Abstract
Plants associate with diverse microbes that exert beneficial, neutral, or pathogenic effects inside the host. During the initial stages of invasion, the plant apoplast constitutes a hospitable environment for invading microbes, providing both water and nutrients. In response to microbial infection, a number of secreted proteins from host cells accumulate in the apoplastic space, which is related to microbial association or colonization processes. However, the molecular mechanisms underlying plant modulation of the apoplast environment and how plant-secreted proteases are involved in pathogen resistance are still poorly understood. Recently, several studies have reported the roles of apoplastic proteases in plant resistance against bacteria, fungi, and oomycetes. On the other hand, microbe-secreted proteins directly and/or indirectly inhibit host-derived apoplastic proteases to promote infection. These findings illustrate the importance of apoplastic proteases in plant–microbe interactions. Therefore, understanding the protease-mediated apoplastic battle between hosts and pathogens is of fundamental importance for understanding plant–pathogen interactions. Here, we provide an overview of plant–microbe interactions in the apoplastic space. We define the apoplast, summarize the physical and chemical properties of these structures, and discuss the roles of plant apoplastic proteases and pathogen protease inhibitors in host–microbe interactions. Challenges and future perspectives for research into protease-mediated apoplastic interactions are discussed, which may facilitate the engineering of resistant crops.
Keywords: apoplast, protease, protease inhibitor, plant immunity, plant–microbe interaction
The apoplast mediates the initial battle between plants and pathogens. This review summarizes the current understanding of plant–microbe interactions in the apoplastic region, with special emphasis on the role of secreted proteases in this process.
Introduction
Plants possess a multi-layered immune system that protects them against infection from pathogens in the phyllosphere (Jones and Dangl, 2006). The recognition of pathogen-associated molecular patterns (PAMPs) by plant plasma membrane-localized pattern recognition receptors (PRRs) leads to the initiation of host immune responses, known as PAMP-triggered immunity (PTI) (Macho and Zipfel, 2014; Bigeard et al., 2015). Both PAMP processing and PAMP-PRR recognition processes take place in the apoplastic region during host–microbe interactions (Mott et al., 2014; Buscaill et al., 2019), suggesting the importance of protein–protein interactions in the apoplast in plant immunity. Host-secreted proteases process PAMPs and other proteins for immune activation (Bohm et al., 2014; Buscaill et al., 2019), and pathogen-secreted proteases directly or indirectly target key components of host immunity (Axtell et al., 2003; Chisholm et al., 2005; Figaj et al., 2019). This protease-mediated communication is required for the crosstalk between hosts and microbes. However, the molecular mechanism by which apoplastic immunity controls pathogen invasion is still largely unknown. In this review, we summarize the current understanding of plant–microbe interactions in the apoplastic region, with special emphasis on the roles of secreted proteases in this process.
Definition of the Apoplast in the Context of Plant–Microbe Interactions
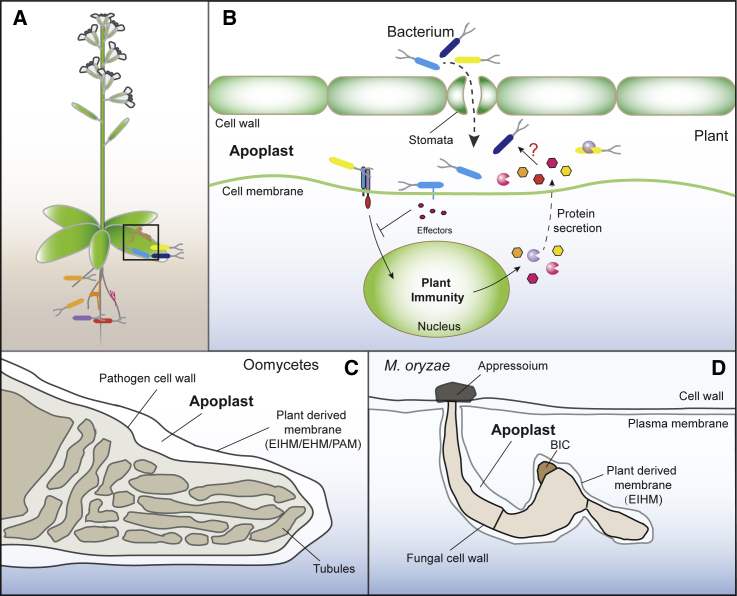
The plant apoplast is an essential environment for signal exchange and nutrient uptake as well as microbe infection and adaptation (Sattelmacher, 2001; Aung et al., 2018). However, due to differences in pathogen lifestyles, apoplast definitions vary. During host–bacterial pathogen interactions, the apoplast defines compartments of the intracellular space beyond the plasma membrane, including the plant cell wall and aqueous intercellular space (Sattelmacher, 2001) (Figure 1). After the successful entry into the host from stomata or wounded regions, bacteria can survive and multiply in this aqueous intercellular space (Bai et al., 2015). Biotrophic or hemibiotrophic fungi and oomycetes can invade host cells using a specialized structure known as the appressorium, which is lacking in bacteria. Furthermore, invasive hyphae or haustoria are surrounded by a host-derived specialized membrane outside the invasive structure, known as the extrainvasive hyphal membrane (EIHM) (Kankanala et al., 2007), extrahaustorial membrane (EHM) (Kwaaitaal et al., 2017), or periarbuscular membrane (PAM) (Ivanov et al., 2019). This narrow space between the pathogen plasma membrane and the host extended membrane is defined as the apoplast (Figure 1). For instance, symbiotic arbuscular mycorrhizal fungi can penetrate the cortical cells of plant roots. After successful infection, a highly branched hyphal structure, known as the arbuscule, is surrounded by the plant-derived PAM (Ivanov et al., 2019). EHM or PAM are similarly formed, after which they surround the invasive haustoria of powdery mildew and the oomycetes Albugo candida, respectively (Soylu et al., 2003; Kwaaitaal et al., 2017). Infection with Magnaporthe oryzae, a hemibiotrophic fungal pathogen, also leads to the formation of EIHM in rice (Kankanala et al., 2007). These findings illustrate that the formation of this extramembrane structure is a general mechanism and essential for the interaction of fungal and oomycete pathogens with plants. Interestingly, M. oryzae delivers fungal effectors into host cells through two distinct secretory systems (Giraldo et al., 2013). In addition to the conventional ER–Golgi secretory system, which mediates the secretion of apoplastic effectors, a specialized structure known as the biotrophic interfacial complex (BIC) is generated. This complex functions in the translocation of cytoplasmic effectors from M. oryzae into the host cytoplasm (Giraldo et al., 2013) (Figure 1). No BIC-like structures have been reported in other fungi or oomycetes. Therefore, it is still unclear how these apoplastic effectors enter host cells.
Figure 1.
The Apoplast as a Site for the Interactions of Plants with Bacteria, Fungi, and Oomycetes.
(A) Plants associate with diverse microbial communities in both their root and leaf compartments.
(B) Bacterial pathogens cause infection in their hosts after entering host cells through stomata. Upon sensing pathogen-derived signals, plants activate immune responses and secrete proteins and metabolites into the plant apoplast.
(C and D) During infection with oomycetes (C) or fungi (D), an additional plant-derived membrane structure is formed outside of the invading microbe, known as the extrainvasive hyphal membrane (EIHM), extrahaustorial membrane (EHM), or periarbuscular membrane (PAM). The space between the oomycete/fungal cell wall and this extramembrane structure is also defined as the apoplast. During infection with Magnaporthe oryzae, an additional biotrophic interfacial complex (BIC) structure is formed to mediate the translocation of cytoplasmic effectors into host cells (D).
Physical and Chemical Properties of the Apoplast during Infection
Plants are surrounded by millions of microbes. Root- and leaf-associated microbiomes are tightly regulated by hosts via the modulation of immunity and nutrient supply. The apoplast, as a site of habitation for the host-associated microbiome, also provides sufficient nutrients for invaders during the early invasion process (Xin et al., 2016; Aung et al., 2018). Therefore, the tight control of key components, including water, nutrients, pH, and reactive oxygen species (ROS), in the apoplast is essential for the regulation of apoplastic interactions between hosts and microbes.
High Levels of Water in the Apoplast Negatively Contribute to Plant Resistance
Rainfall and high humidity enhance plant infection by pathogens, and water soaking is commonly observed at the early stages of infection with phyllosphere pathogens (Aung et al., 2018). This water-soaking phenomenon provides for an extended aqueous environment, which favors pathogen infection of the host. It was reported that Pseudomonas syringae pv. tomato DC3000 (Pst) utilizes the type III effectors AvrE1 and HopM1 to induce water soaking in Arabidopsis leaves (Xin et al., 2016). HopM1 induces water soaking by targeting and degrading the water homeostasis-related protein Arabidopsis thaliana HopM interactor 7 (AtMIN7) (Nomura et al., 2006; Xin et al., 2016). Moreover, it was shown that AtMIN7-mediated immunity can limit the soaking-dependent pathogenesis of Pst (Xin et al., 2016). These findings indicate that the regulation of water availability is essential for the interaction between hosts and microbes.
Plasma membrane intrinsic proteins (PIPs) belong to the aquaporin group of membrane-localized transporter proteins that control water export in plants (Maurel et al., 2008). In Arabidopsis, the pip1;4 mutant is more susceptible, whereas overexpression lines are more resistant to Pst compared with wild-type plants, suggesting a positive role of AtPIP1;4 in plant immunity (Tian et al., 2016). In rice, OsPIP1;3-overexpressing plants are more susceptible to Xanthomonas oryzae, whereas knockout mutants are more resistant to this pathogen (Li et al., 2019). OsPIP1;3 is required for the translocation of bacterial transcription factor-like (TAL) effector AvrXa10-Ni, which induces Xa10-mediated immunity (Tian et al., 2014). Interestingly, OsPIP1;3 is also involved in the translocation of the TAL effector PthXo1, which is required for the pathogenicity of X. oryzae in rice (Zhang et al., 2019). These findings indicate that OsPIP1;3-mediated susceptibility to pathogens is dependent on whether the translocated effector is virulent or avirulent (Tian et al., 2014; Zhang et al., 2019). Taken together, these findings suggest that apoplast water availability is essential for the interaction between hosts and pathogens. However, the link between water soaking, aquaporins, and disease symptoms remains to be addressed.
Sugar Levels in the Apoplast Influence Bacterial Pathogenicity
The apoplast is a nutrient-rich compartment that mediates the translocation of sugars to different tissues during plant development (van Ooij, 2011). Microbial pathogens have evolved strategies to acquire nutrients from this compartment by regulating membrane-localized transporters, thereby promoting their pathogenicity (Bezrutczyk et al., 2018). Xanthomonas employs TAL effectors to activate the expression of Sugars Will Eventually be Exported Transporters (SWEETs), which are plasma membrane-localized sugar transporters, and to increase sugar efflux to the apoplast (Chen et al., 2010; Cox et al., 2017). The oomycete pathogen Phytophthora sojae effector PsXEG1 converts xyloglucan to reduced sugars after secretion into the soybean apoplast (Ma et al., 2015). To counter PsXEG1-mediated pathogenicity, soybean secretes the glucanase inhibitor GmGIP1 into the apoplast, which directly interacts with PsXEG1, to reduce the production of hexose sugars (Ma et al., 2017). Through the secretion of PsXLP1, a paralogous decoy molecule of PsXEG1, P. sojae reduces the interaction between GmGIP1 and PsXEG1, thereby increasing its supply of sugars, which may promote pathogenicity (Ma et al., 2017).
On the other hand, plants can suppress pathogen infection by reducing the apoplastic sugar content through the action of plasma membrane-localized sugar importers, namely sugar transporter proteins (STPs). Genetic evidence has revealed that the Arabidopsis stp1 stp13 double mutant is highly susceptible to Pst (Yamada et al., 2016). Moreover, the exogenous application of flg22 leads to the phosphorylation of STP13 by FLS2 and BAK1. These results suggest that the reduction of apoplastic sugar levels can be considered as an approach to modulate plant innate immunity.
Iron, a Double-Edged Sword in Plant–Microbe Interactions
Iron is an important nutrient not only for plants but also for their surrounding microbes. Competition for iron uptake between hosts and pathogens plays an important role in host-microbe interactions (Skaar, 2010). Bacteria utilize the G-protein-like transporter FeoB or secrete siderophores, extracellular iron chelators, to take up ferrous (Fe2+) and ferric (Fe3+) iron from the environment, respectively (Andrews et al., 2003; Skaar, 2010). In nature, most iron found in the soil is in an insoluble form. Non-grass plants secrete phenolic compounds to increase the solubility of Fe3+ by reducing Fe3+ to Fe2+ via membrane-localized ferric reduction oxidase 2 (FRO2) and then transport Fe2+ into the root epidermis via Arabidopsis iron-regulated transporter 1 (IRT1) (Verbon et al., 2017). The plant iron homeotic system, in particular coumarin-mediated iron uptake, was shown to be involved in the makeup of the root microbiome (Stringlis et al., 2018; Voges et al., 2019). Moreover, transcriptome analysis has revealed that plant immune pathways can suppress the expression of bacterial iron acquisition-related genes in leaves (Nobori et al., 2018). The loss of iron uptake-related genes in the fungal pathogen Ustilago maydis can strongly affect its virulence in plants (Eichhorn et al., 2006). On the other hand, plants can sense siderophores secreted by bacteria and activate immune responses (Aznar et al., 2014), which suggests that plants have evolved direct and indirect mechanisms for the iron-mediated growth suppression of microbial pathogens. However, it is unclear how pathogens are distinguished from beneficial microbes during iron-mediated interactions.
Ferroptosis is a nonapoptotic form of iron-dependent cell death that was first described in mammalian cells (Dixon et al., 2012; Stockwell et al., 2017). It was recently reported that the infection of the blast fungus M. oryzae triggers the accumulation of ferric iron and ROS in the extracellular region in rice, thereby leading to ferroptosis (Dangol et al., 2019). This iron-dependent cell death is involved in the suppression of M. oryzae infection in plants (Dangol et al., 2019). However, ferroptosis is a relatively new concept in plants. Therefore, its role in plant–microbe interactions requires further investigation.
Control of ROS and pH in the Apoplast
ROS production is an indicator of biotic and abiotic stresses but also plant developmental processes (Waszczak et al., 2018). Apoplastic ROS burst is one of the earliest events during PTI in plants (Qi et al., 2017). ROS bursts are triggered by plasma membrane-localized respiratory burst oxidase homologs (RBOHs) and apoplastic peroxidases in response to PAMP signals (Torres et al., 2002; Daudi et al., 2012; Kadota et al., 2014). RbohD is the most important RBOH for apoplastic ROS production during plant–microbe interactions (Torres et al., 2002). In Arabidopsis, trimeric G proteins (XLG2, AGB1, AGG1, AGG2), the calcium-dependent protein kinase 5 (CPK5), and the Botrytis-induced kinase 1 (BIK1) are required for the activation of RBOHD in PTI (Liang et al., 2016). Moreover, it was reported that the apoplast-localized peroxidases PRX33 and PRX34 are involved in apoplastic ROS burst and resistance against P. syringae in Arabidopsis (O'Brien et al., 2012). These results demonstrate that RBOH- and peroxidase-mediated apoplastic ROS accumulation is involved in plant–microbe interactions.
Arabidopsis plasma membrane-localized H+-ATPases (AHAs) function as proton pumps and regulate the apoplastic pH (Haruta et al., 2010). It was shown that ETI triggers stomatal opening by activating AHA1 and AHA2 in guard cells, which increases bacterial entry into leaves (Liu et al., 2009; Lee et al., 2015). The plant apoplast is an acidic compartment with pH 5–6, which is essential for the biochemical function of proteins, especially proteases (Felle, 2006; Barbez et al., 2017). However, the regulation of pH during pathogen infection in plants is unclear. Therefore, further studies on pH dynamics in apoplasts and their role in pathogen resistance may improve our understanding of apoplastic immunity in plants.
Secreted Proteases Are Essential for Plant Resistance to Pathogens
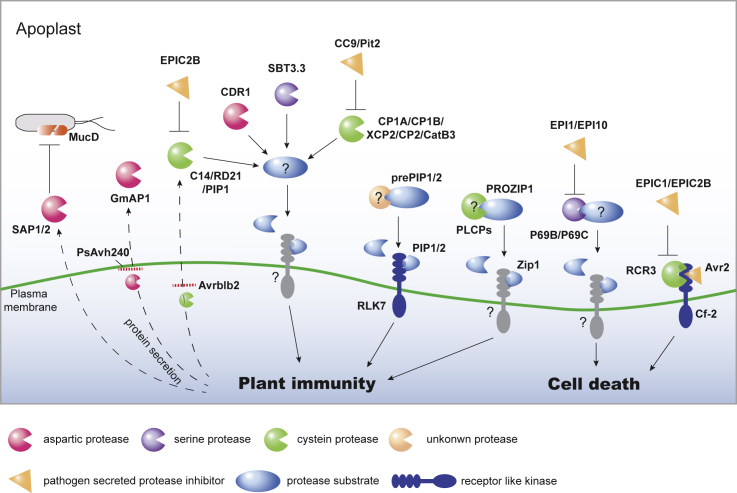
Apoplastic proteins from both plants (Jashni et al., 2015) and animals (Brogden, 2005; Lai and Gallo, 2009) are involved in resistance against pathogens, illustrating that apoplast-mediated pathogen suppression is a widely employed immune strategy. Moreover, the immune-primed apoplastic fluid exerts significant bacterial growth suppression (Wang et al., 2019). However, our knowledge of the molecular mechanisms of plant–pathogen antagonism in the apoplast remains incomplete. Proteases, catabolic enzymes that hydrolyze peptide bonds, are highly conserved in both prokaryotes and eukaryotes (Dunn, 2005). It has been reported that plant-derived proteases are enriched in the apoplastic region during host–microbe interactions where they act to enhance host resistance against different types of pathogens (Kim et al., 2013; Grosse-Holz et al., 2017; Wang et al., 2017). In the following sections, we summarize the current understanding of apoplastic proteases and their importance in immunity (Figure 2). To enhance our understanding of the arms race between hosts and pathogens, pathogen-derived protease inhibitors are also described (Figure 2).
Figure 2.
Secreted Proteases and Protease Inhibitors in Plant–Microbe Interactions.
In response to pathogen infection, plants secrete different types of proteases into the apoplastic region. Plant-derived proteases contribute to plant immune response via different mechanisms. The Arabidopsis secreted protein SAP1/2 suppresses bacterial growth by directly cleaving bacterial growth-related protein MucD. Other proteases, such as P69B/C, RCR3, and PLCPs, also recognize and cleave their substrates in the apoplast. The released peptides can be recognized by host membrane-localized receptor-like kinases, leading to the activation of immune responses. To counter host protease-mediated immunity, pathogens can secrete protease inhibitors to suppress host protease activity via direct interactions or secrete effectors, such as PsAvh240 and Avrblb2, which block the secretion of plant proteases.
Proteases
A protease (also known as a peptidase, proteinase, or proteolytic enzyme) is a type of protein that catalyzes the degradation of other proteins based on its ability to recognize and cleave specific short amino acid sequences (Casem, 2016). The MEROPS database (https://www.ebi.ac.uk/merops) classifies proteases into nine different groups based on catalytic type and homology, namely aspartic, cysteine, glutamic, metalloprotease, asparagine, mixed, serine, threonine, and unknown catalytic type proteases (Rawlings et al., 2018). Plant genomes encode hundreds of proteases that are distributed across all protease families, playing key roles in most aspects of plant physiology and development (Garcia-Lorenzo et al., 2006; Tripathi and Sowdhamini, 2006; van der Hoorn, 2008; Chen et al., 2009; Wang et al., 2018). Aspartic proteases, cysteine proteases, metalloproteases, and serine proteases are the most abundant proteases in plants. Moreover, plant proteases belonging to these families have been reported to participate in plant immune responses and pathogen resistance (van der Hoorn, 2008; Jashni et al., 2015). Interestingly, proteases are highly enriched in the apoplastic region. For instance, among 77 A. thaliana aspartic proteases, 51 were predicted to be extracellularly localized in TAIR10 ( Wang et al., 2019), suggesting that the apoplastic localization of proteases may be essential for their function.
Plant-Secreted Proteases in Plant–Bacteria Interactions
Constitutive disease resistance 1 (AtCDR1), an extracellular aspartic protease, was identified in an activation tagged screening in Arabidopsis (Xia et al., 2004). CDR1-D plants (activation tagging line) exhibit a dwarf phenotype, increased accumulation of pathogenesis-related proteins, and enhanced resistance against Pst in local and systemic tissues (Suzuki et al., 2004; Xia et al., 2004; Simoes et al., 2007). By contrast, enhanced resistance to bacterial and fungal pathogens was observed upon overexpression of an ortholog of AtCDR1, OsCDR1, in rice and Arabidopsis (Prasad et al., 2009).
Subtilases (SBTs) belong to the serine protease family and are especially abundant in plants (Rautengarten et al., 2005; Schaller et al., 2017). It was reported that SBTs are involved in pathogen resistance in plants (Ramirez et al., 2013; Figueiredo et al., 2014). Overexpression of the extracellular subtilase SBT3.3 leads to enhanced mitogen-activated protein kinase activation, defense gene expression, and resistance against bacterial and fungal pathogens (Ramirez et al., 2013). Both CDR1-and SBT3.3-mediated immunity was abolished after crossing with the salicylic acid (SA) signaling mutant npr1 and SA biosynthesis mutant sid2, suggesting that the apoplastic accumulation of CDR1 and SBT3.3 enhances pathogen resistance by amplifying SA immunity in plants. However, the molecular mechanism underlying the amplification of SA immunity by CDR1 and SBT3.3 remains unknown. Interestingly, CDR1 protease activity is necessary for its function in immune activation (Suzuki et al., 2004), suggesting that the protease amplifies the host immune response by cleaving its substrate(s).
PAMP-INDUCED PEPTIDE1 (PIP1) and PIP2 are conserved endogenous peptides derived from the C termini of prePIP family proteins, which trigger immune responses in Arabidopsis (Hou et al., 2014). PrePIPs are secreted and accumulate in the apoplastic region. Moreover, recognition of the PIP peptide is mediated by the plasma membrane-localized receptor-like kinase 7 (RLK7) in Arabidopsis (Hou et al., 2014), suggesting that the processing of prePIP takes place in the apoplast. Indeed, recombinant glutathione S-transferase-prePIP proteins are proteolytically processed after entering the apoplast of Arabidopsis (Hou et al., 2014). Additionally, maize-secreted papain-like cysteine proteases (PLCPs) are required for the release of the bioactive immune signaling peptide Zip1 from its propeptide precursor, which strongly induces SA accumulation in leaves (Ziemann et al., 2018). These findings suggest that apoplastic protease(s) are required for the induction or amplification of immune signals via the cleavage of their substrate(s). Therefore, the identification of immune-related proteases and their specific substrate(s) in the apoplast will contribute to our understanding of the mechanisms by which they mediate pathogen resistance.
The constitutive activation of immunity promotes pathogen resistance. However, it also results in a significant reduction in growth, the so-called immunity-growth trade-off, which is a challenge for the simultaneous optimization of plant growth and pathogen resistance in agricultural settings (Smakowska et al., 2016; Karasov et al., 2017; Scheres and van der Putten, 2017). A recent report showed that overexpression of SAP1 and SAP2, two secreted aspartic proteases of Arabidopsis, can enhance bacterial resistance without constitutive defense activation and growth retardation (Wang et al., 2019), suggesting that SAP1 and SAP2 suppress bacterial growth via a mechanism distinct from that of CDR1 and SBT3.3. Indeed, SAP1 and SAP2 suppress Pst growth by directly targeting the bacterial MucD protein, which is known to be involved in bacterial growth (Wang et al., 2019), thermotolerance (Wood and Ohman, 2006), and resistance against oxidative stress (Yorgey et al., 2001). This is the first evidence that a plant can suppress pathogen infection by direct targeting a pathogen growth-related protein.
A recent study showed that a single Pseudomonas operational taxonomic unit (OTU), which is generally classified as a pathogen, can colonize Arabidopsis plants in the wild (Karasov et al., 2018). Single strains within this OTU diverged from each other at least 300 000 years ago, suggesting that genetic and species diversity may prevent clonal expansions in nature (Karasov et al., 2018). A similar trend was observed for the MucD protein, which is widespread in bacteria and subject to purifying selection. However, site-specific diversity in mucD was detected in Pseudomonas, in contrast to other bacteria where the gene is highly conserved (Wang et al., 2019). Therefore, SAP1-mediated immunity might exert selection pressure on Pseudomonas but not on other bacteria, and the diversity in the MucD sequence may allow Pseudomonas to efficiently colonize plants, which could explain disease outbreaks in wild populations.
Plant-Secreted Proteases in Plant–Oomycete Interactions
Phytophthora infestans, which causes late blight and potato blight, is the major oomycete pathogen for potato and tomato (Nowicki et al., 2012). Secreted proteases derived from different plants play important roles in the suppression of Phytophthora infection. The tomato serine proteases P69B and P69C, which belong to the P69 family of extracellular subtilisin-like proteases, are induced upon pathogen infection and SA treatment (Jorda et al., 1999; Jorda and Vera, 2000), indicating that extracellular serine proteases might be involved in plant resistance against oomycetes. Interestingly, P69B is necessary for matrix metalloproteinase-mediated cell death in tomato (Zimmermann et al., 2016). As no genetic evidence is available, it is still unclear how P69B and P69C contribute to plant immunity. Papain-like proteases, which belong to the cysteine protease family, are also linked with plant immune responses, especially in tomato (Bozkurt et al., 2011; Hou et al., 2014; Ilyas et al., 2015). Overexpression of the apoplastic cysteine proteases C14 and PIP1 reduces the resistance of tomato against P. infestans (Bozkurt et al., 2011; Hou et al., 2014), whereas depletion of PIP1 increases tomato susceptibility to Cladosporium fulvum and P. infestans (Ilyas et al., 2015). A recent study has reported that GmAP1, a secreted aspartic protease from soybean, positively contributes to soybean resistance against P. sojae (Guo et al., 2019). Overexpression of GmAP1 reduces, whereas knockdown of GmAP1 and GmAP2 increases, the biomass of P. sojae in plants (Guo et al., 2019). These findings illustrate that proteolysis by different proteases contributes to plant resistance against pathogen infection in the apoplast.
Plant-Secreted Proteases in Plant–Fungus Interactions
RCR3, an apoplastic cysteine protease from tomato, is required for Cf-2-mediated resistance to Avr2 from the biotrophic fungal pathogen C. fulvum (Dixon et al., 2000; Luderer et al., 2002). RCR3 interacts with and functions as a co-receptor of Cf-2 in recognizing C. fulvum Avr2 (Rooney et al., 2005). Other studies have indicated that Avr2 directly inhibits the extracellular cysteine protease activity of RCR3, as well as its homolog protein PIP1, to promote the infection of C. fulvum in tomato (Shabab et al., 2008; van Esse et al., 2008). Moreover, PIP1-depleted tomato plants are hypersusceptible to bacterial, fungal, and oomycete pathogens, suggesting that PIP1 confers broad resistance against unrelated apoplastic pathogens (Ilyas et al., 2015). However, RCR3-depleted tomato plants do not exhibit altered resistance against fungal and bacterial pathogens but are susceptible to P. infestans (Ilyas et al., 2015), indicating divergent functions of RCR3 and PIP1 in pathogen resistance.
Moreover, the cysteine protease C14 secreted by tomato contributes to the resistance against the oomycete pathogen P. infestans (Kaschani et al., 2010), whereas its ortholog in Arabidopsis, RD21, contributes to the resistance against the fungal pathogen Botrytis cinerea but not the bacterial pathogen Pst and oomycete Hyaloperonospora arabidopsidis (Shindo et al., 2012). Similarly, two cysteine proteases in Nicotiana benthamiana, NbCYP1 and NbCYP2, confer resistance to the hemibiotrophic pathogen Colletotrichum destructivum, but not to the bacterial pathogen Pst (Hao et al., 2006). Therefore, studies of how protease specificity is determined will reveal the molecular mechanisms of proteases in plant–microbe interactions.
Pathogens Counteract Host Protease-Mediated Growth Suppression
As apoplastic proteases suppress bacterial growth, the production of specific or even general protease inhibitors would be a straightforward stratagem of microbes to escape protease-mediated immunity (Figure 2). Oomycetes harbor several extracellular protease inhibitors of cysteine proteases (EPICs) that suppress extracellular cysteine protease-mediated immunity (Tian et al., 2007). EPIC2B directly interacts with and inhibits the cysteine protease activity of tomato C14 (Kaschani et al., 2010). Furthermore, EPIC2B inhibits the protease activity of the tomato cysteine proteases PIP1 and RCR3 through direct interactions (Tian et al., 2007; Song et al., 2009). These examples illustrate that oomycete-secreted extracellular protease inhibitors (EPIs) can target multiple proteases secreted by the host. Moreover, EPIC1 from P. infestans could also bind and inhibit RCR3 (Song et al., 2009), indicating that oomycetes secrete multiple protease inhibitors that target the same proteases to enhance their pathogenicity. Interestingly, specific interactions between protease inhibitors drive the host adaptation of the Phytophthora species. Phytophthora mirabilis is a close relative of P. infestans that has jumped to a different host, namely Mirabilis jalapa. P. mirabilis secretes PmEPIC1, an ortholog of P. infestans EPIC1, which is a strong inhibitor of the RCR3-like protease MRP2 from M. jalapa but a less efficient inhibitor of RCR3 from the non-host plant tomato (Dong et al., 2014). By contrast, P. infestans EPIC1 is less efficient than PmEPIC1 in inhibiting MRP2 activity but more efficient in inhibiting RCR3 homologs from different host species. The specificity of RCR3 inhibitor binding, which was determined using a structural model of the RCR3–EPIC complex, was defined by a 7-amino-acid region that was polymorphic between RCR3 and MRP2 (Dong et al., 2014). Thus, inhibitor specificity allows P. mirabilis to adapt to a new host.
Infection by the biotrophic fungal pathogen U. maydis, which causes smut disease in maize, requires the suppression of host immunity. It was shown that cystatin CC9, a cysteine protease inhibitor, was induced during the infection of U. maydis in maize (van der Linde et al., 2012). Silencing of CC9 led to massive induction of SA-mediated immunity, whereas activated apoplastic cysteine proteases induce SA-associated defense gene expression in naive plants, suggesting that cysteine protease activity in the apoplast is essential for resistance against U. maydis (van der Linde et al., 2012). Consistent with this, the U. maydis-secreted effector protein Pit2, which functions as a cysteine protease inhibitor, is essential for fungal virulence (Mueller et al., 2013). These findings indicate the importance of cysteine proteases in maize defense against biotrophic pathogens. Five different apoplastic cysteine proteases have been identified in the maize apoplast, namely CP1A, CP1B, XCP2, CP2, and CatB3 (van der Linde et al., 2012). Pit2 directly suppresses the enzymatic activity of these proteases, except for CatB3 (Mueller et al., 2013). Recent studies have indicated that a conserved 14-amino-acid motif, PID14, which is conserved in different fungi, functions as the inhibitory core for the suppression of apoplastic cysteine proteases (Misas Villamil et al., 2019).
“Candidatus Liberibacter asiaticus” (CLas), which is transmitted to the citrus species by the Asian citrus psyllid during sap feeding, causes serious Huanglongbing on citrus fruits. During infection, CLas secretes the species-specific effector SDE1 to promote bacterial infection (Clark et al., 2018). SDE1 is associated with multiple citrus PLCPs and inhibits PLCP activity both in vivo and in vitro (Clark et al., 2018). Taken together, these findings illustrate that the inhibition of plant cysteine proteases by secreted effectors is a common strategy used by pathogens to maintain susceptibility.
To counter the P69B- and P69C-mediated immunity, P. infestans secretes a group of Kazal family serine protease inhibitors known as EPIs. In silico analysis revealed that the P. infestans genome contains 14 EPIs (EPI1–EPI14) (Tian et al., 2004). EPI1 and EPI10, which are expressed during infection in tomato, physically bind to and inhibit the protease activity of P69B (Tian et al., 2004, 2005). However, due to a lack of genetic evidence, the importance of P69B/C-EPI1/10 interactions in vivo remains unclear.
Recent studies have reported that oomycetes execute the virulence function by suppressing the secretion of host proteases. For instance, the P. infestans effector AVRblb2 is delivered into host cells where it suppresses the secretion of the C14 protease in tomato (Bozkurt et al., 2011). In soybean, the P. sojae-secreted effector PsAvh240 localizes to the host plasma membrane where it inhibits the secretion of GmAP1 (Guo et al., 2019). Taken together, plant-secreted proteases, including serine proteases, cysteine proteases, and aspartic proteases, are essential for plant immunity against oomycetes. However, oomycetes have established multiple strategies to counter host-secreted proteases, including the suppression of protease secretion and direct inhibition of protease activities.
Concluding Remarks and Future Perspectives
Studying the function of host-secreted proteases during their interaction with different types of microbes will uncover the molecular mechanisms of how plants employ their defense pathways to shape their resident microbial communities during plant–microbe co-evolution. Plant-secreted proteases use two major mechanisms to provide resistance. Firstly, secreted proteases trigger host basal immunity. For instance, maize cysteine proteases trigger host SA immunity via the cleavage of a precursor peptide to generate the bioactive peptide Zip1 (Ziemann et al., 2018). In addition, protease-mediated immune activation can be induced by other types of proteases such as the aspartic protease CDR1 (Xia et al., 2004; Simoes et al., 2007) and serine protease SBT3.3 (Ramirez et al., 2013). Therefore, elucidating how different types of proteases are involved in plant immune activation and how plants sense these signals will deepen our understanding of plant–microbe interactions. Secondly, plant-secreted proteases cleave proteins that are essential for the growth of pathogens, leading to direct suppression of bacterial infection (Wang et al., 2019). It is still unknown whether this mechanism is conserved in other plant species.
Microbes can secrete multiple protease inhibitors to inhibit plant-secreted proteases, which may facilitate their adaptation to different plants. Genome-wide characterization of protease inhibitors from different microbes should advance our understanding of the evolution of these proteins. For instance, the development of a quick screening method, which can be applied to different plant species, would improve our understanding of the outcomes/effects of apoplastic interactions on the co-evolution of plants and their microbial pathogens. Moreover, a recent study has reported that soil humidity is positively correlated with the diversity of the soil microbiota (Taketani et al., 2017). The apoplast, which provides an aqueous environment for the rhizosphere, is also a key barrier conferring plant immunity against microbes. Therefore, an investigation of the roles of secreted proteases in the shaping of microbial communities in roots, leaf rhizosphere, and endosphere would provide fundamental insights into plant–microbe interactions.
Author Contributions
All authors contributed to the writing of the manuscript and production of the figures.
Acknowledgments
We thank Neysan Donnelly for scientific English editing. We apologize to all the colleagues whose original research publications could not be cited due to space limitations. No conflicts of interest are declared.
Published: June 12, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Contributor Information
Yuanchao Wang, Email: wangyc@njau.edu.cn.
Yiming Wang, Email: ymwang@njau.edu.cn.
References
- Andrews S.C., Robinson A.K., Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Aung K., Jiang Y., He S.Y. The role of water in plant-microbe interactions. Plant J. 2018;93:771–780. doi: 10.1111/tpj.13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J., Chisholm S.T., Dahlbeck D., Staskawicz B.J. Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol. Microbiol. 2003;49:1537–1546. doi: 10.1046/j.1365-2958.2003.03666.x. [DOI] [PubMed] [Google Scholar]
- Aznar A., Chen N.W., Rigault M., Riache N., Joseph D., Desmaele D., Mouille G., Boutet S., Soubigou-Taconnat L., Renou J.P. Scavenging iron: a novel mechanism of plant immunity activation by microbial siderophores. Plant Physiol. 2014;164:2167–2183. doi: 10.1104/pp.113.233585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Muller D.B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., Dombrowski N., Munch P.C., Spaepen S., Remus-Emsermann M. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- Barbez E., Dunser K., Gaidora A., Lendl T., Busch W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U S A. 2017;114:E4884–E4893. doi: 10.1073/pnas.1613499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrutczyk M., Yang J., Eom J.S., Prior M., Sosso D., Hartwig T., Szurek B., Oliva R., Vera-Cruz C., White F.F. Sugar flux and signaling in plant-microbe interactions. Plant J. 2018;93:675–685. doi: 10.1111/tpj.13775. [DOI] [PubMed] [Google Scholar]
- Bigeard J., Colcombet J., Hirt H. Signaling mechanisms in pattern-triggered immunity (PTI) Mol. Plant. 2015;8:521–539. doi: 10.1016/j.molp.2014.12.022. [DOI] [PubMed] [Google Scholar]
- Bohm H., Albert I., Fan L., Reinhard A., Nurnberger T. Immune receptor complexes at the plant cell surface. Curr. Opin. Plant Biol. 2014;20:47–54. doi: 10.1016/j.pbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L.M., Jones A.M., Huitema E., van der Hoorn R.A. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. U S A. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K.A. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- Buscaill P., Chandrasekar B., Sanguankiattichai N., Kourelis J., Kaschani F., Thomas E.L., Morimoto K., Kaiser M., Preston G.M., Ichinose Y. Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science. 2019;364:eaav0748. doi: 10.1126/science.aav0748. [DOI] [PubMed] [Google Scholar]
- Casem M.L. Academic Press; Cambridge, MA: 2016. Case Studies in Cell Biology. [Google Scholar]
- Chen J., Ouyang Y., Wang L., Xie W., Zhang Q. Aspartic proteases gene family in rice: gene structure and expression, predicted protein features and phylogenetic relation. Gene. 2009;441:108–118. doi: 10.1016/j.gene.2009.04.021. [DOI] [PubMed] [Google Scholar]
- Chen L.Q., Hou B.H., Lalonde S., Takanaga H., Hartung M.L., Qu X.Q., Guo W.J., Kim J.G., Underwood W., Chaudhuri B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Dahlbeck D., Krishnamurthy N., Day B., Sjolander K., Staskawicz B.J. Molecular characterization of proteolytic cleavage sites for the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. U S A. 2005;102:2087–2092. doi: 10.1073/pnas.0409468102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K., Franco J.Y., Schwizer S., Pang Z., Hawara E., Liebrand T.W.H., Pagliaccia D., Zeng L., Gurung F.B., Wang P. An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun. 2018;9:1718. doi: 10.1038/s41467-018-04140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox K.L., Meng F., Wilkins K.E., Li F., Wang P., Booher N.J., Carpenter S.C.D., Chen L.Q., Zheng H., Gao X. TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 2017;8:15588. doi: 10.1038/ncomms15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangol S., Chen Y., Hwang B.K., Jwa N.-S. Iron- and reactive oxygen species-dependent ferroptosis cell death in rice-Magnaporthe oryzae interactions. Plant Cell. 2019;31:189–209. doi: 10.1105/tpc.18.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daudi A., Cheng Z., O'Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M.S., Golstein C., Thomas C.M., van der Biezen E.A., Jones J.D.G. Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. U S A. 2000;97:8807–8814. doi: 10.1073/pnas.97.16.8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Stam R., Cano L.M., Song J., Sklenar J., Yoshida K., Bozkurt T.O., Oliva R., Liu Z., Tian M. Effector specialization in a lineage of the Irish potato famine pathogen. Science. 2014;343:552–555. doi: 10.1126/science.1246300. [DOI] [PubMed] [Google Scholar]
- Dunn B.M. Determination of protease mechanism. In: Beynon R., Bond J.S., editors. Proteolytic Enzmyes: A Practival Approach. 2nd edn. Oxford Univesity Press; Oxford: 2005. pp. 77–104. [Google Scholar]
- Eichhorn H., Lessing F., Winterberg B., Schirawski J., Kamper J., Muller P., Kahmann R. A ferroxidation/permeation iron uptake system is required for virulence in Ustilago maydis. Plant Cell. 2006;18:3332–3345. doi: 10.1105/tpc.106.043588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaj D., Ambroziak P., Przepiora T., Skorko-Glonek J. The role of proteases in the virulence of plant pathogenic bacteria. Int. J. Mol. Sci. 2019;20:672. doi: 10.3390/ijms20030672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo A., Monteiro F., Sebastiana M. Subtilisin-like proteases in plant-pathogen recognition and immune priming: a perspective. Front. Plant Sci. 2014;5:739. doi: 10.3389/fpls.2014.00739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle H.H. Apoplastic pH during low-oxygen stress in Barley. Ann. Bot. 2006;98:1085–1093. doi: 10.1093/aob/mcl193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lorenzo M., Sjodin A., Jansson S., Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo M.C., Dagdas Y.F., Gupta Y.K., Mentlak T.A., Yi M., Martinez-Rocha A.L., Saitoh H., Terauchi R., Talbot N.J., Valent B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 2013;4:1996. doi: 10.1038/ncomms2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Holz F., Kelly S., Blaskowski S., Kaschani F., Kaiser M., van der Hoorn R.A.L. The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 2017;16:1068–1084. doi: 10.1111/pbi.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Wang H., Yang B., Jiang W., Jing M., Li H., Xia Y., Xu Y., Hu Q., Wang F. Phytophthora sojae Effector PsAvh240 inhibits host aspartic protease secretion ot promote infection. Mol. Plant. 2019;12:552–564. doi: 10.1016/j.molp.2019.01.017. [DOI] [PubMed] [Google Scholar]
- Hao L., Hsiang T., Goodwin P.H. Role of two cysteine proteinases in the susceptible response of Nicotiana benthamiana to Colletotrichum destructivum and the hypersensitive response to Pseudomonas syringae pv. tomato. Plant Sci. 2006;170:1001–1009. [Google Scholar]
- Haruta M., Burch H.L., Nelson R.B., Barrett-Wilt G., Kline K.G., Mohsin S.B., Young J.C., Otegui M.S., Sussman M.R. Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 2010;285:17918–17929. doi: 10.1074/jbc.M110.101733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou S.G., Wang X., Chen D.H., Yang X., Wang M., Turra D., Di Pietro A., Zhang W. The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 2014;10:e1004331. doi: 10.1371/journal.ppat.1004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas M., Horger A.C., Bozkurt T.O., van den Burg H.A., Kaschani F., Kaiser M., Belhaj K., Smoker M., Joosten M.H., Kamoun S. Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 2015;25:2300–2306. doi: 10.1016/j.cub.2015.07.030. [DOI] [PubMed] [Google Scholar]
- Ivanov S., Austin J., Berg R.H., Harrison M.J. Extensive membrane systems at the host-arbuscular mycorrhizal fungus interface. Nat. Plants. 2019;5:194–203. doi: 10.1038/s41477-019-0364-5. [DOI] [PubMed] [Google Scholar]
- Jashni M.K., Mehrabi R., Collemare J., Mesarich C.H., de Wit P.J. The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 2015;6:584. doi: 10.3389/fpls.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jorda L., Coego A., Conejero V., Vera P. A genomic cluster containing four differentially regulated subtilisin-like processing protease genes is in tomato plants. J. Biol. Chem. 1999;274:2360–2365. doi: 10.1074/jbc.274.4.2360. [DOI] [PubMed] [Google Scholar]
- Jorda L., Vera P. Local and systemic induction of two defense-related subtilisin-like protease promoters in transgenic Arabidopsis plants. Luciferin induction of PR gene expression. Plant Physiol. 2000;124:1049–1058. doi: 10.1104/pp.124.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Sklenar J., Derbyshire P., Stransfeld L., Asai S., Ntoukakis V., Jones J.D., Shirasu K., Menke F., Jones A. Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 2014;54:43–55. doi: 10.1016/j.molcel.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., Chae E., Herman J.J., Bergelson J. Mechanisms to mitigate the trade-off between growth and defense. Plant Cell. 2017;29:666–680. doi: 10.1105/tpc.16.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., Almario J., Friedemann C., Ding W., Giolai M., Heavens D., Kersten S., Lundberg D.S., Neumann M., Regalado J. Arabidopsis thaliana and Pseudomonas pathogens exhibit stable associations over evolutionary timescales. Cell Host Microbe. 2018;24:168–179.e4. doi: 10.1016/j.chom.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaschani F., Shabab M., Bozkurt T., Shindo T., Schornack S., Gu C., Ilyas M., Win J., Kamoun S., van der Hoorn R.A. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010;154:1794–1804. doi: 10.1104/pp.110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.G., Wang Y., Lee K.H., Park Z.-Y., Park J., Wu J., Kwon S.J., Lee Y.-H., Agrawal G.K., Rakwal R. In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteomics. 2013;78:58–71. doi: 10.1016/j.jprot.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Kwaaitaal M., Nielsen M.E., Bohlenius H., Thordal-Christensen H. The plant membrane surrounding powdery mildew haustoria shares properties with the endoplasmic reticulum membrane. J. Exp. Bot. 2017;68:5731–5743. doi: 10.1093/jxb/erx403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Gallo R.L. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Bourdais G., Yu G., Robatzek S., Coaker G. Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H(+)-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell. 2015;27:2042–2056. doi: 10.1105/tpc.114.132308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhang L., Mo X., Ji H., Bian H., Hu Y., Majid T., Long J., Pang H., Tao Y. Rice aquaporin PIP1;3 and harpin Hpa1 of bacterial blight pathogen cooperate in a type III effector translocation. J. Exp. Bot. 2019;70:3057–3073. doi: 10.1093/jxb/erz130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Ding P., Lian K., Wang J., Ma M., Li L., Li L., Li M., Zhang X., Chen S. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife. 2016;5:e13568. doi: 10.7554/eLife.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Fuglsang A.T., Palmgren M.G., Staskawicz B.J., Coaker G. RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 2009;7:e1000139. doi: 10.1371/journal.pbio.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luderer R., Takken F.L.W., de Wit P.J.G.M., Joosten M.H.A.J. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol. Microbiol. 2002;45:875–884. doi: 10.1046/j.1365-2958.2002.03060.x. [DOI] [PubMed] [Google Scholar]
- Ma Z., Song T., Zhu L., Ye W., Wang Y., Shao Y., Dong S., Zhang Z., Dou D., Zheng X. A phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell. 2015;27:2057–2072. doi: 10.1105/tpc.15.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Zhu L., Song T., Wang Y., Zhang Q., Xia Y., Qiu M., Lin Y., Li H., Kong L. A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science. 2017;355:710–714. doi: 10.1126/science.aai7919. [DOI] [PubMed] [Google Scholar]
- Macho A.P., Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol. Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- Maurel C., Verdoucq L., Luu D.-T., Santoni V. Plant aquaporins: membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 2008;59:595–624. doi: 10.1146/annurev.arplant.59.032607.092734. [DOI] [PubMed] [Google Scholar]
- Misas Villamil J.C., Mueller A.N., Demir F., Meyer U., Okmen B., Schulze Huynck J., Breuer M., Dauben H., Win J., Huesgen P.F. A fungal substrate mimicking molecule suppresses plant immunity via an inter-kingdom conserved motif. Nat. Commun. 2019;10:1576. doi: 10.1038/s41467-019-09472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott G.A., Middleton M.A., Desveaux D., Guttman D.S. Peptides and small molecules of the plant-pathogen apoplastic arena. Front. Plant Sci. 2014;5:677. doi: 10.3389/fpls.2014.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller A.N., Ziemann S., Treitschke S., Assmann D., Doehlemann G. Compatibility in the Ustilago maydis-maize interaction requires inhibition of host cysteine proteases by the fungal effector Pit2. Plos Pathog. 2013;9:e1003177. doi: 10.1371/journal.ppat.1003177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori T., Velasquez A.C., Wu J., Kvitko B.H., Kremer J.M., Wang Y., He S.Y., Tsuda K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. U S A. 2018;115:E3055–E3064. doi: 10.1073/pnas.1800529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- Nowicki M., Fooled M.R., Nowakowska M., Kozik E.U. Potato and tomato late blight caused by Phytophthora infestans: an overview of pathology and resistance breeding. Plant Dis. 2012;96:4–17. doi: 10.1094/PDIS-05-11-0458. [DOI] [PubMed] [Google Scholar]
- O'Brien J.A., Daudi A., Finch P., Butt V.S., Whitelegge J.P., Souda P., Ausubel F.M., Bolwell G.P. A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 2012;158:2013–2027. doi: 10.1104/pp.111.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad B.D., Creissen G., Lamb C., Chattoo B.B. Overexpression of Rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol. Plant Microbe Interact. 2009;22:1635–1644. doi: 10.1094/MPMI-22-12-1635. [DOI] [PubMed] [Google Scholar]
- Qi J., Wang J., Gong Z., Zhou J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017;38:92–100. doi: 10.1016/j.pbi.2017.04.022. [DOI] [PubMed] [Google Scholar]
- Ramirez V., Lopez A., Mauch-Mani B., Gil M.J., Vera P. An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathog. 2013;9:e1003445. doi: 10.1371/journal.ppat.1003445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Steinhauser D., Bussis D., Stintzi A., Schaller A., Kopka J., Altemann T. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Comput. Biol. 2005;1:297–312. doi: 10.1371/journal.pcbi.0010040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings N.D., Barrett A.J., Thomas P.D., Huang X., Bateman A., Finn R.D. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–D632. doi: 10.1093/nar/gkx1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney H.C.E., van 't Klooster J.W., van der Hoorn R.A.L., Joosten M.H.A.J., Jones J.D.G., de Wit P.J.G.M. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science. 2005;308:1783–1786. doi: 10.1126/science.1111404. [DOI] [PubMed] [Google Scholar]
- Sattelmacher B. Tansley review no. 22—the apoplast and its significance for plant mineral nutrition. New Phytol. 2001;149:167–192. doi: 10.1046/j.1469-8137.2001.00034.x. [DOI] [PubMed] [Google Scholar]
- Schaller A., Stintzi A., Rivas S., Serrano I., Chichkova N.V., Vartapetian A.B., Martinez D., Guiamet J.J., Sueldo D.J., van der Hoorn R.A.L. From structure to function—a family portrait of plant subtilases. New Phytol. 2017;218:901–915. doi: 10.1111/nph.14582. [DOI] [PubMed] [Google Scholar]
- Scheres B., van der Putten W.H. The plant perceptron connects environment to development. Nature. 2017;543:337–345. doi: 10.1038/nature22010. [DOI] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., van der Hoorn R.A.L. Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell. 2008;20:1169–1183. doi: 10.1105/tpc.107.056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo T., Misas-Villamil J.C., Horger A.C., Song J., van der Hoorn R.A.L. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS One. 2012;7:e29317. doi: 10.1371/journal.pone.0029317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes I., Faro R., Bur D., Faro C. Characterization of recombinant CDR1, an Arabidopsis aspartic proteinase involved in disease resistance. J. Biol. Chem. 2007;282:31358–31365. doi: 10.1074/jbc.M702477200. [DOI] [PubMed] [Google Scholar]
- Skaar E.P. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska E., Kong J.X., Busch W., Belkhadir Y. Organ-specific regulation of growth-defense tradeoffs by plants. Curr. Opin. Plant Biol. 2016;29:129–137. doi: 10.1016/j.pbi.2015.12.005. [DOI] [PubMed] [Google Scholar]
- Song J., Win J., Tian M., Schornack S., Kaschani F., Ilyas M., van der Hoorn R.A., Kamoun S. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. U S A. 2009;106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu S., Keshavarzi M., Brown I., Mansfield J.W. Ultrastructural characterisation of interactions between Arabidopsis thaliana and Albugo candida. Physiol. Mol. Plant Pathol. 2003;63:201–211. [Google Scholar]
- Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascon S., Hatzios S.K., Kagan V.E. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis I.A., Yu K., Feussner K., de Jonge R., Van Bentum S., Van Verk M.C., Berendsen R.L., Bakker P., Feussner I., Pieterse C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. U S A. 2018;115:E5213–E5222. doi: 10.1073/pnas.1722335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Xia Y.J., Cameron R., Shadle G., Blount J., Lamb C., Dixon R.A. Signals for local and systemic responses of plants to pathogen attack. J. Exp. Bot. 2004;55:169–179. doi: 10.1093/jxb/erh025. [DOI] [PubMed] [Google Scholar]
- Taketani R.G., Lanconi M.D., Kavamura V.N., Durrer A., Andreote F.D., Melo I.S. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb. Ecol. 2017;73:153–161. doi: 10.1007/s00248-016-0835-4. [DOI] [PubMed] [Google Scholar]
- Tian M., Huitema E., Da Cunha L., Torto-Alalibo T., Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J. Biol. Chem. 2004;279:26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- Tian M., Benedetti B., Kamoun S. A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005;138:1785–1793. doi: 10.1104/pp.105.061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Win J., Song J., van der Hoorn R., van der Knaap E., Kamoun S. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang J., Zeng X. The rice TAL effector–dependent resistance protein Xa10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 2014;26:497–515. doi: 10.1105/tpc.113.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S., Wang X., Li P., Wang H., Ji H., Xie J., Qiu Q., Shen D., Dong H. Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 2016;171:1635–1650. doi: 10.1104/pp.15.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L., Jones J.D. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U S A. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L.P., Sowdhamini R. Cross genome comparasions of serine proteases in Arabidopsis and rice. BMC Genomics. 2006;7:200. doi: 10.1186/1471-2164-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R.A.L. Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- van der Linde K., Hemetsberger C., Kastner C., Kaschani F., van der Hoorn R.A.L., Kumlehn J., Doehlemann G. A maize cystatin suppresses host immunity by inhibiting apoplastic cysteine proteases. Plant Cell. 2012;24:1285–1300. doi: 10.1105/tpc.111.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse H.P., van't Klooster J.W., Bolton M.D., Yadeta K.A., van Baarlen P., Boeren S., Vervoort J., de Wit P.J.G.M., Thomma B.P.H.J. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell. 2008;20:1948–1963. doi: 10.1105/tpc.108.059394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij C. The SWEET life of pathogens. Nat. Rev. Microbiol. 2011;9:4. doi: 10.1038/nrmicro2499. [DOI] [PubMed] [Google Scholar]
- Verbon E.H., Trapet P.L., Stringlis I.A., Kruijs S., Bakker P., Pieterse C.M.J. Iron and immunity. Annu. Rev. Phytopathol. 2017;55:355–375. doi: 10.1146/annurev-phyto-080516-035537. [DOI] [PubMed] [Google Scholar]
- Voges M., Bai Y., Schulze-Lefert P., Sattely E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. U S A. 2019;116:12558–12565. doi: 10.1073/pnas.1820691116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhou X.M., Xiong H.X., Mao W.Y., Zhao P., Sun M.X. Papain-like and legumain-like proteases in rice: genome-wide identification, comprehensive gene feature characterization and expression analysis. BMC Plant Biol. 2018;18:87. doi: 10.1186/s12870-018-1298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gupta R., Song W., Huh H.-H., Lee S.E., Wu J., Agrawal G.K., Rakwal R., Kang K.Y., Park S.R. Label-free quantitative secretome analysis of Xanthomonas oryzae pv. oryzae highlights the involvement of a novel cysteine protease in its pathogenicity. J. Proteomics. 2017;169:202–214. doi: 10.1016/j.jprot.2017.02.012. [DOI] [PubMed] [Google Scholar]
- Wang Y., Garrido-Oter R., Wu J., Winkelmuller T.M., Agler M., Colby T., Nobori T., Kemen E., Tsuda K. Site-specific cleavage of bacterial MucD by secreted proteases mediates antibacterial resistance in Arabidopsis. Nat. Commun. 2019;10:2853. doi: 10.1038/s41467-019-10793-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C., Carmody M., Kangasjarvi J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018;69:209–236. doi: 10.1146/annurev-arplant-042817-040322. [DOI] [PubMed] [Google Scholar]
- Wood L.F., Ohman D.E. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J. Bacteriol. 2006;188:3134–3137. doi: 10.1128/JB.188.8.3134-3137.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y.J., Suzuki H., Borevitz J., Blount J., Guo Z.J., Patel K., Dixon R.A., Lamb C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004;23:980–988. doi: 10.1038/sj.emboj.7600086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X.F., Nomura K., Aung K., Velasquez A.C., Yao J., Boutrot F., Chang J.H., Zipfel C., He S.Y. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524–529. doi: 10.1038/nature20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Saijo Y., Nakagami H., Takano Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science. 2016;354:1427–1430. doi: 10.1126/science.aah5692. [DOI] [PubMed] [Google Scholar]
- Yorgey P., Rahme L.G., Tan M.W., Ausubel F.M. The roles of mucD and alginate in the virulence of Pseudomonas aeruginosa in plants, nematodes and mice. Mol. Microbiol. 2001;41:1063–1076. doi: 10.1046/j.1365-2958.2001.02580.x. [DOI] [PubMed] [Google Scholar]
- Zhang L.Y., Hu Y.Q., Li P., Wang X.B., Dong H.S. Silencing of an aquaporin gene diminishes bacterial blight disease in rice. Australas. Plant Path. 2019;48:143–158. [Google Scholar]
- Ziemann S., van der Linde K., Lahrmann U., Acar B., Kaschani F., Colby T., Kaiser M., Ding Y.Z., Schmelz E., Huffaker A. An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants. 2018;4:172–180. doi: 10.1038/s41477-018-0116-y. [DOI] [PubMed] [Google Scholar]
- Zimmermann D., Gomez-Barrera J.A., Pasule C., Brack-Frick U.B., Sieferer E., Nicholson T.M., Pfannstiel J., Stintzi A., Schaller A. Cell death control by matrix metalloproteinases. Plant Physiol. 2016;171:1456–1469. doi: 10.1104/pp.16.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]