Abstract
Pluripotent stem cells localized in proliferating growth centers, the meristems, are the origin of life-long organ formation and growth in higher plants. In the shoot apical meristem of Arabidopsis thaliana, the closely related ARGONAUTE proteins AGO1 and ZLL/AGO10 bind miR165/166 species to regulate mRNAs of HD-ZIP III transcription factors that are essential to maintaining stem cells. Several genetic studies showed that AGO1 and ZLL/AGO10 act redundantly to maintain stem cells. By contrast, the reported biochemical data suggested antagonistic functions: AGO1 utilizes miR165/166 to slice HD-ZIP III mRNAs, whereas ZLL/AGO10 promotes degradation of miR165/166 and thus stabilizes HD-ZIP III mRNAs. How these different functions are balanced in stem cell regulation has remained enigmatic. Here, we show that autorepression of AGO1 through miR168-mediated slicing of its own RNA is required to maintain the ability of AGO1 to suppress HD-ZIP III mRNAs. Increased AGO1 expression, either in the miR168a-2 mutant or by transgenic expression, inhibits this ability despite the presence of high levels of miR165/166, effectively uncoupling HD-ZIP III and miR165/166 expression. AGO1 activity can be restored, however, by increasing the levels of chaperones SQN and HSP90, which promote assembly of RNA-induced silencing complex (RISC). This suggests that cellular abundance of SQN and HSP chaperones limits AGO1-mediated RNA interference in shoot meristem stem cell regulation. Localized misexpression of AGO1 indicates that the cells surrounding the shoot meristem primordium play a crucial role in stem cell development. Taken together, our study provides a framework that reconciles biochemical and genetic data, showing that restriction of AGO1 levels by miR168-mediated autorepression is key to RISC homeostasis and the function of AGO1 in stem cell regulation.
Key words: Arabidopsis, ARGONAUTE1, ZWILLE (ARGONAUTE10), HD-ZIP III, stem cell, shoot meristem
The restriction of AGO1 levels through miR168-mediated autorepression is essential for RISC homeostasis and AGO1 function in stem cell regulation.
Introduction
The stem cells in the shoot meristem of higher plants give rise to all above-ground organs from germination to maturity. In the model plant Arabidopsis thaliana, the maintenance of the shoot meristem stem cells is regulated by several pathways (Aichinger et al., 2012). Among them, class III homeodomain-leucine zipper (HD-ZIP III) transcription factors promote shoot meristem development. Their activity is negatively regulated by the microRNA (miRNA) species miR165 and miR166 that are used to cleave HD-ZIP III mRNAs (Prigge et al., 2005, Byrne, 2006) by the AGO1 protein, the main slicing activity in plants. AGO1 deficiency leads to upregulation of its miRNA targets, including HD-ZIP III mRNAs, and consequently to pleiotropic developmental phenotypes (Bohmert et al., 1998, Vaucheret et al., 2004). AGO1 also binds miR168 to slice its own mRNA, ensuring correct AGO1 expression levels. Disruption of this negative feedback regulation causing increased AGO1 levels results in pleiotropic phenotypical defects, including abortion of the shoot meristem in severe cases (Vaucheret et al., 2004). Two chaperones, the cyclophilin 40 SQUINT (SQN) and the heat shock protein 90 (HSP90) are required for assembly of an AGO1-containing RNA-induced silencing complex (RISC), the functional unit of RNA interference (Smith et al., 2009, Iki et al., 2012). Mutations in SQN and redundant genes trigger overproliferation of the shoot and floral meristem (Prunet et al., 2008) and suppress shoot meristem termination by CLV3 overexpression (Prunet et al., 2015), suggesting a loss of the stem cell restriction. ZWILLE (ZLL)/AGO10 is the closet paralog of AGO1 and is specifically required for maintaining stem cells of the shoot meristem during embryogenesis, for the initiation of axillary shoot meristems, and for floral meristem regulation (Moussian et al., 1998, Lynn et al., 1999, Tucker et al., 2008).
Genetic and biochemical analysis revealed contradictory results of how AGO1 and ZLL/AGO10 affect shoot meristem maintenance. Several genetic studies suggest that both genes act similarly in stem cell regulation: Double-mutant analysis demonstrated redundant functions of AGO1 and ZLL/AGO10 in the development of bilateral symmetry and the shoot meristem during embryogenesis, including the expression of the stem cell regulator SHOOTMERISTEMLESS (Lynn et al., 1999). In floral meristems, AGO1 and AGO10 redundantly promote stem cell termination (Ji et al., 2011). Finally, several studies showed that AGO1 can complement ZLL/AGO10 function in shoot meristem stem cell maintenance (Mallory et al., 2009, Tucker et al., 2008). In contrast to these genetic data, molecular data suggest opposite functions of AGO1 and ZLL/AGO10: Misexpression of ZLL/AGO10 increases the levels of HDZIP-III mRNAs (Supplemental Figure 1) that are degraded by AGO1 (Liu et al., 2009). Loss-of-function ago1 mutants have opposite effects on miR165/166 and HD-ZIPIII mRNA levels compared with zll/ago1 mutants in flower development (Ji et al., 2011). A detailed biochemical study revealed that ZLL/AGO10 can act as a decoy AGO protein that preferentially binds miR165/166. Unlike AGO1, however, ZLL/AGO10 shows only a weak ability to cleave HD-ZIP III mRNAs in vitro, and in planta this activity seems to be largely inefficient (Ji et al., 2011, Zhu et al., 2011); rather, the bound miRNA is degraded (Yu et al., 2017). As a consequence, ZLL/AGO10 reduces miR165/166 levels and protects HD-ZIP III mRNAs against AGO1-mediated slicing, which in turn promotes stem cell maintenance (Liu et al., 2009).
In this study, we address these different models and show that AGO1 affects stem cell maintenance in different ways depending on its expression levels. We also show that the balanced abundances of AGO1 and the RISC assembly factors SQN and HSP90 are crucial for stem cell maintenance. We reveal a critical role of miR168-mediated autorepression of AGO1 levels to prevent dominant-negative inhibition of AGO1 activity.
Results
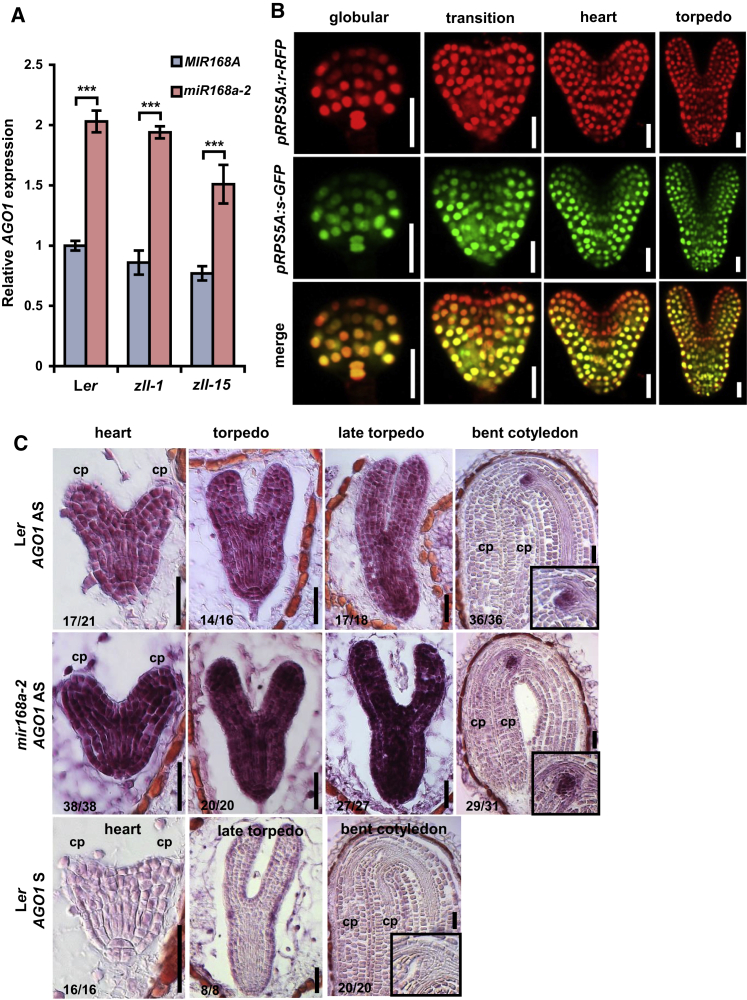
Loss of miR168 Function Restores Shoot Meristem Formation in the zll-1 Knockout Mutant
To address the controversial models of ZLL/AGO10 and AGO1 function in the context of stem cell regulation, we analyzed stem cell maintenance during embryo maturation. We first addressed the consequences of increasing AGO1 level in zll mutants. AGO1 autoregulates its own expression levels in a negative feedback loop by slicing its own mRNA through miR168. We therefore analyzed the effect of the mir168a-2 mutation that causes a severe reduction of miR168 and a two-fold increase in AGO1 mRNA level in wild-type and zll-1 and zll-15 mutants (Vaucheret, 2009) (Figure 1A). Approximately 75% of zll-1 seedlings display a terminated shoot meristem with differentiated organs or structures in place of the stem cells (Figure 2A, 2C, and 2G) (Moussian et al., 1998, Tucker et al., 2008). Introgression of mir168a-2 into zll-1 or the weaker allele zll-15 strongly restored shoot meristem formation (Figure 2D and 2G), while it had no recognizable effect on the shoot meristem in a wild-type background (Figure 2B). As a control, expression of a genomic MIR168A fragment abolished this effect (24 out of 34 transgenic T1 plants; Figure 2E and 2F), confirming that the mutation in the MIR168A gene caused the phenotypes. Similar to the miR168a-2 mutant, reduction of miR168 function through expression of MIMICRY FOR MIR168 (MIM168) (Varallyay et al., 2010) (Supplemental Figure 2A) also caused recovery of shoot meristem formation in zll-1 mutants, albeit to a smaller extent (Supplemental Figure 2B). In an alternative approach, introduction of an additional 8-kb genomic AGO1 fragment (Vaucheret et al., 2004) into zll-1 also strongly recovered shoot meristem formation (Supplemental Table 1). One possible explanation for the suppression of stem cell termination by increasing AGO1 dose could be that AGO1 expression is reduced in the zll-1 mutant. However, qRT–PCR shows no significant difference of AGO1 transcript levels between Ler and zll-1 plants (Figure 1A), arguing against such a mechanism. We did not observe any phenotype suggestive of AGO1 co-suppression in any experiment, such as terminated seedlings as observed by Mallory et al. (2009) when using a different transgene (pZLL:AGO1).
Figure 1.
miR168 Regulates AGO1 Expression in a Dynamic Manner during Embryogenesis.
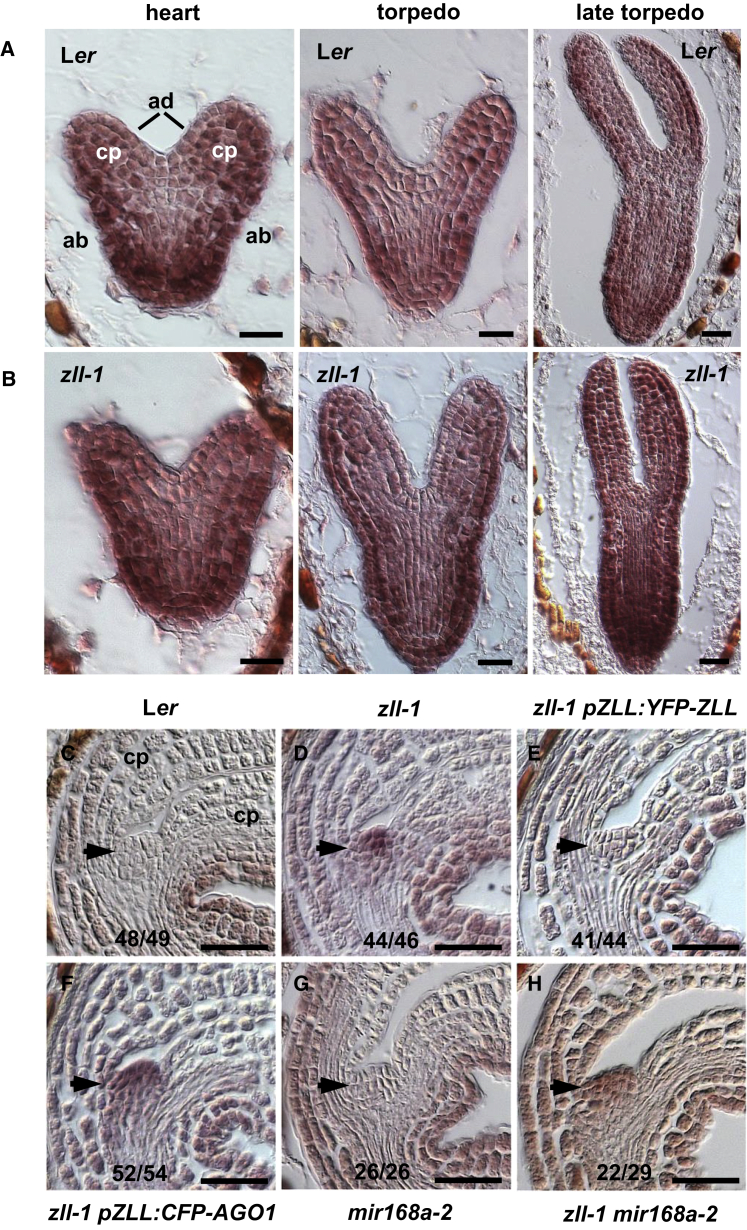
(A) Levels of AGO1 transcript in seedling shoots of the indicated genotypes with or without mir168a-2 mutation. Date represent means ± SD from three biological replicates. ***p < 0.001 by two-sided, non-paired t-test. AGO1 expression levels in Ler, zll-1 and zll-15 carrying the MIR168 wild-type allele are not significantly different.
(B) A tandem reporter co-expressing a miR168-resistant RFP (r-RFP) and a miR168-sensitive GFP (s-GFP) driven by the RPS5A promoter in embryos at the indicated stages. The lower panel shows merged channels. The same pattern was found in 13 out of 14 independent transgenic lines of the tandem reporter.
(C) mRNA pattern of the endogenous AGO1 gene detected by in situ hybridization with antisense AGO1 probe 1 (Supplemental Figure 4). Genotypes, frequencies of observed patterns, and embryo stages are indicated. Insets show higher magnification of the embryonic shoot apical meristems. AS, antisense; S, sense; cp, cotyledon primordium.
Scale bars: 20 μm.
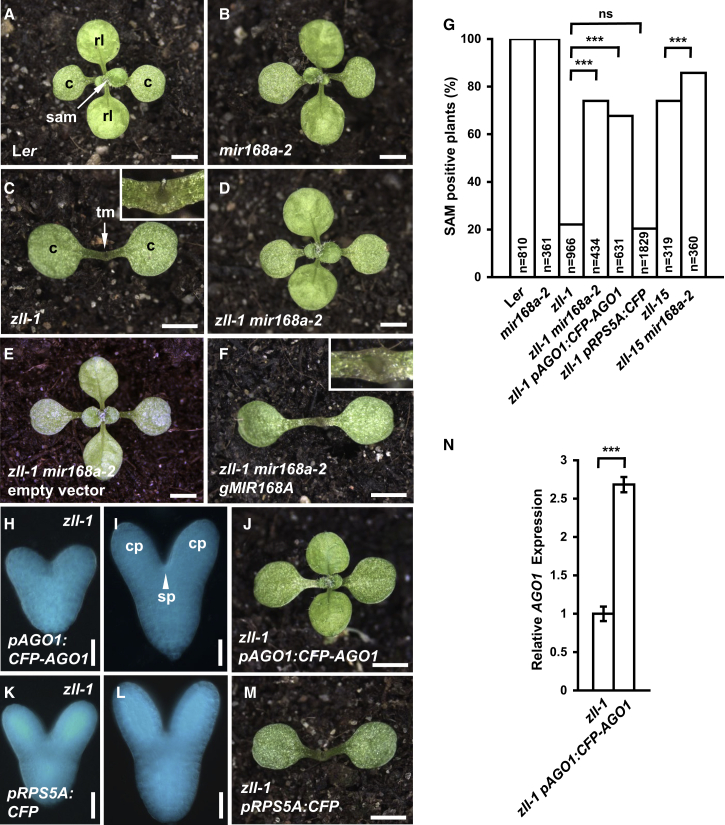
Figure 2.
Increased AGO1 Expression Recovers Shoot Meristem Formation in zll-1.
(A–F) Phenotypes of 14-day-old seedlings of indicated genotypes. The functional shoot apical meristems (sam) in (A), (B), and (D) produce rosette leaves (rl), while the terminated meristem (tm) in (C) only produces a filamentous structure (inset). (E and F)zll-1 mir168a-2 double mutants transformed with empty vector (E) or genomic MIR168A(F).
(G) Frequency of 14-day-old seedlings containing a functional shoot meristem (SAM) for the indicated genotypes. n, number of seedlings. For the transgenic seedlings, data from three independent transgenic lines were pooled. ***p < 0.0001 by two-sided, non-paired Fisher’s exact tests and Bonferroni correction. ns, not significant.
(H and I) Expression patterns of a functional CFP-AGO1 fusion protein driven by the AGO1 promoter in heart (H) and torpedo (I) stage zll-1 embryos. cp, cotyledon primordium; sp, shoot apical meristem primordium.
(J) Phenotype of a 14-day-old transgenic zll-1 seedling expressing pAGO1:CFP-AGO1.
(K–L) Expression patterns of a CFP fluorescent protein driven by the RPS5A promoter in heart (K) and torpedo (L) stage zll-1 embryos. The exposure time of microscopy is one-tenth of that used for pAGO1:CFP-AGO1 embryos.
(M) Phenotype of a 14-day-old transgenic zll-1 seedling expressing pRPS5A:CFP.
(N)AGO1 transcript levels in 14-day-old seedlings of the indicated genotypes.
Data represent means ± SD from three biological replicates. ***p < 0.001 by two-sided, non-paired t-test. Scale bars, 2.5 mm (A–F, J, M) and 20 μm (H, I, K, L).
To elucidate where and when during embryo development AGO1 mRNA is regulated by miR168, we first constructed a fluorescent miR168 sensor. This sensor combines nuclear localized GFP fused to an oligonucleotide carrying the miR168 binding site of AGO1 as the miR168-sensitive reporter and nuclear localized RFP with a mutated version of miR168 binding site as transcriptional control (Supplemental Figure 3). Both GFP and RFP reporters are driven by the RPS5A promoter and arranged on one single T-DNA. The ratio of RFP/GFP, as a direct readout for miR168 activity, indicates that miR168 is active in the apical half of the globular embryo, but subsequently becomes restricted to the shoot apex and the adaxial sides of the cotyledonary primordia (Figure 1B).
To analyze the effect of miR168 on the spatiotemporal AGO1 mRNA pattern, we performed in situ hybridization (Figure 1C and Supplemental Figure 4). In the wild type, AGO1 mRNA is detected throughout the embryo with a slightly higher expression in the vascular and shoot meristem primordia until the late torpedo stage. Thereafter, AGO1 mRNA becomes more pronounced in the shoot apex. In mir168a-2 embryos, AGO1 mRNA signal is stronger than in the wild type throughout the embryo at all stages. Thus, miR168 downregulates AGO1 mRNA in a domain of the embryo that encompasses the developing shoot meristem. Together, release of AGO1 levels from miR168-mediated autorepression complements stem cell defects of the zll-1 mutant, suggesting that AGO1 and ZLL/AGO10 can act synergistically in stem cell maintenance.
Region-Specific AGO1 Expression Restores Shoot Meristem Formation in zll-1
We next determined in which region of the zll-1 embryo upregulation of AGO1 levels restores shoot meristem formation. To this end, we first expressed the CFP-AGO1 fusion protein from the pAGO1 promoter, which mimics the endogenous AGO1 mRNA pattern (Mallory et al., 2009) (Figures 2H, 2I, and 1C) and completely complements the ago1-1 mutant (Supplemental Figure 5), indicating that the fusion protein is functional. Confirming previous findings (Tucker et al., 2008, Mallory et al., 2009), expression of pAGO1:CFP-AGO1 in zll-1 resulted in efficient shoot meristem recovery (Figure 2J and 2G), similar to introducing additional genomic AGO1 copies (Supplemental Table 1), ruling out that the CFP-AGO1 chimeric construct behaves significantly different from the native AGO1. As expected, the AGO1 mRNA level is significantly increased after expressing pAGO1:CFP-AGO1 in zll-1 seedlings, excluding a potential co-suppression effect by the AGO1 transgene (Figure 2N). As a control, overexpression of a CFP protein driven by RPS5A promoter had no effect on shoot meristem recovery in zll-1 background (Figure 2K–2M and 2G), excluding an unexpected effect of the CFP protein.
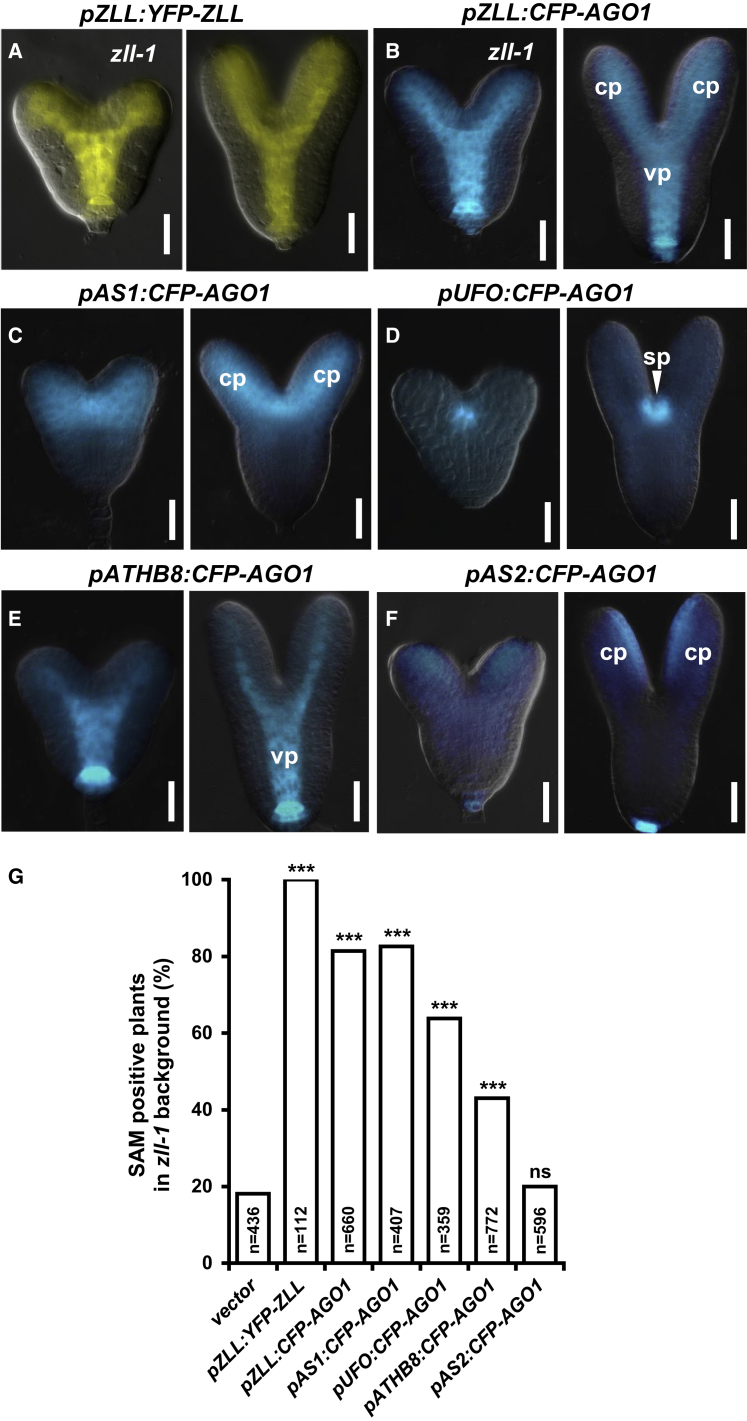
Expression of CFP-AGO1 from the pZLL promoter that is active in the vasculature, the shoot meristem, and the adaxial region of the cotyledons (Tucker et al., 2008) restored stem cells in zll-1 with a slightly reduced efficiency compared with pZLL:YFP-ZLL/AGO10 (Figure 3A, 3B, and 3G). Expression of CFP-AGO1 specifically in the apical embryo domain (pAS1 promoter; Figure 3C) restores stem cells in zll-1 with a similar efficiency (Figure 3G). Expressing CFP-AGO1 in cells surrounding the shoot apical meristem primordium (pUFO; Figure 3D) or in the provasculature (pATHB8; Figure 3E) partially suppresses stem cell termination in zll-1 (Figure 3G), whereas AGO1 expression in the adaxial regions of the cotyledons (pAS2; Figure 3F) had no effect (Figure 3G). Thus, transgenic expression of AGO1 around the shoot apical meristem primordium or in the vasculature is sufficient for shoot meristem stem cell recovery in zll-1.
Figure 3.
Domain-Specific Rescue of the zll-1 Phenotype by AGO1 Expression.
(A–F) Expression patterns of YFP-ZLL and CFP-AGO1 driven by domain-specific promoters as indicated above each panel. For each transgene, a heart-stage embryo (left) and a torpedo-stage embryo (right) are shown. vp, vascular primordium. Scale bars, 20 μm.
(G) Frequencies of 14-day-old zll-1 seedlings with a shoot meristem (SAM) by expressing transgenes as indicated. The numbers were generated by combining data from three independent transgenic lines. ***p < 0.00002 by two-sided, non-paired Fisher’s exact tests and Bonferroni correction. ns, not significant.
Increased AGO1 Expression Restores Stem Cells in zll Mutants by Uncoupling HD-ZIP III Levels from miR165/166 Accumulation
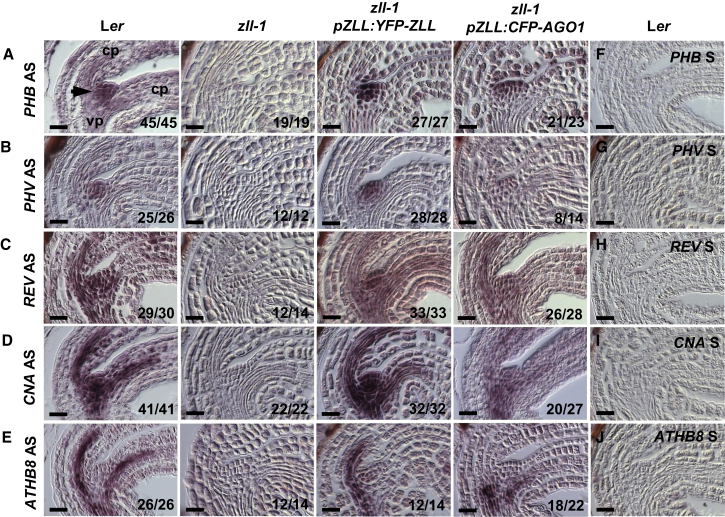
Based on findings that AGO1 and ZLL/AGO10 both can promote stem cell formation despite their opposite biochemical functions, we considered two hypotheses. The first hypothesis holds that because mR165/166 can move from cell to cell (Carlsbecker et al., 2010, Miyashima et al., 2013), increased levels of AGO1 in cells surrounding the shoot meristem primordium might create a sink for miR165/166, thereby draining it from the shoot meristem and consequently increasing HD-ZIP III transcripts. To test this hypothesis, we first compared the levels of HD-ZIP III mRNAs between zll-1 mutants expressing pZLL:YFP-ZLL or pZLL:CFP-AGO1 and the untransformed mutant. The Arabidopsis genome encodes five HD-ZIP III transcription factors, namely PHABULOSA (PHB), PHAVOLUTA (PHV), REVOLUTA (REV), CORONA (CNA), and ARABIDOPSIS THALIANA HOMEOBOX8 (ATHB8). In bent-cotyledon-stage wild-type embryos, the transcripts of PHB, REV, and CNA each are detected in the shoot apical meristem primordia, the adaxial domain of cotyledon primordia, and the provasculature, whereas PHV and ATHB8 transcripts are restricted to the shoot apical meristem primordium and the provasculature, respectively (Figure 4A–4E, first column).
Figure 4.
Increased AGO1 Levels Restore HD-ZIP III mRNA Levels in zll-1 Shoot Apices.
(A–E)In situ hybridization analysis of transcripts of all five HD-ZIP III genes in bent-cotyledon-stage embryos of indicated genotypes using the antisense probes of PHB(A), PHV(B), REV(C), CNA(D), and ATHB8(E).
(F–J) Hybridization using the sense probes of PHB(F), PHV(G), REV(H), CNA(I), and ATHB8(J) in Ler bent-cotyledon-stage embryos.
The position of the embryonic shoot apical meristem is marked by an arrow. The ratio of embryos showing the corresponding pattern of HD-ZIP III transcript is indicated. cp, cotyledon primordium; vp, vascular primordium. Scale bars, 10 μm.
In zll-1 embryos, the expression of all five HD-ZIP III genes in all tissues is strongly reduced (Figure 4A–4E, second column), supporting previous observations (Liu et al., 2009). As expected, expression of pZLL:YFP-ZLL/AGO10 fully restored the expression of all five HD-ZIP III genes in zll-1 (Figure 4A–4E, third column). Likewise, expression of pZLL:CFP-AGO1 also restored the expression of all HD-ZIP III transcripts (Figure 4A–4E, fourth column), albeit to a weaker degree than pZLL:YFP-ZLL/AGO10.
We then asked whether this restoration of HD-ZIP III mRNA levels was due to correspondingly decreased miR165/166 concentrations. In heart- to late-torpedo-stage embryos, miR165/166 accumulation is detected by in situ hybridization with the strongest signal in the abaxial domains of cotyledon primordia and the embryo axis in both wild-type and zll-1 embryos (Figure 5A and 5B). At the bent-cotyledon stage, miR165/166 signal is strongly enhanced in the shoot apical meristem region of zll-1 and to a lesser extent in surrounding tissues, whereas in the wild type, miR165/166 is undetectable in the shoot meristem (Figure 5C and 5D). Reducing the activity of miR165/166 by expressing the miRNA mimicry MIM165/166 in the shoot apex resulted in restoration of a functional shoot meristem (Supplemental Figure 6), confirming that miR165/166 accumulation in the shoot apex is essential for stem cell loss in the zll-1 mutant (Liu et al., 2009). As expected, complementation of the zll-1 mutant by pZLL:YFP-ZLL/AGO10 eliminates miR165/166 accumulation back to the wild-type pattern (Figure 5E). Surprisingly, however, when restoring HD-ZIP III expression and stem cells by expressing pZLL:CFP-AGO1 (Figure 5F) or introducing the miR168a-2 mutation (Figure 5H), miR165/166 accumulation is not reduced.
Figure 5.
Increased AGO1 Levels Do Not Alter miR165/166 Accumulation in the zll-1 Shoot Apical Meristem.
(A and B)In situ hybridization using an antisense miR166 LNA probe in Ler(A) and zll-1(B) embryos at heart, torpedo, and late torpedo stages. Representative patterns for each stage of Ler and zll-1 embryos are shown (n > 20 embryos per stage were examined).
(C–H) miR165/166 accumulation patterns in bent-cotyledon stage embryos of six indicated genotypes. The position of the embryonic shoot apical meristem is marked by an arrow. The ratio of embryos showing the corresponding miR165/166 pattern is indicated.
cp, cotyledon primordium; ad, adaxial region; ab, abaxial region. Scale bars, 20 μm.
In summary, upregulation of AGO1 in the zll-1 mutant increases HD-ZIP III expression in the shoot meristem region but, unlike ZLL/AGO10, does not simultaneously reduce miR165/166 accumulation. This refutes the first hypothesis and suggests that raising AGO1 levels in zll-1 uncouples HD-ZIP III mRNA and miR165/166 levels.
Expression of RISC Chaperones Suppresses the Effects of AGO1 Overexpression
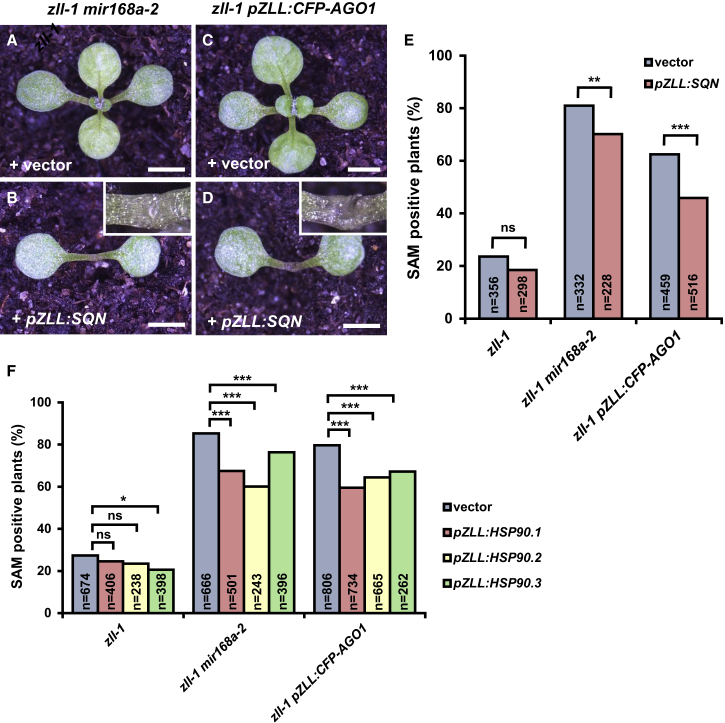
Our second hypothesis holds that increased AGO1 protein levels might sequester co-factors required to assemble a functional RISC and thus cleave HD-ZIP III mRNAs. To address this hypothesis, we compared the effects of expressing SQUINT (SQN) and the Heat Shock Protein 90 (HSP90), which are required for AGO1 function and RISC assembly (Smith et al., 2009, Iki et al., 2012), between zll-1 miR168a-2 or zll-1 pZLL:CFP-AGO1 and the untransformed zll-1 mutant. When expressed from the pZLL promoter, SQN is able to partially suppress the recovery of stem cells in both zll-1 mir168a-2 and zll-1 pZLL:CFP-AGO1, whereas it has no significant effects in the zll-1 control (Figure 6A–6E). To test whether this suppression of AGO1-mediated stem cell recovery is accompanied by downregulation of HD-ZIP III mRNAs, we analyzed the expression levels by qRT–PCR from dissected seedling shoot apices. We found that pZLL:SQN causes downregulation of PHV and REV mRNA levels in zll-1mir168a-2 and of PHB, PHV, and REV mRNA levels in zll-1 pZLL:CFP-AGO1 (Supplemental Figure 7).
Figure 6.
Expression of AGO1 Co-factors SQN and HSP90 Partially Suppresses the Recovery of Shoot Apical Meristem in zll-1.
(A–D) Phenotypes of 14-day-old seedlings of the indicated genotypes expressing either empty vector or pZLL:SQN as indicated. Insets show the magnification of terminated shoot apices.
(E) Shoot apical meristem phenotypes of 14-day-old seedlings of the indicated genotypes in homozygous T3 generations. The numbers from at least two independent transgenic lines were combined. **p < 0.01, ***p < 0.001 by two-sided, non-paired Fisher’s exact tests. ns, not significant.
(F) Shoot apical meristem phenotypes of 14-day-old seedlings of the indicated genotypes in homozygous T3 generations. For the transgenic seedlings, individual counts from two or three independent transgenic lines not significantly different by chi-square test were pooled. *p < 0.0017, ***p < 0.0003 by two-sided, non-paired Fisher’s exact tests and Bonferroni correction. ns, not significant.
Similar to SQN, expression of the HSP90 isoforms HSP90.1–3 (Earley and Poethig, 2011) significantly suppresses the recovery of stem cells in zll-1 mir168a-2 and zll-1 pZLL:CFP-AGO1 (Figure 6F). We found that in these cases, expression of pZLL:HSP90.1 causes downregulation of PHB and PHV mRNA levels in zll-1 mir168a-2, and of PHB in zll-1 pZLL:CFP-AGO1 (Supplemental Figure 8). Thus, elevated HD-ZIP III gene expression and, consequently, stem cell recovery by increased AGO1 levels are suppressed by upregulated levels of SQN, HSP90.1, and HSP90.2.
Discussion
AGO1 and ZLL/AGO10 are central regulators of shoot meristem stem cell maintenance, to a large part through post-transcriptional regulation of HD-ZIP III transcription factors. However, contradictory to their opposite biochemical functions (Zhu et al., 2011, Yu et al., 2017), genetic evidence in several studies suggested that AGO1 and ZLL/AGO10 have overlapping functions in stem cell regulation (Lynn et al., 1999, Tucker et al., 2008, Mallory et al., 2009, Ji et al., 2011). Addressing this discrepancy, we confirmed that increased expression of AGO1 complements the loss of ZLL/AGO10 function, but we find that it does so by protecting HD-ZIP III mRNA via a mechanism different from that of ZLL/AGO10. In the latter case, ZLL/AGO10 promotes degradation of miR165/166 in the shoot meristem and thus protects their targets, HD-ZIP III mRNAs, from AGO1-mediated slicing (Zhu et al., 2011, Yu et al., 2017). By contrast, increased AGO1 expression in zll-1 causes increased HD-ZIP III mRNAs levels without changing the high abundance of miR165/166 in the shoot meristem primordium, thus uncoupling HD-ZIP III mRNA and miR165/166 levels. This suggests that the ability of AGO1 and miR165/166 to catalyze degradation of HD-ZIP III mRNAs is compromised rather than increased, as one might expect, if AGO1 levels increase.
These findings imply that to maintain AGO1 activity in stem cell regulation, its level must be carefully kept in check. We show that mir168a mutation is sufficient to induce dominant-negative effects of increased AGO1 expression, indicating that the role of AGO1 autorepression via miR168 is not only to restrict AGO1 activity but to prohibit increased AGO1 dose that would lead to inactivation. This interpretation is consistent with previous findings that upregulation of AGO1 in a wild-type background increased the levels of several miRNA targets, rather than more efficiently degrading them (Vaucheret et al., 2004, Vaucheret et al., 2006, Deveson et al., 2013).
Why does upregulation of AGO1 levels result in the stabilization of its target HD-ZIP III mRNAs, rather than in their degradation, as one would expect from a Slicer protein? AGO proteins function as part of RISCs (Meister, 2013). We find that upregulation of SQN and HSP90 proteins suppresses AGO1-mediated HD-ZIP III mRNA stabilization and stem cell recovery in zll-1. Previous studies revealed that HSP90 promotes RISC assembly in vitro through a chaperone cycle involving ATP binding and hydrolysis (Iki et al., 2010), and that SQN promotes the activity of AGO1 in vivo and HSP90-mediated RISC assembly in vitro (Smith et al., 2009, Iki et al., 2012).
Therefore, one model to explain how upregulation of AGO1 might stabilize HD-ZIP III mRNAs is that an even moderate excess of AGO1 disturbs the assembly of active RISCs because of limited cellular abundances of SQN and HSP90 co-factors, plausibly by sequestering limited RISC components. In this view, upregulation of SQN and HSP90 levels overcomes this shortage and restores assembly of active AGO1-containing RISC and, consequently, HD-ZIP III degradation and stem cell termination.
The rate-limiting abundance of HSP90 might appear surprising, since HSP90 can be considered a housekeeping gene. However, previous studies showed that HSP90.1 and HSP90.3 in non-stress conditions are mainly expressed in the cotyledons and the root tip of maturing embryos rather than in the shoot meristem (Prasinos et al., 2005), and plants with reduced levels of HSP90 resemble ago1 mutants (Sangster et al., 2007).
We note that increased dosage of SQN and HSP90 only partially suppresses shoot meristem recovery in zll-1 pZLL:CFP-AGO1 or zll-1 mir168a-2, implying that other mechanisms might also contribute. Such mechanisms might involve other miRNAs and/or the phytohormone auxin that were found to act together with ZLL/AGO10 in shoot meristem formation (Ji et al., 2011, Knauer et al., 2013, Roodbarkelari et al., 2015).
A rate-limiting model of SQN and HSP90 for AGO1 activity in stem cell maintenance can also explain differences in ZLL/AGO10 and AGO1 requirements in different Arabidopsis accessions. In the Ler accession that has higher SQN expression levels and thus a higher potential for AGO1-mediated slicing of HD-ZIP III mRNAs, the decoy function of ZLL/AGO10 is crucial for shoot meristem stem cell maintenance during embryogenesis. By contrast, in the Columbia accession that has lower SQN expression, ZLL/AGO10 is dispensable for this process (Tucker et al., 2013).
Transgenic ZLL/AGO10 and AGO1 promote stem cell maintenance not only when expressed in the shoot meristem but also from the underlying vasculature (Tucker et al., 2008). Because miR165/166 has been found to move between embryo cells (Miyashima et al., 2013), the non-cell-autonomous manner of ZLL/AGO10 function can conceivably be explained by its sequestration of miR165/166. In this model, ZLL/AGO10 expression in the vasculature would create a sink for miR165/166, consequently draining it from the shoot meristem primordium (Zhu et al., 2011). Because AGO1 binds to SQN and HSP90 proteins, the non-cell-autonomous function of increased AGO1 levels in the surrounding cells observed here, in analogy, could be explained by creating a sink for SQN or HSP90 co-factors, thus preventing assembly of a functional RISC in the stem cells and consequently enabling HD-ZIPIII function and stem cell survival.
Methods
Plant Materials
The mutant alleles of zll-1 (Ler), zll-15 (Ler), mir168a-2 (Ler), and ago1-1 (Col) have been previously described (Bohmert et al., 1998, Moussian et al., 1998, Tucker et al., 2008, Vaucheret, 2009). The pZLL:YFP-ZLL and pAGO1:ZLL transgenic lines were previously reported (Tucker et al., 2008, Mallory et al., 2009). All plants were grown under 16-h/8-h photoperiod conditions at 19°C. Medium cultivated seedlings were grown in half-strength Murashige and Skoog medium containing vitamins supplemented with 0.5 g/l 2-(N-morpholino)ethanesulfonic acid and 8 g/l agar. The genotyping protocol and primers are listed in Supplemental Tables 2 and 3.
Plasmid Construction
The plasmid carrying the native endogenous AGO1 gene in the form of an 8-kb genomic fragment has already been described (Vaucheret et al., 2004). To generate different Promoter:CFP-AGO1 constructs, we amplified the fragment containing CFP fused to the full-length genomic sequence of AGO1 gene from the original pZLL:CFP-AGO1 construct (A53) (Mallory et al., 2009) by oFD653/oFD654 and cloned it into pBarMAP via AscI/PacI. The promoters of AGO1, ZLL, AS1, UFO, ATHB8, and AS2 were then inserted upstream of the CFP-AGO1 module via AscI. To generate the pRPS5A:CFP construct, we amplified the CFP coding sequence with NOS terminator from ALH007 by oFD653/oFD717 and cloned it into pBarMAP vector via AscI/BamHI. The RPS5A promoter was then inserted upstream of the CFP-NOSt module via AscI.
To generate miR168a-2 complementation constructs, we amplified the genomic fragment of MIR168A gene (−2281 to +1369 bp) from Ler genomic DNA by oFD894/oFD930 and cloned it into the pGreenII vector via AscI/XmaI.
To generate MIM168 constructs, we amplified the MIM168 fragment from 35S:MIM168 (Varallyay et al., 2010) by oFD971/oFD972 and cloned it between pRPS5A and NOSt in the pGreenII vector.
To generate the tandem miR168 sensor, we amplified the NLS-miR168 site-1×GFP-NOSt fragment from STK157 by oFD899/oFD900 and cloned it into pJET1.2 (Thermo Fisher Scientific). The second GFP coding sequence was introduced from MU594 via BsrGI to make NLS-miR168 site-2×GFP-NOSt, which was cloned into the pGreenII vector via SalI/XmaI. The RPS5A promoter was then inserted upstream of the NLS-miR168 site-2×GFP-NOSt module via SalI to make pRPS5A:NLS-miR168 site-2×GFP-NOSt (named pFD205). In parallel, the NLS- miR168-4m site-1×RFP was amplified from STK285 by oFD916/oFD915 and cloned into pJET 1.2. The second RFP coding sequence was introduced from STK286 via BamHI to make NLS-miR168-4m site-2×RFP, which was cloned into pFD205 via KpnI/XhoI. Lastly, pRPS5A and NOSt were introduced upstream and downstream of NLS-miR168-4m site-2×RFP via KpnI and XhoI, respectively.
To generate the pZLL:SQN construct, we amplified the full-length coding sequence (CDS) of SQN from Ler cDNA by oFD1134/oFD1135 and cloned it into pGreenII vector via AscI/NotI. pZLL and NOSt were introduced upstream and downstream of the SQN CDS via AscI and NotI, respectively. To generate the pZLL:HSP90.2 and pZLL:HSP90.3 constructs, we amplified the full-length CDS of HSP90.2 and HSP90.3 from Ler cDNA by oFD1136/oFD1137 and oFD1138/oFD1139 and cloned them into the pGreenII vector via AscI/NotI. pZLL and NOSt were introduced upstream and downstream of each CDS via AscI and NotI, respectively. To generate the pZLL:HSP90.1 construct, we amplified full-length genomic HSP90.1 (from ATG to TAA) from Ler cDNA by oFD1144/oFD1145 and cloned it into the pGreenII vector via AscI/NotI. pZLL and NOSt were introduced upstream and downstream of gHSP90.1 via AscI and NotI, respectively.
To generate the pUFO:MIM165/166 line, we used the LhG4-pOp two-component expression system (Moore et al., 1998). The UFO promoter (cloned in STK195) was introduced into the pGreenII vector STK164b via AscI. pOp:MIM165/166 (SB229) was introduced into pOp:NLS-3×YFP (SB225) via AscI to generate SB233 in the binary vector pBarMap. MIM165/166 sequence was obtained according to Todesco et al. (2010) and was introduced into SB229 by LIC cloning.
Primers and constructs used for cloning are listed in Supplemental Tables 4 and 5.
RNA In Situ Hybridization
For the in situ hybridization experiments, we collected siliques with embryos at different stages from each genotype. To prepare probes, we amplified fragments of protein-coding genes by PCR from Ler cDNA and cloned them into pGEM-T Easy vectors (Promega, catalog no. A1360) for in vitro transcription using the Digoxigenin Labeling Kit (Roche, catalog no. 11277073910). Antisense and sense probes were synthesized using SP6 (Promega, catalog no. P1085) or T7 RNA polymerase (Promega, catalog no. P2075). The long probes were then hydrolyzed to an average length of 150 bp with carbonate buffer (2× buffer: 80 mM NaHCO3, 120 mM Na2CO3) and resuspended in 50% formamide at the desired concentration. The miR166 locked nucleic acid (LNA) probe (zma-miR166a) was obtained from the EXIQON company with 5′ end labeling of digoxigenin: /5′Dig/GGG GAA TGA AGC CTG GTC CGA. Primers for amplifying hybridization probes are listed in Supplemental Table 6.
In situ hybridization experiments were performed as previously described (Zhang et al., 2017). For the miR166 LNA probe, the hybridization and washing were performed at 60°C and staining at 4°C, as previously described (Liu et al., 2009). For other probes, the hybridization and washing were performed at 55°C and staining at room temperature.
RT–PCR and qRT–PCR
Total RNA from aerial parts of randomly collected 14-day-old seedlings was extracted using the RNeasy Plant Mini Kit (Qiagen), and first strand cDNA was synthesized using SuperScript III First Strand Synthesis SuperMix (Invitrogen). For quantitative real-time PCR, LightCycler 480 SYBR Green I Master Mix (Roche) was used and the experiments were performed on the LightCycler 480 II qPCR machine (Roche). qRT–PCR results were analyzed with the LightCycler 480 Software release 1.5.0. Expression levels of target genes were normalized to the levels of ACTIN7 for RT–PCR and PP2AA3 (Czechowski et al., 2005) for qRT–PCR. Primers for RT–PCR and qRT–PCR are listed in Supplemental Table 7.
Image Acquisition
Pictures of seedlings were taken with a Leica DC300 camera on an Olympus SZ-CTV stereozoom microscope. A Zeiss Axioskop microscope was used to take Nomarski pictures of in situ hybridization. For analysis of the expression patterns of YFP-ZLL/AGO10 or CFP-AGO1 driven by different promoters in embryos, ovules were squeezed from siliques, mounted with 10% (v/v) glycerol, and imaged by a Zeiss Axio Imager microscope with AxioVision 4.4 software. For analysis of the patterns of miR168-sensitive GFP and miR168-insensitive RFP, the embryos were imaged by a LSM700 (Zeiss) laser scanning confocal microscope using ZEN 2010 software. All images of independent experiments were taken under the same settings of microscopy.
Funding
This work was funded by grants from the Baden-Württemberg Stiftung and by the German Research Foundation (DFG) under Germany's Excellence Strategy (CIBSS – EXC-2189 – Project ID390939984) and by grants (La606/14, La606/17; ERA-CAPS, to T.L.) and a stipend from the China Scholarship Council to F.D.
Author Contributions
F.D., H.V., and T.L. conceived the project and designed the experiments. F.D., W.G., S.B., M.T., and H.V. performed the experiments. F.D. and T.L. interpreted the data and wrote the manuscript with input from all authors.
Acknowledgments
We thank Zoltán Havelda (Agricultural Biotechnology Center, Hungary) for kindly providing the 35S:MIM168 construct. No conflict of interest declared.
Published: September 3, 2019
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Aichinger E., Kornet N., Friedrich T., Laux T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012;63:615–636. doi: 10.1146/annurev-arplant-042811-105555. [DOI] [PubMed] [Google Scholar]
- Bohmert K., Camus I., Bellini C., Bouchez D., Caboche M., Benning C. AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 1998;17:170–180. doi: 10.1093/emboj/17.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M.E. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS Genet. 2006;2:e89. doi: 10.1371/journal.pgen.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J.Y., Roberts C.J., Dettmer J., Lehesranta S., Zhou J., Lindgren O., Moreno-Risueno M.A., Vaten A., Thitamadee S. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveson I., Li J., Millar A.A. Expression of human ARGONAUTE 2 inhibits endogenous microRNA activity in Arabidopsis. Front. Plant Sci. 2013;4:96. doi: 10.3389/fpls.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Poethig R.S. Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 2011;286:38184–38189. doi: 10.1074/jbc.M111.290130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2012;31:267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Ji L., Liu X., Yan J., Wang W., Yumul R.E., Kim Y.J., Dinh T.T., Liu J., Cui X., Zheng B. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer S., Holt A.L., Rubio-Somoza I., Tucker E.J., Hinze A., Pisch M., Javelle M., Timmermans M.C., Tucker M.R., Laux T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell. 2013;24:125–132. doi: 10.1016/j.devcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Liu Q., Yao X., Pi L., Wang H., Cui X., Huang H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and establishment of leaf polarity by repressing miR165/166 in Arabidopsis. Plant J. 2009;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M.K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- Mallory A.C., Hinze A., Tucker M.R., Bouche N., Gasciolli V., Elmayan T., Lauressergues D., Jauvion V., Vaucheret H., Laux T. Redundant and specific roles of the ARGONAUTE proteins AGO1 and ZLL in development and small RNA-directed gene silencing. PLoS Genet. 2009;5:e1000646. doi: 10.1371/journal.pgen.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- Miyashima S., Honda M., Hashimoto K., Tatematsu K., Hashimoto T., Sato-Nara K., Okada K., Nakajima K. A comprehensive expression analysis of the Arabidopsis MICRORNA165/6 gene family during embryogenesis reveals a conserved role in meristem specification and a non-cell-autonomous function. Plant Cell Physiol. 2013;54:375–384. doi: 10.1093/pcp/pcs188. [DOI] [PubMed] [Google Scholar]
- Moore I., Galweiler L., Grosskopf D., Schell J., Palme K. A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl. Acad. Sci. U S A. 1998;95:376–381. doi: 10.1073/pnas.95.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussian B., Schoof H., Haecker A., Jurgens G., Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasinos C., Krampis K., Samakovli D., Hatzopoulos P. Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J. Exp. Bot. 2005;56:633–644. doi: 10.1093/jxb/eri035. [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Morel P., Thierry A.M., Eshed Y., Bowman J.L., Negrutiu I., Trehin C. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell. 2008;20:901–919. doi: 10.1105/tpc.107.053306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N., Morel P., Champelovier P., Thierry A.M., Negrutiu I., Jack T., Trehin C. SQUINT promotes stem cell homeostasis and floral meristem termination in Arabidopsis through APETALA2 and CLAVATA signalling. J. Exp. Bot. 2015;66:6905–6916. doi: 10.1093/jxb/erv394. [DOI] [PubMed] [Google Scholar]
- Roodbarkelari F., Du F., Truernit E., Laux T. ZLL/AGO10 maintains shoot meristem stem cells during Arabidopsis embryogenesis by down-regulating ARF2-mediated auxin response. BMC Biol. 2015;13:74. doi: 10.1186/s12915-015-0180-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster T.A., Bahrami A., Wilczek A., Watanabe E., Schellenberg K., McLellan C., Kelley A., Kong S.W., Queitsch C., Lindquist S. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS One. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.R., Willmann M.R., Wu G., Berardini T.Z., Moller B., Weijers D., Poethig R.S. Cyclophilin 40 is required for microRNA activity in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2009;106:5424–5429. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Rubio-Somoza I., Paz-Ares J., Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6:e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M.R., Hinze A., Tucker E.J., Takada S., Jurgens G., Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- Tucker M.R., Roodbarkelari F., Truernit E., Adamski N.M., Hinze A., Lohmuller B., Wurschum T., Laux T. Accession-specific modifiers act with ZWILLE/ARGONAUTE10 to maintain shoot meristem stem cells during embryogenesis in Arabidopsis. BMC Genomics. 2013;14:809. doi: 10.1186/1471-2164-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varallyay E., Valoczi A., Agyi A., Burgyan J., Havelda Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010;29:3507–3519. doi: 10.1038/emboj.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. AGO1 homeostasis involves differential production of 21-nt and 22-nt miR168 species by MIR168a and MIR168b. PLoS One. 2009;4:e6442. doi: 10.1371/journal.pone.0006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Mallory A.C., Bartel D.P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell. 2006;22:129–136. doi: 10.1016/j.molcel.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crete P., Bartel D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187–1197. doi: 10.1101/gad.1201404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Ji L., Le B.H., Zhai J., Chen J., Luscher E., Gao L., Liu C., Cao X., Mo B. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017;15:e2001272. doi: 10.1371/journal.pbio.2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tucker E., Hermann M., Laux T. A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell. 2017;40:264–277.e4. doi: 10.1016/j.devcel.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.