The seminal discovery and classification by Mosmann and Coffman of distinct T helper cell subsets set the stage to begin unraveling the diversity of CD4+ T cells and understanding their critical contributions in homeostasis and host defense [1]. It is now appreciated that different CD4+ T helper cell subsets play important roles in clearing intracellular pathogens, parasites and fungal infections, but that a dysregulated CD4+ T helper cell response can lead to allergy, asthma and autoimmunity [2].
In this review, we will highlight recent advances in understanding how distinct T helper (Th) cell subsets develop, which transcription factors regulate their transcriptional signatures and which cytokines contribute to their effector functions. Over the past few years, major themes were metabolism, the microbiome and tissue cues that govern the differentiation of T helper cells and their effector functions, hence these themes find particular emphasis within this review. Although a number of T helper cell subsets have been defined, we will focus on Th1, Th17, Treg and Tr1 cells as they pertain to tissue-specific autoimmunity (Fig. 1). Th1 and Th17 cells are considered the main drivers of T cell-mediated autoimmunity whereas Treg and Tr1 cells are critical for regulation in that they either inhibit induction or mediate resolution of tissue inflammation.
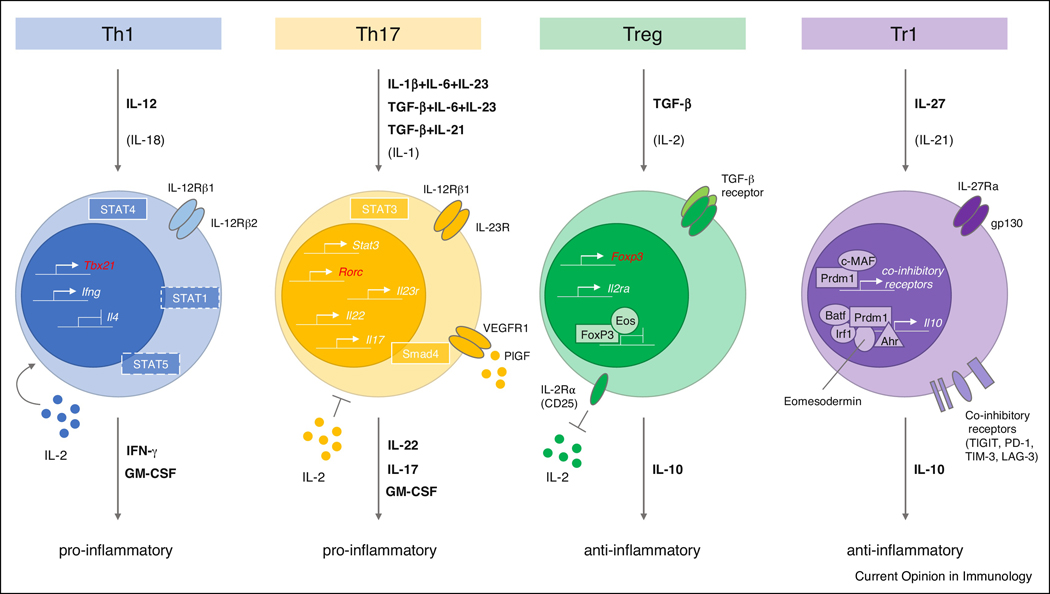
Figure 1.
Overview of Th1, Th17, Treg and Tr1 cell subsets. Cytokines that are critical to their differentiation are highlighted in bold. For Th17 cells, three alternative differentiation conditions are shown. Additional important cytokines contributing to the four subsets are listed in parentheses. Critical genes for each lineage that are actively expressed are highlighted as small promoter symbols with an arrow within the nucleus. In certain instances, protein complexes are highlighted specifically bound to such promoters. Effector cytokines are indicated in bold: IFN-γ and GM-CSF for Th1 cells; IL-17, IL-22 and GM-CSF for Th17 cells; IL-10 for both Tregs and Tr1 cells. IL-2 has been shown to support the Th1 cell lineage whereas it inhibits Th17 cell differentiation. An important aspect of Treg cell function is limiting bio-available IL-2 by expression of IL-2Rα (CD25). The critical importance of IL-23 signaling to the function of Th17 cells is emphasized by depiction of the functional receptor for IL-23 consisting of IL-12Rβ1 and IL-23R. In many instances, master transcription factors suppress the expression of genes important for other T helper cell subsets. This notion is exemplified for Tregs through collaborative repression of genes by Eos and FoxP3 but is an important mechanism of master transcription factors of all lineages so far with the exception of Tr1 cells for which no master transcription factor has been identified yet. The genes encoding the master transcription factors of Th1, Th17 and Treg cells are shown in red, respectively (Tbx21, Rorc and Foxp3). JAK/STAT signaling pathways have particular relevance to T helper cell differentiation as they mediate the instructive signals of the lineage inducing cytokines: STAT1/4 relevant for Th1 cells and STAT3 for Th17 cells, respectively. The critical contribution of TCR signaling for differentiation of T helper cell subsets has been omitted for clarity.
Th1 cells and Th17 cells in Autoimmune Disease
Th1 cells have been shown to play a critical role in a number of human autoimmune diseases including type 1 diabetes, arthritis, multiple sclerosis (MS) and inflammatory bowel disease (IBD) [3]. The signature transcription factor of Th1 cells is T-bet (encoded by the gene Tbx21) and IFN-γ mediates important effector functions of Th1 cells, especially in the setting of host defense against viruses and intracellular pathogens and the promotion of anti-tumor immunity [3,4]. Upon binding of IL-12 to its receptor (IL-12Rβ1/IL-12Rβ2), the JAK/STAT pathway (including STAT1 and STAT4) is activated to induce Tbx21 expression and IFN-γ production [5]. TCR stimulation is critical for induction of Tbx21, which is supported by autocrine signaling via IFN-γ-STAT1 activation. The transcription factors T-bet, STAT4, STAT1 and Runx3 interact, establishing the Th1 cell transcriptional program and at the same time silencing the expression of genes such as Il4 or Gata3 that are important for the development and differentiation of Th2 cells (Fig. 1) [6].
Adoptive transfer of Th1 cells with specificity for self-antigen has been shown to induce tissue inflammation and autoimmunity in multiple experimental autoimmune disease models [7]. However, loss of the signature cytokine IFN-γ, renders mice still susceptible to autoimmunity [8,9]. This observation raised the question of whether Th1 cells are the pathogenic T helper cell subset that induces autoimmunity and provided impetus for the identification of another highly pathogenic T cell subset called Th17 cells.
A hallmark study in 2003 showed that IL-23 is critical for the induction of autoimmunity in the central nervous system (CNS) [10]. Soon thereafter, it was suggested that IL-23 is important for expansion or differentiation of IL-17-producing T cells [11]. We proposed that Th-IL-17 cells (later named Th17 cells) may be a distinct T cell subset, and that IL-23 per se is not the differentiation factor for their generation [12]. This led to the identification of differentiation factors that induce Th17 cells. Three seminal papers defined the conditions that direct the development of Th17 cells from naïve T cell precursors and we proposed that there may be a reciprocal relationship between FoxP3+ Tregs and IL-17-producing Th17 cells [13–15]. It is now realized that there are multiple differentiation pathways by which IL-17-producing cells can be generated [16,17], resulting in development of Th17 cells with different effector phenotypes and functions. Different cytokine combinations, in particular TGF-β + IL-6 + IL-23 or IL-1β + IL-6 + IL-23, lead to the transcriptional program that confers pathogenicity to Th17 cells [18]. The JAK/STAT pathway member STAT3 plays a fundamental role in Th17 cell differentiation programs in that it both transactivates the Rorc gene locus and cooperates with its product, the master transcription factor of the Th17 cell lineage RORγt, to further upregulate signature cytokines and lineage markers of Th17 cells such as Il17a, Il17f and Ccr6 [19,20].
The role of IL-17A and IL-17F, the signature cytokines of Th17 cells, in the induction of experimental autoimmune encephalomyelitis (EAE) has been somewhat controversial in that it was shown that neutralization of IL-17A in Il17f-deficient mice does not have a significant impact on the development of EAE, raising the possibility that IL-17 is not critical for tissue inflammation and that there may be another cytokine critical for the induction of tissue inflammation [21]. This led to the hypothesis that GM-CSF is the cytokine responsible for the induction of tissue inflammation in the absence of canonical Th1 or Th17 cell cytokines [22]. Indeed, GM-CSF, which is part of the pathogenic signature of Th17 cells, has been shown to be important in EAE as early as 2001 and shown to be critical in its expression by myelin-specific, autoreactive T cells [23,24]. GM-CSF received renewed attention more recently [25,26] and two new studies may suggest that a novel pathogenic T cell subset may exist whose signature cytokine is GM-CSF and that this lineage may not depend on IL-6 signaling [27,28]. Furthermore, the authors report that at least in the setting of T cell populations profiled from MS patients, GM-CSF+ cells largely overlap with IFN-γ-secreting cells, hence leaving the possibility that these cells could be in fact Th1 cells [27]. However, the issue remains unresolved whether IL-23 is critical for inducing pathogenicity in bona fide Th1 and Th17 cells by inducing GM-CSF, since IL-23- and IL-23R-deficient mice are completely resistant to the development of EAE [10].
It should be noted that pathogenic Th17 cells, besides expressing GM-CSF, also express IFN-γ and this phenotype is further strengthened when the encephalitogenic T cells enter the CNS and this has also been observed at the single-cell level [29].
However, a recent study showed that IL-17A is critical for driving the development of pathogenic Th17 cells in EAE via the recruitment of myeloid cells that produce IL-1β and IL-23 [30]. Furthermore, loss of both IL-17A and IL-17F results in complete resistance to the development of EAE, even when GM-CSF is actively produced by T cells and when the mice are co-housed to equalize the microbiome differences in the experimental groups (Ho and Kuchroo, unpublished data). This is consistent with the data from Joan Goverman’s group suggesting that indeed GM-CSF is dispensable for induction of EAE and that IL-17 activity compensates for the induction of spinal cord targeted disease [31]. On the other hand, IL-17 and GM-CSF both are required to fully overcome the inhibitory effects of IFN-γ for the induction of inflammation in the brain during EAE [31]. These results suggest that the precise phenotype of pathogenic Th1 or Th17 cells drives distinct patterns of tissue pathology and that heterogeneity within an autoimmune disease phenotype may reflect mechanistic and cellular differences in the T cells inducing the disease phenotype. Indeed, it has been shown in the EAE model that the ratio of infiltrating Th17 to Th1 cells contributes to the location of CNS inflammation and that this ratio is influenced by the myelin antigen-specificity of these T cells [32]. Interestingly, T cells isolated from peripheral blood of MS patients showed also distinct cytokine profiles depending on their antigen-specificity and whether these patients exhibited lesions mostly in the brain or spinal cord [33]. Although clinical trials with anti-GM-CSF have not been conducted in MS yet, a double-blind clinical trial with anti-IL-17A showed a significant decrease in the lesion load and improvement in clinical disease [34], underscoring the role of IL-17 in inducing the human disease. This is further supported by the observation that anti-IL-17 antibody is now approved for treatment of multiple human autoimmune diseases [35].
Fine tuning of Th17 cell responses: transcriptional control and subset diversity
After the discovery of Th17 cells, it became very clear that not all IL-17-producing cells are pathogenic and indeed a vast majority of IL-17-producing cells populate the intestine at homeostasis and do not induce any tissue inflammation [36]. This led to the concept that there might be two different types of IL-17-producing T cells, one that is present at steady-state to promote barrier function and to limit invasion of the microbiome and a second, pathogenic Th17 cell subset, that promotes tissue inflammation and autoimmunity [36–39]. We identified a transcriptional signature that distinguishes pathogenic from non-pathogenic Th17 cells, showing that pathogenic Th17 cells co-express T-bet/IFN-γ and GM-CSF, whereas non-pathogenic Th17 cells express IL-10 and IL-1RN [18,29,40].
An intriguing recent study linked febrile temperatures to an increase in the pathogenicity of Th17 cells through sumoylation of SMAD4 and its nuclear translocation, where it cooperates with RORγt and STAT3 in transactivation of Il17a (Fig. 1) [41]. Hence, CD4+ T cell deficiency for Smad4 protects animals from EAE. Interestingly, expression of Rorc/Rora is not affected by temperature suggesting that the influence of temperature may represent a very fast acting mechanism on the pathogenicity of Th17 cells in vivo through acute upregulation of effector cytokines such as Il17a, Il17f and Il22 and not necessarily through de novo generation of Th17 cells driven by RORγt [41].
IL-21 is an amplification factor that has been known to trigger Th17 differentiation in the absence of IL-6 signaling [17]. Recently, placental growth factor (PlGF) has been shown to induce Th17 cells through the activation of STAT3 signaling (Fig. 1) [42]. Indeed, PlGF is produced by Th17 cells and signals in an autocrine/paracrine manner to strongly upregulate Rorc. The important contribution of PlGF to the pathogenicity of Th17 cells was demonstrated by the observations that mice deficient for Plgf show reduced EAE severity and that Plgf-Tg mice show more severe collagen-induced arthritis (CIA). The authors suggested that PlGF may be able to substitute for the requirement of IL-6 signaling in Th17 cell differentiation in vitro [42]. However, as Il6 deficiency fully protects mice from EAE and Plgf deficiency does not, this suggests that PlGF cannot completely substitute for IL-6 in inducing Th17 cell-dependent CNS disease in vivo [43,44].
IL-23/IL-23R signaling is critical to Th17 cell pathogenicity
IL-23 was found to be a key cytokine in converting non-pathogenic Th17 cells into a pathogenic phenotype. This requirement for IL-23 may be reflected in the genetic linkage to the IL-23:IL-23R pathway in the development of multiple autoimmune diseases [45]. Therefore, transcriptional regulators that control the expression of Il23r have particular relevance for the pathogenicity of Th17 cells and IL23R has been identified as a critical risk gene for multiple human autoimmune diseases including IBD, psoriasis and ankylosing spondylitis [45,46]. IL-23R has been suggested to be important for both the stabilization and the terminal differentiation of Th17 cells [13,47], but the actual signaling pathway and molecular mechanisms by which IL-23 acts on Th17 cells have not been elucidated.
Recently, it was shown that the transcriptional regulator RBPJ can further upregulate the expression of Il23r and at the same time reduces expression of Il10 to control the pathogenicity of Th17 cells. Consequently, genetic deletion of Rbpj protects animals from severe EAE [48]. Furthermore, Blimp-1/PRDM1 co-localizes with RORγt and STAT3 at enhancer regions to control the expression of Il23r, Il17a and Csf2 (encoding the cytokine GM-CSF) in Th17 cells. Similar to deletion of Rbpj, deficiency of Blimp-1/Prdm1 in peripheral T cells ameliorates EAE [49].
A deeper understanding of the mechanism by which IL-23/IL-23R signaling evokes the pathogenicity of Th17 cells and possibly other pathogenic T helper cell subsets will have important therapeutic implications.
Metabolic and microbial regulation of Th1 and Th17 cell responses
Metabolic processes have emerged over the past few years as a critical component of T helper cell function [50–54]. For example, Peng et al. showed that Th1 cells boost aerobic glycolysis through lactase dehydrogenase A and that Ifng expression is driven by acetyl-coenzyme A-augmented histone acetylation (Fig. 2A) [54]. Recent work by the Flavell lab has uncovered an important novel function of mitochondrial metabolism with particular relevance to Th1 cell differentiation [55]. In particular, the authors showed that the tricarboxylic acid (TCA) cycle is important for the terminal effector function of Th1 cells through succinate dehydrogenase. For example, Sdhc-deficient cells, which lack the important subunit C of the succinate dehydrogenase complex, show a defect in upregulation of Tbx21 and Ifng. On the one hand, succinate dehydrogenase activity suppresses Th1 cell proliferation, whereas the malate-aspartate shuttle and mitochondrial citrate export were shown to drive proliferation, including through the production of aspartate [55]. The seemingly opposite functions of components of mitochondrial metabolism may allow tuning of the biological response to allow either terminal effector function or proliferation in response to energy status and nutrient supply. By genetic targeting of key components of the shuttle and mitochondrial citrate export, such as Slc25a11 and Slc25a1, through CRISPR/Cas9 technology, the authors linked these activities to epigenetic histone acetylation. This epigenetic mechanism then fosters the expression of key Th1 cell lineage genes such as Tbx21 (Fig. 2A) [55]. How exactly these metabolic processes are translated mechanistically into locus-specific epigenetic changes of the Th1 cell chromatin landscape remains mostly unknown. Taken together, this work offers a glimpse into how the metabolic state may be tailored to the specific needs of the Th1 cell response and how it can be linked through epigenetic mechanisms to transcriptional control of key Th1 cell genes associated with proliferation or terminal effector functions. We are beginning to understand how metabolic processes intersect with transcriptional control of Th1 or other T helper cell subsets and these metabolic controls will offer new avenues for therapeutic intervention in autoimmunity [56].
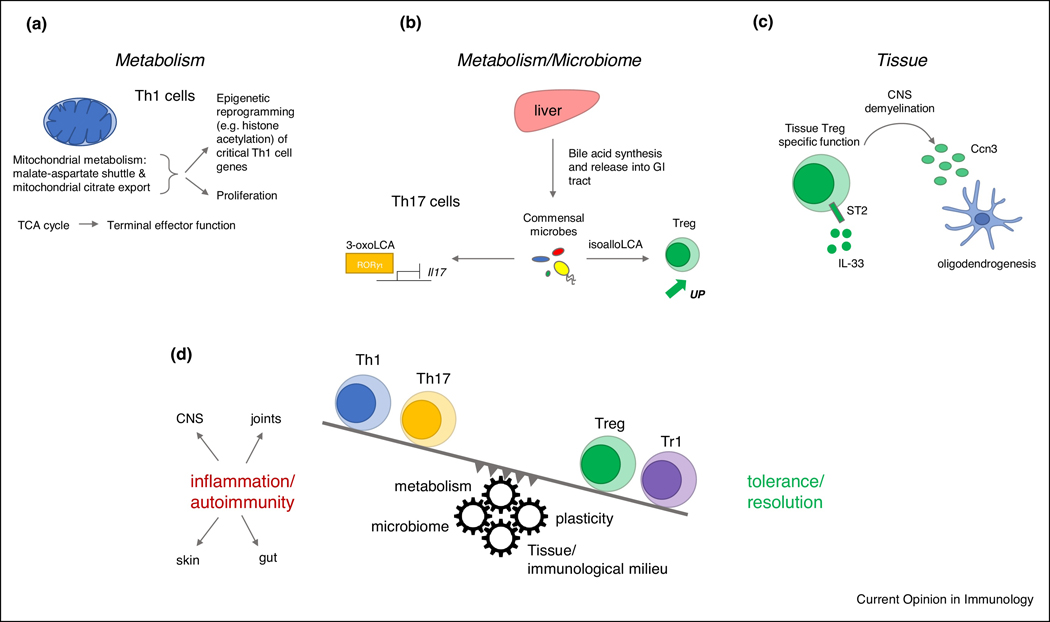
Figure 2.
(A) Mitochondrial metabolism is critical to Th1 cell differentiation and terminal effector function. The malate-aspartate shuttle and mitochondrial citrate export drive proliferation and their activity is translated mechanistically through epigenetic changes influencing the transcription of key Th1 cell genes. The TCA (tricarboxylic acid) cycle is critical to terminal effector function of Th1 cells. (B) Metabolites such as bile acids and their derivatives which are generated through processing by the microbiome are important regulators of Th17 cell differentiation, e.g. 3-oxoLCA can directly bind to the Th17 cell master transcription factor RORγt and inhibit transcription of Il17. In contrast, the bile acid metabolite isoalloLCA can lead to the expansion of Tregs thereby contributing to tolerance. (C) Tregs have been shown to possess very important tissue-resident functions such as the repair of damaged tissue. Indeed, Tregs can secrete oligodendrogenic Ccn3 to enhance oligodendrogenesis in the setting of CNS demyelination. (D) Model showing the interconnected relationship among the pro-inflammatory Th1/Th17 cell subsets and the regulatory Treg/Tr1 cell subsets. The biological outcome of either inflammation/autoimmunity or tolerance/resolution of inflammation is dictated by the expansion and dominance of either pro-inflammatory or regulatory Th cell subsets. T helper cell differentiation and expansion occurs under the influence of a complex network of mechanisms. These mechanisms can dynamically shift the balance between pro-inflammatory and regulatory subsets suggested as cogwheels tipping the balance in a particular direction. For example, metabolites generated or processed by the microbiome can foster Treg cell differentiation. The inherent plasticity of Th17 cells can lead to their transdifferentiation to either Tregs or Tr1 cells. T helper cell differentiation and effector function is critically influenced by the tissue immunological milieu and the interplay with other immune cells.
The microbiome has been shown to be a critical driver of autoimmunity in pre-clinical models of both MS and arthritis [57,58]. Some microbial species such as segmented filamentous bacteria (SFB) are particularly equipped to trigger Th17 cell differentiation [59]. Metabolism and the microbiome are intricately interconnected and a recent study has implicated bile acids in the differentiation of T helper cell subsets and to influence the Th17/Treg cell balance (Fig. 2B). In particular, Hang et al. demonstrated that the bile acid metabolite 3-oxoLCA interferes with Il17a expression through direct binding of RORγt, the critical transcription factor for Th17 cells, and that isoalloLCA leads to an increase in reactive oxygen species (ROS) promoting Treg cell differentiation [60]. Strikingly, bile acid metabolism has been shown to be altered in MS patients and may be amenable to manipulation to curtail neuroinflammation [61].
Even though various studies have identified microbiota and metabolites that can both trigger and expand Th17 cells, the picture is far less clear in regard to the differentiation of Th1 cells. However, germ-free mice orally colonized with microbiota from a Crohn’s disease patient developed a very strong Th1 cell driven intestinal inflammation. In particular, Klebsiella strains were identified to be responsible for triggering this Th1 cell-driven immune response [62]. This pioneering study illustrates that very clear instructive signals are generated by certain microbiota to drive Th1 cell differentiation in the gut and it appears likely that much remains to be learned.
The metabolite tetrahydrobiopterin (BH4) is an important co-factor for enzymatic activity, in particular for the hydroxylation of aromatic amino acids and therefore important for the synthesis of neurotransmitters such as serotonin, dopamine and melatonin as well as for the production of nitric oxide [63]. Interestingly, Cronin et al. discovered a function of BH4 in the proliferation of T cells and their ability to mediate autoimmune tissue inflammation [64]. For example, conditional genetic deletion of the upstream enzyme in BH4 synthesis, GCH1, specifically in T cells, protects animals from severe EAE and adoptive transfer colitis [64].
Serum amyloid A proteins (SAA) have recently been shown to trigger a pathogenic Th17 cell differentiation program independent of TGF-β and the tissue-specific availability of SAA proteins controls the pathogenicity of Th17 cells and the extent of Th17 cell-driven inflammation [65]. In particular, when in vitro differentiated MOG-specific TCR transgenic T cells (2D2) are cultured under pathogenic conditions and adoptively transferred into Saa3-deficient hosts, a marked decrease in EAE severity is observed highlighting the tissue-specific cues that SAA proteins provide to either promote a non-pathogenic/homeostatic response or to foster neuroinflammation [65]. Which factors and signaling pathways may cooperate with SAA proteins to mediate either a homeostatic or pathogenic phenotype in Th17 cells remains incompletely understood but the observation of the highly context-dependent function of SAA proteins suggests they could emerge as strong targets to treat autoimmune disease.
Thus, both host and microbiome metabolic pathways influence the balance between pathogenic Th1 and Th17 responses and non-pathogenic responses that are important for homeostasis and tissue repair.
Lineage Plasticity of Th17 cells
T helper cells, in particular Th17 cells, exhibit a remarkable extent of lineage plasticity. For example, in the course of EAE, Th17 cells have been shown to give rise to IFN-γ+/IL-17A+ double positive Th17 cells and IFN-γ+/IL-17A− ex-Th17 cells [66]. Furthermore, Th17 cells transdifferentiate in the intestine to IL-10+ Tr1-like cells which maintain homeostasis and promote resolution of inflammation in the gut [67]. A recent study connected the plasticity of Th17 cells to their metabolic state in EAE and identified two distinct populations of Th17 cells that are discriminated by TCF-1 and T-bet expression and expression of CD27 [68]. The TCF-1+ Th17 cell population exhibits features of a precursor or stem cell-like state and the authors demonstrated that mTOR signaling enables the progression of these precursor cells to become Th1-like ex-Th17 cells, which is critical for their ability to elicit neuroinflammation [68]. How the plasticity of Th17 cells is regulated on a molecular level and which transcription factors are critical remains under intensive investigation. A recent study from our laboratory demonstrated that the salt-sensing kinase Sgk1, which is induced by IL-23R signaling, regulates the balance between Th17 cells and Tregs [69]. It remains unclear if other pathogenic Th cell subsets such as pathogenic Th1 cells possess a similar plasticity during the course of an inflammatory response. To this end, single-cell studies have been instrumental in elucidating the transcriptional signatures and drivers of Th17 cell differentiation and plasticity including in the settings of autoimmunity and are expected to be similarly insightful in regard to other pathogenic T cell subsets, in particular Th1 cells [29,40].
Tregs and Tr1 cells limit Th1/Th17 cell-driven autoimmunity
Tregs
T regulatory (Treg) cells play a fundamental role in maintaining tolerance and homeostasis which is highlighted by the observation that germline mutations of their master transcription factor FoxP3 lead to multi-organ autoimmunity [70]. The function of FoxP3 is not limited to the activation of the Treg cell transcriptional program but it also silences genes that would destabilize Treg cell function similar to the master transcription factors of other Th cell subsets (Fig. 1) [71]. TGF-β is the critical cytokine inducing peripheral FoxP3+ Tregs in vitro from naïve T cells [13,72]. Tregs have been shown to inhibit effector T cell responses through a host of mechanisms, including regulation of bio-available IL-2 through expression of IL-2Rα (CD25), direct modulation of APC function, secretion of the immunosuppressive cytokine IL-10 and induction of apoptosis of effector T cells [70]. Tregs, in particular, seed to various tissues early during development where they acquire highly specialized transcriptional programs and functions that are tailored to the tissue in which they reside [73].
How these tissue-resident Treg cell subsets are directed transcriptionally remains an area of intense study. It is clear, however, that tissue-specific adaptation of Tregs has critical functions in the control of inflammation which was recently demonstrated in the gut [74]. In the setting of acute brain injury by ischemic stroke, it was shown that brain-specific Tregs depend on IL-2, IL-33, serotonin and TCR activation and that their targeting to the brain is guided by chemokines CCL1 and CCL20 (Fig. 2C) [75]. Furthermore, in a pre-clinical model of acute CNS demyelination, it was shown that these tissue-specific, infiltrating Tregs may foster regeneration of afflicted tissue through secretion of oligodendrogenic Ccn3 (Fig. 2C) [76]. The Teichmann lab used single-cell RNA-sequencing to determine the trajectories of tissue adaptation for skin and colonic Tregs and showed that the acquisition of a tissue-specific transcriptional program proceeds through discrete transitional states with characteristic transcriptional profiles [77]. This observation is consistent with previous findings from the Benoist/Mathis lab regarding visceral adipose tissue (VAT)-Tregs [77,78]. The transcription factors and their timing of action that govern the tissue adaptation of Tregs are largely unknown but recent work by Delacher et al. identified BATF as a transcription factor that plays a critical role in establishing ST2+ populations of tissue Tregs from two distinct precursor stages [79].
Previously, it was shown that distinct intestinal gut symbionts instigate the development of RORγt+FoxP3+ Treg cells [80] and by virtue of co-expression of these two master transcription factors these Treg cells acquire specialized functions in the gut. In addition to the study cited above, describing the influence of bile acids on the intestinal Th17/Treg cell balance [60], Song et al. showed that bile acids play a critical role in regulation of colonic RORγt+ Tregs and demonstrated that Treg cell-specific deficiency of the bile acid receptor Vdr negatively impacts this intestinal population and leads to more severe disease in a pre-clinical model of colitis [81]. Furthermore, genetic manipulation of bile acid metabolic pathways in certain intestinal bacterial strains influences the colonic RORγt+ Treg cell population suggesting that processing of bile acids by the microbiota is important [81].
The Littman lab showed that c-MAF plays an important role in the control of RORγt+FoxP3+ Treg cells and that the function of these cells is critical in maintaining homeostasis and limiting potentially pathogenic microbial species [82]. It was shown that IL-2 produced by ILC3s is implicated in the generation of intestinal Tregs and contributes to their ability to maintain homeostasis [83]. How the transcriptional program of RORγt+FoxP3+ Tregs is influenced by IL-2 made by ILC3s may be informative in the future as a particular relevance of this IL-2/ILC3 axis in Crohn’s disease was proposed [83].
Tr1 cells
T regulatory type 1 (Tr1) cells are characterized by high expression of IL-10, do not express FoxP3 and have been shown to be protective in colitis [84]. Tr1 cells can be differentiated in vitro through IL-27 and Ahr plays a critical role, together with c-MAF, in establishing their transcriptional program (Fig. 1) [85–87]. Unlike for Th1, Th17 and FoxP3+ Treg cells, the master transcription factor for IL-10-producing Tr1 cells has not been identified so far, as no single transcription factor is known to be exclusively directing the development of the Tr1 cell lineage. Indeed, IL-10 is produced by multiple T cell subsets (Th1, Th2, Th17 and Treg cells), raising the possibility that expression of a single master transcription factor for the induction of IL-10 may antagonize differentiation of other T cell subsets and interfere with their development. Therefore, multiple transcription factors may induce IL-10, and they would cooperate with the master transcription factors of other T cell subsets to induce IL-10 together with their respective signature cytokine(s). Recently, the transcription factor Eomesodermin was shown to cooperate with Blimp-1 to activate the Il10 locus [88]. Interestingly, c-MAF possesses important regulatory functions regarding IL-10 production by its ability to regulate IL-2 production which has direct implications for various inflammatory responses ranging from Th1, Th2 to Th17 cell-driven responses [89]. Also, the transcription factors IRF1 and BATF were found to transactivate the Il10 locus in Tr1 cells and to serve as pioneering factors in setting up the chromatin landscape during Tr1 cell differentiation (Fig. 1) [90].
IL-27, which induces Il10 expression, was also shown to regulate the expression of a module of “checkpoint” molecules including PD-1, TIM-3, LAG-3 and TIGIT [91,92] and coexpression of IL-10 with co-inhibitory receptors can be viewed as an indicator of a cellular program to increase regulatory function, both in a cell-intrinsic and cell-extrinsic manner. Blimp-1/PRDM1 and c-Maf were found to be the shared regulators of this module and both of these transcription factors have also been shown to regulate IL-10 production [91]. Expression of both IL-10 and “checkpoint” molecules may be important in the resolution of tissue inflammation and autoimmunity. Indeed, both Tr1 and Treg cells have been shown to express checkpoint molecules, suggesting a role in their function. For example, the co-inhibitory molecule TIGIT has been implicated in Treg-mediated suppression of Th1 and Th17 responses [93]. While undertaking an unbiased expression analysis of PBMCs from autoimmune disease patients, McKinney et al. found that relapse rates correlate with the expression of a gene module in T cells containing co-inhibitory molecules [94]. Based on these studies and studies on the role of IL-10 in regulating tissue inflammation, one would predict that IL-10 and the expression of co-inhibitory molecules in T cells may go hand-in-hand to resolve inflammation and to inhibit autoimmunity [95,96]. It has been shown that Th17 cells transdifferentiate into Tr1-like cells during the resolution of inflammation [67]. Therefore, Tr1 cells may be particularly relevant to the resolution of autoimmune inflammation and preventing a relapse in a particular autoimmune disease whereas Tregs may play a dominant role in the inhibition of auto-reactive T cells.
Control of the balance between pathogenic and regulatory T helper cell subsets
The balance between proinflammatory Th1/Th17 cells and regulatory Treg/Tr1 cells is critical to maintain tolerance yet allow an appropriate response to invading pathogens. The complex balance between proinflammatory and regulatory T helper cell subsets is controlled at multiple layers including metabolic and microbial control, tissue-specific cues and plasticity (Fig. 2D). As this balance can be shifted through a host of mechanisms, autoimmune responses in different tissues such as the CNS, joints, skin and gut may be dependent upon distinct mechanisms that result in inflammation. It is important to emphasize that the specificity of the TCR for its auto-antigen has critical contribution to the expansion of pathogenic Th1/Th17 cells for the development of organ-specific autoimmunity.
As we highlighted in this review, our understanding of how the balance between pathogenic and regulatory responses can be influenced has been deepened in particular in the setting of the gut and the effect of the microbiome. Some microbes and their metabolites can foster the development of pathogenic T helper cell subsets whereas other microbes can foster the development of regulatory T cells. A healthy microbiome promotes the expansion of regulatory T cells which fosters tolerance; for example, a mixture of Clostridia strains has been shown to induce Tregs [97]. If the balance is disrupted through dysbiosis, this will lead to the emergence of pathogenic T cells. Indeed, it has been shown that the transfer of microbiota from MS patients into mice can transfer disease and trigger spontaneous CNS autoimmunity [98].
The knowledge of how, on a mechanistic level, the plasticity of Th cell subsets is controlled remains rather limited at this moment but importantly, not only Th17 cells have been shown to have a significant ability for plasticity but other T cell subsets also have been shown to be plastic [99]. Furthermore, the specific cues that imprint tissue-specific transcriptional programs and function remain poorly understood and are a field of intensive research.
Synthesis and New Frontiers
The classification of T helper cells into different subsets has been extremely useful in facilitating a comprehensive understanding of their diverse functions in homeostasis, host defense and autoimmunity. The application of new technologies has revealed a deeper complexity within the initially defined subsets that likely reflect distinct functional states. A cardinal conclusion of the advent of single-cell technologies is that cellular heterogeneity is far greater than previously appreciated and that this also holds true for T helper cell subsets, in particular in inflammatory settings. Interrogation of the classical Th cell subsets with new technologies such as single-cell genomic technologies and CRISPR/Cas9 systems will further define not only the transcriptional status but also metabolomic and epigenetic states in relation to a variety of other cell types within various autoimmune and inflammatory settings. Greater heterogeneity and plasticity may be embedded in Th cell responses to ensure an appropriate and regulated response to pathogens, the local tissue environment and resolution of inflammation. A very important open question is the individual contributions of pathogenic Th1 and Th17 cells to particular autoimmune diseases and it appears likely that Th1 and Th17 cells contribute differently depending on the organ and/or the type of disease. A deeper understanding of this complexity will allow new approaches to the prevention and treatment of autoimmune disease.
Acknowledgements
This work was supported by grants from the US National Institutes of Health to V.K.K. (R01NS045937, P01AI073748, P01AI039671 and P01AI056299). We are grateful to members of the Kuchroo laboratory for valuable feedback on this review. We would like to thank Mary Collins for critical discussion and advice. Due to the scope of this review, we apologize to investigators whose work and important contributions we could not highlight here.
Footnotes
Declaration of interest
V.K.K. is cofounder of Celsius Therapeutics, Tizona Therapeutics and Bicara Therapeutics. His interests are reviewed and managed by the Brigham and Women’s Hospital and Partners Healthcare in accordance with their conflict of interest policies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
• of special interest
•• of outstanding interest
- 1.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL: Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol 1986, 136:2348–2357. [PubMed] [Google Scholar]
- 2.Bluestone JA, Bour-Jordan H, Cheng M, Anderson M: T cells in the control of organ-specific autoimmunity. J Clin Invest 2015, 125:2250–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH: Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol 2003, 21:713–758. [DOI] [PubMed] [Google Scholar]
- 4.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH: A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000, 100:655–669. [DOI] [PubMed] [Google Scholar]
- 5.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH: Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity 2008, 29:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM: Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol 2007, 8:145–153. [DOI] [PubMed] [Google Scholar]
- 7.Jager A, Dardalhon V, Sobel RA, Bettelli E, Kuchroo VK: Th1, Th17, and Th9 effector cells induce experimental autoimmune encephalomyelitis with different pathological phenotypes. J Immunol 2009, 183:7169–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferber IA, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L, Dalton D,Fathman CG: Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J Immunol 1996, 156:5–7. [PubMed] [Google Scholar]
- 9.Trembleau S, Penna G, Gregori S, Giarratana N, Adorini L: IL-12 administration accelerates autoimmune diabetes in both wild-type and IFN-gamma-deficient nonobese diabetic mice, revealing pathogenic and protective effects of IL-12-induced IFN-gamma. J Immunol 2003, 170:5491–5501. [DOI] [PubMed] [Google Scholar]
- 10.Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B, Lucian L, To W, Kwan S, Churakova T, et al. : Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 2003, 421:744–748. [DOI] [PubMed] [Google Scholar]
- 11.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ: IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 2005, 201:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettelli E, Kuchroo VK: IL-12- and IL-23-induced T helper cell subsets: birds of the same feather flock together. J Exp Med 2005, 201:169–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK: Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006, 441:235–238. [DOI] [PubMed] [Google Scholar]
- 14.Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT: Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 2006, 441:231–234. [DOI] [PubMed] [Google Scholar]
- 15.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B: TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 2006, 24:179–189. [DOI] [PubMed] [Google Scholar]
- 16.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, et al. : Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature 2010, 467:967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK: IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature 2007, 448:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, et al. : Induction and molecular signature of pathogenic TH17 cells. Nat Immunol 2012, 13:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR : The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 2006, 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 20.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, et al. : Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 2010, 32:605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haak S, Croxford AL, Kreymborg K, Heppner FL, Pouly S, Becher B, Waisman A: IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J Clin Invest 2009, 119:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Becher B, Tugues S, Greter M: GM-CSF: From Growth Factor to Central Mediator of Tissue Inflammation. Immunity 2016, 45:963–973. [DOI] [PubMed] [Google Scholar]
- 23.McQualter JL, Darwiche R, Ewing C, Onuki M, Kay TW, Hamilton JA, Reid HH, Bernard CC: Granulocyte macrophage colony-stimulating factor: a new putative therapeutic target in multiple sclerosis. J Exp Med 2001, 194:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ponomarev ED, Shriver LP, Maresz K, Pedras-Vasconcelos J, Verthelyi D, Dittel BN: GM-CSF production by autoreactive T cells is required for the activation of microglial cells and the onset of experimental autoimmune encephalomyelitis. J Immunol 2007, 178:39–48. [DOI] [PubMed] [Google Scholar]
- 25.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A: The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol 2011, 12:568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B: RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol 2011, 12:560–567. [DOI] [PubMed] [Google Scholar]
- 27.Galli E, Hartmann FJ, Schreiner B, Ingelfinger F, Arvaniti E, Diebold M, Mrdjen D, van der Meer F, Krieg C, Nimer FA, et al. : GM-CSF and CXCR4 define a T helper cell signature in multiple sclerosis. Nat Med 2019, 25:1290–1300.• Using Cytof in human T cells from MS patients, the authors described a prominent GM-CSF+ CXCR4+ CD4+ T cell population.
- 28.Komuczki J, Tuzlak S, Friebel E, Hartwig T, Spath S, Rosenstiel P, Waisman A, Opitz L, Oukka M, Schreiner B, et al. : Fate-Mapping of GM-CSF Expression Identifies a Discrete Subset of Inflammation-Driving T Helper Cells Regulated by Cytokines IL-23 and IL-1beta. Immunity 2019, 50:1289–1304 e1286.•• This paper described the generation of GM-CSF fate mapping mice (FROG mice) to elegantly investigate the implication of GM-CSF in neuroinflammation and to profile CNS infiltrating cells by single-cell RNA-seq.
- 29.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. : Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell 2015, 163:1400–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGinley AM, Sutton CE, Edwards SC, Leane CM, DeCourcey J, Teijeiro A, Hamilton JA, Boon L, Djouder N, Mills KHG: Interleukin-17A Serves a Priming Role in Autoimmunity by Recruiting IL-1beta-Producing Myeloid Cells that Promote Pathogenic T Cells. Immunity 2020, 52:342–356 e346.•• This paper demonstrated another important function of IL-17A in tissue inflammation and autoimmunity by attracting myeloid cells that in turn contribute IL-1β and IL-23 to foster development of pathogenic Th17 cells.
- 31.Pierson ER, Goverman JM: GM-CSF is not essential for experimental autoimmune encephalomyelitis but promotes brain-targeted disease. JCI Insight 2017, 2:e92362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stromnes IM, Cerretti LM, Liggitt D, Harris RA, Goverman JM: Differential regulation of central nervous system autoimmunity by T(H)1 and T(H)17 cells. Nat Med 2008, 14:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson MC, Pierson ER, Spieker AJ, Nielsen AS, Posso S, Kita M, Buckner JH, Goverman JM: Distinct T cell signatures define subsets of patients with multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 2016, 3:e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havrdova E, Belova A, Goloborodko A, Tisserant A, Wright A, Wallstroem E, Garren H, Maguire RP, Johns DR: Activity of secukinumab, an anti-IL-17A antibody, on brain lesions in RRMS: results from a randomized, proof-of-concept study. J Neurol 2016, 263:1287–1295. [DOI] [PubMed] [Google Scholar]
- 35.Patel DD, Kuchroo VK: Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity 2015, 43:1040–1051. [DOI] [PubMed] [Google Scholar]
- 36.Esplugues E, Huber S, Gagliani N, Hauser AE, Town T, Wan YY, O’Connor W Jr., Rongvaux A, Van Rooijen N, Haberman, et al. : Control of TH17 cells occurs in the small intestine. Nature 2011, 475:514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Honda K, Littman DR: The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535:75–84. [DOI] [PubMed] [Google Scholar]
- 38.Korn T, Bettelli E, Oukka M, Kuchroo VK: IL-17 and Th17 Cells. Annu Rev Immunol 2009, 27:485–517. [DOI] [PubMed] [Google Scholar]
- 39.Omenetti S, Bussi C, Metidji A, Iseppon A, Lee S, Tolaini M, Li Y, Kelly G, Chakravarty P, Shoaie S, et al. : The Intestine Harbors Functionally Distinct Homeostatic Tissue-Resident and Inflammatory Th17 Cells. Immunity 2019, 51:77–89 e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, et al. : Dynamic regulatory network controlling TH17 cell differentiation. Nature 2013, 496:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Ni L, Wan S, Zhao X, Ding X, Dejean A, Dong C: Febrile Temperature Critically Controls the Differentiation and Pathogenicity of T Helper 17 Cells. Immunity 2020, 52:328–341 e325.• This paper established an interesting link between febrile temperatures and enhanced pathogenicity of Th17 cells through implication of SMAD4-driven Il17 expression.
- 42.Yoo SA, Kim M, Kang MC, Kong JS, Kim KM, Lee S, Hong BK, Jeong GH, Lee J, Shin MG, et al. : Placental growth factor regulates the generation of TH17 cells to link angiogenesis with autoimmunity. Nat Immunol 2019, 20:1348–1359.• This paper identified the angiogenic factor PlGF as a novel driver of Th17 cell differentiation and pathogenicity.
- 43.Samoilova EB, Horton JL, Hilliard B, Liu TS, Chen Y: IL-6-deficient mice are resistant to experimental autoimmune encephalomyelitis: roles of IL-6 in the activation and differentiation of autoreactive T cells. J Immunol 1998, 161:6480–6486. [PubMed] [Google Scholar]
- 44.Eugster HP, Frei K, Kopf M, Lassmann H, Fontana A: IL-6-deficient mice resist myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. Eur J Immunol 1998, 28:2178–2187. [DOI] [PubMed] [Google Scholar]
- 45.Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, Cua DJ: IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med 2015, 21:719–729. [DOI] [PubMed] [Google Scholar]
- 46.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. : A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006, 314:1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ: The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol 2009, 10:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer Zu Horste G, Wu C, Wang C, Cong L, Pawlak M, Lee Y, Elyaman W, Xiao S, Regev A, Kuchroo VK: RBPJ Controls Development of Pathogenic Th17 Cells by Regulating IL-23 Receptor Expression. Cell Rep 2016, 16:392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jain R, Chen Y, Kanno Y, Joyce-Shaikh B, Vahedi G, Hirahara K, Blumenschein WM, Sukumar S, Haines CJ, Sadekova S, et al. : Interleukin-23-Induced Transcription Factor Blimp-1 Promotes Pathogenicity of T Helper 17 Cells. Immunity 2016, 44:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buck MD, Sowell RT, Kaech SM, Pearce EL: Metabolic Instruction of Immunity. Cell 2017, 169:570–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haghikia A, Jorg S, Duscha A, Berg J, Manzel A, Waschbisch A, Hammer A, Lee DH, May C, Wilck N, et al. : Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 2015, 43:817–829. [DOI] [PubMed] [Google Scholar]
- 52.Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, Laidlaw BJ, Araki K, Ahmed R, Kaech SM, et al. : The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity 2015, 43:690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakaya M, Xiao Y, Zhou X, Chang JH, Chang M, Cheng X, Blonska M, Lin X, Sun SC: Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 2014, 40:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng M, Yin N, Chhangawala S, Xu K, Leslie CS, Li MO: Aerobic glycolysis promotes T helper 1 cell differentiation through an epigenetic mechanism. Science 2016, 354:481–484.• This reference showed how Th1 cells boost aerobic glycolysis and how it can mechanistically be coupled through epigenetic regulation to upregulation of Ifng expression.
- 55.Bailis W, Shyer JA, Zhao J, Canaveras JCG, Al Khazal FJ, Qu R, Steach HR, Bielecki P, Khan O, Jackson R, et al. : Distinct modes of mitochondrial metabolism uncouple T cell differentiation and function. Nature 2019, 571:403–407.•• This paper demonstrated how different components of mitochondrial metabolism can be utilized to achieve either Th1 cell proliferation or terminal effector function. In particular, the malate-aspartate shuttle and mitochondrial citrate export are needed for proliferation, including through production of aspartate. In addition, they enhance histone acetylation to influence epigenetically gene expression and therefore differentiation, whereas succinate dehydrogenase antagonizes proliferation to foster terminal effector function.
- 56.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, Snyder SH: Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 2018, 360:449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G: Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011, 479:538–541. [DOI] [PubMed] [Google Scholar]
- 58.Wu HJ, Ivanov, II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D: Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010, 32:815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. : Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, et al. : Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019, 576:143–148.•• This paper showed that bile acids are critical to the Th17/Treg cell balance in the intestine. 3-oxoLCA is shown to directly bind to RORγt to inhibit its ability to activate the expression of Il17 and that another bile acid metabolite, isoalloLCA, drives ROS to foster Treg cell development.
- 61.Bhargava P, Smith MD, Mische L, Harrington EP, Fitzgerald KC, Martin KA, Kim S, Reyes AAA, Gonzalez-Cardona J, Volsko C, et al. : Bile acid metabolism is altered in multiple sclerosis and supplementation ameliorates neuroinflammation. J Clin Invest 2020.•• This paper showed that bile acid metabolism is altered in MS patients and may be amenable to manipulation to achieve therapeutic benefit.
- 62.Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. : Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science 2017, 358:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HK, Han J: Tetrahydrobiopterin in energy metabolism and metabolic diseases. Pharmacol Res 2020:104827. [DOI] [PubMed] [Google Scholar]
- 64.Cronin SJF, Seehus C, Weidinger A, Talbot S, Reissig S, Seifert M, Pierson Y, McNeill E, Longhi MS, Turnes BL, et al. : The metabolite BH4 controls T cell proliferation in autoimmunity and cancer. Nature 2018, 563:564–568.• The authors showed that the metabolite BH4 (important co-factor for the hydroxylation of aromatic amino acids) is important for T cell proliferation and how it could be exploited to treat autoimmunity.
- 65.Lee JY, Hall JA, Kroehling L, Wu L, Najar T, Nguyen HH, Lin WY, Yeung ST, Silva HM, Li D, et al. : Serum Amyloid A Proteins Induce Pathogenic Th17 Cells and Promote Inflammatory Disease. Cell 2020, 180:79–91 e16.•• This paper showed the important effect of serum amyloid A proteins on the pathogenicity of Th17 cells and established a connection of the availability of these proteins to protect from CNS inflammatory disease.
- 66.Hirota K, Duarte JH, Veldhoen M, Hornsby E, Li Y, Cua DJ, Ahlfors H, Wilhelm C, Tolaini M, Menzel U, et al. : Fate mapping of IL-17-producing T cells in inflammatory responses. Nat Immunol 2011, 12:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gagliani N, Amezcua Vesely MC, Iseppon A, Brockmann L, Xu H, Palm NW, de Zoete MR, Licona-Limon P, Paiva RS, Ching T, et al. : Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature 2015, 523:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karmaus PWF, Chen X, Lim SA, Herrada AA, Nguyen TM, Xu B, Dhungana Y, Rankin S, Chen W, Rosencrance C, et al. : Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 2019, 565:101–105.•• This study identified subsets of Th17 cells, one which is characterized by a precursor/stem cell-like state through expression of TCF1 and an effector-like population that is characterized by T-bet and demonstrated how mTOR signaling controls this aspect of Th17 plasticity important for pathogenicity and in particular neuroinflammation.
- 69.Wu C, Chen Z, Xiao S, Thalhamer T, Madi A, Han T, Kuchroo V: SGK1 Governs the Reciprocal Development of Th17 and Regulatory T Cells. Cell Rep 2018, 22:653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakaguchi S, Mikami N, Wing JB, Tanaka A, Ichiyama K, Ohkura N: Regulatory T Cells and Human Disease. Annu Rev Immunol 2020. [DOI] [PubMed] [Google Scholar]
- 71.Pan F, Yu H, Dang EV, Barbi J, Pan X, Grosso JF, Jinasena D, Sharma SM, McCadden EM, Getnet D, et al. : Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science 2009, 325:1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM: Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med 2003, 198:1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Panduro M, Benoist C, Mathis D: Tissue Tregs. Annu Rev Immunol 2016, 34:609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sujino T, London M, Hoytema van Konijnenburg DP, Rendon T, Buch T, Silva HM, Lafaille JJ, Reis BS, Mucida D: Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science 2016, 352:1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ito M, Komai K, Mise-Omata S, Iizuka-Koga M, Noguchi Y, Kondo T, Sakai R, Matsuo K, Nakayama T, Yoshie O, et al. : Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 2019, 565:246–250.•• This study identified brain tissue Tregs that have the ability to suppress astrogliosis and that depend on IL-33/ST2. These brain Tregs can foster neurological recovery.
- 76.Dombrowski Y, O’Hagan T, Dittmer M, Penalva R, Mayoral SR, Bankhead P, Fleville S, Eleftheriadis G, Zhao C, Naughton M, et al. : Regulatory T cells promote myelin regeneration in the central nervous system. Nat Neurosci 2017, 20:674–680.•• This study decribed how tissue Tregs can contribute to neurological repair by secretion of oligodendrogenic Ccn3 in the setting of acute CNS demyelination.
- 77.Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, Whibley N, Tucci A, Chen X, Lindeman I, et al. : Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 2019, 50:493–504 e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, DiSpirito JR, Zemmour D, Spallanzani RG, Kuswanto W, Benoist C, Mathis D: TCR Transgenic Mice Reveal Stepwise, Multi-site Acquisition of the Distinctive Fat-Treg Phenotype. Cell 2018, 174:285–299 e212.• This paper described how tissue Treg phenotype is achieved in a stepwise process.
- 79.Delacher M, Imbusch CD, Hotz-Wagenblatt A, Mallm JP, Bauer K, Simon M, Riegel D, Rendeiro AF, Bittner S, Sanderink L, et al. : Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity 2020, 52:295–312 e211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sefik E, Geva-Zatorsky N, Oh S, Konnikova L, Zemmour D, McGuire AM, Burzyn D, Ortiz-Lopez A, Lobera M, Yang J, et al. : MUCOSAL IMMUNOLOGY. Individual intestinal symbionts induce a distinct population of RORgamma(+) regulatory T cells. Science 2015, 349:993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, et al. : Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature 2020, 577:410–415.•• This paper demonstrated that bile acids are critical to RORγt+FoxP3+ Tregs and contribute vitally to their ability to control intestinal inflammation.
- 82.Xu M, Pokrovskii M, Ding Y, Yi R, Au C, Harrison OJ, Galan C, Belkaid Y, Bonneau R, Littman DR: c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018, 554:373–377.•• This paper decribed the critical implication of c-MAF in intestinal Tregs to homeostasis and the control of tolerance integrating signals from the micobiota.
- 83.Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, Putzel GG, Kabata H, Kelsen JR, Baldassano RN, et al. : Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature 2019, 568:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MG: A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature 1997, 389:737–742. [DOI] [PubMed] [Google Scholar]
- 85.Chihara N, Madi A, Karwacz K, Awasthi A, Kuchroo VK: Differentiation and Characterization of Tr1 Cells. Curr Protoc Immunol 2016, 113:3 27 21–23 27 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D, Burns EJ, Sherr DH, Weiner HL, Kuchroo VK: The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol 2010, 11:854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gandhi R, Kumar D, Burns EJ, Nadeau M, Dake B, Laroni A, Kozoriz D, Weiner HL, Quintana FJ: Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3(+) regulatory T cells. Nat Immunol 2010, 11:846–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang P, Lee JS, Gartlan KH, Schuster IS, Comerford I, Varelias A, Ullah MA, Vuckovic S, Koyama M, Kuns RD, et al. : Eomesodermin promotes the development of type 1 regulatory T (TR1) cells. Sci Immunol 2017, 2 • This study identified Eomesodermin as an important transcription factor for the differentiation of Tr1 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gabrysova L, Alvarez-Martinez M, Luisier R, Cox LS, Sodenkamp J, Hosking C, Perez-Mazliah D, Whicher C, Kannan Y, Potempa K, et al. : c-Maf controls immune responses by regulating disease-specific gene networks and repressing IL-2 in CD4(+) T cells. Nat Immunol 2018, 19:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karwacz K, Miraldi ER, Pokrovskii M, Madi A, Yosef N, Wortman I, Chen X, Watters A, Carriero N, Awasthi A, et al. : Critical role of IRF1 and BATF in forming chromatin landscape during type 1 regulatory cell differentiation. Nat Immunol 2017, 18:412–421.• This study identified IRF1 and BATF as important pioneer factors that enable epigenetic changes to drive the transcriptional program of Tr1 cells.
- 91.Chihara N, Madi A, Kondo T, Zhang H, Acharya N, Singer M, Nyman J, Marjanovic ND, Kowalczyk MS, Wang C, et al. : Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature 2018, 558:454–459.•• The authors demonstrated how IL-27 is important for the induction of an ‘exhaustion’ module exemplified by expression of co-inhibitory receptors TIM-3, LAG-3, PD-1 and TIGIT driven by transcription factors PRDM1 and c-MAF.
- 92.DeLong JH, O’Hara Hall A, Rausch M, Moodley D, Perry J, Park J, Phan AT, Beiting DP, Kedl RM, Hill JA, et al. : IL-27 and TCR Stimulation Promote T Cell Expression of Multiple Inhibitory Receptors. Immunohorizons 2019, 3:13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joller N, Lozano E, Burkett PR, Patel B, Xiao S, Zhu C, Xia J, Tan TG, Sefik E, Yajnik V, et al. : Treg cells expressing the coinhibitory molecule TIGIT selectively inhibit proinflammatory Th1 and Th17 cell responses. Immunity 2014, 40:569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McKinney EF, Lee JC, Jayne DR, Lyons PA, Smith KG: T-cell exhaustion, co-stimulation and clinical outcome in autoimmunity and infection. Nature 2015, 523:612–616.•• This paper decribed how the relapse rate in autoimmune disease patients correlates with the expression of co-inhibitory receptors. The abundance of cells with such a phenotype may therefore be a critical clinical parameter of treatment benefit.
- 95.Okamura T, Fujio K, Shibuya M, Sumitomo S, Shoda H, Sakaguchi S, Yamamoto K: CD4+CD25-LAG3+ regulatory T cells controlled by the transcription factor Egr-2. Proc Natl Acad Sci U S A 2009, 106:13974–13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu C, Sakuishi K, Xiao S, Sun Z, Zaghouani S, Gu G, Wang C, Tan DJ, Wu C, Rangachari M, et al. : An IL-27/NFIL3 signalling axis drives Tim-3 and IL-10 expression and T-cell dysfunction. Nat Commun 2015, 6:6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. : Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500:232–236. [DOI] [PubMed] [Google Scholar]
- 98.Berer K, Gerdes LA, Cekanaviciute E, Jia X, Xiao L, Xia Z, Liu C, Klotz L, Stauffer U, Baranzini SE, et al. : Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci U S A 2017, 114:10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brucklacher-Waldert V, Carr EJ, Linterman MA, Veldhoen M: Cellular Plasticity of CD4+ T Cells in the Intestine. Front Immunol 2014, 5:488. [DOI] [PMC free article] [PubMed] [Google Scholar]