Abstract
Purpose of review:
Alcohol use disorder (AUD) is a complex genetic disorder with very high heritability. This polygenic disorder not only results in increased morbidity and mortality, it is also a substantial social and economic burden on families and the nation. For past three decades, several genetic studies were conducted to identify genes and pathways associated with AUD. This review aims to summarize past efforts and recent advances in genetic association studies of AUD and related traits.
Recent findings:
Initial genetic association studies achieved a limted success and suffered from low power due to small sample sizes. AUD is a polygenic trait and data from several thousands individuals was required to identify the genetic factors of small effect sizes. The scenario changed recently with technological advances and significant reduction in cost of the genome wide association analyses (GWAS). This enabled researchers to generate genomic data on mega biobanks and cohorts with access to extensive clinical and non-clinical phenotypes. Public access to data from biobanks and collaborative efforts of researchers lead to identification of several novel loci associated with AUDs and related traits. Efforts are now underway to identify the causal variants under the GWAS loci to identify target genes and biological mechanisms underpining AUDs. Many GWAS variants occur in promoter or enhancer regions of the genes and are involved in regulation of gene expression of causal genes. This, large amounts of “omics” data from projects such as “ENCODE”, RoadMap and GTEx is also helping researchers to integrate “multi-omics” data to interpret functional significance of GWAS variants.
Summary:
With current review, we aim to present the recent advances in genetic and molecular studies of AUDs. Recent successes in genetic studies of AUDs will definetely motivate researchers and lead to better therapeutic interventions for this complex disorder.
Keywords: alcohol use disorder, GWAS, SNPs, AUDIT scores, ADH1B, multi-omics
Introduction
Alcohol use disorder (AUD) is one of the most common and costly public health problems in the United States and throughout the world[1, 2]. A person with AUD consumes alcohol in quantities that might be injurious to themselves and to people around them[2, 3]. Infect, the physical and mental health issues associated with excessive consumption of alcohol are known for centuries. Worldwide, an estimated 20–30% cases of esophageal cancer, liver cancer, cirrhosis of the liver, homicide, epilepsy, and motor vehicle accidents can be attributed to excessive consumption of alcohol[4].
Factors influencing AUD
AUDs may transmit from one generation to other and have a high degree of familial association[5]. Many twin, adoption and family studies have provided consistent evidence for genetic predispositions to AUDs[6–8]. Heritable influences account for approximately 40% to 60% of the total variance to risk for alcoholism[6, 7]. A host of other social, cultural and personal factors also influence the drinking behaviors of an individual[9]. It is also important to remember that availability of alcohol is the most important factor that influences the outcome of AUD[1, 10]. A person who has never tried alcohol can not become alcohol dependent in spite of his/ her genetic susceptibility to alcoholism. Therefore it is pretty safe to assume that AUD results from a complex interplay of genetic susceptibility (genes associated with risk), environmental influences, and history of alcohol exposure[11, 12]. These combined factors likely contribute to system-wide epigenetic alterations, post-translational modifications, and long-term allostatic changes in brain regions that underlie the alcohol use disorder[12].
Phenotypes/ traits to study AUD
Like many other complex traits, alcoholism appears to be clinically and etiologicaly hetrogenous[13]. This implies that there might be several steps and intermediate conditions in the development of AUD. Information about the underlying genetic factors that influence risk to AUD can be derived from multiple levels of AUD including amounts of drinks (Alcohol consumption), severity and symptoms of alcohol abuse and dependence. Commonly, genome wide association studies (GWAS) of alcoholism have focused on phenotypes based on the Diagnostic & Statistical Manual of Mental Disorders (DSM)[14]. In the 4th edition of the DSM (DSM-IV), alcohol dependence (AD) and abuse were considered as mutually exclusive diagnoses that together made up AUDs. DSM-V[14, 15] on the other hand consolidated AD and abuse as a single disorder as AUD[15],[16]. By considering AD and abuse under single umbrella increased the number of diagnosed subjects, but this number was still not large enough to design powerful GWAS studies. Therefore, many genetic studies of alcoholism also concentrated on nonclinical phenotypes, such as alcohol consumption and Alcohol Use Disorders Identification Test (AUDIT)[17–19], from large population based cohorts. The AUDIT, a 10-item, self-reported test was developed by the World Health Organization as a screen for hazardous and harmful drinking and can be used as a total (AUDIT-T), AUDIT-Consumption (AUDIT-C) and AUDIT-Problems (AUDIT-P) sub-scores.
Genetics of AUD
Given a large heritability for AUDs, sevral studies were conducted to identify the specific genes or genetic variations associated with AUD or related traits[8, 20–22]. The gene identification efforts for AUDs can be divided into pre-GWAS and the GWAS era. The pre-GWAS era mainly focussed on genome-wide linkage and candidate gene studies[23, 23–26]. Although, during this period many genes were nominated as the causal factors, a few genes actually showed consistent evidence of an association with AUD[23, 27]. Even the initial GWAS studies suffered from low power due to smaller sample sizes and failed to identify credible evidence in favor of any particular gene[28–30]. Power of GWAS significantly improved in the past couple of years with the advent of large biobanks (UK Biobank) and the collaborative efforts of large consortiums (Psychiatric Genetics Consortium [PGC]). Following section will briefly describe the most successful and replicated findings in candidate gene studies before moving to recent advances in the field of AUD gene discovery.
Candidate gene studies of AUD and related traits
Most candidate genes selected for AUD genetic association studies can broadly be divided into two categories: 1) genes involved in central nervous system’s (CNS) response to alcohol or other addictive substances (CHRNA5, GABRG1, GABRA2, OPRM1 etc.)[27, 31–35] and 2) genes involved in alcohol metabolism (ADH4, ADH1B, ALDH2)[36–41]. Out of all candidate genes, role of ADH1B in AUD is very well established and replicated, particularly among populations of Asian descent[36, 37, 42]. However, variants in ADH1B are uncommon (~3–5%) in European Americans (EAs) and African Americans (AAs) [36]. This is the reason that a low frequency coding variant of ADH1B gene (rs1229984) was originally identified in Asian samples and was subsequently replicated in EAs and AAs in a large meta-analysis[36]. This single nucleotide change leads to replacement of Arg48 with His48 and results in a “atypical ADH” enzyme that exhibits several times higher catalytic activity than the normal enzyme. Increased accumulation of acetaldehyde from due to higher catalytic activity of “atypical ADH” is responsible for the flushing and severe symptoms of alcohol related sensitivity[37, 40]. The intense aversive reaction to even smaller amounts of alcohol deter individuals from consuming large amounts of alcohol and protects them from developing AUD[36, 37, 40, 42]. Indeed, the His48 allele was also found to be associated with lower alcohol consumption as measured by the subjects’ lifetime maximum alcohol consumption in a 24-hour period (β = −0.28 (95% CI −0.35, − 0.20), p value = 3.24 × 10−13)[36].
GWAS of AUD and related traits
GWASs represent the most recent paradigm shift for the gene discovery[43]. These hypothesis free genome scans allow interrogation of million of SNPs across thousands of genes at relatively modest cost[43]. Many GWAS’s of AUD, AD and alcohol consumption have been completed majorly in the European ancestry[28–30, 44–53]. First reported GWAS study for AD (Treutlein et al. 2009) identified 2 genome-wide significant SNPs (rs7590720 and rs134694) in combined male only sample of 1,460 AD subjects and 2,332 controls[44]. The closest gene to the association signal PECR (peroxisomal trans-2-enoyl-CoA reductase) is involved in the metabolism of fatty acids. Two subsequent AD GWASs did not identify any novel genome-wide significant loci (COGA, SAGE)[29, 54]. Further Heath and colleagues performed GWAS of quantitative indices of excessive alcohol consumption in moderate size cohort of Australian families but failed to identify any genome-wide significant variant[28]. Subsequent family and case control genome-wide efforts met with similar fate of limited success and non replication across different studies[52, 55]. At this point in time researchers already realized that AUD is highly polygenic and sample size of individual cohort is not enough to identify the SNPs with very small effect sizes[28]. Availability of raw genotype data through dbGAP also made it a bit easier to meta-analyze the similarity ascertained cohorts with genome-wide SNP data. Schumann and colleagues[56] meta-analyzed 26,316 population based subjects and a follow-up sample of 21,185 EA subjects and identified variants in the autism susceptibility candidate 2 (AUTS2) gene significantly associated with alcohol consumption (gms/ day/ kg of body weight). Subsequently, Kapoor and colleagues[49] meta-analyzed two large complementary and well-characterized EA cohorts assessed using the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA) and identified rs1229984 SNP in ADH1B to be genome-wide significantly associated with phenotype measuring maximum number of alcoholic drinks in 24 hour. This was the first alcoholism related GWAS that reported genome-wide significance at this locus[49].
The initial genome-wide meta-analysis had a few consistent findings, but the variants identified in these GWASs explained a very small proportion of heritability for alcohol related traits [28, 49, 57, 58]. A very large sample size was needed to account for the missing heritability and it was a great deal of challenge to identify the well characterized large cohorts with AUD phenotypes. To address this challenge, recent genome-wide efforts focussed on larger sample sizes assembled via consortia-led meta-analyses (Figure 1 and table 1). Researchers got access to alcohol consumption data for large number of individuals through UKBiobank and it finally provided necessary boost in gene identification efforts for alcholism. Clarke and colleagues[59] were the first group to take advantage of this cohort and reported genome-wide significant associations at 14 loci including ADH1B gene (Table 1). Soon most of these initial findings were replicated in a very large meta-analysis of alcohol consumption of over 30 datasets (UKBibank, 23andMe and other GWAS) across nearly 1.2 million participants of European ancestry (Table 1; Figure 1)[60]. In this study, Liu and colleagues discovered 566 genetic variants in 406 loci associated with multiple stages of alcohol and tobacco use (initiation, cessation, and heaviness), with 150 loci evidencing pleiotropic association[60]. These results provided a very good starting point to evaluate the effects of these loci in model organisms and more precise measures of AUD.
Figure 1: Relationship among recently published genome-wide association studies related to AUDs.
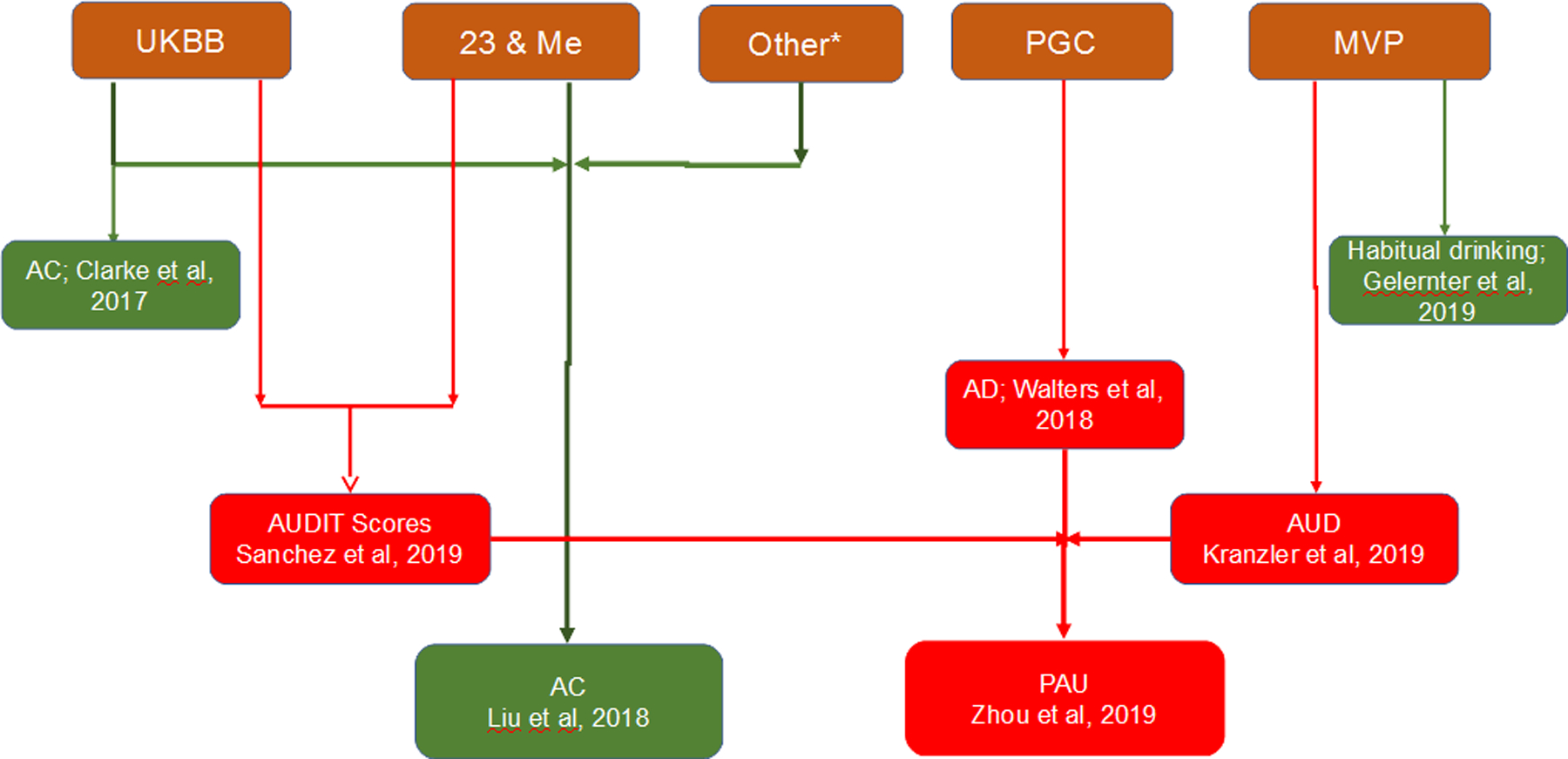
Majority of genomic data for large alcohol consumption and AUD meta-analysis was either from UKBiobank or from Million Veterans Project. Several other cohorts from dbGAP also contributed to large sample size of alcohol consumption GWAS by Liu et al, 2019. Genome-wide data on 14,904 DSM-IV diagnosed AD individuals and 37,944 controls from 28 case/control and family-based studies were meta-analyzed for PGC’s AD GWAS.
Table 1:
List of recently published genome-wide association studies related to AUDs
| GWAS (Cohort) | Year | Phenotype | N | N Loci |
|---|---|---|---|---|
| Zhou et al (MVP, PGC, UKBB) | 2019 | PAU* | 435,563 | 27 |
| Sanchez-Roige et al (UKBB, 23andMe) | 2019 | AUDIT-C, AUDIT-P and AUDIT-T | 141,932 | 7, 3, 8 |
| Liu et al (UKBB, 23andMe, other+) | 2019 | Drinks per week | 941,280 | 81 |
| Kranzler et al (MVP) | 2019 | AUD, AUDIT-C | 55,587 AUD cases, 218,807 controls; 274,424 | 10, 13 |
| Walters et al (PGC) | 2018 | Alcohol dependence | 14,904 AD cases 37,944 controls | 2 |
| Clarke et al (UKBB) | 2017 | Alcohol consumption | 112, 117 | 8 |
Problamatic alchol use (PAU);
Several GWAS datasets from dbGAP with alcohol consumption phenotype;
Despite these advances with the GWASs of alcohol consumption, genetic studies of AUD and AD drew multiple challenges and limited success[45]. In a community based cohort (e.g. UKBiobank), it is relatively easy to derive the measure of alcohol consumption using number of alcoholic drinks consumed by an individual. But these large population based cohorts lack individuals diagnosed with AUD or AD. Substance use disorder working group of PGC (PGCSUD) tried to circumvent this problem by performing genome-wide meta-analysis of well characterized cohorts for DSM-IV diagnosed AD[45]. This meta-analysis identified, genome-wide significant effects of different ADH1B variants in European (rs1229984; P=9.8 ×10–13) and African ancestries (rs2066702; P=2.2 ×10–9). This study also found that the genetic underpinnings of DSM-IV diagnosed AD only partially overlap with those for alcohol consumption, underscoring the genetic distinction between pathological and nonpathological drinking behaviors[45]. Recently, Sanchez and colleagues also reported similar genetic differences among AUD (measured as AUDIT-P) and alcohol consumption (measured as AUDIT-C)[61]. In this meta-analysis (UKBiobank and 23andMe), AUDIT-P score showed a strong genetic correlation with alcohol dependence (PGC-SUD GWAS), while AUDIT-C score showed stronger genetic correlation with alcohol consumption[61]. Although, these genetics differences were not as apparent in African Americans in a recent large GWAS of AUD and AUDIT-C on individuals from Million Veteran Program (MVP)[62]. This GWAS reported many overlapping variants for AUDIT-C and AUD, with moderate-to-high genetic correlation between these traits (0.522 in EAs and 0.930 in AAs). Despite the significant genetic overlap between the AUDIT-C and AUD diagnosis, downstream analyses of MVP data revealed biologically meaningful points of divergence. Kranzler and colleagues[62] found that the polygenic risk score (PRS) calculated from AUD GWAS was significantly associated with tobacco use and multiple psychiatric disorders, whereas the AUDIT-C PRS did not show any association with these traits. These findings further confirmed that the AUD and alcohol consumption (measured by AUDIT-C in MVP) are genetically related but very distinct phenotypes.
Despite a significant boost in the number of genome-wide significant loci, variants identified in these large GWASs still explain a very small proportion of estimated genetic effect (heritability) for AUD and alcohol consumption. The SNP heritability estimates of AUDIT-C scores for all loci in MVP and the meta-analysis of the UKBiobank and 23andMe data ranged from 0.6 to 8 percent respectively[61, 62]. Heritability estimates for AUD were slightly lower and ranged from 0.5 to 5.9 percent in MVP versus UKBiobank and 23andMe meta-analysis respectively[61, 62]. These estimates are still significantly lower than the heritability estimates of AUD from twin and family studies. Given the heterogeneity in the diagnosis and polygenicity of this complex trait it seems that we are still short of required sample size to identify all the variants associated with disease. Some researchers working with other complex psychiatric traits argued that missing heritability can be explained by rare to low frequency variants of relatively large effect sizes. These rare variants can be identified by next generation sequencing in large cohorts. Contrary to expectations, recent sequencing studies for neurological disorders in moderate sample size are not much successful and have not yielded the intended results[63, 64]. These studies concluded that the low frequency disease associated variants generally have low-moderate effect sizes and very large sample size is needed to identify these variants. Sequencing costs are moving down, but still these costs are prohibitive to perform a large whole genome sequencing study of AUD and other complex psychiatric disorders.
Functional significance of GWAS variants
Recent progress in GWAS of AUD has identified several variants across many loci that are significantly associated with alcoholism and related traits (Table 2). Some variants in these loci result in amino acid changes (e.g. rs1229984 in ADH1B) and known to alter the function of the gene to affect outcome of AUD. Most other AUD and alcohol consumption associated variants occur in intergenic or intronic regions and are not directly associated with protein coding changes[65]. Linkage disequilibrium (LD) at many loci span across thousands of variants and further makes it difficult to identify a causal SNP or genes associated with the disorder[66]. Thus, many AUD GWAS just annotated the nearest gene to the lead SNP as the susceptibility loci (Table 2). Furthermore, due to different LD structure across datasets, many times individual study identified different lead SNPs and/ or different nearest genes within the same loci. Recent studies on psychiatric and neurological disorders showed that the most of genome-wide significant variants occur on active enhancers or promoters and might alter the expression level of nearby (cis) or distant (trans) gene[66–68]. These gene expression altering SNPs (expression quantitative loci or eQTLs) can be specific to a particular cell or tissue type. Several post-genomic helper tools such as FUMA[69] are available to functionally annotate the GWAS variants and to predict the functional consequence of a disease associated variant. More recent tools such as PrediXcan[70] and TWAS[71] can impute the genetic component of tissue-specific gene expression in GWAS datasets and help to connect changes in gene expression to trait outcome. Although size of eQTL and transcriptomic datasets can be a limiting factor to detect all functional association. Still initial application of PrediXcan has prioritized several genes (e.g. MAPT, CRHR1, FUT2, ADH1B, ADH4, ADH5, C1QTNF4, GCKR, DRD2) across different tissues [61, 72]. In a recent study some of these genetic targets including ADH1B, GCKR, SLC39A8 and KLB have been shown to play a conserved role in phenotypic responses to alcohol in Caenorhabditis elegans [74]. Researchers are also using the post-mortem brain tissue from alcoholic subjects to understand the effect of long-term alcohol consumption on expression of genes. Recently, Kapoor and colleagues[73] have suggested that genes identified in alcohol related GWASs (AD and alcohol consumption) interact with genes affected by alcohol exposure and results in system wide changes across pathways and networks involved in alcoholism. Better understanding of these pathways will definitely result in better therapeutic interventions for AUD and problematic drinking[73].
Table 2:
Shared loci among GWAS for problematic drinking and alcohol consumption
| Chromosome | Locus | Locus start | Locus end | Closest/ overlapping genes |
|---|---|---|---|---|
| 1 | 1p31.3 | 65907700 | 66907700 | PDE4B |
| 2 | 2p23.3-p23.2 | 27230940 | 28746841 | GCKR* |
| 2 | 2p21 | 43771496 | 45655276 | SLC3A1 |
| 3 | 3p12.1 | 84408785 | 85957240 | CADM2 |
| 4 | 4p14 | 38914993 | 39914993 | KLB |
| 4 | 4q22.3-q24 | 96764066 | 101983024 | ADH1B* |
| 4 | 4q24 | 102688709 | 103688709 | SLC39A8 |
| 7 | 7q36.2 | 152989744 | 153989744 | DPP6 |
| 10 | 10q25.1 | 110007806 | 111007806 | MAPKAPK5P1 |
| 11 | 11p11.2 | 46897353 | 48410823 | SPI1 |
| 11 | 11q23.3-q24.1 | 121044285 | 122044285 | SORL1 |
| 12 | 12q13.12-q13.13 | 51395882 | 52395882 | SLC4A8 |
| 14 | 14q23.1 | 58282779 | 59282779 | ARID4A |
| 11 | 11q23.2 | 112924042 | 113924042 | DRD2* |
Candidate genes
Conclusions
For centuries, it was known that problematic drinking runs in families and genetic factors influence the etiology of AUD. Recent GWAS approaches have started elucidating the genetic loci related to AUD. At many GWAS loci strong LD extend to several hundred megabases and makes it difficult to identify the causal variant and candidate genes associated with alcoholism. Integration of genomic and transcriptomic data has opened the door for fine mapping and molecular genetic investigations into pathways and networks related to AUD. But there is still need of large scale “omics” data from diverse populations to effectively and accurately fine map the loci associated with alcoholism and other complex disorders. Many groups including Collaborative Studies of Genetics of Alcoholism (COGA) are generating “omics” data on African American and other diverse populations. Genomic data in diverse populations will also be useful for accurate disease risk prediction using polygenic risk scores. The future goal of precision medicine cannot be achieved without genetic association studies in diverse populations. Researchers from COGA are also generating transcriptomic and epigenomic data at single nuclei level from many different regions from post-mortem human brains of alcoholics and controls. Single nuclei transcriptomic analysis in human brain will be extremely useful to understand the role of various cellular lineages in development of AUDs. Genetic studies of AUD are advancing in the right direction and insights revealed will elucidate novel therapeutic targets resulting in better understanding of AUD biology.
Funding source:
This work is supported by National Institute on Alcohol Abuse and Alcoholism (R21AA026388 and U10AA008401).
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflicts of interest
The authors declare that they have no conflicts of interest associated with this manuscript.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
References:
- 1.Wang J-C, Foroud T, Hinrichs AL, et al. (2013) A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry 18:1218–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD (2015) 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med 49:e73–e79 [DOI] [PubMed] [Google Scholar]
- 3.Connor JP, Haber PS, Hall WD (2016) Alcohol use disorders. The Lancet 387:988–998 [DOI] [PubMed] [Google Scholar]
- 4.Organization WH (2019) Global Status Report on Alcohol and Health 2018. World Health Organization [Google Scholar]
- 5.Goodwin DW (1985) Alcoholism and Genetics: The Sins of the Fathers. Arch Gen Psychiatry 42:171–174 [DOI] [PubMed] [Google Scholar]
- 6.Verhulst B, Neale MC, Kendler KS (2015) The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med 45:1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schuckit MA (2014) A Brief History of Research on the Genetics of Alcohol and Other Drug Use Disorders. J Stud Alcohol Drugs Suppl 75:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edenberg HJ, Foroud T (2006) The genetics of alcoholism: identifying specific genes through family studies. Addict Biol 11:386–396 [DOI] [PubMed] [Google Scholar]
- 9.Sudhinaraset M, Wigglesworth C, Takeuchi DT (2016) Social and Cultural Contexts of Alcohol Use. Alcohol Res Curr Rev 38:35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chartier KG, Karriker-Jaffe KJ, Cummings CR, Kendler KS (2017) Review: Environmental influences on alcohol use: Informing research on the joint effects of genes and the environment in diverse U.S. populations. Am J Addict 26:446–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dick DM, Kendler KS (2012) The Impact of Gene–Environment Interaction on Alcohol Use Disorders. Alcohol Res Curr Rev 34:318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contet C (2012) Gene Expression Under the Influence: Transcriptional Profiling of Ethanol in the Brain. Curr Psychopharmacol 1:301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloninger CR, Reich T (1983) Genetic heterogeneity in alcoholism and sociopathy. Res Publ - Assoc Res Nerv Ment Dis 60:145–166 [PubMed] [Google Scholar]
- 14.Bell CC (1994) DSM-IV: Diagnostic and Statistical Manual of Mental Disorders. JAMA 272:828–829 [Google Scholar]
- 15.Takahashi T, Lapham G, Chavez LJ, et al. (2017) Comparison of DSM-IV and DSM-5 criteria for alcohol use disorders in VA primary care patients with frequent heavy drinking enrolled in a trial. Addict Sci Clin Pract. 10.1186/s13722-017-0082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grant BF, Goldstein RB, Saha TD, et al. (2015) Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry 72:757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burns E, Gray R, Smith LA (2010) Brief screening questionnaires to identify problem drinking during pregnancy: a systematic review. Addict Abingdon Engl 105:601–614 [DOI] [PubMed] [Google Scholar]
- 18.de Oliveira JB, Kerr-Corrêa F, Lima MCP, Bertolote JM, Santos JLF (2014) Validity of alcohol screening instruments in general population gender studies: an analytical review. Curr Drug Abuse Rev 7:59–65 [DOI] [PubMed] [Google Scholar]
- 19.Reinert DF, Allen JP (2007) The alcohol use disorders identification test: an update of research findings. Alcohol Clin Exp Res 31:185–199 [DOI] [PubMed] [Google Scholar]
- 20.Wang J-C, Kapoor M, Goate AM (2012) The Genetics of Substance Dependence. Annu Rev Genomics Hum Genet 13:241–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal A, Bierut LJ (2012) Identifying genetic variation for alcohol dependence. Alcohol Res Curr Rev 34:274–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edenberg HJ, Gelernter J, Agrawal A (2019) Genetics of Alcoholism. Curr Psychiatry Rep 21:26. [DOI] [PubMed] [Google Scholar]
- 23.Irons DE, Iacono WG, Oetting WS, Kirkpatrick RM, Vrieze SI, Miller MB, McGue M (2014) GABA System Genes – No Evidence for a Role in Alcohol Use and Abuse in a Community-based Sample. Alcohol Clin Exp Res 38:938–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong X, Zhang H (2005) Linkage analysis and association analysis in the presence of linkage using age at onset of COGA alcoholism data. BMC Genet 6:S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X, Cooper R, Kan D, Cao G, Wu X (2005) A genome-wide linkage and association study using COGA data. BMC Genet 6:S128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D, Sulovari A, Cheng C, Zhao H, Kranzler HR, Gelernter J (2014) Association of Gamma-Aminobutyric Acid A Receptor α 2 Gene (GABRA2) with Alcohol Use Disorder. Neuropsychopharmacology 39:907–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dick DM, Foroud T (2003) Candidate Genes for Alcohol Dependence: A Review of Genetic Evidence From Human Studies: Alcohol Clin Exp Res 27:868–879 [DOI] [PubMed] [Google Scholar]
- 28.Heath AC, Whitfield JB, Martin NG, et al. (2011) A Quantitative-Trait Genome-Wide Association Study of Alcoholism Risk in the Community: Findings and Implications. Biol Psychiatry 70:513–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bierut LJ, Agrawal A, Bucholz KK, et al. (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci 107:5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edenberg HJ, Koller DL, Xuei X, et al. (2010) Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 34:840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen RW, Liang J (2017) Role of GABAA receptors in alcohol use disorders suggested by chronic intermittent ethanol (CIE) rodent model. Mol Brain 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA (2008) The CHRNA5/A3/B4 Gene Cluster Variability as an Important Determinant of Early Alcohol and Tobacco Initiation in Young Adults. Biol Psychiatry 63:1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeifer P, Sariyar M, Eggermann T, Zerres K, Vernaleken I, Tüscher O, Fehr C (2015) Alcohol Consumption in Healthy OPRM1 G Allele Carriers and Its Association with Impulsive Behavior. Alcohol Alcohol 50:379–384 [DOI] [PubMed] [Google Scholar]
- 34.Hack LM, Kalsi G, Aliev F, Kuo P-H, Prescott CA, Patterson DG, Walsh D, Dick DM, Riley BP, Kendler KS (2011) Limited Associations of Dopamine System Genes With Alcohol Dependence and Related Traits in the Irish Affected Sib Pair Study of Alcohol Dependence (IASPSAD). Alcohol Clin Exp Res 35:376–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chamorro A-J, Marcos M, Mirón-Canelo J-A, Pastor I, González-Sarmiento R, Laso F-J (2012) Association of μ-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict Biol 17:505–512 [DOI] [PubMed] [Google Scholar]
- 36.Bierut LJ, Goate AM, Breslau N, et al. (2012) ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 17:445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polimanti R, Gelernter J (2018) ADH1B: From alcoholism, natural selection, and cancer to the human phenome. Am J Med Genet B Neuropsychiatr Genet 177:113–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edenberg HJ, Foroud T (2013) Genetics and alcoholism. Nat Rev Gastroenterol Hepatol 10:487–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luczak SE, Glatt SJ, Wall TL (2006) Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull 132:607–621 [DOI] [PubMed] [Google Scholar]
- 40.Edenberg HJ (2007) The Genetics of Alcohol Metabolism: Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase Variants. Alcohol Res Health 30:5–13 [PMC free article] [PubMed] [Google Scholar]
- 41.Almeida OP, Hankey GJ, Yeap BB, Golledge J, Flicker L (2014) The triangular association of ADH1B genetic polymorphism, alcohol consumption and the risk of depression in older men. Mol Psychiatry 19:995–1000 [DOI] [PubMed] [Google Scholar]
- 42.Eng MY, Luczak SE, Wall TL (2007) ALDH2, ADH1B, and ADH1C Genotypes in Asians: A Literature Review. Alcohol Res Health 30:22–27 [PMC free article] [PubMed] [Google Scholar]
- 43.Bush WS, Moore JH (2012) Chapter 11: Genome-Wide Association Studies. PLoS Comput Biol. 10.1371/journal.pcbi.1002822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treutlein J, Cichon S, Ridinger M, et al. (2009) Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 66:773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.**.Walters RK, Polimanti R, Johnson EC, et al. (2018) Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 21:1656–1669 Largest GWAS of alcohol dependent subjects selected on basis of DSMIV diagnosis.
- 46.Wetherill L, Kapoor M, Agrawal A, et al. (2014) Family-based association analysis of alcohol dependence criteria and severity. Alcohol Clin Exp Res 38:354–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai D, Wetherill L, Bertelsen S, et al. (2019) Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav 18:e12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wetherill L, Lai D, Johnson EC, et al. (2019) Genome-wide association study identifies loci associated with liability to alcohol and drug dependence that is associated with variability in reward-related ventral striatum activity in African- and European-Americans. Genes Brain Behav 18:e12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.*.Kapoor M, Wang J-C, Wetherill L, et al. (2013) A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet 132:1141–1151 First GWAS study that reported genome-wide significant association results at ADH locus (rs1229984)
- 50.Kapoor M, Wang J-C, Wetherill L, et al. (2014) Genome-wide survival analysis of age at onset of alcohol dependence in extended high-risk COGA families. Drug Alcohol Depend 142:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai D, Wetherill L, Kapoor M, et al. (2019) Genome-wide association studies of the self-rating of effects of ethanol (SRE). Addict Biol e12800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J-C, Foroud T, Hinrichs AL, et al. (2013) A genome wide association study of alcohol dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. 10.1038/mp.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gelernter J, Kranzler HR, Sherva R, et al. (2014) Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edenberg HJ, Koller DL, Xuei X, et al. (2010) Genome-Wide Association Study of Alcohol Dependence Implicates a Region on Chromosome 11. Alcohol Clin Exp Res 34:840–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV (2011) Association Analysis of Symptoms of Alcohol Dependence in the Molecular Genetics of Schizophrenia (MGS2) Control Sample. Alcohol Clin Exp Res 35:963–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schumann G, Coin LJ, Lourdusamy A, et al. (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci 108:7119–7124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kapoor M, Chou Y-L, Edenberg HJ, et al. (2016) Genome-wide polygenic scores for age at onset of alcohol dependence and association with alcohol-related measures. Transl Psychiatry 6:e761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salvatore JE, Aliev F, Edwards AC, et al. (2014) Polygenic Scores Predict Alcohol Problems in an Independent Sample and Show Moderation by the Environment. Genes 5:330–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.**.Clarke T-K, Adams MJ, Davies G, et al. (2017) Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry 22:1376–1384 This large GWAS of alcohol consumption initiated large scale genome-wide exploration of alcohol related phenotypes.
- 60.**.Liu M, Jiang Y, Wedow R, et al. (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51:237–244 Largest GWAS of alcohol consumption till date.
- 61.Sanchez-Roige S, Palmer AA, Fontanillas P, et al. (2019) Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry 176:107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kranzler HR, Zhou H, Kember RL, et al. (2019) Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10:1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bis JC, Jian X, Kunkle BW, et al. (2018) Whole exome sequencing study identifies novel rare and common Alzheimer’s-Associated variants involved in immune response and transcriptional regulation. Mol Psychiatry 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vardarajan BN, Barral S, Jaworski J, et al. (2018) Whole genome sequencing of Caribbean Hispanic families with late-onset Alzheimer’s disease. Ann Clin Transl Neurol 5:406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallagher MD, Chen-Plotkin AS (2018) The Post-GWAS Era: From Association to Function. Am J Hum Genet 102:717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang K, Marcora E, Pimenova AA, et al. (2017) A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat Neurosci 20:1052–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huckins LM, Dobbyn A, Ruderfer DM, et al. (2019) Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat Genet 51:659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bryois J, Buil A, Evans DM, et al. (2014) Cis and Trans Effects of Human Genomic Variants on Gene Expression. PLOS Genet 10:e1004461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Watanabe K, Taskesen E, Bochoven A van, Posthuma D (2017) Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gamazon ER, Wheeler HE, Shah KP, et al. (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47:1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gusev A, Ko A, Shi H, et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou H, Sealock JM, Sanchez-Roige S, et al. (2019) Meta-analysis of problematic alcohol use in 435,563 individuals identifies 29 risk variants and yields insights into biology, pleiotropy and causality. bioRxiv 738088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.*.Kapoor M, Wang J-C, Farris SP, et al. (2019) Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl Psychiatry 9:1–11 This study integrated transcriptomic and genomics data to prioritize genes and pathways related to alcoholism.
- 74.Thompson A, Cook J, Choquet H, et al. (2020) Functional validity, role, and implications of heavy alcohol consumption genetic loci. Science Advances 6:eaay5034. [DOI] [PMC free article] [PubMed] [Google Scholar]


