Abstract
The genetic identities of Ca2+ channels in root hair (RH) tips essential for constitutive RH growth have remained elusive for decades. Here, we report the identification and characterization of three cyclic nucleotide-gated channel (CNGC) family members, CNGC5, CNGC6, and CNGC9, as Ca2+ channels essential for constitutive RH growth in Arabidopsis. We found that the cngc5-1cngc6-2cngc9-1 triple mutant (designated shrh1) showed significantly shorter and branching RH phenotypes as compared with the wild type. The defective RH growth phenotype of shrh1 could be rescued by either the expression of CNGC5, CNGC6, or CNGC9 single gene or by the supply of high external Ca2+, but could not be rescued by external K+ supply. Cytosolic Ca2+ imaging and patch-clamp data in HEK293T cells showed that these three CNGCs all function as Ca2+-permeable channels. Cytosolic Ca2+ imaging in growing RHs further showed that the Ca2+ gradients and their oscillation in RH tips were dramatically attenuated in shrh1 compared with those in the wild type. Phenotypic analysis revealed that these three CNGCs are Ca2+ channels essential for constitutive RH growth, with different roles in RHs from the conditional player CNGC14. Moreover, we found that these three CNGCs are involved in auxin signaling in RHs. Taken together, our study identified CNGC5, CNGC6, and CNGC9 as three key Ca2+ channels essential for constitutive RH growth and auxin signaling in Arabidopsis.
Video Abstract
Key words: Ca2+ channels, CNGC, root hair, polar growth, Arabidopsis
This study identified and characterized three CNGC family members, CNGC5, CNGC6, and CNGC9, as the long-sought Ca2+ channels required for the regulation of cytosolic Ca2+ signaling in RHs and for constitutive RH growth as compared with the conditional Ca2+ channel CNGC14 in Arabidopsis. In Addition, the three CNGCs are also shown to be involved in auxin signaling in RHs in Arabidopsis.
Introduction
Root hairs (RHs) are tubular-shaped structures resulted from the outgrowth of root epidermal cells. RHs greatly increase the overall surface area of roots and are essential for the acquisition of diverse ion nutrients and water as well as the interaction between plants and rhizosphere microbes (Cui et al., 2017, Ibáñez et al., 2017. The developmental process of RHs can be roughly categorized into three steps: cell-fate determination, RH initiation, and elongating polar growth. The cell fate of root epidermis is determined mainly by an intrinsic transcriptional factor network to be either hair (H) cells or non-hair (N) cells. In this transcriptional factor network, negative regulator GL2 (GLABRA2) is the central component (Rerie et al., 1994, Di Cristina et al., 1996, Berger et al., 1998, Lin and Schiefelbein, 2001. A transcriptional factor complex composed of GL2 (GLABRA2), EGL3 (ENHANCER OF GLABRA3), WER (WEREWOLF), and TTG1 (TRANSPARENT TESTA GLABRA1) positively regulates the expression of GL2, and the accumulation of GL2 protein represses H cell differentiation and leads to N cell fate (Hung et al., 1998, Lee and Schiefelbein, 1999, Walker et al., 1999, Bernhardt et al., 2003, Bernhardt et al., 2005, whereas CPC (CAPRICE) and several redundant players function as negative regulators of GL2 (Wada et al., 1997, Wada et al., 2002, Schellmann et al., 2002, Kirik et al., 2004, Simon et al., 2007. RH initiation is a process of cell-wall loosening and bulge/swelling formation in the epidermal cells adopting H cell fate. External acidification and the production of reactive oxygen species (ROS) facilitate cell-wall loosening during RH initiation and tip growth (Bibikova et al., 1998, Liszkay et al., 2004, Monshausen et al., 2007. The local assembly of F-actin initiates bulge formation, and supports sustained tip growth of RHs (Baluska et al., 2000). A number of components, including ROS, a few small guanosine 5′-triphosphate (GTP)-binding proteins from ROP (Rho-related GTPase from plant) family, receptor-like kinase FERONIA (FER), and guanine nucleotide exchange factors (ROPGEFs), are involved in RH initiation and tip growth (Wymer et al., 1997, Molendijk et al., 2001, Foreman et al., 2003, Duan et al., 2010, Huang et al., 2013. The production of ROS triggers external Ca2+ influx and cytosolic Ca2+ elevation during RH initiation. It is believed that ROS production is required for RH initiation, but ROS-induced cytosolic Ca2+ elevation is not (Wymer et al., 1997, Foreman et al., 2003.
Tip growth of RHs is a process of exocytosis and cytoplasmic restructuring (Ketelaar et al., 2008, Rounds and Bezanilla, 2013, which is regulated by a complex machinery composed of diverse components, including cytoskeleton of F-actin and microtubules, ROP proteins, ROS, and cytosolic Ca2+ signaling (Pei et al., 2012, Grierson et al., 2014. The cytosolic Ca2+ gradient starts from the elevation of cytosolic Ca2+ during RH initiation in epidermis, and the high cytosolic Ca2+ area enters RHs after RH initiation to form a Ca2+ gradient in RH apex. The Ca2+ gradient functions as a key regulator of RH growth and orientation similar to its functions in other tip-growing cells, including pollen tubes, fungal hyphae, and neurons (Konrad et al., 2011, Guan et al., 2013, Akiyama and Kamiguchi, 2015, and the Ca2+-sensitive components of the complex machinery for tip growth regulation can be the targets of Ca2+ signaling, including F-actin binding proteins, microtubule binding proteins, myosin XI motors, and ROPs (Bibikova et al., 1999, Baluska et al., 2000, Molendijk et al., 2001, Sieberer et al., 2005, Tominaga et al., 2012. Early studies revealed that the presence and oscillation of the Ca2+ gradient at RH tips are required for the tip growth and orientation of RHs (Bibikova et al., 1997, Felle and Hepler, 1997, Wymer et al., 1997, Monshausen et al., 2008, and that external Ca2+ influx through inward Ca2+ channels in the plasma membrane at RH tips is the main source of Ca2+ for the establishment, maintenance, and regulation of the Ca2+ gradients (Schiefelbein et al., 1992, Herrmann and Felle, 1995, Felle and Hepler, 1997, Wymer et al., 1997. Therefore, plasma membrane Ca2+ channels at RH tips become key components for the regulation of the Ca2+ gradients, RH tip growth, and orientation. However, the identities of the Ca2+ channels have remained largely unknown for decades. Electrophysiological analysis has detected the activity of three types of Ca2+ channels in Arabidopsis root epidermis and RHs, including Ca2+-permeable non-selective cation channels (NSCCs), hyperpolarization-activated Ca2+ channels (HACCs), and depolarization-activated Ca2+ channel (DACCs) (Kiegle et al., 2000, Véry and Davies, 2000, Demidchik et al., 2002, Miedema et al., 2008). A model has been proposed that NSCCs and HACCs co-exist in root epidermal cells (Demidchik et al., 2002), and HACCs and DACCs co-exist in RHs in Arabidopsis (Miedema et al., 2008). NSCC-mediated Ca2+ influx dominates in mature epidermal cells, whereas HACC-mediated Ca2+ influx predominates in RHs (Demidchik et al., 2002). Further research has detected the activity of hyperpolarization-activated Ca2+ channels in Arabidopsis RHs, which could be activated by NADPH oxidase AtRBOHC (RHD2)-dependent ROS accumulation (Foreman et al., 2003, Liszkay et al., 2004), and tip-localized ROP proteins and their negative regulator RhoGDI (RhoGTPase GDP dissociation inhibitor) function as upstream regulators of RHD2 in RH growth (Molendijk et al., 2001, Jones et al., 2002, Carol et al., 2005, Carol and Dolan, 2006). Efforts have been made to identify the Ca2+ channels essential for RH growth, and progress has been made in the analysis of annexin and cyclic nucleotide-gated channel (CNGC) families in Arabidopsis. Annexin1 is the first Ca2+-permeable channel identified in RHs, which is an ROS-activated non-selective K+- and Ca2+-permeable cation channel in Arabidopsis (Laohavisit et al., 2012). Both hyperpolarized and depolarized voltage-activated channel currents were observed in Arabidopsis RH apical spheroplast, but no significant difference was observed between ann1 mutant and wild type (Laohavisit et al., 2012). Arabidopsis mutant cngc14-1 showed a conditional short-hair phenotype only when RHs grew in solid Murashige and Skoog (MS) medium, and did not show any detectable RH defect as compared with the wild type when RHs were exposed to air (Zhang et al., 2017). Arecent study reported that the cngc5cngc9cngc14 triple mutant did not form any RHs under their experimental conditions (Brost et al., 2019), suggesting that CNGC5/9/14 are required for RH initiation. However, the Ca2+ channels essential for constitutive RH growth have not been identified yet. In this study, we report that three CNGC members, CNGC5, CNGC6, and CNGC9, function redundantly as Ca2+ channels essential for constitutive RH tip growth by maintaining and regulating a sharp cytosolic Ca2+ gradient and its oscillation in RH tips in Arabidopsis, which are different from the roles of the conditional player CNGC14 in RHs. We also found that CNGC5, CNGC6 and CNGC9 are involved in auxin signaling in RHs.
Results
The cngc5 cngc6 cngc9 Triple and cngc5 cngc6 cngc9 cngc14 Quadruple Knockout Mutants Show Strong Defects in RH Growth
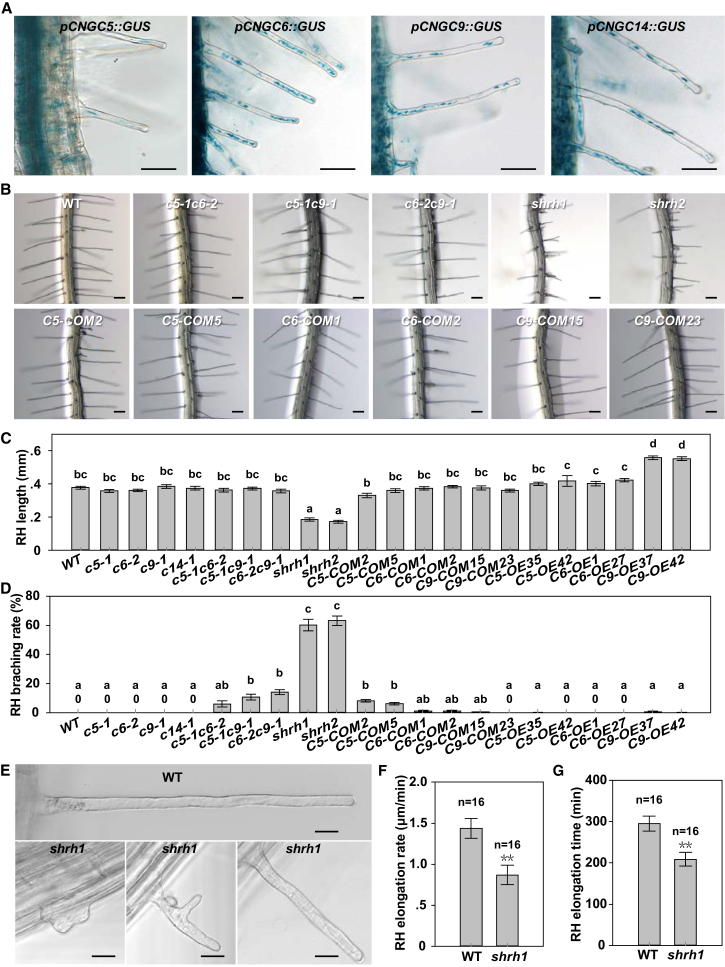
Pollen tubes and RHs are both tip-growing cells. We revealed that CNGC18 was the dominant Ca2+ channel for pollen germination, pollen tube growth, and orientation (Gao et al., 2016, Gu et al., 2017), and other groups recently observed RH-related phenotypes in a few cngc mutants in Arabidopsis (Zhang et al., 2017, Brost et al., 2019). Thus, we hypothesized that the Ca2+ channels essential for RH growth are composed of CNGC members in Arabidopsis. To test the hypothesis, we generated a set of transgenic Arabidopsis lines by introducing a β-glucuronidase (GUS) encoding gene driven by a native promoter of the 20 CNGC members. GUS staining results showed that CNGC5, CNGC6, CNGC9, and CNGC14 were strongly expressed in RHs (Figure 1A), whereas the remaining 16 CNGC members were not obviously expressed in RHs (Supplemental Figure 1). We collected T-DNA insertional single mutants cngc5-1, cngc6-2, cngc9-1, and cngc14-1 and double mutant cngc5-1cngc6-2 (Supplemental Figure 2A) (Wang et al., 2013). cngc5-1 and cngc9-1 were confirmed as knockout mutants (Supplemental Figure 2B and 2C), and cngc6-2 was a knockdown mutant (Supplemental Figure 2D) (Wang et al., 2013). cngc14-1 was already known as a knockout mutant (Zhang et al., 2017). We then generated the double mutants cngc5-1cngc9-1 and cngc6-2cngc9-1 by crossing the related single mutants with each other, triple mutant cngc5-1cngc6-2cngc9-1 by crossing cngc5-1cngc9-1 with cngc5-1cngc6-2, and quadruple mutant cngc5-1cngc6-2cngc9-1cngc14-1 by crossing cngc5-1cngc6-2cngc9-1 with cngc14-1. We analyzed RH-related phenotypes in these single, double, triple, and quadruple mutants by analyzing the RHs exposed to air. We found that the RH lengths in the wild type, four single mutants, and three double mutants were similar to each other without significant difference (Figure 1B and 1C; Supplemental Figure 3A). However, the RHs of triple mutant cngc5-1cngc6-2cngc9-1 and quadruple mutant cngc5-1cngc6-2cngc9-1cngc14-1 were significantly shorter compared with that of the wild type, four single mutants, and three double mutants (Figure 1B and 1C). We thus designated the triple mutant cngc5-1cngc6-2cngc9-1 and quadruple mutant cngc5-1cngc6-2cngc9-1cngc14-1 as shrh1 (short root hair1) and shrh2, respectively. The RHs were significantly shorter by 50.9% for shrh1 and 54% for shrh2 compared with the wild type. We further observed weak RH branching defective phenotype in three double mutants and strong RH branching defective phenotype in shrh1 and shrh2 (Figure 1D and 1E). The RH branching rates were 6.5% for cngc5-1cngc6-2, 14% for cngc5-1cngc9-1, 15% for cngc6-2cngc9-1, 60.2% for shrh1, and 63.2% for shrh2 (Figure 1D). We rarely observed branched RH in the wild type and the four single mutants, which showed RH branching rates of lower than 0.1% (Figure 1D). RH density (RH number per millimeter root length) was not significantly altered in all the mutants compared with the wild type and each other (Supplemental Figure 3B). The diameters of RH trunk close to RH tips and the diameters of RH initiation area were obviously variable and significantly larger by 34% and 43% in shrh1 mutant as compared with that in the wild type, respectively (Figure 1E; Supplemental Figure 3C and 3D). No RH rupturing phenotype was observed in all genotypes. Of note, RHs exposed to the air and located in 2-mm range of the RH zone (RHZ) of roots (Supplemental Figure 3E) were counted in these RH phenotype analyses, including RH length, density, and branching. Next, we mainly focused on CNGC5, CNGC6, and CNGC9 in further investigation, considering that the RH phenotypes of shrh1 and shrh2 were similar to each other.
Figure 1.
CNGC5, CNGC6, CNGC9, and CNGC14 Are Required for RH Growth in Arabidopsis.
(A) GUS staining results of CNGC5, CNGC6, CNGC9, and CNGC14 in RHs.
(B) Images of typical roots and hairs.
(C and D) Statistical analysis of RH length (C) and branching rate (D).
(E) Images of typical RHs.
(F and G) Statistical analysis of elongation rate (F) and elongating time of RHs (G).
WT, wild type. See the figure legend of Supplemental Figure 3 for the annotation of genotypic initials of Arabidopsis lines. No less than 30 roots and 200 RHs (7–10 RHs per root) were counted for each analysis of each line. Error bars depict means ± SEM. Samples with different letters are significantly different with P < 0.05 (Kruskal–Wallis one-way ANOVA) in (C) and (D). Letter n denotes the numbers of RH analyzed, and ** denotes significant difference with P < 0.01 (Mann–Whitney Rank Sum Test) in (F) and (G). Scale bars, 0.05 mm (A), 0.1 mm (B), and 20 μm (E).
To oversee the dynamic growth of RHs, we performed time-lapse experiments to monitor RH growth for no less than 300 min. A set of optical sections under bright field were captured each 1 min, and the set of sectioning images were automatically merged into a single picture by a computer using the software Leica Application Suite X (Leica, Germany). RHs growing at different angles were able to be seen clearly in the merged bright-field pictures. These data showed that shrh1 RHs grew significantly slower and ceased growth earlier than wild type (Figure 1F and 1G; Supplemental Figure 4). The RH initiation zone (RHIZ) of roots (Supplemental Figure 3E) was selected for the time-lapse experiments in bright field for RHs.
To further test whether the defective RH phenotypes resulted from the mutations in CNGC5, CNGC6, and CNGC9, we generated rescued lines by expressing the CNGCs in shrh1 mutant background under their native promoters. We selected two complemented (COM) lines for each of the three CNGCs, six rescued lines in total, namely CNGC5-COM2 (CNGC5-COMPLEMENTED2), CNGC5-COM5, CNGC6-COM1, CNGC6-COM2, CNGC9-COM15, and CNGC9-COM23, for further analysis. The expression of CNGC5, CNGC6, and CNGC9 in the COM lines was confirmed by qRT–PCR experiments (Supplemental Figure 2B–2D). We found that the average RH lengths of the six COM lines were significantly longer than that of shrh1 and shrh2 mutants, but similar to each other and that of wild type, single mutants, and double mutants (Figure 1B and 1C). The RH branching phenotype of shrh1 was also dramatically repressed by the expression of the three CNGCs (Figure 1D). RH densities of the COM lines were similar to each other and that of wild type and all the mutants (Supplemental Figure 3B). These data confirmed that the RH phenotypes of shrh1 mutant resulted from the mutations in CNGC5, CNGC6, and CNGC9.
We next generated overexpression (OE) lines by overexpressing the three CNGCs with an eGFP fused to their N terminus under control of the Arabidopsis Ubiquitin10 promoter in the wild-type background. Two OE lines were selected for each CNGC, and six OE lines in total, namely CNGC5-OE35 (CNGC5-OVEREXPRESSION35), CNGC5-OE42, CNGC6-OE1, CNGC6-OE27, CNGC9-OE37, and CNGC9-OE42, for experiments. The overexpression of CNGC5, CNGC6, and CNGC9 in the OE lines was confirmed by qRT–PCR (Supplemental Figure 2B–2D). We found that the average RH lengths of CNGC5-OE35, CNGC5-OE42, CNGC6-OE1, and CNGC6-OE27 were significantly longer than that of shrh1 and shrh2, but similar to each other and that of wild type, single mutants, double mutants, and COM lines except CNGC5-COM2 (Figure 1C). The average RH lengths were significantly increased in CNGC9-OE37 and CNGC9-OE42 compared with wild type, mutants, COM lines, and the remaining four OE lines (Figure 1C). Considering that CNGC9 rescued the RH phenotype of shrh1 mutant to a similar extent as that of CNGC5 and CNGC6, the longer RHs in CNGC9-OE37 and CNGC9-OE42 implied that CNGC9 could play a slightly dominant role in RH growth relative to CNGC5 and CNGC6. We rarely observed branching RHs in all six OE lines (Figure 1D). The RH densities of the OE lines were not obviously altered relative to wild type, mutants, and COM lines (Supplemental Figure 3B). Together, these data demonstrate that CNGC5, CNGC6, and CNGC9 are required for tip growth of RHs in the air, in which CNGC9 plays a more important role than CNGC5 and CNGC6 in Arabidopsis.
CNGC5, CNGC6, and CNGC9 Are Predominantly Localized in the Periphery of RHs
We analyzed the subcellular localization of the three CNGCs by analyzing the eGFP fluorescent signal in the RHs of three OE lines CNGC5-OE42, CNGC6-OE27, and CNGC9-OE42, in which eGFP-CNGC5, eGFP-CNGC6, and eGFP-CNGC9 were overexpressed. However, we observed a dominant plasma membrane localization of the three CNGC proteins plus some distribution in cytoplasm (data not shown). The cytoplasm distribution of the three CNGC proteins could be a result of the overexpression of the three CNGCs. We thus analyzed the eGFP fluorescence distribution in three weaker OE lines, namely CNGC5-OE4, CNGC6-OE12, and CNGC9-OE18. We found that the eGFP fluorescent signal was distributed randomly in wild-type RHs (Supplemental Figure 5). However, the eGFP fluorescent signal was clearly distributed in the peripheral area of RHs (Supplemental Figure 5). Plasma membrane dye FM4-64 was then used to stain RHs, and an overlap of FM4-64 signal and eGFP fluorescence was clearly observed in the peripheral area in the merged photos, demonstrating a plasma membrane localization of the three CNGC proteins (Supplemental Figure 5).
The Defective RH Phenotypes in Shrh1 Could Be Rescued by the Supply of High External Ca2+ Rather than K+
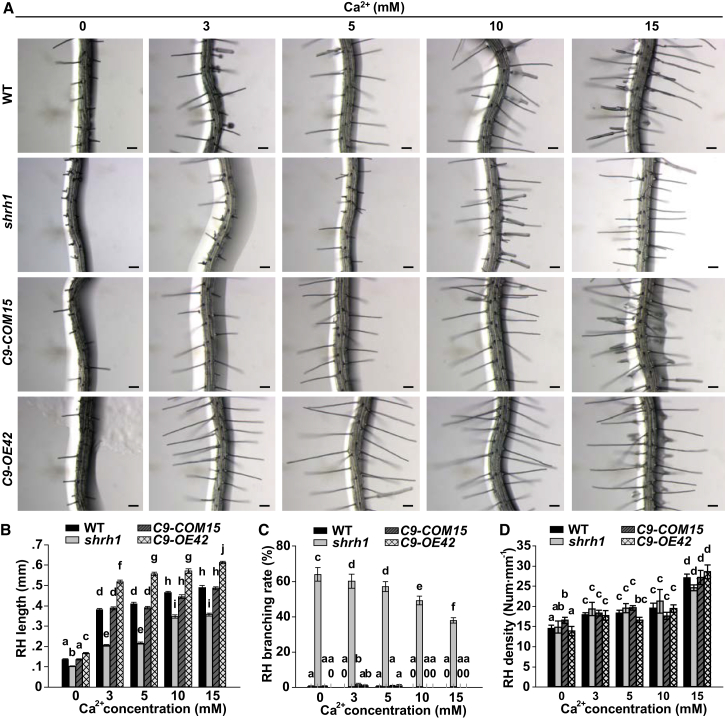
The CNGC family has been predicted as NSCCs. Research from different groups revealed that CNGC2 and CNGC4 are permeable to both K+ and Ca2+ (Leng et al., 1999, Leng et al., 2002, Wang et al., 2017), whereas several other CNGCs (including CNGC7, 8, 9, 10, 11, 12, 14, 16, and 18) are selective to Ca2+ (Urquhart et al., 2011, Gao et al., 2014, Gao et al., 2016, Zhang et al., 2017). The double mutant cngc5-1cngc6-2 showed disrupted Mg2+-permeable channel activity in Arabidopsis guard cells (Wang et al., 2013), suggesting that CNGC5 and CNGC6 are Ca2+-permeable channels because most Ca2+ channels are permeable to diverse divalent cations, including Mg2+, Ca2+, and Ba2+. Thus, we hypothesized that CNGC5, CNGC6, and CNGC9 function as inward Ca2+ channels in RHs. To test the hypothesis, we tested external Ca2+ dependence of RH growth. We found that RH growth was strongly impaired by the absence of external Ca2+ (no Ca2+ added) compared with control condition (3 mM external Ca2+ added), and RHs grew significantly longer when the external Ca2+ concentration ([Ca2+]ext) was increased to a higher concentration in all genotypes tested (Figure 2A and 2B). The RH branching rates were not obviously altered in wild type, CNGC9-COM15, and CNGC9-OE42 upon the increases of [Ca2+]ext, but were significantly reduced in shrh1 mutant relative to wild type, CNGC9-COM15, and CNGC9-OE42 (Figure 2C). These data demonstrate that the shorter and branching RH phenotypes of shrh1 resulted from reduced external Ca2+ influx, consistent with the Ca2+ dependence of RH growth (Schiefelbein et al., 1992). In addition, the promotion of RH growth by high external Ca2+ in wild type, shrh1, CNGC9-COM15, and CNGC9-OE42 (Figure 2A and 2B) suggests the presence of other Ca2+ channels in RHs, and the significantly longer RHs of CNGC9-OE42 relative to CNGC9-COM15 and wild type in multiple external Ca2+ conditions (Figure 2B) support a dominant role of CNGC9 relative to CNGC5 and CNGC6 in RH growth. Moreover, we found that the RH densities were significantly reduced by the absence of external Ca2+ (no Ca2+ added), and significantly increased at the 15 mM [Ca2+]ext condition compared with 3–10 mM [Ca2+]ext condition in all the genotypes tested (Figure 2D), suggesting that RH initiation could be affected by low (0 mM) and high (15 mM) [Ca2+]ext conditions relative to modest external Ca2+ conditions.
Figure 2.
External Ca2+ Dependence of RH Growth in Arabidopsis.
(A) Images of typical roots and hairs. Scale bars, 0.1 mm.
(B–D) Average lengths (B), branching rates (C), and densities (D) of RHs at [Ca2+]ext as indicated. C9-COM15 and C9-OE42 denote CNGC9-COM15 and CNGC9-OE42, respectively. No less than 30 roots and 200 RHs (7-10 RHs per root) were analyzed for each analysis of each line. Error bars depict means ± SEM. Samples with different letters are significantly different with P < 0.05 (two-way ANOVA).
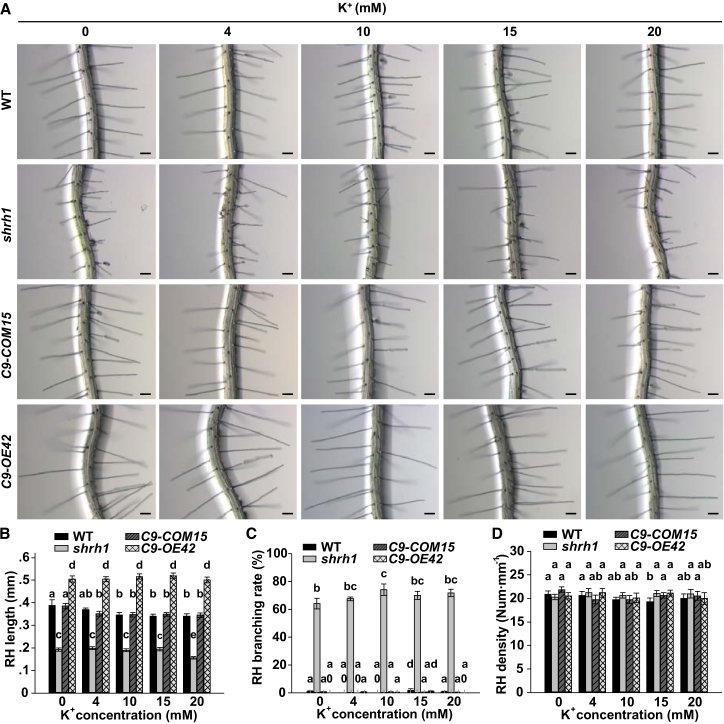
To test whether the three CNGCs were also involved in RH growth as K+ channels by mediating external K+ influx, we fixed external Ca2+ concentration at 3 mM, and analyzed the external K+ dependence of RH length, branching, and density. We found that the average RH length, branching rate, and density were not obviously altered upon the increase of external K+ concentration ([K+]ext) from micromolar level (no external K+ added) (Xu et al., 2006) to 4 mM, 10 mM, 15 mM, and 20 mM in shrh1 mutant, CNGC9-COM15, CNGC9-OE42, and wild type (Figure 3A–3D). However, the RHs were still significantly shorter in shrh1 mutant and significantly longer in CNGC9-OE42 compared with the wild type and CNGC9-COM15 (Figure 3A and 3B), suggesting that the RH-related phenotypes in shrh1 were not obviously related to K+. Together, these data suggest that CNGC5, CNGC6, and CNGC9 function specifically as essential Ca2+ channels rather than K+ channels in RH growth in Arabidopsis.
Figure 3.
Defective RH Growth Phenotypes in Shrh1 Are Independent of External K+.
(A) Images of typical roots and hairs at external [K+] as indicated. Scale bars, 0.1 mm.
(B–D) Statistical analysis of length (B), branching rate (C), and density (D) of RHs. C9-COM15 and C9-OE42 denote CNGC9-COM15 and CNGC9-OE42, respectively. No less than 30 roots and 200 RHs (7-10 RHs per root) were analyzed for each analysis of each line. Error bars depict means ±SEM. Samples with different letters are significantly different with P < 0.05 (two-way ANOVA).
CNGC5, CNGC6, and CNGC9 Are Ca2+-Permeable Channels
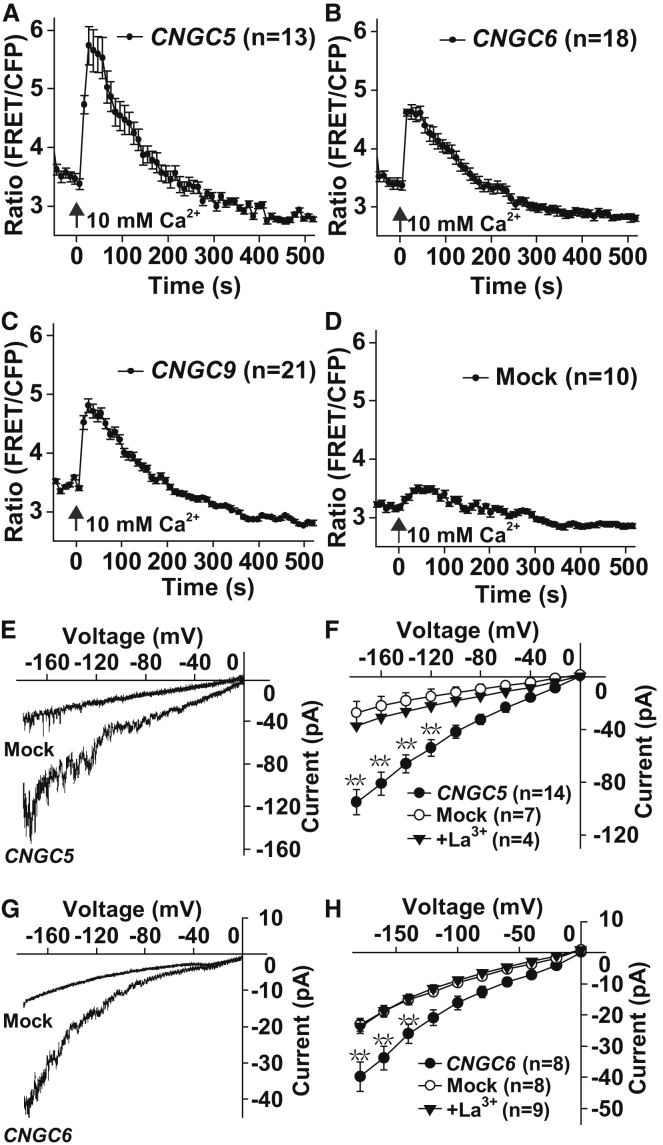
To characterize CNGC5, CNGC6, and CNGC9, we conducted cytosolic Ca2+ imaging experiments by transient expression in HEK293T cells. We observed a large cytosolic Ca2+ increase in HEK293T cells expressing one of the three CNGCs upon the application of 10 mM external Ca2+ (Figure 4A–4C), but only a small cytosolic Ca2+ increase was observed in some of the mock control HEK293T cells expressing eGFP (Figure 4D), suggesting that the three CNGCs are Ca2+-permeable channels. HEK293T cells have an endogenous Ca2+ channel in the plasma membrane called CRACM1, which is specifically activated by the depletion of intracellular Ca2+ stores (Feske et al., 2006). It is not clear whether the small occasional Ca2+ increase in mock control HEK293T cells resulted from CRACM1-mediated external Ca2+ influx. However, CNGC-mediated cytosolic Ca2+ increases were clear and sharp without being obviously affected by the small and occasional background Ca2+ increase in HEK293T cells.
Figure 4.
CNGC5, CNGC6, and CNGC9 Are Ca2+-Permeable Channels.
(A–D) Cytosolic Ca2+ imaging data showing 10 mM Ca2+-triggered cytosolic Ca2+ increases in HEK293T cells expressing CNGC5(A), CNGC6(B), and CNGC9(C) compared with the control HEK293T cells expressing YC3.6(D).
(E–H) Patch-clamp analysis of CNGC5- and CNGC6-mediated channel currents in HEK293T cells. (E and F) Typical whole-cell recordings (E) and average current–voltage curves of steady-state whole-cell currents (F) recorded in HEK293T cells expressing either CNGC5 or eGFP. (G and H) Typical whole-cell recordings (G) and average current–voltage curves of steady-state whole-cell currents (H) recorded in HEK293T cells expressing either CNGC6 or eGFP.
Error bars depict means ± SEM. Letter n denotes the numbers of cells tested, and ** denotes significant difference with P < 0.01 (Kruskal–Wallis one-way ANOVA).
We next conducted patch-clamp experiments in HEK293T cell using Ca2+-based standard bath and pipette solutions (Gao et al., 2014, Gao et al., 2016). HEK293T cells were transformed, and the transformed cells with eGFP fluorescence in the similar size were selected for patch-clamp analysis. We observed large inward Ca2+ channel currents in HEK293T cells expressing either CNGC5 (Figure 4E and 4F) or CNGC6 (Figure 4G and 4H), and these large channel currents were abolished upon the application of the Ca2+ channel blocker La3+ (100 μM) (Figure 4F and 4H). We observed only a small background conductance in mock control cells (Figure 4E–4H). To test whether the inward channel currents were carried by Na+ considering the presence of 120 mM Na+ in the bath solution, we substituted Na+ with the impermeant cation NMDG+ (N-methyl-D-glucamine) in the bath solution, and kept the pipette solution unchanged. We pursued further patch-clamp experiments, and observed obvious inward channel currents in HEK293T cells expressing either CNGC5 (Supplemental Figure 6A and 6B) or CNGC6 (Supplemental Figure 6C and 6D). The inward channel currents were abolished upon the application of the Ca2+ channel blocker Gd3+ (100 μM) (Supplemental Figure 6A–6D). The large inward channel currents recorded in NMDG+-based bath solution were similar to the currents recorded in Na+-based bath solution, and the inhibitory effect of Gd3+ was similar to that of La3+ (Figure 4E–4H and Supplemental Figure 6). These data demonstrate that the inward channel currents recorded in HEK293T cells expressing either CNGC5 or CNGC6 were mainly carried by Ca2+, not Na+.
The cytosolic Ca2+ imaging data and patch-clamp data together demonstrate that CNGC5 and CNGC6 are Ca2+-permeable channels. CNGC9 has been characterized as a Ca2+-permeable channel in HEK293T cells using a patch-clamp technique (Gao et al., 2016). Thus, CNGC5, CNGC6, and CNGC9 are all Ca2+-permeable channels.
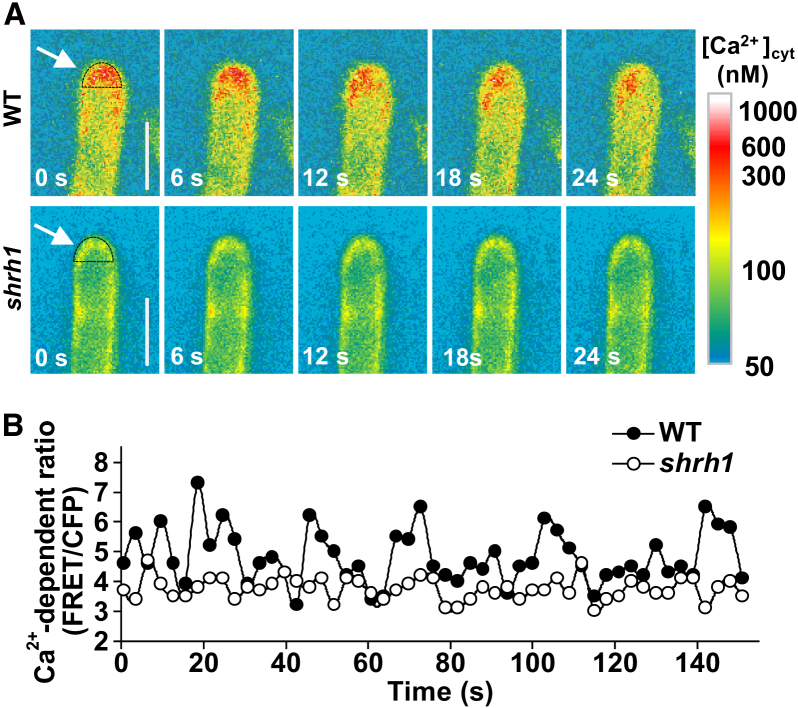
Cytosolic Ca2+ Gradients in RH Tips Are Dramatically Attenuated in Shrh1
To investigate how CNGC5, CNGC6, and CNGC9 are involved in RH growth as Ca2+ channels in vivo, we generated transgenic Arabidopsis lines by introducing a construct containing the Ca2+ indicator encoding gene of Yellow Cameleon version 3.6 (YC 3.6) under the control of Arabidopsis Ubiquitin10 promoter into shrh1 mutant and wild-type plants and performed time-lapse experiments to monitor the Ca2+ gradient and oscillation in growing RH tips. We observed not only a much stronger ratio signal of fluorescence resonance energy transfer (FRET)/CFP, but also a significantly sharper gradient of FRET/CFP in wild-type RH apex as compared with that in shrh1 mutant throughout the whole oscillating growth phase (Figure 5A), suggesting that cytosolic Ca2+ gradients in shrh1 RH tips were significantly reduced compared with those in the wild type. The vacuole regions were observed clearly in both wild-type and shrh1 RHs, confirming the growth of the RHs (Supplemental Video 1).
Figure 5.
Cytosolic Ca2+ Oscillation in RH Tips Is Strongly Attenuated in shrh1.
(A) Typical time-elapses of RH images showing a sharp cytosolic Ca2+ gradient and its strong oscillation in wild type, and an attenuated Ca2+ gradient and its weaker oscillation in shrh1 mutant in RH tips (left). A pseudocolor scale bar for relative cytosolic Ca2+ level calibration is shown on the right. Scale bars, 10 μm.
(B) Typical oscillation of FRET/CFP ratio of YC3.6 in RH tips in wild type and shrh1 mutant. The white arrows in (A) show the regions of interest for Ca2+ oscillation measurement. Fifteen out of 18 wild-type RHs tested showed a sharp Ca2+ gradient and obvious Ca2+ oscillation in RH tips. Twelve out of 12 shrh1 RHs tested showed much weaker Ca2+ gradient and oscillation relative to wild type.
We further analyzed the cytosolic Ca2+ oscillation in RH apex. The wild type and shrh1 mutant RHs showed similar Ca2+ oscillation periods (Figure 5B). However, cytosolic Ca2+ oscillation in shrh1 was much weaker compared with that of wild type (Figure 5B). Results similar to those shown in Figure 5 were observed in 15 out of 18 wild-type RHs tested and 12 out of 12 shrh1 RHs tested. To test whether the weaker Ca2+ gradient and oscillation in shrh1 RHs resulted from a lower expression of YC3.6 relative to wild type, we conducted in situ [Ca2+]cyt calibration (Swanson and Gilroy, 2013). Similar Rmin and Rmax were recorded in wild-type and shrh1 RHs (Supplemental Figure 7A–7D), and the estimated [Ca2+]cyt in shrh1 was similar to that of wild type (Supplemental Figure 7E and 7F), suggesting that the expression of YC3.6 in shrh1 RHs was similar to that of wild type. These data together demonstrate that CNGC5, CNGC6, and CNGC9 are involved in RH growth by establishing and maintaining a sharp Ca2+ gradient and regulating the Ca2+ oscillation in RH apex, both of which are required for RH tip growth.
CNGC5, CNGC6, and CNGC9 Are Essential for Constitutive Growth of RHs
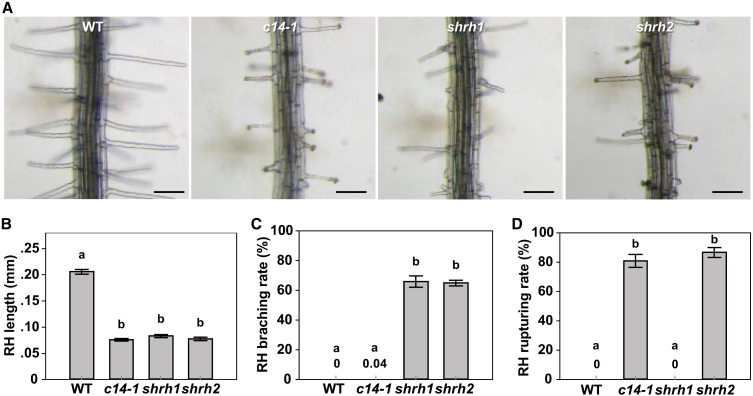
It has been reported that cngc14-1 mutant showed short RH phenotype only when RHs grew in solid medium, not in air (Zhang et al., 2017). The RH phenotypes observed in this research were mainly related to the RHs exposed to air. One possibility is that CNGC5, CNGC6, and CNGC9 function in one signaling pathway while CNGC14 functions in another. A second possibility is that CNGC5, CNGC6, and CNGC9 are Ca2+ channels essential for constitutive RH growth in all different conditions, whereas CNGC14 is only a conditional player. To test these possibilities, we analyzed the RH phenotypes when RHs grew in solid medium, not in air. The experimental results showed that the average RH lengths of cngc14-1, shrh1, and shrh2 were similar to each other without significant difference, but significantly shorter compared with wild type (Figure 6A and 6B), demonstrating that the four CNGC members are all required for polar growth of RHs in solid medium. The RH branching rate was 66% for shrh1 and 65% for shrh2 in this experimental condition, which were similar to each other but significantly higher than that of wild type (0%) and cngc14-1 mutant (0.04%) (Figure 6C), indicating that the RH branching phenotype we observed mainly resulted from the simultaneous mutations in CNGC5, CNGC6, and CNGC9, not CNGC14. We also observed ruptured RHs in solid medium, and the rupturing rates were zero for the wild type and shrh1, 80.9% for cngc14-1, and 86.6% for shrh2 (Figure 6D), suggesting that the RH rupturing phenotype in the condition we used mainly resulted from the mutation in CNGC14, not in CNGC5, CNGC6, and CNGC9. The wild-type, cngc14-1, shrh1, and shrh2 plants showed similar RH density (Supplemental Figure 8), suggesting that all four CNGCs were not obviously involved in RH initiation under these conditions. These data together suggest that CNGC5, CNGC6, and CNGC9 are key Ca2+ channels for constitutive RH growth, while CNGC14ris a conditional Ca2+ channel in RH growth, given that the defective RH growth phenotypes exposed to air mainly result from simultaneous mutations in CNGC5, CNGC6, and CNGC9 (Figure 1; Supplemental Figures 3 and 4).
Figure 6.
Phenotypic Analysis of Arabidopsis RHs Grown in Solid Medium.
(A) Images of typical roots and hairs. Scale bars, 0.1 mm.
(B–D) Statistical analysis of RH length (B), branching rate (C), and rupturing rate (D) of RHs. No less than 30 roots and 200 RHs (7–10 RHs per root) were counted for each analysis of each line. Error bars depict means ±SEM. Samples with different letters are significantly different with P < 0.05 (Kruskal–Wallis one-way ANOVA). See also Supplemental Figure 7 for RH density analysis.
CNGC5, CNGC6, and CNGC9 Are Involved in Auxin Signaling in RHs
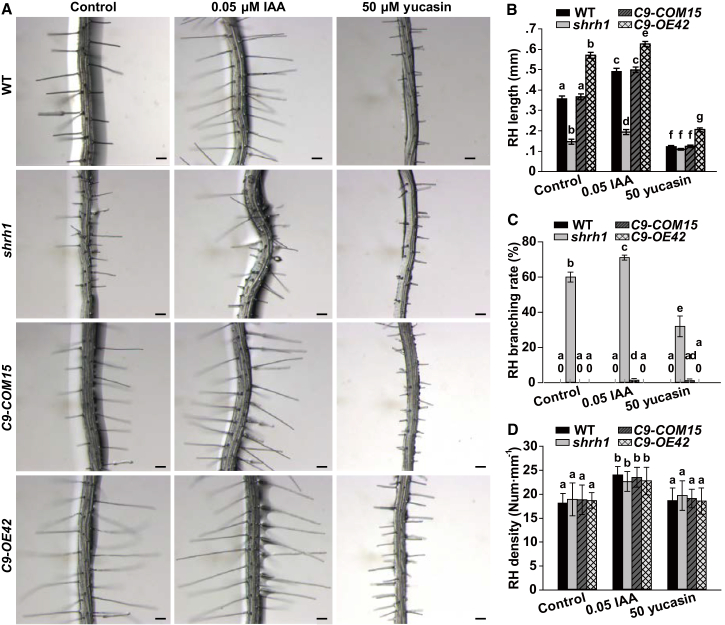
Cyclic nucleotides (cAMP and cGMP) are well characterized as the activators of mammalian cyclic nucleotide-gated channels, which are the orthologs of Arabidopsis CNGCs in mammalian cells. However, it took decades to confirm the presence of cAMP and cGMP and to identify the enzymes responsible for the synthesis and breakdown of the small molecules in plants (Raji and Gehring, 2017). cAMP and cGMP at the femtomolar level have been detected in pollen tubes, which are polar growth plant cells similar to RHs (Tunc-Ozdemir et al., 2013). We thus analyzed the effect of cAMP and cGMP on RH growth by adding either 1 μM cAMP or 1 μM cGMP in medium. However, no obvious effect of the two compounds was observed in RH growth and initiation in shrh1 mutant and wild type (data not shown). It is well known that the initiation and growth of RHs are strongly regulated by auxin, ethylene, and ion nutrient availability. Of these factors, auxin is a dominant stimulating signal for RH initiation and growth (Grierson et al., 2014). To investigate whether CNGC5, CNGC6, and CNGC9 functioned as key components of the auxin signaling pathway in RH growth, we analyzed the effects of indole-3-acetic acid (IAA) and the auxin biosynthesis inhibitor yucasin (Nishimura et al., 2014) on RHs exposed to the air. Either IAA (0.05 μM) or yucasin (50 μM) was added to the solid medium. The experimental results demonstrated that RH growth was significantly promoted upon the application of 0.05 μM external IAA, but was significantly inhibited upon the application of 50 μM external yucasin in shrh1, wild type, CNGC9-COM15, and CNGC9-OE42 to a similar extent without significant difference (Figure 7A and 7B; Supplemental Table 1). The RH branching rate was significantly increased by IAA and significantly reduced by yucasin in shrh1 mutant relative to control conditions (Figure 7C). However, we did not observe an obvious change in the rupturing rate upon the application of either auxin or yucasin in wild type, CNGC9-COM15, and CNGC9-OE42 (data not shown). RH density was significantly increased by auxin compared with control conditions to a similar extent in all genotypes, but was not obviously altered by yucasin relative to control (Figure 7D). These data suggest that CNGC5, CNGC6, and CNGC9 are also involved in RH initiation, and play a role in maintaining the integrity of RHs from bursting. However, it remains unclear whether auxin stimulates RH elongation via CNGC5, CNGC6, and CNGC9.
Figure 7.
CNGC5, CNGC6, and CNGC9 Are Involved in Auxin Signaling in RHs.
(A) Typical roots and hairs. Scale bars, 0.1 mm.
(B–D) Statistical analysis of length (B), branching rate (C), and density (D) of RHs. No less than 30 roots and 200 RHs (7–10 RHs per root) were counted for each analysis of each line. Error bars depict means ±SEM. Samples with different letters are significantly different with P < 0.05 (Kruskal–Wallis one-way ANOVA).
Discussion
CNGC-Mediated External Ca2+ Influx Matters for RH Development
Cytosolic Ca2+ elevation and ROS production can be observed during the bulge formation in epidermis (Wymer et al., 1997, Foreman et al., 2003). ROS production is required, and cytosolic Ca2+ elevation is believed to be not essential for RH initiation (Wymer et al., 1997, Foreman et al., 2003), implying that the Ca2+ channels responsible for Ca2+ influx are not essential for RH initiation. The single mutant cngc14 can form normal RHs in air (Zhang et al., 2017). The triple mutant cngc6cngc9cngc14 can form irregular bulges, but cannot form any RHs in their experimental condition (Brost et al., 2019). We show here that the triple mutant shrh1 and quadruple mutant shrh2 can form normal, but shorter RHs, regardless of whether the roots and hairs are exposed to air or embedded in solid medium (Figures 1 and 6), and RH density is not obviously altered in the triple and quadruple mutant compared with wild type under our experimental conditions (Supplemental Figures 3B and 8). It seems that all the studies from different groups support the conclusion that cytosolic Ca2+ evaluation is not indispensable for RH initiation/bulge or swelling formation. However, RH density was slightly reduced by the absence of external Ca2+ and increased by high external Ca2+ (15 mM) (Figure 2D), implying that CNGC5, CNGC6, and CNGC9 are involved in RH initiation. The irregular bulges in the triple mutant cngc6cngc9cngc14 (Brost et al., 2019) also support a role of CNGC6, CNGC9, and CNGC14 in RH initiation. Swelling formation in epidermis is a process of cytoplasmic restructuring, which may not require the elevation of cytosolic Ca2+. However, cytosolic Ca2+ elevation during RH initiation could be required by RH-elongating growth because the Ca2+ elevation during RH initiation is a start of establishment of the cytosolic Ca2+ gradient and RH-elongating growth. Thus, the two stages of RH development have an overlap. Some of Ca2+-sensitive proteins involved in RH initiation may be affected by the changes in cytosolic Ca2+ level, and bulge formation can be consequently affected. This could be a side effect of Ca2+ on RH initiation under extremely low or high cytosolic Ca2+ conditions because of the overlap between the two stages of RH development. Thus, the three CNGCs as well as their redundant partners could be somehow involved in RH initiation indirectly.
Different RH-related phenotypes were observed in cngc mutants by different groups (Figures 1, 2, 5 and 6) (Zhang et al., 2017, Brost et al., 2019), supporting an indispensable role of the multiple CNGC members for RH-elongating growth, not RH initiation. We revealed here that CNGC5, CNGC6, and CNGC9 are involved in RH growth by establishing and maintaining a sharp cytosolic Ca2+ gradient as well as regulating the oscillation of cytosolic Ca2+ in RH tips in Arabidopsis. Nevertheless, it is clear that the three CNGCs do matter for RH development, including RH initiation and elongating growth. In these processes, the three Arabidopsis CNGCs plus CNGC14 may form heterotetramers in RHs, as reported in Lotus japonicus (Chiasson et al., 2017), to form plastic Ca2+ channels for dynamic regulation of the channel activity at the post-translational level. Further investigations are needed to dissect the molecular mechanism underlying the channel activity regulation in RH growth.
Possible Processes of External Ca2+ Supply for RH Growth
It has been widely believed that the influx of external Ca2+ through Ca2+ channels at RH tips is the main source of Ca2+ for cytosolic Ca2+ gradients and RH growth. When roots are grown in medium or soil, Ca2+ in RHs is presumably absorbed directly from liquid medium or rhizosphere. However, RHs can grow normally in air, where the external Ca2+ supply seems to be absent. Two possibilities have been proposed for the supply of external Ca2+ to RHs (Ryan et al., 2001). First, RHs create their own internal Ca2+ gradients independent of external Ca2+. Ca2+ may be provided by roots through RH's neighboring cells. The Ca2+ could be coated in vesicles. The vesicles travel inside the RHs to the RH apex to release Ca2+ to form a Ca2+ gradient. Second, external Ca2+ diffuses along the hydrated external surface of RHs to the external space close to RH tips, and then enters RHs through the inward Ca2+ channels. There is also a third possibility: the vesicles containing a large amount of Ca2+ may travel inside RHs to RH tips, the vesicle membrane and plasma membrane are fused at RH tips, and Ca2+ is then secreted to the extracellular space by exocytosis. External Ca2+ enters RHs through the inward Ca2+ channels composed of the CNGCs, no matter where the external Ca2+ comes from. The inward rectification of the three CNGCs as Ca2+ channels and the RH-related defects in a series of cngc mutants (Figures 1, 2, 5 and 6; Supplemental Figures 3, 4, and 6) (Gao et al., 2016, Zhang et al., 2017, Brost et al., 2019) strongly supports the last two possibilities, i.e., RHs take in Ca2+ from extracellular space via Ca2+ channel-mediated Ca2+ influx. Further analyses are required to clarify the exact process of external Ca2+ supply for RH growth.
RH development is regulated by an intrinsic sophisticated regulatory network in response to multiple phytohormones and various environmental stimuli(Cui et al., 2017). A large number of components, including transcriptional factors, ROPs, cytoskeleton, and Ca2+, are involved in regulating RH development. This study and previous reports from other groups clearly show that Ca2+ channels composed of CNGCs and annexin1 are critical for RH growth (Laohavisit et al., 2012, Zhang et al., 2017, Brost et al., 2019). Thus, Ca2+ channels could be the core components of the regulatory networks underlying RH development. Further studies are needed to unravel how the multiple Ca2+ channels are integrated with other components of the network in regulation of RH development.
Methods
Plant Growth
Arabidopsis plants (Columbia-0 ecotype) were grown in a growth room with a daily cycle of 16-h light/8-h darkness at 21°C ± 1°C as described by Zhang et al. (2016). Arabidopsis T-DNA insertion lines, SALK_149893 (cngc5-1), SALK_042207 (cngc6-2), SALK_026086 (cngc9-1), and SALK_206460C (cngc14-1), were obtained from the Arabidopsis Biological Resource Center.
Generation of Transgenic Arabidopsis Lines
For the generation of transgenic lines for the GUS staining assay, 20 GUS reporter vectors for CNGC1 to CNGC20 were prepared in pCambia1301P0G. Genomic DNA 2 kb upstream of the CNGCs' coding regions (native promoters) was fused to a GUS reporter gene, and the fused sequences were cloned into pCambia1301P0G vector. Transformation of wild-type Arabidopsis plants was carried out using the Agrobacterium (strain GV3101)-mediated floral dip method (Clough and Bent, 1998). T2 seedlings with hygromycin B resistance were selected for GUS staining assay.
For the generation of rescued lines in the background of shrh1, the full-length cDNAs of CNGC5, CNGC6, and CNGC9 were PCR amplified and cloned in the vector pCambia1305 downstream of their respective native promoters. For the generation of OE lines in wild-type background, eGFP was fused to the N terminus of CNGC5, CNGC6, and CNGC9. The full-length cDNAs of the three fused proteins were cloned downstream of an Ubiquitin10 promoter in the pCambia1305 vector, respectively. These vectors were then transformed into either shrh1 mutant or wild-type plants using the Agrobacterium-mediated floral dip method (Clough and Bent, 1998). Homozygous lines from T3 generation were selected for experiments. eGFP was transformed into shrh1 mutant and wild-type plants, and the transgenic lines selected from T3 generation were used as control plants in experiments. Primers and restriction sites for vector construction are listed in Supplemental Table 2.
GUS Staining Assay
Four-day-old T2 seedlings were incubated overnight in GUS staining buffer containing 100 mM Na2HPO4, 100 mM NaH2PO4, 5 mM EDTA–Na2 (pH 8.0), 2 ml/l Triton X-100, 2.5 mM K3Fe(CN)6, 2.5 mM K4Fe(CN)6, and 0.5 mg/ml X-Gluc (PhytoTech) in a chamber at 37°C, then cleared in 70% ethanol. Images were captured under a stereo microscope (Model M205 FCA, Leica, Germany) using the software Leica Application Suite X (Leica).
RH Phenotype Analysis
Arabidopsis seeds were surface sterilized with 0.5% sodium hypochlorite for 10 min, washed three times with sterile water, and sowed in a Petri dish containing solid medium modified from a previous method (Zhao et al., 2016). The Ca2+-free solid medium contained 5 mM KNO3, 1 mM MgSO4, 1 mM KH2PO4, 0.1 mM NaFeEDTA, 5 mM 2-(N-morpholino)ethanesulfonic acid (MES), 1% (w/v) sucrose, 0.68% (w/v) agarose, MS microelements at full strength, and pH 6.0 adjusted with Tris–Cl. CaCl2 was added to the Ca2+-free medium as indicated for Ca2+-related RH phenotype analysis. K+-free solid medium contained 3 mM NH4NO3, 1.5 mM MgSO4, 1.25 mM NH4H2PO4, 3 mM CaCl2, 5 mM MES, 1% (w/v) sucrose, 0.68% (w/v) agarose, MS microelements at full strength, and pH 6.0 adjusted with Tris–Cl. KCl was added to K+-free medium as indicated for K+-related RH phenotype analysis. Pictures were taken under the Leica stereo microscope 4–5 days after seed sowing.
For the analysis of RHs exposed to air, seeds were sown on the surface of solid medium in the Petri dishes, which were then vertically placed in a growth room as described by Zhang et al. (2016). Roots grew on the surface of the solid medium, and most RHs were exposed to air under these conditions. For the measurement of RH length, roots and RHs were covered by a coverglass to push the RHs down to an angle being basically parallel to the surface of solid medium for the convenience of RH phenotype analysis. For the analysis of RHs embedded in solid medium, seeds were sown on the surface of the solid medium and pushed into the solid medium using a sharp pipette tip. The Petri dishes were placed vertically in a growth room, and all roots and RHs grew in the solid medium under this condition. Pictures were taken under the Leica stereo microscope. The length, diameter, branching, and rupturing phenotypes of RHs were analyzed using the image software Digimizer (https://www.digimizer.com/index.php) after pictures were taken. RHs located within the 2-mm RHZ (Supplemental Figure 3E) were counted in RH-related phenotype analysis. For RH diameter measurements, the RH diameter at the region about 10 μm from RH tips was measured. For the measurements of the diameter of RH initiation area, the bulge-forming region of RHs was measured. RHs or bulges of epidermis less than 10 μm were categorized into initiating RHs, and not counted as RHs in the RH phenotype analysis. No less than 30 roots and 200 RHs (7–10 RHs per root) were analyzed for each Arabidopsis line analyzed. Statistical analysis was performed as described previously (Kolar and Senkova, 2008, Dang et al., 2018).
Time-Lapse Analysis of RH Growth
Time-lapse experiments in bright field in intact growing roots and hairs were conducted to monitor the growth, calculate the growth rate, and measure the growth time of RHs. Four-day-old seedlings grown in a Petri dish placed vertically in a growth room after seeds were sown on the surface of solid medium were used for the experiments. RHIZ of the seedlings with emerging and elongating RHs were selected for the experiments. A Petri dish was placed horizontally on the stage of a stereo microscope (Model M205 FCA; Leica, Germany). A set of optical sectioning images along z axis with a 22.5-μm step was captured each 1 min for no less than 300 min at room temperature (25°C ± 1°C) using a Sequential Frame Scan Mode following manufacturer’s instructions (Leica). Each set of optical sections was composed of 15 sections for shrh1 mutant and 21 sections for wild type, and was merged automatically and immediately to a single bright-field picture after the acquisition of the set of optical sections. Software Leica Application Suite X (Leica) was used for data acquisition and analysis.
RT–PCR and qRT–PCR
Arabidopsis seeds were sown on a plate containing solid medium as described above, and roots were cut off from the four-day-old seedlings and ground using a commercial electro drill in liquid nitrogen. Total RNA was extracted from the ground mix using TRIzol reagent (Invitrogen), and reverse transcriptions were performed with a TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (Transgen). The qRT–PCR analysis was conducted using a TransStart Top Green qPCR SuperMix kit (Transgen) on a Bio-Rad CFX Connect Real-Time PCR System according to the manufacturer's protocols (Wang et al., 2013). Primers are listed in Supplemental Table 2.
Subcellular Localization Assay
Arabidopsis OE lines were used for protein subcellular localization analysis of CNGC5, CNGC6, and CNGC9. Roots of four-day-old seedlings were cut off and placed on a slide. Four-day-old seedlings were immersed in FM4-64 solution (2 μM) for plasma membrane staining for 12 min, and FM4-64 fluorescent pictures were captured 10 min afterward. eGFP and FM4-64 fluorescent images of roots and RHs were taken under a Leica TCS SP8 STED 3X Super-Resolution Confocal Microscope (Leica, Germany). The fluorescence distribution was analyzed using the free software ImageJ2X.
HEK293T Cell Culturing
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Gibco) and 100 IU/ml penicillin–streptomycin (Yeasen) in a water-injected incubator (Thermo, USA) with 5% CO2 at 37°C in a moist atmosphere as described by Gao et al. (2016).
Patch-Clamp Experiments
For patch-clamp experiments in HEK293T cells, the CDS of CNGC5 and CNGC6 were fused upstream of eGFP sequences with an IRES linker (Mizuguchi et al., 2000) and constructed into pCI-neo vector under a T7 promoter, respectively. HEK293T cells were transfected with vectors using a Lipofectamine 2000 Transfection Reagent kit (Invitrogen, USA) in penicillin–streptomycin-free DMEM as described by Gao et al. (2016). Plasmids for HEK293T transfection were extracted from Escherichia coli (DH5α) using a QIAGEN Plasmid Midi kit. HEK293T cells of a similar size showing bright eGFP fluorescence were used for patch-clamp experiments, and the HEK293T cells expressing eGFP were used as mock control.
The standard bath solution contained 120 mM NaCl, 10 mM CaCl2, 10 mM CsCl, 2 mM MgCl2, 10 mM glucose, and 10 mM HEPES (pH 7.2 adjusted with NaOH). NMDG+-based bath solution was derived from standard bath solution by substituting 120 mM NaCl with 120 mM NMDG–Cl, and pH was adjusted to 7.2 with HCl. The standard pipette solution contained 120 mM Cs-glutamate, 8 mM NaCl, 2 mM MgCl2, 3.35 mM CaCl2, 6.7 mM EGTA, and 10 mM HEPES (pH 7.2 adjusted with CsOH). The osmolality of bath and pipette solutions was adjusted to 313 mmol/kg with D-glucose.
Whole-cell patch-clamp experiments were conducted using an Axopatch-200B patch-clamp setup (Axon Instruments, CA, USA) with a Digitata1440A digitizer combined with an inverted microscope (Model A1; Carl Zeiss, Germany). pClamp10.2 software (Axon Instruments) was used for data acquisition and analysis. A glass pipette was prepared with a glass capillary puller (Model PC-10; Narishige, Japan) and polished using a micro-forge (Model MF-830; Narishige). A membrane ramp protocol with 2-s duration from −180 mV to +20 mV was applied each 10 s for whole-cell current recordings in HEK293T cells as described by Gao et al. (2014).
Ca2+ Imaging
[Ca2+]cyt in HEK293T cells and RHs was FRET/CFP imaged (Swanson and Gilroy, 2013) using a 40×, 1.3 numerical aperture, oil-immersion, Fluar objective under an inverted microscope (Model D1; Carl Zeiss, Germany). Samples were excited by a xenon light through a 430-nm excitation filter with 24-nm bandpass. CFP emission through a 470-nm emission filter with 24-nm bandpass and FRET-dependent emission through a 535-nm emission filter with 30-nm bandpass were captured by a Neo sCMOS CCD camera (Andor, UK). The excitation filter and emission filters were mounted in an excitation filter wheel (Lambda XL; Sutter Instrument, USA) and an emission filter wheel (Lambda XL; Sutter Instrument), respectively. Ratio florescence imaging software MetaFluor (version 7.8.0.0; Molecular Devices, USA) was used for data acquisition and analysis. A set of fluorescent and bright-field pictures were captured every 2 s, and the FRET/CFP ratio of interested regions was simultaneously calculated and recorded on the hard drive of a computer using the software MetaFluor. Exposure time to excitation light was 50 ms.
For Ca2+ imaging in HEK293T cells, the coding sequences of Ca2+ indicator YC3.6 were cloned into pCI-neo at the XhoI/NotI site, and were used for HEK293T transfection and cytosolic Ca2+ ([Ca2+]cyt) imaging. HEK293T cells were transfected in penicillin–streptomycin-free DMEM as described by Gao et al. (2014). HEK293T cells were digested with 0.25% trypsin–EDTA, collected by centrifugation at 1000 rpm for 1 min, and resuspended in Ca2+-free incubation buffer (CIB) containing 130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 1.2 mM NaHCO3, 10 mM glucose, and 10 mM HEPES, at pH 7.2 adjusted with NaOH. HEK293T cells showing bright YC3.6 fluorescence were used for [Ca2+]cyt imaging. Cytosolic Ca2+ was analyzed by monitoring the ratio of FRET/CFP in CIB solution at room temperature (25°C ± 1°C) under the Zeiss inverted microscope (Model D1). External Ca2+ (10 mM) was added as indicated.
For [Ca2+]cyt imaging in Arabidopsis RHs, the shrh1 plants were crossed with wild-type plants expressing YC3.6 under a 35S promoter, and homozygous shrh1 mutant with strong YC3.6 fluorescent signal was isolated from progeny for further experiments. FRET/CFP-based [Ca2+]cyt imaging was performed in RHs of four-day-old seedlings adhered to a coverglass with medical adhesive glue (Hollister, USA). The coverglass was immersed in a chamber containing working solution (5 mM KCl, 100 μM CaCl2, 10 mM MES–Tris [pH 5.7]). In situ calibration was performed as described by Swanson and Gilroy (2013). The Ca2+ saturating FRET/CFP ratio for YC3.6 (Rmax) was recorded upon the application of 5 mM CaCl2 and 20 μM Ca2+ ionophore Br-A23187, and the minimum FRET/CFP ratio (Rmin) was recorded upon the application of 20 μM Br-A23187 and 5 mM EGTA. The estimated [Ca2+]cyt value was calculated according to the equation [Ca2+]cyt = (KD[R − Rmin]/[Rmax − R])1/n, where R is the measured ratio value, KD is 250 nM for YC3.6, and the Hill coefficient n is determined as one for YC3.6 as reported by Nagai et al. (2004).
Funding
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB27020102) and the National Natural Science Foundation of China (91635301, 31570262, and 31770292).
Author Contributions
Investigation, Y.-Q.T., Y.Y., L.-L.G., S.-J.S., A.Z., and C.-F.F.; Methodology, W.X., L.W., and H.L.; Funding Acquisition, Supervision, and Writing, Y.-F.W.
Acknowledgments
We thank Yunxiao He for assistance in confocal imaging experiments, Hongjie Wu (Focell) for assistance in cytosolic Ca2+ imaging experiments, and Hong-Wei Xue (Shanghai Jiao Tong University) for providing IAA and yucasin. No conflict of interest declared.
Published: September 4, 2019
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
A video abstract is available at https://doi.org/10.1016/j.xplc.2019.100001#mmc2.
Supplemental Information
References
- Akiyama H., Kamiguchi H. Second messenger networks for accurate growth cone guidance. Dev. Neurobiol. 2015;75:411–422. doi: 10.1002/dneu.22157. [DOI] [PubMed] [Google Scholar]
- Baluska F., Salaj J., Mathur J., Braun M., Jasper F., Samaj J., Chua N.H., Barlow P.W., Volkmann D. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 2000;227:618–632. doi: 10.1006/dbio.2000.9908. [DOI] [PubMed] [Google Scholar]
- Berger F., Haseloff J., Schiefelbein J., Dolan L. Positional information in root epidermis is defined during embryogenesis and acts in domains with strict boundaries. Curr. Biol. 1998;8:421–430. doi: 10.1016/s0960-9822(98)70176-9. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Lee M.M., Gonzalez A., Zhang F., Lloyd A., Schiefelbein J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development. 2003;130:6431–6439. doi: 10.1242/dev.00880. [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Zhao M., Gonzalez A., Lloyd A., Schiefelbein J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development. 2005;132:291–298. doi: 10.1242/dev.01565. [DOI] [PubMed] [Google Scholar]
- Bibikova T.N., Blancaflor E.B., Gilroy S. Microtubules regulate tip growth and orientation in root hairs of Arabidopsis thaliana. Plant J. 1999;17:657–665. doi: 10.1046/j.1365-313x.1999.00415.x. [DOI] [PubMed] [Google Scholar]
- Bibikova T.N., Jacob T., Dahse I., Gilroy S. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development. 1998;125:2925–2934. doi: 10.1242/dev.125.15.2925. [DOI] [PubMed] [Google Scholar]
- Bibikova T.N., Zhigilei A., Gilroy S. Root hair growth in Arabidopsis thaliana is directed by calcium and an endogenous polarity. Planta. 1997;203:495–505. doi: 10.1007/s004250050219. [DOI] [PubMed] [Google Scholar]
- Brost C., Studtrucker T., Reimann R., Denninger P., Czekalla J., Krebs M., Fabry B., Schumacher K., Grossmann G., Dietrich P. Multiple cyclic nucleotide-gated channels coordinate calcium oscillations and polar growth of root hairs. Plant J. 2019;99:910–923. doi: 10.1111/tpj.14371. [DOI] [PubMed] [Google Scholar]
- Carol R.J., Dolan L. The role of reactive oxygen species in cell growth: lessons from root hairs. J. Exp. Bot. 2006;57:1829–1834. doi: 10.1093/jxb/erj201. [DOI] [PubMed] [Google Scholar]
- Carol R.J., Takeda S., Linstead P., Durrant M.C., Kakesova H., Derbyshire P., Drea S., Zarsky V., Dolan L. A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature. 2005;438:1013. doi: 10.1038/nature04198. [DOI] [PubMed] [Google Scholar]
- Chiasson D.M., Haage K., Sollweck K., Brachmann A., Dietrich P., Parniske M. A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. eLife. 2017;6:e25012. doi: 10.7554/eLife.25012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cui S., Suzaki T., Tominaga-Wada R., Yoshida S. Regulation and functional diversification of root hairs. Semi. Cell Dev. Biol. 2017;83:115–122. doi: 10.1016/j.semcdb.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Dang X., Yu P., Li Y., Yang Y., Zhang Y., Ren H., Chen B., Lin D. Reactive oxygen species mediate conical cell shaping in Arabidopsis thaliana petals. PLoS Genet. 2018;14:e1007705. doi: 10.1371/journal.pgen.1007705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V., Bowen H.C., Maathuis F.J.M., Shabala S.N., Tester M.A., White P.J., Davies J.M. Arabidopsis thaliana root non-selective cation channels mediate calcium uptake and are involved in growth. Plant J. 2002;32:799–808. doi: 10.1046/j.1365-313x.2002.01467.x. [DOI] [PubMed] [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 1996;10:393–402. doi: 10.1046/j.1365-313x.1996.10030393.x. [DOI] [PubMed] [Google Scholar]
- Duan Q., Kita D., Li C., Cheung A.Y., Wu H.-M. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl. Acad. Sci. USA. 2010;107:17821–17826. doi: 10.1073/pnas.1005366107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felle H.H., Hepler P.K. The cytosolic Ca2+ concentration gradient of Sinapis alba root hairs as revealed by Ca2+-selective microelectrode tests and fura-dextran ratio imaging. Plant Physiol. 1997;114:39–45. doi: 10.1104/pp.114.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feske S., Gwack Y., Prakriya M., Srikanth S., Puppel S.H., Tanasa B., Hogan P.G., Lewis R.S., Daly M., Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- Foreman J., Demidchik V., Bothwell J.H., Mylona P., Miedema H., Torres M.A., Linstead P., Costa S., Brownlee C., Jones J.D. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Gao Q.-F., Fei C.-F., Dong J.-Y., Gu L.-L., Wang Y.-F. Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant. 2014;7:739–743. doi: 10.1093/mp/sst174. [DOI] [PubMed] [Google Scholar]
- Gao Q.-F., Gu L.-L., Wang H.-Q., Fei C.-F., Fang X., Hussain J., Sun S.-J., Dong J.-Y., Liu H., Wang Y.-F. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2016;113:3096–3101. doi: 10.1073/pnas.1524629113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson C., Nielsen E., Ketelaarc T., Schiefelbein J. Root hairs. The Arabidopsis Book. 2014;12:e0172. doi: 10.1199/tab.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.-L., Gao Q.-F., Wang Y.-F. Cyclic nucleotide-gated channel 18 functions as an essential Ca2+ channel for pollen germination and pollen tube growth in Arabidopsis. Plant Signal Behav. 2017;12:e1197999. doi: 10.1080/15592324.2016.1197999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Guo J., Li H., Yang Z. Signaling in pollen tube growth: crosstalk, feedback, and missing links. Mol. Plant. 2013;6:1053–1064. doi: 10.1093/mp/sst070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A., Felle H.H. Tip growth in root hair cells of Sinapis alba L.: significance of internal and external Ca2+ and pH. New Phytol. 1995;129:523–533. [Google Scholar]
- Huang G.Q., Li E., Ge F.R., Li S., Wang Q., Zhang C.Q., Zhang Y. Arabidopsis RopGEF4 and RopGEF10 are important for FERONIA-mediated developmental but not environmental regulation of root hair growth. New Phytol. 2013;200:1089–1101. doi: 10.1111/nph.12432. [DOI] [PubMed] [Google Scholar]
- Hung C.Y., Lin Y., Zhang M., Pollock S., Marks M.D., Schiefelbein J. A common position-dependent mechanism controls cell-type patterning and GLABRA2 regulation in the root and hypocotyl epidermis of Arabidopsis. Plant Physiol. 1998;117:73–84. doi: 10.1104/pp.117.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez F., Wall L., Fabra A. Starting points in plant-bacteria nitrogen-fixing symbioses: intercellular invasion of the roots. J. Exp. Bot. 2017;68:1905–1918. doi: 10.1093/jxb/erw387. [DOI] [PubMed] [Google Scholar]
- Jones M.A., Shen J.J., Fu Y., Li H., Yang Z., Grierson C.S. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell. 2002;14:763–776. doi: 10.1105/tpc.010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketelaar T., Galway M.E., Mulder B.M., Emons A.M. Rates of exocytosis and endocytosis in Arabidopsis root hairs and pollen tubes. J. Microsc. 2008;231:265–273. doi: 10.1111/j.1365-2818.2008.02031.x. [DOI] [PubMed] [Google Scholar]
- Kiegle E., Gilliham M., Haseloff J., Tester M. Hyperpolarisation-activated calcium currents found only in cells from the elongation zone of Arabidopsis thaliana roots. Plant J. 2000;21:225–229. doi: 10.1046/j.1365-313x.2000.00659.x. [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Kolar J., Senkova J. Reduction of mineral nutrient availability accelerates flowering of Arabidopsis thaliana. J. Plant Physiol. 2008;165:1601–1609. doi: 10.1016/j.jplph.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Konrad K.R., Wudick M.M., Feijo J.A. Calcium regulation of tip growth: new genes for old mechanisms. Curr. Opin. Plant Biol. 2011;14:721–730. doi: 10.1016/j.pbi.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Laohavisit A., Shang Z., Rubio L., Cuin T.A., Véry A.-A., Wang A., Mortimer J.C., Macpherson N., Coxon K.M., Battey N.H. Arabidopsis annexin1 mediates the radical-activated plasma membrane Ca2+- and K+-permeable conductance in root cells. Plant Cell. 2012;24:1522–1533. doi: 10.1105/tpc.112.097881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- Leng Q., Mercier R.W., Yao W., Berkowitz G.A. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 1999;121:753–761. doi: 10.1104/pp.121.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Q., Mercier R.W., Hua B.G., Fromm H., Berkowitz G.A. Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 2002;128:400–410. doi: 10.1104/pp.010832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Schiefelbein J. Embryonic control of epidermal cell patterning in the root and hypocotyl of Arabidopsis. Development. 2001;128:3697–3705. doi: 10.1242/dev.128.19.3697. [DOI] [PubMed] [Google Scholar]
- Liszkay A., van der Zalm E., Schopfer P. Production of reactive oxygen intermediates (O2⋅-, H2O2, and ⋅OH) by maize roots and their role in wall loosening and elongation growth. Plant Physiol. 2004;136:3114–3123. doi: 10.1104/pp.104.044784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedema H., Demidchik V., Véry A.-A., Bothwell J.H.F., Brownlee C., Davies J.M. Two voltage-dependent calcium channels co-exist in the apical plasma membrane of Arabidopsis thaliana root hairs. New Phytol. 2008;179:378–385. doi: 10.1111/j.1469-8137.2008.02465.x. [DOI] [PubMed] [Google Scholar]
- Mizuguchi H., Xu Z., Ishii-Watabe A., Uchida E., Hayakawa T. IRES-dependent second gene expression is significantly lower than cap-dependent first gene expression in a bicistronic vector. Mol. Ther. 2000;1:376–382. doi: 10.1006/mthe.2000.0050. [DOI] [PubMed] [Google Scholar]
- Molendijk A.J., Bischoff F., Rajendrakumar C.S., Friml J., Braun M., Gilroy S., Palme K. Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 2001;20:2779–2788. doi: 10.1093/emboj/20.11.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Bibikova T.N., Messerli M.A., Shi C., Gilroy S. Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA. 2007;104:20996–21001. doi: 10.1073/pnas.0708586104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen G.B., Messerli M.A., Gilroy S. Imaging of the yellow cameleon 3.6 indicator reveals that elevations in cytosolic Ca2+ follow oscillating increases in growth in root hairs of Arabidopsis. Plant Physiol. 2008;147:1690–1698. doi: 10.1104/pp.108.123638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai T., Yamada S., Tominaga T., Ichikawa M., Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T., Hayashi K.-i., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J.-i. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- Pei W., Du F., Zhang Y., He T., Ren H. Control of the actin cytoskeleton in root hair development. Plant Sci. 2012;187:10–18. doi: 10.1016/j.plantsci.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Raji M., Gehring C. In vitro assessment of guanylyl cyclase activity of plant receptor kinases. In: Aalen R.B., editor. Plant Receptor Kinases: Methods and Protocols. Springer New York; New York, NY: 2017. pp. 131–140. [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 1994;8:1388–1399. doi: 10.1101/gad.8.12.1388. [DOI] [PubMed] [Google Scholar]
- Rounds C.M., Bezanilla M. Growth mechanisms in tip-growing plant cells. Ann. Rev. Plant Biol. 2013;64:243–265. doi: 10.1146/annurev-arplant-050312-120150. [DOI] [PubMed] [Google Scholar]
- Ryan E., Steer M., Dolan L. Cell biology and genetics of root hair formation in Arabidopsis thaliana. Protoplasma. 2001;215:140–149. doi: 10.1007/BF01280310. [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jürgens G., Hülskamp M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21:5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J.W., Shipley A., Rowse P. Calcium influx at the tip of growing root-hair cells of Arabidopsis thaliana. Planta. 1992;187:455–459. doi: 10.1007/BF00199963. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Ketelaar T., Esseling J.J., Emons A.M.C. Microtubules guide root hair tip growth. New Phytol. 2005;167:711–719. doi: 10.1111/j.1469-8137.2005.01506.x. [DOI] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 2007;311:566–578. doi: 10.1016/j.ydbio.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Swanson S.J., Gilroy S. Imaging changes in cytoplasmic calcium using the yellow cameleon 3.6 biosensor and confocal microscopy. In: Munnik T., Heilmann I., editors. Plant Lipid Signaling Protocols. Humana Press; Totowa, NJ: 2013. pp. 291–302. [DOI] [PubMed] [Google Scholar]
- Tominaga M., Kojima H., Yokota E., Nakamori R., Anson M., Shimmen T., Oiwa K. Calcium-induced mechanical change in the neck domain alters the activity of plant myosin XI. J. Biol. Chem. 2012;287:30711–30718. doi: 10.1074/jbc.M112.346668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc-Ozdemir M., Tang C., Ishka M.R., Brown E., Groves N.R., Myers C.T., Rato C., Poulsen L.R., McDowell S., Miller G. A cyclic nucleotide-gated channel (CNGC16) in pollen is critical for stress tolerance in pollen reproductive development. Plant Physiol. 2013;161:1010–1020. doi: 10.1104/pp.112.206888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart W., Chin K., Ung H., Moeder W., Yoshioka K. The cyclic nucleotide-gated channels AtCNGC11 and 12 are involved in multiple Ca2+-dependent physiological responses and act in a synergistic manner. J. Exp. Bot. 2011;62:3671–3682. doi: 10.1093/jxb/err074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véry A.A., Davies J.M. Hyperpolarization-activated calcium channels at the tip of Arabidopsis root hairs. Proc. Natl. Acad. Sci. USA. 2000;97:9801–9806. doi: 10.1073/pnas.160250397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- Wada T., Kurata T., Tominaga R., Koshino-Kimura Y., Tachibana T., Goto K., Marks M.D., Shimura Y., Okada K. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development. 2002;129:5409–5419. doi: 10.1242/dev.00111. [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kang Y., Ma C., Miao R., Wu C., Long Y., Ge T., Wu Z., Hou X., Zhang J. CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 2017;173:1342–1354. doi: 10.1104/pp.16.01222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.F., Munemasa S., Nishimura N., Ren H.M., Robert N., Han M., Puzorjova I., Kollist H., Lee S., Mori I. Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 2013;163:578–590. doi: 10.1104/pp.113.225045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymer C.L., Bibikova T.N., Gilroy S. Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 1997;12:427–439. doi: 10.1046/j.1365-313x.1997.12020427.x. [DOI] [PubMed] [Google Scholar]
- Xu J., Li H.D., Chen L.Q., Wang Y., Liu L.L., He L., Wu W.H. A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell. 2006;125:1347–1360. doi: 10.1016/j.cell.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Zhang A., Ren H.-M., Tan Y.-Q., Qi G.-N., Yao F.-Y., Wu G.-L., Yang L.-W., Hussain J., Sun S.-J., Wang Y.-F. S-type anion channels SLAC1 and SLAH3 function as essential negative regulators for K+ channel KAT1 and stomatal opening in Arabidopsis. Plant Cell. 2016;28:949–965. doi: 10.1105/tpc.15.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Pan Y., Tian W., Dong M., Zhu H., Luan S., Li L. Arabidopsis CNGC14 mediates calcium influx required for tip growth in root hairs. Mol. Plant. 2017;10:1004–1006. doi: 10.1016/j.molp.2017.02.007. [DOI] [PubMed] [Google Scholar]
- Zhao S., Zhang M.L., Ma T.L., Wang Y. Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell. 2016;28:3005–3019. doi: 10.1105/tpc.16.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.