Abstract
Dispersal is one of the most important but least understood processes in plant ecology and evolutionary biology. Dispersal of seeds maintains and establishes populations, and pollen and seed dispersal are responsible for gene flow within and among populations. Traditional views of dispersal and gene flow assume models that are governed solely by geographic distance and do not account for variation in dispersal vector behavior in response to heterogenous landscapes. Landscape genetics integrates population genetics with Geographic Information Systems (GIS) to evaluate the effects of landscape features on gene flow patterns (effective dispersal). Surprisingly, relatively few landscape genetic studies have been conducted on plants. Plants present advantages because their populations are stationary, allowing more reliable estimates of the effects of landscape features on effective dispersal rates. On the other hand, plant dispersal is intrinsically complex because it depends on the habitat preferences of the plant and its pollen and seed dispersal vectors. We discuss strategies to assess the separate contributions of pollen and seed movement to effective dispersal and to delineate the effects of plant habitat quality from those of landscape features that affect vector behavior. Preliminary analyses of seed dispersal for three species indicate that isolation by landscape resistance is a better predictor of the rates and patterns of dispersal than geographic distance. Rates of effective dispersal are lower in areas of high plant habitat quality, which may be due to the effects of the shape of the dispersal kernel or to movement behaviors of biotic vectors. Landscape genetic studies in plants have the potential to provide novel insights into the process of gene flow among populations and to improve our understanding of the behavior of biotic and abiotic dispersal vectors in response to heterogeneous landscapes.
Keywords: effective dispersal, cpDNA, ResistanceGA, Circuitscape, geographic information systems (GIS), ecological niche modeling (ENM)
Plant dispersal is affected by both the habitat requirements of the plant and the effects of geographic features on vector behavior. This review discusses the advantages of using plants in landscape genetics studies and provide examples of a novel method for delineating the effects of habitat quality from those of geographic features that influence vector behavior when evaluating patterns of effective dispersal in heterogeneous landscapes.
Introduction
Dispersal is a keystone process that affects fundamental aspects of the ecology and evolution of all organisms. For plants, dispersal is critical for successful reproduction, maintenance of populations, and shifts in geographic range in response to environmental change (Harper, 1977; Howe and Smallwood, 1982; Nathan et al., 2008; Cruzan, 2018). Dispersal of seeds and pollen has important consequences for the biodiversity, conservation, and composition of plant communities (Trakhtenbrot et al., 2005; Jongejans et al., 2008; Damschen et al., 2014; Lohmus et al., 2014). The movement of pollen and seeds contributes to gene flow within and among populations, maintaining genetic variation within populations and enabling the spread of beneficial mutations that confer adaptive responses to environmental challenges (O'Connell et al., 2007; Teplitsky et al., 2014; Chybicki and Oleksa, 2018; Cruzan, 2019; Johnson et al., 2019). Seed dispersal also contributes to demographic and metapopulation processes (Cain et al., 2000; Howe and Miriti, 2004). Given the importance of dispersal for plants, it is crucial to understand the ecological drivers that affect the patterns of seed and pollen movement, as well as the evolutionary consequences of differential rates of dispersal.
The sedentary life form of plants necessitates the exploitation of biotic and abiotic vectors for the movement of seeds and pollen (Holderegger et al., 2010). Effective means of dispersal were apparently important for the early success of flowering plants, as their diversification paralleled the origin and diversification of bees and frugivorous primates in the late Cretaceous and frugivorous birds and bats in the Oligocene and Miocene (Eriksson, 2016). The mechanisms that incentivize effective transport of seeds and pollen often come at an energetic cost. Selection has favored the offering of rewards such as nectar and pollen to manipulate the behavior of pollinators for efficient pollen transfer and the production of fleshy fruits that encourage seed transport by birds and mammals. The potential for seed movement by vectors has been improved by the evolution of morphological modifications of spores, seeds, and fruits that improve their buoyancy (for wind and water dispersal, anemochory, and hydrochory, respectively), attachment to fur and hooves (spines or hooks for ectozoochory), or attraction of frugivorous birds and mammals (fleshy fruits for endozoochory). On the other hand, the large majority of plants lack specific seed and fruit characters and are considered to be gravity dispersed (barochoric), although their seeds and fruits may be subject to inadvertent secondary dispersal by biotic or abiotic vectors (Harper, 1977; Cousens et al., 2008; Grasty et al., 2020). Although mechanisms that improve the potential for dispersal often require the expenditure of resources, this investment is only a small fraction of the ambulatory energetic costs that would be required if plants could transport their own seeds and pollen over a similar diversity of directions and distances (Bonte et al., 2012). Hence, the sedentary life form of plants has significant benefits, as they have evolved to exploit available means of seed and pollen conveyance with a minimal energetic expense. Plants are the major beneficiaries of passive transport by biotic and abiotic vectors and of their coevolutionary relationships with biotic vectors.
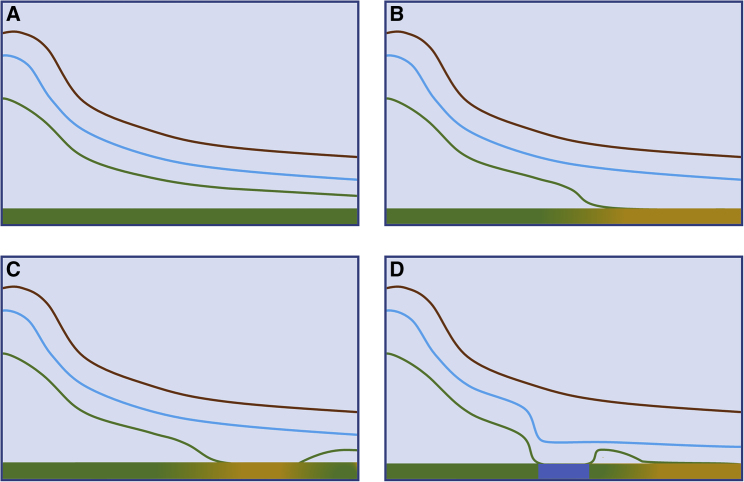
Plants’ lack of direct participation in dispersal increases the difficulty of tracking the movement of their seeds and pollen. Once seeds and pollen are removed from a plant, it is difficult to determine their fates for a variety of reasons. For one, pollen is always—and seeds are almost always—small enough to prevent direct observation of their movements. Moreover, seeds and pollen are often transported by an array of biotic and abiotic vectors whose complex behaviors affect the probability of pickup, the direction and distance of transport, and the probability of deposition at different distances from the source plant (Nathan and Muller-Landau, 2000; Nathan et al., 2008). If the frequency of movement to different distances can be measured, a probability density function can be estimated (dispersal kernel; Figure 1) and used in mathematical models to predict the dispersal potential of seeds and pollen (Okubo and Levin, 1989; Clark, 1998; Nathan et al., 2012). Dispersal kernels indicate the probability of movement to different distances but do not account for success or failure after arrival (Nathan et al., 2012; Klein et al., 2013). Because receptive stigmas and suitable habitats can be exceedingly small targets for dispersal, large proportions of pollen and seeds arrive in locations that are inhospitable for their germination and survival. The small number that does succeed in fertilizing ovules (pollen) or surviving in their new locations (seeds) constitutes the fraction of the dispersal kernel that represents effective dispersal (Auffret et al., 2017; Figure 1).
Figure 1.
The Effects of Geographic Distance, Plant Habitat Quality, and Landscape Features on Dispersal Kernels (Brown Curves), the Behavior of Large Mammals (Dispersal Vectors; Blue Curves), and the Consequences for Patterns of Effective Dispersal (Green Curves).
Bars across the bottom of each graph indicate habitat quality. Green regions of the landscape indicate high habitat quality, and yellow regions indicate low quality.
(A) For dispersal within high-quality habitat, the three curves are parallel such that effective dispersal reflects the dispersal kernel.
(B) The movement of the vector and the dispersal kernel are similar, but plants do not become established in areas of low habitat quality, and effective dispersal declines to zero.
(C) The effective dispersal kernel recovers as vectors move seeds to a separate area of high-quality habitat.
(D) The presence of a river (blue region in the habitat quality bar) reduces vector movement, and consequently, there are lower rates of effective dispersal in a habitat patch on the opposite side.
Ecologists have accumulated substantial amounts of information on the dispersal of both seeds and pollen. For example, we know a great deal about the movement of pollen by biotic vectors such as birds and bumble bees through the estimation of pollen carryover curves based on dye particles and morphologically distinct pollen deposited on stigmas after pollinator visits (Thomson and Thomson, 1989; Cane and Love, 2019), but we know much less about the patterns and distances of pollen movement by wind (Friedman and Barrett, 2009). Estimates of seed dispersal come from release experiments on natural or artificial wind-dispersed seeds (Augspurger, 1986; Damschen et al., 2014), from sticky traps placed at different distances from isolated plants or trees (Harper, 1977; Howe and Smallwood, 1982; Nathan and Katul, 2005; Jones and Muller-Landau, 2008), and from the behaviors of their vectors (Tsoar et al., 2011; Cortes and Uriarte, 2013). These approaches for dispersal quantification have limitations, as they can be applied only under restrictive circumstances and with certain types of vectors and landscapes. For example, wind dispersal of seeds can be accurately measured only for one individual at a time (sticky trap experiments) or under artificial conditions (seed release experiments). However, all of these methods of direct observation chronically underestimate dispersal distance because they do not detect long-distance dispersal (LDD) events (Howe and Smallwood, 1982; Nathan, 2006). The disproportionate importance of LDD is evident from discrepancies between observed and predicted migration rates of understory herbs and trees following the last glacial maximum (Clark, 1998; Clark et al., 1998). Furthermore, dispersal kernels estimated from the direct observation of seeds, pollen, or movement of their vectors may provide misleading information because they do not reflect the effects of population density and other ecological conditions that determine the success of migrant seeds and pollen (Cruzan, 1989; Steinitz et al., 2011; Jansen et al., 2014; Harder et al., 2016; Sullivan et al., 2017).
The use of genetic markers to measure effective dispersal has advantages over direct observation because it provides valuable information on the patterns of seed and pollen movement that are important for ecological, demographic, and evolutionary processes (Ouborg et al., 1999; Cain et al., 2000; Auffret et al., 2017). One method for the estimation of effective dispersal is based on levels of similarity in genetic composition among populations. The use of genetic similarity (or distance) among populations takes advantage of the fact that effective dispersal results in gene flow that affects the genetic composition of populations. Over smaller spatial scales, this method has the advantage of integrating effective dispersal events that occurred over recent history. However, at larger spatial scales, levels of gene flow among populations may have the disadvantage of reflecting effective dispersal over longer time frames that are more important for evolutionary than for ecological processes (Nathan et al., 2003). When populations are sampled at an appropriate spatial scale, estimates of effective dispersal based on genetic marker composition can provide substantial information on ecological processes that influence patterns of seed and pollen movement. Unlike dispersal kernels inferred from observational data, which typically evaluate only the effect of geographic distance, measures of effective dispersal based on genetic marker similarity among populations can be combined with geographic information systems (GIS) methods to test a range of hypotheses about the effects of specific landscape features on gene flow patterns (Manel et al., 2003; McRae et al., 2008; Spear et al., 2010). The field of landscape genetics (genomics) was developed to test specific hypotheses about the influences of landscape features on the patterns of effective dispersal. Landscape genetics has most often been used to evaluate the movement patterns of animals (Storfer et al., 2010; Rissler, 2016; Beninde et al., 2018; Dileo et al., 2018; Cameron et al., 2019; Kunde et al., 2019; Lourenco et al., 2019; Yadav et al., 2019), but more studies on plants are beginning to emerge (e.g., Vandepitte et al., 2007; Arredondo et al., 2018; Morente-López et al., 2018; Alvarado-Serrano et al., 2019; Grasty et al., 2020).
The goal of this review is to describe applications of landscape genetic approaches to the estimation of plant dispersal and to evaluate the challenges and benefits of using plants in landscape genetic studies. We focus on the ecological and evolutionary aspects of dispersal within a contemporary time frame. We explore the advantages of using different types of genetic markers, genetic relatedness measures, geographic scales, plant life forms, and sampling strategies for studies that evaluate the effects of ecological features on patterns of effective dispersal. With the prudent selection of study species, strategic sampling, and advanced analysis methods, plants offer opportunities to evaluate a wide range of hypotheses about the effects of biotic and abiotic behaviors on the patterns of gene flow and dispersal.
An Overview of Landscape Genetics
Our view of pollen and seed dispersal has been dominated by the idea that most transports occur over short distances and that movements become increasingly rare as the distance from the source increases. The resulting dispersal kernel is expected to be a smooth probability curve for deposition as a function of geographic distance. Because it is assumed that effective dispersal is directly related to the dispersal kernel, rates of gene flow are also expected to decrease as distance increases, a model referred to as isolation by distance (IBD; Wright, 1943; Slatkin, 1993; Hutchison and Templeton, 1999; Jenkins et al., 2010). Although there has been much discussion about the shape of dispersal kernels (e.g., leptokurtic or more “fat tailed”) (Rogers et al., 2019), these models consider only geographic distance as the primary driver of dispersal potential. They effectively assume that the landscape is flat and homogeneous and that vectors are well behaved within the confines of the defined density function. In real landscapes, geographic features and spatial variation in habitat quality can have large influences on the behavior of biotic and abiotic vectors. Dispersal kernels that consider only the effects of distance may be adequate over local regions of continuous habitat, but we expect that their predictive power will rapidly erode over larger areas that include changes in topography and vegetation and the presence of geographic features such as streams, rivers, lakes, and various types of human-modified landscapes. Hence, consideration of the effects of heterogeneous landscapes on the behavior of dispersal vectors is particularly important for understanding patterns of dispersal over longer distances.
The recognition that geographic features can inhibit the movement of organisms dates back to early biogeographers such as Augustin-Pyramus de Candolle and Alfred Russell Wallace, who noted that taxonomic discontinuities were often associated with “physical causes” (Crisci, 2001). Through the 1900s, most evaluations of the effects of geographic features on movement focused on species' distributions and were conducted at large scales using biogeographic studies of species' distributions on continents and islands (Cox and Moore, 2010; Schnitzler et al., 2012; Echeverría-Londoño et al., 2018). In particular, MacArthur and Wilson's (1967) Theory of Island Biogeography inspired a number of studies that had a large influence on our understanding of IBD and the effects of habitat fragmentation on community composition and population sizes. Later, genetic marker methods were used to conduct more refined studies of biogeography in the field of phylogeography, which evaluated genetic discontinuities within species to infer historical distributions and patterns of range expansion (Avise et al., 1987; Templeton, 2004; Hickerson et al., 2010). Because population genetic models predicted higher rates of gene flow among neighboring populations, it was widely assumed that patterns of genetic differentiation within species generally followed a model of isolation by geographic distance.
By the late 1990s, there was increasing awareness that IBD may apply to only a limited set of circumstances (Whitlock and McCauley, 1999) and that landscape features could have substantial effects on gene flow patterns (Sork et al., 1999). During this period, there was a growing interest in the effects of spatial distribution (e.g., Cruzan, 2001; Diniz-Filho and De Campos Telles, 2002) and landscape features (e.g., Young et al., 1996; Kudoh and Whigham, 1997; Nason and Hamrick, 1997; Piertney et al., 1998; Keyghobadi et al., 1999; Castric et al., 2001; Gram and Sork, 2001) on genetic variation patterns. The idea that population genetics could be combined with landscape ecology to provide a spatially explicit framework in which to test hypotheses about the effects of geographic features on the patterns of genetic differentiation was first proposed by Manel et al. (2003), who termed this new field “landscape genetics”. Unfortunately, the earliest methods available were limited to evaluating the effects of individual geographic boundaries (Manel et al., 2007; Storfer et al., 2010). With the introduction of circuit theory (McRae and Beier, 2007; McRae et al., 2008; Box 1), the assessment of effective dispersal models expanded to include isolation by resistance (IBR) in addition to IBD.
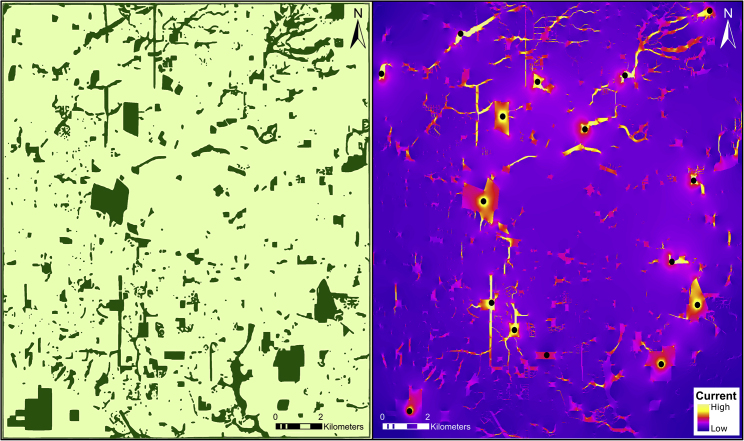
Figure 2.
Patterns of Isolation by Resistance for a Network of Nature Parks (Left Map; Dark Green) Distributed across an Urban/Suburban Landscape (Light Green Matrix), and Consequences for Isolation by Resistance (Right Panel; Cool Colors = High Resistance).
The circuit map was generated by Circuitscape and assumes that only natural areas have low resistance to dispersal. Circuit maps are used to test the hypothesis that they predict dispersal patterns by comparing them with genetic distance matrices among sample locations.
Box 1. Circuit Theory Tools for Estimating Landscape Resistance.
Circuitscape
With the advent of circuit theory, it is possible to test hypotheses of isolation by resistance (IBR), which are more applicable to complex landscapes than isolation by distance (IBD; Figure 2). The popularity of circuit theory in landscape genetics was largely due to the publication of Circuitscape, a program that uses population locations and landscape features to estimate “resistance” to dispersal and, consequently, gene flow for all possible paths across a landscape (McRae et al., 2008; Spear et al., 2010). Circuit theory, more commonly used in electrical fields, operates around concepts of resistance, voltage, and current, where the voltage and resistance determine the current across a circuit. When circuit theory is applied to a biological system, the increase in landscape resistance—and reduction in conductance—restricts the movement of organisms in the current, decreasing gene flow across the landscape (McRae and Beier, 2007; McRae et al., 2008). Circuitscape produces a population-pairwise matrix of landscape resistance, which can be compared with genetic diversity matrices to ascertain which features determine gene flow. Circuitscape and other similar programs (see commuteDistance; van Etten, 2017) have been widely utilized in animal and plant migration studies (Pérez-Espona et al., 2012; Poor et al., 2012; Trumbo et al., 2013; Johnson et al., 2017), as they are easily adaptable to a variety of ecosystems and require minimal input data. Circuitscape analysis requires known population locations and spatially defined landscape features, which are often available through online databases.
ResistanceGA
The main limitation of early versions of circuit analysis was the requirement for a priori knowledge of the resistance values for each landscape feature. For example, one might assume that wind-dispersed seeds travel shorter distances in a closed canopy than in open areas, and a higher resistance value would therefore be assigned to forests than to grasslands. However, assigning resistance values using this approach is arbitrary, based on a best guess or “expert opinion.” Consequently, the resistance values chosen can bias the results and may reinforce preconceived notions of the effects of different features on dispersal (Shirk et al., 2010; Charney, 2012). Moreover, only one landscape layer could be evaluated at a time, making comparisons of different types of landscape features difficult. A number of methods were proposed to optimize resistance values for individual landscape types (e.g., Peterman et al., 2014; Khimoun et al., 2017), but these approaches suffered from limitations, as they would work with only one type of GIS layer, either categorical (e.g., vegetation classifications) or continuous (e.g., environmental gradients). This problem was solved when permutation methods were developed to optimize resistance values for both categorical and continuous landscape layers to predict the patterns of genetic distance among sample locations (ResistanceGA; Peterman, 2018). ResistanceGA has been shown to be superior to alternative methods for the optimization of landscape genetic models of dispersal (Peterman et al., 2019). Given its advantages, ResistanceGA has quickly become the method of choice for the optimization of landscape genetic models. ResistanceGA is an optimization program that addresses the researcher bias introduced in Circuitscape analyses during the assignment of landscape feature values. Using population genetic diversity measurements, ResistanceGA iteratively optimizes feature values in the context of known gene flow (Peterman, 2018). For example, if high gene flow is found in populations in natural parks, ResistanceGA will assign a low resistance value to the natural park feature and a high resistance value to the urban/suburban feature (See Figure 2). Analysis using ResistanceGA also allows for appropriate combinations of multiple features to quantify if more than one aspect of the landscape is determining the patterns of genetic diversity. During model selection by ResistanceGA, IBD, and null hypotheses are also considered. The program determines whether features are resistors or conduits, calculates their optimized resistance values, and identifies features that contribute significantly to the patterns of genetic diversity.
The application of circuit theory to the studies of dispersal permits the testing of explicit hypotheses about the influence of landscape features on patterns of effective dispersal (Box 1). In this approach, landscape layers that represent one or more features are used to test the hypothesis that they predict patterns of effective dispersal among populations (collection sites) by comparing matrices of resistance distance with genetic distance (Spear et al., 2010). These models typically include geographic distance as well as resistance matrices based on a number of landscape layers; they therefore specifically test whether IBD or IBR is a better predictor of dispersal patterns. Although early programs required researchers to decide on resistance values for each landscape feature, ResistanceGA uses permutation methods to optimize resistance values in categorical landscapes (e.g., land-use classifications) and resistance functions for landscapes variables with continuous variation (e.g., elevation; Peterman, 2018). Genetic distance matrices can be based on a variety of measures calculated from nuclear or cytoplasmic genetic markers. This approach permits robust tests of specific hypotheses about the influence of landscape features on effective dispersal patterns.
Advantages of Using Plants in Landscape Genetic Studies
Only a minority of landscape genetic studies have been conducted on plants since the inception of the field in the late 1990s (Storfer et al., 2010), and this trend has continued—especially for studies that use optimized circuit theory methods (e.g., Arredondo et al., 2018; Alvarado-Serrano et al., 2019; Carvalho et al., 2019; Grasty et al., 2020). This is surprising given that plants offer many advantages for evaluating the effects of specific types of landscape features on effective dispersal. First and foremost is the fact that plant populations are stationary, which permits a more accurate evaluation of the effects of individual landscape types on rates of effective dispersal (see below). Furthermore, the separate effects of seed and pollen dispersal can often be determined by comparing the genetic structure based on maternally inherited chloroplast DNA (cpDNA) markers, which is determined by seed dispersal, and the genetic structure based on nuclear markers, which is determined by seed and pollen dispersal (Ennos, 1994; Savolainen et al., 2007; Gerber et al., 2014). Although maternal inheritance of chloroplasts predominates in the flowering plants (Sears, 1980), there can be low levels of paternal inheritance following crosses between divergent populations or species (Cruzan et al., 1993; Ellis et al., 2008), and paternal inheritance of chloroplasts is common in the conifers (Strauss et al., 1989). Evaluating the separate effects of seed and pollen dispersal on gene flow among populations can be important because different dispersal vectors are responsible for pollen and seed movement and are likely to have different responses to landscape features. Although the slow rate of chloroplast genome evolution has historically been an impediment to population genetic studies that use cpDNA markers, this situation has improved through the development of efficient methods for whole chloroplast genome sampling and genotyping (Kohrn et al., 2017; Grasty et al., 2020; Table 1).
Table 1.
Chloroplast Haplotype Diversity within Species Based on Whole-Genome SNP Assays.
| Species | Family | Life form | Sampled populations | Range area (km2) | Discovered haplotypes |
|---|---|---|---|---|---|
| Achyrachaena mollis | Asteraceae | Annual | 46 | 610 | 13 |
| Eriophyllum lanatum | Asteraceae | Perennial | 27 | 2420 | 47 |
| Lasthenia californica | Asteraceae | Annual | 21 | 1.6 | 40 |
| Plagiobothrys nothofulvus | Boraginaceae | Annual | 32 | 1.6 | 16 |
| Plectritis congesta | Caprifoliaceae | Annual | 36 | 920 | 22 |
| Ranunculus occidentalis | Ranunculaceae | Perennial | 32 | 5350 | 18 |
For each species, its life form, number of sampled populations, and size of the sampled region are shown. Because the chloroplast is highly conserved, the rate of mutation and generation of novel haplotypes occurs over thousands of years, resulting in a lower number of discoverable haplotypes, even over large sample areas. Haplotypes were discovered following methods described in Kohrn et al. (2017).
Alternatively, nuclear DNA can be used to detect patterns of effective dispersal in landscape genetics. Relative to the chloroplast, the nuclear genome is less conserved, increasing the level of detectable genetic diversity and providing insights into contemporary dispersal events. Because maternal and paternal contributions are included in the offspring genotype, further delineation is required to identify the effects of pollen and seed dispersal on levels and patterns of gene flow. This can be accomplished by comparing genetic differentiation estimates for maternally inherited markers (e.g., cpDNA) with those for biparentally inherited markers (nuclear markers; Ennos, 1994; Liu et al., 2015) or by conducting parentage analysis of seeds or seedlings (e.g., Gerber et al., 2014; Browne et al., 2018; Melo and Hale, 2019). Separation of parental contributions to gene flow depends upon dispersal mode, as differences in the behavior of dispersal vectors may result in asymmetric pollen and seed dispersal kernels (Bacles et al., 2006; Garcia et al., 2007). For example, many invertebrate pollinators have limited foraging ranges and move pollen relatively short distances (Thomson and Thomson, 1989; Osborne et al., 2008; Danner et al., 2016), resulting in limited pollen-mediated gene flow. By contrast, pollen-mediated gene flow can extend over much larger distances in wind-pollinated species (Dutech et al., 2005; Wang et al., 2012; Wei et al., 2013; Gerber et al., 2014). In pine forests, coniferous pollen grains are equipped with air sacs that facilitate LDD in wind currents (Williams, 2008). The dispersal of pine seeds is often limited because they are either barochoric or have less effective wind dispersal (Benkman, 1995), and local gene flow is therefore likely to be maternally derived (Latta et al., 1998). Whether pollen dispersal occurs by biotic or abiotic vectors, the process of gene flow among established populations is sequential, such that nuclear genomes will be moved first by pollen dispersal and then by the dispersal of fertilized seeds. Consequently, total nuclear gene flow is due to a combination of pollen and seed movement, whereas the gene flow of maternally inherited chloroplast markers occurs solely by seed dispersal.
Landscape genetic studies on plants also have the potential to provide unique information on the behavior of their dispersal vectors. For biotic vectors, evaluation of the movement behavior of animals is generally laborious, data can typically be collected for only a few individuals, and the implementation of tracking technologies is expensive (Nathan, 2008; Katzner and Arlettaz, 2020). Some animals, such as small insect pollinators, are too small to accommodate tracking devices, and direct observation is difficult over larger foraging distances. Similarly, it can be difficult to determine the movement behavior of frugivorous birds (endozoochory) and large mammals (ectozoochory and secondary dispersal of barochoric species) if they travel over large distances. Landscape genetic studies of plant effective dispersal have the advantage of integrating the effects of large numbers of animal movements over time, allowing for the characterization of animals’ responses to specific geographic features such as elevation changes, vegetation types, waterways, and various types of human-modified landscapes.
Abiotic vectors, and wind in particular, are important for plant dispersal, and landscape genetic studies can provide information on the movement patterns of wind- and water-borne spores and propagules (e.g., Wang et al., 2012; Wei et al., 2013; Tumas et al., 2018). Dispersal by wind is notoriously difficult to measure across geographic distances, as the source and destination of individual particles are impossible to track (Nathan et al., 2002). Successful dispersal by wind is subject to variables at both the local scale—such as seed and maternal traits—and the meso and regional scales, where effects of landscape features and meteorological events on spore and seed movement are prevalent. Release height and abscission force (Greene and Quesada, 2011; Treep et al., 2018) from the maternal plant are known to influence the dispersal kernels of anemochoric seeds, and specific morphological attributes of seeds and pollen may affect particle behavior in wind streams. For example, the presence of a pappus on seeds (Nathan et al., 2008) or air bladders in pollen (Di-Giovanni and Kevan, 1991) can change the buoyancy of the particle while aloft. During the long-distance flight of particles, landscape surface roughness influences wind fluid dynamics and induces turbulent fluctuations in flow speed, which may determine dispersal events (Bullock and Clarke, 2000; Bohrer et al., 2008; Damschen et al., 2014). The incorporation of wind data into landscape genetic analyses of wind-dispersed species is a relatively novel approach. However, one study of a wind-dispersed tree found that mean wind speed and direction influenced spatial genetic structure through seed and pollen dispersal (Wang et al., 2016), although another did not detect effects of wind direction on genetic structure in a wind-pollinated species (Dutech et al., 2005). Landscape genetic analyses can quantify the consequences of fluid behavior for plant effective dispersal, which are challenging to predict.
Design of Landscape Genetic Studies
The goal of landscape genetics is to characterize the effects of individual types of geographic features on rates of effective dispersal. Landscape genetics provides an interface between the fields of population genetics and ecology, and it is therefore important to prioritize ecological considerations in the design of landscape genetic studies. In particular, the distribution of sample locations should be guided by an understanding of the dispersal potential of the plant under study (i.e., the morphological characteristics of flowers, seeds, and fruits) relative to the “grain” of landscape features (i.e., the size of features in a spatially variable landscape) that are likely to influence patterns of effective dispersal. Sampling schemes should be developed with some knowledge of the ecology of the plant and the behavioral characteristics of its likely dispersal vectors. It is also important to recognize that, for the large majority of species whose fruits and seeds lack morphological characteristics that aid dispersal (i.e., dispersal is barochoric), primary dispersal may be limited by plant stature, although biotic and abiotic vectors probably contribute to secondary dispersal over longer distances (Harper, 1977; Cousens et al., 2008; Grasty et al., 2020).
In general, the size, life history, distribution, and dispersal ability of the study organism should be considered when developing sampling strategies (Anderson et al., 2010). For larger plants such as trees—and for species whose fruit and seed morphology suggests that they are likely to have good dispersal ability—the scale of analysis must be large enough to capture the effects of the intervening landscape that separates sampling locations, and the sampling region may therefore need to cover tens to hundreds of hectares (e.g., Sork and Smouse, 2006; Born et al., 2008). Conversely, landscape genetic studies of small annuals and species with limited dispersal ability require scaling down to adequately capture the effects of individual landscape types (e.g., Grasty et al., 2020). If the study species is common enough, then sample sites can be strategically placed to evaluate the effects of a homogeneous stretch of high-quality habitat or placed in habitat fragments that are separated by a single type of landscape (Figure 3). Over larger distances, the effects of multiple landscape features may be integrated, making it difficult to evaluate their independent effects on effective dispersal (Anderson et al., 2010; Figures 3 and 4). Hence, the grain of habitat heterogeneity relative to the dispersal ability of the study organism should be considered for the development of sampling strategies that will provide accurate estimates of the effects of individual landscape features on patterns of effective dispersal (O'Connell et al., 2007).
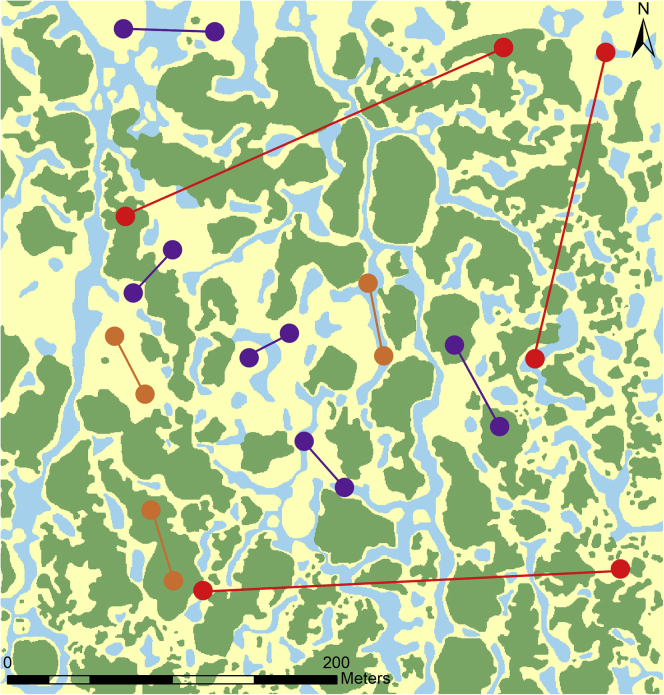
Figure 3.
Sampling Schemes for Landscape Genetics.
This example landscape is a mosaic of vegetation types: shrubs (green), prairie (yellow), and swales/vernal pools (blue). Pairs of orange sample points test for resistance within each type of vegetation, and pairs of purple sample points test for resistance across a single contrasting habitat type. The pairs of red sample points are not optimal because they integrate the effects of many vegetation types across the landscape. Vegetation map modified from Grasty et al. (2020).
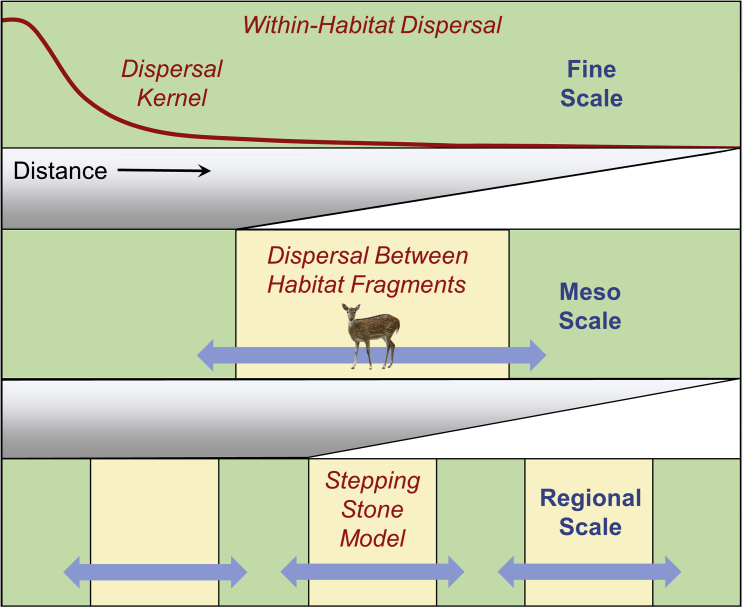
Figure 4.
Dispersal at Different Spatial Scales.
Within contiguous high-quality habitat, we expect rates of dispersal to generally follow a strongly leptokurtic dispersal kernel that is based on the movement behavior of primary and secondary dispersal vectors. Dispersal resistance may be higher within habitat fragments because many dispersal events occur over short distances. Dispersal between pairs of habitat fragments separated by low-quality habitat may appear to have lower resistance because effective dispersal is higher at the tail of the dispersal kernel (Figure 1C) and because there are higher rates of wildlife vector movement between fragments. At regional scales, effective dispersal rates become multigenerational and follow a stepping-stone model, as dispersal is more likely to occur between pairs of neighboring habitat fragments.
Species' distributions are generally aggregated but the scale of aggregation varies widely from sparsely distributed populations that occupy habitat fragments to continuously distributed individuals of the same species that cover tens to thousands of hectares. A complete understanding of dispersal potential requires sampling locations that span areas within continuous habitat and across fragments separated by low-quality habitat. Because humans have modified landscapes across much of the globe, landscape features that separate habitat fragments often include different categories of land use (e.g., agricultural fields and urban and suburban development). These and other features that result from human activity, such as foot trails, roads, and highways, may act as either barriers or conduits for dispersal. When sampling across regions that have been extensively modified, choices for sample locations may be limited by the small number of habitat fragments, and it may be difficult to locate large regions of undisturbed, high-quality habitat. To the extent possible, sample sites should be chosen to evaluate single types of landscape features either within continuous areas of suitable habitat or in pairs of habitat fragments that are separated by a single, homogeneous landscape type (Figure 3).
At larger spatial scales, estimates of landscape resistance are more likely to integrate multiple dispersal events across different types of habitat. This is due to the fact that gene flow over long distances is most likely to occur by a sequence of multiple dispersal events within continuous habitat and between neighboring habitat fragments. Each dispersal event contributes to local and regional patterns of genetic differentiation, and genetic distance measures at larger spatial scales are more likely to reflect “stepping-stone” processes across multiple habitat fragments (Figure 4). As the distance between sample sites increases, the effects of gene flow on genetic similarity decrease and genetic differentiation becomes governed primarily by mutation and genetic drift. Hence, the spatial scale evaluated reflects the temporal scale. Smaller scales are more appropriate for landscape genetic analyses, as they provide information on the effects of contemporary gene flow and dispersal processes. By contrast, larger scales are more appropriate for the study of differentiation patterns that result from phylogeographic and coalescence processes (Avise et al., 1987; Cruzan and Templeton, 2000; Knowles and Maddison, 2002; Wang, 2010). Sampling across larger regions may include groups of populations with unique histories due to differentiation in separate glacial refugia and historical migration patterns, making it impractical to evaluate the effects of contemporary dispersal processes on patterns of genetic differentiation.
From the discussion above, it is clear that the design of landscape genetic studies depends on the life form, dispersal potential, and habitat preferences of the species under study, as well as the questions being addressed. In general, the spatial scale of sampling should be considered carefully to ensure that the patterns of genetic differentiation among sample sites are primarily governed by contemporary dispersal rather than historical processes (Wang, 2010). As mentioned above, the stature, life history, and dispersal ability of the study species should be considered when deciding on an appropriate spatial scale. To the extent possible, it is also desirable to choose sample sites that are separated primarily by single types of landscape features, to sample at multiple spatial scales, and to place multiple samples within undisturbed habitat as well as among habitat fragments. To provide the largest degree of sampling flexibility, more common species should be chosen when possible, and it is important to keep in mind that the population genetic structure of plants with shorter generation times is more likely to be affected by contemporary processes. If the purpose of the study is to measure the cumulative effects of effective dispersal over a number of years, the use of circuit theory analyses is appropriate. If the aim is to detect contemporary dispersal events, other techniques, such as parentage analyses, should be used. Finally, whether the study focuses on pollen or seed dispersal will dictate the type of genetic marker used to estimate genetic distances among sites, but for the most informative studies, both nuclear and cytoplasmic markers should be assayed.
Effects of Habitat Suitability and Vector Behavior
Dispersal within and among plant populations is determined by habitat suitability, as well as the behavior of seed and pollen dispersal vectors. If a seed is dispersed outside the habitat of its parental plant, the probability of encountering high-quality habitat sufficient for germination and seedling recruitment is low (Rey and Alcántara, 2000), and the majority of dispersed propagules will fail to establish in novel locations. Regardless of dispersal distance, the establishment, reproductive success, and fitness of plants may be contingent upon habitat suitability; this dynamic has been observed for plant–soil feedback loops (Sedlacek et al., 2014), temperature and precipitation shifts (Bradley et al., 2016), and light availability (Warren et al., 2012), among other conditions. Hence, dispersal among populations is dependent upon the mosaic of the plant's suitable habitat and landscape-modified vector movement patterns (Figure 1). An optimal design with which to evaluate landscape effects on plant dispersal should delineate features that influence habitat quality (destination effects) and vector movement (path effects).
Movement in response to habitat quality has been observed for some animal species, such as butterflies, which typically migrate quickly through low-quality habitat and linger in regions of high-quality habitat (Albert et al., 2015). If plants are dependent upon animal pollination or zoochory, such animal behavior can significantly influence gene flow. For example, mild disturbances—such as firebreaks and logging roads—have been shown to facilitate the movement of coyote, rabbit, and other seed-dispersing mammals, and ultimately the dispersal of seeds (Suárez-Esteban et al., 2013). Ungulate behavior and range patterns are influenced by the presence of wildlife corridors such as riparian areas, which can serve as dispersal conduits, and by suburban development, which presents a dispersal barrier (Kilpatrick and Spohr, 2000; Vellend et al., 2003). Anemophilous plants may be influenced by disparities in surface roughness (Biswas and Wagner, 2012), such as those caused by tree canopies (Finnigan, 2000) and high-density urbanization (Fernando et al., 2010), that influence wind patterns. Although conscious choice is not involved in plant movement across the landscape, measurements of effective dispersal and landscape genetics analyses should consider path effects due to dispersal vector behavior, as well as the chance that transported seeds are deposited in suitable habitat.
Assessments of plant habitat quality, such as those generated by ecological niche modeling (ENM) (Warren and Seifert, 2011), may provide a method for separating the effects of destination from those of vector behavior (path effects) on the patterns of effective dispersal. ENM considers the distribution of populations of a species and predicts the probability of establishment across the landscape based on the environmental conditions of known occurrences. To generate a “habitat quality” layer for GIS analysis, population location data are analyzed in the context of landscape variables that may influence the growth and survival of the plant species. Hence, variables normally considered in ENM that might affect vector behavior, such as land-use type classifications (e.g., urbanization, agricultural use), tree canopy coverage, elevation, hydraulic features, and roads, are removed. Only variables that affect plant habitat suitability, such as soil (moisture content, composition, pH) and climatic (mean annual and monthly temperature and precipitation) variables, are retained. Site-specific climate data interpolated from weather station records are available from databases such as WorldClim (Fick and Hijmans, 2017) and PRISM (PRISM Climate Group, 2015). The scale of the analysis should also be considered during ENM, as some environmental variables may not change across shorter geographic distances. Many types of climate data are appropriate only when considering a large study range on the order of tens to hundreds of kilometers. When considering niches at a fine scale—on the order of less than 1 km—variables that influence the microhabitat, such as soil variables and vegetation cover, may be more appropriate for habitat quality assessments (e.g., Grasty et al., 2020).
ENM tools such as Maxent (Merow et al., 2013) combined with model selection create a spatially defined habitat suitability map (e.g., an ASCII raster file) that can be used in landscape resistance analyses. Significant landscape features may predict vector behavior in resistance models, whereas an ENM map contains information on plant-specific requirements across the landscape. Habitat quality has been associated with genetic structure (Murphy et al., 2010; Sork et al., 2010), but it is rarely incorporated into resistance analyses. By including an ENM habitat quality map along with layers that may affect vector behavior, we can quantify the individual contributions of plant habitat quality and vector behavior to effective dispersal rates (gene flow) across the geographic range of a plant species. This approach effectively delineates the path from destination effects and can provide a more thorough understanding of dispersal processes across different landscape types.
A Review of Preliminary Results
At present, data are available from three plant landscape genetic studies that attempt to delineate the effects of seed dispersal vector behavior from those of suitable habitat distribution. In all three studies, whole chloroplast genome capture was used prior to Illumina sequencing, and the resulting SNP (single nucleotide polymorphism) data were used to estimate network phylogenies and haplotype frequencies using the methods described in Kohrn et al. (2017); Table 1. All three study species were annuals, maximizing the potential for detecting the effects of landscapes modified by humans over the past 100 years on effective seed dispersal patterns. The studies were conducted at two spatial scales. Plagiobothrys nothofulvus (rusty popcorn flower) is a small, gravity-dispersed annual species that was sampled at a fine scale (10–300 m) across a mosaic of vernal pool, prairie, and shrub vegetation types (Grasty et al., 2020; Figure 3). The area sampled for this species did not show substantial evidence of human disturbance or modification, and in this case, plant density was used as an indicator of habitat suitability. The sea blush (Plectritis congesta) is a gravity-dispersed annual that was sampled at a larger scale (40 m to 20 km) in a region that consisted of a mosaic of natural and human-modified landscapes (agricultural, natural, and urban areas). The wind-dispersed species Achyrachaena mollis (soft blow wives) was sampled across the same region as P. congesta (see Supplemental Information). For the latter two species, ENM was used to generate a GIS layer for habitat quality.
In all three studies, there were effects of landscape layers on patterns of effective seed dispersal, and geographic distance was a poor predictor of genetic similarity patterns among populations (Table 2). Counterintuitively, a positive relationship between habitat quality and dispersal resistance was found in all three species: low-quality habitat appeared to act as a conduit for gene flow, and regions of high habitat quality displayed reduced rates of gene flow. There are two possible explanations for this phenomenon. One explanation considers the differences in the shapes of the dispersal kernel and the distribution of effective dispersal (Figure 1A). The largest proportion of dispersal events occur over short distances, and in a continuous high-quality habitat, most offspring establish close to their maternal parents, reducing the mean effective dispersal distance. Dispersal across fragmented habitats, on the other hand, generates a biased effective dispersal kernel because intermediate distance dispersal events are absent (Figure 1C). Plants are unlikely to grow on low-quality habitat, and seeds may have to travel large distances to germinate in suitable habitat. Due to the nature of resistance model optimization, low-quality habitat appears to be a conduit, as it connects populations of similar genetic composition. Because genetic methods are only able to quantify effective dispersal among established populations, landscape genetic models may be overestimating dispersal rates across low-quality habitat.
Table 2.
Landscape Genetic Analyses for Three Species.
| Species | A. mollis | P. congesta | P. nothofulvus |
|---|---|---|---|
| 1st significant feature | Habitat quality | Habitat quality | Vegetation type + plant density + vole trails |
| Conduit or barrier on 1st feature surface | Barriera | Barrierb | Conduit + barrier + conduit |
| 1st feature marginal R2 | 0.1139 | 0.3307 | 0.1950 |
| 1st feature AICc | −255.597 | −165.738 | −142.126 |
| 2nd significant feature | Elevation | Rivers | Plant density + vole trails |
| Conduit or barrier on 2nd feature surface | Conduitc | Conduit | Barrier + conduit |
| 2nd feature marginal R2 | 0.0988 | 0.0710 | 0.1170 |
| 2nd feature AICc | −253.686 | −162.557 | −140.033 |
| Distance marginal R2 | <0.01 | 0.0307 | 0.079 |
| Distance AICc | −250.974 | −158.023 | −138.765 |
These results describe the two best models to explain genetic distance among populations and include geographic distance models as a comparison. Model selection was conducted using the small-sample corrected Akaike Information Criterion (AICc). Achyrachaena mollis and Plectritis congesta were sampled across a meso-scale range, whereas Plagiobothrys nothofulvus was sampled at a fine scale within a single prairie. Estimates of habitat quality were calculated for A. mollis and P. congesta using the ecological niche modeling program, Maxent. ENM training layers included average annual precipitation, soil content (percentage of clay), maximum annual temperature, mean annual temperature, minimum annual temperature, and percentage of soil moisture. Population occurrence data for ENM were concatenated using our own sampling sites and historical herbarium records. For P. nothofulvus, plant density was used as the best estimate of habitat quality. Although analyses of all three species included interactions among layers, significant interactions among features were found only for P. nothofulvus.
Continuous variable best fit to an inverse-reverse Ricker function.
Continuous variable best fit to an inverse-reverse monomolecular function.
Continuous variable best fit to an inverse Ricker function.
An alternative explanation for the higher rates of effective dispersal across low-quality habitat is the response of biotic dispersal vectors to landscape features, particularly in cases where low-quality habitat corresponds to the presence of human-modified landscapes. For example, white-tailed deer tend to move quickly along wildlife corridors but linger and move in more random patterns within habitat fragments (Kilpatrick and Spohr, 2000). This phenomenon has also been observed in several butterfly and bird species, as mobile organisms may disperse more rapidly through areas of less suitable habitat due to a reduction in available resources (Winker et al., 1995; Haddad and Tewksbury, 2005; Kuefler et al., 2010). Because animals contribute to the seed dispersal of a wide range of plant species, their behavior may increase the rates of effective dispersal across regions of low-quality habitat and increase the chances of seed deposition within habitat fragments. Consequently, the rates of effective dispersal are enhanced in regions where high-quality habitat has been fragmented by human land use.
High-quality habitat appeared to act as a barrier for all three species described above. This would be expected for the two barochoric species if there was significant secondary dispersal by mammals. In the case of P. nothofulvus, which was studied at a fine scale, voles and other small mammals that use rodent runways appeared to be responsible for the majority of longer distance dispersal events (Grasty et al., 2020). Because P. congesta was studied at a meso scale, the higher effective dispersal rates across low-quality habitat probably reflect secondary dispersal and the movement patterns of deer and elk, which are common in the Rogue River Valley where this study was conducted. This hypothesis is supported by the identification of riparian areas (rivers) as conduits for the effective dispersal of P. congesta (Table 2), a pattern consistent with the movement behavior of ungulates (Kilpatrick and Spohr, 2000; Vellend et al., 2003). It is less clear why A. mollis, which has wind-dispersed seeds, should respond to higher quality habitat as if it were a barrier. In this case, the effect of habitat on effective dispersal was much weaker, and it may be that this reflects the bias associated with effective dispersal compared with the dispersal kernel, as discussed above. The seeds of A. mollis are much larger than those of the other two species, and they may have less secondary dispersal by biotic vectors. Based on the results presented here, it appears that both effective dispersal bias due to destination effects and biotic vector behavior in response to landscape features may contribute to limited dispersal distances within high-quality habitat fragments. Further studies with other species may provide additional insights into the effects of landscapes on the patterns and frequencies of LDD.
Recommendations and Future Directions
It is clear from the above discussion that plant landscape genetics studies have much to offer to address a wide range of questions about processes of gene flow and ecological aspects of dispersal and vector behavior. Although whole-genome assays of chloroplasts can provide substantial numbers of haplotypes, the utility of this marker may be limited because there is no recombination, and it effectively acts as a single locus. Genetic distance estimates among populations for haplotype data can be made using parameters such as paired FST, which accounts for haplotype frequencies, or paired NST, which accounts for both frequencies and phylogenetic relatedness among haplotypes (Pons and Petit, 1996). A comparison of these two genetic structure estimates provides information on the relative importance of mutation-drift processes versus dispersal for the determination of genetic differentiation among populations across a study region.
Nuclear markers can provide information on larger numbers of independently segregating genomic regions and consequently provide much greater power for estimating genetic distance than chloroplast assays. In general, one hundred variable SNPs or at least eight variable microsatellite loci are adequate to resolve genetic differentiation levels among populations (Turakulov and Easteal, 2003; Arthofer et al., 2018). A wide range of genetic distance measures based on nuclear markers are available (see reviews by Shirk et al., 2017; Storfer et al., 2018). The disadvantage of using only nuclear markers is that the variation detected may reflect both pollen and seed dispersal. Ideally, both nuclear and cytoplasmic markers would be used to delineate the separate effects of pollen and seed dispersal on patterns of gene flow. However, with the exception of wind-pollinated and perhaps hawkmoth- (Sphingidae) and bat-pollinated species, genetic differentiation measured by nuclear markers at the meso scale may provide accurate estimates of effective seed dispersal processes because most biotic pollinators forage over smaller areas and may be unlikely to move among habitat fragments. A best practice may be to use both maternally inherited cytoplasmic and biparentally inherited nuclear markers when working at scales where average effective dispersal distances are likely to be similar for seeds and pollen. However, nuclear markers should be favored in cases where pollen dispersal is more likely to be limited relative to seed dispersal.
The preliminary results described above indicate that including a GIS layer that acts as an estimate of plant habitat quality may be an effective means of separating path and destination effects on patterns of effective dispersal. Habitat layer (or the plant density layer in the case of P. nothofulvus) explained significant levels of variation in the genetic distance among sample sites in two of the three studies. By using only variables that are unlikely to affect vector behavior for niche modeling, it should be possible to separate the effects of plant establishment and reproduction from those of vector movement. With an adequate number of plant occurrences, the layers generated using this method will reflect habitat conditions independent of human modifications, tree canopy cover, and other features that may affect vector behavior.
Although the methods described above are intended to distinguish path and destination effects on the patterns of effective dispersal, it is important to note that there can be overlap between plant and vector habitat suitability. This may be particularly true in cases where the plant species is limited to habitat fragments in a matrix of human-modified landscapes, such as the example described above for P. congesta and its probable ungulate seed dispersal vectors. On the other hand, some seed vectors such as frugivorous birds and bats may be less restricted to fragments of natural vegetation and are able to move more freely and to utilize food resources in urban landscapes (Tsoar et al., 2011; Grafius et al., 2017). Similarly, insect pollinators may be primarily limited to foraging in natural vegetation fragments in agricultural regions, but many are known to utilize ornamental plants in urban landscapes as food resources (Harrison and Winfree, 2015). Understanding likely dispersal vectors based on flower, seed, and fruit morphologies and considering their possible responses to landscape features in the study region will be important to inform robust sampling designs for landscape genetic studies.
The combination of genome-wide SNP assays, GIS, and ENM provides methods for generating unprecedented levels of information on processes of gene flow and dispersal within and among plant populations. Applications of the approaches described above have the potential to inform studies of genetic structure conducted across heterogeneous landscapes, and studies of conservation ecology and genetics inform prudent management decisions. These applications are particularly relevant as humans strive to mitigate the impacts of their activities on the survival of populations and species. Landscape genetic studies in plants can provide information on dispersal potential across modified landscapes and identify particular situations and species of plants and animals that are candidates for active relocation and restoration efforts.
Funding
This work was supported by funds from a US National Science Foundation MacroSystems Biology award (1340746) and from the Portland State University College of Liberal Arts and Sciences.
Author Contributions
Conceptualization, M.B.C. and E.C.H.; Methodology, M.B.C. and E.C.H.; Formal Analysis, E.C.H.; Investigation, M.B.C. and E.C.H.; Data Curation, E.C.H.; Writing – Original Draft, M.B.C. and E.C.H.; Writing – Review & Editing, M.B.C. and E.C.H.; Visualization, M.B.C. and E.C.H.; Funding Acquisition, M.B.C.
Acknowledgments
We thank T. Arredondo, N. Diaz, K. Gerloff, M. Grasty, A. Hamilton, K. Kline, H. Machiorlete, J. Persinger, A. Pheil, J. Schwoch, and P. Thompson for assistance with field and lab work, and B. Kohrn for assistance with bioinformatics. N. Diaz assisted with ENM analyses, T. Arredondo generated the circuit map, and J. Persinger assembled the chloroplast genomes for the six species listed in Table 1. K. Berg, R. Bice, N. Diaz, and J. Schwoch made helpful comments on the manuscript. No conflict of interest declared.
Published: July 20, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Albert A., Auffret A.G., Cosyns E., Cousins S.A.O., D'hondt B., Eichberg C., Eycott A.E., Heinken T., Hoffmann M., Jaroszewicz B. Seed dispersal by ungulates as an ecological filter: a trait-based meta-analysis. Oikos. 2015;124:1109–1120. [Google Scholar]
- Alvarado-Serrano D.F., Van Etten M.L., Chang S.M., Baucom R.S. The relative contribution of natural landscapes and human-mediated factors on the connectivity of a noxious invasive weed. Heredity. 2019;122:29–40. doi: 10.1038/s41437-018-0106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.D., Epperson B.K., Fortin M.-J., Holdregger R., James P.M.A., Rosenberg M.S., Scribner K.T., Spear S. Considering spatial and temporal scale in landscape-genetic studies of gene flow. Mol. Ecol. 2010;19:3565–3575. doi: 10.1111/j.1365-294X.2010.04757.x. [DOI] [PubMed] [Google Scholar]
- Arredondo T.M., Marchini G.L., Cruzan M.B. Evidence for human-mediated range expansion and gene flow in an invasive grass. Proc. R. Soc. B Biol. Sci. 2018;285:20181125. doi: 10.1098/rspb.2018.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthofer W., Heussler C., Krapf P., Schlick-Steiner B.C., Steiner F.M. Identifying the minimum number of microsatellite loci needed to assess population genetic structure: a case study in fly culturing. Fly (Austin) 2018;12:13–22. doi: 10.1080/19336934.2017.1396400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffret A.G., Rico Y., Bullock J.M., Hooftman D.A.P., Pakeman R.J., Soons M.B., Suarez-Esteban A., Traveset A., Wagner H.H., Cousins S.A.O. Plant functional connectivity - integrating landscape structure and effective dispersal. J. Ecol. 2017;105:1648–1656. [Google Scholar]
- Augspurger C.K. Morphology and dispersal potential of wind-dispersed diaspores of neotropical trees. Am. J. Bot. 1986;73:353–363. [Google Scholar]
- Avise J., Arnold J., Ball R., Eldredge B., Lamb T., Neigel J., Reeb C., Saunders N. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 1987;18:489–522. [Google Scholar]
- Bacles C.F.E., Lowe A.J., Ennos R.A. Effective seed dispersal across a fragmented landscape. Science. 2006;311:628. doi: 10.1126/science.1121543. [DOI] [PubMed] [Google Scholar]
- Beninde J., Feldmeier S., Veith M., Hochkirch A. Admixture of hybrid swarms of native and introduced lizards in cities is determined by the cityscape structure and invasion history. Proc. R. Soc. B Biol. Sci. 2018;285 doi: 10.1098/rspb.2018.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkman C.W. Wind dispersal capacity of pine seeds and the evolution of different seed dispersal modes in pines. Oikos. 1995;73:221–224. [Google Scholar]
- Biswas S.R., Wagner H.H. Landscape contrast: a solution to hidden assumptions in the metacommunity concept? Landscape Ecol. 2012;27:621–631. [Google Scholar]
- Bohrer G., Katul G.G., Nathan R., Walko R.L., Avissar R. Effects of canopy heterogeneity, seed abscission and inertia on wind-driven dispersal kernels of tree seeds. J. Ecol. 2008;96:569–580. [Google Scholar]
- Bonte D., Van Dyck H., Bullock J.M., Coulon A., Delgado M., Gibbs M., Lehouck V., Matthysen E., Mustin K., Saastamoinen M. Costs of dispersal. Biol. Rev. 2012;87:290–312. doi: 10.1111/j.1469-185X.2011.00201.x. [DOI] [PubMed] [Google Scholar]
- Born C., Hardy O.J., Chevalieer M.-H., Ossari S., Atteke C., Wickings E.J., Hossaert-Mickey M. Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation. Mol. Ecol. 2008;17:2041–2050. doi: 10.1111/j.1365-294X.2007.03685.x. [DOI] [PubMed] [Google Scholar]
- Bradley B.A., Curtis C.A., Chambers J.C. Bromus response to climate and projected changes with climate change. In: Germino M.J., Chambers J.C., Brown C.S., editors. Exotic Brome-Grasses in Arid and Semiarid Ecosystems of the Western US: Causes, Consequences, and Management Implications. Springer; New York: 2016. pp. 257–274. [Google Scholar]
- Browne L., Ottewell K., Sork V.L., Karubian J. The relative contributions of seed and pollen dispersal to gene flow and genetic diversity in seedlings of a tropical palm. Mol. Ecol. 2018;27:3159–3173. doi: 10.1111/mec.14768. [DOI] [PubMed] [Google Scholar]
- Bullock J.M., Clarke R.T. Long distance seed dispersal by wind: measuring and modelling the tail of the curve. Oecologia. 2000;124:506–521. doi: 10.1007/PL00008876. [DOI] [PubMed] [Google Scholar]
- Cain M.L., Milligan B.G., Strand A.E. Long-distance seed dispersal in plant populations. Am. J. Bot. 2000;87:1217–1227. [PubMed] [Google Scholar]
- Cameron A.C., Page R.B., Watling J.I., Hickerson C.A.M., Anthony C.D. Using a comparative approach to investigate the relationship between landscape and genetic connectivity among woodland salamander populations. Conserv. Genet. 2019;20:1265–1280. [Google Scholar]
- Cane J.H., Love B. Pollen carryover between sequential foraging trips by a solitary bee: implications for distant outcrossing. J. Pollination Ecol. 2019;24 doi: 10.26786/1920-7603(2018)15. [DOI] [Google Scholar]
- Carvalho C.S., Lanes E.C.M., Silva A.R., Caldeira C.F., Carvalho N., Gastauer M., Imperatriz-Fonseca V.L., Nascimento W., Oliveira G., Siqueira J.O. Habitat loss does not always entail negative genetic consequences. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castric V., Bonney F., Bernatchez L. Landscape structure and hierarchical genetic diversity in the brook char, Salvelinus fontinalis. Evolution. 2001;55:1016–1028. doi: 10.1554/0014-3820(2001)055[1016:lsahgd]2.0.co;2. 1013. [DOI] [PubMed] [Google Scholar]
- Charney N.D. Evaluating expert opinion and spatial scale in an amphibian model. Ecol. Model. 2012;242:37–45. [Google Scholar]
- Chybicki I.J., Oleksa A. Seed and pollen gene dispersal in Taxus baccata, a dioecious conifer in the face of strong population fragmentation. Ann. Bot. 2018;122:409–421. doi: 10.1093/aob/mcy081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.S. Why trees migrate so fast: confronting theory with dispersal biology and the paleorecord. Am. Nat. 1998;152:204–224. doi: 10.1086/286162. [DOI] [PubMed] [Google Scholar]
- Clark J.S., Fastie C., Hurtt G., Jackson S.T., Johnson C., King G.A., Lewis M., Lynch J., Pacala S., Prentice C. Reid's paradox of rapid plant migration. Bioscience. 1998;48:13–24. [Google Scholar]
- Cortes M.C., Uriarte M. Integrating frugivory and animal movement: a review of the evidence and implications for scaling seed dispersal. Biol. Rev. Camb. Philos. Soc. 2013;88:255–272. doi: 10.1111/j.1469-185X.2012.00250.x. [DOI] [PubMed] [Google Scholar]
- Cousens R.D., Dytham C., Law R. Oxford University Press; New York: 2008. Dispersal in Plants - A Population Perspective. [Google Scholar]
- Cox C.B., Moore P.D. John Wiley; New York: 2010. Biogeography: An Ecological and Evolutionary Approach. [Google Scholar]
- Crisci J.V. The voice of historical biogeography. J. Biogeogr. 2001;28:157–168. [Google Scholar]
- Cruzan M.B. Pollen tube attrition in Erythronium grandiflorum. Am. J. Bot. 1989;76:562–570. [Google Scholar]
- Cruzan M.B. Population size and fragmentation thresholds for the maintenance of genetic diversity in the endemic, Scutellaria montana (Lamiaceae) Evolution. 2001;55:1569–1580. doi: 10.1111/j.0014-3820.2001.tb00676.x. [DOI] [PubMed] [Google Scholar]
- Cruzan M.B. Oxford University Press; New York: 2018. Evolutionary Biology - A Plant Perspective. [Google Scholar]
- Cruzan M.B. How to make a weed - the saga of the slender false brome invasion in the North American west and lessons for the future. BioScience. 2019;69:496–507. [Google Scholar]
- Cruzan M.B., Arnold M.L., Carney S.E., Wollenberg K.R. cpDNA inheritance in interspecific crosses and evolutionary inference in Louisiana irises. Am. J. Bot. 1993;80:344–350. [Google Scholar]
- Cruzan M.B., Templeton A.R. Paleoecology and coalescence: phylogeographic analysis of hypotheses from the fossil record. Trends Ecol. Evol. 2000;15:491–496. doi: 10.1016/s0169-5347(00)01998-4. [DOI] [PubMed] [Google Scholar]
- Damschen E.I., Baker D.V., Bohrer G., Nathan R., Orrock J.L., Turner J.R., Brudvig L.A., Haddad N.M., Levey D.J., Tewksbury J.J. How fragmentation and corridors affect wind dynamics and seed dispersal in open habitats. Proc. Natl. Acad. Sci. U S A. 2014;111:3484–3489. doi: 10.1073/pnas.1308968111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danner N., Molitor A.M., Schiele S., Härtel S., Steffan-Dewenter I. Season and landscape composition affect pollen foraging distances and habitat use of honey bees. Ecol. Appl. 2016;26:1920–1929. doi: 10.1890/15-1840.1. [DOI] [PubMed] [Google Scholar]
- Di-Giovanni F., Kevan P.G. Factors affecting pollen dynamics and its importance to pollen contamination: a review. Can. J. For. Res. 1991;21:1155–1170. [Google Scholar]
- Dileo M.F., Husby A., Saastamoinen M. Landscape permeability and individual variation in a dispersal-linked gene jointly determine genetic structure in the Glanville fritillary butterfly. Evol. Lett. 2018;2:544–556. doi: 10.1002/evl3.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diniz-Filho J.A.F., De Campos Telles M.P. Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv. Biol. 2002;16:924–935. [Google Scholar]
- Dutech C., Sork V.L., Irwin A.J., Smouse P.E., Davis F.W. Gene flow and fine-scale genetic structure in a wind-pollinated tree species, Quercus lobata (Fagaceaee) Am. J. Bot. 2005;92:252–261. doi: 10.3732/ajb.92.2.252. [DOI] [PubMed] [Google Scholar]
- Echeverría-Londoño S., Enquist B.J., Neves D.M., Violle C., Boyle B., Kraft N.J.B., Maitner B.S., McGill B., Peet R.K., Sandel B. Plant functional diversity and the biogeography of biomes in North and South America. Front. Ecol. Evol. 2018;6 doi: 10.3389/fevo.2018.00219. [DOI] [Google Scholar]
- Ellis J.R., Bentley K.E., McCauley D.E. Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus. Heredity. 2008;100:574–580. doi: 10.1038/hdy.2008.11. [DOI] [PubMed] [Google Scholar]
- Ennos R.A. Estimating the relative rates of pollen and seed migration among plant populations. Heredity. 1994;72:250–259. [Google Scholar]
- Eriksson O. Evolution of angiosperm seed disperser mutualisms: the timing of origins and their consequences for coevolutionary interactions between angiosperms and frugivores. Biol. Rev. 2016;91:168–186. doi: 10.1111/brv.12164. [DOI] [PubMed] [Google Scholar]
- Fernando H.J.S., Zajic D., Di Sabatino S., Dimitrova R., Hedquist B., Dallman A. Flow, turbulence, and pollutant dispersion in urban atmospheres. Phys. Fluids. 2010;22:051301. [Google Scholar]
- Fick S.E., Hijmans R.J. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017;37:4302–4315. [Google Scholar]
- Finnigan J. Turbulence in plant canopies. Annu. Rev. Fluid Mech. 2000;32:519–571. [Google Scholar]
- Friedman J., Barrett S.C.H. Wind of change: new insights on the ecology and evolution of pollination and mating in wind-pollinated plants. Ann. Bot. 2009;103:1515–1527. doi: 10.1093/aob/mcp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C., Jordano P., Godoy J.A. Contemporary pollen and seed dispersal in a Prunus mahaleb population: patterns in distance and direction. Mol. Ecol. 2007;16:1947–1955. doi: 10.1111/j.1365-294X.2006.03126.x. [DOI] [PubMed] [Google Scholar]
- Gerber S., Chadœuf J., Gugerli F., Lascoux M., Buiteveld J., Cottrell J., Dounavi A., Fineschi S., Forrest L.L., Fogelqvist J. High rates of gene flow by pollen and seed in oak populations across Europe. PLoS One. 2014;9:e85130. doi: 10.1371/journal.pone.0085130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafius D.R., Corstanje R., Siriwardena G.M., Plummer K.E., Harris J.A. A bird’s eye view: using circuit theory to study urban landscape connectivity for birds. Landscape Ecol. 2017;32:1771–1787. doi: 10.1007/s10980-017-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram W.K., Sork V.L. Association between environmental and genetic heterogeneity in forest tree populations. Ecology. 2001;82:2012–2021. [Google Scholar]
- Grasty M.R., Thompson P.G., Pheil A.E., Hendrickson E.C., Cruzan M.B. Fine-scale habitat heterogeneity and vole runways influence seed dispersal in Plagiobothrus nothofulvus. Am. J. Bot. 2020;107:413–422. doi: 10.1002/ajb2.1433. [DOI] [PubMed] [Google Scholar]
- Greene D.F., Quesada M. The differential effect of updrafts, downdrafts and horizontal winds on the seed abscission of Tragopogon dubius. Funct. Ecol. 2011;25:468–472. [Google Scholar]
- Haddad N.M., Tewksbury J.J. Low-quality habitat corridors as movement conduits for two butterfly species. Ecol. Appl. 2005;15:250–257. [Google Scholar]
- Harder L.D., Aizen M.A., Richards S.A. The population ecology of male gametophytes: the link between pollination and seed production. Ecol. Lett. 2016;19:497–509. doi: 10.1111/ele.12596. [DOI] [PubMed] [Google Scholar]
- Harper J.L. Academic Press; London: 1977. Population Biology of Plants. [Google Scholar]
- Harrison T., Winfree R. Urban drivers of plant-pollinator interactions. Funct. Ecol. 2015;29:879–888. [Google Scholar]
- Hickerson M.J., Carstens B.C., Cavender-Bares J., Crandall K.A., Graham C.H., Johnson J.B., Rissler L., Victoriano P.F., Yoder A.D. Phylogeography's past, present, and future: 10 years after Avise, 2000. Mol. Phylogenet. Evol. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Holderegger R., Buehler D., Gugerli F., Manel S. Landscape genetics of plants. Trends Plant Sci. 2010;15:675–683. doi: 10.1016/j.tplants.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Howe H.F., Miriti M.N. When seed dispersal matters. Bioscience. 2004;54:651–660. [Google Scholar]
- Howe H.F., Smallwood J. Ecology of seed dispersal. Annu. Rev. Ecol. Syst. 1982;13:P201–P228. [Google Scholar]
- Hutchison D.W., Templeton A.R. Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the distribution of genetic variability. Evolution. 1999;53:1898–1914. doi: 10.1111/j.1558-5646.1999.tb04571.x. [DOI] [PubMed] [Google Scholar]
- Jansen P.A., Visser M.D., Joseph Wright S., Rutten G., Muller-Landau H.C. Negative density dependence of seed dispersal and seedling recruitment in a neotropical palm. Ecol. Lett. 2014;17:1111–1120. doi: 10.1111/ele.12317. [DOI] [PubMed] [Google Scholar]
- Jenkins D.G., Carey M., Czerniewska J., Fletcher J., Hether T., Jones A., Knight S., Knox J., Long T., Mannino M. A meta-analysis of isolation by distance: relic or reference standard for landscape genetics? Ecography. 2010;33:315–320. [Google Scholar]
- Johnson J.S., Cantrell R.S., Cosner C., Hartig F., Hastings A., Rogers H.S., Schupp E.W., Shea K., Teller B.J., Yu X. Rapid changes in seed dispersal traits may modify plant responses to global change. AoB Plants. 2019;11 doi: 10.1093/aobpla/plz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J.S., Gaddis K.D., Cairns D.M., Konganti K., Krutovsky K.V. Landscape genomic insights into the historic migration of mountain hemlock in response to Holocene climate change. Am. J. Bot. 2017;104:439–450. doi: 10.3732/ajb.1600262. [DOI] [PubMed] [Google Scholar]
- Jones F.A., Muller-Landau H.C. Measuring long-distance seed dispersal in complex natural environments: an evaluation and integration of classical and genetic methods. J. Ecol. 2008;96:642–652. [Google Scholar]
- Jongejans E., Skarpaas O., Shea K. Dispersal, demography and spatial population models for conservation and control management. Perspect. Plant Ecol. Evol. Syst. 2008;9:153–170. [Google Scholar]
- Katzner T.E., Arlettaz R. Evaluating contributions of recent tracking-based animal movement ecology to conservation management. Front. Ecol. Evol. 2020;7 doi: 10.3389/fevo.2019.00519. [DOI] [Google Scholar]
- Keyghobadi N., Roland J., Strobeck C. Influence of landscape on the population genetic structure of the alpine butterfly Parnassius smintheus (Papilionidae) Mol. Ecol. 1999;8:1481–1495. doi: 10.1046/j.1365-294x.1999.00726.x. [DOI] [PubMed] [Google Scholar]
- Khimoun A., Peterman W., Eraud C., Faivre B., Navarro N., Garnier S. Landscape genetic analyses reveal fine-scale effects of forest fragmentation in an insular tropical bird. Mol. Ecol. 2017;26:4906–4919. doi: 10.1111/mec.14233. [DOI] [PubMed] [Google Scholar]
- Kilpatrick H.J., Spohr S.M. Movements of female white-tailed deer in a suburban landscape: a management perspective. Wildl. Soc. Bull. 2000;28:1038–1045. [Google Scholar]
- Klein E.K., Bontemps A., Oddou-Muratorio S. Seed dispersal kernels estimated from genotypes of established seedlings: does density-dependent mortality matter? Methods Ecol. Evol. 2013;4:1059–1069. [Google Scholar]
- Knowles L.L., Maddison W.P. Statistical phylogeography. Mol. Ecol. 2002;11:2623–2635. doi: 10.1046/j.1365-294x.2002.01637.x. [DOI] [PubMed] [Google Scholar]
- Kohrn B.F., Persinger J.M., Cruzan M.B. An efficient pipeline to generate data for studies in plastid population genomics and phylogeography. Appl. Plant Sci. 2017;5:1700053. doi: 10.3732/apps.1700053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh H., Whigham D.F. Microgeographic genetic structure and gene flow in Hibiscus moscheutos (Malvaceae) populations. Am. J. Bot. 1997;84:1285–1293. [PubMed] [Google Scholar]
- Kuefler D., Hudgens B., Haddad N.M., Morris W.F., Thurgate N. The conflicting role of matrix habitats as conduits and barriers for dispersal. Ecology. 2010;91:944–950. doi: 10.1890/09-0614.1. [DOI] [PubMed] [Google Scholar]
- Kunde M.N., Martins R.F., Premier J., Fickel J., Forster D.W. Population and landscape genetic analysis of the Malayan sun bear Helarctos malayanus. Conserv. Genet. 2019 doi: 10.1007/s10592-019-01233-w. [DOI] [Google Scholar]
- Latta R.G., Linhart Y.B., Fleck D., Elliot M. Direct and indirect estimates of seed versus pollen movement within a population of ponderosa pine. Evolution. 1998;52:61–67. doi: 10.1111/j.1558-5646.1998.tb05138.x. [DOI] [PubMed] [Google Scholar]
- Liu M., Compton S.G., Peng F.-E., Zhang J., Chen X.-Y. Movements of genes between populations: are pollinators more effective at transferring their own or plant genetic markers? Proc. Biol. Sci. 2015;282 doi: 10.1098/rspb.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmus K., Paal T., Liira J. Long-term colonization ecology of forest-dwelling species in a fragmented rural landscape - dispersal versus establishment. Ecol. Evol. 2014;4:3113–3126. doi: 10.1002/ece3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco A., Goncalves J., Carvalho F., Wang I.J., Velo-Anton G. Comparative landscape genetics reveals the evolution of viviparity reduces genetic connectivity in fire salamanders. Mol. Ecol. 2019;28:4573–4591. doi: 10.1111/mec.15249. [DOI] [PubMed] [Google Scholar]
- MacArthur R.H., Wilson E.O. Princeton University Press; Princeton, NJ: 1967. The Theory of Island Biogeography. [Google Scholar]
- Manel S., Berthoud F., Bellemain E., Gaudeul M., Luikart G., Swenson J.E., Waits L.P., Taberlet P. A new individual-based spatial approach for identifying genetic discontinuities in natural populations. Mol. Ecol. 2007;16:2031–2043. doi: 10.1111/j.1365-294X.2007.03293.x. [DOI] [PubMed] [Google Scholar]
- Manel S., Schwartz M.K., Luikart G., Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 2003;18:189–197. [Google Scholar]
- McRae B.H., Beier P. Circuit theory predicts gene flow in plant and animal populations. Proc. Natl. Acad. Sci. U S A. 2007;104:19885–19890. doi: 10.1073/pnas.0706568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae B.H., Dickson B.G., Keitt T.H., Shah V.B. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology. 2008;89:2712–2724. doi: 10.1890/07-1861.1. [DOI] [PubMed] [Google Scholar]
- Melo A.T.O., Hale I. ‘Apparent’: a simple and flexible R package for accurate SNP-based parentage analysis in the absence of guiding information. BMC Bioinformatics. 2019;20:108. doi: 10.1186/s12859-019-2662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merow C., Smith M.J., Silander J.A., Jr. A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography. 2013;36:1058–1069. [Google Scholar]
- Morente-López J., García C., Lara-Romero C., García-Fernández A., Draper D., Iriondo J.M. Geography and environment shape landscape genetics of Mediterranean alpine species Silene ciliata Poiret. (Caryophyllaceae) Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M.A., Dezzani R., Pilliod D.S., Storfer A. Landscape genetics of high mountain frog metapopulations. Mol. Ecol. 2010;19:3634–3649. doi: 10.1111/j.1365-294X.2010.04723.x. [DOI] [PubMed] [Google Scholar]
- Nason J.D., Hamrick J.L. Reproductive and genetic consequences of forest fragmentation: two case studies of neotropical canopy trees. J. Hered. 1997;88:264–276. [Google Scholar]
- Nathan R. Long-distance dispersal of plants. Science. 2006;313:786–788. doi: 10.1126/science.1124975. [DOI] [PubMed] [Google Scholar]
- Nathan R. An emerging movement ecology paradigm. Proc. Natl. Acad. Sci. U S A. 2008;105:19050–19051. doi: 10.1073/pnas.0808918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R., Katul G.G. Foliage shedding in deciduous forests lifts up long-distance seed dispersal by wind. Proc. Natl. Acad. Sci. U S A. 2005;102:8251–8256. doi: 10.1073/pnas.0503048102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R., Katul G.G., Horn H.S., Thomas S.M., Oren R., Avissar R., Pacala S.W., Levin S.A. Mechanisms of long-distance dispersal of seeds by wind. Nature. 2002;418:409–413. doi: 10.1038/nature00844. [DOI] [PubMed] [Google Scholar]
- Nathan R., Klein E., Robledo-Arnuncio J.J., Revilla E. Dispersal kernels: review. In: Clobert J., Baguette M., Benton T., Bullock J.M., editors. Dispersal and Spatial Ecology. Oxford University Press; New York: 2012. pp. 187–210. [Google Scholar]
- Nathan R., Muller-Landau H.C. Spatial patterns of seed dispersal, their determinants and consequences for recruitment. Trends Ecol. Evol. 2000;15:278–285. doi: 10.1016/s0169-5347(00)01874-7. [DOI] [PubMed] [Google Scholar]
- Nathan R., Perry G., Cronin J.T., Strand A.E., Cain M.L. Methods for estimating long-distance dispersal. Oikos. 2003;103:261–273. [Google Scholar]
- Nathan R., Schurr F.M., Spiegel O., Steinitz O., Trakhtenbrot A., Tsoar A. Mechanisms of long-distance seed dispersal. Trends Ecol. Evol. 2008;23:638–647. doi: 10.1016/j.tree.2008.08.003. [DOI] [PubMed] [Google Scholar]
- O'Connell L.M., Mosseler A., Rajora O.P. Extensive long-distance pollen dispersal in a fragmented landscape maintains genetic diversity in white spruce. J. Hered. 2007;98:640–645. doi: 10.1093/jhered/esm089. [DOI] [PubMed] [Google Scholar]
- Okubo A., Levin S.A. A theoretical framework for data analysis of wind dispersal of seeds and pollen. Ecology. 1989;70:329–338. [Google Scholar]
- Osborne J.L., Martin A.P., Carreck N.L., Swain J.L., Knight M.E., Goulson D., Hale R.J., Sanderson R.A. Bumblebee flight distances in relation to the forage landscape. J. Anim. Ecol. 2008;77:406–415. doi: 10.1111/j.1365-2656.2007.01333.x. [DOI] [PubMed] [Google Scholar]
- Ouborg N.J., Piquot Y., Van Groenendael J.M. Population genetics, molecular markers and the study of dispersal in plants. J. Ecol. 1999;87:551–568. [Google Scholar]
- Pérez-Espona S., McLeod J.E., Franks N.R. Landscape genetics of a top neotropical predator. Mol. Ecol. 2012;21:5969–5985. doi: 10.1111/mec.12088. [DOI] [PubMed] [Google Scholar]
- Peterman W.E. ResistanceGA: an R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 2018;9:1638–1647. [Google Scholar]
- Peterman W.E., Connette G.M., Semlitsch R.D., Eggert L.S. Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol. Ecol. 2014;23:2402–2413. doi: 10.1111/mec.12747. [DOI] [PubMed] [Google Scholar]
- Peterman W.E., Winiarski K.J., Moore C.E., Carvalho C.D., Gilbert A.L., Spear S.F. A comparison of popular approaches to optimize landscape resistance surfaces. Landscape Ecol. 2019;34:2197–2208. [Google Scholar]
- Piertney S.B., MacColl A.D., Bacon P.J., Dallas J.F. Local genetic structure in red grouse (Lagopus lagopus scoticus): evidence from microsatellite DNA markers. Mol. Ecol. 1998;7:1645–1654. doi: 10.1046/j.1365-294x.1998.00493.x. [DOI] [PubMed] [Google Scholar]
- Pons O., Petit R.J. Measuring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996;144:1237–1245. doi: 10.1093/genetics/144.3.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poor E.E., Loucks C., Jakes A., Urban D.L. Comparing habitat suitability and connectivity modeling methods for conserving pronghorn migrations. PLoS One. 2012;7:e49390. doi: 10.1371/journal.pone.0049390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRISM Climate Group . Oregon State University; Corvallis, OR: 2015. PRISM Gridded Climate Data. [Google Scholar]
- Rey P.J., Alcántara J.M. Recruitment dynamics of a fleshy-fruited plant (Olea europaea): connecting patterns of seed dispersal to seedling establishment. J. Ecol. 2000;88:622–633. [Google Scholar]
- Rissler L.J. Union of phylogeography and landscape genetics. Proc. Natl. Acad. Sci. U S A. 2016;113:8079–8086. doi: 10.1073/pnas.1601073113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H.S., Beckman N.G., Hartig F., Johnson J.S., Pufal G., Shea K., Zurell D., Bullock J.M., Cantrell R.S., Loiselle B. The total dispersal kernel: a review and future directions. AoB Plants. 2019;11 doi: 10.1093/aobpla/plz042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen O., Pyhajarvi T., Knurr T. Gene flow and local adaptation in trees. Annu. Rev. Ecol. Evol. Syst. 2007;38:595–619. [Google Scholar]
- Schnitzler J., Graham C.H., Dormann C.F., Schiffers K., Peter Linder H. Climatic niche evolution and species diversification in the Cape flora, South Africa. J. Biogeogr. 2012;39:2201–2211. [Google Scholar]
- Sears B. Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid. 1980;4:233–255. doi: 10.1016/0147-619x(80)90063-3. [DOI] [PubMed] [Google Scholar]
- Sedlacek J.F., Bossdorf O., Cortés A.J., Wheeler J.A., van Kleunen M. What role do plant–soil interactions play in the habitat suitability and potential range expansion of the alpine dwarf shrub Salix herbacea? Basic Appl. Ecol. 2014;15:305–315. [Google Scholar]
- Shirk A.J., Landguth E.L., Cushman S.A. A comparison of individual-based genetic distance metrics for landscape genetics. Mol. Ecol. Resour. 2017;17:1308–1317. doi: 10.1111/1755-0998.12684. [DOI] [PubMed] [Google Scholar]
- Shirk A.J., Wallin D.O., Cushman S.A., Rice C.G., Warheit K.I. Inferring landscape effects on gene flow: a new model selection framework. Mol. Ecol. 2010;19:3603–3619. doi: 10.1111/j.1365-294X.2010.04745.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and nonequilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Sork V.L., Davis F.W., Westfall R., Flint A., Ikegami M., Wang H., Grivet D. Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Mol. Ecol. 2010;19:3806–3823. doi: 10.1111/j.1365-294X.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- Sork V.L., Nason J., Campbell D.R., Fernandez J.F. Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol. Evol. 1999;14:219–224. doi: 10.1016/s0169-5347(98)01585-7. [DOI] [PubMed] [Google Scholar]
- Sork V.L., Smouse P.E. Genetic analysis of landscape connectivity in tree populations. Landscape Ecol. 2006;21:821–836. [Google Scholar]
- Spear S.F., Balkenhol N., Fortin M.-J., McRae B.H., Scribner K. Use of resistance surfaces for landscape genetic studies: considerations for parameterization and analysis. Mol. Ecol. 2010;19:3576–3591. doi: 10.1111/j.1365-294X.2010.04657.x. [DOI] [PubMed] [Google Scholar]
- Steinitz O., Troupin D., Vendramin G.G., Nathan R. Genetic evidence for a Janzen–Connell recruitment pattern in reproductive offspring of Pinus halepensis trees. Mol. Ecol. 2011;20:4152–4164. doi: 10.1111/j.1365-294X.2011.05203.x. [DOI] [PubMed] [Google Scholar]
- Storfer A., Murphy M.A., Spear S.F., Holderegger R., Waits L.P. Landscape genetics: where are we now? Mol. Ecol. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Storfer A., Patton A., Fraik A.K. Navigating the interface between landscape genetics and landscape genomics. Front. Genet. 2018;9 doi: 10.3389/fgene.2018.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S.H., Neale D.B., Wagner D.B. Genetics of the chloroplast in conifers: biotechnology research reveals some surprises. J. For. 1989;87:11–17. [Google Scholar]
- Suárez-Esteban A., Delibes M., Fedriani J.M. Barriers or corridors? The overlooked role of unpaved roads in endozoochorous seed dispersal. J. Appl. Ecol. 2013;50:767–774. [Google Scholar]
- Sullivan L.L., Li B., Miller T.E.X., Neubert M.G., Shaw A.K. Density dependence in demography and dispersal generates fluctuating invasion speeds. Proc. Natl. Acad. Sci. U S A. 2017;114:5053–5058. doi: 10.1073/pnas.1618744114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A.R. Statistical phylogeography: methods of evaluating and minimizing inference errors. Mol. Ecol. 2004;13:789–809. doi: 10.1046/j.1365-294x.2003.02041.x. [DOI] [PubMed] [Google Scholar]
- Teplitsky C., Robinson M.R., Merilä J. Evolutionary potential and constraints in wild populations. In: Charmantier A., Garant D., Kruuk L.E.B., editors. Quantitative Genetics in the Wild. Oxford University Press; Oxford: 2014. pp. 190–208. [Google Scholar]
- Thomson J.D., Thomson B.A. Dispersal of Erythronium grandiflorum pollen by bumblebees: implications for gene flow and reproductive success. Evolution. 1989;43:657–661. doi: 10.1111/j.1558-5646.1989.tb04261.x. [DOI] [PubMed] [Google Scholar]
- Trakhtenbrot A., Nathan R., Perry G., Richardson D.M. The importance of long-distance dispersal in biodiversity conservation. Divers. Distrib. 2005;11:173–181. [Google Scholar]
- Treep J., de Jager M., Kuiper L.S., Duman T., Katul G.G., Soons M.B. Costs and benefits of non-random seed release for long-distance dispersal in wind-dispersed plant species. Oikos. 2018;127:1330–1343. [Google Scholar]
- Trumbo D.R., Spear S.F., Baumsteiger J., Storfer A. Rangewide landscape genetics of an endemic Pacific northwestern salamander. Mol. Ecol. 2013;22:1250–1266. doi: 10.1111/mec.12168. [DOI] [PubMed] [Google Scholar]
- Tsoar A., Shohami D., Nathan R. A movement ecology approach to study seed dispersal and plant invasion: an overview and application of seed dispersal by fruit bats. In: Richardson D.M., editor. Fifty Years of Invasion Ecology. Wiley-Blackwell; London: 2011. pp. 103–119. [Google Scholar]
- Tumas H.R., Shamblin B.M., Woodrey M., Nibbelink N.P., Chandler R., Nairn C. Landscape genetics of the foundational salt marsh plant species black needlerush (Juncus roemerianus Scheele) across the northeastern Gulf of Mexico. Landscape Ecol. 2018;33:1585–1601. [Google Scholar]
- Turakulov R., Easteal S. Number of SNPS loci needed to detect population structure. Hum. Hered. 2003;55:37–45. doi: 10.1159/000071808. [DOI] [PubMed] [Google Scholar]
- van Etten J. R Package gdistance: distances and routes on geographical grids. J. Stat. Softw. 2017;1 doi: 10.18637/jss.v076.i13. [DOI] [Google Scholar]
- Vandepitte K., Jacquemyn H., Roldan-Ruiz I., Honnay O. Landscape genetics of the self-compatible forest herb Geum urbanum: effects of habitat age, fragmentation and local environment. Mol. Ecol. 2007;16:4171–4179. doi: 10.1111/j.1365-294X.2007.03473.x. [DOI] [PubMed] [Google Scholar]
- Vellend M., Myers J.A., Gardescu S., Marks P.L. Dispersal of Trillium seeds by deer; implications for long-distance migration of forest herbs. Ecology. 2003;84:1067–1072. [Google Scholar]
- Wang I.J. Recognizing the temporal distinctions between landscape genetics and phylogeography. Mol. Ecol. 2010;19:2605–2608. doi: 10.1111/j.1365-294X.2010.04715.x. [DOI] [PubMed] [Google Scholar]
- Wang R., Compton S.G., Shi Y.-S., Chen X.-Y. Fragmentation reduces regional-scale spatial genetic structure in a wind-pollinated tree because genetic barriers are removed. Ecol. Evol. 2012;2:2250–2261. doi: 10.1002/ece3.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-F., Lian J.-Y., Ye W.-H., Cao H.-L., Zhang Q.-M., Wang Z.-M. Pollen and seed flow under different predominant winds in wind-pollinated and wind-dispersed species Engelhardia roxburghiana. Tree Genet. Genomes. 2016;12:19. [Google Scholar]
- Warren D.L., Seifert S.N. Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecol. Appl. 2011;21:335–342. doi: 10.1890/10-1171.1. [DOI] [PubMed] [Google Scholar]
- Warren R.J., II, Bahn V., Bradford M.A. The interaction between propagule pressure, habitat suitability and density-dependent reproduction in species invasion. Oikos. 2012;121:874–881. [Google Scholar]
- Wei X., Meng H., Jiang M. Landscape genetic structure of a streamside tree species Euptelea pleiospermum (Eupteleaceae): contrasting roles of river valley and mountain ridge. PLoS One. 2013;8:e66928. doi: 10.1371/journal.pone.0066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock M.C., McCauley D.E. Indirect measures of gene flow and migration: FST≠ 1/(4Nm + 1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
- Williams C.G. Aerobiology of Pinus taeda pollen clouds. Can. J. For. Res. 2008;38:2177–2188. [Google Scholar]
- Winker K., Rappole J.H., Ramos M.A. The use of movement data as an assay of habitat quality. Oecologia. 1995;101:211–216. doi: 10.1007/BF00317286. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Stow A.J., Dudaniec R.Y. Detection of environmental and morphological adaptation despite high landscape genetic connectivity in a pest grasshopper (Phaulacridium vittatum) Mol. Ecol. 2019;28:3395–3412. doi: 10.1111/mec.15146. [DOI] [PubMed] [Google Scholar]
- Young A., Boyle T., Brown T. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 1996;11:413–418. doi: 10.1016/0169-5347(96)10045-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.