Abstract
Since its discovery as a bacterial adaptive immune system and its development for genome editing in eukaryotes, the CRISPR technology has revolutionized plant research and precision crop breeding. The CRISPR toolbox holds great promise in the production of crops with genetic disease resistance to increase agriculture resilience and reduce chemical crop protection with a strong impact on the environment and public health. In this review, we provide an extensive overview on recent breakthroughs in CRISPR technology, including the newly developed prime editing system that allows precision gene editing in plants. We present how each CRISPR tool can be selected for optimal use in accordance with its specific strengths and limitations, and illustrate how the CRISPR toolbox can foster the development of genetically pathogen-resistant crops for sustainable agriculture.
Key words: CRISPR-Cas9, gene targeting, base editing, prime editing, plant/pathogen interactions, precision crop breeding
The CRISPR-mediated precision breeding toolbox allows researchers and molecular breeders to fine-tune plant genomes with high versatility. Application of these genome-editing tools to genes involved in plant/pathogen interactions can foster the development of sustainable agriculture through the production of genetically pathogen-resistant crops.
Introduction
Primary food production across the globe faces the challenge of sustainably feeding a growing population in an accelerating climate change context, while more than 800 million people suffered from undernourishment worldwide in 2016, particularly in Africa and Asia (http://www.fao.org/state-of-food-security-nutrition/2017/en/). Current agricultural practices mostly rely on the cultivation of a narrow range of plant species, sometimes in poorly suited locations, far away from their area of domestication (Fernie and Yan, 2019). Labor-intensive and time-consuming conventional crop breeding relying on a natural or an induced genetic polymorphism has substantially contributed to plant adaptation to new environments and food availability. Recently, the development of genome engineering in plants has opened new avenues for precision crop breeding, including the improvement of the elite germplasm as well as the molecular domestication of wild species (Zhang et al., 2019).
Plant pathogens, including bacteria, fungi, and viruses, cause substantial economic losses and threaten food security (Savary et al., 2019). Pathogens rely on diverse strategies to bypass plant immunity. For instance, they produce molecular weapons called effectors that act inside or outside of plant cells to target diverse host proteins involved in different cellular processes to promote infection through the successful colonization of the host.
Plants rely on a sophisticated immune system to ward off potential pathogens. Key elements are an arsenal of receptors termed invasion pattern receptors that recognize either microbe- or host-derived signals termed invasion patterns that betray the presence of microbial invaders (Cook et al., 2015). Invasion pattern receptors belong to two main classes: cell surface receptors that are either receptor-like proteins or receptor-like kinases (RLKs) and intracellular receptors that belong to the class of nucleotide-binding leucine-rich repeat domain proteins (NLRs). While NLRs specifically recognize intracellular effectors (Cesari, 2018; Kourelis and van der Hoorn, 2018), receptor-like proteins and RLKs perceive microbe-associated molecular patterns and extracellular effectors originating from the pathogen and damage-associated molecular patterns released by host cells damaged upon pathogen attack (Boutrot and Zipfel, 2017; Kanyuka and Rudd, 2019).
The vast majority of disease resistance (R) genes cloned from plants code for immune receptors (Kourelis and van der Hoorn, 2018) with NLRs being the dominating class. Another successful strategy to confer plant disease resistance relies on a loss of compatibility through mutations of plant susceptibility (S) genes required for pathogen infection and plant susceptibility. As a result, the pathogen will not be able to perform its infectious cycle, resulting in plant disease resistance (van Schie and Takken, 2014).
While conventional resistance breeding can be very successful, it may be associated with linkage drag and the resistance conferred by single R genes may be rapidly bypassed by fast-evolving pathogens. Therefore, the precise engineering of R and S genes constitutes an exciting route for the development of genetically resistant crops (Langner et al., 2018; Tamborski and Krasileva, 2020; van Wersch et al., 2020), thereby limiting the environmental impact of chemical control. Copying mutations across accessions can also circumvent linkage drags associated with classical breeding, as shown for other characters (Li et al., 2017a).
In the last few years, genome-editing tools have evolved very quickly with the development of RNA-guided endonuclease systems (Zhang et al., 2019). Until now, genome editing has been mostly used to generate loss-of-function alleles through DNA error-prone repair of the target site after double-strand cleavage by the classical CRISPR-Cas9 system. For example, this strategy resulted in a powdery mildew-resistant tomato by knocking out the mildew-resistant locus O (Mlo) S gene (Nekrasov et al., 2017), while rice blast resistance increased due to the loss of function of the transcription factor OsERF922 (Wang et al., 2016). However, many traits can be conferred by single or multiple nucleotide substitutions, especially for genes involved in plant/pathogen interactions, where co-evolution exerts a dual selective pressure that favors mutations of pathogen effectors to evade recognition, but also mutations of immune receptors to restore perception (Jones et al., 2016). Therefore, genome-editing tools mediating precise and predictable mutations are highly valuable for the production of gain-of-function mutants, which could lead to a broader perception of the pathogen and/or evasion of the host factor from effectors. Of particular interest is the CRISPR-mediated mimicking of natural alleles conferring pathogen resistance (Bastet et al., 2017), as well as directed in planta evolution to generate new gene variants that are not present in the natural genetic diversity. In the course of this review, we refer to the targeted genome alterations, such as nucleotide changes and small deletions, as precision breeding. This process can involve GM techniques but the resulting plant is devoid of the transgene (Andersen et al., 2015).
In this review, we mostly focus on recent advances in CRISPR technologies used to introduce targeted point mutations in plant genes, including the new “search-and-replace” prime editing technology. We show how the multiple adjustments that have been developed to expand the targeting scope, precision, and efficiency of these CRISPR tools offer complementary strengths and weaknesses that can be mobilized according to specific desired outcomes. The fast adoption and improvement of these precise and versatile genome-editing tools in plants open up new avenues for biotechnology and the development of sustainable agriculture, especially through the development of new genetically resistant crops.
The Basic Machinery for Plant Genome Editing
In the frame of this review, we focus on genome-editing strategies, i.e., approaches that will lead to stable modifications in plant genomic DNA, and result in transgene-free plants through different delivery strategies that have been extensively described in recent reviews (Chen et al., 2019; Kuluev et al., 2019; El-Mounadi et al., 2020). Therefore, we will not cover another important aspect of CRISPR that involves the use of a nuclease that targets RNA for modification such as Cas13. Additional details on this strategy can be found in recent reviews (Wolter and Puchta, 2018; Burmistrz et al., 2020).
The CRISPR-Cas9 System
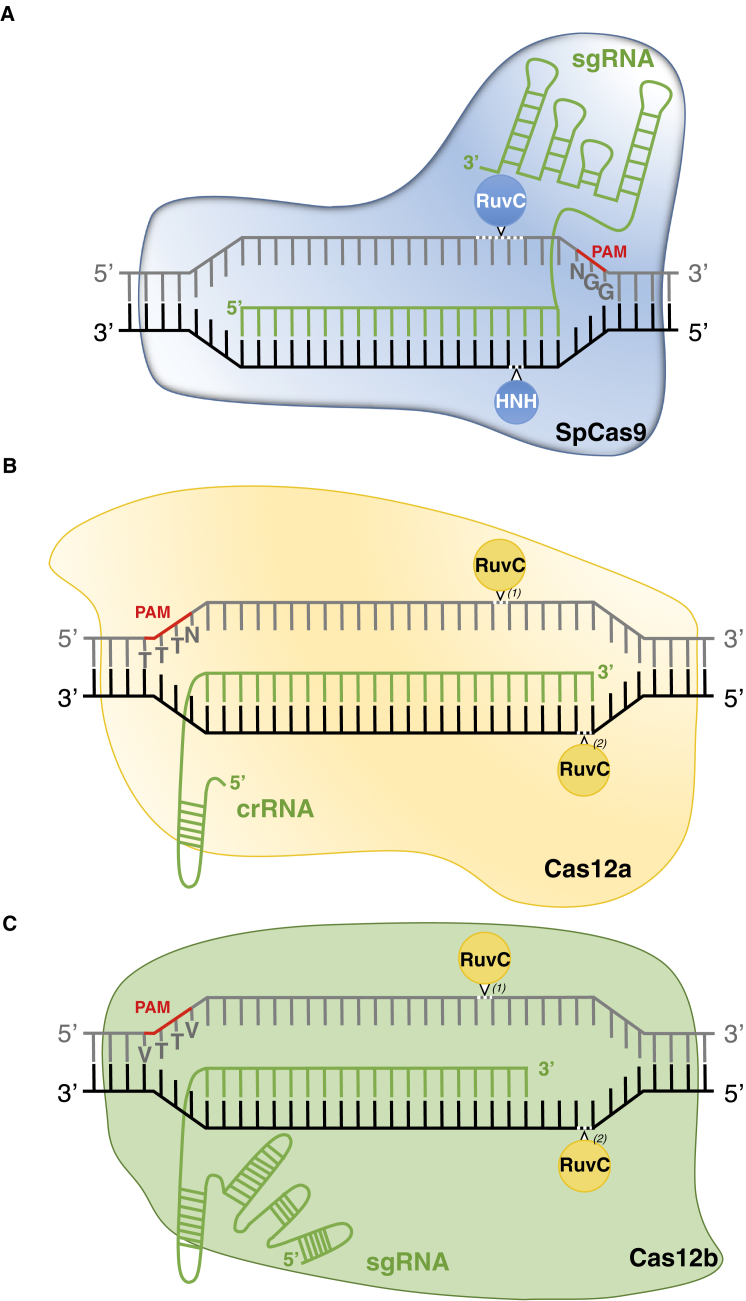
The leading CRISPR-SpCas9 system for genome editing, initially derived from a class 2 type II Streptococcus pyogenes adaptive immune system, consists of a two-component complex made up of the DNA endonuclease SpCas9 (1368 amino acids) and a customizable single-guide RNA (sgRNA) created by the artificial fusion of a crRNA and a trans-activating crRNA (Jinek et al., 2012). The sgRNA is composed of an ∼80-bp scaffold that mediates binding to the Cas9 and a customizable 20-bp sequence at its 5′ end called the spacer sequence that confers DNA targeting specificity to the complex (Figure 1A). Binding of the sgRNA to the SpCas9 triggers the transition of the nuclease from an inactive to a DNA-probing state in search of a canonical 5′-NGG-3′ protospacer adjacent motif (PAM). Natural and engineered Cas9 variants recognizing alternative PAMs have also been extensively used (Zhang et al., 2019). Recognition of a suitable PAM motif leads to quick interrogation of adjacent DNA, followed by local DNA melting and RNA strand invasion (formation of an R-loop structure) for interrogation of the full spacer sequence (Figure 1A). Perfect base pairing between the so-called seed region (10–12 nucleotides from the PAM) of the spacer sequence and target DNA is required for SpCas9-mediated DNA cleavage, while mismatches in the non-seed region can be tolerated, potentially leading to unwanted off-target activity. While the careful design of spacer sequences is generally considered to be sufficient to avoid off-target activity, several Cas9 variants displaying higher specificity have been developed through protein engineering (Zhang et al., 2019). The gradual base pairing triggers SpCas9 conformational changes to an active site, eventually resulting in DNA cleavage by the concerted activity of its HNH and RuvC nuclease domains (Figure 1A). While SpCas9 was thought to only create blunt-ended double-strand DNA breaks (DSBs) about 3-bp upstream of the PAM (Jiang and Doudna, 2017), recent findings demonstrated that SpCas9 nuclease activity results in both blunt and staggered ends, likely because of the RuvC cutting flexibility (Molla and Yang, 2020). The CRISPR-SpCas9 system is now routinely used in numerous species and is considered as the golden tool for genome editing in plants (Manghwar et al., 2019).
Figure 1.
CRISPR-Cas Systems Used for Genome Editing in Plants.
(A) The CRISPR-SpCas9 system comprised of the endonuclease SpCas9, harboring RuvC and HNH catalytic domains, and the sgRNA that guides the complex to an endogenous target sequence upstream of a G-rich PAM (5′-NGG-3′), leading to blunt and/or staggered DNA breaks.
(B) The CRISPR-Cas12a system involves the endonuclease Cas12a that is guided to the target locus, downstream of a T-rich PAM (5′-TTTN-3′), by a short crRNA, leading to a staggered DNA cleavage by a single RuvC domain after conformational changes: (1) and (2).
(C) The CRISPR-Cas12b system relies on a Cas12b endonuclease, harboring a single RuvC catalytic domain that mediates staggered DNA cleavage—(1) and (2)—and an sgRNA that targets the complex to a specific site downstream of a T-rich PAM (5′-VTTV-3′). The schemes are not at scale and are for illustrative purposes only.
The CRISPR-Cas12 Systems
The second leading genome-editing tool, the class 2 type V-A CRISPR-Cas12a system also known as CRISPR-Cpf1, displays unique features and constitutes a relevant alternative to the CRISPR-Cas9 system (Zetsche et al., 2015). Cas12a enzymes (1200–1500 amino acids) mostly recognize T-rich 5′-TTTN-3′ PAMs located upstream of the target sequence. They associate with a short ∼43-bp crRNA and only rely on the RuvC-like domain to cleave both DNA strands in a sequential manner, beginning with the non-target strand and resulting in a cleaved DNA with 4–5-bp overhangs distal to the PAM (Figure 1B) (Zetsche et al., 2015; Zaidi et al., 2017; Alok et al., 2020). Cas12a orthologs from Lachnospiraceae bacterium (LbCas12a), Acidaminococcus sp. (AsCas12a), and Francisella novicida (FnCas12a) have been the most commonly used enzymes in several plant species. They generally display higher specificity and less or no off-targets compared with Cas9 (Endo et al., 2016; Begemann et al., 2017; Kim et al., 2017; Tang et al., 2017, 2018, 2019; Xu et al., 2017, 2019a; Yin et al., 2017; Li et al., 2019a; Lee et al., 2019; Herbert et al., 2020).
The recently established class 2 type V-B CRISPR-Cas12b system uses a smaller Cas12b nuclease (∼1100 amino acids) than CRISPR-SpCas9 and CRISPR-Cas12a systems. Similar to Cas12a, Cas12b prefers T-rich PAMs and induces RuvC-mediated DSBs with staggered ends distal to the PAM (Figure 1C) (Shmakov et al., 2015; Yang et al., 2016). The Cas12b ortholog from Alicyclobacillus acidiphilus (AaCas12b), initially characterized as a high-specificity nuclease with a high optimal temperature in mammalian cells (Teng et al., 2018), was reported to be efficient for rice genome engineering, with a 5′-VTTV-3′ PAM preference (V = A, C, or G) (Ming et al., 2020). In addition, Alicyclobacillus acidoterrestris (AacCas12b) was also successfully used for genome editing in tetraploid cotton plants, displaying an optimal editing efficiency at 45°C and undetectable off-target activity (Wang et al., 2020b). Although promising, further studies are still required to properly assess the strengths and weaknesses associated with Cas12b compared with Cas9 and Cas12a enzymes for genome editing in plants.
Evolving CRISPR-Cas Systems: Going Beyond the Gene Knockout
The three aforementioned CRISPR-Cas systems constitute the base for diversified genome-editing tools. So far, most genome-editing applications in plants have focused on the production of knockout mutants for single or multiple genes (Manghwar et al., 2019). This is due to the predominance of error-prone non-homologous end-joining (NHEJ) mechanisms to repair CRISPR-Cas-mediated DSBs in somatic cells of higher plants (Puchta, 2005). Contrary to homologous recombination (HR), an endogenous DNA repair mechanism that is responsible for crossovers between homologous chromosomes during meiosis, NHEJ mechanisms mediate DSB repair without the need for a homologous template. While the classical NHEJ pathway appears to be mainly error-free, the alternative NHEJ mechanism seems to have a key role in error-prone CRISPR-induced DSB repair (Mara et al., 2019; Atkins and Voytas, 2020). The unfaithful DNA repair eventually creates random small insertion or deletion mutations (indels) at the cleavage site, typically causing frameshift mutations that result in loss-of-function alleles when located in coding sequences. In promoter regions, targeted deletions affecting cis-regulatory elements can result in altered transcriptional regulation.
An interesting feature of the CRISPR-Cas9 system is that the cutting function can be uncoupled from the target recognition. This opens room for repurposing the system and carrying enzymatic domains to a specific locus. Indeed, the inactivation of either the RuvC or HNH catalytic domain by the D10A or H840A substitution produces nickase Cas9 (nCas9), which is only able to cut the targeting and non-targeting strands, respectively, while the introduction of both mutations generates dead Cas9 (dCas9). Similarly, dCas12a and dCas12b enzymes are also available, but nCas12 proteins have yet to be reported. However, the fact that DNA cleavage of Cas12 enzymes is sequentially mediated by a single RuvC-like nuclease domain may prevent the development of nCas12. These impaired Cas proteins keep their DNA-binding properties, thereby allowing targeted applications, such as epigenome editing or transcriptional regulation through the recruitment of the DNA methylation machinery or transcriptional regulators, respectively (Zhang et al., 2019; Gallego-Bartolome, 2020). Of particular interest is the possibility to bring enzymatic domains that specifically replace nucleotides in genomic sequences, thereby directly editing the sequences of genes. In the subsequent sections, we mostly focus on recently developed CRISPR systems that support precise and predictable targeted DNA mutations to confer new traits.
Precision Editing: Refining the Tools?
As many agronomic traits are controlled by single base polymorphisms (Henikoff and Comai, 2003), the introduction of precise base substitutions and/or predictable insertions or deletions could generate plants with new agronomic properties. For example, the targeted substitution of nucleotide(s) could introduce non-synonymous mutations that cause amino acid changes in the encoded protein. Besides, the substitution of nucleotide(s) can broadly affect the gene by creating or correcting premature stop codons or regulating splicing. In the subsequent sections, we summarize current CRISPR tools for precision editing.
CRISPR-Mediated Gene Correction through NHEJ
While NHEJ-mediated DSB repair upon Cas9 cleavage is considered to result in random mutations, it is becoming increasingly obvious that a fraction of Cas9-induced DSB repair outcomes are predictable. User-friendly web tools with machine-learning algorithms have been recently developed to predict repair outcomes in human cells, thereby allowing the selection of suitable guides for the introduction of predictable mutations through NHEJ (Molla and Yang, 2020). The development of such tools in plants would be of great interest, with the possibility of anticipating NHEJ-mediated DSB repair outcomes for predictable mutations in coding or regulatory sequences. While the Cas9 nuclease mainly generates small indels (Figure 2A), Cas12a and Cas12b predominantly produce larger deletions (Bernabe-Orts et al., 2019; Herbert et al., 2020). Whether these different mutation footprints are the result of Cas12 cleavage properties and/or due to the binding time of the nuclease to the broken DNA is still unclear (Chen et al., 2018; Que et al., 2019). Regardless of their mechanisms, the cleavage properties of Cas12 enzymes could have specific practical interest compared with Cas9 such as the removal of larger coding or regulatory motifs (Li et al., 2020e; Herbert et al., 2020).
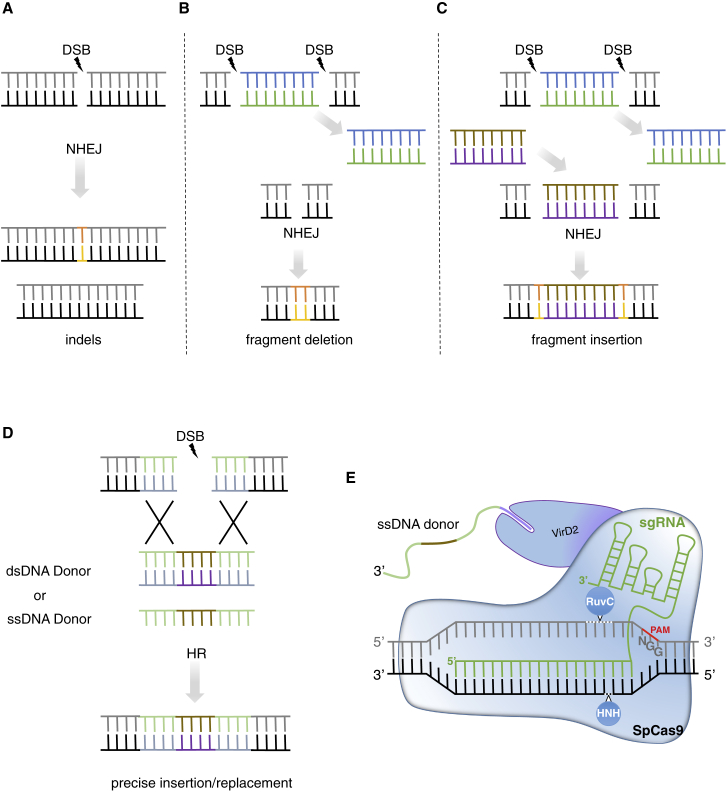
Figure 2.
NHEJ- and HR-Mediated DNA Mutations after CRISPR Cleavage.
(A) CRISPR-mediated gene knockout through the introduction of indel mutations at the cutting site after reparation by the error-prone NHEJ repair mechanism.
(B) CRISPR-mediated fragment deletion after dual sgRNA-induced DSBs, resulting in fragment deletion with associated indels after error-prone NHEJ repair.
(C) CRISPR-mediated fragment replacement after dual sgRNA-induced DSBs, resulting in the replacement of a specific locus by a donor DNA sequence, with associated indels due to error-prone NHEJ repair.
(D) CRISPR-mediated gene targeting (GT) for precise and predictable deletions, insertions, and/or DNA substitutions. HR repair pathway occurs through the introduction of available donor templates (mainly double-stranded RNA [dsDNA] and single-stranded RNA [ssDNA]) harboring homologous sequences with both sides of the CRISPR-induced DSB.
(E) CRISPR–Cas9–VirD2-mediated GT, which repairs the ssDNA template in the vicinity of the cutting site through the interaction between the 5′ specific sequence (purple) of the ssDNA donor template and the VirD2 domain. This spatiotemporal delivery of the repair template may increase the rate of precise repair through the HR pathway.
The schemes are not at scale and are for illustrative purposes only.
With dual sgRNA approaches, larger DNA fragments can be deleted, thereby allowing the removal of complete domains or entire genes (Pauwels et al., 2018) (Figure 2B). The NHEJ-mediated DSB repair approach can also be used for targeted DNA insertion using double-stranded RNA or single-stranded RNA (ssDNA) donors without homologous ends. However, this technique introduces small indels at 5′ and 3′ junctions (Figure 2C) (Wang et al., 2014). This major drawback can be addressed by a strategy where entire exons are replaced by creating DSBs in flanking introns, thereby restricting NHEJ-associated indels to non-coding intron sequences. This NHEJ-mediated exon replacement strategy has been successfully applied to the rice OsEPSPS gene where the introduction of two amino acid changes created glyphosate resistance (Li et al., 2016).
While these NHEJ-mediated editing strategies have proved efficient and reliable in many plant species for gene knockouts, the unpredictable outcomes at the cleavage sites limit their applications for precision editing. This drawback is particularly relevant in vegetatively propagated crops, where desirable or undesirable mutations at the target site cannot be segregated through sexual reproduction. Therefore, the predictable and precise introduction of point mutations or indels through NHEJ-independent pathways is of particular interest.
CRISPR-Mediated Gene Targeting
CRISPR-mediated gene targeting (GT) is a technology relying on HR (Figure 2D) that has been applied for precise nucleotide conversions or precise insertions or deletions in many eukaryotic genomes, including plants. While it is very promising for genome engineering, HR suffers from low efficiency in plant somatic cells (Puchta, 2005), and the delivery of a sufficient amount of donor template in the vicinity of the target site is still challenging, thereby limiting the use of GT in most higher plant species. An illustration of this challenging task is the high number of CRISPR-mediated GT studies that use phenotypic markers, such as herbicide tolerance to facilitate the identification of successful events (Atkins and Voytas, 2020). Nevertheless, a variety of recent improvements substantially enhance GT in plants (Huang and Puchta, 2019). Of particular interest is the use of engineered geminiviral replicon systems, which use the rolling-circle replication to deliver large amounts of the DNA repair template into the plant cell nucleus. The CRISPR-Cas9 GT-geminiviral replicon strategy was successfully applied for large insertions and/or point mutations in tomato, potato, cassava, wheat, and rice (Butler et al., 2016; Cermak et al., 2015; Dahan-Meir et al., 2018; Gil-Humanes et al., 2017; Hummel et al., 2018; Wang et al., 2017). Another interesting approach is the use of Cas12a instead of Cas9 for inducing DSBs. Because Cas12a cuts DNA in the non-seed region distal from the PAM (Figure 1B), allowing multiple rounds of DNA cleavage even after the introduction of NHEJ-mediated indel mutations, and produces sticky ends, HR may be favored (Huang and Puchta, 2019). Consistent with this hypothesis, the CRISPR–Cas12a GT system was successfully applied for targeted insertions or point mutations in rice (Begemann et al., 2017; Li et al., 2018b, 2019c). This system was further improved in tomato using a CRISPR–Cas12a GT–geminiviral multi-replicon strategy, allowing the production of transgene-free salt-tolerant plants due to a single amino acid change (N217D) in the SlHKT1;2 gene (Van Vu et al., 2020). While the geminiviral replicon system allows the delivery of a higher amount of donor template in plants, some improvements for GT are still needed to spatially and temporally bring the CRISPR components and the repair template to the breaking site, as observed in animals (Aird et al., 2018; Savic et al., 2018). This strategy has recently been applied in rice using a fusion between Cas9 and the Agrobacterium VirD2 relaxase (Ali et al., 2020), a key player of ssT-DNA translocation and integration into the plant genome (Gelvin, 2017). The CRISPR–Cas9–VirD2 system facilitated GT likely through the delivery of the ssDNA repair-template in close vicinity to the Cas9-induced DSB (Figure 2E). This enabled the introduction of point mutations in OsALS and OsCCD7 genes to confer herbicide resistance and to engineer plant architecture, respectively, and in-frame insertion of the hemagglutinin epitope at the C terminus of OsHDT (Ali et al., 2020). Together, these recent advances offer new possibilities for precise genome editing, although future improvements to increase the efficiency of CRISPR-mediated GT are still needed for broad and fast adoption in many plant species.
CRISPR-Mediated Base Editing
In contrast to GT-mediated gene correction, CRISPR-mediated base editing is a donor template and DSB-free approach that induces precise base conversion. Cytosine base editors (CBEs) and adenine base editors (ABEs) are fusion proteins made of a catalytically impaired Cas9 and an enzymatic domain mediating cytosine or adenine deamination, respectively. During the formation of the CRISPR-mediated “R-loop” structure, a small window of the non-targeted ssDNA is exposed and can serve as a substrate for deamination (Figure 3A). CBEs catalyze the deamination of cytosine(s) into uracil(s) in the target region. This triggers the base excision repair (BER) pathway that can result in either an error-free or an error-prone repair, thereby leading to a diversification of the edits (C-to-T, C-to-G, and C-to-A), albeit at the cost of indel production at a substantial rate (Figure 3B) (Hess et al., 2017). While varying the edits is interesting for local sequence diversification, predictable targeted base conversions are desirable for precise amino acid changes. The addition of the uracil DNA glycosylase inhibitor (UGI) to the CBE architecture that blocks the BER pathway has been developed as a solution to specifically obtain the C-to-T conversion with generally a low level of by-products (Figure 3C and 3D) (Komor et al., 2017). The deamination of adenine through ABEs (Figure 4A) does not necessitate the use of alkyl adenine DNA glycosylases inhibitors, because BER of inosine intermediates is inefficient for DNA. ABEs therefore create efficient A-to-G conversion with a very low level of by-products (Figure 4B) (Gaudelli et al., 2017). While early BEs harbored dCas9, the incorporation of the edit(s) to the non-deaminated strand was strongly improved by the use of nCas9 with an impaired RuvC domain (D10A), which promoted long-patch BER using the edited strand as a model (Komor et al., 2016). Due to the lack of nCas12, the use of Cas12 enzymes for base-editing applications remains limited.
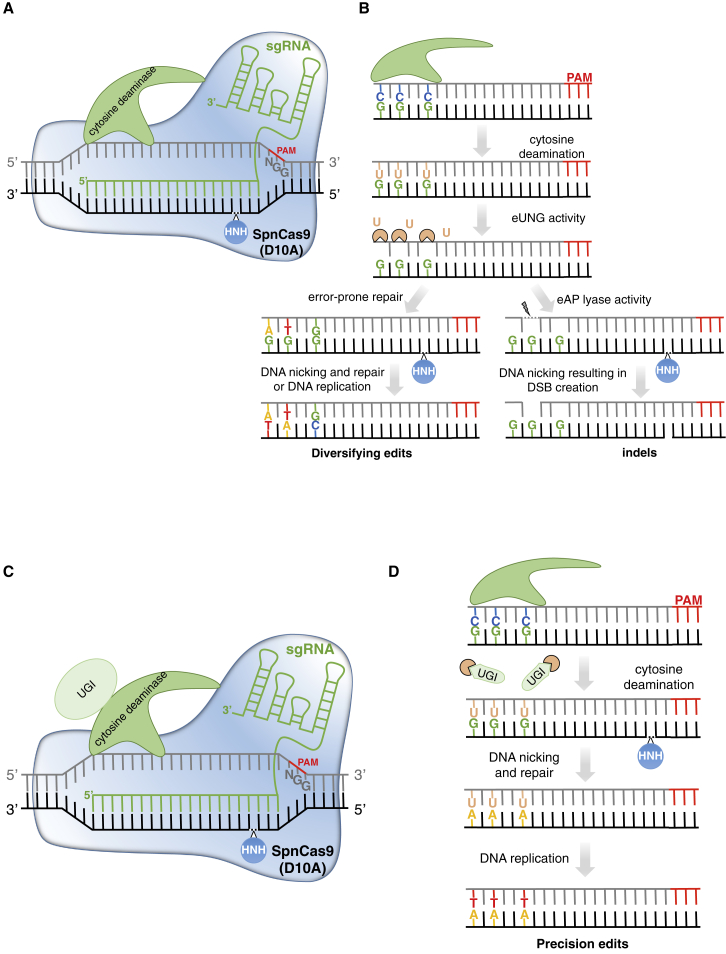
Figure 3.
CRISPR-Mediated Base Editing Using CBEs.
(A) CBEs are composed of nCas9 (D10A) fused to a cytosine deaminase catalytic domain (rAPOBEC1, PmCDA1, hAID, or hA3A) that mediates cytosine deamination in the so-called editing window at the 5′ end of the non-targeted sequence.
(B) After C deamination into U, endogenous uracil DNA glycosylase (eUNG) detects and removes the U, leading to an abasic site, which is further processed through error-free (U-to-C) or error-prone repair, producing different base substitutions, albeit at the cost of indel mutations due to the generation of DSBs through concomitant ssDNA breaks by nCas9 and endogenous AP lyases (eAP lyase). This system allows the production of C-to-T, C-to-G, and C-to-A conversions.
(C) CBE architecture can be upgraded through the fusion of one to several uracil glycosylase inhibitors (UGIs) to the base editor to increase the rate of C-to-T conversion while limiting the formation of by-products.
(D) After C deamination, UGIs protect the U edits from eUNG, thereby preventing the formation of abasic sites and mostly producing C-to-T conversions through the nicking of the non-edited strand and the intervention DNA repair/replication mechanisms, with a low level of by-products, such as indel mutations.
The schemes are not at scale and are for illustrative purposes only.
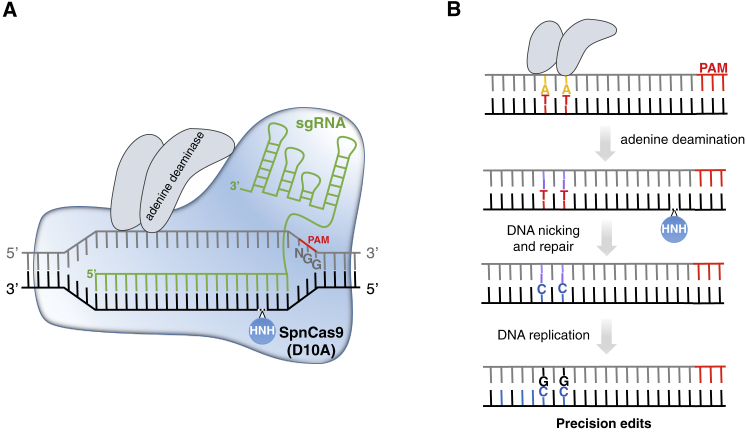
Figure 4.
CRISPR-Mediated Base Editing Using ABEs.
(A) ABEs are composed of nCas9 (D10A) fused to an adenine deaminase catalytic domain (ecTadA-ecTadA∗) that mediates adenine deamination in the so-called editing window at the 5′ end of the non-targeted sequence.
(B) After A deamination into I (inosine), nicking of the non-edited strand and intervention of DNA repair/replication mechanisms produce A-to-G conversions, with very low rates of by-products.
The schemes are not at scale and are for illustrative purposes only.
Soon after their development in animals, CBEs and ABEs were rapidly used in several plant species. The two most frequently used cytosine deaminases, PmCDA1 from Petromyzon marinus and rAPOBEC1 from rat (both devoid of UGI), have been reported to produce C-to-T transitions, but also C-to-G and C-to-A transversions in Arabidopsis, tomato, and potato, albeit with a substantial rate of indels, as discussed above (Li et al., 2017b; Lu and Zhu, 2017; Shimatani et al., 2017; Veillet et al., 2019a, 2019b; Bastet et al., 2019). For an approach requiring a high level of outcome predictability, UGI domain(s) can be added to the CBE architecture, resulting in a higher rate of C-to-T substitutions with lower unwanted mutations (Zong et al., 2017; Qin et al., 2019b). As observed in animals, ABEs produce A-to-G transitions in plants, with a very low rate of indels (Li et al., 2018a, 2019b; Kang et al., 2018; Yan et al., 2018; Hao et al., 2019; Negishi et al., 2019; Hua et al., 2020b). These BEs allowed the production of plants with new agronomic traits, including pathogen resistance (Bharat et al., 2020; Mishra et al., 2020). Recently, dual cytosine and adenine BEs were generated to simultaneously mediate C-to-T and A-to-G transitions in the same editing window, increasing the potential outputs for targeted gene modifications (Li et al., 2020b; Grünewald et al., 2020). Several different deaminases can also be recruited in the target site through sgRNA–protein interactions, thereby increasing the local amount of catalytic domains for the production of diversified outcomes (Zhang et al., 2019; Mishra et al., 2020).
Although base editing constitutes a promising technology, early CBEs and ABEs suffered from several drawbacks. First, their targeting scope is restricted to a sequence harboring a suitable PAM downstream of the targeted sequence, placing the target base in a generally short editing window at the 5' end of the spacer sequence. Much work has been done to use natural Cas9 orthologs with different PAM requirements, such as Staphylococcus aureus and Streptococcus canis Cas9 (Hua et al., 2018; Qin et al., 2019a; Wang et al., 2020a), or to engineer SpCas9 variants with relaxed PAM recognition, expanding the targeting scope of BEs (Wang et al., 2018; Ge et al., 2019; Hua et al., 2019; Niu et al., 2019; Ren et al., 2019; Zhong et al., 2019; Qin et al., 2020; Veillet et al., 2020). Of particular interest is the recent development in animal cells of new SpCas9 variants that recognize non-G PAMs (Miller et al., 2020) or almost any PAM sequence, as illustrated with BEs harboring the SpRY variant that are able to target almost any locus, albeit with a preference for sequences upstream of NRN PAMs (R = A or G) (Walton et al., 2020). Due to almost unrestricted PAM recognition, special attention should be put on limiting sgRNA self-targeting activity when using DNA delivery methods, potentially increasing the off-target risk by introducing mutations into spacer sequences (Qin et al., 2020). Second, the size of the editing window of BEs would benefit from being modular according to the desired editing outcome. The human APOBEC3A cytosine deaminase mediates base conversion inside an extended 17-bp editing window in rice, wheat, and potato, thereby increasing the saturated mutagenesis potential of a targeted locus (Zong et al., 2018). To increase the affinity of CBEs with their ssDNA substrates, Zhang et al. (2020b) fused an ssDNA-binding protein domain between nCas9 and the deaminases and achieved highly efficient cytosine base editing in an expanded editing window. On the contrary, CBEs with narrowed editing windows have been developed to avoid bystander mutations, allowing highly precise base substitutions (Tan et al., 2019, 2020). Third, the CBE harboring the rAPOBEC1 deaminase domain fused to a UGI was shown to induce substantial genome-wide sgRNA–Cas9-independent off-target C-to-T mutations in rice, while the ABE did not result in such unwanted effects (Jin et al., 2019). These single-nucleotide variants were often encountered in genic regions where single-stranded DNA is generated due to active transcription (Jin et al., 2019). To minimize these CBE-mediated unpredictable genome-wide off-target mutations also observed in animals (Zuo et al., 2019; Lee et al., 2020), engineered CBEs have been developed in animals. However, they still need to be validated in plants (Doman et al., 2020).
Combined with sgRNA libraries, the base-editing toolbox holds great promise to drive CRISPR-directed in planta evolution of proteins by generating many targeted mutations in a whole gene or specific sequence-encoding domains, allowing the identification of new key amino acid(s) associated with agronomic traits (Capdeville et al., 2020). So far, CRISPR-directed in planta evolution has been applied to confer herbicide resistance through amino acid substitutions in OsALS1 and OsACC genes (Li et al., 2020b; Kuang et al., 2020; Liu et al., 2020), but there is no doubt that this strategy could be used for ecological-friendly purposes such as the development of pathogen-resistant crops.
The ever-growing base-editing toolbox now includes many CBEs and ABEs that could meet various applications for the development of plants with new traits, such as the precise editing of a particular site or in-vivo-directed evolution. However, in addition to the restricted range of outcomes mediated by current base editors, each application needs a proper and careful selection of the most appropriate tool, limiting the wide adoption of base editing and highlighting the need for more versatility.
CRISPR-Mediated Prime Editing
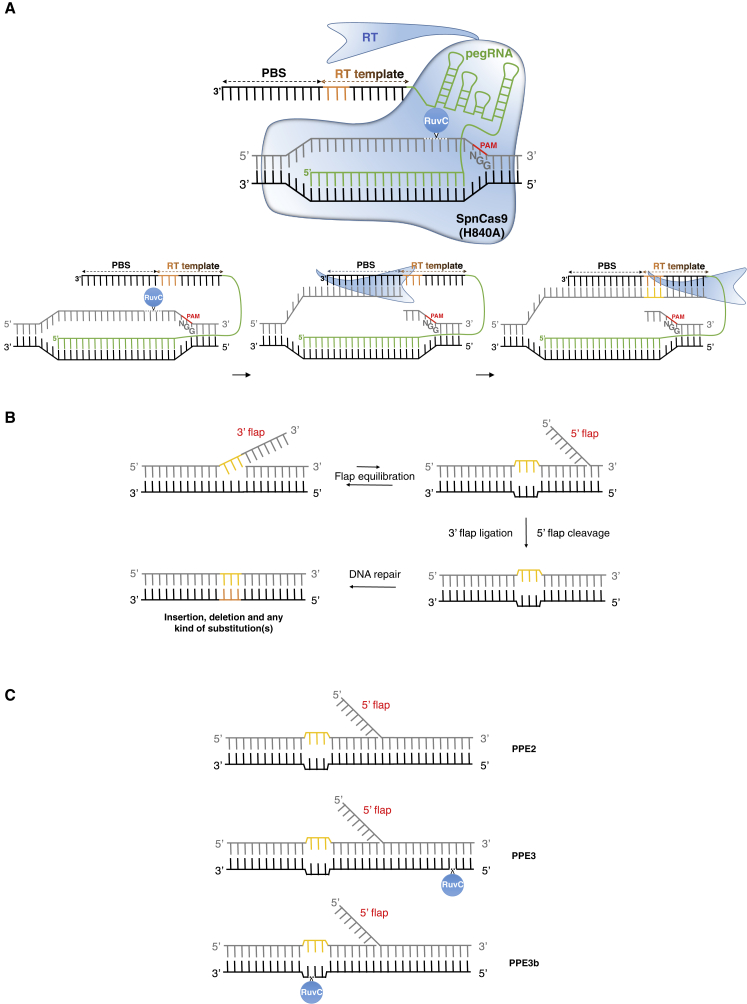
Despite the considerable expansion of the CRISPR toolbox, introducing precise and predictable targeted transversions, insertions, and deletions into eukaryote genomes is still a difficult task. Recently, a new ground-breaking technology that directly mediates the writing of new genetic information into a specific locus has been implemented in mammalian cells, unleashing new possibilities for precise genome editing. This search-and-replace technology, called prime editing, mediates targeted insertions, deletions, and any single or multiple substitutions (transitions and transversions) without requiring a DSB or a DNA donor template (Anzalone et al., 2019). Prime editors (PEs) are composed of a reverse transcriptase (RT) tethered to nCas9 with an impaired HNH domain (H840A) (Figure 5A). The editing protein complex is guided by an engineered prime editing sgRNA named pegRNA and consists of a classical sgRNA fused to a customizable 3′ extension that includes a primer binding sequence (PBS) and an RT template bearing the desired polymorphism (Figure 5A). Site-specific ssDNA breakage of the non-targeted strand and annealing of the PBS to the free 3′ end of the nicked strand result in priming of the reverse transcription of the RT template. This leads to the polymerization of an edited ssDNA at the free 3ʹ end that is complementary to the RT template and called a 3′ edited flap (Figure 5B). Subsequent eukaryotic DNA repair mechanisms favor 5′ flap excision and 3′ edited flap ligation (Keijzers et al., 2015; Liu et al., 2004), thereby producing a heteroduplex between the edited strand and the unmodified strand, which is then resolved to permanently stabilize the desired edit (Figure 5B). Similar to the strategy used for base editing, nicking the non-edited strand substantially increased the efficiency of PEs by favoring the stable incorporation of the edits (Anzalone et al., 2019).
Figure 5.
CRISPR-Mediated Prime Editing.
(A) Plant prime editors (PPEs) are composed of nCas9 (H840A) fused to reverse transcriptase (RT), allowing insertions, deletions, and all kinds of base substitutions. The polymorphism of interest is brought through a pegRNA, containing both an sgRNA for target specificity and a 3′ extension that harbors an RNA template bearing the polymorphism, leading to the targeted writing of new DNA sequences through reverse transcription.
(B) Upon cleavage of the non-targeted strand by the HNH domain of nCas9, the primer binding site (PBS) sequence of the pegRNA hybridizes with the broken ssDNA upstream of the cleavage site. This RNA/DNA structure initiates reverse transcriptase activity, copying the genetic information from the RT template. After the resolution of 3′ flap ligation, DNA repair mechanisms permanently install the mutation.
(C) Different prime editing strategies can be used to increase the rate of desired outcomes. The PPE2 strategy only implies the use of the pegRNA, while the PPE3 strategy requires the use of an additional sgRNA to cut the non-edited strand upstream or downstream of the modified sequence. For the PPE3b strategy, the second sgRNA targets the edited sequence to cut the non-edited strand only after 3′ flap resolution, thereby limiting the risk of indel mutations through the occurrence of DSBs.
The schemes are not at scale and are for illustrative purposes only.
While the successful development of highly versatile and precise PEs in mammalian cells has great potential, the implementation of plant PEs (PPEs) could also contribute to the improvement of food crops (Zhang et al., 2020c). A few months after its application in animals, prime editing has been adopted by several groups working on cereal crops (Butt et al., 2020; Hua et al., 2020a; Li et al., 2020c; Lin et al., 2020; Tang et al., 2020; Xu et al., 2020a, 2020b). Three different PPEs, PPE2, PPE3, and PPE3b, were assayed for their editing efficiency (Figure 5C). While PPE2 only consists of the expression of the nCas9-RT fusion and pegRNA, PPE3 aims to promote favorable repair by nicking the non-edited strand using an additional sgRNA that targets the edited strand upstream or downstream of the editing site. PPE3b also consists of nicking the non-edited strand, but the additional sgRNA targets the newly edited sequence so that nicking is restricted only after 3′ flap resolution, thereby preventing the formation of DSBs that would lead to a higher indel rate (Figure 5C). PPE2, PPE3, and PPE3b systems harboring an engineered version of Moloney murine leukemia virus RT resulted in similar editing efficiencies in rice and wheat protoplasts, as well as in Agrobacterium-mediated transformed rice plants. This indicates that nicking the non-edited strand does not necessarily increase prime editing efficiency in plants (Hua et al., 2020a; Xu et al., 2020a; Butt et al., 2020; Lin et al., 2020). PPEs were shown to specifically allow the introduction of all types of single- or multiple base substitutions, as well as deletions (up to 40 bp) and insertions (up to 15 bp) (Xu et al., 2020b; Li et al., 2020c; Lin et al., 2020; Tang et al., 2020). As observed in mammalian cells, by-products were mainly pegRNA scaffold insertions, which likely originated from the extensive activity of RT, and large deletions due to the paired nicking of both strands (Lin et al., 2020; Tang et al., 2020). Overall, editing efficiencies in rice and wheat were in the low percentage range, although precise 6-bp deletions and single A-to-T transversions were detected in 21.8% and 31.3% of rice plants regenerated from Agrobacterium-mediated transformation, respectively (Xu et al., 2020a; Lin et al., 2020).
The successful proof-of-concept of CRISPR-mediated prime editing in plants opens up exciting perspectives, although some challenges need to be overcome for the broad use of this new tool. The enhancement of the prime editing efficiency constitutes an essential track, especially for polyploids and/or vegetatively propagated species. Because a high variability of prime editing activity was observed among targeted sites, the “copy and replace” mechanism may be enhanced to promote reliable outcome rates. The PPE architecture should be optimized to maximize CRISPR component expression levels (Xu et al., 2020a, 2020b; Tang et al., 2020), and using different RTs that may be more efficient in plant cells is of particular interest, as well as optimizing temperature conditions for reverse transcriptase activity (Lin et al., 2020). The systematic testing of some pegRNA (PBS and RT lengths, esgRNA scaffold) and sgRNA (position of the nicking) designs for new targets is also highly recommended (Li et al., 2020d). While PPEs can accommodate long RT templates and are much less constrained than BEs for PAM availability, the use of Cas9 variants with relaxed PAM recognition may help localize the edit at a putatively favorable position from the ssDNA cutting site. Finally, although prime editing seems to induce lower off-target editing than Cas9 at putative off-target sites in animals (Anzalone et al., 2019), the genome-wide off-target activity of PPEs needs to be carefully evaluated to assess the capacity of RT to cause Cas9-independent unwanted edits.
Precision Breeding, a Matter of Choosing the Right Tool in the Toolbox
Collectively, CRISPR-mediated GT, base editing, and prime editing constitute an extended toolbox for precision editing, offering complementary strengths and weaknesses to edit almost any target site. When large DNA sequences need to be precisely inserted or deleted, the classical GT approach is the most suitable, as prime editing efficiency decreases with increasing length of the desired insertion or deletion (Lin et al., 2020). However, targeted small insertions and deletions can be efficiently mediated by both the prime editing system and the GT strategy. Besides the utility of such modifications for crop improvement, the possibility to label endogenous proteins with specific tags is of particular interest (e.g., cellular localization, purification, immunoprecipitation). Ali et al. (2020) recently managed to insert the hemagglutinin epitope into the C terminus of OsHDT using the CRISPR-Cas9-VirD2 system. It may also be possible to generate such insertion using the prime editing system, provided that the flag length is within the range of possible insertions by PEs.
Base editing appears to be generally more efficient than current PPEs for base substitution(s) (Anzalone et al., 2019; Lin et al., 2020). Therefore, early BEs should be used when bystander mutations are acceptable, whereas new BEs harboring narrowed editing windows should be favored when bystander edits are not desirable. However, when the desired outcome cannot be generated by BEs (e.g., most transversions or multiple base substitutions), PPEs offer much more versatility. For applications requiring targeted local random mutagenesis, such as the directed evolution of proteins, BEs still constitute the most suitable tool. However, PPEs might be modified to randomly insert a polymorphism in the target site through low-fidelity RTs, thereby providing another source of genetic variability. Because prime editing is at an early stage of development, we hope that future improvements will considerably enhance the efficiency and widen the targeting scope of PPEs.
A CRISPR Method for Pathogen Resistance Engineering
Interestingly, CRISPR-Cas can be directly used to target the pathogens’ genome, mainly those of viruses. This can be achieved by either targeting DNA viruses or RNA viruses but requires the transgenic expression of the CRISPR-Cas machinery and specific gRNA, an approach reminiscent of RNAi strategies. Therefore, this falls beyond the scope of precision breeding, but the reader can find details on these strategies as well as their possible caveats in recent reviews (Pyott et al., 2020; Zhao et al., 2020). Now, it is possible to apply precision breeding through the CRISPR technology to improve traits conferred by precise and/or punctual sequence variations, with an extraordinary opportunity to develop genetically resistant crops for sustainable agriculture. CRISPR applications have been predominantly focused on generating loss-of-function alleles, with some successes in the production of pathogen–resistant plants (Langner et al., 2018). However, plant–microorganism interactions result from a long co-evolution involving a complex molecular dialogue with several key players. As a result, CRISPR-mediated gain-of-function mutations appear to be highly relevant for developing crops with improved resistance to pathogens. In the subsequent sections, we review the current literature on CRISPR-mediated precision editing for pathogen resistance and provide interesting avenues that are now within CRISPR reach.
Immune Receptor Engineering
Considerable progress has been made in recent years regarding the molecular mechanisms of action, structural properties, and evolution of NLR receptors (Kourelis and van der Hoorn, 2018; Burdett et al., 2019; Tamborski and Krasileva, 2020). This enables novel strategies to improve the capacity of NLRs to induce immune responses, broaden their pathogen recognition spectrum, or even create new recognition specificities. However, there are currently very few examples of immune receptors that have been improved in this way (Cesari, 2018; Grund et al., 2019; Tamborski and Krasileva, 2020). Besides, current NLR engineering strategies essentially rely on either testing modified NLR genes in transient expression systems (e.g., by agroinfiltration in Nicotiana tabacum or Nicotiana benthamiana) or complementing susceptible varieties using stable transformation. The use of a CRISPR-based system for engineering NLR genes has not been reported. However, this represents a promising strategy to create new disease resistances directly in elite varieties. The development and rapid improvement of a wide range of CRISPR tools pave the way toward these new strategies.
One approach for NLR engineering relies on the editing of residues required for the regulation of these receptors to enhance their activation potential and, by this, enlarge their pathogen recognition spectrum. This strategy has been used for the wheat powdery mildew resistance gene Pm3, which forms an allelic series mediating the specific recognition of Blumeria graminis f. sp. tritici (Bgt) isolates. By comparing several alleles of Pm3 that exhibit a broad (a and b alleles) or narrow (f allele) resistance spectrum, Stirnweis et al. (2014) identified two polymorphisms in the nucleotide binding domain that are responsible for enhanced signaling activity and extended resistance spectrum. CRISPR-mediated prime editing of such regulatory residues in NLRs could create artificial “trigger happy” variants with a broadened resistance spectrum directly in elite cultivars. However, the misregulation of NLRs carries the risk of pleiotropic phenotypes, and such a potential trade-off phenomenon must be taken into consideration in this approach.
Alternatively, the recognition spectrum of NLRs can be broadened or modified by changing residues responsible for effector recognition specificity. In allelic NLRs series where distinct alleles exhibit different pathogen recognition specificities (e.g., barley MLA, wheat Pm3, flax L, or rice Pi-2/Piz-t/Pi50), the LRR domain plays a crucial role in effector recognition specificities (Dodds et al., 2006; Saur et al., 2019). In these cases, an attractive application of the CRISPR technology is to provide an elite cultivar with a recognition specificity already existing in other varieties by mutating the specific residues or sequences in the LRR domain that determine specificity. This would enable the adaptation of the pathogen recognition specificities of elite cultivars according to pathogen populations without going through tedious crossing and selection steps. The potential for this type of approach is illustrated by the historical example of the flax NLRs L2, L6, and L10 for which swaps of LRR domains have enabled changes in flax-rust recognition specificities (Ellis et al., 1999).
Knowledge-guided engineering of completely new recognition specificities by targeted mutagenesis of specific residues in the LRR domain is not yet possible. For this, one would require much better insight into the molecular mechanism of NLR activation as well as specific and precise knowledge on the LRR residues mediating effector recognition and specificity. The investigation of the allelic diversity coupled with the structural modeling of LRR domains may help in the identification of polymorphic surface residues that are likely involved in effector binding. Filling this knowledge gap is therefore a priority. Indeed, for the moment, novel recognition specificities by mutations in the LRR domain were only generated by random mutagenesis approaches. For example, in the potato NLR Rx, which confers resistance to potato virus X, point mutations in the LRR domain were found to extend the recognition spectrum (Farnham and Baulcombe, 2006). CRISPR-mediated introduction of such mutations identified by random mutagenesis approaches in high-throughput screening systems promises to create novel or broadened resistances.
Another strategy based on genome-editing techniques consists in reactivating pseudogenized NLR genes in elite varieties of agronomic interest. This would allow the "resuscitation" of resistance without laborious cloning and complementation steps and, in many countries, issues related to GMO regulations. This strategy is relevant for NLRs where the loss of function is due to a limited number of polymorphisms that can be “repaired” through base editing. This strategy has been tested using transcription activator-like effector nucleases editing on the wheat Lr21 gene, which provides race-specific resistance to leaf rust disease caused by Puccinia triticina (Luo et al., 2019). The inactive lr21Ψ allele differs from Lr21 by three non-synonymous polymorphisms and a single base deletion that disrupts the ORF. By editing the single base deletion, Luo et al. (2019) restored the lr21Ψ ORF but this did not reconstitute a functional resistance gene. CRISPR-mediated base editing has been successfully used in rice to reactivate the RLK coding gene Pi-d2, which confers resistance to blast disease (Ren et al., 2018). Rapid progress in the fields of comparative genomics, population genomics, and the intraspecific detection of NLRs (e.g., by resistance gene enrichment sequencing), which enable the identification of polymorphisms in NLR genes associated with disease resistance or susceptibility, will benefit these NLR engineering approaches.
Some NLRs contain unconventional integrated domains (IDs) that interact with pathogen effectors (Cesari et al., 2014; Le Roux et al., 2015; Sarris et al., 2015, 2016; Kroj et al., 2016; Bailey et al., 2018). Precise engineering of these IDs could result in enhanced and/or broader resistance (Cesari, 2018). Recently, the 3D structures of two IDs in complex with the effectors they recognize have been resolved, enabling precise identification of the residues for effector binding (Maqbool et al., 2015; Guo et al., 2018). This allowed, in the case of Pikp-1, which recognizes the Magnaporthe oryzae effector AVR-PikD, the structure-informed editing of the ID leading to the recognition of the previously not recognized effector allele AVR-PikE (De la Concepcion et al., 2019). This gain of specificity was shown in vitro and in transient assays in N. benthamiana. Whether the mutations lead to an extended resistance in the homologous rice/Magnaporthe oryzae system remains to be demonstrated. A CRISPR-mediated base-editing strategy in the true host plant would be a real asset in this type of experiment. Although extremely powerful, these approaches remain complicated because of gaps in our knowledge on the mode of action and structure of NLRs, in particular those that operate in pairs. When these gaps are filled, it will be virtually possible to create engineered NLR receptors capable of recognizing a wide variety of biotrophic or hemibiotrophic pathogens.
In the future, CRISPR-mediated directed evolution of NLR domains using BEs or even PEs, followed by screening for gain-of-resistance mutants, promises to become a powerful strategy for the development of new resistance in crops through completely new effector recognition specificities. However, its development awaits a better molecular understanding of NLR function to precisely target the right motifs and will require special attention to preserve agronomic traits by avoiding improper regulation of NLRs that can result in autoimmunity. This highlights the need to find a balance between pathogen detection and fitness (van Wersch et al., 2020).
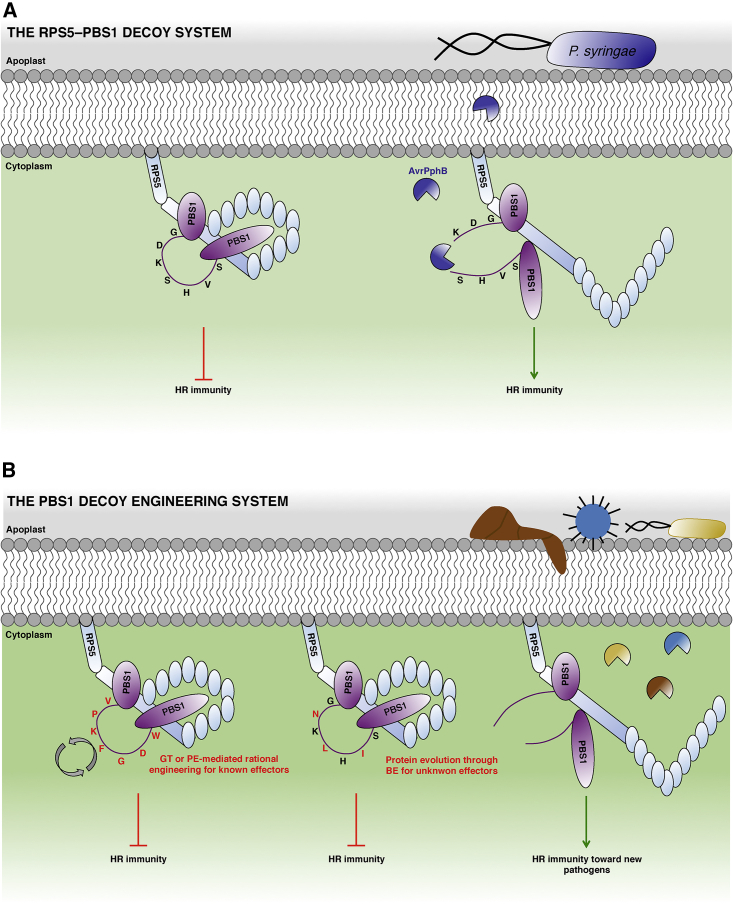
In many cases, the recognition of effectors by NLRs is indirect and occurs through the detection of effector-mediated modifications of plant proteins, called guardees or decoys (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008). A promising strategy for resistance engineering consists in modifying such decoys or guardees to trap novel pathogen effectors. A proof for this concept was provided in Arabidopsis thaliana using the serine-threonine kinase PBS1, whose cleavage by the bacterial effector AvrPphB is monitored by the NLR RPS5. Transforming RPS5 plants with a PBS1 mutant carrying the cleavage sites of other bacterial or viral proteases resulted in the recognition of these proteases and novel bacterial or virus resistance (Kim et al., 2016) (Figure 6A). Using genome-editing tools, such as CRISPR-mediated GT or prime editing, the endogenous locus encoding the seven-residue cleavage site of PBS1 could be readily modified into the cleavage sites of other pathogen proteases (Figure 6B), resulting in RPS5-mediated surveillance of these novel effectors (Pottinger and Innes, 2020). PBS1 is highly conserved among flowering plants and NLR-mediated surveillance of its cleavage emerged repeatedly in evolution, making it a versatile decoy system in corresponding crops (Carter et al., 2019; Pottinger and Innes, 2020). More generally, similar trap systems for proteases or other effectors can probably be engineered with other decoys or guardess in a large spectrum of crops even if they do not possess a PBS1 surveillance system (Giannakopoulou et al., 2016; Kim et al., 2016; Pottinger et al., 2020).
Figure 6.
Representative Model of the Natural and Engineered RPS5-PBS1 Decoy Systems.
(A) RPS5 and PBS1 form an inactive preactivation complex at the plasma membrane. Upon cleavage of the GDKSHVS motif in the activation loop of PBS1 by the Pseudomonas syringae AvrPphB type III protease, RPS5 senses the conformational change of PBS1, leading to the activation of RPS5-mediated HR.
(B) Using CRISPR precision editing tools, it is possible to replace the AvrPphB target cleavage sequence of PBS1 by a motif recognized by another secreted protease, such as the AvrRpt2 effector that cleaves the VPKFGDW sequence. GT or prime editing (PE) tools can be used to replace the initial target cleavage sequence to confer immunity toward pathogens (fungi, bacteria, and viruses) that secrete proteases with known cleavage recognition motifs. Alternatively, protein evolution using base editing (BE) can generate punctual amino acid shifts to generate potential new cleavage sequences. The functionality of these PBS1 variants can be screened toward pathogens that secrete proteases with unknown molecular characteristics, potentially conferring new sources of crop resistance.
The schemes are not at scale and are for illustrative purposes only.
Host Factor Engineering
Because NLR-mediated resistance is often quickly bypassed by pathogens, S gene engineering constitutes an exciting alternative for diversifying the sources of resistance. S genes, which can be targeted by pathogen effectors or act independently, facilitate pathogen infection, and can either encode proteins involved in host recognition, penetration or metabolism, or act as regulators of plant immunity (Langner et al., 2018). Contrary to R genes that are generally dominant, the loss of susceptibility conferred by engineering S factors is mainly recessive, meaning that all alleles should be altered to achieve resistance. This is a substantial challenge, especially for polyploid plants. To date, most genome-editing applications aiming at conferring pathogen resistance involved the knock out of S genes (Langner et al., 2018; Zaidi et al., 2018). However, this strategy may be associated with deleterious side effects as the host S genes may encode essential proteins (see below).
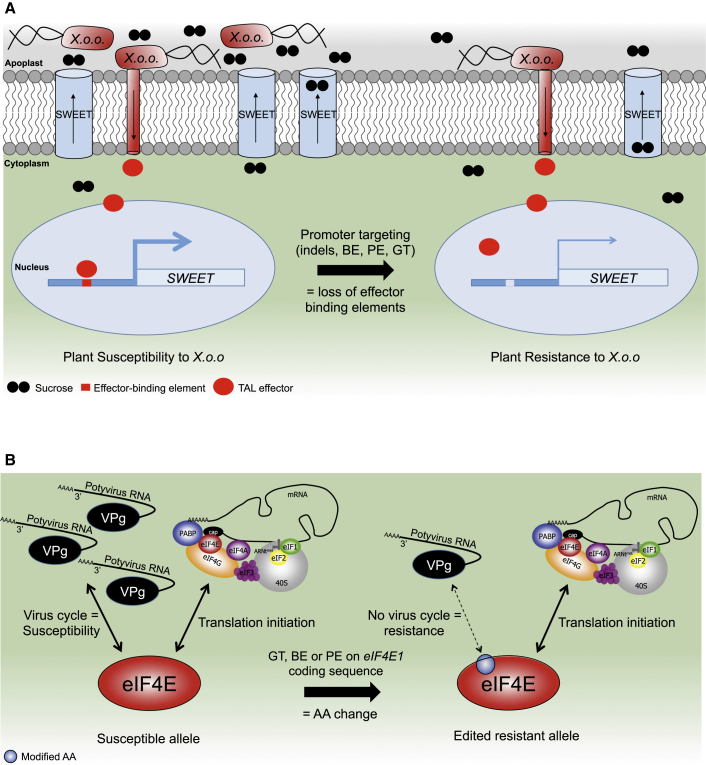
For example, bacterial and fungal infections lead to a competition for carbon resources at the plant/pathogen interface, in which host sugar transporters are essential for the outcome of the interaction (Lemoine et al., 2013). To increase sugar supply in the apoplast, the bacterium Xanthomonas oryzae pv. oryzae activates the transcription of members of the rice SWEET gene family, which encode proteins that mediate the passive diffusion of sucrose across the plasma membrane (Figure 7A). This is achieved through the expression of the so-called transcription activator-like effectors that bind specific regions of the SWEET promoters to activate their transcription, resulting in an enhanced export of sucrose to the apoplast that sustains bacterial growth. Because SWEET proteins are key components of phloem loading for long-distance sucrose transport (Lemoine et al., 2013), CRISPR-mediated loss-of-function approaches may result in unwanted developmental effects (Chen et al., 2012). In this regard, promoter targeting is an attractive alternative to introduce random indel mutations into transcription activator-like effector binding elements. This strategy was performed by targeting several OsSWEET genes, thereby preventing OsSWEET induction by bacterial effectors and conferring bacterial blight broad-spectrum resistance (Xu et al., 2019b; Oliva et al., 2019; Li et al., 2020a). Similarly, CRISPR-Cas9/Cas12a-mediated promoter editing of the CsLOB1 gene, which is specifically targeted by bacterial effectors for transcription activation, resulted in the generation of canker-resistant citrus cultivars (Peng et al., 2017; Jia et al., 2019). Because Cas9 nuclease mostly induces small deletions, we postulate that this strategy can be improved by using Cas12a and Cas12b nucleases, which introduce larger deletions at a higher rate, as previously discussed. The use of Cas variants with relaxed PAM recognition may also be valuable in precisely targeting cis-regulatory elements.
Figure 7.
Representative Model of Editing Resistance by Loss of Susceptibility.
(A) Resistance to bacteria through editing the SWEET promoter. During infection leading to susceptibility (left side), Xanthomonas oryzae pv. oryzae (Xoo) bacteria express transcription activator-like effectors (TAL effector) in the plant cell. These effectors bind effector-binding elements (EBE) located in the promoters of the SWEET genes that encode sucrose transporters. The binding triggers the activation of SWEET genes, and of the encoded sucrose transporter, and results in an increase in sucrose content in the apoplast. The excess of sucrose benefits to the bacteria and contribute to its multiplication. Genetic resistance can be engineered (right side) by removing the EBE region(s) from the SWEET promoter region: the SWEET gene is no longer activated by the TAL effector, such that sucrose content stays low in the apoplast, resulting in resistance.
(B) Resistance to potyvirus through base editing of the translation initiation factor eIF4E. In susceptible plants (left side), the translation initiator eIF4E is necessary for the infection cycle of potyviruses, represented by their ssRNA+ genome linked in 5′ to the viral protein genome-linked (VPg). At the same time, eIF4E proteins are involved in translation initiation of the host mRNA for protein synthesis. Base editing of the eIF4E coding sequence (right side) can be used to introduce non-synonymous mutations associated with amino acid changes usually found in resistance alleles from the natural diversity of plants. This mutation does not affect the translation initiation function of eIF4E while suppressing its interaction with potyvirus, leading to resistance. This allows resistance enhancement at no developmental cost. The translation initiation complex depiction is adapted from Robaglia and Caranta (2006).
The schemes are not at scale and are for illustrative purposes only.
With the recent expansion of the CRISPR toolbox, it is now possible to edit specific bases that can lead to predetermined punctual amino acid changes to develop new or mimick natural resistance-conferring alleles. The eukaryotic Initiation Factor 4E (eIF4E) genes are key elements of eukaryotic protein synthesis. At the same time, they are also important susceptibility factors to members of the large Potyviridae family, which rely on these factors to complete their infectious cycle in the plant (Bastet et al., 2017) (Figure 7B). Natural resistance found in various plant species often relies on functional resistance eIF4E alleles that contain non-synonymous mutations in the coding sequence. These alleles are devoid of associated fitness costs or developmental defects that are associated with loss-of-function alleles. Moreover, it has been shown that the deployment of these functional alleles can reduce the risk of resistance-breaking (Bastet et al., 2017). As a result, conversion of the Arabidopsis eIF4E1 susceptibility allele into a resistant allele through CBE-mediated single amino acid mutation (N176K) was recently achieved with no yield penalty (Bastet et al., 2019). It is expected that this approach could be generalized to any crops that are devoid of the natural eIF4E resistance allele to potyviruses and related single-strand positive RNA viruses. However, current base-editing tools are quite limited in generating a large range of amino acid changes associated with resistance that could be copied across species. Therefore, it is expected that prime editing could greatly facilitate the design of new resistance alleles to mimic more accurately natural resistance alleles that can gather up to five independent non-synonymous amino acid changes compared with the susceptible allele. The larger number of mutations will help to increase the resistance spectrum as well as the resistance durability associated with this allele (Moury et al., 2014).
Besides translation initiation factors, a large number of S genes are available to design new sources of resistance (van Schie and Takken, 2014; Hashimoto et al., 2016). The precise modification of other host factors to prevent their recognition by pathogen effectors, such as auxin response factors that are targeted by Fijiviruses proteins, will provide additional resistance mechanisms for crop molecular breeding toward viruses (Zhang et al., 2020a). We expect that several other host factors could be precisely edited in the coming years, providing new means for developing elite crops with improved genetic resistance toward a broad spectrum of pathogens.
Bottlenecks and Perspectives
The CRISPR toolbox for precision breeding in plants greatly expanded in the last few years, allowing the precise and predictable editing of almost any locus in the genome, at least in theory. While improvements of the new prime editing system are needed, plant scientists now have access to a highly versatile genome-editing toolbox for both functional genomics and molecular crop breeding.
However, in addition to the CRISPR system, delivery methods of genome-editing reagents into plant cells constitute the main technical limitation. While the transformation of major plant crops, such as rice, wheat, tomato and potato, is well established, some bottlenecks still stand in the way for the broad use of CRISPR in crop precision breeding. First, classical delivery methods, such as Agrobacterium-mediated transformation, protoplast transfection, and biolistic transformation, mostly target somatic cells and therefore involve subsequent regenerative steps that are time-consuming and highly genotype-dependent (Atkins and Voytas, 2020). Furthermore, delivery and tissue culture methods can cause unwanted changes to the genome, as recently demonstrated for protoplast transfection and Agrobacterium-mediated transformation in the tetraploid potato (Fossi et al., 2019), and for biolistic transformation in rice and maize (Liu et al., 2019). Second, most current delivery methods involve the stable integration of foreign DNA into plant genomes. While these sequences can be segregated out following Mendelian inheritance, it would be advantageous to minimize their expression window to avoid off-target effects, especially for base editors. Furthermore, the introduction of DNA intermediates into the plant nucleus may result in genome-wide random insertions, pointing out the necessity to use DNA-free delivery methods. As a result, while we are now able to precisely edit target sites through highly specific CRISPR tools, a special focus should be put on minimizing CRISPR-independent side effects, highlighting the need to develop alternative delivery methods into plant cells to avoid or limit such undesirable effects (Demirer et al., 2019; Toda et al., 2019; Maher et al., 2020), thereby unlocking the full potential of the CRISPR technology.
Finally, it is evident that the CRISPR technology has great potential for both plant biology research and precision crop breeding. The CRISPR precision toolbox, which is expanding and disseminating at an extraordinary speed, will help to decipher plant immune responses upon pathogen infection. However, while we are now able to mimic or evolve immune molecular mechanisms that confer genetic resistance to a broad range of pathogens, with the potential to support food security and safety in a sustainable way through the reduction of chemical use, regulatory frameworks constitute the main obstacle to CRISPR application in food crops, especially in Europe (Zhang et al., 2020c). We expect that a product-based regulatory framework could provide a rational balance between human/environment safety concerns and plant breeding innovation.
Funding
Our plant genome-editing research is supported by the Investissement d’Avenir program of the French National Agency of Research for the project GENIUS (ANR-11-BTBR-0001_GENIUS) and the Institut Carnot Plant2Pro program for the project POTATOCRISP. Our research on plant/pathogen interactions is supported by the ANR project Immunereceptor (ANR-15-CE20-0007).
Author Contributions
F.V., T.K., S.C., and J.-L.G. jointly wrote the original manuscript draft. F.V. and M.D. prepared the figures. F.V. and J.-L.G. planned the review outline. All authors contributed to the reviewing and editing of the manuscript.
Acknowledgments
We apologize to our colleagues whose work was not cited in this review due to limited space. We thank Fabien Nogué (INRAE Versailles) for inspiring and constructive discussions about CRISPR systems. We thank Diana Ortiz (INRAE Avignon) for discussions about engineered decoy systems. No conflict of interest declared.
Published: July 25, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
References
- Aird E.J., Lovendahl K.N., St Martin A., Harris R.S., Gordon W.R. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol. 2018;1:54. doi: 10.1038/s42003-018-0054-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z., Shami A., Sedeek K., Kamel R., Alhabsi A., Tehseen M., Hassan N., Butt H., Kababji A., Hamdan S.M. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun. Biol. 2020;3:44. doi: 10.1038/s42003-020-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alok A., Sandhya D., Jogam P., Rodrigues V., Bhati K.K., Sharma H., Kumar J. The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front. Plant Sci. 2020;11:264. doi: 10.3389/fpls.2020.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M.M., Landes X., Xiang W., Anyshchenko A., Falhof J., Østerberg J.T., Olsen L.I., Edenbrandt A.K., Vedel S.E., Thorsen B.J. Feasibility of new breeding techniques for organic farming. Trends Plant Sci. 2015;20:426–434. doi: 10.1016/j.tplants.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins P.A., Voytas D.F. Overcoming bottlenecks in plant gene editing. Curr. Opin. Plant Biol. 2020;54:79–84. doi: 10.1016/j.pbi.2020.01.002. [DOI] [PubMed] [Google Scholar]
- Bailey P.C., Schudoma C., Jackson W., Baggs E., Dagdas G., Haerty W., Moscou M., Krasileva K.V. Dominant integration locus drives continuous diversification of plant immune receptors with exogenous domain fusions. Genome Biol. 2018;19:23. doi: 10.1186/s13059-018-1392-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet A., Robaglia C., Gallois J.L. eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 2017;22:411–419. doi: 10.1016/j.tplants.2017.01.008. [DOI] [PubMed] [Google Scholar]
- Bastet A., Zafirov D., Giovinazzo N., Guyon-Debast A., Nogué F., Robaglia C., Gallois J.-L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019;17:1736–1750. doi: 10.1111/pbi.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann M.B., Gray B.N., January E., Gordon G.C., He Y., Liu H., Wu X., Brutnell T.P., Mockler T.C., Oufattole M. Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci. Rep. 2017;7:11606. doi: 10.1038/s41598-017-11760-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabe-Orts J.M., Casas-Rodrigo I., Minguet E.G., Landolfi V., Garcia-Carpintero V., Gianoglio S., Vazquez-Vilar M., Granell A., Orzaez D. Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol. J. 2019;17:1971–1984. doi: 10.1111/pbi.13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat S.S., Li S., Li J., Yan L., Xia L. Base editing in plants: current status and challenges. Crop J. 2020;8:384–395. [Google Scholar]
- Boutrot F., Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- Burdett H., Kobe B., Anderson P.A. Animal NLRs continue to inform plant NLR structure and function. Arch. Biochem. Biophys. 2019;670:58–68. doi: 10.1016/j.abb.2019.05.001. [DOI] [PubMed] [Google Scholar]
- Burmistrz M., Krakowski K., Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N.M., Baltes N.J., Voytas D.F., Douches D.S. Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. Plant Sci. 2016;7:1045. doi: 10.3389/fpls.2016.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt H., Rao G.S., Sedeek K., Aman R., Kamel R., Mahfouz M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol. J. 2020 doi: 10.1111/pbi.13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capdeville N., Schindele P., Puchta H. Application of CRISPR/Cas-mediated base editing for directed protein evolution in plants. Sci. China Life Sci. 2020;63:613–616. doi: 10.1007/s11427-020-1655-9. [DOI] [PubMed] [Google Scholar]
- Carter M.E., Helm M., Chapman A.V.E., Wan E., Restrepo Sierra A.M., Innes R.W., Bogdanove A.J., Wise R.P. Convergent evolution of effector protease recognition by Arabidopsis and barley. Mol. Plant Microbe Interact. 2019;32:550–565. doi: 10.1094/MPMI-07-18-0202-FI. [DOI] [PubMed] [Google Scholar]
- Cermak T., Baltes N.J., Cegan R., Zhang Y., Voytas D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S. Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 2018;219:17–24. doi: 10.1111/nph.14877. [DOI] [PubMed] [Google Scholar]
- Cesari S., Bernoux M., Moncuquet P., Kroj T., Dodds P.N. A novel conserved mechanism for plant NLR protein pairs: the "integrated decoy" hypothesis. Front. Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Chen L.-Q., Qu X.-Q., Hou B.-H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science. 2012;335:207. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- Cook D.E., Mesarich C.H., Thomma B.P. Understanding plant immunity as a surveillance system to detect invasion. Annu. Rev. Phytopathol. 2015;53:541–563. doi: 10.1146/annurev-phyto-080614-120114. [DOI] [PubMed] [Google Scholar]
- Dahan-Meir T., Filler-Hayut S., Melamed-Bessudo C., Bocobza S., Czosnek H., Aharoni A., Levy A.A. Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 2018;95:5–16. doi: 10.1111/tpj.13932. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- De la Concepcion J.C., Franceschetti M., MacLean D., Terauchi R., Kamoun S., Banfield M.J. Protein engineering expands the effector recognition profile of a rice NLR immune receptor. eLife. 2019;8:e47713. doi: 10.7554/eLife.47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirer G.S., Zhang H., Matos J.L., Goh N.S., Cunningham F.J., Sung Y., Chang R., Aditham A.J., Chio L., Cho M.J. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019;14:456–464. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.-M., Teh T., Wang C.-I.A., Ayliffe M.A., Kobe B., Ellis J.G. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U S A. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doman J.L., Raguram A., Newby G.A., Liu D.R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 2020;38:620–628. doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mounadi K., Morales-Floriano M.L., Garcia-Ruiz H. Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 2020;11:56. doi: 10.3389/fpls.2020.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J.G., Lawrence G.J., Luck J.E., Dodds P.N. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Masafumi M., Kaya H., Toki S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 2016;6:38169. doi: 10.1038/srep38169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnham G., Baulcombe D.C. Artificial evolution extends the spectrum of viruses that are targeted by a disease-resistance gene from potato. Proc. Natl. Acad. Sci. U S A. 2006;103:18828–18833. doi: 10.1073/pnas.0605777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie A.R., Yan J. De novo domestication: an alternative route toward new crops for the future. Mol. Plant. 2019;12:615–631. doi: 10.1016/j.molp.2019.03.016. [DOI] [PubMed] [Google Scholar]
- Fossi M., Amundson K.R., Kuppu S., Britt A.B., Comai L. Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 2019;180:78–86. doi: 10.1104/pp.18.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolome J. DNA methylation in plants: mechanisms and tools for targeted manipulation. New Phytol. 2020;227:38–44. doi: 10.1111/nph.16529. [DOI] [PubMed] [Google Scholar]
- Gaudelli N.M., Komor A.C., Rees H.A., Packer M.S., Badran A.H., Bryson D.I., Liu D.R. Programmable base editing of A∗T to G∗C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z., Zheng L., Zhao Y., Jiang J., Zhang E.J., Liu T., Gu H., Qu L.J. Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol. J. 2019;17:1865–1867. doi: 10.1111/pbi.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 2017;51:195–217. doi: 10.1146/annurev-genet-120215-035320. [DOI] [PubMed] [Google Scholar]
- Giannakopoulou A., Bialas A., Kamoun S., Vleeshouwers V.G. Plant immunity switched from bacteria to virus. Nat. Biotechnol. 2016;34:391–392. doi: 10.1038/nbt.3538. [DOI] [PubMed] [Google Scholar]
- Gil-Humanes J., Wang Y., Liang Z., Shan Q., Ozuna C.V., Sanchez-Leon S., Baltes N.J., Starker C., Barro F., Gao C. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017;89:1251–1262. doi: 10.1111/tpj.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grund E., Tremousaygue D., Deslandes L. Plant NLRs with integrated domains: unity makes strength. Plant Physiol. 2019;179:1227–1235. doi: 10.1104/pp.18.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J., Zhou R., Lareau C.A., Garcia S.P., Iyer S., Miller B.R., Langner L.M., Hsu J.Y., Aryee M.J., Joung J.K. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 2020;38:861–864. doi: 10.1038/s41587-020-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Cesari S., de Guillen K., Chalvon V., Mammri L., Ma M., Meusnier I., Bonnot F., Padilla A., Peng Y.-L. Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces. Proc. Natl. Acad. Sci. U S A. 2018;115:11637. doi: 10.1073/pnas.1810705115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao L., Ruiying Q., Xiaoshuang L., Shengxiang L., Rongfang X., Jianbo Y., Pengcheng W. CRISPR/Cas9-mediated adenine base editing in rice genome. Rice Sci. 2019;26:125–128. [Google Scholar]
- Hashimoto M., Neriya Y., Yamaji Y., Namba S. Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Front. Microbiol. 2016;7:1695. doi: 10.3389/fmicb.2016.01695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Comai L. Single-nucleotide mutations for plant functional genomics. Annu. Rev. Plant Biol. 2003;54:375–401. doi: 10.1146/annurev.arplant.54.031902.135009. [DOI] [PubMed] [Google Scholar]
- Herbert L., Meunier A.C., Bes M., Vernet A., Portefaix M., Durandet F., Michel R., Chaine C., This P., Guiderdoni E. Beyond seek and destroy: how to generate allelic series using genome editing tools. Rice (N Y) 2020;13:5. doi: 10.1186/s12284-020-0366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess G.T., Tycko J., Yao D., Bassik M.C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell. 2017;68:26–43. doi: 10.1016/j.molcel.2017.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Jiang Y., Tao X., Zhu J.K. Precision genome engineering in rice using prime editing system. Plant Biotechnol. J. 2020 doi: 10.1111/pbi.13395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Tao X., Han P., Wang R., Zhu J.K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant. 2019;12:1003–1014. doi: 10.1016/j.molp.2019.03.009. [DOI] [PubMed] [Google Scholar]
- Hua K., Tao X., Liang W., Zhang Z., Gou R., Zhu J.K. Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol. J. 2020;18:770–778. doi: 10.1111/pbi.13244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua K., Tao X., Zhu J.K. Expanding the base editing scope in rice by using Cas9 variants. Plant Biotechnol. J. 2018;17:499–504. doi: 10.1111/pbi.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T.K., Puchta H. CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep. 2019;38:443–453. doi: 10.1007/s00299-019-02379-0. [DOI] [PubMed] [Google Scholar]
- Hummel A.W., Chauhan R.D., Cermak T., Mutka A.M., Vijayaraghavan A., Boyher A., Starker C.G., Bart R., Voytas D.F., Taylor N.J. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 2018;16:1275–1282. doi: 10.1111/pbi.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Orbovic V., Wang N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019;17:1928–1937. doi: 10.1111/pbi.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Doudna J.A. CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Jin S., Zong Y., Gao Q., Zhu Z., Wang Y., Qin P., Liang C., Wang D., Qiu J.-L., Zhang F. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Vance R.E., Dangl J.L. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Kang B.C., Yun J.Y., Kim S.T., Shin Y., Ryu J., Choi M., Woo J.W., Kim J.S. Precision genome engineering through adenine base editing in plants. Nat. Plants. 2018;4:427–431. doi: 10.1038/s41477-018-0178-x. [DOI] [PubMed] [Google Scholar]
- Kanyuka K., Rudd J.J. Cell surface immune receptors: the guardians of the plant's extracellular spaces. Curr. Opin. Plant Biol. 2019;50:1–8. doi: 10.1016/j.pbi.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keijzers G., Bohr V.A., Rasmussen L.J. Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci. Rep. 2015;35:e00206. doi: 10.1042/BSR20150058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Kim S.T., Ryu J., Kang B.C., Kim J.S., Kim S.G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017;8:14406. doi: 10.1038/ncomms14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Qi D., Ashfield T., Helm M., Innes R.W. Using decoys to expand the recognition specificity of a plant disease resistance protein. Science. 2016;351:684. doi: 10.1126/science.aad3436. [DOI] [PubMed] [Google Scholar]
- Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor A.C., Zhao K.T., Packer M.S., Gaudelli N.M., Waterbury A.L., Koblan L.W., Kim Y.B., Badran A.H., Liu D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 2017;3:eaao4774. doi: 10.1126/sciadv.aao4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J., van der Hoorn R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018;30:285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroj T., Chanclud E., Michel-Romiti C., Grand X., Morel J.B. Integration of decoy domains derived from protein targets of pathogen effectors into plant immune receptors is widespread. New Phytol. 2016;210:618–626. doi: 10.1111/nph.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang Y., Li S., Ren B., Yan F., Spetz C., Li X., Zhou X., Zhou H. Base-editing-mediated artificial evolution of OsALS1 in planta to develop novel herbicide-tolerant rice germplasms. Mol. Plant. 2020;13:565–572. doi: 10.1016/j.molp.2020.01.010. [DOI] [PubMed] [Google Scholar]
- Kuluev B.R., Gumerova G.R., Mikhaylova E.V., Gerashchenkov G.A., Rozhnova N.A., Vershinina Z.R., Khyazev A.V., Matniyazov R.T., Baymiev A.K., Baymiev A.K. Delivery of CRISPR/Cas components into higher plant cells for genome editing. Russ. J. Plant Physiol. 2019;66:694–706. [Google Scholar]
- Langner T., Kamoun S., Belhaj K. CRISPR crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 2018;56:479–512. doi: 10.1146/annurev-phyto-080417-050158. [DOI] [PubMed] [Google Scholar]
- Le Roux C., Huet G., Jauneau A., Camborde L., Tremousaygue D., Kraut A., Zhou B., Levaillant M., Adachi H., Yoshioka H. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Smith H.E., Liu C., Willi M., Hennighausen L. Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun. Biol. 2020;3:19. doi: 10.1038/s42003-019-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Zhang Y., Kleinstiver B.P., Guo J.A., Aryee M.J., Miller J., Malzahn A., Zarecor S., Lawrence-Dill C.J., Joung J.K. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019;17:362–372. doi: 10.1111/pbi.12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine R., La Camera S., Atanassova R., Dedaldechamp F., Allario T., Pourtau N., Bonnemain J.L., Laloi M., Coutos-Thevenot P., Maurousset L. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013;4:272. doi: 10.3389/fpls.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Rui H., Li Y., Wang Q., Alariqi M., Qin L., Sun L., Ding X., Wang F., Zou J. Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum) Plant Biotechnol. J. 2019;17:1862–1864. doi: 10.1111/pbi.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li W., Zhou Z., Chen H., Xie C., Lin Y. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 2020;18:313–315. doi: 10.1111/pbi.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Liu C., Qi X., Wu Y., Fei X., Mao L., Cheng B., Li X., Xie C. RNA-guided Cas9 as an in vivo desired-target mutator in maize. Plant Biotechnol. J. 2017;15:1566–1576. doi: 10.1111/pbi.12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Zhang R., Meng X., Chen S., Zong Y., Lu C., Qiu J.L., Chen Y.H., Li J., Gao C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020;38:875–882. doi: 10.1038/s41587-019-0393-7. [DOI] [PubMed] [Google Scholar]
- Li C., Zong Y., Wang Y., Jin S., Zhang D., Song Q., Zhang R., Gao C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018;19:59. doi: 10.1186/s13059-018-1443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Li J., Chen J., Yan L., Xia L. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol. Plant. 2020;13:671–674. doi: 10.1016/j.molp.2020.03.011. [DOI] [PubMed] [Google Scholar]
- Li J., Li H., Chen J., Yan L., Xia L. Toward precision genome editing in crop plants. Mol. Plant. 2020;13:811–813. doi: 10.1016/j.molp.2020.04.008. [DOI] [PubMed] [Google Scholar]
- Li J., Meng X., Zong Y., Chen K., Zhang H., Liu J., Li J., Gao C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants. 2016;2:16139. doi: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- Li J., Qin R., Zhang Y., Xu S., Liu X., Yang J., Zhang X., Wei P. Optimizing plant adenine base editor systems by modifying the transgene selection system. Plant Biotechnol. J. 2019;18:1495–1497. doi: 10.1111/pbi.13304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sun Y., Du J., Zhao Y., Xia L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant. 2017;10:526–529. doi: 10.1016/j.molp.2016.12.001. [DOI] [PubMed] [Google Scholar]
- Li Q., Sapkota M., van der Knaap E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: unlocking the neglected potential for crop improvement. Hortic. Res. 2020;7:36. doi: 10.1038/s41438-020-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li J., He Y., Xu M., Zhang J., Du W., Zhao Y., Xia L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019;37:445–450. doi: 10.1038/s41587-019-0065-7. [DOI] [PubMed] [Google Scholar]
- Li S., Li J., Zhang J., Du W., Fu J., Sutar S., Zhao Y., Xia L. Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J. Exp. Bot. 2018;69:4715–4721. doi: 10.1093/jxb/ery245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Zong Y., Xue C., Wang S., Jin S., Zhu Z., Wang Y., Anzalone A.V., Raguram A., Doman J.L. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020;38:582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- Liu J., Nannas N.J., Fu F.F., Shi J., Aspinwall B., Parrott W.A., Dawe R.K. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell. 2019;31:368–383. doi: 10.1105/tpc.18.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Qin R., Li J., Liao S., Shan T., Xu R., Wu D., Wei P. A CRISPR-Cas9-mediated domain-specific base-editing screen enables functional assessment of ACCase variants in rice. Plant Biotechnol. J. 2020 doi: 10.1111/pbi.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Kao H.I., Bambara R.A. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- Lu Y., Zhu J.K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant. 2017;10:523–525. doi: 10.1016/j.molp.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Luo M., Li H., Chakraborty S., Morbitzer R., Rinaldo A., Upadhyaya N., Bhatt D., Louis S., Richardson T., Lahaye T. Efficient TALEN-mediated gene editing in wheat. Plant Biotechnol. J. 2019;17:2026–2028. doi: 10.1111/pbi.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher M.F., Nasti R.A., Vollbrecht M., Starker C.G., Clark M.D., Voytas D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020;38:84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar H., Lindsey K., Zhang X., Jin S. CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 2019;24:1102–1125. doi: 10.1016/j.tplants.2019.09.006. [DOI] [PubMed] [Google Scholar]
- Maqbool A., Saitoh H., Franceschetti M., Stevenson C.E., Uemura A., Kanzaki H., Kamoun S., Terauchi R., Banfield M.J. Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife. 2015;4:e08709. doi: 10.7554/eLife.08709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mara K., Charlot F., Guyon-Debast A., Schaefer D.G., Collonnier C., Grelon M., Nogue F. POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens. New Phytol. 2019;222:1380–1391. doi: 10.1111/nph.15680. [DOI] [PubMed] [Google Scholar]
- Miller S.M., Wang T., Randolph P.B., Arbab M., Shen M.W., Huang T.P., Matuszek Z., Newby G.A., Rees H.A., Liu D.R. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat. Biotechnol. 2020;38:471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming M., Ren Q., Pan C., He Y., Zhang Y., Liu S., Zhong Z., Wang J., Malzahn A.A., Wu J. CRISPR-Cas12b enables efficient plant genome engineering. Nat. Plants. 2020;6:202–208. doi: 10.1038/s41477-020-0614-6. [DOI] [PubMed] [Google Scholar]
- Mishra R., Joshi R.K., Zhao K. Base editing in crops: current advances, limitations and future implications. Plant Biotechnol. J. 2020;18:20–31. doi: 10.1111/pbi.13225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla K.A., Yang Y. Predicting CRISPR/Cas9-induced mutations for precise genome editing. Trends Biotechnol. 2020;38:136–141. doi: 10.1016/j.tibtech.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Moury B., Charron C., Janzac B., Simon V., Gallois J.L., Palloix A., Caranta C. Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): a game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 2014;27:472–480. doi: 10.1016/j.meegid.2013.11.024. [DOI] [PubMed] [Google Scholar]
- Negishi K., Kaya H., Abe K., Hara N., Saika H., Toki S. An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol. J. 2019;17:1476–1478. doi: 10.1111/pbi.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Wang C., Win J., Lanz C., Weigel D., Kamoun S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017;7:482. doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Q., Wu S., Yang X., Liu P., Xu Y., Lang Z. Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid. J. Integr. Plant Biol. 2019;62:398–402. doi: 10.1111/jipb.12886. [DOI] [PubMed] [Google Scholar]
- Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.S., Li C., Nguyen H., Liu B. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019;37:1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L., De Clercq R., Goossens J., Inigo S., Williams C., Ron M., Britt A., Goossens A. A dual sgRNA approach for functional genomics in Arabidopsis thaliana. G3 (Bethesda) 2018;8:2603–2615. doi: 10.1534/g3.118.200046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng A., Chen S., Lei T., Xu L., He Y., Wu L., Yao L., Zou X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017;15:1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottinger S.E., Bak A., Margets A., Helm M., Tang L., Casteel C., Innes R.W. Optimizing the PBS1 decoy system to confer resistance to potyvirus infection in Arabidopsis and soybean. Mol. Plant Microbe Interact. 2020;33:932–944. doi: 10.1094/MPMI-07-19-0190-R. [DOI] [PubMed] [Google Scholar]
- Pottinger S.E., Innes R.W. RPS5-Mediated disease resistance: fundamental insights and translational applications. Annu. Rev. Phytopathol. 2020;58 doi: 10.1146/annurev-phyto-010820-012733. [DOI] [PubMed] [Google Scholar]
- Puchta H. The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 2005;56:1–14. doi: 10.1093/jxb/eri025. [DOI] [PubMed] [Google Scholar]
- Pyott D.E., Fei Y., Molnar A. Potential for gene editing in antiviral resistance. Curr. Opin. Virol. 2020;42:47–52. doi: 10.1016/j.coviro.2020.04.005. [DOI] [PubMed] [Google Scholar]
- Qin R., Li J., Li H., Zhang Y., Liu X., Miao Y., Zhang X., Wei P. Developing a highly efficient and wildly adaptive CRISPR-SaCas9 toolset for plant genome editing. Plant Biotechnol. J. 2019;17:706–708. doi: 10.1111/pbi.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin R., Li J., Liu X., Xu R., Yang J., Wei P. SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat. Plants. 2020;6:197–201. doi: 10.1038/s41477-020-0603-9. [DOI] [PubMed] [Google Scholar]
- Qin R., Liao S., Li J., Li H., Liu X., Yang J., Wei P. Increasing fidelity and efficiency by modifying cytidine base-editing systems in rice. Crop J. 2019;8:396–402. [Google Scholar]
- Que Q., Chen Z., Kelliher T., Skibbe D., Dong S., Chilton M.D. Plant DNA repair pathways and their applications in genome engineering. Methods Mol. Biol. 2019;1917:3–24. doi: 10.1007/978-1-4939-8991-1_1. [DOI] [PubMed] [Google Scholar]
- Ren B., Liu L., Li S., Kuang Y., Wang J., Zhang D., Zhou X., Lin H., Zhou H. Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol. Plant. 2019;12:1015–1026. doi: 10.1016/j.molp.2019.03.010. [DOI] [PubMed] [Google Scholar]
- Ren B., Yan F., Kuang Y., Li N., Zhang D., Zhou X., Lin H., Zhou H. Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant. Mol. Plant. 2018;11:623–626. doi: 10.1016/j.molp.2018.01.005. [DOI] [PubMed] [Google Scholar]
- Robaglia C., Caranta C. Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 2006;11:40–45. doi: 10.1016/j.tplants.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Sarris P.F., Cevik V., Dagdas G., Jones J.D., Krasileva K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016;14:8. doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P.F., Duxbury Z., Huh S.U., Ma Y., Segonzac C., Sklenar J., Derbyshire P., Cevik V., Rallapalli G., Saucet S.B. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Saur I.M., Bauer S., Kracher B., Lu X., Franzeskakis L., Muller M.C., Sabelleck B., Kummel F., Panstruga R., Maekawa T. Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife. 2019;8:e44471. doi: 10.7554/eLife.44471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savary S., Willocquet L., Pethybridge S.J., Esker P., McRoberts N., Nelson A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- Savic N., Ringnalda F.C., Lindsay H., Berk C., Bargsten K., Li Y., Neri D., Robinson M.D., Ciaudo C., Hall J. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife. 2018;7:e33761. doi: 10.7554/eLife.33761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimatani Z., Kashojiya S., Takayama M., Terada R., Arazoe T., Ishii H., Teramura H., Yamamoto T., Komatsu H., Miura K. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017;35:441–443. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweis D., Milani S.D., Brunner S., Herren G., Buchmann G., Peditto D., Jordan T., Keller B. Suppression among alleles encoding nucleotide-binding-leucine-rich repeat resistance proteins interferes with resistance in F1 hybrid and allele-pyramided wheat plants. Plant J. 2014;79:893–903. doi: 10.1111/tpj.12592. [DOI] [PubMed] [Google Scholar]
- Tamborski J., Krasileva K.V. Evolution of plant NLRs: from natural history to precise modifications. Annu. Rev. Plant Biol. 2020;71:355–378. doi: 10.1146/annurev-arplant-081519-035901. [DOI] [PubMed] [Google Scholar]
- Tan J., Zhang F., Karcher D., Bock R. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 2019;10:439. doi: 10.1038/s41467-018-08034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J., Zhang F., Karcher D., Bock R. Expanding the genome-targeting scope and the site selectivity of high-precision base editors. Nat. Commun. 2020;11:629. doi: 10.1038/s41467-020-14465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Liu G., Zhou J., Ren Q., You Q., Tian L., Xin X., Zhong Z., Liu B., Zheng X. A large-scale whole-genome sequencing analysis reveals highly specific genome editing by both Cas9 and Cpf1 (Cas12a) nucleases in rice. Genome Biol. 2018;19:84. doi: 10.1186/s13059-018-1458-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Lowder L.G., Zhang T., Malzahn A.A., Zheng X., Voytas D.F., Zhong Z., Chen Y., Ren Q., Li Q. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants. 2017;3:17018. doi: 10.1038/nplants.2017.18. [DOI] [PubMed] [Google Scholar]
- Tang X., Ren Q., Yang L., Bao Y., Zhong Z., He Y., Liu S., Qi C., Liu B., Wang Y. Single transcript unit CRISPR 2.0 systems for robust Cas9 and Cas12a mediated plant genome editing. Plant Biotechnol. J. 2019;17:1431–1445. doi: 10.1111/pbi.13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Sretenovic S., Ren Q., Jia X., Li M., Fan T., Yin D., Xiang S., Guo Y., Liu L. Plant prime editors enable precise gene editing in rice cells. Mol. Plant. 2020;13:667–670. doi: 10.1016/j.molp.2020.03.010. [DOI] [PubMed] [Google Scholar]
- Teng F., Cui T., Feng G., Guo L., Xu K., Gao Q., Li T., Li J., Zhou Q., Li W. Repurposing CRISPR-Cas12b for mammalian genome engineering. Cell Discov. 2018;4:63. doi: 10.1038/s41421-018-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda E., Koiso N., Takebayashi A., Ichikawa M., Kiba T., Osakabe K., Osakabe Y., Sakakibara H., Kato N., Okamoto T. An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants. 2019;5:363–368. doi: 10.1038/s41477-019-0386-z. [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A., Kamoun S. From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie C.C., Takken F.L. Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 2014;52:551–581. doi: 10.1146/annurev-phyto-102313-045854. [DOI] [PubMed] [Google Scholar]
- Van Vu T., Sivankalyani V., Kim E.J., Doan D.T.H., Tran M.T., Kim J., Sung Y.W., Park M., Kang Y.J., Kim J.Y. Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol. J. 2020 doi: 10.1111/pbi.13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch S., Tian L., Hoy R., Li X. Plant NLRs: the whistleblowers of plant immunity. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2019.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet F., Chauvin L., Kermarrec M.P., Sevestre F., Merrer M., Terret Z., Szydlowski N., Devaux P., Gallois J.L., Chauvin J.E. The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 2019;38:1065–1080. doi: 10.1007/s00299-019-02426-w. [DOI] [PubMed] [Google Scholar]
- Veillet F., Perrot L., Chauvin L., Kermarrec M.-P., Guyon-Debast A., Chauvin J.-E., Nogué F., Mazier M. Transgene-free genome editing in tomato and potato plants using Agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillet F., Perrot L., Guyon-Debast A., Kermarrec M.P., Chauvin L., Chauvin J.E., Gallois J.L., Mazier M., Nogue F. Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in solanaceae crops. Int. J. Mol. Sci. 2020;21:1024. doi: 10.3390/ijms21031024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton R.T., Christie K.A., Whittaker M.N., Kleinstiver B.P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Wang C., Liu P., Lei C., Hao W., Gao Y., Liu Y.G., Zhao K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11:e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Meng X., Hu X., Sun T., Li J., Wang K., Yu H. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol. J. 2018;17:709–711. doi: 10.1111/pbi.13053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Lu Y., Botella J.R., Mao Y., Hua K., Zhu J.K. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant. 2017;10:1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- Wang M., Xu Z., Gosavi G., Ren B., Cao Y., Kuang Y., Zhou C., Spetz C., Yan F., Zhou X. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J. 2020;18:1645–1647. doi: 10.1111/pbi.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Alariqi M., Wang F., Li B., Ding X., Rui H., Li Y., Xu Z., Qin L., Sun L. The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 2020 doi: 10.1111/pbi.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Wolter F., Puchta H. The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J. 2018;94:767–775. doi: 10.1111/tpj.13899. [DOI] [PubMed] [Google Scholar]
- Xu R., Li J., Liu X., Shan T., Qin R., Wei P. Development of a plant prime editing system for precise editing in the rice genome. Plant Commun. 2020;1 doi: 10.1016/j.xplc.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Qin R., Li H., Li D., Li L., Wei P., Yang J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 2017;15:713–717. doi: 10.1111/pbi.12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Qin R., Li H., Li J., Yang J., Wei P. Enhanced genome editing in rice using single transcript unit CRISPR-LbCpf1 systems. Plant Biotechnol. J. 2019;17:553–555. doi: 10.1111/pbi.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Zhang C., Yang Y., Zhao S., Kang G., He X., Song J., Yang J. Versatile nucleotides substitution in plant using an improved prime editing system. Mol. Plant. 2020;13:675–678. doi: 10.1016/j.molp.2020.03.012. [DOI] [PubMed] [Google Scholar]
- Xu Z., Xu X., Gong Q., Li Z., Li Y., Wang S., Yang Y., Ma W., Liu L., Zhu B. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant. 2019;12:1434–1446. doi: 10.1016/j.molp.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Yan F., Kuang Y., Ren B., Wang J., Zhang D., Lin H., Yang B., Zhou X., Zhou H. Highly efficient A.T to G.C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant. 2018;11:631–634. doi: 10.1016/j.molp.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Yang H., Gao P., Rajashankar K.R., Patel D.J. PAM-dependent target DNA recognition and cleavage by C2c1 CRISPR-Cas endonuclease. Cell. 2016;167:1814–1828.e12. doi: 10.1016/j.cell.2016.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Biswal A.K., Dionora J., Perdigon K.M., Balahadia C.P., Mazumdar S., Chater C., Lin H.C., Coe R.A., Kretzschmar T. CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep. 2017;36:745–757. doi: 10.1007/s00299-017-2118-z. [DOI] [PubMed] [Google Scholar]
- Zaidi S.S., Mahfouz M.M., Mansoor S. CRISPR-Cpf1: a new tool for plant genome editing. Trends Plant Sci. 2017;22:550–553. doi: 10.1016/j.tplants.2017.05.001. [DOI] [PubMed] [Google Scholar]
- Zaidi S.S., Mukhtar M.S., Mansoor S. Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 2018;36:898–906. doi: 10.1016/j.tibtech.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li L., He Y., Qin Q., Chen C., Wei Z., Tan X., Xie K., Zhang R., Hong G. Distinct modes of manipulation of rice auxin response factor OsARF17 by different plant RNA viruses for infection. Proc. Natl. Acad. Sci. U S A. 2020;117:9112–9121. doi: 10.1073/pnas.1918254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen L., Zhu B., Wang L., Chen C., Hong M., Huang Y., Li H., Han H., Cai B. Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat. Cell Biol. 2020;22:740–750. doi: 10.1038/s41556-020-0518-8. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Malzahn A.A., Sretenovic S., Qi Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants. 2019;5:778–794. doi: 10.1038/s41477-019-0461-5. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Pribil M., Palmgren M., Gao C. A CRISPR way for accelerating improvement of food crops. Nat. Food. 2020;1:200–205. [Google Scholar]
- Zhao Y., Yang X., Zhou G., Zhang T. Engineering plant virus resistance: from RNA silencing to genome editing strategies. Plant Biotechnol. J. 2020;18:328–336. doi: 10.1111/pbi.13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Sretenovic S., Ren Q., Yang L., Bao Y., Qi C., Yuan M., He Y., Liu S., Liu X. Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol. Plant. 2019;12:1027–1036. doi: 10.1016/j.molp.2019.03.011. [DOI] [PubMed] [Google Scholar]
- Zong Y., Song Q., Li C., Jin S., Zhang D., Wang Y., Qiu J.L., Gao C. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. 2018;36:950–953. doi: 10.1038/nbt.4261. [DOI] [PubMed] [Google Scholar]
- Zong Y., Wang Y., Li C., Zhang R., Chen K., Ran Y., Qiu J.L., Wang D., Gao C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017;35:438–440. doi: 10.1038/nbt.3811. [DOI] [PubMed] [Google Scholar]
- Zuo E., Sun Y., Wei W., Yuan T., Ying W., Sun H., Yuan L., Steinmetz L.M., Li Y., Yang H. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science. 2019;364:289. doi: 10.1126/science.aav9973. [DOI] [PMC free article] [PubMed] [Google Scholar]