Abstract
Angiosperms (flowering plants) are the most diverse and species-rich group of plants. The vast majority (∼99.95%) of angiosperms form a clade called Mesangiospermae, which is subdivided into five major groups: eudicots, monocots, magnoliids, Chloranthales, and Ceratophyllales. The relationships among these Mesangiospermae groups have been the subject of long debate. In this study, we assembled a phylogenomic dataset of 1594 genes from 151 angiosperm taxa, including representatives of all five lineages, to investigate the phylogeny of major angiosperm lineages under both coalescent- and concatenation-based methods. We dissected the phylogenetic signal and found that more than half of the genes lack phylogenetic information for the backbone of angiosperm phylogeny. We further removed the genes with weak phylogenetic signal and showed that eudicots, Ceratophyllales, and Chloranthales form a clade, with magnoliids and monocots being the next successive sister lineages. Similar frequencies of gene tree conflict are suggestive of incomplete lineage sorting along the backbone of the angiosperm phylogeny. Our analyses suggest that a fully bifurcating species tree may not be the best way to represent the early radiation of angiosperms. Meanwhile, we inferred that the crown-group angiosperms originated approximately between 255.1 and 222.2 million years ago, and Mesangiospermae diversified into the five extant groups in a short time span (∼27 million years) at the Early to Late Jurassic.
Key words: Mesangiospermae, gene tree conflict, phylogenomics, phylogenetic signal, divergence times
Angiosperms (flowering plants) are the most diverse and species-rich group of plants. The relationships among the early divergent lineages of angiosperms have been the subject of long debate. By assembling a phylogenomic dataset of 1594 genes from 151 angiosperm taxa, this study investigates the angiosperm phylogeny and reveals that a fully bifurcating species tree may not be the best way to represent the early radiation of angiosperms.
Introduction
Flowering plants (angiosperms) are among the largest and most structurally and functionally diverse plant groups on Earth (Judd et al., 1999). Angiosperms have crucial roles in current terrestrial ecosystems (Foster, 2016) and provide food for humans and domestic animals and other materials important to human society (Tilman et al., 2002). Among angiosperms, Mesangiospermae account for ∼99.95% of extant species and include five lineages: eudicots, monocots, magnoliids, Chloranthales, and Ceratophyllales (Cantino et al., 2007). Eudicots and monocots are the two largest and most diverse clades of Mesangiospermae, accounting for ∼75% and ∼20% of angiosperm species (http://www.theplantlist.org/). Magnoliids, with over 10 000 species, form the third major clade, and comprise four orders: Canellales, Laurales, Magnoliales, and Piperales (Cantino et al., 2007). The other two Mesangiospermae groups, Chloranthales and Ceratophyllales, are relatively small lineages, but are evolutionarily significant with macrofossil records dating back to the Early Cretaceous (Friis et al., 2010). Chloranthales encompass four genera and about 77 extant species, and Ceratophyllales comprise four species of the single extant genus Ceratophyllum (Maarten et al., 2016).
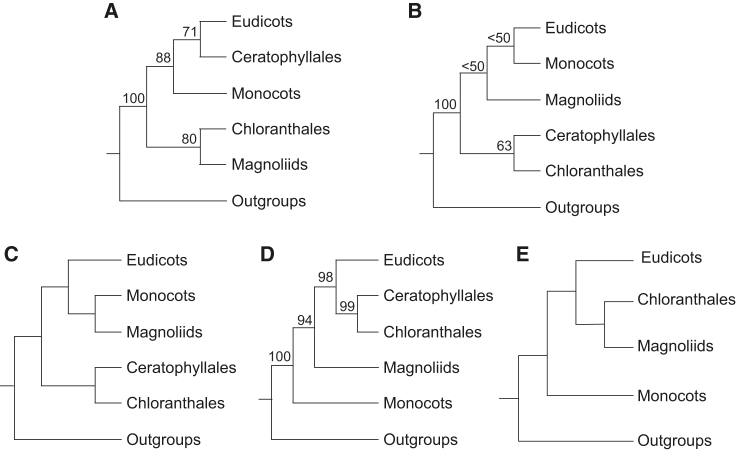
A fully resolved and well-supported phylogeny is important for understanding the evolutionary history of angiosperms, and provides a foundation for research on gene function and phenotypic evolution. The angiosperm phylogeny has been greatly improved in recent years (e.g., Moore et al., 2007, Soltis et al., 2011, Ruhfel et al., 2014, Wickett et al., 2014, Zeng et al., 2014, Byng et al., 2016, Zhong and Betancur-R, 2017, Gitzendanner et al., 2018, Leebens-Mack et al., 2019, Li et al., 2019), but the branching order of early divergent lineages remains contentious, especially among Mesangiospermae (Figure 1). The latest version of the Angiosperm Phylogeny Group classification, APG IV (Byng et al., 2016), placed an uncertain relationship among Chloranthales, magnoliids, and a monocots–Ceratophyllales–eudicots clade. Of particular note is that molecular data from the different genomic sources within angiosperms often carry different phylogenetic signals. Previous analyses based on plastid genes suggest that a clade comprising Chloranthales + magnoliids is sister to monocots + (Ceratophyllales + eudicots) (Figure 1A; Moore et al., 2007, Soltis et al., 2011, Ruhfel et al., 2014, Gitzendanner et al., 2018). Despite sampling nearly 3000 chloroplast genomes across the angiosperm phylogeny, Li et al. (2019) have shown that the relationships among Mesangiospermae remained poorly resolved and that the rapid radiations may have occurred during the early evolutionary history of Mesangiospermae. Analyses based on mitochondrial genes recover different relationships within Mesangiospermae, with eudicots + monocots forming the sister group to magnoliids, and these three lineages being the sister group to Ceratophyllales + Chloranthales (Figure 1B; Qiu et al., 2010). Endress and Doyle (2009) combined plastid and morphological data, and recovered eudicots + (monocots + magnoliids) as sister to the clade including Ceratophyllales and Chloranthales (Figure 1C). The relationships among these clades remain discrepancies in different analyses using nuclear datasets. Zeng et al. (2014) analyzed 59 low-copy nuclear genes of 60 angiosperms, and suggested that a clade comprising Chloranthales + Ceratophyllales is closely related to eudicots, with magnoliids and monocots as successive sister groups (Figure 1D). Wickett et al. (2014) used large number of nuclear genes (674 or fewer genes) to estimate the relationships among land plants and recovered a similar angiosperm topology to that reported by Zeng et al. (2014), but included few angiosperms (37 species) and did not sample Ceratophyllales. Puttick et al. (2018) used expanded data from Wickett et al. (2014) with optimized phylogenetic analyses, and found a close relationship between eudicots and Chloranthales + magnoliids, with monocots being the sister lineage to these three groups (Figure 1E). Recently Leebens-Mack et al. (2019) showed that the deep-branching relationships of angiosperm remain unresolved even using thousands of transcriptomes of green plants. Overall, despite being a widely studied topic, the relationships along the backbone of angiosperm phylogeny remain elusive.
Figure 1.
Representative Published Topologies among the Five Major Groups of Mesangiospermae: Eudicots, Monocots, Magnoliids, Chloranthales, and Ceratophyllales.
Shown are topologies derived from: (A) plastid genes (Moore et al., 2007, Moore et al., 2010, Soltis et al., 2011, Ruhfel et al., 2014, Gitzendanner et al., 2018); (B) mitochondrial genes (Qiu et al., 2010); (C) combined plastid and morphological data (Endress and Doyle, 2009); (D) nuclear data (Wickett et al., 2014, Zeng et al., 2014); and (E) nuclear data (Puttick et al., 2018).
Fossil evidence suggests that angiosperms arose in the Early Cretaceous, approximately 140 million years ago (Ma) (Doyle, 2012, Gomez et al., 2015), and underwent an extremely rapid radiation in their early evolutionary history (Lidgard and Crane, 1988, Lidgard and Crane, 1990, Crane and Lidgard, 1989, Crane and Lidgard, 1990, Friis et al., 2010, Herendeen et al., 2017). Consequently, angiosperms quickly came to dominate terrestrial environments (Magallón and Castillo, 2009, Friis et al., 2011). This rise and subsequent apparently rapid diversification of flowering plants in the Mid-Cretaceous was famously described as an “abominable mystery” in Charles Darwin's letter to Joseph Hooker in 1879 (Darwin et al., 1903, Friedman, 2009). Despite new fossil discoveries and considerable advances in methods, the timing of the origin of angiosperms, and implicitly Mesangiospermae, remains elusive. In contrast to the purely fossil-based estimates of angiosperms arising in the Early Cretaceous, which might represent the radiation of Mesangiospermae, recent molecular dating studies often recover highly variable divergence times for Mesangiospermae (Smith et al., 2010, Zeng et al., 2014, Foster et al., 2017, Barba-Montoya et al., 2018, Li et al., 2019). For example, Zeng et al. (2014) reported that the onset of Mesangiospermae was estimated to have occurred between 191 and 151 Ma in the Jurassic, consistent with an early origin of Mesangiospermae proposed by Foster et al. (2017) (195–157 Ma) and Li et al. (2019) (193–146 Ma). However, in a study with very strong maximum age constraints, Magallón et al. (2015) indicated that the diversification of Mesangiospermae begun at 137–135 Ma in the Cretaceous.
In this study, our main objective is to investigate the causes of the lack of resolution surrounding the backbone of angiosperm phylogeny. We sampled 1594 protein-coding nuclear genes from 151 angiosperm taxa, including the earliest diverging angiosperm lineages and representatives of all five major groups of Mesangiospermae. The large nuclear dataset allows us to dissect whether lack of phylogenetic signal or gene tree conflict can result in low resolution of phylogenetic relationship of five major lineages of angiosperms, and whether the angiosperm evolution is strictly bifurcating. We employed both coalescent and concatenation approaches to infer angiosperm phylogenetic trees. Additionally, as a comparison with recent mainly plastome-based molecular dating studies, we also infer the evolutionary timescale of angiosperms.
Results
Datasets
We searched transcriptome datasets we previously generated and a newly generated transcriptomic dataset for a third species of Chloranthales (Hedyosmum orientale), as well as additional public sources for orthologous nuclear sequence data from angiosperms and gymnosperm outgroup taxa. Our initial dataset contained 4180 orthologous genes (OGs) from 151 angiosperm species, including 98 eudicots, 24 monocots, 19 magnoliids, four Chloranthales species, two Ceratophyllales species, and four species of ANA grade, along with two gymnosperm outgroups (Ginkgo biloba and Pinus taeda) (Supplemental Table 1). After filtering out putatively spurious and/or paralogous sequences with extremely long branches, 1696 OGs were retained for further analyses. We calculated the average bootstrap support (ABS) value and the “Tree Certainty all” (TCA) score to quantify the accuracy of gene tree estimation (Salichos and Rokas, 2013, Salichos et al., 2014). We retained only genes that had lengths of a minimum of 600 nucleotides, with both ABS ≥50% and TCA >0.3, resulting in a working dataset of 1594 genes. These genes had taxon coverage ranging from 70% to 100% (average of 87.6%) and an average length of 1080 nucleotides.
Phylogenetic Analyses Based on 1594 Nuclear Genes
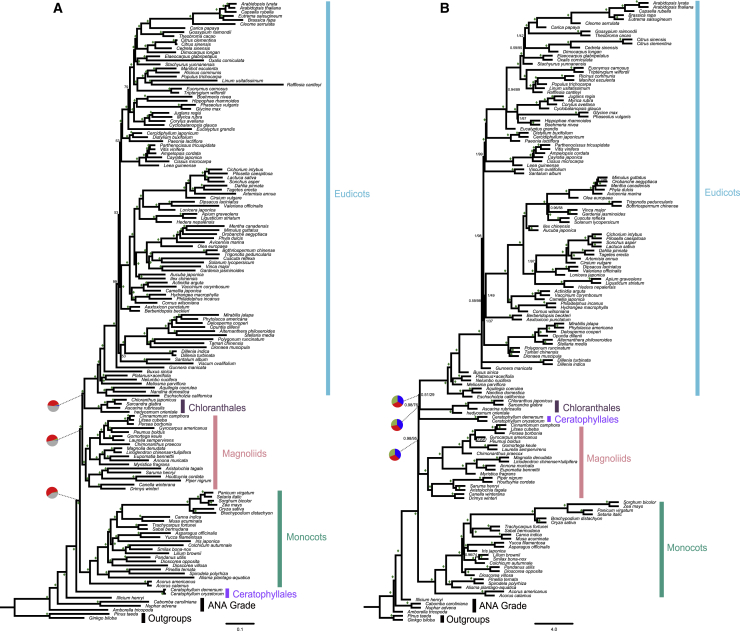
We applied both coalescent and concatenation methods to reconstruct the angiosperm phylogeny using the 1594-gene dataset. Our phylogenomic analyses recovered full support for Amborella being sister to all other extant angiosperms, followed successively by Nymphaeales and Austrobaileyales (ANA grade: Amborella/Nymphaeales/Austrobaileyales), in agreement with previous studies (e.g., The Amborella Genome Project, 2013, Wickett et al., 2014, Zeng et al., 2014, Zhong and Betancur-R, 2017, Gitzendanner et al., 2018, Leebens-Mack et al., 2019, Li et al., 2019, Zhang et al., 2020). All five major lineages of the remaining angiosperms (Mesangiospermae) were recovered as monophyletic groups with maximum support, consistent with previous analyses (Ruhfel et al., 2014, Zeng et al., 2014, Gitzendanner et al., 2018, Leebens-Mack et al., 2019, Li et al., 2019). However, the relationships among the five lineages of Mesangiospermae were less robustly resolved (Figure 2), especially the position of Ceratophyllales. In the concatenation analyses with ultrafast bootstrapping support (UFboot), eudicots and Chloranthales were recovered as a sister group with full support (Figure 2A). Magnoliids and monocots were recovered as the next two successive sister lineages with maximum support, and Ceratophyllales as sister to the rest of Mesangiospermae with 100% UFboot support. The coalescent-based phylogeny was inferred with ASTRAL, which accounts for gene tree heterogeneity due to incomplete lineages sorting (ILS), and the node support was estimated by local posterior probability (PP) and multilocus bootstrapping (MLBS). The coalescent-based analyses revealed that eudicots and Chloranthales were sisters with low support (0.51 PP and 29% MLBS), with Ceratophyllales being sister to eudicots + Chloranthales with 0.98 PP and 76% MLBS (Figure 2B). Magnoliids and monocots were recovered as the next two successive sister lineages, with strong support (0.98 PP/95% MLBS and 1.0 PP/100% MLBS, respectively).
Figure 2.
The Species Trees Inferred from the Dataset of 1594 Nuclear Gene.
(A) The concatenation-based species tree inferred by IQ-TREE. Numbers associated with nodes are the ultrafast bootstrapping support (UFboot); diamonds indicate UFboot support of 100%. The branch lengths in substitutions per site were estimated by IQ-TREE. The pie charts at three backbone nodes of angiosperms present the proportion of gene tree concordance and conflict in the 1594-gene dataset. Pie chart color coding: blue, fraction of gene trees that are concordant with the species tree; green, fraction of gene trees supporting the second most common conflicting topology; red, fraction of gene trees supporting all other alternative conflicting partitions; gray, fraction of gene trees with <50% bootstrap support at that node.
(B) The coalescent-based species tree was inferred by ASTRAL. Numbers associated with nodes are support values obtained by the posterior probability (PP, on the left) and multilocus bootstrapping (MLBS, on the right). Diamonds indicate PP of 1.0 and MLBS of 100%. The branch lengths in coalescent units were estimated by ASTRAL. Pie charts show relative quartet support for the current tree (blue) and the two (green and red) alternative quartets.
Evaluation of Gene Tree Conflict and Phylogenetic Signal
The conflicts between gene trees and concatenation-based species tree were prevalent at the nodes along the backbone of the angiosperm phylogeny (Figure 2A and Supplemental Figure 2), although the concatenation analyses yielded highly supported topology. The percentage of conflicting bipartitions for the three backbone nodes among Mesangiospermae ranged from 45.55% to 59.66%, and the topological concordance for these nodes ranged only from 0.63% to 2.70% (pie chart in Figure 2A). Importantly, the quartet score calculated by ASTRAL was low at these three internal nodes (e.g., 34%, with alterative quartet support 34% and 30%; Figure 2B and Supplemental Figure 3) based on the coalescent-based species tree. The uninformative bipartitions with bootstrap values lower than 50% were prevalent at most internal nodes, specifically at the three internal nodes representing the relationships among five major Mesangiosperm lineages (39.71%–51.76% uninformative bipartitions for concatenation-based topology; Figure 2A). To further gauge the phylogenetic signal present in our phylogenomic dataset, for each gene tree, we used the approximately unbiased (AU) test (Shimodaira, 2002) to compare the likelihoods of the coalescent-based species tree based on 1594-gene dataset (Figure 2B) and four alternative hypotheses for the five major lineages of Mesangiosperm (Figure 1A–1D). We found that many among the 1594 genes lacked phylogenetic signals to resolve the relationships among five Mesangiosperm lineages (Supplemental Figure 4): 53% of genes could not reject any of the five topologies at the 5% confidence level, and 29% of genes rejected one of the five hypotheses. Only 12% of genes could significantly reject two hypotheses. Therefore, we filtered out the genes with low phylogenetic signal to obtain two subsets (756 genes and 296 genes) for subsequent phylogenomic inferences.
Inferring the Species Tree of Five Major Groups of Mesangiospermae
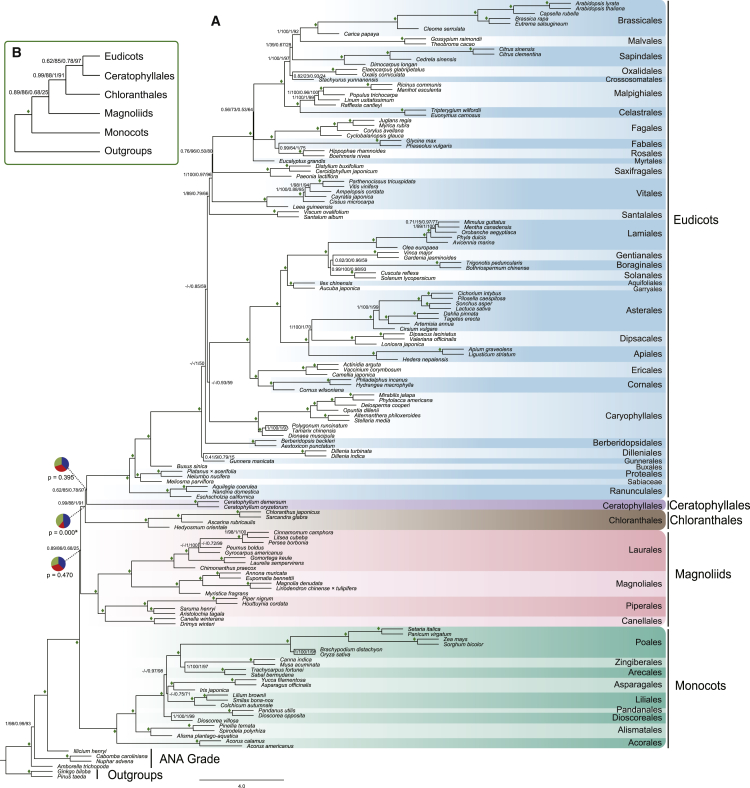
Coalescent-based analyses of the two smaller datasets (756 genes and 296 genes) produced well-supported results for the backbone nodes of the angiosperm phylogeny (Figure 3 and Supplemental Figure 5). The monophyly of angiosperms received maximal support, with Amborella sister to all other angiosperms, followed by Nymphaeales and Austrobaileyales with strong support. Mesangiospermae and all of its five major lineages were recovered as monophyletic with maximum support. We found that eudicots, Ceratophyllales, and Chloranthales constituted a clade with high support. Within the clade, eudicots and Ceratophyllales as a sister group were supported with moderate support (0.62 PP and 85% MLBS for 756 genes; 0.78 PP and 97% MLBS for 296 genes). Magnoliids and monocots were successive next sister group to this clade with 0.89 PP and 86% MLBS for 756 genes but 0.68 PP and 25% MLBS for 296 genes (Figure 3B). The inferences based on the 756-gene and 296-gene datasets were largely congruent with that of the 1594-gene dataset, except for the placement of Ceratophyllales and Chloranthales (cf. the relationship of (Ceratophyllales (Chloranthales + eudicots)) in the 1594-gene dataset).
Figure 3.
The Coalescent-Based Species Tree of Angiosperms Is Supported by the Trees Inferred from Two Subsets of Nuclear Genes.
(A) The tree inferred by ASTRAL using 296 genes. The detailed phylogeny of 756 genes is presented in Supplemental Figure 4. Numbers associated with nodes are support values: posterior probability and multilocus bootstrapping (PP and MLBS). The first two are PP and MLBS values from 756 genes, and the last two are PP and MLBS values from 296 genes. Diamonds indicate PP of 1.0 and MLBS of 100% in both datasets. The branch lengths are in coalescent units, as estimated by ASTRAL. The pie charts at three backbone nodes of angiosperms show relative quartet support for the current tree (blue) and the two (green and red) alternative quartets. p values for the polytomy test are given for four backbone nodes of angiosperm below the respective pie charts for those nodes, and significance (p ≤ 0.05) is indicated with an asterisk.
(B) An abbreviated tree showing the relationship of the five lineages of Mesangiospermae with associated support values (PP/MLBS).
Within the eudicots, Ranunculales, Proteales, and Buxales were successive sisters to all other eudicots (also referred to as core eudicots) with maximal support (Figure 3A). The positions of Dilleniaceae remain uncertain, with 0.41 and 0.79 PP but only 9% and 15% MLBS values for its placement as sister to Gunnerales using 756- and 296-gene sets, respectively. The relationships among remaining eudicots lineages were broadly consistent with those inferred in Leebens-Mack et al. (2019), but different from APG IV and Li et al. (2019). We consistently recovered core rosids and Saxifragales in a clade (0.76 PP and 96% MLBS for 756 genes; 0.5 PP and 80% MLBS for 296 genes), with Vitales and Santalales being moderately supported successive next clades. However, the position of Berberidopsidales was uncertain (Figure 4A and Supplemental Figure 7). Berberidopsidales was either sister to Asterids (756 genes) or sister of the most core eudicots, except Gunnerales (296 genes).
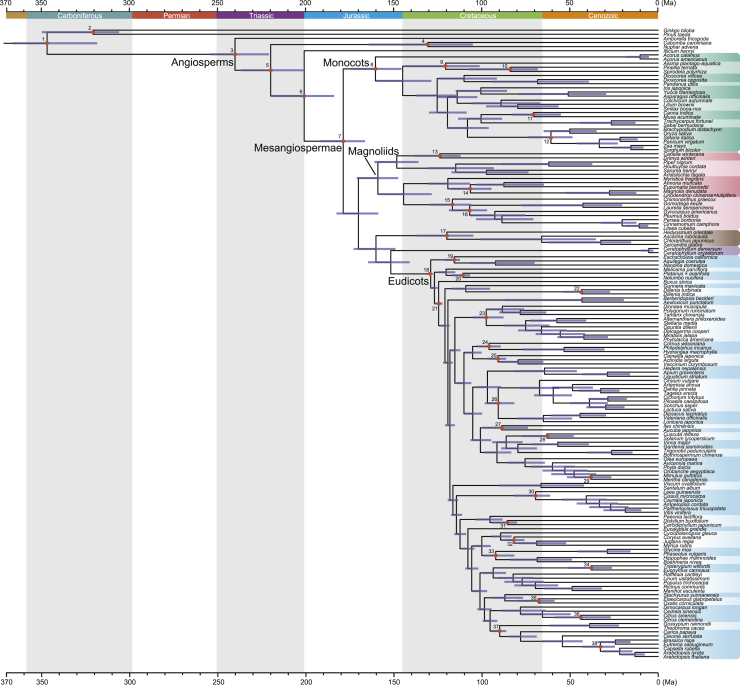
Figure 4.
Chronogram Depicting the Evolutionary Timescale of 151 Angiosperms and Two Gymnosperm Outgroups.
Divergence times were estimated using Bayesian inference of 296 genes with 38 calibration points in MCMCTree. Fossil constraints are shown by red dots, and the details of fossils are available in Supplemental Information. Horizontal bars represent 95% credibility intervals.
Relationships within the monocots were largely consistent with published hypotheses, with Acorales, followed by Alismatales, as successive sisters to the rest of monocots with maximal support (Zeng et al., 2014, Hertweck et al., 2015, Gitzendanner et al., 2018). However, the relationships of Dioscoreales and Liliales to other monocot orders were different here. Dioscoreales were recovered as paraphyletic, with Dioscorea opposita close to the order Pandanales, although there were only two taxa of Dioscoreales and one of Pandanales. Thus, additional samplings of these two orders are likely needed to better resolve their relationships to other monocots. Among the remaining monocots, we recovered Poales as sister to a clade of Zingiberales + Arecales. The relationships of Asparagales and Liliales remain uncertain, either being successive sister clades of (Poales (Zingiberales + Arecales)) (756 genes), or as sister groups (296 genes). The topology within magnoliids was largely congruent with recent analyses (Massoni et al., 2014). There was strong support for a clade with Magnoliales and Laurales, which are in turn sister to a clade of Piperales and Canellales with maximal support using both 756 and 296 genes.
Evolutionary Timescales of Angiosperms
The evolutionary timescale of angiosperms is one of the most contentious questions in evolutionary biology. Recent investigations to estimate the angiosperm timescale have been predominantly based on datasets of chloroplast markers (e.g., Magallón et al., 2015, Foster et al., 2017, Barba-Montoya et al., 2018, Li et al., 2019), with limited analysis using only dozens of nuclear genes (Zeng et al., 2014). Although not the primary aim of this study, we inferred the evolutionary timescale of angiosperms using our 296-gene dataset as a comparison with plastome-based estimates. Recent studies investigating the evolutionary timescale of angiosperms have comprehensively assessed the impact of many potential biases on inferred ages (e.g., Foster et al., 2017, Barba-Montoya et al., 2018). We carefully chose values for all parameters of our analysis, such as by selecting fossil calibrations according to contemporary gold standards (Parham et al., 2012).
Our results did not appear to be affected by life-history-associated rate heterogeneity, or by our choice of maximum age calibration (see Supplemental Information). Here, we report 95% credibility intervals for estimated divergence times for crown groups, as obtained through analysis with MCMCTree (Figure 4). The estimated timeline suggests that crown-group angiosperms diverged in the interval 255.1–222.2 Ma (Late Permian to Late Triassic), which is similar to those estimates in some of the recent studies (Foster et al., 2017, Barba-Montoya et al., 2018, Morris et al., 2018, Li et al., 2019). The diversification of Mesangiospermae was inferred to begin at 192.2–166.4 Ma. Within Mesangiospermae, both magnoliids and monocots originated almost contemporaneously. We inferred crown-group magnoliids to have arisen in the Middle and Late Jurassic (170.7–147.1 Ma), and the divergence time of monocots was dated between 174.6 and 145.7 Ma. The onset of crown-group eudicot diversification occurred between 131.1 and 127.8 Ma. Intriguingly, the five major groups of Mesangiospermae diverged from one another mainly during the Early to Late Jurassic (178.8–151.8 Ma, Figure 4). Therefore, over a time span as short as 27 million years (Myr), angiosperms underwent a rapid radiation with explosive diversification.
Discussion
Many recent analyses have focused on resolving the angiosperm phylogeny with large genome-scale datasets (Gitzendanner et al., 2018, Leebens-Mack et al., 2019, Li et al., 2019). Yet, the relationships among Mesangiospermae remained poorly resolved in these studies, and the taxon sampling of Chloranthales and Ceratophyllales was still rather limited. These examples demonstrate that for problematic deep nodes of the angiosperm phylogeny, such as the relationships among major lineages of Mesangiospermae, simply increasing taxon sampling is unlikely to lead to accurate inference or increase bootstrap support, but is likely to increase computational difficulties. Considering this, we tailored our taxon sampling to target the relationships at the order level and above, specifically focusing on the relationships among the major clades of Mesangiospermae. For three major lineages (eudicots, monocots, and magnoliids), we sampled the representative species for each order covering 45 of 59 orders of APG IV (for details see Methods). For two smaller groups, Ceratophyllales and Chloranthales, we maximized representative sampling and newly sequenced one transcriptome of the genus Hedyosmum (Chloranthales). In total, we sampled two species of Ceratophyllales and four species of Chloranthales.
Incomplete Lineage Sorting during the Early Angiosperm Evolution
In our phylogenomic analyses, we recovered two inconsistent topologies for the five Mesangiospermae lineages using coalescent- and concatenation-based approaches. We calculated topological concordance and discordance between 1594 gene trees and the species tree, showing low percentage of topological concordance but a large degree of gene tree heterogeneity at deep internal branches, especially along the backbone of the angiosperm phylogeny (Figure 2; Supplemental Figures 2 and 3). Importantly, given the nearly identical quartet frequencies for alternative topologies for the backbone of angiosperm with our datasets (Figures 2B and 3; Supplemental Figure 6), ILS is likely to impede phylogenetic resolution for the backbone of the angiosperm phylogeny. The concatenation method assumes that all genes have the same or similar evolutionary histories and implicitly ignore several complicated evolutionary realities, including gene tree conflict due to ILS and hybridization. Analyses of simulated and empirical data have demonstrated that concatenation approaches yield inconsistent results in the presence of ILS (Kubatko and Degnan, 2007, Zhong et al., 2013, Jarvis et al., 2014, Xi et al., 2014, Roch and Steel, 2015). Therefore, the concatenation approach may not be suitable for resolution of relationships among the five Mesangiospermae lineages.
Lack of phylogenetic signal may be part of the cause for the contentious relationship among these five groups of angiosperms. A large fraction of bipartitions of 1594 gene trees was uninformative at deep internal branches of the angiosperm tree of life (Figure 2A and Supplemental Figure 2). We used these uninformative genes (53% of 1594 genes) to infer a coalescent-based species tree, and found that support for relationships among the five main Mesangiospermae clades was negligible (Supplemental Figure 8). After filtering out these genes, the phylogenomic analyses based on the two subsets produced a largely congruent species tree, and the backbone of angiosperm phylogeny was well supported (Figure 3). The clade consisting of eudicots, Ceratophyllales, and Chloranthales was recovered in all coalescent-based analyses. The sister-group relationship between eudicots and Ceratophyllales was moderately supported, but the polytomy for the relationships among eudicots, Ceratophyllales, and Chloranthales could not be rejected (Figure 3). The magnoliids and monocots, being successive sister lineages of the eudicots–Ceratophyllales–Chloranthales clade, were recovered in the coalescent-based analyses. However, the support values of these nodes were moderate, and the polytomy test could not be rejected among these two lineages and the eudicots–Ceratophyllales–Chloranthales clade in the analyses of two subsets. These results suggested that a fully bifurcating tree may be an inadequate representation of the early radiation of angiosperms.
Rapid Radiation of Mesangiospermae
We inferred that five major clades of Mesangiospermae arose during a rapid radiation (∼27 Myr), similar to the findings of recent studies (Foster et al., 2017: 27 Myr; Salomo et al., 2017: 34 Myr; Li et al., 2019: 5 Myr). During this radiation, the diverse morphological and functional attributes of angiosperms were established (Magallón et al., 2015). Our estimated timeline showed that the earliest angiosperms originated in the Middle Triassic and the emergence of Mesangiospermae was in the Early Jurassic period. The similarity of our results to recent plastome-based studies reinforces suggest that more accurate and precise estimates of the evolutionary timescale of angiosperms will arise through improved models of molecular evolution and through the discovery of more early-diverging crown-group angiosperm fossils, rather than through increased gene sampling (Foster et al., 2017).
It is intriguing that the radiation of the crown groups of the major Mesangiospermae lineages is inferred to have occurred nearly 100 Myr after the origin of crown-group angiosperms. Possible explanations can be derived from paleoclimatic records. Climate simulations by Chaboureau et al. (2014) suggest that the Middle Triassic and Early Jurassic were characterized by extensive continuous latitudinal desert zones and a dry environment, which might have limited the potential migration and radiation of angiosperms. From the Early Jurassic (180 Ma) to the Early Cretaceous (120 Ma), the breakup of Pangea led to sharply increased continental rainfall and decreased desertic belts, possibly providing a geographical space and suitable environment for the major radiations of angiosperms. Our estimates for the timing of diversification of the crown groups of Mesangiospermae appears to coincide with global climatic changes associated with the decrease of desertic belts and the subsequent expansion of temperate zones during the Triassic to Cretaceous period, and might be an example of range expansion promoting diversification.
Methods
Taxon Sampling and Data Collection
We sampled 151 species representing the major lineages of angiosperms: 98 eudicots, 24 monocots, 19 magnoliids, four Chloranthales, two Ceratophyllales, and four early-diverging angiosperms. Our dataset includes representatives of most orders of Mesangiospermae: 32/44 eudicots orders, 9/11 monocots orders, 4/4 magnoliids orders, as well as Chloranthales and Ceratophyllales. Two gymnosperms (G.biloba and P. taeda) were used as outgroups. We source our data from 31 genomes from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html), 72 transcriptomes from GenBank (http://www.ncbi.nlm.nih.gov/genbank/), 18 transcriptomes retrieved from the “1000 plants” project database (https://db.cngb.org/blast4onekp), and 31 previously generated transcriptomes (Zeng et al., 2014, Huang et al., 2016a, Huang et al., 2016b, Zeng et al., 2017; C.-H.H. and H.M., unpublished). We newly sequenced the transcriptome of H. orientale following the protocols outlined in Zeng et al. (2014) (details provided in Supplemental Table 1). The complete list of taxa and information of data sources are provided in Supplemental Table 2.
Identification of Candidate Orthologous Genes
To identify reliable OGs for phylogenomic analyses, we obtained an initial dataset of 4180 putative orthologous genes (pOGs), which are shared by nine angiosperm genomes (Arabidopsis thaliana, Populus trichocarpa, Glycine max, Medicago truncatula, Vitis vinifera, Solanum lycopersicum, Oryza sativa, Sorghum bicolor, and Zea mays) available from the Deep Metazoan Phylogeny website (Ebersberger et al., 2009; http://www.deep-phylogeny.org/hamstr/). The 4180 pOGs were used as queries to retrieve homologous sequences from 153 species using HMMER v3.1b2 (Eddy, 2011) with default parameters. Amino acid sequences of each pOG were aligned by MAFFT v.7.3 (Katoh and Standley, 2013) using the L-INS-I algorithm, after which corresponding nucleotide alignments of each pOG were generated using PAL2NAL v14 (Suyama et al., 2006). Ambiguously aligned regions were excluded using Gblocks v.0.91b (Castresana, 2000) with the “codon” model and half gaps allowed. In addition, genes shorter than 600 nucleotides and sequences shorter than 50% of the alignment length were culled to reduce the amount of missing data, resulting in 2435 pOGs.
To identify putatively spurious or paralogous sequences, we applied a paralog-filtering work flow (Supplemental Figure 1; Simion et al., 2017). First, we estimated gene trees for the 2435 pOGs using maximum likelihood (ML) in RAxML v8.2.4 (Stamatakis, 2014) with the GTR+G substitution model and 100 rapid bootstrap replicates. We then inferred a reference species tree using a coalescent approach in ASTRAL-III v5.5.9 (Zhang et al., 2018) (hereafter ASTRAL), with all 2435 ML gene trees as the input. The branch lengths of the reference tree were optimized by analyzing a concatenated dataset of all 2435 pOGs using RAxML. We further estimated the branch lengths of each pOG based on constraint search of the reference topology, and removed putatively spurious or paralogous sequences if a ratio of the terminal branch length on the constrained gene tree and the reference tree was greater than 5 times. In addition, we calculated the Pearson correlation coefficient r between branch lengths on the constrained gene tree and the reference tree, and removed genes whose r values were outliers. Outlier genes were defined as those whose r values were greater than the upper whisker or smaller than the lower whisker of a box plot in the R programming environment (Ihaka and Gentleman, 1996):
| Upper whisker = min(max(x), Q3 + 1.5 IQR), | (Equation 1) |
| Lower whisker = max(min(x), Q1 − 1.5 IQR), | (Equation 2) |
where max(x) and min(x) are the maximum and minimum value for a set of r values, respectively. Q1 and Q3 are the first quartile and the third quartile, and IQR (interquartile range) is the difference in values between Q3 and Q1 (Q3 − Q1). To relax the assumption of a constrained gene tree, we further compared the branch lengths of each ML gene tree (unconstrained gene tree) and the reference tree, and sequences were removed if a ratio was greater than 5 times.
The final remaining sequences of each pOG were realigned and trimmed, and genes were discarded if the length was below 600 nucleotides and species coverage was lower than 70%. As Ceratophyllales and Chloranthales have smaller numbers of species, it is necessary to maintain all samples of these two clades (six species in total). Therefore, to identify OGs with sufficient taxonomic coverage of these two small clades, genes with less than six species of Ceratophyllales and Chloranthales were removed, ending up with a dataset of 1696 OGs.
Evaluation of Gene Tree Incongruence
To quantify the incongruence of gene trees, we examined the ABS values and the TCA scores of individual ML gene trees. The ABS of each gene tree was calculated using a custom R script. The relative TCA score was calculated using RAxML based on the best ML gene tree and 100 bootstrap replicates (Salichos et al., 2014). The TCA score is the sum of “Internode Certainty all” values across all internodes of a phylogeny. If gene tree incongruence is rare, most bipartitions across the tree are consistently recovered in much higher frequencies than conflicting bipartitions and the TCA score is near 1. Alternatively, TCA is close to 0 if many internodes have a low resolution, suggesting that a high frequency of conflicting bipartitions have been inferred across bootstrap replicates. We removed the genes that had ABS <50% and TCA ≤0.3 from the 1696 pOGs, resulting in a working dataset of 1594 OGs.
Phylogenetic Inferences
Coalescent and concatenation approaches were used to construct phylogenetic trees. For the coalescent approach, single ML gene trees were inferred using RAxML with the GTR+G model and 100 rapid bootstrap replicates. Because the summary coalescent methods are sensitive to gene tree estimation error (Gatesy and Springer, 2014, Bayzid et al., 2015), we collapsed low support branches (<20% bootstrap support) in gene trees to minimize potential impacts of gene tree error for species tree reconstruction. The species tree was inferred using ASTRAL (Zhang et al., 2018) with node support estimated by PP and MLBS. The quartet score was estimated for each node showing quartet support for the species tree and two alternative topologies. The polytomy test was calculated by ASTRAL (Sayyari and Mirarab, 2018) to evaluate whether a polytomy can be rejected for the relationships among five Mesangiospermae lineages, where a p value of <0.05 is considered to reject the null hypothesis of a polytomy. For the concatenation approach, nucleotide sequences of OGs were concatenated by Geneious v10.2.3 (Kearse et al., 2012). The ML tree was estimated using IQ-TREE 1.16.11(Nguyen et al., 2015) under the GTR+G model and supports were evaluated with ultrafast bootstrapping testing (1000 replicates) (Minh et al., 2013, Nguyen et al., 2015).
Dissecting Phylogenetic Signal among Orthologous Genes
Phylogenetic signal among gene trees was quantified by mapping 1594 rooted gene trees onto the concatenation-based species tree of the 1594-gene dataset using Phyparts (Smith et al., 2015) and ETE3 (Huerta-Cepas et al., 2016) implemented in PhyPartsPieCharts (https://github.com/mossmatters/MJPythonNotebooks; last accessed June 24, 2017). Gene trees were rooted on G. biloba if present, using P. taeda or Amborella trichopoda if Ginkgo was absent, and with Illicium henryi if the first three species were not present. Gene tree bootstrap cutoff was applied as 50% using -s parameter in Phyparts.
To further assess the phylogenetic signal present in our phylogenomic dataset, we examined five topological hypotheses, including our new coalescent-based phylogeny (Figure 2B) and four published representative topologies (Figure 1A–1D). For each topology, the constrained ML gene tree and per-site log-likelihood scores were estimated for 1594-gene dataset using RAxML. For each gene, we performed the AU test to statistically test the five topologies among the major lineages of Mesangiospermae in the program CONSEL (Shimodaira and Hasegawa, 2001). If a gene could not significantly reject any of the five hypotheses, the gene was considered to be uninformative for resolving the backbone of the angiosperm phylogeny (lack of phylogenetic signal). If a gene could significantly reject at least one of the five hypotheses, the gene was considered to have phylogenetic signal.
Divergence-Time Estimation
We estimated divergence times for our angiosperm-wide, 296-gene dataset using Bayesian inference in MCMCTree v.4.9h package (Yang, 2007; version released on March 31, 2018) with the uncorrelated relaxed clock model (clock = 2). To specify the prior on the overall substitution rate, we first ran BASEML (Yang, 2007) under a strict molecular clock with the root age (crown Spermatophyta) set to 340 Ma. The ML estimates of the branch lengths were then calculated using BASEML using the GTR+G nucleotide substitution model. We set the prior on the overall substitution rate across loci (μ) to G(1, 16.5), meaning 6 × 10−10 substitutions per site per year. The prior for the degree of rate variation across branches (σ2) was set to G(1, 3.4). The time unit was set to 100 Ma, and the parameters for the birth-death process were set as λ = μ = 1 and ρ = 0.0004. The sampling proportion (ρ) of 0.04% was based on our sample size (151 taxa) compared with the number of angiosperm species (∼352 000, http://www.theplantlist.org/). We also ran the program without sequence data to examine the effective priors and compared the shape of specified prior and effective prior distributions on all 38 nodes (Supplemental Figure 13). Our assessment showed that the paleontological constraints are reflected in the effective time prior. The MCMC analyses were run for 9 million generations sampled every 900 generations after a burn-in of 900 000 iterations. The chain convergence was assessed by running MCMC analyses twice, and the effective sample size of all parameters was confirmed to be >200 using Tracer v.1.6 (Rambaut et al., 2014). All calibration constraints were represented as uniform distributions with soft bounds, allowing 2.5% of the probability distribution to exceed the specified limit. The details of 38 selected fossil calibrations following contemporary standards (Parham et al., 2012) are available in Supplemental Information. Given the recent concerns that molecular dating of angiosperms might be biased by shifts in rates of molecular evolution, particularly among early-diverging lineages (Beaulieu et al., 2015), we tested for the impacts of rate heterogeneity among lineages on our estimates. To do so, we followed the methods of Foster et al. (2017), and tested for the impact of any life history-associated rate heterogeneity. We also tested the impact of alternative maximum age calibrations. Each of these analyses is described in Supplemental Information.
Funding
This study is supported by the National Natural Science Foundation of China (no. 31970229, no. 31570219, and no. 91531301), the Young Elite Scientists Sponsorship Program of Jiangsu Province, the State Key Laboratory of Paleobiology and Stratigraphy (Nanjing Institute of Geology and Paleontology, CAS) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author Contributions
B.Z. designed the study and managed the project. L.Y., D.S., X.C., C.S.P.F., L.S., and X.Z. performed phylogenetic and divergence-time analyses. C.-H.H., L.Z., and H.M. collected plant materials and prepared RNA samples. L.Y. and D.S. drafted the manuscript and C.S.P.F., H.M., and B.Z. revised the manuscript. All authors contributed and approved the final manuscript.
Acknowledgments
The authors thank Liang Liu, Ling Fang, Zhenhua Zhang, Yuan Nie, Xiaoya Ma, and Ning Zhang for assistance with analyses. No conflict of interest declared.
Published: February 4, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Accession Numbers
The dataset in this study have been deposited in the figshare (https://figshare.com/s/27c41bba65a30dbfd3c7).
Supplemental Information
References
- Barba-Montoya J., Reis M.D., Schneider H., Donoghue P.C.J., Yang Z. Constraining uncertainty in the timescale of angiosperm evolution and the veracity of a cretaceous terrestrial revolution. New Phytol. 2018;218:819–834. doi: 10.1111/nph.15011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayzid M.S., Mirarab S., Boussau B., Warnow T. Weighted statistical binning: enabling statistically consistent genome-scale phylogenetic analyses. PLoS One. 2015;10:e0129183. doi: 10.1371/journal.pone.0129183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J.M., O'Meara B.C., Crane P., Donoghue M.J. Heterogeneous rates of molecular evolution and diversification could explain the Triassic age estimate for angiosperms. Syst. Biol. 2015;64:869–878. doi: 10.1093/sysbio/syv027. [DOI] [PubMed] [Google Scholar]
- Byng J.W., Chase M., Christenhusz M., Fay M.F., Judd W.F., Mabberley D., Sennikov A., Soltis D.E., Soltis P.S., Stevens P. An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016;181:1–20. [Google Scholar]
- Cantino P.D., Doyle J.A., Graham S.W., Judd W.S., Olmstead R.G., Soltis D.E., Soltis P.S., Donoghue M.J. Towards a phylogenetic nomenclature of Tracheophyta. Taxon. 2007;56:822–846. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chaboureau A.C., Sepulchre P., Donnadieu Y., Franc A. Tectonic-driven climate change and the diversification of angiosperms. Proc. Natl. Acad. Sci. U S A. 2014;111:14066–14070. doi: 10.1073/pnas.1324002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane P.R., Lidgard S. Angiosperm diversification and paleolatitudinal gradients in Cretaceous floristic diversity. Science. 1989;246:675–678. doi: 10.1126/science.246.4930.675. [DOI] [PubMed] [Google Scholar]
- Crane P.R., Lidgard S. Angiosperm radiation and patterns of Cretaceous palynological diversity. In: Taylor P.D., Larwood G.P., editors. Major Evolutionary Radiations. Clarendon Press; Oxford: 1990. pp. 377–407. [Google Scholar]
- Darwin C., Darwin F., Seward A.C. Murray; London: 1903. More Letters of Charles Darwin: A Record of His Work in a Series of hitherto Unpublished Letters. [Google Scholar]
- Doyle J.A. Molecular and fossil evidence on the origin of angiosperms. Annu. Rev. Earth Planet. Sci. 2012;40:301–326. [Google Scholar]
- Ebersberger I., Strauss S., von Haeseler A. HaMStR: profile hidden markov model based search for orthologs in ESTs. BMC Evol. Biol. 2009;9:157. doi: 10.1186/1471-2148-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress P.K., Doyle J.A. Reconstructing the ancestral angiosperm flower and its initial specializations. Am. J. Bot. 2009;96:22–66. doi: 10.3732/ajb.0800047. [DOI] [PubMed] [Google Scholar]
- Eddy S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman W.E. The meaning of Darwin’s ‘abominable mystery’. Am. J. Bot. 2009;96:5–21. doi: 10.3732/ajb.0800150. [DOI] [PubMed] [Google Scholar]
- Friis E.M., Pedersen K.R., Crane P.R. Diversity in obscurity: fossil flowers and the early history of angiosperms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:369–382. doi: 10.1098/rstb.2009.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis E.M., Crane P.R., Pedersen K.R. Cambridge University Press; Cambridge: 2011. Early Flowers and Angiosperm Evolution. [Google Scholar]
- Foster C.S.P. The evolutionary history of flowering plants. J. Proc. R. Soc. New South Wales. 2016;149:65–82. [Google Scholar]
- Foster C.S.P., Sauquet H., van der Merve M., McPherson H., Rossetto M., Ho S.Y.W. Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Syst. Biol. 2017;66:338–351. doi: 10.1093/sysbio/syw086. [DOI] [PubMed] [Google Scholar]
- Gatesy J., Springer M.S. Phylogenetic analysis at deep timescales: unreliable gene trees, bypassed hidden support, and the coalescence/concatalescence conundrum. Mol. Phylogenet. Evol. 2014;80:231–266. doi: 10.1016/j.ympev.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Gomez B., Daviero-Gomez V., Coiffard C., Martin-Closas C., Dilcher D.L. Montsechia, an ancient aquatic angiosperm. Proc. Natl. Acad. Sci. U S A. 2015;112:10985–10988. doi: 10.1073/pnas.1509241112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitzendanner M.A., Soltis P.S., Wong G.K., Ruhfel B.R., Soltis D.E. Plastid phylogenomic analysis of green plants: a billion years of evolutionary history. Am. J. Bot. 2018;105:291–301. doi: 10.1002/ajb2.1048. [DOI] [PubMed] [Google Scholar]
- Herendeen P.S., Friis E.M., Pedersen K.R., Crane P.R. Palaeobotanical redux: revisiting the age of the angiosperms. Nat. Plants. 2017;3:17015. doi: 10.1038/nplants.2017.15. [DOI] [PubMed] [Google Scholar]
- Hertweck K.L., Kinney M.S., Stuart S.A., Maurin O., Mathews S., Chase M.W., Gandolfo M.A., Pires J.C. Phylogenetics, divergence times and diversification from three genomic partitions in monocots. Bot. J. Linn. Soc. 2015;178:375–393. [Google Scholar]
- Huang C., Sun R., Hu Y., Zeng L., Zhang N., Cai L., Zhang Q., Koch M.A., Al-Shehbaz I., Edger P.P. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 2016;33:394–412. doi: 10.1093/molbev/msv226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-H., Zhang C., Liu M., Hu Y., Gao T., Qi J., Ma H. Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol. Biol. Evol. 2016;33:2820–2835. doi: 10.1093/molbev/msw157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Serra F., Bork P. ETE3: reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R.R. A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Judd W.S., Campbell C.S., Kellogg E.A., Stevens P.F., Donoghue M.J. Sinauer Associates; Sunderland: 1999. Plant Systematics: A Phylogenetic Approach. [Google Scholar]
- Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubatko L.S., Degnan J.H. Inconsistency of phylogenetic estimates from concatenated data under coalescence. Syst. Biol. 2007;56:17–24. doi: 10.1080/10635150601146041. [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J.H., Barker M.S., Carpenter E.J., Deyholos M.K., Gitzendanner M.A., Graham S.W., Grosse I., Li Z., Melkonian M., Mirarab S. One thousand plant transcriptomes and the phylogenomics of green plants. Nature. 2019;574:679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.T., Yi T.S., Gao L.M., Ma P.F., Zhang T., Yang J.B., Gitzendanner M.A., Fritsch P.W., Cai J., Luo Y. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants. 2019;5:461–470. doi: 10.1038/s41477-019-0421-0. [DOI] [PubMed] [Google Scholar]
- Lidgard S., Crane P.R. Quantitative analyses of the early angiosperm radiation. Nature. 1988;331:344–346. [Google Scholar]
- Lidgard S., Crane P.R. Angiosperm diversification and Cretaceous floristic trends: a comparison of palynofloras and leaf macrofloras. Paleobiology. 1990;16:77–93. [Google Scholar]
- Maarten J.M., Christenhuse, James W.B. The number of known plants species in the world and its annual increase. Phytotaxa. 2016;261:201–217. [Google Scholar]
- Massoni J., Forest F., Sauquet H. Increased sampling of both genes and taxa improves resolution of phylogenetic relationships within Magnoliidae, a large and early-diverging clade of angiosperms. Mol. Phylogenet. Evol. 2014;70:84–93. doi: 10.1016/j.ympev.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Moore M.J., Bell C.D., Soltis P.S., Soltis D.E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. U S A. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. U S A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magallón S., Castillo A. Angiosperm diversification through time. Am. J. Bot. 2009;96:349–365. doi: 10.3732/ajb.0800060. [DOI] [PubMed] [Google Scholar]
- Magallón S., Gómez-Acevedo S., Sánchez-Reyes L.L., Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207:437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Nguyen M.A., von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.L., Puttick M.N., Clark J.W., Edwards D., Kenrick P., Pressel S., Wellman C.H., Yang Z., Schneider H., Donoghue P.C.J. The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U S A. 2018;115:E2274–E2283. doi: 10.1073/pnas.1719588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham J.F., Donoghue P.C., Bell C.J., Calway T.D., Head J.J., Holrooyd P.A., Irmis R.B., Joyce W.G., Ksepka D.T., Patané J.S. Best practices for justifying fossil calibrations. Syst. Biol. 2012;61:346–359. doi: 10.1093/sysbio/syr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick M.N., Morris J.L., Williams T.A., Cox C.J., Edwards D., Kenrick P., Pressel S., Wellman C.H., Schneider H., Pisani D. The interrelationships of land plants and the nature of the ancestral embryophyte. Curr. Biol. 2018;28:733–745. doi: 10.1016/j.cub.2018.01.063. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li L., Wang B., Xue J., Hendry T.A., Li R., Brown J.W., Liu Y., Hudson G.T., Chen Z. Angiosperm phylogeny inferred from sequences of four mitochondrial genes. J. Syst. Evol. 2010;48:391–425. [Google Scholar]
- Ruhfel B., Gitzendanner M., Soltis P., Soltis D., Burleigh J. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014;14:23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Suchard M., Drummond A. Tracer, version 1.6.0. 2014. http://tree.bio.ed.ac.uk/software/tracer/
- Roch S., Steel M. Likelihood-based tree reconstruction on a concatenation of aligned sequence data sets can be statistically inconsistent. Theor. Popul. Biol. 2015;100:56–62. doi: 10.1016/j.tpb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Salichos L., Rokas A. Inferring ancient divergences requires genes with strong phylogenetic signals. Nature. 2013;497:327–331. doi: 10.1038/nature12130. [DOI] [PubMed] [Google Scholar]
- Salichos L., Stamatakis A., Rokas A. Novel information theory-based measures for quantifying incongruence among phylogenetic trees. Mol. Biol. Evol. 2014;31:1261–1271. doi: 10.1093/molbev/msu061. [DOI] [PubMed] [Google Scholar]
- Sayyari E., Mirarab S. Testing for polytomies in phylogenetic species trees using quartet frequencies. Genes. 2018;9:132. doi: 10.3390/genes9030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H., Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Smith S.A., Beaulieu J., Donoghue M. An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proc. Natl. Acad. Sci. U S A. 2010;107:5897–5902. doi: 10.1073/pnas.1001225107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., Moore M.J., Brown J.W., Yang Y. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evol. Biol. 2015;15:150. doi: 10.1186/s12862-015-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Smith S.A., Cellinese, Wurdack K.J., Tank D.C., Brockington S.F., Refulio-Rodriguez N.F., Walker J.B., Moore M.J., Carlsward B.S. Angiosperm phylogeny: 17 genes, 640 taxa. Am. J. Bot. 2011;98:704–730. doi: 10.3732/ajb.1000404. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML Version 8: a tool for phylogenetic analysis and post- analysis of large phylogenies. Bioinformatics. 2014;30:1312–1315. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simion P., Philippe H., Baurain D., Jager M., Richter D.J., Di Franco A., Roure B., Satoh N., Quéinnec É., Ereskovsky A. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 2017;27:958–967. doi: 10.1016/j.cub.2017.02.031. [DOI] [PubMed] [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Amborella Genome Project The Amborella genome and the evolution of flowering plants. Science. 2013;342:1241089. doi: 10.1126/science.1241089. [DOI] [PubMed] [Google Scholar]
- Tilman D., Cassman K.G., Matson P.A., Naylor R., Polasky S. Agricultural sustainability and intensive production practices. Nature. 2002;418:671–677. doi: 10.1038/nature01014. [DOI] [PubMed] [Google Scholar]
- Wickett N.J., Mirarab S., Nguyen N., Warnow T., Carpenter E., Matasci N., Ayyampalayam S., Barker M.S., Burleigh J.G., Gitzendanner M.A. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. U S A. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Liu L., Rest J.S., Davis C.C. Coalescent versus concatenation methods and the placement of Amborella as sister to water lilies. Syst. Biol. 2014;63:919–932. doi: 10.1093/sysbio/syu055. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zeng L., Zhang Q., Sun R., Kong H., Zhang N., Ma H. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014;5:4956. doi: 10.1038/ncomms5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Zhang N., Zhang Q., Endress P.K., Huang J., Ma H. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017;214:1338–1354. doi: 10.1111/nph.14503. [DOI] [PubMed] [Google Scholar]
- Zhong B., Liu L., Yan Z., Penny D. Origin of land plants using the multispecies coalescent model. Trends Plant Sci. 2013;18:492–495. doi: 10.1016/j.tplants.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Zhong B., Betancur-R R. Expanded taxonomic sampling coupled with gene genealogy interrogation provides unambiguous resolution for the evolutionary root of angiosperms. Genome Biol. Evol. 2017;9:3154–3161. [Google Scholar]
- Zhang C., Sayyari E., Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19:153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen F., Zhang X. The water lily genome and the early evolution of flowering plants. Nature. 2020;577:79–84. doi: 10.1038/s41586-019-1852-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomo K., Smith J., F., Feild T., S. The Emergence of Earliest Angiosperms may be Earlier than Fossil Evidence Indicates. Syst. Bot. 2017;42:607–619. doi: 10.1600/036364417X696438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.