2020; 4: pkaa002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sharma P, Klemp JR, Kimler BF, et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat 2014; 145: 707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Massihnia D, Galvano A, Fanale D, et al. Triple negative breast cancer: shedding light onto the role of PI3K/AKT/mTOR pathway. Oncotarget 2016; 7: 60712–60722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oualla K, El-Zawahry HM, Arun B, et al. Novel therapeutic strategies in the treatment of triple-negative breast cancer. Ther Adv Med Oncol 2017; 9: 493–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hahnen E, Hauke J, Engel C, et al. Germline mutations in triple-negative breast cancer. Breast Care 2017; 12: 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis SL, Eckhardt SG, Tentler JJ, et al. Triple-negative breast cancer: bridging the gap from cancer genomics to predictive biomarkers. Ther Adv Med Oncol 2014; 6: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Prokunina-Olsson L, Larsen MJ, Kruse TA, et al. Classifications within molecular subtypes enables identification of BRCA1/BRCA2 mutation carriers by RNA tumor profiling. PLoS One 2013; 8: e64268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shah PD, Patil S, Dickler MN, et al. Twenty-one-gene recurrence score assay inBRCA-associated versus sporadic breast cancers: Differences based on germline mutation status. Cancer 2016; 122: 1178–1184. [DOI] [PubMed] [Google Scholar]
- 21. Rebbeck TR, Mitra N, Wan F, et al. Association of type and location of BRCA1 and BRCA2 mutations with risk of breast and ovarian cancer. JAMA 2015; 313: 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thompson D, Easton D. Variation in cancer risks, by mutation position, in BRCA2 mutation carriers. Am J Hum Genet 2001; 68: 410–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kondov B, Milenkovikj Z, Kondov G, et al. Presentation of the molecular subtypes of breast cancer detected by immunohistochemistry in surgically treated patients. Open Access Maced J Med Sci 2018; 6: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mazzola E, Blackford A, Parmigiani G, et al. Recent enhancements to the genetic risk prediction model BRCAPRO. Cancer Inform 2015; 14s2: CIN.S17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hoheisel JD, Simbolo M, Gottardi M, et al. DNA qualification workflow for next generation sequencing of histopathological samples. PLoS One 2013; 8: e62692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fanale D, Iovanna JL, Calvo EL, et al. Germline copy number variation in theYTHDC2gene: does it have a role in finding a novel potential molecular target involved in pancreatic adenocarcinoma susceptibility? Expert Opin Ther Targets 2014; 18: 841–850. [DOI] [PubMed] [Google Scholar]
- 27. Incorvaia L, Fanale D, Badalamenti G, et al. Hereditary breast and ovarian cancer in families from Southern Italy (Sicily)—prevalence and geographic distribution of pathogenic variants in BRCA1/2 genes. Cancers 2020; 12: 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008; 29: 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data: figure 1. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Szabo C, Masiello A, Ryan JF, et al. The breast cancer information core: database design, structure, and scope. Hum Mutat 2000; 16: 123–131. [DOI] [PubMed] [Google Scholar]
- 32. den Dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 2016; 37: 564–569. [DOI] [PubMed] [Google Scholar]
- 33. Finch A, Wang M, Fine A, et al. Genetic testing for BRCA1 and BRCA2 in the Province of Ontario. Clin Genet 2016; 89: 304–311. [DOI] [PubMed] [Google Scholar]
- 34. Russo A, Calò V, Agnese V, et al. BRCA1 genetic testing in 106 breast and ovarian cancer families from southern Italy (Sicily): a mutation analyses. Breast Cancer Res Treat 2007; 105: 267–276. [DOI] [PubMed] [Google Scholar]
- 35. Nielsen FC, van Overeem Hansen T, Sørensen CS. Hereditary breast and ovarian cancer: new genes in confined pathways. Nat Rev Cancer 2016; 16: 599–612. [DOI] [PubMed] [Google Scholar]
- 36. Giannini G, Capalbo C, Ristori E, et al. Novel BRCA1 and BRCA2 germline mutations and assessment of mutation spectrum and prevalence in Italian breast and/or ovarian cancer families. Breast Cancer Res Treat 2006; 100: 83–91. [DOI] [PubMed] [Google Scholar]
- 37. Wallace AJ. New challenges for BRCA testing: a view from the diagnostic laboratory. Eur J Med Genet 2016; 24: S10–S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gori S, Barberis M, Bella MA, et al. Recommendations for the implementation of BRCA testing in ovarian cancer patients and their relatives. Crit Rev Oncol Hematol 2019; 140: 67–72. [DOI] [PubMed] [Google Scholar]
- 39. Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014; 371: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tung N, Lin NU, Kidd J, et al. Frequency of germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol 2016; 34: 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. LaDuca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2,000 patients. Genet Med 2014; 16: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russo A, Calò V, Bruno L, et al. Is BRCA1-5083del19, identified in breast cancer patients of Sicilian origin, a Calabrian founder mutation? Breast Cancer Res Treat 2008; 113: 67–70. [DOI] [PubMed] [Google Scholar]
- 43. Manchanda R, Loggenberg K, Sanderson S, et al. Population testing for cancer predisposing BRCA1/BRCA2 Mutations in the Ashkenazi-Jewish community: a randomized controlled trial. J Natl Cancer Inst 2015; 107: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rowley SM, Mascarenhas L, Devereux L, et al. Population-based genetic testing of asymptomatic women for breast and ovarian cancer susceptibility. Genet Med 2018; 21: 913–922. [DOI] [PubMed] [Google Scholar]
- 45. Toss A, Molinaro E, Venturelli M, et al. BRCA detection rate in an Italian cohort of luminal early-onset and triple-negative breast cancer patients without family history: when biology overcomes genealogy. Cancers 2020; 12: 1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bayraktar S, Jackson M, Gutierrez-Barrera AM, et al. Genotype-phenotype correlations by ethnicity and mutation location inBRCA mutation carriers. Breast J 2015; 21: 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Santonocito C, Rizza R, Paris I, et al. Spectrum of germline BRCA1 and BRCA2 variants identified in 2351 ovarian and breast cancer patients referring to a reference cancer hospital of Rome. Cancers 2020; 12: 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ellsworth DL, Turner CE, Ellsworth RE. A review of the hereditary component of triple negative breast cancer: high- and moderate-penetrance breast cancer genes, low-penetrance loci, and the role of nontraditional genetic elements. J Oncol 2019; 2019: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gayther SA, Mangion J, Russell P, et al. Variation of risks of breast and ovarian cancer associated with different germline mutations of the BRCA2 gene. Nat Genet 1997; 15: 103–105. [DOI] [PubMed] [Google Scholar]
- 50. Dine J, Deng C-X. Mouse models of BRCA1 and their application to breast cancer research. Cancer Metastasis Rev 2012; 32: 25–37. [DOI] [PubMed] [Google Scholar]
- 51. Fanale D, Incorvaia L, Filorizzo C, et al. Detection of germline mutations in a cohort of 139 patients with bilateral breast cancer by multi-gene panel testing: impact of pathogenic variants in other genes beyond BRCA1/2. Cancers 2020; 12: 2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers 2019; 5. [DOI] [PubMed] [Google Scholar]
- 53. Tung NM, Garber JE. BRCA1/2 testing: therapeutic implications for breast cancer management. Br J Cancer 2018; 119: 141–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dines JN, Shirts BH, Slavin TP, et al. Systematic misclassification of missense variants in BRCA1 and BRCA2 “coldspots”. Genet Med 2020; 22: 825–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Clark SL, Rodriguez AM, Snyder RR, et al. Structure-function of the tumor suppressor BRCA1. Comput Struct Biotechnol J 2012; 1: e201204005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tam-10.1177_1758835920975326 for BRCA1/2 pathogenic variants in triple-negative versus luminal-like breast cancers: genotype–phenotype correlation in a cohort of 531 patients by Lorena Incorvaia, Daniele Fanale, Marco Bono, Valentina Calò, Alessia Fiorino, Chiara Brando, Lidia Rita Corsini, Sofia Cutaia, Daniela Cancelliere, Alessia Pivetti, Clarissa Filorizzo, Maria La Mantia, Nadia Barraco, Stefania Cusenza, Giuseppe Badalamenti, Antonio Russo and Viviana Bazan in Therapeutic Advances in Medical Oncology