Abstract
Birds, notably wild ducks, are reservoirs of pathogenic and zoonotic viruses such as influenza viruses and coronaviruses. In the current study, we used metagenomics to detect and characterise avian DNA and RNA viruses from wild Pacific black ducks, Chestnut teals and Grey teals collected at different time points from a single location. We characterised a likely new species of duck aviadenovirus and a novel duck gyrovirus. We also report what, to the best of our knowledge, is the first finding of an avian orthoreovirus from Pacific black ducks and a rotavirus F from Chestnut teals. Other viruses characterised from the samples from these wild ducks belong to the virus families Astroviridae, Caliciviridae and Coronaviridae. Some of the viruses may have potential cross-species transmissibility, while others indicated a wide genetic diversity of duck viruses within a genus. The study also showed evidence of potential transmission of viruses along the East Asian—Australasian Flyway; potentially facilitated by migrating shorebirds. The detection and characterisation of several avian viruses not previously described, and causing asymptomatic but potentially also symptomatic infections suggest the need for more virus surveillance studies for pathogenic and potential zoonotic viruses in wildlife reservoirs.
Subject terms: Microbiology, Virology
Introduction
Birds, notably wild ducks, can be reservoirs for zoonotic viruses including, but not limited to, influenza viruses1,2 and coronaviruses3,4. These viruses could become potential threats to other wild animals, poultry and possibly humans under the right set of circumstances2. Virome (virus community) analysis of potential reservoir bird hosts is expected to unveil the role of viruses causing acute or long-term asymptomatic infections and the diversity and range of viruses that infect wild birds. Virome analysis is also expected to aid in understanding the transmission and possible role of viruses on host health in the wild. Metagenomics of viruses, utilises the power of NGS for the discovery, analysis and characterisation of viruses, thereby enabling virome analysis and providing insight into viruses for which these birds act as a natural reservoir. The use of the metagenomic technique in bird samples can lead to the determination of the virome from an individual bird or a population, and the identification of various novel avian viruses3,5,6. Nevertheless, only a few studies have explored the viruses circulating among wild ducks in Australia3–6.
In the current study, we collected fresh duck faecal samples from a single location at various time points. Metagenomics of viruses was carried out to detect and characterise avian viruses present in the sample of Australian wild ducks using a protocol optimised in our laboratory and described earlier3. We previously focused on avian parvoviruses and picornaviruses from these duck samples as they were found to be abundant compared to other avian viruses6. We also reported a reassorted low pathogenicity H9N2 avian influenza virus with intercontinental gene segments in Chestnut teal samples collected on August 20181. It was the first report of an H9N2 avian influenza virus in resident wild birds in Australia1. In the current study, we focus on the sequence characterisation of additional avian DNA and RNA viruses from multiple virus families that were present in the Australian wild ducks. We aimed to determine the abundance of the different viruses in Australian wild ducks and the factors that might influence the ecology of these viruses in relation to host species.
Results
We have previously described viruses belonging to the virus families Parvoviridae and Picornaviridae from our duck samples as they had the most abundant virus reads generated6. Avian viruses belonging to virus families other than Parvoviridae and Picornaviridae are described here and were detected and characterised in samples from Pacific black ducks (PBD12.16, PBD05.18 and PBD08.18), Chestnut teals (CT08.18 and CT11.18) and Grey teals (GT11.18) (Table 1). From the Chestnut teal (CT05.18) and Wood duck (WD08.18) samples collected in May and August 2018, respectively, no other avian viruses were detected other than those previously described viruses belonging to Parvoviridae and Picornaviridae6.
Table 1.
Avian viruses detected and characterised from the duck samples.
| Sample | Avian viruses detected and characterised |
|---|---|
| Pacific black duck | |
| PBD12.16 | Aviadenovirus (PBDAdV/PBD12.16) |
| Calicivirus (PBDCV/PBD12.16) | |
| PBD05.18 | Avastrovirus (PBDAstV/PBD05.18) |
| PBD08.18 | Avian orthoreovirus (PBDORV/PBD08.18) |
| Rotavirus G (PBDRVG/PBD08.18) | |
| Chestnut teal | |
| CT08.18 | Gammacoronavirus |
| Rotavirus F (CTRVF/CT08.18) | |
| CT11.18 | Avastrovirus (CTAstV/CT11.18) |
| Grey teal | |
| GT11.18 | Gyrovirus (GTGV/GT11.18) |
DNA viruses characterised from the duck samples
Two DNA viruses, in addition to the parvoviruses already described6, were detected and characterised from the duck samples, one belonging to the virus family Adenoviridae and another to Anelloviridae.
Aviadenovirus from Pacific black duck (PBDAdV/PBD12.16)
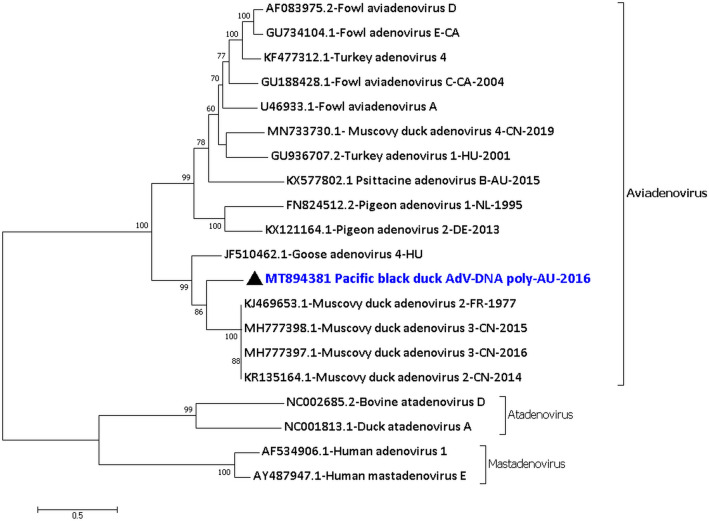
A brief account of this avian virus detected in this sample was provided earlier3. However, more nucleotide sequences were generated by resequencing the sample at a greater depth. A total of 37,832 nucleotides (out of ~ 43 kb) of adenovirus sequence was generated from seven consensus sequences of the virus with a coverage ranging from 2 to 4456 (Supplementary material 1 Row 1–7 and Supplementary material 2 Fig. S1). The seven consensus sequences of the Pacific black duck aviadenovirus encoded the dUTPase, ORF52, ORF2, ORF14, ORF12, IVa2, DNA polymerase, pTP, partial 52 K, complete pIIIa, penton base protein, pVII, pX, minor capsid protein VI, hexon, protease, DNA binding protein, 100 K, 22 K, 33 K, pVIII, U-exon, partial fibre, complete ORF22, ORF20, ORF56, ORF19 (lipase), partial ORF54, partial ORF19B and complete ORF53. The amino acid sequences of the DNA polymerase showed that the PBDAdV/PBD12.16 was only 81.9% (1028/1255 amino acids) identical to the closest sequence in NCBI which is a Muscovy duck adenovirus 2 from France in the year 19777 (KJ469653.1), and only 81.5% (1024/1255 amino acids) identical to JF510462.1 Goose adenovirus 4 from Hungary8 (Fig. 1). The species demarcation criteria of the genus Aviadenovirus by ICTV is based on DNA polymerase phylogenetic distance (> 5–15% distance matrix), genomic organisation, RFLP analysis, host range, pathogenicity, cross-neutralization and ability to recombine9. From the PBDAdV/PBD12.16 consensus sequences generated here and based on the DNA polymerase phylogenetic analysis and host species; this virus can be considered potentially as a new species. BLASTP analysis of these protein sequences showed that some of them are more identical but still distant to their counterparts expressed by the KJ469653.1 Muscovy duck adenovirus 2 or the KR135164.1 Muscovy duck adenovirus 2, while others to that of JF510462.1 Goose adenovirus 4 (Supplementary Material 1 Column J).
Figure 1.
Phylogenetic analysis of the amino acid sequences of the DNA polymerase of Pacific black duck aviadenovirus (PBDAdV/PBD12.16). The amino acid sequences were aligned and analysed by using the maximum likelihood method based on the LG + G model72 in MEGA771 with a bootstrapping of 1000 replicates. The analysis involved 20 amino acid sequences and all positions containing gaps and missing data were eliminated. The final dataset contained a total of 936 amino acid positions. The numbers at the nodes represent bootstrap values and only bootstrap values at or above 60% are shown. Pacific black aviadenovirus is shown in blue and marked with a black triangle.
From the generated consensus sequences, 18 genes have been identified (IVa2–fibre) to be located in the central region of the Pacific black duck adenovirus genome. Thirty genes or ORFs (out of 34–36 genes) were found in total from the seven consensus sequences of the virus. The motifs and other sequence or amino acid features found such as bipartite nuclear localisation signals have been given in the Supplementary material 1 Column K.
Gyrovirus from Grey teal (GTGV/GT11.18)
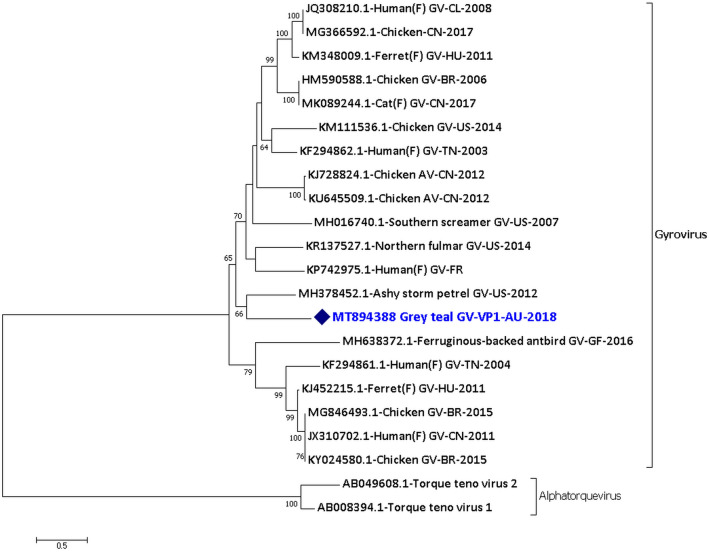
A novel gyrovirus was found in the single Grey teal sample (GT11.18). Real-time PCR amplification and Sanger sequencing were carried out and were successful for the Grey teal gyrovirus for the region from 264th to 761st nucleotide. A near-complete genome of 1549 nucleotides long consensus sequences encoding complete VP1, VP2 and hypothetical VP3 protein of GTGV/GT11.18 was generated from the NGS reads of the sample with a coverage ranging from 2 to 105 (Supplementary material 1 Row 47). The phylogenetic analysis of the VP1 protein sequence of the Grey teal gyrovirus shows that it is distantly related to other currently known gyroviruses (Fig. 2). Gyroviruses were recently classified into the family Anelloviridae, and the species demarcation criteria of some of the genera in the family Anelloviridae are based on ORF1/VP1 (capsid) protein with a nucleotide divergence of > 35%10. Hence, based on the phylogenetic analysis of the VP1 protein sequence the Grey teal gyrovirus can be considered as a new virus species. The GTGV/GT11.18 was relatively most identical, but still distant, to MH378452.1 gyrovirus from Ashy storm petrel faecal sample from the US in 2012 with 55.2% (159/288 amino acids) identity only. The GTGV/GT11.18 was also distantly related to the Southern screamer gyrovirus (MH016740.1) from the US in 200711 with 52.4% identity only in the VP1 protein sequence.
Figure 2.
Phylogenetic analysis of the amino acid sequences of the VP1 protein of Grey teal gyrovirus (GTGV/GT11.18). The amino acid sequences were aligned and analysed using the maximum likelihood method based on the WAG + G + I + F model73 in MEGACC74 with a bootstrapping of 1000 replicates. The analysis involved 22 amino acid sequences and all positions containing gaps and missing data were eliminated. The final dataset contained a total of 233 amino acid positions. The numbers at the nodes represent bootstrap values and only bootstrap values at or above 60% are shown. Grey teal gyrovirus is shown in blue and marked with a blue diamond. (F) shows faecal samples.
The VP1 protein of the GTGV/GT11.18 contained the motif 141WWRWA145, and also had a high number of basic amino acids, especially arginines in its N-terminal region. The minimal signature motif for the protein tyrosine phosphatases (PTPase) superfamily CX5R (46CSCGNFR52) and the motif 34WX7HX3CXCX5H54 was found in the VP2 protein, suggesting that this protein may exhibit PTPase function12,13. It is also to be noted that the VP3 protein did not have any apoptin conserved protein domains.
RNA viruses characterised from the duck samples
The RNA viruses, in addition to the picornaviruses already described6, detected and characterised from the duck samples belonged to the virus families Reoviridae, Astroviridae, Caliciviridae and Coronaviridae.
Avian orthoreovirus from Pacific black duck (PBDORV/PBD08.18)
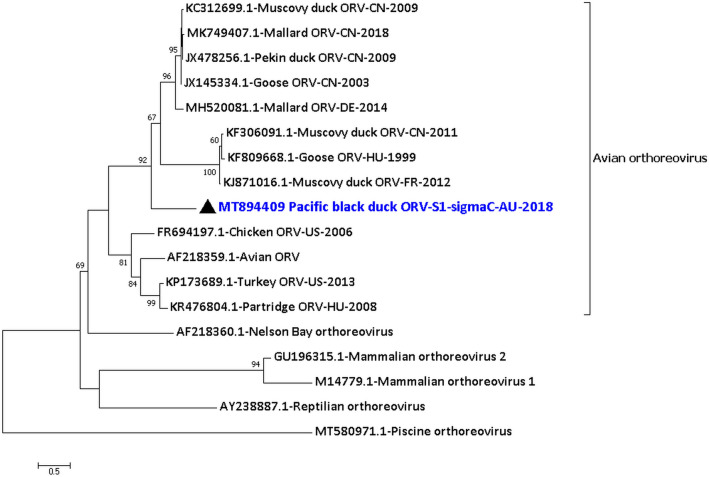
From the Pacific black duck 08.18 sample, an orthoreovirus, family Reoviridae, was detected and characterised. All the 10 segments were detected and a total of 21,170 nucleotides (out of ~ 23 kb) of the consensus sequence of the Pacific black duck orthoreovirus genome were generated with a coverage from 2 to 336 (Supplementary material 1 Row 16–29). Table 2 provides the details on the avian orthoreovirus from the PBD08.18 sample along with the percentage identity to the closest relative of the nucleotide sequences and the proteins encoded by each segment. The analysis of the protein sequences showed that they were closely related to other avian orthoreoviruses with > 96% identity (for example, Fig. S2 of Supplementary material 2), except for the proteins p10, p17 and sigma C encoded by segment S1. However, the nucleotide sequences of the 10 segments of the PBDORV/PBD08.18 were about 84–96% identity to its closest relative, except for segment S1 (Table 2). The phylogenetic analysis of the assembled consensus sequence of the sigma C protein from segment S1 was most identical, but still distant, to the JX145334.1 Goose orthoreovirus sigma C protein from China in the year 200314 and JX478256.1 Pekin duck orthoreovirus sigma C protein from China in the year 200915 with 54.8% identity (187/341 amino acids) than to any other orthoreovirus sigma C proteins (Fig. 3). The partial p10 protein and complete p17 protein were most identical but still distant to MH520081.1 Mallard orthoreovirus p10 protein from Germany16 with 67.2% identity and to JX478256.1 Pekin duck orthoreovirus p17 protein from China15 with 52.1% identity, respectively (Table 2).
Table 2.
Details on the avian orthoreovirus from Pacific black duck 08.18 sample (PBDORV/PBD08.18).
| Segment|protein encoded | Length of the generated nucleotide sequence (nt) | Protein function | Percentage identity to its closest relative using MEGA | |
|---|---|---|---|---|
| Amino acid | Nucleotide | |||
| L1|partial lambda (λ) A | 3860 | Core shell scaffold42 | KC312700.1 Muscovy duck ORV75 with 99.0% identity | MH520075.1 Mallard duck ORV16 with 91.9% identity |
| L2|partial lambda (λ) B | 3661 | Putative transcriptase42 | MH520077.1 Mallard ORV16 with 98.6% identity | MH520077.1 Mallard ORV16 with 93.1% identity |
| L3|partial lambda (λ) C | 3624 | Capping enzyme42 | MK955820.1 Cherry valley duck ORV14 with 97.3% identity | JX145330.1 Goose ORV14 with 92.8% identity |
| M1|partial mu (µ) A | Two consensus sequences of 939 and 1199nt long | Putative transcriptase co-factor42 | MH520078.1 Mallard ORV16 with 97.4% and 96.7% identity, respectively | KF306085.1 Muscovy duck ORV76 with 90.9% and 91.9% identity, respectively |
| M2|partial mu (µ) B | Two consensus sequences of 497 and 1066nt long | Part of the outer capsid and may help in the penetration to the host cell42 | KR476802.1 Partridge ORV77 with 97.5% and 99.1% identity, respectively | KR476802.1 Partridge ORV77 with 84.1% and 85.8% identity, respectively |
| M3|partial mu (µ) NS | Two consensus sequences of 983 and 542nt long | Formation of virus factories and protein recruitment42 | MH520080.1 Mallard ORV16 with 96.6% and 98.1% identity respectively | MH520080.1 Mallard ORV16 with 90.9% and 91.6% identity respectively |
| S1|partial p10, complete p17 and sigma (σ) C | Two consensus sequences of 211 and 1328nt long | σC forms part of the outer capsid that helps in host cell attachment of the virus particle42 | p10: MH520081.1 Mallard ORV16 with 67.2% identity p17: JX478256.1 Pekin duck ORV15 with 52.1% identity σC: JX145334.1 Goose ORV14 and JX478256.1 Pekin duck ORV15 with 54.8% identity | 1328nt sequence: JX478256.1 Pekin duck ORV15 and KC312699.1 Muscovy duck ORV with 67.6% identity |
| S2|sigma (σ) A | 1269 | Part of the inner core of the capsid42 | KC508653.1 Muscovy duck ORV78 with 97.8% identity | KF306088.1 Muscovy duck ORV76 with 91.1% identity |
| S3|partial sigma (σ) B | 1107 | Part of the outer capsid42 | MH520083.1 Mallard ORV16 with 96.6% identity | JX145336.1 Goose ORV14 with 85.5% identity |
| S4|partial sigma (σ) NS | 884 | ssRNA binding42 | KJ871025.1 Muscovy duck ORV with 99.6% identity | KJ871015.1Muscovy duck ORV with 95.7% identity |
Figure 3.
Phylogenetic analysis of the amino acid sequences of the sigma C protein of Pacific black duck orthoreovirus segment S1 (PBDORV/S1/PBD08.18). The amino acid sequences were aligned and analysed by using the maximum likelihood method based on the WAG + G + F model73 in MEGACC74 with a bootstrapping of 1000 replicates. The analysis involved 18 amino acid sequences and all positions containing gaps and missing data were eliminated. The final dataset contained a total of 257 amino acid positions. The numbers at the nodes represent bootstrap values and only bootstrap values at or above 60% are shown. Pacific black orthoreovirus is shown in blue and marked with a black triangle. No orthologous sigma C protein was found in Broome ORV, Baboon ORV and Mahlapitsi ORV.
Rotavirus G from Pacific black duck (PBDRVG/PBD08.18)
Rotavirus G also belongs to the virus family Reoviridae and was detected and characterised from the PBD08.18 sample. All the 11 segments of the virus were detected and characterised. A total of 17,808 nucleotides (out of ~ 18 kb) of the consensus sequence of the Pacific black duck rotavirus G genome were generated with a coverage of 2–512 (Supplementary material 1 Row 30–40). It should be noted that all the segments have been named as per the rotavirus G NCBI RefSeq sequence (NC021580.1 to NC021590.1). Table 3 provides the details on the PBDRVG/PBD08.18 along with the percentage identity to the closest relative for each segment. Out of the 11 proteins encoded by the 11 segments of Pacific black duck rotavirus G, seven proteins were most closely related to their orthologous proteins of Grey teal rotavirus G from south west Victoria, Australia from 20175 with 87.2–99.6% identity (Table 3). Segment 7, encoding the NSP3 protein, was most closely related to the MK204407.1 Grey teal partial NSP3 protein (only 152 amino acids given) from Australia in the year 20175 with 64.9% identity. The complete NSP3 protein of the PBDRVG/PBD08.18 was most identical but still distant to KY689682.1 Turkey rotavirus G from the US in the year 201617 with 44.3% identity only. It should also be noted that the NSP4 protein was not generated from the Grey teal sample from Australia5 and hence was not available for comparison. The VP4 and the VP7 protein of the Pacific black duck rotavirus G were also distantly related to the orthologous proteins of Grey teal rotavirus G from Australia5. The nucleotide sequences of all the segments of the PBDRVG/PBD08.18 were only about 56–98% identity to its closest relative.
Table 3.
Details on the rotavirus G from Pacific black duck 08.18 sample (PBDRVG/PBD08.18).
| Segment|protein encoded | Length of the generated nucleotide sequence (nt) | Protein function | Percentage identity to its closest relative using MEGA | |
|---|---|---|---|---|
| Amino acid | Nucleotide | |||
| S1|partial VP1 | 3479 | RNA polymerase45,46 | MK204401.1 Grey teal RVG5 with 98.6% identity | MK204401.1 Grey teal RVG5 with 92.6% identity |
| S2|partial VP2 | 2832 | Part of the inner layer of the capsid and is associated with the RNA polymerase and the VP318 | MK204402.1 Grey teal RVG VP25 with 98.5% identity | MK204402.1 Grey teal RVG5 with 79.8% identity |
| S3|partial VP4 | 2322 | Part of the outer capsid18 | KY689679.1 Turkey RVG17 with 54.3% identity | KY689679.1 Turkey RVG17 with 60.3% identity |
| S4|partial VP3 | 2277 | A multifunctional protein which may be associated with guanylyltransferase79 | MK204403.1 Grey teal RVG5 with 98.2% identity | MK204403.1 Grey teal RVG5 with 95.1% identity |
| S5|partial NSP1-1 and complete NSP1 | 1198 | Innate host immune response regulator80,81 | NSP-1 protein: MK204405.1 Grey teal RVG5 with 98.0% identity | MK204405.1 Grey teal RVG5 with 96.6% identity |
| S6|VP6 | 1272 | Part of the intermediate layer of the capsid44 | MK204406.1 Grey teal RVG5 with 87.2% identity | MK204406.1 Grey teal RVG5 with 73.8% identity |
| S7|NSP3 | 1276 | Regulates the virus mRNA translation18 | MK204407.1 Grey teal RVG partial NSP3 protein5 with 88.1% identity; KY689682.1 Turkey RVG17 with 60.4% identity | MK204407.1 Grey teal RVG partial S75 with 89.1% identity; KY689682.1 Turkey RVG17 with 70.4% identity |
| S8|NSP2 | 997 | Aids in the virus particle assembly82 | MK204408.1 Grey teal RVG5 with 99.6% identity | MK204408.1 Grey teal RVG5 with 97.8% identity |
| S9|partial VP7 | 745 | Part of the outer capsid18 | MT025062.1 Gentoo penguin RVG with 66.7% identity and MK204409.1 Grey teal RVG with 66.4% identity | MT025062.1 Gentoo penguin RVG with 58.6% identity and MK204409.1 Grey teal RVG with 56.5% identity |
| S10|partial NSP4 | 772 | Acts as viral enterotoxin83 | MH453872.1 Ruddy turnstone RVG19 with 53.7% identity | MH453872.1 Ruddy turnstone RVG19 with 67.8% identity |
| S11|NSP5 | 613 | Aids in the virus particle assembly82 | MK204410.1 Grey teal RVG NSP55 with 92.9% identity | MK204410.1 Grey teal RVG5 with 80.0% identity |
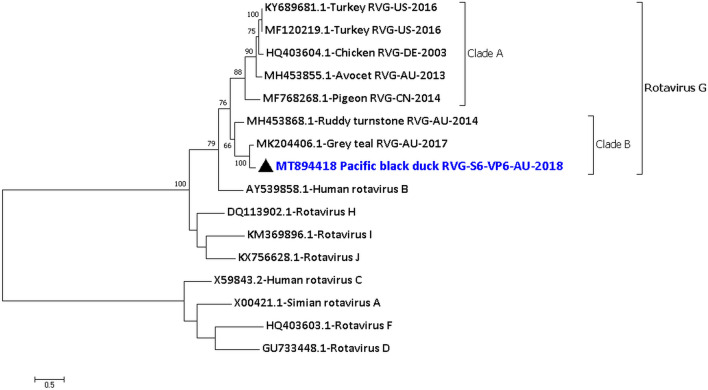
From the phylogenetic analysis of the VP1 and the VP6 protein (Fig. 4 and Fig. S3 of Supplementary Material 2), it is clear that the rotavirus G forms two different clades. Clade A includes the rotavirus G from chicken18, turkey17, avocet19 and pigeon while Clade B includes the rotavirus G from the Pacific black duck, Grey teal5 and Ruddy turnstone19.
Figure 4.
Phylogenetic analysis of the amino acid sequences of the VP6 protein of Pacific black duck rotavirus G segment S6 (PBDRVG/PBD08.18). The amino acid sequences were aligned and analysed by using the maximum likelihood method based on the LG + G + F model72 in MEGACC74 with a bootstrapping of 1000 replicates. The analysis involved 16 amino acid sequences and all positions containing gaps and missing data were eliminated. The final dataset contained a total of 371 amino acid positions. The numbers at the nodes represent bootstrap values and only bootstrap values at or above 60% are shown. Pacific black rotavirus G is shown in blue and marked with a black triangle.
Various motifs found in the PBDRVG/PBD08.18 partial VP3 protein of segment 4 varied to those found in VP3 protein of the other rotavirus G. For example, in lieu of an EXXFXXXN motif in the N-terminal region18, the PBDRVG/PBD08.18 has an EXXFXXXS motif; and in lieu of an ALYXLSNXXN motif in the central region18, the PBDRVG/PBD08.18 has an ALYXISNXXN motif in the central region. Also, the putative guanylyl transferase KXTAMDXEXP and KXXGNNH motifs found in rotavirus A and C20 were not found in the Pacific black duck rotavirus G, as observed for other rotavirus G sequences18. The conserved domain HGXGHXRXV at 229 to 237 amino acid and histidine triad His-X-His-X-His-XX at 229–234 amino acid of the NSP2 protein that aid in the binding of nucleoside triphosphate21 was found in the NSP2 protein. The five fully conserved cysteine residues were found in the partial VP7 of the PBDRVG/PBD08.18 at the expected location, which in turns forms the disulphide bonds that is thought to stabilise the protein structure18.
Rotavirus F from Chestnut teal (CTRVF/CT08.18)
From the Chestnut teal 08.18 sample, a few good quality NGS reads of Chestnut teal rotavirus F were detected and characterised. The NGS reads were mapped to segment 7 of the virus encoding partial NSP3 protein. Further analysis led to the generation of a 647 nucleotides long consensus sequences with a coverage of 2–11 with a minimum mapping quality of 80 or higher (Supplementary material 1 Row 42). The partial NSP3 protein, which regulates the virus mRNA translation18, was relatively closely identical to the JQ920001.1 Chicken rotavirus F from Germany in the year 200318 with 88.8% (191/215 amino acids) identity.
Avastrovirus from Pacific black duck (PBDAstV/PBD05.18) and Chestnut teal (CTAstV/CT11.18)
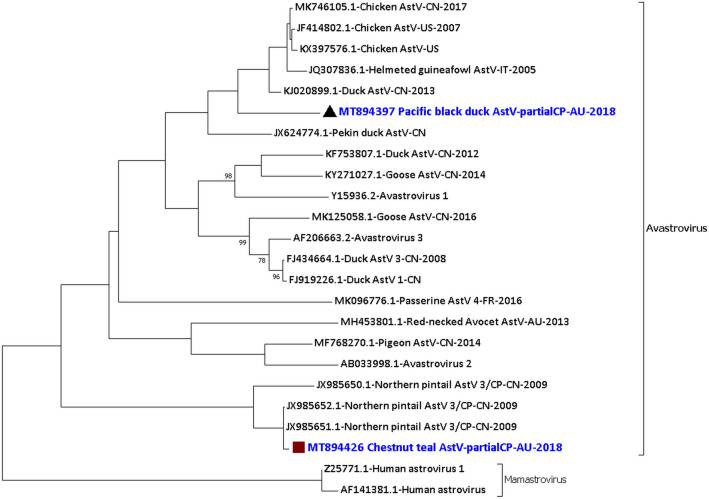
From the Pacific black duck 05.18 sample and Chestnut teal 11.18 sample we detected and characterised two astroviruses that belong to the virus family Astroviridae and the genus Avastrovirus. A total of 3443 nucleotides (out of ~ 7 kb) of the consensus sequence of the Pacific black duck avastrovirus genome was generated. In comparison, a total of 4185 nucleotides of the consensus sequence for the Chestnut teal avastrovirus genome was generated (Supplementary material 1 Row 12–15 and 43–46 and Supplementary material 2 Fig. S4). The -1 ribosomal frameshift signal was found to be from the 79th to 85th nucleotides of the 738 nucleotides long consensus sequence encoding partial ORF1a and ORF1b of the PBDAstV/PBD05.18 and from the 1026th to 1032nd nucleotides of the 1,554 nucleotides long consensus sequence encoding partial ORF1a and ORF1b of the CTAstV/CT11.18. The phylogenetic analysis of the partial capsid protein (ORF2) encoding consensus sequence (PBDAstV/1297nt/PBD05.18) of the PBDAstV showed that it was most identical, but still distant to KJ020899.1 Duck astrovirus from China in 201322 with 83.1% (356/428 amino acids) identity only (Fig. 5). The phylogenetic analysis of the partial capsid protein (ORF2) encoding consensus sequence (CTAstV/1564nt/CT11.18) of the CTAstV showed that it was closely related to JX985715.1 Northern pintail astrovirus from China in the year 200923 with 98.0% (251/256 amino acids) identity (Fig. 5) while having only 85.2% identity (655/768 nucleotides) at the nucleotide level. The amino acid sequence of the partial capsid region of Chestnut teal astrovirus (CTAstV/1564nt/CT11.18) is only 41.4% identical to the Pacific black duck astrovirus partial capsid region (PBDAstV/1297nt/PBD05.18). Analysis of other proteins encoded by the consensus sequences of the duck avastroviruses showed differences in the identity to their closest relative (Supplementary material 1 Column J).
Figure 5.
Phylogenetic analysis of the amino acid sequences of the partial capsid protein (ORF2) of Pacific black duck avastrovirus (PBDAstV/1297nt/PBD05.18) and Chestnut teal avastrovirus (CTAstV/1564nt/CT11.18). The amino acid sequences were aligned and analysed by using the maximum likelihood method based on the LG + G model72 in MEGACC74 with a bootstrapping of 1000 replicates. The analysis involved 24 amino acid sequences and all positions containing gaps and missing data were eliminated. The final dataset contained a total of 224 amino acid positions. The numbers at the nodes represent bootstrap values and only bootstrap values at or above 60% are shown. Pacific black avastrovirus is shown in blue and marked with a black triangle and Chestnut teal avastrovirus is shown in blue and marked with a brown square.
Calicivirus from Pacific black duck (PBDCV/PBD12.16)
From the Pacific black duck December 2016 sample, we detected and characterised a duck calicivirus, belonging to the virus family Caliciviridae. A total of 4,710 nucleotides (out of ~ 8.3 kb) of the consensus sequence of the virus genome were generated. Five consensus sequences of 305 nucleotides, 1596 nucleotides, 331 nucleotides, 1129 nucleotides, and 1112 nucleotides encoding partial ORF1 were generated with a coverage from 2–90 (Supplementary material 1 Row 6–10). Another consensus sequence of 237 nucleotides long was generated encoding partial VP2 protein (Supplementary material 1 Row 11). These sequences had the characteristic motifs of a calicivirus at the expected location in the amino acid sequence (Supplementary material 1 Column K). The Pacific black duck calicivirus consensus sequences showed 96.3% to 97.2% identity at the nucleotide level (blastn) to the MK204392.1 Grey teal calicivirus from south west Victoria, Australia in the year 20175. Phylogenetic analysis of the amino acid sequence of the partial ORF1 containing Helicase motif (PBDCV/1596nt/PBD12.16) and the RdRp region (PBDCV/1129nt/PBD12.16) showed these to be 100% identical to the MK204392.1 Grey teal calicivirus ORF1 protein. In comparison, the VP1 region (PBDCV/1112nt/PBD12.16) was 99.4% (368/370 amino acids) identical to the MK204392.1 Grey teal calicivirus ORF1 protein (Fig. S5 of Supplementary material 2), while being distantly related to the other caliciviruses with 36.2–45.6% (134–169/370 amino acid) identity only. However, based on the phylogenetic analysis of the VP1 region and genus demarcation criteria of the family Caliciviridae24, the calicivirus from the PBD12.16 sample belongs to the genus Nacovirus.
Gammacoronavirus from Chestnut teal
From the Chestnut teal 08.18 sample, a few good quality reads of gammacoronavirus were detected and characterised. Further analysis led to the generation of a short 382 nucleotides long consensus sequence with a coverage of 2–9 (Supplementary material 1 Row 41). The BLASTN analysis of the short consensus sequences shows that it is 100% identical to the MK204393.1 Grey teal gammacoronavirus from Australia in the year 20175. Further analysis of this virus was not carried out due to the low quantity and short consensus sequence.
Discussion
Previously, we described in detail the metagenomics methodology that can be used to detect and characterise both DNA and RNA viruses from bird faecal samples3. Using this method, we have now conducted a surveillance study of Australian wild ducks from which faecal samples were collected at different time points from a single location. As reported earlier1,3,4,6 and also shown in the current study, we were able to detect and characterise both DNA and RNA viruses. We were also able to characterise novel viruses from these ducks6.
In the current study, details on the genome sequences of a duck aviadenovirus genetically different to currently known waterfowl aviadenoviruses and a novel gyrovirus are provided. We also report what, to the best of our knowledge, is the first finding of an avian orthoreovirus from Pacific black ducks and a rotavirus F from Chestnut teals, albeit being a short partial sequence from segment 7 of the rotavirus genome.
The Pacific black duck aviadenovirus characterised here belongs to the genus Aviadenovirus. Adenoviruses belonging to the genus Aviadenovirus have been found in, but not limited to, chickens25, turkeys26, geese8, pigeons and ducks7. The abundance of the Pacific black duck aviadenovirus in the juvenile Pacific black ducks faecal sample (PBD12.16) might indicate active infection and shedding of the virus from these birds. Analysis of the virus proteins showed that some of them were more identical but still distant to their counterparts expressed by the KJ469653.1 Muscovy duck adenovirus 2, while others to that of JF510462.1 Goose adenovirus 4. Nevertheless, the Pacific black duck aviadenovirus was almost equidistant from both the viruses in the maximum likelihood tree generated. This analysis is consistent with our previous analysis of the PBDAdV/PBD12.16 virus, as reported from the very short sequences generated3 before resequencing of the sample. From the central region of the partial Pacific black duck aviadenovirus genome, 18 genes (IVa2–fibre) were found in total which are common to all adenoviruses and said to be inherited from a common ancestor of all known adenoviruses8. However, only a partial fibre gene sequence was generated from the NGS data, and hence at this point, it is uncertain if the PBDAdV/PBD12.16 have one fibre gene like the Fowl adenovirus 825 or two fibre genes like the Goose adenovirus 48. Among the ORFs, ORF2 contained Parvovirus NS1 superfamily domain at 2nd–191st amino acids, as seen in some of the other waterfowl aviadenoviruses8. As reported earlier6, in the PBD12.16 sample, we detected an adeno-associated virus which may utilise the Pacific black duck aviadenovirus as a helper virus for its replication. However, currently, how the ORF2 protein assists the adeno-associated virus replication is uncertain.
A novel gyrovirus was found in the Grey teal 11.18 sample and was distantly related to other gyroviruses. Gyroviruses have been recently reclassified in the Anelloviridae family12. Gyroviruses have been found in cloacal, oral, and blood samples from birds27–29 and faecal samples of carnivorous mammals such as ferrets, cats and also humans30–34. However, the natural host of gyroviruses is thought to be birds. The finding of the GTGV/GT11.18 contributes to the wide genetic diversity of the gyroviruses. The Southern screamer gyrovirus (MH016740.1)11, the only other gyrovirus from a bird belonging to the order Anseriformes yet characterised, was distantly related to the Grey teal gyrovirus. Despite being very different to currently known gyroviruses, genetic analysis revealed that the N-terminal region of the VP1 protein is rich in basic amino acids and is a characteristic shared by all members of the Anelloviridae family12. The motif 173WWRWA177 in the VP1 protein of the GTGV/GT11.18 also appears to be conserved for all gyroviruses30. The VP2 of the GTGV/GT11.18 has the characteristic motif of protein tyrosine phosphatases which is the CX5R motif and the WX7HX3CXCX5H motif, that is conserved in other gyroviruses and anelloviruses12,13. A minimal signature motif with the configuration CX5R is highly conserved in all PTPases. PTPases catalyse the removal of phosphate from phosphotyrosine35 and may be important for the replication of the virus13. The VP3 protein of many gyroviruses, such as chicken anaemia virus, has some important motifs related to the induction of apoptosis in infected cells29,36–38. However, similar to some other gyroviruses from zoo and wild birds, no such related motif was found in the GTGV/GT11.18.
Reoviruses were detected and characterised in both Pacific black ducks and Chestnut teals collected in August 2018, which suggest that these reovirus infections may be more common in winter compared to other seasons. Other reovirus seasonal distribution studies in humans and chickens also show reovirus infections to be high during colder months of the year39–41.
The sigma C protein of the Pacific black duck orthoreovirus that forms part of the outer capsid and helps in host cell attachment of the virus particle42 was only 54.5% identical to its nearest homologous avian virus protein counterpart. This may be because of a specific host adaptation and to the best of our knowledge, this is the first report of an avian orthoreovirus from Pacific black ducks and more widely from a wild duck in Australia. It is also to be noted that sigma C is the most variable protein of avian orthoreoviruses and antibodies to this protein can neutralise the virus infection43. Nevertheless, further studies are required to determine whether the Pacific black duck orthoreovirus can infect other closely related host species that can lead to reassortment. However, the analysis of the other encoded proteins of the virus suggests that this virus may infect closely related host species leading to reassortment.
All the 11 segments of a Pacific black duck rotavirus G were detected and characterised from the PBD08.18 sample. The phylogenetic analysis of the VP6 protein, one of the most conserved proteins that form the intermediate layer of the capsid44, and the phylogenetic analysis of the VP1 protein that functions as the RdRp45,46 shows that rotavirus G forms two different clades. Clade B includes the rotavirus G from the Pacific black duck, Grey teal5 and Ruddy turnstone19 (all three viruses from Australia). In contrast, clade A includes the rotavirus G from chicken18, turkey17, avocet19 (from Australia) and pigeon (unpublished). The most diverse segments of PBDRVG/PBD08.18 were S3 (VP4 protein), S9 (VP7 protein) and S10 (NSP4 protein). VP4 and VP7 form part of the outer capsid and the genetic diversity may be due to host-related immune selection pressure. However, the NSP4, which act as a viral enterotoxin, is only 53.7% identical to its closest relative and could indicate the presence of a diverse group of rotavirus G in this wildlife population that may exhibit different patterns of virulence in these hosts.
The complete genome of rotavirus F has only been molecularly characterised from chickens18, although partial sequences of the same have been detected and characterised from a pig (unpublished) and partridge47. The current study is the first to detect and characterise rotavirus F from a duck species, albeit only a partial segment being identified. This is due to the low amount of virus (0.0001% of NGS reads) present in the sample. The genetic diversity and evolution of rotavirus F are less understood due to the limited characterisation of the virus, and further studies involving more infected duck samples may help in its elucidation.
The phylogenetic analysis of the avastrovirus from Pacific black ducks and Chestnut teals suggest that these viruses are quite different to one another with ~ 60% difference albeit being detected from the same location at different time points. This indicates the wide genetic diversity of the avastroviruses.
The Pacific black duck orthoreovirus (except for the segment S1) and the Pacific black duck rotavirus G from the PBD08.18 sample, and the Chestnut teal avastrovirus from the CT11.18 sample showed conservation at the amino acid sequences while being more distantly related at the nucleotide level to their closest relatives, respectively. Taken together, this may indicate that there is significant evolutionary distance/time between a common ancestor of the viruses, but that conservation at the amino acid sequence of the protein is subjected to a strong selective pressure. The conservation of protein structure across different avian hosts may also indicate that these viruses may be capable of cross-species infection or that cross-species infection is part of their evolutionary history. The close relatedness of the Northern pintail avastrovirus detected from China23 to the Chestnut teal avastrovirus suggests that the migration of birds (likely migratory shorebirds) along the East Asian—Australasian Flyway could be important in the evolution of these viruses, as similarly observed for an avian paramyxovirus 6 characterised earlier from a Pacific black duck sample collected from the same site 18 months prior to this sample3.
The Pacific black duck calicivirus is very closely related to the MK204392.1 Grey teal calicivirus from south west Victoria, Australia5. The Pacific black duck collection site is about 375 km away from the MK204392.1 Grey teal calicivirus collection site and was taken three months prior to the sample collected from the Grey teal. Both Pacific black duck and Grey teal are dabbling ducks found in Australia that may share habitat at any particular time point. Dabbling ducks can travel about 25 km to hundreds of kilometres per day48,49. Caliciviruses are thought to be transmitted through direct contact with an infected host or indirectly via oral-faecal route50. Thus, the calicivirus detected in the current study may indicate cross-species transmission and infection occurs between these related duck species.
It should be noted that at the collection time, no clinical signs of disease were observed in the ducks and no close physical or extended examination such as post mortem or histopathological examination, was carried out. However, the high abundance of virus reads for some of the viruses from the bird samples may indicate an active infection and shedding of the virus. Aviadenoviruses can cause gizzard erosion or severe, often fatal cases of hepatitis-hydropericardium syndrome or inclusion body hepatitis depending on the virus and the host species25,51,52. For viruses in the genus Gyrovirus, Chicken anaemia virus is so far known to be the only pathogenic member of that genus12 causing an economically important clinical and subclinical disease in young chickens, with a worldwide distribution53. Nonetheless, a gyrovirus was also sequenced from Northern fulmar (Fulmarus glacialis) and Southern screamers (Chauna torquata) with neurological clinical signs11,28, as well as from healthy birds11.
Although avian orthoreoviruses have been associated with various disease conditions, a direct link between the virus and disease has been demonstrated only for viral arthritis syndrome or tenosynovitis in chickens42. However, avian orthoreoviruses were sequenced from diseased Muscovy ducks that had severe haemorrhagic and necrotic lesions in the liver and spleen54 and from Pekin ducks that had spleen necrosis55. Rotavirus G and rotavirus F were found in chickens with the runting-stunting syndrome; however, its role in the disease remains unclear56.
Among birds, astroviruses have been associated with various diseases such as enteritis, hepatitis, nephritis and runting-stunning syndrome57–59. However, the pathogenicity of the viruses described in the current study is currently unknown, and further studies involving histopathological examinations are required for its elucidation. Nevertheless, all of the viruses described in the current study may spread through the faecal-oral route27,42,60 and may cause asymptomatic infections as observed of the wild ducks in the current study.
The current study was carried out by sampling several representative bird species from a bird community at specific time points from a single location in an effort to comprehend virus diversity, abundance and seasonal prevalence. The Grey teal gyrovirus was the most abundant virus (414 reads at Q ≥ 20) from the GT11.18 sample and exceeded the two parvoviruses previously reported from this sample with 141 and 222 reads at Q ≥ 20 respectively6. The generation of the partial sequences of the other avian viruses from the duck samples described here may be due to various factors, including but not limited to, virus particle quantity, stage of infection and library preparation. Also, as stated earlier, the most abundant avian virus from each of the duck samples, except for the GT11.18 sample, was either an avian parvovirus or an avian picornavirus6. Future studies involving more extensive sample collection may lead to the complete genome elucidation of the viruses described here. However, the presence of motifs at the expected location in the generated virus consensus sequences suggests that they form part of a functional virus genome, although the description of each motif functions is beyond the scope of the manuscript. Furthermore, taken together with the previously described viruses1,3,4,6, the detection of more than one virus from each sample indicates co-infection, which could have a more significant health impact on the host when compared to infection with an individual virus.
Detection and characterisation of several potentially pathogenic avian viruses, some genetically novel while others similar to those detected in other avian hosts, indicates further virus surveillance studies of wild ducks are warranted. Ducks being a natural reservoir of pathogenic and zoonotic viruses should be monitored continuously to detect any potential threat to other wildlife and humans. Future pathological studies on the pathogenicity of the characterised viruses will also aid in comprehending the host–virus relationship.
Materials and methods
Sample collection
The current study uses the samples reported earlier6. Fresh wild duck faecal samples of Pacific black ducks (PBD), Chestnut teals (CT), Grey teals (GT) and Wood ducks (WD) were collected in May 2018 (late autumn), August 2018 (late winter), November 2018 and December 2016 (late spring/early summer) from Wallington, south-eastern Victoria, Australia. Pacific black ducks were captured in May 2018 (PBD05.18), August 2018 (PBD08.18) and December 2016 (PBD12.16 (described as MAD previously3)). Chestnut teal samples were collected in May (CT05.18), August (CT08.18) and November 2018 (CT11.18). Grey teal samples were collected in November 2018 (GT11.18) only, while Wood duck samples were collected in August 2018 (WD08.18) only. All samples were stored at − 80 °C within 1–3 h of collection until processing.
Bird sample collection was approved under Deakin University’s Animal Ethics Committee project number B43–2016 and Department of Environment, Land, Water and Planning permit number 1008206. The current study involving these samples were performed in accordance with relevant guidelines and regulations.
Virus enrichment from samples
Virus particles enrichment and virus nucleic acid extraction was carried out as per a previously optimised protocol in our laboratory3. Briefly, the faecal samples were subjected to various biophysical methods such as homogenisation (25 Hz for 2 min), centrifugation (17000×g for 3 min) and filtration using a 0.8 µm PES filter (17000×g for 1 min). The sample was then divided into two aliquots (aliquot A—ultracentrifuged, aliquot B—non-ultracentrifuged) as described earlier3. Both aliquots were then nuclease treated using benzonase and micrococcal nuclease (37 °C for 2 h), followed by nucleic acid extraction using the QIAamp Viral RNA mini kit (Qiagen), as previously described3.
Next-generation sequencing (NGS)
cDNA synthesis and amplification was carried out using the SeqPlex RNA Amplification Kit (Sigma) as per the manufacturer’s instructions using the extracted virus nucleic acids from both aliquots of the samples. Next generation sequencing library preparation was performed using the Ion Fragment Library kit (Life Technologies) followed by quantification of libraries, as described earlier3. Libraries were then pooled and were loaded onto Ion 530 or 540 chips using the Ion Chef Instrument. Template preparation was carried out and then the chips were run on an Ion Torrent S5XL System (Thermo Fisher Scientific) as per company protocols and as described earlier3.
NGS data analyses
NGS data analyses were carried out as described earlier3,6,61,62. Two local BLAST datasets were created for all the available virus sequences and all RefSeq virus sequences from the NCBI GenBank genetic sequence database (Dec 2018), as described earlier6. BLASTN and TBLASTX query against the two virus reference sequence databases was performed with an e-value cut-off of 1 × 10−10 and 1 × 10–30. The query results files were converted into spreadsheet files, sorted by virus matches, a list of potential virus targets created and then viruses of interest were identified for each sample, as described earlier3,6.
The NGS reads were mapped against reference virus genomes of interest using the TMAP plugin on the Ion Torrent server. The mapped reads were used to obtain full or partial consensus sequences of the viruses using Integrative Genomics Viewer software (IGV) (Broad Institute, MA, USA), as described earlier3,6,61,62. Contigs were generated from the AssemblerSPAdes 5.6.0 plugin on the Ion Torrent Server by using the NGS reads of each sample. The contigs were bioinformatically analysed to identify virus sequences as described above using the same e-value cut-offs. Virus contigs (> 500 nucleotides) were then used as references in TMAP plugin and trimmed to regions with a mapping quality of 80 or higher and a coverage depth of at least 2 unless specified, as described earlier3,6. Virus sequences were also generated by assembling overlapping sequences from contigs and consensus sequences by using MEGA7 and magicblast63. Final virus consensus sequences were then generated after subjecting the sequences again to TMAP and mapping using IGV, as described above.
Grey teal gyrovirus PCR and Sanger sequencing
Real-time PCR amplification and Sanger sequencing were carried out for Grey teal gyrovirus from the GT11.18 sample for the region from 264th to 761st nucleotide, as a potential indication of this virus being different from other gyroviruses was observed during NGS analysis. The GTGV/GT11.18 was the most abundant virus from the sample compared to the two parvoviruses previously reported6, and the region of PCR analysis had NGS reads with very low abundance. Primer-BLAST64 software was used to design two sets of primers (Micormon—Monash University, Victoria, Australia). FP1-GTGV-347nt (5′-CCCGGAAACCTGTACGAGTC-3′) RP1-GTGV-761nt (5′-TCTCCTGTAGTGGCGTCTGA-3′) and FP2-GTGV-264nt (5′-TCCTGTGGCAACTTTCGGAG-3′) RP2-GTGV-756nt (5′-TGTAGTGGCGTCTGAATCGG-3′) were used as the forward and reverse primers. The PCR master mix was prepared with 2X AmpliTaq Gold™ 360 Master Mix with 1 mM of each primer and 2 µl of DNA in a total reaction volume of 10 µl. The PCR reaction was carried out in QuantStudio™ Flex 6 real-time thermal cycler (Applied Biosystems) using the following conditions: 95 °C for 10 min, 40 cycles of 95 °C for 30 s, 54 °C for 30 s, 72 °C for 30 s, followed by a final step of 72 °C for 3 min. This is followed by a melt curve stage of 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s and a final hold at 4 °C. The two PCR products of ~ 434 bp size and ~ 512 bp size, respectively, were purified using the 2% Size Select E-Gel System (Thermo Fisher, USA). These products were sequenced using the Big Dye Terminator Cycle v3.1 on a Hitachi 3500XL Genetic Analyzer (Applied Biosystems, USA) as per the manufacture instructions.
ORF prediction and motif analysis
Basic Local Alignment Search Tool65,66 (BLASTN, BLASTX and BLASTP), NCBI ORFfinder67, ScanProsite68 and PSORT II69 were used for identifying open reading frames (ORFs), and features of the virus genomes such as promotors, motifs and nuclear-locating signals, as described earlier6. The presence of these features in their expected location was used as an additional confirmation of the generated consensus sequences.
Phylogenetic analysis of virus sequences
Relevant related sequences were selected from the NCBI GenBank database. Nucleotide and protein sequences were aligned using Clustal-W70 in MEGA 771 software. MEGA7/MEGACC software was used to identify the best evolutionary model and the selected model was then used for generating the maximum likelihood tree with a bootstrapping of 1000 replicates. The distance data between sequences were calculated using the same software.
Supplementary Information
Acknowledgements
The work was supported by funding from the Australian Government Research Training Program Scholarship through Deakin University; NHMRC Equipment Grant GNT9000413 and NHMRC CRE APPRISE APP1116530 through Deakin University and funding provided by Deakin University, Barwon Health and CSIRO to Soren Alexandersen; BirdLife Australia funding to Jessy Vibin; and ARC funding to Marcel Klaassen. We also acknowledge Dr Jason Hodge, laboratory manager of the GCEID laboratory, and Mr Tarka Raj Bhatta for the laboratory work support.
Author contributions
S.A. coordinated the work carried out at GCEID. M.K. collected bird samples. J.V. performed the laboratory work, including nucleic acid sequencing. J.V. performed sanger sequencing under the guidance of A.C. NGS data analysis and Sanger sequencing data analysis was carried out by J.V., A.C. and S.A. J.V. drafted the initial manuscript together with S.A., and later versions were based on input and suggestions from all. All authors contributed to the final submitted version.
Data availability
All sequences analysed have been deposited in NCBI GenBank under accession numbers MT894381-MT894428. Other datasets generated or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jessy Vibin, Email: jessyatall@gmail.com.
Soren Alexandersen, Email: soren.alexandersen@deakin.edu.au.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-79413-9.
References
- 1.Bhatta TR, Chamings A, Vibin J, Klaassen M, Alexandersen S. Detection of a reassortant H9N2 avian influenza virus with intercontinental gene segments in a resident australian chestnut teal. Viruses. 2020;12:88. doi: 10.3390/v12010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JK, Negovetich NJ, Forrest HL, Webster RG. Ducks: The ‘Trojan Horses’ of H5N1 influenza. Influenza Other Respir. Viruses. 2009;3:121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vibin J, et al. Metagenomics detection and characterisation of viruses in faecal samples from Australian wild birds. Sci. Rep. 2018;8:8686. doi: 10.1038/s41598-018-26851-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamings A, et al. Detection and characterisation of coronaviruses in migratory and non-migratory Australian wild birds. Sci. Rep. 2018;8:5980. doi: 10.1038/s41598-018-24407-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wille M, Shi M, Klaassen M, Hurt AC, Holmes EC. Virome heterogeneity and connectivity in waterfowl and shorebird communities. ISME J. 2019 doi: 10.1038/s41396-019-0458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vibin J, Chamings A, Klaassen M, Bhatta TR, Alexandersen S. Metagenomic characterisation of avian parvoviruses and picornaviruses from Australian wild ducks. Sci. Rep. 2020;10:12800. doi: 10.1038/s41598-020-69557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marek A, et al. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology. 2014;462–463:107–114. doi: 10.1016/j.virol.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 8.Kaján GL, Davison AJ, Palya V, Harrach B, Benko M. Genome sequence of a waterfowl aviadenovirus, goose adenovirus 4. J. Gen. Virol. 2012;93:2457–2465. doi: 10.1099/vir.0.042028-0. [DOI] [PubMed] [Google Scholar]
- 9.Benko M, Harrach B, Both G, Russell W, Adair B. ‘Adenoviridae’ Virus Taxonomy: VIIIth Report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier; 2005. [Google Scholar]
- 10.Anelloviridae - ssDNA Viruses (2011) International Committee on Taxonomy of Viruses (ICTV). https://talk.ictvonline.org/ictv-reports/ictv_9th_report/ssdna-viruses-2011/w/ssdna_viruses/139/anelloviridae. Accessed 16 Nov 2020.
- 11.Goldberg TL, Clyde VL, Gendron-Fitzpatrick A, Sibley SD, Wallace R. Severe neurologic disease and chick mortality in crested screamers (Chauna torquata) infected with a novel Gyrovirus. Virology. 2018;520:111–115. doi: 10.1016/j.virol.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Rosario K, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017;162:1447–1463. doi: 10.1007/s00705-017-3247-y. [DOI] [PubMed] [Google Scholar]
- 13.Peters MA, Jackson DC, Crabb BS, Browning GF. Chicken anemia virus VP2 is a novel dual specificity protein phosphatase. J. Biol. Chem. 2002;277:39566–39573. doi: 10.1074/jbc.M201752200. [DOI] [PubMed] [Google Scholar]
- 14.Yun T, et al. Complete genomic sequence of goose-origin reovirus from China. J. Virol. 2012;86:10257. doi: 10.1128/JVI.01692-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma G, et al. Complete genomic sequence of a reovirus isolate from Pekin ducklings in China. J. Virol. 2012;86:13137. doi: 10.1128/JVI.02512-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farkas SL, et al. Genomic sequence and phylogenetic analyses of two novel orthoreovirus strains isolated from Pekin ducks in 2014 in Germany. Virus Res. 2018;257:57–62. doi: 10.1016/j.virusres.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, et al. Genome characterization of Turkey rotavirus G strains from the United States identifies potential recombination events with human rotavirus B strains. J. Gen. Virol. 2017;98:2931–2936. doi: 10.1099/jgv.0.000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kindler E, Trojnar E, Heckel G, Otto PH, Johne R. Analysis of rotavirus species diversity and evolution including the newly determined full-length genome sequences of rotavirus F and G. Infect. Genet. Evol. 2013;14:58–67. doi: 10.1016/j.meegid.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Wille M, et al. Virus-virus interactions and host ecology are associated with RNA virome structure in wild birds. Mol. Ecol. 2018;27:5263–5278. doi: 10.1111/mec.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook JP, McCrae MA. Sequence analysis of the guanylyltransferase (VP3) of group A rotaviruses. J. Gen. Virol. 2004;85:929–932. doi: 10.1099/vir.0.19629-0. [DOI] [PubMed] [Google Scholar]
- 21.Kumar M, et al. Crystallographic and biochemical analysis of rotavirus NSP2 with nucleotides reveals a nucleoside diphosphate kinase-like activity. J. Virol. 2007;81:12272–12284. doi: 10.1128/JVI.00984-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N, Wang F, Zhang D. Complete sequence of a novel duck astrovirus. Arch. Virol. 2014;159:2823–2827. doi: 10.1007/s00705-014-2141-0. [DOI] [PubMed] [Google Scholar]
- 23.Chu DKW, et al. A novel group of avian astroviruses in wild aquatic birds. J. Virol. 2012;86:13772–13778. doi: 10.1128/JVI.02105-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vinjé J, et al. ICTV virus taxonomy profile: Caliciviridae. J. Gen. Virol. 2019;100:1469–1470. doi: 10.1099/jgv.0.001332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grgić H, Yang DH, Nagy É. Pathogenicity and complete genome sequence of a fowl adenovirus serotype 8 isolate. Virus Res. 2011;156:91–97. doi: 10.1016/j.virusres.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Marek A, et al. Whole-genome sequences of two Turkey adenovirus types reveal the existence of two unknown lineages that merit the establishment of novel species within the genus Aviadenovirus. J. Gen. Virol. 2014;95:156–170. doi: 10.1099/vir.0.057711-0. [DOI] [PubMed] [Google Scholar]
- 27.Truchado DA, et al. A novel and divergent gyrovirus with unusual genomic features detected in wild passerine birds from a remote rainforest in French Guiana. Viruses. 2019;11:1148. doi: 10.3390/v11121148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, et al. A gyrovirus infecting a sea bird. Arch. Virol. 2015;160:2105–2109. doi: 10.1007/s00705-015-2468-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao S, et al. Novel characteristics of the avian gyrovirus 2 genome. Sci. Rep. 2017;7:41068. doi: 10.1038/srep41068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehér E, Pazár P, Lengyel G, Phan TG, Bányai K. Sequence and phylogenetic analysis identifies a putative novel gyrovirus 3 genotype in ferret feces. Virus Genes. 2015;50:137–141. doi: 10.1007/s11262-014-1128-y. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, et al. Identification of a chicken anemia virus variant-related gyrovirus in stray cats in china, 2012. Biomed. Res. Int. 2014;2014:313252. doi: 10.1155/2014/313252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu DKW, et al. Characterization of a novel gyrovirus in human stool and chicken meat. J. Clin. Virol. 2012;55:209–213. doi: 10.1016/j.jcv.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gia Phan T, et al. Divergent gyroviruses in the feces of Tunisian children. Virology. 2013;446:346–348. doi: 10.1016/j.virol.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fehér E, et al. Molecular detection and characterization of human gyroviruses identified in the ferret fecal virome. Arch. Virol. 2014;159:3401–3406. doi: 10.1007/s00705-014-2203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W-Q, Sun J-P, Zhang Z-Y. An overview of the protein tyrosine phosphatase superfamily. Curr. Top. Med. Chem. 2005;3:739–748. doi: 10.2174/1568026033452302. [DOI] [PubMed] [Google Scholar]
- 36.Bullenkamp J, et al. Human Gyrovirus apoptin shows a similar subcellular distribution pattern and apoptosis induction as the chicken anaemia virus derived VP3/Apoptin. Cell Death Dis. 2012;3:e296. doi: 10.1038/cddis.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noteborn MH, et al. A single chicken anemia virus protein induces apoptosis. J. Virol. 1994;68:346–351. doi: 10.1128/JVI.68.1.346-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Danen-Van Oorschot AAAM, et al. Importance of nuclear localization of apoptin for tumor-specific induction of apoptosis. J. Biol. Chem. 2003;278:27729–27736. doi: 10.1074/jbc.M303114200. [DOI] [PubMed] [Google Scholar]
- 39.Patel MM, et al. Global seasonality of rotavirus disease. Pediatr. Infect. Dis. J. 2013;32:1. doi: 10.1097/INF.0b013e31826fb32d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapikian AZ, Shope RE. Rotaviruses, Reoviruses, Coltiviruses, and Orbiviruses Medical Microbiology. Galveston: University of Texas Medical Branch; 1996. [PubMed] [Google Scholar]
- 41.Nham EG, et al. Flock-level prevalence, geographical distribution, and seasonal variation of avian reovirus among broiler flocks in Ontario. Can. Vet. J. 2017;58:828–834. [PMC free article] [PubMed] [Google Scholar]
- 42.Benavente J, Martínez-Costas J. Avian reovirus: Structure and biology. Virus Res. 2007;123:105–119. doi: 10.1016/j.virusres.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Goldenberg D, et al. Genetic and antigenic characterization of sigma C protein from avian reovirus. Avian Pathol. 2010;39:189–199. doi: 10.1080/03079457.2010.480969. [DOI] [PubMed] [Google Scholar]
- 44.Johne R, et al. Sequence analysis of the VP6-encoding genome segment of avian group F and G rotaviruses. Virology. 2011;412:384–391. doi: 10.1016/j.virol.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Ogden KM, Johne R, Patton JT. Rotavirus RNA polymerases resolve into two phylogenetically distinct classes that differ in their mechanism of template recognition. Virology. 2012;431:50–57. doi: 10.1016/j.virol.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu X, et al. Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1. Structure. 2008;16:1678–1688. doi: 10.1016/j.str.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Falcone E, et al. Molecular characterization of avian rotaviruses circulating in Italian poultry flocks. Avian Pathol. 2015;44:509–515. doi: 10.1080/03079457.2015.1096011. [DOI] [PubMed] [Google Scholar]
- 48.Bengtsson D, et al. Movements, home-range size and habitat selection of mallards during autumn migration. PLoS ONE. 2014;9:e100265. doi: 10.1371/journal.pone.0100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roshier D, Klomp N, Asmus M. Movements of a nomadic waterfowl, Grey Teal Anas gracilis, across inland Australia: Results from satellite telemetry spanning fifteen months. Ardea. 2006;94:461–475. [Google Scholar]
- 50.Smith AW, Skilling DE, Cherry N, Mead JH, Matson DO. Calicivirus emergence from ocean reservoirs: Zoonotic and interspecies movements. Emerg. Infect. Dis. 1998;4:13–20. doi: 10.3201/eid0401.980103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yugo DM, Hauck R, Shivaprasad HL, Meng X-J. Hepatitis virus infections in poultry. Avian Dis. 2016;60:576–588. doi: 10.1637/11229-070515-Review.1. [DOI] [PubMed] [Google Scholar]
- 52.Schachner A, Matos M, Grafl B, Hess M. Fowl adenovirus-induced diseases and strategies for their control: a review on the current global situation. Avian Pathol. 2018;47:111–126. doi: 10.1080/03079457.2017.1385724. [DOI] [PubMed] [Google Scholar]
- 53.Fatoba AJ, Adeleke MA. Chicken anemia virus: A deadly pathogen of poultry. Acta Virol. 2019;63:19–25. doi: 10.4149/av_2019_110. [DOI] [PubMed] [Google Scholar]
- 54.Zhang XL, et al. Molecular characterization of two novel reoviruses isolated from Muscovy ducklings in Guangdong. China. BMC Vet. Res. 2019;15:1. doi: 10.1186/s12917-018-1758-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H, Gao B, Chen H, Diao Y, Tang Y. Isolation and characterization of a variant duck orthoreovirus causing spleen necrosis in Peking ducks, China. Transbound. Emerg. Dis. 2019;66:2033–2044. doi: 10.1111/tbed.13252. [DOI] [PubMed] [Google Scholar]
- 56.Otto P, et al. Detection of rotaviruses and intestinal lesions in broiler chicks from flocks with runting and stunting syndrome (RSS) Avian Dis. 2006;50:411–418. doi: 10.1637/7511-020106R.1. [DOI] [PubMed] [Google Scholar]
- 57.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. Astrovirus infections in humans and animals: Molecular biology, genetic diversity, and interspecies transmissions. Infect. Genet. Evol. 2011;11:1529–1544. doi: 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Donato C, Vijaykrishna D. The broad host range and genetic diversity of mammalian and avian astroviruses. Viruses. 2017;9:102. doi: 10.3390/v9050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang K, et al. Chicken astrovirus as an aetiological agent of runting-stunting syndrome in broiler chickens. J. Gen. Virol. 2018;99:512–524. doi: 10.1099/jgv.0.001025. [DOI] [PubMed] [Google Scholar]
- 60.Kohn A. Gallus adeno-like virus in chickens–studies on infection, excretion, and immunity. Am. J. Vet. Res. 1962;23:562–568. [PubMed] [Google Scholar]
- 61.Nelson TM, et al. An outbreak of severe infections among Australian infants caused by a novel recombinant strain of human parechovirus type 3. Sci. Rep. 2017;7:44423. doi: 10.1038/srep44423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexandersen S, Nelson TM, Hodge J, Druce J. Evolutionary and network analysis of virus sequences from infants infected with an Australian recombinant strain of human parechovirus type 3. Sci. Rep. 2017;7:3861. doi: 10.1038/s41598-017-04145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang W, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol. 2015;16:133. doi: 10.1186/s13059-015-0694-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye J, et al. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 66.Mount, D. W. Using the basic local alignment search tool (BLAST). Cold Spring Harb. Protoc.2007, pdb.top17 (2007). [DOI] [PubMed]
- 67.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Castro E, et al. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakai K, Horton P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/S0968-0004(98)01336-X. [DOI] [PubMed] [Google Scholar]
- 70.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 71.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 73.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 74.Kumar S, Stecher G, Peterson D, Tamura K. MEGA-CC: Computing core of molecular evolutionary genetics analysis program for automated and iterative data analysis. Bioinformatics. 2012;28:2685–2686. doi: 10.1093/bioinformatics/bts507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen S-Y, et al. The isolation and identification of novel duck reovirus. Chinese J. Virol. 2012;28:224–230. [PubMed] [Google Scholar]
- 76.Yun T, et al. Isolation and genomic characterization of a classical Muscovy duck reovirus isolated in Zhejiang, China. Infect. Genet. Evol. 2013;20:444–453. doi: 10.1016/j.meegid.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 77.Kugler R, et al. Phylogenetic analysis of a novel reassortant orthoreovirus strain detected in partridge (Perdix perdix) Virus Res. 2016;215:99–103. doi: 10.1016/j.virusres.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 78.Wang D, Shi J, Yuan Y, Zheng L, Zhang D. Complete sequence of a reovirus associated with necrotic focus formation in the liver and spleen of Muscovy ducklings. Vet. Microbiol. 2013;166:109–122. doi: 10.1016/j.vetmic.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 79.Pizarro JL, Sandino AM, Pizarro JM, Fernandez J, Spencer E. Characterization of rotavirus guanylyltransferase activity associated with polypeptide VP3. J. Gen. Virol. 1991;72:325–332. doi: 10.1099/0022-1317-72-2-325. [DOI] [PubMed] [Google Scholar]
- 80.Barro M, Patton JT. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 2007;81:4473–4481. doi: 10.1128/JVI.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graff JW, Ettayebi K, Hardy ME. Rotavirus NSP1 inhibits NFκB activation by inducing proteasome-dependent degradation of β-TrCP: A novel mechanism of IFN antagonism. PLoS Pathog. 2009;5:e100028. doi: 10.1371/journal.ppat.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fabbretti E, Afrikanova I, Vascotto F, Burrone OR. Two non-structural rotavirus proteins, NSP2 and NSP5, form viroplasm-like structures in vivo. J. Gen. Virol. 1999;80:333–339. doi: 10.1099/0022-1317-80-2-333. [DOI] [PubMed] [Google Scholar]
- 83.Ball JM, Mitchell DM, Gibbons TF, Parr RD. Rotavirus NSP4: A multifunctional viral enterotoxin. Viral Immunol. 2005;18:27–40. doi: 10.1089/vim.2005.18.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences analysed have been deposited in NCBI GenBank under accession numbers MT894381-MT894428. Other datasets generated or analysed during the current study are available from the corresponding author on reasonable request.