Abstract
In the late 19th century, it was discovered that legumes can establish a root nodule endosymbiosis with nitrogen-fixing rhizobia. Soon after, the question was raised whether it is possible to transfer this trait to non-leguminous crops. In the past century, an ever-increasing amount of knowledge provided unique insights into the cellular, molecular, and genetic processes controlling this endosymbiosis. In addition, recent phylogenomic studies uncovered several genes that evolved to function specifically to control nodule formation and bacterial infection. However, despite this massive body of knowledge, the long-standing objective to engineer the nitrogen-fixing nodulation trait on non-leguminous crop plants has not been achieved yet. In this review, the unsolved questions and engineering strategies toward nitrogen-fixing nodulation in non-legume plants are discussed and highlighted.
Key words: nodulation, legumes, actinorhizal plants, Parasponia, engineering nitrogen fixation
Engineering the nitrogen-fixing nodulation trait into crop plants is a longstanding research objective. Recent phylogenomic studies uncovered several genes that evolved to function specifically to control nodulation. Supported by the increase in molecular tools to work in non-model non-crop plants, research is now moving towards hands-on engineering.
Introduction
Since ancient times, legumes (Fabaceae) have been known as “nitrogen accumulators” important for soil fertility (Hirsch, 2009). The anatomy of legume plants, including nodules, was already well known in the 17th century. However, it took until the late 19th century to discover that nodules provide a unique trait. Namely, nodule cells host endosymbiotic nitrogen-fixing (diazotrophic) bacteria, which are now known as rhizobia and which provide the plant with an additional source of fixed nitrogen (Beijerinck, 1888, Hellriegel and Wilfarth, 1888, Hirsch, 2009). These findings have set off research programs aiming to reveal how legumes can establish such “profitable” symbiosis, whereas most other plant species cannot. It was questioned as early as 1917 whether “a symbiosis is possible between legume bacteria and non-legume plants” (Burrill and Hansen, 1917). Over the past decades, this enigma has come back over and over again (Beringer and Hirsch, 1984, de Bruijn et al., 1995, Saikia and Jain, 2007, Charpentier and Oldroyd, 2010, Beatty and Good, 2011, Untergasser et al., 2012, Mus et al., 2016). Up until now, the research on biological nitrogen fixation has mainly focused on understanding the nitrogen-fixing nodule symbiosis in legumes, in particular in two model species. This has led to significant advances in our understanding of the morphology, molecular genetics, biochemistry, and physiology of nodulation and the variability and conservation between different nodulating species. Supported by the increase in molecular tools to work in non-model non-crop plants, the research field is now moving from discovery-driven research toward hands-on engineering. Here, we will review the molecular genetics of the nodulation trait relevant for such engineering approaches and explore step-by-step strategies that may lead to engineering nitrogen-fixing nodule symbiosis in non-leguminous crop plants.
Signposts on the Road to Engineering Root Nodule Symbiosis
To summarize the major achievements of the field over the past decades, we break the past research down into four themes that represent signposts on the road to engineering a nitrogen-fixing nodule symbiosis: (i) the variety of blueprints that are available for nitrogen-fixing nodule symbiosis; (ii) the use of non-model non-crop models to identify core symbiosis genes; (iii) the recruitment of the ancient arbuscular mycorrhizal (AM) signaling pathway for nodulation; (iv) the cross talk between generic plant developmental programs and nodulation. Not only do these themes represent major breakthroughs in our understanding of the symbiosis, they also provide essential clues for the possibilities and constraints of the engineering strategies.
There Is a Variety of Blueprints for Nitrogen-Fixing Nodule Symbiosis
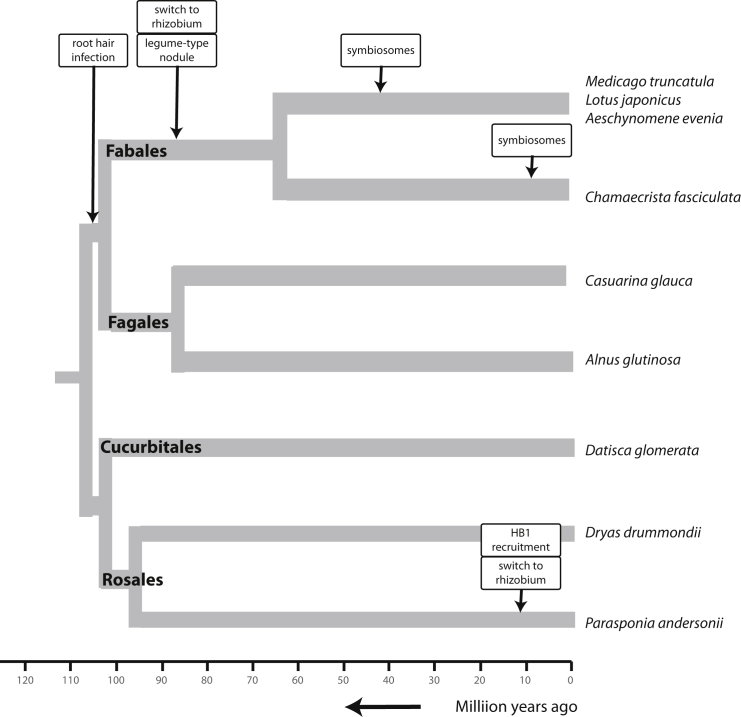
Instead of starting from scratch, the probably most achievable way to engineer a nitrogen-fixing root nodule symbiosis is to mimic an existing symbiosis by transferring genes from nodulating to non-nodulating plant species. For this, it should be appreciated that there is a wide variety of nitrogen-fixing symbioses that can provide several blueprints for nodulation, requiring the transfer of different sets of genes. Plant species that possess the nitrogen-fixing nodulation trait are distributed across 10 lineages in the related taxonomic orders Fabales, Fagales, Cucurbitales, and Rosales (Soltis et al., 1995, Doyle, 2011, Li et al., 2015) (Figure 1). Together these orders are known as the nitrogen-fixing clade, despite the occurrence of many lineages of non-nodulating species in this clade. This distribution of nodulating and non-nodulating plant species can be explained by a shared evolutionary origin of the nitrogen-fixing nodulation trait in the root of the nitrogen-fixing clade (∼110 million years ago) followed by multiple parallel losses of this trait (Soltis et al., 1995, van Velzen et al., 2019). We take this “single gain–parallel loss” hypothesis as the basis for this review.
Figure 1.
The Nitrogen-Fixing Clade Depicting the Phylogenetic Relation of Nodulating Plant Species Selected for Comparative Studies.
The nitrogen-fixing nodulation trait finds its origin about 110 million years ago prior to the diversification into four orders: Fabales, Fagales, Cucurbitales, and Rosales. The major lineage-specific adaptations are indicated.
Species of eight lineages in the orders Fagales, Cucurbitales, and Rosales establish a nitrogen-fixing nodule symbiosis with diazotrophic Actinobacteria of the genus Frankia and are named accordingly actinorhizal plants (Pawlowski and Demchenko, 2012, Li et al., 2015). The remaining two lineages, legumes (Fabaceae, Fabales) and Parasponia (Cannabaceae, Rosales), establish a nodule symbiosis with diazotrophic ɑ- and β-proteobacteria that are collectively known as rhizobia (Masson-Boivin et al., 2009, Li et al., 2015). In line with the single gain–massive loss hypothesis, the occurrence of two distinct classes of diazotrophic microsymbionts suggests that at least two major switches in microsymbiont partner have occurred. Frankia species have intrinsic characteristics to protect nitrogenase from oxidation and can fix nitrogen in a free-living form (Sellstedt and Richau, 2013). In contrast, rhizobia are dependent on the mechanisms provided by the plant. In line with this, it is hypothesized that Frankia was the ancestral microsymbiont in the nitrogen-fixing clade (van Velzen et al., 2018, van Velzen et al., 2019).
Of all nitrogen-fixing plants, legumes are most prominent. The legume family massively expanded soon after the Cretaceous–Paleogene boundary (60 million years ago) and now encompasses six subfamilies and over 19 500 species (Lewis et al., 2005, Lavin et al., 2005, Azani et al., 2017, Koenen et al., 2019). Nodulation is prominent in two subfamilies—Caesalpinioideae and Papilionoideae—that represent the vast majority of legume species (Sprent, 2007, Azani et al., 2017). In contrast to the large amount of nodulating legumes, there are only five Parasponia and a not-well-defined number of actinorhizal species that range in estimates from fewer than 200 to more than 400 species divided over 25 genera in eight taxonomic families (Becking, 1992, Soltis et al., 1995, Pawlowski and Demchenko, 2012, Li et al., 2015). Whereas Parasponia and actinorhizal species do not include economically important crop species, several nitrogen-fixing legumes, especially from the Papilionoideae subfamily, are cultivated for their relatively high nitrogen/protein and/or oil content, e.g., pea (Pisum sativum), soybean (Glycine max), common bean (Phaseolus vulgaris), cowpea (Vigna unguiculata), groundnut (Arachis hypogaea), lentil (Lens esculenta), chickpea (Cicer arietinum), pigeon pea (Cajanus cajan), alfalfa (Medicago sativa), and clovers (Trifolium spp). Consequently, most research on nodulation has been done on papilionoid legumes, initially focusing on crops, but for the last two decades mainly on the models Medicago truncatula and Lotus japonicus, resulting in a rather legume-biased knowledge of nodulation. The nitrogen-fixing nodulation trait in these legumes is rather advanced in several aspects compared with most non-legume nodulating species. Five of these advances will be discussed here.
-
(i)
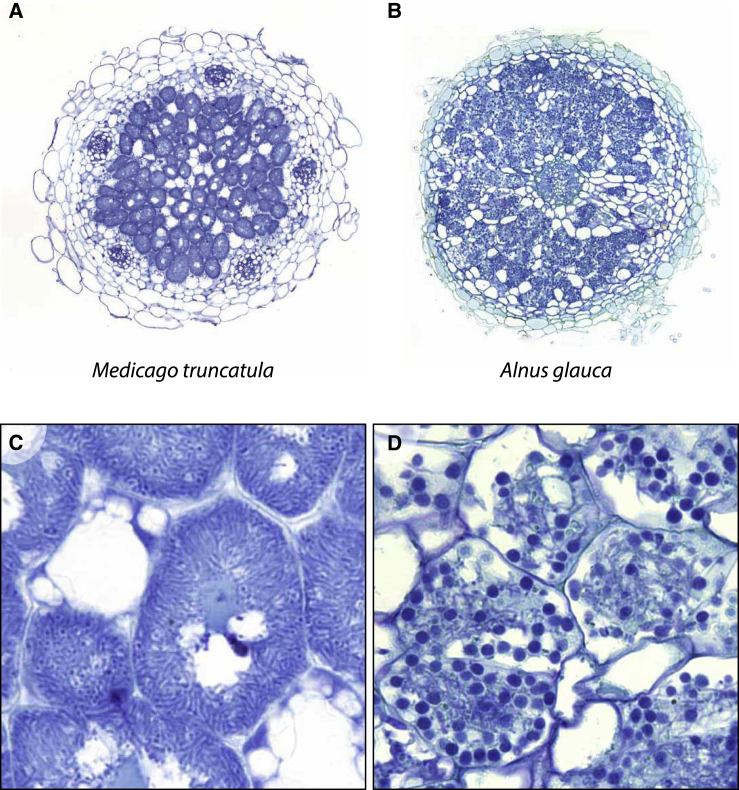
Legume nodules have a unique ontogeny. Legume nodules have a large central zone containing the rhizobium-infected cells, surrounded by an endodermis. The peripheral cortex tissue of a legume nodule contains several vascular bundles allowing an efficient flux of nutrients fueling the enzymatic conversion of molecular dinitrogen into ammonia by the nitrogenase enzyme complex (Figure 2A). Furthermore, this nodule architecture allows maintenance of a low-oxygen environment in the central zone of the nodule to allow optimal functioning of the nitrogenase enzyme complex, without affecting the respiration in the nodule vascular bundles. In contrast to legumes, Parasponia and actinorhizal plant species have a single central vascular bundle. This gives nodules of these species a lateral rootlike appearance, having infected cells organized in lobes of cortex cells (Figure 2B).
-
(ii)
Most legumes select their rhizobial microsymbiont by capturing a single bacterium in a curled root hair. From there, clonally propagating rhizobia can penetrate the root hair cell via a tubular infection thread, which guides the microbe toward the newly formed nodule primordium (Gage, 2004). In contrast, in Parasponia and actinorhizal plants of the orders Rosales and Cucurbitales, the microsymbionts enter the root apoplastically via epidermal cracks made by emerging lateral roots or formed due to induced epidermal cell divisions. Root hair-based infection increases the chance that a single bacterial strain colonizes the nodule (Gage, 2002), which allows plants to sanction cheaters and allocate more resources to fixing nodules. Root hair-based infection is also found in actinorhizal plants of the Fagales order (Svistoonoff et al., 2014). Since recent phylogenetic studies indicated that Fagales and Fabales are sister lineages (Koenen et al., 2019), root hair-based infection could have a single evolutionary origin (Figure 1).
-
(iii)
Most legumes host rhizobia as transient nitrogen-fixing organelle-like structures (known as symbiosomes) in the cells of the nodule central zone (Figure 2C). In this way, the bacteria are fully dependent on their host plant, which likely contributes to optimized nutrient exchange between both partners. In contrast, Frankia is a filamentous bacterium, which obstructs the release of single bacterial cells into nodule cells. Consequently, nitrogen-fixing Frankia inside cells of an actinorhizal nodule remains in contact with their extranodular mycelium, which may complicate plant control of nutrient fluxes. Likewise, Parasponia species host nitrogen-fixing rhizobia in thread-like structures (named “persistent infection threads” or “fixation threads”), rather than releasing bacteria as symbiosomes into the cytoplasm of nodule cells (Trinick, 1979, Lancelle and Torrey, 1984, Lancelle and Torrey, 1985). Such fixation threads are found also in nodules of some basal legumes, e.g., Andira and some Chamaecrista species, and are considered more primitive compared with hosting rhizobia as symbiosomes (de Faria et al., 1986, de Faria et al., 1987, Naisbitt et al., 1992). This suggests that symbiosomes have evolved multiple times in parallel in the Fabaceae (Figure 1).
-
(iv)
Engaging with rhizobia as diazotrophic microsymbionts requires active modulation of the oxygen balance by the plant in infected cells to avoid oxidation of the bacterial nitrogenase enzyme complex. Legumes and Parasponia independently recruited hemoglobin genes to commit this task, as they recruited different paralogs (Ott et al., 2005, Sturms et al., 2010, Kakar et al., 2011, van Velzen et al., 2018, Wang et al., 2019). Also, some actinorhizal plant species in the Fagales clade use hemoglobin genes to function in root nodules (e.g., Casuarina and Myrica species); however, most actinorhizal species do not (Jacobsen-Lyon et al., 1995, Huss-Danell, 1997, Heckmann et al., 2006). This is because Frankia can rely on intrinsic mechanisms to protect its nitrogenase enzyme complex from oxidation, for example, by the formation of rigid infection structures, known as vesicles, that form a physical oxygen barrier (Figure 2D). Also, it can produce hopanoid derivatives that can form protective laminar lipid layers and/or express the truncated hemoglobin HbO to control its oxygen homeostasis (Silvester et al., 2007, Huss-Danell, 1997, Coats et al., 2009).
-
(v)
In ecological settings, legumes seem more dynamic in regulating the nitrogen-fixing symbiosis in response to exogenous nitrogen sources compared with actinorhizal plants (Andrews et al., 2011). This suggests that legumes evolved a more efficient mechanism to lower their nitrogen-fixation capacity in response to the occurrence of exogenous nitrogen sources or in case other nutrients become growth limiting (Andrews et al., 2011). Such mechanisms are relevant, as the carbon cost for full reliance on a nitrogen-fixing nodule symbiosis is estimated to be more than 30% of the total photosynthates (Pate and Layzell, 1990).
Figure 2.
Cytoarchitecture of a Legume (Medicago truncatula) and an Actinorhizal Nodule (Alnus glutinosa).
(A) A cross section of an M. truncatula nodule visualizing a large central zone of infected cells and five peripheral vascular bundles.
(B) A cross section of an A. glutinosa nodule visualizing a central vascular bundle surrounded by infected cortical cells.
(C) Infected M. truncatula nodule cells containing hundreds of elongated rhizobia as transient nitrogen-fixing organelle-like structures.
(D) Infected A. glutinosa nodule cells containing fixation threads and tens of densely stained nitrogen-fixing Frankia vesicles.
Taken together, these differences between legumes, Parasponia, and actinorhizal plants demonstrate that there have been lineage-specific adaptations in the nitrogen-fixing nodulation trait. As nodulation in the nitrogen-fixing clade shares a single evolutionary origin (Griesmann et al., 2018, van Velzen et al., 2018, van Velzen et al., 2019), each nodulating species as found today is the result of ∼110 million years of evolution over which it may have accumulated complexity. Some of these interactions seem less intricate and could represent a more accessible starting point for an engineering approach than the legume–rhizobium symbiosis. Thus, studying actinorhizal plants and Parasponia can provide alternative and simpler blueprints for the nitrogen-fixing nodulation trait. Further, comparing the genomes and transcriptomes of the full diversity of nodulating plants will allow us to pinpoint which genes are associated with different aspects of nodulation.
Comparative Studies of Different Nodulating Plant Species Can Be Used to Identify Core Symbiosis Genes
Genetic and experimental studies in legumes identified over a hundred genes that function in nitrogen-fixing nodule symbiosis (Supplemental Table 1), whereas a multitude of genes have been found to possess a nodule-enhanced expression profile (Colebatch et al., 2004, Küster et al., 2007, Tadege et al., 2008, Benedito et al., 2008, Høgslund et al., 2009, Maunoury et al., 2010, Takanashi et al., 2012, Demina et al., 2013, Limpens et al., 2013, Breakspear et al., 2014, Roux et al., 2014). Many of these genes evolved upon genome and/or segmental duplication events that allowed sub- and/or neofunctionalization, resulting in diverged expression profiles of paralogous genes (Schmutz et al., 2010, Young et al., 2011, Schranz et al., 2012, Young and Bharti, 2012, De Mita et al., 2014, Cannon et al., 2015). Selecting which of these genes represent targets in an engineering approach seems a daunting task. Therefore, it is vital to consider the nitrogen-fixing nodule symbiosis in other clades. By comparing the different nitrogen-fixing nodule symbioses and the genes that are involved, we can distinguish shared features and identify a core set of genes, which must have been recruited early in the evolution of the trait. Features and genes that are specific for a particular clade likely represent later adaptations to the symbiosis trait that are not absolutely required. Knowledge of these genes may nevertheless still be useful, as they may be associated with processes such as root hair infection, nodule ontogeny, and symbiosome formation, allowing for a mix-and-match approach to engineering that incorporates features from multiple symbioses.
To define a core set of symbiosis genes that are essential for nodulation, as well as to identify lineage-specific adaptations, representative species of distinct clades can be selected for comparative analysis. Ideally, such species should be suited to laboratory experimentation having established protocols for synchronized seed germination and quantitative nodulation assays, have a relatively small diploid genome that is fairly homozygotic, and be able to be transformed. The last especially is a pro, since it will allow CRISPR–Cas9-based reverse genetics to study gene function.
We have listed seven non-model non-crop species that have been used for experimentation in several studies, that represent significant diversity in the nodulation trait, and whose genomes have been sequenced (Table 1). These include two legumes, namely, Aeschynomene evenia (Papilionoideae) and Chamaecrista fasciculata (Caesalpinioideae). The latter species does not share the genome duplication event that occurred in the root of the papilionoid clade and that may have been an important driver of further adaptations of the nitrogen-fixing nodulation trait (Singer et al., 2009, Young et al., 2011, Cannon et al., 2015, Koenen et al., 2019). Furthermore, the use of a Chamaecrista species as a comparative system may afford insights into basal adaptations in the Fabaceae, e.g., the formation of legume-type nodules. However, C. fasciculata, which has been proposed as a research system (Singer et al., 2009), is mainly outcrossed with a complex fertilization strategy depending on long-tongued bees (Lee and Bazzaz, 1982). As a consequence, C. fasciculata is difficult to reproduce under laboratory conditions. This complicates the generation of homozygous lines and will affect genomic studies. The Chamaecrista genus represents ∼330 species, many of which are confirmed nodulators (Naisbitt et al., 1992, Lewis et al., 2005). Alternative species that are more suitable as experimental systems could be selected, e.g., Chamaecrista mimosoides, which can be propagated efficiently due to its self-compatible nature, has a genome of ∼460 Mb and is amenable to genetic analyses (W. Kohlen, unpublished results). The second legume listed, A. evenia, represents one of the few legume species that nodulate with Bradyrhizobium strains that do not produce the canonical lipochitooligosaccharide (LCO) signal molecules known as nodulation (Nod) factors (see below), but depend on the secretion of effectors to trigger symbiotic signaling (Arrighi et al., 2012, Arrighi and Cartieaux, 2015, Teulet et al., 2019). In that respect, it is more similar to actinorhizal plant species—like Casuarina glauca and Alnus glutinosa—that are nodulated by Frankia species that also lack the potential to produce LCO signal molecules in response to their host plants (Normand et al., 2007). Studying these plants and their corresponding microsymbionts may provide a blueprint of LCO-independent nodulation. The remaining three species listed—Datisca glomerata, Dryas drummondii, and Parasponia andersonii—can be used to identify the core gene set for LCO-driven nodulation. P. andersonii is of particular interest as it is postulated that the genus Parasponia only recently switched its microsymbiont from Frankia to rhizobia (van Velzen et al., 2018, van Velzen et al., 2019).
Table 1.
Non-model–Non-crop Nodulating Species with Sequenced Genomes Used for Comparative Analysis.
| Species | Symbiont | Genome (Mb) | Seed-to-seed time | Transformation |
|---|---|---|---|---|
| Fabales, Fabaceae, Papilionoideae | ||||
| Aeschynomene evenia | rhizobia1,2 | 4601,2 | 2–3 months1,2 | Agrobacterium rhizogenes; Agrobacterium tumefaciens1,2 |
| Fabales, Fabaceae, Caesalpinioideae | ||||
| Chamaecrista fasciculata | rhizobia3 | 5504 | unknown | A. rhizogenes3 |
| Rosales, Cannabaceae | ||||
| Parasponia andersonii | rhizobia5,6 | 5637 | 6 months6 | A. rhizogenes; A. tumefaciens6,8,9 |
| Rosales, Rosaceae | ||||
| Dryas drummondii | Frankia cluster II10−12 | 2534 | >1 year12 | A. rhizogenes12 |
| Fagales, Casuarinaceae | ||||
| Casuarina glauca | Frankia cluster I9,12 | 3144 | 3–4 years13 | A. rhizogenes, A. tumefaciens13 |
| Fagales, Betulaceae | ||||
| Alnus glutinosa | Frankia cluster I10 | 4614 | 3 years14 | n.d. |
| Cucurbitales; Datiscaceae | ||||
| Datisca glomerata | Frankia cluster II10,11 | 8274 | 6 months15 | A. rhizogenes; A. tumefaciens16,17 |
n.d., not developed.
1Arrighi et al., 2012; 2Arrighi and Cartieaux, 2015; 3Singer et al., 2009; 4Griesmann et al., 2018: 5Op den Camp et al., 2012; 6Wardhani et al., 2019; 7van Velzen et al., 2018; 8Op den Camp et al., 2011; 9van Zeijl et al., 2018; 10Pawlowski and Demchenko, 2012; 11Normand et al., 2017; 12Billault-Penneteau et al., 2019; 13Zhong et al., 2013; 14McVean, 1955; 15Claessens et al., 2010; 16Wang and Berry, 1996; 17Markmann et al., 2008.
At present, two different strategies have been applied to distill a core set of nodulation genes. First, comparative analyses of nodule transcriptomes are conducted (Hocher et al., 2011, Battenberg et al., 2018, van Velzen et al., 2018, Salgado et al., 2018). These studies identified core sets of symbiosis genes with a nodule-enhanced expression profile. Among these are genes that in legumes have shown to be essential for nodulation, like the coiled-coil protein-encoding gene RHIZOBIUM POLAR GROWTH (RPG) that is essential for intracellular rhizobium infection and the transcription factor module NODULE INCEPTION (NIN)–NUCLEAR FACTOR-YA1 (NF-YA1) controlling nodule organogenesis and rhizobial infection (Schauser et al., 1999, Borisov et al., 2003, Combier et al., 2006, Marsh et al., 2007, Arrighi et al., 2008, Laporte et al., 2014, Laloum et al., 2014, Hossain et al., 2016, Rípodas et al., 2019). As these genes have a conserved nodule-enhanced expression profile in all species studied, it is most probable that they have conserved functions in nodulation. Nevertheless, it remains essential to prove that this is indeed the case, as functions can have diverged in different nodulating lineages. For NIN, such studies have been done in C. glauca. By using RNA interference (RNAi) it was demonstrated that knockdown of CgNIN resulted in a reduced nodulation efficiency (Clavijo et al., 2015). However, it would be relevant to extend these studies, preferably by creating knockout mutants. This would enable more in-depth phenotypic analysis as well as trans-complementation studies with engineering constructs (see below).
In the second strategy of comparative analyses, the gene contents of nodulating and non-nodulating species in the nitrogen-fixing clade were compared (Griesmann et al., 2018, van Velzen et al., 2018). These phylogenomics approaches identified NIN and RPG as correlated to the nitrogen-fixing nodulation trait. These genes accumulated mutations and have become pseudogenes, or are even completely lost, in most non-nodulating relatives in the nitrogen-fixing clade. In contrast, orthologs of both genes occur outside the nitrogen-fixing clade, where they must carry out other functions that currently remain unknown. This suggests that NIN and RPG have experienced adaptations allowing both genes to function in the nitrogen-fixing nodulation trait. As both genes have a conserved nodule-specific expression profile, such adaptations most probably have occurred in the root of the nitrogen-fixing clade and are most probably essential for nodulation. Therefore, NIN and RPG represent primary targets for engineering.
In addition to identifying genes that are essential for nitrogen-fixing nodule symbiosis, successful engineering also requires the correct transcriptional regulation of these genes in other plant species. It will probably be hard to unravel the transcriptional networks that govern root nodule symbiosis by just comparing the genomes and transcriptomes of nodulating plants. Therefore, it is essential to experimentally uncover these networks, of which the progress will be discussed in the next two signposts.
Nitrogen-Fixing Nodule Symbiosis Co-opted the Arbuscular Mycorrhizal Symbiosis Signaling Pathway
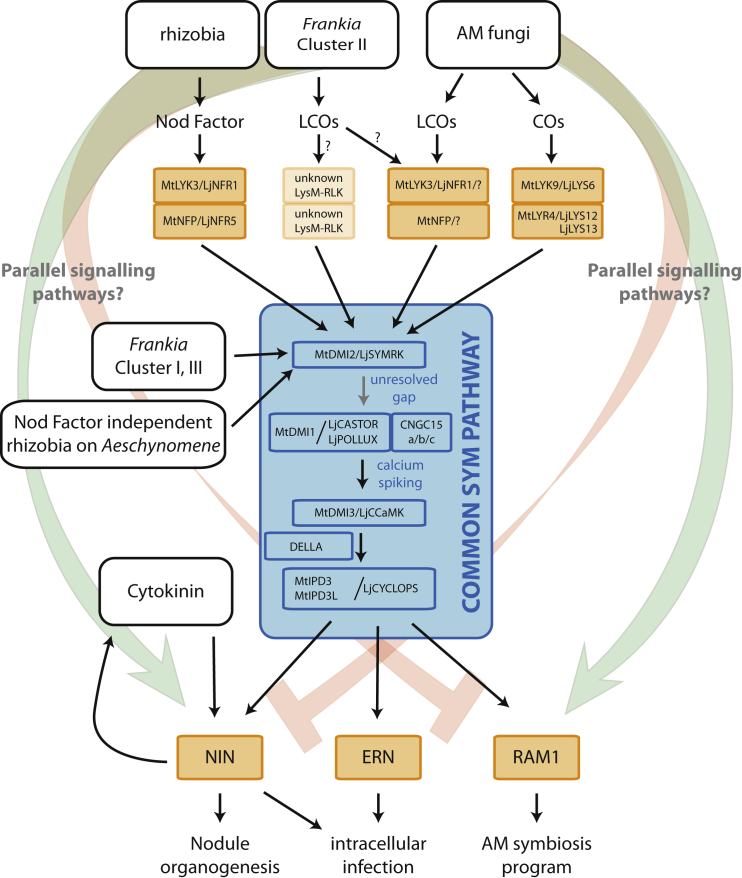
To start a symbiotic engagement between a plant and nitrogen-fixing bacteria, the plant needs to recognize its endosymbiont to initiate nodule organogenesis and to allow bacteria to infect plant cells. To transfer the nitrogen-fixing nodulation trait to other plant species, it is crucial to know which genes are responsible for this communication and how they translate the perception of bacterial signal molecules into transcriptional and cellular responses. As mentioned above, most symbiotic rhizobia secrete LCOs, which are known as Nod factors (Lerouge et al., 1990). These molecules have a core structure of three to five N-acetyl glucosamines with an acyl chain attached to the non-reducing residue. Further modifications can be present, such as sulfation, acetylation, fucosylation, glycosylation, and methylation, depending on the rhizobial species or strain. These decorations are important determinants of host specificity (Mergaert et al., 1997). Rhizobium Nod factors are perceived by legumes and Parasponia species and trigger a symbiotic signaling cascade (Granqvist et al., 2015). Many genes of this signaling cascade are also required for the symbiosis between plants and AM fungi (Duc et al., 1989, Parniske, 2008, Kouchi et al., 2010). These genes are therefore referred to as the common symbiotic signaling pathway or common SYM pathway (Figure 3). AM symbiosis is much older (∼450 million years) and widespread (∼72% of all land plants) than nitrogen-fixing nodule symbioses (Brundrett and Tedersoo, 2018). Therefore, a key step in the evolution of the nitrogen-fixing nodulation trait was the recruitment of the already existing AM symbiosis signaling pathway. AM fungi trigger the common SYM pathway by the release of chitin oligomers and LCOs, which are collectively called Myc factors (Maillet et al., 2011, Genre et al., 2013). Since Nod factors are structurally very similar—or even identical—to some Myc factors, it is most probable that the first symbiotic nitrogen-fixing bacteria recruited the AM signaling pathway by mimicking AM signaling molecules.
Figure 3.
Schematic Representation of the Common SYM Pathway.
Lipochitooligosaccharides (LCOs) of rhizobia (Nod factors), Frankia cluster II, and arbuscular mycorrhizal (AM) fungi (Myc factors), as well as chitin oligomers (COs) of AM fungi, are perceived by different heterodimeric complexes of LysM-RLKs. These complexes interact also with the leucine-rich repeat-type SYMBIOSIS RECEPTOR KINASE (LjSYMRK/MtDMI2), thereby activating the common SYM pathway. In the case of LCO-independent Frankia clusters I and III as well as Aeschynomene legumes, SYMRK is activated by an as-yet unknown mechanism. Nuclear Ca2+ oscillations are a hallmark of symbiotic signaling induced by rhizobia, Frankia, and AM fungi. Ca2+ oscillations are decoded by calcium-/calmodulin-dependent kinase (CCaMK), leading to activation of MtIPD3/LjCYCLOPS. Downstream of this transcription factor a symbiosis-specific transcriptional network is activated, which includes ERN1 and the NIN–cytokinin feed-forward loop, in the case of nodule formation in legumes, and RAM1 in the case of AM symbiosis. To allow such symbiosis-specific transcriptional activation, parallel pathways are predicted, which can include activating (green) or inhibiting (red) activity.
In this light, it is also worth considering the symbiotic signals secreted by Frankia species. There are four taxonomic clusters of Frankia, of which three are able to engage in a root nodule symbiosis: clusters I, II, and III (Pawlowski and Demchenko, 2012). Genome sequencing revealed that only Frankia cluster II species harbor nodulation (nod) genes essential for LCO biosynthesis and secretion (Persson et al., 2011, Ghodhbane-Gtari et al., 2014, Nguyen et al., 2016, Tisa et al., 2016, Gueddou et al., 2018, Van Nguyen et al., 2019). As Frankia cluster II species represent the most basal taxonomic Frankia clade, it is conceivable that the common ancestor of Frankia microsymbionts did produce LCOs, allowing it to activate the common SYM pathway by mimicking the AM signal (Markmann et al., 2008). However, Frankia species of cluster I and cluster III generally lack the canonical nod genes, and therefore it is most probable that these do not produce LCOs and gained an alternative signal to communicate to the host plant. Initial characterization showed that these molecules are hydrophilic, thermoresistant, and resistant to chitinase digestion, indicating structural differences from LCOs (Cissoko et al., 2018). Even though the nature of this signal remains elusive, RNAi experiments on the common SYM genes CALCIUM AND CALMODULIN-DEPENDENT PROTEIN KINASE (CCAMK) in C. glauca and SYMBIOTIC RECEPTOR KINASE (SYMRK) in C. glauca and D. glomerata demonstrated that the symbiotic signaling triggered by Frankia depends on the common SYM pathway (Figure 3) (Gherbi et al., 2008, Svistoonoff et al., 2013). Similarly, it was shown that Nod factor-independent nodulation in the legume A. evenia depends on the common SYM pathway (Giraud et al., 2007, Fabre et al., 2015). Thus, all nitrogen-fixing symbioses require the activation of the common SYM pathway.
Both Myc and Nod factors are perceived by lysine-motif domain-containing receptor-like kinases (LysM-RLKs) on the plasma membrane of plant cells (Madsen et al., 2003, Radutoiu et al., 2003, Limpens et al., 2003, Mulder et al., 2006, Zhukov et al., 2008, Indrasumunar et al., 2011, Maillet et al., 2011, Broghammer et al., 2012, Miyata et al., 2014, Carotenuto et al., 2017, Bozsoki et al., 2017). Pairs of LysM-RLKs are proposed to be drawn into each other’s proximity by ligand binding to their extracellular domain, enabling the intracellular-kinase domains to phosphorylate target proteins (Madsen et al., 2011, Hayafune et al., 2014). In M. truncatula and L. japonicus, Nod factor perception requires two distinct LysM-RLK receptors that can heterodimerize, namely, M. truncatula LYSM DOMAIN CONTAINING RECEPTOR KINASE3 and L. japonicus NOD FACTOR RECEPTOR1 (MtLYK3/LjNFR1) and M. truncatula NOD FACTOR PERCEPTION (MtNFP)/LjNFR5 (Madsen et al., 2011, Moling et al., 2014). Upon Nod factor binding, these receptors recruit other proteins into an active signaling complex (Haney et al., 2011, Liang et al., 2018), including LjSYMRK/M. truncatula DOES NOT MAKE INFECTIONS2 (MtDMI2), the first component of the common SYM pathway that interacts with LjNFR5 (Antolín-Llovera et al., 2014). If the ancestor of symbiotic nitrogen-fixing bacteria recruited the AM signaling pathway by mimicking Myc factors, this most likely depended on the same pair of receptors that perceived mycorrhizal LCOs. However, at present the perception of Nod factor and Myc-LCOs in M. truncatula and L. japonicus is only partially dependent on the same LysM-RLKs: MtNFP is required for Myc-LCO-dependent calcium spiking (Sun et al., 2015). However, in Ljnfr1/Mtlyk3 mutants, Myc-LCOs are still perceived, albeit in a reduced number of cells (Zhang et al., 2015). Most probably, in these species, gene duplication events allowed subneofunctionalization of receptor functioning. Studies in Parasponia species revealed that the NFP/NFR5-type Nod factor receptor originates from a duplication that occurred in the root of the nitrogen-fixing clade. The NFP/NFR5 ortholog in Parasponia species (named PanNFP2 in P. andersonii) has been conserved, whereas this gene has been lost or pseudogenized in Rosales species that lost nodulation (van Velzen et al., 2018). This suggests that within the Rosales, genes in the MtNFP/LjNFR5/PanNFP2 orthogroup function specifically in Nod factor-induced nodulation. For P. andersonii, it was initially claimed that a single LysM-RLK, PanNFP1, was responsible for the detection of both AM fungi and rhizobia, based on Agrobacterium rhizogenes-mediated RNAi of PanNFP1 (Op den Camp et al., 2011). However, with the sequencing of the P. andersonii genome and recent analysis of mutants generated with CRISPR–Cas9, it became clear that this RNAi resulted in several off-targets and that also, in Parasponia, different receptor pairs may be required for AM and rhizobium symbiosis (van Velzen et al., 2018) (R.G., unpublished results). Taken together, this suggests that MtNFP/LjNFR5/PanNFP2 represents a third target gene in an engineering approach.
A driving force for the subfunctionalization of the symbiotic LysM-RLKs may be that it allowed adaptations to the Nod factor receptors without affecting AM symbiosis. This resulted in the evolution of Nod factor receptors that have a high affinity and stringency for Nod factors (Sun et al., 2015) and allowed co-evolution of the Nod factor receptors with the Nod factor structure of the preferred rhizobial partner (Radutoiu et al., 2007). In addition, Nod factor receptors may have acquired additional functions beyond activating the common SYM pathway. In L. japonicus mutants having a constitutively active CCaMK, spontaneous nodules are formed that can be infected by rhizobia through root hair infection. In contrast, when LjNFR1 or LjNFR5 is knocked out in this CCaMK autoactive background, nodules are rarely infected, since root hair infection is abolished (Madsen et al., 2010). This suggests a role for Nod factor receptors in infection thread growth independent of the common SYM pathway. Further, it was shown in M. truncatula that SYMBIOTIC REMORIN (MtSYMREM), one of the components of the Nod factor signaling complex, is expressed only after Nod factor perception, playing a role in infection thread growth and release of bacteria in the nodule (Lefebvre et al., 2010, Liang et al., 2018). This suggests that the nanodomains containing Nod factor receptors may have a role, such as providing a spatial cue for proteins that are required for rhizobial infection (Haney et al., 2011, Liang et al., 2018). Such a role could require additional adaptations to Nod factor receptors that would distinguish them from Myc factor receptors.
Despite intensive research, there are still gaps in our knowledge of the common SYM pathway. After perception of Nod or Myc factors, the first observed response is an oscillation of the nuclear calcium concentration that is produced by the nuclear envelope-localized cation channels LjCASTOR/LjPOLLUX/MtDMI1 and CYCLIC NUCLEOTIDE GATED CHANNEL 15 (CNGC15) (Peiter et al., 2007, Charpentier et al., 2008, Charpentier et al., 2016). At present, it is still unknown which signal connects the SYMRK receptor complex at the plasma membrane to the cation channels at the nuclear envelope. The nuclear calcium oscillation is decoded by CCaMK, which phosphorylates the transcription factors LjCYCLOPS/M. truncatula INTERACTOR OF DMI3 (MtIPD3), and MtIPD3-LIKE (MtIPD3L) (Yano et al., 2008, Miller et al., 2013, Jin et al., 2018). The direct downstream targets of LjCYCLOPS/MtIPD3 that are currently identified are the transcription factor-encoding genes REDUCED ARBUSCULAR MYCORRHIZA1 (RAM1), NIN, and ETHYLENE RESPONSIVE FACTOR REQUIRED FOR NODULATION1 (ERN1) (Singh et al., 2014, Pimprikar et al., 2016, Cerri et al., 2017). In legumes, induction of RAM1 is highly specific for AM symbiosis, while NIN and ERN1 expression is specific for nodulation (Schauser et al., 1999, Marsh et al., 2007, Gobbato et al., 2012, Cerri et al., 2017) (Figure 2). Since RAM1 and NIN are the master regulators required for the vast majority of transcriptional responses to AM fungi and rhizobia, respectively, the transcriptional responses to rhizobia and AM fungi are markedly different (Pimprikar et al., 2016, Liu et al., 2019b, Liu et al., 2019c, Schiessl et al., 2019). This suggests that there are other signaling pathways parallel to the common SYM pathway that allow plants to discriminate between different microbial partners. Alternatively, specificity is maintained throughout the common SYM pathway, by a still unknown mechanism (Genre and Russo, 2016). For engineering approaches to the nitrogen-fixing symbiosis trait, resolving these issues is relevant, as it will provide insights into the mechanisms by which plants discriminate between two distinct microsymbionts.
Symbiotic Signaling and Nodule Organogenesis Are Interlinked with Hormonal Pathways Regulating Plant Development
When the nitrogen-fixing nodule symbiosis is to be introduced into non-nodulating plants it is essential to identify and engineer cross talk—or the lack thereof—between symbiotic signaling and existing plant processes. As is evident from the nodule morphology of plant species interacting with Frankia (Figure 2), these nodules are morphologically somewhat similar to lateral roots. Also in legumes, there is a high transcriptomic overlap between developing nodules and lateral roots (Schiessl et al., 2019). Further, regulatory networks that govern the maintenance and identity of lateral root and nodule meristems are shared (Franssen et al., 2015). Thus, nodule organogenesis likely depends on the lateral root developmental program. Consequently, the mechanisms regulating the initiation of lateral roots under different ecological conditions may affect nodule formation. Even if this regulation is disconnected in nodulating species, it has to be considered when engineering root nodules on plants outside of the nitrogen-fixation clade. Further, the common SYM pathway, rhizobium-specific signaling, and nodule organogenesis are tightly interlinked with hormonal signaling: ethylene, gibberellin, cytokinin, and auxin (Breakspear et al., 2014, Van Zeijl et al., 2015, Fonouni-Farde et al., 2016, Jin et al., 2016).
The formation of a nodule primordium and meristem draws on generic plant hormone signaling, including the formation of a local auxin maximum (De Smet et al., 2007, Deinum et al., 2012, Ryu et al., 2012, Buhian and Bensmihen, 2018). In legumes, the first cell divisions that give rise to the nodule primordium occur in the inner cell layers, pericycle, and cortex, while rhizobia are infecting the epidermal cells (Timmers et al., 1999, Xiao et al., 2014). At this stage, Nod factor signaling in M. truncatula is limited to the epidermis (Vernié et al., 2015). Therefore, there must be a mobile signal from the epidermis to the pericycle to start nodule organogenesis (Ng et al., 2015, Deinum et al., 2016). The nature of this signal remains elusive. It is speculated it may be cytokinin, as the vast majority of rhizobium-induced responses in M. truncatula are dependent on cytokinin-induced signaling, and in L. japonicus it was shown that cytokinin biosynthesis genes are transcriptionally activated upon LCO signaling (Van Zeijl et al., 2015, Deinum et al., 2016, Reid et al., 2017, Schiessl et al., 2019). This cytokinin dependence is mainly related to the transcriptional regulation of NIN in the inner cell layers of the root, for which this gene in legumes gained a cluster of cytokinin-responsive cis-regulatory elements in its promoter (Yoro et al., 2014, Liu et al., 2019a). This cytokinin-responsive promoter element, together with a cytokinin signaling feed-forward loop, is essential for rhizobium LCO-induced nodule organogenesis in legumes. This suggests that such cytokinin–NIN signaling circuit may be a relevant element in an engineering approach. However, to date, it remains elusive whether this circuit is conserved in Parasponia and/or actinorhizal plants. This may not be the case, as comparative studies using exogenously applied cytokinin on A. glutinosa, legumes, and non-nodulating plants indicated that only nodulating legumes respond by the formation of nodule-like structures (Gauthier-Coles et al., 2018).
The importance of NIN activation in legume nodule formation is highlighted by ectopic expression studies that showed that NIN is sufficient to induce the formation of nodule-like structures (Soyano et al., 2013). Therefore, understanding the genetic adaptations that have occurred in NIN regulation is relevant for any engineering strategy. NIN is part of a small family of NIN LIKE PROTEINS (NLPs). The NLP family consists of three clades (orthogroups), one of which includes symbiotic NIN genes of nodulating plants (Mu and Luo, 2019). Species outside the nitrogen-fixing clade possess putative orthologous genes in this clade, e.g., Arabidopsis thaliana AtNLP1, AtNLP2, and AtNLP3, but these genes have not been functionally analyzed. NLP genes of a sister clade, namely AtNLP6 and AtNLP7, encode transcription factors that relocalize from the cytoplasm to the nucleus upon nitrate perception (Marchive et al., 2013, Liu et al., 2017), where they can bind to nitrate-responsive cis-regulatory elements (Konishi and Yanagisawa, 2013). Studies in L. japonicus show that legume NIN can also bind to nitrate-responsive cis-regulatory elements (Suzuki et al., 2013). However, in contrast to NLPs, legume NIN lacks the putative nitrate-binding site, by which it is non-responsive (Suzuki et al., 2013). The question remains whether these adaptations were relevant for its recruitment in nodulation. Functional and comparative studies of NIN in Parasponia and/or actinorhizal plants may provide such insights.
A Step-by-Step, Species by Species Engineering Approach
Can we engineer a nitrogen-fixing rhizobium symbiosis before knowing all the genetic adaptations that are associated with this trait? We strongly believe that an engineering strategy inspired by an “agile approach” popular in software development is the most efficient way of identifying gaps in our knowledge. In software development, “agile” refers to methodologies based on iterative development, where requirements and solutions evolve over time.
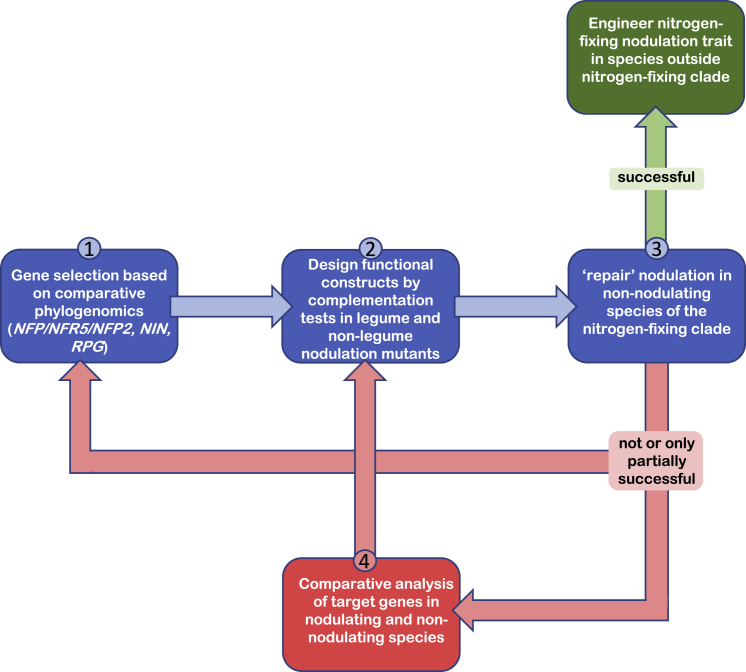
Obviously, nitrogen-demanding crops, like cereals, would be ideal targets for engineering the nitrogen-fixing nodulation trait (Charpentier and Oldroyd, 2010, Beatty and Good, 2011). However, Poaceae (Poales) and the nitrogen-fixing orders Fabales, Fagales, Cucurbitales, and Rosales have diverged significantly, and their root systems display several developmental adaptations (Hochholdinger and Zimmermann, 2008, Orman-Ligeza et al., 2013). The more distantly related a target species is from the nitrogen-fixing clade, the higher the chances are that the endogenous genes and processes that are to be recruited have diverged too much to function in a symbiotic context. Therefore, a stepwise approach seems more strategic, in which a proof of concept is provided in species that are evolutionarily more related to the nitrogen-fixing clade. We anticipate that even in closely related plant species the engineering will be an iterative process in which more than the currently identified target genes have to be introduced before a functional symbiosis is engineered (Figure 4). When all the genes that are required to engineer nitrogen-fixing symbiosis in these species are identified, these will provide a more solid basis for the engineering in more distantly related crops.
Figure 4.
A Roadmap for Engineering the Nitrogen-Fixing Nodulation Trait Based on an Evolutionary Blueprint from Nodulating Legumes or Non-legumes.
Step 1: Selection of engineering targets based on comparative phylogenomics using nodulating and non-nodulating species of the nitrogen-fixing clade. Currently, these studies have provided three potential targets: the LysM-RLK LCO receptor NFP/NFR5/NFP2, the transcription factor NIN, and the coiled-coil protein RPG. Step 2: Design constructs that can functionally complement legume and non-legume nodulation mutants. The nature of the gene constructs can depend on the selected target species for engineering. Step 3: Repair nodulation in non-nodulating species of the nitrogen-fixing clade. If this turns out not—or only partially—successful, then additional engineering targets should be found, focusing on genes that can be associated with the phenotype of the unsuccessful complementation attempt (step 1). Alternatively, it can be determined whether downstream targets of, e.g., NIN, are conserved (step 4). In case nodulation can be successfully repaired, the obtained knowledge can guide an engineering strategy for species outside the nitrogen-fixing clade.
Engineering Within the Nitrogen-Fixation Clade: A Matter of Repairing
As it is hypothesized that the nitrogen-fixing nodulation trait has a single evolutionary origin, many species within the nitrogen-fixing clade have lost nodulation (van Velzen et al., 2018, Griesmann et al., 2018, van Velzen et al., 2019). A straightforward strategy is the reintroduction of mutated symbiosis genes into a non-nodulating species of the nitrogen-fixing clade. Several such species represent crops for which transformation protocols are developed, for example, strawberry (Fragaria × ananassa and Fragaria vesca), cucumber (Cucumis sativus), and melon (Cucumis melo) (Barceló et al., 1998, Akasaka-Kennedy et al., 2004, Oosumi et al., 2006, Nanasato et al., 2013). Alternatively, a species that represents a sister lineage of a nodulating clade can be targeted. Such species are also described, for example, Trema species (Cannabaceae) as a non-nodulating sister of the Parasponia lineage (Cannabaceae), Dryas octopetala as non-nodulating sister of D. drummondii (Rosaceae), and Nissolia schotii as non-nodulating sister of Papilionoid legumes (Fabaceae) (Werner et al., 2014, Griesmann et al., 2018, van Velzen et al., 2018, Billault-Penneteau et al., 2019). The disadvantage of these non-model non-crop species is that transformation protocols and growth conditions need to be established. However, this may be outweighed by the fact that these species only relatively recently diverged from their nodulating relatives. Therefore, these species may have experienced fewer genetic adaptations that could hinder reengineering of the nitrogen-fixing nodulation trait compared with more distantly related crop species. For example, nodulation-specific cis-regulatory elements in genes that also function in a non-symbiotic context can be lost, as transcriptional networks are rewired.
Engineering nodulation in non-nodulating plant species will have to rely on the endogenous genes of the target species as much as possible, but a number of transgenes will have to be introduced. As a starting point, NFP/NFR5/NFP2, NIN, and RPG can be used. The question remains whether these genes represent the only genes that adopted a nodule-specific function and consequently are lost, or pseudogenized, in non-nodulating species in the nitrogen-fixing clade. This low number is in stark contrast to the hundreds of genes that correlate with AM symbiosis and subsequently are lost in species that lost this fungal symbiosis trait (Delaux et al., 2014, Bravo et al., 2016). Most probably, additional proteins have undergone small, but essential, adaptations that are easily missed in comparative omics studies, such as leghemoglobin (van Velzen et al., 2018). However, not knowing all genes that need to be transferred should not discourage us from starting to engineer nodulation in non-nodulation plants right now. Analyzing the phenotypes of our “failures” may be most informative in identifying missing components.
Both the transgene constructs that will be introduced and the endogenous genes of the target species should be functional in a nitrogen-fixing symbiosis context. For transgenes, functional promoters should be identified. For the endogenous genes that are expected to be important for symbiosis, both the proteins that they encode and their promoters should be tested in a symbiotic context. These aspects can be tested in mutant complementation studies in nodulating plant species (Figure 4). Especially proteins that are heavily connected by protein–protein interactions or transcription factors that recognize a specific DNA motif may not be functional outside of their native plant species. These issues can be identified by testing trans-complementation of the interactors and targets of all transgenes. Further, it will be useful to test trans-complementation in a wide range of nodulating plant species. Therefore, it is essential to have symbiosis mutants in an actinorhizal or Parasponia species. In this respect, protocols for CRISPR–Cas9-mediated mutagenesis as developed for P. andersonii are a valuable tool in optimizing an engineering strategy (van Zeijl et al., 2018). Based on interactions and functionality of endogenous genes and their promoters, the number of transgenes to be introduced likely has to be expanded. This expansion may also include endogenous genes driven by a different promoter to enable expression in a symbiotic context. We do not expect that a fully functional symbiosis will be achieved in a single attempt. Instead, the engineering will be an iterative process. In addition, partial repair of symbiosis will help to identify additional processes and genes that need to be introduced until the full set of genes is identified, and the nitrogen-fixing symbiosis can be restored.
Engineering Outside of the Nitrogen-Fixation Clade
Knowledge obtained from the “repairing” of nodulation can inspire an engineering approach to crops outside the nitrogen-fixing clade. Again, these species can be related more closely to the nitrogen-fixing clade—like Populus species and cassava (Manihot esculenta)—or are more distinct, like cereals. In any case, these species may have close homologs—or even orthologs—of essential symbiosis genes. For example, a putative ortholog of RPG can be found in Rosid species, whereas the origin of NIN can be traced back to the origin of eudicots (Schauser et al., 2005, van Velzen et al., 2018). As the function of these genes in a non-nodulating context remains largely unknown, it is unclear whether the functioning of these genes can be adapted such that they can support nodulation or, alternatively, have to be replaced by engineered versions.
Concluding Remarks
Phylogenomic studies, driven by de novo genome sequencing of non-model species, have provided new insights into the origin of the nodulation trait. Also, these studies provided leads for the design of an engineering strategy. Although these comparative phylogenomic studies were not yet exhaustive, and most probably more target genes can be identified, they inspired researchers to increase efforts to provide a proof of concept of engineering the nitrogen-fixing symbiosis trait. Even though it will be a long and iterative path, we are convinced that an approach based on an evolutionary blueprint can be successful. By comparing legumes to Parasponia and actinorhizal plant species, the simplest blueprint of the nitrogen-fixing nodulation trait can be derived and a core symbiosis gene set can be defined. However, an untouched question remains as to what the trade-off will be of having such an engineered symbiotic interaction. Currently, there is no widely accepted theory explaining why nodulation was lost in several lineages of the nitrogen-fixing clade. Hypotheses vary from the occurrence of pathogenic microbes that exploited plant nodules to a gradual decrease of atmospheric CO2 that hampered the photosynthetic potential to fuel biological nitrogen fixation (Griesmann et al., 2018, van Velzen et al., 2019). In our opinion, it is highly relevant to substantiate research in this direction because, if the latter hypothesis is correct, crops with engineered nitrogen-fixing nodules may have a bright future in the Anthropocene epoch.
Funding
This work was supported by the ENSA project funded by the Bill & Melinda Gates Foundation to the University of Cambridge to R.G. No conflict of interest declared.
Acknowledgments
We thank Katharina Pawlowski (Stockholm University) for her input on Actinorhiza–Frankia symbiosis, Wouter Kohlen (Wageningen University) for sharing information on Chamaecrista mimosoides, Defeng Shen (Wageningen University) for providing images for Figure 2, and Arjan van Zeijl for critical reading of the manuscript.
Published: January 13, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and IPPE, CAS.
Supplemental Information is available at Plant Communications Online.
Supplemental Information
References
- Akasaka-Kennedy Y., Tomita K.-O., Ezura H. Efficient plant regeneration and Agrobacterium-mediated transformation via somatic embryogenesis in melon (Cucumis melo L.) Plant Sci. 2004;166:763–769. [Google Scholar]
- Andrews M., James E.K., Sprent J.I., Boddey R.M., Gross E., dos Reis F.B. Nitrogen fixation in legumes and actinorhizal plants in natural ecosystems: values obtained using 15N natural abundance. Plant Ecol. Divers. 2011;4:131–140. [Google Scholar]
- Antolín-Llovera M., Ried M.K., Parniske M. Cleavage of the SYMBIOSIS RECEPTOR- LIKE KINASE ectodomain promotes complex formation with NOD FACTOR RECEPTOR 5. Curr. Biol. 2014;24:422–427. doi: 10.1016/j.cub.2013.12.053. [DOI] [PubMed] [Google Scholar]
- Arrighi J.-F., Cartieaux F. In: de Bruijn F.J., editor. Vol. 1. John Wiley & Sons, Inc.; Hoboken, New Jersey: 2015. Out of water of a new model legume: the Nod-independent Aeschynomene evenia; pp. 447–454. (Biological Nitrogen Fixation). [Google Scholar]
- Arrighi J.-F., Godfroy O., de Billy F., Saurat O., Jauneau A., Gough C. The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. U S A. 2008;105:9817–9822. doi: 10.1073/pnas.0710273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.-F., Cartieaux F., Brown S.C., Rodier-Goud M., Boursot M., Fardoux J., Patrel D., Gully D., Fabre S., Chaintreuil C., et al. Aeschynomene evenia, a model plant for studying the molecular genetics of the nod-independent rhizobium-legume symbiosis. Mol. Plant Microbe Interact. 2012;25:851–861. doi: 10.1094/MPMI-02-12-0045-TA. [DOI] [PubMed] [Google Scholar]
- Azani N., Babineau M., Bailey C.D., Banks H. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny the Legume Phylogeny Working Group (LPWG) Taxon. 2017;66:44–77. [Google Scholar]
- Barceló M., El-Mansouri I., Mercado J.A., Quesada M.A., Pliego Alfaro F. Regeneration and transformation via Agrobacterium tumefaciens of the strawberry cultivar Chandler. Plant Cell Tissue Organ Cult. 1998;54:29–36. [Google Scholar]
- Battenberg K., Potter D., Tabuloc C., Chiu J.C., Berry A.M. Comparative transcriptomics of two actinorhizal plants and the legume Medicago truncatula support the homology of root nodule symbioses and is congruent with a two-step process of evolution in the nitrogen-fixing clade of angiosperms. Front. Plant Sci. 2018;9:1256. doi: 10.3389/fpls.2018.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty P.H., Good A.G. Future prospects for cereals that fix nitrogen. Science. 2011;333:416–417. doi: 10.1126/science.1209467. [DOI] [PubMed] [Google Scholar]
- Becking J.H. In: Biological Nitrogen Fixation. Stacey G., Burris R.H., Evans H.J., editors. Routledge, Chapman and Hall; New York: 1992. The Rhizobium symbiosis of the nonlegume Parasponia; pp. 497–559. [Google Scholar]
- Beijerinck M.W. Rhizobial systematic. Botany Zeitung. 1888;46:796–804. [Google Scholar]
- Benedito V.A., Torres-Jerez I., Murray J.D., Andriankaja A., Allen S., Kakar K., Wandrey M., Verdier J., Zuber H., Ott T., et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- Beringer J.E., Hirsch P.R. Genetic engineering and nitrogen fixation. Biotechnol. Genet. Eng. Rev. 1984;1:65–88. [Google Scholar]
- Billault-Penneteau B., Sandré A., Folgmann J., Parniske M., Pawlowski K. Dryas as a model for studying the root symbioses of the Rosaceae. Front. Plant Sci. 2019;10:661. doi: 10.3389/fpls.2019.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov A.Y., Madsen L.H., Tsyganov V.E., Umehara Y., Voroshilova V.A., Batagov A.O., Sandal N., Mortensen A., Schauser L., Ellis N., et al. The Sym35 gene required for root nodule development in pea is an ortholog of Nin from Lotus japonicus. Plant Physiol. 2003;131:1009–1017. doi: 10.1104/pp.102.016071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozsoki Z., Cheng J., Feng F., Gysel K., Vinther M., Andersen K.R., Oldroyd G., Blaise M., Radutoiu S., Stougaard J. Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc. Natl. Acad. Sci. U S A. 2017;114:E8118–E8127. doi: 10.1073/pnas.1706795114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., York T., Pumplin N., Mueller L.A., Harrison M.J. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants. 2016;2:15208. doi: 10.1038/nplants.2015.208. [DOI] [PubMed] [Google Scholar]
- Breakspear A., Liu C., Roy S., Stacey N., Rogers C., Trick M., Morieri G., Mysore K.S., Wen J., Oldroyd G.E.D., et al. The Root Hair “Infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell. 2014;26:4680–4701. doi: 10.1105/tpc.114.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., Vinther M., Lorentzen A., Madsen E.B., Jensen K.J., et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. U S A. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F.J., Jing Y., Dazzo F.B. Potential and pitfalls of trying to extend symbiotic interactions of nitrogen-fixing organisms to presently non-nodulated plants, such as rice. Plant Soil. 1995;174:225–240. [Google Scholar]
- Brundrett M.C., Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol. 2018;220:1108–1115. doi: 10.1111/nph.14976. [DOI] [PubMed] [Google Scholar]
- Buhian W.P., Bensmihen S. Mini-review: nod factor regulation of phytohormone signaling and homeostasis during rhizobia-legume symbiosis. Front. Plant Sci. 2018;9:1247. doi: 10.3389/fpls.2018.01247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrill T.J., Hansen R. University of Illinois Agricultural Experiment Station; 1917. Is Symbiosis Possible between Legume Bacteria and Non-legume Plants? Bulletin 202. [Google Scholar]
- Cannon S.B., McKain M.R., Harkess A., Nelson M.N., Dash S., Deyholos M.K., Peng Y., Joyce B., Stewart C.N., Jr., Rolf M., et al. Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol. Biol. Evol. 2015;32:193–210. doi: 10.1093/molbev/msu296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H., Shibuya N., Nakagawa T., Barker D.G., Genre A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- Cerri M.R., Wang Q., Stolz P., Folgmann J., Frances L., Katzer K., Li X., Heckmann A.B., Wang T.L., Downie J.A., et al. The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol. 2017;215:323–337. doi: 10.1111/nph.14547. [DOI] [PubMed] [Google Scholar]
- Charpentier M., Oldroyd G. How close are we to nitrogen-fixing cereals? Curr. Opin. Plant Biol. 2010;13:556–564. doi: 10.1016/j.pbi.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 2008;20:3467–3479. doi: 10.1105/tpc.108.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Sun J., Martins T.V., Radhakrishnan G.V., Findlay K., Soumpourou E., Thouin J., Véry A.-A., Sanders D., Morris R.J., et al. Nuclear-localized cyclic nucleotide–gated channels mediate symbiotic calcium oscillations. Science. 2016;352:1102–1105. doi: 10.1126/science.aae0109. [DOI] [PubMed] [Google Scholar]
- Claessens H., Oosterbaan A., Savill P., Rondeux J. A review of thecharacteristics of black alder (Alnus glutinosa (L.) Gaertn.) and theirimplications for silvicultural practices. Forestry. 2010;83:163–175. [Google Scholar]
- Cissoko M., Hocher V., Gherbi H., Gully D., Carré-Mlouka A., Sane S., Pignoly S., Champion A., Ngom M., Pujic P., et al. Actinorhizal signaling molecules: Frankia root hair deforming factor shares properties with NIN inducing factor. Front. Plant Sci. 2018;9:1494. doi: 10.3389/fpls.2018.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo F., Diedhiou I., Vaissayre V., Brottier L., Acolatse J., Moukouanga D., Crabos A., Auguy F., Franche C., Gherbi H., et al. The Casuarina NIN gene is transcriptionally activated throughout Frankia root infection as well as in response to bacterial diffusible signals. New Phytol. 2015;208:887–903. doi: 10.1111/nph.13506. [DOI] [PubMed] [Google Scholar]
- Coats V., Schwintzer C.R., Tjepkema J.D. Truncated hemoglobins in Frankia CcI3: effects of nitrogen source, oxygen concentration, and nitric oxide. Can. J. Microbiol. 2009;55:867–873. doi: 10.1139/w09-042. [DOI] [PubMed] [Google Scholar]
- Colebatch G., Desbrosses G., Ott T., Krusell L., Montanari O., Kloska S., Kopka J., Udvardi M.K. Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 2004;39:487–512. doi: 10.1111/j.1365-313X.2004.02150.x. [DOI] [PubMed] [Google Scholar]
- Combier J.-P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernié T., Ott T., Gamas P., Crespi M., et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum E.E., Geurts R., Bisseling T., Mulder B.M. Modeling a cortical auxin maximum for nodulation: different signatures of potential strategies. Front. Plant Sci. 2012;3:1–19. doi: 10.3389/fpls.2012.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinum E.E., Kohlen W., Geurts R. Quantitative modelling of legume root nodule primordium induction by a diffusive signal of epidermal origin that inhibits auxin efflux. BMC Plant Biol. 2016;16:254. doi: 10.1186/s12870-016-0935-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaux P.-M., Varala K., Edger P.P., Coruzzi G.M., Pires J.C., Ané J.-M. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014;10:e1004487. doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demina I.V., Persson T., Santos P., Plaszczyca M., Pawlowski K. Comparison of the nodule vs. root transcriptome of the actinorhizal plant Datisca glomerata: actinorhizal nodules contain a specific class of defensins. PLoS One. 2013;8:e72442. doi: 10.1371/journal.pone.0072442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J. Phylogenetic perspectives on the origins of nodulation. Mol. Plant Microbe Interact. 2011;24:1289–1295. doi: 10.1094/MPMI-05-11-0114. [DOI] [PubMed] [Google Scholar]
- Duc G., Trouvelot A., Gianinazzi-Pearson V., Gianinazzi S. First report of non-mycorrhizal plant mutants (Myc−) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.) Plant Sci. 1989;60:215–222. [Google Scholar]
- Fabre S., Gully D., Poitout A., Patrel D., Arrighi J.-F., Giraud E., Czernic P., Cartieaux F. Nod factor-independent nodulation in Aeschynomene evenia required the common plant-microbe symbiotic toolkit. Plant Physiol. 2015;169:2654–2664. doi: 10.1104/pp.15.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Faria S.M., Sutherland J.M., Sprent J.I. A new type of infected cell in root nodules of Andira spp. (Leguminosae) Plant Sci. 1986;45:143–147. [Google Scholar]
- de Faria S.M., McInroy S.G., Sprent J.I. The occurrence of infected cells, with persistent infection threads, in legume root nodules. Can. J. Bot. 1987;65:553–558. [Google Scholar]
- Fonouni-Farde C., Tan S., Baudin M., Brault M., Wen J., Mysore K.S., Niebel A., Frugier F., Diet A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016;7:12636. doi: 10.1038/ncomms12636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H.J., Xiao T.T., Kulikova O., Wan X., Bisseling T., Scheres B., Heidstra R. Root developmental programs shape the Medicago truncatula nodule meristem. Development. 2015;142:2941–2950. doi: 10.1242/dev.120774. [DOI] [PubMed] [Google Scholar]
- Gage D.J. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 2002;184:7042–7046. doi: 10.1128/JB.184.24.7042-7046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage D.J. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier-Coles C., White R.G., Mathesius U. Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Front. Plant Sci. 2018;9:1901. doi: 10.3389/fpls.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Russo G. Does a common pathway transduce symbiotic signals in plant–microbe interactions? Front. Plant Sci. 2016;7:96. doi: 10.3389/fpls.2016.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- Gherbi H., Markmann K., Svistoonoff S., Estevan J., Autran D., Giczey G., Auguy F., Péret B., Laplaze L., Franche C., et al. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc. Natl. Acad. Sci. U S A. 2008;105:4928–4932. doi: 10.1073/pnas.0710618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodhbane-Gtari F., Hurst S.G., Oshone R., Morris K., Abebe-Akele F., Thomas W.K., Ktari A., Salem K., Gtari M., Tisa L.S. Draft genome sequence of Frankia sp. strain BMG5. 23, a salt-tolerant nitrogen-fixing actinobacterium isolated from the root nodules of Casuarina glauca grown in Tunisia. Genome Announc. 2014;2 doi: 10.1128/genomeA.00520-14. e00520–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E., Moulin L., Vallenet D., Barbe V., Cytryn E., Avarre J.-C., Jaubert M., Simon D., Cartieaux F., Prin Y., et al. Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science. 2007;316:1307–1312. doi: 10.1126/science.1139548. [DOI] [PubMed] [Google Scholar]
- Gobbato E., Marsh J.F., Vernié T., Wang E., Maillet F., Kim J., Miller J.B., Sun J., Bano S.A., Ratet P., et al. A GRAS-type transcription factor with a specific function in mycorrhizal signaling. Curr. Biol. 2012;22:2236–2241. doi: 10.1016/j.cub.2012.09.044. [DOI] [PubMed] [Google Scholar]
- Granqvist E., Sun J., Op Den Camp R., Pujic P., Hill L., Normand P., Morris R.J., Downie J.A., Geurts R., Oldroyd G.E.D., et al. Bacterial-induced calcium oscillations are common to nitrogen-fixing associations of nodulating legumes and non-legumes. New Phytol. 2015;207:551–558. doi: 10.1111/nph.13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesmann M., Chang Y., Liu X., Song Y., Haberer G., Crook M.B., Billault-Penneteau B., Lauressergues D., Keller J., Imanishi L., et al. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361:eaat1743. doi: 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- Gueddou A., Swanson E., Hezbri K., Nouioui I., Ktari A., Simpson S., Morris K., Kelley Thomas W., Ghodhbane-Gtari F., Gtari M., et al. Draft genome sequence of the symbiotic Frankia sp. strain BMG5.30 isolated from root nodules of Coriaria myrtifolia in Tunisia. Antonie Van Leeuwenhoek. 2018 doi: 10.1007/s10482-018-1138-1. [DOI] [PubMed] [Google Scholar]
- Haney C.H., Riely B.K., Tricoli D.M., Cook D.R., Ehrhardt D.W., Long S.R. Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell. 2011;23:2774–2787. doi: 10.1105/tpc.111.086389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayafune M., Berisio R., Marchetti R., Silipo A., Kayama M., Desaki Y., Arima S., Squeglia F., Ruggiero A., Tokuyasu K., et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U S A. 2014;111:E404–E413. doi: 10.1073/pnas.1312099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Hebelstrup K.H., Larsen K., Micaelo N.M., Jensen E.Ø. A single hemoglobin gene in Myrica gale retains both symbiotic and non-symbiotic specificity. Plant Mol. Biol. 2006;61:769–779. doi: 10.1007/s11103-006-0048-1. [DOI] [PubMed] [Google Scholar]
- Hellriegel H., Wilfarth H. Buchdruckerei der “Post” Kayssler; Berlin: 1888. Untersuchungen über die Stickstoffnahrung der Gramineen und Leguminosen. [Google Scholar]
- Hirsch A.M. Brief history of the discovery of nitrogen-fixing organisms. 2009. http://www.mcdb.ucla.edu/Research/Hirsch/imagesb/HistoryDiscoveryN2fixing Organisms.pdf
- Hocher V., Alloisio N., Auguy F., Fournier P., Doumas P., Pujic P., Gherbi H., Queiroux C., Da Silva C., Wincker P., et al. Transcriptomics of actinorhizal symbioses reveals homologs of the whole common symbiotic signaling cascade. Plant Physiol. 2011;156:700–711. doi: 10.1104/pp.111.174151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F., Zimmermann R. Conserved and diverse mechanisms in root development. Curr. Opin. Plant Biol. 2008;11:70–74. doi: 10.1016/j.pbi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Høgslund N., Radutoiu S., Krusell L., Voroshilova V., Hannah M.A., Goffard N., Sanchez D.H., Lippold F., Ott T., Sato S., et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS One. 2009;4:e6556. doi: 10.1371/journal.pone.0006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.S., Shrestha A., Zhong S., Miri M., Austin R.S., Sato S., Ross L., Huebert T., Tromas A., Torres-Jerez I., et al. Lotus japonicus NF-YA1 plays an essential role during nodule differentiation and targets members of the SHI/STY gene family. Mol. Plant Microbe Interact. 2016;29:950–964. doi: 10.1094/MPMI-10-16-0206-R. [DOI] [PubMed] [Google Scholar]
- Huss-Danell K. Tansley Review No. 93. Actinorhizal symbioses and their N2 fixation. New Phytol. 1997;136:375–405. doi: 10.1046/j.1469-8137.1997.00755.x. [DOI] [PubMed] [Google Scholar]
- Indrasumunar A., Searle I., Lin M.-H., Kereszt A., Men A., Carroll B.J., Gresshoff P.M. Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr) Plant J. 2011;65:39–50. doi: 10.1111/j.1365-313X.2010.04398.x. [DOI] [PubMed] [Google Scholar]
- Jacobsen-Lyon K., Jensen E.O., Jørgensen J.E., Marcker K.A., Peacock W.J., Dennis E.S. Symbiotic and nonsymbiotic hemoglobin genes of Casuarina glauca. Plant Cell. 1995;7:213–223. doi: 10.1105/tpc.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Liu H., Luo D., Yu N., Dong W., Wang C., Zhang X., Dai H., Yang J., Wang E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016;7:12433. doi: 10.1038/ncomms12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Chen Z., Yang J., Mysore K.S., Wen J., Huang J., Yu N., Wang E. IPD3 and IPD3L function redundantly in rhizobial and mycorrhizal symbioses. Front. Plant Sci. 2018;9:267. doi: 10.3389/fpls.2018.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakar S., Sturms R., Tiffany A., Nix J.C., Dispirito A.A., Hargrove M.S. Crystal structures of Parasponia and Trema hemoglobins: differential heme coordination is linked to quaternary structure. Biochemistry. 2011;50:4273–4280. doi: 10.1021/bi2002423. [DOI] [PubMed] [Google Scholar]
- Koenen E.J.M., Ojeda D.I., Steeves R., Migliore J. The origin and early evolution of the legumes are a complex paleopolyploid phylogenomic tangle closely associated with the Cretaceous-Paleogene (K-Pg) bioRxiv. 2019 doi: 10.1101/577957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Yanagisawa S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013;4:1617. doi: 10.1038/ncomms2621. [DOI] [PubMed] [Google Scholar]
- Kouchi H., Imaizumi-Anraku H., Hayashi M., Hakoyama T., Nakagawa T., Umehara Y., Suganuma N., Kawaguchi M. How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 2010;51:1381–1397. doi: 10.1093/pcp/pcq107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küster H., Vieweg M.F., Manthey K., Baier M.C., Hohnjec N., Perlick A.M. Identification and expression regulation of symbiotically activated legume genes. Phytochemistry. 2007;68:8–18. doi: 10.1016/j.phytochem.2006.09.029. [DOI] [PubMed] [Google Scholar]
- Laloum T., Baudin M., Frances L., Lepage A., Billault-Penneteau B., Cerri M.R., Ariel F., Jardinaud M.-F., Gamas P., de Carvalho-Niebel F., et al. Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 2014;79:757–768. doi: 10.1111/tpj.12587. [DOI] [PubMed] [Google Scholar]
- Lancelle S.A., Torrey J.G. Early development of Rhizobium-induced root nodules of Parasponia rigida. I. Infection and early nodule initiation. Protoplasma. 1984;123:26–37. [Google Scholar]
- Lancelle S.A., Torrey J.G. Early development of Rhizobium-induced root nodules of Parasponia rigida. II. Nodule morphogenesis and symbiotic development. Can. J. Bot. 1985;63:25–35. [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M.-F., Mun J.-H., Larrainzar E., Cook D.R., Gamas P., et al. The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 2014;65:481–494. doi: 10.1093/jxb/ert392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M., Herendeen P.S., Wojciechowski M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lee T.D., Bazzaz F.A. Regulation of fruit and seed production in an annual legume, Cassia fasciculata. Ecology. 1982;63:1363–1373. [Google Scholar]
- Lefebvre B., Timmers T., Mbengue M., Moreau S., Hervé C., Tóth K., Bittencourt-Silvestre J., Klaus D., Deslandes L., Godiard L., et al. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U S A. 2010;107:2343–2348. doi: 10.1073/pnas.0913320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Lewis G., Schrire B., MacKinder B., Lock M. Royal Botanic Gardens; Kew, UK: 2005. Legumes of the World. [Google Scholar]
- Li H.-L., Wang W., Mortimer P.E., Li R.-Q., Li D.-Z., Hyde K.D., Xu J.-C., Soltis D.E., Chen Z.-D. Large-scale phylogenetic analyses reveal multiple gains of actinorhizal nitrogen-fixing symbioses in angiosperms associated with climate change. Sci. Rep. 2015;5:14023. doi: 10.1038/srep14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Stratil T.F., Popp C., Marín M., Folgmann J., Mysore K.S., Wen J., Ott T. Symbiotic root infections in Medicago truncatula require remorin-mediated receptor stabilization in membrane nanodomains. Proc. Natl. Acad. Sci. U S A. 2018;115:5289–5294. doi: 10.1073/pnas.1721868115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E., Franken C., Smit P., Willemse J., Bisseling T., Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science. 2003;302:630–633. doi: 10.1126/science.1090074. [DOI] [PubMed] [Google Scholar]
- Limpens E., Moling S., Hooiveld G., Pereira P.A., Bisseling T., Becker J.D., Küster H. cell- and tissue-specific transcriptome analyses of Medicago truncatula root nodules. PLoS One. 2013;8:e64377. doi: 10.1371/journal.pone.0064377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.-H., Niu Y., Konishi M., Wu Y., Du H., Sun Chung H., Li L., Boudsocq M., McCormack M., Maekawa S., et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature. 2017;545:311–316. doi: 10.1038/nature22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Rutten L., Limpens E., van der Molen T., van Velzen R., Chen R., Chen Y., Geurts R., Kohlen W., Kulikova O., et al. A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. Plant Cell. 2019;31:68–83. doi: 10.1105/tpc.18.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-W., Breakspear A., Guan D., Cerri M.R., Jackson K., Jiang S., Robson F., Radhakrishnan G.V., Roy S., Bone C., et al. NIN acts as a network Hub controlling a growth module required for rhizobial infection. Plant Physiol. 2019;179:1704–1722. doi: 10.1104/pp.18.01572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Soyano T., Yano K., Hayashi M., Kawaguchi M. ERN1 and CYCLOPS coordinately activate NIN signaling to promote infection thread formation in Lotus japonicus. J. Plant Res. 2019 doi: 10.1007/s10265-019-01122-w. [DOI] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E.B., Antolín-Llovera M., Grossmann C., Ye J., Vieweg S., Broghammer A., Krusell L., Radutoiu S., Jensen O.N., Stougaard J., et al. Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signalling in cooperation with Nod factor receptor 5. Plant J. 2011;65:404–417. doi: 10.1111/j.1365-313X.2010.04431.x. [DOI] [PubMed] [Google Scholar]
- Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A., et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- Marchive C., Roudier F., Castaings L., Bréhaut V., Blondet E., Colot V., Meyer C., Krapp A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- Markmann K., Giczey G., Parniske M. Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 2008;6:0497–0506. doi: 10.1371/journal.pbio.0060068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive Calcium/Calmodulin-Dependent Protein Kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Boivin C., Giraud E., Perret X., Batut J. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 2009;17:458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Maunoury N., Redondo-Nieto M., Bourcy M., Van de Velde W., Alunni B., Laporte P., Durand P., Agier N., Marisa L., Vaubert D., et al. Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS One. 2010;5:e9519. doi: 10.1371/journal.pone.0009519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean D.N. Ecology of Alnus Glutinosa (L.) Gaertn.: I. Fruit formation. J. Ecol. 1955;43:46–60. [Google Scholar]
- Miller J.B., Pratap A., Miyahara A., Zhou L., Bornemann S., Morris R.J., Oldroyd G.E.D. Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell. 2013;25:5053–5066. doi: 10.1105/tpc.113.116921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S., Streng A., Bisseling T., Geurts R. Evolution of a symbiotic receptor through gene duplications in the legume–rhizobium mutualism. New Phytol. 2014;201:961–972. doi: 10.1111/nph.12549. [DOI] [PubMed] [Google Scholar]
- Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E., Okabe Y., Umehara Y., Miyamoto A., Kobae Y., et al. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- Mergaert P., Van Montagu M., Holsters M. Molecular mechanisms of Nod factor diversity. Mol. Microbiol. 1997;25:811–817. doi: 10.1111/j.1365-2958.1997.mmi526.x. [DOI] [PubMed] [Google Scholar]
- Moling S., Pietraszewska-Bogiel A., Postma M., Fedorova E., Hink M.A., Limpens E., Gadella T.W.J., Bisseling T. Nod factor receptors form heteromeric complexes and are essential for intracellular infection in medicago nodules. Plant Cell. 2014;26:4188–4199. doi: 10.1105/tpc.114.129502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X., Luo J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell. Mol. Life Sci. 2019 doi: 10.1007/s00018-019-03164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder L., Lefebvre B., Cullimore J., Imberty A. LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling and docking of chitooligosaccharides and Nod factors. Glycobiology. 2006;16:801–809. doi: 10.1093/glycob/cwl006. [DOI] [PubMed] [Google Scholar]
- Mus F., Crook M.B., Garcia K., Garcia Costas A., Geddes B.A., Kouri E.D., Paramasivan P., Ryu M.-H., Oldroyd G.E.D., Poole P.S., et al. Symbiotic nitrogen fixation and the challenges to its extension to Nonlegumes. Appl. Environ. Microbiol. 2016;82:3698–3710. doi: 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naisbitt T., James E.K., Sprent J.I. The evolutionary significance of the legume genus Chamaecrista, as determined by nodule structure. New Phytol. 1992;122:487–492. doi: 10.1111/j.1469-8137.1992.tb00077.x. [DOI] [PubMed] [Google Scholar]
- Nanasato Y., Konagaya K.-I., Okuzaki A., Tsuda M., Tabei Y. Improvement of Agrobacterium-mediated transformation of cucumber (Cucumis sativus L.) by combination of vacuum infiltration and co-cultivation on filter paper wicks. Plant Biotechnol. Rep. 2013;7:267–276. doi: 10.1007/s11816-012-0260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J.L.P., Hassan S., Truong T.T., Hocart C.H., Laffont C., Frugier F., Mathesius U. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. Plant Cell. 2015;27:2210–2226. doi: 10.1105/tpc.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.V., Wibberg D., Battenberg K., Blom J., Vanden Heuvel B., Berry A.M., Kalinowski J., Pawlowski K. An assemblage of Frankia Cluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. BMC Genomics. 2016;17:796. doi: 10.1186/s12864-016-3140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nguyen T., Wibberg D., Vigil-Stenman T., Berckx F., Battenberg K., Demchenko K.N., Blom J., Fernandez M.P., Yamanaka T., Berry A.M., et al. Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: they come in teams. Genome Biol. Evol. 2019;11:2273–2291. doi: 10.1093/gbe/evz153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Lapierre P., Tisa L.S., Gogarten J.P., Alloisio N., Bagnarol E., Bassi C.A., Berry A.M., Bickhart D.M., Choisne N., et al. Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 2007;17:7–15. doi: 10.1101/gr.5798407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Van Nguyen T., Battenberg K., Berry A.M., Heuvel B.V., Fernandez M.P., Pawlowski K. Proposal of 'Candidatus Frankia californiensis', the uncultured symbiont in nitrogen-fixing root nodules of a phylogenetically broad group of hosts endemic to western North America. Int. J. Syst. Evol. Microbiol. 2017;67:3706–3715. doi: 10.1099/ijsem.0.002147. [DOI] [PubMed] [Google Scholar]
- Oosumi T., Gruszewski H.A., Blischak L.A., Baxter A.J., Wadl P.A., Shuman J.L., Veilleux R.E., Shulaev V. High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta. 2006;223:1219–1230. doi: 10.1007/s00425-005-0170-3. [DOI] [PubMed] [Google Scholar]
- Op den Camp R., Streng A., De Mita S., Cao Q., Polone E., Liu W., Ammiraju J.S.S., Kudrna D., Wing R., Untergasser A., et al. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- Op den Camp R.H.M., Polone E., Fedorova E., Roelofsen W., Squartini A., Op den Camp H.J.M., Bisseling T., Geurts R. Nonlegume Parasponia andersonii deploys a broad rhizobium host range strategy resulting in largely variable symbiotic effectiveness. Mol. Plant Microbe Interact. 2012;25:954–963. doi: 10.1094/MPMI-11-11-0304. [DOI] [PubMed] [Google Scholar]
- Orman-Ligeza B., Parizot B., Gantet P.P., Beeckman T., Bennett M.J., Draye X. Post-embryonic root organogenesis in cereals: branching out from model plants. Trends Plant Sci. 2013;18:459–467. doi: 10.1016/j.tplants.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Ott T., van Dongen J.T., Günther C., Krusell L., Desbrosses G., Vigeolas H., Bock V., Czechowski T., Geigenberger P., et al. Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 2005;15:531–535. doi: 10.1016/j.cub.2005.01.042. [DOI] [PubMed] [Google Scholar]
- Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- Pate J.S., Layzell D.B. Energetics and biological costs of nitrogen assimilation. Biochem. Plants. 1990;16:1–42. [Google Scholar]
- Pawlowski K., Demchenko K.N. The diversity of actinorhizal symbiosis. Protoplasma. 2012;249:967–979. doi: 10.1007/s00709-012-0388-4. [DOI] [PubMed] [Google Scholar]
- Peiter E., Sun J., Heckmann A.B., Venkateshwaran M., Riely B.K., Otegui M.S., Edwards A., Freshour G., Hahn M.G., Cook D.R., et al. The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 2007;145:192–203. doi: 10.1104/pp.107.097261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson T., Benson D.R., Normand P., Vanden Heuvel B., Pujic P., Chertkov O., Teshima H., Bruce D.C., Detter C., Tapia R., et al. Genome sequence of “Candidatus Frankia datiscae” Dg1, the uncultured microsymbiont from nitrogen-fixing root nodules of the Dicot Datisca glomerata. J. Bacteriol. 2011;193:7017–7018. doi: 10.1128/JB.06208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P., Carbonnel S., Paries M., Katzer K., Klingl V., Bohmer M.J.J., Karl L., Floss D.S.S., Harrison M.J.J., Parniske M., et al. A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr. Biol. 2016;26:987–998. doi: 10.1016/j.cub.2016.01.069. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M.H., Albrektsen A.S., James E.K., Thirup S., Stougaard J. LysM domains mediate lipochitin–oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D., Nadzieja M., Novák O., Heckmann A.B., Sandal N., Stougaard J. Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 2017;175:361–375. doi: 10.1104/pp.17.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rípodas C., Castaingts M., Clúa J., Villafañe J., Blanco F.A., Zanetti M.E. The PvNF-YA1 and PvNF-YB7 subunits of the heterotrimeric NF-Y transcription factor influence strain preference in the Phaseolus vulgaris–Rhizobium etli symbiosis. Front. Plant Sci. 2019;10:221. doi: 10.3389/fpls.2019.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B., Rodde N., Jardinaud M.F., Timmers T., Sauviac L., Cottret L., Carr??re S., Sallet E., Courcelle E., Moreau S., et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77:817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- Ryu H., Cho H., Choi D., Hwang I. Plant hormonal regulation of nitrogen-fixing nodule organogenesis. Mol. Cells. 2012;34:117–126. doi: 10.1007/s10059-012-0131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S.P., Jain V. Biological nitrogen fixation with non-legumes: an achievable target or a dogma? Curr. Sci. 2007;92:317–322. [Google Scholar]
- Salgado M.G., van Velzen R., Van Nguyen T., Battenberg K., Berry A.M., Lundin D., Pawlowski K. Comparative analysis of the nodule transcriptomes of Ceanothus thyrsiflorus (Rhamnaceae, Rosales) and Datisca glomerata (Datiscaceae, Cucurbitales) Front. Plant Sci. 2018;9:1629. doi: 10.3389/fpls.2018.01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- Schauser L., Wieloch W., Stougaard J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005;60:229–237. doi: 10.1007/s00239-004-0144-2. [DOI] [PubMed] [Google Scholar]
- Schiessl K., Lilley J.L.S., Lee T., Tamvakis I., Kohlen W., Bailey P.C., Thomas A., Luptak J., Ramakrishnan K., Carpenter M.D., et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019 doi: 10.1016/j.cub.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S.B., Schlueter J., Ma J., Mitros T., Nelson W., Hyten D.L., Song Q., Thelen J.J., Cheng J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schranz M.E., Mohammadin S., Edger P.P. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012;15:147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Sellstedt A., Richau K.H. Aspects of nitrogen-fixing Actinobacteria, in particular free-living and symbiotic Frankia. FEMS Microbiol. Lett. 2013;342:179–186. doi: 10.1111/1574-6968.12116. [DOI] [PubMed] [Google Scholar]
- Silvester W.B., Berg R.H., Schwintzer C.R., Tjepkema J.D. In: Nitrogen-Fixing Actinorhizal Symbioses. Pawlowski K., Newton W.E., editors. Springer; Dordrecht: 2007. Oxygen responses, hemoglobin, and the structure and function of vesicles; pp. 105–146. [Google Scholar]
- Singer S.R., Maki S.L., Farmer A.D., Ilut D., May G.D., Cannon S.B., Doyle J.J. Venturing beyond beans and peas: what can we learn from Chamaecrista? Plant Physiol. 2009;151:1041–1047. doi: 10.1104/pp.109.144774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe. 2014;15:139–152. doi: 10.1016/j.chom.2014.01.011. [DOI] [PubMed] [Google Scholar]
- De Smet I., Tetsumura T., De Rybel B., Frei dit Frey N., Laplaze L., Casimiro I., Swarup R., Naudts M., Vanneste S., Audenaert D., et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Morgan D.R., Swensen S.M., Mullin B.C., Dowd J.M., Martin P.G. Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. U S A. 1995;92:2647–2651. doi: 10.1073/pnas.92.7.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013;9:e1003352. doi: 10.1371/journal.pgen.1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J.I. Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol. 2007;174:11–25. doi: 10.1111/j.1469-8137.2007.02015.x. [DOI] [PubMed] [Google Scholar]
- Sturms R., Kakar S., Trent J., Hargrove M.S. Trema and parasponia hemoglobins reveal convergent evolution of oxygen transport in plants. Biochemistry. 2010;49:4085–4093. doi: 10.1021/bi1002844. [DOI] [PubMed] [Google Scholar]
- Sun J., Miller J.B., Granqvist E., Wiley-Kalil A., Gobbato E., Maillet F., Cottaz S., Samain E., Venkateshwaran M., Fort S., et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki W., Konishi M., Yanagisawa S. The evolutionary events necessary for the emergence of symbiotic nitrogen fixation in legumes may involve a loss of nitrate responsiveness of the NIN transcription factor. Plant Signal. Behav. 2013;8 doi: 10.4161/psb.25975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S., Benabdoun F.M., Nambiar-Veetil M., Imanishi L., Vaissayre V., Cesari S., Diagne N., Hocher V., de Billy F., Bonneau J., et al. The independent acquisition of plant root nitrogen-fixing symbiosis in Fabids recruited the same genetic pathway for nodule organogenesis. PLoS One. 2013;8:e64515. doi: 10.1371/journal.pone.0064515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S., Hocher V., Gherbi H. Actinorhizal root nodule symbioses: what is signalling telling on the origins of nodulation? Curr. Opin. Plant Biol. 2014;20:11–18. doi: 10.1016/j.pbi.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Tadege M., Wen J., He J., Tu H., Kwak Y., Eschstruth A., Cayrel A., Endre G., Zhao P.X., Chabaud M., et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- Takanashi K., Takahashi H., Sakurai N., Sugiyama A., Suzuki H., Shibata D., Nakazono M., Yazaki K. Tissue-specific transcriptome analysis in nodules of Lotus japonicus. Mol. Plant Microbe Interact. 2012;25:869–876. doi: 10.1094/MPMI-01-12-0011-R. [DOI] [PubMed] [Google Scholar]
- Teulet A., Busset N., Fardoux J., Gully D., Chaintreuil C., Cartieaux F., Jauneau A., Comorge V., Okazaki S., Kaneko T., et al. The rhizobial type III effector ErnA confers the ability to form nodules in legumes. Proc. Natl. Acad. Sci. U S A. 2019 doi: 10.1073/pnas.1904456116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- Tisa L.S., Oshone R., Sarkar I., Ktari A., Sen A., Gtari M. Genomic approaches toward understanding the actinorhizal symbiosis: an update on the status of the Frankia genomes. Symbiosis. 2016;70:5–16. [Google Scholar]
- Trinick M.J. Structure of nitrogen-fixing nodules formed by Rhizobium on roots of Parasponia andersonii Planch. Can. J. Microbiol. 1979;25:565–578. doi: 10.1139/m79-082. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Bijl G.J.M., Liu W., Bisseling T., Schaart J.G., Geurts R. One-step Agrobacterium mediated transformation of eight genes essential for rhizobium symbiotic signaling using the novel binary vector system pHUGE. PLoS One. 2012;7:e47885. doi: 10.1371/journal.pone.0047885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen R., Holmer R., Bu F., Rutten L., van Zeijl A., Liu W., Santuari L., Cao Q., Sharma T., Shen D., et al. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. U S A. 2018;115:E4700–E4709. doi: 10.1073/pnas.1721395115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen R., Doyle J.J., Geurts R. A resurrected scenario: single gain and massive loss of nitrogen-fixing nodulation. Trends Plant Sci. 2019;24:49–57. doi: 10.1016/j.tplants.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Vernié T., Kim J., Frances L., Ding Y., Sun J., Guan D., Niebel A., Gifford M.L., de Carvalho-Niebel F., Oldroyd G.E.D. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell. 2015;27:3410–3424. doi: 10.1105/tpc.15.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Berry A.M. Plant regeneration from leaf segments of Datisca glomerata. Acta Bot. Gallica. 1996;143:609–612. [Google Scholar]
- Wang L., Rubio M.C., Xin X., Zhang B., Fan Q., Wang Q., Ning G., Becana M., Duanmu D. CRISPR/Cas9 knockout of leghemoglobin genes in Lotus japonicus uncovers their synergistic roles in symbiotic nitrogen fixation. New Phytol. 2019 doi: 10.1111/nph.16077. [DOI] [PubMed] [Google Scholar]
- Wardhani T.A.K., Roswanjaya Y.P., Dupin S., Li H., Linders S., Hartog M., Geurts R., van Zeijl A. Transforming, genome editing and phenotyping the nitrogen-fixing tropical Cannabaceae tree Parasponia andersonii. J. Vis. Exp. 2019 doi: 10.3791/59971. [DOI] [PubMed] [Google Scholar]
- Werner G.D.A., Cornwell W.K., Sprent J.I., Kattge J., Kiers E.T. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat. Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. Fate map of Medicago truncatula root nodules. Development. 2014;141:3517–3528. doi: 10.1242/dev.110775. [DOI] [PubMed] [Google Scholar]
- Yano K., Yoshida S., Müller J., Singh S., Banba M., Vickers K., Markmann K., White C., Schuller B., Sato S., et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. U S A. 2008;105:20540–20545. doi: 10.1073/pnas.0806858105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E., Suzaki T., Toyokura K., Miyazawa H., Fukaki H., Kawaguchi M. A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 2014;165:747–758. doi: 10.1104/pp.113.233379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N.D., Bharti A.K. Genome-enabled insights into legume biology. Annu. Rev. Plant Biol. 2012;63:283–305. doi: 10.1146/annurev-arplant-042110-103754. [DOI] [PubMed] [Google Scholar]
- Young N.D., Debellé F., Oldroyd G.E.D., Geurts R., Cannon S.B., Udvardi M.K., Benedito V.A., Mayer K.F.X., Gouzy J., Schoof H., et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480:520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zeijl A., Op den Camp R.H.M., Deinum E.E.E., Charnikhova T., Franssen H., Op den Camp H.J.M., Bouwmeester H., Kohlen W., Bisseling T., Geurts R., et al. Rhizobium Lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant. 2015;8:1213–1226. doi: 10.1016/j.molp.2015.03.010. [DOI] [PubMed] [Google Scholar]
- van Zeijl A., Wardhani T.A.K., Seifi Kalhor M., Rutten L., Bu F., Hartog M., Linders S., Fedorova E.E., Bisseling T., Kohlen W., et al. CRISPR/Cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Front. Plant Sci. 2018;9:284. doi: 10.3389/fpls.2018.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., Oldroyd G.E.D., Wang E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- Zhong C., Mansour S., Nambiar-Veetil M., Bogusz D., Franche C. Casuarina glauca: a model tree for basic research in actinorhizal symbiosis. J. Biosci. 2013;38:815–823. doi: 10.1007/s12038-013-9370-3. [DOI] [PubMed] [Google Scholar]
- Zhukov V., Radutoiu S., Madsen L.H., Rychagova T., Ovchinnikova E., Borisov A., Tikhonovich I., Stougaard J. The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol. Plant Microbe Interact. 2008;21:1600–1608. doi: 10.1094/MPMI-21-12-1600. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.