Abstract
Blood production is essential to maintain human health, and even small perturbations in hematopoiesis can cause disease. Hematopoiesis has therefore been the focus of much research for many years. Experiments determining the lineage potentials of hematopoietic stem and progenitor cells (HSPCs) in vitro and after transplantation revealed a hierarchy of progenitor cell states, where differentiating cells undergo lineage commitment – a series of irreversible changes that progressively restrict their potential. New technologies have recently been developed that allow for a more detailed analysis of the molecular states and fates of differentiating HSPCs. Proteomic and lineage-tracing approaches, alongside single cell transcriptomic analyses have recently helped to reveal the biological complexity underlying lineage commitment during hematopoiesis. Recent insights from these new technologies were presented by Drs. Marjorie Brand and Allon Klein in the Summer 2019 ISEH Webinar, and are discussed in this Perspective.
Keywords: Hematopoiesis, hematopoietic stem cell, hematopoietic stem and progenitor cell, lineage commitment, lineage tracing, single cell biology, proteomics
Introduction:
The hematopoietic system plays a major role in human health and disease, supplying oxygen and nutrients to tissues, supporting healing, and fighting infections. These essential functions are carried out by the various types of mature hematopoietic cells, including red blood cells, platelets, myeloid immune cells (macrophages, neutrophils), and lymphocytes (T cells, B cells, natural killer [NK] cells). Most of the mature cells lack the ability to proliferate and have limited lifespans, so they must be constantly replenished by a process called hematopoiesis (Eaves, 2015; Orkin and Zon, 2008). Disturbance in the homeostasis of this process results in hematological diseases such as leukemias, lymphomas, anemias, thrombocytopenias, and immunodeficiencies. Hematopoiesis has therefore been the focus of considerable experimental research for many years.
Hematopoiesis is sustained by rare hematopoietic stem cells (HSCs) that have two definitive characteristics (Eaves, 2015; Orkin and Zon, 2008). HSCs can self-renew, dividing to produce new HSC daughter cells to maintain lifelong hematopoiesis. HSCs are also multipotent, i.e. they have the ability to differentiate into any of the adult hematopoietic cell lineages. To produce mature blood cells, the progeny of HSCs undergo lineage commitment, a process of differentiation where the potential to produce all hematopoietic cell types is progressively lost until they become restricted to forming one type of blood cell. The molecular mechanisms, cellular relationships and timing of lineage commitment are fundamental to the regulation of blood production in homeostasis and in disease.
Recently developed technologies including lineage tracing, single-cell transcriptomics, and proteomics, have provided important new insights into lineage commitment during hematopoiesis. “Changing concepts in hematopoietic lineage commitment” was the focus of the Summer 2019 International Society for Experimental Hematology (ISEH) New Investigator Committee Webinar, presented by Drs. Marjorie Brand and Allon Klein, and moderated by Dr. Stephen Loughran. Dr. Brand discussed recent progress in using proteomic approaches to trace lineage commitment while Dr. Klein covered how novel lineage tracing methods in combination with single-cell transcriptomics were uncovering new cell fate trajectories. In this Perspective, we provide a brief summary of the classical view of hematopoiesis and a discussion of the topics covered by this recent webinar, which can also be watched online (https://www.youtube.com/watch?v=RqUsYsXqFfA).
Hierarchical lineage commitment revealed by measuring the potential of isolated HSPCs
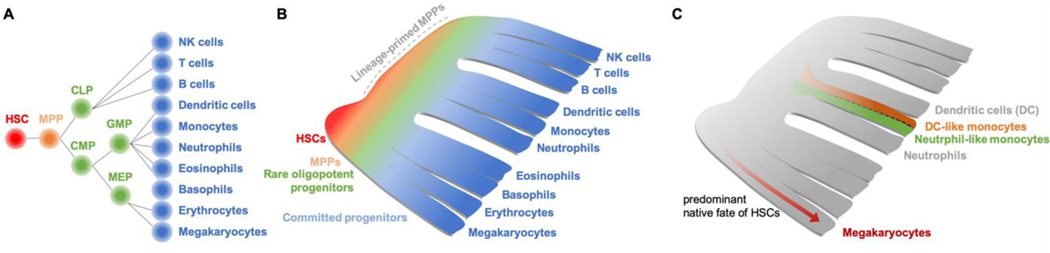
The differentiation potential of various HSPCs was determined over many years of experimentation, using in vitro colony assays and transplantation of prospectively isolated cells into myeloablated mice (Eaves, 2015; Seita and Weissman, 2010). This allowed HSPCs with varying potentials to be fitted into a cellular hierarchy with HSCs at the apex and mature blood cell types at the base. Hematopoiesis is therefore often depicted as a process of branching transitions between phenotypically identifiable cell states, with lineage commitment occurring during these transitions (Weissman et al., 2001) (Figure 1A).
Figure 1: Models of hematopoietic stem cell lineage commitment.
(A) The classical hematopoietic tree model, which suggested lineage commitment associated with branching transitions between cells with distinct lineage potentials.
(B) A schematic representation of hematopoiesis based on recent findings discussed within the review, which suggest that HSC differentiation occurs via continuum with phenotypically defined progenitor populations representing heterogeneous lineage potential.
(C) Lineage tracing in combination with single cell analyses have suggested that (1) the predominant fate of HSCs in native hematopoiesis is megakaryopoiesis and (2) there may be multiple lineage trajectories that generate equivalent mature hematopoietic cell types. For example, monocytes may be generated via dendritic (DC)-like or neutrophil-like trajectories.
Studies in the early and late 2000s revealed further complexity, demonstrating considerable functional and molecular variability between cells with similar cell surface marker phenotypes, and alternative lineage commitment pathways. These included single-cell transplantation, label-retention assays, and molecular analyses, which identified tremendous heterogeneity within the HSC pool, including variability in long-term reconstitution capacity, lineage biases, cell cycle activity and proliferative history of individual HSCs (Adolfsson et al., 2005; Beerman et al., 2010; Benz et al., 2012; Challen et al., 2010; Copley et al., 2012; Dykstra et al., 2007; Månsson et al., 2007; Morita et al., 2010; Muller-Sieburg et al., 2004; Müller-Sieburg et al., 2002; Wilson et al., 2008). Moreover, paired-daughter transplant experiments revealed that a subpopulation of HSCs could produce one multipotent daughter cell, and one lineage committed daughter, indicating that some multipotent stem cells can undergo lineage commitment within a single division, ‘by-passing’ certain differentiation stages (Yamamoto et al., 2013). However, it is worth noting that lineage-tracing using Flk2-Cre mice suggested that at steady state (non-transplantation settings), the majority of RBCs and platelets derive from a Flk2-expressing MPP precursors, rather than from the Flk2-negative HSC pool (Boyer et al., 2011).
New insights from lineage-tracing and single-cell approaches:
More recently, single-cell and lineage tracing approaches have provided additional insights into HSC lineage commitment, and have also been extensively reviewed elsewhere (Haas et al., 2018; Jacobsen and Nerlov, 2019; Laurenti and Göttgens, 2018; Yamamoto et al., 2018a). In line with observations by Yamamoto et al., recent single-cell transcriptomic and functional studies have identified a considerable fraction of the murine and human HSC compartment that exclusively adopt a lineage-restricted megakaryocyte fate in vivo (Haas et al., 2015; Notta et al., 2016; Roch et al., 2015; Rodriguez-Fraticelli et al., 2018). It also appears that this fraction expands during aging in mice (Yamamoto et al., 2018b). Megakaryocyte-primed HSPCs are efficiently driven into maturation by inflammatory signals, resulting in enhanced platelet generation, and it has been suggested that these might serve as an emergency pool for rapid platelet generation in scenarios of high platelet demand, such as upon blood loss or during infection (Boettcher and Manz, 2017; Haas et al., 2015; Hirche et al., 2017).
It is worth noting that a recent study, performing quantitative analyses of the absolute production of mature cells in clonal in vivo assays, revealed a strong erythroid potential of HSCs and MPPs. (Boyer et al., 2019). Additionally, at least two studies have now identified plasticity in the lineage-restriction of certain self-renewing stem cell types; stem cells displaying platelet-restriction in primary transplantation recipients were capable of additional myeloid and lymphoid fates when differentiated in vitro (Carrelha et al., 2018), or in the case of aged-specific latent-HSCs, serial transplantation (Yamamoto et al., 2018b). Notably, this is different to the previously observed cell-autonomous lineage bias of HSCs and progenitors, where cells are predisposed towards one lineage, but not fully committed to it (Dykstra et al., 2007; Sanjuan-Pla et al., 2013).
Recent fate mapping studies of native hematopoiesis have also identified megakaryocyte-restricted output from HSCs (Rodriguez-Fraticelli et al., 2018). While early native hematopoiesis fate mapping studies suggested that a large proportion of HSCs were not contributing to steady-state hematopoiesis, these studies did not measure megakaryocyte output (Busch et al., 2015; Pei et al., 2017; Sun et al., 2014). In contrast, more recent in vivo lineage-tracing experiments that analysed megakaryocytic output have found that a large proportion of HSCs only produce megakaryocytic cells, suggesting that this is a predominant native fate of many HSCs that were previously thought to be dormant (Rodriguez-Fraticelli et al., 2018). One limitation of fate mapping studies is that it is not possible to distinguish a lineage committed cell from a lineage primed cell, or from a fully multipotent cell that is located in a microenvironment that only permits differentiation into a single lineage.
In the last four years, large-scale single-cell transcriptomics of the mouse, human and zebrafish hematopoietic stem and progenitor cell (HSPC) compartments have provided detailed insights into the transcriptomic landscape of hematopoiesis (Macaulay et al., 2016; Nestorowa et al., 2016; Pellin et al., 2019; Tusi et al., 2018; Velten et al., 2017; Zheng et al., 2018). These studies have suggested that at the mRNA level, hematopoiesis occurs as a continuum rather than by the acquisition of discrete transcriptional states (Figure 1B). Using these methods, discrete transcriptional patterns were only observed at the level of mature cell types (Macaulay et al., 2016; Nestorowa et al., 2016; Tusi et al., 2018; Velten et al., 2017). In trajectory analyses, early transcriptional lineage priming gradually separates erythroid-megakaryocyte-eosinophil-basophil-primed progenitors from lympho-myeloid-primed progenitors in mouse and human (Pellin et al., 2019; Tusi et al., 2018; Velten et al., 2017; Zheng et al., 2018). In later stages, lineage-specific gene expression programs are acquired, coinciding with functional lineage commitment (Velten et al., 2017). The early separation between erythroid-megakaryocyte-eosinophil-basophil and the lympho-myeloid lineages is also supported by single-cell transcriptomic and single-cell functional assays of downstream progenitor compartments in mouse and human (Drissen et al., 2016, 2019; Görgens et al., 2013).
However, these single cell transcriptomic datasets only represent snapshots of distinct stages of HSC commitment rather than a time-resolved picture of hematopoiesis. To overcome this limitation, new tools combining lineage-tracing and single-cell transcriptomics have recently been developed to address the question how accurately single cell RNA sequencing (scRNA-seq)-inferred hierarchies reflect actual fate choices, and to determine transcriptional states upstream of lineage commitment branchpoints (Weinreb et al., 2020). A new method, developed by Klein and colleagues, to overlay lineage relationships from barcoding analysis with single cell transcriptomics data is providing a powerful approach to interrogate how single HSCs (and their progeny) move through the continuous lineage differentiation models provided by single cell transcriptomics (Rodriguez-Fraticelli et al., 2018; Weinreb et al., 2020). First, these data reveal that hematopoietic differentiation is not a strict tree-like branching process; instead, some cell types appear to reflect more than one possible sequence of molecular events, leading to ‘loops’ as different branches of the tree converge (Figure 1C). This was most apparent for monocytes. Second, using sister cell experiments they reveal that cells with very similar gene expression profiles can nonetheless be pre-committed to different fates, suggesting that transcriptional circuits alone do not encode the potential of cells towards different fates. Combining CRISPR/Cas9 perturbation with large-scale single readouts (Dixit et al., 2016; Gundry et al., 2017; Jaitin et al., 2016) ex vivo and in vivo, is also likely to provide novel insights into the molecular mechanisms driving HSC lineage commitment.
Quantification of lineage-specific transcription factors in single cells during lineage commitment
The studies described above focused on linking cellular differentiation with patterns of mRNA expression. These transcriptional changes are orchestrated by transcription factor proteins that regulate gene expression (Wilkinson and Göttgens, 2013). The control of myeloid progenitor fate choice by GATA1 and PU.1 (SPI1) is a paradigmatic example of transcription factor levels instructing lineage commitment (Graf and Enver, 2009). GATA1 promotes the expression of the erythroid/megakaryocyte programme, and PU.1 promotes the granulocyte/monocyte programme (Ferreira et al., 2005; Rosmarin et al., 2005). Furthermore, GATA1 and PU.1 each upregulate their own expression, and inhibit the expression of the other transcription factor (Okuno et al., 2005; Tsai et al., 1991; Yu et al., 2002). These properties led to a model where low levels of GATA1 and PU.1 maintain myeloid progenitors in a metastable multipotential state until a developmental switch occurs when either GATA1 or PU.1 expression dominates, leading to the dominant transcription factor suppressing the expression of the other, while it forms an autoregulatory loop to further activate its own expression and drive lineage commitment (Cantor and Orkin, 2001; Graf, 2002). This model is supported by the levels of GATA1 and PU.1 mRNA detected in populations of cells during differentiation, and by studies where GATA1 or PU.1 overexpression were able to drive expression of erythroid/megakaryocyte or granulocyte/monocyte programs respectively in cells of the other lineage (Heyworth, 2002; Kulessa et al., 1995; Nerlov and Graf, 1998; Visvader et al., 1992). Other pairs of mutually antagonistic transcription factor switches that similarly control binary lineage commitment decisions during hematopoiesis have been identified, including KLF1 and FLI1, which promote erythroid or megakaryocyte commitment respectively (Bouilloux et al., 2008; Frontelo et al., 2007; Graf and Enver, 2009; Siripin et al., 2015).
Recent studies measuring dynamic changes in transcription factor abundance at the protein level within single cells have challenged these binary fate models. For example, Hoppe et al. quantified fluorescently tagged GATA1 and PU.1 in a large number of single differentiating HSCs and their progeny for several days (Hoppe et al., 2016). PU.1 was detected in all HSCs and uncommitted progenitors. GATA1 expression was not detected at any time during lineage commitment to the granulocyte/macrophage lineage. In granulocyte/macrophage differentiation events, PU.1 protein levels increased steadily during about half of the events, and in the other half PU.1 levels dipped transiently before undergoing a similar steady increase. Cells in which GATA1 was detected, even at low levels, invariably continued to express GATA1 and differentiated into GATA1+ PU.1– megakaryocytic and/or erythroid cells. During the majority of these megakaryocyte/erythroid differentiation events, downregulation of PU.1 occurred before detection of GATA1. These findings are incompatible with an abrupt GATA1 versus PU.1 binary switching event driving lineage commitment: uncommitted progenitors contained only PU.1, GATA1 detection was always associated with megakaryocyte/erythroid commitment, and changes in GATA1 and PU.1 protein levels during differentiation occurred gradually.
Another recent study by Brand and colleagues was the first to quantify the levels of endogenous lineage-specific transcription factors in single cells during hematopoietic differentiation. Mass cytometry time of flight (CyTOF) was used to simultaneously measure 11 cell surface proteins and 16 transcription factors in single human hematopoietic stem and progenitor cells and their progeny at 13 stages of in vitro erythroid development (Palii et al., 2019). Using this technique, KLF1 and FLI1 co-expression was detected in the majority of single MEPs, and GATA1 and PU.1 co-expression in the majority of CMPs. These data conflict with the lack of co-expression reported by Hoppe et al. and highlight potential limitations in the ability of live imaging of fluorescent labels to detect of proteins present at low levels. As MEPs underwent erythroid differentiation, KLF1 levels gradually increased, and FLI1 gradually decreased. Co-expression of KLF1 and FLI1 persisted for 14 days, until the pro-erythroblast stage of differentiation. This demonstrates that lineage commitment is not an abrupt transition from a metastable uncommitted state to one of two distinct lineage committed states initiated by the rapid switching of a pair of cross-antagonistic, autoregulatory transcription factors. Instead, these findings are consistent with lineage commitment occurring during a continuous process of differentiation where cells gradually transition along an ordered series of states. Changes in lineage-specific transcription factor levels during lineage commitment were gradual and continuous.
Under the same conditions, artificially increasing FLI1 protein levels in progenitors was sufficient to divert cells from their preferred erythroid trajectory to take on a megakaryocytic fate (Palii et al., 2019). This highlights the ability of quantitative changes in transcription factor protein levels to determine cell fate decisions, and strongly supports them being the main process that initiates lineage commitment, a finding also compatible with single cell RNA-seq data. However, because it is difficult to precisely determine when a cell undergoes lineage commitment, it is difficult to distinguish which changes in transcription factor levels initiate lineage commitment, and which are downstream events that reinforce a prior commitment decision. The granulocyte-macrophage versus erythroid-megakaryocyte lineage commitment decision timepoint, computationally inferred from the genealogy of single-cell-tracked differentiating HSCs, occurred much earlier than changes in the level of fluorescently-tagged PU.1 (Strasser et al., 2018). This suggests that PU.1 protein levels do not initiate this lineage commitment decision, and demonstrates that further work is required to determine which transcription factors are responsible.
Conclusions:
The development and application of new technologies to study hematopoiesis are creating a more complex but more complete understanding of hematopoietic lineage commitment. This hematopoietic lineage commitment landscape has now been mapped by different technologies in several distinct, but related, ways. One type of map shows the lineage potential of HSPCs – what lineages can isolated cells differentiate into if placed in appropriate conditions? Another is a map of lineage fates – what lineages do HSPCs produce in situ? The third maps the molecular state of individual HSPCs in particular tissues at snapshots in time. Recent single cell analyses of hematopoiesis at both the mRNA and protein levels have revealed a continuum of states in hematopoiesis, suggesting that the molecular changes that underlie lineage commitment occur on a time scale comparable to the lifetime of mRNA molecules - hours or days. However, little is known about how rapidly individual cells move between these molecular states during steady state hematopoiesis, and it is not yet possible to accurately predict the lineage potential of a single cell from its transcriptome. To better understand the regulation of lineage commitment, links between the three types of map must be discovered – associating the molecular profile of individual cells to their potential, and to the fate they are most likely to take in the bone marrow. Further characterization of transcription factor protein levels and genomic binding in single cells across different stages of differentiation, and new methods combining cell fate tracking and single cell transcriptomics, will provide new information to help address this, and allow the development of more accurate models of the molecular regulation of hematopoietic lineage commitment. These findings have important implications for how we interpret the perturbations in hematopoiesis that underlie hematological diseases as well as our efforts to develop new therapies for these diseases.
Highlights:
Review of cellular lineage commitment within hematopoiesis
Discusses insights from novel lineage tracing and single cell transcriptomics studies
Describes how single cell proteomics have resolved cell state transitions in hematopoiesis
Acknowledgements:
We thank the ISEH staff and New Investigator Committee for their support. We thank the ISEH New Investigator Committee and the ISEH staff for their support. ACW is supported by the Leukemia & Lymphoma Society (3385-19) and the NIH (K99HL150218).
Footnotes
Authorship statement:
All authors contributed to writing and editing the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge O-J, Thoren LAM, et al. (2005). Identification of Flt3+ Lympho-Myeloid Stem Cells Lacking Erythro-Megakaryocytic Potential. Cell 121, 295–306. [DOI] [PubMed] [Google Scholar]
- Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, and Rossi DJ (2010). Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc. Natl. Acad. Sci. 107, 5465–5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C, Copley MR, Kent DG, Wohrer S, Cortes A, Aghaeepour N, Ma E, Mader H, Rowe K, Day C, et al. (2012). Hematopoietic Stem Cell Subtypes Expand Differentially during Development and Display Distinct Lymphopoietic Programs. Cell Stem Cell 10, 273–283. [DOI] [PubMed] [Google Scholar]
- Boettcher S, and Manz MG (2017). Regulation of Inflammation- and Infection-Driven Hematopoiesis. Trends Immunol. 38, 345–357. [DOI] [PubMed] [Google Scholar]
- Bouilloux F, Juban G, Cohet N, Buet D, Guyot B, Vainchenker W, Louache F, and Morlé F. (2008). EKLF restricts megakaryocytic differentiation at the benefit of erythrocytic differentiation. Blood 112, 576–584. [DOI] [PubMed] [Google Scholar]
- Boyer SW, Schroeder AV, Smith-Berdan S, and Forsberg EC (2011). All Hematopoietic Cells Develop from Hematopoietic Stem Cells through Flk2/Flt3-Positive Progenitor Cells. Cell Stem Cell 9, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer SW, Rajendiran S, Beaudin AE, Smith-Berdan S, Muthuswamy PK, Perez-Cunningham J, Martin EW, Cheung C, Tsang H, Landon M, et al. (2019). Clonal and Quantitative In Vivo Assessment of Hematopoietic Stem Cell Differentiation Reveals Strong Erythroid Potential of Multipotent Cells. Stem Cell Reports 12, 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, and Rodewald H-R (2015). Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518, 542–546. [DOI] [PubMed] [Google Scholar]
- Cantor AB, and Orkin SH (2001). Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11, 513–519. [DOI] [PubMed] [Google Scholar]
- Carrelha J, Meng Y, Kettyle LM, Luis TC, Norfo R, Alcolea V, Grasso F, Gambardella A, Grover A, Högstrand K, et al. (2018). Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 1–26. [DOI] [PubMed] [Google Scholar]
- Challen GA, Boles NC, Chambers SM, and Goodell MA (2010). Distinct Hematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-β1. Cell Stem Cell 6, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley MR, Beer PA, and Eaves CJ (2012). Hematopoietic Stem Cell Heterogeneity Takes Center Stage. Cell Stem Cell 10, 690–697. [DOI] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Buza-Vidas N, Woll P, Thongjuea S, Gambardella A, Giustacchini A, Mancini E, Zriwil A, Lutteropp M, Grover A, et al. (2016). Distinct myeloid progenitor–differentiation pathways identified through single-cell RNA sequencing. Nat. Immunol. 17, 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R, Thongjuea S, Theilgaard-Mönch K, and Nerlov C. (2019). Identification of two distinct pathways of human myelopoiesis. Sci. Immunol. 4, eaau7148. [DOI] [PubMed] [Google Scholar]
- Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, Lee SJ, Brinkman R, and Eaves C. (2007). Long-Term Propagation of Distinct Hematopoietic Differentiation Programs In Vivo. Cell Stem Cell 1, 218–229. [DOI] [PubMed] [Google Scholar]
- Eaves CJ (2015). Hematopoietic stem cells: Concepts, definitions, and the new reality. Blood 125, 2605–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R, Ohneda K, Yamamoto M, and Philipsen S. (2005). GATA1 Function, a Paradigm for Transcription Factors in Hematopoiesis. Mol. Cell. Biol. 25, 1215–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frontelo P, Manwani D, Galdass M, Karsunky H, Lohmann F, Gallagher PG, and Bieker JJ (2007). Novel role for EKLF in megakaryocyte lineage commitment. Blood 110, 3871–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görgens A, Radtke S, Möllmann M, Cross M, Dürig J, Horn PA, and Giebel B. (2013). Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. Cell Rep. 3, 1539–1552. [DOI] [PubMed] [Google Scholar]
- Graf T. (2002). Differentiation plasticity of hematopoietic cells. Blood 99, 3089–3101. [DOI] [PubMed] [Google Scholar]
- Graf T, and Enver T. (2009). Forcing cells to change lineages. Nature 462, 587–594. [DOI] [PubMed] [Google Scholar]
- Gundry MC, Dever DP, Yudovich D, Bauer DE, Haas S, Wilkinson AC, and Singbrant S. (2017). Technical Considerations for the Use of CRISPR/Cas9 in Hematology Research. Exp. Hematol. 54, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S, Hansson J, Klimmeck D, Loeffler D, Velten L, Uckelmann H, Wurzer S, Prendergast ÁM, Schnell A, Hexel K, et al. (2015). Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell 17, 422–434. [DOI] [PubMed] [Google Scholar]
- Haas S, Trumpp A, and Milsom MD (2018). Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell 22, 627–638. [DOI] [PubMed] [Google Scholar]
- Heyworth C. (2002). Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 21, 3770–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirche C, Frenz T, Haas SF, Döring M, Borst K, Tegtmeyer P-K, Brizic I, Jordan S, Keyser K, Chhatbar C, et al. (2017). Systemic Virus Infections Differentially Modulate Cell Cycle State and Functionality of Long-Term Hematopoietic Stem Cells In Vivo. Cell Rep. 19, 2345–2356. [DOI] [PubMed] [Google Scholar]
- Hoppe PS, Schwarzfischer M, Loeffler D, Kokkaliaris KD, Hilsenbeck O, Moritz N, Endele M, Filipczyk A, Gambardella A, Ahmed N, et al. (2016). Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature 535, 299–302. [DOI] [PubMed] [Google Scholar]
- Jacobsen SEW, and Nerlov C. (2019). Haematopoiesis in the era of advanced single-cell technologies. Nat. Cell Biol. 21, 2–8. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, and Amit I. (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- Kulessa H, Frampton J, and Graf T. (1995). GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9, 1250–1262. [DOI] [PubMed] [Google Scholar]
- Laurenti E, and Göttgens B. (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Svensson V, Labalette C, Ferreira L, Hamey F, Voet T, Teichmann SA, and Cvejic A. (2016). Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep. 14, 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thorén L, Adolfsson J, et al. (2007). Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity 26, 407–419. [DOI] [PubMed] [Google Scholar]
- Morita Y, Ema H, and Nakauchi H. (2010). Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J. Exp. Med. 207, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Sieburg CE, Cho RH, Karlsson L, Huang J-F, and Sieburg HB (2004). Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood 103, 4111–4118. [DOI] [PubMed] [Google Scholar]
- Müller-Sieburg CE, Cho RH, Thoman M, Adkins B, and Sieburg HB (2002). Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood 100, 1302–1309. [PubMed] [Google Scholar]
- Nerlov C, and Graf T. (1998). PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, Wilson NK, Kent DG, and Göttgens B. (2016). A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 128, e20–e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Zandi S, Takayama N, Dobson S, Gan OI, Wilson G, Kaufmann KB, McLeod J, Laurenti E, Dunant CF, et al. (2016). Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science (80-. ). 351, aab2116–aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Huang G, Rosenbauer F, Evans EK, Radomska HS, Iwasaki H, Akashi K, Moreau-Gachelin F, Li Y, Zhang P, et al. (2005). Potential Autoregulation of Transcription Factor PU.1 by an Upstream Regulatory Element. Mol. Cell. Biol. 25, 2832–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, and Zon LI (2008). Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell 132, 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palii CG, Cheng Q, Gillespie MA, Shannon P, Mazurczyk M, Napolitani G, Price ND, Ranish JA, Morrissey E, Higgs DR, et al. (2019). Single-Cell Proteomics Reveal that Quantitative Changes in Co-expressed Lineage-Specific Transcription Factors Determine Cell Fate. Cell Stem Cell 24, 812–820.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rössler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, et al. (2017). Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellin D, Loperfido M, Baricordi C, Wolock SL, Montepeloso A, Weinberg OK, Biffi A, Klein AM, and Biasco L. (2019). A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 10, 2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch A, Trachsel V, and Lutolf MP (2015). Brief Report: Single-Cell Analysis Reveals Cell Division-Independent Emergence of Megakaryocytes From Phenotypic Hematopoietic Stem Cells. Stem Cells 33, 3152–3157. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, and Camargo FD (2018). Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmarin AG, Yang Z, and Resendes KK (2005). Transcriptional regulation in myelopoiesis: Hematopoietic fate choice, myeloid differentiation, and leukemogenesis. Exp. Hematol. 33, 131–143. [DOI] [PubMed] [Google Scholar]
- Sanjuan-Pla A, Macaulay IC, Jensen CT, Woll PS, Luis TC, Mead A, Moore S, Carella C, Matsuoka S, Jones TB, et al. (2013). Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature 502, 232–236. [DOI] [PubMed] [Google Scholar]
- Seita J, and Weissman IL (2010). Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripin D, Kheolamai P, U-Pratya Y, Supokawej A, Wattanapanitch M, Klincumhom N, Laowtammathron C, and Issaragrisil S. (2015). Transdifferentiation of erythroblasts to megakaryocytes using FLI1 and ERG transcription factors. Thromb. Haemost. 114, 593–602. [DOI] [PubMed] [Google Scholar]
- Strasser MK, Hoppe PS, Loeffler D, Kokkaliaris KD, Schroeder T, Theis FJ, and Marr C. (2018). Lineage marker synchrony in hematopoietic genealogies refutes the PU.1/GATA1 toggle switch paradigm. Nat. Commun. 9, 2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho Y-JJ, Klein A, Hofmann O, and Camargo FD (2014). Clonal dynamics of native haematopoiesis. Nature 514, 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Strauss E, and Orkin SH (1991). Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 5, 919–931. [DOI] [PubMed] [Google Scholar]
- Tusi BK, Wolock SL, Weinreb C, Hwang Y, Hidalgo D, Zilionis R, Waisman A, Huh JR, Klein AM, and Socolovsky M. (2018). Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten L, Haas SF, Raffel S, Blaszkiewicz S, Islam S, Hennig BP, Hirche C, Lutz C, Buss EC, Nowak D, et al. (2017). Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 19, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Elefanty AG, Strasser A, and Adams JM (1992). GATA-1 but not SCL induces megakaryocytic differentiation in an early myeloid line. EMBO J. 11, 4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreb C, Rodriguez-Fraticelli A, Camargo FD, and Klein AM (2020). Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Anderson DJ, and Gage F. (2001). Stem and Progenitor Cells: Origins, Phenotypes, Lineage Commitments, and Transdifferentiations. Annu. Rev. Cell Dev. Biol. 17, 387–403. [DOI] [PubMed] [Google Scholar]
- Wilkinson AC, and Göttgens B. (2013). Transcriptional Regulation of Haematopoietic Stem Cells. pp. 187–212. [DOI] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. (2008). Hematopoietic Stem Cells Reversibly Switch from Dormancy to Self-Renewal during Homeostasis and Repair. Cell 135, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, and Nakauchi H. (2013). Clonal Analysis Unveils Self-Renewing Lineage-Restricted Progenitors Generated Directly from Hematopoietic Stem Cells. Cell 154, 1112–1126. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Wilkinson AC, and Nakauchi H. (2018a). Changing concepts in hematopoietic stem cells. Science (80-. ). 362, 895–896. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Wilkinson AC, Ooehara J, Lan X, Lai C-Y, Nakauchi Y, Pritchard JK, and Nakauchi H. (2018b). Large-Scale Clonal Analysis Resolves Aging of the Mouse Hematopoietic Stem Cell Compartment. Cell Stem Cell 22, 600–607.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, and Orkin SH (2002). Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 195, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Papalexi E, Butler A, Stephenson W, and Satija R. (2018). Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol. Syst. Biol. 14, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]