Abstract
Genetic diversity provides the foundation for plant breeding and genetic research. Over 3000 rice genomes were recently sequenced as part of the 3K Rice Genome (3KRG) Project. We added four additional Indian rice accessions to create a panel of 3004 accessions. However, such a large collection of germplasm is difficult to preserve and evaluate. The construction of core and mini-core collections is an efficient method for the management of genetic resources. In this study, we developed a mini-core comprising 520 accessions that captured most of the SNPs and represented all of the phenotypes and geographic regions from the original panel. The mini-core was validated using different statistical analyses and contained representatives from all major rice groups, including japonica, indica, aus/boro, and aromatic/basmati. Genome-wide association analyses of the mini-core panel efficiently reproduced the marker–trait associations identified in the original panel. Haplotype analysis validated the utility of the mini-core panel. In the current era with many ongoing large-scale sequencing projects, such a strategy for mini-core design should be useful in many crops. The rice mini-core collection developed in this study would be valuable for agronomic trait evaluation and useful for rice improvement via marker-assisted molecular breeding.
Keywords: rice, mini-core, SNPs, GWAS, 3KRG, agronomic trait
The handling and evaluation of a large germplasm panel is very challenging. This study designs a mini-core collection that effectively represents the phenotypic and genotypic diversity of 3004 rice accessions from multiple geographic regions of the world. The mini-core can be used for different marker–trait association analyses.
Introduction
Rice (Oryza sativa) is among the primary staple crops that fulfil the nutritional requirements of more than half of the world's population. Improvements in global rice production will have a direct impact on meeting the world’s growing food demand. More than 90% of global rice production is contributed by Asian countries, particularly China and India (FAOSTAT, 2017). India is the second largest producer of rice (165.3 million tons) after China (208.4 million tons) and accounts for ∼22% of total global rice production. Increases in rice yield are achieved mainly through improved cropping methods, fertilizer use, and—in many areas—intensive irrigation. However, the outcome of these strategies is now reaching saturation and becoming limited, and there is a demand for alternative means of yield improvement. The genetic improvement of rice cultivars and varieties can be an effective strategy in this regard. Yield improvement can be realized through breeding programs that incorporate marker-assisted selection and genetic methods to identify new sources of genetic variation that may help to increase productivity (McCouch et al., 2016). Productivity/yield is a complex trait that is governed by multiple genes and depends on both genetic composition and environmental factors. Variability arises due to segregating alleles at multiple loci whose individual effects on the phenotypic trait are relatively small, and the overall expression is also influenced by environmental conditions. Single-nucleotide polymorphisms (SNPs), present throughout the genome, are one of the major causes of allelic variation that underlie genetic variability in a population. Genetic variation leads to a multitude of phenotypes, which form the basis for selection of improved cultivars for breeding and agricultural purposes. Identification of loci that govern quantitative traits is critical for the maintenance of variation within and among populations. Identification of quantitative trait loci (QTLs) by the conventional method of linkage mapping or QTL mapping involves the development of a mapping population, a time-consuming process that captures a limited number of recombination events based on parental combinations. This methodology forms a part of the marker-assisted selection and biotechnological approach that has been used in a large number of crops for the identification of genes that govern complex traits (Edgerton, 2009, Morrell et al., 2012).
With advances in high-throughput genome sequencing and phenotyping methods, genome-wide association studies (GWAS) have been initiated. GWAS analysis has proved to be very effective for crop improvement. It is a very efficient approach for the identification of marker–trait associations and has been used to identify genes or loci that govern complex traits (Gupta et al., 2005, Breseghello and Sorrells, 2006, Huang et al., 2010, Huang et al., 2012, Famoso et al., 2011, Ingvarsson and Street, 2011, Kump et al., 2011, Zhao et al., 2011, Morrell et al., 2012). The advantage of GWAS is that it does not require a mapping population. It explores the genomic and phenotypic diversity present in the available population to assess marker–trait associations. It also captures a large number of historical recombination events that are prevalent in the population. The basic requirement for GWAS is a diverse panel that harbors historical recombination events for greater genetic resolution (Morrell et al., 2012). This purpose is best served by a core collection that is designed to capture the maximum available/possible diversity (genetic, phenotypic, and geographic) of the entire population, with a limited number of individuals that share low or no kinship (Korte et al., 2012). Core collections have been used as association panels for GWAS in different studies (El Bakkali et al., 2013, Zhang et al., 2014, Perseguini et al., 2015, Ambreen et al., 2018). In the case of rice, attempts have been made to generate core collections and use them as association panels. The US Department of Agriculture MC collection consists of 217 accessions that represent the genotypic and phenotypic diversity of the rice core subset of 1794 accessions, but it is based on a small number of simple sequence repeats (SSRs) and InDel markers (Agrama et al., 2009). More recently, a Rice Diversity Panel was developed that consisted of different collections: Rice Diversity Panel 1 (RDP1), Rice Diversity Panel 2 (RDP2), and a collection from the Institute of Agrobiological Sciences, NARO (Eizenga et al., 2014, Ebana et al., 2008, McCouch et al., 2016). However, the accessions in these panels were genotyped with a fixed array of 700K SNPs (Liakat Ali et al., 2011, Eizenga et al., 2014, McCouch et al., 2016).
Recently, with the availability of a resequencing dataset for 3000 diverse rice accessions that generated 32 million SNPs, a deep and robust platform has been provided to promote marker-associated breeding efforts for various agronomic traits (Li et al., 2014, Alexandrov et al., 2015, Mansueto et al., 2016). Follow-up studies have explored the detailed structural variation and introgression patterns in the 3KRG dataset, further strengthening our understanding of diverse genomes and trait domestication (Wang et al., 2018, Fuentes et al., 2019). Although this panel of 3000 accessions represents the core collection of global rice accessions, it is still relatively large and may present difficulties in management and phenotypic evaluation (Brown, 2011). Therefore, there is a need for a smaller subset that mirrors this large germplasm panel for convenient breeding efforts. In this study, we have developed a mini-core collection (520 accessions) from the original collection of 3004 rice accessions and have used it as an association panel for GWAS analysis with >2 million genome-wide SNPs. In designing the mini-core collection, we considered genotypic data (SNPs), phenotypic data (18 agronomic traits), and representation from various regional gene pools (geographic diversity) to preserve the maximum possible diversity. The comparatively small size of the association panel designed in this study will be useful and convenient for various phenotype–genotype relationship studies, which currently remain a major limitation in plant-breeding programs. We demonstrate that such subset formulations and analyses can lead to the identification of both existing and novel trait associations that are important for increasing crop yield.
Results and Discussion
Generation of the SNP Dataset
We used publically available SNP data from the 3000 rice accessions in the 3K Rice Genome (3KRG) project (Alexandrov et al., 2015, Mansueto et al., 2017). In addition, we resequenced four Indian rice accessions: LGR, PB 1121, Sonasal, and Bindli. After filtering and alignment, we obtained 3 564 117 high-quality SNPs for these four genotypes with reference to the Nipponbare genome. The SNP read depth varied from 10 to more than 11 000, and the overall sequencing depth for the four rice accessions ranged from 42× to 48×. In the present study, we combined the new Indian rice dataset with the 3KRG SNP dataset. Overall, 18.9 million SNPs were identified among the 3000 sequenced genomes with an average depth of ∼14×, ranging from ∼4× to 60×. To bring the 3KRG dataset to the same level of quality as the new data, we considered the filtered dataset (∼4 800 000 SNPs) corrected for excess of heterozygosity and linkage disequilibrium (LD). Finally, we merged both datasets and identified their common SNPs (2 081 521). The common SNPs were non-uniformly distributed over different rice chromosomes. The greatest number of SNPs were located on chromosomes 1, 11, and 2, whereas the smallest number of SNPs was found on chromosome 9.
Development of the Mini-Core Collection
To create a representative mini-core group, we used genotypic data (SNPs) from 3000 rice accessions and phenotypic data on 18 agronomic traits from 2266 rice accessions (Mansueto et al., 2017). We intitially chose to develop independent mini-cores from phenotypic data and genotypic data to avoid tradeoffs and capture the maximum possible phenotypic and genotypic variability present in the original collection. For the phenotype-based subset, scanning of 2266 accessions resulted in a mini-core collection (CC1) of 227 accessions that represented 10% of the initial collection. We added the four Indian accessions (LGR, PB 1121, Sonasal, and Bindli) to this panel because of their notable genomic and phenotypic diversity. Mini-core collection CC1 therefore consisted of 231 accessions representing diversity in phenotypic traits. The 3000 accessions with their SNP data were analyzed separately for the development of a second mini-core collection (CC2) consisting of 300 accessions that represented 10% of the original collection and also included the four Indian accessions sequenced in our laboratory.
Mini-core collections CC1 and CC2 were assessed for their coverage of phenotypic variation with reference to the original panel (Supplemental Table 1). Neither of the two mini-cores captured the entire range of phenotypic traits present in the original collection. Traits that could not be captured in the mini-cores included days to 80% heading (DEH), 100 grain weight (HGW), days to first flowering (DFF), grain width (GW), panicle length (PL), and seedling height (SH). The two mini-core collections were further assessed with various evaluation criteria such as Shannon's diversity index, Nei's gene diversity, mean difference percentage (MD%), variance difference percentage (VD%), variable rate of coefficient of variance (VR%), and coincidence rate of range (CR%) to assess their efficiency in capturing the maximum diversity present in the original collection. The MD% of the mini-core collections ranged from 2.8% to 4.08%, well below the prescribed value of 20% (Supplemental Table 2). VD%, which represents the variance captured in the mini-core collections, ranged from 19.78% to 39.77%. The VR% ranged from 86% for CC1 to 107.68% for CC2. CC1 had a higher CR% value of 92%, whereas that of CC2 was 91.1%. The value of the Shannon–Weaver index (H) ranged from 1.98 for CC1 to 2.25 for CC2. The value of Nei's genetic diversity (I) was higher for CC2 (0.79) than for CC1 (0.77) (Supplemental Table 2).
The mini-core collections were also assessed for their representation of all the varietal groups and regional gene pools present in the original panel (Supplemental Tables 3 and 4). The most prevalent group in mini-core CC1 was indica (129 accessions), followed by Temperate japonica (38), Intermediate (19), Tropical japonica (15), japonica (14), aus/boro (11), and Aromatic (5). The most prevalent group in mini-core CC2 was indica (171), followed by Intermediate (45), aus/boro (42), Aromatic and japonica (12 each), Tropical japonica (10), and Temperate japonica (8) (Supplemental Table 3). We also compared the distribution of accessions from different varietal groups in the mini-cores and found that CC2 had a higher proportion of accessions from the aus/boro (19.5% of the original collection), Intermediate (33.3%), and Aromatic (16.9%) groups. On the other hand, CC1 had only 5.1% of the original representation from aus/boro, 14% from Intermediate, and 7% from Aromatic (Supplemental Table 3). Accessions from the Temperate japonica group were highly represented in CC1 (11.9% of the original representation), whereas only a small proportion (2.5%) was represented in CC2 (Supplemental Table 3). Comparable portions of accessions from the indica (7.4% in CC1 and 9.8% in CC2), Tropical japonica (3.8% in CC1 and 2.5% in CC2), and japonica (10.6% in CC1 and 9% in CC2) groups were present in both CC1 and CC2 mini-cores. Thus, neither of the two mini-cores developed here contained 10% of the representatives from all varietal groups (Supplemental Table 3).
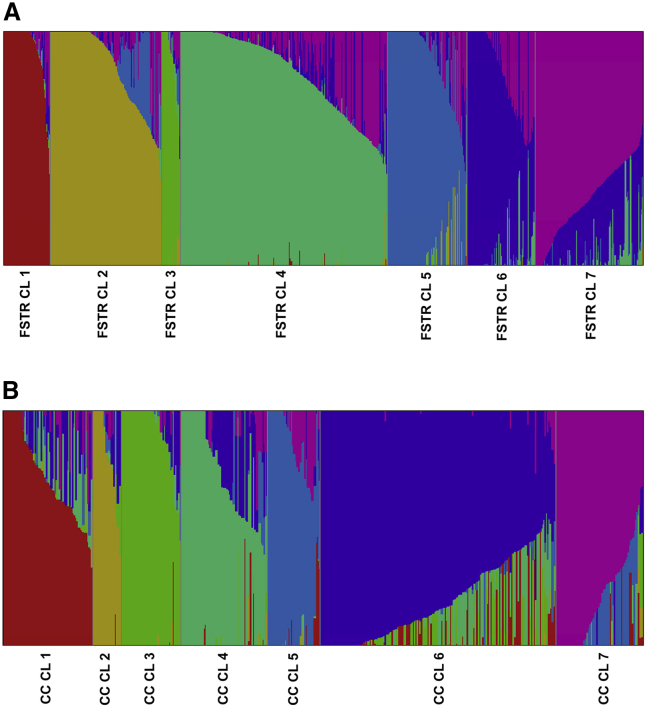
Figure 4.
Population Structure Analysis of the Rice Accessions from the Original Collection and the Mini-Core Panel.
(A) Population structure of the 3004 accessions from the original collection. Each sub-population is represented by a different color code (FSTR CL1–FSTR CL7).
(B) Population structure of the 520 rice accessions from mini-core CC3. Each sub-population is represented by a different color code (CC CL1–CC CL7.) Each vertical bar represents a single rice accession.
The mini-core collections were then assessed for their distribution of accessions from different regional gene pools (Supplemental Table 4). Mini-core CC1 contained 55 accessions from South Asia (6.9% of the original collection), followed by 52 accessions from South East Asia (5.1%), 52 accessions from China (10.8%), 18 accessions from Europe (15.2%), 17 accessions from America (10.2%), 15 accessions each from East Asia and Africa (11.4% and 5.9%, respectively), four accessions from Oceania (23.5%), and three accessions of unknown origin (8.8%). CC2 contained 122 accessions from South Asia (15.5% of the original collection), followed by 70 accessions from South East Asia (6.9%), 55 accessions from China (11.4%), 23 accessions from Africa (9.1%), 13 accessions from America (7.8%), eight accessions from East Asia (6%), six accessions of unknown origin (17.4%), two accessions from Europe (1%), and one accession from Oceania (5.9%; Supplemental Table 4).
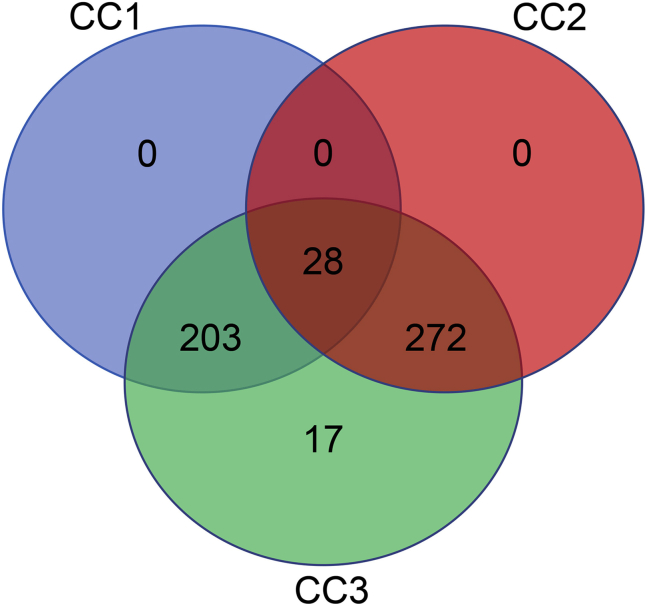
Only 28 accessions were shared between CC1 and CC2, showing that different accessions were selected on the basis of phenotypic and genotypic variation and justifying our concern about designing the mini-cores independently using only phenotypic or genotypic data. An ideal mini-core should represent the maximum possible diversity present in the original collection. However, different evaluation criteria such as phenotypic range (Supplemental Table 1), MD%, VD%, VR%, CR%, Shannon’s and Nei's indices (Supplemental Table 2), and varietal (Supplemental Table 3) and geographic coverage (Supplemental Table 4) revealed that neither of the mini-cores (CC1 and CC2) captured sufficient diversity from the original collection to be considered an ideal representative subset. Therefore, we merged CC1 and CC2 to develop mini-core collection CC3, comprising 520 non-redundant accessions (503 accessions from the merging of CC1 and CC2 and 17 accessions that captured the extreme values of the phenotypic traits discussed below), in order to capture the maximum possible allele/trait diversity and prevent any tradeoffs between the two datasets (phenotypic and genotypic) when used in conjunction (Figure 1). The 520 accessions of CC3 represented 17.3% of the original collection (3004 accessions) and fulfilled the initial size requirement for an ideal core collection, which should range between 5% and 20% of the original collection (Brown and Spilllane, 1999). CC3 was assessed for its representation of the original collection and various traits under consideration by different evaluation criteria (Supplemental Tables 1–4). CC3 covered the entire range of traits from the original collection, including traits not completely covered by CC1 and CC2, such as DEH, HGW, DFF, GW, PL, and SH (Supplemental Table 1). The MD% of CC3 was 2.9% and was within the range of 2.8%–4.08% observed for CC1 and CC2 (Supplemental Table 2). The value of VD% representing the variance captured by the CC3 accessions was 18.9%, which was lower than the VD% values of CC1 and CC2. The value of VR% captured by the CC3 accessions was 109.3%, the highest of the three mini-cores. Furthermore, CC3 had the highest value of CR% (96.2%) of the three mini-cores. The values of Shannon–Weaver H and Nei I for CC3 were 2.17 and 0.79, respectively (Supplemental Table 2).
Figure 1.
Venn Diagram Showing the Distribution of Accessions in Different Mini-Core Collections Developed in This Study.
CC1 represents the mini-core designed using phenotypic data. CC2 represents the mini-core designed using SNP data. CC3 represents the merged (CC1 + CC2 + 17 accessions) mini-core collection.
Mini-core CC3 was also assessed for its representation of the varieties and regional gene pools present in the original collection (Supplemental Tables 3 and 4). All varieties had at least 10% representation from the original collection except for the Tropical japonica group, which had only 6.9% representation from the original collection (27 accessions; Supplemental Table 3). The number of accessions and the percentage representation of all varietal groups from the original collection in mini-core CC3 are presented in Supplemental Table 3. Representation of accessions from different regional gene pools in the original collection varied from 12% to 29.4% in mini-core CC3 (Supplemental Table 4). The most prevalent region in CC3 was South Asia (176 accessions; 22.4% of its original representation), followed by China (101 accessions; 20.95%), Africa (38 accessions; 15%), America (28 accessions; 16.9%), East Asia (21 accessions; 15.9%), Europe (19 accessions; 16.1%), unknown origin (9 accessions; 25.5%), and Oceania (5 accessions; 29.4%; Supplemental Table 4). Thus, mini-core collection CC3 more successfully fulfilled the criteria for capturing the maximum possible diversity from the original panel than did mini-cores CC1 and CC2, and it was considered further for its utility as an association panel.
Distance-Based Cluster Analysis and Principal Component Analysis
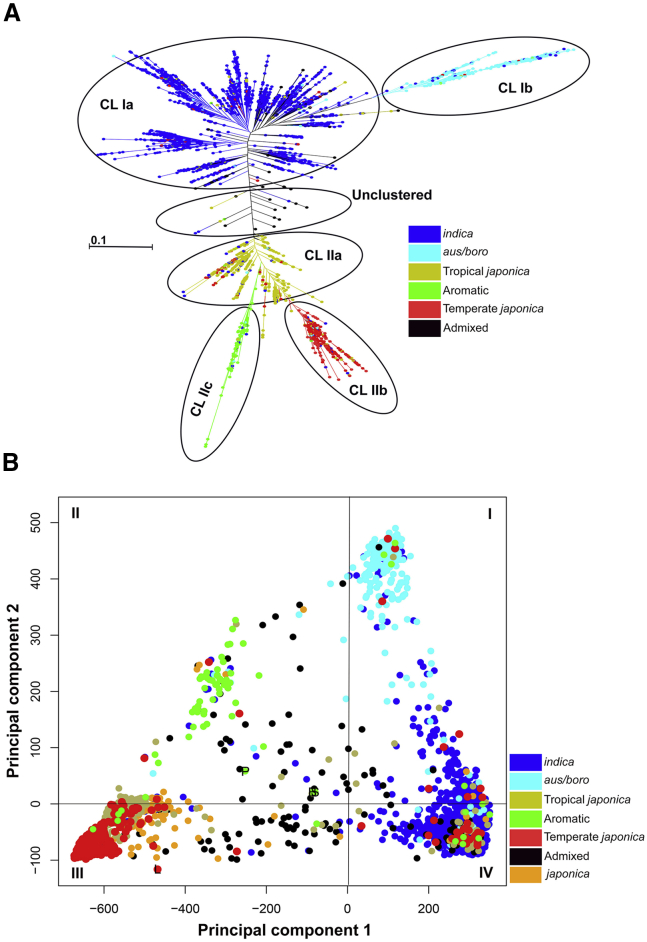
Distance-based cluster analysis was performed to assess the grouping of accessions from the original collection of rice genotypes. Analysis of the SNP data (2 081 521 SNPs) using the maximum-likelihood method grouped the 3004 accessions into two major clusters (CL I and CL II) with internal subgroupings (Figure 2A). The CL I cluster contained the greatest number of accessions (66%) from the original collection, and CL II contained approximately 32% of the original accessions (Supplemental Table 5). Approximately 1.4% of the accessions did not belong to either of the clusters (Figure 2A). These unclustered accessions (43) were mainly Intermediate (21) and indica (15) genotypes. In addition, some of the japonica (3), Tropical japonica (2), Temperate japonica (1), and Aromatic (1) genotypes also remained unclustered. The 1987 accessions of Cluster CL I were further grouped into two subclusters, CL Ia and CL Ib. The larger subcluster, CL Ia, consisted of 1771 accessions and was mainly dominated by indica (1641) genotypes, whereas cluster CL Ib consisted of 216 accessions with major contributions from aus/boro (172) and indica (25) genotypes. The 974 accessions of CL II were divided into three subclusters, CL IIa, IIb, and IIc. The largest subcluster, CL IIa, contained 519 accessions and was mainly dominated by Tropical japonica (329) and japonica (80) genotypes. Subcluster CL IIb contained 358 accessions and was dominated by Temperate japonica (250) genotypes. The Indian genotype LGR was also part of subcluster CL IIb. CL IIc was the smallest subcluster of CL II and contained 97 accessions, with major representation from Aromatic (50) and Intermediate (23) genotypes. Notably, Bindli, PB 1121, and Sonasal grouped together in subcluster CL IIc (Figure 2A and Supplemental Table 5).
Figure 2.
Grouping of the 3004 Rice Accessions Based on Polymorphic SNP Markers.
(A) Maximum-likelihood dendrogram illustrating the genetic relationships among accessions. The two clusters were designated CL I (CL Ia, b) and CL II (C IIa–c) with further subclustering shown.
(B) Principal component analysis of the 3004 accessions from the original collection, showing principal component axes 1 and 2. The distribution of accessions in different quadrants (I–IV) is shown. Varietal group color codes are provided. Color codes representing different varietal groups are given on the right.
In the principal component analysis (PCA), the 3004 original accessions were evenly distributed along coordinate axes 1 and 2, which accounted for 45.6% and 26% of the total variance, respectively (Figure 2B). The indica accessions clustered together in the PCA, consistent with the results of the distance-based analysis. They formed the largest group in the original collection and were mainly present in Cluster Ia of the distance-based analysis and quadrants I and IV of the PCA. The japonica, Temperate japonica, and Tropical japonica accessions were part of Cluster II in the distance-based analysis and were found in quadrants III and IV of the PCA. Accessions from the aus/boro group were present in quadrant I, and Aromatic accessions were present in quadrant II. Accessions of the Intermediate type were spread across all quadrants of the PCA, consistent with the maximum-likelihood dendrogram in which they were present in all clusters in even proportions (Figure 2A and 2B).
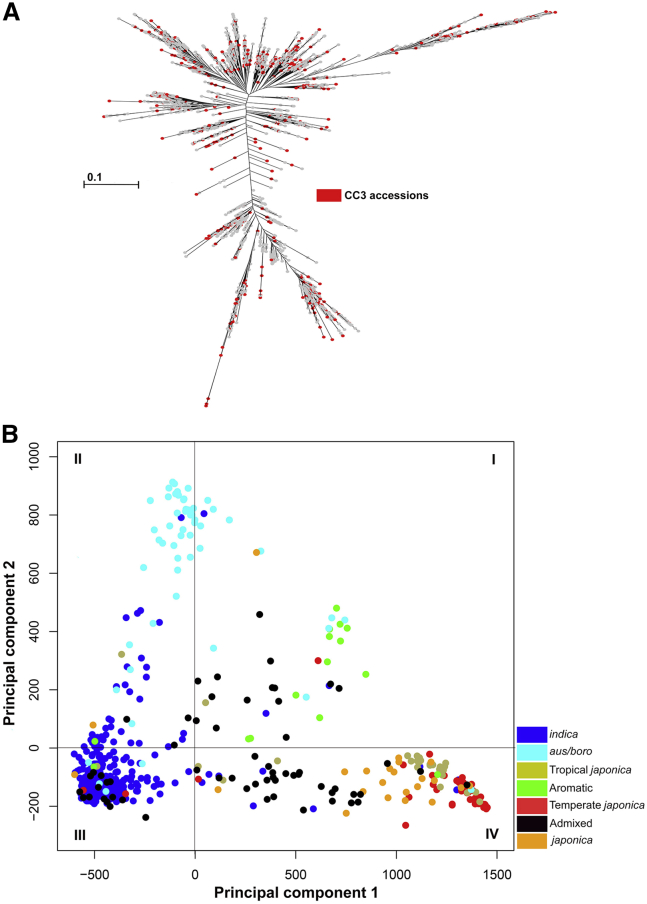
Next, we checked the distribution of the CC3 accessions on the distance-based maximum-likelihood dendrogram and the PCA of the original collection to assess their distribution in all clusters and quadrants. CC3 showed balanced representation (10.5%–25.7%) of all clusters of the maximum-likelihood dendrogram (Figure 3A and Supplemental Table 6). It contained 18.1% of the accessions from subcluster CL Ia, 19.4% of the accessions from subcluster CL Ib, 25% of the unclustered accessions, 10.5% of the accessions from subcluster CL IIa, 18% of the accessions from subcluster CL IIb, and 25.7% of the accessions from subcluster CL IIc (Figure 3A and Supplemental Table 6). Similarly, accessions from all quadrants of the PCA were present in CC3 (Figure 3B). Thus, CC3 had contained representative accessions from all clusters of the maximum-likelihood dendrogram and all quadrants of the PCA, capturing the maximum possible genotypic diversity.
Figure 3.
Distribution of the 520 Accessions of Mini-Core Collection CC3.
(A) Distribution of rice accessions in different clusters of the maximum-likelihood dendrogram of the original collection of 3004 rice accessions (Figure 2A). CC3 accessions are indicated by red dots.
(B) Principal component analysis of the 520 accessions of mini-core collection CC3, showing principal component axes 1 and 2. The distribution of the accessions in different quadrants (I–IV) is shown. Varietal group color codes are provided. Color codes representing different varietal groups are given on the right.
Population Structure Analysis of the Original Collection Using FastSTRUCTURE
Population structure analysis of the original 3004 rice accessions was performed using the FastSTRUCTURE program (Raj et al., 2014). The best clustering was observed at K = 7, and the clusters obtained were named FSTR CL 1–7 (Figure 4A). FSTR CL 1 consisted of 219 accessions and was mainly dominated by aus/boro (179 accessions) and indica genotypes (28; Supplemental Table 7); it showed congruence with CL Ib from the maximum-likelihood analysis. FSTR CL 2 consisted of 522 accessions, with highest representation from the Tropical japonica (310) and japonica groups (94); its accessions were similar to those of CL IIa from the maximum-likelihood analysis. Some indica (47), Temperate japonica (35), and Intermediate (27) accessions were also found in FSTR CL 2. The smallest cluster was FSTR CL 3, whose 90 accessions were dominated by the Aromatic group (50), followed by the Intermediate group (19); it showed congruence with CL IIc from the maximum-likelihood analysis. The largest cluster was FSTR CL 4, which contained 973 accessions and was dominated by the indica group (885), with minor contributions from the Tropical japonica (26), Intermediate (21), Temperate japonica (17), and aus/boro (14) groups. FSTR CL 5 contained 372 accessions and was dominated by the Temperate japonica group (248), with minor contributions from the Tropical japonica (35), Intermediate (29), indica (26), and japonica (25) groups; it was similar to CL IIb from the maximum-likelihood analysis. FSTR CL 6 consisted of 323 accessions and was dominated by indica varieties (297), and FSTR CL 7 consisted of 505 accessions with major contributions from indica (451) and Intermediate varieties (27). FSTR CLs 4, 6, and 7 together corresponded to CL Ia from the maximum-likelihood dendrogram, suggesting that the CL Ia accessions could be further divided into three subgroups. The numbers of accessions that constituted different clusters in the FastSTRUCTURE analysis are provided in Supplemental Table 7.
Next, we looked for admixed genotypes in all groups and found that 41% (1242) of the accessions were admixed in nature (Supplemental Table 8). Among all the clusters, the 505 accessions of FSTR CL 7 contained more admixed individuals (351) than pure individuals (154 accessions), followed by FSTR CL 6 (145 admixed and 148 pure accessions; Supplemental Table 8). Assessment of admixtures within varietal groups revealed that the Intermediate category contained more admixtures (94) than pure (41) accessions, whereas the indica population had 821 admixed individuals (47%) out of 1743 accessions. Analysis of regional gene pools revealed that only the European region had more admixed (65) than pure individuals (53) (Supplemental Table 8). Distribution of the 520 CC3 accessions in different clusters of the FastSTRUCTURE analysis (FSTR CL 1–7) was assessed to determine the representation of individuals from each cluster in the mini-core collection. CC3 captured 50 (40 pure individuals with Q value >80%) of the 219 FSTR CL 1 accessions (Supplemental Table 9), 42 (23 pure individuals) of the 522 FSTR CL 2 accessions, 24 (13 pure individuals) of the FSTR CL 3 accessions, 185 (109 pure individuals) of the 973 FSTR CL 4 accessions, 74 (37 pure individuals) of the 372 FSTR CL 5 accessions, 61 (28 pure individuals) of the 323 FSTR CL 6 accessions, and 84 (25 pure individuals) of the 505 FSTR CL 7 accessions (Supplemental Table 9). Thus, CC3 contained representatives of both pure and admixed accessions from all seven clusters of the population structure analysis derived from the original collection of rice accessions. We were therefore able to fulfil the initial objective of developing a mini-core collection (CC3) that represented the maximum phenotypic, genotypic, varietal, and geographic variability present in the original collection of 3004 rice accessions.
Assessment of Mini-Core Collection CC3 for Its Utility as an Association Panel
To avoid spurious marker–trait associations, an association panel should contain nucleotide diversity (π) equivalent to that of a larger panel, as well as low population structure and low kinship among its members (Yu and Buckler, 2006, Zhu et al., 2008, Yang et al., 2010, Nachimuthu et al., 2015). We therefore performed nucleotide diversity, population structure, and kinship analyses for the mini-core CC3 collection to assess its utility as an association panel.
Nucleotide Diversity and Population Structure Analysis of Mini-Core CC3
To determine whether mini-core C3 recapitulated the nucleotide diversity of the original collection, we tested important genes known to be associated with various traits, including GW, cooking quality, grain color, grain size, flowering time, and panicle development. We found that the nucleotide diversity within these important genes was comparable in both panels (Supplemental Table 10). This result suggests that mini-core CC3, despite being a smaller subset, captures the essential nucleotide diversity of the larger panel. Next, the underlying population structure of the 520 mini-core CC3 accessions was estimated using FastSTRUCTURE, which grouped them into seven clusters (K = 7) named CC CL1–CC CL7 (Figure 4B and Supplemental Table 11). CC CL1 contained 73 accessions and was dominated by the indica (57) and Intermediate (12) groups. CC CL2 was the smallest of the clusters; it contained 23 accessions and was dominated by the Aromatic (9) and Intermediate (8) groups. CC CL3 contained 49 accessions and was dominated by aus/boro (41). CC CL4 contained 70 accessions and had representatives from the indica (58 accessions) and Intermediate (8) groups. CC CL5 contained 43 accessions from different varietal groups, including the Tropical japonica (17), japonica (10), and Intermediate (8) groups. CC CL6 was the largest cluster and contained 191 accessions predominately from the indica (165) and Intermediate (11) groups. CC CL7 contained 77 accessions mainly from the Temperate japonica (36), Intermediate (14), and japonica (9) groups. A detailed distribution of the accessions from different varietal groups in the seven clusters of mini-core collection CC3 is presented in Supplemental Table 11.
We found that 47% (245) of the accessions in the CC3 mini-core collection were admixtures (Supplemental Table 12). CC CL1 contained 73 accessions and had more admixed individuals (48) than pure individuals (25), followed by CC CL4 with 70 accessions (42 admixed and 28 pure individuals) (Supplemental Table 12). Clusters CC CL2, 5, and 7 had approximately equal numbers of pure and admixed accessions, whereas CC CL3 and CC CL6 had more pure individuals than admixed individuals. Admixture assessment of the varietal groups in CC3 revealed that the Intermediate group had more admixtures (47) than pure (14) individuals, followed by japonica with 12 admixed and 11 pure individuals. The indica group had 144 admixed genotypes (49%) out of 295 accessions, and the Tropical japonica group had 13 admixed accessions out of 27 individuals. Analysis of rice accessions from different regional gene pools in CC3 revealed that South East Asia (63 admixed and 60 pure accessions), China (54 admixed and 47 pure accessions), America (23 admixed and 15 pure accessions), Europe (13 admixed and 6 pure accessions), and Oceania (3 admixed and 2 pure accessions) gene pools contained more admixed individuals than pure individuals (Supplemental Table 12). A detailed distribution of admixed and pure accessions from the different groups present in CC3 is provided in Supplemental Table 12. An increased number of admixed individuals in the clusters derived from population structuring (CC CL1–CC CL7), varietal, and geographic identities confirms that CC3 contained more unrelated individuals than the original collection and validates its suitability as an association panel.
Kinship Analysis of Mini-Core CC3 Individuals
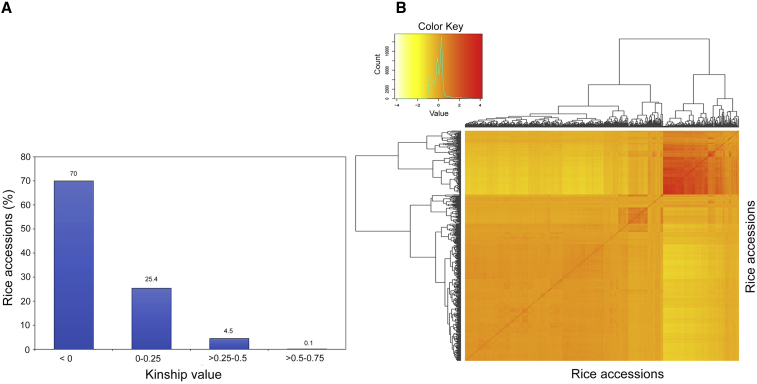
Kinship analysis between individuals from mini-core collection CC3 was performed to estimate their co-ancestry. Seventy percent of the possible pairs of CC3 accessions had kinship values less than zero, whereas 25.4% of the accession pairs had kinship values ranging between 0% and 0.25% (Figure 5). Approximately 4.5% of the CC3 accession pairs showed kinship values in the range of 0.25%–0.50%, and only 0.1% of accession pairs had kinship values in the range of 0.5%–0.75% (Figure 5). Thus, the kinship values for most CC3 accessions exhibited an absence or a weak level of genetic relatedness, fulfilling the primary requirement for utilization of the CC3 mini-core collection as an association panel.
Figure 5.
Kinship Analysis of the 520 Accessions from Mini-Core CC3.
(A) Histogram showing the kinship status of rice accessions from mini-core CC3.
(B) Kinship matrix showing the relatedness of rice accessions from mini-core CC3.
GWAS of Mini-Core Collection CC3
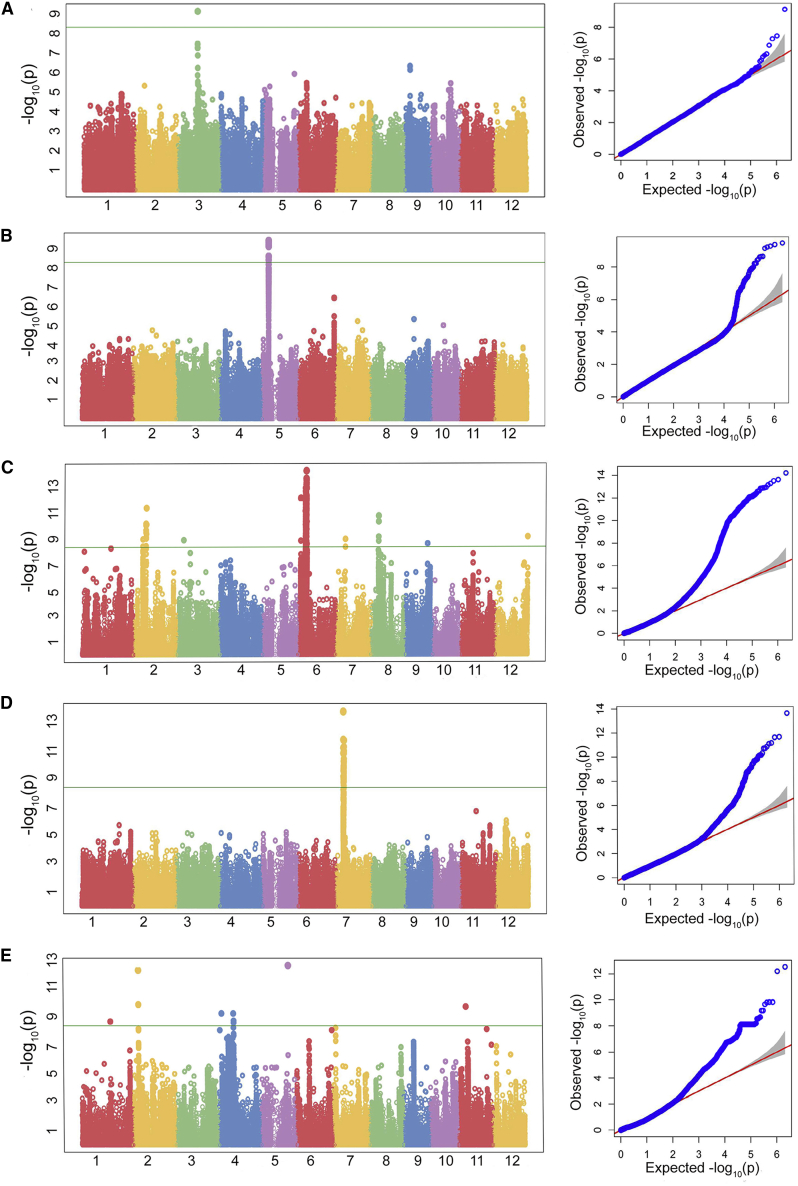
Because mini-core collection CC3 showed low population structure and low kinship values, we proceeded to study its utility for GWAS in rice. GWAS was performed on 520 CC3 mini-core accessions using a compressed mixed linear model (MLM) with 2 081 521 SNPs (MAF >0.02) and 18 yield-related traits of agronomic importance. Association between markers and traits was considered to be significant at P < 1 × 10−8, with a false discovery rate (FDR) adjusted P value of <0.05 and a correlation value (R2) of ≥10%. Six of the 18 traits showed significant marker–trait associations, namely endosperm type (ET), grain length (GL), GW, panicle axis (PA), secondary branching (SB), and seed coat color (SCC) (Table 1). In all, 5924 SNPs were found to be significantly associated with the aforementioned six traits, explaining between 10.4% and 61.6% of their phenotypic variation.
Table 1.
List of SNPs that Showed Significant Associations with Different Traits Identified in Mini-Core Collection CC3 and in the Original Collection of 3004 Rice Accessions.
| Trait | Chr | Position | Major allele | Minor allele | Minor allele frequency | Nipp. allele | FDR-adjusted P value | R2 value (%) | Known loci |
|---|---|---|---|---|---|---|---|---|---|
| ∗Grain length | 3 | 16 733 441 | G | T | 0.36 | G | 1.4 × 10−3 | 32.3 | GS3 |
| #Grain length | 3 | 16 733 441 | G | T | 0.36 | G | 3.4 × 10−43 | 43.5 | GS3 |
| ∗Grain width | 5 | 5 371 949 | C | G | 0.46 | C | 2.8 × 10−4 | 34.2 | qSW5 |
| #Grain width | 5 | 5 371 686 | C | T | 0.49 | C | 9.3 × 10−34 | 51.4 | qSW5 |
| ∗Endosperm type | 6 | 1 765 761 | T | G | 0.13 | T | 6.4 × 10−8 | 25 | Waxy |
| #Endosperm type | 6 | 1 731 808 | G | C | 0.20 | G | 1.03 × 10−29 | 20.2 | Waxy |
| ∗Endosperm type | 6 | 6 294 468 | G | T | 0.07 | G | 1.2 × 10−8 | 29 | |
| #Endosperm type | 6 | 6 830 286 | G | A | 0.21 | G | 3.4 × 10−8 | 15.6 | |
| $Endosperm type | 2 | 7 413 964 | C | G | 0.24 | C | 8.8 × 10−6 | 20.3 | |
| ∗Seed coat color | 7 | 6 124 457 | T | C | 0.456 | T | 4.5 × 10−8 | 61.6 | Rc |
| #Seed coat color | 7 | 6 133 394 | G | A | 0.26 | G | 6.6 × 10−11 | 7.2 | Rc |
| ∗Seed coat color | 7 | 6 660 825 | T | G | 0.454 | T | 1.6 × 10−6 | 59.7 | |
| #Seed coat color | 7 | 6 656 052 | T | C | 0.43 | T | 1.8 × 10−8 | 6.8 | |
| $Secondary branching | 2 | 5 032 535 | C | T | 0.013 | C | 6.4 × 10−7 | 32 | |
| $Secondary branching | 4 | 2 521 459 | A | G | 0.052 | A | 1.6 × 10−4 | 23.5 | |
| $Secondary branching | 4 | 12 427 420 | G | A | 0.208 | G | 1.6 × 10−4 | 23.4 | |
| $Panicle axis | 4 | 1 075 655 | A | C | 0.013 | A | 3.7 × 10−4 | 24 | |
| $Panicle axis | 6 | 28 676 456 | G | A | 0.0078 | G | 3.7 × 10−4 | 23.1 | |
| $Panicle axis | 10 | 14 829 875 | C | A | 0.0078 | C | 3.7 × 10−4 | 23 |
Nipp, Nipponbare, Chr, chromosome.
∗ and # represent the mini-core and original collection association markers, respectively. $ represents the association markers found exclusively in the mini-core subset CC3.
Three SNPs on chromosome 3 were significantly associated with GL. The most significant SNP (G/T) associated with GL on chromosome 3 was located at position 16 733 441. It had an FDR-adjusted P value of 1.4 × 10−3 and explained 32.2% of the phenotypic variation (Figure 6A and Table 1). This was previously reported as GS3, a well-known QTL for GL (Fan et al., 2006). Another important trait, GW, showed significant association with 64 SNPs on chromosome 5. The most significant SNP (C/G) associated with GW on chromosome 5 was located at position 5 371 949, had an FDR-adjusted P value of 2.8 × 10−4, and explained 34.2% of the phenotypic variation (Figure 6B and Table 1). This SNP was associated with the gene qSW5/GW5, which has a well-established correlation with GW (Shomura et al., 2008). ET showed significant associations with 3651 SNPs on chromosomes 2, 4, 6, 8, 11, and 12. The most significant SNP (G/T) was located on chromosome 6 at position 6 294 468, had an FDR-adjusted P value of 1.2 × 10−8, and explained 29% of the phenotypic variation (Figure 6C and Table 1). The other significant SNP (T/G) associated with ET was located on chromosome 6 at position 1 765 761, had an FDR-adjusted P value of 6.4 × 10−8, and accounted for 25% of the phenotypic variation. This was also previously reported by various researchers as the locus of the Waxy gene (GAO, 2003, Tian et al., 2009, Huang et al., 2010). Another SNP showing significant association with ET was located on chromosome 2 at position 7 413 964 (C/G), had an FDR-adjusted P value of 8.8 × 10−6, and explained 20.3% of the phenotypic variation (Table 1). SCC was associated with 306 SNPs on chromosome 7. The most significant SNP (T/C) was located at position 6 124 457, had an FDR-adjusted P value of 4.5 × 10−8, and explained 61.6% of the phenotypic variation (Figure 6D and Table 1). This SNP was associated with the Rc gene described in a previous report as an important locus for SCC (Sweeney et al., 2006). Another SNP (T/G) significantly associated with SCC was located at position 6 660 825 on chromosome 7, had an FDR-adjusted P value of 1.6 × 10−6, and explained 59.7% of the phenotypic variation. SB was associated with 1779 SNPs on chromosomes 2, 4, 6, 7, 9, and 11. The most significant SNP (C/T) associated with SB was located on chromosome 2 at position 5 032 535, had an FDR-adjusted P value of 6.4 × 10−7, and explained 32% of the phenotypic variation (Figure 6E and Table 1). Two SNPs significantly associated with SB were identified on chromosome 4 at positions 2 521 459 (A/G) and 12 427 420 (G/A). They had an FDR-adjusted P value of 1.6 × 10−4 and explained 23.5% and 23.4% of the phenotypic variation, respectively. PA was associated with 121 SNPs on chromosomes 2, 4, 6, and 10. The most significant SNP (A/C) was located on chromosome 4 at position 1 075 655, had an FDR-adjusted P value of 3.7 × 10−4, and explained 24% of the phenotypic variation (Supplemental Figure 1 and Table 1). SNPs on chromosomes 6 and 10 at positions 28 676 456 (G/A) and 14 829 875 (C/A) also showed an association with PA. They had an FDR-adjusted P value of 3.7 × 10−4 and explained 23.1% and 23% of the phenotypic variation, respectively.
Figure 6.
Genome-wide Mapping of SNPs Associated with Different Yield-Related Traits in Accessions from Mini-Core CC3.
Manhattan (left) and Q–Q (right) plots of compressed MLM for (A) grain length, (B) grain width, (C) endosperm type, (D) seed coat color, and (E) secondary branching. Negative log10-transformed P values (y axis) from the compressed MLM are plotted against the positions of SNPs (x axis) on different chromosomes. The green line in each figure represents the genome-wide cutoff for significant association. Red and blue lines in the Q–Q plot represent the trajectory for the null hypothesis and the observed values, respectively.
Next, we evaluated the utility of the mini-core collection for a trait other than yield. Fortunately, salt-tolerance data are now available for the original panel accessions. We therefore performed GWAS for salt injury (EC18) using the 520 mini-core CC3 accessions and identified seven SNPs that showed significant association with salt tolerance. These SNPs were distributed on chromosomes 1, 5, 6, 8, 9, 11, and 12. Of the seven SNPs, five had previously reported associations with salt-tolerance- or abiotic-stress-linked QTLs (saltol, qSNC1, qClLV-8.1a, qSSISFH-8.1, qSSIGY5.1, and qSSIGY6.2) (Pandit et al., 2010, Tiwari et al., 2016, Naveed et al., 2018). The details of the SNPs associated with salt stress are provided in Supplemental Table 13. One recent study evaluated the salinity tolerance of 191 Temperate japonica accessions from the 3KRG panel and identified one overlapping QTL, qPD18_11.1 & qSES18_11.1 (Batayeva et al., 2018). There were 24 accessions in common between this panel of 191 Temperate japonica accessions and mini-core CC3 designed in the present study (Supplemental Table 14). This result suggests that mini-core CC3 is also suitable for studying other traits. The identification of previously characterized QTLs for yield traits confirms the utility and importance of mini-core CC3. A detailed description of the SNPs that showed significant associations with ET, GL, GW, PA, SB, and SCC in the CC3 GWAS analysis is presented in Table 1.
GWAS Using the Original Panel of 3004 Accessions
To further validate the efficiency of the CC3 mini-core collection in capturing the maximum number of marker–trait associations, we performed GWAS analyses for the same yield traits using the original collection of 3004 rice accessions covering genome-wide SNPs. The number of SNPs was reduced due to limitations on matrix size in the R program. For the original collection, 1790 SNPs were significantly associated with different traits and explained from 5.6% to 51.4% of phenotypic variation. Notably, four of the six traits (ET, GL, GW, and SCC) showed significant marker–trait association. However, two traits, 100 grain weight (HGW) and panicle threshability (PT), showed associations in the analysis of the original collection but were missing in the analysis of the CC3 mini-core (Supplemental Table 15). GL was associated with 325 SNPs on chromosomes 3 and 5. The most significant SNP (G/T) associated with GL was located on chromosome 3 at position 16 733 441, had an FDR-adjusted P value of 3.4 × 10−43, and explained 43.5% of the phenotypic variation (Supplemental Figure 2A and Supplemental Table 15). Another SNP associated with GL was located on chromosome 5 (G/A) at position 5 361 894, had an FDR-adjusted P value of 1.03 × 10−9, and explained 38.8% of the phenotypic variation. GW was associated with 737 SNPs on chromosome 5. Consistent with the earlier studies, the most significant SNP on chromosome 5 (C/T) was located at position 5 371 686, had an FDR-adjusted P value of 9.3 × 10−34, and explained 51.4% of the phenotypic variation (Supplemental Figure 2B and Supplemental Table 15). The second SNP (T/C) associated with GW on chromosome 5 was located at position 28 019 687, had an FDR-adjusted P value of 8.4 × 10−6, and explained 48% of the phenotypic variation. HGW was significantly associated with 54 SNPs on chromosomes 3 and 5. The most significant SNP (G/T) was located on chromosome 3 at position 16 733 441, had an FDR-adjusted P value of 7.9 × 10−5, and explained 35.2% of the phenotypic variation. Another SNP (T/C) was present on chromosome 5 at position 5 375 201, had an FDR-adjusted P value of 7.9 × 10−5, and explained 35.2% of the phenotypic variation (Supplemental Figure 2C and Supplemental Table 15). ET was associated with 503 SNPs on chromosome 6. The most significant SNP (G/C) was located at position 1 731 808, had an FDR-adjusted P value of 1.03 × 10−29, and explained 20.2% of the phenotypic variation (Supplemental Figure 2D). The next significant SNP (G/A) was identified at position 6 830 286, had an FDR-adjusted P value of 3.4 × 10−8, and explained 15.6% of the phenotypic variation. Several SNPs significantly associated with SCC were identified on chromosomes 2 and 7. The most significant SNP was located on chromosome 7 (G/A) at position 6 133 394, had an FDR-adjusted P value of 6.6 × 10−11, and explained 7.2% of the phenotypic variation (Supplemental Figure 2E). Three additional SNPs were associated with SCC: SNP (G/T, 6 417 000) and SNP (T/C, 6 656 052) on chromosome 7 and SNP (A/G, 32 431 463) on chromosome 2. These three associations explained between 5.6% and 7.1% of the phenotypic variation. One SNP (C/T) on chromosome 2 showed a significant association with PT; it had an FDR-adjusted P value of 6.8 × 10−3 and explained 16.4% of the phenotypic variation (Supplemental Figure 2F). A detailed distribution of the SNPs associated with traits such as ET, GL, GW, HGW, PT, and SCC in the original panel of 3004 accessions is provided in Supplemental Table 15.
Linkage Disequilibrium and Haplotype Analysis
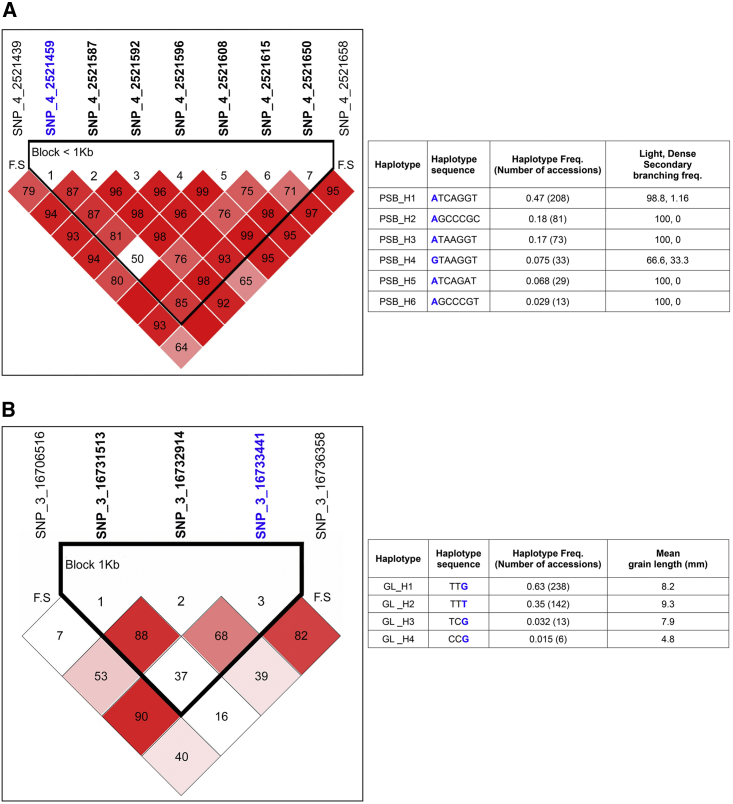
To gain further insight into some of the less well-characterized marker–trait associations identified in the CC3 panel, we studied the associated SNPs for LD pattern and haplotype block formation using their flanking nucleotides. In the case of trait SB, 100 SNPs flanking the most significantly associated SNP (A/G, 2 521 459) on chromosome 4 were used, and an LD block containing the associated SNP was identified. The formulated block showed strong LD in a span of 2 kb that contained seven neighboring SNPs, including the A/G at 2 521 459 (Figure 7A). Haplotype analysis of this block revealed that the PSB_H1 (ATCAGGT) haplotype had the highest frequency (f = 0.45). Distribution of haplotypes between light and dense panicle secondary branching revealed that all but the H4 haplotype had significant associations with light-level branching (Figure 7A). By contrast, the H4 haplotype showed an inclination toward a dense branching trait. One recent study has demonstrated the association of elevated haplotype diversity in SHORT PANICLE 1 (SP1) with phenotype in the japonica rice group (Jang et al., 2018). Similarly, a GL-associated SNP (G/T, 16 733 441) on chromosome 3 was also analyzed for haplotype mining. LD analysis was performed to identify a block that contained the associated SNP. This block showed strong LD within a span of 1 kb that contained three neighboring SNPs, including the associated one (Figure 7B). Haplotype analysis of this block revealed that the GL_H1 (TTG) haplotype had the highest frequency (f = 0.631) of all the haplotypes (GL_H2; TTT = 0.315, GL_H3; TCG = 0.034, GL_H4; CCG = 0.013). Distribution of these haplotypes between long- and short-grain accessions revealed that GL_H2, in addition to being the second most frequent haplotype, was also maximally associated with long grains, with an average of 9.33 mm GL. Similarly, haplotypes GL_H1 and GL_H3 were linked to intermediate GL, with mean values of 8.2 and 7.9 mm, respectively (Figure 7B). On the other hand, the H4 haplotype showed an inclination toward the short-grain trait, with the lowest mean GL of 4.8 mm. This haplotyping observation was similar to that of a previous study that dissected the separate clustering of grain-length haplotypes for varying size and different rice groups (Singh et al., 2017).
Figure 7.
Linkage Disequilibrium and Haplotype Analysis.
(A) Depiction of strong linkage disequilibrium (LD) on chromosome 4 and the haplotype block containing the GWAS-identified SNP for panicle secondary branching (PSB). The table shows the distribution of various haplotypes for the PSB trait in the mini-core CC3 population.
(B) Depiction of strong LD on chromosome 3 and the haplotype block containing the GWAS-identified SNP for grain length (GL). The table shows the distribution of various haplotypes for the GL trait in the mini-core CC3 population. The GWAS-identified SNP ID is highlighted in blue. The SNP ID in black bold format depicts the block comprising SNP. F. S denotes the block flanking SNP. Red blocks, D′ (normalized LD measure or D) ≤1.0, with logarithm of odds (LOD) score ≥2.0; white blocks, D′ < 1.0 with LOD < 2.0; blue blocks, D′ = 1.0 with LOD < 2.0. Numbers in blocks denote D′ values. The genomic organization is described above the LD plot. LOD was defined as log10(L1/L0), where L1 = likelihood of the data under LD, and L0 = likelihood of the data under linkage equilibrium.
Concluding Remarks
Despite tremendous efforts, the resolution of QTLs responsible for yield-related traits and their causative genes has remained limited due to their complex, multifactorial nature. QTL mapping using a diverse panel and GWAS analysis have been proven to be effective tools for understanding the genetic basis of any trait. For GWAS analysis, estimation of the underlying population structure of the panel under consideration is important and helps to avoid spurious associations between phenotypes and genotypes (Pritchard and Rosenberg, 1999, Pritchard et al., 2000, Pritchard and Donnelly, 2001). Most earlier studies in rice have considered a particular population (Huang et al., 2010, Lu et al., 2015) that may have had a high level of structure and kinship affecting the GWAS analysis and resulting in spurious marker–trait associations. This study represents the first time that a large set of diversified rice germplasms (3004) was used, providing complete coverage of the global rice gene pool. The mini-core was developed using more than 2 million genome-wide SNPs, 18 different phenotypes, and 89 country locations. The mini-core accounted for 17.3% of the original collection and captured the maximum SNP polymorphism. All the original phenotypes and geographic regions were represented in the mini-core. The mini-core showed nucleotide diversity equivalent to that of the original panel, as well as low population structure and low or no kinship among individuals, thereby avoiding spurious marker–trait associations. Furthermore, an increase in the number of admixed individuals in different clusters of the CC3 structure analysis showed that the panel was unstructured and diverse in nature, appropriate for use in association analysis.
On the utility front, GWAS with the mini-core panel identified various novel marker–trait associations and validated earlier reported associations. This analysis also provided a tool for comparison between the CC3 mini-core and the original collection. We were able to show that CC3 captured the associations prevalent in the original collection and was therefore a representative subset. In conclusion, we were able to generate and validate mini-core CC3 as a robust, diversified, non-redundant, and manageable association panel that efficiently mirrored the large collection of 3004 diverse rice accessions. We suggest that this relatively small subset can be used effectively for efficient agronomic trait evaluation, which in turn will be useful for marker-assisted breeding programs for rice crop improvement.
Methods
Genotypic and Phenotypic Data of the Rice Germplasm Collection
We used SNP data from 3004 rice accessions (hereafter referred to as the original collection) and phenotypic data for 18 yield-related traits (DEH, HGW, ET, DFF, GL, GW, leaf senescence, PA, PL, panicle shattering, PT, SB, SCC, SH, spikelet fertility, culm length, culm number, and culm diameter) to develop mini-core collections and perform association analyses. The 3000 Rice Genome Project (3K RGP) data for 18.9 million polymorphic SNPs and associated phenotypic data were retrieved from the SNP-Seek database (http://snp-seek.irri.org) (Alexandrov et al., 2015, Mansueto et al., 2017). In addition, we performed whole-genome sequencing of four Indian accessions (LGR, PB-1121, Sonasal, and Bindli) at a depth of 45× and collected phenotypic data for the aforementioned traits during the 2016 and 2017 growing seasons. The original collection of rice accessions came from 89 countries and represented all the regional pools and varieties of rice grown throughout the world.
Isolation of Genomic DNA, Genome Sequencing, and SNP Calling
The four Indian rice accessions (long grain: LGR [LG] and PB 1121 [PB]; short grain: Sonasal [SN] and Bindli [BN]) were grown in a research field at the National Institute of Plant Genome Research in 2016. Ten-day-old rice seedlings were used for the isolation of genomic DNA with the Sigma GenElute Plant genomic DNA kit. The integrity of the genomic DNA was analyzed using a 2100 Bioanalyzer (Agilent Technologies, Singapore). Samples for sequencing were prepared using the Illumina TruSeq DNA sample preparation kit (Illumina, USA). Sequencing was performed with 90-bp paired-end chemistry on an Illumina HiSeq 2000 instrument.
Raw reads were quality-checked, and low-quality bases (Phred score < Q30) were removed. The filtered reads were then mapped to the rice Nipponbare reference genome (IRGSP-1.0 pseudomolecule/MSU7) using the BWA program with the –q20 setting. The Picard program was used to remove duplicate reads.
Variant calling of SNPs and InDels was performed using the Genome Analysis TKLite-2.3-9 Unified Genotyper (GATK) (McKenna et al., 2010). SNPs and InDels with a polymorphism call rate of <90% were eliminated. After calling, total variants were stringently filtered based on a read depth threshold of ≥10 and a quality score threshold of ≥30 to eliminate low-quality variants; only good-quality variants were retained for subsequent analysis. All SNPs consecutive and adjacent to indels were also eliminated. The Ensembl Plants database was used to obtain gene models for annotation. All identified SNPs and IndDels were annotated using customized VariMAT (SciGenome, India).
Development of the Mini-core Collections
The program Core Hunter 3 (De Beukelaer et al., 2018) was used to develop independent mini-core collections based on phenotypic and genotypic data. More than 2 million genome-wide SNPs and 18 phenotypic traits for 3004 rice accessions were used. A cutoff value of 10% of the initial collection was used to design the mini-core collections in Core Hunter 3 with default parameters. The mini-cores were also assessed for coverage of the entire range of all quantitative traits with reference to the initial collection. The diversity captured in the mini-core collections relative to the initial collection was assessed using multiple evaluation indices, such as Shannon's diversity index H, Nei's gene diversity I, MD%, VD%, VR%, and CR% (Hu et al., 2000). The Pearson correlation coefficient (r) was used to determine correlations between different quantitative traits using PAST version 3.10 (Hammer et al., 2001).
Phylogenetic and Population Structure Analysis
The SNP data were used to construct a distance-based dendrogram with the maximum-likelihood method in the SNPhylo program (Lee et al., 2014). Principal component analysis was performed to estimate the overall relationships among accessions. Bayesian analysis of the population structure was performed using FastSTRUCTURE (Raj et al., 2014), which estimated the optimal K value for the dataset. Pairwise kinship coefficients were estimated using SPAGeDi (Hardy and Vekemans, 2002). To estimate the proportion of ancestral contribution for each accession, we followed the admixture model. The analysis was performed independent of the geographic and varietal origin of the accessions. Accessions with a Q value (membership proportion) ≥80% were considered to be pure and assigned to a particular cluster, whereas accessions with Q < 80% were considered to be admixtures.
Genome-wide Association Studies
All GWAS analyses were performed using GAPIT (Lipka et al., 2012) based on a compressed MLM for 18 rice agronomic traits. For the original panel (3004 accessions) and the CC3 mini-core (520 accessions), 520 381 and 2 081 521 SNP markers were used, respectively. Due to a computational bottleneck in the R program, SNPs (520 381) for the original panel association study were filtered from 2 081 521 by selecting every fourth SNP. The phenotyping data for 18 traits in 2266 rice accessions were obtained from the SNP-Seek-II repository (http://snp-seek.irri.org) (Alexandrov et al., 2015, Mansueto et al., 2017). Phenotyping of the four Indian accessions was performed at two different locations (New Delhi and Chennai) in two consecutive years (2016 and 2017). The SNP data (filtered with a minor allele frequency of >0.02) and various phenotypic data for 3004 rice accessions (including the four accessions sequenced in the current study) were combined with their relative kinship matrix (K) and PCA information using a P3D/compressed MLM as described elsewhere (Lipka et al., 2012, Upadhyaya et al., 2015). The inflation factor (λ) and test statistics were evaluated using a quantile–quantile (Q–Q) plot. An FDR-corrected P value threshold of 0.05 was used for the analysis. The 100-kb genomic region (based on accepted LD decay in different rice populations) on both sides of the most significantly associated SNP was identified as the QTL region (McNally et al., 2009).
LD and Haplotype Analyses
Haplotypes were generated from the genotype data. The LD and haplotype analyses were performed using Haploview 4.2 (Barrett et al., 2005) with default parameters (MAF < 0.001), the Hardy–Weinberg equilibrium test (<0.001), and the percent genotype test (cutoff value = 75%). The four-gamete-rule method was employed to identify the more refined genomic block that contained the associated SNPs.
Funding
This study was financially supported by the grants BT/AB/NIPGR/SEED BIOLOGY/2012 and BT/BI/04/069/2006 for the establishment of Distributed Information Sub-Centre from the Department of Biotechnology, Government of India.
Author Contributions
A.K. performed all the analyses and contributed to the writing; S.K. contributed to the analyses and wrote the initial draft with contributions from all the authors; K.B.M.S. and M.P. provided technical assistance; J.K.T. conceived and supervised the project, complemented the writing, and secured funding for the project.
Acknowledgments
A.K. acknowledges the University Grant Commission, Government of India and NIPGR for the Research Fellowships. S.K. acknowledges a National Postdoctoral Fellowship from the Science and Engineering Research Board, Department of Science and Technology, Government of India and a Short-Term Research Fellowship from NIPGR. K.B.M.S. acknowledges the Council of Scientific and Industrial Research, Government of India for the Junior Research Fellowship. The authors are grateful to the DBT-eLibrary Consortium for providing access to literature. No conflict of interest declared.
Published: April 24, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information can be found online at Plant Communications Online.
Supplemental Information
References
- Agrama H.A., Yan W., Lee F., Fjellstrom R., Chen M.-H., Jia M., McClung A. Genetic assessment of a mini-core subset developed from the USDA rice genebank. Crop Sci. 2009;49:1336. [Google Scholar]
- Alexandrov N., Tai S., Wang W., Mansueto L., Palis K., Fuentes R.R., Ulat V.J., Chebotarov D., Zhang G., Li Z. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43:D1023–D1027. doi: 10.1093/nar/gku1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambreen H., Kumar S., Kumar A., Agarwal M., Jagannath A., Goel S. Association mapping for important agronomic traits in safflower (Carthamus tinctorius L.) core collection using microsatellite markers. Front. Plant Sci. 2018;9:402. doi: 10.3389/fpls.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Batayeva D., Labaco B., Ye C., Li X., Usenbekov B., Rysbekova A., Dyuskalieva G., Vergara G., Reinke R., Leung H. Genome-wide association study of seedling stage salinity tolerance in temperate japonica rice germplasm. BMC Genet. 2018 doi: 10.1186/s12863-017-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello F., Sorrells M.E. Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci. 2006;46:1323. [Google Scholar]
- Brown A.H.D. Core collections: a practical approach to genetic resources management. Genome. 2011;31:818–824. [Google Scholar]
- Brown A.H.D., Spilllane C. Implementing core collections—principles, procedures, progress, problems and promise. In: Johnson R.C., Hodgkin T., editors. Core Collections for Today and Tomorrow. IPGRI; Rome: 1999. pp. 7–17. [Google Scholar]
- De Beukelaer H., Davenport G.F., Fack V. Core Hunter 3: flexible core subset selection. BMC Bioinformatics. 2018;19:203. doi: 10.1186/s12859-018-2209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebana K., Kojima Y., Fukuoka S., Nagamine T., Kawase M. Development of mini core collection of Japanese rice landrace. Breed. Sci. 2008;58:281–291. [Google Scholar]
- Edgerton M.D. Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol. 2009;149:7–13. doi: 10.1104/pp.108.130195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizenga G.C., Ali M.L., Bryant R.J., Yeater K.M., McClung A.M., McCouch S.R. Registration of the rice diversity panel 1 for genomewide association studies. J. Plant Regist. 2014;8:109. [Google Scholar]
- Eizenga G.C., Ali M.L., Bryant R.J., Yeater K.M., McClung A.M., McCouch S.R. Registration of the Rice Diversity Panel 1 for Genomewide Association Studies. J. Plant Regist. 2014;8(1):109–116. [Google Scholar]
- El Bakkali A., Haouane H., Moukhli A., Costes E., Van Damme P., Khadari B. Construction of core collections suitable for association mapping to optimize use of mediterranean olive (Olea europaea L.) genetic resources. PLoS One. 2013;8:e61265. doi: 10.1371/journal.pone.0061265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famoso A.N., Zhao K., Clark R.T., Tung C.-W., Wright M.H., Bustamante C., Kochian L.V., McCouch S.R. Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. Plos Genet. 2011;7:e1002221. doi: 10.1371/journal.pgen.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., Li X., Zhang Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organization of the United Nations. FAOSTAT Database. 2017.
- Fuentes R.R., Chebotarov D., Duitama J., Smith S., De la Hoz J.F., Mohiyuddin M., Wing R.A., McNally K.L., Tatarinova T., Grigoriev A. Structural variants in 3000 rice genomes. Genome Res. 2019;29:870–880. doi: 10.1101/gr.241240.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAO Z. Map-based cloning of the ALK gene, which controls the gelatinization temperature of rice. Sci. China Ser. C. 2003;46:661. doi: 10.1360/03yc0099. [DOI] [PubMed] [Google Scholar]
- Gupta P.K., Rustgi S., Kulwal P.L. Linkage disequilibrium and association studies in higher plants: present status and future prospects. Plant Mol. Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- Hammer Ø., Harper D., Ryan P. Past: paleontological statistics software package for education and data analysis. Paleontol. Electron. 2001;4 http://palaeo-electronica.org/2001_1/past/issue1_01.htm [Google Scholar]
- Hardy O.J., Vekemans X. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes. 2002;2:618–620. [Google Scholar]
- Hu J., Zhu J., Xu H.M. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor. Appl. Genet. 2000;101(1–2):264–268. [Google Scholar]
- Huang X., Wei X., Sang T., Zhao Q., Feng Q., Zhao Y., Li C., Zhu C., Lu T., Zhang Z. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Wei X., Li C., Wang A., Zhao Q., Li W., Guo Y., Deng L., Zhu C. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–39. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- Ingvarsson P.K., Street N.R. Association genetics of complex traits in plants. New Phytol. 2011;189:909–922. doi: 10.1111/j.1469-8137.2010.03593.x. [DOI] [PubMed] [Google Scholar]
- Jang S., Lee Y., Lee G., Seo J., Lee D., Yu Y., Chin J.H., Koh H.-J. Association between sequence variants in panicle development genes and the number of spikelets per panicle in rice. BMC Genet. 2018;19:5. doi: 10.1186/s12863-017-0591-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A., Vilhjálmsson B.J., Segura V., Platt A., Long Q., Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 2012;44:1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump K.L., Bradbury P.J., Wisser R.J., Buckler E.S., Belcher A.R., Oropeza-Rosas M.A., Zwonitzer J.C., Kresovich S., McMullen M.D., Ware D. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- Lee T.-H., Guo H., Wang X., Kim C., Paterson A.H. SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics. 2014;15:162. doi: 10.1186/1471-2164-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Rutger J., Yu S., Xu W., Vijayakumar C., Ali J., Fu B., Xu J., Marghirang R., Domingo J. The 3,000 rice genomes project. Gigascience. 2014;3:7. [Google Scholar]
- Liakat Ali M., McClung A.M., Jia M.H., Kimball J.A., McCouch S.R., Eizenga G.C. A rice diversity panel evaluated for genetic and agro-morphological diversity between subpopulations and its geographic distribution. Crop Sci. 2011 doi: 10.2135/cropsci2010.11.0641. [DOI] [Google Scholar]
- Lipka A.E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P.J., Gore M.A., Buckler E.S., Zhang Z. GAPIT: genome association and prediction integrated tool. Bioinformatics. 2012;28:2397–2399. doi: 10.1093/bioinformatics/bts444. [DOI] [PubMed] [Google Scholar]
- Lu Q., Zhang M., Niu X., Wang S., Xu Q., Feng Y., Wang C., Deng H., Yuan X., Yu H. Genetic variation and association mapping for 12 agronomic traits in indica rice. BMC Genomics. 2015;16:1067. doi: 10.1186/s12864-015-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansueto L., Fuentes R.R., Chebotarov D., Borja F.N., Detras J., Abriol-Santos J.M., Palis K., Poliakov A., Dubchak I., Solovyev V. SNP-Seek II: a resource for allele mining and analysis of big genomic data in Oryza sativa. Curr. Plant Biol. 2016;7–8:16–25. [Google Scholar]
- Mansueto L., Fuentes R.R., Borja F.N., Detras J., Abrio-Santos J.M., Chebotarov D., Sanciangco M., Palis K., Copetti D., Poliakov A. Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res. 2017;45:D1075–D1081. doi: 10.1093/nar/gkw1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch S.R., Wright M.H., Tung C.-W., Maron L.G., McNally K.L., Fitzgerald M., Singh N., DeClerck G., Agosto-Perez F., Korniliev P. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016;7:10532. doi: 10.1038/ncomms10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.L., Childs K.L., Bohnert R., Davidson R.M., Zhao K., Ulat V.J., Zeller G., Clark R.M., Hoen D.R., Bureau T.E. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl. Acad. Sci. U S A. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell P.L., Buckler E.S., Ross-Ibarra J. Crop genomics: advances and applications. Nat. Rev. Genet. 2012;13:85–96. doi: 10.1038/nrg3097. [DOI] [PubMed] [Google Scholar]
- Nachimuthu V.V., Muthurajan R., Duraialaguraja S., Sivakami R., Pandian B.A., Ponniah G., Gunasekaran K., Swaminathan M., Suji K., Sabariappan R. Analysis of population structure and genetic diversity in rice germplasm using SSR markers: an initiative towards association mapping of agronomic traits in Oryza sativa. Rice. 2015;8:30. doi: 10.1186/s12284-015-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed S.A., Zhang F., Zhang J., Zheng T.Q., Meng L.J., Pang Y.L., Xu J.L., Li Z.K. Identification of QTN and candidate genes for salinity tolerance at the germination and seedling stages in rice by genome-wide association analyses. Sci. Rep. 2018;8:6505. doi: 10.1038/s41598-018-24946-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit A., Rai V., Bal S., Sinha S., Kumar V., Chauhan M., Gautam R.K., Singh R., Sharma P.C., Singh A.K. Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rice (Oryza sativa L.) Mol. Genet. Genomics. 2010;284:121–136. doi: 10.1007/s00438-010-0551-6. [DOI] [PubMed] [Google Scholar]
- Perseguini J.M.K.C., Silva G.M.B., Rosa J.R.B.F., Gazaffi R., Marçal J.F., Carbonell S.A.M., Chiorato A.F., Zucchi M.I., Garcia A.A.F., Benchimol-Reis L.L. Developing a common bean core collection suitable for association mapping studies. Genet. Mol. Biol. 2015;38:67–78. doi: 10.1590/S1415-475738120140126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Donnelly P. Case-control studies of association in structured or admixed populations. Theor. Popul. Biol. 2001;60:227–237. doi: 10.1006/tpbi.2001.1543. [DOI] [PubMed] [Google Scholar]
- Pritchard J.K., Rosenberg N.A. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Stephens M., Pritchard J.K. FastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shomura A., Izawa T., Ebana K., Ebitani T., Kanegae H., Konishi S., Yano M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- Singh N., Singh B., Rai V., Sidhu S., Singh A.K., Singh N.K. Evolutionary insights based on SNP haplotypes of red pericarp, grain size and starch synthase genes in wild and cultivated rice. Front. Plant Sci. 2017;8:972. doi: 10.3389/fpls.2017.00972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M.T., Thomson M.J., Pfeil B.E., McCouch S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell. 2006;18:283–294. doi: 10.1105/tpc.105.038430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Tan G., Liu Y., Rong T., Huang Y. Origin and evolution of Chinese waxy maize: evidence from the Globulin-1 gene. Genet. Resour. Crop Evol. 2009;56:247–255. [Google Scholar]
- Tiwari S., SL K., Kumar V., Singh B., Rao A., Mithra SV A., Rai V., Singh A.K., Singh N.K. Mapping QTLs for salt tolerance in rice (Oryza sativa L.) by bulked segregant analysis of recombinant inbred lines using 50K SNP chip. PLoS One. 2016;11:e0153610. doi: 10.1371/journal.pone.0153610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya H.D., Bajaj D., Das S., Saxena M.S., Badoni S., Kumar V., Tripathi S., Gowda C.L.L., Sharma S., Tyagi A.K. A genome-scale integrated approach aids in genetic dissection of complex flowering time trait in chickpea. Plant Mol. Biol. 2015;89:403–420. doi: 10.1007/s11103-015-0377-z. [DOI] [PubMed] [Google Scholar]
- Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., Li M., Zheng T., Fuentes R.R., Zhang F. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Yan J., Shah T., Warburton M.L., Li Q., Li L., Gao Y., Chai Y., Fu Z., Zhou Y. Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 2010;121:417–431. doi: 10.1007/s00122-010-1320-y. [DOI] [PubMed] [Google Scholar]
- Yu J., Buckler E.S. Genetic association mapping and genome organization of maize. Curr. Opin. Biotechnol. 2006;17:155–160. doi: 10.1016/j.copbio.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Zhang P., Liu X., Tong H., Lu Y., Li J. Association mapping for important agronomic traits in core collection of rice (Oryza sativa L.) with SSR markers. PLoS One. 2014;9:e111508. doi: 10.1371/journal.pone.0111508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Tung C.-W., Eizenga G.C., Wright M.H., Ali M.L., Price A.H., Norton G.J., Islam M.R., Reynolds A., Mezey J. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Gore M., Buckler E.S., Yu J. Status and prospects of association mapping in plants. Plant Genome J. 2008;1:5. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.