Abstract
The actin-related protein 2/3 complex (Arp2/3 complex), a key regulator of actin cytoskeletal dynamics, has been linked to multiple cellular processes, including those associated with response to stress. Herein, the Solanum habrochaites ARPC3 gene, encoding a subunit protein of the Arp2/3 complex, was identified and characterized. ShARPC3 encodes a 174-amino acid protein possessing a conserved P21-Arc domain. Silencing of ShARPC3 resulted in enhanced susceptibility to the powdery mildew pathogen Oidium neolycopersici (On-Lz), demonstrating a role for ShARPC3 in defence signalling. Interestingly, a loss of ShARPC3 coincided with enhanced susceptibility to On-Lz, a process that we hypothesize is the result of a block in the activity of SA-mediated defence signalling. Conversely, overexpression of ShARPC3 in Arabidopsis thaliana, followed by inoculation with On-Lz, showed enhanced resistance, including the rapid induction of hypersensitive cell death and the generation of reactive oxygen. Heterologous expression of ShARPC3 in the arc18 mutant of Saccharomyces cerevisiae (i.e., Δarc18) resulted in complementation of stress-induced phenotypes, including high-temperature tolerance. Taken together, these data support a role for ShARPC3 in tomato through positive regulation of plant immunity in response to O. neolycopersici pathogenesis.
Keywords: actin cytoskeleton, Arp2/3 complex, ARPC3, powdery mildew, resistance, Solanum habrochaites
1 |. INTRODUCTION
Tomato powdery mildew, Oidium neolycopersici, is an obligate biotrophic fungus that can parasitize more than 60 plant species in 13 families, including members of the Solanaceae (Jones, Whipps, & Guu, 2001). Upon infection of a susceptible host, O. neolycopersici causes powdery white lesions on the adaxial tomato leaf surface, abaxial surfaces, petioles, and the calyx; only the fruit remains uninfected. Pathogen infection typically affects leaves of the host plant, causing up to 50% yield losses (fruit) as a result of loss of vigour (Roberts, Momol, & Pernezny, 2002), and has caused devastating epidemics from Europe to North and South America, as well as in Asia (Lebeda et al., 2014). At present, chemical control remains the primary method to manage tomato powdery mildew in greenhouse production; however, chemical fungicide application introduces numerous inherent risks, including the development of pathogen resistance and the accumulation of toxic residues in fruit, both of which pose potential risks to the environment (Nakajima & Akutsu, 2014).
Plants defence signalling in response to bacterial and fungal pathogen infection is mediated by at least two primary nodes of the host innate immune system (Jones & Dangl, 2006). The first, pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), is often sufficient to protect plants against most pathogens and is often described as a mechanism of nonhost (basal) resistance (Marcel et al., 2008). In response to PTI, pathogens evolved mechanisms counter host defences, a process that functions via the delivery of virulence molecules which target a variety of host functions, including immune signalling. To counter this pathogen virulence function, plants evolved to recognize and respond to effector delivery, a process known as effector-triggered immunity (ETI). In short, ETI results in the activation of robust immune signalling, often characterized by localized cell death (i.e., the hypersensitive response [HR]) and the subsequent halt of pathogen growth and proliferation (Chisholm, Coaker, Day, & Staskawicz, 2006).
To date, most of the cloned resistance (R) genes encode proteins with an N-terminal nucleotide-binding site (NBS) and C-terminal leucine-rich repeats (LRRs; Takken, Albrecht, & Tameling, 2006). In wild tomato species, several R genes have been identified, including six monogenic genes comprising five dominant (Ol-1, Ol-3, Ol-4, Ol-5, and Ol-6), one recessive (ol-2) loci, and three polygenic resistance quantitative trait loci (QTLs; Bai et al., 2005; Bai, Huang, Van Der Hulst, Meijer-Dekens, & Bonnema, 2003). In brief, Ol-4-mediated resistance relies on the salicylic acid (SA) pathway, whereas Ol-1 and Ol-qtls require ethylene (ET) to promote delayed cell death during powdery mildew resistance, and jasmonic acid (JA) deficiency can compromise resistance mediated by ol-2 (Bai et al., 2005; Bai et al., 2003; Ciccarese, Amenduni, Ambrico, & Cirulli, 2000; Lindhout, Pet, & van der Beek, 1994; Lindhout, van der Beek, & Pet, 1994; Pei et al., 2011). In all instances, resistance to O. neolycopersici is associated with the induction of the HR, with a high frequency of necrosis in epidermal cells accompanying H2O2 accumulation induced by the fungal haustoria (Bai et al., 2005). Interestingly, when cells undergo the HR, uninfected neighbouring cells show an accumulation of focal actin microfilaments (AFs; Kobayashi, Kobayashi, & Hardham, 1994).
The eukaryotic cytoskeleton forms a contiguous network within all cells and includes microtubules (MT) and AF systems, both of which are associated with the function of numerous cellular processes (Day, Henty, Porter, & Staiger, 2011; Kim, Park, Kim, & Hwang, 2005; Li & Day, 2019; Sparkes, Runions, Hawes, & Griffing, 2009; Yokota et al., 2009). In short, MTs are hypothesized to play a role in maintaining cell polarity, whereas AFs ensure the targeted delivery of vesicles that carry plasma membrane and cell wall components to the site of growth (Mathur & Hülskamp, 2002). Additionally, AFs support the formation of penetration barriers by recruiting defence-related products to the subcellular site of fungal attack (Opalski, Schultheiss, Kogel, & Hückelhoven, 2005). As a putative mechanism underpinning actin-associated defence signalling, work by Henty-Ridilla et al. (2013) and Shimono, Higaki, et al. (2016) noted that early transient accumulation of actin (i.e., increased density and decreased filament bundling) is associated with the activation of PTI. Additional evidence points to the requirement for a suite actin-binding proteins (ABPs), including the actin-related protein 2 and 3 (Arp2/3) complex, profilin, and actin depolymerizing factors (ADF; reviewed in Porter & Day, 2015).
A key step in AF organization is actin nucleation, the key rate-limiting step necessary to ensure proper filaments formation (Campellone & Welch, 2010); this process is regulated by the Arp2/3 complex (Chesarone & Goode, 2009). The Arp2/3 complex contains seven subunits, including two actin-related proteins (ARP2 and ARP3) and five unrelated subunits (Machesky et al., 1999; Machesky, Atkinson, Ampe, Vandekerckhove, & Pollard, 1994). In eukaryotes, this complex facilitates branched actin network nucleation associated with regions of the plasma membrane (Pollard & Borisy, 2003; Weaver, Young, Lee, & Cooper, 2003) and is involved in actin polymerization-mediated motility of organelles (Machesky et al., 1999; Welch, Holtzman, & Drubin, 2002). Interestingly, Mathur, Mathur, Kernebeck, and Hülskamp (2003) found that expansion growth in Arabidopsis requires ARP2/3 activity, and its loss results in inefficient fine F-actin formation, leading to enhanced F-actin aggregation and bundling.
At a fundamental level, the Arp2/3 complex polymerizes new AFs in response to suite of signals, including those associated with growth, development, and response to external stimuli (Pantaloni, le Clainche, & Carlier, 2001; Pollard, Blanchoin, & Mullins, 2000; Takenawa & Miki, 2001). Interestingly, the seven subunits serve distinct roles in numerous developmental processes. For example, Arp2 is essential for membrane association (Kotchoni et al., 2009), whereas Arp3 is primarily associated with sites of actin nucleation (Maisch, Fiserova, Fischer, & Nick, 2009). Further research also demonstrated that deletion of ARPC1 or ARPC2 resulted in lethality and severe reductions in viability in Saccharomyces cerevisiae. In plants, Mathur, Mathur, Kernebeck et al. (2003); Mathur, Mathur, Kirik, et al. (2003) observed that loss-of-function mutants in Arabidopsis ARP3 or ARPC5 leads to shorter and sometimes sinuous root hairs. As a function of plant immunity, recent work by Qi et al. (2017) demonstrated that TaARPC3 is a key subunit of the Arp2/3 complex that is required for wheat resistance against Puccinia striiformis f. sp. tritici.
In the current study, we found that expression of ShARPC3 was significantly up-regulated during an incompatible host-pathogen interaction, suggestive of a role for ShARPC3 in plant defence signalling and immunity. This is significant, as a defined role for Arp2/3 signalling during plant pathogenesis by fungal pathogens has not been described. Using the model plant tomato and infection by the powdery mildew pathogen O. neolycopersici, we describe a function for ShARPC3 during infection and defence signalling in tomato. In total, the current study contributes to a broader understanding of the role of the Arp2/3 complex in pathogen defence signalling, and moreover, sheds light on the regulation of this complex in growth, development, and response to stress.
2 |. RESULTS
2.1 |. Identification and sequence analysis of tomato ARPC3
A tomato 525-bp homologue of actin-related protein was isolated from tomato LA1777 by homology-based cloning, and the obtained cDNA was designated as ShARPC3. Blast analysis of ShARPC3 nucleotide sequence in the Tomato Genome CDS (ITAG release 3.20) database revealed that one copy was present, localized on chromosome 7. The predicted ORF of ShARPC3 encodes a protein of 174 amino acid with a predicted molecular weight of 21 kDa. As shown in Figure 1a, phylogenetic analysis of ShARPC3 with other tomato ARP2/3 complex members and other species ARP2/3 complex members revealed that ShARPC3 clusters with AtARPC3 (AT1G60430) and NtARPC3 (XM_009610279), all of which are members of actin-related proteins in dicotyledons. Amino acid sequences alignment of ShARPC3 with AtARPC3, NsARPC3 (XP_009791878), NtARPC3 (XP_009608574), BnARPC3 (XP_022548276), SbARPC3 (XP_002451839), VrARPC3 (XP_014510301), and OsARPC3 (XP_015626361) predicts that ShARPC3 encodes a protein with unique conserved domains also found in the ARP2/3 complex-containing subunit ARPC3 (Figure 1b). In silico-based sequence analysis revealed that ShARPC3 has high sequence similarity to AtARPC3 and NtARPC3, with identities of 86% and 93%, respectively. SMART and RCSB PDB analyses revealed that ShARPC3 possesses a conserved Pfam:P21-Arc domain (amino acids 1–173), one glycyl lysine isopeptide (Lys-Gly; interchain with G-Cter in SUMO2)-cross-link site (amino acid 14), one phosphotyrosine-modified residue (amino acid 47), and two N6-acetyllysine-modified residues (amino acids 56 and 61). These data reveal that ShARPC3 is closely related to AtARPC3 and NtARPC3.
FIGURE 1.
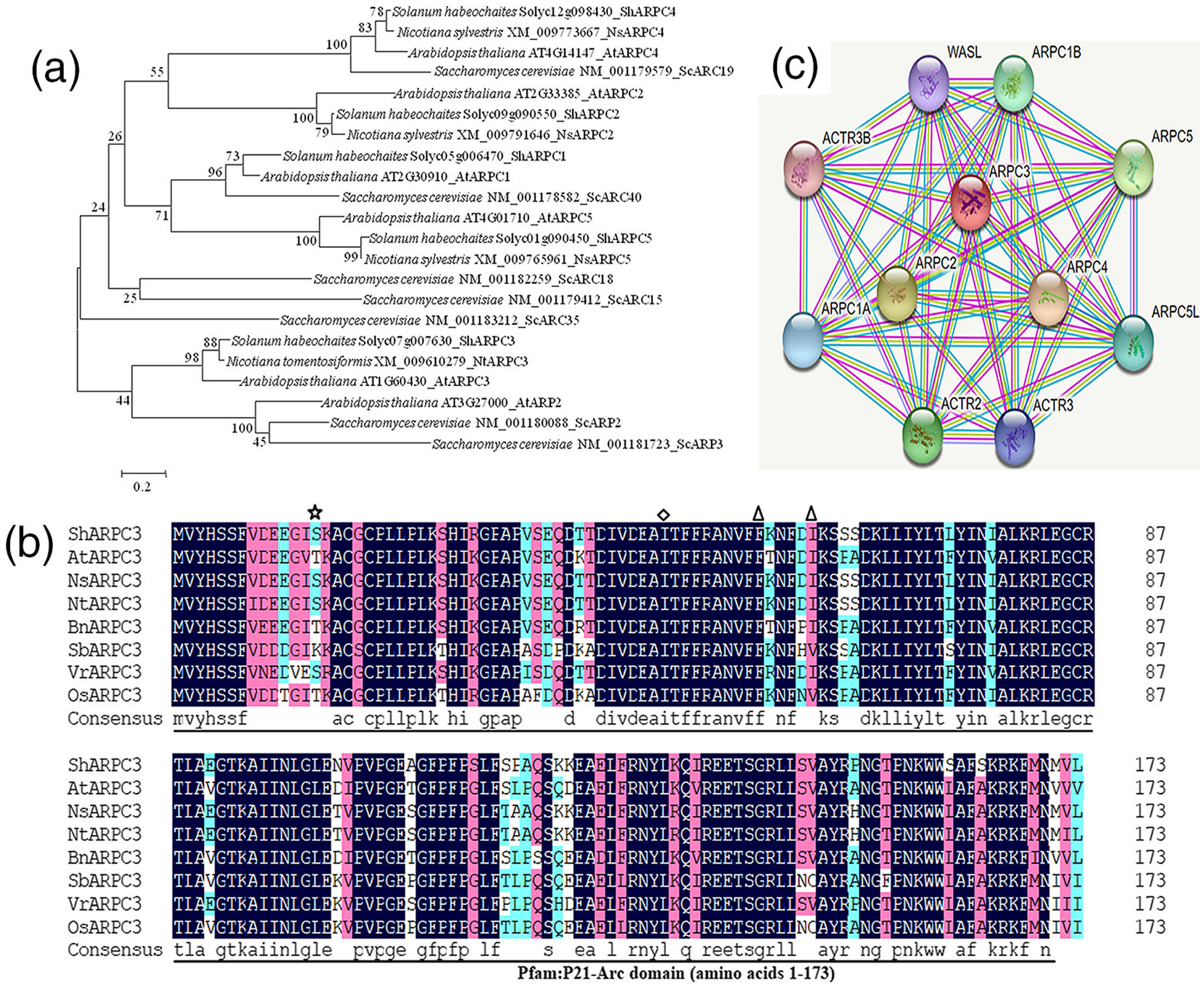
Sequence analysis and predicted interaction network of tomato actin-related protein 2 and 3 (Arp2/3) complex subunit 3 (ShARPC3). (a) Evolutionary analysis of ShARPC3. Analysis was conducted using the neighbour-joining method in MEGA6.0. Representative phylogenetic tree of ShARPC3 and ARP2/3 complex sequences from Solanum habrochaites (ShARP2/3), Arabidopsis thaliana (AtARP2/3), Nicotiana sylvestris (NsARP2/3), Nicotiana tomentosiformis (NtARP2/3), and Saccharomyces cerevisiae (ScARP2/3). Accession numbers are shown after the gene names. ShARPC3, encoded by the tomato gene Solyc07g007630, is considered to be the tomato ortholog of NtARPC3 and AtARPC3. (b) Multiple protein sequence alignment of ShARPC3 and characterized members of ARPC3 proteins. Sequence alignment of ShARPC3 (Solyc07g007630), AtARPC3 (AT1G60430), NsARPC3 (XP_009791878), NtARPC3 (XP_009608574), BnARPC3 (XP_022548276), SbARPC3 (XP_002451839), VrARPC3 (XP_014510301), and OsARPC3 (XP_015626361). The ShARPC3 sequence encodes a 174-amino acid protein containing one P21-Arc domain (amino acids 1–173), one glycyl lysine isopeptide (Lys-Gly; interchain with G-Cter in SUMO2)-cross-link site (amino acid 14) is highlighted with asterisk, one phosphotyrosine-modified residue (amino acid 47) is highlighted with a diamond, two N6-acetyllysine-modified residues (amino acids 56 and 61) are highlighted with a triangle. Amino acid similarity are shown within the Pfam:P21-Arc domain. Black boxes indicate regions of 100% homology, pink boxes indicate ≥75% homology, and cyan boxes highlight ≥50% homology. (c) The network map showed the interaction of ShARPC3 with other tomato proteins according to STRING database analysis. Here, the ARPC1A node in the figure represents the unknown three-dimensional (3D) structure, whereas other nodes indicate the availability of known or predicted 3D structures. The red coloured node represents the query protein. The edge, coloured in turquoise, yellow, purple, and white, indicates that the prediction was made on the basis of curated databases, text-mining, experimentally determined, and/or protein homology, respectively
Using an in silico-based approach (e.g., STRING database analysis), we generated a predicted protein network map (Figure 1c; high confidence score = 0.982), which revealed interactions between tomato ARPC3 and numerous additional tomato proteins. As shown, tomato ARPC3 is predicted to interact with actin-related protein 2/3 complex subunit 1A (ARPC1A), ARP2/3 complex subunit 1B (ARPC1B), ARP2/3 complex subunit 2 (ARPC2), ARP2/3 complex subunit 4 (ARPC4), ARP2/3 complex subunit 5 (ARPC5), ARP2/3 complex subunit 5-like protein (ARPC5L), ARP2 (ACTR2), ARP3 (ACTR3), ARP3B (ACTR3B), and Neural Wiskott-Aldrich syndrome protein (WASL). In total, the interaction map contains a total 11 nodes with 54 edges, and further analysis indicates that all of these tomato proteins are involved in actin cytoskeletal organization, mitosis, and cytokinesis. Based on this, we hypothesize that ShARPC3 functions through dynamic interactions with other ARP2/3 complex subunits to regulate cellular morphology, as well as abiotic and biotic signalling responses.
2.2 |. ShARPC3 is localized in the cytoplasm and plasma membrane in tomato protoplast
Based on the analysis above, we hypothesized that ShARPC3 is localized within multiple subcellular environments, including plasma membrane, extracellular spaces, cytoplasm, mitochondria, endoplasmic reticulum, peroxisomes, Golgi, and chloroplast (Table S4). To further determine the precise subcellular localization of ShARPC3, the ShARPC3 ORF was fused to GFP and placed under the control of the constitutive Cauliflower mosaic virus 35S promoter and transiently expressed in tomato protoplasts. As shown in Figure 2, pCaMV35S:GFP (negative control) revealed a diffuse, non-specific cellular address, whereas cells expressing GFP-tagged ShARPC3 was localized predominantly in the cytoplasm and nucleus.
FIGURE 2.
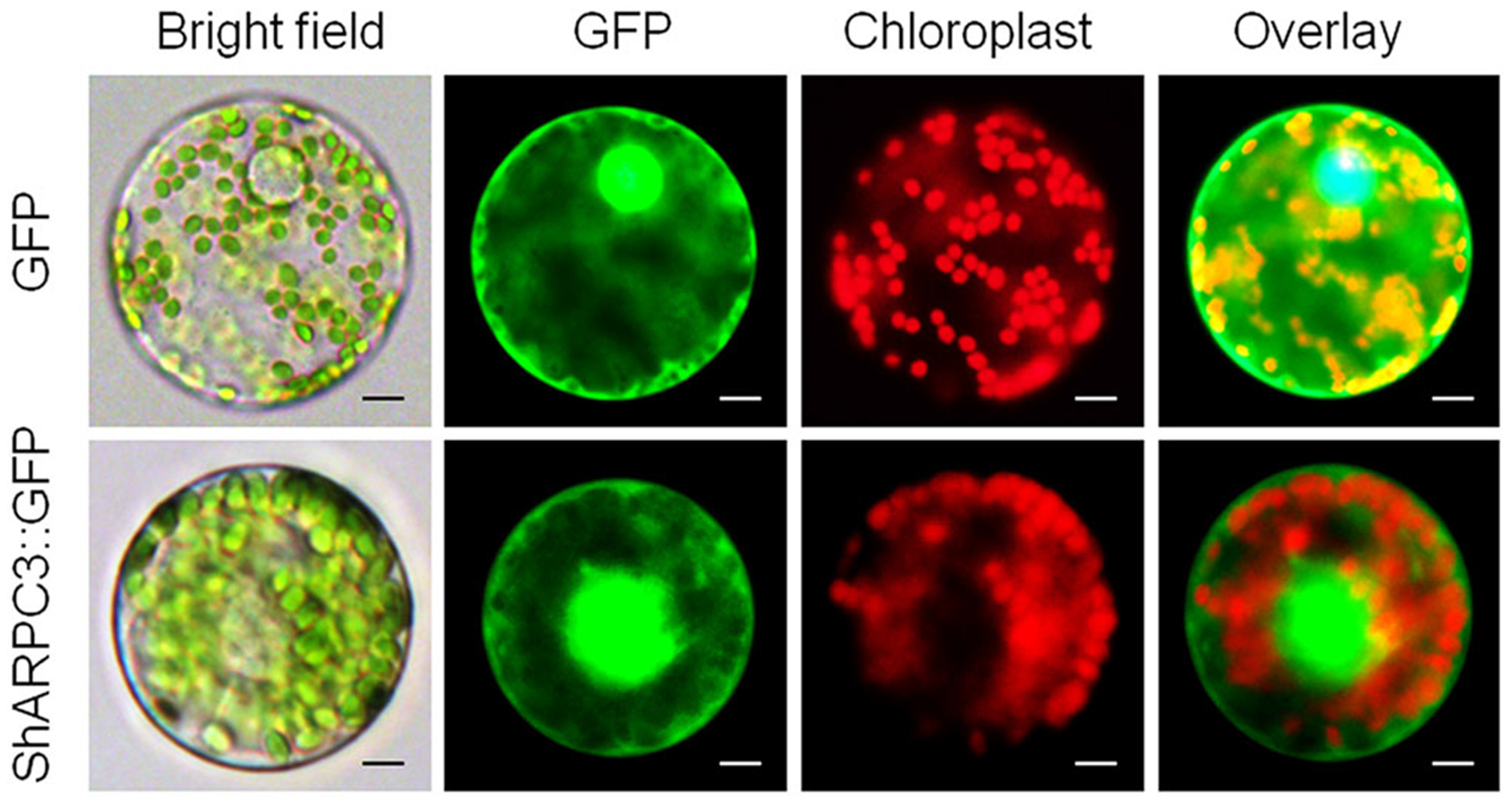
Subcellular localization of ShARPC3. Protoplast transient expression using GFP-ShARPC3 fusion constructs was used to determine the subcellular localization. The 35S:GFP-ShARPC3 constructs were transformed into a tomato protoplast cell. Fluorescence images of GFP and chlorophyll autofluorescence (Chl) were captured by laser confocal scanning microscopy, as indicated by green and red emission signals, respectively (scale bars, 20 μm)
2.3 |. ShARPC3 does not induce the hypersensitive cell death response nor suppress BAX-induced necrosis
To further define ShARPC3 function, we next employed a PVX-based high-throughput transient plant overexpression system under the regulation of the 35S promoter, using Nicotiana benthamiana, to evaluate ARPC3 function and activity during cell death elicitation. As shown in Figure S1, infiltration of N. benthamiana leaves with Agrobacterium expressing pGR106:GFP (site “a,” CK), pGR106: ShARPC3 (site “b”), or buffer alone (site “c”) did not result in the induction of cell death symptoms. These data indicate that ARPC3 does not possess cell death-inducing activity. To determine if the overexpression of ARPC3 can positively, or negatively, influence BAX-induced cell death, N. benthamiana leaves were infiltrated with Agrobacterium cells harbouring ShARPC3 gene 24 hr prior to infiltration with the pGR106:BAX-harbouring cells produced cell death symptoms (Figure S1, site “e”). As expected, cell death was observed in N. benthamiana leaves infiltrated with Agrobacterium cells expressing pGR106:GFP:BAX (Figure S1, site “d”). As shown, the BAX-induced cell death response, in the presence of ARPC3, was no different than overexpression of BAX alone (Figure S1, site “f”).
2.4 |. ShARPC3 is differentially induced by On-Lz infection
To determine the expression profile of ShARPC3, the mRNA accumulation of ShARPC3 was monitored during the interaction between tomato and On-Lz using qRT-PCR. LA1777 plants showed resistance against On-Lz, whereas obvious powdery mildew disease lesions appeared in MM plants (Figure S2a). The disease indexes of MM plants were significantly higher, with lesion indices reaching 18.5 and 26.4 at 7 and 14 dpi, respectively (Figure S2b). As shown in Figure 3, during an incompatible interaction, ShARPC3 transcripts were significantly up-regulated at 12–24 hpi, and mRNA accumulation peaked at 18 hpi, 16 times (P < .05) than that in control (uninoculated) plants. Additionally, ShARPC3 transcript levels were much higher during an incompatible interaction than during a compatible interaction at all time points evaluated, except at 96 hpi. Based on this, we hypothesize that ShARPC3 is associated with resistance of tomato against On-Lz infection.
FIGURE 3.
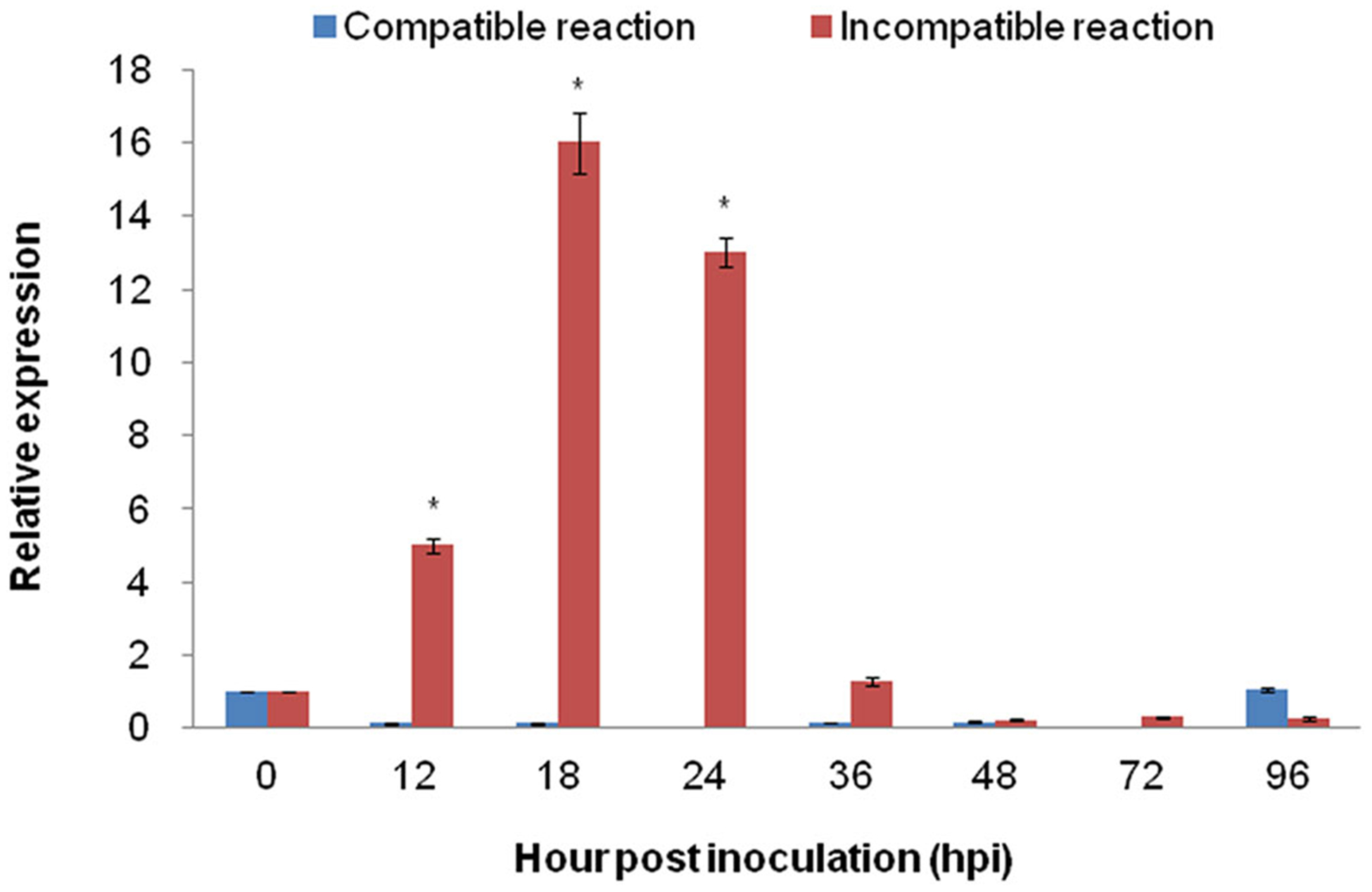
Quantitative real-time PCR expression analysis of ARPC3 in tomato leaves. The expression level of ARPC3 in tomato leaves was induced in incompatible reaction between tomato-On-Lz interaction. Error bars represent standard deviation from three independent replicates. Asterisks (*) indicate significant difference (P < .05) from 0 hpi using Student’s t test
2.5 |. Silencing of ShARPC3 results in host susceptibility to On-Lz
To determine the role of ShARPC3 during tomato infection by On-Lz, we used a tobacco rattle virus-induced gene silencing (TRV-VIGS)-based approach to silence ShARPC3 expression in LA1777. As shown in Figure 4, TRV2 (CK), TRV2:ShPDS (phytoene desaturase), and TRV2:ShARPC3 plasmids were inoculated onto tomato leaves for silencing. As expected, plants inoculated with TRV2:ShPDS showed a photobleaching phenotype at ~30 days post-inoculation (Figure 4 a), indicating the induction of TRV-VIGS-mediated silencing. At this point (i.e., 30 dpi), all plants were inoculated with On-Lz, and infection phenotypes were recorded. Compared with control plants, plants carrying TRV2:ShARPC3 showed obvious powdery mildew disease spot lesions (Figure 4a), and the disease indexes of ShARPC3-silenced plants were significantly higher, with lesion indices reaching 10.7 and 21.1 at 7 and 14 dpi, respectively (Figure 4b). In parallel, samples were collected to assess the efficiency of gene silencing using qRT-PCR analysis. The expression of ShARPC3 was reduced by 60–85% at 0–72 hpi compared with control-inoculated leaves (Figure S3); ShARPC3-silenced efficiency peaked at 24 hpi. Based on these data, we conclude that ShARPC3 is required for resistance to On-Lz.
FIGURE 4.
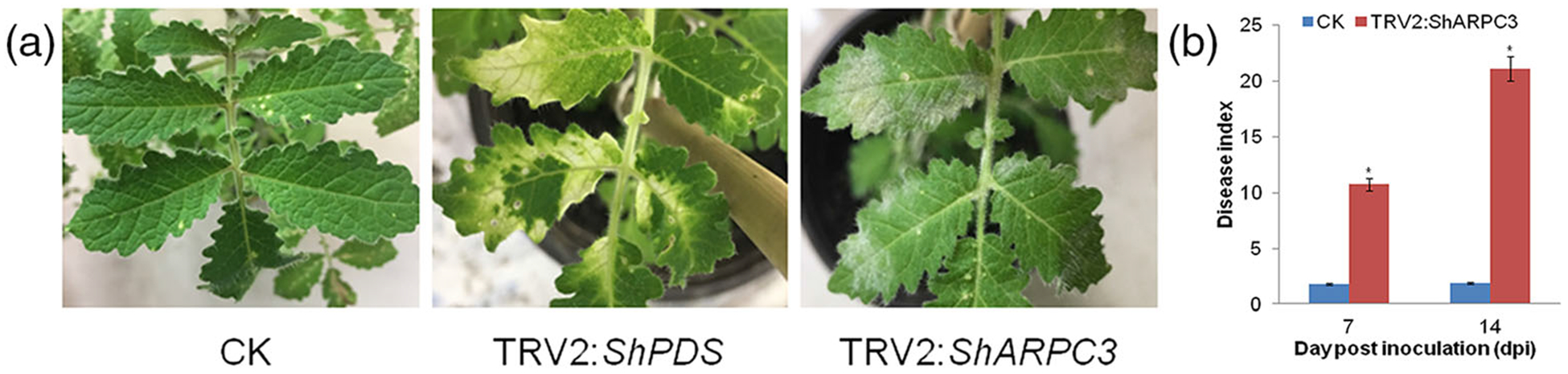
Silencing of ShARPC3 in tomato LA1777 renders plants susceptible to On-Lz. (a) Infection phenotypes of CK (TRV2), TRV2:ShPDS, and TRV2:ShARPC3 tomato leaves at 7 dpi. (b) Disease indexes of CK and TRV2:ShARPC3 plants at 7 and 14 dpi, respectively. Error bars represent the variations among three independent replicates. The asterisk (*) indicates statistically significant differences in disease index between CK (TRV2) and TRV2:ShARPC3 plants according to independent samples t test (P < .05)
2.6 |. Silencing of ShARPC3 results in reduced defence responses following On-Lz inoculation
To further investigate how ShARPC3 participates in tomato resistance to On-Lz, we measured the accumulation of H2O2 and the development of the HR at 6, 18, 24, 48, and 72 hpi via a microscopic examination of the infection process (Figure 5). As shown, a histological examination of On-Lz growth was performed in both control and ShARPC3-silenced plants by analysing the HR production rate (Figure 5a,c). We found that HR production in the ShARPC3-silenced plants was much less compared with the control plants at 48 and 72 hpi (P < .05). There were no obvious differences between TRV2-treated and ShARPC3-silenced plants in HR production at 18 and 24 hpi. As the infection progressed, we also observed a significant decrease in the amount of H2O2 accumulated in ShARPC3-silenced plants (leaves) at 72 hpi (P < .05); H2O2 was 0.45 times that in TRV2-treated leaves (CK; Figure 5b,d). Additionally, with the increase of haustoria formation, more invasive growth in ShARPC3-silenced plants was observed than in control plants. Taken together, these results indicate that a reduction in ShARPC3 expression compromises resistance signalling in response to On-Lz infection.
FIGURE 5.
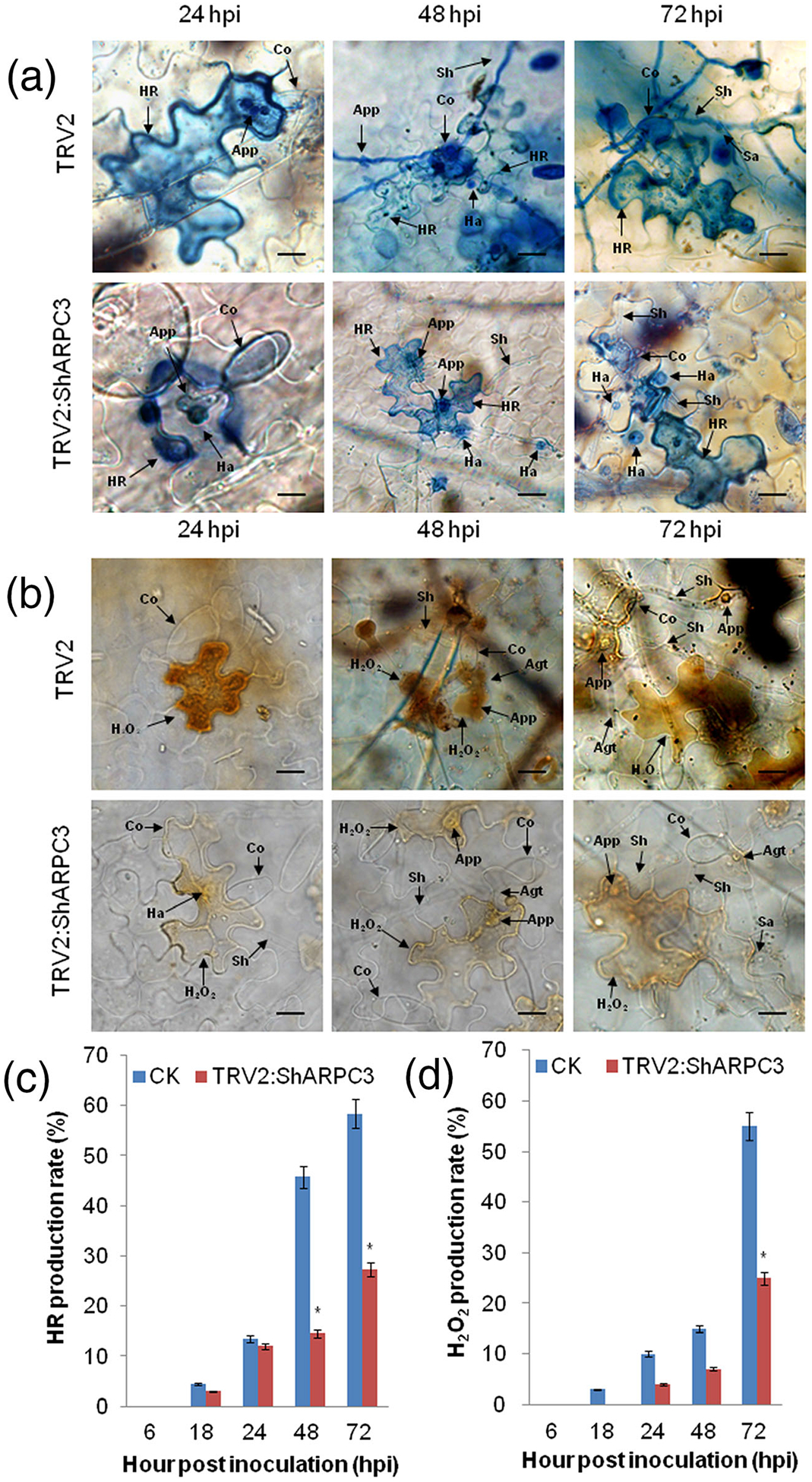
Silencing of ShARPC3 in tomato reduced defence responses and increased On-Lz infection. (a) Histological observation of hypersensitive cell death in CK (TRV2) and TRV2:ShARPC3 tomato leaves inoculated with On-Lz by microscopy. Blue (trypan) staining indicates hypersensitive cell death. (b) Histological observation of H2O2 accumulation in CK and TRV2:ShARPC3 tomato leaves inoculated with On-Lz by microscopy. (c) HR production rate of tomato leaves carrying TRV2 (CK) or TRV2:ShARPC3 at 6, 18, 24, 48, and 72 hpi, respectively. (d) H2O2 production in CK or TRV2:ShARPC3 tomato leaves at 6, 18, 24, 48, and 72 hpi, respectively. Agt, appressorium germ tube; App, appressorium; Co, conidium; Ha, haustorium; HR, hypersensitive response; Pa, papilla; Sa, secondary appressorium; Sh, secondary hyphae. Bar, 50 μm. Error bars represent the variations among three independent replicates. The asterisk (*) indicates statistically significant differences between CK (TRV2) and TRV2:ShARPC3 plants (P < .05)
2.7 |. Effects of ShARPC3 silencing on phytohormone accumulation and PR transcript levels
To confirm the relevance of ShARPC3 expression as a function of the activity of SA signalling pathway, we measured SA level in ShARPC3-silenced plants at 0–24 hpi with On-Lz. As shown in Figure 6a, in control plants (CK), SA content reached its highest level of 1,818.0 μg·g−1 at 12 hpi, >2.5 times that in ShARPC3-silenced plants. Additionally, SA levels were significantly lower in ShARPC3-silenced plants than in the control plants at all time points and higher than that at 0 hpi. We further studied pathogenesis-related (PR) PR1b1 (PR1), Glucanase A (PR2), Glucanase B (PR2-like), Chitinase 3 (PR3), and Chitinase 9 (PR3-like) gene expression after inoculation with On-Lz. In ShARPC3-silenced plants at 24 hpi, the expressions of PR1, PR2, and PR3 genes (SA pathway-specific gene expression markers) were significantly reduced (Figure 6b). Taken together, we posit that ShARPC3 is required for the positive regulation of host defences, mediated in part by SA signalling.
FIGURE 6.
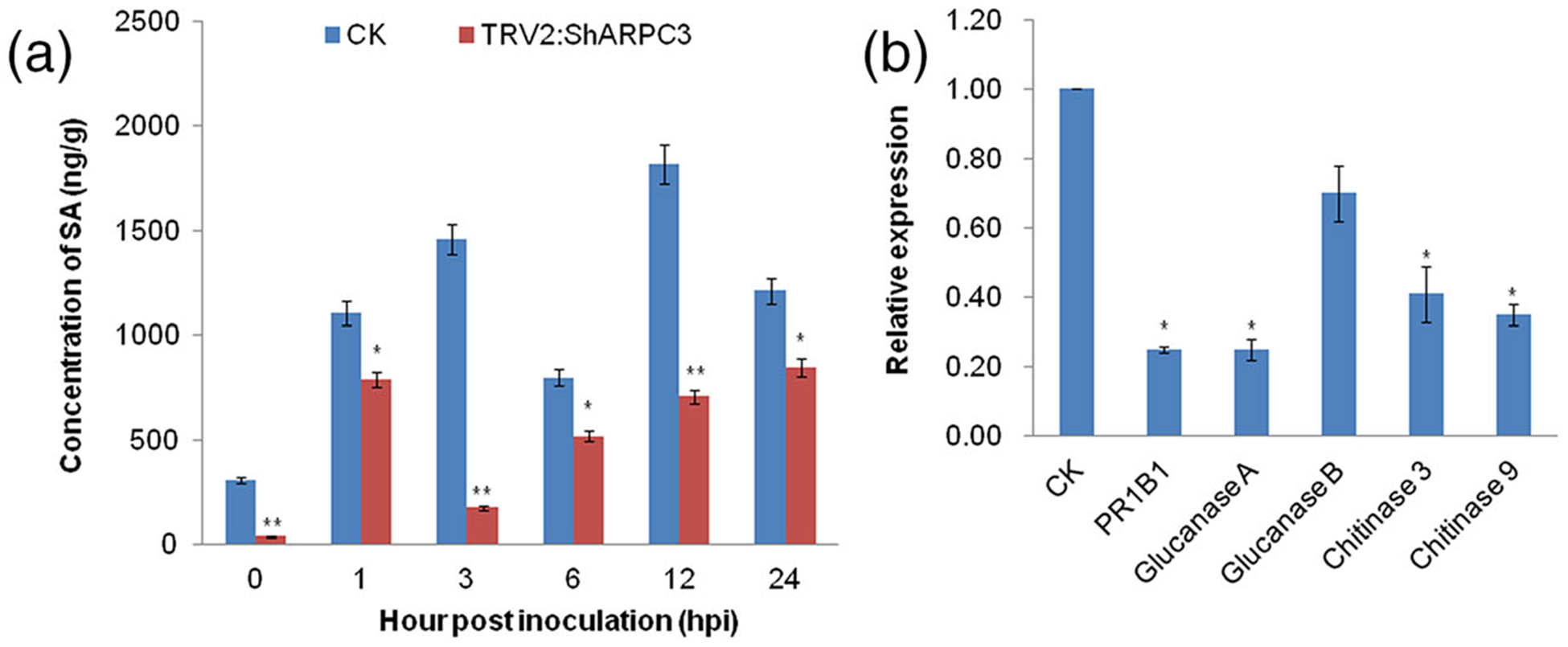
SA content and relative mRNA transcript levels of PRs genes are reduced in On-Lz-inoculated ShARPC3-silenced plants. (a) Changes in SA levels in tomato leaves carrying TRV2 (CK) or TRV2:ShARPC3 at 0, 1, 3, 6, 12, and 24 hpi, respectively. (b) Relative mRNA transcript levels of PR1B1 (PR1), β−1,3-glucanase (PR2), and chitinase (PR3; SA pathway-specific gene expression markers) in CK and TRV2:ShARPC3 tomato leaves at 24 hpi as determined by quantitative real-time PCR. Error bars represent the variations among three independent replicates. Number of asterisks indicate statistically degree of significance between CK (TRV2) and TRV2: ShARPC3 plants (Student’s t test, *P < .05; **P < .01)
2.8 |. Overexpression of ShARPC3 functionally complements the arpc3 mutation
To test whether ShARPC3-based resistance occurs in Arabidopsis, we evaluated a homozygous arpc3 mutant, an arpc3 complementation line, and WT Col-0 plants overexpressing ShARPC3 under the regulation of the 35S promoter and challenged them with On-Lz. As expected, macroscopic inspection revealed that arpc3 plants were susceptible to On-Lz (Figure 7b). However, arpc3 complementation plants, like the control plants (i.e., WT Col-0), showed only slight disease symptoms following infection. Conversely, WT Col-0 plants carrying 35S:pCAMBIA3301-ShARPC3 showed no obvious symptoms, possibly due to the additive effect of native + transgene overexpression of ShARPC3. Microscopic examination revealed that resistance in arpc3 plants carrying 35S:pCAMBIA3301-ShARPC3 and WT Col-0 plants carrying 35S:pCAMBIA3301-ShARPC3 was incomplete and characterized by reduced conidiophore and haustorium formation (Figure 7c–e). Compared with control plants (i.e., WT Col-0), conidiophore production was significantly higher in arpc3 mutant plants (P < .05). Additionally, the growth ability of the complemented arpc3 plants containing 35S:pCAMBIA3301-ShARPC3 was stronger than that in the arpc3 mutant yet weaker than that in WT Col-0 plants (Figure 7a).
FIGURE 7.
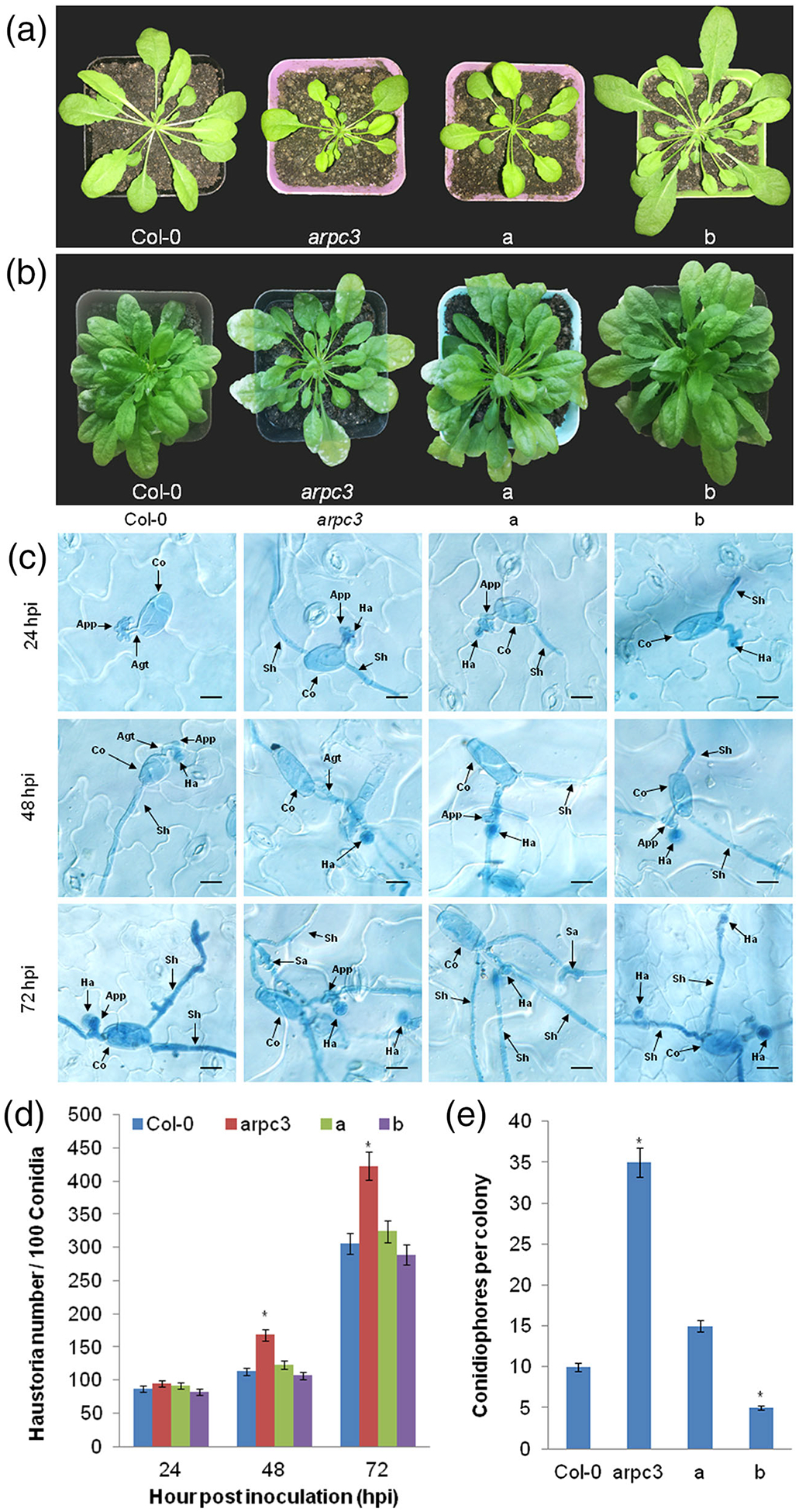
Complementation of ShARPC3 function confers On-Lz resistance in A. thaliana. (a) Macroscopic phenotypes of representative unchallenged plants at 8 weeks. (b) Infection phenotypes of WT Col-0 plants, arpc3 mutants, arpc3 plants transgenic plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 transgenic plants expressing pCAMBIA3301-ShARPC3. Images were taken at 7 dpi. (c) Infection process of conidiation in WT Col-0 plants, arpc3 mutants, arpc3 plants transgenic plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 transgenic plants expressing pCAMBIA3301-ShARPC3 by microscopy. Images were taken at 24, 48, and 72 hpi. (d) Quantitative assessment of haustoria/100 conidia on WT Col-0 plants, the arpc3 mutant, the arpc3 mutant expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Images were taken at 24, 48, and 72 hpi. (e) Quantitative assessment of conidiation on WT Col-0 plants, the arpc3 mutant, the arpc3 mutant expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Images were taken at 7 dpi. a: arpc3 plants transgenic expressing pCAMBIA3301-ShARPC3; b: WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Error bars represent the variations among three independent replicates. Asterisks (*) indicate a significant difference between WT Col-0 plants, arpc3 mutants, arpc3 plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 (P < .05)
2.9 |. Resistance mechanisms revealed in histological and biochemical studies
To assess the impact of ARPC3 on the response of Arabidopsis to On-Lz, we next investigated the host response via a microscopic examination of the infection process. To do this, we measured the development of the HR (Figure 8a,c) and the accumulation of H2O2 (Figure 8b,d) at 6, 18, 24, 48, and 72 hpi. As shown, we observed no significant differences in either HR development or H2O2 accumulation among four treatments at 6–24 hpi. However, when compared with ShARPC3-overexpression plants (i.e., arpc3 complementation lines or WT Col-0 overexpression), cell death elicitation was significantly reduced in control and arpc3 plants at 48 hpi. Moreover, we observed that HR production in arpc3 plants was much less than that in the control plants at 72 hpi (P < .05). Consistent with this observation, H2O2 production rate was significantly lower in arpc3 plants than that of other treatments at 48 hpi (P < .05). Indeed, H2O2 accumulation was frequently observed at levels significantly higher in mesophyll cells of arpc3 plants carrying pCAMBIA3301-ShARPC3 and WT Col-0 plants carrying pCAMBIA3301-ShARPC3 than in control plants and arpc3 mutant plants at 72 hpi. In addition to altered H2O2 accumulation and cell death, qRT-PCR analyses revealed that accumulation of AtARPC3 mRNA was rapidly induced and increased significantly over time in ShARPC3-overexpression plants compared with AtARPC3 mRNA levels in WT Col-0 plants (Figure S4). Conversely, the expression of AtARPC3 was significantly lower in arpc3 plants than that in WT Col-0 plants at 0–72 hpi. Taken together, we posit that an increase in ShARPC3 mRNA accumulation is required for robust defence signalling in response to On-Lz infection.
FIGURE 8.
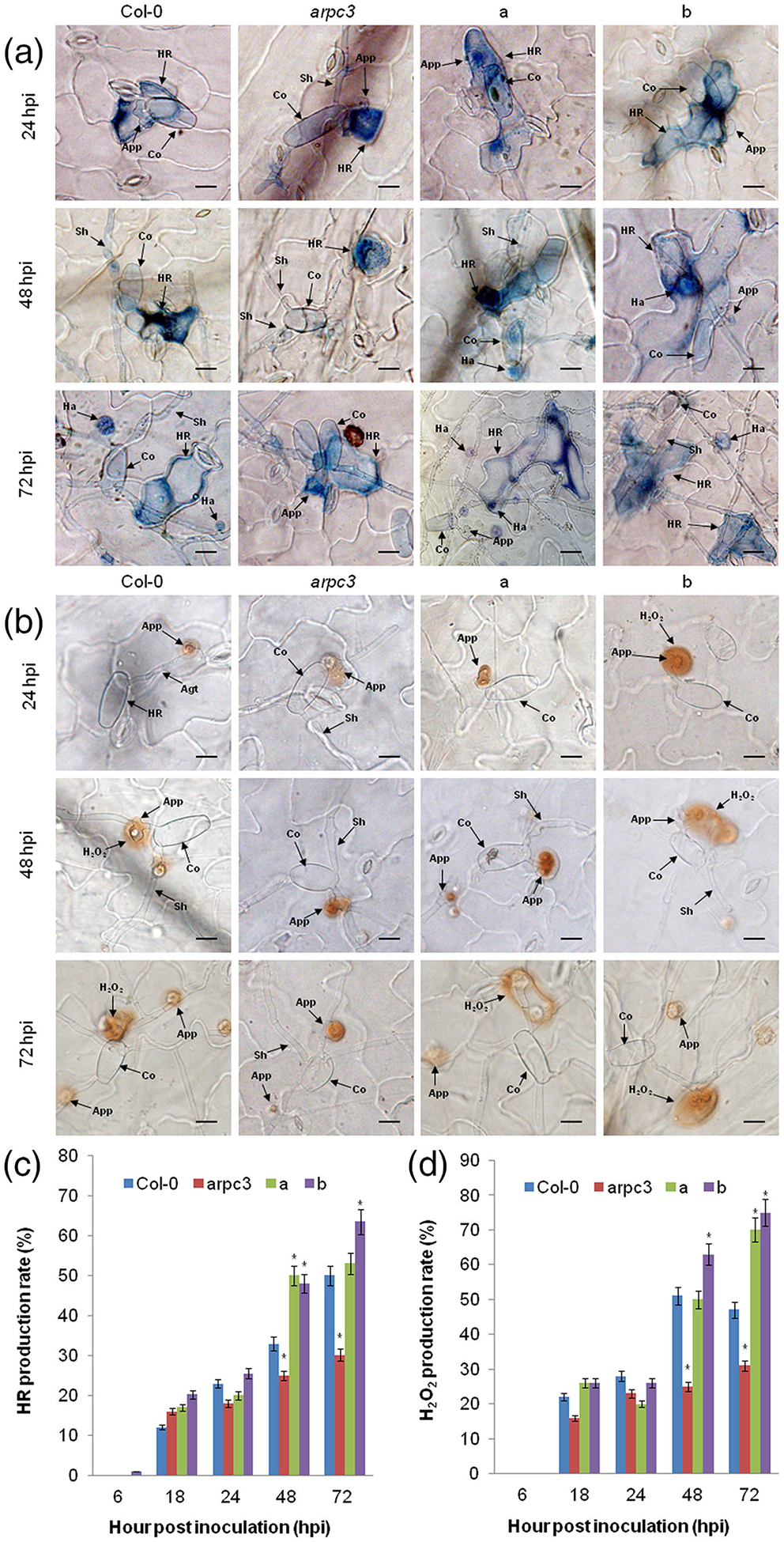
Overexpression of ShARPC3 in Arabidopsis thaliana leads to enhanced defence responses. (a) Histological observation of hypersensitive cell death in WT Col-0 plants, the arpc3 mutant, the arpc3 mutant expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 inoculated with On-Lz by microscopy. Blue (trypan) staining indicates hypersensitive cell death. (b) Histological observation of H2O2 accumulation in WT Col-0 plants, the arpc3 mutant, the arpc3 mutant expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 inoculated with On-Lz by microscopy. (c) HR production rate in WT Col-0 plants, arpc3 mutants, arpc3 transgenic plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 at 6, 18, 24, 48, and 72 hpi, respectively. (d) H2O2 production rate of Col-0 plants, arpc3 mutants, arpc3 plants transgenic for pCAMBIA3301-ShARPC3, and WT Col-0 plants transgenic for pCAMBIA3301-ShARPC3 at 6, 18, 24, 48, and 72 hpi, respectively. Agt, appressorium germ tube; App, appressorium; Co, conidium; Ha, haustorium; HR, hypersensitive response; Pa, papilla; Sa, secondary appressorium; Sh, secondary hyphae. Bar, 50 μm. a: arpc3 plants transgenic for pCAMBIA3301-ShARPC3; b: WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Error bars represent the absolute variations in conidia number among three independent biological replicates. Asterisks (*) indicate a significant difference between WT Col-0 plants and arpc3 mutants and arpc3 plants transgenic expressing pCAMBIA3301-ShARPC3, as well as WT Col-0 plants expressing pCAMBIA3301-ShARPC3 (P < .05)
2.10 |. Heterologous expression of ShARPC3 partially rescues the arc18 mutant in S. cerevisiae
S. cerevisiae growth is sensitive to high temperatures, with optimum growth at 30°C. To further explore the function of ShARPC3 in response to temperature stress, ShARPC3 was expressed in the S. cerevisiae arc18 mutant strain, and transformants were grown under different temperature conditions (e.g., 24°C, 30°C, 36°C, and 42°C). As shown in Figure S5, the growth S. cerevisiae strain Y06714 harbouring pDR195-ShARPC3 was more robust than that of the arpc3 S. cerevisiae mutant carrying the empty expression vector pDR195 but was weaker than that of the WT yeast strain, BY4741. Taken together, these data demonstrate that heterologous expression of ShARPC3 in yeast partially restores the growth at elevated temperatures, suggestive of a functional conservation between ShARPC3 and yeast ARC18 and a role in abiotic stress signalling and tolerance.
3 |. DISCUSSION
Previous work demonstrated that the Arp2/3 complex is required for actin polymerization and the formation of branched actin networks (Welch, Iwamatsu, & Mitchison, 1997). In the current study, and as a function of plant defence signalling, our data reveal a new role for ShARPC3 during tomato infection by the powdery mildew pathogen On-Lz. This is significant, as the current study provides insight into the network of processes that are activated upon pathogen perception and immune signalling. Indeed, we observed that accumulation of ShARPC3 was rapidly induced during an incompatible interaction, suggesting that ShARPC3 accumulation might be an early defence activation signal which associated with the stimulation of a large number of primary haustoria. Conversely, and in support of this hypothesis, in the compatible interaction, the expression of ShARPC3 was down-regulated at all time points—with the exception of 96 hpi—as compared with 0 hpi. In total, these data support the hypothesis that ShARPC3 is a key regulator of the disease resistance response in tomato against O. neolycopersici.
Using a VIGS-based approach, further support for this was revealed by silencing of ShARPC3 in LA1777, followed by infection with On-Lz, led to the development of susceptibility, indicating that ShARPC3 is a key component of the Arp2/3 complex-mediated disease resistance response in tomato against O. neolycopersici. Although it remains to be investigated, we suggest that this process is likely associated with actin polymerization catalysed by the Arp2/3 complex, and moreover, functions as a pivotal component in the plant defence signalling cascade (Tian et al., 2009; Welch & Mullins, 2002; Zhang et al., 2017).
In the current study, we provide evidence for a role for ShARPC3 as a resistance-associated gene activated in response to On-Lz infection. We observed that arpc3 mutant complementation with a 35S-expressing ShARPC3 construct restores On-Lz resistance; in WT Col-0 plants carrying pCAMBIA3301-ShARPC3, we observed enhanced resistance to On-Lz compared with arpc3 plants carrying pCAMBIA3301-ShARPC3 (Figure 7b). Based on this, it seems plausible that in the case of complementation of arpc3 by ShARPC3, there is also a quantitative effect due to differing levels of ShARPC3 expression in individual transformants, leading to the observation that ShARPC3 only partially restores resistance. In this study, compared with nonsilenced resistant LA1777 plants, which show a rapid induction of the HR and H2O2 accumulation, ShARPC3-silenced plants showed a markedly slower HR and H2O2 accumulation, responses that coincide with a loss of On-Lz resistance. These observations support our previous findings that transcriptomic differences between compatible and incompatible interactions of tomato and O. neolycopersici are mainly in timing (Li et al., 2006; Pei et al., 2011). Furthermore, it also indicates that the expression level of ShARPC3 correlates with the level of On-Lz induced HR and H2O2 accumulation in tomato, implying that the Arp2/3 complex plays a role in the tomato-O. neolycopersici interaction through adjusting the level, and amplitude, of the HR and H2O2 accumulation.
As a possible, yet undefined, mechanism underpinning this response, numerous reports have observed that changes in cytoskeletal organization lead to reactive oxygen bursts and subsequent cell death (Gourlay & Ayscough, 2005; Fu et al., 2014; Shimono, Lu, et al., 2016; Zhang et al., 2017). Indeed, rearrangement of the plant actin cytoskeleton has been correlated with the activation of defence signalling; most notably, the generation of ROS, changes in gene expression and formation of papillae at the site of penetration (Gourlay & Ayscough, 2005; Huot et al., 1998; Wang et al., 2009). Taken together with our observation of the impact of ShARPC3 overexpression in Arabidopsis, we posit a role for ShARPC3 as a positive regulator of immunity in response to On-Lz infection.
Our data describe a role for ShARPC3 associated with SA signalling (Lan et al., 2013; Li & Zou, 2017). Indeed, we observed that SA level was elevated in WT plants following inoculation and was reduced in ShARPC3-silenced plants. And tomato can receive the stimuli from On-Lz quickly and trigger SA-inducible PR proteins (PR1, PR2, and PR3) to protect themselves from being invaded, because the strength of PR gene expression has been linked to penetration resistance (Molitor et al., 2011; Peterhänsel, Freialdenhoven, Kurth, Kolsch, & Schulze-Lefert, 1997; Van Verk, Pappaioannou, Neeleman, Bol, & Linthorst, 2008). Based on these observations, it is reasonable to assume that SA is involved in the development of resistance to On-Lz in tomato, and that modulation of this defence hormone induces the expression of several defence-related genes (e.g., β−1,3-glucanase and chitinase) as observed in other host-pathogen systems (e.g., Mouekouba et al., 2014; Sun et al., 2017). In agreement with our findings, Sun et al. (2017) reported that Glucanase-A, Glucanase-B, Chitinase-3, Chitinase-9, and PR-1 transcripts significantly changed in the ε-poly-L-lysine-treated + Botrytis cinerea infected tomato plants. Interestingly, no significant fitness costs in terms of reduced growth were observed via ShARPC3 silencing, thus making this gene a promising target for mutagenesis to obtain suitable ShARPC3 alleles for disease resistance breeding in tomato. However, the growth ability of Arabidopsis arpc3 mutant was weaker than that in WT Col-0 plants. We speculate that the decrease of disease resistance in arpc3 mutant is due to blocked SA signalling pathways rather than changes in plant size. Rops, as upstream regulators of ARP2/3 complex, can affect the polymerization of microfilament skeleton (Jaffe & Hall, 2005). Poraty-Gavra et al. (2013) indicated that the morphological aberration of Arabidopsis rop6 mutant results from perturbations that are independent from the SA-associated response. These perturbations uncouple SA-dependent defence signalling from disease resistance execution. Based on the sum of the data presented herein, we hypothesize that ShARPC3 may function in a similar manner, and moreover, the current study points to a role for the Arp2/3 signalling platform in the control of multiple intercellular signalling processes that are required for immune activation and general stress response signalling.
4 |. MATERIALS AND METHODS
4.1 |. Strains and plant growth
O. neolycopersici Lanzhou strain (On-Lz) was isolated from tomato plants in Gansu Province, China, and was kindly provided by the College of Plant Protection, Northwest A&F University, Yangling, China. The isolate was preserved and propagated on tomato (cv. Money-maker [MM]) in an environmentally controlled growth chamber at 20 ± 3°C with 70 ± 5% humidity and a 16 hr photoperiod.
The S. cerevisiae diploid mutant strain Y06714 (MATa; ura310; leu210; his311; met1510; YLR370c::kanMX4) was grown on SC-U medium at 30°C, and the WT strain BY4741 (MATa; his311; leu210; met1510; ura310) was grown on yeast peptone dextrose (YPD) medium at 30°C. Yeast strains were obtained from the EUROSCARF collection.
Escherichia coli strain DH5α was grown on Luria-Bertani (LB) medium containing antibiotics, at 37°C. Agrobacterium tumefaciens strain GV3101 harbouring binary vector constructs was grown on antibiotic-containing LB media at 28°C.
Two tomato genotypes were used in this study: Solanum lycopersicum MM and Solanum habrochaites cv. LA1777. LA1777 and On-Lz constitute an incompatible interaction, whereas MM is highly susceptible to On-Lz. Tomato seeds were surface sterilized with 3% sodium hypochlorite for 3 min and immediately rinsed with sterile distilled water three times, as previously described (Sun et al., 2018). Seeds were germinated in vermiculite and grown in a growth chamber for 7 days under the following environmental conditions: 25 ± 0.5°C, 95–100% relative humidity (RH), and continuous illumination of 3,500 lux provided by fluorescent lamps. Once the cotyledon leaves had unfolded, the plants were transplanted to soil in 15 cm pots and grown in a temperature-controlled glasshouse (25 ± 3°C) for 1 month before they were used for pathogen inoculation experiments.
N. benthamiana plants were grown in a growth chamber at 20°C under a 16-hr light/8-hr dark cycle with 60% relative humidity and a light intensity of 120-mmol photons m−2 s−1.
Arabidopsis thaliana seeds included ecotype Columbia-0 (WT Col-0) and the T-DNA insertion mutant arpc3 (SALK_099449) were obtained from the Arabidopsis Biological Resource Center (ABRC; The Ohio State University). Seeds were sterilized in 50% bleach (v/v) containing 0.05% Triton X-100 (v/v) for 10 min, rinsed five times with sterile water, and incubated at 4°C for 3 days. For germination, 10 seeds were transferred to 1 ml ½-strength Murashige–Skoog (MS) liquid medium (MS salt supplemented with 0.5% sucrose [w/v], pH 5.5 [pH adjusted to 5.7 by KOH, pH 5.5 after autoclaving], in each well of a six-well plate; Wu et al., 2014). Germination and growth took place in a growth chamber at 21°C and 19°C during the 8 hr day and 16 hr night periods, respectively. Relative humidity was 70% and light intensity was 100 W m−2.
4.2 |. Sequencing and phylogenetic analysis of ShARPC3
Tomato ARPC3 (Accession number Solyc07g007630) amino acid sequence was used as a query in a TBLASTN program against the SGN Tomato Combined database (http://solgenomics.net/tools/blast/), in a BLASTP program against an Arabidopsis protein database (http://www.arabidopsis.org/Blast/index.jsp), or GenBank using BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to identify homologous sequences. Conserved and homologous tomato expressed sequence tag (EST) sequences were extracted and assembled. The cDNA sequence of ShARPC3 was analysed in silico using BLAST and the Open Reading Frame (ORF) Finder software in NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Multiple sequence alignments were performed using CLUSTALX2.0 (DNASTAR; http://www.dnastar.com/) and DNAMAN6.0 (Lynnon BioSoft). Phylogenetic analysis was performed using the MEGA software package (v.6.0; http://www.megasoftware.net/) using the neighbour-joining method. The protein domain predictions were assigned on the basis of analysis using the SMART database (http://smart.embl-heidelberg.de) and RCSB PDB protein databank (https://www.rcsb.org/).
Predicted protein-protein interactions between tomato ARPC3 and other tomato proteins were determined using STRING (http://string-db.org/; Franceschini et al., 2013). To reduce the number of hits (i.e., potential false positives), the minimum required interaction score was set to the highest confidence interval (0.900), and the maximum number of interactors was set to not more than 10 interactors. The subcellular localization of ShARPC3 protein was predicted from the amino acid sequence using Procomp (v.9.0; http://linux1.softberry.com/berry.phtml).
4.3 |. Plasmid construction
The coding sequence (cDNAs) of ShARPC3 was amplified from reverse-transcribed RNA and cloned using gene-specific DNA primers (Table S1). For subcellular localization analysis of ShARPC3, the full-length cDNA of ShARPC3 was cloned into the pCaMV35S::GFP vector via BamHI digestion and ligation. For transient expression assays in tobacco, the ShARPC3 ORF was amplified with DNA primers (Table S1) designed to introduce a SalI restriction enzyme site. The amplified product was digested and ligated into the SalI site of the binary vector pGR106 using SmartSeamless Cloning kit. The VIGS vectors were constructed using tobacco rattle virus (TRV1 and TRV2) according to the methods of Dong, Burch-Smith, Liu, Mamillapalli, and Dinesh-Kumar (2007). The target sequence for ShARPC3 VIGS started from the non-coding 5′ region and included 427 nucleotides of the ShARPC3 ORF. Selected gene fragments of ShARPC3 were amplified by PCR from tomato cDNA using the primers BamHI with restriction enzyme site (Table S1). The resultant PCR products were ligated into the TRV2 vector using the SmartSeamless Cloning Kit, yielding TRV2:ShARPC3. DNA constructs were extracted using the plasmid extracting kit from Qiagen (Shanghai, China) and sequenced to confirm the presence of the intended inserts. For the Arabidopsis arpc3 mutant complementation assays, PCR-amplified cDNAs of ShARPC3 (Table S1) was subcloned with BamHI digestion site and inserted between 35S promoter and NOS terminator sequences of the binary vector pCAMBIA3301-ShARPC3 using the SmartSeamless Cloning Kit. For yeast arc18 mutant complementation assays, PDR195-ShARPC3 expression constructs were created by cloning PCR-amplified ShARPC3 cDNA into the NotI restriction enzyme (Promega, Madison, WI, USA) site of the binary vector PDR195 using the SmartSeamless Cloning Kit (Jieyi Biotech, Shanghai, China).
4.4 |. Subcellular localization analysis
Protoplast isolation from tomato leaves was performed as previously described (Yu et al., 2015), with minor modifications. In brief, leaves of ~8-week-old tomato plants were used for subcellular localization analyses. Leaves were dissected into 0.5–1-mm strips using a razor blade and incubated at room temperature (~22°C) in 1% cellulase R10 (Yakult Pharmaceutical Ind. Co., Ltd., Tokyo, Japan), 0.25% macerozyme R10 (Yakult Pharmaceutical Ind. Co., Ltd.), 0.4 M mannitol, 20 mM KCl, 10 mM CaCl2, and 20 mM 2-(N-morpholino) ethanesulfonic acid (MES) for 3–4 hr, with shaking at 40–50 rpm. Following incubation at room temperature, the protoplast preparation was filtered through a metal sieve, and the eluate was centrifuged at 100 g for 5 min. Pelleted protoplasts were suspended in 5 ml of W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES/KOH; pH 5.7) and centrifuged for 5 min at 100 g. The protoplasts were transferred to a tube containing 5 ml of the W5 solution. The protoplasts were pelleted again by centrifugation at 100 g for 5 min and resuspended in 5 ml of the W5 solution. Protoplasts were incubated on ice for 30 min and then resuspended in 5 ml of MMg buffer (400 mM mannitol, 15 mM MgCl2, and 4 mM MES/KOH; pH 5.7). For cotransformation, 10 μg of recombinant plasmid pCaMV35S:ShARPC3-GFP, or empty vector pCaMV35S:GFP was added to 100 μl of the protoplast suspension. An equal volume of 40% (w/v) PEG3350, freshly prepared with 0.1 M CaCl2 and 0.8 M mannitol solution, was added. Then, the mixture was incubated at room temperature for 30 min. After incubation, the mixture was diluted with 500 μl of the W5 solution. The solution containing the protoplasts was gently mixed and the protoplasts were pelleted by centrifugation at 100 g for 5 min. Protoplasts were washed twice using the W5 solution, gently resuspended in 1 ml of W5 solution, and incubated in 12-well plates at room temperature for 18–36 hr in the dark. GFP fluorescence was detected using an Olympus FV1000 laser confocal microscope equipped with a 488 nm filter. The experiment was repeated three times.
4.5 |. Agrobacterium-mediated transient expression in N. benthamiana
The ShARPC3 gene was amplified using combinations of primers for the PVX (pGR106) assay (Lu et al., 2003). Recombinant plasmid pGR106-ShARPC3 or pGR106-GFP prepared from E. coli DH5α was transformed into A. tumefaciens strain GV3101 via electroporation. Transformants were grown at 28°C with 50 μg ml−1 of each of rifampicin and kanamycin until cultures reached stationary growth phase. Cultures of Agrobacterium transformed with pGR106-GFP or pGR106-ShARPC3 were centrifuged and the resultant bacterial pellets resuspended in infiltration buffer (10 mM MgCl2, 150 μM acetosyringone, 10 mM MES pH 5.6) to a final OD 600 nm of 0.1. After incubation for 2–3 hr in the dark, A. tumefaciens cells carrying ShARPC3, GFP, or buffer were infiltrated. After 24 hr, the same infiltration site was challenged with A. tumefaciens cells carrying the BAX gene. BAX, a death-promoting member of the Bcl-2 family of proteins, triggered cell death when expressed in plants (Lacomme & Santa, 1999). A. tumefaciens strains carrying GFP gene alone were infiltrated in parallel as negative control. Symptom development was monitored visually 5 to 7 days after infiltration. This experiment was repeated three times with each assay consisting of three plants each, with three leaves inoculated.
4.6 |. TRV2-mediated silencing of ShARPC3 in tomato
VIGS TRV1 and TRV2-ShARPC3 were introduced into A. tumefaciens strain GV3101 by heat shock. A total of 5 ml of an overnight culture was grown at 28°C in the appropriate antibiotic selection medium in a 15-ml glass tube for 24 hr, after which time cultures were spun down and cells were resuspended in infiltration medium (10 mM MES, 10 mM MgCl2, 200 μM acetosyringone), adjusted to an OD 600 nm of 1.0, and incubated at room temperature for 3 hr. Agroinfiltration was performed as previously described (Liu, Schiff, Marathe, & Dinesh-Kumar, 2002). The first and second leaves of four-leaf stage LA1777 plants were infiltrated with a 1:1 mixture of TRV1 and TRV2-ShARPC3 fragment for the clone. The empty vector TRV2 was used as negative control. To monitor silencing efficiency, plants were inoculated in parallel with TRV2 expressing phytoene desaturase (i.e., TRV2:ShPDS; accession number NM_001247166). Following virus inoculation, seedlings were transferred to an environmentally controlled growth chamber (25°C, 16-hr light/8-hr dark photoperiod). Photobleaching symptoms in the PDS control plants were observed ~30 days after virus inoculation. The fourth leaves of plants were inoculated with On-Lz; sampled at 6, 18, 24, 48, and 72 hpi; and processed for histological observation. Five plants were inoculated per trial, and three biological replicates were performed.
4.7 |. A. thaliana transgenic complementation analysis
Arabidopsis genomic DNA preparation was performed as previously described (Qian et al., 2001). SALK_099449 T-DNA insertions in the AtARPC3 gene were confirmed by PCR and sequencing using the following DNA primers: SALK_099449LP (5′-AAGGTAGAGGCTCAAACGCTC-3′), SALK_099449RP (5′-AGAATCGCCACCTTTAGCTTC-3′), and the T-DNA-specific primer LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′). The complete ORF of ShARPC3 was cloned into the binary expression vector pCAMBIA3301 carrying CaMV35S promoter. pCAMBIA3301-ShARPC3 was transformed into A. tumefaciens GV3101 and was introduced into WT Col-0 and arpc3 by the floral dip method (Clough & Bent, 1998). Transgenic T1 plants were selected with kanamycin (25 μg ml−1) on ½-strength MS medium containing 1% Phytoagar. After 2 weeks, kanamycin-resistant seedlings were transplanted to soil. Three- month-old kanamycin-resistant T2 plants from individual T1 lines were pooled (three or four individuals per sample) and assayed for resistance to On-Lz as described below. Kanamycin-resistant siblings of these T2 individuals were confirmed as transgenic by PCR using the following DNA primers: (LP-5′-CGGTGTTCTCTCCAAATGAAATGAACTT-3′ and RP-5′-AATTGAGACTTTTCAACAAAGGGTAATA-3′) specific to the 35S promoter of the transgene expression construct. The leaves of plants were inoculated with On-Lz; sampled at 6, 18, 24, 48, and 72 hpi; and processed for histological observation. Five plants were inoculated per trial and three biological replicates were performed.
4.8 |. Quantitative real-time PCR analysis (qRT-PCR)
To evaluate the expression levels of ShARPC3 in response to On-Lz infection, MM, and LA1777 plant leaves were harvested at 0, 12, 18, 24, 36, 48, 72, and 96 hpi, respectively. For TRV2-mediated silencing of ShARPC3 assay efficiency the relative expression of PR1B1 (PR1), Glucanase A (PR2), Glucanase B (PR2-like), Chitinase 3 (PR3), and Chitinase 9 (PR3-like; SA pathway-specific gene expression markers; Schenk et al., 2000) were assessed by sampling the fourth leaves of SlARPC3-silenced tomato plants at 0, 24, 48, and 72 hpi following infection with On-Lz. The GAPDH gene was used as the internal control for qRT-PCR analysis (Chalupowicz et al., 2010). DNA primers for qRT-PCR are listed in Table S2. For Arabidopsis transgenic complementation assays, leaves were inoculated with On-Lz; sampled at 0, 24, 48, and 72 hpi; and processed for RNA isolation (below). The UBQ10 gene was used as the control for qRT-PCR reactions. DNA primers used for qRT-PCR analysis are listed in Table S3. Total RNA was prepared using the BIOZOL total RNA extraction kit (Gentaur, Belgium) following the manufacturer’s instructions. RNA integrity was evaluated by gel electrophoresis, and the RNA concentration was determined using a Qubit 2.0 Fluorometer (Life Technologies, Thermo Fisher, Waltham, MA, USA). Total RNA (1 μg) was used for first strand cDNA synthesis using the single strand cDNA synthesis kit (GeneCopoeia, Rockville, MD, USA) with oligo (dT)18 primer (MBI Fermentas/Thermo Fisher, Waltham, MA, USA).
The IQ™5 Real-Time PCR System (BioRad, Hercules, CA, USA) was employed for quantitative PCR amplification. For real-time PCR reactions, 1 μl total cDNA was added to a 20 μl PCR reaction mixture containing 10 μl of 2X ultra SYBR Mixture (CWBIO, Beijing, China), 0.4 μl of each primer, 2 μl template, and 7.2 μl of water. qRT-PCR conditions were as follows: denaturing at 95°C for 10 min, followed by 40 cycles of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Each reaction included a nontemplate control. All analyses were performed in biological triplicate. Relative transcript quantification was calculated by the comparative 2−ΔΔCT method (Livak & Schmittgen, 2001). Statistical analysis was performed using two-tailed analysis of variance (ANOVA) with the SPSS 17.0 program (IBM SPSS Statistics; https://www.ibm.com/products/spss-statistics). A value of P < .05 indicates a significant difference between groups.
4.9 |. Pathogen inoculation, phenotypic, and histological observations
The fresh On-Lz conidia were collected from infected plant leaves with sterile water. Four-week plants were used for inoculation by spraying a spore suspension of 5 × 104 conidia/ml on the whole plants for the histological study. The infection phenotypes of tomato and A. thaliana leaves were observed at 7 dpi. For VIGS assay, disease index (DI) was evaluated at 7–14 dpi (Sun et al., 2017). Disease severity was scored according to the following: 0 = no diseased leaves; 1 = 0–5% of leaves having lesions; 3 = leaves with infection lesions up to 6–10%; 5 = leaves with infection lesions up to 11–20%; 7 = leaves with infection lesions up to 21–40%; and 9 = leaves with infection lesions up to 41–100%.
Disease index was calculated using the following equation:
Disease index (DI) = [Σ (number of diseased plant leaves at a given disease severity × the disease severity)/(total plant leaves analysed × 9)] × 100%. An average DI was calculated at three independent times for each infected plant.
To quantify fungal growth, the number of conidiophores per colony was counted at 7 dpi as previously described (Consonni et al., 2006). At least 25 colonies were counted for each genotype in each experiment. Infection assays and disease severity evaluations were repeated three times. To quantify the accumulation of H2O2 (H2O2 production rate = H2O2 numbers per 100 penetration sites) and the induction of HR cell death (HR production rate = HR numbers per 100 penetration sites) during On-Lz infection, 3,3-diaminobenzidine (DAB) and trypan blue staining were performed following the method of Thordal-Christensen, Zhang, Wei, and Collinge (1997). All microscopic examinations were performed using a Nikon Eclipse 80i microscope equipped with DIC (Nikon Corporation, Tokyo, Japan). At least 50 penetration sites on each of four-leaf samples were observed at each time point. Standard errors of deviation were calculated using Microsoft Excel. Statistical significance was assessed by a Student’s t test (P < .05) using SPSS software.
4.10 |. Quantification of phytohormone content
Tomato leaves from VIGS experiments were used for phytohormone analysis. Leaves were separately collected at 0, 1, 3, 6, 12, and 24 hr after inoculation with On-Lz according to Gong et al. (2017). Three biological replicates were performed. Standard errors of deviation were calculated using Microsoft Excel. Statistical significance was assessed by a Student’s t test (P < .05) using SPSS software.
4.11 |. Yeast mutant complementation assay
The complete ORF of ShARPC3 was cloned into pDR195 (Addgene, plasmid #36028). The transformed cell (Δarc18 + ShARPC3) was obtained by transforming the reconstructed vector (pDR195- ShARPC3) into the mutant strain Y06714. The transformed cell containing the empty vector pDR195 (Δarc18 + empty) was used as a control. To investigate yeast cell survival under stress, transformed cells (initial optical density [OD] of 0.6–1.0 at 600 nm) were cultured in yeast medium. The concentration of pDR195 (Δarc18 + empty) and pDR195-ShARPC3 (Δarc18 + ShARPC3) transformed cells, as well as WT yeast cells (BY4741) were serially diluted to 107, 106, 105, 104, and 103 (cells ml−1) using an Olympus BX-51 microscope (Olympus) coupled with a blood counting chamber. This experiment was repeated three times.
Supplementary Material
Figure S1. Agrobacterium-mediated transient expression in Nicotiana benthamiana. (A) Leaf phenotypes 7 days post-inoculation (dpi) with Agrobacterium tumefaciens-expressing constructs. (B) Leaf inoculation phenotypes after clearing in ethanol/acetic acid (1:1 v/v) for 2 days. a: pGR106-GFP (CK); b: pGR106-ShARPC3; c: Buffer; d: pGR106-GFP-BAX; e: pGR106-ShARPC3-BAX; f: Buffer-BAX. Agroinfiltration sites in N. benthamiana leaves expressing ShARPC3 were challenged with A. tumefaciens expressing the BAX elicitin. BAX-induced cell death was scored at 3- and 4-days post-inoculation. BAX, a death-promoting member of the Bcl-2 family of proteins, triggered cell death when expressed in plants. A. tumefaciens strain carrying pGR106-GFP was used as a negative control.
Figure S2. Infection level of On-Lz in MM and LA1777 tomato plants.
(A) Infection phenotypes of MM and LA1777 tomato leaves at 7 dpi. (B) Disease indexes of MM and LA1777 plants at 7 and 14 dpi, respectively. Error bars represent the variations among three independent biological replicates. The asterisk (*) indicates statistically significant differences in disease index between MM and LA1777 plants (P < 0.05).
Figure S3. Relative expression levels of ShARPC3 mRNA in TRV2: ShARPC3 plants challenged with On-Lz.
Figure S4. Relative expression levels of AtARPC3 in WT Col-0, the arpc3 and arpc3 plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 challenged with On-Lz. The relative transcript level of AtARPC3 was calculated by the comparative threshold method (2−ΔΔCT). (A) arpc3 plants expressing pCAMBIA3301-ShARPC3. (B) WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Error bars represent the standard deviation among three independent replicates. Asterisks (*) indicate a significant difference from WT Col-0 plants, arpc3 mutants, arpc3 plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 (P < 0.05).
Figure S5. Functional characterization of ShARPC3 in the Saccharomyces cerevisiae arc18 mutant. Survival of the mutant yeast strain Δarc18 expressing pDR195 (Δarc18 + vec) or pDR195-ShARPC3 (Δarc18 + ShARPC3) were spotted on SC-URA medium at 24°C, 30°C, 36°C, and 42°C. The final densities were diluted to 107, 106, 105, 104, and 103 (cell ml−1) with sterile water using a blood counting chamber.
Table S1. DNA primers used to amplify ShARPC3 gene and recombination. The primer sequences are composed of two parts. The lowercase letters represent the sequences of corresponding vector homologous arm and restriction enzyme site, and the capital letters indicate the gene-specific nucleotide sequences.
Table S2. qRT-PCR genes and DNA primers used in VIGS experiment.
Table S3. qRT-PCR genes and primers used in Arabidopsis thaliana transgenic complementation assay.
Table S4. The predicted subcellular localizations of ShARPC3 using ProtComp 9.0. The predicted localization of tomato ARPC3 protein supported by ProtComp 9.0 (http://linux1.softberry.com/berry.phtml). Neural Nets, Pentamers, and Integral predictions show ShARPC3 protein localizes predominantly in the extracellular space.
ACKNOWLEDGMENTS
The research presented herein was supported by the National Natural Science Foundation of China (Grant 31571960), Key Industrial Chain Projects of Shaanxi (2019ZDLNY03-07), Northwest A&F University extension Project (2018-10), and the 111 Project from the Ministry of Education of China (Grant B07049). Research in the laboratory of B.D. is supported by the U.S. National Science Foundation (IOS-1557437) and the National Institutes of General Medical Sciences (1R01GM125743).
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
REFERENCES
- Bai Y, Huang CC, Van Der Hulst R, Meijer-Dekens F, & Bonnema G (2003). Lindhout P QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Molecular Plant-Microbe Interactions, 16, 169–176. 10.1094/MPMI.2003.16.2.169 [DOI] [PubMed] [Google Scholar]
- Bai Y, Van Der Hulst R, Bonnema G, Marcel TC, Meijer-Dekens F, Niks R, & Lindhout P (2005). Tomato defense to Oidium neolycopersici: Dominant Ol genes confer isolate-dependent resistance via a different mechanism than recessive ol-2. Molecular Plant-Microbe Interactions, 18, 354–362. 10.1094/MPMI-18-0354 [DOI] [PubMed] [Google Scholar]
- Campellone KG, & Welch MD (2010). A nucleator arms race: Cellular control of actin assembly. Nature Reviews Molecular Cell Biology, 11, 237–251. 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalupowicz L, Cohen-Kandli M, Dror O, Eichenlaub R, Gartemann KH, Sessa G, … Manulis-Sasson S (2010). Sequential expression of bacterial virulence and plant defense genes during infection of tomato with Clavibacter michiganensis subsp. michiganensis. Phytopathology, 100, 252–261. 10.1094/PHYTO-100-3-0252 [DOI] [PubMed] [Google Scholar]
- Chesarone MA, & Goode BL (2009). Actin nucleation and elongation factors: Mechanisms and interplay. Current Opinion in Cell Biology, 21, 28–37. 10.1016/j.ceb.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S, Coaker G, Day B, & Staskawicz BJ (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell, 124, 1–12. [DOI] [PubMed] [Google Scholar]
- Ciccarese F, Amenduni M, Ambrico A, & Cirulli M (2000). The resistance to Oidium lycoeprsici conferred by ol-2 gene in tomato. Acta Physiologiae Plantarum, 22, 266. [Google Scholar]
- Clough SJ, & Bent AF (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal, 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, … Panstruga R (2006). Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nature Genetics, 38, 716–720. 10.1038/ng1806 [DOI] [PubMed] [Google Scholar]
- Day B, Henty J, Porter K, & Staiger C (2011). The pathogen-actin connection: A platform for defense signaling in plants. Annual Review of Phytopathology, 49, 483–506. 10.1146/annurev-phyto-072910-095426 [DOI] [PubMed] [Google Scholar]
- Dong YY, Burch-Smith TM, Liu YL, Mamillapalli P, & Dinesh-Kumar SP (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NBMADS4–1 and −2 in floral development. Plant Physiology, 145, 1161–1170. 10.1104/pp.107.107391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Simonovic M, Roth A, Von Mering C, Szklarczyk D, Pletscher-Frankild S, … Bork P (2013). STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research, 41, D808–D815. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Duan X, Tang C, Li X, Voegele RT, Wang X, … Kang Z (2014). TaADF7, an actin-depolymerizing factor, contributes to wheat resistance against Puccinia striiformis f. sp. tritici. The Plant Journal, 78, 16–30. 10.1111/tpj.12457 [DOI] [PubMed] [Google Scholar]
- Gong C, Liu Y, Liu SY, Cheng MZ, Zhang Y, Wang RH, … Wang AX (2017). Analysis of Clonostachys rosea-induced resistance to grey mould disease and identification of the key proteins induced in tomato fruit. Postharvest Biology and Technology, 123, 83–93. 10.1016/j.postharvbio.2016.08.004 [DOI] [Google Scholar]
- Gourlay CW, & Ayscough KR (2005). The actin cytoskeleton: A key regulator of apoptosis and ageing. Nature Reviews Molecular Cell Biology, 6, 583–589. 10.1038/nrm1682 [DOI] [PubMed] [Google Scholar]
- Henty-Ridilla JL, Shimono M, Li J, Chang J, Day B, & Staiger CJ (2013). The plant actin cytoskeleton responds to signals from microbe-associated molecular patterns. PLoS Pathogens, 9, e1003290 10.1371/journal.ppat.1003290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot J, Houle F, Rousseau S, Deschesnes RG, Shah GM, & Landry J (1998). SAPK2/p38-dependent F-actin reorganization regulates early membrane blebbing during stress-induced apoptosis. Journal of Cell Biology, 143, 1361–1373. 10.1083/jcb.143.5.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, & Hall A (2005). Rho GTPases: Biochemistry and biology. Annual Review of Cell and Developmental Biology, 21, 247–269. 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jones H, Whipps JM, & Guu SJ (2001). The tomato powdery mildew fungus Oidium neolycopersici. Molecular Plant Pathology, 2, 303–309. 10.1046/j.1464-6722.2001.00084.x [DOI] [PubMed] [Google Scholar]
- Jones JD, & Dangl JL (2006). The plant immune system. Nature, 444, 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, & Hwang I (2005). Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell, 17, 888–902. 10.1105/tpc.104.028829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I, Kobayashi Y, & Hardham AR (1994). Dynamic reorganization of microtubules and microfilaments in flax cells during the resistance response to flax rust infection. Planta, 195, 237–247. [Google Scholar]
- Kotchoni SO, Zakharova T, Mallery EL, Le J, El-Assal SE, & Szymanski DB (2009). The association of the Arabidopsis actin-related protein2/3 complex with cell membranes is linked to its assembly status but not its activation. Plant Physiology, 151, 2095–2109. 10.1104/pp.109.143859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme C, & Santa CS (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proceedings of the National Academy of Sciences, 96, 7956–7961. 10.1073/pnas.96.14.7956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan A, Huang J, Zhao W, Peng Y, Chen Z, & Kang D (2013). A salicylic acid-induced rice (Oryza sativa L.) transcription factor OsWRKY77 is involved in disease resistance of Arabidopsis thaliana. Plant Biology, 15, 452–461. 10.1111/j.1438-8677.2012.00664.x [DOI] [PubMed] [Google Scholar]
- Lebeda A, Mieslerová B, Petřivalský M, Luhová L, Špundová M, Sedlářová M, … Pink DAC (2014). Resistance mechanisms of wild tomato germplasm to infection of Oidium neolycopersici. European Journal of Plant Pathology, 138(SI), 569–596. 10.1007/s10658-013-0307-3 [DOI] [Google Scholar]
- Li CW, Bai YL, Jacobsen E, Visser R, Lindhout P, & Bonnema G (2006). Tomato defense to the powdery mildew fungus: Differences in expression of genes in susceptible, monogenic- and polygenic resistance responses are mainly in timing. Plant Molecular Biology, 62, 127–140. 10.1007/s11103-006-9008-z [DOI] [PubMed] [Google Scholar]
- Li L, & Zou Y (2017). Induction of disease resistance by salicylic acid and calcium ion against Botrytis cinerea in tomato (Lycopersicon esculentum). Emirates Journal of Food Agriculture, 29, 78–82. 10.9755/ejfa.2016-10-1515 [DOI] [Google Scholar]
- Li P, & Day B (2019). Battlefield cytoskeleton: Cytoskeletal regulation and pathogen targeting of plant immunity. Mol Plant-Microbe Interact., 32, 25–34. 10.1094/MPMI-07-18-0195-FI [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindhout P, Pet G, & van der Beek H (1994). Screening wild Lycopersicon species for resistance to powdery mildew (Oidium lycopersicum). Euphytica, 72, 43–49. [DOI] [PubMed] [Google Scholar]
- Lindhout P, van der Beek H, & Pet G (1994). Wild Lycopersicon species as sources for resistance to powdery mildew (Oidium lycopersicum): Mapping of resistance gene Ol-1 on chromosome 6 of Lycopersicon hirsutum. Acta Horticulturae, (376), 387–394. [DOI] [PubMed] [Google Scholar]
- Liu YL, Schiff M, Marathe R, & Dinesh-Kumar SP (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. The Plant Journal, 30, 415–429. 10.1046/j.1365-313X.2002.01297.x [DOI] [PubMed] [Google Scholar]
- Livak KJ, & Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, … Baulcombe DC (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO Journal, 22, 5690–5699. 10.1093/emboj/cdg546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, & Pollard TD (1994). Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. Journal of Cell Biology, 127, 107–116. 10.1083/jcb.127.1.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, … Pollard TD (1999). Scar, a WASP-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proceedings of the National Academy of Science of the United States of America, 96, 3739–3744. 10.1073/pnas.96.7.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch J, Fiserova J, Fischer L, & Nick P (2009). Tobacco Arp3 is localized to actin-nucleating sites in vivo. Journal of Experimental Botany, 60, 603–614. 10.1093/jxb/ern307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel TC, Gorguet B, Ta MT, Kohutova Z, Vels A, & Niks RE (2008). Isolate specificity of quantitative trait loci for partial resistance of barley to Puccinia hordei confirmed in mapping populations and near-isogenic lines. New Phytologist, 177, 743–755. 10.1111/j.1469-8137.2007.02298.x [DOI] [PubMed] [Google Scholar]
- Mathur J, & Hülskamp M (2002). Microtubules and microfilaments in cell morphogenesis in higher plants. Current Biology, 12, R669–R676. 10.1016/S0960-9822(02)01164-8 [DOI] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kernebeck B, & Hülskamp M (2003). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. The Plant Cell, 15, 1632–1645. 10.1105/tpc.011676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J, Mathur N, Kirik V, Kernebeck B, Srinivas BP, & Hulskamp M (2003). Arabidopsis, CROOKED encodes for the smallest subunit of the ARP2/3 complex and controls cell shape by region specific fine F-actin formation. Development, 130, 3137–3146. 10.1242/dev.00549 [DOI] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, Pons KHJ, Samans B, Kogel KH, & Waller F (2011). Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Molecular Plant-Microbe Interactions, 24, 1427–1439. 10.1094/MPMI-06-11-0177 [DOI] [PubMed] [Google Scholar]
- Mouekouba LD, Zhang L, Guan X, Chen X, Chen H, Zhang J, … Wang A (2014). Analysis of Clonostachys rosea-induced resistance to tomato gray mold disease in tomato leaves. PLoS One, 9, e102690 10.1371/journal.pone.0102690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M, & Akutsu K (2014). Virulence factors of Botrytis cinerea. Journal of General Plant Pathology, 80, 15–23. 10.1007/s10327-013-0492-0 [DOI] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, & Hückelhoven R (2005). The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. The Plant Journal, 41, 291–303. 10.1111/j.1365-313X.2004.02292.x [DOI] [PubMed] [Google Scholar]
- Pantaloni D, le Clainche C, & Carlier MF (2001). Mechanism of actin-based motility. Science, 292, 1502–1506. 10.1126/science.1059975 [DOI] [PubMed] [Google Scholar]
- Pei DL, Ma HZ, Zhang Y, Ma YS, Wang WJ, Geng HX, … Li CW (2011). Silencing a putative cytosolic NADP-malic enzyme gene compromised tomato resistance to Oidium neolycopersici. Life Science Journal, 8, 652–657. [Google Scholar]
- Peterhänsel C, Freialdenhoven A, Kurth J, Kolsch R, & Schulze-Lefert P (1997). Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. The Plant Cell, 9, 1397–1409. 10.1105/tpc.9.8.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, & Mullins RD (2000). Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annual Review of Biophysics and Biomolecular Structure, 29, 545–576. [DOI] [PubMed] [Google Scholar]
- Pollard TD, & Borisy GG (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell, 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Poraty-Gavra L, Zimmermann P, Haigis S, Bednarek P, Hazak O, Stelmakh OR, … Yalovsky S (2013). The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiology, 161, 1172–1188. 10.1104/pp.112.213165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter K, & Day B (2015). From filaments to function: The role of the plant actin cytoskeleton in pathogen perception, signaling, and immunity. Journal of Integrative Plant Biology, 58, 299–311. [DOI] [PubMed] [Google Scholar]
- Qi T, Wang J, Sun Q, Day B, Guo J, & Ma Q (2017). TaARPC3, contributes to wheat resistance against the stripe rust fungus. Frontiers in Plant Science, 8 10.3389/fpls.2017.01245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Li YH, Zeng D, Teng S, Wang Z, Li X, … Li J (2001). Isolation and genetic characterization of a fragile plant mutant rice. (Oryza sativa L.). Chinese Science Bulletin, 46, 2082–2085. 10.1007/BF02901137 [DOI] [Google Scholar]
- Roberts P, Momol T, & Pernezny K (2002). Powdery mildew on tomato, document PP-191 of Plant Pathology Department, Florida Cooperative Extension Service, Institute of Food and Agricultural Sciences, University of Florida. [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, & Manners JM (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proceedings of the National Academy of Sciences, 97, 11655–11660. 10.1073/pnas.97.21.11655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Higaki T, Kaku H, Shibuya N, Hasezawa S, & Day B (2016). Quantitative evaluation of stomatal cytoskeletal patterns during the activation of immune signaling in Arabidopsis thaliana. PLoS One, 11, e0159291 10.1371/journal.pone.0159291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono M, Lu Y, Porter K, Kvitko B, Creason A, Henty-Ridilla J, … Day B (2016). The Pseudomonas syringae type III effector HopG1 induces actin filament remodeling in Arabidopsis in association with disease symptom development. Plant Physiology, 171, 2239–2255. 10.1104/pp.16.01593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, & Griffing L (2009). Movement and remodeling of the endoplasmic reticulum in non-dividing cells of tobacco leaves. The Plant Cell, 21, 3937–3949. 10.1105/tpc.109.072249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun GZ, Wang H, Shi BB, Shangguan NN, Wang Y, & Ma Q (2017). Control efficiency and expressions of resistance genes in tomato plants treated with ε-poly-l-lysine against Botrytis cinerea. Pesticide Biochemistry and Physiology, 143, 191–198. 10.1016/j.pestbp.2017.07.007 [DOI] [PubMed] [Google Scholar]
- Sun GZ, Yang QC, Zhang AC, Guo J, Liu XJ, Wang Y, & Ma Q (2018). Synergistic effect of the combined bio-fungicides ε-poly-L-lysine and chitooligosaccharide in controlling grey mould (Botrytis cinerea) in tomatoes. International Journal of Food Microbiology, 276, 46–53. 10.1016/j.ijfoodmicro.2018.04.006 [DOI] [PubMed] [Google Scholar]
- Takenawa T, & Miki H (2001). WASP and WAVE family proteins: Key molecules for rapid rearrangement of cortical actin filaments and cell movement. Journal of Cell Science, 114, 1801–1809. [DOI] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, & Tameling WI (2006). Resistance proteins: Molecular switches of plant defence. Current Opinion in Plant Biology, 9, 383–390. 10.1016/j.pbi.2006.05.009 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang ZG, Wei YD, & Collinge DB (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. The Plant Journal, 11, 1187–1194. 10.1046/j.1365-313X.1997.11061187.x [DOI] [Google Scholar]
- Tian M, Chaudhry F, Ruzicka DR, Meagher RB, Staiger CJ, & Day B (2009). Arabidopsis actin-depolymerizing factor AtADF4 mediates defense signal transduction triggered by the Pseudomonas syringae effector AvrPphB. Plant Physiology, 150, 815–824. 10.1104/pp.109.137604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Verk MC, Pappaioannou D, Neeleman L, Bol JF, & Linthorst HJ (2008). A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiology, 146, 1983–1995. 10.1104/pp.107.112789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tang C, Zhang G, Li Y, Wang C, Liu B, … Kang ZS (2009). cDNA-AFLP analysis reveals differential gene expression in compatible interaction of wheat challenged with Puccinia striiformis f. sp. tritici. BMC Genomics, 10, 289 10.1186/1471-2164-10-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Young ME, Lee WL, & Cooper JA (2003). Integration of signals to the Arp2/3 complex. Current Opinion in Cell Biology, 15, 23–30. 10.1016/S0955-0674(02)00015-7 [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, & Mitchison TJ (1997). Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature, 385, 265–268. 10.1038/385265a0 [DOI] [PubMed] [Google Scholar]
- Welch MD, & Mullins RD (2002). Cellular control of actin nucleation. Annual Review of Cell Developmental Biology, 18, 247–288. 10.1146/annurev.cellbio.18.040202.112133 [DOI] [PubMed] [Google Scholar]
- Wu HY, Liu KH, Wang YC, Wu JF, Chiu WL, Chen CY, … Lai EM (2014). AGROBEST: An efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant Methods, 10, 19 10.1186/1746-4811-10-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota K, Fukai E, Madsen LH, Jurkiewicz A, Rueda P, Radutoiu S, … Rusek AM (2009). Rearrangement of actin cytoskeleton mediates invasion of Lotus japonicus roots by Mesorhizobium loti. Plant Cell, 21, 267–284. 10.1105/tpc.108.063693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Jiang W, Liu Q, Zhang H, Piao MX, Chen ZD, & Bian MD (2015). Expression pattern and subcellular localization of the ovate protein family in rice. PLoS One, 10, e0118966 10.1371/journal.pone.0118966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Yuan H, Wang J, Huo Y, Shimono M, Day B, & Ma Q (2017). TaADF4, an actin-depolymerizing factor from wheat, is required for resistance to the stripe rust pathogen Puccinia striiformis f. sp. tritici. The Plant Journal, 89, 1210–1224. 10.1111/tpj.13459 [DOI] [PubMed] [Google Scholar]
- Zhang R, Xia X, Lindsey K, & Da RP (2012). Functional complementation of dwf4 mutants of Arabidopsis by overexpression of CYP724A1. Journal of Plant Physiology, 169, 421–428. 10.1016/j.jplph.2011.10.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Agrobacterium-mediated transient expression in Nicotiana benthamiana. (A) Leaf phenotypes 7 days post-inoculation (dpi) with Agrobacterium tumefaciens-expressing constructs. (B) Leaf inoculation phenotypes after clearing in ethanol/acetic acid (1:1 v/v) for 2 days. a: pGR106-GFP (CK); b: pGR106-ShARPC3; c: Buffer; d: pGR106-GFP-BAX; e: pGR106-ShARPC3-BAX; f: Buffer-BAX. Agroinfiltration sites in N. benthamiana leaves expressing ShARPC3 were challenged with A. tumefaciens expressing the BAX elicitin. BAX-induced cell death was scored at 3- and 4-days post-inoculation. BAX, a death-promoting member of the Bcl-2 family of proteins, triggered cell death when expressed in plants. A. tumefaciens strain carrying pGR106-GFP was used as a negative control.
Figure S2. Infection level of On-Lz in MM and LA1777 tomato plants.
(A) Infection phenotypes of MM and LA1777 tomato leaves at 7 dpi. (B) Disease indexes of MM and LA1777 plants at 7 and 14 dpi, respectively. Error bars represent the variations among three independent biological replicates. The asterisk (*) indicates statistically significant differences in disease index between MM and LA1777 plants (P < 0.05).
Figure S3. Relative expression levels of ShARPC3 mRNA in TRV2: ShARPC3 plants challenged with On-Lz.
Figure S4. Relative expression levels of AtARPC3 in WT Col-0, the arpc3 and arpc3 plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 challenged with On-Lz. The relative transcript level of AtARPC3 was calculated by the comparative threshold method (2−ΔΔCT). (A) arpc3 plants expressing pCAMBIA3301-ShARPC3. (B) WT Col-0 plants expressing pCAMBIA3301-ShARPC3. Error bars represent the standard deviation among three independent replicates. Asterisks (*) indicate a significant difference from WT Col-0 plants, arpc3 mutants, arpc3 plants expressing pCAMBIA3301-ShARPC3, and WT Col-0 plants expressing pCAMBIA3301-ShARPC3 (P < 0.05).
Figure S5. Functional characterization of ShARPC3 in the Saccharomyces cerevisiae arc18 mutant. Survival of the mutant yeast strain Δarc18 expressing pDR195 (Δarc18 + vec) or pDR195-ShARPC3 (Δarc18 + ShARPC3) were spotted on SC-URA medium at 24°C, 30°C, 36°C, and 42°C. The final densities were diluted to 107, 106, 105, 104, and 103 (cell ml−1) with sterile water using a blood counting chamber.
Table S1. DNA primers used to amplify ShARPC3 gene and recombination. The primer sequences are composed of two parts. The lowercase letters represent the sequences of corresponding vector homologous arm and restriction enzyme site, and the capital letters indicate the gene-specific nucleotide sequences.
Table S2. qRT-PCR genes and DNA primers used in VIGS experiment.
Table S3. qRT-PCR genes and primers used in Arabidopsis thaliana transgenic complementation assay.
Table S4. The predicted subcellular localizations of ShARPC3 using ProtComp 9.0. The predicted localization of tomato ARPC3 protein supported by ProtComp 9.0 (http://linux1.softberry.com/berry.phtml). Neural Nets, Pentamers, and Integral predictions show ShARPC3 protein localizes predominantly in the extracellular space.


