Abstract
Purpose of Review.
We describe and contrast the strengths of precision medicine with Western medicine, and complex trait genetics with Mendelian genetics. Classic genetics focuses on highly penetrant pathogenic variants in a single gene believed to cause or confer a high risk for well-defined phenotypes. However, a minority of disorders have a single gene etiology. Further, even individuals with identical Mendelian disease-associated genotypes may exhibit substantial phenotypic variability indicative of genetic and environmental modifiers. Still, most diseases are considered complex traits (or complex diseases).
Recent findings:
New insights into the genetic underpinnings of complex traits provide opportunities for advances in diagnosis and management. Precision medicine provides the framework for integrating complex trait knowledge into clinical care through a sophisticated analysis pipeline. Multidimensional modeling of acquired diseases includes multiple genetic risks scattered over many genes and gene regulators that must be interpreted based on functional evidence (e.g. genomics, transcriptomics) with structured models and expert systems; strengthened with machine learning and artificial intelligence. The choice of genotyping approaches (shot-gun sequencing, SNP-chips, targeted panels) is discussed.
Summary:
The result of a good precision medicine tool is clinical decision support and guidance to tackle complex disorders such as pancreatitis, diabetes and pancreatic cancer oncogenesis.
Keywords: Precision Medicine, Genetics, Genomics, chronic pancreatitis, modeling, artificial intelligence
Introduction.
Precision Medicine is an alternative paradigm to traditional Western medicine (based on the Germ Theory of disease) that focuses on complex mechanisms causing cell-level disorders in an individual patient before the disorder progresses to disease. These different paradigms are contrasted in a recent primer on precision medicine for complex disorders.[1] Complex diseases, such as acute pancreatitis (AP), recurrent acute pancreatitis (RAP), chronic pancreatitis (CP), diabetes (DM) types 1–3c (T1D, T2D, T3cD) and pancreatic ductal adenocarcinoma (PDAC) have multiple risk and complex etiologies.[2–5] Thus, the traditional approach of evaluating and diagnosing complex disease requires the new paradigm of precision medicine.
Managing complex diseases requires the use of genetic information integrated with biomarkers and knowledge of biology and medicine integrated into brief, understandable reports that can guide the caretaker and patient. The focus must be on underlying mechanistic disorders and treatments that assist dysfunctional cells to compensate for ongoing stress (e.g. early chronic pancreatitis, glucose intolerance) or block disease processes that cannot be reversed (e.g. CP, DM, PDAC).
This purpose of this review is to describe differences between clinical precision medicine reports and traditional genetic reports. Precision medicine provide valuable new insights, noting that it is impossible to consider and integrate all variables causing complex diseases in an individual person. Thus, the reports should serve as clinical decision support (CDS) tools in prioritizing and improving diagnostic evaluations, choosing first lines of therapies, selecting biomarkers for more accurate disease monitoring, and minimizing the development of high-risk complications.
Human Genetic Data: Fun facts and frightening features
Precision medicine integrates the effects of genetic variants on disease models to predict or replicate observed conditions. Determining the functional effect of a variant requires many lines of evidence, noting that subtle effects are typically compensated for by the second allele and the fantastic ability of the human body to adapt to significant problems for years without obvious consequences.
In 2015, the 1000 Genomes Project consortium published summary genetic data on 2,504 individuals.[6] They reported 84.7 million single nucleotide polymorphisms / variants (SNPs, or SNVs), 3.6 million short insertions/deletions (indels), and 60,000 structural variants. However, this sample represented only 0.00003% of all the people on the planet, and several populations were not represented. The Genome Aggregation Database (gnomAD) consortium recently analyzed the DNA sequence on 71,702 people and identified 602 million SNPs and 105 million indels.[7] Expanded testing indicates that millions of unrecognized variants are yet to be discover.
A typical genome differs from the reference human genome at 4.1 million to 5.0 million sites, or about 0.15% of the genome.[6] About 99.9% of variants consist of SNPs and short indels. The rest includes large deletions, copy-number variants (CNV) and large structural variants. Among the SNPs in each person, the majority were common—only 1–4% had a frequency <0.5%. About 2,000 variants per genome were previously associated with complex traits found in genome-wide association studies (GWAS) and 24–30 variants per genome were implicated in rare diseases. Yet, most people do not have hundreds of common or multiple rare diseases at the same time – or ever. Thus, there must be other factors, including compensatory mechanisms that protect most people from most diseases for a lifetime – with occasional failures that cause disease.
Mendelian Disorders.
Mendelian genetics focuses on very strong “pathogenic” mutation in a single gene with inheritance patterns such as autosomal dominant (e.g. hereditary pancreatitis) or autosomal recessive (e.g. cystic fibrosis). These “one gene - one disease” Mendelian disorders are “diagnosed” with the combination of clinical features and genetic data. This is a well-established field (Figure 1A), and guidelines for reporting pathogenic variants are well established.[8, 9] However, Mendelian genetics does not capture the impact of multiple common genetic variants (e.g. risk factors) in common diseases, and guidelines on reporting risk factors in complex disorders are needed.
Figure 1. Illustrative pipeline for Mendelian genetic reports and precision medicine reports.
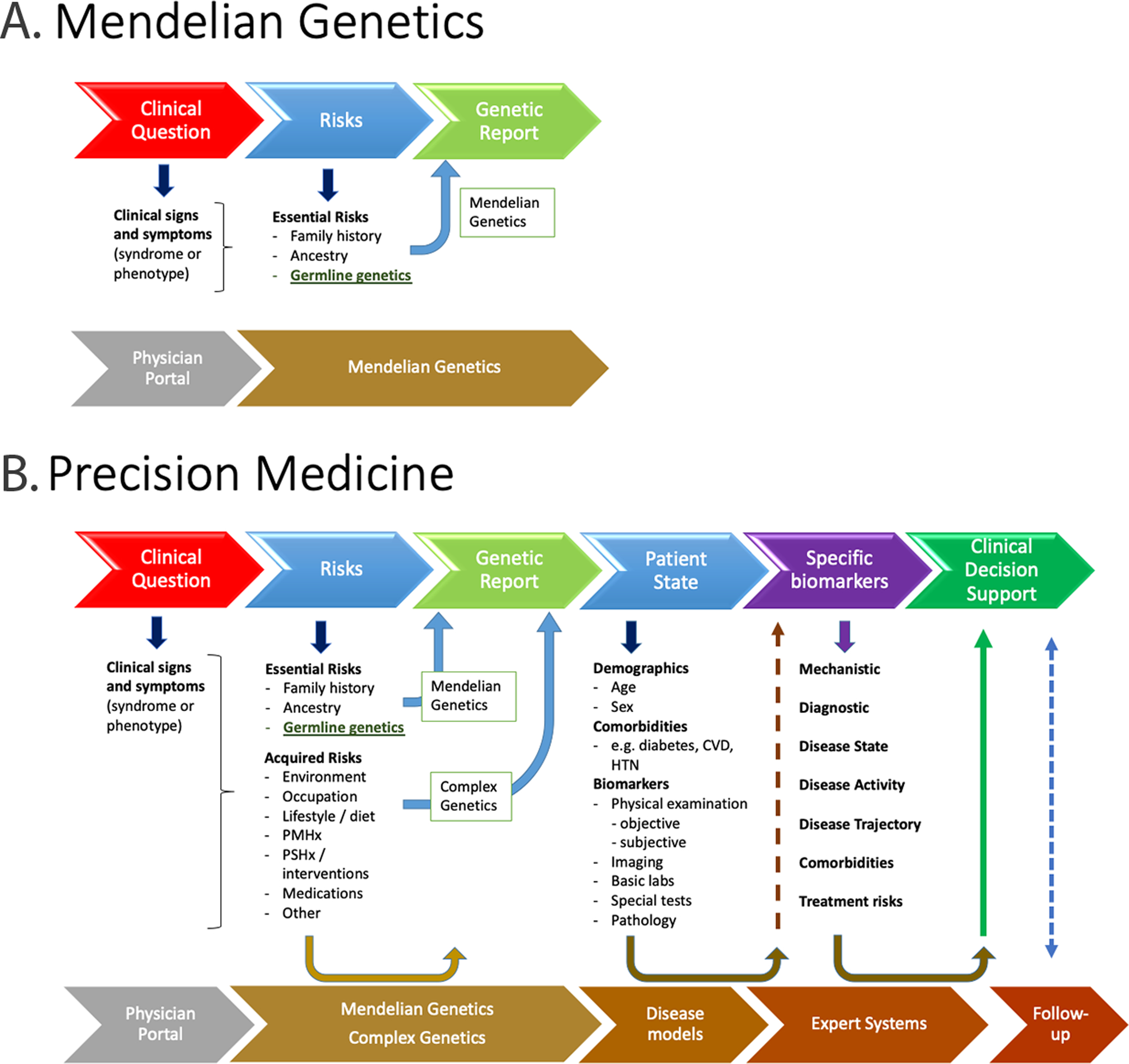
A. Mendelian genetic analysis is initiated with clinical suspicion of a genetic disorder. Top row, physician/patient activities. Bottom row, precision medicine analysis pipeline/expert systems activities. The report focuses on the genetic variants identified and whether they support the clinical diagnosis. B. Precision medicine goes beyond pathogenic genetic variants to include genetic risk factors and acquired risks, then clinical state and evidences of disease and complications, then recommendations for further specific biomarkers to establish the patient’s pathophysiology, measures of disease trajectory and treatment options for clinical decision support. Patient follow-up is recommended to continually refine the care and improve the accuracy of future predictions. The data for precision medicine do not need to be gathered or analyzed in a strictly sequential timeline. PMHx, personal medical history; PSHx, personal surgical history; CVD, cardiovascular disease; HTN, hypertension.
Genomics and Complex Disorders
Genomics is the study of the genome, including interactions of genes with each other and the person’s environment. This includes complex diseases such as heart disease, asthma, diabetes, and cancer—diseases typically caused by a combination of genetic and environmental factors.[10]
Many SNPs and haplotypes (groups of SNPs on the same allele) that are associated with complex diseases alter gene expression by altering transcription factor binding sites (TFBS). They are designated as expression quantitative train loci (eQTL). There are many TFBS for each gene allowing for controlled up- or down-regulation depending on the circumstances and the combination of transcription factors at any time. Gene expression is therefore determined by a complex, multi-level, tightly controlled regulatory system (see Encyclopedia of DNA elements (ENCODE) project (https://www.encodeproject.org/).[11, 12] Furthermore, transcription factors regulate the expression of multiple genes at the same time so the risk of dysfunction in a cellular mechanism may be caused by TFBS variants causing a supply deficit of one or more proteins. Epigenetic effects such as methylation of promoter sites, histone function, etc. are also important.
Although genomics is critical to precision medicine, it is not sufficient. Rich information without contest is uninterpretable, and the promised advances in precision medicine by genomics can be “lost in translation”.[13, 14] A new approach for interpretation and reporting the patient’s condition is needed to accurately assess the underlying disorder, to integrate biomarkers of disease state and activity, and to choose the right treatment in the right amount at the right time to minimize disease, suffering and cost of care.
Disease Models for Complex Pancreatic Diseases.
Disease models are critical to precision medicine because they direct an explicit reduction in information complexity, and both define and order variables in precise, rule-based (e.g. evidence based) programs. They also allow for multiple hypothetical conditions to be considered to understand emerging concepts.
Disease models.
A model is a simplified representation of a thing that is used to explain the relationship between factors that result in observed phenomenon. A disease model includes one or more factors that, when present, explain a pathologic process that leads to a disease. Western Medicine is designed to consider one variable at a time in case-control statistical approaches. Randomized, controlled treatment trials using this approach for complex disorders either fail or result in a high number needed to treat (NNT).[1] For complex disorders, new approaches such as mechanism-based n-of-1 studies are needed, with individualized disease models prioritizing the treatment.[15–17]
Precision medicine must rely on models, but the models must be both sophisticated in biological principles and dynamic so that they can be modified by mechanistic and quantitative variables specific to an individual patient. There is tremendous interest in “big data” and data-driven models, but these are association-based and limited by the complexity of the disease (heterogeneity), the number of people who have the disease (study power), the use of traditional disease classification and the accuracy of phenotyping (specificity), biomarkers /outcome (endpoints) fidelity, and correction of big data from multiple testing (false discovery). In contrast, precision medicine must be based on mechanistic (predictive) modeling, integrating risk variants and patient status into the model.[18]
Data-driven models can identify common disease drivers, risk factors, protective factors, response mechanisms, covariates and other features. But big data alone has proven to be insufficient to define and deliver precision medicine. However, this data can inform the model builders on key pathways and variables needed in mechanistic disease models. Fortunately, disease models for pancreatic disease are orders of magnitude easier than Crohn’s disease or liver diseases since the pancreas only has a few cell types (acinar, duct, islet), and they each have one primary function.
A Focus on the Function of Specialized Cells.
Precision medicine models focus on the function of specialized cells and their response to stimulation, stress and injury.[1, 19] Living cells respond to both external and internal signals that up-regulate or down-regulate specific cell functions that are executed by protein molecules that form machines. Only subsets of genes are expressed at any time, and focusing on these genes informs us on active machines and reduces genomic complexity.
The repertoire of genes being expressed in a tissue under defined conditions can be determined by sequencing RNA and is called the expressome. A new tool of single cell RNA-sequencing allows the repertoire of gene expression in individual cells to be determined. These technologies are important for modeling single cells and obtaining evidences of the impact of genetic variants in health and disease.
The goal of developing mechanistic disease models is to determine where within a complex system it is likely that something will go wrong under defined conditions. The “where” is defined by the components of a machine and/or process, and the “when” is defined by the condition such as stress or other perturbations of the specialized cell.
Artificial Intelligence (AI) is the use of a computer to mimic intelligence human in performing specific tasks that may include “the ability to reason, discover meaning, generalize, or learn from past experience and to achieve goals without being explicitly programmed for specific action”.[20] AI gained fame in playing games such as chess or Jeopardy and proving to be useful in optical recognition of polyps in gastroenterology. However, in precision medicine AI programs such as IBM Watson have largely failed. The reason can be explained by a principal that I learned in my first computer course in 1977- GIGO (garbage in, garbage out). In this case, the garbage is clinical information that is defined by syndromes and pathologic appearance based on an inadequate paradigm designed over a century ago. Thus, the idea of AI is great, but the success in medicine, as in chess and Jeopardy, requires explicit rules, exact definitions and reproduceable and predictable patterns. However, AI does have a bright future, especially as the same mechanism-linked features that are critical for understanding precision medicine replace outdated terminology and symptom-based disease definitions. Furthermore, key insights are now being made through sophisticated advances in artificial intelligence, including machine learning. Thus, AI is not in conflict with precision medicine. These new tools provide added valuable for generating testable models, particularly when the models are constrained by biological principles and established mechanisms.
Mathematical Duct Cell Model.
There are many different types of models, and the selection of the type and sophistication depends on the process or phenomenon that is being modeled. For example, the pancreatic duct cell has a function of secreting bicarbonate ions. Ion movement is determined by known electro-chemical forces (e.g. the Nernst equation), so it is possible to build a model of the duct cell that has physical dimensions and features, with known ion transporters, pumps and exchangers organized in their anatomical location (basolateral or apical surface). Linking the concentration of various ions in the various compartments (plasma, intracellular, duct) and changing the parameters of key molecules (e.g. lowering the permeability of CFTR to chloride or bicarbonate) it is possible to simulate the function of duct cell secretion.[21] Furthermore, since we know the effect of genetic variants on the function of CFTR[22], we can simulate what will happen to chloride or bicarbonate secretion with various combinations of variants (e.g. using differential equations and a modeling program). Furthermore, since we know the effect of therapeutic agents on CFTR[23], the degree of improvement of function with therapy can be predicted.
Multi-dimensional models.
Further consideration of the duct model is warranted. Basic physiology reminds us that duct cells go from minimal secretion to high volume secretion with a meal. The mechanism includes stimulation from the vagus nerve (cholinergic) and hormonal stimulation from secretin released from the duodenum upon acidification. Pancreatic juice secretion is a plumbing issue, with flow dependent on upstream hydrostatic pressure (generated by the upstream duct cells) and downstream resistance, such as the sphincter of Oddi. The duct cell can also be activated by high calcium concentrations in the duct via a calcium-sensing receptor. Patients with pancreatitis but without cystic fibrosis often have mild CFTR variants that are predicted to reduce, but not eliminate duct function. A multidimensional model may also incorporate epidemiologic information into specific aspects of a cell-based model, such as cigarette smoking.[24] In this case, the context becomes important as confounding factors lower the function of the duct cell below the threshold needed to protect the duct from intraductal trypsin activation. Thus, a mechanistic, mathematical model of the duct can be a component of a larger physiologic, rule-based model.
Integration of genetic risk factors into complex duct models.
Genetic risk factors are important for determining likelihood of CFTR dysfunction in the duct cell model. First, some CFTR variants were thought to be benign because they do not cause lung disease. Instead, they cause diseases of the pancreas, male reproductive system and chronic sinusitis by altering the ability of CFTR to secrete bicarbonate despite continued chloride conductance.[25–27] In other cases, diminished CFTR function may cause pancreatitis in in the context of diminished trypsin inhibition capacity linked the SPINK1 p.N34S or CTRC p.G60G risk haplotypes.[28–32] Genetic risk variants in CFTR in combination with risk variants in the calcium sensing receptor gene (CASR) also increase risk through diminished signaling to the defective CFTR to open when there is high duct calcium concentrations (which protects active trypsin from degradation). Worse pancreatic disease is also seen with combined CFTR and SLC26A9 variants, with the latter molecule being a chloride channel that may help rescue CFTR function to some degree.[33] Finally, when duct cells are under stress they upregulate GGT1, an extracellular enzyme that provides substrates for intracellular glutathione-linked detoxification.[34] Genetic variants in the regulatory region of GGT1 were previously been associated with risk of pancreatitis and pancreatic cancer.[35, 36] New approaches to understanding complex disorders suggest that minor CFTR variants that are not sufficient to cause disease alone can be associated with pancreatitis in combination with GGT1 variants – suggesting that lower level injury with failure of cell-specific compensatory or protective mechanisms will also cause disease.[37]
Disease stage.
Understanding the underlying mechanism of injury or dysfunction is only one step. Next, the expert will need to consider where the patient is in the disease process. The considerations for management are totally different for a person with early clinical signs and symptoms and another person with end-stage disease (Figure 2). For example, if the patient has had an episode of idiopathic acute pancreatitis, the likelihood and mechanisms of recurrence is very important for risk management. Furthermore, knowledge of risk for diabetes, pain syndromes or cancer may be important. Early knowledge of injury and inflammation in the pancreas thereby enhances the accuracy of risk assessments of specific types of diabetes (T1D, T2D, T3cD, other)[38–40] or likelihood of developing PDAC.[41–43]
Figure 2. Progressive stage model of CP with multiple complications.
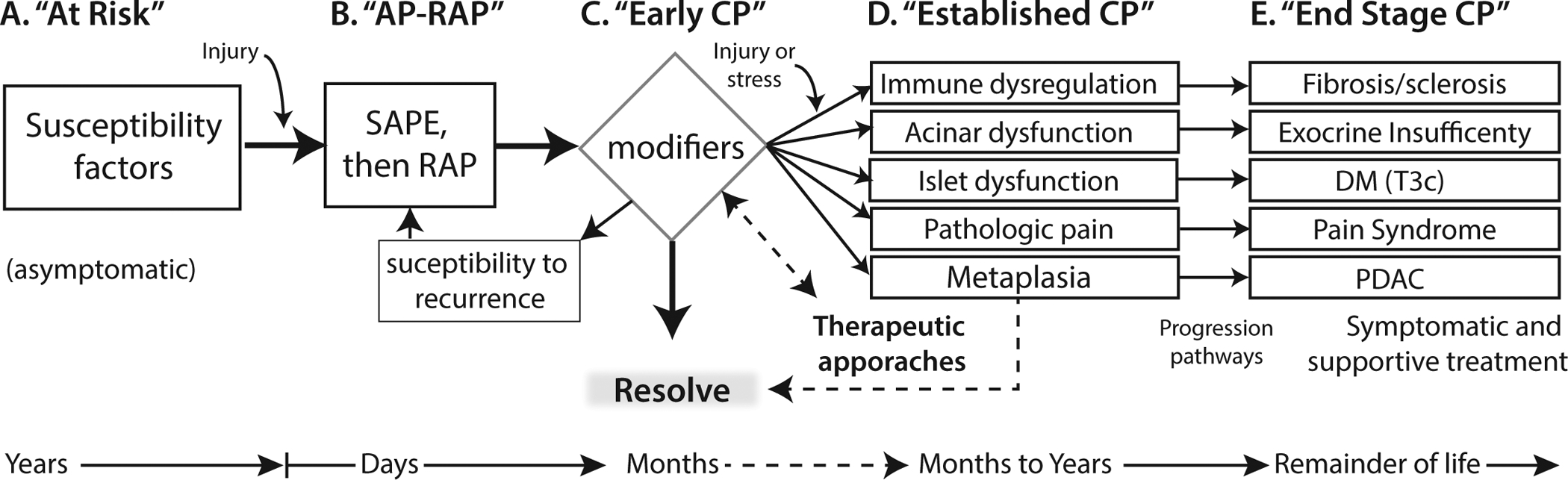
Stage A is asymptomatic risk. Stage B indicates the initiation of the pathogenic process toward chronic pancreatitis that is usually caused by AP and especially RAP. Stage C includes the detection of features of later stages (D & E) but they are not persistent or have not progressed to the stage of established CP. Preventative and therapeutic approaches are aimed at Stages C and D (dashed lines), whereas symptomatic and function replacement therapies (e.g. pancreatic enzyme replacement, insulin replacement) are the mainstay of Stage E. The features in D and E are not surrogates of each other and may need to be monitored independently. AP-RAP, acute pancreatitis and recurrent acute pancreatitis; CP, chronic pancreatitis; DM (T3c), diabetes mellitus Type IIIc or pancreatogenic diabetes mellitus; PDAC, pancreatic ductal adenocarcinoma; SAPE sentinel acute pancreatitis event. From Whitcomb [3]
Choosing genotyping data and biomarkers.
Physicians and health care workers often have trouble choosing between different tests, which come with very different price tags. The choice reverts to the old adage: Price, Quality, Service - choose two of the three. For genotyping, the price depends on the breadth, depth and method of genotyping. Thus, one gene can be evaluated by traditional Sanger DNA sequencing, which is highly accurate but is relatively inefficient, expensive and only covers several hundred base pairs. For complex disorders, multiple genes and regulatory regions must be sequenced.
Shot-gun sequencing.
If multiple genes must be evaluated, then large-scale “shot-gun” sequencing may be used, where it is possible to sequence all of the exons (whole exome sequencing, WES) or the entire genome (whole genome sequencing, WGS). Shot-gun sequencing involves shearing DNA into fragments that can be sequenced in parallel, resulting in a huge number of sequenced reads ~50–300 base pairs in length. A computer aligns these reads to a reference genome to determine where the subject’s DNA mismatches with the reference sequence.
The good news is that it is possible to get accurate DNA sequences from most of the genome, but this approach lacks accuracy. Errors occur in highly homologous regions such as between genes and pseudogenes, and between isoforms of families of genes (e.g. trypsin genes, including PRSS1). Also, shot-gun sequencing is a biochemical reaction, so some areas sequence very well, while other areas sequence poorly. To compensate for unevenness, sequencing depth may be increased. In this approach, the entire exome or genome is sequenced on the average of 30, 40 or more times to improve coverage of poorly sequenced areas. Errors also come from next-generation sequencing bioinformatics pipelines, with different algorithms often generating slightly different results. WES and especially WGS are more expensive per person, both in the cost of raw materials, computational cost and data storage. Tremendous effort continues to be exerted to improve the results of shot-gun sequencing.
SNP-chips.
SNP-chips (or SNP arrays) were designed to tag haplotypes at specific locations across the genome for GWAS studies. A tremendous number of disease-associated loci were discovered, but the association does not indicate that the SNP associated with the disease causes the disease. Furthermore, by design, the SNPs on earlier chips were common (so that variants would be seen at some frequency, such as >5%, in both cases and controls). To a large extent, the disease-associated haplotypes are used as risk indicators rather than mechanistic determinants. Also, SNP-chips typically represent only the most common disease-associated haplotypes, missing almost all uncommon or novel variants.
Limitations of comparing SNP-chips are often addressed by imputation, a technique that makes assumptions about the probability of other SNPs on a haplotype to obtain broader coverage.[44] SNP-chips are also used for ancestry, and they can have pathogenic variants added to help classify patients into risk groups. One popular approach with SNP-chip data is the polygenic risk score (PRS), a method of taking a dozen or more SNPs from a GWAS study that are independently associated with disease, and testing how common a combination of these are seen in a population with and without disease (a type of burden test).
We recently used a Type 2 diabetes (T2D) PRS to demonstrate that patients with CP and diabetes have a T2D PRS that matched a T2D populations without pancreatitis. Furthermore, CP patients without diabetes have a PRS that matched a population without DM.[39] However, common demographic risks for T2D also increase risk for DM in CP patients[38] indicating the importance of both common genetic and environmental factors in causing this secondary disease.
Gene panels.
A gene panel is a carefully curated and well-designed group of genes and other DNA loci that are known to contribute to specific diseases. They integrate insights from Sanger sequencing (with carefully designed primers to get exactly the right locus), shot-gun sequencing (massive parallel sequencing) and selected SNPs (capturing key haplotypes). The advantage is outstanding coverage of targeted genes and SNPs at a fraction of the cost. This approach has been popularized with cancer risk panels, and multiple gene testing is recommended for early evaluation of idiopathic pancreatitis patients by national and international societies.[45–47]
Precision Medicine Report.
The goal of a precision medicine report is to provide clinical decision support for clinicians and patients. Clinical decision support is defined by the National Coordinator for Health Information Technology (ONC, HealthIT.gov) as a service that “provides clinicians, staff, patients or other individuals with knowledge and person-specific information, intelligently filtered or presented at appropriate times, to enhance health and health care”. In this context, knowledge represents the integrated scientific and clinical facts and experience of expert physicians. The person-specific information represents the optimal or available data that is uploaded into analysis system. The appropriate time should be “on demand” beginning with initial evaluation, through the various phases and into longer term follow up. This type of analysis requires a complex pipeline (Figure 1B)
Underlying Disorder.
The “knowledge” to generate a precision medicine report includes the information that an “expert” would need for integration of the patient’s signs and symptoms, predictions from disease models utilizing genetics and other risks, and supporting biomarker evidence of cell or system dysfunction. Considerations must also include environment, lifestyle, injuries, metabolic stresses, other diseases – and even the microbiome. For pancreatic diseases, the major, well-defined etiology/risk factors have been organized into lists, including the recently updated Toxic-Metabolic, Idiopathic, Genetic, Autoimmune, Recurrent or severe acute pancreatitis and Obstruction (TIGAR-O).[2] Important considerations include demographic features such as ancestry, family history, sex, age, past medical history, past surgical history, lifestyle, diet, medications and other information. All of these are patient-specific factors considered by experts, and therefore need to be considered by expert systems. Precision medicine must include expert systems, providing the advantage of capturing and systematizing the insights of multiple experts (like a multidisciplinary team). Through automation, a physician is supported to make better decisions based on considerations of large amounts of information in a very short period of time.
There are important limits to precision medicine and precision medicine reports. The analysis has many steps in the pipeline, and missing or inaccurate data reduces prediction accuracy. More importantly, the exact effect of most genetic variants must be estimated, many variants have been incorrectly classified as benign based on limited data, and more damaging variants are yet to be discovered. Thus, the precision medicine report is a CDS tool rather than a definitive solution to every individual patient. It frames the patient within a population of patients with similar features but somewhat variable outcomes.
Conclusion.
Precision medicine is needed to advance the care of people who are developing pancreatic diseases through complex genetic and environmental interactions. The complexity of human cell and systems biology in combination with the human genome requires caregivers to use new genomic models and accurate biomarkers of disease state and activity to optimize personalized patient management. With high throughput ‘omic’ technologies and high-performance computing, models with expert systems can be leveraged to provide practical clinical decision support and guidance to tackle complex disorders such as pancreatitis, diabetes and pancreatic cancer.
Key Points:
Mendelian genetics focuses on pathogenic genetic variants causing monogenic diseases. This approach is inadequate for precision medicine.
Precision medicine reports must incorporate pathogenic genetic variants, genetic risk factors, genomic effect predictions, environmental factors, biomarkers and clinical context within disease models and linked to expert systems to provide clinical decision support recommendations.
Multiple methods are available for germline genotyping that differ in quality and quantity, as well as price and utility.
Continued genetic epidemiology research continues to discover new variants; machine learning and AI are improving data mining and improved disease modeling is providing more accurate, patient-specific insights into pancreatic diseases.
Financial support and sponsorship
Support for this work includes funding from the National Institutes of Health, DK061451 (DCW) and DK063922 (CAS)
Disclosures: Support for this work includes funding from the National Institutes of Health, DK061451 (DCW).
Footnotes
Conflicts of interest
Celeste Shelton CGC, PhD is employed by Ariel Precision Medicine. David Whitcomb MD PhD is a consultant for AbbVie, Abbott, Ariel Precision Medicine, BioNTech, Regeneron and Samsung. He is a cofounder of Ariel Precision Medicine and may have equity.
References.
- 1.Whitcomb DC. Primer on Precision Medicine for Complex Chronic Disorders. Clin Transl Gastroenterol. 2019;10(7):e00067. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper is a brief discussion of the difference between traditional Western medicine (taught in all medical schools) and precision medicine, and why a completely new perspective is needed to move forward in medicine.
- 2.Whitcomb DC, North American Pancreatitis Study G. Pancreatitis: TIGAR-O Version 2 Risk/Etiology Checklist With Topic Reviews, Updates, and Use Primers. Clin Transl Gastroenterol. 2019;10(6):e00027. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper is a catelogue of all of the major known risk factors for recurrent acute pancreatitis and chronic pancreatitis. It is an update of the original TIGAR-O classification from 20 years ago, based on new findings over the past two decades
- 3.Whitcomb DC, Frulloni L, Garg P, Greer JB, Schneider A, Yadav D, et al. Chronic pancreatitis: An international draft consensus proposal for a new mechanistic definition. Pancreatology. 2016;16:218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This paper redefines chronic pancreatitis as an inflammatory process rather than an end-stage disease so that early diagnois and intervention is possible. This has bee accepted as the preferred definition by all of the major pancreas societies.
- 4.American Diabetes A. Standards of medical care in diabetes--2014. Diabetes Care. 2014;37 Suppl 1:S14–80. [DOI] [PubMed] [Google Scholar]
- 5.Sharma A, Kandlakunta H, Nagpal SJS, Feng Z, Hoos W, Petersen GM, et al. Model to Determine Risk of Pancreatic Cancer in Patients With New-Onset Diabetes. Gastroenterology. 2018;155(3):730–9 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francioli L, Macarthur DG. gnomAD v3.0: MacArthur Lab; 2019. [cited 2020 May 3, 2020] Available from: https://macarthurlab.org/2019/10/16/gnomad-v3-0/.
- 8.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The standard method of defiing pathogenic variants for Mendelian genetic disorders.
- 9.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–55. [DOI] [PubMed] [Google Scholar]
- 10.Anonymous. Genetics vs. Genomics Fact Sheet: National Human Genome Research Institute; 2018. [updated September 7, 2018; cited 2020 April 3, 2020] Available from: https://www.genome.gov/about-genomics/fact-sheets/Genetics-vs-Genomics.
- 11.Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krawczyk M, Liebe R, Lammert F. Toward Genetic Prediction of Nonalcoholic Fatty Liver Disease Trajectories: PNPLA3 and Beyond. Gastroenterology. 2020;158(7):1865–80 e1. [DOI] [PubMed] [Google Scholar]
- 14.Whitcomb DC, Shelton C, Brand RE. Genetics and Genetic Testing in Pancreatic Cancer. Gastroenterology. 2015. [DOI] [PubMed] [Google Scholar]
- 15.Lowe ME, Goodman MT, Cote GA, Glesby MJ, Haupt M, Schork NJ, et al. Accelerating the Drug Delivery Pipeline for Acute and Chronic Pancreatitis: Summary of the Working Group on Drug Development and Trials in Recurrent Acute Pancreatitis at the National Institute of Diabetes and Digestive and Kidney Diseases Workshop. Pancreas. 2018;47(10):1193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; * An important conference marking the transition from traditional approaches to study pancreatic disease in humans to new approaches based on the mechanistic definitio of pancreatitis.
- 16.Schork NJ. Personalized medicine: Time for one-person trials. Nature. 2015;520(7549):609–11. [DOI] [PubMed] [Google Scholar]
- 17.Schork NJ, Goetz LH. Single-Subject Studies in Translational Nutrition Research. Annu Rev Nutr. 2017;37:395–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitcomb DC, Aoun E, Vodovotz Y, Clermont G, Barmada MM. Evaluating disorders with a complex genetics basis: The future role of meta-analysis and systems biology. Dig Dis Sci. 2005;50(12):2195–202. [DOI] [PubMed] [Google Scholar]
- 19.Whitcomb DC. What is personalized medicine and what should it replace? Nat Rev Gastroenterol Hepatol. 2012;9(7):418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bali J, Garg R, Bali RT. Artificial intelligence (AI) in healthcare and biomedical research: Why a strong computational/AI bioethics framework is required? Indian J Ophthalmol. 2019;67(1):3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitcomb DC, Ermentrout GB. A mathematical model of the pancreatic duct cell generating high bicarbonate concentrations in pancreatic juice. Pancreas. 2004;29(2):E30–E40. [DOI] [PubMed] [Google Scholar]
- 22.Raraigh KS, Han ST, Davis E, Evans TA, Pellicore MJ, McCague AF, et al. Functional Assays Are Essential for Interpretation of Missense Variants Associated with Variable Expressivity. Am J Hum Genet. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han ST, Rab A, Pellicore MJ, Davis EF, McCague AF, Evans TA, et al. Residual function of cystic fibrosis mutants predicts response to small molecule CFTR modulators. JCI Insight. 2018;3(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hegyi P, Wilschanski M, Muallem S, Lukacs GL, Sahin-Toth M, Uc A, et al. CFTR: A New Horizon in the Pathomechanism and Treatment of Pancreatitis. Rev Physiol Biochem Pharmacol. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider A, Larusch J, Sun X, Aloe A, Lamb J, Hawes R, et al. Combined Bicarbonate Conductance-Impairing Variants in CFTR and SPINK1 Variants Are Associated With Chronic Pancreatitis in Patients Without Cystic Fibrosis. Gastroenterology. 2011;140(1):162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaRusch J, Jung J, General IJ, Lewis MD, Park HW, Brand RE, et al. Mechanisms of CFTR functional variants that impair regulated bicarbonate permeation and increase risk for pancreatitis but not for cystic fibrosis. PLoS Genetics. 2014;10(7):e1004376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y, Jun I, Shin DH, Yoon JG, Piao H, Jung J, et al. Regulation of CFTR Bicarbonate Channel Activity by WNK1: Implications for Pancreatitis and CFTR-Related Disorders. Cell Mol Gastroenterol Hepatol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohn JA, Noone PG, Jowell PS. Idiopathic pancreatitis related to CFTR: complex inheritance and identification of a modifier gene. J Investig Med. 2002;50(5):247S–55S. [DOI] [PubMed] [Google Scholar]
- 29.Tzetis M, Kaliakatsos M, Fotoulaki M, Papatheodorou A, Doudounakis S, Tsezou A, et al. Contribution of the CFTR gene, the pancreatic secretory trypsin inhibitor gene (SPINK1) and the cationic trypsinogen gene (PRSS1) to the etiology of recurrent pancreatitis. Clin Genet. 2007;71(5):451–7. [DOI] [PubMed] [Google Scholar]
- 30.Masson E, Chen JM, Audrezet MP, Cooper DN, Ferec C. A conservative assessment of the major genetic causes of idiopathic chronic pancreatitis: data from a comprehensive analysis of PRSS1, SPINK1, CTRC and CFTR genes in 253 young French patients. PLoS One. 2013;8(8):e73522. [DOI] [PMC free article] [PubMed] [Google Scholar]; * One of the first studies to demonstrate the complexity of chronic pancreatitis as a disorder with multiple risk factors rather than from the effect of one mutated gene.
- 31.Zou WB, Tang XY, Zhou DZ, Qian YY, Hu LH, Yu FF, et al. SPINK1, PRSS1, CTRC, and CFTR Genotypes Influence Disease Onset and Clinical Outcomes in Chronic Pancreatitis. Clin Transl Gastroenterol. 2018;9(11):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaRusch J, Lozano-Leon A, Stello K, Moore A, Muddana V, O’Connell M, et al. The Common Chymotrypsinogen C (CTRC) Variant G60G (C.180T) Increases Risk of Chronic Pancreatitis But Not Recurrent Acute Pancreatitis in a North American Population. Clin Transl Gastroenterol. 2015;6:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam AN, Aksit MA, Vecchio-Pagan B, Shelton CA, Osorio DL, Anzmann AF, et al. Increased expression of anion transporter SLC26A9 delays diabetes onset in cystic fibrosis. J Clin Invest. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanigan MH. Gamma-glutamyl transpeptidase: redox regulation and drug resistance. Adv Cancer Res. 2014;122:103–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brand H, Diergaarde B, O’Connell MR, Whitcomb DC, Brand RE. Variation in the gamma-glutamyltransferase 1 gene and risk of chronic pancreatitis. Pancreas. 2013;42(5):836–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diergaarde B, Brand R, Lamb J, Cheong SY, Stello K, Barmada MM, et al. Pooling-Based Genome-Wide Association Study Implicates Gamma-Glutamyltransferase 1 (GGT1) Gene in Pancreatic Carcinogenesis. Pancreatology. 2010;10(2–3):194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellison MA, Spagnolo DM, Shelton CA, O’rlova E, Larusch J, Whitcomb DC, et al. Complex Genetics in Pancreatitis: Insights Gained from a New Candidate Locus Panel. Pancreas. 2020;(in press). [DOI] [PubMed] [Google Scholar]
- 38.Bellin MD, Whitcomb DC, Abberbock J, Sherman S, Sandhu BS, Gardner TB, et al. Patient and Disease Characteristics Associated With the Presence of Diabetes Mellitus in Adults With Chronic Pancreatitis in the United States. Am J Gastroenterol. 2017;112(9):1457–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodarzi MO, Nagpal T, Greer P, Cui J, Chen YI, Guo X, et al. Genetic Risk Score in Diabetes Associated With Chronic Pancreatitis Versus Type 2 Diabetes Mellitus. Clin Transl Gastroenterol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]; * This paper shows that inflammation of the exocrine pancreas can drive islet dysfunction and type 2 diabetes without requiring the islets to be destroyed as in type 3c diabetes.
- 40.Zeggini E, Gloyn AL, Barton AC, Wain LV. Translational genomics and precision medicine: Moving from the lab to the clinic. Science. 2019;365(6460):1409–13. [DOI] [PubMed] [Google Scholar]
- 41.Maisonneuve P, Lowenfels AB. Risk factors for pancreatic cancer: a summary review of meta-analytical studies. Int J Epidemiol. 2015;44(1):186–98. [DOI] [PubMed] [Google Scholar]; * An authoative review of the major risk factors for pancreatic cancer.
- 42.Zhan W, Shelton CA, Greer PJ, Brand RE, Whitcomb DC. Germline Variants and Risk for Pancreatic Cancer: A Systematic Review and Emerging Concepts. Pancreas. 2018;47(8):924–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Childs EJ, Mocci E, Bracci P, Gallinger S, Li D, et al. Analysis of Heritability and Genetic Architecture of Pancreatic Cancer: A PanC4 Study. Cancer Epidemiol Biomarkers Prev. 2019;28(7):1238–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hegyi P, Parniczky A, Lerch MM, Sheel ARG, Rebours V, Forsmark CE, et al. International Consensus Guidelines for Risk Factors in Chronic Pancreatitis. Recommendations from the working group for the international consensus guidelines for chronic pancreatitis in collaboration with the International Association of Pancreatology, the American Pancreatic Association, the Japan Pancreas Society, and European Pancreatic Club. Pancreatology. 2020. [DOI] [PubMed] [Google Scholar]
- 46.Gardner TB, Adler DG, Forsmark CE, Sauer BG, Taylor JR, Whitcomb DC. ACG Clinical Guideline: Chronic Pancreatitis. Am J Gastroenterol. 2020. [DOI] [PubMed] [Google Scholar]
- 47.Vivian E, Cler L, Conwell D, Cote GA, Dickerman R, Freeman M, et al. Acute Pancreatitis Task Force on Quality: Development of Quality Indicators for Acute Pancreatitis Management. Am J Gastroenterol. 2019;114(8):1322–42. [DOI] [PubMed] [Google Scholar]


