Abstract
The establishment of symbiotic nitrogen fixation requires the coordination of both nodule development and infection events. Despite the evolution of a variety of anatomical structures, nodule organs serve a common purpose in establishing a localized area that facilitates efficient nitrogen fixation. As in all plant developmental processes, the establishment of a new nodule organ is regulated by plant hormones. During nodule initiation, regulation of plant hormone signaling is one of the major targets of symbiotic signaling. We review the role of major developmental hormones in the initiation of the nodule organ and argue that the manipulation of plant hormones is a key requirement for engineering nitrogen fixation in non-legumes as the basis for improved food security and sustainability.
Key words: nitrogen fixation, symbiosis, hormones, nodule, legume
Legume nodules are specialized symbiotic organs that develop to facilitate efficient nitrogen fixation. This review discusses how plant hormones regulate the expression of symbiotic genes and the process of nodule development. Manipulating these hormone-symbiosis interactions is essential for improving legume crops to support nitrogen fixation in various environments and for engineering nitrogen fixation in cereal crops.
Introduction
Nitrogen Fixing Symbioses
Nitrogen limits the production of food in all agroecosystems. Besides synthetic fertilizer, the second greatest source of nitrogen in agriculture is biological nitrogen fixation, such as that which occurs in the symbiotic relationship between legume crops and soil bacteria called rhizobia. Legumes are members of the FaFaCuRo clade (Fabales, Fagales, Cucurbitales, Rosales), which contains the only plant families that engage in this type of symbiosis (Soltis et al., 1995). A common feature of this symbiosis is the formation of a specialized organ (known as a nodule) that enables the reciprocal transfer of carbon from the plant and fixed nitrogen from the bacteria.
Importance of the Nodule Organ for Symbiotic Nitrogen Fixation
The nodule organ provides a number of favorable conditions for efficient nitrogen fixation. It is likely that no single function is sufficient to explain the requirement for a nodule, but together they provide an advantageous environment that necessitates the formation of such an organ. These functions include the facilitation of infection by host cell division (Breakspear et al., 2014), the development of a sink in which localized high gene expression levels facilitate nutrient and metabolite exchange (Clarke et al., 2014), the establishment of a low-oxygen region that facilitates nitrogenase activity (Layzell and Hunt, 1990), and the local relaxation of defense responses (Benezech et al., 2020).
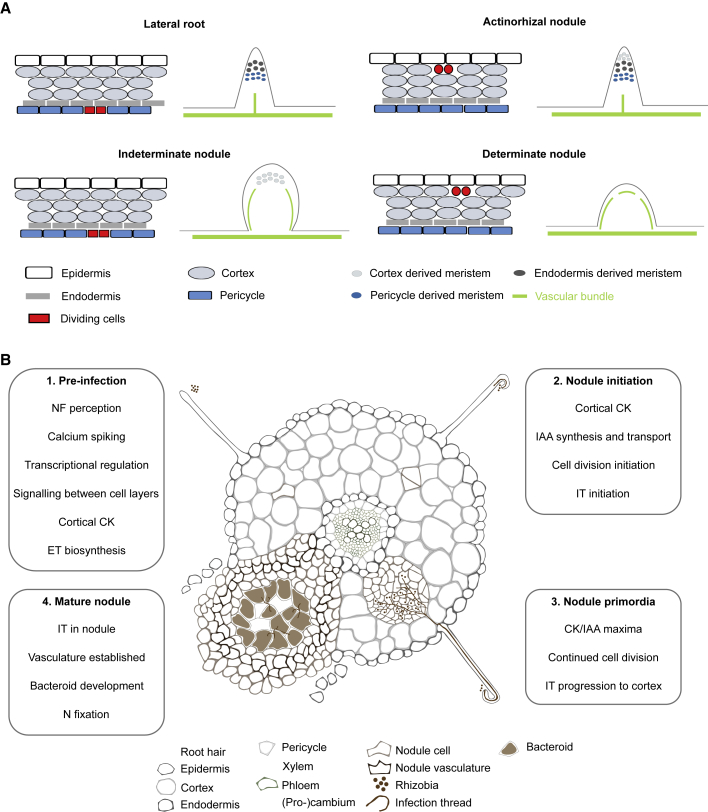
There is some flexibility in the allowed structure of a nodule, as a number of related developmental programs are observed (Figure 1A). These differences are principally in the nodule initiation site (the cell layer varies from the pericycle to the outer cortex), the vascular arrangement, and the source and persistence of meristematic cells (Szczyglowski et al., 1998; Xiao et al., 2014, 2019; Shen et al., 2020). Although there are numerous determinants of nitrogen fixation efficiency, it is not clear whether any are directly related to the varied organ structures that have evolved (Terpolilli et al., 2012).
Figure 1.
Nodule Development and Signaling Pathways.
(A) Comparison of the initiation sites and structures of lateral roots and several major nodule types. The first cell division (red) can vary within and between nodule types from the pericycle to the outer cortex. Vascular patterning and persistence of meristematic cells also differ.
(B) Major events during nodule organogenesis. Development of a determinate-type nodule as found in the model legume Lotus japonicus is exemplified. The establishment of cortical cytokinin (CK) and auxin signaling domains are major requirements for pre-infection signaling to stimulate nodule initiation and growth. NF, Nod factor; ET, ethylene; IT, Infection thread; IAA, indole-3-acetic acid (auxin).
Plant Hormones Involved in Nodule Organogenesis
Plant hormones regulate all developmental processes, including nodule formation (Ferguson and Mathesius, 2014). The ubiquity of hormones means that they are often hubs that integrate plant environmental signals with developmental processes, including nodulation.
Here, we review the role of developmental hormones in the initiation of the nodule organ in legumes (Figure 1B). We pay particular attention to developmental hormones for which the greatest mechanistic insights have been uncovered: cytokinin (CK), auxin, ethylene, gibberellins (GA), and peptide hormones. Finally, the challenge of engineering nodule organogenesis in non-legumes is discussed in the context of plant hormone signaling as a key target.
Initiation of Nodulation as a Target of Symbiotic Signaling
Nodule initiation in legumes occurs in response to the perception of Nod factors by the LysM receptors (LjNFR1/LjNFR5 and orthologs; Madsen et al., 2003; Radutoiu et al., 2003). Following receptor activation, perinuclear calcium spiking activates the common symbiosis pathway components, named for their common role in both nodulation and arbuscular mycorrhizal symbioses (2020) (for further details, see recent review by Roy et al., 2020). Transcriptional regulators are crucial for this early signaling and include CCAMK (MtDMI3) (Lévy et al., 2004), CYCLOPS (MtIPD3) (Messinese et al., 2007; Yano et al., 2008; Singh et al., 2014b), NSP1 (Smit et al., 2005), and NSP2 (Kaló et al., 2005). Additional transcriptional regulators that are specific to nodulation include ERN1 (Andriankaja et al., 2007), NIN (Schauser et al., 1999), and members of the NF-Y heterocomplex (Laloum et al., 2014; Laporte et al., 2014; Baudin et al., 2015). These transcriptional regulators are critical for the initiation of normal infection and organogenesis processes and are interlinked with many hormone signaling pathways (Buhian and Bensmihen, 2018). Transcriptome studies indicate that the expression of many, if not all, of these transcription factors is modulated by hormones. A key role of this early signaling is also to initiate hormone signaling, including that of cytokinin and auxin, which are the two major hormones that influence the cell-cycle initiation and progression required for nodule initiation. Further information is given on the mechanisms by which plant hormones interact with the nodulation pathway in each section below.
Cytokinins
Cytokinin Synthesis, Metabolism, and Signaling
CKs were named after their ability to regulate cell division (Miller et al., 1955, 1956) and have been implicated in nearly all aspects of plant growth and development (reviewed in Werner and Schmülling, 2009; Kieber and Schaller, 2014; Cortleven et al., 2019).
Structurally, natural CKs are adenine derivatives that are either isoprenylated or carry an aromatic side chain at the N6-residue (Sakakibara, 2006). Studies, mainly in Arabidopsis, suggest that N6-isopentenyladenine (iP) and trans-zeatin (tZ) are the most biologically relevant CKs, with iP being primarily important for root development and tZ being crucial for most developmental processes in the shoot (Miyawaki et al., 2006; Kiba et al., 2013). They are initially synthesized by isopentenyltransferases (IPTs), forming iP nucleotides (Kakimoto, 2001; Takei et al., 2001), which may be converted further to tZ nucleotides by cytochrome P450 monooxygenases (CYP735As) (Takei et al., 2004; Kiba et al., 2013). Bioactive CKs are formed via dephosphoribosylation of CK riboside monophosphate precursors by LONELY GUYs (LOGs) (Kurakawa et al., 2007). They can be irreversibly degraded by either N-glucosylation or side-chain cleavage.
Bioactive CKs activate signaling through a histidine/aspartate phosphorylation cascade that bears similarities to the two-component signaling which is frequently seen in bacteria to couple membrane sensors with cellular responses (reviewed in Werner and Schmülling, 2009; Kieber and Schaller, 2014). Central components are the histidine kinase (HK) receptors (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001), which in turn phosphorylate the histidine residues of histidine phosphotransfer proteins (HPs) (Hutchison et al., 2006). The phosphorylation signal is transferred from HPs to type-B response regulator (RRB) transcription factors, activating them and ultimately leading to CK-dependent gene expression (Imamura et al., 1999). Acting antagonistically to these, RRAs act as negative regulators of signaling, establishing a feedback mechanism (Brandstatter and Kieber, 1998; Sakai et al., 2001).
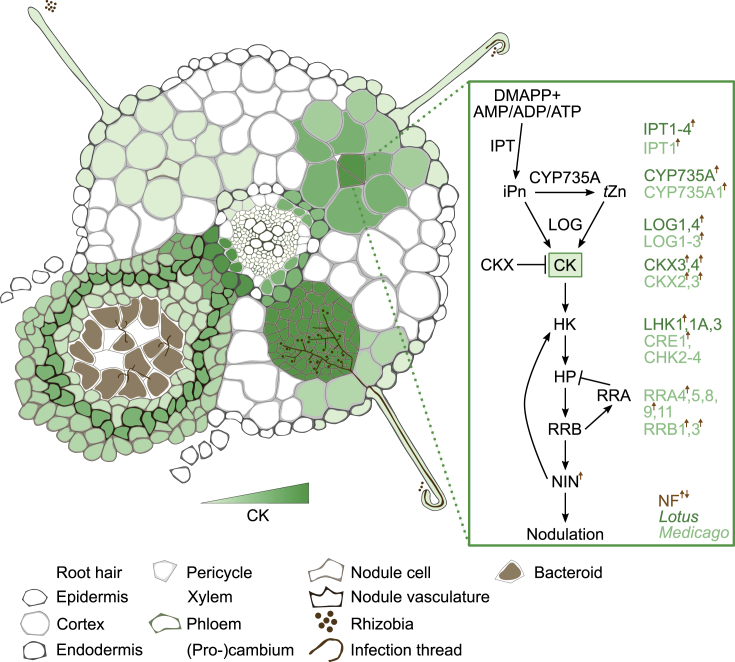
As well as their roles in many aspects of plant development, CKs are central regulators of nodule organogenesis and root hair infection (Figure 2; reviewed in Frugier et al., 2008; Miri et al., 2016; Gamas et al., 2017).
Figure 2.
CK Biosynthesis, Metabolism, and Signaling.
Intensity of signaling output at different developmental stages is depicted based on reporter studies, transcriptome experiments, and signaling mutant phenotypes. Following the perception of rhizobia, CK level and signaling is rapidly induced in the cortex. Cortical CK remains increased during nodule initiation and growth, while surrounding tissue and epidermal responses are restricted. Mature nodules retain CK signaling in the vasculature, with reduced signaling in infected cells. Core biosynthesis, metabolism, and signaling components are depicted at the right, along with species-specific components, where known. NF-induced/repressed components are indicated with a brown arrow (up or down, respectively).
Cytokinin in Nodule Organogenesis
Among the first indications that CK was of major importance for nodule organogenesis were the reports that CK application led to increased nodule formation in Sesbania rostrata (tZ) and alfalfa (BAP) (Dehio and de Bruijn, 1992; Cooper and Long, 1994; Bauer et al., 1996; Fang and Hirsch, 1998). A critical molecular link was established with the demonstration in Lotus japonicus and Medicago truncatula that the CK receptor LjLHK1 was necessary for nodulation and, when activated, sufficient to initiate nodules spontaneously in the absence of bacteria (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007).
All isoprenoid class CKs, including the most relevant iP-type and tZ-type CKs, are induced upon infection with rhizobia or Nod factor (NF) treatment in M. truncatula and L. japonicus (van Zeijl et al., 2015; Reid et al., 2016). Multiple IPT-encoding genes have been identified as rapidly induced by rhizobia or NF treatment (Chen et al., 2014; van Zeijl et al., 2015; Reid et al., 2017; Mens et al., 2018). In L. japonicus, mutants in Ipt4 or knockdown of Ipt3 caused a reduced number of nodules, whereas Ipt2, which is the most significantly induced at the early stage, has not been characterized by reverse genetics (Chen et al., 2014; Reid et al., 2017). Intriguingly, nodulation of ipt3ipt4 plants is indistinguishable from that of wild type (Reid et al., 2017), suggesting that the CK pool is sufficient for nodulation but that the balance with active forms may be altered. Active CKs are also rapidly formed, and several LOG family genes increase in abundance within 24 h of inoculation and throughout nodule primordia establishment (Mortier et al., 2014; van Zeijl et al., 2015; Reid et al., 2017). Although the functional relevance of these genes has not been demonstrated by analysis of loss-of-function mutants, overexpression of LjLog4 leads to spontaneous nodule formation (Reid et al., 2017), and both knockdown and overexpression of MtLog1 result in reduced nodule numbers compared with wild-type M. truncatula (Mortier et al., 2014). The relative activity of different L. japonicus LOG enzymes is unknown, although it would appear likely that overexpression of other LOG enzymes would achieve a similar accumulation of active cytokinin and thereby trigger organogenesis. Together, these studies suggest a key role for CK biosynthesis in the establishment of an early burst of CK to initiate cell division. Aside from the role of CKs in nodule organogenesis, biosynthesis of iP-type CKs by IPT3 in the shoot has also been suggested to play a role in the long-distance (systemic) negative regulation of nodule initiation (Sasaki et al., 2014). A similar shoot induction of CK biosynthesis has been found for soybean GmIpt5 (Mens et al., 2018). The role of CK in the autoregulation of nodulation (AON) pathway is discussed further in later sections.
Whether iP- or tZ-type CKs play particular roles in nodulation remains unclear. An increased abundance of LjCyp735a transcripts and higher tZR levels occur within 24 h of rhizobia infection in L. japonicus (Reid et al., 2016, 2017). In M. truncatula, MtCyp735a1 is induced and tZ levels are increased 3 h after NF treatment (van Zeijl et al., 2015) or predicted by transcriptional networks in the epidermis 24 h after NF treatment (Jardinaud et al., 2016). However, only one predicted Cyp735a gene exists in L. japonicus, and Ljcyp735a mutants do not show a nodulation phenotype, indicating that tZ-type CK is not critical for nodulation in L. japonicus (Reid et al., 2017).
The regulation of CK biosynthesis appears complex: MtIpt1 and MtCyp735a1 are induced in a partially CRE1-dependent manner, whereas LjIpt2 and LjLog4 show increased expression in Ljlhk1 mutants, indicating that regulation may differ between species (van Zeijl et al., 2015; Reid et al., 2017). Future studies to identify which combinations of Ipt and Log genes are required for nodule organogenesis and to characterize the regulation of these genes will be critical for defining how the initial burst of CK required for nodule induction is regulated by the symbiotic pathway.
The importance of CK degradation during nodule formation has been described in L. japonicus. LjCkx3 is specifically expressed in xylem-pole pericycle cells and dividing cortical cells during early nodule development (Reid et al., 2016), and in M. truncatula, MtCkx1 is a primary target of cytokinin signaling (Ariel et al., 2012). Mutants in LjCkx3 have reduced infection and nodule numbers, which results at least in part from crosstalk with ethylene signaling (see below; Reid et al., 2016).
The active pool of CK triggers the CK signaling pathway, including the primary CK response by the RRBs. The CK-responsive TCS/TCSn promoters, which carry repetitive elements targeted by these response regulators, have been used to define the signaling domains of CK (Müller and Sheen, 2008; Zürcher et al., 2013). These promoters revealed that CK signaling in L. japonicus is activated in cortical cells within 24 h and is highly active throughout the development of young nodules, whereas the signaling is limited to vascular bundles of mature nodules (Held et al., 2014; Reid et al., 2017). A similar pattern in early nodule development is seen in M. truncatula, with the strongest CK signaling occurring in endodermal and pericycle cells (van Zeijl et al., 2015). There remains some uncertainty over the timing and relative signaling intensity of CK in the epidermis. In experiments with rhizobia inoculation in L. japonicus, early epidermal signaling activity was not detected, and later epidermal activity was mutually exclusive with infection (Reid et al., 2017). In M. truncatula, strong epidermal and outer cortical activity was detected in response to NF or Sinorhizobium meliloti inoculation through both tissue-specific transcriptomics and TCSn analyses (Jardinaud et al., 2016), and activity in the cortex, endodermis, and pericycle was detected in response to NF treatment using TCS (van Zeijl et al., 2015). The regulation of this process is highly dynamic and therefore difficult to compare between experimental systems. Whether CK accumulation and signaling differ between species with different nodule types and initiation locations will be key questions to resolve. Studies to directly identify CK accumulation rather than signaling at a cellular resolution may be key to resolving this question.
Most studies of CK signaling components in nodulation have focused on the role of CK receptors, and the current understanding is that three L. japonicus CK receptors act partially redundantly, with LjLHK1 playing the predominant role in nodulation (Held et al., 2014). In hyperinfection thread1 (hit1-1 = lhk1 har1) roots, nodule primordia are aborted despite hyperinfection of root hairs, highlighting the different roles of CK in nodule organogenesis and root hair infection (Murray et al., 2007). Similarly, the LjLHK1 homolog MtCRE1 functions as the main CK receptor during nodulation in M. truncatula. MtCre1 knockdown or loss-of-function mutants have a reduced nodule number but, in contrast to lhk1-1, no increase in infection events (Gonzalez-Rizzo et al., 2006; Plet et al., 2011), and the other three receptors (MtCHK2–4) have partially redundant functions (Boivin et al., 2016). Gain-of-function alleles of LjLhk1 named spontaneous nodule formation2/5 (snf2/snf5) also demonstrated that increased CK signaling is sufficient to induce nodulation, independent of rhizobia infection (Tirichine et al., 2007; Liu et al., 2018b). Similarly, the application of CK or overexpression of its biosynthetic pathway achieves similar results (Heckmann et al., 2011; Reid et al., 2017). LjLhk1 expression mostly overlaps with the expression pattern of TCS/TCSn during nodulation (Held et al., 2014), and its expression is induced upon CK treatment, suggesting a positive feedback loop during nodule organogenesis (Murray et al., 2007). In M. truncatula, the transcriptional regulator MtNIN plays a key role in this positive feedback mechanism (Vernié et al., 2015). The cortical expression of LjLhk1 is sufficient for both organogenesis initiation and infection regulation, highlighting the key role played by CK in linking these two processes (Miri et al., 2019). This cortex-dependent infection regulation may require ethylene as an intermediary and is discussed in more detail below.
Downstream of the well-studied CK receptors, the high degrees of redundancy in the core signaling pathway have hindered detailed study. Of the core components, several Rrs in M. truncatula are induced upon rhizobia infection and CK treatment (Gonzalez-Rizzo et al., 2006; van Zeijl et al., 2015). Among these, MtRrb3 is most strongly expressed in nodules, and mutants exhibit fewer nodules, indicating that redundancy in signaling components is only partial (Tan et al., 2020). This work also identified a direct regulatory relationship with Nsp2 and control of endoreduplication by CK in nodulation.
Interconnection between CK and Nodulation Signaling
Given the central role of CK in nodule organogenesis, the question arises: how do CK synthesis, metabolism, and signaling interact with other factors that are crucial for nodulation? Studies in L. japonicus suggest that CK acts downstream of the NF receptors LjNFR1 and LjNFR5 and the calcium signaling decoded by LjCCaMK (Tirichine et al., 2007; Madsen et al., 2010). At the same time, CK biosynthesis and signaling act upstream of LjNIN to positively regulate its expression (Murray et al., 2007; Tirichine et al., 2007; Madsen et al., 2010; Heckmann et al., 2011; Chen et al., 2014). This regulation of LjNin by CK occurs through a distal cis-regulatory region in the LjNin promoter that renders Ljdaphne (Yoro et al., 2014) and Mtdaphne-like mutants (Liu et al., 2019) unable to form nodules despite competence for infection events, as the distal promoter regions are not required for Nin expression in the epidermis (Liu et al., 2019). The exact factors that control these distant regulatory motifs remain to be identified. The contribution of CK to early symbiotic signaling in the epidermis remains a key point to resolve, as MtCRE1 was reported to be required for the vast majority of symbiotic signaling (Schiessl et al., 2019), and null mutants or RNAi targeting of MtCre1 results in reduced or aborted infection threads (Gonzalez-Rizzo et al., 2006; Plet et al., 2011). On the other hand, Ljlhk1 mutants remain highly sensitive to NF and rhizobia inoculation, with hyperactive CK biosynthesis and hyperinfection (Murray et al., 2007; Reid et al., 2017). The precise genetic regulation of CK biosynthesis and signaling response by the symbiosis pathways is likely to be key to identifying how signals are propagated between the epidermis and cortex to initiate nodulation, and the analysis of signaling components such as RRs (as in RRB3; Tan et al., 2020) is likely to identify mechanisms that link CK with organ initiation and identity.
Auxin
Auxin Biosynthesis, Metabolism, and Signaling
Auxins regulate essentially all plant developmental processes (Teale et al., 2006). The main natural auxin, indole-3-acetic acid (IAA), is predominantly synthesized through the tryptophan-dependent indole-3-pyruvic acid (IPA) pathway (reviewed in Zhao, 2012; Korasick et al., 2013). Initially, TRYPTOPHAN AMIDOTRANSFERASE OF ARABIDOPSIS1 (TAA1) and related (TAR) proteins convert tryptophan to IPA (Stepanova et al., 2008), which is subsequently converted to bioactive free IAA by YUCCA proteins (Zhao et al., 2001). IAA may be modified/inactivated in many ways, including oxidation or amino acid and carbohydrate conjugation. Intracellular IAA activates the auxin signaling pathway after perception by the TRANSPORT INHIBITOR RESISTANT1 (TIR1)/AUXIN signaling F-BOX (AFB) receptors (Dharmasiri et al., 2005a, 2005b; Parry et al., 2009). In the absence of IAA, TIR/AFBs are inactive, which allows AUX/IAA proteins to inhibit the function of AUXIN RESPONSE FACTOR (ARF) transcription factors through direct interaction (Ulmasov et al., 1997b, 1999b). Upon perception of IAA by TIR1/AFB receptors, a Skp1-Cullin-F-box (SCF)-TIR1/AFB complex is formed that inhibits Aux/IAA function by mediating their ubiquitination and thus their proteasome-dependent degradation. Consequently, ARFs are released from Aux/IAA repression and can form dimers to positively or negatively regulate the transcription of output genes through binding to auxin response elements (Ulmasov et al., 1997a, 1999a, 1999b).
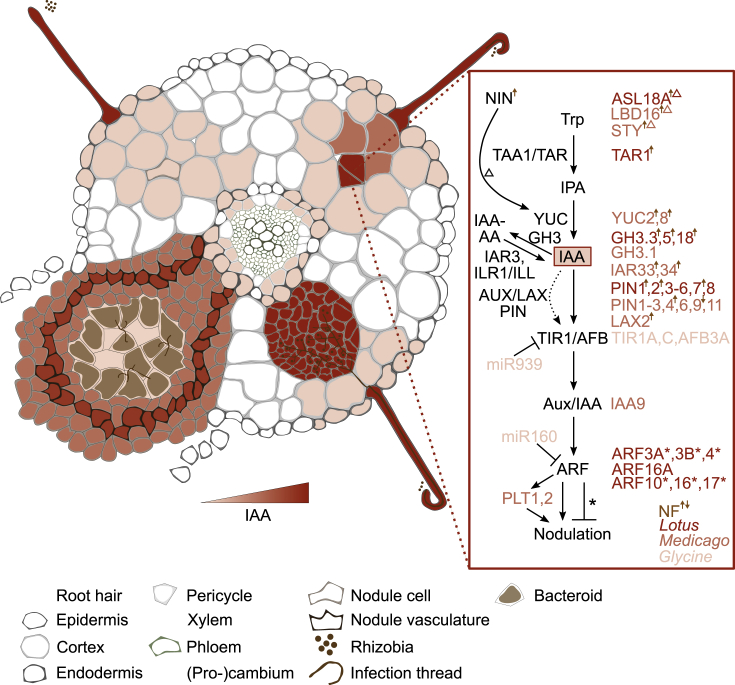
The role of auxin in nodule organogenesis has been studied extensively, and greater mechanistic insights are now emerging (Figure 3; reviewed in Kohlen et al., 2018).
Figure 3.
Auxin Biosynthesis, Metabolism, and Signaling.
Intensity of signaling output at different developmental stages is depicted based on reporter studies, transcriptome experiments, and signaling mutant phenotypes. Following rhizobia perception, auxin levels increase, and signaling output is initiated in the epidermis. Auxin biosynthesis and transport regulation contribute to increased signaling associated with nodule initiation and growth and are maintained until nodule maturity. Generally known biosynthesis, metabolism, and signaling components are depicted at the right with species-specific components, where known. A dotted line indicates IAA transport by AUX/LAX/PIN proteins. Factors through which NIN induces YUC are marked with a triangle. ARFs acting as negative regulators of nodulation are depicted with asterisks. NF-induced/repressed components are indicated with a brown arrow (up or down, respectively).
Auxin in Nodule Development
The importance of auxin for nodule development is perhaps best illustrated by the ability of auxin transport inhibitors to induce organogenesis and Enod gene expression (Hirsch et al., 1989; Scheres et al., 1992; Fang and Hirsch, 1998; Rightmyer and Long, 2011). In M. truncatula, auxin transport inhibitor application is also sufficient to partially rescue the organogenesis defect of the cre1 mutant (Ng et al., 2015).
Auxin production and response in root hairs occur rapidly in response to NF through both enhanced biosynthesis and transport regulation (Breakspear et al., 2014; Nadzieja et al., 2018). During nodule organogenesis, auxin response occurs in an LjLHK1- and LjNIN-dependent manner (Suzaki et al., 2012). In M. truncatula, this link is established by MtNIN-dependent induction of MtYuc2,8 expression via MtLOB-DOMAIN PROTEIN 16 (MtLBD16) (Schiessl et al., 2019). The L. japonicus ortholog of Lbd16 (Asl18a) is regulated by LjNIN through cis elements in the intron, as well as by auxin during lateral root formation (Soyano et al., 2019). Shi/Sty genes, which in Arabidopsis are known to regulate Yuc expression, are also regulated by NF-YA1 in L. japonicus (Hossain et al., 2016). One of the major roles of these transcriptional regulators is therefore to establish connections between symbiotic signaling and the production and localization of an auxin maximum required for cell division and nodule initiation. This interconnection between auxin and organogenesis is also evident in other plant developmental processes, in particular lateral root initiation, where LBD16 and SHI/STYs are also central to the establishment of auxin signaling domains.
Induction of IAA conjugation by GH3s is a common feedback feature of auxin response in many developmental contexts, including nodulation, and is therefore commonly used as a marker for auxin signaling. Expression of Gh3s during both infection and nodule development highlights the importance of metabolic feedback on auxin signaling in these processes (Mathesius et al., 1998; van Noorden et al., 2007; Singh et al., 2014a; Breakspear et al., 2014). In addition, regulation of MtIAR33 and MtIAR34 during nodulation may indicate that IAA-Asp, IAA-Glc, and IBA-Ala are hydrolyzed during nodule organogenesis (Campanella et al., 2008).
Compared with auxin metabolism, more attention has been paid to the role of auxin transport during nodule organogenesis. Rhizobia inoculation of M. truncatula leads to a substantial reduction in auxin transport over time, correlating with a reduction in nodule formation (van Noorden et al., 2006). Gravitropic stimulation of roots, which results in asymmetric auxin distribution, is also sufficient to alter nodule positioning in L. japonicus (Nadzieja et al., 2019), and application of auxin import inhibitors before rhizobia infection reduced the number of nodules in M. truncatula (Roy et al., 2017). The requirement for auxin transport regulation is supported by the fact that several MtPins show enhanced expression during nodulation (Huo et al., 2006; Plet et al., 2011; Sańko-Sawczenko et al., 2016). Moreover, knockdown of MtPin2, MtPin3, or MtPin4 results in a reduced number of nodules (Huo et al., 2006). In L. japonicus, all eight LjPin genes are expressed in young nodules and the vasculature of mature nodules (Sańko-Sawczenko et al., 2016). Auxin importers MtLAX1 and MtLAX2 are expressed during early stages of nodule formation and in the vasculature and the apex of mature nodules (de Billy et al., 2001; Roy et al., 2017), and lax2 mutants form fewer nodules than wild type and have a reduced auxin response within nodules (Roy et al., 2017). The high degree of redundancy in the PIN family has hindered the establishment of additional mechanistic links between symbiotic signaling and auxin transport regulation (Ng et al., 2020).
Together, these transport studies indicate that auxin must accumulate during different stages of nodule development and subsequently activate auxin signaling. Studies with the auxin reporter constructs pDR5::Gus, DII-Venus, and pGH3::Gus in L. japonicus and M. truncatula demonstrated that auxin signaling starts to emerge in cell layers underneath infected root hairs and is strongest in the cortex and pericycle of young nodules (Pacios-Bras et al., 2003; Suzaki et al., 2012; Breakspear et al., 2014) and in the vasculature and apex of mature nodules (Takanashi et al., 2011; Suzaki et al., 2012; Guan et al., 2013; Breakspear et al., 2014; Franssen et al., 2015; Nadzieja et al., 2019). These observations support a requirement for auxin signaling in both infection and nodule organogenesis and may relate to the requirement for cell-cycle initiation and progression that occurs in both these processes.
Although the locations where auxin signaling occurs are relatively well established, few downstream signaling components have been demonstrated to have specific roles in nodulation. In soybean, six auxin receptor genes encoding either GmTIR1 or GmAFB3 are important for proper nodule development, and their degradation depends on miRNA393 (Cai et al., 2017). S. meliloti infection of M. truncatula leads to increased transcript abundance of the auxin-responsive genes MtIaa9 and MtArf16a, and arf16a mutants show a reduced number of infection events (Breakspear et al., 2014). In soybean, GmARF8a/8b, GmARF10, GmARF16, and GmARF17 are negative regulators of auxin signaling and are targets of miR167/miR160, which reduces their expression to increase the sensitivity to auxin (Turner et al., 2013; Nizampatnam et al., 2015; Wang et al., 2015). In L. japonicus, LjARF3A, LjARF3B, and LjARF4 can potentially act as negative regulators of nodulation. Their expression is increased in rel3 plants, which are impaired in the biogenesis of trans-acting small interfering RNAs (siRNAs), have a reduced nodule number, and exhibit insensitivity to auxins but greater sensitivity to auxin transport inhibitors during nodulation (Li et al., 2014). Taken together, these results show that miRNAs and siRNAs contribute to the maintenance of robust auxin signaling during nodule formation. Additional tools, such as conditional knockouts of signaling components or transporters without pleiotropic phenotypes, may provide mechanistic insights into auxin signaling and help to clarify the relative contributions of biosynthesis, transport, and signaling components during nodule initiation.
Interaction of Auxin and Other Signaling Pathways during Nodule Organogenesis
During many developmental processes, auxin and CK interact with and mostly counteract each other by regulating each other's metabolism, signaling, and transport (Coenen and Lomax, 1997; El-Showk et al., 2013). In nodule development, auxin and CK have more synergistic activity, as the presence of both is required for nodule initiation.
Several studies have investigated how auxin and CK interact to regulate nodule development and suggest that LHK1/CRE1-dependent CK signaling acts upstream of auxin biosynthesis and transport. In M. truncatula, inhibition of polar auxin transport (PAT) upon rhizobia infection is MtCRE1 dependent and accomplished through the regulation of MtPin expression and protein accumulation (Plet et al., 2011). The lack of PAT inhibition is causative for the reduced the nodulation of cre1 plants and can be rescued by N-1-naphthylphthalamic acid (NPA) or 2,3,5-triiodobenzoic acid (TIBA) supplementation. Moreover, MtCRE1-dependent flavonoid accumulation is required to inhibit PAT (Ng et al., 2015). As discussed earlier, MtCRE1-dependent CK signaling is also required to induce MtNin expression. This CRE1-dependent induction is required to induce the expression of MtSty, MtLbd16, MtYuc2, and MtYuc8, resulting in auxin accumulation (Schiessl et al., 2019). Interestingly, miR160 regulates the sensitivity to auxin but also to CK during nodule organogenesis in soybean, and the nodulation phenotypes of miR160 mimicry and overexpressing lines can be rescued by application of one or both hormones (Turner et al., 2013; Nizampatnam et al., 2015). In L. japonicus, a connection was also established whereby, during root hair infection, LjLHK1-dependent signaling restricts auxin accumulation after inoculation with rhizobia in the epidermal cell layer. It remains to be seen whether this requires LHK1 signaling in the cortex or epidermis. At the same time, auxin may counteract the inhibitory role of CK signaling in the epidermis by inhibiting the expression of the CK biosynthesis genes LjCyp735a and LjLog4 (Nadzieja et al., 2018). These observations suggest a finely balanced antagonism between auxin promoting infection progression and CK inhibiting it in the epidermis, while both hormones positively influence the initiation and maintenance of cell division for nodule organogenesis.
Ethylene
Ethylene Biosynthesis and Signaling
Ethylene is a gaseous hormone involved in the regulation of numerous developmental processes, such as seedling growth, fruit ripening, leaf and root development, and senescence (Schaller and Kieber, 2002; Schaller, 2012).
Ethylene synthesis is a multistep process in which ACC synthase (ACS) (Liang et al., 1995; Yamagami et al., 2003) and ACC oxidase (ACO) (Linkies et al., 2009) are often considered the limiting steps, and their regulation is thus key for plant development.
In the absence of ethylene, ETHYLENE RESPONSE (ETR)/ETHYLENE RESPONSE SENSOR (ERS)/ETHYLENE INSENSITIVE4 (EIN4) receptors act as negative regulators of ethylene signaling (Ecker, 1995; Hua et al., 1995; Sakai et al., 1998), together with CONSTITUTIVE TRIPLE RESPONSE1 (CTR1), a Ser/Thr kinase (Kieber et al., 1993). CTR1 phosphorylates EIN2 at its C terminus, causing its inactivation (Ju et al., 2012). In addition, it may be degraded in an F-box EIN2 TARGETING PROTEINs (ETPs)-dependent manner (Qiao et al., 2009). Without active EIN2, EIN3/EIN3-LIKE (EIL) transcription factors are degraded by EIN3-BINDING F-BOX PROTEINs (EBFs), resulting in an absence of ethylene response gene transcription (Guo and Ecker, 2003; Potuschak et al., 2003). Ethylene perception inactivates ETR/ERS/EIN4 and CTR1, enabling a phosphorylation-dependent interaction of EIN2 with ETR/ERS/EIN4 that protects EIN2 from degradation (Bisson and Groth, 2010). Cleavage and nuclear migration of the EIN2 C terminus stabilize EIN3/EIL proteins (An et al., 2010; Wen et al., 2012), resulting in the transcriptional regulation of ethylene response genes (Chao et al., 1997; Guo and Ecker, 2003).
Role of Ethylene in Nodule Organogenesis
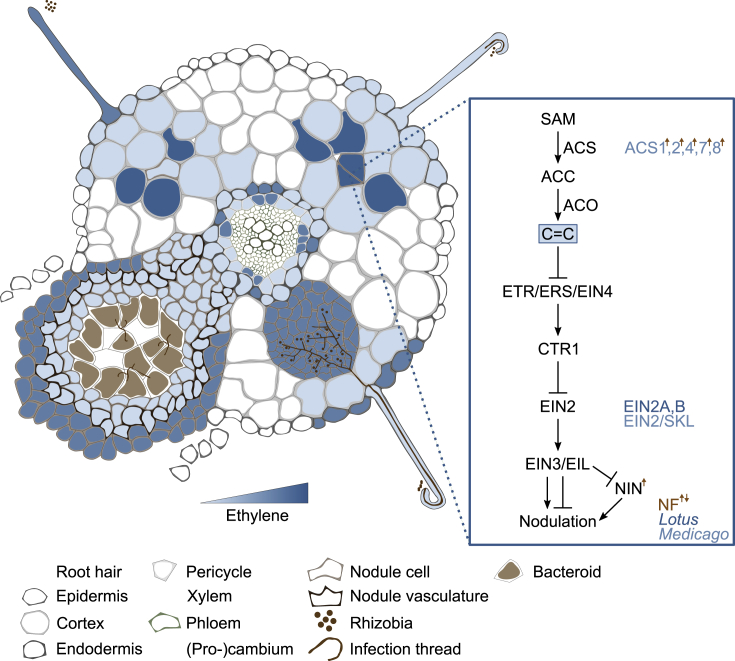
Ethylene appears to function primarily as a negative regulator of nodulation in most legumes (Figure 4). Application of the ethylene precursor ACC reduces nodule number, whereas inhibition of ethylene biosynthesis, e.g., through the application of AVG or Ag+, enhances nodulation in Vicia sativa (Heidstra et al., 1997), M. truncatula (Peters and Crist-Estes, 1989; Penmetsa and Cook, 1997), pea (Goodlass and Smith, 1979; Lorteau et al., 2001), and L. japonicus (Heckmann et al., 2011). This inhibitory effect acts very early in the signaling pathway, as ACC treatment not only inhibits nodule formation but also inhibits Ca2+ spiking in M. truncatula root hairs in response to NF (Oldroyd et al., 2001). Transcriptional studies in ethylene-insensitive mutants show a massive impact of ethylene on essentially all symbiosis-induced transcription (Larrainzar et al., 2015). The symbiotic induction of ethylene occurs rapidly, with rhizobia inoculation stimulating an increase in ethylene within six hpi (Reid et al., 2018), and MtAcs1 and MtAcs2 are induced upon NF application in M. truncatula (van Zeijl et al., 2015). MtAcs4, MtAcs7, and MtAcs8 are expressed in the periphery of young nodules and in the meristem of mature nodules (Larrainzar et al., 2015). This rapid synthesis and inhibitory effect of ethylene thus create a rapid feedback mechanism to restrict symbiotic gene expression and nodulation.
Figure 4.
Ethylene Biosynthesis, Metabolism, and Signaling.
Following perception of rhizobia, ethylene levels and signaling are rapidly induced; however, precise localization has not been achieved, and signaling intensity at different developmental stages is depicted based on mutant phenotypes, indicating roles at all stages of nodule development and in nodule positioning. Core biosynthesis, metabolism, and signaling components are depicted in black at the right, together with species-specific components, where known. NF-induced/repressed components are indicated with a brown arrow (up or down, respectively).
In addition to its negative regulatory role, ethylene may act as a positional cue for nodule initiation, exerting stronger inhibitory effects at some positions within a tissue. For example, in Vicia sativa, Aco expression was found to accumulate in cells opposite phloem poles and in the first cortex layers adjacent to them (Heidstra et al., 1997). These observations were further supported by ethylene signaling mutants in M. truncatula: skl (ein2) mutants form nodules opposite xylem and phloem poles, in contrast to wild-type plants that only form nodules opposite xylem poles (Penmetsa et al., 2003; 2008). This suggests that ethylene signaling before rhizobia infection is required to suppress excessive nodule formation in general and specifically in cell layers over phloem-pole pericycle cells. L. japonicus possesses two EIN2 encoding genes, LjEin2a and LjEin2b, and the respective double mutant forms more nodules, akin to the skl/ein2 mutant (Reid et al., 2018).
Although some ethylene responsive markers have been described, none so far developed offers the high-resolution and dynamic range available for CK and auxin (DR5/DII-VENUS reporter; Nadzieja et al., 2019), making it challenging to identify the precise location and signaling of ethylene responses during nodule organogenesis. Novel approaches such as single-cell sequencing might circumvent this restriction or lead to the development of such ethylene markers in the future.
Interaction between Ethylene and Symbiotic Signaling
Recent studies suggest that ethylene and CK balance each other's responses during nodule organogenesis in a feedback loop. In M. truncatula, induction of MtAcs1 and MtAcs2 expression upon NF application is MtCRE1-dependent (as is regulation of CK biosynthesis/signaling genes), and cre1 mutants are insensitive to the AVG-dependent enhancement of this NF effect (van Zeijl et al., 2015), suggesting that CK acts as a positive regulator of ethylene biosynthesis and signaling. The increased ethylene emission of uninoculated L. japonicus gain-of-function lhk1 (snf2) plants provides additional support for this view (Reid et al., 2018) and is consistent with the CK-dependent induction of ethylene biosynthesis in Arabidopsis (Dugardeyn and Van Der Straeten, 2008). The question remains, where is ethylene active in this context? Given the inhibitory effect of ethylene on infection thread formation and the fact that reduced infection thread formation in L. japonicus ckx3 mutants can be rescued by AVG supplementation (Reid et al., 2016), it is highly likely that CK-dependent ethylene synthesis inhibits infection thread formation, as proposed by Miri et al. (2016).
Evidence for the influence of ethylene on CK biosynthesis or signaling was provided recently. In M. truncatula skl/ein2 plants, iP concentrations are strongly increased due to both a compensatory effect and an inhibitory effect of ethylene signaling on symbiotic signaling and CK biosynthesis (van Zeijl et al., 2015). In support of the latter effect, the skl/ein2 nodulation phenotype is almost completely suppressed by the introduction of the cre1 allele (Plet et al., 2011), implying that MtEIN2 may act as a negative regulator of MtCRE1-dependent signaling. The identity of the molecular targets of ethylene-dependent inhibition of the symbiotic signaling pathway remains a key outstanding question. Studies in Arabidopsis suggest that ethylene and CK signaling may be interconnected through direct phosphorylation of AHPs by ethylene receptors (Zdarska et al., 2019). It will be interesting to determine whether and how the interconnection of these pathways has been recruited in nodule development or regulation.
Gibberellins
Gibberellin Biosynthesis and Signaling
Gibberellins (GAs) are a large group of compounds formed through multiple consecutive reactions from geranylgeranyl diphosphate (GGDP); they are involved in many aspects of plant growth and development, including seed germination and the development of leaves and seeds (Sun, 2008; Yamaguchi, 2008). GGDP is converted further by a number of GA oxidases, ultimately leading to the formation of bioactive GAs (Mitchum et al., 2006; Hu et al., 2008; Sun, 2008).
Bioactive GAs activate a nucleus-localized signaling pathway that involves GIBBERELLIN INSENSITIVE DWARF1 (GID1) receptors, DELLA proteins GIBBERELLIC ACID INSENSITIVE (GAI)/REPRESSOR-OF-GA1-3 (RGA)/RGA-LIKE (RGL), F-box proteins SLEEPY1 (SLY1) and SNEEZY (SNE), and transcription factors, e.g., PHYTOCHROME-INTERACTING FACTORs (PIFs) (Sun, 2008; Schwechheimer, 2012; Davière and Achard, 2013). In the absence of bioactive GA, GID1 receptors are inactive (Ueguchi-Tanaka et al., 2005; Griffiths et al., 2006), allowing DELLA proteins to inhibit the GA signaling pathway by inhibiting the function of transcription factors such as PIFs (Sun, 2008; Schwechheimer, 2012; Davière and Achard, 2013). GA perception by GID1 receptors results in complex formation with F-box proteins SLY and SNE, mediating DELLA degradation (McGinnis et al., 2003; Dill et al., 2004; Ariizumi et al., 2011). As a result, formerly repressed transcription factors can mediate the transcription of output genes. A summary of GA-dependent nodulation in legumes has been given by Hayashi et al. (2014). We highlight recent advances in the mechanistic understanding of GA in nodule organogenesis (Figure 5).
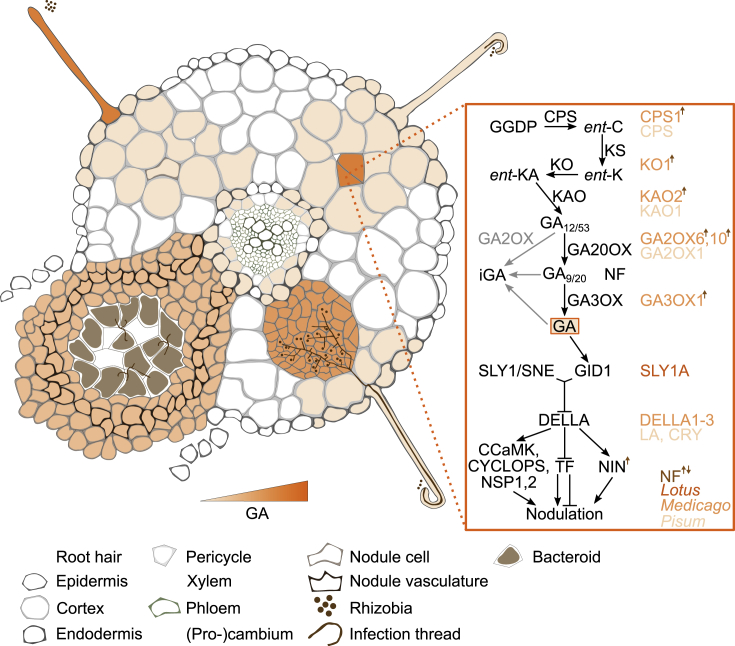
Figure 5.
GA Biosynthesis, Metabolism, and Signaling.
Intensity of signaling output at different developmental stages is depicted based on transcriptome experiments and signaling mutant phenotypes; however, it has not been precisely localized using reporter studies. Following rhizobia perception, GA level/signaling accompanies the activation of symbiotic signaling. Core biosynthesis, metabolism, and signaling components are depicted in black/gray at the right with species-specific components shown where they have been investigated. NF-induced/repressed components are indicated with a brown arrow (up or down, respectively).
Gibberellin Signaling in Nodulation
Our current understanding of the role of GA in nodulation is mainly based on research in pea and M. truncatula and suggests that levels of active GA must be precisely coordinated. Although there are contradictory reports that GA application either induces (Kawaguchi et al., 1996) or inhibits (Maekawa et al., 2009) the formation of nodules (and infection threads) in L. japonicus, nodulation promoting effect is seen in pea at low GA concentrations, whereas an inhibitory effect is seen at high concentrations (Ferguson et al., 2005). In M. truncatula, GA application inhibits nodule formation (Fonouni-Farde et al., 2016a; Jin et al., 2016). However, the GA antagonist paclobutrazol reduces nodule formation in pea (McAdam et al., 2018) but stimulates nodulation at low concentrations in M. truncatula (Fonouni-Farde et al., 2016a). These somewhat contradictory results may be explained by the sensitive GA balance during nodule development and the fact that GA is likely to cause artifacts when applied at high concentrations outside the normal range. Another possible explanation could be a bell-shaped dose–response curve similar to that described, e.g., for IAA/NAA-dependent protoplast swelling (Yamagami et al., 2004).
Extensive studies with pea mutants defective in GA biosynthesis and degradation have been conducted and show the importance of GA biosynthesis during nodule organogenesis. Generally, these mutants exhibit reduced nodule formation and also display pleiotropic effects on root and shoot development (Ferguson et al., 2005). Four of the mutants (ls, na, lh, and sln) with reduced nodule numbers (Ross et al., 1993; Yaxley et al., 2001) are loss-of-function mutants of PsCPS, PsKAO1 (Davidson et al., 2003), PsKO1 (Davidson et al., 2004), and PsGA2OX1 (Lester et al., 1999). Moreover, grafting experiments with lh/ko1 and wild type revealed that normal GA synthesis in the shoot is sufficient to rescue the nodulation phenotype of lh/ko1 (Ferguson et al., 2005). Crosses of na-1 with the hypernodulation mutants sym28, sym29, or nod3 show additivity of the nodulation phenotypes (Ferguson et al., 2011). Although biosynthesis mutants are not yet characterized in L. japonicus, Medicago ga2ox10 plants display a reduced number of nodules (Kim et al., 2019). Moreover, rhizobia infection induces MtGa3ox1, MtGa2ox6, MtGa2ox10, MtCps1, MtKo1, and MtKao2 expression between 1 and 12 dpi in M. truncatula (Breakspear et al., 2014; Kim et al., 2019) and LjGa2ox expression between 4 and 12 dpi in L. japonicus (Kouchi, 2004).
GA signaling components involved in the regulation of nodulation identified to date encode SLY1 and DELLA proteins (Maekawa et al., 2009; Fonouni-Farde et al., 2016b; Jin et al., 2016). In L. japonicus, overexpression of LjSly1a and a gain-of-function LjSly1a allele (SLY1A-d) reduce the number of nodules and also reduce LjGa20ox expression, indicating that constitutive GA signaling negatively regulates nodule organogenesis (Maekawa et al., 2009). This idea is further supported by the observation that GA application inhibits the expression of LjNin and LjNsp1/2 (Maekawa et al., 2009). In pea, loss of the DELLA proteins LA and CRY leads to a reduction in nodule number, also indicating the repressive function of GA signaling in nodulation (Ferguson et al., 2011). Similar to pea, MtDELLA1,2,3 are positive regulators of nodule organogenesis in M. truncatula. Expression of MtDella1,2,3 is induced 1 d after inoculation with rhizobia in all root tissues, and these genes are also expressed in the nodule meristematic zone (Fonouni-Farde et al., 2016a, 2016b). Triple della1,2,3 mutants display a strong reduction in nodule formation, and all three MtDELLAs can physically interact with MtNSP2, MtNF-YA1, and MtIPD3 to positively regulate the expression of nodulation genes such as MtErn1, MtNin, or MtRip (Fonouni-Farde et al., 2016a, 2016b; Jin et al., 2016).
All in all, DELLA accumulation, which promotes the transcriptional activity of multiple symbiotic signaling components and thus nodule initiation, is antagonized by increased GA levels. Therefore, the precise regulation of GA content and signaling is necessary for nodule initiation.
Interaction between GA and Other Hormones during Nodule Organogenesis
A number of interactions between GA signaling components and other nodulation factors have been demonstrated recently. A potential link between CK and GA was shown by GA supplementation experiments in L. japonicus. Both wild-type and snf2 plants respond to GA supplementation with a decrease in nodule number caused by GA-dependent inhibition of LjNsp1/2 and LjNin expression (Maekawa et al., 2009). In M. truncatula, the introduction of della2 della3 into CRE1 gain-of-function plants suppresses their spontaneous nodule formation (Jin et al., 2016). As MtDELLAs are positive regulators of MtNin and MtErn1 expression (Jin et al., 2016), one possible explanation is that they act downstream of CK signaling to regulate MtNin and MtErn1 expression during nodule organogenesis. Indeed, MtDELLAs also antagonize cytokinin in the regulation of Nsp2, with dominant active DELLA forms triggering spontaneous nodule organogenesis (Fonouni-Farde et al., 2017).
There is also some evidence that GA and ethylene interact during nodulation. In pea, GA acts as a negative regulator of ethylene biosynthesis, as na plants show higher endogenous ethylene concentrations, which may result from increased PsAcs1 and PsAco1 expression. Moreover, treating na plants with AVG can partially rescue the nodulation phenotype (Ferguson et al., 2011). This may indicate that a proper GA/ethylene balance is required for nodule organogenesis. However, crosses of na-1 with ein2 suggest that both hormones may act independently (McAdam et al., 2018).
GA signaling is therefore positioned as a key regulator of symbiotic signaling, with DELLA interacting with core members of the symbiosis pathway to positively regulate transcription of NF signaling genes (Liu et al., 2018a).
Peptide Hormones
It is becoming increasingly clear that peptide hormones perform myriad plant developmental functions alongside the classical small-molecule plant hormones (reviewed in Matsubayashi and Sakagami, 2006; Hirakawa and Sawa, 2019). In legume nodulation, several peptide hormones mediate systemic regulation of nodulation to ensure that organogenesis is balanced with internal and environmental signals (Figure 6).
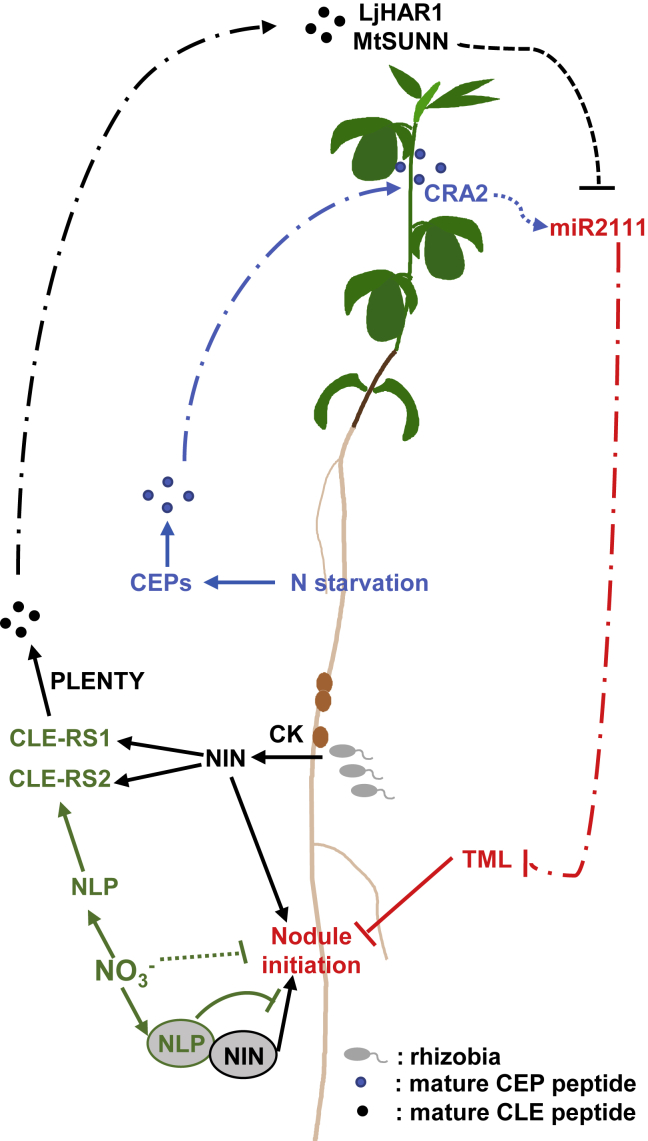
Figure 6.
Peptide Hormones as Systemic Regulators of Nodulation.
CK associated with nodule initiation induces NIN and production of CLE-RS peptides. CLE-RS2 is also induced by nitrate via NLP4. Although CLE-RS2 participates in systemic regulation, NLP1 locally inhibits nodulation through competition with NIN for downstream regulatory elements. Processed CLE-RS peptides are transported to, and perceived in, the shoot by HAR1. HAR1 then inhibits miRNA2111 abundance to prevent negative regulation of TML, establishing AON. Nitrogen starvation responses are integrated into this pathway by CEP/CRA2 signaling, which acts antagonistically to HAR1 on miRNA2111 levels.
Negative Regulators of Nodulation
The most studied plant peptide hormones in the CLAVATA (CLV)/EMBRYO SURROUNDING REGION (ESR)-RELATED PROTEIN (CLE) family play roles in the regulation of cell division and differentiation in various developmental contexts (Ito et al., 2006; Hirakawa and Sawa, 2019). After processing, CLE peptides typically consist of 12–13 amino acids (aa) and exert diverse effects on the shoot, inflorescence, vascular, and root meristem development in both monocots and dicots (Clark et al., 1996; Hirakawa et al., 2008; Xu et al., 2015; Je et al., 2018).
In legumes, several Cle genes are upregulated by rhizobia or NF to activate the AON pathway (see recent review by Ferguson et al., 2019). LjCle-RS1/2/3, MtCle12/13, and GmRic1/2 are upregulated by rhizobia inoculation, and overexpression of these genes is sufficient to inhibit nodulation (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011; Nishida et al., 2016). LjNIN can directly bind to the LjCle-RS1/2 promoter to induce gene expression, and MtNIN is also critical for MtCle12/13 expression (Mortier et al., 2010; Soyano et al., 2014). In contrast to the early induction of LjCle-RS1/2, LjCle-RS3 is upregulated at later stages of nodule organogenesis, implying that its regulation may differ from that of LjCle-RS1/2 (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011; Nishida et al., 2016). CLE peptides also participate in nitrate regulation of nodulation, with nitrate inducing the expression of LjCle-RS2, LjCle-RS3, and GmNic1 (Okamoto et al., 2009; Mortier et al., 2010; Reid et al., 2011; Nishida et al., 2016).
After translation, CLEs are processed into 12–13-aa peptides and modified by tri-arabinosylation, which is essential for receptor binding and signal transduction (Okamoto et al., 2013). HYPERNODULATION RELATED GENE1 (MtRDN1/LjPLENTY) is a homolog of the Arabidopsis hydroxyproline O-arabinosyltransferases (HPAT1/2/3), which are required for this modification (Ogawa-Ohnishi et al., 2013; Kassaw et al., 2017; Yoro et al., 2019). Tri-arabinosylation of MtCLE13 (CLE13-TaP) is sufficient to inhibit nodulation and is dependent on MtRDN1 (Kassaw et al., 2017). Intriguingly, LjCle-RS3 overexpression reduces nodule number in an LjPLENTY-dependent manner, whereas LjCle-RS1/2 does not, indicating differences in substrate specificity, peptide stability, or functional redundancy, as in the case of MtRND2 (Kassaw et al., 2017; Yoro et al., 2019).
Mature peptides are transported from root to shoot, where they are perceived by LjHAR1 (Okamoto et al., 2013). Genetic evidence suggests that MtSUNN, GmNARK, PsSYM29, and PvNARK function as equivalent receptors in other legumes, exhibiting hypernodulation and nitrate tolerant nodulation phenotypes (Carroll et al., 1985; Wopereis et al., 2000; Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003; Schnabel et al., 2005; Ferguson et al., 2014). Mutants with similar phenotypes in other Leucine Rich Repeat (LRR) receptor-like kinases indicate that these likely function as part of an AON signaling complex (Miyazawa et al., 2010; Krusell et al., 2011). Activation of HAR1 leads to downregulation of the shoot-to-root mobile miR2111, which is expressed in the shoot but targets the TML mRNA for cleavage in the root. Subsequent increased TML levels negatively regulate nodule initiation and infection in the root (Tsikou et al., 2018). TML is a putative Kelch repeat-containing F-box protein and is predicted to target proteins for degradation by E3 ligase activity, but its target remains unknown (Takahara et al., 2013).
In summary, AON functions to systemically control nodule number. Before inoculation, roots remain susceptible to rhizobia infection due to high miR2111 levels and relatively low levels of TML mRNA and protein. When nodule organogenesis events commence, the CLE-dependent induction of the negative regulatory loop is initiated (Figure 6).
Positive Regulators of Nodulation
C-TERMINALLY ENCODED PEPTIDEs (CEPs) are 14- or 15-aa peptides first found to regulate root development in response to nitrogen starvation in Arabidopsis (Ohyama et al., 2008). In M. truncatula, nitrogen deprivation can also induce the expression of CEPs (Imin et al., 2013). In contrast to AtCEP, MtCEP1 overexpression or peptide application does not affect root length but instead increases nodule formation, reduces lateral root number, and triggers periodic circumferential cell proliferation (Ohyama et al., 2008; Imin et al., 2013). Processing and modification are crucial for CEP peptides; for instance, hydroxylation or arabinosylation of prolines in MtCEP1 increases or reduces its activity, respectively (Mohd-Radzman et al., 2015; Patel et al., 2018; Taleski et al., 2018).
In Arabidopsis, mature AtCEP1 is transported from root to shoot to be perceived by the LRR receptor kinase AtCEPR1/2 (Tabata et al., 2014). Similarly, MtCRA2, the homolog of AtCEPR1, plays a vital role in systemic promotion of nodule formation while acting locally to control root architecture (Huault et al., 2014). Perception of CEP peptides in Arabidopsis shoots leads to the production of the shoot-to-root mobile signals AtCEPD1/2 and AtCEPDL1/2, which regulate nitrate uptake and assimilation (Ohkubo et al., 2017; Ota et al., 2020). In M. truncatula, MtCEPD1 and MtCEPD2 are regulated in the shoot in an MtCRA2-dependent manner, and unlike their Arabidopsis orthologs, they also show root expression (Gautrat et al., 2020).
Integration of Systemic Signaling Pathways
Integration of environmental signals into the nodulation program is critical for growth optimization under available conditions, balancing cheap soil nitrate or comparatively expensive fixed nitrogen with the carbon budget. The maintenance of numerous positively and negatively acting pathways is likely to allow finer control over this integration. Both negative feedback through CLE/HAR1 and positive regulation through the CEP/CRA2 pathways can be induced by cytokinin and NIN (Laffont et al., 2020) and appear to converge on a common regulator in miR2111 (Figure 6; Laffont et al., 2019; Gautrat et al., 2020). HAR1-dependent CK synthesis in the shoot also appears to play a role in systemic nodule regulation (Sasaki et al., 2014), although it has not yet been clearly established whether this also acts through the regulation of miR2111 or through additional, parallel pathways. Regulation of auxin transport is also linked to both CLE and CEP signaling, as sunn and cra2 mutants show increased auxin transport from the shoot (van Noorden et al., 2006; Mathesius et al., 2019; Chapman et al., 2020). Identification of the molecular players that act downstream of CRA2 and HAR1 in the shoot is likely to help clarify these relationships.
Other Hormones
In addition to the four major hormones discussed here, all plant hormones appear to play roles in fine-tuning the nodulation program in response to environmental conditions or through crosstalk with the hormones discussed above. Although further mechanistic insight into their roles will be critical, physiological studies have shown that strigolactone, abscisic acid, brassinosteroids, salicylic acid, and jasmonic acid all play roles in nodule development and have been reviewed elsewhere (Ferguson and Mathesius, 2014).
Manipulation of Plant Hormones to Engineer Nodule Organogenesis in Non-legumes
The massive environmental impact of agriculture demands the development of more sustainable farming systems. Excessive nitrogen use has already exceeded the safe limits for humanity and requires urgent attention (Rockström et al., 2009; Springmann et al., 2018). While associative nitrogen fixation can provide substantial benefits and is already a part of agricultural practice, the efficiencies derived from symbiotic nitrogen fixation in a specialized organ make engineering an analogous symbiosis in cereals highly desirable.
Induction of lateral root organs (both Lateral roots - LRs, and nodules) by plant hormones, particularly auxin, is a conserved feature of vascular plants. Legume nodules share a transcriptional and developmental program with lateral roots (Xiao et al., 2014, 2019; Bensmihen, 2015; Schiessl et al., 2019; Soyano et al., 2019), and some actinorhizal plants accommodate actinorhizal bacteria in structures that are somewhat intermediate between lateral roots and legume nodules (Hirsch et al., 1997; Pawlowski and Bisseling, 1997). Auxin is critical to both nodule and lateral root processes, with NIN found to directly target LjAsl18a/MtLbd16, which coordinates auxin signaling in both nodule and LR initiation (Schiessl et al., 2019; Soyano et al., 2019). In addition, LjNF-YA1 directly targets members of the SHI/STY family, which in Arabidopsis regulate auxin biosynthesis and lateral root development (Hossain et al., 2016). The existence of a common developmental program is further supported by the ability of cereals to develop nodule-like structures, which are presumably modified lateral root primordia, when treated with high levels of auxin (Hiltenbrand et al., 2016; Thomas et al., 2018, 2020). In legumes, similar responses can be triggered by auxin transport inhibitors (Rightmyer and Long, 2011; Li et al., 2014; Ng et al., 2015). The existing lateral root developmental program in cereals may therefore be considered a developmental blueprint for engineering a new lateral organ capable of supporting nitrogen fixation if manipulation of auxin signaling can be restricted to a small subset of cells in the root, thereby avoiding detrimental impacts on general root development. Further understanding of the similarities between lateral roots, actinorhizal nodules, and legume nodule development may help to clarify the requirements for a nodule developmental program and inform the engineering of cereals.
Cell division of the indeterminate nodule occurs first in the pericycle before extending to inner cortical cells, which develop into the meristem (Xiao et al., 2014; Bensmihen, 2015; Kohlen et al., 2018). On the other hand, cell division of the determinate nodule occurs first in the cortical cells, without the establishment of a persistent meristem (Xiao et al., 2019). This flexibility indicates that the site of induction is not critical per se, but rather the acquisition of nodule identity factors. The requirement for acquiring and maintaining nodule identity is perhaps best exemplified by the Noot/Cochleata regulators, loss of which causes meristem identity and vascular arrangement to alter periodically between lateral root and nodule characteristics (Ferguson and Reid, 2005; Couzigou et al., 2012; Magne et al., 2018; Shen et al., 2020). This indicates that, together with the initiation of a new lateral root organ, manipulation of legume regulators required for nodule identity (e.g., NIN, NOOT) (Griesmann et al., 2018; van Velzen et al., 2018) should be a priority in nodule engineering. A key adaptation of legumes appears to be the susceptibility of the cortical cells to cell division induction by CK (Gauthier-Coles et al., 2019). In addition to their role in nodule initiation, CKs appear to play a key role in the coordination of nodule identity. Gain-of-function mutations in symbiotic (CCamK) or cytokinin (Lhk1) signaling components give nodule-like structures with characteristics closer to those of true nodules (Gleason et al., 2006; Tirichine et al., 2006, 2007), whereas manipulation of other hormones, including auxin, gibberellin, and regulators such as Nin and Nf-ya1, induces organogenesis without conferring all nodule characteristics (Soyano et al., 2013; Fonouni-Farde et al., 2017; Schiessl et al., 2019).
In our view, successful engineering of a nodule organ in cereals will therefore require: initiation of lateral root organs through precise manipulation of auxin; establishment of nodule identity on the newly initiated organ, which in legumes is achieved with cytokinin signaling and specific nodule identity regulators; and organ-specific gene expression to support the unique requirements for hosting symbiotic bacteria.
Concluding Remarks and Future Perspectives
Plant hormones are universal regulators of development, yet they play a number of specific roles during the initiation of a nodule organ. Ongoing efforts to identify the symbiotic pathway components that regulate, and are regulated by, plant hormones together with the timing and location of each hormone's action are important next steps. Establishing the profiles of transcripts, proteins, and metabolites at cellular resolution is rapidly emerging as a feasible means to address these challenges. These efforts may help to unravel the nature of signaling between the root surface and initiation of a nodule in deeper cell layers, as well as the underlying characteristics that render a specific root zone susceptible to organ initiation. In our opinion, parallel studies are now needed in multiple species and within ecotypes of a species to identify the degree of flexibility that exists in these regulatory pathways. This understanding of the core nodule initiation program will be critical not only for our ability to improve legume cultivation but also for ongoing efforts to engineer an organ that is competent for nitrogen fixation in diverse non-legume crop species.
Although the engineering of cereal crops for efficient nitrogen fixation is a desirable goal, it may remain out of reach for some time. Ongoing research in nodule development must also focus on meeting demands for sustainable food and feed production. Adverse environmental conditions for crop production, such as insufficient or excess water, mineral toxicities and deficiencies, and varying nutrient conditions, all affect nodule development. To develop improved legume crops that maintain robust nodule development and nitrogen fixation in varied conditions, we must understand how these environmental signals are perceived and integrated with hormone signaling pathways and the nodule development program. This work will also necessitate a switch to greater focus on research in legume crops rather than model legumes for our understanding of nodule development. Improved genomic resources and targeted mutagenesis capabilities in many legumes are already facilitating such a transition.
Funding
We are grateful for support from the project Engineering Nitrogen Symbiosis for Africa (ENSA), currently supported through a grant to the University of Cambridge by the Bill & Melinda Gates Foundation and the UK government's Department for International Development (DFID). This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and innovaWon program (grant agreement no. 834221).
Acknowledgments
We thank Jens Stougaard for his helpful comments on the manuscript.
Published: August 21, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
References
- An F., Zhao Q., Ji Y., Li W., Jiang Z., Yu X., Zhang C., Han Y., He W., Liu Y. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. AP2-ERF transcription factors mediate Nod factor-dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell. 2007;19:2866–2885. doi: 10.1105/tpc.107.052944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Brault-Hernandez M., Laffont C., Huault E., Brault M., Plet J., Moison M., Blanchet S., Ichanté J.L., Chabaud M. Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. Plant Cell. 2012;24:3838–3852. doi: 10.1105/tpc.112.103267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Lawrence P.K., Steber C.M. The role of two F-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling. Plant Physiol. 2011;155:765–775. doi: 10.1104/pp.110.166272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin M., Laloum T., Lepage A., Rípodas C., Ariel F., Frances L., Crespi M., Gamas P., Blanco F.A., Zanetti M.E. A phylogenetically conserved group of nuclear factor-Y transcription factors interact to control nodulation in legumes. Plant Physiol. 2015;169:2761–2773. doi: 10.1104/pp.15.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P., Ratet P., Crespi M.D., Schultze M., Kondorosi A. Nod factors and cytokinins induce similar cortical cell division, amyloplast deposition and MsEnod12A expression patterns in alfalfa roots. Plant J. 1996;10:91–105. [Google Scholar]
- Benezech C., Berrabah F., Jardinaud M.-F., Le Scornet A., Milhes M., Jiang G., George J., Ratet P., Vailleau F., Gourion B. Medicago-Sinorhizobium-Ralstonia co-infection reveals legume nodules as pathogen confined infection sites developing weak defenses. Curr. Biol. 2020;30:351–358.e4. doi: 10.1016/j.cub.2019.11.066. [DOI] [PubMed] [Google Scholar]
- Bensmihen S. Hormonal control of lateral root and nodule development in legumes. Plants. 2015;4:523–547. doi: 10.3390/plants4030523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisson M.M.A., Groth G. New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol. Plant. 2010;3:882–889. doi: 10.1093/mp/ssq036. [DOI] [PubMed] [Google Scholar]
- Boivin S., Kazmierczak T., Brault M., Wen J., Gamas P., Mysore K.S., Frugier F. Different cytokinin histidine kinase receptors regulate nodule initiation as well as later nodule developmental stages in Medicago truncatula. Plant Cell Environ. 2016;39:2198–2209. doi: 10.1111/pce.12779. [DOI] [PubMed] [Google Scholar]
- Brandstatter I., Kieber J.J. Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell. 1998;10:1009–1019. doi: 10.1105/tpc.10.6.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E.D. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell. 2014;26:4680–4701. doi: 10.1105/tpc.114.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhian W.P., Bensmihen S. Mini-review: Nod factor regulation of phytohormone signaling and homeostasis during Rhizobia-legume symbiosis. Front. Plant Sci. 2018;9:1247. doi: 10.3389/fpls.2018.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z., Wang Y., Zhu L., Tian Y., Chen L., Sun Z., Ullah I., Li X. GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 2017;215:672–686. doi: 10.1111/nph.14632. [DOI] [PubMed] [Google Scholar]
- Campanella J.J., Smith S.M., Leibu D., Wexler S., Ludwig-Müller J. The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 2008;27:26–38. [Google Scholar]
- Carroll B.J., McNeil D.L., Gresshoff P.M. A supernodulation and nitrate-tolerant symbiotic (nts) soybean mutant. Plant Physiol. 1985;78:34–40. doi: 10.1104/pp.78.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Q., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- Chapman K., Ivanovici A., Taleski M., Sturrock C.J., Ng J.L.P., Mohd-Radzman N.A., Frugier F., Bennett M.J., Mathesius U., Djordjevic M.A. CEP receptor signalling controls root system architecture in Arabidopsis and Medicago. New Phytol. 2020;226:1809–1821. doi: 10.1111/nph.16483. [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen W., Li X., Jiang H., Wu P., Xia K., Yang Y., Wu G. Knockdown of LjIPT3 influences nodule development in Lotus japonicus. Plant Cell Physiol. 2014;55:183–193. doi: 10.1093/pcp/pct171. [DOI] [PubMed] [Google Scholar]
- Clark S.E., Jacobsen S.E., Levin J.Z., Meyerowitz E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- Clarke V.C., Loughlin P.C., Day D.A., Smith P.M.C. Transport processes of the legume symbiosome membrane. Front. Plant Sci. 2014;5:699. doi: 10.3389/fpls.2014.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenen C., Lomax T.L. Auxin–cytokinin interactions in higher plants: old problems and new tools. Trends Plant Sci. 1997;2:351–356. doi: 10.1016/S1360-1385(97)84623-7. [DOI] [PubMed] [Google Scholar]
- Cooper J.B., Long S.R. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. Plant Cell. 1994;6:215–225. doi: 10.1105/tpc.6.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortleven A., Leuendorf J.E., Frank M., Pezzetta D., Bolt S., Schmülling T. Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 2019;42:998–1018. doi: 10.1111/pce.13494. [DOI] [PubMed] [Google Scholar]
- Couzigou J.-M., Zhukov V., Mondy S., Abu el Heba G., Cosson V., Ellis T.H.N., Ambrose M., Wen J., Tadege M., Tikhonovich I. NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. Plant Cell. 2012;24:4498–4510. doi: 10.1105/tpc.112.103747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S.E., Elliott R.C., Helliwell C.A., Poole A.T., Reid J.B. The pea gene NA encodesent-kaurenoic acid oxidase. Plant Physiol. 2003;131:335–344. doi: 10.1104/pp.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S.E., Smith J.J., Helliwell C.A., Poole A.T., Reid J.B. The pea gene LH encodes ent-kaurene oxidase. Plant Physiol. 2004;134:1123–1134. doi: 10.1104/pp.103.032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–1151. doi: 10.1242/dev.087650. [DOI] [PubMed] [Google Scholar]
- de Billy F., Grosjean C., May S., Bennett M., Cullimore J.V. Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Mol. Plant Microbe Interact. 2001;14:267–277. doi: 10.1094/MPMI.2001.14.3.267. [DOI] [PubMed] [Google Scholar]
- Dehio C., de Bruijn F.J. The early nodulin gene SrEnod2 from Sesbania rostrata is inducible by cytokinin. Plant J. 1992;2:117–128. doi: 10.1046/j.1365-313x.1992.t01-51-00999.x. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Weijers D., Lechner E., Yamada M., Hobbie L., Ehrismann J.S., Jürgens G., Estelle M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell. 2005;9:109–119. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Dill A., Thomas S.G., Hu J., Steber C.M., Sun T.-P. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugardeyn J., Van Der Straeten D. Ethylene: fine-tuning plant growth and development by stimulation and inhibition of elongation. Plant Sci. 2008;175:59–70. [Google Scholar]
- Ecker J.R. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- El-Showk S., Ruonala R., Helariutta Y. Crossing paths: cytokinin signalling and crosstalk. Development. 2013;140:1373–1383. doi: 10.1242/dev.086371. [DOI] [PubMed] [Google Scholar]
- Fang Y., Hirsch A.M. Studying early nodulin gene ENOD40 expression and induction by nodulation factor and cytokinin in transgenic alfalfa. Plant Physiol. 1998;116:53–68. doi: 10.1104/pp.116.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Mathesius U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 2014;40:770–790. doi: 10.1007/s10886-014-0472-7. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Reid J.B. Cochleata: getting to the root of legume nodules. Plant Cell Physiol. 2005;46:1583–1589. doi: 10.1093/pcp/pci171. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Ross J.J., Reid J.B. Nodulation phenotypes of gibberellin and brassinosteroid mutants of pea. Plant Physiol. 2005;138:2396–2405. doi: 10.1104/pp.105.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Foo E., Ross J.J., Reid J.B. Relationship between gibberellin, ethylene and nodulation in Pisum sativum. New Phytol. 2011;189:829–842. doi: 10.1111/j.1469-8137.2010.03542.x. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Li D., Hastwell A.H., Reid D.E., Li Y., Jackson S.A., Gresshoff P.M. The soybean (Glycine max) nodulation-suppressive CLE peptide, GmRIC1, functions interspecifically in common white bean (Phaseolus vulgaris), but not in a supernodulating line mutated in the receptor PvNARK. Plant Biotechnol. J. 2014;12:1085–1097. doi: 10.1111/pbi.12216. [DOI] [PubMed] [Google Scholar]
- Ferguson B.J., Mens C., Hastwell A.H., Zhang M., Su H., Jones C.H., Chu X., Gresshoff P.M. Legume nodulation: the host controls the party. Plant Cell Environ. 2019;42:41–51. doi: 10.1111/pce.13348. [DOI] [PubMed] [Google Scholar]
- Fonouni-Farde C., Tan S., Baudin M., Brault M., Wen J., Mysore K.S., Niebel A., Frugier F., Diet A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016;7:12636. doi: 10.1038/ncomms12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonouni-Farde C., Diet A., Frugier F. Root development and endosymbioses: DELLAs lead the orchestra. Trends Plant Sci. 2016;21:898–900. doi: 10.1016/j.tplants.2016.08.012. [DOI] [PubMed] [Google Scholar]
- Fonouni-Farde C., Kisiala A., Brault M., Emery R.J.N., Diet A., Frugier F. DELLA1-mediated gibberellin signaling regulates cytokinin-dependent symbiotic nodulation. Plant Physiol. 2017;175:1795–1806. doi: 10.1104/pp.17.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H.J., Xiao T.T., Kulikova O., Wan X., Bisseling T., Scheres B., Heidstra R. Root developmental programs shape the Medicago truncatula nodule meristem. Development. 2015;142:2941–2950. doi: 10.1242/dev.120774. [DOI] [PubMed] [Google Scholar]
- Frugier F., Kosuta S., Murray J.D., Crespi M., Szczyglowski K. Cytokinin: secret agent of symbiosis. Trends Plant Sci. 2008;13:115–120. doi: 10.1016/j.tplants.2008.01.003. [DOI] [PubMed] [Google Scholar]
- Gamas P., Brault M., Jardinaud M.-F., Frugier F. Cytokinins in symbiotic nodulation: when, where, what for? Trends Plant Sci. 2017;22:792–802. doi: 10.1016/j.tplants.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Gauthier-Coles C., White R.G., Mathesius U. Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Front. Plant Sci. 2019;9:1901. doi: 10.3389/fpls.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautrat P., Laffont C., Frugier F. Compact root architecture 2 promotes root competence for nodulation through the miR2111 systemic effector. Curr. Biol. 2020;30:1339–1345.e3. doi: 10.1016/j.cub.2020.01.084. [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Muñoz A., Poovaiah B.W., Oldroyd G.E.D. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell. 2006;18:2680–2693. doi: 10.1105/tpc.106.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodlass G., Smith K.A. Effects of ethylene on root extension and nodulation of pea (Pisum sativum L.) and white clover (Trifolium repens L.) Plant Soil. 1979;51:387–395. [Google Scholar]
- Griesmann M., Chang Y., Liu X., Song Y., Haberer G., Crook M.B., Billault-Penneteau B., Lauressergues D., Keller J., Imanishi L. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361:eaat1743. doi: 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.-L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.-P. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell. 2006;18:3399–3414. doi: 10.1105/tpc.106.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D., Stacey N., Liu C., Wen J., Mysore K.S., Torres-Jerez I., Vernié T., Tadege M., Zhou C., Wang Z.-Y. Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 2013;162:107–115. doi: 10.1104/pp.113.215111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Ecker J.R. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2- dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Hayashi S., Gresshoff P.M., Ferguson B.J. Mechanistic action of gibberellins in legume nodulation. J. Integr. Plant Biol. 2014;56:971–978. doi: 10.1111/jipb.12201. [DOI] [PubMed] [Google Scholar]
- Heckmann A.B., Sandal N., Bek A.S., Madsen L.H., Jurkiewicz A., Nielsen M.W., Tirichine L., Stougaard J. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 2011;24:1385–1395. doi: 10.1094/MPMI-05-11-0142. [DOI] [PubMed] [Google Scholar]
- Heidstra R., Yang W.C., Yalcin Y. Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development. 1997;124:1781–1787. doi: 10.1242/dev.124.9.1781. [DOI] [PubMed] [Google Scholar]
- Held M., Hou H., Miri M., Huynh C., Ross L., Hossain M.S., Sato S., Tabata S., Perry J., Wang T.L. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell. 2014;26:678–694. doi: 10.1105/tpc.113.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltenbrand R., Thomas J., McCarthy H., Dykema K.J., Spurr A., Newhart H., Winn M.E., Mukherjee A. A developmental and molecular view of formation of auxin-induced nodule-like structures in land plants. Front. Plant Sci. 2016;7:1692. doi: 10.3389/fpls.2016.01692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Sawa S. Diverse function of plant peptide hormones in local signaling and development. Curr. Opin. Plant Biol. 2019;51:81–87. doi: 10.1016/j.pbi.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U S A. 2008;105:15208–15213. doi: 10.1073/pnas.0808444105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A.M., Bhuvaneswari T.V., Torrey J.G., Bisseling T. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. U S A. 1989;86:1244–1248. doi: 10.1073/pnas.86.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch A.M., Larue T.A., Doyle J. Is the legume nodule a modified root or stem or an organ sui generis? CRC Crit. Rev. Plant Sci. 1997;16:361–392. [Google Scholar]
- Hossain M.S., Shrestha A., Zhong S., Miri M., Austin R.S., Sato S., Ross L., Huebert T., Tromas A., Torres-Jerez I. Lotus japonicus NF-YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Mol. Plant Microbe Interact. 2016;29:950–964. doi: 10.1094/MPMI-10-16-0206-R. [DOI] [PubMed] [Google Scholar]
- Hu J., Mitchum M.G., Barnaby N., Ayele B.T., Ogawa M., Nam E., Lai W.-C., Hanada A., Alonso J.M., Ecker J.R. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell. 2008;20:320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Chang C., Sun Q., Meyerowitz E.M. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Huault E., Laffont C., Wen J., Mysore K.S., Ratet P., Duc G., Frugier F. Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet. 2014;10:e1004891. doi: 10.1371/journal.pgen.1004891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X., Schnabel E., Hughes K., Frugoli J. RNAi phenotypes and the localization of a protein::GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. J. Plant Growth Regul. 2006;25:156–165. doi: 10.1007/s00344-005-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison C.E., Li J., Argueso C., Gonzalez M., Lee E., Lewis M.W., Maxwell B.B., Perdue T.D., Schaller G.E., Alonso J.M. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell. 2006;18:3073–3087. doi: 10.1105/tpc.106.045674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura A., Hanaki N., Nakamura A., Suzuki T., Taniguchi M., Kiba T., Ueguchi C., Sugiyama T., Mizuno T. Compilation and characterization of Arabidopsis thaliana response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 1999;40:733–742. doi: 10.1093/oxfordjournals.pcp.a029600. [DOI] [PubMed] [Google Scholar]
- Imin N., Mohd-Radzman N.A., Ogilvie H.A., Djordjevic M.A. The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 2013;64:5395–5409. doi: 10.1093/jxb/ert369. [DOI] [PubMed] [Google Scholar]
- Inoue T., Higuchi M., Hashimoto Y., Seki M., Kobayashi M., Kato T., Tabata S., Shinozaki K., Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 2006;313:842–845. doi: 10.1126/science.1128436. [DOI] [PubMed] [Google Scholar]
- Jardinaud M.-F., Boivin S., Rodde N., Catrice O., Kisiala A., Lepage A., Moreau S., Roux B., Cottret L., Sallet E. A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiol. 2016;171:2256–2276. doi: 10.1104/pp.16.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Je B.I., Xu F., Wu Q., Liu L., Meeley R., Gallagher J.P., Corcilius L., Payne R.J., Bartlett M.E., Jackson D. The CLAVATA receptor FASCIATED EAR2 responds to distinct CLE peptides by signaling through two downstream effectors. Elife. 2018;7:e35673. doi: 10.7554/eLife.35673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Liu H., Luo D., Yu N., Dong W., Wang C., Zhang X., Dai H., Yang J., Wang E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016;7:12433. doi: 10.1038/ncomms12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju C., Yoon G.M., Shemansky J.M., Lin D.Y., Ying Z.I., Chang J., Garrett W.M., Kessenbrock M., Groth G., Tucker M.L. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kaló P., Gleason C., Edwards A., Marsh J., Mitra R.M., Hirsch S., Jakab J., Sims S., Long S.R., Rogers J. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- Kassaw T., Nowak S., Schnabel E., Frugoli J. ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiol. 2017;174:2445–2456. doi: 10.1104/pp.17.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi M., Imaizumi-Anraku H., Fukai S., Syono K. Unusual branching in the seedlings of Lotus japonicas–gibberellins reveal the nitrogen-sensitive cell divisions within the pericycle on roots. Plant Cell Physiol. 1996;37:461–470. [Google Scholar]
- Kiba T., Takei K., Kojima M., Sakakibara H. Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell. 2013;27:452–461. doi: 10.1016/j.devcel.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Kieber J.J., Schaller G. Cytokinins. Arabidopsis Book. 2014;12:e0168. doi: 10.1199/tab.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. CTRI, a negative regulator of the ethylene pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Kim G.-B., Son S.-U., Yu H.-J., Mun J.-H. MtGA2ox10 encoding C20-GA2-oxidase regulates rhizobial infection and nodule development in Medicago truncatula. Sci. Rep. 2019;9:5952. doi: 10.1038/s41598-019-42407-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlen W., Ng J.L.P., Deinum E.E., Mathesius U. Auxin transport, metabolism, and signalling during nodule initiation: indeterminate and determinate nodules. J. Exp. Bot. 2018;69:229–244. doi: 10.1093/jxb/erx308. [DOI] [PubMed] [Google Scholar]
- Korasick D.A., Enders T.A., Strader L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouchi H. Large-scale analysis of gene expression profiles during early stages of root nodule formation in a model legume, Lotus japonicus. DNA Res. 2004;11:263–274. doi: 10.1093/dnares/11.4.263. [DOI] [PubMed] [Google Scholar]
- Krusell L., Madsen L.H., Sato S., Aubert G. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420:422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- Krusell L., Sato N., Fukuhara I., Koch B.E.V., Grossmann C., Okamoto S., Oka-Kira E., Otsubo Y., Aubert G., Nakagawa T. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011;65:861–871. doi: 10.1111/j.1365-313X.2010.04474.x. [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., Sakakibara H., Kyozuka J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- Laffont C., Huault E., Gautrat P., Endre G., Kalo P., Bourion V., Duc G., Frugier F. Independent regulation of symbiotic nodulation by the SUNN negative and CRA2 positive systemic pathways. Plant Physiol. 2019 doi: 10.1104/pp.18.01588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffont C., Ivanovici A., Gautrat P., Brault M., Djordjevic M.A., Frugier F. The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically. Nat. Commun. 2020;11:3167. doi: 10.1038/s41467-020-16968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloum T., Baudin M., Frances L., Lepage A., Billault-Penneteau B., Cerri M.R., Ariel F., Jardinaud M.-F., Gamas P., de Carvalho-Niebel F. Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 2014;79:757–768. doi: 10.1111/tpj.12587. [DOI] [PubMed] [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M.-F., Mun J.-H., Larrainzar E., Cook D.R., Gamas P. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 2014;65:481–494. doi: 10.1093/jxb/ert392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrainzar E., Riely B.K., Kim S.C., Carrasquilla-Garcia N., Yu H.-J., Hwang H.-J., Oh M., Kim G.B., Surendrarao A.K., Chasman D. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 2015;169:233–265. doi: 10.1104/pp.15.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layzell D.B., Hunt S. Oxygen and the regulation of nitrogen fixation in legume nodules. Physiol. Plant. 1990;80:322–327. [Google Scholar]
- Lester D.R., MacKenzie-Hose A.K., Davies P.J., Ross J.J., Reid J.B. The influence of the null le-2 mutation on gibberellin levels in developing pea seeds. Plant Growth Regul. 1999;27:83–89. [Google Scholar]
- Lévy J., Bres C., Geurts R., Chalhoub B., Kulikova O., Duc G., Journet E.-P., Ané J.-M., Lauber E., Bisseling T. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science. 2004;303:1361–1364. doi: 10.1126/science.1093038. [DOI] [PubMed] [Google Scholar]
- Li X., Lei M., Yan Z., Wang Q., Chen A., Sun J., Luo D., Wang Y. The REL3-mediated TAS3 ta-siRNA pathway integrates auxin and ethylene signaling to regulate nodulation in Lotus japonicus. New Phytol. 2014;201:531–544. doi: 10.1111/nph.12550. [DOI] [PubMed] [Google Scholar]
- Liang X., Oono Y., Shen N.F., Köhler C., Li K., Scolnik P.A., Theologis A. Characterization of two members (ACS1 and ACS3) of the 1-aminocyclopropane-1-carboxylate synthase gene family of Arabidopsis thaliana. Gene. 1995;167:17–24. doi: 10.1016/0378-1119(95)00694-x. [DOI] [PubMed] [Google Scholar]
- Linkies A., Muller K., Morris K., Tureckova V., Wenk M., Cadman C.S.C., Corbineau F., Strnad M., Lynn J.R., Finch-Savage W.E. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell. 2009;21:3803–3822. doi: 10.1105/tpc.109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang C., Yang J., Yu N., Wang E. Hormone modulation of legume-rhizobial symbiosis. J. Integr. Plant Biol. 2018;60:632–648. doi: 10.1111/jipb.12653. [DOI] [PubMed] [Google Scholar]
- Liu H., Sandal N., Andersen K.R., James E.K., Stougaard J., Kelly S., Kawaharada Y. A genetic screen for plant mutants with altered nodulation phenotypes in response to rhizobial glycan mutants. New Phytol. 2018;220:526–538. doi: 10.1111/nph.15293. [DOI] [PubMed] [Google Scholar]
- Liu J., Rutten L., Limpens E., van der Molen T., van Velzen R., Chen R., Chen Y., Geurts R., Kohlen W., Kulikova O. A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. Plant Cell. 2019;31:68–83. doi: 10.1105/tpc.18.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorteau M.-A., Ferguson B.J., Guinel F.C. Effects of cytokinin on ethylene production and nodulation in pea (Pisum sativum) cv. Sparkle. Physiol. Plant. 2001;112:421–428. doi: 10.1034/j.1399-3054.2001.1120316.x. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Maekawa-Yoshikawa M., Takeda N., Imaizumi-Anraku H., Murooka Y., Hayashi M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009;58:183–194. doi: 10.1111/j.1365-313X.2008.03774.x. [DOI] [PubMed] [Google Scholar]
- Magne K., Couzigou J.-M., Schiessl K., Liu S., George J., Zhukov V., Sahl L., Boyer F., Iantcheva A., Mysore K.S. MtNODULE ROOT1 and MtNODULE ROOT2 are essential for indeterminate nodule identity. Plant Physiol. 2018;178:295–316. doi: 10.1104/pp.18.00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U., Schlaman H.R., Spaink H.P., Of Sautter C., Rolfe B.G., Djordjevic M.A. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998;14:23–34. doi: 10.1046/j.1365-313X.1998.00090.x. [DOI] [PubMed] [Google Scholar]
- Mathesius U., van Noorden G.E., Jin J. The autoregulation gene SUNN mediates changes in nodule and lateral root formation in response to nitrogen through changes of shoot-to-root auxin transport. In: de Bruijn F., editor. The Model Legume Medicago Truncatula. Wiley Blackwell; Hoboken, NJ: 2019. pp. 811–816. [Google Scholar]
- Matsubayashi Y., Sakagami Y. Peptide hormones in plants. Annu. Rev. Plant Biol. 2006;57:649–674. doi: 10.1146/annurev.arplant.56.032604.144204. [DOI] [PubMed] [Google Scholar]
- McAdam E.L., Reid J.B., Foo E. Gibberellins promote nodule organogenesis but inhibit the infection stages of nodulation. J. Exp. Bot. 2018;69:2117–2130. doi: 10.1093/jxb/ery046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis K.M., Thomas S.G., Soule J.D., Strader L.C., Zale J.M., Sun T.-P., Steber C.M. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell. 2003;15:1120–1130. doi: 10.1105/tpc.010827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mens C., Li D., Haaima L.E., Gresshoff P.M., Ferguson B.J. Local and systemic effect of cytokinins on soybean nodulation and regulation of their isopentenyl transferase (IPT) biosynthesis genes following rhizobia inoculation. Front. Plant Sci. 2018;9:1150. doi: 10.3389/fpls.2018.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messinese E., Mun J.-H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.-J., Cook D.R. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 2007;20:912–921. doi: 10.1094/MPMI-20-8-0912. [DOI] [PubMed] [Google Scholar]
- Miller C.O., Skoog F., Von Saltza M.H., Strong F.M. Kinetin, a cell division factor from deoxyribonucleic acid1. J. Am. Chem. Soc. 1955;77:1392. [Google Scholar]
- Miller C.O., Skoog F., Okumura F.S., Von Saltza M.H., Strong F.M. Isolation, structure and synthesis of kinetin, a substance promoting cell Division1,2. J. Am. Chem. Soc. 1956;78:1375–1380. [Google Scholar]
- Miri M., Janakirama P., Held M., Ross L., Szczyglowski K. Into the root: how cytokinin controls rhizobial infection. Trends Plant Sci. 2016;21:178–186. doi: 10.1016/j.tplants.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Miri M., Janakirama P., Huebert T., Ross L., McDowell T., Orosz K., Markmann K., Szczyglowski K. Inside out: root cortex-localized LHK1 cytokinin receptor limits epidermal infection of Lotus japonicus roots by Mesorhizobium loti. New Phytol. 2019;222:1523–1537. doi: 10.1111/nph.15683. [DOI] [PubMed] [Google Scholar]
- Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.-P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., Tabata S., Sandberg G., Kakimoto T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U S A. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa H., Oka-Kira E., Sato N., Takahashi H., Wu G.-J., Sato S., Hayashi M., Betsuyaku S., Nakazono M., Tabata S. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development. 2010;137:4317–4325. doi: 10.1242/dev.058891. [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman N.A., Binos S., Truong T.T., Imin N., Mariani M., Djordjevic M.A. Novel MtCEP1 peptides produced in vivo differentially regulate root development in Medicago truncatula. J. Exp. Bot. 2015;66:5289–5300. doi: 10.1093/jxb/erv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V., Den Herder G., Whitford R., Van De Velde W., Rombauts S., D’haeseleer K., Holsters M., Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortier V., Wasson A., Jaworek P., De Keyser A., Decroos M., Holsters M., Tarkowski P., Mathesius U., Goormachtig S. Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytol. 2014;202:582–593. doi: 10.1111/nph.12681. [DOI] [PubMed] [Google Scholar]
- Müller B., Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315:101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- Nadzieja M., Kelly S., Stougaard J., Reid D. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 2018;95:101–111. doi: 10.1111/tpj.13934. [DOI] [PubMed] [Google Scholar]
- Nadzieja M., Stougaard J., Reid D. A toolkit for high resolution imaging of cell division and phytohormone signaling in legume roots and root nodules. Front. Plant Sci. 2019;10:1000. doi: 10.3389/fpls.2019.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.L.P., Hassan S., Truong T.T., Hocart C.H., Laffont C., Frugier F., Mathesius U. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell. 2015;27:2210–2226. doi: 10.1105/tpc.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.L.P., Welvaert A., Wen J., Chen R., Mathesius U. The Medicago truncatula PIN2 auxin transporter mediates basipetal auxin transport but is not necessary for nodulation. J. Exp. Bot. 2020;71:1562–1573. doi: 10.1093/jxb/erz510. [DOI] [PubMed] [Google Scholar]
- Nishida H., Handa Y., Tanaka S., Suzaki T., Kawaguchi M. Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. J. Plant Res. 2016;129:909–919. doi: 10.1007/s10265-016-0842-z. [DOI] [PubMed] [Google Scholar]
- Nishimura R., Hayashi M., Wu G.-J., Kouchi H., Imaizumi-Anraku H., Murakami Y., Kawasaki S., Akao S., Ohmori M., Nagasawa M. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- Nizampatnam N.R., Schreier S.J., Damodaran S., Adhikari S., Subramanian S. microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015;84:140–153. doi: 10.1111/tpj.12965. [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsushita W., Matsubayashi Y. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 2013;9:726–730. doi: 10.1038/nchembio.1351. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M., Matsubayashi Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants. 2017;3:17029. doi: 10.1038/nplants.2017.29. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 2008;55:152–160. doi: 10.1111/j.1365-313X.2008.03464.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Engstrom E.M., Long S.R. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell. 2001;13:1835–1849. doi: 10.1105/TPC.010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota R., Ohkubo Y., Yamashita Y., Ogawa-Ohnishi M., Matsubayashi Y. Shoot-to-root mobile CEPD-like 2 integrates shoot nitrogen status to systemically regulate nitrate uptake in Arabidopsis. Nat. Commun. 2020;11:641. doi: 10.1038/s41467-020-14440-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacios-Bras C., Schlaman H.R.M., Boot K., Admiraal P., Mateos Langerak J., Stougaard J., Spaink H.P. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 2003;52:1169–1180. doi: 10.1023/b:plan.0000004308.78057.f5. [DOI] [PubMed] [Google Scholar]
- Parry G., Calderon-Villalobos L.I., Prigge M., Peret B., Dharmasiri S., Itoh H., Lechner E., Gray W.M., Bennett M., Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proc. Natl. Acad. Sci. U S A. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Mohd-Radzman N.A., Corcilius L., Crossett B., Connolly A., Cordwell S.J., Ivanovici A., Taylor K., Williams J., Binos S. Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome. Mol. Cell. Proteomics. 2018;17:160–174. doi: 10.1074/mcp.RA117.000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski K., Bisseling T. Legume and actinorhizal root nodule formation. In: Anderson H.M., Barlow P.W., Clarkson D.T., Jackson M.B., Shewry P.R., editors. Plant Roots - from Cells to Systems. Springer; 1997. pp. 137–142. [Google Scholar]
- Penmetsa R.V., Cook D.R. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science. 1997;275:527–530. doi: 10.1126/science.275.5299.527. [DOI] [PubMed] [Google Scholar]
- Penmetsa R.V., Frugoli J.A., Smith L.S., Long S.R., Cook D.R. Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 2003;131:998–1008. doi: 10.1104/pp.015677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmetsa R.V., Uribe P., Anderson J., Lichtenzveig J., Gish J.-C., Nam Y.W., Engstrom E., Xu K., Sckisel G., Pereira M. The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 2008;55:580–595. doi: 10.1111/j.1365-313X.2008.03531.x. [DOI] [PubMed] [Google Scholar]
- Peters N.K., Crist-Estes D.K. Nodule formation is stimulated by the ethylene inhibitor aminoethoxyvinylglycine. Plant Physiol. 1989;91:690–693. doi: 10.1104/pp.91.2.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 2011;65:622–633. doi: 10.1111/j.1365-313X.2010.04447.x. [DOI] [PubMed] [Google Scholar]
- Potuschak T., Lechner E., Parmentier Y., Yanagisawa S., Grava S., Koncz C., Genschik P. EIN3-Dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Qiao H., Chang K.N., Yazaki J., Ecker J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009;23:512–521. doi: 10.1101/gad.1765709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Reid D.E., Ferguson B.J., Gresshoff P.M. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- Reid D.E., Heckmann A.B., Novák O., Kelly S., Stougaard J. CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiol. 2016;170:1060–1074. doi: 10.1104/pp.15.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D., Nadzieja M., Novák O., Heckmann A.B., Sandal N., Stougaard J. Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 2017;175:361–375. doi: 10.1104/pp.17.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D.E., Liu H., Kelly S. Dynamics of Lotus japonicus ethylene production in response to compatible Nod factor. Plant Physiol. 2018;176:1764–1772. doi: 10.1104/pp.17.01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rightmyer A.P., Long S.R. Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Mol. Plant Microbe Interact. 2011;24:1372–1384. doi: 10.1094/MPMI-04-11-0103. [DOI] [PubMed] [Google Scholar]
- Rockström J., Steffen W., Noone K., Persson A. A safe operating space for humanity. Nature. 2009;461:e475. doi: 10.1038/461472a. [DOI] [PubMed] [Google Scholar]
- Ross J.J., Reid J.B., Swain S.M. Control of stem elongation by gibberellin A1: evidence from genetic studies including the slender mutant sln. Funct. Plant Biol. 1993;20:585–599. [Google Scholar]
- Roy S., Robson F., Lilley J., Liu C.-W., Cheng X., Wen J., Walker S., Sun J., Cousins D., Bone C. MtLAX2, a functional homologue of the Arabidopsis auxin influx transporter AUX1, is required for nodule organogenesis. Plant Physiol. 2017;174:326–338. doi: 10.1104/pp.16.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Liu W., Nandety R.S., Crook A., Mysore K.S., Pislariu C.I., Frugoli J., Dickstein R., Udvardi M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell. 2020;32:15–41. doi: 10.1105/tpc.19.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Hua J., Chen Q.G., Chang C., Medrano L.J., Bleecker A.B., Meyerowitz E.M. ETR2 is an ETR1-like gene involved in ethylene signaling in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., Honma T., Aoyama T., Sato S., Kato T., Tabata S., Oka A. ARR1, a transcription factor for genes immediately responsive to cytokinins. Science. 2001;294:1519–1521. doi: 10.1126/science.1065201. [DOI] [PubMed] [Google Scholar]
- Sakakibara H. Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- Sańko-Sawczenko I., Łotocka B., Czarnocka W. Expression analysis of PIN genes in root tips and nodules of Medicago truncatula. Int. J. Mol. Sci. 2016;17:1197. doi: 10.3390/ijms17081197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 2014;5:4983. doi: 10.1038/ncomms5983. [DOI] [PubMed] [Google Scholar]
- Schaller G.E. Ethylene and the regulation of plant development. BMC Biol. 2012;10:9. doi: 10.1186/1741-7007-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller G.E., Kieber J.J. Ethylene. Arabidopsis Book. 2002;1:e0071. doi: 10.1199/tab.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Scheres B., McKhann H.I., Zalensky A., Löbler M., Bisseling T., Hirsch A.M. The PsENOD12 gene is expressed at two different sites in Afghanistan pea pseudonodules induced by auxin transport inhibitors. Plant Physiol. 1992;100:1649–1655. doi: 10.1104/pp.100.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K., Lilley J.L.S., Lee T., Tamvakis I., Kohlen W., Bailey P.C., Thomas A., Luptak J., Ramakrishnan K., Carpenter M.D. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019;29:3657–3668.e5. doi: 10.1016/j.cub.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E., Journet E.-P., de Carvalho-Niebel F., Duc G., Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C. Gibberellin signaling in plants—the extended version. Front. Plant Sci. 2012;2:107. doi: 10.3389/fpls.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- Shen D., Xiao T.T., van Velzen R., Kulikova O., Gong X., Geurts R., Pawlowski K., Bisseling T. A homeotic mutation changes legume nodule ontogeny into actinorhizal-type ontogeny. Plant Cell. 2020;32:1868–1885. doi: 10.1105/tpc.19.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V.K., Jain M., Garg R. Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front. Plant Sci. 2014;5:789. doi: 10.3389/fpls.2014.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15:139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Morgan D.R., Swensen S.M., Mullin B.C., Dowd J.M., Martin P.G. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. U S A. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Hirakawa H., Sato S., Hayashi M., Kawaguchi M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. U S A. 2014;111:14607–14612. doi: 10.1073/pnas.1412716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Shimoda Y., Kawaguchi M., Hayashi M. A shared gene drives lateral root development and root nodule symbiosis pathways in Lotus. Science. 2019;366:1021–1023. doi: 10.1126/science.aax2153. [DOI] [PubMed] [Google Scholar]
- Springmann M., Clark M., Mason-D’Croz D., Wiebe K., Bodirsky B.L., Lassaletta L., de Vries W., Vermeulen S.J., Herrero M., Carlson K.M. Options for keeping the food system within environmental limits. Nature. 2018;562:519–525. doi: 10.1038/s41586-018-0594-0. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Doležal K., Schlereth A., Jürgens G., Alonso J.M. TAA1-Mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Sun T.-P. Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book. 2008;6:e0103. doi: 10.1199/tab.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development. 2012;139:3997–4006. doi: 10.1242/dev.084079. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Miwa K., Ishikawa K., Yamada H., Aiba H., Mizuno T. The Arabidopsis sensor His-kinase, AHk4, can respond to cytokinins. Plant Cell Physiol. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Szczyglowski K., Shaw R.S., Wopereis J., Copeland S., Hamburger D., Kasiborski B., Dazzo F.B., de Bruijn F.J. Nodule organogenesis and symbiotic mutants of the model legume Lotus japonicus. Mol. Plant Microbe Interact. 1998;11:684–697. [Google Scholar]
- Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014;346:343–346. doi: 10.1126/science.1257800. [DOI] [PubMed] [Google Scholar]
- Takahara M., Magori S., Soyano T., Okamoto S., Yoshida C., Yano K., Sato S., Tabata S., Yamaguchi K., Shigenobu S. Too much love, a novel Kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 2013;54:433–447. doi: 10.1093/pcp/pct022. [DOI] [PubMed] [Google Scholar]
- Takanashi K., Sugiyama A., Yazaki K. Involvement of auxin distribution in root nodule development of Lotus japonicus. Planta. 2011;234:73–81. doi: 10.1007/s00425-011-1385-0. [DOI] [PubMed] [Google Scholar]
- Takei K., Sakakibara H., Sugiyama T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001;276:26405–26410. doi: 10.1074/jbc.M102130200. [DOI] [PubMed] [Google Scholar]
- Takei K., Yamaya T., Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J. Biol. Chem. 2004;279:41866–41872. doi: 10.1074/jbc.M406337200. [DOI] [PubMed] [Google Scholar]
- Taleski M., Imin N., Djordjevic M.A. CEP peptide hormones: key players in orchestrating nitrogen-demand signalling, root nodulation, and lateral root development. J. Exp. Bot. 2018;69:1829–1836. doi: 10.1093/jxb/ery037. [DOI] [PubMed] [Google Scholar]
- Tan S., Sanchez M., Laffont C., Boivin S., Le Signor C., Thompson R., Frugier F., Brault M. A cytokinin signaling type-B response regulator transcription factor acting in early nodulation. Plant Physiol. 2020;183:1319–1330. doi: 10.1104/pp.19.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006;7:847–859. doi: 10.1038/nrm2020. [DOI] [PubMed] [Google Scholar]
- Terpolilli J.J., Hood G.A., Poole P.S. What determines the efficiency of N(2)-fixing Rhizobium-legume symbioses? Adv. Microb. Physiol. 2012;60:325–389. doi: 10.1016/B978-0-12-398264-3.00005-X. [DOI] [PubMed] [Google Scholar]
- Thomas J., Bowman M.J., Vega A., Kim H.R., Mukherjee A. Comparative transcriptome analysis provides key insights into gene expression pattern during the formation of nodule-like structures in Brachypodium. Funct. Integr. Genomics. 2018;18:315–326. doi: 10.1007/s10142-018-0594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., Hiltenbrand R., Bowman M.J., Kim H.R., Winn M.E., Mukherjee A. Time-course RNA-seq analysis provides an improved understanding of gene regulation during the formation of nodule-like structures in rice. Plant Mol. Biol. 2020;103:113–128. doi: 10.1007/s11103-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L., Imaizumi-Anraku H., Yoshida S., Murakami Y., Madsen L.H., Miwa H., Nakagawa T., Sandal N., Albrektsen A.S., Kawaguchi M. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science. 2007;315:104–107. doi: 10.1126/science.1132397. [DOI] [PubMed] [Google Scholar]
- Tsikou D., Yan Z., Holt D.B., Abel N.B., Reid D.E., Madsen L.H., Bhasin H., Sexauer M., Stougaard J., Markmann K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science. 2018;362:233–236. doi: 10.1126/science.aat6907. [DOI] [PubMed] [Google Scholar]
- Turner M., Nizampatnam N.R., Baron M., Coppin S., Damodaran S., Adhikari S., Arunachalam S.P., Yu O., Subramanian S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013;162:2042–2055. doi: 10.1104/pp.113.220699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C., Sato S., Kato T., Tabata S. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 2001;42:751–755. doi: 10.1093/pcp/pce094. [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., Chow T.Y., Hsing Y.-I.I., Kitano H., Yamaguchi I. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. ARF1, a transcription factor that binds to auxin response elements. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- Ulmasov T., Murfett J., Hagen G., Guilfoyle T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. U S A. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999;19:309–319. doi: 10.1046/j.1365-313x.1999.00538.x. [DOI] [PubMed] [Google Scholar]
- van Noorden G.E., Ross J.J., Reid J.B., Rolfe B.G., Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noorden G.E., Kerim T., Goffard N., Wiblin R., Pellerone F.I., Rolfe B.G., Mathesius U. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 2007;144:1115–1131. doi: 10.1104/pp.107.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen R., Holmer R., Bu F., Rutten L., van Zeijl A., Liu W., Santuari L., Cao Q., Sharma T., Shen D. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing Rhizobium symbioses. Proc. Natl. Acad. Sci. U S A. 2018;115:E4700–E4709. doi: 10.1073/pnas.1721395115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A., Op den Camp R.H.M., Deinum E.E., Charnikhova T., Franssen H., Op den Camp H.J.M., Bouwmeester H., Kohlen W., Bisseling T., Geurts R. Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant. 2015;8:1213–1226. doi: 10.1016/j.molp.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Vernié T., Kim J., Frances L., Ding Y., Sun J., Guan D., Niebel A., Gifford M.L., de Carvalho-Niebel F., Oldroyd G.E.D. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li K., Chen L., Zou Y., Liu H., Tian Y., Li D., Wang R., Zhao F., Ferguson B.J. MicroRNA167-Directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 2015;168:984–999. doi: 10.1104/pp.15.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., An F., Jiang L., Guo H. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T., Schmülling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Wopereis J., Pajuelo E., Dazzo F.B., Jiang Q., Gresshoff P.M., De Bruijn F.J., Stougaard J., Szczyglowski K. Short root mutant of Lotus japonicus with a dramatically altered symbiotic phenotype. Plant J. 2000;23:97–114. doi: 10.1046/j.1365-313x.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. Fate map of Medicago truncatula root nodules. Development. 2014;141:3517–3528. doi: 10.1242/dev.110775. [DOI] [PubMed] [Google Scholar]
- Xiao T.T., van Velzen R., Kulikova O., Franken C., Bisseling T. Lateral root formation involving cell division in both pericycle, cortex and endodermis is a common and ancestral trait in seed plants. Development. 2019;146:dev182592. doi: 10.1242/dev.182592. [DOI] [PubMed] [Google Scholar]
- Xu C., Liberatore K.L., MacAlister C.A., Huang Z., Chu Y.-H., Jiang K., Brooks C., Ogawa-Ohnishi M., Xiong G., Pauly M. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- Yamada H., Suzuki T., Terada K., Takei K., Ishikawa K., Miwa K., Yamashino T., Mizuno T. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 2001;42:1017–1023. doi: 10.1093/pcp/pce127. [DOI] [PubMed] [Google Scholar]
- Yamagami T., Tsuchisaka A., Yamada K., Haddon W.F., Harden L.A., Theologis A. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 2003;278:49102–49112. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- Yamagami M., Haga K., Napier R.M., Iino M. Two distinct signaling pathways participate in auxin-induced swelling of pea epidermal protoplasts. Plant Physiol. 2004;134:735–747. doi: 10.1104/pp.103.031294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., Markmann K., White C., Schuller B., Sato S. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U S A. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaxley J.R., Ross J.J., Sherriff L.J., Reid J.B. Gibberellin biosynthesis mutations and root development in pea. Plant Physiol. 2001;125:627–633. doi: 10.1104/pp.125.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E., Suzaki T., Toyokura K., Miyazawa H., Fukaki H., Kawaguchi M. A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 2014;165:747–758. doi: 10.1104/pp.113.233379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E., Nishida H., Ogawa-Ohnishi M., Yoshida C., Suzaki T., Matsubayashi Y., Kawaguchi M. PLENTY, a hydroxyproline O-arabinosyltransferase, negatively regulates root nodule symbiosis in Lotus japonicus. J. Exp. Bot. 2019;70:507–517. doi: 10.1093/jxb/ery364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdarska M., Cuyacot A.R., Tarr P.T., Yamoune A., Szmitkowska A., Hrdinová V., Gelová Z., Meyerowitz E.M., Hejátko J. ETR1 integrates response to ethylene and cytokinins into a single multistep phosphorelay pathway to control root growth. Mol. Plant. 2019;12:1338–1352. doi: 10.1016/j.molp.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant. 2012;5:334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zürcher E., Tavor-Deslex D., Lituiev D., Enkerli K., Tarr P.T., Müller B. A robust and sensitive synthetic sensor to monitor the transcriptional output of the cytokinin signaling network in planta. Plant Physiol. 2013;161:1066–1075. doi: 10.1104/pp.112.211763. [DOI] [PMC free article] [PubMed] [Google Scholar]