Abstract
Background
Rhinovirus (RV) is the most common pathogen involved in asthma, and COVID-19, caused by SARS-COV-2, may be more severe in asthma patients. Here, we applied integrated bioinformatics to identify potential key genes and cytokine pathways after RV infection in asthma, and analyzed changes in angiotensin-converting enzyme 2 (ACE2), the cellular receptor of SARS-COV-2.
Material/Methods
The gene expression profile dataset GSE149273 was downloaded from NCBI-GEO, which included 90 samples of non-infected, RVA, and RVC. Differentially expressed genes (DEGs) were identified using t tests in the limma R package, and subsequently investigated by GO, KEGG, and DO analysis. Moreover, the expression of ACE2 and the proportion of immune cells were further analyzed to determine the effects of RV on cytokines.
Results
A total of 555 DEGs of RVA and 421 of RVC were identified. There were 415 DEGs in RVA and RVC, of which 406 were upregulated and 9 were downregulated. The functional enrichment analysis showed that most DEGs were obviously enriched in cytokines, and were mainly enriched in “influenza” and “hepatitis C, chronic”. In addition, the expression of ACE2 increased significantly and the proportion of immune cytokines significantly changed after RV infection. Our results suggest that RV can activate the cytokine pathway associated with COVID-19 by increasing ACE2.
Conclusions
The DEGs and related cytokine pathways after asthma RV infection identified using integrated bioinformatics in this study elucidate the potential link between RV and COVID-19.
MeSH Keywords: Computational Biology, COVID-19, Rhinovirus
Background
Respiratory virus infections are closely associated with the exacerbation of asthma, in which rhinovirus (RV) is the most common pathogen [1]. Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-COV-2), may be more serious in patients with chronic lung diseases, including asthma [2]. In asthma patients, biological factors may affect the susceptibility to SARS-COV-2 or the severity of COVID-19. Kim et al. found that RV infection was also the most common additional respiratory pathogen in 116 SARS-COV-2-positive specimens [3]. It is well known that there is a great diversity in RV, in which RV-B infection usually causes mild or asymptomatic symptoms, and RV-A or RV-C are usually associated with more severe illness or exacerbation of asthma [4]. RV infection induces the expression of interferon (IFN)-stimulated genes and subsequent production of cytokines [5].
Angiotensin-converting enzyme 2 (ACE2), a transmembrane protein with an extracellular carboxypeptidase domain located on the cell membrane of a variety of epithelial cells, was originally described as playing a role in the renin-angiotensin system [6]. The various roles of ACE2 have gradually been discovered [7]. It participates in the regulation of cardiovascular physiology, dietary amino acid homeostasis, innate immunity, and intestinal microbial ecology, and plays an important role in heart failure, hypertension, myocardial infarction, and cardiovascular complications of diabetes [8]. It is also a cellular receptor for SARS-COVs, including SARS-COV-2 which caused the COVID-19 epidemic [9]. The spike protein of virus binds to the ACE2 of the host cell, mediating viral invasion of cell membranes and transmission to other cells [10]. Therefore, we speculated that there are differences in ACE2 gene expression in RV-infected asthma patients, and it may be associated with severe COVID19 infections.
Gene expression microarray, as an efficient and large-scale technique to obtain genetic data, is widely used to study the gene expression profile of diseases and plays an important role in screening differentially expressed genes (DEGs) [11]. With the wider application of this technology, a large quantity of microarray and sequencing data has been collated and stored in the public database platforms, which are available for free download. Further analysis and integration of the data through relevant statistical software or online analysis tools can lead to more in-depth study of molecular mechanisms of disease, providing a new method for the study of disease-related genes. However, the results of different studies on the identification of DEGs are not completely consistent, and there are still some limitations in single-cohort studies. Many studies have identified the DEGs of different diseases, including ovarian cancer [12], leukemia [13], and gastric cancer [14]. The identification of DEGs has been widely used in the analysis of various types of diseases and can provide effective therapeutic targets for the disease.
In this study, the original microarray dataset GSE149273 [15] was downloaded from the NCBI-Gene Expression Omnibus (GEO), which contained a total of 90 samples. DEGs were identified in RV-A16 infections (RVA) or RV-C15 infections (RVC) compared to non-infection (control) in patients with asthma. Additionally, gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and Disease ontology (DO) analysis were performed to identify the intersecting DEGs of RVA and RVC. The gene expression of ACE2 and the proportion of 22 kinds of immune cells in each sample were further analyzed to determine the effects of FVA or FVC infection on cytokines. Overall, the findings of the present study highlighted key candidate genes and cytokine pathways in RV infection in people with asthma. These may provide a reliable molecular marker to better understand RV infection and analyze its relationship with COVID-19.
Material and Methods
Dataset sources
The GSE149273 profile dataset was downloaded from the NCBI-GEO database (https://www.ncbi.nlm.nih.gov/geo/). The platform for GSE149273 is GPL21290 Illumina HiSeq 3000 (Homo sapiens), which includes 90 samples from 30 asthma patients with non-infection, RVA, and RVC. Platform and series matrix file(s) were downloaded as TXT files.
Expression analysis of DEGs
The original data were converted into an expression matrix, and then the mRNA expressions of 90 samples were normalized using the R software package with the robust multi-array average algorithm. The t test method in the limma R package was subsequently used to identify DEGs between RVA and RVC infected samples and non-infected samples, respectively. The log fold change >2 and P<0.05 were considered as the screening criteria for DEGs identification. The intersection of the analysis results of DEGs of RVA and RVC was demonstrated using VennDiagram in the R package, and the heatmap.2 of R package was used to draw the heatmaps of RVC, RVA, and control groups.
Functional analyses
To further explore the biological processes and signal pathways in which the intersecting DEGs of RVA and RVC might be involved, functional analyses of the DEGs at the intersection of RVA and RVC were performed. GO analysis was performed to demonstrate the dominant function of the intersecting DEGs from molecular function and biological process using the clusterProfiler R package. KEGG pathway analysis was performed to find the correlation between the intersecting DEGs and signal pathways using the clusterprofiler R package. DO analysis was performed to explore the relationship between the intersecting DEGs and diseases using DisGeNET data (https://www.disgenet.org/). All the results were visualized, and P<0.05 and gene counts >5 were considered to show a statistically significant difference in this functional enrichment analysis.
Analysis of ACE2 and immune cells
The t test was used to analyze the differences in gene expression of ACEs between RVA infection or RVC infection and control. P<0.05 was statistically significant. Moreover, the proportion of 22 immune cells in each sample was estimated to analyze the infiltration of immune cells after infection using cibersortx (https://cibersortx.stanford.edu/index.php), and a heatmap was drawn. The vioplot in the R package was used to visualize the change in the proportion of 22 immune cells relative to the control after RVA or RVC infection. P<0.05 was considered statistically significant.
Results
Data information and identification of DEGs
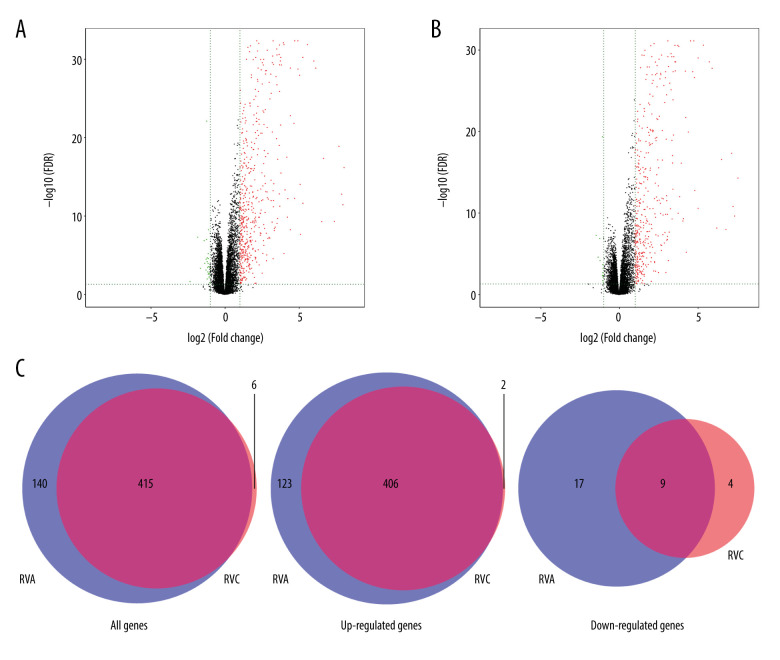
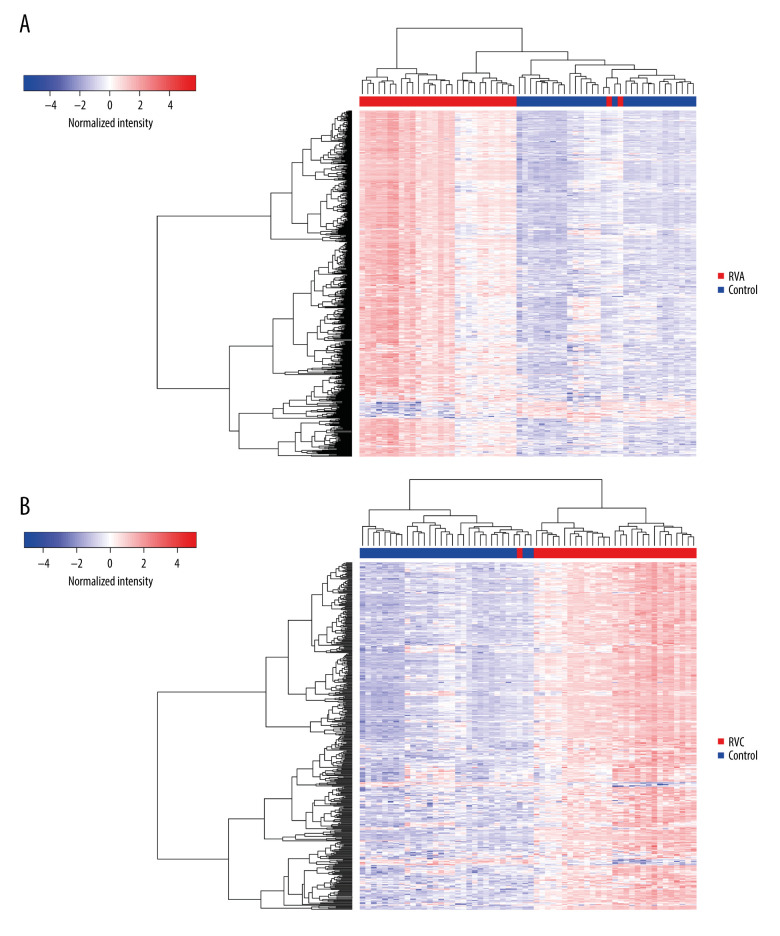
The GSE149273 profile dataset consisted of 30 patients with a median age of 35 years and a balanced sex ratio, 60% of whom were non-Hispanic white individuals. Thirty individuals were treated with non-infection, RVA, and RVC infection respectively, and 90 samples were obtained. The mRNA expression levels of the 90 samples in the microarray dataset GSE149273 were standardized, and the results are shown in Figure 1. The GSE149273 dataset was screened using the limma R package (logFC >2 and P<0.05), and 555 DEGs were obtained between RVA and control and 421 DEGs between RVC and control. Among them, 529 upregulated genes and 26 downregulated genes were identified in RVA, while 408 upregulated genes and 13 downregulated genes were classified in RVC. In addition, there were a total of 415 DEGs both in RVA and RVC, among which 406 were upregulated and 9 were downregulated (Table 1). The differential expression of multiple genes from FVA or FVC and control in the microarray is shown in Figure 2A and 2B, and the intersection of RVA and RVC DEGs is shown in Figure 2C. The cluster heatmaps of FVA and FVC are shown in Figure 3.
Figure 1.
Standardization of gene expression.
Table 1.
415 DEGs in the intersection of FVA and FVC.
| DEGs | Genes |
|---|---|
| Upregulated | IRF7, MX2, CMPK2, TRIM25, OAS3, IFIT5, DDX60L, RSAD2, MX1, IFI16, ADAR, USP18, STAT1, PNPT1, TRIM14, OAS1, OAS2, XAF1, IFIT1, DHX58, DDX58, CMTR1, RTP4, PARP12, SP100, EPSTI1, IFIT2, PARP9, IFI35, HERC6, LAMP3, UBE2L6, SAMD9L, PLSCR1, HERC5, OASL, IFIT3, C19orf66, IFI44, TRANK1, SP110, HSH2D, DTX3L, ISG15, IFIH1, ZC3HAV1, PARP10, TRIM21, DDX60, PARP14, IFITM1, NMI, SLC25A28, ISG20, ETV7, STAT2, NLRC5, TRIM5, MYD88, TDRD7, SMCHD1, XRN1, TRIM22, PHF11, SAMD9, GTPBP2, RNF213, NT5C3A, LAP3, ZCCHC2, FAM46A, IFI44L, TREX1, SCO2, GTPBP1, PML, FBXO6, TOR1B, PLEKHA4, UBA7, AIM2, TNFSF10, TAP1, NUB1, GBP4, LMO2, MOV10, APOL6, TAPSAR1, GBP1, SCAMP1-AS1, STARD5, EIF2AK2, N4BP1, BST2, TYMP, IFITM3, IRF9, HELZ2, RNF19B, HLA-E, MOB3C, LGALS9, CNP, SPATS2L, ZNFX1, GMPR, OPTN, CXCL10, C5orf56, RBCK1, APOBEC3G, HDX, CSRNP1, SECTM1, PRKD2, PSMB8, FBXO39, TTC39B, PSMB9, WARS, IRF1, BATF2, TRIM38, LGALS17A, LY6E, IFI6, JAK2, CLEC7A, SAMHD1, SP140L, EXOC3L1, PI4K2B, NUPR1, SIDT1, PCGF5, IFNL3, CASP1, ACSL1, EHD4, CD68, TAP2, SLFN5, TLR3, LOC101927027, TRIB2, LIFR, GRIP2, ACE2, MASTL, BCL2L14, TRAFD1, PRICKLE4, APOL2, BCL3, GBP5, BTC, PIK3AP1, SLC15A3, NFE2L3, ZBP1, ZFYVE26, RASGRP3, TMEM140, ZC3H12A, KIAA1551, CTRL, MLKL, NFKBIA, LYSMD2, MAB21L2, GNB4, C17orf67, GBP1P1, DUOX2, PMAIP1, USP30-AS1, SOD2, CIITA, OGFR, LYPD5, APOBEC3A, IFNL1, AKAP7, ACKR4, TNFAIP3, RBMS2, HESX1, HTR2B, MXD1, IL4I1, BLZF1, DUOXA2, NCOA7, RBM11, HRASLS2, SUSD3, FGD2, CCL5, IFI27, TICAM1, CCDC109B, APOBEC3F, GCH1, IL22RA1, TNFSF13B, C19orf38, IKBKE, IFNL2, CASP10, LRRN2, PPM1K, CD7, APOL3, IRAK2, NFKBIZ, UBQLNL, C21orf91, IL7, HLA-F, CD38, RELB, GCNT4, RHBDF2, CX3CL1, CD83, TNFRSF6B, PPM1J, TAGAP, BTN3A3, TMEM229B, FFAR2, CARD16, SOCS1, STX11, OVOL1, IL19, BIRC3, GLRX, CPNE5, MAFF, CXorf49, CXorf49B, CEACAM1, SEC16B, PRDM1, ADCY4, ATF3, LINC00623, TLR2, IFITM2, NRG2, BBC3, SOCS3, KIAA0226, CXCL11, CXCL9, IL15RA, LAG3, STARD4, IDO1, SOCS2, ICOSLG, C1GALT1, FANCA, KCNQ4, NCF1, C10orf105, AP5B1, TMEM171, GBP2, HK2, BRIP1, RAB43, MUC13, IFI30, NCF1C, SP140, HAPLN3, ARID5A, SLC16A1, CD274, KLHDC7B, CACNA1A, CSF3, MAP2, CCL20, TMEM106A, SERPING1, TNFAIP2, CLCA3P, TNIP3, DLG2, CXCL8, MAPK8IP2, IL17C, PLAUR, NEURL3, PROX1, APOBEC3B, TNF, MB21D1, THEMIS2, FRMD3, HBEGF, BRCA2, DUXAP10, TMEM92, DUSP5, EXOC3L4, APOL4, C11orf82, RIMS2, C15orf48, C2CD4B, NOV, ADAM8, SLFN12, MICB, GBP3, GRIN3A, C1R, LOC102724163, SEMA3D, IL2RG, LOC645638, FGF2, FAM169B, LOC100294362, MT2A, ABCD1, ZNF618, LRRC3, SAA4, CYP21A1P, SLC26A4-AS1, SERPINB9, SERPINB9P1, BATF, LOC388692, TLDC2, IL36G, PRRX2, TGM1, ELOVL7, CXCL3, TNFRSF10A, RTKN2, CCL22, HDAC9, ATP10A, IL6, SBK1, BCL2A1, DOC2B, C2CD4A, C1S, ADM, HCG4B, PTPRH, HCAR3, PDCD1LG2, C8orf4, CXCL2, STS, RET, NOS2, SLC26A4, SLCO4A1, SELL, NTNG2, HLA-J, RND1, CFB, XDH, MMP13, TRIM31, SAA2, SPRR2A, PTGS2, MTNR1A, AKAP2, ACHE, DEFB4A, ICAM1, EREG, SPRR2F, CH25H, IL1RN, NEXN, CSF1, HSPA6, LOC554223, APOL1, SLAMF7, LIF, MEF2C, SSC5D, TGM2, ZDHHC14, CXCL13, ERAP2, ANGPTL4, SH2D1B, THSD1, PLA2G4E |
| Downregulated | C20orf195, FAM13C, HPX, INHBB, GPATCH11, PTGFR, PRR18, SLC16A14, SLC47A2 |
Figure 2.
Differential expression of data between infection and control. (A) Differential expression of data between RVA and control; (B) Differential expression of data between RVC and control; (C) The intersection of RVA and RVC DEGs. The red points in A and B represent upregulated genes, the green points represent downregulated genes, and the black points represent genes with no significant difference.
Figure 3.
The heatmap of RVA or RVC infection compared with control. (A) The heatmap of RVA and control; (B) The heatmap of RVC and control.
Functional enrichment analysis
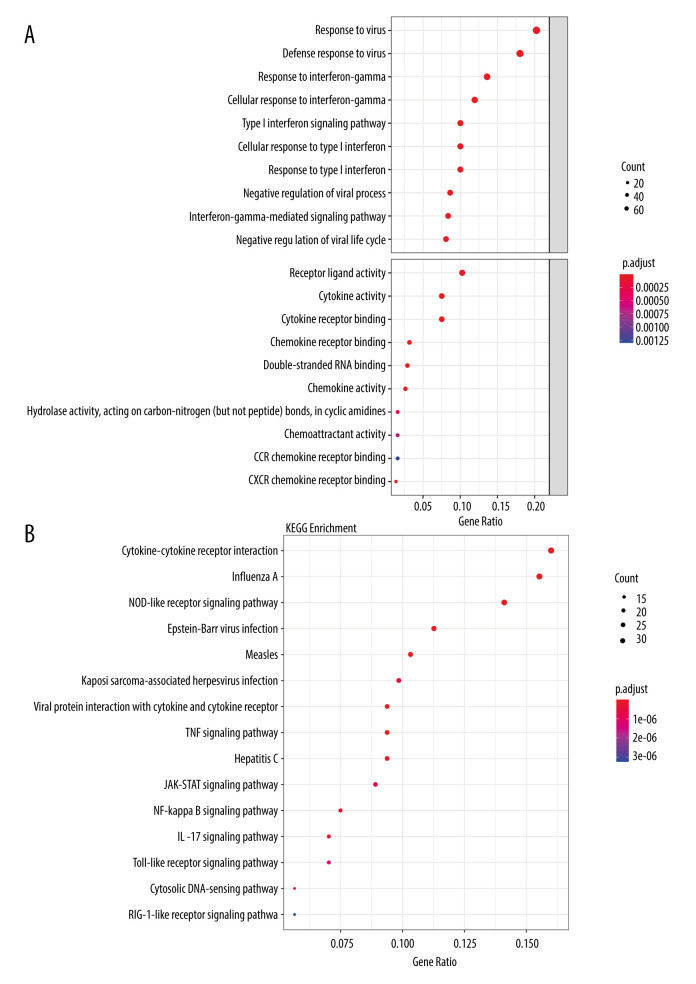
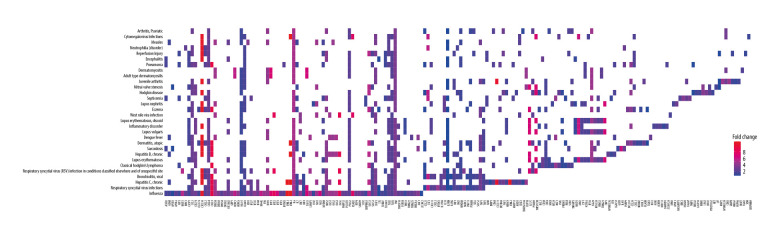
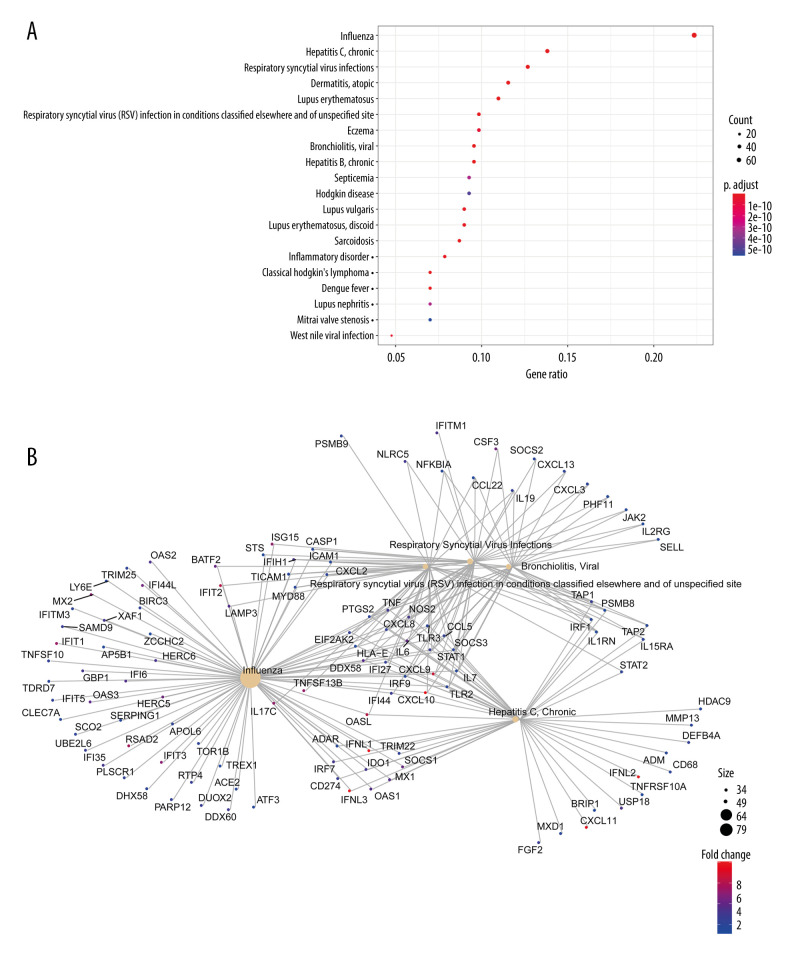
To identify which biological functions and pathways were most significantly associated with the DEGs, a total of 415 genes in the intersection of RVA and RVC were analyzed for GO and KEGG pathways. GO analysis of DEGs in this study was divided into 2 functional groups: a biological processes group and a molecular function group (Figure 4A). In the biological processes group, the DEGs were mainly enriched in “response to virus”, “defense response to virus”, and “response to interferon-gamma”. In the molecular function group, the DEGs were mainly enriched in “receptor ligand activity”, “cytokine activity”, and “cytokine receptor binding”. The top 10 GO terms for each category are listed in Table 2. KEGG pathway analysis demonstrated that DEGs were significantly enriched in “Cytokine-cytokine receptor interaction”, “Influenza A”, and “NOD-like receptor signaling pathway” (Figure 4B). All KEGG pathway terms are listed in Table 3. These results indicate that most DEGs were significantly enriched in virus and cytokine. To further clarify the relationship between DEGs and various diseases, 415 DEGs were subjected to DO analysis. The specific conditions of diseases enriched by each gene are shown in Figure 5. After enrichment analysis, it was found that the DEGs were mainly enriched in “Influenza”, “Hepatitis C, Chronic”, and “Respiratory Syncytial Virus Infections”. The enrichment results are shown in Figure 6A, and the interaction network between diseases and DEGs is shown in Figure 6B.
Figure 4.
Functional enrichment analysis of DEGs in the intersection of RVA and RVC. (A) GO analysis revealed that DEGs were significantly enriched in biological processes (BP) and molecular function (MF) terms; (B) Significantly enriched KEGG terms obtained from KEGG analysis. GO – gene ontology; KEGG – Kyoto Encyclopedia of Genes and Genomes.
Table 2.
GO functional annotation for the most significantly related targets of 415 DEGs.
| GO ID | GO Term | Count | P-value |
|---|---|---|---|
| BP | |||
| GO: 0009615 | Response to virus | 75 | 9.27E-60 |
| GO: 0051607 | Defense response to virus | 67 | 1.20E-59 |
| GO: 0034341 | Response to interferon-gamma | 51 | 5.09E-43 |
| GO: 0060337 | Type I interferon signaling pathway | 38 | 1.25E-40 |
| GO: 0071357 | Cellular response to type i interferon | 38 | 1.25E-40 |
| GO: 0034340 | Response to type i interferon | 38 | 8.46E-40 |
| GO: 0071346 | Cellular response to interferon-gamma | 45 | 1.81E-37 |
| GO: 0048525 | Negative regulation of viral process | 33 | 2.80E-32 |
| GO: 0060333 | Interferon-gamma-mediated signaling pathway | 32 | 3.39E-32 |
| GO: 1903901 | Negative regulation of viral life cycle | 31 | 8.43E-32 |
| MF | |||
| GO: 0005125 | Cytokine activity | 29 | 1.03E-15 |
| GO: 0048018 | Receptor ligand activity | 39 | 1.62E-13 |
| GO: 0005126 | Cytokine receptor binding | 29 | 9.81E-13 |
| GO: 0042379 | Chemokine receptor binding | 13 | 5.95E-10 |
| GO: 0008009 | Chemokine activity | 11 | 2.92E-09 |
| GO: 0045236 | CXCR chemokine receptor binding | 6 | 2.79E-08 |
| GO: 0003725 | Double-stranded RNA binding | 12 | 3.25E-08 |
| GO: 0016814 | Hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines | 7 | 4.39E-06 |
| GO: 0042056 | Chemoattractant activity | 7 | 9.61E-06 |
| GO: 0048020 | CCR chemokine receptor binding | 7 | 2.25E-05 |
Table 3.
KEGG functional annotation for 415 DEGs.
| KEGG ID | KEGG Term | Count | P-value |
|---|---|---|---|
| hsa05164 | Influenza A | 33 | 4.44E-20 |
| hsa04621 | NOD-like receptor signaling pathway | 30 | 2.20E-16 |
| hsa04060 | Cytokine-cytokine receptor interaction | 34 | 1.28E-13 |
| hsa04061 | Viral protein interaction with cytokine and cytokine receptor | 20 | 7.62E-13 |
| hsa04668 | TNF signaling pathway | 20 | 7.04E-12 |
| hsa05162 | Measles | 22 | 7.43E-12 |
| hsa05169 | Epstein-Barr virus infection | 24 | 3.95E-10 |
| hsa05160 | Hepatitis C | 20 | 3.30E-09 |
| hsa04064 | NF-kappa B signaling pathway | 16 | 9.32E-09 |
| hsa04657 | IL-17 signaling pathway | 15 | 1.65E-08 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | 21 | 1.77E-08 |
| hsa04630 | JAK-STAT signaling pathway | 19 | 3.65E-08 |
| hsa04623 | Cytosolic DNA-sensing pathway | 12 | 6.12E-08 |
| hsa04620 | Toll-like receptor signaling pathway | 15 | 6.73E-08 |
| hsa04622 | RIG-I-like receptor signaling pathway | 12 | 2.08E-07 |
| hsa04625 | C-type lectin receptor signaling pathway | 14 | 4.49E-07 |
| hsa04217 | Necroptosis | 17 | 7.52E-07 |
| hsa05134 | Legionellosis | 10 | 1.77E-06 |
| hsa04062 | Chemokine signaling pathway | 18 | 1.94E-06 |
| hsa05133 | Pertussis | 11 | 3.86E-06 |
| hsa05161 | Hepatitis B | 16 | 4.60E-06 |
| hsa05168 | Herpes simplex virus 1 infection | 29 | 2.77E-05 |
| hsa05323 | Rheumatoid arthritis | 11 | 2.78E-05 |
| hsa05170 | Human immunodeficiency virus 1 infection | 16 | 0.000129 |
| hsa05145 | Toxoplasmosis | 11 | 0.000155 |
| hsa05140 | Leishmaniasis | 9 | 0.000166 |
| hsa05144 | Malaria | 7 | 0.000289 |
| hsa05143 | African trypanosomiasis | 6 | 0.00035 |
| hsa05235 | PD-L1 expression and PD-1 checkpoint pathway in cancer | 9 | 0.000498 |
| hsa04612 | Antigen processing and presentation | 8 | 0.000928 |
| hsa04930 | Type II diabetes mellitus | 6 | 0.00116 |
| hsa05142 | Chagas disease (American trypanosomiasis) | 9 | 0.001339 |
Figure 5.
DGEs-enriched diseases.
Figure 6.
Disease enrichment analysis of DEGs in the intersection of RVA and RVC. (A) Disease enrichment analysis; (B) The interaction network between diseases and DEGs.
Expression of ACE2 gene and immune cells
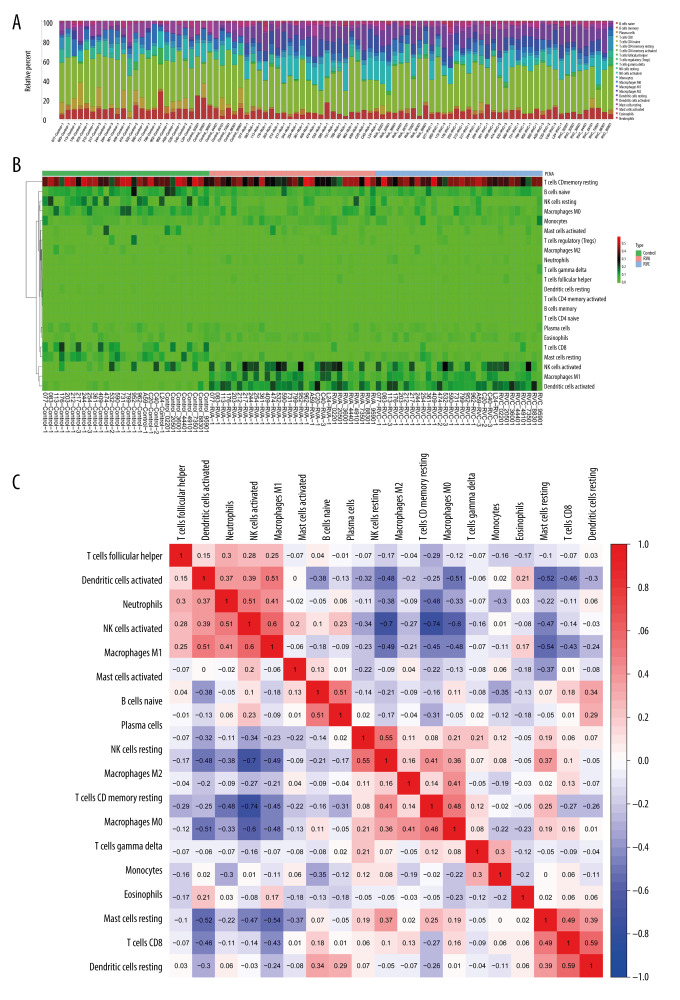
The ACE2 gene is one of the 415 DGEs, and this study further confirmed that the expression of ACE was significantly increased after RVA or RVC infection (P<0.01), along with upregulated DGEs (Figure 7). Functional enrichment analysis showed that most DEGs were significantly related to cytokines, so we analyzed the content and proportion of 22 kinds of immune cells in 90 samples. The content of immune cells is shown in Figure 8A and the heatmap of cell proportions is shown in Figure 8B. The results showed that the proportion of “NK cells activated”, “Macrophages M1”, and “Dendritic cells activated” was increased after infection with RVA or RVC, while that of “T cells CD8” and “Mast cells resting” was decreased. Based on the correlation analysis of the immune cells, we found that “T cells follicular helper”, “Dendritic cells activated”, “Neutrophils”, and “NK cells activated” had a positive correlation with “Macrophages M1”, and “T cells CD8” was also positively correlated with “Dendritic cells resting” (Figure 8C).
Figure 7.
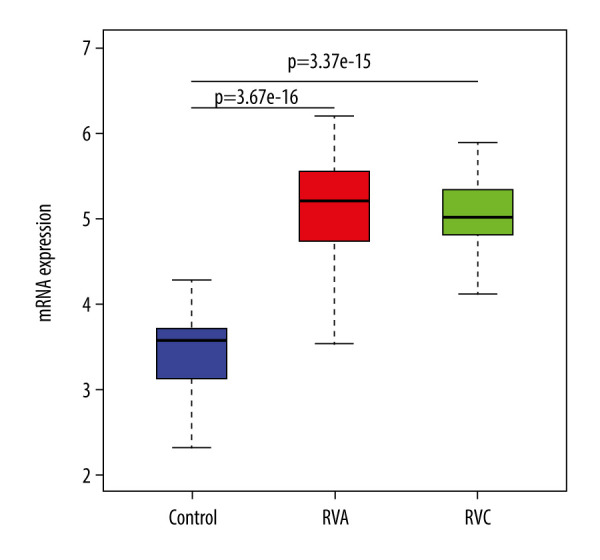
The expression of ACE gene in different treatments.
Figure 8.
Expression of 22 kinds of immune cells in the 90 samples. (A) The content of 22 kinds of immune cells; (B) The heatmap of 22 kinds of immune cells proportional; (C) Correlation between different immune cells.
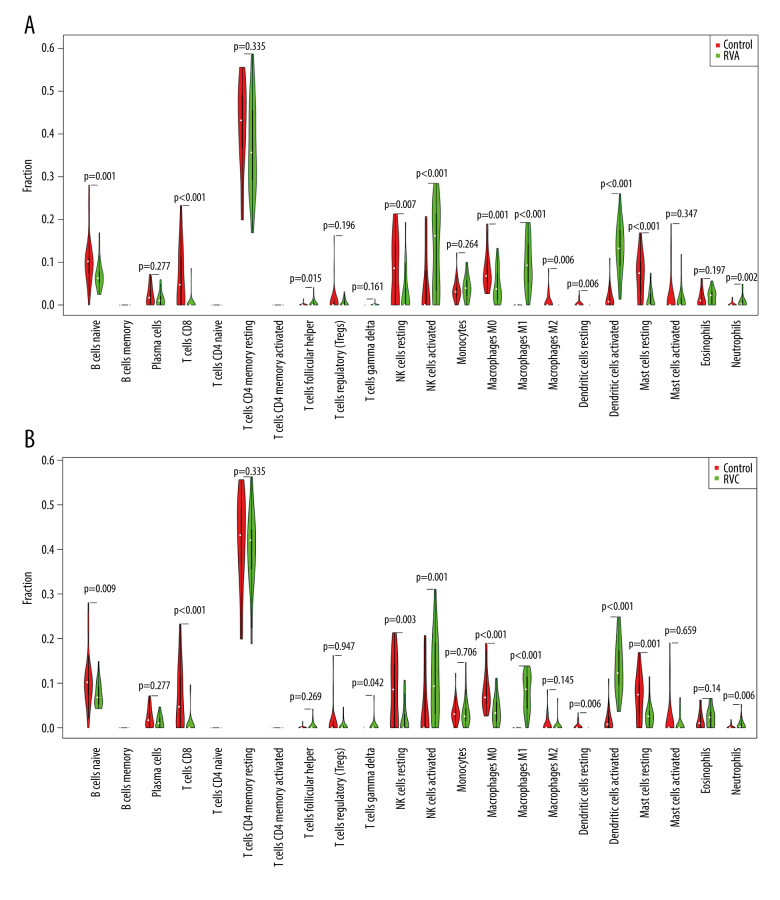
To identify changes in the expression of individual immune cells after infection, we visualized the proportion of 22 kinds of immune cells relative to control after RVA or RVC infection. The results showed that the proportions of “B cells naïve”, “T cells CD8”, “NK cells resting”, “Macrophages M0”, “Macrophages M2”, “Dendritic cells resting”, and “Mast cells resting” were significantly decreased, while that of “T cells follicular helper”, “NK cells activated”, “Macrophages M1”, “Dendritic cells activated” and “Neutrophils” increased significantly after RVA infection (Figure 9A). And the proportion of “B cells naïve”, “T cells CD8”, “NK cells resting”, “Macrophages M0”, “Dendritic cells resting”, and “Mast cells resting” was significantly decreased, while “T cells gamma delta”, “NK cells activated”, “Macrophages M1”, “Dendritic cells activated”, and “Neutrophils” was significantly increased after RVC infection (Figure 9B). In general, RV infection significantly activated the expression of NK cells, dendritic cells, and neutrophils, and reduced the expression of CD8 T cells.
Figure 9.
Expression of 22 kinds of immune cells after RVA or RVC infection. (A) Expression of 22 kinds of immune cells after RVA infection; (B) Expression of 22 kinds of immune cells after RVC infection.
Discussion
SARS-COV-2, causing COVID-19, is a novel virus first detected in December 2019 in Wuhan, China and the disease is thought to be more severe in patients with chronic lung disease [16]. RV airway infection is the most common virus causing the common cold and is a major contributor to the development and exacerbation of asthma [17]. RV infection induces the expression of IFN-stimulated genes and subsequent cytokines [18], and ACE2, the receptor of SARS-COV-2, has recently been identified as an IFN-stimulated gene [19]. Therefore, RV infection may be associated with severe COVID-19 infection through key candidate genes and cytokine pathways. Studies have shown that there are large clinical variations due to differences in autoantibodies among individuals in the course of SARS-COV-2 infection, and type I IFNs in protective immunity against SARS-Cov-2 are crucial [20]. There are many immunological tests for SARS-COV-2 infection, but the protective immune genes for SARS-CoV-2 have not yet been fully clarified, and the study of genes and cytokines can provide a unique opportunity to control the infection and disease [21]. Microarrays and high-throughput sequencing have been widely used to detect the expressions of various genes in the human genome and to predict potential targets for related diseases, which can help identify target genes for diagnosis or treatment of diseases [22].
In the present study, the genomic dataset was obtained from GEO, including 90 samples with non-infection, RVA infection, and RVC infection. Bioinformatics analysis was conducted, identifying 555 DEGs between RVA and control and 421 DEGs between RVC and control. A total of 415 DEGs were involved both in RVA and RVC, among which 406 were upregulated and 9 were downregulated. Functional enrichment analysis showed that DEGs were mainly enriched in “response to virus”, “response to interferon-gamma”, “receptor ligand activity”, “cytokine activity”, “cytokines-cytokine receptor interaction”, “NOD-like receptor signaling pathway”, “Influenza A”, and “respiratory syncytial virus infection”. These results indicate that most DEGs were significantly enriched in virus and cytokine, which is consistent with normal expression after virus infection.
SARS-COV-2 infection is mediated by a Spike virus protein and ACE2, and fuses with target cells through a type I mechanism [23]. ACE2 is expressed in the alveoli, heart, esophagus, kidney, bladder, and ileum, suggesting that all of these organs may be affected by SARS-COV-2 [24,25]. The expression of ACE2 mediates SARS-COV-2 infection in host lung cells [10], so it can be reasonably concluded that increased expression of ACE2 in lung cells will increase susceptibility to SARS-COV-2 infection or lead to more severe COVID19 disease. Generally, after cellular detection of viral entry into a host cell, IFN-induced IFN-stimulated genes are essential for host antiviral defense, and studies have shown that ACE2 is an INF-stimulated gene in human airway epithelial cells [19]. Inducers of the IFN pathway, including influenza infection, can lead to increased expression of ACE2 in cells, thereby increasing susceptibility to COVID-19. Our study found that ACE2 was one of the 415 DEGs in RV and control, and there was a significant increase in the expression of ACE2 in asthma patients infected with RV, indicating that RV infection can increase susceptibility to SARS-COV-2 or lead to more severe COVID19.
Immune cells are the cells involved in or related to the immune response, and they play an important role in the human body. Studies have shown that NK cells are activated in COVID-19 [26], and can also be activated in humans after influenza virus infection, while dendritic cells are necessary for NK cell activation [27]. Dendritic cells play an important role in defense against lung infections [28]. In human dendritic cells derived from monocytes infected by SARS-COV, the chemokines are upregulated [29]. Furthermore, lymphocytosis may be due to the direct effects of cytokines on T cell populations and/or the indirect effects on other cell types such as dendritic cells [30] and neutrophils [31,32] in severe disease. Our research found that NK cells, dendritic cells, and neutrophils were activated after RV infection in asthma, which was consistent with the previous description. In addition, we found a significant decrease in CD8 T cells of people with asthma infected with RV, which was similar to earlier observations about SARS-CoV-1 infection. Some studies reported that lymphopenia was caused by a sharp decrease in the number of T cells CD4 and CD8 in COVID-19 patients [33]. T cells play an important role in viral infections, in which CD4 T cells help to produce antibodies and coordinate the response of other immune cells, while CD8 kills infected cells to reduce the viral burden, and lower numbers of CD8 cells have been shown to be correlated with severity of COVID-19 [34]. The hyperactivation of monocytes, macrophages, and dendritic cells is the driving factor of human immunopathology. Our study found a significant correlation between CD8 and dendritic cells after RV infection, indicating that asthma patients infected with RV would be more susceptible to COVID-19, which was also consistent with the higher incidence of COVID-19 among young asthmatics, accounting for 27% of the hospitalized 18–49 age group in the United States [35]. All results showed that viral infections associated with asthma exacerbations and SARS-COV-2 infections showed synergistic biomolecular interactions. RV infection can aggravate COVID-19 infection by increasing key gene and cytokine pathways associated with COVID-19.
Conclusions
Using integrated analysis of microarray datasets downloaded from the NCBI-GEO database, the present study identified 415 DEGs that may be involved in the progression of RV infection in people with asthma. These DEGs were mainly related to cytokines. Our results contribute to a better understanding of the molecular mechanism of RV infection. We found that there was significant activation of NK cells, dendritic cell, and neutrophils, and a significant decrease in CD8 T cells after RV infection in people with asthma. Moreover, the expression of ACE2, which is closely related to COVID-19, was significantly increased after RV infection. The evidence suggests that RV infection activates the cytokine pathways and increases the expression of ACE2, thus exacerbating COVID-19. These results enhance our understanding of RV infection and its correlation with COVID-19. However, further molecular biological experiments are required to confirm the results of this study.
Footnotes
Conflicts of interest
None.
Source of support: This work was supported by a special COVID-19 project initiated by Shanxi Provincial Education Department
References
- 1.Kennedy JL, Pham S, Borish L. Rhinovirus and asthma exacerbations. Immunol Allergy Clin North Am. 2019;39:335–44. doi: 10.1016/j.iac.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–43. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim D, Quinn J, Pinsky B, et al. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323:2085–86. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee WM, Lemanske RF, Jr, Evans MD, et al. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186:886–91. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansel TT, Tunstall T, Trujillo-Torralbo MB, et al. A comprehensive evaluation of nasal and bronchial cytokines and chemokines following experimental rhinovirus infection in allergic asthma: increased interferons (IFN-γ and IFN-λ) and type 2 inflammation (IL-5 and IL-13) EBioMedicine. 2017;19:128–38. doi: 10.1016/j.ebiom.2017.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donoghue M, Hsieh F, Baronas E, et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. 2000;87:E1–9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 7.Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: Celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126:1456–74. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel VB, Zhong JC, Grant MB, Oudit GY. Role of the ACE2/angiotensin 1–7 axis of the renin-angiotensin system in heart failure. Circ Res. 2016;118:1313–26. doi: 10.1161/CIRCRESAHA.116.307708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–48. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffmann M, Kleine-Weber H, Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petryszak R, Burdett T, Fiorelli B, et al. Expression Atlas update – a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res. 2014;42:D926–32. doi: 10.1093/nar/gkt1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang X, Zhu S, Li L, et al. Identification of differentially expressed genes and signaling pathways in ovarian cancer by integrated bioinformatics analysis. Onco Targets Ther. 2018;11:1457–74. doi: 10.2147/OTT.S152238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Y, Wang YP, Cao H, et al. Integrated computational biology analysis to evaluate target genes for chronic myelogenous leukemia. Mol Med Rep. 2018;18:1766–72. doi: 10.3892/mmr.2018.9125. [DOI] [PubMed] [Google Scholar]
- 14.Zhu Y, Sun X, Lin J, et al. Investigating potential molecular mechanisms of carcinogenesis and genes as biomarkers for prognosis of gastric cancer based on integrated bioinformatics analysis. Pathol Oncol Res. 2019;25:1125–33. doi: 10.1007/s12253-018-0523-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang EH, Willis AL, Romanoski CE, et al. Rhinovirus infections in individuals with asthma increase ACE2 expression and cytokine pathways implicated in COVID-19. Am J Respir Crit Care Med. 2020;202:753–55. doi: 10.1164/rccm.202004-1343LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busse WW, Lemanske RF, Jr, Gern JE. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet. 2010;376:826–34. doi: 10.1016/S0140-6736(10)61380-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaheer RS, Proud D. Human rhinovirus-induced epithelial production of CXCL10 is dependent upon IFN regulatory factor-1. Am J Respir Cell Mol Biol. 2010;43:413–21. doi: 10.1165/rcmb.2009-0203OC. [DOI] [PubMed] [Google Scholar]
- 19.Ziegler CGK, Allon SJ, Nyquist SK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–35. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bastard P, Rosen LB, Zhang Q, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casanova JL, Su HC. A global effort to define the human genetics of protective immunity to SARS-CoV-2 infection. Cell. 2020;181:1194–99. doi: 10.1016/j.cell.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng T, Wang A, Hu D, Wang Y. Molecular mechanisms of breast cancer metastasis by gene expression profile analysis. Mol Med Rep. 2017;16:4671–77. doi: 10.3892/mmr.2017.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross LZF, Sacerdoti M, Piiper A, et al. ACE2, the receptor that enables infection by SARS-CoV-2: Biochemistry, structure, allostery and evaluation of the potential development of ACE2 modulators. ChemMedChem. 2020;15:1682–90. doi: 10.1002/cmdc.202000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou X, Chen K, Zou J, et al. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020;14:185–92. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Zhao Z, Wang Y, et al. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. bioRxiv. 2020;2020 doi: 10.1164/rccm.202001-0179LE. 919985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maucourant C, Filipovic I, Ponzetta A, et al. Natural killer cell activation related to clinical outcome of COVID-19. medRxiv. 2020;2020 20148478. [Google Scholar]
- 27.Draghi M, Pashine A, Sanjanwala B, et al. NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. J Immunol. 2007;178:2688–98. doi: 10.4049/jimmunol.178.5.2688. [DOI] [PubMed] [Google Scholar]
- 28.Guilliams M, Lambrecht BN, Hammad H. Division of labor between lung dendritic cells and macrophages in the defense against pulmonary infections. Mucosal Immunol. 2013;6:464–73. doi: 10.1038/mi.2013.14. [DOI] [PubMed] [Google Scholar]
- 29.Law HK, Cheung CY, Ng HY, et al. Chemokine up-regulation in SARS-coronavirus-infected, monocyte-derived human dendritic cells. Blood. 2005;106:2366–74. doi: 10.1182/blood-2004-10-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waal Malefyt R, Haanen J, Spits H, et al. Interleukin 10 (IL-10) and viral IL-10 strongly reduce antigen-specific human T cell proliferation by diminishing the antigen-presenting capacity of monocytes via downregulation of class II major histocompatibility complex expression. J Exp Med. 1991;174:915–24. doi: 10.1084/jem.174.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Du X, Chen J, et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Shi N, Fan Y, et al. Predictive value of the neutrophil-to-lymphocyte ratio (NLR) for diagnosis and worse clinical course of the COVID-19: Findings from ten provinces in China. SSRN Electronic Journal. 2020 [Google Scholar]
- 33.Vabret N, Britton GJ, Gruber C, et al. Immunology of COVID-19: Current state of the science. Immunity. 2020;52:910–41. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie S, Zhao X, Zhao K, et al. Metabolic disturbances and inflammatory dysfunction predict severity of coronavirus disease 2019 (COVID-19): A retrospective study. medRxiv. 2020;2020 20042283. [Google Scholar]
- 35.Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 – COVID-NET, 14 states, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:458–64. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]