Abstract
Introduction:
Some studies suggest that the accuracy of Helicobacter pylori diagnostic tests is decreased in peptic ulcer bleeding (PUB). We aimed to assess the accuracy of diagnostic tests for H. pylori in patients with PUB in a diagnostic test accuracy (DTA) network meta-analysis.
Methods:
A systematic search was carried out in seven databases until November 2019. We collected or calculated true and false positive and negative values, and constructed 2×2 diagnostic contingency tables with reference standards including histology, rapid urease test, urea breath test, serology, stool antigen test, culture, and polymerase chain reaction. We ranked the index tests by the superiority indices (SI) and calculated pooled sensitivity and specificity of each test.
Discussion:
Our search yielded 40 eligible studies with 27 different diagnostic strategies for H. pylori. In 32 articles, the reference standard was a combination of multiple tests. In 12 studies, the index tests were compared with a single testing method. We analyzed seven networks with the reference standards against a single or a combination of diagnostic index tests. None of the index tests had better diagnostic accuracy (SI between 9.94 and 2.17) compared with the individual index tests as all the confidence intervals included 1. Combined testing strategies had higher sensitivities (0.92–0.62) and lower specificities (0.85–0.46) while single tests proved to have higher specificities (0.83–0.77) and lower sensitivities (0.73–0.42).
Conclusion:
Use of combined tests may have a rationale in clinical practice due to their higher sensitivities. The differences between the included DTA studies limited the comparison of the testing strategies.
Keywords: accuracy, bleeding, diagnostic, Helicobacter, histology, peptic, pylori, tests, UGIB, ulcer
Introduction
Peptic ulcer bleeding (PUB) is the most frequent cause of acute nonvariceal upper gastrointestinal (GI) bleeding,1–4 and has a reported mortality of between 11% and 13.1%.5,6 Helicobacter pylori infection (HPI) remains one of the common causes of peptic ulcer disease.7,8 In the case of underlying HPI, untoward outcomes of the PUB episode such as rebleeding, need for repeat endoscopy and transfusion are better if the bacteria are eradicated.9 Gisbert et al. found that eradication of HPI can reduce rebleeding episodes from 23.7% to 4.5%.10
An optimal testing strategy would be desirable; however, the international guidelines on PUB do not provide clear guidance for clinicians concerning HPI testing in the acute setting. The American College of Gastroenterology (ACG) guidelines recommend biopsy-based testing,4 whereas the European Society of Gastroenterological Endoscopy (ESGE)1,2 does not specify a diagnostic method. The American Society of Gastroenterological Endoscopy (ASGE) recommends the eradication of HPI, but it does not determine the method of testing.3
There is a lack of guidance in the Maastricht/Florence V guideline on the ideal timing of eradication therapy after an acute episode of PUB,11 and there are multiple logistical factors that hamper early testing and eradication.
Multiple previous studies and reviews proved a decreased diagnostic performance of HPI tests in PUB. The reasons include recent proton pump inhibitor (PPI) use, which can change the number and load of detectable organisms. Intragastric blood contains albumin and human plasma with killing factors, which can interfere with bacteria.11–13 A meta-regression reported that testing on the index admission underestimates the true prevalence of HPI in PUB, likely due to the decreased diagnostic performance.14 Therefore, an optimal strategy or preference for the available tests is needed.
The meta-analysis of Gisbert et al. in 2006 on the diagnostic accuracy of H. pylori testing in the setting of acute PUB drew many conclusions on the diagnostic performance of single tests compared with ‘gold standards’.15 They found that biopsy-based tests had low sensitivity and high specificity in PUB. The urea breath test remained highly accurate, but the stool antigen test was less reliable. They did not recommend serology as the first test. Since then, multiple studies have been completed, the genre of diagnostic test accuracy (DTA) meta-analysis improved significantly, with detailed recommendations in the Cochrane Handbook for DTA reviews.16 Also, the method of network meta-analysis for DTA studies was developed.17
We aimed to assess the diagnostic performance of all HPI testing strategies in PUB in a diagnostic test accuracy network meta-analysis.
Materials and methods
Protocol
A diagnostic accuracy meta-analysis and systematic review were planned using the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) protocols for DTA studies.18 The analysis was registered in advance on PROSPERO with registration number CRD42019113083, and the protocol was later updated as a network meta-analysis due to the significant variation between the comparisons of testing strategies.19
Data sources and searches
We included studies from adult populations with PUB where index tests were compared with reference tests for identifying HPI. The outcomes were the diagnostic performance measures of the different diagnostic tests.
A systematic search was conducted in seven databases: Medline via PubMed, Embase, Cochrane Central Register of Controlled Trials, ClinicalTrials.gov, WHO Global Health Library, Web of Science, Scopus, from inception to 1 November 2019. There were no restrictions or filters imposed on the search strategy apart from human studies.
Keywords for the computer-aided search were (bleed* OR haemorrhage OR hemorrhage OR haematemesis OR hematemesis OR melaena OR melena) AND (‘upper gastrointestinal’ OR ‘upper GI’ OR nonvariceal OR peptic OR gastric OR duodenal OR gastroduodenal OR antrum OR antral OR pylorus OR pyloric OR GU OR DU OR PU OR ulcer OR stomach OR curling) AND (helicobacter OR pylori). Additional articles were identified from the reference lists of primarily eligible studies.
Study selection
Records were managed by EndNote X7.4, software (Clarivate Analytics, Philadelphia, PA, USA). After exclusion of duplicates, the remaining studies were screened by title, abstract and finally by full text by two independent authors (NV, ME). Additional articles were searched manually and identified from the reference lists of primarily eligible studies. We calculated Cohen’s kappa coefficient to measure the agreement between two raters (NV, ME) in three levels (title, abstract and full text) of the selection process.20 Disagreements were resolved by consensus and the involvement of the senior reviewer (BE).
Eligibility criteria
All prospective, cross-sectional DTA studies with relevant information about the accuracy of any HPI diagnostic test without language restriction were included in our analysis. Articles without direct or indirect information on the true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN) were excluded from the analysis. Conference abstracts were excluded after we discovered they did not contain enough information.
Data extraction
Data were extracted independently by two investigators (NV, ME) and populated manually into a purpose-designed Excel 2016 sheet (Office 365, Microsoft, Redmond, WA, USA). Data were collected on the year of publication, geographical location, study type, number of enrolled patients, and basic demographics (age, sex ratio). Most importantly, the raw data (TP, TN, FP, FN), the name, manufacturer, cut off value, biopsy site and timing of both index tests and reference standards were collected. Data about therapy after admission before a diagnosis of HPI, the timing of endoscopic examination, the bleeding source and the risk factors (smoking, alcohol consumption, nonsteroidal anti-inflammatory drug and aspirin use, history of GI bleeding and PUB) were also collected. Other relevant findings were mentioned in an additional column as free text. Disagreements were resolved by consensus and the involvement of the corresponding author.
Risk of bias and applicability
A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) was used for the quality assessment of the DTA studies, and the result of the assessment was graphically demonstrated.21 Risk of bias was assessed independently by two investigators (NV, ME). Disagreements were resolved by consensus and the involvement of the corresponding author.
Statistical analysis
We performed a DTA network meta-analysis to investigate which diagnostic method can be the best choice to detect HPI in PUB. This method allows us to make direct as well as indirect comparisons through a common comparator (i.e. the reference standard).
We collected the raw data of diagnostic tests, TP, TN, FP, and FN values and created 2×2 tables for each study. If raw data on the diagnostic accuracy were not provided, but detailed indirect data on the diagnostic performance were available, TP, TN, FP, FN were calculated. To assess the relative performance of a diagnostic test, we calculated pooled sensitivity and specificity for the index tests compared with the reference standard, and ranked them according to superiority indices (SI). The larger the SI, the more accurately a test is expected to predict the targeted condition compared with other tests. The network meta-analytical calculations were performed by the R programming language (R Core Team 2019, Vienna, Austria, R version 3.6.1) developed by Nyaga et al.,17 with the use of the ANOVA arm-based model. Publication bias was not assessed as the Handbook for DTA Reviews of Cochrane Methods argues that conventional ways of assessment are not reliable and can lead to misinterpretation of the publication bias.16
To display the network, we constructed a graph where nodes represent different screening methods, and edges represent head-to-head comparisons. The size of nodes correlates with the number of studies. The thickness of edges represents the number of comparisons between the two tests. The potential nodes of the network were the single tests that had enough connections with other tests to allow statistical analysis in networks.
Since we performed a network meta-analysis of diagnostic accuracy studies in R software, inconsistency should have been calculated instead of heterogeneity. However, since the conditions of node splitting analysis were not met, we were unable to assess inconsistency.
Results
Study selection
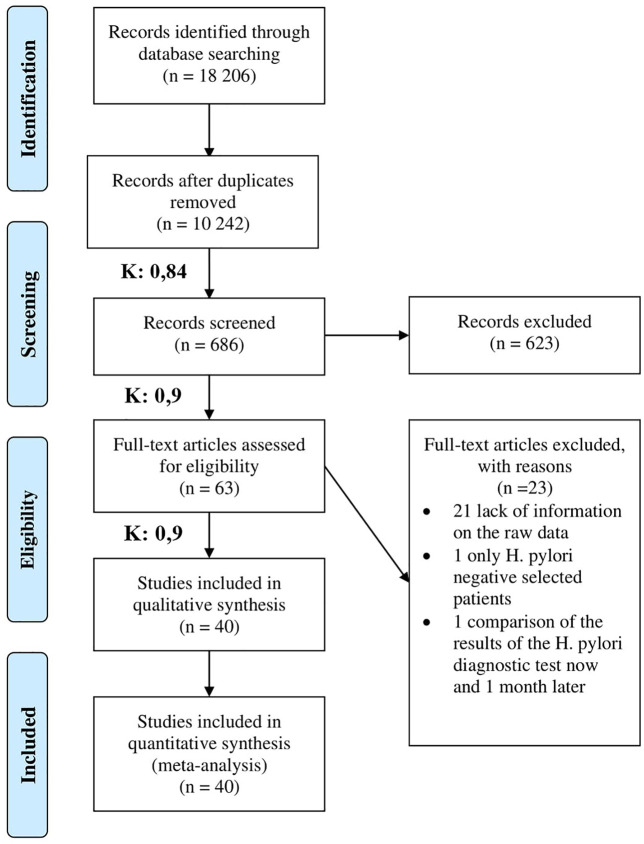
Our final statistical analyses included 40 observational cross-sectional studies.12,22–60 The study selection process with the Cohen’s kappa values is shown in Figure 1. Eligible studies were reported between 1998 and 2016 from four continents. The number of study participants ranged between 32 and 324. Characteristics of the studies included in our analysis are shown in Table 1.
Figure 1.
PRISMA flow chart for the study selection procedure. K value is the Cohen’s kappa coefficient, if K is between 0.81 and 1.00 it means an almost perfect or perfect agreement.
PRISMA, preferred reporting items for systematic review and meta-analysis.
Table 1.
Main characteristics of included studies.
| Author | Country | Index tests | Definition of reference standards | No of patients |
|---|---|---|---|---|
| Archimandritis et al.22 | Greece | Rapid urease test | Positive histology | 72 |
| Bravo Paredes et al.23 | Peru | Rapid urease test | Positive histology | 93 |
| Castro-Fernandez et al.24 | Spain | Rapid urease test | Assumed 100% infection | 120 |
| Castro-Fernandez et al.25 | Spain | Rapid urease test, histology | Positive at least one of the followings: rapid urease test and histology or urea breath test | 173 |
| Chandrasakha et al. 26 | Thailand | Polymerase chain reaction, rapid urease test, histology, culture | Positive culture or a positive rapid urease test and histology | 64 |
| Choi et al.27 | Korea | Culture, rapid urease test, histology | Positive culture or any two of the followings: rapid urease test, histology, serology | 157 |
| Chung et al.28 | Korea | Histology, rapid urease test, urea breath test, serology | At least two positive of the followings: rapid urease test, histology, urea breath test, serology | 32 |
| Chung et al.29 | Korea | Polymerase chain reaction | Positive in two of the followings: histology, rapid urease test, serology | 170 |
| Colin et al.30 | France | Rapid urease test, histology, culture | Positive serology | 182 |
| Demiray et al.31 | Turkey | Stool antigen test | Positive urea breath test or histology and/or rapid urease test | 22 |
| Garcia-Diaz et al.32 | Spain | Serology | Positive in for at least one of the followings: rapid urease test, histology, culture or urea breath test | 214 |
| Gisbert et al.34 | Spain | Stool antigen test | Positive rapid urease test or histology or urea breath test | 34 |
| Gisbert et al.33 | Spain | Urea breath test | Positive urea breath test | 131 |
| Grino et al.35 | Spain | Rapid urease test, urea breath test serology, histology | Positive histology or positive serology and urea breath test | 78 |
| Grino et al.36 | Spain | Stool antigen test, rapid urease test, histology, urea breath test, serology | Positive histology or positive two of the followings: rapid urease test, serology, urea breath test | 68 |
| Hanvivatong et al.37 | Taiwan | Stool antigen test, urea breath test serology, rapid urease test, histology | Positive culture or two of histology, rapid urease test, serology, urea breath test | 56 |
| Lahmidani et al.38 | Marocco | Rapid urease test | Positive histology | 116 |
| Lee et al.39 | Taiwan | Rapid urease test | Positive culture or histology | 55 |
| Lee et al.40 | Taiwan | Rapid urease test | Positive urea breath test | 116 |
| Liao et al.41 | China | Rapid urease test, histology, urea breath test | At least two positive of the followings: rapid urease test, histology, urea breath test | 57 |
| Lin et al.42 | Taiwan | Stool antigen test | Positive culture or positive histology and rapid urease test | 93 |
| Lin et al.43 | Taiwan | Polymerase chain reaction | Positive culture or positive histology and rapid urease test | 88 |
| Lo et al.44 | Taiwan | Polymerase chain reaction, rapid urease test, histology, culture, serology, urea breath test | Positive culture or at least three of the followings: rapid urease test, histology, urea breath test, serology | 55 |
| López Peñas et al.45 | Spain | Stool antigen test, rapid urease test, histology, urea breath test, serology | Positive in at least two of the followings: rapid urease test, histology, serology, urea breath test | 32 |
| Manguso et al.46 | Italy | Serology, culture, rapid urease test, histology | Positive culture or positive rapid urease test and histology | 80 |
| Pascual et al.47 | Spain | Rapid urease test, histology | Positive rapid urease test or histology or urea breath test | 73 |
| Peitz et al.48 | Germany | Stool antigen test | Positive culture or positive rapid urease test and histology | 114 |
| Peitz et al.49 | Germany | Serology | Positive culture or positive rapid urease test and histology | 106 |
| Ramírez-Lázaro et al.50 | Spain | Histology | Positive polymerase chain reaction | 52 |
| Romero Gómez et al.51 | Spain | Rapid urease test, histology, culture | Positive culture or positive rapid urease test and histology | 55 |
| Saez et al.52 | Spain | Polymerase chain reaction | Positive culture or at least two of the followings: histology, rapid urease test, stool antigen test, urea breath test | 79 |
| Shilling et al.53 | Germany | Rapid urease test | Positive histology and urea breath test | 96 |
| Sfarti et al.54 | Romania | Urea breath test | Positive urea breath test | 39 |
| Tang et al.55 | Taiwan | Rapid urease test | Positive histology | 324 |
| Tu et al.12 | Taiwan | Rapid urease test, culture, histology, urea breath test, serology | Positive histology and culture or any two of the followings: rapid urease test, urea breath test, serology | 77 |
| van Leerdam et al.56 | The Netherlands | Stool antigen test | Positive culture or histology and rapid urease test | 36 |
| Velayos et al.57 | Spain | Urea breath test | Positive histology or rapid urease test or urea breath test | 70 |
| Wildner-Christensen et al.58 | Denmark | Rapid urease test, urea breath test, serology | Positive histology | 95 |
| Winiarski et al.59 | Poland | Urea breath test | Positive serology | 81 |
| Wong et al.60 | Japan | Serology | Positive rapid urease test or culture or histology | 59 |
Results of meta-analysis
The included 40 studies used 27 different definitions of the reference standard. In 32 articles, the reference standard was a combination of multiple tests. In 12 studies, the index tests were compared with a single testing method. We could form seven networks with the single tests (histology, rapid urease test, urea breath test, serology, stool antigen test, culture, polymerase chain reaction) serving as gold standards of the following networks.
In the seven networks, the top three index tests based on their SI are shown in Table 2. None of the index tests had better diagnostic accuracy (SI between 9.94 and 2.17) compared with the individual index tests as all the confidence intervals included 1. Combined testing strategies had higher sensitivities (0.92–0.62) and lower specificities (0.85–0.46), while single tests proved to have higher specificities (0.83–0.77) and lower sensitivities (0.73–0.42). Out of the single tests, only the urea breath test against histology, and culture against polymerase chain reaction, had SI values ranked within the top three. The graphically displayed networks, results of the full analysis and ranking are detailed in Supplementary Files S1–S7. When we ranked the index tests based on their pooled sensitivity, only combinations of tests ranked in the top three in all seven networks. Ranking of index tests based on their pooled specificity identified nine single tests among all 21 top three ranks. However, all sensitivity and specificity values had wide 95% confidence intervals (CIs) ranging between 0.0 and 1. The pooled specificity values with a corresponding CIs and top three specificity and sensitivity values highlighted are shown in Supplementary Files S1–S7.
Table 2.
Summary of results.
| Gold standards | Index tests | No of patients | Superiority index (95% CI) | Pooled sensitivity (95% CI) | Pooled specificity (95% CI) |
|---|---|---|---|---|---|
| Histology | Positive culture or two of the followings: histology, rapid urease test, serology, urea breath test | 41 | 7.51 (0.05–25) | 0.79 (0.33–1.00) | 0.70 (0.22–0.96) |
| Positive culture or two of the followings: rapid urease test, histology, serology | 157 | 6.52 (0.05–23) | 0.81 (0.29–1.00) | 0.67 (0.17–0.96) | |
| Positive urea breath test | 95 | 5.98 (0.05–21) | 0.73 (0.23–0.97) | 0.77 (0.27–0.99) | |
| Rapid urease test | Positive histology and/or rapid urease test | 81 | 9.94 (0.05–33) | 0.84 (0.32–1.00) | 0.59 (0.14–0.95) |
| Positive histology and urea breath test | 96 | 7.89 (0.04–31) | 0.81 (0.33–1.00) | 0.61 (0.14–0.95) | |
| Positive at least two of the followings: rapid urease test, histology, urea breath test | 57 | 7.24 (0.04–33) | 0.86 (0.36–1.00) | 0.46 (0.10–0.76) | |
| Urea breath test | Positive histology and rapid urease test | 81 | 7.56 (0.07–23) | 0.81 (0.34–1.00) | 0.73 (0.23–0.99) |
| Positive at least two of the followings: rapid urease test, histology, urea breath test, serology | 64 | 6.53 (0.08–21) | 0.89 (0.48–1.00) | 0.61 (0.16–0.97) | |
| Positive at least two of the followings: rapid urease test, histology, urea breath test | 57 | 6.48 (0.07–21) | 0.76 (0.24–0.99) | 0.79 (0.31–1.00) | |
| Serology | Positive at least two of the followings: rapid urease test, histology, urea breath test, serology | 32 | 7.81 (0.08–21) | 0.72 (0.35–0.97) | 0.80 (0.23–1.00) |
| Positive histology or positive serology and urea breath test | 68 | 5.69 (0.06–19) | 0.83 (0.36–0.99) | 0.57 (0.12–0.90) | |
| Positive histology or two of the followings: rapid urease test, serology, urea breath test | 78 | 5.06 (0.06–17) | 0.82 (0.29–0.99) | 0.52 (0.10–0.88) | |
| Stool antigen test | Positive culture or positive rapid urease test and histology | 93 | 3.87 (0.33–9) | 0.76 (0.56–0.89) | 0.73 (0.49–0.89) |
| Positive culture or two of the followings: histology, rapid urease test, serology, urea breath test | 34 | 3.77 (0.11–11) | 0.80 (0.32–1.00) | 0.55 (0.13–0.93) | |
| Positive histology or two of the followings: rapid urease test, serology, urea breath test | 68 | 2.66 (0.11–9.05) | 0.72 (0.23–0.98) | 0.56 (0.12–0.96) | |
| Culture | Positive culture or positive rapid urease test and histology | 199 | 7.35 (0.71–11) | 0.94 (0.83–1.00) | 0.77 (0.45–0.94) |
| Positive culture or two of the followings: rapid urease test, histology, serology | 157 | 2.28 (0.11–9) | 0.84 (0.35–1.00) | 0.46 (0.08–0.91) | |
| Positive culture or two of the followings: rapid urease test, histology, serology | 55 | 2.17 (0.11–9) | 0.82 (0.36–1.00) | 0.49 (0.10–0.88) | |
| Polymerase chain reaction | Positive culture or rapid urease test and histology | 152 | 4.99 (0.11–13) | 0.62 (0.34–0.88) | 0.85 (0.48–0.96) |
| Positive culture | 79 | 4.47 (0.07–11) | 0.42 (0.07–0.79) | 0.83 (0.32–1.00) | |
| Positive culture or positive three of the followings: rapid urease test, histology, urea breath test, serology | 55 | 4.44 (0.14–15) | 0.68 (0.34–1.00) | 0.80 (0.27–0.98) |
CI, confidence interval.
Risk of bias and applicability assessment
With the use of QUADAS-2 assessment tool, overall, only two studies56,58 proved to be free of an unclear or high risk of bias. Five studies42,43,45,47,51 were found to carry an unclear risk of bias. The remaining 33 articles carried a high risk of bias. Detailed results of the risk assessment are shown in Table 3.
Table 3.
Results of the study quality assessment.
| Study | Risk of bias | Applicability concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient selection | Index test | Reference standard | Flow and timing | Patient selection | Index test | Reference standard | |
| Archimandritis et al.22 |

|

|

|

|

|

|

|
| Bravo Paredes et al.23 |

|

|

|

|

|

|

|
| Castro-Fernandez et al.24 |

|

|

|

|

|

|

|
| Castro-Fernandez et al.25 |

|

|

|

|

|

|

|
| Chandrasakha et al.26 |

|

|

|

|

|

|

|
| Choi et al.27 |

|

|

|

|

|

|

|
| Chung et al.28 |

|

|

|

|

|

|

|
| Chung et al.29 |

|

|

|

|

|

|

|
| Colin et al.30 |

|

|

|

|

|

|

|
| Demiray et al.31 |

|

|

|

|

|

|

|
| Garcia-Diaz et al.32 |

|

|

|

|

|

|

|
| Gisbert et al.34 |

|

|

|

|

|

|

|
| Gisbert et al.33 |

|

|

|

|

|

|

|
| Grino et al.35 |

|

|

|

|

|

|

|
| Grino et al.36 |

|

|

|

|

|

|

|
| Hanvivatong et al.37 |

|

|

|

|

|

|

|
| Lahmidani et al.38 |

|

|

|

|

|

|

|
| Lee et al.39 |

|

|

|

|

|

|

|
| Lee et al.40 |

|

|

|

|

|

|

|
| Liao et al.41 |

|

|

|

|

|

|

|
| Lin et al.42 |

|

|

|

|

|

|

|
| Lin et al.43 |

|

|

|

|

|

|

|
| Lo et al.44 |

|

|

|

|

|

|

|
| López Peñas et al.45 |

|

|

|

|

|

|

|
| Manguso et al.46 |

|

|

|

|

|

|

|
| Pascual et al.47 |

|

|

|

|

|

|

|
| Peitz et al.48 |

|

|

|

|

|

|

|
| Peitz et al.49 |

|

|

|

|

|

|

|
| Ramírez-Lázaro et al.50 |

|

|

|

|

|

|

|
| Romero Gómez et al.51 |

|

|

|

|

|

|

|
| Saez et al.52 |

|

|

|

|

|

|

|
| Shilling et al.53 |

|

|

|

|

|

|

|
| Sfarti et al.54 |

|

|

|

|

|

|

|
| Tang et al.55 |

|

|

|

|

|

|

|
| Tu et al.12 |

|

|

|

|

|

|

|
| van Leerdam et al.56 |

|

|

|

|

|

|

|
| Velayos et al.57 |

|

|

|

|

|

|

|
| Wildner-Christensen et al.58 |

|

|

|

|

|

|

|
| Winiarski et al.59 |

|

|

|

|

|

|

|
| Wong et al.60 |

|

|

|

|

|

|

|
 low risk.
low risk.
 high risk.
high risk.
 unclear risk of bias.
unclear risk of bias.
Discussion
Our results from the network meta-analysis of the eligible DTA studies could order the index tests for the detection of HPI in PUB based on their superiority index. Still, their wide confidence intervals could not prove this order beyond doubt. Combined index tests showed a tendency of higher sensitivity, while single index tests had higher specificity values when ranked.
Reasons for combined tests as diagnostic gold standards in the included studies
The majority of the included DTA studies (28 of 40) used a combination of multiple testing methods for HPI as a gold standard. None of these studies gave specific reasons for the approach above. This seems to be an established strategy across the studies without sufficient evidence. With the use of the combined tests, one can increase the sensitivity of the testing. The use of this strategy in DTA studies is controversial, as it identifies more patients with HPI but compromises the validity of the results of the DTA – even more so, as 15 of 30 DTA studies included the index test in their combined method of the gold standard.
We believe that HPI in PUB should be detected as soon as possible and it is a preferable strategy over delayed testing. With this approach, clinicians can maximise the number of patients who genuinely need eradication and, at the same time, a small but increased proportion of patients will receive eradication unnecessarily (FNs). Given the significant risks of untreated HPI after PUB and the low risks of potential side effects (diarrhoea 1.6%; bloating or abdominal pain 1.3%; nausea or vomiting 0.4%) from the unnecessary antibiotic therapy,61 the clinical approach of combined testing seems reasonable.
Another reason for the combined and more aggressive testing approach can be the risk of loss to follow up after hospitalisation for PUB. The previous Maastricht/Florence IV guideline recommended the initiation of eradication at the time of introduction of oral feeding,62 arguing that a proportion of patients would be lost to follow up. In the study of Yoon et al., results showed that 13.3% of the patients who were lost to follow up before the confirmation of HPI, and 41.4% of the patients confirmed to have HPI after discharge, did not receive eradication.63 In 2014, as part of a retrospective analysis, Kim et al. reported that only 47% of PUB patients had HPI testing during their index admission, less than 10% had any testing after discharge, and 15% were lost to follow up.64 Given the findings above, the identification of HPI in the acute setting is of utmost importance.
Reasons for missing the opportunity to test HPI during admission for PUB
Invasive tests requiring tissue sampling during endoscopy
The endoscopic procedures for patients presenting with PUB are often stressful and are done out of hours, when access to diagnostic tools may be limited. Endoscopic management of acute GI bleeding is likely associated with patient and operator fatigue, and results in poorer adherence to guidelines. After the successful endoscopic termination of an acute PUB, the endoscopist may feel that histological sampling could contribute to a recurrent episode of bleeding. Finally, poor visualisation due to residual blood during the intervention may prevent safe sampling.
Another essential clinical problem leading to reluctance to take biopsies is PUB aggravated by anticoagulant and antiplatelet treatment. A recent multicentric retrospective study from France on upper GI bleeding found that 475 of 2498 patients (19%) took oral anticoagulants, either Vitamin K antagonist or direct oral anticoagulant.65 A French prospective multicenter study in 2011 reported 8.1% antiplatelet use in upper GI bleeding.66
Non-invasive tests
The urea breath test has very low feasibility in the acute setting of PUB, as patients have to fast before and often after the index endoscopy.
Stool antigen testing has a similar problem concerning feasibility; the opportunity of stool sampling for HPI testing in the admission department is often missed. Also, a Dutch study revealed a high rate of false-positive results in PUB patients explained by a cross-reaction with the blood.56
Also, patients with PUB receive, and are committed to, long-term PPI treatment before a urea breath test and stool antigen test is performed.1 The current guideline suggests a 14 day PPI free period before a urea breath test can be performed.11 Even 3 days of high dose PPI treatment reduces the detection of H. pylori significantly, and patients presenting with PUB would often receive PPI treatment before the index endoscopy.67
Serology would seem the most feasible out of all non-invasive tests, but the presence of antibodies may only indicate a previous infection instead of an acute one.68
Strengths of the study
The update of the most recent DTA meta-analysis on the same topic was published in 2006,15 with the then most recent study published in 2004; therefore, an update was necessary. We used a comprehensive and rigorous search strategy in seven databases. Detailed data extraction covered 70 items, which are shown in Supplementary file S8. We used a new statistical method of network meta-analysis developed for DTA studies. Assessment of the risks of bias was performed by the purpose-designed tool of QUADAS-2. Due to the clear study designs of the included reports, the patient population matched the review question.
Limitations of the study
As detailed in Table 3, the overall quality of the included studies was suboptimal, with high and unclear risks of bias. In many studies, it was unclear whether enrollment was consecutive, and inappropriate exclusion of subjects often occurred. Blinding the interpretation of the index tests and threshold of cut off value were not defined in many studies. Blinding the interpretation of the reference tests were not pre-specified. Another significant limitation was the unclear or prolonged interval between the index and reference tests. Not all patients received the same reference standard test when a combination was used. In some studies, a few participants were excluded from the final analysis.
Not only the combination of tests and index tests, but also the actual tests differed (manufacturer, methodology, etc.). In some articles, PPI use preceded either or both index and reference tests. The tissue sampling was not uniform across studies: some used antral, others antral and gastric body mucosal samples.
Implications for clinical practice
Combined tests may have a role in HPI testing in PUB as they have higher sensitivities. Endoscopic and gastroenterology units should have a tailored approach based on the availability of the individual tests.
Implication for research
Future DTA studies should use uniform gold standards. Also, they should focus on the feasibility and cost-effectiveness of the combined testing strategies.
In conclusion, our network meta-analysis demonstrated that none of the individual tests or the strategy of combined tests is superior in the detection of HPI. The combined tests have an increased sensitivity, which can translate to an optimized eradication strategy as it can result in the identification of most patients needing eradication therapy.
Supplemental Material
Supplemental material, sj-pdf-1-tag-10.1177_1756284820965324 for Accuracy of the Helicobacter pylori diagnostic tests in patients with peptic ulcer bleeding: a systematic review and network meta-analysis by Nóra Vörhendi, Alexandra Soós, Marie Anne Engh, Benedek Tinusz, Zsolt Szakács, Dániel Pécsi, Alexandra Mikó, Patrícia Sarlós, Péter Hegyi and Bálint Eröss in Therapeutic Advances in Gastroenterology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data, sj-xlsx-2-tag-10.1177_1756284820965324 for Accuracy of the Helicobacter pylori diagnostic tests in patients with peptic ulcer bleeding: a systematic review and network meta-analysis by Nóra Vörhendi, Alexandra Soós, Marie Anne Engh, Benedek Tinusz, Zsolt Szakács, Dániel Pécsi, Alexandra Mikó, Patrícia Sarlós, Péter Hegyi and Bálint Eröss in Therapeutic Advances in Gastroenterology
Footnotes
Author contributions: NV, BT, AS and PH conceptualized and designed the study in cooperation with BE; DP, SP, and ZS constructed the search query. NV, AM, and ZS carried out the search process. NV and ME screened the articles for eligibility. NV, ME, and BE performed the data extraction; NV, ME and BE conducted the quality assessment. NV, BE, BT and ZS wrote the article. AS carried out the statistical analysis. DP, PS, AM, BT, ME, and PH provided valuable feedback after critically reviewing the first drafts of the manuscript. All authors contributed and approved the final manuscript for publication.
Conflict of interest statement: The authors declare that there is no conflict of interest.
Ethical approval: No ethical approval was required for this review as all data were already published in peer reviewed journals. No patients were involved in the design, conduct or interpretation of our review.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research was supported by Project Grants (K131996 to PH and FK131864 to MA), an Economic Development and Innovation Operative Programme Grant (GINOP 2.3.2-15-2016-00048 to PH), The Grant of the Hungarian Science Foundation (FK 132834 to PS) and a Human Resources Development Operational Programme Grant (EFOP-3.6.2-16-2017-00006 to PH) of the National Research, Development and Innovation Office.
Guarantor: Bálint Eröss
ORCID iD: Bálint Eröss  https://orcid.org/0000-0003-3658-8427
https://orcid.org/0000-0003-3658-8427
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Nóra Vörhendi, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary.
Alexandra Soós, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary.
Marie Anne Engh, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary.
Benedek Tinusz, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary; Division of Gastroenterology, First Department of Medicine, Medical School, University of Pécs, Pécs, Hungary.
Zsolt Szakács, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary.
Dániel Pécsi, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary.
Alexandra Mikó, Institute for Translational Medicine, University of Pécs, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary; Division of Gastroenterology, First Department of Medicine, Medical School, University of Pécs, Pécs, Hungary.
Patrícia Sarlós, Division of Gastroenterology, First Department of Medicine, Medical School, University of Pécs, Pécs, Hungary.
Péter Hegyi, Institute for Translational Medicine, Medical School, Szentágothai Research Centre, University of Pécs, Pécs, Hungary; Division of Gastroenterology, First Department of Medicine, Medical School, University of Pécs, Pécs, Hungary.
Bálint Eröss, Institute for Translational Medicine, University of Pécs, 12 Szigeti út. II. floor, PÉCS, 7624, Hungary; Division of Gastroenterology, First Department of Medicine, Medical School, University of Pécs, Pécs, Hungary.
References
- 1. Gralnek IM, Dumonceau J-M, Kuipers EJ, et al. Diagnosis and management of nonvariceal upper gastrointestinal hemorrhage: European Society of Gastrointestinal Endoscopy (ESGE) guideline. Endoscopy 2015; 47: a1–a46. [DOI] [PubMed] [Google Scholar]
- 2. Karstensen JG, Ebigbo A, Aabakken L, et al. Nonvariceal upper gastrointestinal hemorrhage: European Society of Gastrointestinal Endoscopy (ESGE) cascade guideline. Endosc Int Open 2018; 6: E1256–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adler DG, Leighton JA, Davila RE, et al. ASGE guideline: the role of endoscopy in acute non-variceal upper-GI hemorrhage. Gastrointest Endosc 2004; 60: 497–504. [DOI] [PubMed] [Google Scholar]
- 4. Laine L, Jensen DM. Management of patients with ulcer bleeding. Am J Gastroenterol 2012; 107: 345–360; quiz 61. [DOI] [PubMed] [Google Scholar]
- 5. Rosenstock SJ, Moller MH, Larsson H, et al. Improving quality of care in peptic ulcer bleeding: nationwide cohort study of 13,498 consecutive patients in the Danish Clinical Register of Emergency Surgery. Am J Gastroenterol 2013; 108: 1449–1457. [DOI] [PubMed] [Google Scholar]
- 6. Crooks C, Card T, West J. Reductions in 28-day mortality following hospital admission for upper gastrointestinal hemorrhage. Gastroenterology 2011; 141: 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Leerdam ME. Epidemiology of acute upper gastrointestinal bleeding. Best Pract Res Clin Gastroenterol 2008; 22: 209–224. [DOI] [PubMed] [Google Scholar]
- 8. Hearnshaw SA, Logan RF, Lowe D, et al. Acute upper gastrointestinal bleeding in the UK: patient characteristics, diagnoses and outcomes in the 2007 UK audit. Gut 2011; 60: 1327–1335. [DOI] [PubMed] [Google Scholar]
- 9. Leodolter A, Kulig M, Brasch H, et al. A meta-analysis comparing eradication, healing and relapse rates in patients with Helicobacter pylori-associated gastric or duodenal ulcer. Aliment Pharmacol Ther 2001; 15: 1949–1958. [DOI] [PubMed] [Google Scholar]
- 10. Gisbert JP, Khorrami S, Carballo F, et al. Meta-analysis: Helicobacter pylori eradication therapy vs. Antisecretory non-eradication therapy for the prevention of recurrent bleeding from peptic ulcer. Aliment Pharmacol Ther 2004; 19: 617–629. [DOI] [PubMed] [Google Scholar]
- 11. Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection-the Maastricht V/Florence consensus report. Gut 2017; 66: 6–30. [DOI] [PubMed] [Google Scholar]
- 12. Tu TC, Lee CL, Wu CH, et al. Comparison of invasive and noninvasive tests for detecting Helicobacter pylori infection in bleeding peptic ulcers. Gastrointest Endosc 1999; 49: 302–306. [DOI] [PubMed] [Google Scholar]
- 13. Huang T-C, Lee C-L. Diagnosis, treatment, and outcome in patients with bleeding peptic ulcers and Helicobacter pylori infections. Biomed Res Int 2014; 2014: 658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sánchez-Delgado J, Gené E, Suárez D, et al. Has H. Pylori prevalence in bleeding peptic ulcer been underestimated? A meta-regression. Am J Gastroenterol 2011; 106: 398–405. [DOI] [PubMed] [Google Scholar]
- 15. Gisbert JP, Abraira V. Accuracy of Helicobacter pylori diagnostic tests in patients with bleeding peptic ulcer: a systematic review and meta-analysis. Am J Gastroenterol 2006; 101: 848–863. [DOI] [PubMed] [Google Scholar]
- 16. Group CDTAW. Handbook for DTA reviews. London: The Cochrane Collaboration, 2009. [Google Scholar]
- 17. Nyaga VN, Aerts M, Arbyn M. ANOVA model for network meta-analysis of diagnostic test accuracy data. Stat Methods Med Res 2018; 27: 1766–1784. [DOI] [PubMed] [Google Scholar]
- 18. McInnes MDF, Moher D, Thombs BD, et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies: the PRISMA-DTA statement. JAMA 2018; 319: 388–396. [DOI] [PubMed] [Google Scholar]
- 19. Hutton B, Salanti G, Caldwell DM, et al. The PRISMA extension statement for reporting of systematic reviews incorporating network meta-analyses of health care interventions: checklist and explanations. Ann Intern Med 2015; 162: 777–784. [DOI] [PubMed] [Google Scholar]
- 20. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012; 22: 276–282. [PMC free article] [PubMed] [Google Scholar]
- 21. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- 22. Archimandritis A, Tzivras M, Sougioultzis S, et al. Rapid urease test is less sensitive than histology in diagnosing Helicobacter pylori infection in patients with non-variceal upper gastrointestinal bleeding. J Gastroenterol Hepatol 2000; 15: 369–373. [DOI] [PubMed] [Google Scholar]
- 23. Bravo Paredes E, Guzmán Rojas P, Gallegos López R, et al. [Utility of urease rapid test for detection of Helicobacter pylori in patients with upper gastrointestinal bleeding from peptic ulcer]. Rev Gastroenterol Peru 2011; 31: 17–20. [PubMed] [Google Scholar]
- 24. Castro Fernández M, Sánchez Muñoz D, García Díaz E, et al. Diagnosis of Helicobacter pylori infection using urease rapid test in patients with bleeding duodenal ulcer: influence of endoscopic signs and simultaneous corporal and antral biopsies. Rev Esp Enferm Dig 2004; 96: 599–605. [DOI] [PubMed] [Google Scholar]
- 25. Castro-Fernandez M, Sanchez-Munoz D, Garcia-Diaz E, et al. Diagnosis of Helicobacter pylori infection in patients with bleeding ulcer disease: rapid urease test and histology. Rev Esp Enferm Dig 2004; 96: 395–401. [DOI] [PubMed] [Google Scholar]
- 26. Chandrasakha S, Hansombun P, Sirinawasatien A, et al. Gastric juice PCR for the diagnosis of Helicobacter pylori infection in patients with upper gastrointestinal bleeding. J Gastroenterol Hepatol 2015; 30: 116. [Google Scholar]
- 27. Choi YJ, Kim N, Lim J, et al. Accuracy of diagnostic tests for Helicobacter pylori in patients with peptic ulcer bleeding. Helicobacter 2012; 17: 77–85. [DOI] [PubMed] [Google Scholar]
- 28. Chung IK, Hong SJ, Kim EJ, et al. What is the best method to diagnose Helicobacter infection in bleeding peptic ulcers?: a prospective trial. Korena J Intern Med 2001; 16: 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung WC, Jeon EJ, Oh JH, et al. Dual-priming oligonucleotide-based multiplex PCR using tissue samples from the rapid urease test kit for the detection of Helicobacter pylori in bleeding peptic ulcers. Dig Liver Dis 2016; 48: 899–903. [DOI] [PubMed] [Google Scholar]
- 30. Colin R, Czernichow P, Baty V, et al. Low sensitivity of invasive tests for the detection of Helicobacter pylori infection in patients with bleeding ulcer. Gastroenterol Clin Biol 2000; 24: 31–35. [PubMed] [Google Scholar]
- 31. Demiray E, Yilmaz Ö, Șarkiș C, et al. Comparison of invasive methods and two different stool antigen tests for diagnosis of H pylori infection in patients with gastric bleeding. World J Gastroenterol 2006; 12: 4206–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia-Diaz E, Castro-Fernandez M, Romero-Gomez M, et al. The effectiveness of (IgG-ELISA) serology as an alternative diagnostic method for detecting Helicobacter pylori infection in patients with gastro-intestinal bleeding due to gastro-duodenal ulcer. Rev Esp Enferm Dig 2002; 94: 725–736. [PubMed] [Google Scholar]
- 33. Gisbert JP, Esteban C, Jimenez I, et al. 13C-urea breath test during hospitalization for the diagnosis of Helicobacter pylori infection in peptic ulcer bleeding. Helicobacter 2007; 12: 231–237. [DOI] [PubMed] [Google Scholar]
- 34. Gisbert JP, Trapero M, Calvet X, et al. Evaluation of three different tests for the detection of stool antigens to diagnose Helicobacter pylori infection in patients with upper gastrointestinal bleeding. Aliment Pharmacol Ther 2004; 19: 923–929. [DOI] [PubMed] [Google Scholar]
- 35. Grino P, Pascual S, Such J, et al. Comparison of diagnostic methods for Helicobacter pylori infection in patients with upper gastrointestinal bleeding. Scand J Gastroenterol 2001; 36: 1254–1258. [DOI] [PubMed] [Google Scholar]
- 36. Griñó P, Pascual S, Such J, et al. Comparison of stool immunoassay with standard methods for detection of Helicobacter pylori infection in patients with upper-gastrointestinal bleeding of peptic origin. Eur J Gastroenterol Hepatol 2003; 15: 525–529. [DOI] [PubMed] [Google Scholar]
- 37. Hanvivatvong O, Thong-Ngam D, Kuakarn S, et al. Evaluation of Helicobacter pylori stool antigen test in Thai patients with upper gastrointestinal bleeding. J Med Assoc Thai 2006; 89(Suppl. 3): S98–S103. [PubMed] [Google Scholar]
- 38. Lahmidani N, Aqodad N, ElYousfi M, et al. Accuracy of rapid urease test in the diagnosis of H. Pylori during bleeding period. Acta Endosc 2013; 43: 14–18. [Google Scholar]
- 39. Lee JM, Breslin NP, Fallon C, et al. Rapid urease tests lack sensitivity in Helicobacter pylori diagnosis when peptic ulcer disease presents with bleeding. Am J Gastroenterol 2000; 95: 1166–1170. [DOI] [PubMed] [Google Scholar]
- 40. Lee TH, Lin CC, Chung CS, et al. Increasing biopsy number and sampling from gastric body improve the sensitivity of rapid urease test in patients with peptic ulcer bleeding. Dig Dis Sci 2015; 60: 454–457. [DOI] [PubMed] [Google Scholar]
- 41. Liao CC, Lee CL, Lai YC, et al. Accuracy of three diagnostic tests used alone and in combination for detecting Helicobacter pylori infection in patients with bleeding gastric ulcers. Chin Med J 2003; 116: 1821–1826. [PubMed] [Google Scholar]
- 42. Lin HJ, Lo WC, Perng CL, et al. Helicobacter pylori stool antigen test in patients with bleeding peptic ulcers. Helicobacter 2004; 9: 663–668. [DOI] [PubMed] [Google Scholar]
- 43. Lin HJ, Lo WC, Perng CL, et al. Mucosal polymerase chain reaction for diagnosing Helicobacter pylori infection in patients with bleeding peptic ulcers. World J Gastroenterol 2005; 11: 382–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lo CC, Lai KH, Peng NJ, et al. Polymerase chain reaction: a sensitive method for detecting Helicobacter pylori infection in bleeding peptic ulcers. World J Gastroenterol 2005; 11: 3909–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. López Peñas D, Naranjo Rodríguez A, Muñoz Molinero J, et al. Efficacy of fecal detection of Helicobacter pylori with the HpSA technique in patients with upper digestive hemorrhage. Gastroenterol Hepatol 2001; 24: 5–8. [PubMed] [Google Scholar]
- 46. Manguso F, Riccio E, de Nucci G, et al. Helicobacter pylori infection in bleeding peptic ulcer patients after non-steroidal antiinflammatory drug consumption. World J Gastroenterol 2011; 17: 4509–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pascual S, Grino P, Casellas JA, et al. [Etiology of upper gastrointestinal bleeding of peptic origin: role of Helicobacter pylori and NSAIDs]. Gastroenterol Hepatol 2003; 26: 630–634. [DOI] [PubMed] [Google Scholar]
- 48. Peitz U, Leodolter A, Kahl S, et al. Antigen stool test for assessment of Helicobacter pylori infection in patients with upper gastrointestinal bleeding. Aliment Pharmacol Ther 2003; 17: 1075–1084. [DOI] [PubMed] [Google Scholar]
- 49. Peitz U, Leodolter A, Wex T, et al. Diagnostics of Helicobacter pylori infection in patients with peptic ulcer bleeding. Z Gastroenterol 2004; 42: 141–146. [DOI] [PubMed] [Google Scholar]
- 50. Ramirez-Lazaro MJ, Lario S, Casalots A, et al. Real-time PCR improves Helicobacter pylori detection in patients with peptic ulcer bleeding. PLoS One 2011; 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Romero Gómez M, Vargas J, Utrilla D, et al. Prospective study on the influence of gastroduodenal ulcer hemorrhage on the diagnostic methods in Helicobacter pylori infection. Gastroenterol Hepatol 1998; 21: 267–271. [PubMed] [Google Scholar]
- 52. Saez J, Belda S, Santibanez M, et al. Real-time PCR for diagnosing Helicobacter pylori infection in patients with upper gastrointestinal bleeding: comparison with other classical diagnostic methods. J Clin Microbiol 2012; 50: 3233–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schilling D, Demel A, Adamek HE, et al. A negative rapid urease test is unreliable for exclusion of Helicobacter pylori infection during acute phase of ulcer bleeding. A prospective case control study. Dig Liver Dis 2003; 35: 217–221. [DOI] [PubMed] [Google Scholar]
- 54. Sfarti C, Stanciu C, Cojocariu C, et al. 13C-urea breath test for the diagnosis of Helicobacter pylori infection in bleeding duodenal ulcer. Rev Med Chir Soc Med Nat Iasi 2009; 113: 704–709. [PubMed] [Google Scholar]
- 55. Tang JH, Liu NJ, Cheng HT, et al. Endoscopic diagnosis of Helicobacter pylori infection by rapid urease test in bleeding peptic ulcers: a prospective case-control study. J Clin Gastroenterol 2009; 43: 133–139. [DOI] [PubMed] [Google Scholar]
- 56. Van Leerdam ME, Van der Ende A, Ten Kate FJW, et al. Lack of accuracy of the noninvasive Helicobacter pylori stool antigen test in patients with gastroduodenal ulcer bleeding. Am J Gastroenterol 2003; 98: 798–801. [DOI] [PubMed] [Google Scholar]
- 57. Velayos B, Fernandez-Salazar L, Pons-Renedo F, et al. Accuracy of urea breath test performed immediately after emergency endoscopy in peptic ulcer bleeding. Dig Dis Sci 2012; 57: 1880–1886. [DOI] [PubMed] [Google Scholar]
- 58. Wildner-Christensen M, Lassen AT, Lindebjerg J, et al. Diagnosis of Helicobacter pylori in bleeding peptic ulcer patients, evaluation of urea-based tests. Digestion 2002; 66: 9–13. [DOI] [PubMed] [Google Scholar]
- 59. Winiarski M, Bielanski W, Plonka M, et al. The usefulness of capsulated 13C-urea breath test in diagnosis of Helicobacter pylori infection in patients with upper gastrointestinal bleeding. J Clin Gastroenterol 2003; 37: 34–38. [DOI] [PubMed] [Google Scholar]
- 60. Wong RM, Ota S, Bamba H, et al. Accuracy of endoscopic diagnosis of Helicobacter pylori in patients with hemorrhagic peptic ulcers. Dig Endosc 2003; 15: 25–29. [Google Scholar]
- 61. Kim SE, Park MI, Park SJ, et al. Trends in Helicobacter pylori eradication rates by first-line triple therapy and related factors in eradication therapy. Korean J Intern Med 2015; 30: 801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Malfertheiner P, Megraud F, O’Morain CA, et al. Management of Helicobacter pylori infection-the Maastricht IV/ Florence Consensus Report. Gut 2012; 61: 646–664. [DOI] [PubMed] [Google Scholar]
- 63. Yoon H, Lee DH, Jang ES, et al. Optimal initiation of Helicobacter pylori eradication in patients with peptic ulcer bleeding. World J Gastroenterol 2015; 21: 2497–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim JJ, Lee JS, Olafsson S, et al. Low adherence to Helicobacter pylori testing in hospitalized patients with bleeding peptic ulcer disease. Helicobacter 2014; 19: 98–104. [DOI] [PubMed] [Google Scholar]
- 65. UEG week 2019 oral presentations. United European Gastroenterol J 2019; 7(Suppl. 8): 10–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nahon S, Hagege H, Latrive JP, et al. Epidemiological and prognostic factors involved in upper gastrointestinal bleeding: results of a French prospective multicenter study. Endoscopy 2012; 44: 998–1008. [DOI] [PubMed] [Google Scholar]
- 67. Udd M, Miettinen P, Palmu A, et al. Effect of short-term treatment with regular or high doses of omeprazole on the detection of Helicobacter pylori in bleeding peptic ulcer patients. Scand J Gastroenterol 2003; 38: 588–593. [PubMed] [Google Scholar]
- 68. Wang WM, Chen CY, Jan CM, et al. Long-term follow-up and serological study after triple therapy of Helicobacter pylori-associated duodenal ulcer. Am J Gastroenterol 1994; 89: 1793–1796. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tag-10.1177_1756284820965324 for Accuracy of the Helicobacter pylori diagnostic tests in patients with peptic ulcer bleeding: a systematic review and network meta-analysis by Nóra Vörhendi, Alexandra Soós, Marie Anne Engh, Benedek Tinusz, Zsolt Szakács, Dániel Pécsi, Alexandra Mikó, Patrícia Sarlós, Péter Hegyi and Bálint Eröss in Therapeutic Advances in Gastroenterology
This article is distributed under the terms of the Creative Commons Attribution 4.0 License (http://www.creativecommons.org/licenses/by/4.0/) which permits any use, reproduction and distribution of the work without further permission provided the original work is attributed as specified on the SAGE and Open Access pages (https://us.sagepub.com/en-us/nam/open-access-at-sage).
Research Data, sj-xlsx-2-tag-10.1177_1756284820965324 for Accuracy of the Helicobacter pylori diagnostic tests in patients with peptic ulcer bleeding: a systematic review and network meta-analysis by Nóra Vörhendi, Alexandra Soós, Marie Anne Engh, Benedek Tinusz, Zsolt Szakács, Dániel Pécsi, Alexandra Mikó, Patrícia Sarlós, Péter Hegyi and Bálint Eröss in Therapeutic Advances in Gastroenterology