Abstract
Effector proteins delivered inside plant cells are powerful weapons for bacterial pathogens, but this exposes the pathogen to potential recognition by the plant immune system. Therefore, the effector repertoire of a given pathogen must be balanced for a successful infection. Ralstonia solanacearum is an aggressive pathogen with a large repertoire of secreted effectors. One of these effectors, RipE1, is conserved in most R. solanacearum strains sequenced to date. In this work, we found that RipE1 triggers immunity in N. benthamiana, which requires the immune regulator SGT1, but not EDS1 or NRCs. Interestingly, RipE1-triggered immunity induces the accumulation of salicylic acid (SA) and the overexpression of several genes encoding phenylalanine-ammonia lyases (PALs), suggesting that the unconventional PAL-mediated pathway is responsible for the observed SA biosynthesis. Surprisingly, RipE1 recognition also induces the expression of jasmonic acid (JA)-responsive genes and JA biosynthesis, suggesting that both SA and JA may act cooperatively in response to RipE1. We further found that RipE1 expression leads to the accumulation of glutathione in plant cells, which precedes the activation of immune responses. R. solanacearum secretes another effector, RipAY, which is known to inhibit immune responses by degrading cellular glutathione. Accordingly, RipAY inhibits RipE1-triggered immune responses. This work shows a strategy employed by R. solanacearum to counteract the perception of its effector proteins by plant immune system.
Key words: ETI, SGT1, effector, immunity, Ralstonia, PAL
The type-III secreted effector RipE1, from Ralstonia solanacearum, triggers immune responses in Arabidopsis and Nicotiana benthamiana. Such immune responses correlate with an activation of signaling mediated by salicylic acid and jasmonic acid. RipE1-triggered immunity is suppressed by another effector in R. solanacearum, RipAY, showing a bacterial strategy to counteract effector-triggered immunity.
Introduction
Ralstonia solanacearum is considered one of the most destructive plant pathogens, and is able to cause disease in more than 250 plant species (Mansfield et al., 2012, Jiang et al., 2017). As a soil-borne bacterial pathogen, R. solanacearum enters plants through the roots, reaches the vascular system, and spreads through xylem vessels, colonizing the plant systemically (Mansfield et al., 2012). This is followed by massive bacterial replication and the disruption of the plant vascular system, leading to eventual plant wilting (Turner et al., 2009, Digonnet et al., 2012).
Most bacterial pathogens deliver proteins inside plant cells via a type-III secretion system (T3SS); such proteins are thus called type-III effectors (T3Es) (Galán et al., 2014). T3Es have been reported to mediate the suppression of basal defenses and the manipulation of plant physiological functions to support bacterial proliferation (Macho and Zipfel, 2015, Macho, 2016). Resistant plants have evolved intracellular receptors defined by the presence of nucleotide-binding sites (NBS) and leucine-rich repeat domains (LRRs), thus termed NLRs (Cui et al., 2015). Specific NLRs can detect the activities of specific T3Es, leading to the activation of immune responses, which effectively prevent pathogen proliferation (Chiang and Coaker, 2015). The outcome of these responses is named effector-triggered immunity (ETI), and, in certain cases, may cause a hypersensitive response (HR) that involves the collapse of plant cells. Hormone-mediated signaling plays an essential role in plant immunity. Salicylic acid (SA) is considered the most important hormone in plant immunity against biotrophic pathogens (Vlot et al., 2009, Burger and Chory, 2019); Jasmonic acid (JA), on the other hand, is considered the main mediator of immune responses against necrotrophic pathogens (Burger and Chory, 2019). In most cases, both hormones are considered as antagonistic, balancing the effects of each other (Burger and Chory, 2019).
In an evolutionary response to ETI, successful pathogens have acquired T3E activities to suppress this phenomenon (Jones and Dangl, 2006), although reports characterizing T3E suppression of ETI remain scarce, particularly among T3Es within the same strain. While the development of additional T3E activities is a powerful virulence strategy, it also exposes the pathogen to further events of effector recognition. Therefore, the benefits and penalties of T3E secretion need to be finely and dynamically balanced in specific hosts to ensure the appropriate manipulation of plant functions while evading or suppressing ETI. This balance may be particularly important for R. solanacearum, which secretes a larger number of T3Es in comparison with other bacterial plant pathogens (e.g., the reference GMI1000 strain is able to secrete more than 70 T3Es) (Peeters et al., 2013a, Peeters et al., 2013b).
Plants have evolved to recognize immune elicitors from R. solanacearum (Jayaraman et al., 2016, Wei et al., 2018). In terms of mechanism of T3E recognition, the most studied case in R. solanacearum is RipP2 (also known as PopP2), which is perceived in Arabidopsis by the RRS1–RPS4 NLR pair (Gassmann et al., 2002, Deslandes et al., 2002, Tasset et al., 2010, Williams et al., 2014, Le Roux et al., 2015, Sarris et al., 2015). Additionally, several R. solanacearum T3Es were shown to induce cell death in different plant species (Peeters et al., 2013a, Peeters et al., 2013b, Clarke et al., 2015), although, in most cases, it is unclear whether these are due to toxic effects caused by effector overexpression or a host immune response. Some R. solanacearum T3Es have also been shown to cause a restriction of host range; such is the case for RipAA and RipP1 (also known as AvrA and PopP1, respectively), which are perceived and restrict host range in Nicotiana species (Poueymiro et al., 2009). RipP1 also triggers resistance in petunia (Lavie et al., 2002). Similarly, RipB-triggered immunity has been reported as the major cause of avirulence of R. solanacearum RS1000 in Nicotiana species (Nakano and Mukaihara, 2019), RipAX2 (also known as Rip36) have been shown to induce resistance in eggplant and its wild relative Solanum torvum (Nahar et al., 2014, Morel et al., 2018a), and several T3Es from the AWR family (also known as RipA) restrict bacterial growth in Arabidopsis (Solé et al., 2012). Although the utilization of these recognition systems to generate disease-resistant crops is tantalizing, it is imperative to understand the mechanisms underlying the activation of plant immunity and their potential suppression by other T3Es within R. solanacearum.
The ripE1 gene encodes a protein secreted by the type-III secretion system in the R. solanacearum GMI1000 strain (phylotype I) (Mukaihara et al., 2010), and is conserved across R. solanacearum strains from different phylotypes (Peeters et al., 2013a, Peeters et al., 2013b). Based on sequence analysis, RipE1 is homologous to other T3Es in Pseudomonas syringae (HopX) and Xanthomonas spp (XopE) (Supplemental Figure 1; Peeters et al., 2013a, Peeters et al., 2013b), belonging to the HopX/AvrPphB T3E family (Nimchuk et al., 2007). This family is characterized by the presence of a putative catalytic triad consisting of specific cysteine, histidine, and aspartic acid residues, which are conserved in RipE1 (Nimchuk et al., 2007; Supplemental Figure 1), and is similar to several enzyme families from the transglutaminase protein superfamily, such as peptide N-glycanases, phytochelatin synthases, and cysteine proteases (Makarova et al., 1999). AvrPphB, from P. syringae pv. phaseolicola, the original member of the HopX/AvrPphB family, was identified on the basis of its ability to activate immunity in certain bean cultivars (Mansfield et al., 1994). Divergent members from this family in other strains also trigger immunity, and this requires the putative catalytic cysteine (Nimchuk et al., 2007). Previous sequence analysis of T3Es from the HopX family also identified a conserved domain (domain A) required for HopX induction of immunity in bean and Arabidopsis, which was hypothesized to represent a host–target interaction domain or a novel nucleotide/cofactor binding domain (Nimchuk et al., 2007).
In this work, we studied the impact of RipE1 in plant cells and found that RipE1 is recognized by the plant immune system in both Nicotiana benthamiana and Arabidopsis, leading to the activation of immune responses. We further investigated the immune components and signaling pathways associated with this effector recognition. Finally, we found that another effector in R. solanacearum GMI1000 is able to inhibit RipE1-triggered immune responses in N. benthamiana, explaining the fact that RipE1 does not seem to be an avirulence determinant in this plant species.
Results
RipE1 Triggers Cell Death upon Transient Expression in N. benthamiana
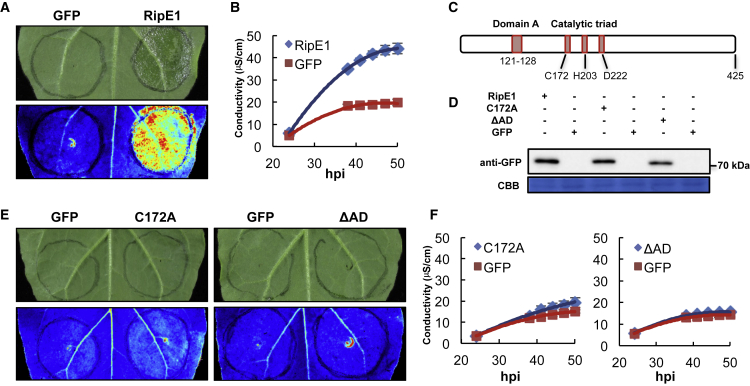
To understand the impact of RipE1 in plant cells, we first used an Agrobacterium tumefaciens (hereafter Agrobacterium)-mediated expression system in N. benthamiana leaves to transiently express RipE1 that is fused to a carboxyl-terminal green fluorescent protein (GFP) tag (RipE1-GFP). Two days after Agrobacterium infiltration, we noticed the collapse of infiltrated tissues expressing RipE1-GFP, but not a GFP control (Figure 1A). This tissue collapse correlated with a release of ions from plant cells (Figure 1B), and cell death was confirmed by trypan blue staining (Supplemental Figure 2). Mutation of the catalytic cysteine to an alanine residue has been shown to disrupt the catalytic activity of enzymes with a catalytic triad similar to that conserved in RipE1 (Gimenez-Ibanez et al., 2014; Figure 1C). To determine whether the putative catalytic activity is required for RipE1 induction of cell death, we generated an equivalent mutant in RipE1 (C172A; Figure 1C). We also generated an independent mutant with a deletion on the eight amino acids that constitute the conserved domain A (Nimchuk et al., 2007; Figure 1C). These mutations did not affect the accumulation of RipE1 (Figure 1D), but abolished the induction of tissue collapse and the ion leakage caused by RipE1 expression (Figure 1E and 1F), indicating that RipE1 requires both the catalytic cysteine and the conserved domain A for the induction of cell death in plants.
Figure 1.
RipE1 Triggers Cell Death in Nicotiana benthamiana.
(A) RipE1-GFP or GFP (as control) were expressed in the same leaf of N. benthamiana using Agrobacterium with an OD600 of 0.5. Photos were taken 2 days post inoculation with a CCD camera (upper panel) or a UV camera (lower panel). UV signal corresponds to the development of cell death (not GFP fluorescence). UV images were taken from the abaxial side and flipped horizontally for representation.
(B) Ion leakage measured in leaf discs taken from N. benthamiana tissues expressing RipE1-GFP or GFP (as control), representative of cell death, at the indicated time points. Values indicate mean ± SE (n = 3 biological replicates).
(C) Simplified diagram of RipE1, including the residues comprising the domain A and the predicted catalytic triad.
(D) Western blot showing the accumulation of RipE1 mutant variants. ΔAD corresponds to a deletion mutant of the domain A (residues 121–128). Molecular weight (kDa) marker bands are indicated for reference.
(E) Cell death triggered by RipE1 mutant variants (conditions as in A).
(F) Ion leakage measured in leaf discs taken from N. benthamiana tissues expressing RipE1 mutant variants (conditions as in B). Each experiment was repeated at least three times with similar results.
hpi, hours post inoculation.
Interestingly, RipE1 was also identified in a systematic screen performed in our laboratory to identify R. solanacearum T3Es that suppress immune responses triggered by bacterial elicitors. In this screen we found that RipE1 expression suppresses the burst of reactive oxygen species (ROS) and the activation of mitogen-activated protein kinases (MAPKs) triggered upon treatment with the bacterial flagellin epitope flg22, which acts as an immune elicitor (Supplemental Figure 3A and 3B). RipE1 requires both the catalytic cysteine and the conserved domain A for this activity (Supplemental Figure 3C). However, we considered the possibility that these responses are abolished by the death of plant cells rather than an active immune suppression. Time-course experiments showed that the suppression of flg22-triggered ROS correlated with the appearance of cell death (Supplemental Figure 3A and 3D), making it difficult to uncouple these observations.
RipE1 Activates Salicylic Acid-Dependent Immunity in N. benthamiana
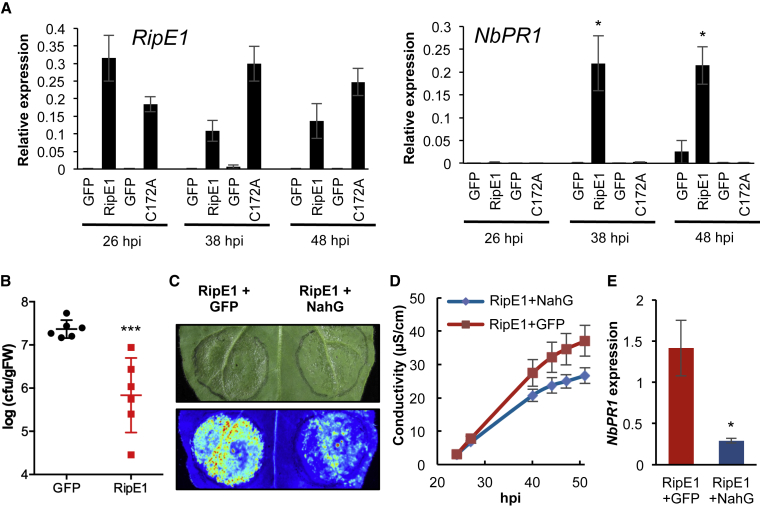
The induction of cell death by pathogen effectors may reflect toxicity in plant cells or the activation of immune responses that lead to an HR. SA plays a major role in the activation of immune responses after the perception of different types of invasion patterns (Vlot et al., 2009). To determine whether RipE1 activates immune responses, we first measured the expression of the N. benthamiana ortholog of the Arabidopsis gene PATHOGENESIS-RELATED-1 (PR1), which is a hallmark of SA-dependent immune responses (Ward et al., 1991, Vlot et al., 2009). Expression of RipE1-GFP (but not the C172A catalytic mutant) significantly enhanced the accumulation of NbPR1 transcripts (Figure 2A). In keeping with the notion that RipE1 activates a defense response against R. solanacearum, RipE1 expression in N. benthamiana leaves enhanced resistance against subsequently inoculated R. solanacearum Y45, which is otherwise pathogenic in N. benthamiana (Li et al., 2011) (Figure 2B). The bacterial salicylate hydroxylase NahG converts SA to catechol, which leads to the suppression of SA-dependent responses (Delaney et al., 1994). The expression of NahG-GFP in N. benthamiana slightly enhanced the accumulation of RipE1 fused to a carboxyl-terminal N-luciferase tag (Nluc) (Supplemental Figure 4), consistent with the reported role of SA in hindering Agrobacterium-mediated transformation (Rosas-Díaz et al., 2017); despite this, NahG expression partially suppressed RipE1-triggered cell death, ion leakage, and NbPR1 expression (Figure 2C–2E). Altogether, these data suggest that RipE1 induces SA-dependent immune responses in plant cells, which cause the development of an HR.
Figure 2.
RipE1 Triggers SA-Dependent Immune Responses in N. benthamiana.
(A) Quantitative RT–PCR (qRT–PCR) to determine the expression of RipE1 and NbPR1 in N. benthamiana tissues expressing GFP, RipE1, or RipE1 C172A, using Agrobacterium with an OD600 of 0.1. Samples were taken at the indicated times (hours post infiltration; hpi) after Agrobacterium infiltration. In each case, the RipE1 variants and their respective GFP control were expressed in the same leaf, and values are represented side by side. Expression values are relative to the expression of the housekeeping gene NbEF1a. Values indicate mean ± SE (n = 3 biological replicates).
(B) RipE1-GFP or GFP (as control) were expressed in the same leaf of N. benthamiana using Agrobacterium with an OD600 of 0.5. Twenty-four hours after Agrobacterium infiltration, before the appearance of cell death, a 105 CFU/ml inoculum of R. solanacearum Y45 was infiltrated into the same tissues. Samples were taken 1 day post inoculation to determine Y45 CFU per gram of tissue. Values indicate mean ± SE (n = 6 biological replicates).
(C–E) RipE1-Nluc was expressed 24 h after expression of GFP (as control) or with NahG-GFP in the same leaf. Protein accumulation is shown in Supplemental Figure 4. (C) Photos were taken 2.5 days post inoculation with a CCD camera (upper panel) or a UV camera (lower panel). UV signal corresponds to the development of cell death (not GFP fluorescence). UV images were taken from the abaxial side and flipped horizontally for representation. (D) Ion leakage measured in leaf discs taken from N. benthamiana tissues expressing RipE1 together with GFP or NahG-GFP, representative of cell death, at the indicated time points. Values indicate mean ± SE (n = 3 biological replicates). (E) qRT–PCR to determine the expression of NbPR1 in N. benthamiana tissues 48 h after Agrobacterium infiltration. Expression values are relative to the expression of the housekeeping gene NbEF1a. Values indicate mean ± SE (n = 3 biological replicates). Asterisks indicate significant differences compared with the mock control according to a Student's t-test (*p < 0.05; ***p < 0.001). Each experiment was repeated at least three times with similar results.
RipE1 Enhances the Expression of PAL Genes and the Biosynthesis of Salicylic Acid and Jasmonic Acid
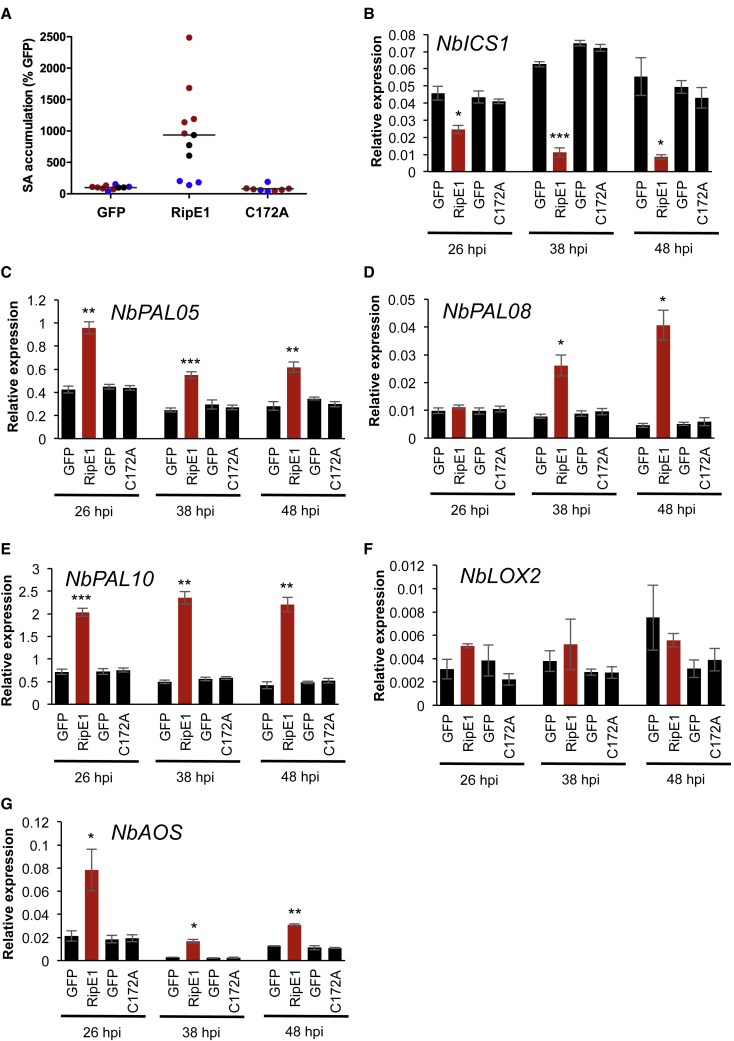
The expression of RipE1 led to a dramatic increase in SA accumulation in N. benthamiana (Figure 3A), consistent with the observed overexpression of NbPR1 (Figure 2A). This reinforces the idea that RipE1 is perceived by the plant immune system, and this leads to the activation of SA biosynthesis and SA-dependent immune responses. In Arabidopsis, the chloroplastic pathway mediated by isochorismate synthetase 1 (ICS1) plays a predominant role in the pathogen-induced SA biosynthesis (Wildermuth et al., 2001, Garcion et al., 2008). However, gene expression analysis showed that the expression of the N. benthamiana ortholog of the Arabidopsis ICS1, NbICS1, was significantly reduced upon RipE1 expression (Figure 3B), despite the simultaneous high NbPR1 transcript accumulation (Figure 2A). SA can also be synthesized from phenylalanine in a pathway mediated by phenylalanine-ammonia lyases (PALs). In contrast with the expression of NbICS1, several genes encoding NbPALs were upregulated upon expression of RipE1, but not the catalytic mutant version (Figure 3C–3E), suggesting that this pathway may mediate the enhancement of SA biosynthesis upon perception of RipE1 activity. SA and JA are considered antagonistic hormones in plant immune responses. Surprisingly, instead of a reduction of the expression of genes associated to JA biosynthesis, we found an increase in the accumulation of transcripts of NbLOX2 and NbAOS upon expression of catalytically active RipE1 (Figure 3F). In Arabidopsis, LOX2 and AOS contribute to the biosynthesis of JA (Bell et al., 1995, Laudert et al., 1996). Accordingly, we detected an increase in JA contents upon RipE1 expression (Supplemental Figure 5), indicating that RipE1 perception does not inhibit JA signaling, but rather leads to an enhancement of JA biosynthesis and associated gene expression.
Figure 3.
Perception of RipE1 Results in Enhanced Expression of PAL Genes and SA Biosynthesis in N. benthamiana.
(A) Measurement of SA accumulation in N. benthamiana tissues expressing GFP, RipE1, or RipE1 C172A, using Agrobacterium with an OD600 of 0.5. Samples were taken 42 h after Agrobacterium infiltration. Three independent biological repeats were performed, and the different colors indicate values from different replicates. Values are represented as percentage of the GFP control in each replicate.
(B–G)qRT–PCR to determine the expression of NbICS1(B), NbPAL05(C), NbPAL08(D), NbPAL10(E), NbLOX2(F), and NbAOS(G), in N. benthamiana tissues expressing GFP, RipE1, or RipE1 C172A, using Agrobacterium with an OD600 of 0.5. Samples were taken at the indicated times (hours post infiltration; hpi) after Agrobacterium infiltration. In each case, the RipE1 variants and their respective GFP control were expressed in the same leaf, and values are represented side by side. Expression values are relative to the expression of the housekeeping gene NbEF1a. Values indicate mean ± SE (n = 3 biological replicates). Asterisks indicate significant differences compared with the mock control according to a Student's t-test (*p < 0.05; **p < 0.01; ***p < 0.001). Each experiment was repeated at least three times with similar results.
RipE1-Triggered Immunity Requires SGT1, but Not EDS1 or NRC Proteins
The suppressor of the G2 allele of skp1 (SGT1) plays an essential role in ETI, and is required for the induction of disease resistance mediated by most NLRs (Azevedo et al., 2002, Kadota et al., 2010). Virus-induced gene silencing (VIGS) of NbSGT1 abolished RipE1-triggered cell death, ion leakage, and NbPR1 expression (Figure 4A–4D), indicating that RipE1-triggered immunity requires SGT1. While most NLRs require SGT1 to function, a specific group of NLRs containing an N-terminal Toll-like interleukin-1 receptor (TIR) domain also requires EDS1 (Wiermer et al., 2005, Schultink et al., 2017). N. benthamiana plants carrying a stable knockout mutation in EDS1 (Schultink et al., 2017) displayed clear RipE1-triggered cell death (Figure 4E), suggesting that RipE1-triggered immunity is not mediated by a TIR-NLR. Other NLRs contain a C-terminal coiled-coil (CC) domain, and a specific subset of CC-NLRs requires a network of helper NLRs termed NRC proteins (Wu et al., 2016). Interestingly, silencing of NRC proteins did not affect RipE1-triggered cell death (Supplemental Figure 6), suggesting that RipE1-triggered immunity is not mediated by an NLR within the NRC network.
Figure 4.
RipE1-Triggered Immune Responses Require SGT1 but Not EDS1.
(A–D) RipE1-GFP or GFP (as control) were expressed in the same leaf of N. benthamiana undergoing VIGS of NbSGT1 or VIGS with an empty vector (EV) construct (as control), using Agrobacterium with an OD600 of 0.5. (A) Western blot showing the accumulation of GFP, RipE1-GFP, and endogenous NbSGT1. Molecular weight (kDa) marker bands are indicated for reference. (B) Photos were taken 2 days post inoculation with a CCD camera (upper panel) or a UV camera (lower panel). UV signal corresponds to the development of cell death (not GFP fluorescence). UV images were taken from the abaxial side and flipped horizontally for representation. (C) Ion leakage measured in leaf discs taken from N. benthamiana tissues expressing RipE1-GFP or GFP (as control), representative of cell death, 48 h after Agrobacterium infiltration. Values indicate mean ± SE (n = 3 biological replicates). (D) qRT–PCR to determine the expression of NbPR1 in N. benthamiana tissues 48 h after Agrobacterium infiltration. Expression values are relative to the expression of the housekeeping gene NbEF1a. Values indicate mean ± SE (n = 3 biological replicates).
(E) RipE1-GFP or GFP (as control) were expressed in the same leaf of N. benthamiana wild type or a stable eds1 knockout mutant, using Agrobacterium with an OD600 of 0.5. Photos were taken 2 days post inoculation with a CCD camera (upper panel) or a UV camera (lower panel). UV signal corresponds to the development of cell death (not GFP fluorescence). UV images were taken from the abaxial side and flipped horizontally for representation. Asterisks indicate significant differences compared with the mock control according to a Student's t-test (***p < 0.001). Each experiment was repeated at least three times with similar results.
RipE1 Triggers Immune Responses in Arabidopsis Plants
Because transgenic Arabidopsis plants expressing RipE1-GFP from an inducible 35S promoter died after germination, we generated Arabidopsis transgenic plants expressing RipE1-GFP and RipE1C172A-GFP from an estradiol (EST)-inducible promoter. Five-week-old plants expressing RipE1-GFP, but not RipE1C172A-GFP, showed reduced growth in soil upon EST treatment for 14 days (Figure 5A). To determine whether RipE1-triggered growth reduction in Arabidopsis correlates with the activation of immunity, we first monitored the expression of defense-related genes. Similar to the result observed upon expression in N. benthamiana, expression of RipE1 in Arabidopsis triggered the overexpression of AtPR1 (Figure 5B). However, in Arabidopsis, the enhanced PR1 expression correlated with an overexpression of AtICS1, but not AtPAL1, upon RipE1 expression (Figure 5B). As observed in N. benthamiana, RipE1 expression led to the overexpression of the JA marker genes AtVSP2 and AtPDF1.2 (Figure 5B). This indicates that, as observed in N. benthamiana, RipE1 activates SA- and JA-dependent signaling in Arabidopsis. To determine whether the activation of defense-related genes in Arabidopsis leads to an efficient immune response against R. solanacearum, we inoculated RipE1-expressing plants by soil drenching with R. solanacearum after EST treatment for 2 days. As shown in Figure 5C, RipE1-expressing plants displayed weaker and delayed disease symptoms upon R. solanacearum inoculation, reflecting an enhanced disease resistance upon RipE1 expression. RipE1-expressing plants also showed a moderate reduction in bacterial growth after R. solanacearum infiltration in the leaves (Supplemental Figure 7A), suggesting that the immune response is not exclusively associated with invasion or proliferation in the root. However, RipE1-expressing plants did not display enhanced resistance against the leaf-borne pathogen Pseudomonas syringae pv. tomato DC3000 (Supplemental Figure 7B and 7C).
Figure 5.
RipE1 Triggers Immunity in Arabidopsis Plants.
(A)Arabidopsis Col-0 wild type or independent stable transgenic lines expressing RipE1 or RipE1 C172A from an estradiol (EST)-inducible promoter were grown for 3 weeks and then sprayed with 100 μM EST daily. Photographs were taken 2 weeks after beginning the EST treatment.
(B)Arabidopsis 4-day-old seedlings were treated with 25 μM EST and samples were taken 1, 2, 3, or 4 days after EST treatment. qRT–PCR was used to determine the expression of RipE1, AtPR1, AtPAL1, AtICS1, AtVSP2, and AtPDF1.2. Expression values are relative to the expression of the housekeeping gene AtACT2. Values indicate mean ± SE (n = 3 biological replicates).
(C)Arabidopsis Col-0 wild type or EST-RipE1 transgenic plants were grown for 4 weeks and then treated with 100 μM EST for 2 days before inoculation with R. solanacearum GMI1000 by soil drenching. Plants showed no difference in root or shoot size at the time of inoculation. The results are represented as disease progression, showing the average wilting symptoms on a scale from 0 to 4 (mean ± SE). n = 20 plants per genotype. Asterisks indicate significant differences compared with the mock control according to a Student's t-test (*p < 0.05; **p < 0.01; ***p < 0.001). Each experiment was repeated at least three times with similar results.
RipE1-Triggered Immune Responses Are Suppressed by RipAY
RipE1 expression activates immunity in Arabidopsis and N. benthamiana, although both plant species are susceptible hosts for R. solanacearum GMI1000 (or a derivative strain carrying mutations in ripP1 and ripAA, in the case of N. benthamiana; Poueymiro et al., 2009), which carries RipE1. Therefore, we reasoned that other T3Es in GMI1000 may be able to suppress RipE1-triggered immunity in the context of infection. We recently identified an R. solanacearum T3E, RipAY, which is able to suppress SA-dependent immune responses through the degradation of glutathione (Mukaihara et al., 2016, Sang et al., 2016); however, the ability of RipAY to suppress immunity triggered by other R. solanacearum T3Es remained unknown. Interestingly, the expression of RipE1 in N. benthamiana leads to an increase in glutathione accumulation in plant tissues, which precedes the onset of immune responses (Figure 6A). Considering that both RipAY and RipE1 are present in GMI1000, we sought to determine whether RipAY has the ability to suppress RipE1-triggered immunity. Indeed, expression of RipAY in N. benthamiana did not affect the accumulation of RipE1 (Supplemental Figure 8) but inhibited the tissue collapse and ion leakage caused by RipE1 expression (Figure 6B and 6C). Moreover, RipAY was able to suppress the overexpression of several SA-related genes triggered by RipE1 (Figure 6D and Supplemental Figure 9), indicating that RipAY suppresses RipE1-triggered immune responses. RipAY did not significantly suppress the expression of NbLOX2 or NbAOS (Supplemental Figure 9). This could reflect a predominant role of RipAY in the suppression of RipE1-triggered SA responses, and may be responsible for the absence of a full suppression of RipE1-triggered HR (Figure 6B and 6C). Interestingly, however, a RipAY point mutant unable to degrade glutathione (RipAYE216Q; Sang et al., 2016) did not suppress RipE1-triggered responses (Figure 6B–6D), suggesting that RipAY suppresses RipE1-triggered immunity through the degradation of cellular glutathione.
Figure 6.
RipE1-Triggered Immune Responses Are Suppressed by RipAY.
(A) RipE1-GFP or GFP (as control) were expressed in the same leaf of N. benthamiana using Agrobacterium with an OD600 of 0.5, and samples were taken at the indicated time points to measure the accumulation of glutathione (GSH).
(B–D) RipE1-Nluc was expressed 24 h after expression of GFP (as control), RipAY-GFP, or RipAY-E216Q-GFP, respectively, in the same leaf. Protein accumulation is shown in Supplemental Figure 8. (B) Photos were taken 2.5 days post inoculation with a CCD camera (upper panel) or a UV camera (lower panel). UV signal corresponds to the development of cell death (not GFP fluorescence). UV images were taken from the abaxial side and flipped horizontally for representation. (C) Ion leakage measured in leaf discs taken from N. benthamiana tissues expressing RipE1 together with GFP or RipAY-GFP, representative of cell death, at the indicated time points. Values indicate mean ± SE (n = 3 biological replicates). (D) qRT–PCR to determine the expression of NbPR1 in N. benthamiana tissues 48 h after Agrobacterium infiltration. Expression values are relative to the expression of the housekeeping gene NbEF1a. Values indicate mean ± SE (n = 3 biological replicates). Asterisks indicate significant differences compared with the mock control according to a Student's t-test (*p < 0.05; ***p < 0.001). Each experiment was repeated at least three times with similar results.
Discussion
Expression of T3Es in plant cells may either induce cell death because of cell toxicity or lead to the activation of an immunity-associated HR. Overexpression of RipE1 in N. benthamiana leads to an HR that: (1) is dependent on the immune regulator SGT1; (2) activates SA accumulation and PR1 expression; (3) restricts growth of R. solanacearum Y45; and (4) is suppressed by the NahG and other R. solanacearum effectors, indicating that RipE1-mediated cell death is due to the activation of immunity in the host. It is, however, noteworthy that cell death induced by RipE1 develops slower than that triggered by other HR-inducing T3Es (i.e., RipAA; Supplemental Figure 2). Several T3Es within the HopX/AvrPphB family are predicted enzymes that are associated with activation of host immunity, although the association of the predicted catalytic activity with the activation of immunity seems to differ among them. While the ability of AvrPphB and several other family members to trigger immunity requires the putative catalytic cysteine (Mansfield et al., 1994, Nimchuk et al., 2007), other members with the predicted catalytic activity, such as HopX from P. syringae pv. tabaci or P. syringae pv. phaseolicola race 6, do not trigger immunity in the same hosts (Stevens et al., 1998, Nimchuk et al., 2007). In the case of RipE1, the putative catalytic cysteine is required for the induction of immunity, which suggests that RipE1 is an active enzyme and that this catalytic activity leads to perception by the host immune system. Moreover, the conserved domain A (Nimchuk et al., 2007) is also required for the activation of immunity by RipE1. In addition, we found that RipE1 is able to suppress elicitor-triggered immune responses in N. benthamiana. However, since this activity correlates with the induction of cell death, it is difficult to uncouple both observations, and further studies on the virulence activity of RipE1 will require the utilization of a host plant that is unable to recognize it.
The fact that RipE1 is recognized, and activates immune responses, in both N. benthamiana and Arabidopsis suggests at least two scenarios: it is possible that the NLR responsible for this recognition is conserved in both species; on the other hand, it is also possible that both species have independently developed NLRs that recognize RipE1. Although we did not identify the NLR involved, we determined that, at least in N. benthamiana, RipE1 recognition does not rely on EDS1 or the NRC network, pointing to a CC-NRC-independent NLR. Interestingly, although RipE1 perception leads to the accumulation of SA in both plant species, the associated gene expression patterns seem to differ. The ICS pathway plays a predominant role in the pathogen-induced SA biosynthesis in Arabidopsis (Wildermuth et al., 2001, Garcion et al., 2008). In agreement with this, the RipE1-triggered overexpression of AtPR1 in Arabidopsis correlates with an enhanced expression of AtICS1 but not AtPAL1. However, it seems that the RipE1-induced increase in SA content in N. benthamiana correlates with a reduction in NbICS1 gene expression and an increase in the expression of several NbPAL genes. Considering that ICS1 is normally regulated at the transcriptional level upon pathogen perception (Wildemurth et al., 2001), our results suggest that the PAL pathway is more relevant than the ICS pathway for the induction of RipE1-triggered immunity in N. benthamiana, indicating that both pathways are differentially required for distinct immune responses in different plant species. Similarly, both the ICS and PAL pathways have been reported to be required for pathogen-induced SA biosynthesis in soybean (Shine et al., 2016). The reduction in ICS1 expression in N. benthamiana may reflect a compensatory effect between the ICS and PAL pathway. In addition to different gene expression patterns, the physiological output in both plant species may be different. Although RipE1 expression caused an inhibition of Arabidopsis growth, we did not observe any signs of cell death (data not shown), which contrasts with our observation in N. benthamiana. However, this may be caused by differences in the expression system used in both plants (Agrobacterium-mediated transient expression in N. benthamiana versus EST-induced expression in Arabidopsis stable transgenic plants).
Another surprising aspect of RipE1-triggered immunity is the fact that it leads to the simultaneous accumulation of SA and JA, and to a strong and moderate SA- and JA-triggered gene expression, respectively, in both N. benthamiana and Arabidopsis. This suggests that, in the case of RipE1-triggered immunity, SA and JA may play a cooperative role, possibly reflecting the complexity of the R. solanacearum infection process compared with other pathogens. In keeping with this notion, although RipE1-expressing Arabidopsis plants displayed enhanced resistance against R. solanacearum and upregulation of SA-related genes, they did not show enhanced resistance against the leaf-borne pathogen P. syringae pv tomato DC3000 (Supplemental Figure 6). Since the enhancement of JA signaling has been associated with a promotion of virulence by this pathogen (Gimenez-Ibanez et al., 2016), the observed upregulation of JA-related genes may underlie this phenomenon.
If RipE1 triggers immunity in N. benthamiana, why is it that a GMI1000 strain without RipP1 and RipAA (but having RipE1) can cause a successful infection in N. benthamiana without triggering immunity (Poueymiro et al., 2009)? Here, we found that another effector within GMI1000, RipAY, is able to inhibit RipE1-triggered immunity. Since RipE1 perception correlates with an enhancement of cellular glutathione, and RipAY requires its γ-glutamyl cyclotransferase activity to inhibit RipE1-triggered HR, the degradation of glutathione or other γ-glutamyl compounds (Fujiwara et al., 2016, Mukaihara et al., 2016, Sang et al., 2016) is the most likely mechanism for this inhibition. Besides RipAY, other T3Es within GMI1000 contribute to the suppression of RipE1-triggered HR by targeting other immune functions (Yu et al., 2019b; Keke Wang and A.P.M., unpublished data), playing a redundant role that likely leads to the robust suppression of RipE1-triggered immunity in GMI1000. This reflects bacterial adaptation: RipE1 could be important for virulence, but also triggers immunity. In this context, instead of losing RipE1, R. solanacearum has developed other effectors to suppress the induction of immunity while keeping RipE1 virulence activity. This is reminiscent of what has been shown for P. syringae pv syringae B728a, where several effectors within the same strain suppress the HR triggered by HopZ3, which otherwise acts as a virulence factor (Rufián et al., 2018). Similarly, although transient expression of HopX from P. syringae pv. tomato (Pto) triggers HR in specific Arabidopsis accessions, it does not trigger HR in the context of Pto infection (Nimchuk et al., 2007). It is possible that, as in the case of RipE1, the immune responses triggered by HopX are masked during Pto infection (as suggested in Nimchuk et al., 2007), likely due to the suppression by other effectors within the same strain.
Methods
Plant Materials and Growth Conditions
N. benthamiana plants were grown on soil at one plant per pot in an environmentally controlled growth room at 25°C under a 16-h light/8-h dark photoperiod with a light intensity of 130 mE m−2 s−1. A. thaliana plants were grown under the same conditions as N. benthamiana for collection of seeds. For bacterial virulence and ROS burst assays, A. thaliana plants were grown in a growth chamber controlled at 22°C with a 10-h photoperiod and a light intensity of 100–150 mE m−2 s−1. After R. solanacearum inoculation, Arabidopsis plants were transferred to a growth chamber at 27°C with 75% humidity under a 12-h light/12-h dark photoperiod.
Chemicals
The flg22 peptide (TRLSSGLKINSAKDDAAGLQIA) was purchased from Abclonal (USA). All other chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Plasmids, Bacterial Strains, and Cultivation Conditions
R. solanacearum GMI1000 was grown on solid BG-11 medium plates or cultivated overnight in liquid BG-11 medium at 28°C (Morel et al., 2018b). The ripE1 gene from R. solanacearum GMI1000 cloned in pDONR207 (donated by Nemo Peeters and Anne-Claire Cazale) was subcloned into pGWB505 by LR reaction (Thermo Fisher, USA) to generate a fusion protein with eGFP tag at the C terminus (Nakagawa et al., 2007). RipE1 and ripE1 mutants were inserted between BamHI and XhoI restriction sites on sXVE:GFPc:Bar estradiol-inducible vector using enzyme digestion (Schlücking et al., 2013). These generated binary vectors were transformed into Agrobacterium tumefaciens (Agrobacterium) GV3101 for transient or stable gene expression in N. benthamiana and A. thaliana plants. Agrobacterium carrying pGWB505 vectors were grown at 28°C and 220 rpm in Luria–Bertani medium supplemented with 50 mg/l rifampicin, 25 mg/l gentamycin, and 50 mg/l spectinomycin, while those carrying estradiol-inducible vectors were grown in 50 mg/l rifampicin, 25 mg/l gentamycin, and 50 mg/l kanamycin.
Site-Directed Mutagenesis
RipE1C172A and RipE1 ΔAD mutant variants were generated using the QuickChange Lightning Site-Directed Mutagenesis Kit (Life Technologies, USA) following the manufacturer's instructions. RipE1/pDONR207 plasmid was used as template. Primers used for the mutagenesis are listed in Supplemental Table 1.
Agrobacterium-Mediated Gene Expression in A. thaliana and N. benthamiana
Stable transgenic Arabidopsis plants with RipE1 and RipE1 mutated variants driven by estradiol-inducible promoter were obtained using the floral dip method (Zhang et al., 2006). Homozygous T3 lines were used for all the experiments. Agrobacterium-mediated transient expression in N. benthamiana was performed as described by Li (2011). Agrobacterium carrying the resultant plasmids was suspended in infiltration buffer to a final OD600 of 0.1–0.5 and infiltrated into the abaxial side of the leaves using a 1-ml needless syringe. Leaf samples were taken at 1–3 days post infiltration for analysis based on experimental requirements.
Protein Extraction and Western Blots
Plant tissues were collected into 2-ml tubes with metal beads and frozen in liquid nitrogen. After grinding with a tissue lyser (Qiagen, Germany) for 1 min at 30 rpm/s, proteins were extracted using protein extraction buffer (100 mM Tris–HCl [pH 8], 150 mM NaCl, 10% glycerol, 5 mM EDTA, 2 mM dithiothreitol, 1× plant protease inhibitor cocktail, 1% NP-40, 2 mM phenylmethylsulfonyl fluoride, 10 mM Na2MoO4, 10 mM NaF, 2 mM Na3VO4) and incubating for 5 min. After centrifugation, the supernatants were mixed with SDS loading buffer, incubated at 70°C for 10 min, and resolved using SDS–PAGE. Proteins were transferred to a polyvinylidene fluoride membrane and monitored by western blot using anti-GFP (Abicode, M0802-3a) and anti-luciferase (Sigma, L0159) antibodies.
Measurement of ROS Generation and MAPK Activation
PAMP-triggered ROS burst and MAPK activation in plant leaves were measured as described previously (Segonzac et al., 2011, Sang and Macho, 2017). ROS was elicited with 50 nM flg22. MAPK activation assays were performed using 4- to 5-week-old N. benthamiana. Two days after Agrobacterium infiltration at OD600 of 0.1, the intact leaves were elicited for 15 min after vacuum infiltration of 100 nM flg22. Leaf discs were taken to monitor MAPK activation by western blot with Phospho-p44/42 MAPK (Erk1/2; Thr-202/Tyr-204) antibodies.
Cell Death Measurement
Cell death in plant leaves was quantified as previously described (Yu et al., 2019b) by measuring the electrolyte leakage using a conductivity meter (Thermo Fisher) or by observing the autofluorescence using the Bio-Rad Gel Imager (Bio-Rad, USA). In brief, 1 day after Agrobacterium infiltration in N. benthamiana, one 13-mm leaf disc was immersed in 4 ml of distilled water for 1 h with gentle shaking and then transferred to a 6-well culture plate containing 4 ml of distilled water in each well. The ion conductivity was then measured at different time intervals. Autofluorescence in intact N. benthamiana leaves was measured at 2.5 days post infiltration. Trypan blue staining was performed as previously described (Lv et al., 2019).
RNA Isolation and qRT–PCR
Five- to 8-day-old Arabidopsis seedlings were grown in sterile conditions and 8–10 seedlings grown on an independent plate were collected as one biological sample. For N. benthamiana tissues, three leaf discs were taken from each leaf from different plants and collected as one biological sample. Total RNA was extracted using the E.Z.N.A. Plant RNA kit with DNA digestion on column (Biotek, China) according to the manufacturer's instructions. RNA samples were quantified with a Nanodrop spectrophotometer (Thermo Fisher). First-strand cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). qRT–PCR was performed using the iTaq Universal SYBR Green Supermix (Bio-Rad) and CFX96 Real-time system (Bio-Rad), and the qPCR data were analyzed as previously described (Livak and Schmittgen, 2001, Wang et al., 2019). The identifiers of the genes analyzed by qRT–PCR are: NbPR1 (Niben101Scf03376g03004); NbICS1 (Niben101Scf00593g04010); NbPAL05 (Niben101Scf05617g00005); NbPAL08 (Niben101Scf03712g01008); NbPAL10 (Niben101Scf12881g00010); NbLOX2 (Niben101Scf06364g00003); NbAOS (Niben101Scf05799g02010); NbEF1a (Niben101Scf08618g01001); AtPR1 (AT2G14610); AtICS1 (AT1G74710); AtPAL1 (AT2G37040); AtPDF1.2 (AT5G44420); AtVSP2 (AT5G24770); and AtACTIN2 (AT3G18780). Primer sequences are listed in Supplemental Table 1.
Measurements of SA and JA Content in Plant Leaves
SA and JA content were quantified using the method described by Forcat et al. (2008) with the following modifications. Leaves (50 mg fresh weight) were collected 42 h after Agrobacterium infiltration and frozen in liquid nitrogen before grinding into fine powder with the Qiagen tissue lyser. SA and JA were extracted at 10°C for 1 h using 70% methanol extraction solvent spiked with d4-SA as internal standard. Supernatant was taken after centrifugation at 20 000 g for 10 min and analyzed on an ACQUITY UPLC I-class coupled with AB SCIEX TripleTOF 5600+ apparatus. The analytical column used was an ACQUITY UPLC BECH C18 1.7-μm, 2.1 × 150-mm column. The JA concentration was calculated based on the calibration curve created by running a JA standard solution. The results were analyzed by Peakview1.2.
Measurements of Total Cellular Glutathione in N. benthamiana Leaves
Total cellular glutathione was measured as previously described (Sang et al., 2016). In brief, 10 mg of N. benthamiana leaves was collected and glutathione was measured using a Glutathione Assay Kit (Beyotime, China) according to the manufacturer's instructions.
Virus-Induced Gene Silencing (VIGS) in N. benthamiana
VIGS in N. benthamiana plants was performed using TRV vectors as described by Senthil-Kumar and Mysore (2014). VIGS of NbSGT1 was performed with several modifications described by Yu et al. (2019a). Cultures of Agrobacterium carrying pTRV2:NbSGT1 plasmids or pTRV2 plasmids were mixed at 1:1 ratio and co-infiltrated into the lower leaves of 3-week-old N. benthamiana plants. The upper leaves were used for experimental assay within 7–10 days after VIGS application. Silencing of NRCs (NLR required for cell death) in N. benthamiana and subsequent expression of T3Es was performed as described by Wu et al. (2017).
Pseudomonas syringae Virulence Assays
For leaf infiltration with P. syringae, Arabidopsis plants were treated with 100 μM EST for 2 days before inoculation. Plants showed no difference in root or shoot size at the time of inoculation. Pto DC3000 was resuspended in water at 105 colony-forming units (CFU)/ml. The bacterial suspensions were then infiltrated into 4- to 5-week-old Arabidopsis leaves using a needleless syringe. For spray inoculation, Pto DC3000 was resuspended in water at 108 CFU/ml, and silwet-L77 was added to a final concentration of 0.02% before spraying onto 3-week-old Arabidopsis seedlings. Bacterial numbers were determined 3 days post inoculation as previously described (Macho et al., 2012, Wang et al., 2019).
Ralstonia solanacearum Virulence Assays
For standard R. solanacearum virulence assays, 4-week-old A. thaliana plants, grown in Jiffy pots, were inoculated with R. solanacearum without wounding by soil drenching. For experiments using inducible transgenic lines, all the plants were treated with 100 μM EST for 2 days before inoculation. Plants showed no difference in root or shoot size at the time of inoculation. An overnight-grown bacterial suspension was diluted to obtain an inoculum of 5 × 107 CFU/ml. Once the Jiffy pots were completely drenched, the plants were removed from the bacterial solution and placed back on a bed of potting mixture soil. The genotypes to be tested were placed in a random order to allow an unbiased analysis of the wilting. Daily scoring of the visible wilting on a scale ranging from 0 to 4 (or 0%–100% leaves wilting) led to an analysis using Kaplan–Meier survival analysis, log-rank test, and HR calculation as previously described (Morel et al., 2018b).
To determine For determination of R. solanacearum growth in Arabidopsis leaves, a 107 CFU/ml inoculum was infiltrated into leaves of 4-week-old Arabidopsis plants 2 days after EST treatment, and samples were taken 2 days after inoculation. To determine R. solanacearum growth in N. benthamiana leaves, a 105 CFU/ml inoculum of R. solanacearum Y45 was infiltrated into N. benthamiana leaves expressing RipE1-GFP or a GFP control. RipE1-GFP was expressed using Agrobacterium, and R. solanacearum Y45 was infiltrated in leaf tissues 24 h after Agrobacterium infiltration, before the development of cell death. R. solanacearum Y45 is a strain originally isolated from tobacco (Li et al., 2011), which is pathogenic in N. benthamiana (unpublished data).To determine bacterial numbers, leaf discs (3 leaf discs from Arabidopsis plants and four leaf discs from N. benthamiana plants) were taken and weighed. The plant tissue was ground and homogenized in distilled water before serial dilutions were plated to determine CFU per gram of fresh weight.
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB27040204), the National Natural Science Foundation of China (grant 31571973), the Chinese 1000 Talents Program, and the Shanghai Center for Plant Stress Biology (Chinese Academy of Sciences).
Author Contributions
Y.S., W.Y., L.D., and A.P.M. designed the experiments. Y.S., W.Y., H.Z., Y.W., L.D., G.Y., and J.L. performed the experiments and analyzed the data. A.P.M. conceived the project, analyzed the data, and wrote the manuscript with input from all of the authors.
Acknowledgments
We thank Nemo Peeters and Anne-Claire Cazale for sharing unpublished biological materials, Sophien Kamoun for helpful discussions, Longjiang Fan, Yong Liu, Chanhong Kim, Alex Schultink, and Brian Staskawicz for sharing biological materials, Rosa Lozano-Duran for helpful discussions and critical reading of the manuscript, and Xinyu Jian for technical and administrative assistance during this work. We thank the PSC Cell Biology, Proteomics, and Metabolomics core facilities for assistance with confocal microscopy and mass spectrometry analysis, respectively. No conflict of interest declared.
Published: January 21, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Azevedo C., Sadanandom A., Kitagawa K., Freialdenhoven A., Shirasu K., Schulze-Lefert P. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- Bell E., Creelman R.A., Mullet J.E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger M., Chory J. Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe. 2019;26:163–172. doi: 10.1016/j.chom.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.-H., Coaker G. Effector triggered immunity: NLR immune perception and downstream defense responses. Arabidopsis Book. 2015;13:e0183. [Google Scholar]
- Clarke C.R., Studholme D.J., Byron H., Brendan R., Alexandra W., Rongman C., Tadeusz W., Marie-Christine D., Emmanuel W., Castillo J.A. Genome-enabled phylogeographic investigation of the quarantine pathogen Ralstonia solanacearum Race 3 Biovar 2 and screening for sources of resistance against its core effectors. Phytopathology. 2015;105:597–607. doi: 10.1094/PHYTO-12-14-0373-R. [DOI] [PubMed] [Google Scholar]
- Cui H., Tsuda K., Parker J.E. Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E. A central role of salicylic acid in plant disease resistance. Science. 1994;266:1247–1250. doi: 10.1126/science.266.5188.1247. [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Theulieres F. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digonnet C., Martinez Y., Denancé N. Deciphering the route of Ralstonia solanacearum colonization in Arabidopsis thaliana roots during a compatible interaction: focus at the plant cell wall. Planta. 2012;236:1419. doi: 10.1007/s00425-012-1694-y. [DOI] [PubMed] [Google Scholar]
- Forcat S., Bennett M., Mansfield J., Grant M. A rapid and robust method for simultaneously measuring changes in the phytohormones ABA, JA and SA in plants following biotic and abiotic stress. Plant Methods. 2008;4:16. doi: 10.1186/1746-4811-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S., Kawazoe T., Ohnishi K., Kitagawa T., Popa C., Valls M., Genin S., Nakamura K., Kuramitsu Y., Tanaka N. RipAY, a plant pathogen effector protein exhibits robust γ-glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 2016;291:6813–6830. doi: 10.1074/jbc.M115.678953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J.E., Lara-Tejero M., Marlovits T.C., Wagner S. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcion C., Lohmann A., Lamodière E., Catinot J., Buchala A., Doermann P., Métraux J.-P. Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 2008;147:1279–1287. doi: 10.1104/pp.108.119420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W., Hinsch M.E., Staskawicz B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 2002;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Boter M., Fernández-Barbero G., Chini A., Rathjen J.P., Solano R. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014;12:e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S., Chini A., Solano R. How microbes twist jasmonate signaling around their little fingers. Plants (Basel) 2016;5:9. doi: 10.3390/plants5010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman J., Segonzac C., Cho H., Jung G., Sohn K.H. Effector-assisted breeding for bacterial wilt resistance in horticultural crops. Hortic. Environ. Biotechnol. 2016;57:415–423. [Google Scholar]
- Jiang G., Wei Z., Xu J., Chen H., Zhang Y., She X., Macho A.P., Ding W., Liao B. Bacterial wilt in China: history, current status, and future perspectives. Front. Plant Sci. 2017;8:1549. doi: 10.3389/fpls.2017.01549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Guerois R. NLR sensors meet at the SGT1–HSP90 crossroad. Trends Biochem. Sci. 2010;35:199–207. doi: 10.1016/j.tibs.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Laudert D., Pfannschmidt U., Lottspeich F., Hollander-Czytko H., Weiler E.W. Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol. Biol. 1996;31:323–335. doi: 10.1007/BF00021793. [DOI] [PubMed] [Google Scholar]
- Lavie M., Shillington E., Eguiluz C., Grimsley N., Boucher C. PopP1, a new member of the YopJ/AvrRxv family of type III effector proteins, acts as a host-specificity factor and modulates aggressiveness of Ralstonia solanacearum. Mol. Plant Microbe Interact. 2002;15:1058–1068. doi: 10.1094/MPMI.2002.15.10.1058. [DOI] [PubMed] [Google Scholar]
- Le Roux C., Huet G., Jauneau A., Camborde L., Trémousaygue D., Kraut A., Zhou B., Levaillant M., Adachi H., Yoshioka H. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Li Z., Wu S., Bai X., Liu Y., Lu J., Liu Y., Xiao B., Lu X., Fan L. Genome sequence of the tobacco bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 2011;193:6088–6089. doi: 10.1128/JB.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio Protoc. 2011;1:e95. [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lv R., Li Z., Li M., Dogra V., Lv S., Liu R., Lee K.P., Kim C. Uncoupled expression of nuclear and plastid photosynthesis-associated genes contributes to cell death in a lesion mimic mutant. Plant Cell. 2019;31:210–230. doi: 10.1105/tpc.18.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P., Boutrot F., Rathjen J.P., Zipfel C. ASPARTATE OXIDASE plays an important role in Arabidopsis stomatal immunity. Plant Physiol. 2012;159:1845–1856. doi: 10.1104/pp.112.199810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho A.P. Subversion of plant cellular functions by bacterial type-III effectors: beyond suppression of immunity. New Phytol. 2016;210:51–57. doi: 10.1111/nph.13605. [DOI] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr. Opin. Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Aravind L., Koonin E.V. A superfamily of archaeal, bacterial, and eukaryotic proteins homologous to animal transglutaminases. Protein Sci. 1999;8:1714–1719. doi: 10.1110/ps.8.8.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J., Genin S., Magori S., Citovsky V., Sriariyanum M., Ronald P., Dow M., Verdier V., Beer S.V., Machado M.A. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield J., Jenner C., Hockenhull R., Bennett M.A., Stewart R. Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol. Plant Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- Morel A., Guinard J., Lonjon F., Sujeeun L., Barberis P., Genin S., Vailleau F., Daunay M.C., Dintinger J., Poussier S. The eggplant AG91-25 recognizes the Type III-secreted effector RipAX2 to trigger resistance to bacterial wilt (Ralstonia solanacearum species complex) Mol. Plant Pathol. 2018;19:2459–2472. doi: 10.1111/mpp.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel A., Peeters N., Vailleau F., Barberis P., Jiang G., Berthomé R., Guidot A. Plant pathogenicity phenotyping of Ralstonia solanacearum strains. In: Medina C., López-Baena F.J., editors. Host-Pathogen Interactions: Methods and Protocols. Springer; New York: 2018. pp. 223–239. [DOI] [PubMed] [Google Scholar]
- Mukaihara T., Hatanaka T., Nakano M., Oda K. Ralstonia solanacearum Type III effector RipAY is a glutathione-degrading enzyme that is activated by plant cytosolic thioredoxins and suppresses plant immunity. MBio. 2016;7:e00359. doi: 10.1128/mBio.00359-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaihara T., Tamura N., Iwabuchi M. Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 2010;23:251–262. doi: 10.1094/MPMI-23-3-0251. [DOI] [PubMed] [Google Scholar]
- Nahar K., Matsumoto I., Taguchi F., Inagaki Y., Yamamoto M., Toyoda K., Shiraishi T., Ichinose Y., Mukaihara T. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 2014;15:297–303. doi: 10.1111/mpp.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki T., Murata S., Nakamura S., Hino T., Maeo K., Tabata R., Kawai T., Tanaka K., Niwa Y. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. J. Agric. Chem. Soc. Jpn. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Nakano M., Mukaihara T. The type III effector RipB from Ralstonia solanacearum RS1000 acts as a major avirulence factor in Nicotiana benthamiana and other Nicotiana species. Mol. Plant Pathol. 2019;20:1237–1251. doi: 10.1111/mpp.12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimchuk Z.L., Fisher E.J., Desveaux D., Chang J.H., Dangl J.L. The HopX (AvrPphE) family of Pseudomonas syringae type III effectors require a catalytic triad and a novel N-terminal domain for function. Mol. Plant Microbe Interact. 2007;20:346–357. doi: 10.1094/MPMI-20-4-0346. [DOI] [PubMed] [Google Scholar]
- Peeters N., Carrère S., Anisimova M., Plener L., Cazalé A.C., Genin S. Repertoire, unified nomenclature and evolution of the Type III effector gene set in the Ralstonia solanacearum species complex. BMC Genomics. 2013;14:859. doi: 10.1186/1471-2164-14-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Guidot A., Vailleau F., Valls M. Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 2013;14:651–662. doi: 10.1111/mpp.12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poueymiro M., Cunnac S., Barberis P., Deslandes L., Peeters N., Cazale-Noel A.C., Boucher C., Genin S. Two type III secretion system effectors from Ralstonia solanacearum GMI1000 determine host-range specificity on tobacco. Mol. Plant Microbe Interact. 2009;22:538–550. doi: 10.1094/MPMI-22-5-0538. [DOI] [PubMed] [Google Scholar]
- Rosas-Díaz T., Cana-Quijada P., Amorim-Silva V., Botella M.A., Lozano-Durán R., Bejarano E.R. Arabidopsis NahG plants as a suitable and efficient system for transient expression using Agrobacterium tumefaciens. Mol. Plant. 2017;10:353–356. doi: 10.1016/j.molp.2016.11.005. [DOI] [PubMed] [Google Scholar]
- Rufián J.S., Lucía A., Rueda-Blanco J., Zumaquero A., Guevara C.M., Ortiz-Martín I., Ruiz-Aldea G., Macho A.P., Beuzón C.R., Ruiz-Albert J. Suppression of HopZ effector-triggered plant immunity in a natural pathosystem. Front. Plant Sci. 2018;9:977–981. doi: 10.3389/fpls.2018.00977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y., Macho A.P. Analysis of PAMP-triggered ROS burst in plant immunity. Methods Mol. Biol. 2017;1578:143–153. doi: 10.1007/978-1-4939-6859-6_11. [DOI] [PubMed] [Google Scholar]
- Sang Y., Wang Y., Ni H., Cazalã A.C., She Y.M., Peeters N., Macho A.P. The Ralstonia solanacearum type III effector RipAY targets plant redox regulators to suppress immune responses. Mol. Plant Pathol. 2016;19:129–142. doi: 10.1111/mpp.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarris P., Duxbury Z., Huh S.U., Ma Y., Segonzac C., Sklenar J., Derbyshire P., Cevik V., Rallapalli G., Saucet S. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- Schlücking K., Kai H.E., Köster P., Drerup M.M., Eckert C., Steinhorst L., Waadt R., Batistič O., Kudla J. A new β-estradiol-inducible vector set that facilitates easy construction and efficient expression of transgenes reveals CBL3-dependent cytoplasm to tonoplast translocation of CIPK5. Mol. Plant. 2013;6:1814–1829. doi: 10.1093/mp/sst065. [DOI] [PubMed] [Google Scholar]
- Schultink A., Qi T., Lee A., Steinbrenner A.D., Staskawicz B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017;92:787–795. doi: 10.1111/tpj.13715. [DOI] [PubMed] [Google Scholar]
- Segonzac C., Feike D., Gimenez Ibanez S., Hann D.R., Zipfel C., Rathjen J.P. Hierarchy and roles of pathogen-associated molecular pattern-induced responses in Nicotiana benthamiana. Plant Physiol. 2011;156:687–699. doi: 10.1104/pp.110.171249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M., Mysore K.S. Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 2014;9:1549–1562. doi: 10.1038/nprot.2014.092. [DOI] [PubMed] [Google Scholar]
- Shine M.B., Yang J.W., El-Habbak M., Nagyabhyru P., Fu D.Q., Navarre D., Ghabrial S., Kachroo P., Kachroo A. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016;212:627–636. doi: 10.1111/nph.14078. [DOI] [PubMed] [Google Scholar]
- Solé M., Popa C., Mith O., Sohn K.H., Jones J., Deslandes L., Valls M. The awr gene family encodes a novel class of Ralstonia solanacearum type III effectors displaying virulence and avirulence activities. Mol. Plant Microbe Interact. 2012;25:941–953. doi: 10.1094/MPMI-12-11-0321. [DOI] [PubMed] [Google Scholar]
- Stevens C., Bennett M.A., Athanassopoulos E., Tsiamis G., Taylor J.D., Mansfield J.W. Sequence variations in alleles of the avirulence gene avrPphE.R2 from Pseudomonas syringae pv. phaseolicola lead to loss of recognition of the AvrPphE protein within bean cells and a gain in cultivar-specific virulence. Mol. Microbiol. 1998;29:165–177. doi: 10.1046/j.1365-2958.1998.00918.x. [DOI] [PubMed] [Google Scholar]
- Tasset C., Bernoux M., Jauneau A., Pouzet C., Brière C., Kieffer-Jacquinod S., Rivas S., Marco Y., Deslandes L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Jauneau A., Genin S., Tavella M.J., Vailleau F., Gentzbittel L., Jardinaud M.F. Dissection of bacterial wilt on Medicago truncatula revealed two type III secretion system effectors acting on root infection process and disease development. Plant Physiol. 2009;150:1713–1722. doi: 10.1104/pp.109.141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.M.A., Klessig D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Wang Y., Li Y., Rosas-Diaz T., Caceres-Moreno C., Lozano-Durán R., Macho A.P. The IMMUNE-ASSOCIATED NUCLEOTIDE-BINDING 9 protein is a regulator of basal immunity in Arabidopsis thaliana. Mol. Plant Microbe Interact. 2019;32:65–75. doi: 10.1094/MPMI-03-18-0062-R. [DOI] [PubMed] [Google Scholar]
- Ward E.R., Uknes S.J., Williams S.C., Dincher S.S., Wiederhold D.L., Alexander D.C., Ahlgoy P., Métraux J.P., Ryals J.A. Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell. 1991;3:1085–1094. doi: 10.1105/tpc.3.10.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Caceres-Moreno C., Jimenez-Gongora T., Wang K., Sang Y., Lozano-Duran R., Macho A.P. The Ralstonia solanacearum csp22 peptide, but not flagellin-derived peptides, is perceived by plants from the Solanaceae family. Plant Biotechnol. J. 2018;16:1349–1362. doi: 10.1111/pbi.12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Feys B.J., Parker J.E. Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature. 2001;414:562–565. doi: 10.1038/35107108. [DOI] [PubMed] [Google Scholar]
- Williams S.J., Kee Hoon S., Li W., Maud B., Sarris P.F., Cecile S., Thomas V., Yan M., Saucet S.B., Ericsson D.J. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Wu C.-H., Abd-El-Haliem A., Bozkurt T.O., Belhaj K., Terauchi R., Vossen J.H., Kamoun S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. U S A. 2017;114:8113–8118. doi: 10.1073/pnas.1702041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Belhaj K., Bozkurt T.O., Birk M.S., Kamoun S. Helper NLR proteins NRC2a/b and NRC3 but not NRC1 are required for Pto-mediated cell death and resistance in Nicotiana benthamiana. New Phytol. 2016;209:1344–1352. doi: 10.1111/nph.13764. [DOI] [PubMed] [Google Scholar]
- Yu G., Xian L., Sang Y., Macho A.P. Cautionary notes on the use of Agrobacterium-mediated transient gene expression upon SGT1 silencing in Nicotiana benthamiana. New Phytol. 2019;222:14–17. doi: 10.1111/nph.15601. [DOI] [PubMed] [Google Scholar]
- Yu G., Xian L., Xue H., Yu W., Rufian J., Sang Y., Morcillo R., Wang Y., Macho A.P. A bacterial effector protein prevents MAPK-mediated phosphorylation of SGT1 to suppress plant immunity. bioRxiv. 2019:641241. doi: 10.1371/journal.ppat.1008933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.-S., Niu Q.-W., Chua N.-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.