Abstract
Chlorophyll (Chl) is essential for photosynthetic reactions and chloroplast development. While the enzymatic pathway for Chl biosynthesis is well established, the regulatory mechanism underlying the homeostasis of Chl levels remains largely unknown. In this study, we identified CBD1 (Chlorophyll Biosynthetic Defect1), which functions in the regulation of chlorophyll biosynthesis. The CBD1 gene was expressed specifically in green tissues and its protein product was embedded in the thylakoid membrane. Furthermore, CBD1 was precisely co-expressed and functionally correlated with GUN5 (Genome Uncoupled 5). Analysis of chlorophyll metabolic intermediates indicated that cbd1 and cbd1gun5 mutants over-accumulated magnesium protoporphyrin IX (Mg-Proto IX). In addition, the cbd1 mutant thylakoid contained less Mg than the wild type not only as a result of lower Chl content, but also implicating CBD1 in Mg transport. This was supported by the finding that CBD1 complemented a Mg2+ uptake-deficient Salmonella strain under low Mg conditions. Taken together, these results indicate that CBD1 functions synergistically with CHLH/GUN5 in Mg-Proto IX processing, and may serve as a Mg-transport protein to maintain Mg homeostasis in the chloroplast.
Keywords: chlorophyll biosynthesis, magnesium, thylakoid membrane, GUN5, magnesium protoporphyrin
Magnesium (Mg) is not only a structural element of chlorophyll but also a cofactor for a number of enzymes in the chloroplast. This study shows that a thylakoid membrane protein, CBD1, functions synergistically with CHLH/GUN5 in chlorophyll biosynthesis, and serves as a Mg-transport protein to maintain Mg homeostasis in the chloroplast.
Introduction
Chlorophyll (Chl) is a tetrapyrrole macrocycle critical for the photosynthetic process that underpins life on earth. As the most abundant pigment, Chl plays vital roles in harvesting light energy in the antenna system and driving electron transfer in the reaction center for the initiation of photosynthesis in all green lineages. Chl in algae and plants is synthesized from a branched tetrapyrrole biosynthetic pathway localized entirely within plastids (Larkin, 2016). This pathway begins with the synthesis of 5-aminolevulinic acid (ALA), a universal precursor of all tetrapyrroles (Tanaka et al., 2011; Brzezowski et al., 2015). In the subsequent enzymatic steps, eight molecules of ALA are converted into protoporphyrin IX (Proto IX), which is then allocated into two distinct branches, namely, the heme-synthesizing Fe branch and the Chl-synthesizing Mg branch.
In the Fe branch, Fe2+ is inserted into Proto IX by ferrochelatase (FeCh) to form heme, a cofactor for cytochromes and other redox proteins (Chow et al., 1998; Tanaka et al., 2011; Scharfenberg et al., 2015). The Mg branch results in Chl biosynthesis and begins with ATP-dependent Mg2+ insertion into Proto IX, which is catalyzed by magnesium chelatase (MgCh) to produce Mg-Proto IX. Unlike FeCh, MgCh is a complex composed of three subunits, CHLI, CHLD, and CHLH (Gibson et al., 1995; Jensen et al., 1998, 1999). Catalysis by MgCh involves a two-step reaction, namely, an enzyme activation step, followed by an Mg2+ insertion step (Masuda, 2008; Tanaka et al., 2011). In the first step, the CHLD hexamer and CHLI hexamer interact to form a CHLI–CHLD–Mg-ATP complex without ATP hydrolysis. This complex then binds to the substrate-binding CHLH subunit to form a transient holoenzyme complex. Mg chelation occurs with ATP hydrolysis to generate Mg-Proto IX, followed by the disassembly and turnover of the MgCh complex (Walker and Willows, 1997; Jensen et al., 1998; Masuda, 2008; Tanaka et al., 2011). Subsequently, the propionate side chain of Mg-Proto IX is methylated by Mg-Proto IX methyltransferase (MgMT) to generate Mg-Proto IX monomethyl ester (Mg-Proto IX ME). The enzyme MgMT, which is encoded by the CHLM gene, has been reported to form a complex with MgCh (Alawady et al., 2005; Shepherd et al., 2005). In the next step, a Mg-Proto IX ME cyclase encoded by the CHL27/CRD1 gene in Arabidopsis catalyzes the conversion of Mg-Proto IX ME into protochlorophyllide, the key precursor of mature Chl (Bang et al., 2008; Tanaka et al., 2011).
As the catalytic subunit of MgCh, CHLH has also been proposed to interact with and enhance MgMT catalysis by accelerating the formation and breakdown of the catalytic intermediate. Interactions between MgMT and MgCh facilitate the methylation step by channeling Mg-Proto IX, the product of MgCh and substrate of MgMT (Alawady et al., 2005; Shepherd et al., 2005; Pontier et al., 2007). CHLH (also known as GENOMES UNCOUPLED 5 [GUN5]) also functions in other processes, including plastid-to-nucleus retrograde signaling and the ABA response (Mochizuki et al., 2001; Shang et al., 2010). Absence of CHLH/GUN5 resulted in a lethal phenotype in Chlamydomonas reinhardtii (Chekounova et al., 2001); however, several chlh/gun5 mutants, including the T-DNA insertional gun5 mutant, are viable in Arabidopsis (Chen et al., 2017), implying that additional components might function collaboratively with CHLH/GUN5 in Arabidopsis. In this study, we identified a previously unknown protein that functionally interacts with CHLH/GUN5 in Chl biosynthesis.
Results
CBD1 Is Co-expressed with CHLH/GUN5 and Localized in the Thylakoid Membrane
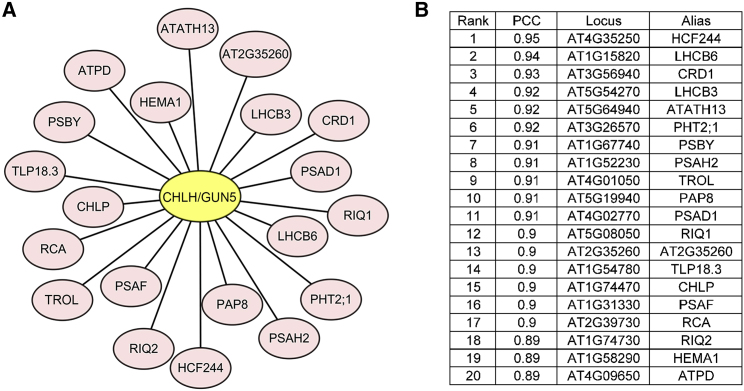
Genes involved in related biological pathways are often co-expressed in space and time to establish their common functions (Eisen et al., 1998; Obayashi et al., 2011). To identify potential proteins working together with CHLH/GUN5 in Chl biosynthesis, we performed co-expression analysis with CHLH/GUN5 as a query gene using the Genevestigator online tool (Zimmermann et al., 2004). Genes with relatively high Pearson's correlation coefficients under perturbation conditions (Supplemental Figure 1) were isolated (Figure 1A). The 20 most correlated genes included known genes of the Chl biosynthetic pathway, such as CHL27/CRD1, CHLP (encoding the geranylgeranyl pyrophosphate reductase), and HEMA1 (encoding the glutamyl-tRNA reductase), as well as genes encoding the subunits of photosynthetic complexes, such as LHCB3, LHCB6, PSBY, PSAH2, PSAD1, and PSAF. Many other co-expressed genes in the list were also reported to function in the chloroplast (Figure 1B). However, the co-expressed gene AT2G35260 encodes a novel protein of unknown function. We hypothesized that this gene may play a role in Chl biosynthesis, and designated it as CBD1 (Chlorophyll Biosynthetic Defect1) based on our research described in this report.
Figure 1.
CBD1 is Co-expressed with CHLH/GUN5.
(A) Genes co-expressed with CHLH/GUN5. Data were collected from the Genevestigator database (www.genevestigator.com), and the figure was processed using Cytoscape 3.6.1.
(B) List of co-expressed genes; genes were ranked by their Pearson's correlation coefficients.
Genes in the Chl biosynthetic pathway were grouped into four categories from cluster 1 (c1) to cluster 4 (c4) (Matsumoto et al., 2004). The genes in the c1 cluster (HEMA1, CHLH/GUN5, CHL27/CRD1, CAO, GUN4, and CHLP), which are coordinately regulated by light and the circadian rhythm, form a tight co-expression network with each other and are closely correlated (Kobayashi and Masuda, 2016; Masuda and Fujita, 2008), and CBD1 was co-expressed with most of the genes in this cluster. The expression level of CBD1, similar to that of CHLH/GUN5, rapidly increased after etiolated seedlings were exposed to light (Figure 2A). This light-inducible expression pattern is typical for genes involved in Chl biosynthesis (Matsumoto et al., 2004). To determine the rhythmic expression profile of CBD1, samples were harvested from 3-week-old Arabidopsis seedlings grown under 12-h light/12-h dark conditions. After data processing, we found that the phases and amplitudes of CBD1 and CHLH/GUN5 were synchronized (Figure 2B). This again supported the finding from our initial co-expression analysis of CBD1, pointing to a tight correlation between CBD1 and the Chl biosynthetic genes (Masuda and Fujita, 2008; Kobayashi and Masuda, 2016).
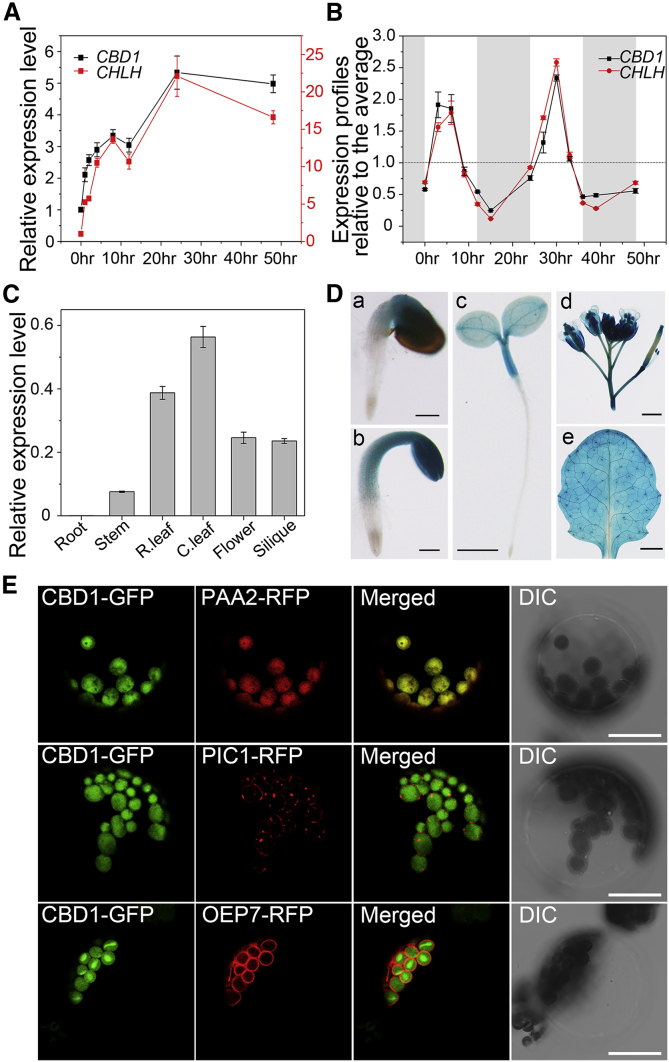
Figure 2.
Expression Profiles of CBD1 and Subcellular Localization of CBD1 in the Thylakoid Membrane.
(A) Three-day-old dark-grown etiolated WT seedlings were placed under continuous light exposure for indicated periods of time on the bottom. Relative expression levels of CBD1 at different time points were detected by quantitative real-time PCR.
(B) Rhythmic regulation of CBD1 and CHLH/GUN5 in mature plants. In the diurnal cycle, 3-week-old seedlings were maintained on a 12-h light/12-h dark (gray) cycle. Hours after the onset of the experiment are indicated at the bottom. The expression level relative to the average (dotted line) was plotted. Data are means ± SD. n = 4.
(C) Relative expression levels of CBD1 in various tissues as revealed by qRT–PCR analysis.
(D) GUS staining of (a–c) 1- to 3-day-old seedlings after germination, respectively, (d) inflorescence and silique from 4-week-old plants, and (e) the rosette leaf from 3-week-old plants grown in soil. Scale bars correspond to 200 μm in (a) and (b), 1 mm in (c) and (f), and 2 mm in (d) and (e). Data are means ± SD. n = 4.
(E) The 35S::CBD1-GFP construct was co-transformed with the marker PAA2-RFP (thylakoid membrane), PIC1-RFP (inner envelope), or OEP7-RFP (outer envelope) into Arabidopsis leaf protoplasts for fluorescence observation by confocal laser scanning microscopy after 16 h of expression. Green fluorescence signals, chlorophyll red auto-fluorescence, merged signals from GFP and chlorophyll channels, and bright-field images showing the intactness of the protoplasts are shown separately from left to right in each lane. Scale bars correspond to 20 μm.
To confirm the expression pattern of CBD1 in Arabidopsis, we conducted quantitative real-time PCR (qRT–PCR) analysis and found that the CBD1 mRNA was specifically expressed in green tissues, but not detected in roots (Figure 2C). Consistent with the qRT–PCR results, the GUS staining of transgenic plants expressing the proCBD1::GUS construct revealed that CBD1 promoter activity was restricted to green tissues, including leaves, stems, flowers, and immature siliques, and it was non-detectable in roots (Figure 2D), supporting the initial in silico expression analysis and the tight correlation between CBD1 expression and the Chl biosynthetic pathway.
The subcellular localization of a protein is a critical indicator of its function. Among the top 20 candidates co-expressed with CHLH/GUN5, all known proteins are located and function in the chloroplast. We speculated that CBD1 may also be localized in the chloroplast. We tested this possibility by transiently expressing the CBD1::GFP fusion protein in Arabidopsis mesophyll protoplasts and examined the subcellular localization of the fusion protein by confocal microscopy. The GFP signal from the CBD1::GFP fusion protein was specifically associated with chloroplast clusters, whereas the signal from the control GFP vector was detected in the cytosolic portions of the cell, except for areas covered with chloroplasts and vacuoles (Supplemental Figure 2A and 2B).
The chloroplast is surrounded by a dual-membrane envelope composed of the outer and inner envelope, and contains an internal membrane system known as the thylakoid (Sandelius and Aronsson, 2009). Because CBD1 is a multi-transmembrane domain protein, we expected it to be embedded into one of these membrane systems in the chloroplast. To determine the exact location of CBD1 within the chloroplast, we used marker proteins (fused to GFP) with known sub-chloroplast locations to serve as references (Supplemental Figure 2C–2F). For example, PAA2 resides in the thylakoid membrane (Abdel-Ghany et al., 2005), whereas PIC1 and OEP7 are located in the inner and outer envelopes, respectively (Duy et al., 2007). RBCS1, a small subunit of the Rubisco enzyme, is present in the stroma (Lee et al., 2006). We found that CBD1 showed the same localization pattern as PAA2, a thylakoid membrane protein. To further confirm this localization, we co-transformed CBD1::GFP with PAA2::RFP, PIC1::RFP, or OEP7::RFP into Arabidopsis protoplasts, respectively (Figure 2E). The results demonstrated that CBD1::GFP was co-localized with PAA2::RFP, but not with other markers. Thus, we concluded that CBD1 is localized in the thylakoid membrane of chloroplasts.
Cbd1 Mutant Plants Display Lower Chl Content
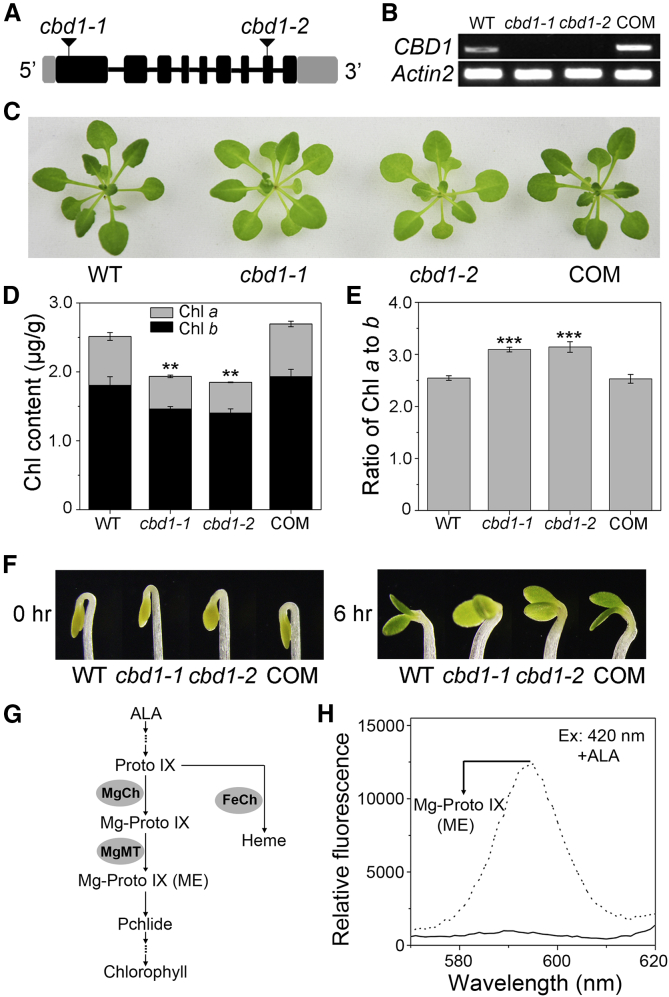
The putative function of CBD1 was explored using two T-DNA insertion mutant alleles of the gene. The first allele, referred to as cbd1-1 (SALK_031802), contained a T-DNA insertion in the first exon, whereas the second allele, cbd1-2 (SALK_058830), contained a T-DNA insertion in the eighth exon of the coding region (Figure 3A). The full-length CBD1 transcript was not detected by RT–PCR in either mutant (Figure 3B). The two different cbd1 mutant lines both exhibited a consistent pale-green phenotype and a 40% reduction in total Chl content compared with the wild type (WT), which indicated possible defects in Chl synthesis (Figure 3C and 3D). There was a stronger reduction in Chl b content than Chl a, leading to a higher Chl a/b ratio in the cbd1 mutant than in the WT (Figure 3E). To further confirm that the phenotype of the cbd1 mutant resulted from the loss-function of CBD1, a 3.4-kb genomic fragment of CBD1 (including the putative promoter, untranslated region, and coding sequence) was cloned into pCAMBIA1300, and the construct was used to transform the cbd1-1 mutant line by Agrobacterium-mediated gene transformation. Both the Chl content and Chl a/b ratio were recovered to WT levels in transgenic plants that we referred to as complementation (COM) lines (Figure 3C–3E). Taken together, these results suggest that CBD1 is required for Chl accumulation in Arabidopsis plants. To evaluate the contribution of CBD1 in Chl biosynthesis, we turned to the de-etiolation process in which Chl is initially synthesized after the first exposure of dark-grown plants to light. When 3-day-old WT, cbd1-1, cbd1-2, and COM etiolated seedlings were exposed to light, they gradually turned green reflecting Chl synthesis (Supplemental Figure 3). Compared with WT and COM plants, cbd1 mutants turned green more slowly, as it became visually evident 6 h after light exposure (Figure 3F). This finding, together with typical lower Chl content in adult plants, supported our hypothesis that CBD1 plays a role in Chl biosynthesis. As the two null alleles showed the same phenotype, we selected cbd1-1 for subsequent experiments.
Figure 3.
Mutation of CBD1 Leads to Impaired Chlorophyll Biosynthesis.
(A) The structure of the CBD1 gene and the positions of T-DNA insertion sites of two mutant lines. Locations of the two T-DNA insertions are indicated by the solid triangle. Exons are shown as black boxes, introns as black lines, and untranslated regions as gray boxes.
(B) Semi-quantitative RT–PCR analysis of the accumulation of CBD1 transcripts in the wild type (WT), the two mutant lines (cbd1-1 and cbd1-2), and the complementation (COM) line transformed with CBD1 genomic DNA fragment. Actin2 was used as an internal control.
(C) Three-week-old WT, cbd1-1, cbd1-2, and COM grown horizontally in half-strength MS plates in a growth chamber. Scale bar corresponds to 1 cm.
(D) Chl content in plants in (C).
(E) Ratio of Chl a to b calculated from results in (D). Data are means ± SD. n = 4.
(F) Three-day-old dark-grown etiolated WT, cbd1-1, cbd1-2, and COM seedlings exposed to continuous light for 6 h. The scale bar corresponds to 1 mm. Asterisks indicate statistically significant differences compared with the WT (Student's t-test, ∗∗P < 0.01, ∗∗∗P < 0.001).
(G) Schematic representation of a summarized tetrapyrrole biosynthesis pathway. Only representative intermediates and enzymes (gray sphere) are shown.
(H) Levels of Chl intermediates in dark-grown WT and cbd1 seedlings treated with 10 mM ALA, as determined by spectrofluorometry. Three-week-old WT and cbd1 seedlings were dark adapted for 3 days before measurement. Excitation at 420 nm produces an emission peak (arrow) at 595 nm corresponding to Mg-Proto IX and Mg-Proto IX (ME).
Another gene, CBD2 (AT4G17840), was homologous to CBD1 in Arabidopsis. CBD2 encodes a protein of 422 amino acids that shared a 58% amino acid sequence identity with CBD1 (Supplemental Figure 4A). Similar to CBD1, CBD2 was also highly expressed in green tissues, as detected by qRT–PCR and GUS reporter analysis (Supplemental Figure 4B and 4C). We also examined the subcellular localization of CBD2 using the GFP fusion construct and found that CBD2::GFP was co-localized with RBCS::RFP, indicating a stromal localization of CBD2 (Supplemental Figure 4D). To study the function of CBD2, we generated cbd2 single and cbd1cbd2 double mutants using CRISPR/Cas9-mediated mutagenesis. Under normal growth conditions, the cbd2 single mutant showed essentially the same phenotype as the WT. However, the cbd1cbd2 double mutant was much more necrotic and consequently showed a more severe decrease in Chl content and a more increased Chl a/b ratio compared with the single mutants or WT (Supplemental Figure 5A, 5C, and 5D). After 5 days of dark incubation, cbd2 and cbd1cbd2 mutants became more chlorotic and wilting, whereas the cbd1 mutant retained its pale-green phenotype (Supplemental Figure 5B). In addition, we also compared the relative expression levels of CBD1 and CBD2 in leaves at different developmental stages (Supplemental Figure 5E). The expression level of CBD1 was higher in young and mature leaves. By contrast, the expression level of CBD2 was low in young and mature leaves but high in senescent leaves. These results suggested that, unlike CBD1, CBD2 might participate in a different metabolic pathway related to dark-adapted Chl degradation. We focused on a more detailed analysis of CBD1 function in this study.
Specific Defects Associated with the Chl Biosynthetic Pathway in the cbd1 Mutant
Because CHLH/GUN5 is an essential component in the Chl biosynthetic pathway and CBD1 was co-expressed with CHLH/GUN5, we hypothesized that CBD1 might participate in the same pathway as CHLH/GUN5. Moreover, the decreased Chl content and the slower rate of “greening” of cbd1 seedlings also supported a function of CBD1 in Chl biosynthesis. To explore the mechanism of action of CBD1, we surveyed the expression profiles of genes involved in the Chl biosynthesis pathway in mutant and WT plants during the dark-to-light switch. When plants were transferred from light to dark, the expression levels of these genes were downregulated but the down-turn was slower in the cbd1 mutant (Supplemental Figure 7A). While most of the genes in this pathway were upregulated upon the dark-to-light switch, they were induced to a higher level in the cbd1 mutant than in the WT (Supplemental Figure 7B). The perturbed expression levels of Chl biosynthetic genes also supported the idea that CBD1 was functionally involved in this pathway.
We next assessed the levels of Chl intermediates in 3-week-old WT and cbd1 seedlings by spectrofluorometry. In dark-adapted seedlings, the content of Proto IX was increased in the cbd1 mutant compared with the WT (Supplemental Figure 8A), implying that loss-of-function of CBD1 may hinder fluxes through the Chl biosynthesis pathway (Figure 3G). In spectrofluorometry, excitation at 420 nm produces an emission peak at 595 nm, corresponding to Mg-Proto IX and Mg-Proto IX ME (Pontier et al., 2007). However, there was no discernable 595-nm peak in dark-adapted WT and cbd1 seedlings (Supplemental Figure 8B), indicating that the content of Mg-Proto IX and Mg-Proto IX ME was too low to be detected by spectrofluorometry under these conditions. To overcome this problem, we fed dark-adapted WT and cbd1 seedlings with the early tetrapyrrole precursor ALA. After ALA feeding, the contents of intermediates consistently increased (Supplemental Figure 8A). Notably, a strong 595-nm peak was detected in cbd1 but not in WT plants, suggesting that Mg-Proto IX and/or Mg-Proto IX ME were over-accumulated in cbd1 mutant (Figure 3H).
CBD1 Functionally Interact with GUN5 in Chl Biosynthesis and Chloroplast Development
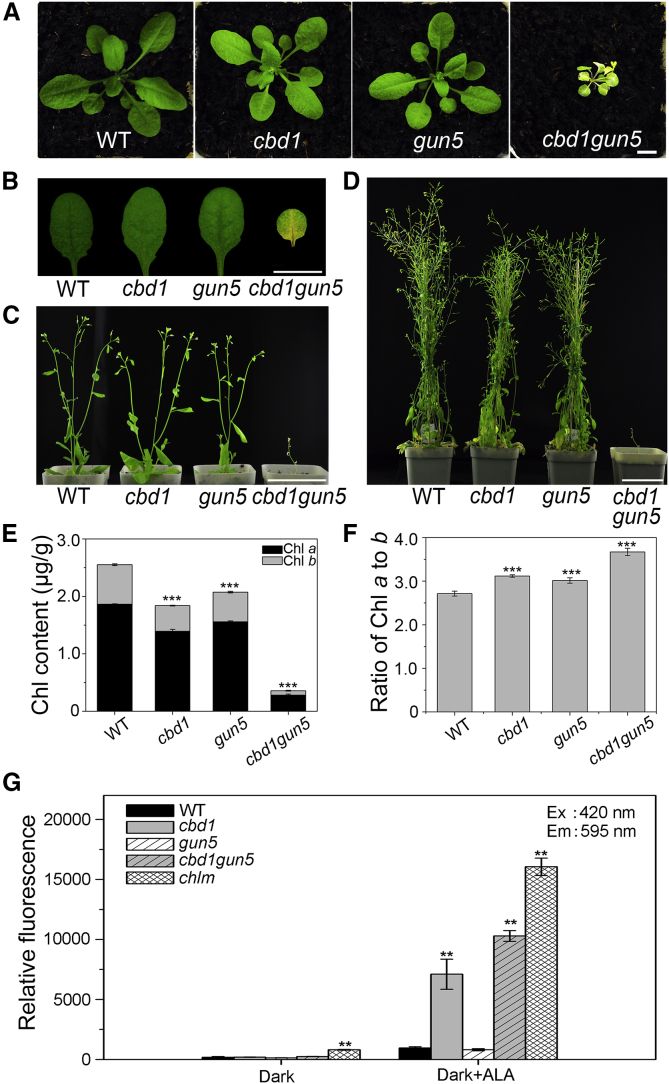
Based on the aforementioned results, CBD1 and CHLH/GUN5 appear to work together at the step of Mg-Proto IX conversion in the Chl biosynthesis pathway. To further investigate the functional connection between CBD1 and CHLH/GUN5, we generated double mutants of cbd1 and chlh/gun5. The gun5-1 mutant (hereafter called gun5) with a point mutation (Ala990→Val) in CHLH/GUN5 (Mochizuki et al., 2001) was used in this experiment. In the F2 generation from a ♂cbd1 x ♀gun5-1 cross, we found several pale-yellow seedlings that were homozygous cbd1gun5 double mutants, as verified by genomic PCR and sequencing. The homozygous cbd1gun5 F2 seedlings showed a more chlorotic phenotype compared with the two single mutants (Figure 4A and 4B). After being transferred to the soil, cbd1gun5 plants showed stunted growth; however, they were able to produce seeds (Figure 4C and 4D). After assessing the Chl content, we found that the cbd1gun5 double mutant showed an additive effect of the two single mutants and retained a dramatically lower Chl content and a higher Chl a/b ratio compared with its parental lines (Figure 4E and 4F). We also used cch (conditional chlorina), another gun5 allele that has a Pro990→Leu point mutation (Mochizuki et al., 2001), to generate a cbd1cch double mutant and found that the cbd1cch double mutant phenocopied the cbd1gun5 double mutant (Supplemental Figure 9).
Figure 4.
Arabidopsis cbd1gun5 Double Mutant Exhibited Variable Chlorotic Phenotypes and Mg-Proto IX levels.
(A–D) (A) Three-week-old, (C) 4-week-old, and (D) 7-week-old WT, cbd1, gun5, and cbd1gun5 seedlings grown in soil in a greenhouse. (B) Detached leaves from (A).
(E) Chlorophyll content in plants in (A). Scale bars correspond to 1 cm in (A) and (B) and 7 cm in (C) and (D).
(F) Ratio of chlorophyll a to b in plants in (A). Data are means ± SD. n = 4.
(G) Three-week-old WT and cbd1, gun5, cbd1gun5, and chlm mutants were dark-adapted for 3 days before Mg-Proto IX (ME) level was determined by spectrofluorometry under different treatments. Asterisks indicate statistically significant differences compared with the WT (Student's t-test, ∗∗P < 0.01, ∗∗∗P < 0.001).
CHLH/GUN5 functions not only as the catalytic subunit of MgCh but also as an activator of MgMT. To assess the steps at which CBD1 and CHLH/GUN5 function in Chl biosynthesis, we analyzed the Chl intermediates in dark-adapted WT, cbd1, gun5, and cbd1gun5 seedlings under ALA feeding conditions described earlier. The Mg-Proto IX content was reduced in gun5, consistent with the enzymatic activity of CHLH/GUN5 in MgCh (Figure 4G) (Mochizuki et al., 2001). By contrast, the 595-nm peak, which reflects the content of Mg-Proto IX and Mg-Proto IX ME, was considerably higher in cbd1gun5 mutants compared with the WT or cbd1 mutant (Figure 4G), indicating that CBD1 and GUN5 work together to regulate steps downstream of the Mg chelation. We also assessed the 595-nm peak in the chlm mutant as a reference. As reported earlier, the Mg-Proto IX content, but not the Mg-Proto IX ME content, was increased in chlm (Figure 4G) (Pontier et al., 2007; Mochizuki et al., 2008). Furthermore, the cbd1chlm double mutant showed the same phenotype as the chlm single mutant (Supplemental Figure 10), supporting the notion that CBD1 functions upstream of CHLM. Therefore, we suggested that CBD1 and CHLH/GUN5 work collaboratively in trafficking Mg-Proto IX to the next step in the pathway.
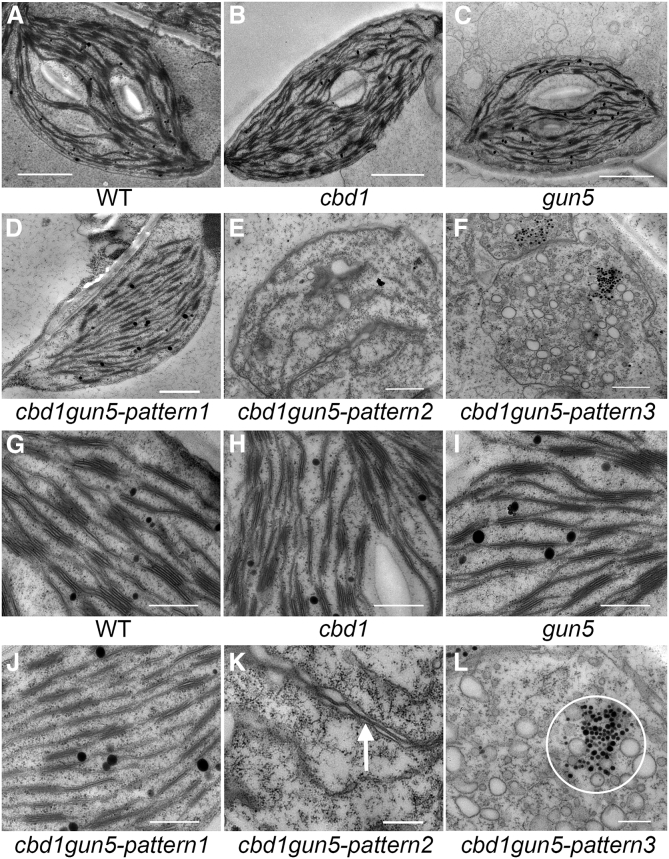
To assess chloroplast development in the mutants, we examined the ultrastructure of chloroplasts and thylakoids in 4-week-old plants by transmission electron microscopy. Chloroplasts and thylakoid stacks in cbd1 (Figure 5B and 5H) and gun5 (Figure 5C and 5I) were approximately the same size and shape as the WT (Figure 5A and 5G), however, fewer starch grains were observed in the two single mutants. By contrast, the development of chloroplasts (Figure 5D–5F) and thylakoids (Figure 5J–5L) was severely defective in the cbd1gun5 double mutant. Three types of chloroplasts with varying degrees of defects were observed: (1) well-developed but not fully developed chloroplasts with smaller and less stacked thylakoids (Figure 5D and 5J), (2) a proplastid-like morphology in which only rudimentary thylakoids were present (Figure 5E and 5K), and (3) a lack of thylakoids (Figure 5F) with only vesicles possibly formed by undeveloped thylakoid membrane. In addition, osmiophilic granules were accumulated in the cbd1gun5 double mutant, which may have resulted from the degradation of the thylakoid membrane (Figure 5L).
Figure 5.
Transmission Electron Microscopy Analysis of Chloroplasts in WT, cbd1, gun5, and cbd1gun5.
(A–C) Chloroplasts containing starch grains and thylakoids from the leaves of 4-week-old WT, cbd1, and gun5.
(D–F) Chloroplasts from the leaves of 4-week-old cbd1gun5.
(G–I) Thylakoids containing grana and stroma thylakoids from the leaves of 4-week-old WT, cbd1, and gun5.
(J–L) Thylakoids from the leaves of 4-week-old cbd1gun5. The white arrow in (K) indicates the rudimentary thylakoids and the white circle in (L) indicates the aggregated osmiophilic granules.
Scale bars correspond to 2 μm in (A–C), 1 μm in (D–F), and 500 nm in (G–L).
The decrease in Chl content may also affect the assembly of photosynthetic apparatus. We examined the abundance of thylakoid protein complexes using blue native (BN) gel electrophoresis (Supplemental Figure 11A). The cbd1 and gun5 single mutants showed a consistently altered pattern in photosynthetic complexes. Notably, the mutants exhibited a pronounced increase in the bands representing monomeric photosystem I (PSI-M) and CF0–CF1 complex (CF1). The band corresponding to monomeric PSII (PSII-M) also showed an increase compared with the WT. Furthermore, the cbd1gun5 double mutant was almost devoid of the PSII super-complexes (PSII SC). In addition, the cbd1gun5 double mutant, but not the single mutants, exhibited a reduced level in the PSI–light-harvesting complex I (PSI–LHCI) and PSII-dimer (PSI–LHCI, PSII-D) complex compared with the WT. These results indicated a possible role of CBD1 and CHLH/GUN5 in the assembly of photosynthetic super-complexes. Analyses of 2D SDS–PAGE silver-stained gels and western blots confirmed that the large majority of protein subunits were associated with lower-molecular-weight complexes, suggesting a defect in the assembly of super-complexes (especially PSI) in the mutants, especially in the cbd1gun5 double mutants (Supplemental Figure 11B and 12). We also examined these mutants using Chl fluorescence and found a reduced minimal fluorescence (Fo) (Supplemental Figure 13A) but increased PSII maximum efficiency (Fvʹ/Fmʹ) and non-photochemical quenching (NPQ) in both the single and double mutants (Supplemental Figure 13B and 13C). The similar photosynthetic parameters in cbd1, gun5, and cbd1gun5 mutants implied that CBD1 and CHLH/GUN5 function in a similar mechanism in regulating photosynthesis.
CBD1 Regulates Mg Homeostasis in Chloroplast
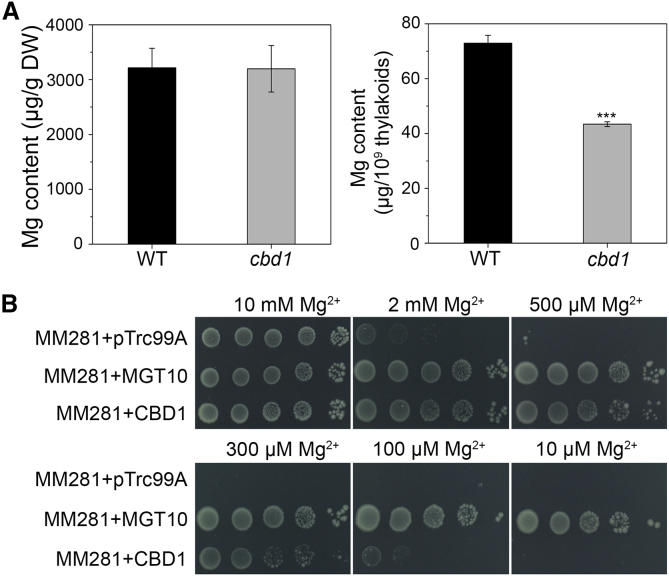
The CBD1 protein had 382 amino acid residues with six putative transmembrane domains (Supplemental Figure 14A). Based on multiple sequence alignments, homologs of CBD1 were evolutionarily conserved in diverse plant species, including moss, ferns, and flowering plants (Supplemental Figure 14B), although none of these proteins has been functionally characterized. A sequence alignment based on Position-Specific Iterated-BLAST identified a C-terminal region of 100 amino acids in CBD1 containing three conserved motifs characteristic for type II CAAX prenyl proteases that were conserved in diverse prokaryotic and eukaryotic species (Pei and Grishin, 2001) (Supplemental Figure 14C). In these motifs, the conserved amino acids required for chelating or binding metal ions were also present in CBD1, suggesting a possible involvement of CBD1 in metal homeostasis. To test this possibility, we conducted an elemental analysis using inductively coupled plasma-mass spectrometry (ICP–MS). Although no significant differences in the contents of metals were found in the leaves of the WT and cbd1 mutant (Supplemental Table 1), the Mg content in intact thylakoids of cbd1 showed a 40% decrease compared with the WT (Figure 6A), whereas the contents of Fe, Mn, Cu, and Ca were not significantly different (Supplemental Table 1). As the total Chl content was reduced by 19% in the cbd1 mutant compared with the WT, the 40% reduction in thylakoid Mg content was not simply attributed to the decrease in Chl levels.
Figure 6.
CBD1 Regulates Mg Homeostasis In Vivo and In Vitro.
(A) Mg content in the leaves and intact thylakoids of 3-week-old WT and cbd1 seedlings grown on half-strength MS medium. Data are means ± SD. n = 4. Asterisks indicate statistically significant differences compared with the WT (Student's t-test, ∗∗∗P < 0.001).
(B) Growth of different bacterial cells on the N-minimal medium containing 10 mM, 2 mM, 500 μM, 300 μM,100 μM, and 10 μM MgSO4. The strains were MM281 transformed with the pTrc99A vector only (MM281 + pTrc99A) as the negative control, MGT10 cDNA in the pTrc99A vector (MM281 + MGT10) as the positive control, and CBD1 cDNA in pTrc99A vector (MM281 + CBD1). From left to right is a 10-fold dilution series of bacterial cultures.
Considering the presence of six putative transmembrane domains in the CBD1 protein, we speculated that CBD1 may have Mg transport activity and tested this idea using the Salmonella mutant strain MM281, which lacks Mg2+ transport systems. We cloned the full-length cDNA of CBD1 into the pTrc99A vector and then transformed it into MM281. While the MM281 strain transformed with the empty pTrc99A vector barely grew in medium containing less than 2 mM Mg2+, MM281 expressing AtMGT10, a high-affinity Mg2+ transporter, grew well in medium containing only 10 μM Mg2+. MM281 transformed with the CBD1-containing construct grew in medium with 100 μM Mg2+ (Figure 6B), suggesting that CBD1 may mediate Mg2+ transport in the high μM range. This affinity of CBD1 for Mg binding is in the physiological range relevant to chloroplast Mg levels. Taken collectively, these results indicated that CBD1 may bind/translocate Mg2+ and regulate its incorporation into Chl biosynthesis processes in synergy with CHLH/GUN5 and other enzymes.
Discussion
Despite the indispensable role of Chl in photosynthetic light harvesting, photo-oxidative damage to cells can result from Chl and its highly photoreactive intermediates, that is, if they are not rapidly integrated into the protein complexes. Therefore, the biosynthesis and homeostasis of Chl are precisely controlled to fine-tune the metabolic flow and protect plants from photodamage (Triantaphylidès and Havaux, 2009). For this purpose, enzymes involved in Chl biosynthesis often form protein complexes to enable the efficient channeling of intermediates (Tanaka and Tanaka, 2007). In the Mg-chelating reaction, protoporphyrinogen IX oxidase (PPOX), the enzyme that produces Proto IX in chloroplasts, is in close contact with CHLH/GUN5 to form a super-complex that enables efficient reactions to occur (Chen et al., 2015). Thereafter, the association of CHLH to the chloroplast membrane is prompted by the GUN4-porphyrin complexes (Adhikari et al., 2009, 2011). The PPOX-CHLH/GUN5-GUN4 complex ensures the transformation of Proto IX to Mg-Proto IX. In the following step, a tight complex formed between MgCh and MgMT enables the direct channeling of Mg-Proto IX from CHLH/GUN5 to CHLM for the methylation (Gorchein, 1972). In fact, the direct interaction of CHLH/GUN5 with the CHLM protein enhances the overall activity of MgMT (Jensen et al., 1999; Alawady et al., 2005; Alawady and Grimm, 2005; Shepherd et al., 2005; Chen et al., 2014).
In this study, we uncovered a thylakoid membrane localized protein, CBD1, as a critical component in Chl biosynthesis. Detailed analysis indicated that CBD1 may work synergistically with CHLH/GUN5 in promoting the formation and activity of the MgCh–MgMT complex. CBD1 was identified by co-expression analysis using CHLH/GUN5 as a query gene (Figure 1). The cbd1 mutant in Arabidopsis showed a pale-green phenotype, reduced total Chl content, and an increased Chl a/b ratio, reminiscent of the gun5 mutant (Figure 3C–3E). Furthermore, the cbd1gun5 double mutant had more severe chlorotic leaves and more stunted growth (Figure 4A–4F), suggesting that CBD1 and CHLH/GUN5 might participate in similar biological processes.
While preparing this study for publication, a report (Wang et al., 2020) described the analysis of BCM1 (Balance of Chlorophyll Metabolism1) that is identical to CBD1. BCM1 was also identified as a thylakoid membrane protein playing a role in Chl biosynthesis. It is important to compare several points between the two studies, which may help to better understand the mechanism underlying BCM1/CBD1 function in Chl biosynthesis. Both studies addressed the step in Chl biosynthesis at which BCM1/CBD1 may play a role. In the bcm1 mutant (Wang et al., 2020), the authors observed a decrease in the ALA synthesis rate and reduced accumulation of Proto IX, Mg-Proto IX, and Mg-Proto IX ME. It is difficult to pinpoint which step of Chl biosynthesis is affected based on this profile of metabolites. The fact that BCM1 physically and genetically interacts with GUN4 prompted the conclusion that BCM1 facilitates the biosynthesis of Mg-Proto IX. We took a different approach to examine the later steps in Chl biosynthesis by feeding adequate ALA to the cbd1 mutant and WT plants and investigated the intermediate accumulation. We observed Mg-Proto IX/ME over-accumulation in cbd1, and this accumulation was more dramatic in the cbd1gun5 double mutant (Figures 3H and 4G). Therefore, we suggest that CBD1 likely functions in synergy with CHLH/GUN5 in Mg-Proto IX trafficking to the methylation step. This is consistent with the idea that CHLH/GUN5 not only serves as the catalytic subunit in the Mg chelation step, but also as the “activator” of CHLM in the methylation step. With regard to the mechanism of action, BCM1 is shown to interact with GUN4 and by doing so serves as an important “scaffold” to promote the organization and targeting of a MgCh–GUN4–CHLM enzyme complex at the thylakoid membrane (Wang et al., 2020). We also speculated that the thylakoid membrane-localized CBD1 may serve as an anchor protein of MgCh for its subsequent interaction with MgMT. Consistent with this hypothesis, double mutation of CBD1 and CHLH/GUN5 resulted in severely disturbed MgMT function as reflected by the over-accumulation of Mg-Proto IX/ME upon ALA feeding. In addition to the functional interaction between CBD1 and GUN5, we also provided evidence that CBD1 may bind/translocate Mg2+ at the thylakoid membrane and affect Mg2+ homeostasis (Figure 6), thus leading to Mg2+ accumulation in the stroma. Although adequate free Mg2+ is required for the Mg-chelating reaction in Chl biosynthesis, excessive Mg2+ in the stroma is known to inhibit MgMT activity due to the aggregation of the enzyme at a high free-Mg2+ concentration (Sawicki and Willows, 2007, 2010). The exact mechanism of how CBD1 affects the Mg balance between the stroma and thylakoid warrants further study.
Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana WT (ecotype Columbia-0) and T-DNA insertion line cbd1-1 (SALK_031802) and cbd1-2 (SALK_058830) were ordered from the Arabidopsis Biological Resource Center. The homozygous mutants were obtained from heterozygous plants and screened by genomic PCR using the primers listed in Supplemental Table 2. Homozygous individuals were further identified using RT–PCR analysis. Seeds were surface sterilized in 75% ethanol three times before being sown on half-strength Murashige and Skoog (MS) medium. To ensure synchronized germination, seeds were incubated in darkness for 2 days at 4°C before transferring to a growth chamber. For culture on agar plates in the growth chamber, WT and mutant plants used in this study were grown in half-strength MS medium containing 1% (w/v) sucrose (Sigma) and 0.8% (w/v) phytagel (Caisson Labs) in the growth chamber under long-day conditions (16-h light/8-h dark cycle with a photon flux density of 90 μmol m−2 s−1) at 22°C. For soil culture in a greenhouse, 10-day-old seedlings germinated on half-strength MS agar plates were transferred to soil for further growth in a greenhouse under long-day conditions (16-h light/8-h dark cycle with a photon flux density of 90 μmol m−2 s−1) at 22°C.
Plasmid Construction and Plant Transformation
For the construction of the genetic complementation vector, the 3.4-kb full-length genomic DNA fragment (including the 979-bp promoter and the 2434-bp genomic region) of CBD1 was amplified and cloned into the pCAMBIA-1300 binary vector. To generate GUS fusion expression constructs (CBD1pro::GUS), a 979-bp DNA fragment between CBD1 and its upstream contiguous gene was amplified to generate GUS fusion expression constructs (CBD2pro::GUS), and a 1803-bp DNA fragment between CBD2 and its upstream contiguous gene was amplified. The fragments were then ligated to the modified pCAMBIA1300 vector with a GUS reporter gene. Transgenic plants were generated by the flora dip method with the Agrobacterium tumefaciens strain GV3101 containing the recombinant binary plasmids mentioned above. Transformants were screened on half-strength MS agar plates with 25 mg/l hygromycin. To analyze the subcellular localization in the transient expression assay, the CDS fragments of CBD1, CBD2, PAA2 (AT5G21930), PIC1 (AT2G15290), OEP7 (AT3G52420), and RBCS1 (AT1G67090) deprived of stop codons were amplified and cloned into the PEZS-NL-GFP vector. The CDS fragments of PAA2, PIC1, OEP7, and RBCS1 were further cloned into the PEZS-NL-RFP vector, respectively, for co-localization experiments. In the MM281 complementation assay, the full-length CDS of CBD1 was amplified and cloned into the pTrc99A vector. All primers used are listed in Supplemental Table 2.
Chl Content Determination
To analyze Chl content, plants (∼20 mg fresh weight) were harvested. Leaf samples were ground in 1 ml 80% acetone and placed in the dark at 4°C for 30 min before centrifugation. The supernatants were used for spectrometric measurements at 645 and 663 nm, respectively, using a spectrometer (Biomate 3S, Thermo Scientific).
Subcellular Localization of CBD1
Arabidopsis mesophyll protoplasts were prepared from 4-week-old rosette leaves by soaking leaf slices with an enzymatic mixture containing 1% (w/v) cellulase R10 and 0.4% (w/v) macerozyme R10 (Yakult Pharmaceutical) for 2–3 h. The digested protoplasts were resuspended in the W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES [pH 5.8]), and transfected in combination with 20 μg recombinant plasmids (CBD1-PEZSNL, PAA2-PEZSNL, PIC1-PEZSNL, OEP7-PEZSNL, and RBCS1-PEZSNL) using a PEG-mediated transformation protocol. The transformed protoplasts were incubated in the dark at 22°C overnight before imaging using a laser scanning confocal microscope (Lei TCS SP8-MaiTai MP and Leica TCS-SL).
Histochemical GUS Analysis
Approximately 70 transformants harboring the CBD1pro::GUS fusion protein were selected and a representative line was used in this assay. For histochemical analysis, plant materials were fixed in 90% ice-cold acetone for 30 min and then washed with the X-Gluc free staining buffer (0.5 mM K4Fe(CN)6, 0.5 mM K3Fe(CN)6, 0.1% Triton X-100, 10 mM EDTA, and 100 mM sodium phosphate). Samples were then immersed in the staining buffer supplemented with 1 mM X-Gluc. After a short vacuuming period, samples were incubated at 37°C for 12 h. To clear the background, the stained materials were then rinsed in 75% ethanol overnight to remove Chl. Representative seedlings were photographed under a microscope (Olympus, SZX16).
Generation of a cbd2 Knockout Mutant by CRISPR/Cas9
For the identification of CRISPR/Cas9 target sites in CBD2, the online CRISPR-P software (http://cbi.hzau.edu.cn/crispr/) was used. Secondary structure analysis of target single-guide (sgRNA) sequences was carried out with the program RNA Folding Form (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form2.3). After screening, three sgRNAs were selected. For each target sequence, primer pairs were designed to form a dimer with two BsaI overhangs. The three sgRNA expression cassettes were generated by adaptor-ligation and PCR amplification, and subsequently ligated into the pYLCRISPR/Cas9 vector by Golden Gate cloning. The recombinant constructs were then transformed into the WT or cbd1 mutant using the flora dip method with the Agrobacterium tumefaciens strain EHA105. To check for mutations in CBD2, genomic DNA was extracted, and PCR amplifications were carried out using primer pairs flanking the target sites. These PCR products were sequenced to detect frameshift or deletion mutations in CBD2. All primers used are listed in Supplemental Table 2.
Functional Complementation of Mg2+ Transport by Salmonella typhimurium Mutant MM281
The competent MM281 cells were transformed with the empty pTrc99A vector, recombinant MGT10-pTrc99A, and CBD1-pTrc99A by electroporation (Gene Pulser Xcell, Bio-Rad). Transformed cells were plated onto LB medium containing 10 mM Mg2+ and indicated antibiotics (17 μg/ml chloramphenicol and 100 μg/ml ampicillin). After overnight incubation at 37°C, transformants were selected and confirmed by PCR. Positive ones were selected and further grown in liquid LB medium containing 10 mM Mg2+ and antibiotics as indicated above; 50 μM IPTG was applied to induce protein expression. The liquid cultures were adjusted to an optical density at 600 nm of 1.0, 5 μl of a 10-fold serial dilution was spotted onto the N-minimal medium plates as indicated above, further diluted in a 10-fold series, and 4 μl spotted onto N-minimal medium (5 mM KCl, 0.5 mM K2SO4, 1 mM KH2PO4, 7.5 mM (NH4)2SO4, 0.1 mM Tris, 0.02% [w/v] glucose, and 0.1% [w/v] casein hydrolysate) supplemented with indicated MgSO4 and antibiotics. The plates were incubated at 37°C for 48 h before being photographed.
Isolation of Total RNA, RT–PCR, and qRT–PCR Analysis
Total RNA was extracted using the TRIzol reagent (Invitrogen) following the manufacturer's instructions. The first-strand cDNA was synthesized by M-MLV Reverse Transcriptase (Promega). Semi-quantitative RT–PCR analysis of gene expression using cDNA of WT, two mutant lines (cbd1-1 and cbd1-2), and the COM line, followed by 26 cycles of PCR, was carried out. qRT–PCR analysis was performed using the FastStart Universal SYBR Green Master mix (Roche) on a CFX Connect real-time system (Bio-Rad). Target quantifications were performed with specific primer pairs designed using the Primer designing tool in NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Actin2 (AT3G18780) was used as an internal reference in both RT–PCR and qRT–PCR analysis. All primers used are listed in Supplemental Table 2.
Analysis of Intermediates in the Chl Biosynthetic Pathway
Wild-type, cbd1, gun5, cbd1gun5, and chlm were grown horizontally in a growth chamber for 3 weeks, and plates were dark adapted for another 3 days, after which ALA feeding solution (10 mM ALA, 5 mM MgCl2, and 10 mM HEPES in 10 mM NaPO4 [pH 7.0]) was added to each plate. The seedlings were incubated overnight and harvested under dim green light. Approximately 20 mg of leaf material was homogenized in extraction solution (acetone:NH4OH = 9:1 [v/v]). Samples were centrifuged for 10 min at 12 000 g. The supernatants were then transferred to fresh microcentrifuge tubes and extracted three times with hexane to remove carotenoids and Chls. Fluorescence emission spectra were recorded from 560 to 700 nm using an F7000 luminescence spectrophotometer (HITACHI) at room temperature. The samples were excited at 400 and 420 nm for the detection of ProtoIX and Mg-ProtoIX (ME), respectively.
BN Gel, 2D SDS–PAGE Electrophoresis and Immunoblot Analysis
BN gel electrophoresis was performed as described previously (Zhang et al., 2018). Thylakoid membrane samples containing equal amounts of Chl (10 μg) were loaded onto a BN gradient gel. Electrophoresis was performed at 100 V at 4°C with a 1–8 mA current (PowerSac Universal, Bio-Rad). The cathode buffer initially contained 0.01% (w/v) Serva Blue G dye and was replaced with cathode buffer free of dye halfway through the run. For 2D SDS–PAGE analysis, the excised BN gel lanes were soaked for 10 min in SDS sample buffer (12.5 mM Tris, 4% [w/v] SDS, 20% [v/v] glycerol, and 0.02% [w/v] bromophenol blue) supplied with 5% (v/v) β-mercaptoethanol. Each lane with denatured proteins was placed on top of 12% SDS–PAGE and electrophorated at 100 V at 4°C at 20–58 mA. After electrophoresis, gels were stained using a protein silver stain kit (CWBIO) and photographed (Gel Doc XR+, Bio-Rad). Protein samples corresponding to equal amounts of Chl (1 μg) were separated on 15% SDS–PAGE gel and transferred to polyvinylidene fluoride membrane. After blocking non-specific binding with 5% (w/v) milk, the blot was subsequently incubated with antibodies generated against the indicated proteins and detected using a SuperSignal West Pico Trial kit (Thermo Scientific).
Chl Fluorescence Measurement
Intact leaves of 3-week-old WT and mutant plants grown on half-strength MS agar plates were used in this assay. Plants were dark adapted for 40 min before each measurement. To assess the photosynthetic performance of PSII, basic parameters Fo, Foʹ, Fm, Fmʹ, and Fs were measured with an FMS2 fluorometer (Hansatech, Norfolk, UK). Fvʹ/Fmʹ, and NPQ were calculated according to a previous study (Fu et al., 2007).
Transmission Electron Microscopy
For transmission electron microscopy processing, rosette leaves were collected from 3-week-old WT and mutant plants grown in half-strength MS medium in a growth chamber. The leaves were cut into slices and fixed in 2.5% (v/v) glutaraldehyde under mild vacuuming. The fixed samples were used to make ultrathin sections using a diamond knife on an ultramicrotome (PowerTome-XL, RMC). Micrographs were taken under a transmission electron microscope (H7650, HITACHI).
Isolation of Intact Chloroplasts and Thylakoids
Leaves of 3-week-old WT and mutant plants grown on half-strength MS agar plates were harvested in the daytime. To isolate intact chloroplasts, leaf samples (at least 10 g fresh weight) were homogenized in ice-cold grind buffer (0.33 M sorbitol, 10 mM EDTA, 50 mM HEPES, and 0.5 g/L BSA [pH was adjusted to 8.0 with KOH]). The homogenate was filtered through two layers of Miracloth (Calbiochem) and concentrated by centrifugation for 5 min at 700 g at 4°C using a swing-out rotor (Centrifuge 5810R, Eppendorf). The pellets were gently resuspended in grinding buffer using a brush and then layered onto a discontinuous 40% (w/v)/80% (w/v) Percoll (GE Healthcare) gradient. After centrifugation for 15 min at 2000 g at 4°C, intact chloroplasts were isolated from the interface between the two layers. The interface was gently sucked into a new tube and washed in SH (0.33 M sorbitol and 50 mM HEPES [pH adjusted to 8.0 with KOH]) to remove the remaining Percoll. For the further isolation of thylakoids, intact chloroplasts were lysed in lysis buffer (10 mM HEPES [pH adjusted to 8.0 with KOH]) for 10 min on ice. After centrifugation at 700 g at 4°C for 10 min, the transparent supernatant containing stroma fractions was discarded, and the pellets were intact thylakoids.
Metal Ion Content Determination
Leaves, intact chloroplasts, and thylakoids were collected and evaporated for further analysis. Dried samples were digested with concentrated HNO3 (69%) at 80°C in a digester (DigiBlock ED16, LabTech). The ion concentration was measured by ICP–MS (NexION 300, PerkinElmer).
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31900220 to C.Z., and 31770267 to W.L.) and the National Science Foundation (MCB-1714795 to S.L.).
Author Contributions
C.Z., B.Z., A.F., W.L., and S.L. designed the research. C.Z., B.Z., and B.M. carried out the experiments. C.Z., B.Z., B.M., A.F., W.L., and S.L. analyzed the data. X.Z. and F.Z. provided technical assistance. C.Z., B.Z., and S.L. wrote the manuscript.
Acknowledgments
We thank Dr. Nobuyoshi Mochizuki for the gun5-1, cch, and chlm mutant seeds used in this work, Dr Yaoguang Liu for the CRISPR/Cas9 vectors, and Dr. Fangjie Zhao for advice on ICP–MS analysis. No conflict of interest declared.
Published: July 3, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information can be found online at Plant Communications Online.
Contributor Information
Wenzhi Lan, Email: lanw@nju.edu.cn.
Aigen Fu, Email: aigenfu@nwu.edu.cn.
Sheng Luan, Email: sluan@berkeley.edu.
Supplemental Information
References
- Abdel-Ghany S.E., Muller-Moule P., Niyogi K.K., Pilon M., Shikanai T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell. 2005;17:1233–1251. doi: 10.1105/tpc.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari N.D., Froehlich J.E., Strand D.D., Buck S.M., Kramer D.M., Larkin R.M. GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell. 2011;23:1449–1467. doi: 10.1105/tpc.110.082503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari N.D., Orler R., Chory J., Froehlich J.E., Larkin R.M. Porphyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J. Biol. Chem. 2009;284:24783–24796. doi: 10.1074/jbc.M109.025205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alawady A., Reski R., Yaronskaya E., Grimm B. Cloning and expression of the tobacco CHLM sequence encoding Mg protoporphyrin IX methyltransferase and its interaction with Mg chelatase. Plant Mol. Biol. 2005;57:679–691. doi: 10.1007/s11103-005-1427-8. [DOI] [PubMed] [Google Scholar]
- Alawady A.E., Grimm B. Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. Plant J. 2005;41:282–290. doi: 10.1111/j.1365-313X.2004.02291.x. [DOI] [PubMed] [Google Scholar]
- Bang W.Y., Jeong I.S., Kim D.W., Im C.H., Ji C., Hwang S.M., Kim S.W., Son Y.S., Jeong J., Shiina T. Role of Arabidopsis CHL27 protein for photosynthesis, chloroplast development and gene expression profiling. Plant Cell physiol. 2008;49:1350–1363. doi: 10.1093/pcp/pcn111. [DOI] [PubMed] [Google Scholar]
- Brzezowski P., Richter A.S., Grimm B. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta. 2015;1847:968–985. doi: 10.1016/j.bbabio.2015.05.007. [DOI] [PubMed] [Google Scholar]
- Chekounova E., Voronetskaya V., Papenbrock J., Grimm B., Beck C.F. Characterization of Chlamydomonas mutants defective in the H subunit of Mg-chelatase. Mol. Genet. Genomics. 2001;266:363–373. doi: 10.1007/s004380100574. [DOI] [PubMed] [Google Scholar]
- Chen S.T., He N.Y., Chen J.H., Guo F.Q. Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5-mediated retrograde pathway in Arabidopsis. Plant J. 2017;89:1106–1118. doi: 10.1111/tpj.13447. [DOI] [PubMed] [Google Scholar]
- Chen X., Wang X., Feng J., Chen Y., Fang Y., Zhao S., Zhao A., Zhang M., Liu L. Structural insights into the catalytic mechanism of Synechocystis magnesium protoporphyrin IX O-methyltransferase (ChlM) J. Biol. Chem. 2014;289:25690–25698. doi: 10.1074/jbc.M114.584920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Pu H., Fang Y., Wang X., Zhao S., Lin Y., Zhang M., Dai H.E., Gong W., Liu L. Crystal structure of the catalytic subunit of magnesium chelatase. Nat. plants. 2015;1:15125. doi: 10.1038/nplants.2015.125. [DOI] [PubMed] [Google Scholar]
- Chow K.S., Singh D.P., Walker A.R., Smith A.G. Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and subcellular targeting of the precursor proteins. Plant J. 1998;15:531–541. doi: 10.1046/j.1365-313x.1998.00235.x. [DOI] [PubMed] [Google Scholar]
- Duy D., Wanner G., Meda A.R., von Wiren N., Soll J., Philippar K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell. 2007;19:986–1006. doi: 10.1105/tpc.106.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A., He Z., Cho H.S., Lima A., Buchanan B.B., Luan S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 2007;104:15947–15952. doi: 10.1073/pnas.0707851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson L.C., Willows R.D., Kannangara C.G., Von W.D., Hunter C.N. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. U S A. 1995;92:1941–1944. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorchein A. Magnesium protoporphyrin chelatase activity in Rhodopseudomonas spheroides. Studies with whole cells. Biochem. J. 1972;127:97–106. doi: 10.1042/bj1270097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.E., Gibson L.C., Hunter C.N. Determinants of catalytic activity with the use of purified I, D and H subunits of the magnesium protoporphyrin IX chelatase from Synechocystis PCC6803. Biochem. J. 1998;334(Pt 2):335–344. doi: 10.1042/bj3340335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen P.E., Gibson L.C., Hunter C.N. ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem. J. 1999;339(Pt 1):127–134. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Masuda T. Transcriptional regulation of tetrapyrrole biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 2016;7:1811. doi: 10.3389/fpls.2016.01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin R.M. Tetrapyrrole signaling in plants. Front. Plant Sci. 2016;7:1586. doi: 10.3389/fpls.2016.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.W., Lee S., Lee G.J., Lee K.H., Kim S., Cheong G.W., Hwang I. Functional characterization of sequence motifs in the transit peptide of Arabidopsis small subunit of rubisco. Plant Physiol. 2006;140:466–483. doi: 10.1104/pp.105.074575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth. Res. 2008;96:121–143. doi: 10.1007/s11120-008-9291-4. [DOI] [PubMed] [Google Scholar]
- Masuda T., Fujita Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008;7:1131–1149. doi: 10.1039/b807210h. [DOI] [PubMed] [Google Scholar]
- Matsumoto F., Obayashi T., Sasakisekimoto Y., Ohta H., Takamiya K., Masuda T. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 2004;135:2379. doi: 10.1104/pp.104.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.a., Larkin R., Nagatani a., Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U S A. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Tanaka R., Tanaka A., Masuda T., Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Nishida K., Kasahara K., Kinoshita K. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J., Grishin N.V. Type II CAAX prenyl endopeptidases belong to a novel superfamily of putative membrane-bound metalloproteases. Trends Biochem. Sci. 2001;26:275–277. doi: 10.1016/s0968-0004(01)01813-8. [DOI] [PubMed] [Google Scholar]
- Pontier D., Albrieux C., Joyard J., Lagrange T., Block M.A. Knock-out of the magnesium protoporphyrin IX methyltransferase gene in Arabidopsis. Effects on chloroplast development and on chloroplast-to-nucleus signaling. J. Biol. Chem. 2007;282:2297–2304. doi: 10.1074/jbc.M610286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelius A.S., Aronsson H. Springer-Verlag; Berlin Heidelberg: 2009. The Chloroplast: Interactions with the Environment. [Google Scholar]
- Sawicki A., Willows Robert D. S-Adenosyl-L-methionine:magnesium-protoporphyrin IX O-methyltransferase from Rhodobacter capsulatus: mechanistic insights and stimulation with phospholipids. Biochem. J. 2007;406:469–478. doi: 10.1042/BJ20070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki A., Willows R.D. BchJ and BchM interact in a 1:1 ratio with the magnesium chelatase BchH subunit of Rhodobacter capsulatus. FEBS J. 2010;277:4709–4721. doi: 10.1111/j.1742-4658.2010.07877.x. [DOI] [PubMed] [Google Scholar]
- Scharfenberg M., Mittermayr L., E V.O.N.R.-L., Schlicke H., Grimm B., Leister D., Kleine T. Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 2015;38:280–298. doi: 10.1111/pce.12248. [DOI] [PubMed] [Google Scholar]
- Shang Y., Yan L., Liu Z.-Q., Cao Z., Mei C., Xin Q., Wu F.-Q., Wang X.-F., Du S.-Y., Jiang T. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell. 2010;22:1909–1935. doi: 10.1105/tpc.110.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd M., McLean S., Hunter C.N. Kinetic basis for linking the first two enzymes of chlorophyll biosynthesis. FEBS J. 2005;272:4532–4539. doi: 10.1111/j.1742-4658.2005.04873.x. [DOI] [PubMed] [Google Scholar]
- Tanaka R., Kobayashi K., Masuda T. Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis book. 2011;9:e0145. doi: 10.1199/tab.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R., Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Bio. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C., Havaux M. Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 2009;14:219–228. doi: 10.1016/j.tplants.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Walker C.J., Willows R.D. Mechanism and regulation of Mg-chelatase. Biochem. J. 1997;327(Pt 2):321–333. doi: 10.1042/bj3270321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Richter A.S., Kleeberg J.R.W., Geimer S., Grimm B. Post-translational coordination of chlorophyll biosynthesis and breakdown by BCMs maintains chlorophyll homeostasis during leaf development. Nat. Commun. 2020;11:1254. doi: 10.1038/s41467-020-14992-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang C., Liu C., Jing Y., Wang Y., Jin L., Yang L., Fu A., Shi J., Zhao F. Inner envelope CHLOROPLAST MANGANESE TRANSPORTER 1 supports manganese homeostasis and phototrophic growth in Arabidopsis. Mol. Plant. 2018;11:943–954. doi: 10.1016/j.molp.2018.04.007. [DOI] [PubMed] [Google Scholar]
- Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.