Abstract
The nucleotide-binding domain and leucine-rich repeat (NLR) gene family is highly expanded in the plant lineage with extensive sequence and structure polymorphisms. To survey the landscape of NLR expansion, we mined the published long-read data generated by the resistance gene enrichment sequencing of 64 diverse Arabidopsis thaliana accessions. We found that the hot spots of massive multi-gene NLR cluster expansion did not typically span the whole cluster; instead, they were restricted to a handful of, or only one, dominant radiation(s). All sequences in such a radiation were distinct from other genes in the cluster but not from each other in the clade, making it difficult to assign trustworthy reference-based orthologies when multiple reference genes were present in the radiation. Consequently, NLR genes can be broadly divided into two types: radiating or high-fidelity, where high-fidelity genes are well conserved and well separated from other clades. A similar distinction could be made for NLR clusters, depending on whether cluster size was determined primarily by extensive radiation or the presence of numerous high-fidelity genes. We also identified groups of well-conserved NLR clades that were missing from the Columbia-0 reference genome. This suggests that the classification of NLRs using gene IDs from a single reference accession can rarely capture all major paralogs in a cluster accurately and representatively and that a reference-agnostic perspective is required to properly characterize these additional variations. Finally, we present a quantitative visualization method for differentiating these situations in a given clade of interest.
Keywords: NLR, cluster, evolution, phylogenetics, disease resistance, plant immunity
Analysis of the recently published RenSeq data of 64 Arabidopsis thaliana accessions revealed extensive copy-number variation in several large multi-gene NLR clusters involved in hybrid necrosis. This was mainly driven by the asymmetric expansion of the clusters, where some members within the cluster were more prone to duplication and diversification than others.
Introduction
Plant genomes encode a variety of disease resistance (R) proteins critical for triggering and mounting immune responses against pathogens. The majority of these R proteins are intracellular nucleotide-binding domain and leucine-rich repeat (NLR) receptors (Kourelis and Van Der Hoorn, 2018). This class of R genes is characterized by the presence of both a nucleotide-binding domain (found in APAF-1 [apoptotic protease-activating factor 1], R proteins, and CED-4 [Caenorhabditis elegans death-4 protein] [van der Biezen and Jones, 1998]) (NB-ARC) and a leucine-rich repeat domain (LRR) (Dangl and Jones, 2001; Meyers et al., 2003). Depending on the presence or absence of the Toll/interleukin-1 receptor (TIR) N-terminal domain, NLRs can be broadly divided into TIR and non-TIR types, and the latter can be further subdivided based on the presence of the N-terminal coiled-coil (CC) or RPW8 domain (Shao et al., 2016; Gao et al., 2018). NLRs are known to participate in a broad spectrum of molecular interactions, including effector recognition, signal transduction, and immune response (Dangl et al., 2013; Kourelis and Van Der Hoorn, 2018). Consequently, NLRs are a highly diversified and abundant group of R genes typically found in the hundreds in the genomes of flowering plants (Jacob et al., 2013; Sarris et al., 2016).
NLRs, either individually or in combination, are known to contribute to immunity against specific pathogens, making their transgenic introduction into cash and food crops highly desirable (Dong and Ronald, 2019). Rpi-amr3i, an NLR-encoding gene isolated from Solanum americanum, conferred full resistance to Phytophthora infestans (potato late blight disease) when transiently expressed in Nicotiana benthamiana and stably expressed in potato, the pathogen's natural host (Witek et al., 2016). Although most NLR studies have focused on functional R genes in representative genomes (Krattinger and Keller, 2016; Periyannan et al., 2017) with minimal insight into the history of diversification and population-level dynamics, this limitation can be overcome by comprehensively surveying the NLR repertoires of one or multiple species using recent improvements in high-throughput sequencing (Giolai et al., 2017; Yang et al., 2017).
A recent survey of the NLR inventory of 64 accessions of the model plant Arabidopsis thaliana using resistance gene enrichment sequencing (RenSeq) revealed an astonishing diversity within the species alone, uncovering 75 novel domain architectures that were absent from the Col-0 reference genome (Van de Weyer et al., 2019). Of great interest are the highly polymorphic clusters of tandem NLR repeats that are postulated to be hotspots of diversification where new NLRs are generated, diversified, and possibly pruned at accelerated rates (Michelmore and Meyers, 1998; Borrelli et al., 2018; van Wersch and Li, 2019; Jiao and Schneeberger, 2020). Although gene clustering in eukaryotes is rare (Lee and Sonnhammer, 2003), it is significantly more common among NLRs (van Wersch and Li, 2019). In A. thaliana, these clusters can be small, consisting of as few as two genes, such as the head-to-head pair RPS4-RRS1 (Narusaka et al., 2009), or extremely large, such as the B5 cluster on chromosome 1 that consists of 11 NLRs (Holub, 2001) and the RPP4/RPP5 cluster on chromosome 4 with eight members spanning ~77 kb (Meyers et al., 2003). Although clustered genes can be co-expressed and functionally dependent, for example, all known examples of head-to-head genes from various plant groups feature one typical NLR and one NLR with an integrated domain (van Wersch and Li, 2019), genes in other clusters may be physically linked as a consequence of tandem duplication (Parker et al., 1997; Botella et al., 1998; Meyers et al., 1998).
A single reference genome cannot capture the diversity of NLR genes and haplotypes present in clusters of tandem arrays, as they tend to be highly polymorphic and vary extensively in the number of members within the same species (Noël et al., 1999; Kuang et al., 2004; Christopoulou et al., 2015; MacQueen et al., 2019) and even within the same population (Stam et al., 2016, 2019). For example, the three genes that confer partially overlapping resistance to distinct Hyaloperonospora arabidopsidis (Hpa; formerly Peronospora parasitica) avirulence determinants in the A. thaliana accession Ws-0 were all mapped to the RPP1 cluster (Botella et al., 1998). The topology of the RPP1 cluster is known to exist in various combinations of RPP1-like NLRs in a handful of accessions, hinting at a complex evolutionary process that generates distinct alleles (Alcázar et al., 2009; Chae et al., 2014; Goritschnig et al., 2016). These highly variable clusters are thought to promote recognition specificity and sensitivity by facilitating the duplication and functional diversification of beneficial NLRs into a collection of related genes with an expanded range of resistance specificities (Hall et al., 2009; Seeholzer et al., 2010; Lu et al., 2016).
However, this comes at a cost. Loci of clusters with more than two members are much more likely to be resistant to crossing-over events (Rowan et al., 2019), resulting in minimal exchange of paralogs between the same clusters of different lineages. This increases the risk of accumulating mutations within the cluster, making it incompatible with the same cluster of another lineage when combined in a single hybrid individual (Smith et al., 2011; Todesco et al., 2014; Shen et al., 2017). In addition to intracluster incompatibility, breaking the extended co-evolution of physically distant loci in a hybrid can also lead to unfavorable or incompatible combinations of genes, resulting in necrosis and reduced fitness (Alcázar et al., 2009; Chae et al., 2014; Guerrero et al., 2017; Tran et al., 2017). Loci predisposed to hybrid incompatibility in plants disproportionately encode immune genes (Chen et al., 2016), and Dangerous Mix (DM) combinations for hybrid necrosis in A. thaliana often involve NLR pairs from physically unlinked loci (Chae et al., 2014). Therefore, it is important to characterize such multi-gene clusters.
The panNLRome resource published by Van de Weyer et al. (2019) was generated by long-read sequencing, which overcomes the long-standing limitations innate to short reads that prevented the proper assessment of copy-number variation (CNV) and diversification of duplicated sequences. Using this dataset, we aim to address the degree of structural variation within multi-NLR clusters by inferring cluster expansion and contraction events within A. thaliana as well as relative to the reference Arabidopsis lyrata genome. To highlight potential risk factors for genetic incompatibility, we tested whether cluster size is correlated with geographic or demographic history and assessed the presence of any asymmetric contribution to cluster expansion by members within each cluster. Our analysis of the dataset covering species-wide NLR variations reveals distinct NLR cluster variation patterns and demonstrates that cluster members contribute unequally to cluster expansion and diversification.
Results
Size Distribution of Major NLR Multi-gene Clusters in 64 A. thaliana Accessions
We first defined clusters by setting a maximum distance of 50 kb between adjacent NLRs. Allowances were made for genes located far away from clusters that were previously annotated to be highly similar to any cluster member. According to these criteria, we identified 36 clusters using the Columbia-0 (Col-0) genome as a reference (Supplemental Table 1). Clusters without existing names or well-known members, of which there were 16, were named by prefixing “c” for “cluster” to the gene ID of the first gene in the cluster and were referred to as “minor clusters” hereafter. Although NLR genes usually contain an N-terminal domain, a catalytic NB-ARC domain, and a variable number of LRR domains (Meyers et al., 1999), there exist functional R genes deficient in one or a combination of these domains (Xiao et al., 2001; Zhao et al., 2015; Nishimura et al., 2017; Roth et al., 2017). Nevertheless, as the NB-ARC domain is the most conserved (and therefore most amenable to alignment) and usually present as a single copy in almost all R genes (Meyers et al., 2002), we used the NB-ARC domain as a proxy for gene counts.
To discover homologs in the Van de Weyer et al. (2019) RenSeq dataset, we first used rpsblast+ to define and extract the genomic sequences encoding the NB-ARC domains in 164 reference Col-0 NLR genes and identified 166 NB-ARC domains in 162 genes (Supplemental Table 1). These sequences were then used as baits in a discontiguous BLAST search against the RenSeq dataset to search for homologous stretches. We assigned a Col-0 gene ID to each predicted homolog based on the closest reference gene in the NB-ARC tree (see Methods).
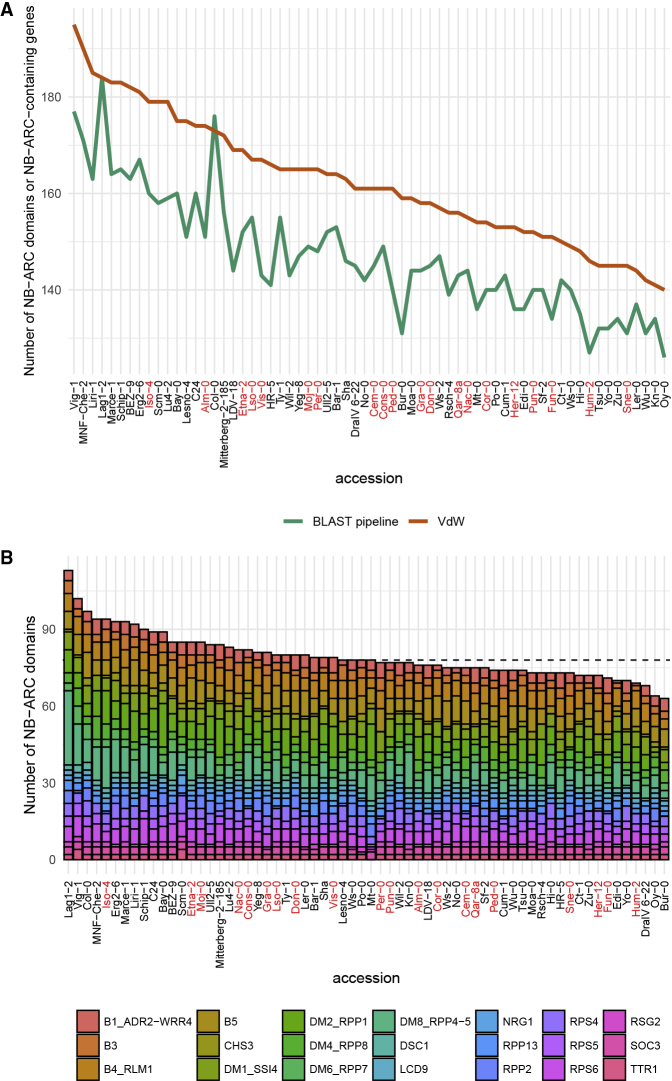
In general, the number of domains identified in different accessions by this BLAST pipeline conforms to the trend reported by Van de Weyer et al. (2019) (Figure 1A). Except for Col-0, the overall ranking of accessions by the number of domains discovered by our BLAST pipeline did not conflict with the number of NB-ARC-containing genes (VdW) identified by Van de Weyer et al. (2019). Compared with the number of genes Van de Weyer et al. (2019) assigned to Col-0 orthogroups (OGs), our BLAST pipeline was more sensitive to paralogs but nevertheless stringent, such that it did not pick up spurious sequences when tested on Col-0 in the RenSeq dataset as a control (Supplemental Table 2). Of the novel, non-bait sequences discovered by our pipeline, two were NLR-like genes (AT4G16930, AT5G45440) and three were pseudogenes (AT1G43180, AT1G50210, AT5G40920).
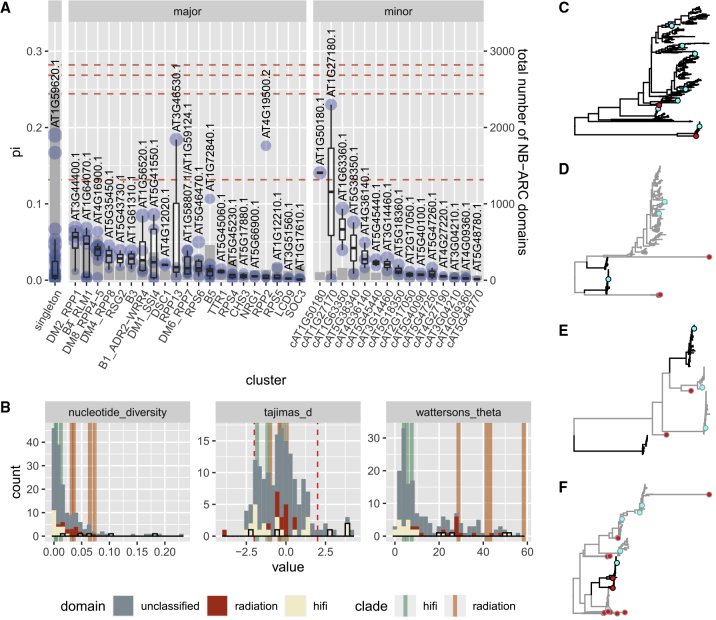
Figure 1.
Summary of the NB-ARC Domain Repertoire in 64 A. thaliana Accessions.
(A) Number of NB-ARC-containing genes identified by Van de Weyer et al. (2019) (VdW) and NB-ARC domains discovered by the pipeline in this study (BLAST pipeline).
(B) Size of major cluster NB-ARC domain repertoires identified in this study, grouped by accession number, and sorted by total cluster repertoire size. The median repertoire size is indicated by a dashed line. Relict accessions are indicated in red.
We are aware that the overall number of NLRs in non-reference accessions may be consistently underestimated as some of the baits used in this pipeline may be Col-0-specific, as suggested by the peak of genes in Col-0 identified by our pipeline (Figure 1A). However, when we plotted the predicted NB-ARC inventory sizes of major clusters and sorted by total cluster inventory size (Figure 1B), relict and European accessions were evenly distributed. This is also true when all identified NB-ARCs were analyzed (Supplemental Figure 1). Although European accessions are presumably more closely related to Col-0 than relict accessions (Van de Weyer et al., 2019), there appeared to be a limited discovery bias toward them (Figure 1B and Supplemental Table 3).
We found that deviations from the median NB-ARC repertoire size among major clusters were more likely to be toward larger than smaller inventories (Figure 1B and Supplemental Figure 1A). When restricted to major clusters analyzed in this paper, the largest NB-ARC repertoire (113 domains found in Lag1-2) nearly doubled the size of the smallest (63 domains found in Oy-0) (Figure 1B and Supplemental Table 3). This could be largely attributed to the DM2/RPP1 and DM8/RPP4/RPP5 clusters, which were highly expanded in Lag1-2 but not in Oy-0 (Figure 1B and Supplemental Table 3).
Cluster Size Is Not Strongly Correlated with Geography or Hierarchy
Although R gene expression is only weakly associated with latitudinal cline (Macqueen and Bergelson, 2016), at least one NLR, CHS3, has a presence/absence pattern strongly associated with altitude. CHS3 responds to chilling stress and appears to be fixed in high-altitude A. thaliana populations, but segregates in low-altitude populations (Günther et al., 2016). Therefore, as the CNV of NLRs may be affected by environmental or evolutionary history, we investigated whether cluster size is correlated with variables affecting demographic history.
To test whether geography contributes to cluster expansion, we fitted a generalized linear regression model for 41 of the 64 accessions that had altitude, latitude, and longitude data. We also calculated Moran's I for each cluster using the geographical distance between 63 of the 64 accessions with reliable latitude and longitude data. We found minimal evidence for the correlation between cluster size and altitude, latitude, longitude, or any combination of interactions between these three geographical predictors (Supplemental Table 4). Additionally, there was no obvious relationship between the cluster sizes of accessions in geographical proximity to each other (Moran's I p value >0.05 for all clusters; Supplemental Figure 2 and Supplemental Table 5). Moran's I was also calculated using the number of pairwise SNPs between 56 accessions for which these data were available. However, no correlation between cluster size and genetic distance was observed (Moran's I p value >0.05 for all clusters; Supplemental Table 5).
Unequal Expansion of Clusters across Accessions and Species
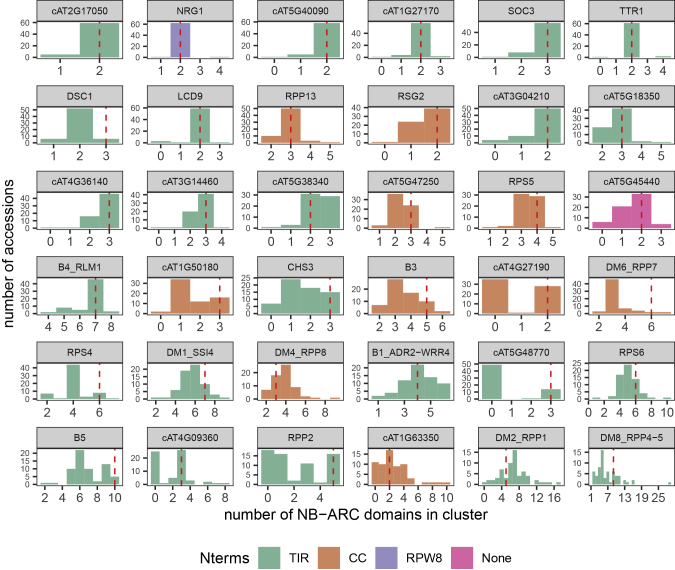
As depicted in Figure 1, the pattern of cluster size variation was not the same for all clusters. In Figure 2, we show a histogram of cluster size for each cluster in all 64 A. thaliana accessions studied. In terms of the range and median of cluster sizes, as well as the CNV pattern, the clusters differed drastically from each other. Some small clusters had a highly conserved copy number that held steady at ~2 for most accessions (NRG1, TTR1, and LCD9), while others were more likely to vary in copy number (RSG2 and CHS3). A similar pattern was observed for larger clusters as well, where a few had extremely dominant copy numbers (B4/RLM1, DM6/RPP7, and RPS4) and others had notable dominant copy numbers (RPP2 and RPS5). Nevertheless, a majority of larger clusters did not appear to have such tight restrictions on cluster size (B1/ADR2/WRR4, B3, DM1/SSI4, DM2/RPP1, DM4/RPP8, and DM8/RPP4/RPP5).
Figure 2.
Patterns of NB-ARC Copy-Number Variation by Cluster.
Histogram of cluster sizes in 64 different A. thaliana accessions. Clusters are colored by the presence of TIR, CC, or RPW8 domains present in their NLR genes and sorted by the SD of cluster sizes. Red dashed lines indicate the size of each cluster in the Col-0 reference genome.
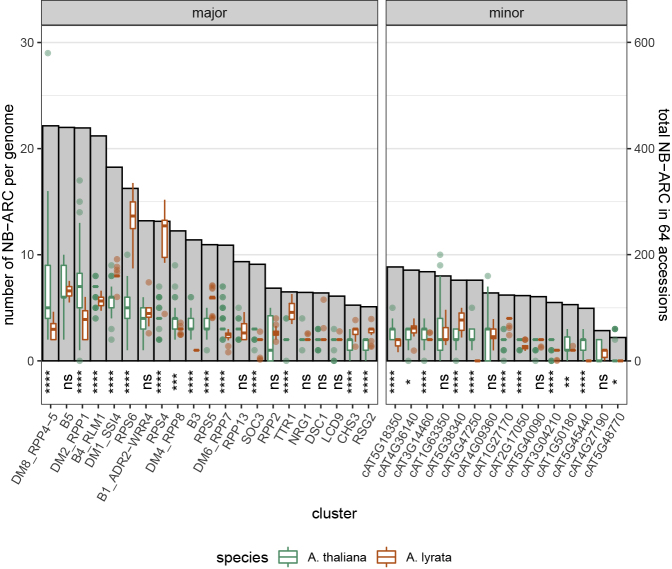
Given that CNV can be massive within clusters (Figures 1B and 2; Supplemental Figure 1), we sought to compare the variations within A. thaliana with the copy numbers in its sister species Arabidopsis lyrata. To do so, we identified the A. lyrata homologs of each A. thaliana NLR (Supplemental Table 6) and estimated the copy number of each A. lyrata homolog in 17 A. lyrata accessions using the normalized read depth of short reads from the whole-genome shotgun sequencing data generated by Novikova et al. (2016) (Supplemental Table 7). Of the 21 major clusters examined in this paper, several stood out as having significantly fewer NB-ARC homologs in A. lyrata than expected (Wilcoxon rank-sum test p value <0.0001) (Figure 3). This suggests that these clusters may have emerged from lineage-specific expansions in A. thaliana after the sister species diverged. Two of these clusters (DM2/RPP1 and DM8/RPP4/RPP5) ranked among the top three clusters with the most members among all 64 A. thaliana accessions and exhibited the highest variation in the number of cluster members (Figures 2 and 3). The number of DM8/RPP4/RPP5 members ranged from two in eight accessions to as many as 29 in Lag1-2, whereas DM2/RPP1 appeared to be entirely missing from IP-Moa-0 (Figure 3; Supplemental Tables 3, 8, and 9). Perhaps even more strikingly, the smaller B3 cluster had only one member in all A. lyrata accessions while all A. thaliana accessions consistently displayed two or more NB-ARC homologs (Figure 3), implying that this cluster is relatively young and entirely A. thaliana-specific. In Col-0, these clusters generally maintained physical clustering within 50 kb from other cluster members with very few, if any, ectopic members, although this may not be the case for other accessions. By contrast, other clusters, such as RPS4, RPS6, and TTR1, were significantly larger in A. lyrata (Figure 3).
Figure 3.
Copy-Number Variation of Cluster NB-ARC Domains across 64 A. thaliana Accessions and 17 A. lyrata Accessions.
Box plot (y-axis left): the number of cluster members distributed across accessions, with hinges corresponding with the 25th and 75th quartiles, and whiskers extending to the smallest and largest values within 1.5 times the interquartile range from the lower and upper hinges, respectively. Bar plot (gray; y-axis right): total number of NB-ARC domains assigned to each cluster across all accessions. Wilcoxon rank-sum test (two-sided) for interspecies comparison: ∗∗∗∗p ≤ 0.0001, ∗∗∗p ≤ 0.001, ∗∗p ≤ 0.01, ∗p ≤ 0.05; ns: p ≥ 0.05.
Interestingly, the number of TTR1 cluster NB-ARCs was always strictly a multiple of two, with an equal number of each gene's NB-ARC homolog in all 64 A. thaliana accessions and several A. lyrata genomes (Figure 3 and Supplemental Figure 3). Although this cluster is highly expanded in the A. lyrata reference genome, there were exactly five copies of each TTR1 cluster NB-ARC for a total of ten (Supplemental Tables 8 and 9). Similarly, the two NRG1 cluster NB-ARC domains existed in a 1:1 ratio in NRG1 clusters across all A. thaliana (except Po-0, which lacks AT5G66900) and A. lyrata accessions (Supplemental Tables 8 and 9). Unlike the TTR1 cluster, however, the size of the NRG1 cluster appeared well conserved in both sister species (Figure 3).
With the exception of IP-Moa-0's lack of a DM2/RPP1 cluster, the presence/absence of the entire cluster was largely restricted to clusters with a median of two members per accession (RPP2, TTR1, LCD9, CHS3, RSG2). The NB-ARC domains of many of these clusters appeared to be descended from a single ancestral pair of domains that diversified into different paired clusters in some common ancestors of A. thaliana and A. lyrata (Supplemental Figure 4). On the other hand, although minor clusters typically contained no more than two or three NB-ARC domains in most accessions, cluster cAT1G63350 had as many as ten members (Figure 3) in DraIV 6-22. This is in contrast with the absence of the large DM2/RPP1 cluster in IP-Moa-0.
Cluster Expansion and Paralog Maintenance Is Often Asymmetric
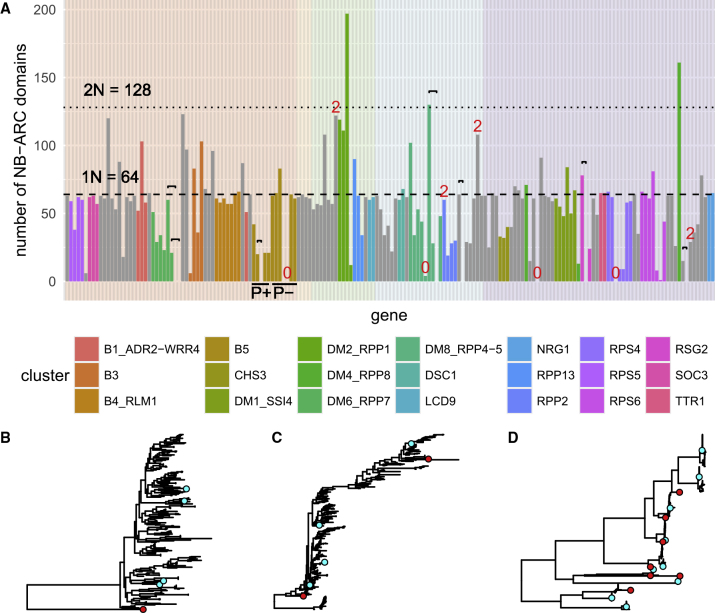
We next set out to determine whether members within each cluster contribute equally to cluster expansion, which would be the case with whole cluster duplication, or if some sequences are more predisposed toward duplication than others. In Figure 4A, we show the number of homologous NB-ARC domains discovered per gene in the 64 A. thaliana accessions, along with their relative genomic positions in Col-0, which were colored by cluster.
Figure 4.
Expansion in High CNV Clusters Can Be Attributed to a Single Massive Radiation.
(A) The number of NB-ARC homologs assigned to each gene was plotted, ordered by position in the genome, and colored by cluster. The background is colored to distinguish the five chromosomes. Singletons are in gray. Black brackets connect genes with identical NB-ARC sequences (including intervening introns), and red numbers indicate the number of NB-ARC domains in genes with more than one or fewer than one NB-ARC domain as detected by rpsblast+. P+ and P− indicate genes containing functional and degenerate P loops, respectively, in the B5 cluster.
(B–D) NB-ARC trees of B3 (B), DM2/RPP1(C), and B5 (D) cluster NB-ARCs showing various degrees of conservation and radiation. Non-B5 cluster sequences that form a monophyletic clade with the B5 cluster are not included in the tree in (D). Col-0 and A. lyrata sequences are shown in cyan and red, respectively.
We found that NB-ARC homologs contributed unequally to cluster size in many major clusters, with only one or two NB-ARC domains responsible for generating most of the clusters' members among the 64 accessions analyzed (Figure 4A). These NB-ARC domains had a disproportionate number of homologs compared with other members of the same cluster, both across all accessions (Figure 4A) and within individual accessions (Supplemental Figure 5A). Therefore, it is clear that some NB-ARC variants have a propensity to spread throughout a cluster. In fact, these NB-ARC domains may contribute remarkably to expansion in some accessions so that there can be up to ten copies in a single individual (Supplemental Figure 5A). Four of the five NB-ARCs with the greatest number of homologs across all accessions belonged to major clusters (Figure 4A and Supplemental Figure 5). Breaking it down further, two of these NB-ARCs (from AT3G44400 and AT3G44630) were found in the DM2/RPP1 cluster, one in the DM4/RPP8 cluster (AT5G48620), and one in the B3 cluster (AT1G61190).
This unequal contribution suggests that duplication events required for generating massive tandem arrays appear to be derived from the radiation of a small number of ancestral sequences. The B3 cluster is an extreme example, where the entire cluster evolved from a single radiation event in the A. thaliana lineage (Figure 4B). In a slightly less extreme example, radiations are also responsible for much of the diversity in both DM2/RPP1 (Figure 4C) and DM4/RPP8 clusters. In other clusters, NB-ARC domains belonging to different genes in the same cluster tend to have high-fidelity. In other words, they are characterized by clear separation from each other, while retaining high sequence conservation, as observed for the B5 cluster (Figure 4D).
Of the major clusters, the B5 cluster stands out by having genes that can be sorted into two groups that are physically separated by a non-NLR gene (AT1G72880). Interestingly, this cluster also neatly segregates along this same divide into genes containing fully functional NB-ARC domains (the former group) from those with degenerate P-loop motifs (the latter group) (Bonardi et al., 2012). These groups have distinct presence/absence patterns among the 64 accessions. The first group, which consisted of AT1G72840, AT1G72850, AT1G72860, and AT1G72870, was characterized by the absence of at least two of these conserved NB-ARC paralogs in most accessions (the genes are indicated with ‘P+’ in Figure 4A and Supplemental Figure 5). On the other hand, the NB-ARCs of genes in the second group, consisting of AT1G72890, AT1G72910, AT1G72940, and AT1G72950, were consistently present in almost all accessions (indicated with “P−” in Figure 4A and Supplemental Figure 5). This copy-number fidelity was even more pronounced in A. lyrata (Supplemental Table 7).
We also found different rates of duplication and spreading between the NB-ARC and TIR domains of the same gene (Supplemental Figure 5), which was not unexpected given the frequency of gene-conversion events within the NLR clusters (Kuang et al., 2004).
Sequence Conservation in NB-ARC Is Rarely Relaxed in All Members of a Cluster
After determining that genes contribute asymmetrically to the NB-ARC repertoire of a cluster, we set out to investigate whether the contribution to cluster sequence diversity is also asymmetric. More specifically, we aimed to determine whether high-copy-number genes contribute the most to cluster sequence diversity. As there is evidence that gene duplication and cluster expansion is associated with sequence and, consequently, functional diversification (Botella et al., 1998; Seeholzer et al., 2010; Goritschnig et al., 2016; Lu et al., 2016), we sought to verify this by calculating the nucleotide diversity (π) in the coding region of each NB-ARC within each NLR.
In general, π scaled weakly with the number of members in a cluster (Figure 5A), the number of domain homologs (Supplemental Figure 6A), and CNV (Supplemental Figure 6B). However, the B5 cluster was a notable exception to this, despite being the second largest cluster that exhibited a fair range of CNV (Figure 3). The π values of its NB-ARCs were comparable to those of much smaller clusters with well-conserved copy numbers such as TTR1 (Figure 5A). The main reason is that it is composed of high-fidelity clades (Figure 4D), which generally exhibit lower nucleotide diversity and Watterson's theta (Wilcoxon rank-sum test [one-sided] p value <0.01 for both) (Figure 5B). By contrast, clusters with the greatest median π values (DM2/RPP1, DM4/RPP8, and DM8/RPP4/RPP5) were mostly populated by members derived from radiations (Figures 4C, 5C, and 5D). High Tajima's D is an indicator of balancing selection, whereas low Tajima's D suggests an excess of rare variants in a population. Based on Tajima's D, an elevated Watterson's theta and sequence diversity in radiating clades compared with high-fidelity clades did not appear to be driven by strong selection pressures in the NB-ARC domain (Figure 5B).
Figure 5.
Cluster Sequence Diversity Is Often Asymmetric.
(A) Nucleotide diversity of NB-ARC domains in each gene in each major cluster calculated per domain from coding sequences only. Refer to Figure 3 for descriptions on boxplot hinges and whiskers. The gene with the highest nucleotide diversity in the NB-ARC domain in each cluster is shown. “.1” and “.2” denote the first and second NB-ARC domains in the gene from the N terminus, respectively. The sizes of blue circles are proportional to the total number of homologs for each gene across all 64 accessions. Red dashed lines denote the nucleotide diversity of NB-ARCs from four A. thaliana genes with two NB-ARC domains when nucleotide diversity is calculated for both domains together.
(B) Histogram of the nucleotide diversity (calculated as in A), Tajima's D, and Watterson's theta of the CDS region of all A. thaliana NLR NB-ARC domains, with selected NB-ARCs belonging to radiating and high-fidelity clades colored in red and beige, respectively. In the back, each statistic was calculated as a whole for three high-fidelity (green) and four radiating (orange) representative clades. See Supplemental Table 10 for domains and clades classified as radiating or high-fidelity. Red dashed lines indicate Tajima's D values of −2 and 2, which are generally considered significant.
(C–F) NB-ARC trees of the DM8/RPP4/RPP5(C), DM4/RPP8(D), RPP13(E), and RPS6(F) clusters showing examples of a cluster with high sequence diversity (C) and clusters with conserved clades missing in Col-0 (D–F). Col-0 and A. lyrata sequences are shown in cyan and red, respectively. Conflated clades are shown in black in (D) to (F), while the rest of the cluster is shown in gray.
In some clusters, there were genes with extremely high π values that were greater than the 75th quartile +1.5 interquartile (Figure 5A). These π values often (but not always) fell approximately around the range defined when π was calculated for two NB-ARC domains belonging to the same gene as if they were a single domain (red dashed lines in Figure 5A). In Col-0, there were four genes with two NB-ARC domains, including AT4G19500 (RPP2A) in cluster RPP2. By inspecting the genes that fell within this range, we could detect instances where two vastly different NB-ARC domains with clear separation were assigned to a single gene present in Col-0. This should only occur when one of the NB-ARC domains in non-reference accessions is absent in Col-0 and has to be assigned to the closest Col-0 homolog, which was evident when we analyzed the trees for RPP13 and RPS6 clusters (Figure 5E and 5F). The existence of such clades (marked in black in Figure 5D–5F) exemplifies the limitation of the field-wide convention of assigning gene names from a single reference genome to non-reference sequences and demonstrates the usefulness of our pipeline for identifying NLR clades not present in reference genomes.
On the other extreme are clusters such as DM2/RPP1, DM4/RPP8, and DM8/RPP4/RPP5, with radiations that exhibit massive sequence diversification and limited sequence conservation. Multiple NB-ARCs in these clusters had relatively high π values, some of which even exceeded the π value of the conflated NB-ARC domain in RPS6 (AT5G46470) (Figure 5A). These patterns could also be observed in their trees (Figures 4C, 5C, and 5D). In fact, these genes with high sequence diversity were nearly indistinguishable from each other in the tree. Their homologs exhibited a relatively continuous range of diversity from each other (Figure 4B and 4C), and were assigned different gene names only by virtue of being slightly more similar to one reference Col-0 gene than another. Interestingly, the DM4/RPP8 cluster also contained a pair of conflated clades (Figure 5D), but the sequence diversity in the DM4/RPP8 radiation was high enough to prevent them from being easily detectable in Figure 5A, unlike the conflated clades in RPP13 and RPS6.
When conflated sequences are excluded as outliers, contribution to cluster sequence diversity is rarely (if ever) dominated by a single “gene” in high-fidelity clusters. Instead, most members typically have comparable π values within a fairly narrow range. Nevertheless, for most large clusters, there was at least one relatively well conserved NB-ARC domain with a π value similar to those of smaller clusters (Figure 5A), which suggests that not all members of a cluster undergo duplication and diversification.
Visualization and Quantification of Cluster Radiation and Problematic Homologs
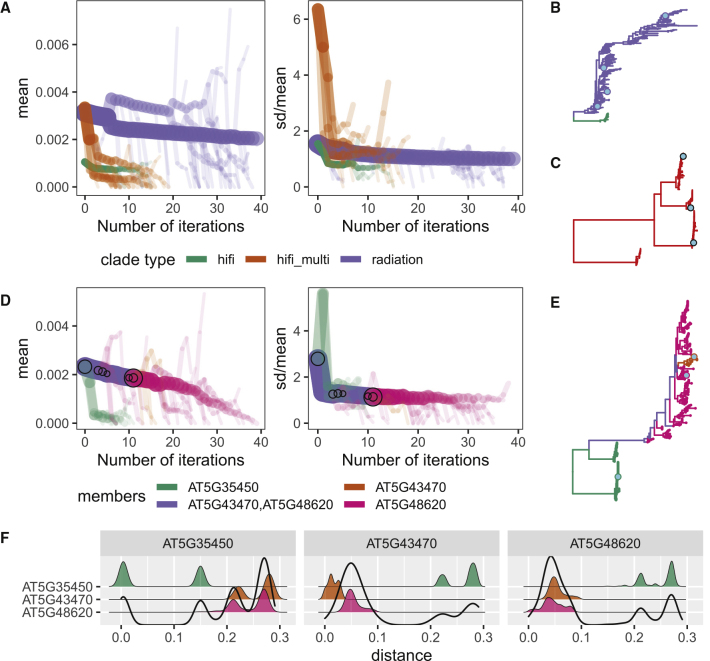
Thus far, we have used “radiating” and “high-fidelity” to describe clades based on the visual inspection of tree shapes. In this section, we present a more quantifiable visualization alternative that captures the essence of this classification scheme using two branch length measurements: the mean and the ratio of standard deviation (SD) to mean. By tracking how these two metrics change when a clade is recursively split at its longest branch, we can identify several distinct patterns to help in the classification of clades.
In Figure 6A, three representative clades were selected to demonstrate these patterns: one radiating clade from the DM2/RPP1 cluster (purple), one high-fidelity clade from the DM2/RPP1 cluster (green), and one clade consisting of multiple high-fidelity clades from the RPP13 cluster (orange), which served as a control. The trees of these clades are shown in Figure 6B (DM2/RPP1) and Figure 6C (RPP13), and are colored by bootstrap confidence in Supplemental Figure 7. According to Figure 6A, three trends helped to distinguish different types of clades. (1) If a clade could be clearly divided into multiple subclades (orange), both the mean and SD/mean ratio started out high and decreased rapidly before abruptly transitioning into a much gentler gradient due to the presence of multiple long branches separating distinct subclades that only contained much shorter branches. As this type of clade was successively split at the longest branches, the resultant clades lacked distinct subclades, giving rise to (2) If a clade could not be clearly divided into multiple subclades (green and purple; orange as well after three iterations), both the mean and SD/mean ratio decreased gradually. This was due to the absence of extremely long branches separating distinct subclades. The SD/mean ratio was generally between 1 and 2. Lastly, (3) if a clade was radiating (purple), it had a much higher mean than a high-fidelity clade. This was because the branches in rapidly evolving radiating clades were longer in general. Due to branch length heterogeneity, fluctuations in mean may occur in smaller clades.
Figure 6.
Visualization and Quantification of Clade Types.
(A and D) Bifurcating plots of the decay of mean and SD as representative clades (A) or the DM4/RPP8 cluster (D) progressively split at their longest branch for 40 iterations, colored by clade type (A) and the set of reference gene ID(s) assigned to clade members (D), respectively. Line width and opacity are proportional to the number of branches in the clade. Clades with only one terminal leaf are not shown. In (A), “hifi” represents the high-fidelity clade and “hifi_multi” denotes the clade containing multiple distinct high-fidelity sub-clades. In (D), circles mark splits that alter gene membership within clades from one iteration to the next, and their sizes are proportional to the number of branches in each resulting clade.
(B, C, and E) NB-ARC trees of the DM2/RPP1(B) and RPP13(C) clusters analyzed in (A) and the DM4/RPP8 cluster (E) analyzed in (D). Clades are colored according to the legends of the relevant decay plots. Col-0 sequences are shown in cyan.
(F) Density plots of intracluster pairwise distances between terminal nodes in the DM4/RPP8 cluster, grouped by assigned reference gene homolog and colored along the y-axis according to the same legend as (D). The combination of facet title and y-axis label indicates the pairwise comparisons included in each density plot. The unfilled distribution at the forefront is the overall distribution of all distances between homologs assigned to the gene in the facet title and all sequences in the cluster.
In Figure 6D, we applied the quantification and visualization method to the NB-ARC tree of cluster DM4/RPP8, which contained both a radiating clade (consisting of AT5G43470 and AT5G48620 NB-ARC homologs) and two well-separated high-fidelity clades (consisting of AT5G35450 NB-ARC homologs) (tree shown in Figure 6E, bootstrap confidence shown in Supplemental Figure 7). The DM4/RPP8 cluster additionally demonstrated both types of problems encountered when using a single reference genome as described previously. As cryptic conflations in the first scenario and rampant diversification in the second scenario had relatively similar π values (Figure 5A), they could not be distinguished by π values alone. However, this visualization method can help to overcome this issue. The cryptic conflation of high-fidelity AT5G35450 subclades could be detected by the fact that both the mean and SD/mean ratio remained high even after AT5G35450 homologs (green in Figure 6D) split off from the rest of the cluster, and required an additional iteration to split these distinct subclades from each other before dropping to the expected mean and SD/mean ratio of well-defined, high-fidelity clades. Rampant diversification leading to a radiating clade of AT5G43470 and AT5G48620 homologs that could not be clearly subdivided along gene boundaries was also detected by the high mean branch length, low SD/mean ratio, as well as the negligible change in mean and SD/mean ratio, when AT5G43470 and AT5G48620 finally split from each other (purple to pink transition from iteration 11 to 12 in Figure 6D).
Figure 6F takes a different approach to visualizing clade separation by plotting the distribution of intracluster pairwise distances between all terminal leaves in the DM4/RPP8 cluster. The extent of the separation between the two distinct clades assigned to AT5G35450 was obvious from the distance between the two distinct peaks in the distribution of pairwise distances between sequences assigned to AT5G35450 (green distribution in the first facet of Figure 6F), where small pairwise distances marked intraclade comparisons and large pairwise distances marked interclade comparisons. On the other hand, the lack of separation between AT5G43470 and AT5G48620 sequences was demonstrated by the overlap between the AT5G43470-AT5G43470 and AT5G43470-AT5G48620 comparisons (orange and pink distributions in the second facet) as well as the AT5G43470-AT5G48620 and AT5G48620-AT5G48620 comparisons (orange and pink distributions in the third facet). The smoothness of their combined distributions (unfilled distributions in the second and third facets) further suggests that sequences assigned to AT5G43470 and AT5G48620 were derived from a single radiation event.
Discussion
Copy-Number and Cluster-Expansion Patterns
We have observed several interesting copy-number patterns for various clusters. For example, the exact 1:1 ratio of the NB-ARC domain abundance in the TTR1 cluster in all 64 A. thaliana accessions and the near 1:1 ratio in the NRG1 cluster (Supplemental Table 8) suggest that the gene pairs in these two clusters operate as functionally linked genes with NB-ARC domains that either work together or not at all. This, combined with the low sequence diversity (Figure 5), implies conservation of sequence and function among physically paired NLRs, which has been shown for the RPS4B/RRS1B cluster (Saucet et al., 2015), as well as negative pressure against the rampant duplication of individual genes without their partners.
Using the NB-ARC domain as a proxy for gene counts, we also noticed that the DM2/RPP1 cluster was the only large cluster that was not present in all 64 accessions (Figure 3). This cluster features prominently in Hpa resistance (Botella et al., 1998; Goritschnig et al., 2016). If IP-Moa-0, the only accession lacking this cluster, also faces Hpa challenge, it would be interesting to investigate whether it has evolved a different mechanism for Hpa resistance in the absence of the major Hpa resistance cluster. By contrast, a handful of small clusters, such as cAT1G63350, which has a median size of two members, can have up to ten NB-ARC homologs in some accessions. A closer investigation may determine whether there is any functional reason for these expansions such as the evolution of a particularly beneficial mutation.
Radiating clades are generally associated with high sequence diversity and can contribute disproportionately to cluster size and diversity (Figures 4 and 5). Many clusters with radiating clades, such as the DM2/RPP1 and DM4/RPP8 clusters, are associated with hybrid incompatibility. This result is in line with the findings of Jiao and Schneeberger (2020), who reported that R gene clusters are enriched among loci that experience limited meiotic recombination due to extensive structural variations between accessions. Massive radiation also appears to be associated with genetic incompatibility that leads to autoimmunity (annotated with DM), suggesting that potentially deleterious mutations are maintained in the cluster along with a resistance trait. Although the NB-ARC domain is considered to be extremely well conserved, especially compared with the highly variable LRR domain in NLRs (Meyers et al., 2002), the fact that the range of sequence diversity is so appreciable, even with the NB-ARC domain alone, suggests a capacity for the NB-ARC domain to adapt rapidly in response to changes in other domains within the same gene. Nevertheless, most clusters with radiating clades are not composed only of members from the radiation and typically possess one or more well-conserved members, which potentially corroborates the proposed trend that a subset of conserved genes flank and anchor relatively large clusters while intervening cluster members are allowed to diversify extensively (Kuang et al., 2004).
Asymmetric Expansion and Sequence Diversification in Large NLR Clusters
Using existing panNLRome data from 64 A. thaliana accessions, we have conducted a comprehensive survey of the number and distribution of NB-ARC domains of known NLR clusters and revealed asymmetric patterns of expansion. By quantifying the number of gene homologs and calculating various diversity statistics, we demonstrated that not all sequences in highly expanded clusters are equally prone to spreading and diversification, as most of the distinct clades within a single cluster are often relatively well conserved in terms of both sequence and copy number. A brief comparison between the number of homologs of NB-ARC and TIR domains belonging to the same reference NLR (Supplemental Figure 5) further showed that domains found in the same reference gene can be present in drastically different abundance in A. thaliana. This supports previous observations showing that an NLR gene unit can be readily broken up to allow for domain recombination (Kuang et al., 2004; MacQueen et al., 2019), providing another path for adaptive immunity to generate potentially different functions.
Clear subclade separation and a high level of conservation may suggest that the high-fidelity genes participate in network interactions that make their conservation indispensable to some pathways or cellular functions (Adachi et al., 2019), although this cannot be concluded without further experimental investigation. A subset of genes in the B5 cluster was particularly interesting, as the NB-ARC domains in the cluster that were present in almost all A. thaliana and A. lyrata accessions contained degenerate P loops, while the NB-ARC domains with intact P loops were absent in various combinations in most accessions (Supplemental Figure 7 and Supplemental Table 7). High levels of sequence fidelity relative to other clusters of a similar size further reinforce the idea that these catalytic null NB-ARC domains may have more to offer to gene function than merely ATP hydrolysis, although how and why they were maintained remains to be uncovered. Curiously, we also discovered that a few of these high-fidelity clades were missing in the reference accession Col-0. As these clades are well conserved, it is likely that the NLRs exert important functions under certain pressures. However, based on what is known about growth–defense trade-offs in R genes (Tian et al., 2003; Harris et al., 2013), the fitness cost for maintaining these genes may be very high. As a result, accessions may have dispensed entirely with the gene when the benefits maintaining it contrasted by the exacted cost. This can be verified by introducing the missing genes into Col-0 and testing the fitness cost.
On the other hand, many well-documented NLR alleles that confer resistance to a range of biotrophic pathogens are known to map to the same highly expandable clusters. For example, multiple Hpa-resistance alleles in various accessions have been traced to DM2/RPP1 (Botella et al., 1998; Goritschnig et al., 2016), a cluster mainly comprised of alleles derived from a single massive radiation (Figure 4C). This suggests the possible existence of qualitative differences in the sequences, functions, or neighboring regions of these sources of proliferation, making them good candidates for adaptive diversification. In particular, transposable elements (TEs) that facilitate chromosomal rearrangement are known to be extremely common around immune gene clusters (Kawakatsu et al., 2016). How TE-associated NLRs respond to changing pathogen stresses and whether a similar radiating phenomenon in the R genes can be induced in other plant species that show insufficient resistance against destructive pathogens await further investigation. In doing so, it may be possible to kick-start unsupervised adaptive radiation to chance upon a winning combination of mutations that confers broader sensitivity either alone or together. Interestingly, although the DM2/RPP1 cluster appears almost indispensable for Hpa resistance in many accessions, it is entirely absent from IP-Moa-0, suggesting that this accession is either no longer facing strong pressures from pathogens or has evolved alternative pathways for addressing this problem.
In summary, we discovered a number of key patterns associated with NLR clusters and identified a few intriguing questions worthy of further exploration. There is still much to be discovered with regard to NLR cluster variations in A. thaliana. This research serves as a starting point and provides candidates for future studies.
Reference-Agnostic R Gene Classification
When using a single reference genome to assign gene IDs to non-reference sequences, we encountered two primary sources of complications: (1) cryptic paralogs in non-reference accessions were forced to be assigned to the closest gene present in Col-0 and (2) sequences belonging to the same radiation were inappropriately sorted to different reference gene IDs simply because reference sequences in the radiation were classified as distinct genes in the reference genome.
These problems caused by using a single reference genome could be solved by the methods we used to visualize the relationship and relatedness of members in the cluster by plotting the decay of the mean and SD of branch length (Figure 6A and 6D) and the density distribution of pairwise distances between sequences (Figure 6E). Using these methods, we were able to clearly distinguish both types of inappropriate groupings despite the limitations of working with a non-representative reference genome. For the more statistically inclined, the density distributions can even be used to formally quantify the conservation, divergence, and number of conflated NB-ARC domains by applying statistical methods to detect multimodal distributions to the density distributions of pairwise distances. For example, the dip test can be used to identify bimodal distributions, and the bimodality coefficient can be used to quantify the magnitude of separation between subclades. Additionally, the level of sequence conservation within a subclade can be quantified by calculating the SD or variance of discrete peaks.
This can also be applied to quantify the patterns of CNV and presence/absence variations in important crops. In fact, one may even expect the patterns to be more prominent in crops, whose traits have been rigorously selected for by humans. Given the known trade-offs between growth and defense (Todesco et al., 2010; Chae et al., 2016), it is more than likely that a number of immune genes have been selectively pruned from crop genomes to engineer larger and more fecund individuals. A closer look into the immune repertoires of crops (Hubner et al., 2019) may shed light on whether crop genomes have evolved ways to mitigate defense trade-offs due to high yield selection, and, if so, whether these genes are capable of conferring a similar level of resistance against pathogens while maintaining growth.
Van de Weyer et al. (2019) previously pointed out that many genes found in non-reference accessions are absent in the reference genome. Combined with what our analysis has revealed about the presence of high-fidelity clades that are absent in Col-0, as well as the smear of relatedness within a radiation, genes in the reference genome should not always be treated as equally distinct. Some of them, such as those generated by a single bush-like radiation (Figure 4B), are more likely to be variations on a theme rather than their own discrete category (Figure 4D). Therefore, one should keep this in mind when surveying R gene repertoires. When possible, information from multiple diverse genomes should be used to obtain a clearer and more representative understanding of immune genes to avoid blindly assigning all non-reference genes to their closest or most well-known reference homolog (as may potentially be the case with the RPP13 cluster) or dividing a radiation into too many discrete units of genes (such as the radiating clade of the DM2/RPP1 cluster).
Methods
Dataset
Analysis of the A. thaliana intraspecific inventory was conducted using the contig-level RenSeq data of 64 accessions, including Col-0, generated by Van de Weyer et al. (2019). The data were retrieved from http://ftp.tuebingen.mpg.de/ebio/alkeller/pan_NLRome/. Of the accessions included in the dataset, 20 were relict accessions sampled from the Iberian Peninsula (Supplemental Table 11), in which the ancestor of A. thaliana was predicted to have evolved. They can therefore be expected to contain significantly more diverse NLR sequences than populations descending from small founding populations that initiated the colonization in Europe (Van de Weyer et al., 2019). This dataset is referred to as the RenSeq dataset. Both the latitude and longitude data for 57 of the 64 accessions, as well as the latitude data for the six remaining accessions, were retrieved from http://1001genomes.org/accessions.html. The SNP dataset used in this paper was previously described by Exposito-Alonso et al. (2018).
Extraction of NLR Domains from Reference Assemblies of A. thaliana (Col-0) and A. lyrata
The coding sequences of all isoforms of 164 NLRs (Supplemental Table 1) annotated in the reference accession Col-0 (TAIR10 assembly; GenBank assembly accession GCA_000001735.2; retrieved from https://www.ncbi.nlm.nih.gov/assembly/GCF_000001735.4) and 189 A. lyrata NLRs identified by Guo et al. (2011) (GenBank assembly accession GCA_000004255.1; retrieved from https://genome.jgi.doe.gov/portal/Araly1/download/Araly1_assembly_scaffolds.fasta.gz) were extracted based on CDS annotations in GFF3 files (retrieved from https://www.arabidopsis.org/download_files/Genes/TAIR10_genome_release/TAIR10_gff3/TAIR10_GFF3_genes.gff (A. thaliana) and https://genome.jgi.doe.gov/portal/Araly1/download/Araly1_GeneModels_FilteredModels6.gff.gz (A. lyrata)). They were translated into amino acid sequences using Biopython (Cock et al., 2009). The resulting amino acid sequences were queried against the CDD database (version CDD.v.3.17, retrieved from ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/cdd.tar.gz) using rpsblast+ (Camacho et al., 2009) to define the boundaries of the NB-ARC (PSSM-Id 307194) and TIR (PSSM-Id 214587) domain(s) in each protein isoform. The corresponding nucleotide sequences encoding these NB-ARC residues (with and without intervening introns, henceforth referred to as “complete” and “CDS-only”, respectively) were extracted by mapping the amino acid positions back to the reference genome using GFF3 annotations.
Identification of Homologous NLR Domains in the RenSeq Dataset
Discontiguous BLAST (Altschul et al., 1990; Camacho et al., 2009) was conducted using complete domains identified in Col-0 as queries and non-reference sequences from the RenSeq dataset as subjects. BLAST hits within 150 bp were merged to account for the introduction of small insertions. Merged regions that did not contain one or more hits >85% identity to any reference domains were dropped. The 85% threshold was set after the manual inspection of general intracluster sequence similarity to ensure sufficient sensitivity so that new cluster genes could be discovered without losing too much specificity. The lowest detected TIR and NB-ARC domain lengths in all reference protein isoforms were converted to their corresponding nucleotide lengths, rounded down to the nearest multiple of 10 (240 bp and 200 bp for the TIR and NB-ARC domains, respectively), and set as the minimum number of bases each merged sequence must overlap with any combination of CDS-only reference sequences to be retained for downstream analysis. The retained nucleotide sequences constituted the set of predicted genomic NLR domain homologs in A. thaliana, and thus, were referred to as RenSeq homologs.
Identification by Discontiguous BLAST of Non-bait Col-0 RenSeq Homologs
Predicted NLR domains for Col-0 (accession ID 6909) in RenSeq homologs were queried against the TAIR10 reference genome using megablast (Morgulis et al., 2008; Camacho et al., 2009), and only the top two hits by bit score for each predicted domain were retained. Hit ranges were extracted and intersected with the TAIR10 GFF3 file using bedtools intersect (Quinlan and Hall, 2010). The results were filtered for gene and pseudogene entries with 100% identity only. To determine whether the domains identified in genes encode polypeptides, we merged CDS entries for each query–subject combination separately and intersected them with the hit range of the query given by the megablast output previously generated. The sum of intersected overlaps for each query–subject combination was calculated, and predicted domains with no overlapping CDS ranges were designated non-coding.
Alignment and Inference of Phylogenetic Relationships among Predicted NLR Domains
The protein sequences of additional genes with predicted coding domains that were discovered in the previous section were extracted from the TAIR10 reference assembly, fed into rpsblast+, and processed as previously described to obtain CDS-only and complete sequences. These were included in the existing list of CDS-only and complete Col-0 sequences. Col-0 RenSeq homologs that were mapped to pseudogenes, non-coding regions within genes, or failed to be identified by rpsblast+ were included in the list of complete Col-0 sequences only. For a final list of reference genes and pseudogenes included in this analysis, see Supplemental Table 1. Sequences were then aligned in discrete steps in increasing order of diversity. Using MAFFT (Katoh and Standley, 2013), CDS-only Col-0 and A. lyrata sequences were aligned, after which complete Col-0 and A. lyrata sequences were added to the alignment, followed by the non-reference sequences from the RenSeq dataset and raising the–adjustdirectionaccurately flag for this last step. The nucleotide alignment was finally fed into FastTree 2 (Price et al., 2010) to construct a maximum-likelihood tree using the General Time Reversible model. All other settings, such as the number of bootstrap iterations (1000), were left as default.
Assignment of A. lyrata and Non-reference A. thaliana Domains to the Closest Col-0 Homolog
Each non-reference domain was assigned to a Col-0 gene/pseudogene (and, consequently, assigned singleton status or to a cluster based on their Col-0 homolog) according to which complete reference gene/pseudogene was the closest in distance in the phylogenetic tree. The TIR tree was rooted using the midpoint using Biopython's Phylo module, while the NB-ARC tree was manually rooted using Dendroscope (Huson and Scornavacca, 2012) based on the assumption that the most basal division was between NLRs with non-TIR N-terminal domains and NLRs with TIR domains. All subsequent steps for homolog assignment were also performed using Biopython's Phylo module. Traversing from each complete Col-0 sequence inward to the root, the distances between all complete Col-0 sequences and internal nodes that lead directly to the root (including the root itself) were stored. To identify the closest Col-0 homologs, we traced the direct path leading from each non-reference A. thaliana sequence and A. lyrata sequence. Whenever an internal node was encountered, if the node was along a direct path between any complete Col-0 sequence and the root, the sum of the distance already traversed and the distance from the node to any complete Col-0 sequence were calculated. The Col-0 sequence(s) that result in the smallest sum was then assigned to each predicted and A. lyrata sequence.
Calculation of Moran's I for Geographical and Genetic Distance
Pairwise geographical distances were calculated based on distances between accession sampling coordinates. To calculate genetic distance, we first filtered the VCF file of SNP data using VCFtools (v.0.1.17) (Danecek et al., 2011) for accessions in the dataset and positions that are called in all of these accessions. The number of SNPs between each pair of accessions was tabulated and used to represent genetic distance. Moran's I was calculated using the R package ape (Paradis and Schliep, 2019).
Estimation of NLR Copy Number in 17 A. lyrata Accessions
Whole-genome shotgun sequencing data for 17 diploid A. lyrata accessions (including two from the subspecies lyrata and 15 from the subspecies petraea) were retrieved from ENA (accession number PRJNA284572) (Novikova et al., 2016) and aligned to the A. lyrata reference genome using BWA MEM (Li, 2013) with default parameters. Using SAMtools (Li et al., 2009), SAM files were converted to BAM files and then sorted and indexed. The indexed BAM files were passed into CNVnator (Abyzov et al., 2011) using a bin size of 500. The genomic ranges of regions determined by CNVnator to exhibit CNV were extracted and intersected with the positions of NB-ARC domains in canonical A. lyrata NLRs in the reference genome. The normalized read depth (RD) of these intersected regions were extracted. Domains that were not identified by CNVnator to exhibit CNV (and therefore were not included in the normalized read depth data output by CNVnator) were assigned a normalized RD of 1.
Calculation of Nucleotide Diversity, Tajima's D, and Watterson's Theta
Nucleotide diversity, Tajima's D, and Watterson's theta were calculated for each domain using the Biopython's Dendropy module based on alignments extracted from the master alignment.
Analysis of the Decay of Mean and SD to Mean Ratio
The NB-ARC phylogenetic tree was imported into Python using Biopython's Phylo module, and all A. lyrata sequences were removed. For a given clade of interest, the lengths of all branches within the clade were obtained by traversing from the root to leaves and storing the lengths of each branch along the way. The mean and SD of branch lengths were calculated using the Python module NumPy (Van Der Walt et al., 2011). At every iteration, the longest branch in a clade was identified and the clade was split by removing the monophyletic clade, for which this branch was the root, from the rest of the clade. This yielded two trees, one of which was the monophyletic clade that had been removed and the other was either a monophyletic or paraphyletic clade depending on whether the longest branch was from the most basal split or not. The process of mean and SD calculation and tree splitting was repeated with each resultant tree until a tree containing only one leaf was achieved or a recursion depth of 40 was reached, whichever came first.
Calculation of Pairwise Distances between Homologs in a Cluster
The distances between NB-ARC homologs within each cluster were calculated for each combination of pairs. The method described above in the section detailing the assignment of gene IDs to homologs was adapted for this purpose by identifying the inner node that all homologs were children of. This node was designated the root. For the step that calculated inner node distances up until the “root,” all cluster homologs were treated if they were Col-0 complete sequences. Finally, the pairwise distance was calculated for each homolog by tracing from each homolog toward the “root” and recording the minimum distance to each of the other homologs.
Funding
This work was supported by the Academic Research Fund (MOE 2019-T2-1-134) from Singapore Ministry of Education and the Intramural Research Fund (R-154-000-B20-114 and R-154-000-B33-114) from National University of Singapore.
Author Contributions
E.C. and R.L. conceptualized and designed the work; R.L. performed the analysis; E.C. and R.L. wrote the paper.
Acknowledgments
We thank Dr. Sang-Tae Kim for the discussion on NLR evolution and data analysis. The authors have no conflict of interest to declare.
Published: June 24, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Accession Numbers
The code generated during this study is available at https://github.com/rlrq/nlr_cluster_survey (in the process of uploading).
Supplemental Information
References
- Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi H., Derevnina L., Kamoun S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 2019;50:121–131. doi: 10.1016/j.pbi.2019.04.007. [DOI] [PubMed] [Google Scholar]
- Alcázar R., García A.V., Parker J.E., Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc. Natl. Acad. Sci. U S A. 2009;106:334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- van der Biezen E.A., Jones J.D. The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998;8:R226–R227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- Bonardi V., Cherkis K., Nishimura M.T., Dangl J.L. A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr. Opin. Immunol. 2012;24:41–50. doi: 10.1016/j.coi.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli G.M., Mazzucotelli E., Marone D., Crosatti C., Michelotti V., Valè G., Mastrangelo A.M. Regulation and evolution of NLR genes: a close interconnection for plant immunity. Int. J. Mol. Sci. 2018;19:1662. doi: 10.3390/ijms19061662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella M.A., Parker J.E., Frost L.N., Bittner-Eddy P.D., Beynon J.L., Daniels M.J., Holub E.B., Jones J.D.G. Three genes of the arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell. 1998;10:1847–1860. doi: 10.1105/tpc.10.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:1–9. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., Bomblies K., Kim S.T., Karelina D., Zaidem M., Ossowski S., Martín-Pizarro C., Laitinen R.A.E., Rowan B.A., Tenenboim H. Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell. 2014;159:1341–1351. doi: 10.1016/j.cell.2014.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E., Tran D.T.N., Weigel D. Cooperation and conflict in the plant immune system. PLOS Pathog. 2016;12:e1005452. doi: 10.1371/journal.ppat.1005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhiguo E., Lin H.X. Evolution and molecular control of hybrid incompatibility in plants. Front. Plant Sci. 2016;7:1–10. doi: 10.3389/fpls.2016.01208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulou M., Wo S.R.C., Kozik A., McHale L.K., Truco M.J., Wroblewski T., Michelmore R.W. Genome-wide architecture of disease resistance genes in lettuce. G3 (Bethesda) 2015;5:2655–2669. doi: 10.1534/g3.115.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. Defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Dangl J.L., Horvath D.M., Staskawich B.J. Pivoting the plant immune system. Science. 2013;341:745–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Walt S., Colbert S.C., Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011;13:22–30. [Google Scholar]
- Dong O.X., Ronald P.C. Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 2019;180:26–38. doi: 10.1104/pp.18.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exposito-Alonso M., Vasseur F., Ding W., Wang G., Burbano H.A., Weigel D. Genomic basis and evolutionary potential for extreme drought adaptation in Arabidopsis thaliana. Nat. Ecol. Evol. 2018;2:352–358. doi: 10.1038/s41559-017-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Wang W., Zhang T., Gong Z., Zhao H., Han G.Z. Out of water: the origin and early diversification of plant R-genes. Plant Physiol. 2018;177:82–89. doi: 10.1104/pp.18.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giolai M., Paajanen P., Verweij W., Witek K., Jones J.D.G., Clark M.D. Comparative analysis of targeted long read sequencing approaches for characterization of a plant’s immune receptor repertoire. BMC Genomics. 2017;18:1–15. doi: 10.1186/s12864-017-3936-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goritschnig S., Steinbrenner A.D., Grunwald D.J., Staskawicz B.J. Structurally distinct Arabidopsis thaliana NLR immune receptors recognize tandem WY domains of an oomycete effector. New Phytol. 2016;210:984–996. doi: 10.1111/nph.13823. [DOI] [PubMed] [Google Scholar]
- Guerrero R.F., Muir C.D., Josway S., Moyle L.C. Pervasive antagonistic interactions among hybrid incompatibility loci. Plos Genet. 2017;13:1–19. doi: 10.1371/journal.pgen.1006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther T., Lampei C., Barilar I., Schmid K.J. Genomic and phenotypic differentiation of Arabidopsis thaliana along altitudinal gradients in the North Italian Alps. Mol. Ecol. 2016;25:3574–3592. doi: 10.1111/mec.13705. [DOI] [PubMed] [Google Scholar]
- Guo Y.-L., Fitz J., Schneeberger K., Ossowski S., Cao J., Weigel D. Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol. 2011;157:757–769. doi: 10.1104/pp.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S.A., Allen R.L., Baumber R.E., Baxter L.A., Fisher K., Bittner-Eddy P.D., Rose L.E., Holub E.B., Beynon J.L. Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions. Mol. Plant Pathol. 2009;10:449–457. doi: 10.1111/j.1364-3703.2009.00544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C.J., Slootweg E.J., Goverse A., Baulcombe D.C. Stepwise artificial evolution of a plant disease resistance gene. Proc. Natl. Acad. Sci. U. S. A. 2013;110:21189–21194. doi: 10.1073/pnas.1311134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001;2:516–527. doi: 10.1038/35080508. [DOI] [PubMed] [Google Scholar]
- Hubner S., Bercovich N., Todesco M., Mandel J.R., Odenheimer J., Ziegler E., Lee J.S., Baute G.J., Owens G.L., Grassa C.J. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants. 2019;5:54–62. doi: 10.1038/s41477-018-0329-0. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Jacob F., Vernaldi S., Maekawa T. Evolution and conservation of plant NLR functions. Front. Immunol. 2013;4:1–16. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W.B., Schneeberger K. Chromosome-level assemblies of multiple Arabidopsis genomes reveal hotspots of rearrangements with altered evolutionary dynamics. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-14779-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Huang S., Shan C., Jupe F., Sasaki E., Schmitz R.J.J., Urich M.A.A., Castanon R., Nery J.R.R., Barragan C., He Y. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J., Van Der Hoorn R.A.L. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018;30:285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krattinger S.G., Keller B. Molecular genetics and evolution of disease resistance in cereals. New Phytol. 2016;212:320–332. doi: 10.1111/nph.14097. [DOI] [PubMed] [Google Scholar]
- Kuang H., Woo S.S., Meyers B.C., Nevo E., Michelmore R.W. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell. 2004;16:2870–2894. doi: 10.1105/tpc.104.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Sonnhammer E.L.L. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13:875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 1303.3997. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Kracher B., Saur I.M.L., Bauer S., Ellwood S.R., Wise R., Yaeno T., Maekawa T., Schulze-Lefert P. Allelic barley MLA immune receptors recognize sequence-unrelated avirulence effectors of the powdery mildew pathogen. Proc. Natl. Acad. Sci. U S A. 2016;113:E6486–E6495. doi: 10.1073/pnas.1612947113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen A., Bergelson J. Modulation of R-gene expression across environments. J. Exp. Bot. 2016;67:2093–2105. doi: 10.1093/jxb/erv530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A., Tian D., Chang W., Holub E., Kreitman M., Bergelson J. Population genetics of the highly polymorphic RPP8 gene family. Genes (Basel). 2019;10:691. doi: 10.3390/genes10090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Chin D.B., Shen K.A., Sivaramakrishnan S., Lavelle D.O., Zhang Z., Michelmore R.W. The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell. 1998;10:1817–1832. doi: 10.1105/tpc.10.11.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Dickerman A.W., Michelmore R.W., Sivaramakrishnan S., Sobral B.W., Young N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999;20:317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Morgante M., Michelmore R.W. TIR-X and TIR-NBS proteins: two new families related to disease resistance TIR-NBS-LRR proteins encoded in Arabidopsis and other plant genomes. Plant J. 2002;32:77–92. doi: 10.1046/j.1365-313x.2002.01404.x. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore R.W., Meyers B.C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Morgulis A., Coulouris G., Raytselis Y., Madden T.L., Agarwala R., Schaffer A.A. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Nishimura M.T., Anderson R.G., Cherkis K.A., Law T.F., Liu Q.L., Machius M., Nimchuk Z.L., Yang L., Chung E.H., El Kasmi F. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2017;114:E2053–E2062. doi: 10.1073/pnas.1620973114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël L., Moores T.L., Van Der Biezen E.A., Parniske M., Daniels M.J., Parker J.E., Jones J.D.G. Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell. 1999;11:2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Novikova P.Y., Hohmann N., Nizhynska V., Tsuchimatsu T., Ali J., Muir G., Guggisberg A., Paape T., Schmid K., Fedorenko O.M. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat. Genet. 2016;48:1077–1082. doi: 10.1038/ng.3617. [DOI] [PubMed] [Google Scholar]
- Paradis E., Schliep K. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- Parker J.E., Coleman M.J., Szabò V., Frost L.N., Schmidt R., Van Der Biezen E.A., Moores T., Dean C., Daniels M.J., Jones J.D.G. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periyannan S., Milne R.J., Figueroa M., Lagudah E.S., Dodds P.N. An overview of genetic rust resistance: from broad to specific mechanisms. PLoS Pathog. 2017;13:1–6. doi: 10.1371/journal.ppat.1006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M.N., Dehal P.S., Arkin A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C., Lüdke D., Klenke M., Quathamer A., Valerius O., Braus G.H., Wiermer M. The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner required for plant immunity. Plant J. 2017;92:808–821. doi: 10.1111/tpj.13717. [DOI] [PubMed] [Google Scholar]
- Rowan B.A., Heavens D., Feuerborn T.R., Tock A.J., Henderson I.R., Weigel D. An ultra high-density arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics. 2019;213:771–787. doi: 10.1534/genetics.119.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P.F., Cevik V., Dagdas G., Jones J.D.G., Krasileva K.V. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016;14 doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015;6:6338. doi: 10.1038/ncomms7338. [DOI] [PubMed] [Google Scholar]
- Seeholzer S., Tsuchimatsu T., Jordan T., Bieri S., Pajonk S., Yang W., Jahoor A., Shimizu K.K., Keller B., Schulze-Lefert P. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact. 2010;23:497–509. doi: 10.1094/MPMI-23-4-0497. [DOI] [PubMed] [Google Scholar]
- Shao Z.Q., Xue J.Y., Wu P., Zhang Y.M., Wu Y., Hang Y.Y., Wang B., Chen J.Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016;170:2095–2109. doi: 10.1104/pp.15.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Wang L., Liu X., Wu J., Jin W., Zhao X., Xie X., Zhu Q., Tang H., Li Q. Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat. Commun. 2017;8:1310. doi: 10.1038/s41467-017-01400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L.M., Bomblies K., Weigel D. Complex evolutionary events at a tandem cluster of Arabidopsis thaliana genes resulting in a single-locus genetic incompatibility. PLoS Genet. 2011;7:e1002164. doi: 10.1371/journal.pgen.1002164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R., Scheikl D., Tellier A. Pooled enrichment sequencing identifies diversity and evolutionary pressures at NLR resistance genes within a wild tomato population. Genome Biol. Evol. 2016;8:1501–1515. doi: 10.1093/gbe/evw094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam R., Silva-Arias G.A., Tellier A. Subsets of NLR genes show differential signatures of adaptation during colonization of new habitats. New Phytol. 2019;224:367–379. doi: 10.1111/nph.16017. [DOI] [PubMed] [Google Scholar]
- Tian D., Traw M.B., Chen J.Q., Kreitman M., Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T.T., Traw M.B., Horton M., Epple P., Kuhns C., Sureshkumar S., Schwartz C., Lanz C. Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana. Nature. 2010;465:632–636. doi: 10.1038/nature09083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Kim S.T., Chae E., Bomblies K., Zaidem M., Smith L.M., Weigel D., Laitinen R.A.E. Activation of the Arabidopsis thaliana immune system by combinations of common ACD6 alleles. PLoS Genet. 2014;10:e1004459. doi: 10.1371/journal.pgen.1004459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D.T.N., Chung E.H., Habring-Müller A., Demar M., Schwab R., Dangl J.L., Weigel D., Chae E. Activation of a plant NLR complex through heteromeric association with an autoimmune risk variant of another NLR. Curr. Biol. 2017;27:1148–1160. doi: 10.1016/j.cub.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wersch S., Li X. Stronger when together: clustering of plant NLR disease resistance genes. Trends Plant Sci. 2019;24:688–699. doi: 10.1016/j.tplants.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Van de Weyer A.L., Monteiro F., Furzer O.J., Nishimura M.T., Cevik V., Witek K., Jones J.D.G., Dangl J.L., Weigel D., Bemm F. A species-wide inventory of NLR genes and alleles in Arabidopsis thaliana. Cell. 2019;178:1260–1272.e14. doi: 10.1016/j.cell.2019.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witek K., Jupe F., Witek A.I., Baker D., Clark M.D., Jones J.D.G. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. 2016;34:656–660. doi: 10.1038/nbt.3540. [DOI] [PubMed] [Google Scholar]
- Xiao S., Ellwood S., Calis O., Patrick E., Li T., Coleman M., Turner J.G. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- Yang R.C., Peng F.Y., Hu Z. Inferring defense-related gene families in Arabidopsis and wheat. BMC Genomics. 2017;18:1–13. doi: 10.1186/s12864-017-4381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Rui L., Li J., Nishimura M.T., Vogel J.P., Liu N., Liu S., Zhao Y., Dangl J.L., Tang D. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015;11:1–28. doi: 10.1371/journal.pgen.1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.