Abstract
Purpose of review:
This review summarizes mechanisms by which Porphyromonas gingivalis interacts with community members and the host so that it can persist in the periodontium under inflammatory conditions that drive periodontal disease.
Recent findings:
Recent advances indicate that, in great part, the pathogenicity of P. gingivalis is dependent upon its ability to establish residence in the subgingival environment and to subvert innate immunity in a manner that uncouples the nutritionally favorable (for the bacteria) inflammatory response from antimicrobial pathways. While the initial establishment of P. gingivalis is dependent upon interactions with early colonizing bacteria, the immune subversion strategies of P. gingivalis in turn benefit co-habiting species.
Summary:
Specific interspecies interactions and subversion of the host response contribute to the emergence and persistence of dysbiotic communities and are thus targets of therapeutic approaches for the treatment of periodontitis.
Keywords: P. gingivalis, inflammation, dysbiosis, immune subversion, periodontitis
Introduction
Perhaps the best-studied oral microorganism, Porphyromonas gingivalis, is a Gram-negative anaerobic and asaccharolytic bacterium that is implicated as a contributor in human periodontitis and is also suspected to play a role in systemic disorders, such as atherosclerosis, aspiration pneumonia, rheumatoid arthritis and Alzheimer’s disease [1–9]. P. gingivalis expresses an array of virulence factors, such as cysteine proteinases (gingipains), lipopolysaccharide (LPS), hemagglutinins, and adhesive hair-like appendages known as fimbriae [10–14]. These molecules are thought to contribute to the ability of P. gingivalis to colonize, secure nutrients, and persist within the inflammatory environment of the periodontal pocket while evading host immunity [15, 16, 12, 17–21]. However, the potential pathogenicity of P. gingivalis is better appreciated through interactions with certain partner species and in the context of the greater microbial community. In this regard, recent advances from microbiome and mechanistic studies suggest a new model of periodontal disease pathogenesis, in which disease arises not from individual pathogens but from polymicrobial synergy and dysbiosis, which perturbs the ecologically balanced microbiota associated with periodontal health [22–31]. Here we review how P. gingivalis, as a community member, interacts with the host and other microbes leading to dysbiotic inflammation that contributes to periodontal disease development. This work is dedicated to Dr. Robert (Bob) J. Genco, a former mentor of one of the co-authors (GH), who is commemorated in this issue for his pivotal role in periodontal research and his inspiring influence on countless scientists in the field. Much of the progress achieved on the biology of P. gingivalis and its fimA-encoded fimbriae is based on pioneering studies by Bob and his group [32–41, 10].
P. gingivalis in heterotypic bacterial communities
The predominant habitat of P. gingivalis is in the subgingival crevice which develops into a periodontal pocket in periodontitis. Although P. gingivalis is indigenous to the oral cavity of humans, it can only be detected in a small fraction of periodontally healthy subjects. At a given time, P. gingivalis can be detected via a sensitive and specific PCR assay in 25% of subjects presenting with periodontal health or mild periodontitis (with pocket probing depths 5 mm or less). In contrast, it is detected in 79% of subjects with deep pockets [42]. The interactions with other microbial species in subgingival biofilms and the inflammatory products released by the host are likely to determine whether P. gingivalis can successfully establish in the subgingival environment.
The development of subgingival microbial communities is a complex process dependent upon stable attachment to the substratum, interspecies adhesive interactions, metabolic compatibility, and capacity of the community to resist host immunity while maintaining an inflammatory environment [31, 2, 43]. P. gingivalis depends on early colonizers, such as oral streptococci, to support its initial attachment and biofilm formation [44]. For instance, P. gingivalis engages in multivalent co-adherence interactions with Streptococcus gordonii, in which the major (FimA) and the minor (Mfa1) fimbrial subunit proteins of the former interact, respectively, with the surface proteins glyceraldehyde-3-phosphate dehydrogenase and SspA/B of the latter [45, 46]. Dual-species communities of P. gingivalis and S. gordonii display mutualistic growth in in vitro models [47, 48]. Numerous interactions between co-adhered S. gordonii and P. gingivalis have been characterized including the elucidation that an S. gordonii gene, cbe, is essential for P. gingivalis accumulation on the streptococcal surface [49]. Later work revealed that the product of cbe, the folate precursor 4-aminobenzoate/para-amino benzoic acid (pABA) is responsible for enhancing P. gingivalis biofilm accumulation [50]. Exogenously added pABA was shown to alter the transcriptome and metabolome of P. gingivalis, increasing the expression of the adhesive fimbrial components FimA and Mfa1, among other effects. Consistent with this finding, pABA was seen to facilitate colonization of P. gingivalis in the mouse oral cavity. However, despite increased numbers, pABA-treated P. gingivalis induced significantly less bone loss than vehicle-treated cells. These findings show that the interaction of P. gingivalis and early-colonizing partners could modulate in a different manner colonization and pathogenicity.
P. gingivalis interactions with viridans-group streptococci may also be antagonistic. For instance, Streptococcus cristatus-derived arginine deiminase inhibits the expression of the FimA fimbrial protein in P. gingivalis leading to impaired periodontal colonization and pathogenicity of P. gingivalis in mice, consistent with observations that the distribution of P. gingivalis and S. cristatus is negatively correlated in the human subgingival biofilm [51–53]. Thus, distinct streptococcal species can potentially promote or suppress P. gingivalis colonization. Conversely, P. gingivalis has been shown to induce cell death and DNA fragmentation of the health-associated commensal Streptococcus mitis, although the mechanism behind this interaction is still unknown [54].
Other early colonizers and ubiquitous components of health-associated biofilms, such as Veillonella and Fusobacterium species, have been shown to interact synergistically with P. gingivalis. Veillonella atypica coaggregates with P. gingivalis via the Veillonella hemagglutinin protein Hag1 [55]. Veillonella species are also proposed to be important for the colonization of P. gingivalis as they possess the gene repertoire for de novo biosynthesis of heme, which is the iron form required by P. gingivalis for growth. Indeed, V. atypica supports the in vitro growth of P. gingivalis when heme is excluded from the growth medium and deletion of hemE in V. atypica, which abolishes heme synthesis, results in an abrogation of the growth-supporting effect [56]. Since P. gingivalis is an anaerobe, its colonization requires interactions with other community members able to reduce the environment to oxygen levels it can tolerate. Fusobacterium nucleatum, a ubiquitous oxygen-tolerant species has been shown to be important for the survival of other anaerobes, including P. gingivalis, in continuous culture chemostat models under aerated conditions [57, 58]. Although F. nucleatum is also an anaerobe, it possesses enzymatic activities such as that of NADH oxidase, which contribute to the rapid metabolism of oxygen and the reduction of the environment [57, 59]. In addition, F. nucleatum supports the growth of P. gingivalis by supplying it with carbon dioxide [57].
P. gingivalis engages in synergistic interactions with other pathogenic organisms in subgingival microbial communities. For example, isobutyric acid production by P. gingivalis stimulates the growth of Treponema denticola, and reciprocally T. denticola produces succinic acid that promotes P. gingivalis growth [60]. Moreover, upon contact with T. denticola, P. gingivalis upregulates the expression of adhesins and proteases [61]. Consistent with these in vitro findings, P. gingivalis and T. denticola are synergistically pathogenic in vivo [62, 63]. With regard to cross-kingdom interactions, the internalin family surface protein InlJ of P. gingivalis binds to the candidal hyphal protein Als3 of Candida albicans leading to the upregulation of genes encoding components of the Type IX section system (T9SS) of P. gingivalis [64]. Given that important virulence factors of P. gingivalis, including its gingipains, are secreted through the T9SS, communities of P. gingivalis and C. albicans may exhibit increased pathogenicity. In this regard, C. albicans colonizes the periodontal pockets of approximately 15–20% of chronic periodontitis patients and its presence in periodontal pockets is associated with the severity of chronic periodontitis [65–68].
In vivo interactions are bound to be more complex than those elucidated using the reductionist model systems utilized by most of the studies described above. Simplified models, however, clearly show that the interaction of P. gingivalis with other subgingival species is a determinant of its colonization and pathogenic potential. While reductionist models do not capture the complex emergent characteristics of in vivo communities, they represent a proof of the concept that the community context plays a role in modulating the establishment of P. gingivalis.
P. gingivalis subversion of innate immunity
Progress in the past 20 years has increased our understanding of how P. gingivalis resists immune elimination and persists in periodontal tissues. It would not be possible to discuss all relevant research and the present review will focus primarily on how P. gingivalis manipulates Toll-like receptors (TLRs) and co-receptors as well as the complement system which intimately interacts with TLRs. Other evasive mechanisms of this oral pathogen, such as, to inhibit production of the IL-8 chemokine by gingival epithelial cells [69], to degrade secreted cytokines [70] as well as many other tactics have been covered in earlier reviews [71, 12, 72, 73].
TLRs, a major family of pattern-recognition receptors (PRRs), are strategically located at the host-microbe interface [74, 75]. They recognize conserved microbial structures known as microbe-associated molecular patterns (MAMPs) and play a central role in inducing innate immune responses for controlling infection [74, 75]. Different TLRs respond to distinct MAMPs (e.g., TLR2 responds to lipoteichoic acid and TLR4 to LPS) thus endowing the innate immune response with relative specificity [76]. TLRs do not function in isolation but cooperate with other PRRs in multireceptor complexes within membrane lipid rafts [76–78]. In this regard, TLR4 alone is not sufficient for inducing a vigorous innate response to LPS and requires MD-2 and CD14 [79, 80]. TLR2 responds to its ligands in association with TLR1 or TLR6 as signaling partners, and with CD14 or CD36 as important coreceptors for robust activation of TLR2/1 or TLR2/6 complexes [76]. Moreover, TLRs engage in signaling crosstalk with the complement system, apparently to coordinate innate immunity and inflammation via either synergistic or antagonistic interactions [81, 82]. However, it seems that P. gingivalis has evolved mechanisms whereby it can exploit TLRs or their crosstalk interactions in a manner that increases its adaptive fitness as well as the pathogenicity (nososymbiocity) of the microbial community where it resides [83]. Indeed, P. gingivalis evades leukocyte killing by exploiting signaling crosstalk between TLRs and other receptors, such as CXC-chemokine receptor 4 (CXCR4) [84–86], complement receptor 3 (CR3; Mac1) [87, 88], and complement C5a receptor 1 (C5aR1) [89–91]. Although first shown for P. gingivalis, some of these evasion mechanisms were later shown to be exploited by other pathogens, such as Bacillus anthracis and Francisella tularensis [92, 93] or even malignant tumors [94]. Strikingly, the manipulation of the periodontal host response by P. gingivalis benefits the entire microbial community, which becomes dysbiotic (altered composition and increased total counts) and causes inflammatory periodontal bone loss [26].
The ability of P. gingivalis to orchestrate experimental periodontitis in mice by promoting the emergence of a dysbiotic microbial community [91, 26], while being a quantitatively minor constituent of the microbiota has prompted its characterization as a keystone pathogen (Figure 1), by analogy to the crucial role of a keystone holding an entire arch together [26, 95, 27].
Figure 1. Keystone pathogen-induced dysbiosis in periodontal disease.
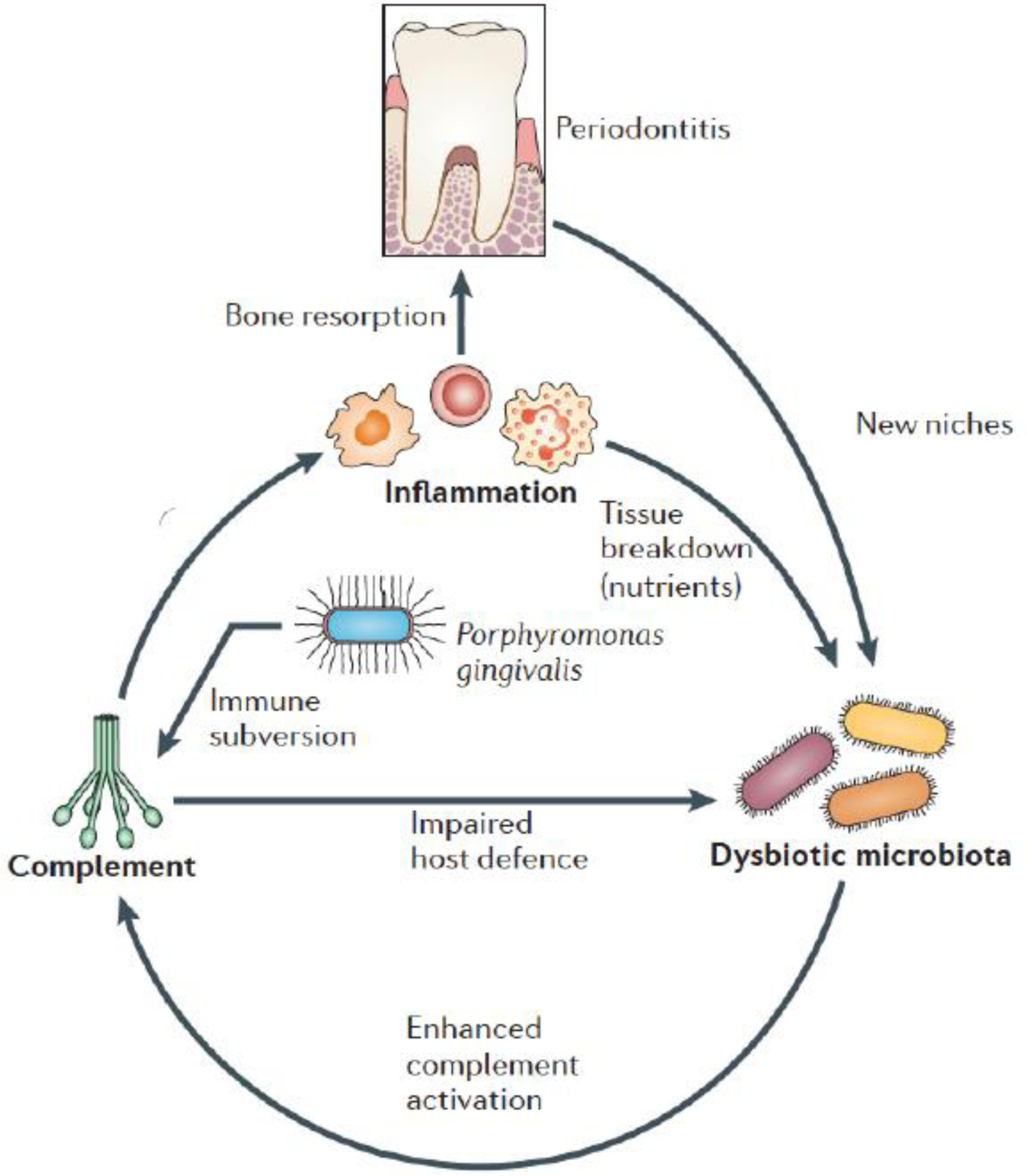
P. gingivalis subverts complement in a manner that compromises antimicrobial defense while enhancing inflammation. These effects contribute to dysbiotic changes of the periodontal microbiota (altered composition, increased total counts), which causes further inflammation, in great part through complement activation. Inflammatory tissue destruction fuels further bacterial growth by generating a nutrient-rich gingival inflammatory exudate (containing degraded host proteins and hemin, sources of amino acids and iron, respectively). These environmental changes favor proteolytic and asaccharolytic bacteria, thus explaining, at least in part, why inflammation causes compositional changes in the bacterial community. Moreover, inflammatory bone loss provides new niches for colonization by the dysbiotic microbiota. Overall, these changes create a self-sustained ‘vicious cycle’, where inflammation and dysbiosis are reciprocally reinforced. It should be noted, however, that whereas P. gingivalis can initiate dysbiosis, it is not an obligatory condition for dysbiosis. From reference [27]. Used by permission.
TLR4 antagonism
P. gingivalis expresses heterogeneous and atypical LPS molecules that act as potent TLR4 antagonists, weak agonists or are immunologically inert [96]. Specifically, P. gingivalis can enzymatically modify the lipid A moiety of its lipopolysaccharide and the shifting of lipid A activity from TLR4-agonistic to TLR4-antagonistic depends on endogenous lipid A phosphatases [96]. These enzymes are controlled by growth phase or environmental factors, such as temperature and hemin availability [97, 98]. Different lipid A structures produced by P. gingivalis include non-phosphorylated tetra-acylated lipid A (inert for TLR4 activation), mono-phosphorylated penta-acylated lipid A (weak TLR4 agonist), and mono-phosphorylated tetra-acylated lipid A (TLR4 antagonist). Genetic inactivation of 4′-phosphatase activity (or bacterial growth at ≥39°C) results in the synthesis of TLR4 agonist lipid A, while ablation of 1-phosphatase activity (or growth in hemin-replete conditions) results in TLR4 antagonist lipid A [96, 99]. The ability of this oral pathogen to manipulate the biological activity of its lipid A allows it to evade or proactively inhibit a variety of potential TLR4-mediated antimicrobial functions, such as inhibition of expression of antimicrobial peptides (β defensins) in human epithelial cells [28]. Moreover, the production of inert or antagonistic lipid A enhances the resistance of P. gingivalis to cationic antimicrobial peptides, owing to changes in the outer surface charge of the bacterial surface that affect the binding of cationic peptides [96, 97, 99, 100]. Since P. gingivalis releases LPS-bearing membrane vesicles that can readily diffuse in the crevice or even penetrate gingival tissue [13], the TLR4 antagonistic LPS of P. gingivalis might inhibit TLR4-dependent antimicrobial responses against other bacteria in the same mixed-species biofilm [28].
Exploitation of TLR2 and integrins
In contrast to TLR4, P. gingivalis does not appear to antagonize TLR2 at the receptor level. However, this oral bacterium can block TLR2 antimicrobial responses by inducing subversive signaling crosstalk between TLR2 and other receptors, such as, C5aR1, CXCR4 and the β2 integrin CD11b/CD18 (also known as Mac1 or complement receptor 3). This subsection will focus on CD11b/CD18. A major TLR2 ligand of P. gingivalis involved in these subversive interactions appears to be its fimA gene-encoded fimbriae. The fimbriae extend to a significant distance (up to 3 μm) from the bacterial cell wall [11], suggesting that they might be the first P. gingivalis molecule to interact with host cells and initiate intracellular signaling. The interaction of P. gingivalis fimbriae with TLR2 is promoted by an initial binding event between the fimbriae and CD14, which greatly facilitates TLR2-induced NF-κB activation and production of proinflammatory cytokines [14, 101, 102] (Figure 2). In addition to this pro-inflammatory signaling pathway, the interaction of P. gingivalis and its fimbriae with CD14/TLR2 in monocytes/macrophages also stimulates proadhesive signaling that activates the ligand-binding capacity of the CD11b/CD18 integrin [103, 104] (Figure 2). The regulation of the binding affinity of β2 integrin from within the cell through proadhesive signaling is referred to as ‘inside-out signaling’ [105]. Inside-out signaling may target cytoplasmic proteins to the integrin cytoplasmic tails causing high-affinity conformational changes on the ligand-binding domains of integrins [105].
Figure 2. P. gingivalis instigates TLR2-CXCR4 and TLR2-CD11b/CD18 crosstalk interactions to subvert macrophages.
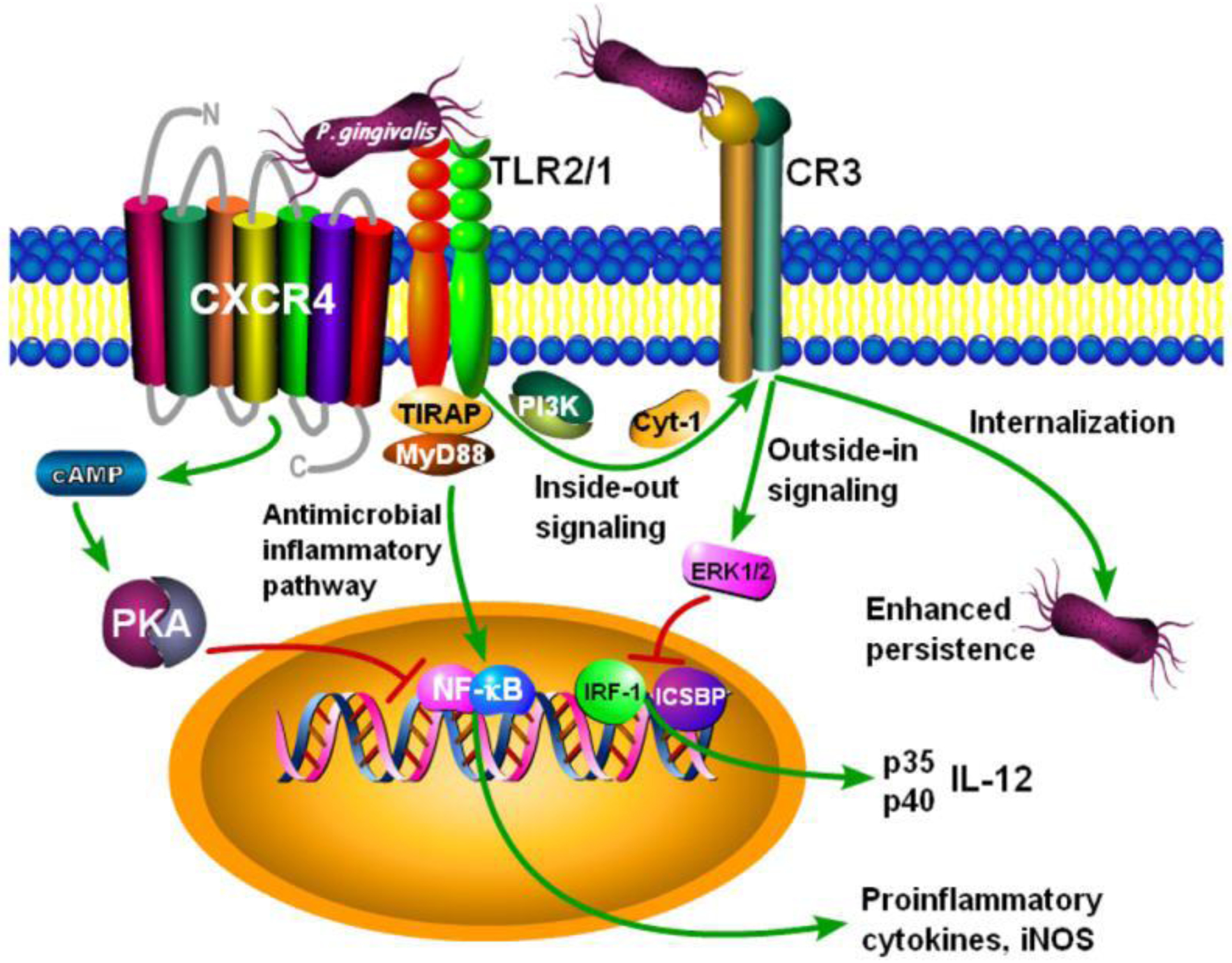
Through its FimA fimbriae, P. gingivalis can bind TLR2 (specifically the TLR2/TLR1 heterodimer; TLR2/1). Through a different structural component of the same molecule, P. gingivalis interacts with CXCR4 which cross-talks with and inhibits the TLR2/1-induced TIRAP/MyD88-mediated antimicrobial pathway. The underlying mechanism involves CXCR4-mediated stimulation of cAMP-dependent protein kinase A (PKA) signaling which limits NF-κB activation and induction of the inducible nitric oxide synthase (iNOS) that generates nitric oxide. The inhibitory effect on the production of nitric oxide, a potent antimicrobial mechanism for intracellular killing, promotes P. gingivalis survival in vitro and in vivo. By activating TLR2/1, P. gingivalis initiates inside-out signaling, which proceeds via phosphatidylinositol 3-kinase (PI3K) and cytohesin-1 to induce the high-affinity conformation of CD11b/CD18 (a β2 integrin also known as complement receptor 3). P. gingivalis binds activated CD11b/CD18 and is thereby internalized in a relatively safe manner as CD11b/CD18 is not linked to potent microbicidal mechanisms. Moreover, the P. gingivalis-CD11b/CD18 interaction activates extracellular signal-related kinase 1/2 (ERK1/2) signaling which downregulates IL-12 p35 and p40 mRNA expression. From reference [16]. Used by permission.
Following interaction with the CD14/TLR2 recognition complex, P. gingivalis FimA fimbriae induce inside-out signaling involving Rac1 phosphatidylinositol-3-kinase (PI3K), and cytohesin-1 that activates the ligand-binding capacity of CD11b/CD18 [103, 104, 106] (Figure 2). This mechanism was shown to promote CD11b/CD18-dependent monocyte adhesion and transendothelial migration [104]. Interestingly, however, P. gingivalis has co-opted this TLR2/CD11b/CD18 proadhesive pathway for CD11b/CD18 binding and entry into macrophages in a relatively safe manner that promotes the fitness of this pathogen [77] (Figure 2). Specifically, the interaction of CD11b/CD18 with P. gingivalis leads to suppression of bioactive IL-12, increased intracellular survival of the pathogen in macrophages, and enhanced in vivo persistence and capacity to cause periodontal bone loss [88, 87]. Therefore, CD11b/CD18 may represent an ‘Achilles’ heel’ exploited by P. gingivalis to promote its persistence in the mammalian host, probably because CD11b/CD18 is not linked to vigorous microbicidal mechanisms [107–110].
Subversion of CXCR4-TLR2 crosstalk
The outcome of TLR2 stimulation by microbial pathogens may be influenced by differential association of TLR2 with co-receptors. In this regard, by means of its FimA fimbriae, which can induce co-association of CXCR4 with TLR2 in lipid rafts, P. gingivalis induces CXCR4/TLR2 signaling crosstalk in human monocytes or mouse macrophages that compromises their killing function [84]. Specifically, the P. gingivalis-induced crosstalk between CXCR4 and TLR2 leads to enhanced cAMP-dependent protein kinase A (PKA) signaling, which in turn suppresses the generation of nitric oxide, a potent antimicrobial molecule for intracellular bacterial killing [84] (Figure 2). Consistent with this mechanism, mice subjected to P. gingivalis-induced periodontitis and treated with a CXCR4 antagonist are protected against periodontal tissue colonization by P. gingivalis and development of periodontal disease [86]. Intriguingly, it was subsequently shown that the soluble protein pancreatic adenocarcinoma upregulated factor (PAUF) also interacts with the CXCR4-TLR2 complex and elevates intracellular cAMP levels for inhibiting cellular activation [94]. The fact that both P. gingivalis fimbriae and PAUF induce the formation of the same receptor complex and exploit it to control cell signaling [84, 94] suggests that microbes and tumors may share common immune-evasive strategies.
Manipulation of TLR2 and complement
An intriguing question has been how P. gingivalis manages to selectively inhibit immune elimination without overall inhibiting inflammation. This is a crucial issue: If P. gingivalis would simply cause immune suppression (as many other pathogens do in other parts of the body [111]), this would spare P. gingivalis from immune clearance but it would starve it to death. This is because the survival and growth of this asaccharolytic organism (and other periodontitis-associated bacteria) critically depends on inflammatory tissue breakdown products (e.g., degraded proteins and hemin, a source of essential iron) [112, 2]. Since neutrophils are heavily involved in human periodontitis and comprise ≥95% of total leukocytes in the gingival crevice (or periodontal pocket), where they constantly encounter P. gingivalis [113–116], this question was addressed in the context of P. gingivalis-neutrophil interactions. The gingival crevicular neutrophils form what looks like a “defense wall” against the periodontal bacteria; however, in periodontitis, the neutrophils largely fail to control the bacteria despite being viable and capable of immune and inflammatory responses [114, 117–122]. At least one major mechanism by which P. gingivalis can manipulate neutrophil responses is via instigating a subversive complement-TLR cross-talk.
In this regard, the arginine-specific gingipains (HRgpA, RgpB) of P. gingivalis can cleave C5 to generate high local concentrations of biologically active C5a [123, 89] independently of the immunological activation of the complement system [124]. Thus, given that neutrophils can recognize P. gingivalis via TLR2 [125], this oral bacterium can coactivate C5aR1 and TLR2 [91]. P. gingivalis-induced C5aR1-TLR2 crosstalk signaling leads to ubiquitylation and proteasomal degradation of the TLR2 adapter MyD88, thereby preventing a host-protective antimicrobial response [91] (Figure 3). Furthermore, the C5aR1-TLR2 crosstalk induces the activation of Pl3K, which inhibits RhoA GTPase and actin polymerization and consequently the phagocytic uptake of P. gingivalis (as well as bystander bacteria) (Figure 3). Importantly, moreover, this TLR2-PI3K signaling pathway induces inflammation that is nutritionally beneficial to the bacteria. Therefore, P. gingivalis subverts intracellular signaling in human or mouse neutrophils in ways that uncouple inflammation from bactericidal activity. Importantly, the local pharmacological inhibition of C5aR1, TLR2 or PI3K in the gingiva of P. gingivalis-colonized mice leads to immune clearance of P. gingivalis, reverses microbial dysbiosis effected by P. gingivalis colonization and suppresses periodontal inflammation [91]. These findings not only are consistent with the keystone pathogen concept but suggested that complement inhibition may be an effective strategy to inhibit periodontal disease, especially since complement is required for a persisting periodontal inflammatory response (mice deficient in the central complement component C3 are protected from inflammatory bone loss) [126]. Indeed, local complement inhibition in non-human primates, using the C3 inhibitor Cp40 (AMY-101), protected the animals against inducible or naturally-occurring periodontal disease [127, 126, 128]. In 2019, AMY-101 received Investigational New Drug approval by the U.S. Food and Drug Administration for the first clinical trial to assess its efficacy in humans with periodontal inflammation (gingivitis) (ClinicalTrials.gov Identifier: NCT03694444).
Figure 3. P. gingivalis dissociates immune clearance from inflammatory responses in neutrophils.
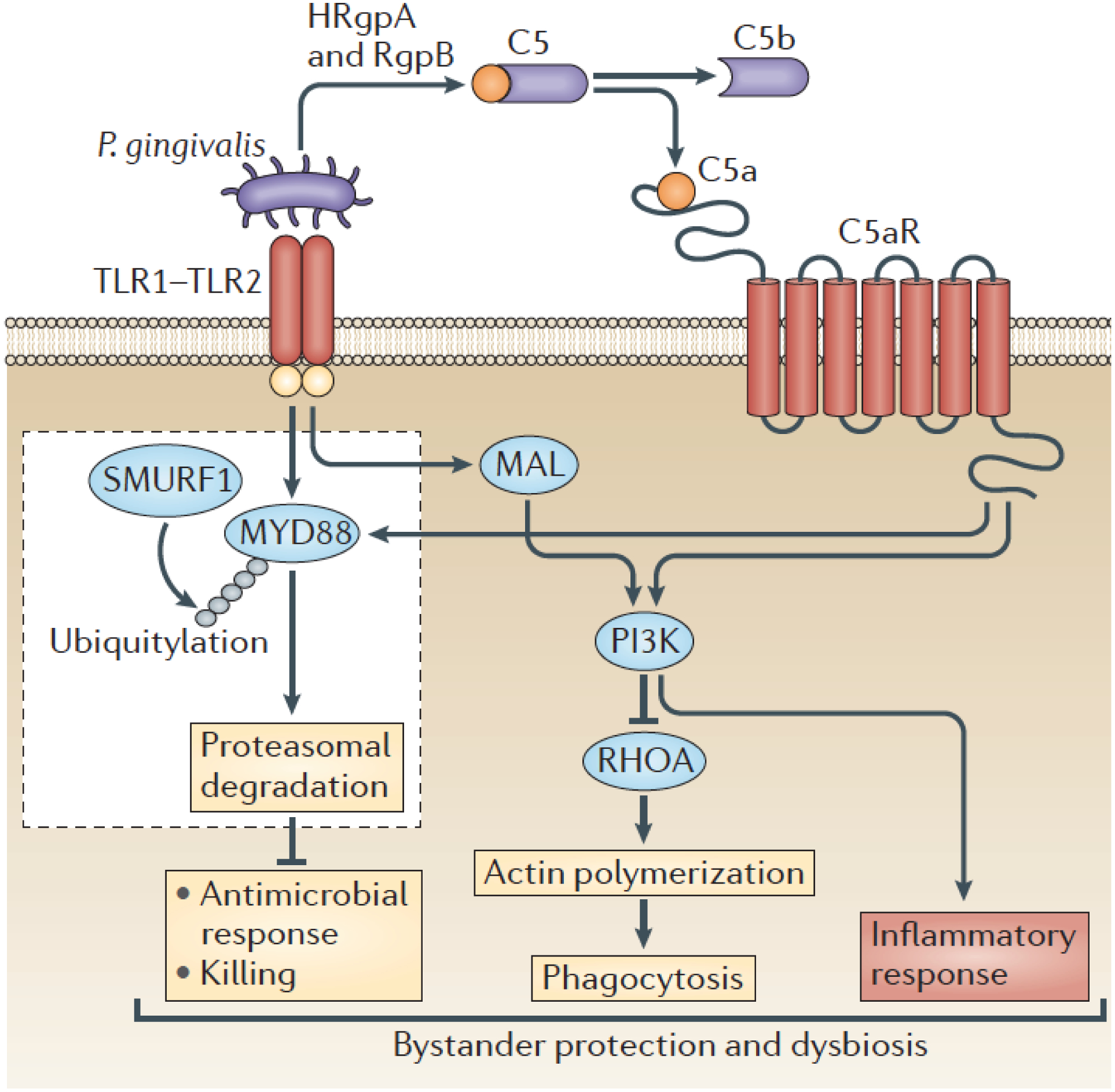
P. gingivalis activates the TLR1-TLR2 heterodimer and C5aR1, the latter through the generation of C5a by its gingipains. Specifically, these enzymes (HRgpA and RgpB) can cleave C5 to release biologically active C5a. The co-activation of TLR2 and C5aR1 by P. gingivalis and the resulting signaling crosstalk leads to the ubiquitylation (via SMURF, an E3 ubiquitin-protein ligase) and proteasomal degradation of the TLR2 adaptor MYD88, thereby blocking a host-protective antimicrobial mechanism. Moreover, the TLR2/C5aR1 crosstalk activates PI3K, which limits phagocytosis through the inhibition of the GTPase RHOA and hence actin polymerization. On the other hand, PI3K stimulates the production of inflammatory cytokines. Contrary to MYD88, another TLR2 adaptor, MYD88-like adaptor protein (MAL), participates in immune subversion by acting upstream of PI3K. These functionally integrated pathways offer ‘bystander’ protection to otherwise susceptible periodontal organisms and enhance polymicrobial dysbiotic inflammation in vivo. From reference [6]. Used by permission.
Summary and conclusions
P. gingivalis requires interactions with other subgingival species, which are likely to represent the first determinants of its successful establishment in the subgingival environment. Once P. gingivalis stably colonizes a site, it interacts with the host creating favorable conditions for its long-term persistence. In so doing, P. gingivalis can favorably promote the greater microbial community. Although P. gingivalis can influence gene and protein expression in other community members by direct interbacterial interactions [54, 43, 72], the major keystone-related function of this oral organism is likely via interference with innate immunity (Figs. 1–3). This interference not only compromises the ability of the host to control the periodontal microbial community but also leads to a dysregulated inflammatory response that contributes to tissue damage and bone resorption. Tissue destruction will also release peptides and heme-containing compounds which stimulate the growth of P. gingivalis and bystander species in the community. Although based on studies in the mouse model, the keystone pathogen concept is also consistent with P. gingivalis often being a quantitatively minor constituent of human periodontitis-associated biofilms, despite its increased prevalence and association with progressive bone loss in periodontal patients [129–133, 22]. However, an intervention study targeting specifically P. gingivalis would be required to test whether P. gingivalis exerts keystone function in human periodontitis. In this regard, in non-human primates, in which P. gingivalis is a natural member of their periodontal microbiota, a gingipain-based vaccine was shown to cause reduction in both the counts of P. gingivalis and the total subgingival bacterial burden [134], suggesting that the entire microbial community benefits from the presence of P. gingivalis. The discussed literature suggests that therapeutic targeting of host immune responses may be an effective way to control P. gingivalis and periodontal disease. In this regard, anti-inflammatory approaches, such as complement inhibition [127, 126, 135], to block harmful inflammation would also have indirect antimicrobial effects (limitation of inflammatory exudate-derived nutrients for the bacteria). Moreover, targeting complement pathways in the gingival tissues could render the host non-responsive to the subversive action of P. gingivalis, thereby neutralizing its promoting effect on periodontitis.
Acknowledgments
GH is supported by U.S. Public Health Service grants from the National Institutes of Health (DE015254, DE024153, DE024716, and DE026152).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflict of Interest:
George Hajishengallis is an inventor of a patent that describes the use of complement inhibitors for therapeutic purposes in periodontal disease.
Patricia I. Diaz declares that she has no conflict of interest.
Human and Animal Rights and Informed Consent: This article does not contain any primary research studies with human or animal subjects performed by any of the authors.
References
- 1.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–44. [DOI] [PubMed] [Google Scholar]
- 2.Diaz PI, Hoare A, Hong BY. Subgingival Microbiome Shifts and Community Dynamics in Periodontal Diseases. Journal of the California Dental Association. 2016;44(7):421–35. [PubMed] [Google Scholar]
- 3.Kebschull M, Demmer RT, Papapanou PN. “Gum bug leave my heart alone”: Epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent Res. 2010;89:879–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, Konradi A et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study identified the presence of gingipains from the keystone pathogen Porphyromonas gingivalis in the brain of Alzheimer’s patients, and showed that inhibition of gingipains by small-molecule inhibitors reduced the P. gingivalis load in a preclinical brain infection model.
- 5.Potempa J, Mydel P, Koziel J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nature reviews Rheumatology. 2017;13(10):606–20. [DOI] [PubMed] [Google Scholar]
- 6.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews Immunology. 2015;15(1):30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kholy KE, Genco RJ, Van Dyke TE. Oral infections and cardiovascular disease. Trends Endocrinol Metab. 2015;26(6):315–21. [DOI] [PubMed] [Google Scholar]
- 8.Barth K, Remick DG, Genco CA. Disruption of immune regulation by microbial pathogens and resulting chronic inflammation. Journal of cellular physiology. 2013;228:1413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinane DF, Stathopoulou PG, Papapanou PN. Periodontal diseases. Nat Rev Dis Primers. 2017;3:17038. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson DP, Kubiniec MA, Yoshimura F, Genco RJ. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170(4):1658–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamont RJ, Jenkinson HF. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atanasova KR, Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Molecular oral microbiology. 2014;29(2):55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amano A, Takeuchi H, Furuta N. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes and infection / Institut Pasteur. 2010;12(11):791–8. [DOI] [PubMed] [Google Scholar]
- 14.Hajishengallis G, Ratti P, Harokopakis E. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. The Journal of biological chemistry. 2005;280(47):38902–13. [DOI] [PubMed] [Google Scholar]
- 15.Lamont RJ, Jenkinson HF. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol Immunol. 2000;15(6):341–9. [DOI] [PubMed] [Google Scholar]
- 16.Hajishengallis G Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes and infection / Institut Pasteur. 2009;11(6–7):637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajishengallis G, Sojar H, Genco RJ, DeNardin E. Intracellular signaling and cytokine induction upon interactions of Porphyromonas gingivalis fimbriae with pattern-recognition receptors. Immunological investigations. 2004;33(2):157–72. [DOI] [PubMed] [Google Scholar]
- 18.Gibson FC 3rd, Hong C, Chou HH, Yumoto H, Chen J, Lien E et al. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation. 2004;109(22):2801–6. [DOI] [PubMed] [Google Scholar]
- 19.Hajishengallis G, Martin M, Sojar HT, Sharma A, Schifferle RE, DeNardin E et al. Dependence of bacterial protein adhesins on toll-like receptors for proinflammatory cytokine induction. Clinical and diagnostic laboratory immunology. 2002;9(2):403–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eskan MA, Hajishengallis G, Kinane DF. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infection and immunity. 2007;75(2):892–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jotwani R, Cutler CW. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infection and immunity. 2004;72(3):1725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME journal. 2013;7(5):1016–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal. 2012;6(6):1176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorth P, Turner KH, Gumus P, Nizam N, Buduneli N, Whiteley M. Metatranscriptomics of the human oral microbiome during health and disease. mBio. 2014;5(2):e01012–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10(5):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nature reviews Microbiology. 2012;10(10):717–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature reviews Microbiology. 2010;8(7):481–90. [DOI] [PubMed] [Google Scholar]
- 29.Rosier BT, de Jager M, Zaura E, Krom BP. Historical and contemporary hypotheses on the development of oral diseases: are we there yet? Frontiers in cellular and infection microbiology. 2014;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The Polymicrobial Synergy and Dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27(6):409–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nature reviews Microbiology. 2018;16(12):745–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma A, Honma K, Evans RT, Hruby DE, Genco RJ. Oral immunization with recombinant Streptococcus gordonii expressing Porphyromonas gingivalis FimA domains. Infection and immunity. 2001;69(5):2928–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sojar HT, Han Y, Hamada N, Sharma A, Genco RJ. Role of the amino-terminal region of Porphyromonas gingivalis fimbriae in adherence to epithelial cells. Infect Immun. 1999;67(11):6173–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Njoroge T, Genco RJ, Sojar HT, Hamada N, Genco CA. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect Immun. 1997;65(5):1980–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amano A, Fujiwara T, Nagata H, Kuboniwa M, Sharma A, Sojar HT et al. Porphyromonas gingivalis fimbriae mediate coaggregation with Streptococcus oralis through specific domains. J Dent Res. 1997;76(4):852–7. [DOI] [PubMed] [Google Scholar]
- 36.Amano A, Sharma A, Lee JY, Sojar HT, Raj PA, Genco RJ. Structural domains of Porphyromonas gingivalis recombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64(5):1631–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sojar HT, Lee J-Y, Genco RJ. Fibronectin binding domain of P. gingivalis fimbriae. Biochem Biophys Res Commun. 1995;216(3):785–92. [DOI] [PubMed] [Google Scholar]
- 38.Amano A, Sojar HT, Lee JY, Sharma A, Levine MJ, Genco RJ. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994;62(8):3372–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amano A, Sharma A, Sojar HT, Kuramitsu HK, Genco RJ. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sojar HT, Lee JY, Bedi GS, Cho M-I, Genco RJ. Purification, characterization and immunolocalization of fimbrial protein from Porphyromonas (Bacteroides) gingivalis. Biochem Biophys Res Commun. 1991;175(2):713–9. [DOI] [PubMed] [Google Scholar]
- 41.Klausen B, Evans RT, Ramamurthy NS, Golub LM, Sfintescu C, Lee JY et al. Periodontal bone level and gingival proteinase activity in gnotobiotic rats immunized with Bacteroides gingivalis. Oral microbiology and immunology. 1991;6(4):193–201. [DOI] [PubMed] [Google Scholar]
- 42.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and periodontal health status. Journal of clinical microbiology. 1998;36(11):3239–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yost S, Duran-Pinedo AE, Teles R, Krishnan K, Frias-Lopez J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slots J, Gibbons RJ. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infection and immunity. 1978;19(1):254–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda K, Nagata H, Nonaka A, Kataoka K, Tanaka M, Shizukuishi S. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes and Infection. 2004;6(13):1163–70. [DOI] [PubMed] [Google Scholar]
- 46.Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infection and immunity. 2005;73(7):3983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191(22):6804–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamont RJ, El-Sabaeny A, Park Y, Cook GS, Costerton JW, Demuth DR. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology. 2002;148(6):1627–36. [DOI] [PubMed] [Google Scholar]
- 49.Kuboniwa M, Tribble GD, James CE, Kilic AO, Tao L, Herzberg MC et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Molecular microbiology. 2006;60(1):121–39. [DOI] [PubMed] [Google Scholar]
- 50.Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2017:doi: 10.1038/s41564-017-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; ••This study has demonstrated multidimensional communication between Porphyromonas gingivalis and Streptococcus gordonii in the oral microbial community that can either enhance or suppress the pathogenicity of the community.
- 51.Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. 2008;190(12):4367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. Journal of clinical microbiology. 2009;47(12):3902–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. Journal of periodontal research. 2012;47(5):578–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duran-Pinedo AE, Baker VD, Frias-Lopez J. The periodontal pathogen Porphyromonas gingivalis Induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infection and immunity. 2014;82(8):3374–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou P, Liu J, Merritt J, Qi F. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis, and human oral buccal cells. Molecular oral microbiology. 2015;30(4):269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou P, Li X, Qi F. Identification and characterization of a haem biosynthesis locus in Veillonella. Microbiology. 2016;162(10):1735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diaz PI, Zilm PS, Rogers AH. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 2002;148(Pt 2):467–72. [DOI] [PubMed] [Google Scholar]
- 58.Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infection and immunity. 1998;66(10):4729–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diaz PI, Zilm PS, Rogers AH. The response to oxidative stress of Fusobacterium nucleatum grown in continuous culture. FEMS Microbiol Lett. 2000;187(1):31–4. [DOI] [PubMed] [Google Scholar]
- 60.Grenier D Nutritional interactions between two suspected periodontopathogens, Treponema denticola and Porphyromonas gingivalis. Infection and immunity. 1992;60(12):5298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meuric V, Martin B, Guyodo H, Rouillon A, Tamanai-Shacoori Z, Barloy-Hubler F et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Molecular oral microbiology. 2013;28(1):40–53. [DOI] [PubMed] [Google Scholar]
- 62.Kesavalu L, Sathishkumar S, Bakthavatchalu V, Matthews C, Dawson D, Steffen M et al. Rat model of polymicrobial infection, immunity, and alveolar bone resorption in periodontal disease. Infection and immunity. 2007;75(4):1704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kesavalu L, Holt SC, Ebersole JL. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral microbiology and immunology. 1998;13(6):373–7. [DOI] [PubMed] [Google Scholar]
- 64.Sztukowska MN, Dutton LC, Delaney C, Ramsdale M, Ramage G, Jenkinson HF et al. Community Development between Porphyromonas gingivalis and Candida albicans Mediated by InlJ and Als3. mBio. 2018;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]; •This paper has characterized inter-kingdom interactions between Candida albicans and Porphyromonas gingivalis that regulate gene expression by P. gingivalis in a manner that increases its pathogenic potential.
- 65.Reynaud AH, Nygaard-Ostby B, Boygard GK, Eribe ER, Olsen I, Gjermo P. Yeasts in periodontal pockets. Journal of clinical periodontology. 2001;28(9):860–4. [DOI] [PubMed] [Google Scholar]
- 66.Urzua B, Hermosilla G, Gamonal J, Morales-Bozo I, Canals M, Barahona S et al. Yeast diversity in the oral microbiota of subjects with periodontitis: Candida albicans and Candida dubliniensis colonize the periodontal pockets. Med Mycol. 2008;46(8):783–93. [DOI] [PubMed] [Google Scholar]
- 67.Jarvensivu A, Hietanen J, Rautemaa R, Sorsa T, Richardson M. Candida yeasts in chronic periodontitis tissues and subgingival microbial biofilms in vivo. Oral diseases. 2004;10(2):106–12. [DOI] [PubMed] [Google Scholar]
- 68.Canabarro A, Valle C, Farias MR, Santos FB, Lazera M, Wanke B. Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. Journal of periodontal research. 2013;48(4):428–32. [DOI] [PubMed] [Google Scholar]
- 69.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infection and immunity. 1998;66(4):1660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Calkins CC, Platt K, Potempa J, Travis J. Inactivation of tumor necrosis factor-a by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J Biol Chem. 1998;273(12):6611–4. [DOI] [PubMed] [Google Scholar]
- 71.Hajishengallis G, Lamont RJ. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. European journal of immunology. 2014;44(2):328–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. Porphyromonas gingivalis: keeping the pathos out of the biont. Journal of oral microbiology. 2013;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333(1):1–9. [DOI] [PubMed] [Google Scholar]
- 74.Medzhitov R Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–45. [DOI] [PubMed] [Google Scholar]
- 75.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature immunology. 2010;11(5):373–84. [DOI] [PubMed] [Google Scholar]
- 76.Beutler B, Jiang Z, Georgel P, Crozat K, Croker B, Rutschmann S et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev Immunol. 2006;24:353–89. [DOI] [PubMed] [Google Scholar]
- 77.Hajishengallis G, Wang M, Harokopakis E, Triantafilou M, Triantafilou K. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infection and immunity. 2006;74(10):5658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, Hartung T et al. Membrane sorting of toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281(41):31002–11. [DOI] [PubMed] [Google Scholar]
- 79.Triantafilou K, Triantafilou M, Dedrick RL. A CD14-independent LPS receptor cluster. Nat Immunol. 2001;2(4):338–45. [DOI] [PubMed] [Google Scholar]
- 80.Latz E, Visintin A, Lien E, Fitzgerald KA, Monks BG, Kurt-Jones EA et al. Lipopolysaccharide rapidly traffics to and from the Golgi apparatus with the toll-like receptor 4-MD-2-CD14 complex in a process that is distinct from the initiation of signal transduction. J Biol Chem. 2002;277(49):47834–43. [DOI] [PubMed] [Google Scholar]
- 81.Hajishengallis G, Lambris JD. More than complementing Tolls: complement-Toll-like receptor synergy and crosstalk in innate immunity and inflammation. Immunological reviews. 2016;274(1):233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nature reviews Immunology. 2011;11(3):187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hajishengallis G, Lamont RJ. Dancing with the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends in microbiology. 2016;24(6):477–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(36):13532–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pierce DL, Nishiyama S, Liang S, Wang M, Triantafilou M, Triantafilou K et al. Host adhesive activities and virulence of novel fimbrial proteins of Porphyromonas gingivalis. Infection and immunity. 2009;77(8):3294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McIntosh ML, Hajishengallis G. Inhibition of Porphyromonas gingivalis-induced periodontal bone loss by CXCR4 antagonist treatment. Molecular oral microbiology. 2012;27(6):449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179(4):2359–67. [DOI] [PubMed] [Google Scholar]
- 88.Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F et al. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179(4):2349–58. [DOI] [PubMed] [Google Scholar]
- 89.Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186(2):869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P et al. Microbial hijacking of complement-toll-like receptor crosstalk. Science signaling. 2010;3(109):ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maekawa T, Krauss JL, Abe T, Jotwani R, Triantafilou M, Triantafilou K et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe. 2014;15(6):768–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Oliva C, Turnbough CL Jr., Kearney JF. CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dai S, Rajaram MV, Curry HM, Leander R, Schlesinger LS. Fine tuning inflammation at the front door: macrophage complement receptor 3-mediates phagocytosis and immune suppression for Francisella tularensis. PLoS pathogens. 2013;9(1):e1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park HD, Lee Y, Oh YK, Jung JG, Park YW, Myung K et al. Pancreatic adenocarcinoma upregulated factor promotes metastasis by regulating TLR/CXCR4 activation. Oncogene. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91(9):816–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT et al. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4’- phosphatase activities. Cellular microbiology. 2009;11:1587–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Curtis MA, Percival RS, Devine D, Darveau RP, Coats SR, Rangarajan M et al. Temperature dependent modulation of Porphyromonas gingivalis lipid A structure and interaction with the innate host defences. Infection and immunity. 2011;79:1187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infection and immunity. 2006;74(8):4474–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zenobia C, Hasturk H, Nguyen D, Van Dyke TE, Kantarci A, Darveau RP. Porphyromonas gingivalis lipid A phosphatase activity is critical for colonization and increasing the commensal load in the rabbit ligature model. Infection and immunity. 2014;82(2):650–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Slocum C, Coats SR, Hua N, Kramer C, Papadopoulos G, Weinberg EO et al. Distinct lipid A moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS pathogens. 2014;10(7):e1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S, Ratti P, Schifferle RE et al. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cellular microbiology. 2006;8(10):1557–70. [DOI] [PubMed] [Google Scholar]
- 102.Asai Y, Ohyama Y, Gen K, Ogawa T. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect Immun. 2001;69:7387–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harokopakis E, Hajishengallis G. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. European journal of immunology. 2005;35(4):1201–10. [DOI] [PubMed] [Google Scholar]
- 104.Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176(12):7645–56. [DOI] [PubMed] [Google Scholar]
- 105.Shimaoka M, Takagi J, Springer TA. Conformational regulation of integrin structure and function. Annu Rev Biophys Biomol Struct. 2002;31:485–516. [DOI] [PubMed] [Google Scholar]
- 106.Hajishengallis G, Wang M, Liang S. Induction of distinct TLR2-mediated proinflammatory and proadhesive signaling pathways in response to Porphyromonas gingivalis fimbriae. J Immunol. 2009;182(11):6690–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lowell CA. Rewiring phagocytic signal transduction. Immunity. 2006;24:243–5. [DOI] [PubMed] [Google Scholar]
- 108.Wright SD, Silverstein SC. Receptors for C3b and C3bi promote phagocytosis but not the release of toxic oxygen from human phagocytes. J Exp Med. 1983;158(6):2016–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282(5394):1717–21. [DOI] [PubMed] [Google Scholar]
- 110.Hellwig SM, van Oirschot HF, Hazenbos WL, van Spriel AB, Mooi FR, van De Winkel JG. Targeting to Fcg receptors, but not CR3 (CD11b/CD18), increases clearance of Bordetella pertussis. J Infect Dis. 2001;183(6):871–9. [DOI] [PubMed] [Google Scholar]
- 111.Cyktor JC, Turner J. Interleukin-10 and immunity against prokaryotic and eukaryotic intracellular pathogens. Infection and immunity. 2011;79(8):2964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hajishengallis G The inflammophilic character of the periodontitis-associated microbiota. Molecular oral microbiology. 2014;29(6):248–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nussbaum G, Shapira L. How has neutrophil research improved our understanding of periodontal pathogenesis? J Clin Periodontol. 2011;38:49–59. [DOI] [PubMed] [Google Scholar]
- 114.Delima AJ, Van Dyke TE. Origin and function of the cellular components in gingival crevice fluid. Periodontology 2000. 2003;31:55–76. [DOI] [PubMed] [Google Scholar]
- 115.Eskan MA, Jotwani R, Abe T, Chmelar J, Lim JH, Liang S et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nature immunology. 2012;13(5):465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hajishengallis G, Chavakis T, Hajishengallis E, Lambris JD. Neutrophil homeostasis and inflammation: novel paradigms from studying periodontitis. Journal of leukocyte biology. 2015;98(4):539–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lange D, Schroeder HE. Cytochemistry and ultrastructure of gingival sulcus cells. Helv Odontol Acta. 1971;15:65–86. [PubMed] [Google Scholar]
- 118.Newman HN. Neutrophils and IgG at the host-plaque interface on children’s teeth. Journal of periodontology. 1980;51(11):642–51. [DOI] [PubMed] [Google Scholar]
- 119.Schroeder HE, Listgarten MA. The gingival tissues: the architecture of periodontal protection. Periodontology 2000. 1997;13(1):91–120. [DOI] [PubMed] [Google Scholar]
- 120.Ryder MI. Comparison of neutrophil functions in aggressive and chronic periodontitis. Periodontology 2000. 2010;53:124–37. [DOI] [PubMed] [Google Scholar]
- 121.Vitkov L, Klappacher M, Hannig M, Krautgartner WD. Neutrophil fate in gingival crevicular fluid. Ultrastruct Pathol. 2010;34(1):25–30. [DOI] [PubMed] [Google Scholar]
- 122.Chapple IL, Matthews JB. The role of reactive oxygen and antioxidant species in periodontal tissue destruction. Periodontology 2000. 2007;43:160–232. [DOI] [PubMed] [Google Scholar]
- 123.Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J Biol Chem. 1992;267(26):18902–7. [PubMed] [Google Scholar]
- 124.Hajishengallis G, Reis ES, Mastellos DC, Ricklin D, Lambris JD. Novel mechanisms and functions of complement. Nature immunology. 2017;18(12):1288–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J Immunol. 2006;177(12):8296–300. [DOI] [PubMed] [Google Scholar]
- 126.Maekawa T, Briones RA, Resuello RR, Tuplano JV, Hajishengallis E, Kajikawa T et al. Inhibition of pre-existing natural periodontitis in non-human primates by a locally administered peptide inhibitor of complement C3. Journal of clinical periodontology. 2016;43:238–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kajikawa T, Briones RA, Resuello RRG, Tuplano JV, Reis ES, Hajishengallis E et al. Safety and Efficacy of the Complement Inhibitor AMY-101 in a Natural Model of Periodontitis in Non-human Primates. Mol Ther Methods Clin Dev. 2017;6:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]; ●●This study has established the safety and efficacy of a small-molecule inhibitor of complement C3 in non-human primate periodontitis and paved the way to a clinical trial in humans.
- 128.Maekawa T, Abe T, Hajishengallis E, Hosur KB, DeAngelis RA, Ricklin D et al. Genetic and intervention studies implicating complement C3 as a major target for the treatment of periodontitis. J Immunol. 2014;192 (12):6020–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chaves ES, Jeffcoat MK, Ryerson CC, Snyder B. Persistent bacterial colonization of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in periodontitis and its association with alveolar bone loss after 6 months of therapy. Journal of clinical periodontology. 2000;27(12):897–903. [DOI] [PubMed] [Google Scholar]
- 130.Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. Journal of clinical periodontology. 1991;18(10):729–39. [DOI] [PubMed] [Google Scholar]
- 131.Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infection and immunity. 1982;38(3):1137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. Journal of clinical microbiology. 2006;44(10):3665–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Doungudomdacha S, Rawlinson A, Douglas CW. Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method. Journal of medical microbiology. 2000;49(10):861–74. [DOI] [PubMed] [Google Scholar]
- 134.Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral microbiology and immunology. 2007;22(3):162–8. [DOI] [PubMed] [Google Scholar]
- 135.Hajishengallis G, Hajishengallis E, Kajikawa T, Wang B, Yancopoulou D, Ricklin D et al. Complement inhibition in pre-clinical models of periodontitis and prospects for clinical application. Seminars in immunology. 2016;28(3):285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]


