Abstract
Hybridization is a creative evolutionary force, increasing genomic diversity and facilitating adaptation and even speciation. Hybrids often face significant challenges to establishment, including reduced fertility that arises from genomic incompatibilities between their parents. Whole-genome duplication in hybrids (allopolyploidy) can restore fertility, cause immediate phenotypic changes, and generate reproductive isolation. Yet the survival of polyploid lineages is uncertain, and few studies have compared the performance of recently formed allopolyploids and their parents under field conditions. Here, we use natural and synthetically produced hybrid and polyploid monkeyflowers (Mimulus spp.) to study how polyploidy contributes to the fertility, reproductive isolation, phenotype, and performance of hybrids in the field. We find that polyploidization restores fertility and that allopolyploids are reproductively isolated from their parents. The phenotype of allopolyploids displays the classic gigas effect of whole-genome duplication, in which plants have larger organs and are slower to flower. Field experiments indicate that survival of synthetic hybrids before and after polyploidization is intermediate between that of the parents, whereas natural hybrids have higher survival than all other taxa. We conclude that hybridization and polyploidy can act as sources of genomic novelty, but adaptive evolution is key in mediating the establishment of young allopolyploid lineages.
Keywords: allopolyploid, Erythranthe, Mimulus, polyploidy, speciation, whole-genome duplication
Hybridization and whole-genome duplication can be creative evolutionary forces and can fuel adaptation and speciation. This study uses natural and synthetically produced hybrid and polyploid monkeyflowers (Mimulus spp.) to study how polyploidy contributes to the fertility, reproductive isolation, phenotype, and performance of hybrids in the field.
Introduction
“Hybrids are more paths to the future than dead ends.”
Although the role of hybridization in plant evolution has been recognized for a long time (Anderson, 1948; Stebbins, 1959; Rieseberg, 1995; Arnold, 1997), recent evolutionary and genomic analyses have catapulted hybridization to the forefront of evolutionary biology in both plants and animals (Schwenk et al., 2008; Soltis and Soltis, 2009; Abbott et al., 2013; Mallet et al., 2016; Elgvin et al., 2017; Grant and Grant, 2019; Runemark et al., 2019; Taylor and Larson, 2019). Hybrids are considered to play an important role in the evolution of different taxa, as genomic introgression across species boundaries can increase genetic and phenotypic variation (Rieseberg and Carney, 1998), facilitate adaptive evolution (Dasmahapatra et al., 2012; Suarez-Gonzalez et al., 2018) and adaptive radiations (Seehausen, 2004), and even fuel speciation (Mallet, 2007; Abbott et al., 2010). However, the contribution of hybridization to evolution may be modulated initially by the fitness of early-generation hybrids. Hybrids between genetically divergent lineages often suffer from reduced fitness, including low viability and fertility (Grant, 1966; Arnold and Hodges, 1995; Arnold et al., 2001). Extreme examples of this phenomenon are inter-ploidy hybrids, in which genetic incompatibilities are compounded by differences in parental chromosome numbers. The absence of a homologous chromosome to perform pairing in the hybrid interferes with the normal segregation of chromosomes during meiosis and often results in sexual sterility (Vallejo-Marin and Hiscock, 2016). If sterility is complete, these hybrids are effectively an evolutionary dead-end.
One mechanism that has come to the rescue of evolutionarily dead-end hybrid lineages is whole-genome duplication (WGD). WGD leads to polyploidy (Soltis et al., 2015), which in turn is associated with a restoration of sexual fertility in initially sterile hybrid lineages (Abbott and Lowe, 2004; Vallejo-Marin and Hiscock, 2016). WGD in a hybrid background provides homologous chromosome copies for each individual chromosome, restoring meiosis I bivalent formation, balanced (i.e., disomic) chromosome segregation, and euploid spore formation in the allopolyploid line. This results in the restoration of F1 hybrid fertility. Surprisingly, there is little direct experimental evidence of the extent to which WGD restores the fertility of initially sterile hybrids. However, hybridization and polyploidy are often linked in the evolution of new lineages (Stebbins, 1985; Soltis and Soltis, 1989; Mandakova et al., 2013). The association of hybridization and polyploidy evidenced by allopolyploids of both ancient (Renny-Byfield et al., 2015; Alix et al., 2017; Liston et al., 2020) and recent origin (Ashton and Abbott, 1992; Ainouche et al., 2003) provides circumstantial evidence for the hypothesis that WGD plays an important role in the evolutionary stabilization and establishment of some hybrid lineages.
The relationship between hybridization and polyploidy may be related to the causal effect of hybridization on the formation and establishment of polyploids. Polyploids can be formed through either somatic WGD or by the mating of unreduced gametes with somatic chromosome numbers (Madlung, 2013). Although such gametes are produced at low rates in many plant species (Ramsey, 2007; Mason and Pires, 2015), hybrids seem to have increased rates of unreduced gamete production (Ramsey and Schemske, 1998, 2002; Van de Peer et al., 2017). In F1 hybrids that result from distant inter-species crosses (no homology between parental genomes), there is a high incidence of meiotic restitution, and the resulting 2n gametes have a selective advantage compared with other aneuploid spores, which are often lethal. Moreover, it has been suggested that hybridization may facilitate adaptation to WGD (Baduel et al., 2018). In Arabidopsis lyrata, adaptive introgression from tetraploid Arabidopsis arenosa has helped to stabilize the meiosis of autotetraploid populations (Marburger et al., 2019). By contrast, polyploidy may provide a mechanism by which hybrid lineages can become reproductively isolated and established, even when they occur with their parental taxa (Husband, 2004; Bomblies and Madlung, 2014). For example, WGD may erect reproductive barriers between taxa with different ploidy levels. In Chamerion angustifolium, differences in chromosome number and associated changes in phenology contribute to reproductive isolation between diploid and autotetraploid individuals (Husband and Schemske, 2000; Husband and Sabara, 2004; Ramsey, 2011). Polyploidy is expected to affect phenotypic traits (Otto and Whitton, 2000; Porturas et al., 2019), partly due to its promotion of cell expansion, sometimes called the gigas effect (Stebbins, 1971; Levin, 2002; Soltis et al., 2014). These polyploid-induced changes may compound differentiation between hybrids and parental taxa and increase the potential for hybrids to become established in the parental environment or to colonize new ecological niches.
One of the challenges associated with the study of allopolyploids is that it becomes extremely difficult to disentangle the separate contributions of hybridization and polyploidy to the phenotype (Madlung, 2013). Moreover, in older allopolyploid lineages, the effect of subsequent evolution following hybridization and WGD events becomes increasingly important and difficult to identify (Otto and Whitton, 2000; Otto, 2007). In part for this reason, recently formed hybrids and polyploids (neopolyploids) hold extraordinary promise as biological model systems for understanding the early stages of allopolyploid evolution (Ramsey and Schemske, 2002). The very recent (<200 years) evolution of allopolyploid lineages in non-native and invasive taxa is particularly useful, as often the parental and intermediate taxa are known and available for study. This is the case, for example, for allopolyploid Spartina anglica (Ainouche et al., 2009), Senecio cambrensis (Hegarty et al., 2012), and Mimulus peregrinus (Vallejo-Marin et al., 2015). Moreover, in many cases, hybrids can be artificially produced (synthesized) with relative ease (Hegarty et al., 2013), and polyploidy can be chemically induced in both hybrid and non-hybrid taxa (Tate et al., 2009; Castro et al., 2018; Van Drunen and Husband, 2018; Wei et al., 2020). Colchicine treatment is a common procedure used to generate synthetic polyploids. Colchicine interferes with microtubule formation during cell division and can cause endomitosis (i.e., chromosome segregation without cell division), thereby producing cell lineages with two or more times the typical number of chromosomes (Castro et al., 2018). The application of colchicine to meristematic tissue or young plants (seeds) allows one to obtain polyploid individuals that can then be compared with their non-polyploid relatives (Husband et al., 2008).
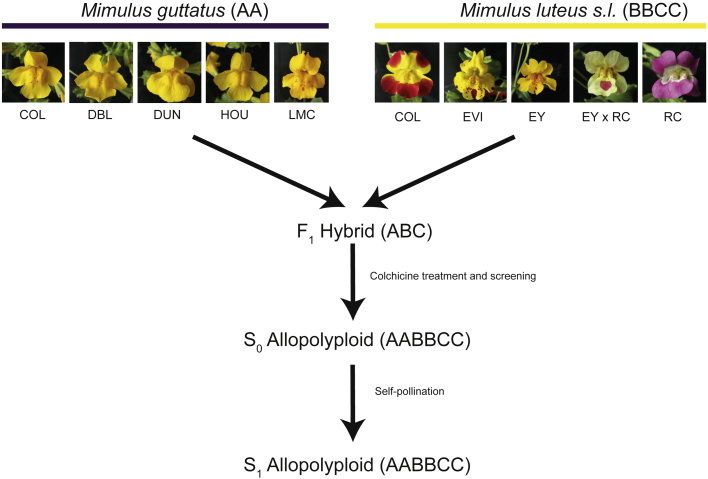
Here, we used natural and synthetic hybrids and polyploid monkeyflowers (Mimulus spp., Phrymaceae; Figure 1) as model systems to investigate the contribution of WGD to the phenotype and fertility of hybrid lineages. Both hybridization and polyploidy have played an important role in the evolutionary history of Mimulus (Vickery, 1995; Beardsley et al., 2004; Benedict et al., 2012). The evolution of chromosome number in Mimulus captures ploidy variation both among (Beardsley et al., 2004) and within species complexes (Vickery, 1978; Coughlan et al., 2020), and aneuploidy seems to be common in some groups (Vickery, 1995). Furthermore, closely related species show porous reproductive barriers, which enable experimental studies of hybridization (Vickery, 1964; Vickery and Mukherjee, 1966; Vickery and Anderson, 1967; Martin and Willis, 2007; Cooley and Willis, 2009; Stanton et al., 2016). The discovery in the British Isles of natural populations of recently formed (<150 years) allohexaploid Mimulus that coexist with triploid hybrids and both diploid and tetraploid parental taxa, together with the ability to resynthesize these taxa in the laboratory, provides a special opportunity to study the effects of WGD on the early evolutionary trajectory of hybrids. The allohexaploid M. peregrinus (2n = 6x) (Vallejo-Marin, 2012) is the WGD product of sterile triploid hybrids (2n = 3x; Mimulus × robertsii). These hybrids have been produced through inter-specific hybridization between two species that are allopatric in their native range but come into contact in the introduced range: diploid Mimulus guttatus (2n = 2x) and ancient allotetraploid Mimulus luteus (2n = 4x) (Vallejo-Marin and Lye, 2013; Da Re et al., 2020). In this study, we address four specific questions. (1) To what extent do differences in ploidy result in reproductive isolation between taxa? (2) What is the effect of WGD on the restoration of fertility in sterile triploid hybrids? (3) To what extent does WGD affect the phenotype of inter-specific hybrids? (4) What is the relative survival of allopolyploids under field conditions compared with related taxa?
Figure 1.
Diagram of the Experimental Design Used to Generate Synthetic Hybrid (Triploid, ABC) and Allohexaploid (AABBCC) Monkeyflowers (Mimulus spp.).
Five populations were used for each of the parental species, diploid M. guttatus (AA) and the ancient allotetraploid M. luteus sensu lato (BBCC). For M. guttatus, we included three introduced populations in the British Isles (COL, DBL, and HOU) and two native populations (DUN and LMC). For M. luteus s.l. we included two introduced populations from the British Isles (COL and EVI), two native populations of two varieties (M. luteus var. luteus [EY] and M. luteus var. variegatus [RC]), and an experimental hybrid between the two varieties (EY × RC). F1 seeds were treated with colchicine and screened using flow cytometry to identify experimental polyploids, which were then brought to flower and self-fertilized to generate S1 seeds. F1 and S1 seeds were used in all subsequent experiments. Population details are given in Supplemental Table 2.
Results
Hybrid Inviability in Inter-ploidy and Inter-specific Crosses
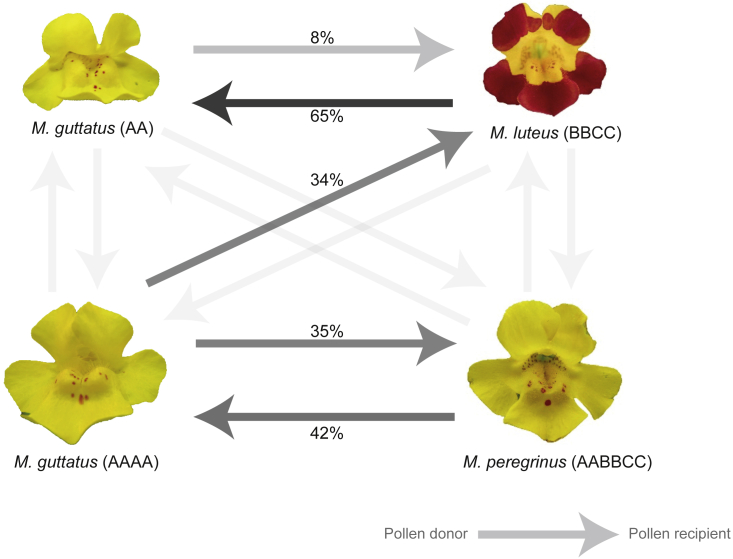
We found significant variation in levels of hybrid inviability among inter-ploidy and inter-specific crosses measured as failure to germinate (seed inviability; Figure 2; statistical significance assessed in a binomial model with two categorical factors—inter-ploidy cross and inter-species cross—as explanatory variables). Regardless of ploidy level, intra-specific crosses (within M. guttatus, M. luteus, or M. peregrinus) had higher viability (0.58 ± 0.09; proportion of germinated seeds, mean ± SE) than inter-specific crosses (0.21 ± 0.06) (inter-species effect, P < 0.001). In general, inter-ploidy crosses produced seeds with lower average viability (0.18 ± 0.08) than intra-ploidy crosses (0.62 ± 0.05) (Supplemental Table 1; inter-ploidy effect, P < 0.001). For M. guttatus, we could further assess the strength of inter-ploidy barriers within an individual species, as we had both diploid and tetraploid individuals. As expected, seeds from intra-ploidy crosses (2x or 4x) had a higher viability (0.75 ± 0.06) than seeds from inter-ploidy crosses (0.01 ± 0.01; within M. guttatus Tukey contrasts, P < 0.001). In inter-specific crosses that yielded viable seed, the direction of the cross, i.e., which taxon was used as the maternal or paternal parent, also affected hybrid inviability. As shown previously, M. guttatus (2x) × M. luteus (4x) crosses with M. guttatus as the maternal parent produced seeds with higher germination (0.65 ± 0.01) than crosses with M. guttatus as the paternal parent (0.08 ± 0.08; cross direction within this cross type, P < 0.001). Furthermore, we found weaker viability barriers between M. peregrinus (6x) and M. guttatus when the crosses involved tetraploid M. guttatus (0.51 ± 0.14) than when they involved diploid M. guttatus (0.02 ± 0.02) (Figure 2; contrast of diploid versus tetraploid M. guttatus parentage, P < 0.001). By contrast, M. peregrinus and M. luteus (4x) crosses produced very few viable seeds, regardless of the mating direction (0.02 ± 0.02 in total).
Figure 2.
Diagram Illustrating the Viability of Seeds Obtained from Crosses between Monkeyflowers (Mimulus spp.) with Different Ploidy and Genomic Composition (Subgenomes Shown with Different Capital Letters).
Arrowheads indicate the recipient of pollen in each cross type. Seed viability was assessed as the proportion of germinated seeds and is qualitatively indicated with different degrees of arrow shading. The lightest shading corresponds to germination proportions less than or equal to 2%. Quantitative values for seed germination are given in Table 1.
Genome-Size Estimation
Table 1 shows the estimated genome size (2C) of intra- and inter-taxon crosses of the Mimulus taxa studied here. We were able to estimate genome size in 116 individuals from all taxa and cross types using 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei, but in only 18 individuals from a subset of taxa using propidium iodide (PI)-stained nuclei. Because genome-size estimates based on DAPI-stained nuclei are affected by potential differences in the AT/GC ratio between the sample and the standard, these genome-size estimates should not be treated as absolute estimates of DNA content or as directly comparable with other methods such as PI staining. However, we found that PI estimates of genome size, which are not affected by AT/GC ratio differences between sample and standard, were similar (3%–8% higher) to estimates from DAPI-stained nuclei (Table 1). With these caveats in mind, DAPI-stained nuclei can be used as an approximation of genome size and, because we used the same size standard for all samples, can be directly compared among samples in our experiment. The DAPI genome size of tetraploid M. guttatus crosses was twice that of diploid M. guttatus (1.598 ± 0.023 versus 0.814 ± 0.026 pg, respectively; Table 1). The genome size of the ancient tetraploid M. luteus was 86% that of an M. guttatus tetraploid (Table 1). As expected, M. × robertsii, the triploid hybrid product of an M. guttatus and M. luteus cross, had a genome size approximately equal to the arithmetic mean of both parents (1.120 ± 0.004 pg), whereas the allohexaploid M. peregrinus had a genome size approximately double that of M. × robertsii (2.121 ± 0.017 pg). The offspring of crosses between tetraploid M. guttatus and M. luteus had a DAPI genome size approximately 20% lower than the arithmetic mean expectation (expected = 1.490 versus observed = 1.205 ± 0.097 pg). By contrast, the product of crosses between diploid M. guttatus and hexaploid M. peregrinus had a genome size closer to the arithmetic mean expectation (expected = 1.467 versus observed = 1.539 ± 0.010 pg). Finally, the product of crosses between tetraploid M. guttatus and hexaploid M. peregrinus had a genome size close to the arithmetic mean expectation (expected = 1.859 versus the observed average of the reciprocal crosses = 1.890, Table 1).
Table 1.
Genome Size (2C Values in Picograms) Estimates of Individuals Produced by Intra- and Inter-ploidy Crosses in Several Taxa of Mimulus spp. (Phrymaceae).
| Maternal parent | Paternal parent | Expected ploidy | 2C genome size (pg) |
||||
|---|---|---|---|---|---|---|---|
| DAPI | N | PI | n | ||||
| M. guttatus (2x) | × | M. guttatus (2x) | 2x | 0.814 ± 0.026 | 6 | 0.841 ± 0.004 | 5 |
| M. guttatus (2x) | × | M. luteus (4x) | 3x | 1.120 ± 0.004 | 29 | 1.160 ± 0.017 | 3 |
| M. luteus (4x) | × | M. luteus (4x) | 4x | 1.382 ± 0.029 | 9 | – | |
| M. guttatus (4x) | × | M. guttatus (4x) | 4x | 1.598 ± 0.023 | 12 | – | |
| M. guttatus (4x) | × | M. luteus (4x) | 4x | 1.205 ± 0.097 | 2 | – | |
| M. peregrinus (6x) | × | M. guttatus (2x) | 4x | 1.539 ± 0.010 | 10 | 1.673 ± 0.021 | 3 |
| M. guttatus (4x) | × | M. peregrinus (6x) | 5x | 1.853 | 1 | 1.967 | 1 |
| M. peregrinus (6x) | × | M. guttatus (4x) | 5x | 1.928 ± 0.006 | 29 | 2.078 ± 0.044 | 6 |
| M. peregrinus (6x) | × | M. peregrinus (6x) | 6x | 2.121 ± 0.017 | 18 | – | |
| Total | 116 | 18 | |||||
n, number of individuals. Genome size estimates are presented separately for nuclei stained with DAPI (relative genome size) and PI (absolute genome size). Note that not all samples were analyzed with PI and that DAPI estimates are affected by potential differences in AT/GC ratios between the sample and the size standard.
Pollen Viability in Parental Taxa and Synthetic Allopolyploids
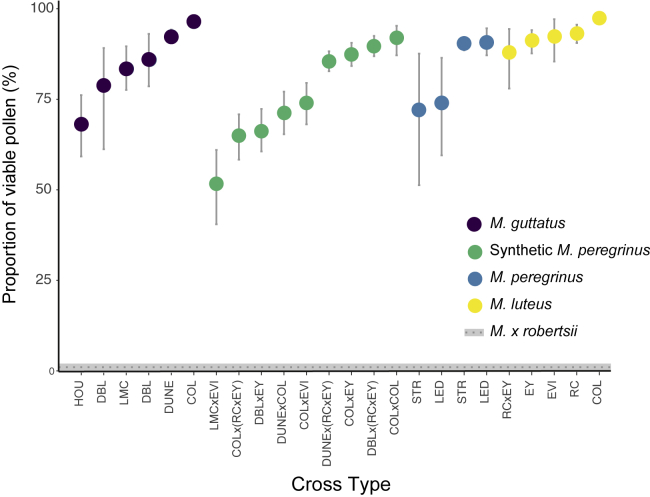
We found that polyploidization of colchicine-treated triploid hybrids resulted in a strong recovery of pollen viability from the near complete pollen sterility documented in triploid M. × robertsii (Figure 3). The nine lines of synthetic allohexaploids varied significantly in average pollen viability, ranging from 51% to 92% viable pollen (P < 0.001; Figure 3). This range in pollen viability of synthetic allohexaploids was similar to that observed within two natural populations of M. peregrinus (72%–91%), although some synthetic lines showed comparatively low pollen viability (e.g., synthetic allopolyploid line LMCxEVI, 51%; Figure 3). The average fertility of the analyzed M. guttatus parental lines ranged from 68% to 96%, and that of the M. luteus parental lines ranged from 88% to 97% (Figure 3).
Figure 3.
Pollen Viability in Individual Products of Intra- or Inter-taxon Crosses.
The nine synthetic allopolyploid lines (“M. peregrinus”) are shown in green. Natural allopolyploids (M. peregrinus) are shown in blue (populations LED and STR). The parental species are shown in purple (M. guttatus) and yellow (M. luteus). The dotted line represents the average fertility reported for M. × robertsii (Vallejo-Marin, 2012). Each data point represents the mean of individuals derived from the same cross (full-sib family). The labels on the x-axis show the codes of populations used in each cross and are detailed in Supplemental Table 2. Vertical bars indicate the 95% confidence intervals (CIs) estimated using bootstrapping.
Phenotypic Variation in a Common Garden
We found clear phenotypic differences between the five studied taxa (M. guttatus, M. luteus, synthetic M. × robertsii, synthetic M. peregrinus, and natural M. peregrinus) in a common garden (Supplemental Figure 1). The first linear discriminant axis distinguished M. guttatus from the other taxa, and the second axis partially separated the other four taxa. A multivariate analysis of variance (MANOVA) of 16 morphological traits indicated significant differences associated with taxon (P < 0.001 for each of the individual traits). In general, M. guttatus had the shortest time to flowering, the largest number of flowers, the shortest pedicles, and the smallest bracts of all taxa (Supplemental Figure 2). The synthetic triploids and derivative allohexaploids showed intermediate values between the parental taxa for several of the 16 measured traits but exceeded both parents in corolla height and bract width (Supplemental Figure 2). The random forest analysis showed a classification error of 9.05% (out-of-bag error), indicating a generally high ability to correctly assign individuals to their true taxon based only on their phenotypic values. The lowest classification error (the proportion of individuals assigned to the wrong taxa) was for M. guttatus (1.8%). The classification error for synthetic M. × robertsii was 9.56%, with most incorrectly assigned individuals (17/23) placed in synthetic M. peregrinus and the rest in M. guttatus (2/23) or M. luteus (1/23). Synthetic M. peregrinus had a low classification error (5.9%), with most (15/17) of the erroneously classified individuals assigned to M. × robertsii and the rest (2/17) to M. guttatus. Natural M. peregrinus was correctly assigned in 82% of the cases, with erroneous classifications mostly assigned to synthetic M. peregrinus (11/13). Finally, M. luteus had the highest classification error (21%), with 12 of 16 erroneous assignments to M. × robertsii, 3 of 16 to synthetic M. peregrinus, and 1 of 16 to M. guttatus. The analysis of variable importance as measured by the random forest shows that the traits with the largest contribution to phenotypic classification are plant height and floral throat width, whereas anther-stigma distance contributes the least (Supplemental Figure 3).
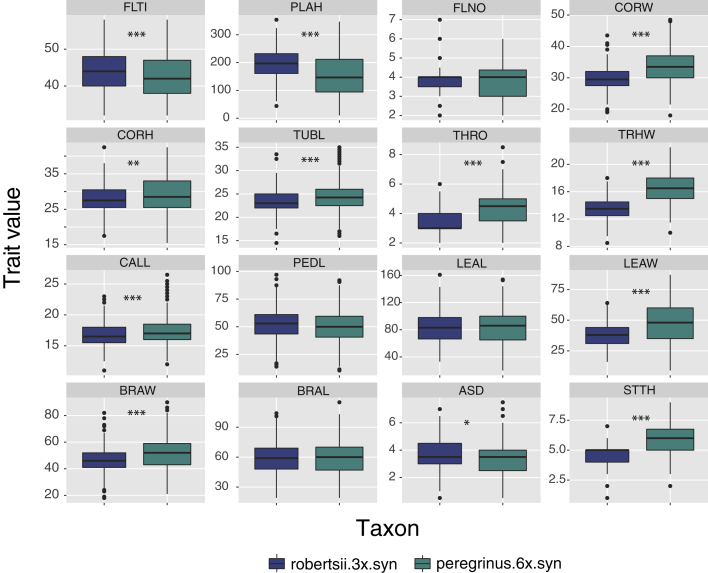
We further compared the phenotypic effects of polyploidization using M. × robertsii synthetic lines and their colchicine-induced allopolyploid counterparts. This comparison allowed us to determine the effect of polyploidization on individual traits. We found statistically significant differences, as assessed by MANOVA (P < 0.05), in 12 of the 16 phenotypic traits analyzed (Figure 4). In general, the effect of WGD on the phenotype of the hybrids was to increase the size of the flower, the width of leaves and bracts, and the thickness of the main stem, while reducing plant height, anther-stigma distance, and the number of days to flowering (Figure 4).
Figure 4.
Comparison of 16 Phenotypic Traits in Synthetic Triploid Hybrid (Robertsii.3x.syn) Monkeyflowers (Mimulus spp.) and Their Synthetic Allohexaploid Derivatives (Peregrinus.6x.syn) Measured in a Common Garden in the Greenhouse.
The traits measured were: days to flowering (FLTI), plant height (PLAH), flowering node of the first flower (FLNO), corolla width (CORW), corolla height (CORH), corolla tube length (TUBL), throat opening (THRO), throat width (THRW), calyx length (CALL), pedicle length (PEDL), the width (LEAW) and length (LEAL) of the largest leaf, bract width (BRAW) and length (BRAL), anther-stigma distance (ASD), and stem thickness (STTH). All units are in millimeters except FLTI (days) and FLNO (flower number). Statistical significance of differences between the triploid and the allohexaploids was assessed by MANOVA. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
Field Experiments
The first field experiment was performed in an experimental plot at the University of Stirling, where we compared the phenotype and survival of four taxa (M. guttatus, M. luteus, synthetic triploid M. × robertsii, and synthetic allohexaploid M. peregrinus) over 2 years. We observed relatively little phenotypic differentiation between triploids and hexaploids, compared with the differences observed among parental taxa in seven measured phenotypic traits (Supplemental Figure 4). The only traits that differed significantly between triploids and hexaploids in the field were related to flower size (corolla width, corolla height, and corolla tube length; MANOVA, P < 0.05) (Supplemental Figure 5).
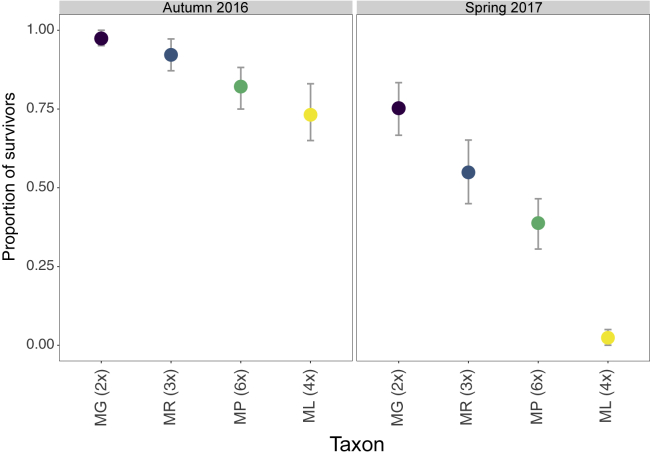
We found no statistical differences in the probability of flowering within the first year among the four taxa studied (taxon effect, likelihood ratio test [LRT] = 3.090, P = 0.3778). By contrast, the probability of survival to the end of the first flowering season (September 2016) and to the following spring (May 2017) varied among taxa (taxon effect, P < 0.001 in both cases; Figure 5). The difference was more marked in the May 2017 survival data and was driven exclusively by the significantly lower survival of M. luteus (probability of survival = 2%) compared with all other taxa (Figure 5).
Figure 5.
Proportion of Mimulus spp. Individuals of Mimulus spp.Surviving in a Field Plot in Central Scotland to the End of the First Growing Season (Autumn 2016) and Over Winter to the Following Spring (Spring 2017).
Triploid (3x) individuals of M. × robertsii (MR) were created synthetically by crossing M. guttatus (MG) (2x) and M. luteus (ML) (4x). Allohexaploid (6x) individuals of M. peregrinus (MP) were synthesized by treating the seeds from synthetic M. × robertsii with colchicine. The experiment included 843 individuals and 1796 flowers; detailed sample sizes are provided in Supplemental Table 3. The symbols indicate mean survival, and the vertical bars show 95% CIs obtained by bootstrapping.
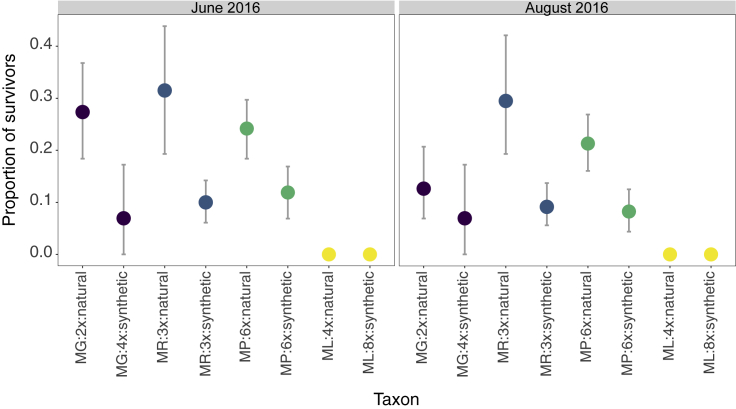
The second field experiment was performed in the southern uplands of Scotland near the town of Leadhills and provided an opportunity to assess survivorship of different Mimulus taxa under more realistic conditions. This area is home to natural populations of M. × robertsii and M. peregrinus. We observed a relatively low average survival through the winter season (<40%), as assessed in both the June 2016 and the August 2016 census (Figure 6). Moreover, this experiment allowed us to compare both auto- and allopolyploids, as well as natural and synthetic triploids and hexaploids. Again, M. luteus performed worst among all taxa, with every natural M. luteus and its synthetic autopolyploid derivative (octoploid M. luteus) failing to survive over the first winter. A statistical analysis of the remaining taxa (defined as a combination of species, ploidy, and natural or synthetic origin) showed different levels of survival among taxa (August census: LRT = 27.06, df = 5, P < 0.001). Diploid M. guttatus had a survival probability of 27.6% by the beginning of the summer, but this decreased to 12.6% toward the end of the summer (Figure 6). Tetraploid M. guttatus had a low survival (6.9%) at both censuses. The synthetic M. × robertsii and synthetic M. peregrinus had similar survival probabilities (10.1% versus 11.8% by June and 9.1% versus 8.1% by August for the triploid and hexaploid, respectively). By contrast, both natural M. × robertsii and natural M. peregrinus had the highest survival probabilities of all taxa (31.6% versus 24.1% in June and 29.8% versus 21.22% in August for the triploid and hexaploid, respectively) (Figure 6). A post hoc pairwise Tukey test showed significant survivorship differences between natural and synthetic individuals of both M. × robertsii and M. peregrinus but detected no significant differences between triploids and hexaploids (P < 0.05) (Figure 6).
Figure 6.
Survival of Mimulus spp. in a Field Plot in the Southern Uplands of Scotland Near Leadhills.
Individuals were planted as cuttings in the autumn of 2015, and survivorship was assessed at the beginning of the following summer (June 2016) and at the end of the summer (August 2016). The experiment contained 828 individuals (ramets) from 298 genotypes (genets) of eight taxa and encompassed parents (M. guttatus:2x and M. luteus:4x) and their autopolyploid derivatives (M. guttatus:4x and M. luteus:8x), as well as inter-specific hybrids before (M. × robertsii:3x) and after WGD (M. peregrinus:6x) (for sample size information per taxon, see Supplemental Table 4). Both triploid and allohexaploid hybrids were represented by both natural and synthetically created individuals. The symbols indicate mean survival, and the vertical bars show 95% CIs obtained by bootstrapping.
Discussion
Crossing Barriers among Ploidy Levels
Our results provide further evidence that differences in ploidy result in strong, but leaky, reproductive barriers both within and between species. Our experiment focused on the viability of seeds produced in controlled crosses and therefore provides a conservative estimate of the strength of intrinsic reproductive barriers (Coyne and Orr, 2004), as they do not include potential reductions in seed number or subsequent reductions in fertility. Within M. guttatus, we found that diploids and tetraploids are significantly reproductively isolated by the low viability of inter-ploidy hybrids (approximately 1% germination success). This finding contrasts with previous work from the native range, which found that crosses between one diploid and two different tetraploid populations produced viable hybrids, although these were sterile (Vickery and Mia, 1967; Vickery et al., 1968). In our experiment, the crossing barriers among taxa of different ploidy levels are clearly imperfect, and inter-ploidy hybrids can be viable. A particularly interesting result is that an increase in ploidy level can change the level and even the direction of crossing barriers between species. For instance, M. guttatus × M. luteus hybrids are much more likely to be viable when diploid M. guttatus is the maternal parent (65.5% viability) than when it is the paternal parent (8.5%) (cf. Vallejo-Marin et al., 2016). Yet the result is reversed with tetraploid M. guttatus, and in this case, the cross in which M. luteus is the maternal parent produces more viable hybrids (46%) than the opposite cross (0.3%). The asymmetry in hybrid formation and its response to the ploidy level of the parents suggests that inviability in hybrid seeds is mediated by parental conflict resulting in endosperm development failure (Lafon-Placette and Kohler, 2014; Kinser et al., 2018; Coughlan et al., 2020). The inter-ploidy barrier between M. guttatus and M. peregrinus also varies strongly depending on whether M. guttatus is diploid (strong barrier, 0%–1% viability) or tetraploid (weaker barrier, 35%–42% viability), independent of the crossing direction. A similar weakening of reproductive isolation at higher ploidy levels has been observed in other systems (Sonnleitner et al., 2013; Sutherland and Galloway, 2017) and may represent a general phenomenon.
Genome-Size Variation
Genome size in early-generation hybrids shows a moderate departure from additivity. The genome sizes of the synthetic inter-ploidy hybrids (triploids, tetraploids, and pentaploids) were close to the arithmetic mean expectation (Table 1). Similarly, the synthetic allohexaploid had a value approximately, if somewhat smaller, than the expected genome size of double the hybrid value. Crosses between tetraploid M. guttatus and tetraploid M. luteus yielded a smaller than expected genome size (20% lower). In addition, the variation in genome size in crosses between diploid M. guttatus and M. peregrinus depended on the direction of the cross. However, these differences remain to be confirmed with larger sample sizes. The biggest difference in relative (DAPI-stained) genome size was between the recently derived autotetraploid M. guttatus (2C = 1.598 ± 0.023 pg) and the ancient allotetraploid M. luteus (2C = 1.382 ± 0.029 pg). This difference in size is opposite to the difference in chromosome number (2n = 56 versus 2n = 60–64, for tetraploid M. guttatus and M. luteus, respectively (Mukherjee and Vickery, 1962; Vickery et al., 1968; Simón-Porcar et al., 2017), but it also probably reflects different genome sizes in the parental taxa of M. luteus, as well as subsequent evolution in genome size (Leitch et al., 2008).
Fertility Restoration
A major effect of WGD on inter-specific triploid hybrids is the restoration of sexual fertility (Dobzhansky, 1937; Grant, 1971). This instantaneous recovery of fertility strongly suggests that sterility in the hybrids is mainly due to unbalanced chromosome numbers (Husband, 2004; Kohler et al., 2010). Synthetic allohexaploids showed a major increase in pollen viability from about 1% in triploids (Vallejo-Marin, 2012) to 50%–90% among different lines of allohexaploids. This variation in fertility may further indicate that even after genome doubling, other reproductive barriers remain in the allohexaploids. Even within species, Mimulus shows a large amount of variation in fertility among inter-population hybrids (Vickery, 1959), and reproductive barriers are common among closely related species (Sweigart et al., 2006; Kenney and Sweigart, 2016; Kerwin and Sweigart, 2020). The variation in the pollen fertility of synthetic allopolyploids is similar to that in natural populations of M. peregrinus (70–90%). To the extent that this genetic variation in fertility is genetically based (Bomblies et al., 2015), we expect that selection in natural populations of M. peregrinus will cause the evolution of higher pollen fertility. The consistent effect of WGD on the restoration of sexual fertility in hybrids is an important mechanism that facilitates the repeated formation of allopolyploid lineages, as has occurred in M. peregrinus and many other polyploids (Wyatt et al., 1988; Soltis and Soltis, 1991; Ashton and Abbott, 1992; Mavrodiev et al., 2015; Vallejo-Marin et al., 2015; Soltis et al., 2016). Importantly, independently originated lineages of polyploids are often interfertile (Vallejo-Marín et al., 2017), supporting the idea that the same biological hybrid species can arise rapidly and repeatedly.
Phenotypic Effects of WGD
The effect of WGD on hybrids could be clearly detected in the greenhouse common garden, as 12 of 16 traits were significantly different between triploids and their derivative allopolyploids. The phenotypic changes induced by WGD in hybrid Mimulus are primarily organ-size increases (flowers, leaves, stem thickness), shorter plants with flowers that exhibit reduced herkogamy (anther-stigma distance), and a developmental delay in flowering. Although colchicine itself can affect the phenotype independent of its effects on chromosome doubling (Husband et al., 2008) even after two generations (Munzbergova, 2017), our results are consistent with polyploid-dependent effects. The phenotypic changes caused by increases in DNA per cell are called nucleotypic effects (Levin, 1983; Snodgrass et al., 2017; Doyle and Coate, 2019), and they can cause immediate increases in the size of cells (the gigas effect, Soltis et al., 2014), including stomatal guard cells and pollen grains (Stebbins, 1971; Levin, 2002). These expectations are met in natural M. peregrinus, which shows larger stomata and pollen grains than its parents (Vallejo-Marin, 2012). In some cases, these nucleotypic effects seem to be directly associated with physiological changes. For example, in autopolyploid Chamerion angustifolium, synthetic tetraploids have larger stomata and increased drought tolerance than diploids (but less than natural tetraploids) (Maherali et al., 2009). Increases in DNA content and cell size can lead to larger organs (Doyle and Coate, 2019) but slower growth rates, as developmental rate seems to be negatively correlated with DNA content across angiosperms (Levin, 1983). Our results of larger flowers and leaves but slightly delayed flowering following WGD in hybrids are consistent with these general patterns. Delay in flowering time in polyploids compared with their diploid relatives has been observed in several plant groups and can range from a few days to more than a month (Levin, 1983). The mechanistic connection between WGD and phenotypic changes is not well understood at the genetic and cellular levels (Doyle and Coate, 2019). However, previous work on WGD in hybrids has established that although hybridization itself triggers changes in gene expression patterns, polyploidy has an additional effect on the transcriptome (Hegarty and Hiscock, 2005; Hegarty et al., 2006), and expression patterns continue to change in subsequent generations (Edger et al., 2017). The general consensus from studies of recently formed polyploids, including our study on neo-allopolyploids, is that WGD affects the phenotype. The extent to which phenotypically different hybrids and allopolyploids differ in their field performance remains to be established.
Hybrid Survival in the Field
To our knowledge, this is the first study to combine synthetic hybridization and WGD to compare the survival of parental taxa and both natural and synthetic hybrids and allopolyploids in the field. Evolutionary experiments with synthetic hybrids in the field, excluding crops, are relatively rare. For instance, a recent study following homoploid Helianthus hybrids (Helianthus annuus × Helianthus debilis) over seven generations showed that hybrid lines adapted at a faster rate than non-hybrid lines (Mitchell et al., 2019). In contrast to the Helianthus experiment, here, we followed only a single generation of hybrids and allopolyploids, however, we performed comparisons not only with their parental taxa but also with their autopolyploid parents. In the first field experiment in central Scotland, synthetic triploid and allohexaploid Mimulus had intermediate survivorship compared with the parental taxa, and synthetic M. peregrinus performed slightly worse than M. × robertsii. These results are consistent with the second field experiment in the harsher environment of the Lowther Hills in the southern uplands of Scotland that included natural hybrids as well as both parental taxa and their autopolyploid derivatives. Here, survivorship was generally lower than in the first experiment, but synthetic triploids and allohexaploids again had intermediate survival between the two parents. By contrast, natural M. × robertsii and M. peregrinus had better survival than all other taxa, including their synthetic counterparts, both parents, and their autopolyploids. It is important to note that the lower survival of the synthetic hybrids and allopolyploids compared with the natural ones could not be due to colchicine treatment alone, as the triploid hybrids included in the experiment were not treated with colchicine. The increased survival of natural hybrids and allopolyploids compared with synthetic ones raises the possibility that this difference has evolved through natural selection despite the recent origin of these taxa. Even asexual taxa such as M. × robertsii can evolve through clonal (genotypic) selection (Pan and Price, 2001; Dalrymple et al., 2015). Reciprocal transplant experiments on M. × robertsii populations from opposite ends of the British Isles show the home advantage of local populations and are consistent with selection shaping local performance in asexual lineages (Simon-Porcar, Silva and Vallejo-Marin, unpublished data). In sexual M. peregrinus, natural selection could continue to shape the superior performance of hybrids already seen in M. × robertsii, with the added advantage of sexual recombination. It has been suggested that the establishment of polyploids may be contingent on being near new adaptive peaks when they are formed (Mallet, 2007). Natural selection on the widespread triploid hybrid from which polyploid M. peregrinus evolved may have given this “hopeful monster” (Goldschmidt, 1933; Jenczewski, 2013) a vantage point from which to seek an adaptive peak.
Methods
Study System
We focused on two hybridizing species of Mimulus Section Simiolus (Nesom, 2012): M. guttatus DC. and M. luteus L. (Grant, 1924). M. guttatus is native to western North America, and M. luteus comes from the Andean region of Chile and Argentina (Vickery, 1959; Medel et al., 2003). Both M. guttatus and M. luteus were introduced into the British Isles in the 19th century and became naturalized (Stace, 2010; Vallejo-Marin and Lye, 2013; Stace and Crawley, 2015; Pantoja et al., 2017). Most populations of M. guttatus are diploid (2n = 2x = 28) (Mukherjee and Vickery, 1962) in both native and introduced ranges, although tetraploids (2n = 4x = 56) occur in North America (Vickery et al., 1968) and have recently evolved in the introduced range from local populations in the Shetland Isles (Simón-Porcar et al., 2017). M. luteus is an ancient tetraploid (2n = 4x = 60–64) (Vickery et al., 1968), and recent genomic analysis suggests that it originated from an allopolyploidization event (Edger et al., 2017). M. luteus is part of an interfertile species complex that includes M. luteus var. luteus, M. luteus var. variegatus, and Mimulus naiandinus (Cooley and Willis, 2009; Stanton et al., 2016). M. guttatus and M. luteus hybridize in the British Isles to produce the hybrid M. × robertsii Silverside (Silverside, 1990; Stace et al., 2015), which has a chromosome number intermediate between those of its parents (2n = 3x = 44–46). M. × robertsii is highly sexually sterile (Vickery and Mukherjee, 1966; Stace, 2010) but is capable of vigorous clonal reproduction via lateral stems and has become widely distributed in the British Isles (Preston et al., 2002; Vallejo-Marin and Lye, 2013; Da Re et al., 2020). M. × robertsii has given rise through WGD to a new, sexually fertile and vegetatively reproducing species, the allohexaploid M. peregrinus (2n = 6x = 90–92) (Vallejo-Marin, 2012). M. peregrinus has evolved at least twice from local populations in two regions of Scotland, the Lowther Hills in the southern highlands and the Orkney Isles in the north (Vallejo-Marin, 2012; Vallejo-Marin et al., 2015).
Generation of Synthetic Hybrids and Polyploids
We grew plants of M. guttatus (2n = 2x) and M. luteus (2n = 4x) from seeds of five populations per species (Supplemental Table 2 and Figure 1) in a controlled environment facility (18 h light at 24°C and 6 h dark at 18°C, 70% relative humidity) at the University of Stirling in the autumn of 2014. The two taxa in this parental generation were crossed reciprocally to produce inter-specific triploid (2n = 3x) hybrid seeds, using all five populations per species and performing up to five replicate crosses per combination of populations. All flowers were emasculated the day before performing the inter-specific crosses. To induce polyploidy and generate synthetic allohexaploids (2n = 6x), a subset of the seeds generated in inter-specific crosses (2n = 3x) were treated with colchicine (Dermen, 1940; Eng and Ho, 2019). Seeds were incubated in 1 ml of 0.1% (w/v) aqueous colchicine solution in the dark at room temperature for 24 h. After the incubation period, seeds were germinated in the controlled environment facility as described above.
Assessment of Ploidy Level of Colchicine-Treated Plants
The ploidy level of colchicine-treated plants was assessed by flow cytometry (Doležel et al., 2007). In brief, cell nuclei were isolated from fresh leaf tissue in Woody Plant Buffer (Loureiro et al., 2007) together with an internal standard. The solution was passed through a 50-μm nylon filter, stained with a PI and RNase solution (both at a final concentration of 50 μg/ml), and analyzed in a Guava EasyCyte flow cytometer (Millipore Limited, UK), using a diploid M. guttatus as an internal standard (relative 2C genome size = 2x). Successfully transformed plants were identified by having one fluorescence peak that corresponded to the expected genome size of the allohexaploid (relative 2C genome size = 6x) (Vallejo-Marin, 2012). Transformed plants (S0) were often mixoploid, showing two peaks at approximately 3x and 6x, indicating a mosaic of transformed and untransformed cells. To obtain fully transformed allopolyploid individuals, we grew these S0 plants until flowering and self-pollinated them (Figure 1). The resulting S1 generation was germinated as described above and their ploidy level was assessed again, revealing a single 2C DNA peak at around 6x. More precise genome-size estimates were obtained in a separate experiment (see Genome-Size Estimation of Inter-ploidy Crosses). Chromosome counts were performed on a subset of individuals as an additional confirmation of successful polyploidization (2n = 90–92 chromosomes).
Inter-ploidy Crosses
To assess the inter-fertility of plants of different ploidy levels, we performed a series of controlled crosses in June 2018. Natural and synthetic plants of M. guttatus (2x), M. guttatus (4x), M. luteus (4x), and M. peregrinus (6x) were grown from seed as described above. At the flowering stage, we emasculated a flower and cross-pollinated it with fresh pollen. All four taxa were crossed with each other in both directions (16 cross types, including within-taxon crosses). Seeds were collected approximately 17–21 days after pollination and stored at 4°C. As it was not possible to standardize the number of viable pollen grains used in cross-pollinations, we estimated seed viability as the proportion of seeds that germinated. Germination was assessed for 48 crosses from which we obtained and planted 10 or more seeds per cross (10–110, average = 69.27 seeds per cross).
Genome-Size Estimation of Inter-ploidy Crosses
To obtain more precise estimates of absolute and relative (DNA-ploidy level) genome size, we performed flow-cytometry analysis of the seeds that resulted from the inter-ploidy cross-hybridizations (Doležel et al., 2007). This time, we used Solanum pseudocapsicum as the internal standard (2C = 2.59 pg [Temsch et al., 2010]), chopped both sample and standard in 0.5 ml of ice-cold Otto I buffer (0.1 M citric acid, 0.5% Tween 20), and passed the mixture through a 42-μm nylon filter. For ploidy-level estimation (relative genome size), 1 ml of staining solution containing Otto II buffer (0.4 M Na2HPO4·12H2O), 4 μg/ml DAPI, and 2 μl/ml β-mercaptoethanol was added to the nuclei suspension. The stained nuclei were analyzed in a Partec Space cytometer (Partec, Münster, Germany) equipped with a 365-nm UV-LED light for DAPI excitation. In the case of absolute genome-size estimation, we used PI and RNase IIA (both at a final concentration of 50 μg/ml). The fluorescence intensity of 5000 isolated nuclei was analyzed using a Partec Cyflow cytometer equipped with a 532-nm diode-pumped solid-state green laser (Cobolt Samba, 100 mW output power). For absolute genome-size estimation, samples were analyzed at three different days, and measurements that deviated from the between-day variance by more than 3% (= machine error rate) were reanalyzed. The absolute genome size was calculated as the mean of the three measurements × standard genome size. The same procedure was used for the estimation of relative genome size, with the caveat that differences in AT/GC composition between the sample and the standard can affect the estimates and cause differences between PI- and DAPI-based estimates. The resulting estimates should therefore be considered relative approximations of true genome size. More samples were analyzed with DAPI than with PI, as our initial goal was to detect ploidy-level differences rather than absolute genome size.
Fertility Measurements
To assess pollen size and viability, we collected anthers from the fifth flower of each plant in 1 ml of 70% ethanol and kept them in the fridge at 4°C to fix and conserve the pollen. Pollen viability was determined with a Multisizer III (Beckman Coulter) using the method described in De Storme et al. (2013). This method relies on physical differences between viable and inviable pollen (Kelly et al., 2002). Following vortexing, centrifugation, and the removal of the supernatant, the pollen in the pellet was suspended in a weak isotonic electrolyte (Isoton II) and immediately run through the Multisizer to prevent a change in pollen volume (De Storme et al., 2013). On average, 2658 pollen grains per sample were counted within the lower and upper size threshold of the pollen (viable and inviable) size range. For each ploidy level, we manually determined the pollen size cutoff value between viable and inviable pollen.
Phenotypic Variation in a Common Garden
We conducted a common garden experiment to assess phenotypic variation in M. guttatus and M. luteus, synthetic F1 M. guttatus × M. luteus hybrids, and allopolyploids (both natural and synthetic) (Supplemental Table 3). From each seed family, 20 seedlings were transplanted to separate 0.37-l pots, placed in greenhouses at the University of Stirling, and maintained with supplemental lighting (16 h per day). We measured 16 phenotypic traits: plant height (PLAH), stem thickness (STTH), the width (LEAW) and length (LEAL) of the largest leaf, bract width (BRAW) and length (BRAL), days to flowering (FLTI), flowering node of the first flower (FLNO) and pedicel length (PEDL), corolla width (CORW), corolla height (CORH), corolla tube length (TUBL), throat opening (THRO), throat width (THRW), calyx length (CALL), and anther-stigma distance (ASD). All traits were measured to the nearest millimeter with digital calipers (Mitutoyo, Japan) on the first day of flowering of each plant. Vegetative traits were measured when the first flower on a given plant had opened, whereas floral trait measurements were obtained from the first two flowers of each plant.
Field Experiments
We conducted two field experiments to measure the performance of plants of different ploidy levels, both natural and synthetic. The first experiment involved a subset of populations from four taxa: the two parental taxa, M. guttatus and M. luteus, as well as the synthetic triploids and allohexaploids (Supplemental Table 3; full plant IDs available at http://hdl.handle.net/11667/155). Only crosses in which M. guttatus was the maternal parent were included in this experiment. To minimize maternal effects, all seeds in this experiment were produced from parental plants grown from seed in the greenhouse. In total, we used 455 individuals. In late spring 2016, we planted and germinated seeds as described above and transferred them to the greenhouse for acclimation. Between 30 June and 5 July 2016, when plants had between four and six true leaves, they were transplanted to a 4 m × 16 m plot in the experimental gardens of the University of Stirling. Plants were arranged in a 13 × 84 rectangular grid, with individuals equally spaced and separated by 40 cm. Prior to transplanting, the plot was rototilled and fenced with galvanized metal netting to exclude rabbits. Due to a severe infestation of common slugs, we also had to deploy beer slug-traps to avoid the experiment being consumed overnight. We scored the probability of flowering up to the end of the growing season in 2016 and measured the following eight traits at the onset of flowering: (1) plant height (PLAH), (2) stem thickness (STTH), (3) flowering node of the first flower (FLNO), (4) pedicel length (PEDL), (5) corolla width (CORW), (6) corolla height (CORH), (7) corolla tube length (TUBL), and (8) calyx length (CALL). Floral traits were measured in two flowers per individual in all individuals that flowered. We also recorded survival to the end of the summer in 2016 (September) and survival to the end of the experiment in May 2017.
The goal of the second experiment was to assess the survival of different taxa and ploidy levels under more realistic field conditions. In this experiment, we tried to analyze as many taxa and ploidy levels as possible, including both natural and artificially produced (through inter-specific crosses and colchicine treatment, hereafter synthetic) individuals. We studied the following taxa: M. guttatus (2n = 2x), M. × robertsii (2n = 3x), M. guttatus (2n = 4x), M. luteus (2n = 4x), M. peregrinus (2n = 6x), and M. luteus (2n = 8x). Full IDs of all plants, including cross type and population of origin, are provided in the supplemental files available at http://hdl.handle.net/11667/155. To minimize maternal effects, all material used in this experiment was derived from clonal replicates obtained from plants that were first grown from seeds and propagated in the greenhouse. Our experiment included 298 genotypes (genets) (Supplemental Table 4). Each of these genotypes was clonally replicated up to three times, and these individuals (ramets) were transplanted to the field (Supplemental Table 4). In total, we worked with 298 genotypes and 828 individuals (ramets) from eight taxon-ploidy combinations.
A field plot was established in the southern uplands of Scotland near the town of Leadhills, South Lanarkshire, in 2015 (latitude 55.4188°, longitude −3.7378°). This site is approximately 550 m from the holotype locality of M. peregrinus (Vallejo-Marin, 2012) and 30 m from a stream (Shortcleuch Waters). Both M. × robertsii and M. peregrinus co-occur in the vicinity of this area (Vallejo-Marin et al., 2015). Because we wanted to avoid the introduction of exotic material to this natural population, we collected any fruits that formed in the plants of the experimental plots before they were dispersed and ensured that all experimental plants had been destroyed by the end of the experiment. The experimental plot consisted of a 12 m × 15 m area that was rotovated in October 2015 and enclosed with a wire fence that excluded deer and sheep but not smaller herbivores such as rabbits. All the material used in this experiment was grown and clonally propagated in the University of Stirling research greenhouses. In October 2015, ramets of approximately 10 cm in length were potted in individual 0.37-l pots to encourage rooting and leaf growth for about 3 weeks. Plants were transplanted to the field plot at the beginning of November 2015. Plants were arranged in a rectangular grid (26 × 32) with rows and columns spaced about 40 cm apart. A preliminary survey in early December 2015 indicated that the vast majority of plants (99.5%) had survived transplant and become established in the field. Survivorship in this experimental plot was assessed in the following summer on 4 June 2016 and again on 24 August 2016 when the experiment was terminated.
Statistical Analysis
Phenotypic Variation in the Common Garden
We conducted a linear discriminant analysis (LDA) to summarize the phenotypic differences among natural and synthetic taxa in the common garden. Prior to LDA, a Box-Cox transformation was applied to all variables. To investigate the extent to which the measured variables could be used to distinguish between the five taxa (M. guttatus, M. luteus, M. × robertsii, and both natural and synthetic M. peregrinus), we performed a random forest classification using the R package randomForest (Liaw and Wiener, 2002). The random forest was tuned using the function tuneRF with 1000 trees and a step factor of 0.5. The classification error (confusion matrix) and the importance of each variable were estimated using stratified sampling with equal sample sizes for all variables, and the mean decrease accuracy and mean Gini index were calculated. To determine the effect of WGD on the phenotype, we compared the synthesized M. × robertsii and M. peregrinus using MANOVA.
Field Experiments
In the experiment carried out at the University of Stirling experimental gardens, we performed an LDA and a MANOVA on the set of eight phenotypic variables after Box-Cox transformation. The probability of flowering, survival to the end of the summer, and next year’s survival were analyzed using generalized linear mixed-effects models with a binomial error implemented in lme4 (Bates et al., 2014) with taxon as a fixed effect and population (or population cross) as a random effect. Pairwise comparisons between taxa were performed with a Tukey test using the package multcomp (Hothorn et al., 2008).
In the field experiment carried out at Leadhills, we analyzed the probability of surviving by classifying each individual as dead or alive (0,1) at census time and using generalized linear mixed-effects models with a binomial error implemented in lme4. We used the combination of species, ploidy level, and origin (natural or synthetic) to produce a single factor (taxon) and used this as a fixed effect in the models. We used clonal membership (clone) as a random effect. The survivorship of both tetraploid and octoploid M. luteus was zero on both census dates, and we excluded these two taxa from subsequent analyses, as a preliminary investigation showed that including these taxa resulted in a lack of convergence of the statistical models. Pairwise comparisons between taxa were performed with a Tukey test using the package multcomp.
Variation in Pollen Viability among Synthetic Allohexaploid Lines
We tested for differences in pollen viability among nine synthetic allohexaploid lines using a generalized linear model with a binomial error and line as the explanatory variable.
Funding
This work was supported by a Plant Fellows Postdoctoral Fellowship to S.M. (FP7, Marie Curie Actions, COFUND-University of Stirling; 2014–2016) and research grants from The Carnegie Trust (Research Incentive Grant 70158) and the Botanical Society of Britain and Ireland (BSBI-2015) to M.V.-M.
Author Contributions
M.V.-M. and S.M. conceived and designed the experiments. S.M. generated the hybrid and polyploid lines and conducted greenhouse and field work with help from M.V.-M. M.V.-M. performed inter-ploidy crosses and assessed their germination. K.S. and S.M. performed flow-cytometry analyses. S.M. and N.D.S. analyzed pollen fertility with the equipment and consumables from D.G. S.M. and M.V.-M. performed the analyses. M.V.-M. and S.M. wrote the manuscript with input from all authors.
Acknowledgments
We dedicate this paper to Robert K. Vickery Jr. for his pioneering and inspiring work on Mimulus hybrids and polyploids over many decades. We thank Sophie Webster, Mariah Wuerges, David Real, Patricia Gonzalez, Pauline Pantoja, Karen Zapata, Violeta Simon-Porcar, and others in the Vallejo-Marín lab for help with plant growth and field assistance, and Jacqueline McKenna and the Gardens and Grounds staff for support with experimental setup at Stirling and Leadhills. We are grateful to Billy Steel and the Leadhills Estate for providing support and access to set up and execute the field experiment at the birthplace of M. peregrinus in the Lowther Hills. Joao Loureiro and Silvia Castro are thanked for early advice on flow cytometry to M.V.-M., without which this study would not have been possible. Arielle Cooley kindly shared her seed material for M. l. luteus and M. l. variegatus. The authors have no conflicts of interest to declare.
Published: July 3, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information can be found online at Plant Communications Online.
Supplemental Information
References
- Abbott R., Albach D., Ansell S., Arntzen J.W., Baird S.J., Bierne N., Boughman J., Brelsford A., Buerkle C.A., Buggs R. Hybridization and speciation. J. Evol. Biol. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Abbott R.J., Hegarty M.J., Hiscock S.J., Brennan A.C. Homoploid hybrid speciation in action. Taxon. 2010;59:1375–1386. [Google Scholar]
- Abbott R.J., Lowe A.J. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biol. J. Linn. Soc. 2004;82:467–474. [Google Scholar]
- Ainouche M.L., Baumel A., Salmon A., Yannic G. Hybridization, polyploidy and speciation in Spartina (Poaceae) New Phytol. 2003;161:165–172. [Google Scholar]
- Ainouche M.L., Fortune P.M., Salmon A., Parisod C., Grandbastien M.A., Fukunaga K., Ricou M., Misset M.T. Hybridization, polyploidy and invasion: lessons from Spartina (Poaceae) Biol. Inv. 2009;11:1159–1173. [Google Scholar]
- Alix K., Gerard P.R., Schwarzacher T., Heslop-Harrison J.S.P. Polyploidy and interspecific hybridization: partners for adaptation, speciation and evolution in plants. Ann. Bot. 2017;120:183–194. doi: 10.1093/aob/mcx079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. Hybridization of the habitat. Evolution. 1948:1–9. [Google Scholar]
- Arnold M.L. Oxford University Press; Oxford: 1997. Natural Hybridization and Evolution. [Google Scholar]
- Arnold M.L., Hodges S.A. Are natural hybrids fit or unfit relative to their parents? Trends Ecol. Evol. 1995;10:67–71. doi: 10.1016/S0169-5347(00)88979-X. [DOI] [PubMed] [Google Scholar]
- Arnold M.L., Kentner E.K., Johnston J.A., Cornman S., Bouck A.C. Natural hybridisation and fitness. Taxon. 2001;50:93–104. [Google Scholar]
- Ashton P.A., Abbott R.J. Multiple origins and genetic diversity in the newly arisen allopolyploid species, Senecio cambrensis Rosser (Compositae) Heredity. 1992;68:25–32. [Google Scholar]
- Baduel P., Bray S., Vallejo-Marin M., Kolář F., Yant L. The “polyploid hop”: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front. Ecol. Evol. 2018;6:117. [Google Scholar]
- Bates D., Maechler M., Bolker B. 2014. lme4: Linear Mixed-Effects Models Using Eigen and S4. R Package Version 1.1-7.http://CRAN.R-project.org/package=lme4 [Google Scholar]
- Beardsley P.M., Schoenig S.E., Whittall J.B., Olmstead R.G. Patterns of evolution in western North American Mimulus (Phrymaceae) Am. J. Bot. 2004;91:474–489. doi: 10.3732/ajb.91.3.474. [DOI] [PubMed] [Google Scholar]
- Benedict B.G., Modliszewski J.L., Sweigart A.L., Martin N.H., Ganders F.R., Willis J.H. Mimulus sookensis (Phrymaceae), a new allotetraploid species derived from Mimulus guttatus and Mimulus nasutus. Madroño. 2012;59:29–43. [Google Scholar]
- Bomblies K., Higgins J.D., Yant L. Meiosis evolves: adaptation to external and internal environments. New Phytol. 2015;208:306–323. doi: 10.1111/nph.13499. [DOI] [PubMed] [Google Scholar]
- Bomblies K., Madlung A. Polyploidy in the Arabidopsis genus. Chromosome Res. 2014;22:117–134. doi: 10.1007/s10577-014-9416-x. [DOI] [PubMed] [Google Scholar]
- Castro M., Castro S., Loureiro J. Production of synthetic tetraploids as a tool for polyploid research. Web Ecol. 2018;18:129–141. [Google Scholar]
- Cooley A.M., Willis J.H. Genetic divergence causes parallel evolution of flower color in Chilean Mimulus. New Phytol. 2009;183:729–739. doi: 10.1111/j.1469-8137.2009.02858.x. [DOI] [PubMed] [Google Scholar]
- Coughlan J.M., Wilson Brown M., Willis J.H. Patterns of hybrid seed inviability in the Mimulus guttatus sp. complex reveal a potential role of parental conflict in reproductive isolation. Curr. Biol. 2020;30:83–93 e85. doi: 10.1016/j.cub.2019.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J.A., Orr H.A. Sinauer Associates; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Da Re D., Olivares A.P., Smith W., Vallejo-Marín M. Global analysis of ecological niche conservation and niche shift in invasive and hybrid populations of monkeyflowers (Mimulus guttatus, M. luteus and M. × robertsii) Plant Ecol. Div. 2020 doi: 10.1080/17550874.2020.1750721. [DOI] [Google Scholar]
- Dalrymple R.L., Buswell J.M., Moles A.T. Asexual plants change just as often and just as fast as do sexual plants when introduced to a new range. Oikos. 2015;124:196–205. [Google Scholar]
- Dasmahapatra K.K., Walters J.R., Briscoe A.D., Davey J.W., Whibley A., Nadeau N.J., Zimin A.V., Hughes D.S., Ferguson L.C., Martin S.H. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487:94. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N., Zamariola L., Mau M., Sharbel T.F., Geelen D. Volume-based pollen size analysis: an advanced method to assess somatic and gametophytic ploidy in flowering plants. Plant Reprod. 2013;26:65–81. doi: 10.1007/s00497-012-0209-0. [DOI] [PubMed] [Google Scholar]
- Dermen H. Colchicine polyploidy and technique. Biol. Rev. 1940;6:599–635. [Google Scholar]
- Dobzhansky T.G. Columbia University Press; New York: 1937. Genetics and the Origin of Species. [Google Scholar]
- Doležel J., Greilhuber J., Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007;2:2233. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Coate J.E. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 2019;180:1–52. [Google Scholar]
- Edger P.P., Smith R., McKain M.R., Cooley A.M., Vallejo-Marin M., Yuan Y., Bewick A.J., Ji L., Platts A.E., Bowman M.J. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017;29:2150–2167. doi: 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgvin T.O., Trier C.N., Torresen O.K., Hagen I.J., Lien S., Nederbragt A.J., Ravinet M., Jensen H., Saetre G.P. The genomic mosaicism of hybrid speciation. Sci. Adv. 2017;3:e1602996. doi: 10.1126/sciadv.1602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng W.H., Ho W.S. Polyploidization using colchicine in horticultural plants: a review. Sci. Hort. 2019;246:604–617. [Google Scholar]
- Goldschmidt R. Some aspects of evolution. Science. 1933;78:539–547. doi: 10.1126/science.78.2033.539. [DOI] [PubMed] [Google Scholar]
- Grant A.L. A monograph of the genus Mimulus. Ann. Miss. Bot. Gard. 1924;11:99–380. [Google Scholar]
- Grant P.R., Grant B.R. Hybridization increases population variation during adaptive radiation. Proc. Natl. Acad. Sci. U S A. 2019;116:23216–23224. doi: 10.1073/pnas.1913534116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. The origin of a new species of Gilia in a hybridization experiment. Genetics. 1966;54:1189–1199. doi: 10.1093/genetics/54.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V. Columbia University Press; New York: 1971. Plant Speciation. [Google Scholar]
- Hegarty M., Coate J., Sherman-Broyles S., Abbott R., Hiscock S., Doyle J. Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 2013;140:204–225. doi: 10.1159/000353361. [DOI] [PubMed] [Google Scholar]
- Hegarty M.J., Abbott R.J., Hiscock S.J. Allopolyploid speciation in action: the origins and evolution of Senecio cambrensis. In: Soltis P.S., Soltis D.E., editors. Polyploidy and Genome Evolution. Springer-Verlag; Berlin: 2012. pp. 245–270. [Google Scholar]
- Hegarty M.J., Barker G.L., Wilson I.D., Abbott R.J., Edwards K.J., Hiscock S.J. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- Hegarty M.J., Hiscock S.J. Hybrid speciation in plants: new insights from molecular studies. New Phytol. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Hothorn T., Bretz F., Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- Husband B.C. The role of triploid hybrids in the evolutionary dynamics of mixed-ploidy populations. Biol. J. Linn. Soc. 2004;82:537–546. [Google Scholar]
- Husband B.C., Ozimec B., Martin S.L., Pollock L. Mating consequences of polyploid evolution in flowering plants: Current trends and insights from synthetic polyploids. Int. J. Plant Sci. 2008;169:195–206. [Google Scholar]
- Husband B.C., Sabara H.A. Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae) New Phytol. 2004;161:703–713. doi: 10.1046/j.1469-8137.2004.00998.x. [DOI] [PubMed] [Google Scholar]
- Husband B.C., Schemske D.W. Ecological mechanisms of reproductive isolation between diploid and tetraploid Chamerion angustifolium. J. Ecol. 2000;88:689–701. [Google Scholar]
- Jenczewski E. Evolution: he who grabs too much loses all. Curr. Biol. 2013;23:R961–R963. doi: 10.1016/j.cub.2013.09.023. [DOI] [PubMed] [Google Scholar]
- Kelly J.K., Rasch A., Kalisz S. A method to estimate pollen viability from pollen size variation. Am. J. Bot. 2002;89:1021–1023. doi: 10.3732/ajb.89.6.1021. [DOI] [PubMed] [Google Scholar]
- Kenney A.M., Sweigart A.L. Reproductive isolation and introgression between sympatric Mimulus species. Mol. Ecol. 2016;25:2499–2517. doi: 10.1111/mec.13630. [DOI] [PubMed] [Google Scholar]
- Kerwin R.E., Sweigart A.L. Rampant misexpression in a Mimulus (monkeyflower) introgression line caused by hybrid sterility, not regulatory divergence. Mol. Biol. Evol. 2020;37:2084–2098. doi: 10.1093/molbev/msaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinser T.J., Smith R.D., Lawrence A.H., Cooley A.M., Vallejo-Marín M., Smith G.C., Puzey J.R. Mechanisms driving endosperm-based hybrid incompatibilities: insights from hybrid monkeyflowers. bioRxiv. 2018 doi: 10.1101/461939. [DOI] [Google Scholar]
- Kohler C., Mittelsten Scheid O., Erilova A. The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet. 2010;26:142–148. doi: 10.1016/j.tig.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Lafon-Placette C., Kohler C. Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 2014;17:64–69. doi: 10.1016/j.pbi.2013.11.008. [DOI] [PubMed] [Google Scholar]
- Leitch I.J., Hanson L., Lim K.Y., Kovarik A., Chase M.W., Clarkson J.J., Leitch A.R. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Ann. Bot. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D.A. Polyploidy and novelty in flowering plants. Am. Nat. 1983;122:1–25. [Google Scholar]
- Levin D.A. Oxford University Press; Oxford: 2002. The Role of Chromosomal Change in Plant Evolution. [Google Scholar]
- Liaw A., Wiener M. Classification and regression by randomForest. R. News. 2002;2:18–22. [Google Scholar]
- Liston A., Wei N., Tennessen J.A., Li J., Dong M., Ashman T.L. Revisiting the origin of octoploid strawberry. Nat. Genet. 2020;52:2–4. doi: 10.1038/s41588-019-0543-3. [DOI] [PubMed] [Google Scholar]
- Loureiro J., Rodriguez E., Dolezel J., Santos C. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann. Bot. 2007;100:875–888. doi: 10.1093/aob/mcm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity. 2013;110:99–104. doi: 10.1038/hdy.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H., Walden A.E., Husband B.C. Genome duplication and the evolution of physiological responses to water stress. New Phytol. 2009;184:721–731. doi: 10.1111/j.1469-8137.2009.02997.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Mallet J., Besansky N., Hahn M.W. How reticulated are species? BioEssays. 2016;38:140–149. doi: 10.1002/bies.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandakova T., Kovarik A., Zozomova-Lihova J., Shimizu-Inatsugi R., Shimizu K.K., Mummenhoff K., Marhold K., Lysak M.A. The more the merrier: recent hybridization and polyploidy in cardamine. Plant Cell. 2013;25:3280–3295. doi: 10.1105/tpc.113.114405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburger S., Monnahan P., Seear P.J., Martin S.H., Koch J., Paajanen P., Bohutinska M., Higgins J.D., Schmickl R., Yant L. Interspecific introgression mediates adaptation to whole genome duplication. Nat. Commun. 2019;10:5218. doi: 10.1038/s41467-019-13159-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N.H., Willis J.H. Ecological divergence associated with mating system causes nearly complete reproductive isolation between sympatric Mimulus species. Evolution. 2007;61:68–82. doi: 10.1111/j.1558-5646.2007.00006.x. [DOI] [PubMed] [Google Scholar]
- Mason A.S., Pires J.C. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 2015;31:5–10. doi: 10.1016/j.tig.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Mavrodiev E.V., Chester M., Suarez-Santiago V.N., Visger C.J., Rodriguez R., Susanna A., Baldini R.M., Soltis P.S., Soltis D.E. Multiple origins and chromosomal novelty in the allotetraploid Tragopogon castellanus (Asteraceae) New Phytol. 2015;206:1172–1183. doi: 10.1111/nph.13227. [DOI] [PubMed] [Google Scholar]
- Medel R., Botto-Mahan C., Kalin-Arroyo M. Pollinator-mediated selection on the nectar guide phenotype in the Andean monkey flower, Mimulus luteus. Ecology. 2003;84:1721–1732. [Google Scholar]
- Mitchell N., Owens G.L., Hovick S.M., Rieseberg L.H., Whitney K.D. Hybridization speeds adaptive evolution in an eight-year field experiment. Sci. Rep. 2019;9:6746. doi: 10.1038/s41598-019-43119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B.B., Vickery R.K. Chromosome counts in the section Simiolus of the genus Mimulus (Scrophulariaceae). V. The chromosomal homologies of M. guttatus and its allied species and varieties. Madroño. 1962;16:141–172. [Google Scholar]
- Munzbergova Z. Colchicine application significantly affects plant performance in the second generation of synthetic polyploids and its effects vary between populations. Ann. Bot. 2017;120:329–339. doi: 10.1093/aob/mcx070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesom G.L. Taxonomy of Erythranthe Sect. Simiola (Phrymaceae) in the USA and Mexico. Phytoneuron. 2012;40:1–123. [Google Scholar]
- Otto S.P. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Otto S.P., Whitton J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Pan J.J., Price J.S. Fitness and evolution in clonal plants: the impact of clonal growth. Evol. Ecol. 2001;15:583–600. [Google Scholar]
- Pantoja P.O., Simón-Porcar V.I., Puzey J.R., Vallejo-Marín M. Genetic variation and clonal diversity in introduced populations of Mimulus guttatus assessed by genotyping at 62 single nucleotide polymorphism loci. Plant Ecol. Div. 2017;10:5–15. [Google Scholar]
- Porturas L.D., Anneberg T.J., Curé A.E., Wang S., Althoff D.M., Segraves K.A. A meta-analysis of whole genome duplication and the effects on flowering traits in plants. Am. J. Bot. 2019;106:469–476. doi: 10.1002/ajb2.1258. [DOI] [PubMed] [Google Scholar]
- Preston C.D., Pearman D.A., Dines T.D. Oxford University Press; Oxford: 2002. New Atlas of the British and Irish Flora. [Google Scholar]
- Ramsey J. Unreduced gametes and neopolyploids in natural populations of Achillea borealis (Asteraceae) Heredity. 2007;98:143–150. doi: 10.1038/sj.hdy.6800912. [DOI] [PubMed] [Google Scholar]
- Ramsey J. Polyploidy and ecological adaptation in wild yarrow. Proc. Natl. Acad. Sci. U S A. 2011;108:7096–7101. doi: 10.1073/pnas.1016631108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J., Schemske D.W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Ann. Rev. Ecol. Syst. 1998;29:467–501. [Google Scholar]
- Ramsey J., Schemske D.W. Neopolyploidy in flowering plants. Ann. Rev. Ecol. Syst. 2002;33:589–639. [Google Scholar]
- Renny-Byfield S., Gong L., Gallagher J.P., Wendel J.F. Persistence of subgenomes in paleopolyploid cotton after 60 my of evolution. Mol. Biol. Evol. 2015;32:1063–1071. doi: 10.1093/molbev/msv001. [DOI] [PubMed] [Google Scholar]
- Rieseberg L.H. The role of hybridization in evolution: old wine in new skins. Am. J. Bot. 1995;82:944–953. [Google Scholar]
- Rieseberg L.H., Carney S.E. Plant hybridization. New Phytol. 1998;140:599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- Runemark A., Vallejo-Marin M., Meier J.I. Eukaryote hybrid genomes. Plos Genet. 2019;15:e1008404. doi: 10.1371/journal.pgen.1008404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk K., Brede N., Streit B. Introduction. Extent, processes and evolutionary impact of interspecific hybridization in animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:2805–2811. doi: 10.1098/rstb.2008.0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol. Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Silverside A.J. A new binomial in Mimulus. Watsonia. 1990;18:210–212. [Google Scholar]
- Simón-Porcar V.I., Silva J.L., Meeus S., Higgins J.D., Vallejo-Marín M. Recent autopolyploidization in a naturalized population of Mimulus guttatus (Phrymaceae) Bot. J. Linn. Soc. 2017;185:189–207. [Google Scholar]
- Snodgrass S.J., Jareczek J., Wendel J.F. An examination of nucleotypic effects in diploid and polyploid cotton. AoB Plants. 2017;9 doi: 10.1093/aobpla/plw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S. Allopolyploid speciation in Tragopogon - insights from chloroplast DNA. Am. J. Bot. 1989;76:1119–1124. [Google Scholar]
- Soltis D.E., Visger C.J., Marchant D.B., Soltis P.S. Polyploidy: Pitfalls and paths to a paradigm. Am. J. Bot. 2016;103:1146–1166. doi: 10.3732/ajb.1500501. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Liu X., Marchant D.B., Visger C.J., Soltis D.E. Polyploidy and novelty: Gottlieb's legacy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2013.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P.S., Marchant D.B., Van de Peer Y., Soltis D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015;35:119–125. doi: 10.1016/j.gde.2015.11.003. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. Multiple origins of the allotetraploid Tragopogon mirus (Compositae): rDNA Evidence. Syst. Bot. 1991;16:407–413. [Google Scholar]
- Soltis P.S., Soltis D.E. The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Sonnleitner M., Weis B., Flatscher R., Garcia P.E., Suda J., Krejcikova J., Schneeweiss G.M., Winkler M., Schonswetter P., Hulber K. Parental ploidy strongly affects offspring fitness in heteroploid crosses among three cytotypes of autopolyploid Jacobaea carniolica (Asteraceae) PLoS One. 2013;8:e78959. doi: 10.1371/journal.pone.0078959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stace C.A. Cambridge University Press; Cambridge: 2010. New Flora of the British Isles. [Google Scholar]
- Stace C.A., Crawley M.J. William Collins; London: 2015. Alien Plants. [Google Scholar]
- Stace C.A., Preston C.D., Pearman D.A. Botanical Society of Britain and Ireland; Bristol: 2015. Hybrid Flora of the British Isles. [Google Scholar]
- Stanton K., Valentin C.M., Wijnen M.E., Stutstman S., Palacios J.J., Cooley A.M. Absence of postmating barriers between a selfing versus outcrossing Chilean Mimulus species pair. Am. J. Bot. 2016;103:1030–1040. doi: 10.3732/ajb.1600079. [DOI] [PubMed] [Google Scholar]
- Stebbins G.L. The role of hybridization in evolution. Proc. Am. Phil. Soc. 1959;103:231–251. [Google Scholar]
- Stebbins G.L. Edward Arnold; London: 1971. Chromosomal Evolution in Higher Plants. [Google Scholar]
- Stebbins G.L. Polyploidy, hybridization, and the invasion of new habitats. Ann. Miss. Bot. Gard. 1985;72:824–832. [Google Scholar]
- Suarez-Gonzalez A., Lexer C., Cronk Q.C.B. Adaptive introgression: a plant perspective. Biol. Lett. 2018;14:20170688. doi: 10.1098/rsbl.2017.0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B.L., Galloway L.F. Postzygotic isolation varies by ploidy level within a polyploid complex. New Phytol. 2017;213:404–412. doi: 10.1111/nph.14116. [DOI] [PubMed] [Google Scholar]
- Sweigart A.L., Fishman L., Willis J.H. A simple genetic incompatibility causes hybrid male sterility in Mimulus. Genetics. 2006;172:2465–2479. doi: 10.1534/genetics.105.053686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J.A., Symonds V.V., Doust A.N., Buggs R.J., Mavrodiev E., Majure L.C., Soltis P.S., Soltis D.E. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 Years after Ownbey's discovery. Am. J. Bot. 2009;96:979–988. doi: 10.3732/ajb.0800299. [DOI] [PubMed] [Google Scholar]
- Taylor S.A., Larson E.L. Insights from genomes into the evolutionary importance and prevalence of hybridization in nature. Nat. Ecol. Evol. 2019;3:170–177. doi: 10.1038/s41559-018-0777-y. [DOI] [PubMed] [Google Scholar]
- Temsch E.M., Greilhuber J., Krisai R. Genome size in liverworts. Preslia. 2010;82:63–80. [Google Scholar]
- Vallejo-Marin M. Mimulus peregrinus (Phrymaceae): a new British allopolyploid species. PhytoKeys. 2012;14:1–14. doi: 10.3897/phytokeys.14.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marin M., Buggs R.J.A., Cooley A.M., Puzey J.R. Speciation by genome duplication: repeated origins and genomic composition of the recently formed allopolyploid species Mimulus peregrinus. Evolution. 2015;69:1487–1500. doi: 10.1111/evo.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marin M., Cooley A.M., Lee M.Y., Folmer M., McKain M.R., Puzey J.R. Strongly asymmetric hybridization barriers shape the origin of a new polyploid species and its hybrid ancestor. Am. J. Bot. 2016;103:1272–1288. doi: 10.3732/ajb.1500471. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marin M., Hiscock S.J. Hybridization and hybrid speciation under global change. New Phytol. 2016;211:1170–1187. doi: 10.1111/nph.14004. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marin M., Lye G.C. Hybridisation and genetic diversity in introduced Mimulus (Phrymaceae) Heredity. 2013;110:111–122. doi: 10.1038/hdy.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo-Marín M., Quenu M., Ritchie S., Meeus S. Partial interfertility between independently originated populations of the neo-allopolyploid Mimulus peregrinus. Plant Syst. Evol. 2017;303:1081–1092. [Google Scholar]
- Van de Peer Y., Mizrachi E., Marchal K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017;18:411–424. doi: 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Van Drunen W.E., Husband B.C. Immediate versus evolutionary consequences of polyploidy on clonal reproduction in an autopolyploid plant. Ann. Bot. 2018;122:195–205. doi: 10.1093/aob/mcy071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickery R.K. Barriers to gene exchange within Mimulus guttatus (Scrophulariaceae) Evolution. 1959;13:300–310. [Google Scholar]
- Vickery R.K. Barriers to gene exchange between members of Mimulus guttatus complex ( Scrophulariaceae ) Evolution. 1964;18:52. [Google Scholar]
- Vickery R.K. Springer; New York: 1978. Case Studies in the Evolution of Species Complexes in Mimulus. [Google Scholar]
- Vickery R.K. Speciation by aneuploidy and polyploidy in Mimulus (Scrophulariaceae) Great Basin Nat. 1995;55:174–176. [Google Scholar]
- Vickery R.K., Anderson D.G. Experimental hybridizations in the genus Mimulus. VI. Section Erythranthe. Proc. Utah Acad. Sci. 1967;44:321–333. [Google Scholar]
- Vickery R.K., Crook K., Lindsay D., Mia M., Tai W. Chromosome counts in section Simiolus of the genus Mimulus (Scrophulariaceae). VII. New numbers for M. guttatus, M. cupreus, and M. tilingii. Madroño. 1968;19:211–218. [Google Scholar]
- Vickery R.K., Mia M. Experimental hybridizations in the genus Mimulus (Scrophulariaceae). III. Utah Acad. Proc. 1967;43:105–114. [Google Scholar]
- Vickery R.K., Mukherjee B.B. Experimental hybridizations in the genus Mimulus. I. Proc. Utah Acad. Sci. 1966;43:83–113. [Google Scholar]
- Wei N., Du Z., Liston A., Ashman T.L. Genome duplication effects on functional traits and fitness are genetic context and species dependent: studies of synthetic polyploid Fragaria. Am. J. Bot. 2020;107:262–272. doi: 10.1002/ajb2.1377. [DOI] [PubMed] [Google Scholar]
- Wyatt R., Odrzykoski I.J., Stoneburner A., Bass H.W., Galau G.A. Allopolyploidy in bryophytes: multiple origins of Plagiomnium medium. Proc. Natl. Acad. Sci. U S A. 1988;85:5601–5604. doi: 10.1073/pnas.85.15.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.